Abstract
INTRODUCTION:
Circadian clocks provide an internal representation of local time inside cells and control the timing of gene expression in anticipation of sunrise and sunset. In cyanobacteria, timekeeping is achieved by way of an oscillator comprising three Kai (meaning “cycle” in Japanese) proteins, KaiA, KaiB, and KaiC, which relay temporal information downstream through two sensor histidine kinases, SasA and CikA, to regulate the transcription factor RpaA. Identifying the specific mechanisms by which circadian clocks exert temporal control over gene expression has proven challenging in the complex milieu of cells. Thus, we reassembled an intact clock, including the core oscillator and signal transduction components, under defined conditions in vitro. Together with structural studies and biochemical analyses of partial clock reactions, we acquired insights into mechanisms by which the cyanobacterial circadian clock functions to control gene expression.
RATIONALE:
Although the core oscillator can be reconstituted in vitro with just the Kai proteins, these reactions assayed by SDS–polyacrylamide gel electrophoresis report only on the phosphorylation status of KaiC and have therefore not offered mechanistic insight into clock protein interactions or signal transduction. Thus, we developed a fluorescence polarization–based in vitro whole-clock system that includes the KaiABC oscillator, SasA and/or CikA, RpaA, and a clock promoter–bearing DNA fragment. This in vitro clock (IVC) oscillates autonomously for days and allows monitoring of each component individually in real time, revealing the formation and phase of macromolecular assemblies from the timekeeper to DNA binding by RpaA.
RESULTS:
The IVC was used to examine the sufficiency of SasA or CikA to maintain clock output under constant conditions and identify how each contributes to the phase of DNA binding by RpaA; it even allowed the dissection of the breakdown in signaling of an arrhythmic mutant of RpaA. We challenged a long-standing paradigm that timekeeping by the oscillator depends solely on the Kai proteins, with SasA and CikA providing only input-output signaling. The in vitro oscillator is known to function under a relatively narrow set of Kai protein concentrations; we show here that a KaiABC-only mixture that fails to oscillate in a sustained manner owing to limiting levels of KaiB can be rescued by SasA, which acts to recruit KaiB to the KaiC hexamer through heterotropic cooperativity. Cooperativity is based on structural mimicry between SasA and KaiB, and mutations that eliminate heterocooperativity in vitro profoundly affect circadian rhythms in vivo. CikA also rescues period defects under low levels of KaiA. Together, our data help to explain how the clock compensates in vivo for changes in concentrations of oscillator components that occur as part of the transcription-translation feedback loop and protein turnover. The intimate coupling between oscillator and input-output components blurs their distinction, although the extent to which this coupling can be generalized to eukaryotic clocks remains an open question.
CONCLUSION:
We developed the reconstituted IVC to establish causal links between clock biochemistry and in vivo phenotypes. Our system provides a powerful platform to explore, for example, how changes in factors such as temperature or adenosine triphosphate levels are compensated to allow generation of reliable circadian rhythms of transcription.
In fact, this IVC is just a starting point, one with many possibilities. For example, the real-time data presented here will allow mathematical models of the cyanobacterial clock to extend beyond the KaiABC oscillator to address the events that effect temporal control of physiology and metabolism. As a next step, transplanting this IVC to artificial cells could enable single-cell microscopy experiments of this streamlined clock under conditions that are much closer to physiological. In an artificial cellular system, transcriptional and translational networks could be included, allowing higher-complexity customized frequencies and outputs to be engineered, opening the door to synthetic biology applications. ■
Graphical Abstract
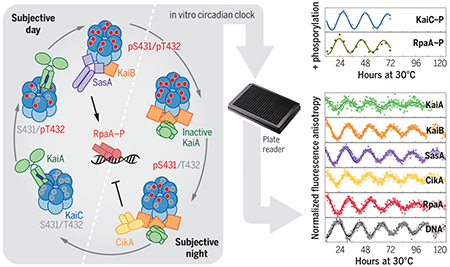
Reconstituting an intact clock in vitro. Cyanobacterial circadian proteins KaiA, KaiB, KaiC, SasA, CikA, RpaA, and a promoter-bearing DNA fragment were reconstituted into an in vitro clock. Together with structural studies and biochemical assays, this system allowed dissection of a circadian clock system to demonstrate that the concept of the core oscillator should be extended to include output proteins SasA and CikA. The dashed line separates subjective day and subjective night, which refer to the halves of a circadian period under constant conditions that correspond to day and night in a light-dark cycle. S, serine; T, threonine; p, phosphorylated.
Circadian clocks control gene expression to provide an internal representation of local time. We report reconstitution of a complete cyanobacterial circadian clock in vitro, including the central oscillator, signal transduction pathways, downstream transcription factor, and promoter DNA. The entire system oscillates autonomously and remains phase coherent for many days with a fluorescence-based readout that enables real-time observation of each component simultaneously without user intervention. We identified the molecular basis for loss of cycling in an arrhythmic mutant and explored fundamental mechanisms of timekeeping in the cyanobacterial clock. We find that SasA, a circadian sensor histidine kinase associated with clock output, engages directly with KaiB on the KaiC hexamer to regulate period and amplitude of the central oscillator. SasA uses structural mimicry to cooperatively recruit the rare, fold-switched conformation of KaiB to the KaiC hexamer to form the nighttime repressive complex and enhance rhythmicity of the oscillator, particularly under limiting concentrations of KaiB. Thus, the expanded in vitro clock reveals previously unknown mechanisms by which the circadian system of cyanobacteria maintains the pace and rhythmicity under variable protein concentrations.
Circadian clocks are intracellular systems that provide organisms with an internal representation of local time and have profound consequences to health (1). The clock in the cyanobacterium Synechococcus elongatus generates circadian rhythms of genetic, physiological, and metabolic activities that enhance fitness (2, 3). The core circadian clock genes of cyanobacteria, kaiA, kaiB, and kaiC, are essential for rhythmic gene expression (4), and their proteins generate an autonomous ~24-hour rhythm of KaiC phosphorylation in vivo (5), which can be recapitulated in vitro (6). Ordered phosphorylation of KaiC at residues S431 and T432 (7, 8), stimulated by KaiA during the day (9, 10 and suppressed by KaiB at night (11, 12), helps set the circadian pace and phase of the oscillator (13–15). The input-output sensor histidine kinases, Synechococcus adaptive sensor A, SasA (16), and circadian input kinase A, CikA (17), interact with Kai proteins (14, 18) to regulate the master transcription factor, RpaA (19), and generate circadian rhythms of gene expression in cyanobacteria (20, 21).
Although the reconstituted KaiABC oscillator has been a powerful tool to study mechanisms of circadian timekeeping in cyanobacteria (6, 15, 22–24), it does not include the signal transduction machinery necessary to provide a readout of timing encoded by the biochemical oscillator. To explore how each component interacts in the network and how output proteins integrate with the core oscillator, we reconstituted clock-controlled DNA binding in a cell-free system that enables real-time reporting from different partners. In addition to the core oscillator components KaiA, KaiB, and KaiC, this in vitro clock (IVC) includes SasA and/or CikA, RpaA, and a DNA duplex that carries a clock-controlled promoter, demonstrating that these components are sufficient to form the fundamental regulatory network for autonomous timekeeping and temporal relay to downstream events. We then use this IVC to identify how a loss-of-function allele in RpaA disrupts circadian rhythms, highlighting its power to provide mechanistic insight for in vivo studies. Moreover, through careful deconstruction of our expanded IVC, we discovered that SasA helps to cooperatively recruit KaiB to the KaiC hexamer, thus defining a previously unknown role for SasA in the core oscillator that it is required for proper circadian rhythms in vivo.
Signal transduction in a reconstituted IVC
To monitor protein interactions in the IVC, we attached fluorescent probes (25) to each of the individual clock protein components, KaiA, KaiB, SasA, CikA, and RpaA (see methods in the supplementary materials and tables S1 to S3) using sortase A–mediated ligation (26, 27) and monitored their association into larger complexes using fluorescence anisotropy (FA) (Fig. 1, A to C, and fig. S1).
Fig. 1. Reconstitution of an intact clock that controls rhythmic DNA binding in vitro.
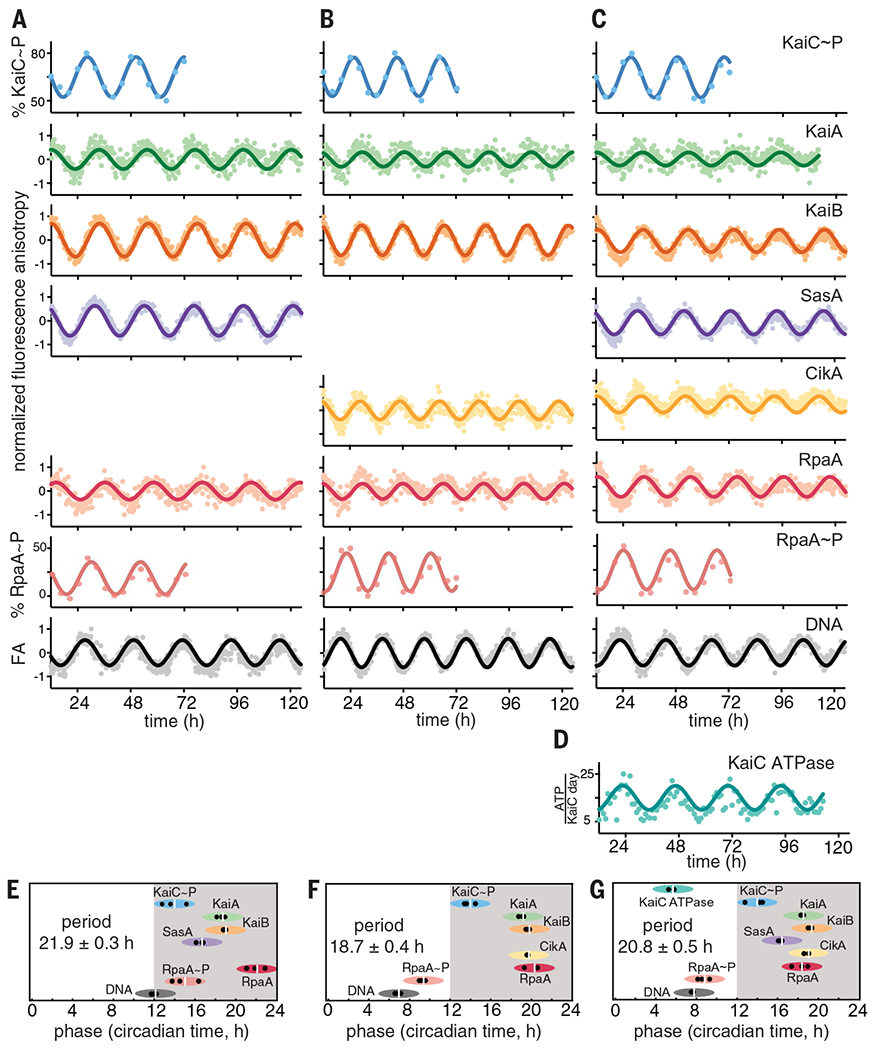
(A to C) IVC reactions containing KaiA, KaiB, KaiC, RpaA, PkaiBC DNA, 1 mM ATP, 5 mM MgCl2, and SasA and/or CikA. 50 nM fluorescently labeled KaiA (green), KaiB (orange), SasA (purple), CikA (yellow), RpaA (red), or 100 nM PkaiBC (gray) were included in separate reactions, and their fluorescence anisotropies (FA) were measured in a microplate reader at 30°C. For phosphorylation experiments, KaiC~P (blue) and RpaA~P (salmon) were quantified by gel electrophoresis on SDS-PAGE and Zn2+-Phos-tag gels respectively. After truncating the first 12 hours of data collection during which samples approached stable limit cycles, data were baseline-corrected and fit to a damped cosine function to calculate periods and phases (see materials and methods in the supplementary materials). FA values were normalized before fitting. (D) KaiC-ATPase was measured by real-time 1H-NMR in a KaiABC-only reaction to isolate the KaiC ATPase activity. (E to G) Relative phases for each component in (A) to (C), determined from three separate experiments, are shown by black dots, with the average denoted by a white vertical line, and the peak of KaiC~P is referenced to CT14 (9). The width of each ellipse is ±2 hours centered at the peak. Full dataset is available in data S1.
Fluorescently labeled synthetic double-stranded DNA representing the promoter sequence of the kaiBC operon, PkaiBC, was used as a representative RpaA target (20), and each labeled species was monitored in separate but parallel clock reactions (table S4). Control experiments were performed to verify the specific binding events that cause changes in FA of each labeled species (figs. S2 and S3), assigning their corresponding peaks to known interactions and circadian properties.
We used a multimode microplate reader to monitor in vitro oscillations in parallel, with individual reactions containing 50 to 100 nM of a single fluorescently labeled species for each protein or DNA monitored, as well as physiological concentrations (20) of each unlabeled clock protein. This method allowed measurement of FA for each clock component in a single plate and in real time over a duration of several days.
The data from these experiments were fit to a damped cosine function to extract period and phase relative to maximal KaiC phosphorylation, which was referenced to its peak at circadian time (CT) 14 in vivo (9), with CT 0 and CT 12 representing subjective dawn and dusk, respectively, under constant lab conditions. Combining FA measurements with traditional time courses of RpaA and KaiC phosphorylation rhythms measured in parallel by gel electrophoresis (Fig. 1 and figs. S4 and S5), we present a comprehensive view of a fully reconstituted IVC.
KaiC adenosine triphosphatase (ATPase) activity was monitored in real time by proton nuclear magnetic resonance (1H-NMR) imaging (Fig. 1D and fig. S6) (28) using the core oscillator to avoid conflation with ATP consumption by the output kinases. We found that the ATPase activity of KaiC peaks during subjective midday with the hydrolysis of ~13 to 18 ATPs per KaiC monomer per day but decreases to a nadir of ~7 or 8 ATPs per KaiC monomer per day during subjective night, which is consistent with earlier studies (29). Using this information, we next established the phase relationships of all the clock components under each IVC condition (Fig. 1, E to G). We found that the FA of RpaA increased when it was dephosphorylated or interacting with SasA in the absence of KaiC and that it decreased when RpaA was phosphorylated by CikA or a SasA-KaiC complex (fig. S3), which explains the observed phase reversal between peak RpaA anisotropy and RpaA phosphorylation.
Exploring clock output by SasA and CikA
Transcriptional output in cyanobacterial circadian rhythms is based on the antagonistic activity of two sensor histidine kinases; SasA is a KaiC-dependent kinase that phosphorylates RpaA to stimulate DNA binding and activate gene expression at dusk, whereas CikA is stimulated to act as a phosphatase that dephosphorylates RpaA when it associates with the Kai complex at night (16, 17, 19, 21, 30). To better understand the activities of each sensor histidine kinase in clock output, we first studied the reconstituted clock under conditions where only one was present at a time. In an IVC that contained KaiA, KaiB, KaiC, and SasA alone with RpaA and DNA (Fig. 1A), the relative phases of the protein interaction rhythms (Fig. 1E) were largely consistent with those reported in vivo (31). For example, FA rhythms of KaiA, KaiB, and SasA peaked from CT 15 to 22, indicating the formation of large protein complexes during subjective night owing to interaction with KaiC. These proteins also form large complexes in vivo around Zeitgeber time (ZT) 16 to 22 (32), where ZT 0 and ZT 12 correspond to dawn and dusk, respectively, under a light-dark cycle. Although SasA and KaiB bind to overlapping sites on KaiC (33), we observed that the rhythms of SasA binding to KaiC peak ~3 hours earlier than those of KaiB (Fig. 1, A and E). In principle, this phase difference could provide a temporal window for the KaiC-dependent activation of SasA autophosphorylation (20), allowing the phosphoryl transfer from SasA to RpaA (19) that is observed around subjective dusk in the IVC (Fig. 1E). RpaA dephosphorylation in the SasA-only IVC is likely due to a combination of the inherent autophosphatase activity typically seen in response regulators (34, 35) and weak phosphatase activity of SasA (21).
When SasA was replaced by CikA in the IVC, we still saw circadian rhythms in the interaction of KaiA and KaiB with KaiC in the same relative phase with KaiC phosphorylation (Fig. 1, B and F), although the period was ~3 hours shorter. The FA of CikA peaked in phase with KaiB, consistent with recruitment of the pseudo-receiver (PsR) domain of CikA to the KaiB-KaiC complex in the subjective night (36). We verified that formation of this complex stimulates the phosphatase activity of CikA toward RpaA and that CikA can act as an RpaA kinase when not associated with the KaiB-KaiC complex (figs. S7 and S8), as previously reported (21, 37). The bifunctional switching of CikA between phosphatase and kinase in the IVC likely causes the phase advance in the rhythms of RpaA phosphorylation and DNA binding relative to the SasA-only IVC by ~5 hours (Fig. 1F and fig. S9). CikA levels in vivo are normally low under high-intensity light (38) (fig. S10, B and C), suggesting that it may be limited to acting predominantly as a phosphatase at night. However, ΔsasA strains generate circadian rhythms in low light (16), suggesting that environmental conditions that favor CikA stability can rescue circadian rhythms. We demonstrated robust CikA-dependent circadian rhythms in strains that lack SasA but still express CikA and in which KaiB and KaiC were boosted to wild-type (WT) levels by circumventing autoregulation; notably, these rhythms occurred with a phase advance similar to that of the CikA-only IVC (fig. S10A). Taken with the observation that ΔsasAΔcikA strains are arrhythmic (39), these data suggest that the circadian rhythms observed in ΔsasA strains arise from a CikA-mediated regulatory circuit on RpaA. When CikA is present constitutively in the IVC, it can drive rhythms of RpaA phosphorylation both in the absence (Fig. 1B) and presence of SasA (Fig. 1C) to push the phase of peak DNA binding to midday instead of dusk (Fig. 1, F and G). Thus, the IVC recapitulates to a reasonable extent the relative in vivo phases of the circadian clock of S. elongatus and enables aspects of the clock—input (40, 41), oscillator (6), and output (20, 21, 39, 42)—to be measured under defined conditions with multiple readouts simultaneously in real time, including the phosphorylation-dependent interactions of RpaA (fig. S11) with a cognate promoter element in DNA.
Leveraging the extended oscillator to study clock mechanisms
Reconstitution of the cyanobacterial clock offers the possibility to ask questions about how downstream signal transduction components are coupled to the KaiABC oscillator to provide deeper mechanistic insight into circadian biology. First, we sought to explore the mechanistic origins of clock disruption in an arrhythmic S. elongatus strain that was originally reported as a crm1 transposon-insertion allele (43). However, in addition to disruption of crm1, we also noted a single-nucleotide polymorphism in the rpaA gene located immediately downstream, resulting in an Arg121→Gln (R121Q) amino acyl substitution in RpaA. When we reconstructed the allele of rpaA-R121Q in a WT reporter strain by markerless CRISPR-Cas12a editing (44), it displayed the same arrhythmic phenotype (Fig. 2A), demonstrating that mutation of rpaA, not crm1, disrupted clock function. Despite having a WT RpaA phosphorylation pattern (Fig. 2B and fig. S12), the mutant strain did not produce rhythmic gene expression (Fig. 2A). We used the IVC to test the hypothesis that the RpaA-R121Q variant is deficient in DNA binding, even in its phosphorylated form. Although we observed circadian rhythms of FA (fig. S13) and phosphorylation (Fig. 2B and fig. S14) in the mutant that were similar to WT RpaA, the R121Q mutant bound poorly to the PkaiBC promoter, as indicated by the low amplitude of DNA FA (Fig. 2C). Thus, the IVC helped to identify the mechanism of clock disruption by the R121Q amino acyl substitution in RpaA—it does not hinder regulatory interactions with SasA or CikA but instead prevents the phosphorylated sensor domain from regulating its DNA binding domain.
Fig. 2. The IVC reveals a defect in DNA binding by the RpaA-R121Q mutant.
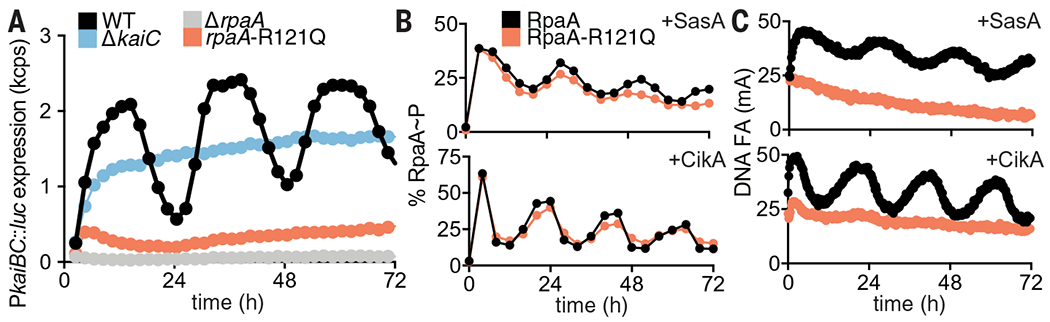
(A) Rhythms of bioluminescence generated by PkaiBC::luc expression were monitored for entrained WT (black), ΔkaiC (blue), ΔrpaA (gray), and rpaA-R121Q (salmon) cyanobacterial strains. Each plot represents an average of six biological replicates. kcps, kilocounts per second. (B) Rhythms of RpaA phosphorylation or (C) FA of PkaiBC DNA in IVC reactions containing either SasA or CikA, as in Fig. 1, A and B.
Effect of protein concentrations on rhythmicity and period of the clock
The core oscillator has a well-established, albeit relatively narrow, range of concentrations of KaiA, KaiB, and KaiC under which sustained biochemical rhythms can be observed in vitro (45). Using our newly developed fluorescence-based readout, we next measured how rhythms in the core oscillator depend on concentrations of KaiA and KaiB (Fig. 3). We found a notable convergence with previous studies monitoring rhythms of KaiC phosphorylation by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (45). Because the fluorescence-based oscillator can be monitored noninvasively in a 384-well format, we could follow rhythms for many days longer than is practical for the KaiC phosphorylation assay and quantify sensitivity of oscillation amplitude and period to concentrations of the oscillator components. In general, the oscillator functions well with substoichiometric concentrations of KaiA, although the period lengthened as KaiA concentration was decreased (Fig. 3, A to C, and table S5). By contrast, oscillator period was not affected by titrating KaiB, although there was a clear requirement for KaiB to be present at concentrations at least equivalent to that of KaiC (Fig. 3D).
Fig. 3. Rhythmicity and period of the oscillator depend in different ways on the concentrations of KaiA and KaiB.
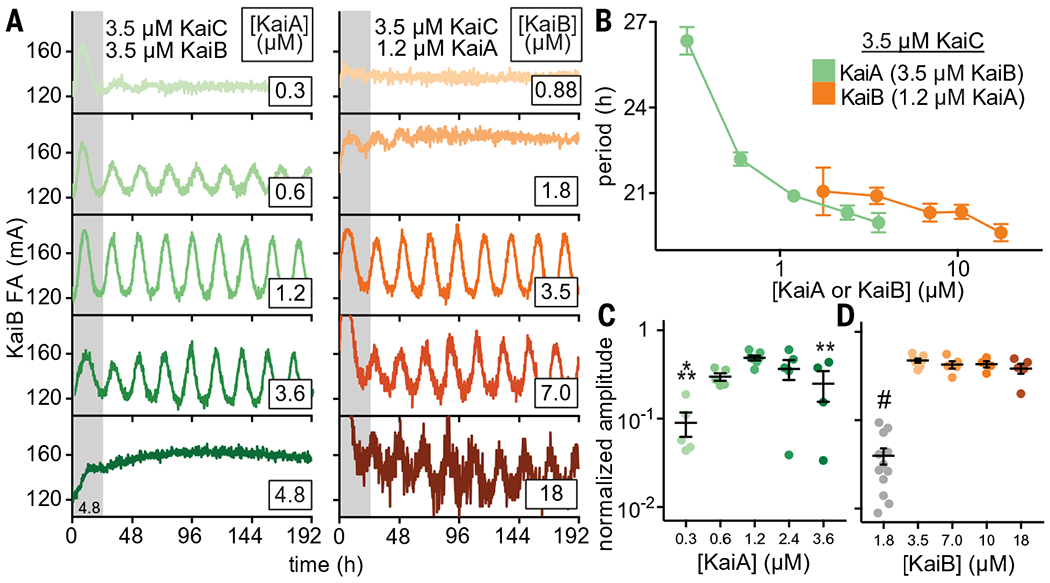
(A) In vitro oscillator reactions containing 3.5 μM KaiC, 3.5 μM KaiB (including 50 nM fluorescently labeled KaiB as a probe), and titrations of KaiA from 0.3 to 6.0 μM (green scale, left column). Representative assay from n = 3 are shown; the first 24 hours of the reactions are shaded gray. Assays were also performed with 3.5 μM KaiC, 1.2 μM KaiA, and titrations of KaiB from 0.7 to 18 μM (orange scale, right panel). FA data were fit to a cosine function to extract period (B) and amplitude (C and D) as a function of KaiA or KaiB concentration. Amplitudes for KaiB titrations were normalized to account for an increasing ratio of unlabeled KaiB to fluorescently labeled probe KaiB. Reactions that did not oscillate were not included. Data are shown as mean ± SD (n ≥ 3). One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test was used for comparison of each suboptimal condition with the 1.2 μM KaiA, 3.5 μM KaiB condition: *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.0001. Where no symbol is shown, differences were determined not to be statistically significant. Full datasets are available in data S2 and S3.
At one-half stoichiometry of KaiB relative to KaiC (1.75 μM KaiB and 3.5 μM KaiC), the oscillator functioned at low amplitude for only a few days with a normal circadian period before significantly damping to arrhythmicity (Fig. 3A). As KaiB is found in slight excess over KaiC in vivo (11, 46), it may be essential for the KaiC hexamer to recruit a full stoichiometric complement of six molecules of KaiB to stably sequester KaiA and facilitate the KaiC dephosphorylation phase in subjective night. Consistent with this model, rhythms abruptly diminished as the concentration of KaiA exceeded that of KaiC (Fig. 3A) (45, 47).
CikA rescues oscillator period under limiting concentrations of KaiA
Although CikA is primarily considered an input-output protein (17, 21, 39), it is known to affect circadian period in vivo and was recently shown to regulate the period of the core oscillator in vitro (37, 48). We previously determined that the PsR domain of CikA competes for the KaiA binding site on the KaiB-KaiC night-time complex (36). This competition likely underlies period control by CikA; by displacing KaiA from the repressive complex, CikA promotes the activating potential of KaiA to stimulate KaiC phosphorylation (48). Using our extended oscillator system, we found that increasing concentrations of CikA led to progressive period shortening, particularly under limiting concentrations of KaiA, bringing it in line with the period found under idealized oscillator conditions (Fig. 4A). This result demonstrates one way that competitive mechanisms can compensate for fluctuating in vivo concentrations of oscillator proteins to influence timekeeping by the cyanobacterial clock.
Fig. 4. Output proteins compensate for period and amplitude defects in suboptimal in vitro oscillators.
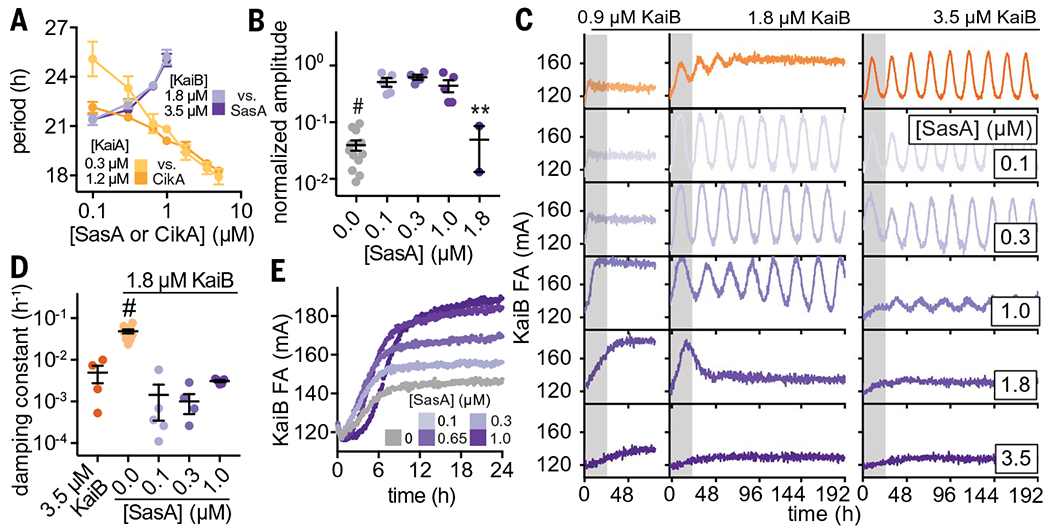
(A) Period of clock reactions with varied concentrations of KaiA (light and dark yellow) or KaiB (light and dark purple) supplemented as a function of CikA or SasA concentration, respectively. (B) Amplitudes of rhythms with probe KaiB + 1.8 μM unlabeled KaiB in the presence of SasA (gray for KaiB alone; light to dark purple for SasA). Reactions that could not be fit to a sine function because they did not oscillate were not plotted. Statistical analysis was performed exactly as described in Fig. 3, C and D. (C) In vitro oscillator assays with 3.5 μM KaiC, 1.2 μM KaiA, and the indicated concentrations of KaiB as well as various amounts of full-length SasA ranging from 0.1 to 3.5 μM. Representative assay from n ≥ 3 shown; the first 24 hours of the time course are shaded gray. (D) Damping constants derived from least-squares fitting of the FA data from (C). Full datasets are available in data S2 and S3. (E) “Partial oscillator” reactions containing 50 nM KaiB, 3.5 μM KaiC, and 1.2 uM KaiA with various amounts of SasA.
SasA rescues oscillations under limiting concentrations of KaiB
SasA was originally described as an amplifier of circadian rhythms needed to maintain robust, high-amplitude rhythms, presumably by controlling rhythmic transcription of the kaiBC cluster (16). In contrast to the effect we observed with CikA, the addition of SasA led to progressive lengthening of the period (Fig. 4A and table S5). The isolated thioredoxin-like domain of SasA was much less effective at lengthening the period than was the full-length SasA dimer, consistent with the avidity observed in binding studies between KaiC and full-length SasA (fig. S15). It should be noted that addition of the isolated PsR domain or full-length CikA caused concentration-dependent changes in period length in vitro that are apparently balanced out by period changes imparted by SasA (Fig. 4 and fig. S16). These data support a model in which the proteins that impart output contribute substantively to the timing of the clock through interactions with the Kai proteins.
In exploring the combined effects of KaiB and SasA concentrations on oscillator period, we unexpectedly observed that amplitude and damping in a weak oscillator with suboptimal concentrations of KaiB could be rescued through the addition of SasA (Figs. 4, B to D). To better understand this phenomenon, we set up assays with increasing concentrations of SasA under typical in vitro oscillator conditions (6) as well as two concentrations where KaiB was limiting relative to KaiC. When KaiB concentrations were too low to support oscillations, we observed low FA values for KaiB, consistent with incomplete recruitment of labeled KaiB to KaiC hexamers (Fig. 4C, top left column). Addition of SasA up to 1 μM increased KaiB FA, demonstrating that SasA enhances KaiB binding to KaiC. However, as the concentration of SasA approached or exceeded that of KaiC, FA values for KaiB dropped, demonstrating that SasA eventually outcompetes KaiB for binding to the KaiC hexamer, as previously observed (36, 49).
Using a KaiB concentration equal to half that of KaiC, we saw rapidly damping, low-amplitude rhythms (Fig. 4C, middle column). We also found that addition of substoichiometric amounts of SasA restored full amplitude and eliminated damping of the oscillations. Increasing SasA concentrations from 0.1 to 1 μM under these conditions did not further influence amplitude, and higher concentrations of SasA attenuated oscillations by outcompeting KaiB for binding to KaiC, as reflected by low anisotropy levels of the labeled KaiB. Under standard oscillator conditions with stoichiometric KaiB and KaiC, the amplitude of oscillation was affected by addition of SasA only at 1.0 μM and higher (Fig. 4, B and C, right column). The sharp drop in oscillator amplitude and KaiB FA levels demonstrates that a functional switch from recruitment to competition by SasA occurs at around ~0.65 to 1 μM, depending on the exact oscillator conditions. This setpoint is close to the concentration of SasA in vivo estimated by quantitative Western blotting to be around 0.6 μM (21).
We hypothesized that SasA expands the range of permissive KaiB concentrations for biochemical oscillation by directly enhancing the interaction of KaiB and KaiC. As a test, we used a partial oscillator experiment in which unlabeled KaiB was omitted from the reaction, and association of the 50 nM labeled KaiB probe was measured as phosphorylated KaiC accumulated upon stimulation by KaiA (Fig. 4E). We observed a concentration-dependent enhancement in KaiB association as SasA concentration approached the physiological value of ~0.65 μM. This finding suggests that, aside from its role in regulating transcriptional output from the oscillator (16), SasA also directly modulates KaiBC association to control formation of the nighttime repressive state and stability of the clock itself. Moreover, we observed a delay in KaiB association with KaiC when SasA was present at higher concentrations, consistent with the period-lengthening effects observed with SasA in the IVC (Fig. 4A and fig. S16).
Structural mimicry in the KaiC-binding region of SasA and KaiB
During subjective day, when KaiB is free, it exists in a distinctive ground-state fold, but the fold-switched monomer that binds KaiC at subjective night is thioredoxin-like, which is the same fold adopted by the KaiC-binding domain of SasA (SasAtrx) (37). A high-resolution crystal structure of the subcomplex comprising a single KaiC-CI domain and a fold-switch-locked mutant KaiB I88A from the related thermophilic species T. elongatus (36) (I87A in S. elongatus, referred to herein as fsKaiB) illustrates how KaiB docks onto the exposed B-loop of the CI domain (Fig. 5, A and B). To better understand the relationship between SasA and KaiB recruitment to KaiC, we solved a crystal structure of SasAtrx bound to the KaiC-CI domain from T. elongatus to reveal that SasA binds the B-loop in a similar orientation to KaiB (Fig. 5C and table S6). To investigate the importance of this interface, we probed substitutions in several KaiC residues in the B-loop using affinity assays to KaiC hexamer (Fig. 5D). We performed equilibrium binding assays using the KaiC phosphomimetic mutants KaiC-EE and KaiC-EA, where X and Y in KaiC-XY represent amino acyl residues at positions 431 and 432, respectively. Glutamyl/glutamyl (EE) and glutamyl/alanyl (EA) substitutions at these positions are widely used to approximate the dusk-like phosphoserine/phosphothreonine (pS/pT) and nighttime-like phosphoserine (pS/T) states (8). Using these mimetics, we found that KaiB has similar affinity for both states, whereas SasA has a preference for the pS/pT state (fig. S17), consistent with prior results (50) and its earlier phase of binding in the IVC (Fig. 1, A and E). Furthermore, although the isolated SasAtrx domain is necessary and sufficient for binding to KaiC (16), avidity effects in the full-length dimer enhanced affinity for KaiC-EE by at least two orders of magnitude compared with the isolated, monomeric SasAtrx domain (fig. S17).
Fig. 5. SasA and KaiB bind to the same site on KaiC.
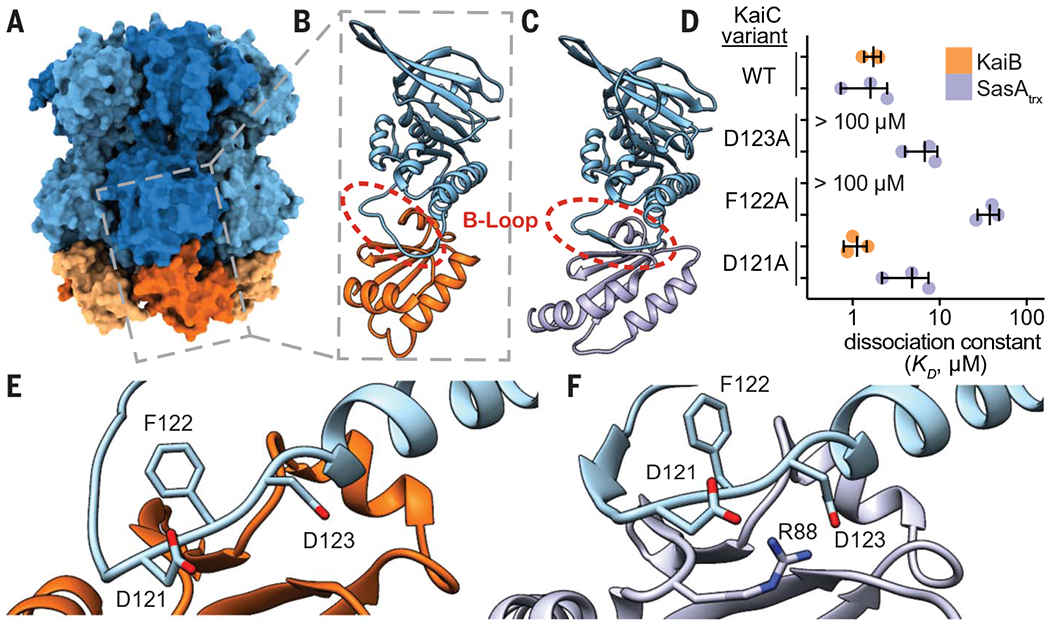
(A) The KaiB-KaiC heterododecamer [Protein Data Bank (PDB) ID 5JWQ] with subunits of the KaiC ring depicted in alternating light and dark blue, and fsKaiB subunits in alternating light and dark orange. The gray box indicates the position of (B) the isolated KaiC-CI domain-fsKaiB subcomplex (PDB ID 5JWO). (C) The KaiC-CI domain–SasAtrx subcomplex (PDB ID 6X61), with KaiC (blue) and SasA (purple). (D) Equilibrium dissociation constants (KD) for KaiB (orange) or SasAtrx (purple) binding to KaiC-EE mutants (mean ± SD, n = 3). Where indicated, binding was too weak for curve fitting, and KD is reported as >100 μM. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (E and F) The interface of KaiC-CI with fsKaiB (E) or SasAtrx (F) with key residues highlighted.
Comparing the KaiB and SasA binding interfaces on KaiC, we found that substitutions at sites conserved between S. elongatus and T. elongatus (KaiC F122 and D123) decrease affinity for both KaiB and SasAtrx (Fig. 5, D to F). We then examined how differences in the structures of KaiB and SasA or their orientations on the B-loop might influence binding to adjacent subunits of the KaiC hexamer (fig. S18). Small changes in the length and orientation of the C-terminal helix between the SasAtrx and KaiB could lead to steric clashes of SasAtrx with a neighboring subunit. Consistent with this idea, saturation binding experiments showed that SasAtrx domains cannot fully occupy all six binding sites on the KaiC hexamer (fig. S18).
SasA enhances KaiBC interaction through heterotropic cooperativity
Six monomers of the active, thioredoxin-like form of KaiB assemble onto the KaiC hexamer of S. elongatus (36, 51) to nucleate formation of the nighttime repressive state. Prior native mass spectrometry analyses demonstrated that KaiB binds to the KaiC hexamer with positive homotropic cooperativity (52, 53). Given the similarity between the SasA-KaiC and KaiB-KaiC interactions, we speculated that SasA could facilitate KaiB-KaiC interactions through positive heterotropic cooperativity. We developed an assay to test directly for cooperativity by monitoring 50 nM fluorescently labeled KaiB probe with KaiC-EE using unlabeled SasA or fsKaiB as a secondary titrant (Fig. 6, A and B). Although these assay conditions do not assess the degree of homotropic cooperativity for KaiC-EE with the KaiB probe alone, we found that low concentrations (50 to 100 nM) of SasA or fsKaiB significantly increased binding of KaiB probe, demonstrating more efficient recruitment to the KaiC hexamer. Higher concentrations of SasA or fsKaiB resulted in competition with the probe, reducing its binding to the KaiC hexamer; an equivalence point was reached when the concentration of KaiC-EE matched that of the competitor. We observed a considerable overshoot of KaiB probe anisotropy values around this point, consistent with formation of a higher-molecular-weight complex consisting of both KaiB probe and SasA bound to the KaiC hexamer. Association of the KaiB probe to the WT KaiC-CI monomer was not significantly affected by the addition of 300 nM SasA or fsKaiB (fig. S19), nor did SasA interact directly with KaiB in the absence of KaiC, demonstrating that these competitors only enhance KaiB association in the context of the KaiC hexamer, as shown using phosphomimetics, which is consistent with our cooperativity model.
Fig. 6. The thioredoxin-like fold of SasA and fsKaiB cooperatively recruits KaiB to the KaiC hexamer.
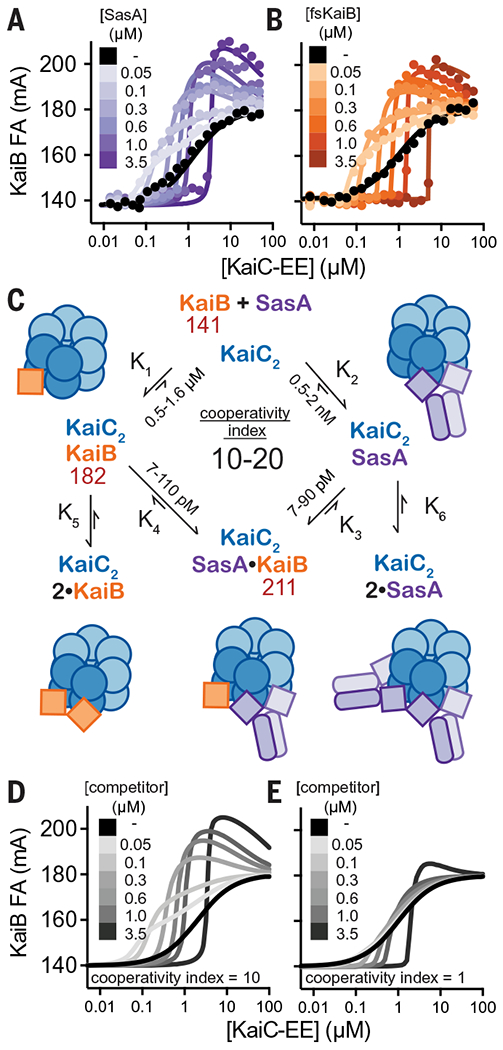
(A and B) Equilibrium binding assays of fluorescently labeled KaiB with KaiC-EE in the absence (black) or presence of (A) unlabeled SasA (light to dark purple) or (B) fsKaiB (light to dark orange). Each 2D titration is shown from representative assays with best fit lines from global curve-fitting. (C) Summary of a two-site thermodynamic model for SasA heterocooperativity, and mean equilibrium constants derived from least-squares fitting of SasA 2D titration assays (n = 3); values in red are mean peak anisotropy values observed from fitting of all SasA 2D titration datasets using DynaFit software. The cooperativity index is defined as the ratio K1/K3 = K2/K4. This model was used to generate simulated 2D titrations with a cooperativity index of either 10 (D) or 1 (E). (See data S6 to S8 for DynaFit scripts and full datasets.)
To quantitatively represent the heterotropic cooperativity observed in these two-dimensional (2D) titrations, a simplified two-site thermodynamic model was found by least-squares fitting to be sufficient to account for the data (Fig. 6C and fig. S19). In this model, we defined the heterotropic “cooperativity index” as the fold-increase in KaiB affinity given by the ratio of equilibrium constants K1/K3 (= K2/K4). Comparison of 2D titrations with SasA or fsKaiB to simulated data representing heterotropic cooperative binding (Fig. 6, A and B), to a model with competition and no cooperativity (Fig. 6, D and E), demonstrates that SasA and fsKaiB similarly influence cooperative recruitment of the KaiB probe on KaiC.
To explore the role of intermolecular KaiB-KaiB interactions in cooperative KaiB-KaiC binding, we characterized KaiB-R22A, a mutant originally identified in Anabaena (54) that reduces the apparent affinity of KaiB for KaiC although it is not located at the KaiB-KaiC interface (36). R22 has been implicated in KaiB cooperativity using native mass spectrometry (52) and is situated at the KaiB-KaiB interface in the KaiBC hexamer structure. We tested this mutant in our cooperativity assay and observed a small decrease in the cooperative recruitment of the KaiB probe relative to that of fsKaiB (fig. S19), demonstrating that intersubunit KaiB-KaiB interactions play a role in the cooperative recruitment process and opening the possibility that mutations at the SasA-KaiB interface could also disrupt cooperativity.
SasA-KaiB complementarity is required for proper circadian timing
To identify whether the enhancement of KaiBC association is important for the function of SasA in vivo, we set out to identify point mutations that would separate the cooperativity and output signaling functions of SasA. To this end, we first showed that the catalytically inactive SasA mutant H161A (19) is as effective as WT SasA in stimulating cooperative recruitment of KaiB to KaiC in vitro (fig. S19). Next, we constructed a structural model by overlaying crystal structures of SasAtrx-CI and KaiB-CI onto the lower-resolution structure of the full-length KaiBC hexamer complex (36) and used it to predict potential SasAtrx-KaiB interactions along the clockwise (CW) and counterclockwise (CCW) interfaces of the hybrid complex (Fig. 7A). Within this framework, we investigated a number of SasA substitutions near the CW and CCW interface (fig. S20) and estimated their ability to cooperatively recruit KaiB in simplified 2D titration assays comparing a single SasA concentration to the no-SasA control. The individual mutants H28A (CCW interface) or Q94A (CW interface) showed decreases in their cooperativity indices that were further decreased in the H28A-Q94A double mutant (Fig. 7B). It should be noted that the estimates of cooperativity in wild type were slightly lower than those determined in the full 2D titrations (Fig. 6, A and C), likely owing to differences in experimental setup. While the H28A-Q94A double mutant did not strongly affect other SasA functions in vitro, such as KaiC association and the KaiC-dependent phosphotransfer to RpaA (fig. S21), it lacked the ability to stimulate KaiB binding in the 2D titration assay (compare Fig. 7C and Fig. 6E) demonstrating that both interfaces contribute to SasA heterotropic cooperativity.
Fig. 7. SasA-KaiB interactions mediate cooperative recruitment of KaiB in vitro and sustain robust circadian rhythms in vivo.
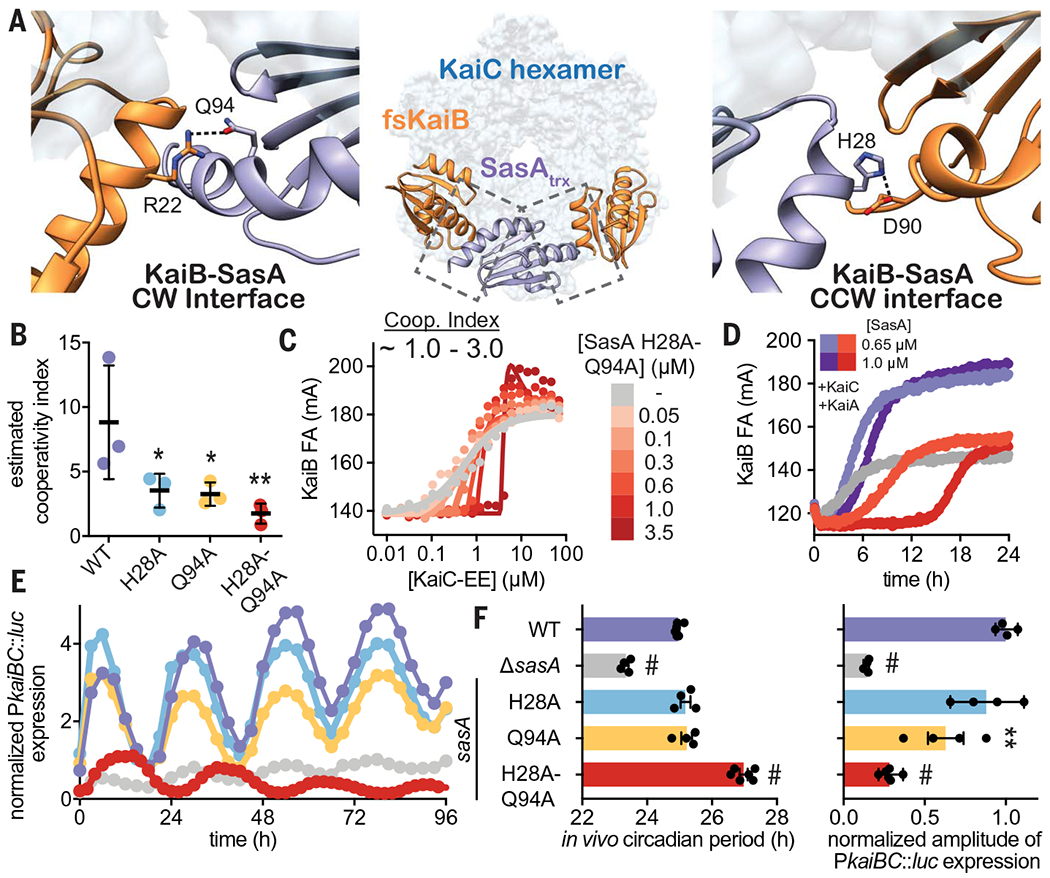
(A) Structural model of KaiB:SasA interactions on KaiC using KaiC-CI subcomplexes with fsKaiB (orange, PDB ID 5JWO) and SasAtrx (purple, PDB ID 6X61) modeled onto adjacent subunits of KaiC (light blue) of the KaiBC heterododecamer (PDB ID 5JWQ). Key residues at clockwise (CW) and counterclockwise (CCW) interfaces are modeled as S. elongatus variants on the basis of the alignment in fig. S20. Polar contacts predicted by our hybrid structural model are indicated by dashed lines. (B) Cooperativity indices for SasA variants extracted from KaiC titrations done in the presence of 0.3 μM SasA (mean SD, n = 3). Statistical significance of differences between WT and mutant SasA variants was tested using one-way ANOVA with Dunnett’s multiple comparisons as described in Fig. 3. (C) Representative 2D titration of fluorescently labeled KaiB with KaiC-EE in the absence (gray) or presence (light to dark red) of SasA-H28A-Q94A. (D) Time-dependent FA trajectories of 50 nM KaiB in the presence of 3.5 μM KaiC and 1.2 uM KaiA with 0.65 or 1.0 μM SasA WT (purple shades) or H28A-Q94A mutant (red shades). (E) Representative time course of bioluminescence driven by PkaiBC from S. elongatus cultures entrained under 12-hour light-dark cycles for 48 hours and subsequently allowed to free run in LL. (F) Raw luminescence curves were fit to a sine function to extract amplitude of PkaiBC::luc expression as well as free-running circadian period for each strain. WT control was included for each experiment, and the luminescence of each mutant was normalized to the amplitude of luminescence oscillation for the WT control in that run. Error bars depict SEM among replicate cultures (n = 4 to 6); when error bars are not visible, they were smaller than could be depicted. Symbols indicate significance determined from one-way ANOVA with Dunnett’s multiple comparisons between mutant and WT S. elongatus with symbols to depict significance as summarized in Fig. 3 legend. Normal distribution was confirmed through quantile-quantile plots showing predicted versus actual residual values in addition to Anderson-Darling, D’Agostino-Pearson omnibus, and Shapiro-Wilk tests, which were all performed in GraphPad PRISM. Equal variance of the data was verified with Brown-Forsythe tests yielding P values above 0.05. Color scheme for data in (E) is consistent with and indicated on left vertical axis of (F).
To better understand these mutations in a more native context, we tested the ability of the H28A-Q94A variant to nucleate formation of the repressive complex using a partial oscillator experiment to directly measure the kinetics of KaiBC formation under limiting concentrations of KaiB with WT or H28A-Q94A SasA (Fig. 7D and fig. S21C). Notably, enhancement of KaiB recruitment was essentially nonexistent with the SasA H28A-Q94A mutant, instead showing a lengthened competitive phase that occurs even at physiological concentrations of SasA. When concentration of the double mutant was increased beyond physiological levels, the delay in KaiB association became even more exaggerated, suggesting that complementarity between the SasA-KaiB interfaces is critical for timely replacement of SasA with KaiB on the KaiC hexamer.
With this separation-of-function mutant in hand, we set out to test whether the SasA-induced cooperativity we observed for the expanded oscillator in vitro could influence circadian rhythms in vivo. We introduced the double mutation, as well as several single alanine substitutions, into SasA using markerless CRISPR-Cas12a and monitored bioluminescence from a PkaiBC luciferase reporter in constant light (LL) after synchronization in 12-hours light:12-hours dark cycles (Fig. 7E and fig. S22). Small effects on amplitude were seen with the individual H28 or Q94 alanine substitutions on their own, but when these were combined in the H28A-Q94A mutant, the amplitude of circadian rhythms decreased to an extent that was similar to the SasA knockout strain (Fig. 7F, ΔsasA), even though the mutant proteins were expressed to a similar level in vivo (fig. S22D), and the double mutant strain exhibited a ~2-hour-longer period (Fig. 7F and fig. S22). Taken together, these results provide a clear link between loss of complementarity at SasA-KaiB interfaces and the long-period phenotype observed in vivo, further supporting our conclusion that cooperative interactions between SasA and KaiB are crucial for proper circadian timing.
Discussion
This extended IVC is an autonomously oscillating system that remains phase coherent for several days without user intervention, allows real-time observation of rhythmic behaviors of several clock components in parallel, and rhythmically activates the interaction of a transcription factor with a promoter element on DNA. Complementing this assay with parallel measurements of phosphorylation and ATPase activities rounds out a movie of an intact cyanobacterial clock. As implemented here, the IVC has revealed a set of minimal components to execute clock-controlled gene expression, helped to dissect the mechanistic origins of a clock-disrupting mutant, and provided insight into mechanisms behind the narrow stoichiometric ratios that are tolerated in the in vitro oscillator (45). In particular, our data demonstrate that SasA, previously regarded solely as an output protein, plays a critical role in the enhancing the oscillator amplitude through heterocooperative regulation of KaiB association with KaiC. This newly developed IVC system offers an unparalleled opportunity to explore how the clock maintains consistency in vivo despite rhythmic changes in the concentration of clock components that result from the associated transcription-translation feedback (11, 55) and protein turnover (56, 57) and provides an experimental platform for integrating the oscillator with the upstream and downstream components with which it interacts. By reconstructing, and subsequently deconstructing, this elegant system, we have shown that a dichotomous view of timekeeping and signaling components provides an insufficient representation of the complexity and synergy that has evolved in this ancient circadian clock.
Supplementary Material
ACKNOWLEDGMENTS
We thank staff at the 23-ID-D beamline of the Advanced Photon Source, Argonne National Laboratory, for their help with data collection. The Advanced Photon Source (contract DE-AC02-06CH11357) is supported by the US Department of Energy. Molecular graphics and analyses performed with UCSF Chimera and ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (UCSF), with support from National Institutes of Health P41-GM103311(Chimera) and R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases (ChimeraX). We thank G. Chow, Y.-G. Chang, M. Rust, L. Hong, S. Sukenik, and M. Zoghbi for helpful discussions. We also thank D. Rice and S. Grimaldi for maintaining the NMR facility and NMR cryoplatform, respectively, at UC Merced.
Funding:
This work was supported by US National Institutes of Health grants R01 GM107521 (to A.L.), R01 GM121507 (to C.L.P.), and R35 GM118290 (to S.S.G.), and NSF-CREST: Center for Cellular and Biomolecular Machines at the University of California, Merced (NSF-HRD-1547848). J.H. was supported by NSF-CREST CCBM HRD-1547848. J.G.P. was supported by NIH IMSD grant R25 GM058903-20. P.C. is supported by EMBO long-term fellowship 57-2019.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All data are available in the main text or the supplementary materials.
REFERENCES AND NOTES
- 1.Dunlap JC, Loros JJ, DeCoursey PJ, Eds., Chronobiology: Biological Timekeeping (Sinauer Associates, 2004). [Google Scholar]
- 2.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH, The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol 14,1481–1486 (2004). doi: 10.1016/j.cub.2004.08.023 [DOI] [PubMed] [Google Scholar]
- 3.Kondo T et al. , Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 90, 5672–5676 (1993). doi: 10.1073/pnas.90.12.5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishiura M et al. , Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 (1998). doi: 10.1126/science.281.5382.1519 [DOI] [PubMed] [Google Scholar]
- 5.Tomita J, Nakajima M, Kondo T, Iwasaki H, No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254 (2005). doi: 10.1126/science.1102540 [DOI] [PubMed] [Google Scholar]
- 6.Nakajima M et al. , Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005). doi: 10.1126/science.1108451 [DOI] [PubMed] [Google Scholar]
- 7.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK, Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318, 809–812 (2007). doi: 10.1126/science.1148596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiwaki T et al. , A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 26, 4029–4037 (2007). doi: 10.1038/sj.emboj.7601832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T, KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 99, 15788–15793 (2002). doi: 10.1073/pnas.222467299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams SB, Vakonakis I, Golden SS, LiWang AC, Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc. Natl. Acad. Sci. U.S.A 99, 15357–15362 (2002). doi: 10.1073/pnas.232517099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T, KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22, 2127–2134 (2003). doi: 10.1093/emboj/cdg212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Mori T, Johnson CH, Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 22, 2117–2126 (2003). doi: 10.1093/emboj/cdg168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y-G, Tseng R, Kuo N-W, LiWang A, Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. U.S.A 109, 16847–16851 (2012). doi: 10.1073/pnas.1211508109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Y-G, Kuo N-W, Tseng R, LiWang A, Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 108, 14431–14436 (2011). doi: 10.1073/pnas.1104221108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe J et al. , Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316 (2015). doi: 10.1126/science.1261040 [DOI] [PubMed] [Google Scholar]
- 16.Iwasaki H et al. , A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101, 223–233 (2000). doi: 10.1016/S0092-8674(00)80832-6 [DOI] [PubMed] [Google Scholar]
- 17.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS, CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289, 765–768 (2000). doi: 10.1126/science.289.5480.765 [DOI] [PubMed] [Google Scholar]
- 18.Ivleva NB, Gao T, LiWang AC, Golden SS, Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A 103,17468–17473 (2006). doi: 10.1073/pnas.0606639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takai N et al. , A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 103, 12109–12114 (2006). doi: 10.1073/pnas.0602955103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markson JS, Piechura JR, Puszynska AM, O’Shea EK, Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell 155, 1396–1408 (2013). doi: 10.1016/j.cell.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutu A, O’Shea EK, Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol. Cell 50, 288–294 (2013). doi: 10.1016/j.molcel.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito-Miwa K, Furuike Y, Akiyama S, Kondo T, Tuning the circadian period of cyanobacteria up to 6.6 days by the single amino acid substitutions in KaiC. Proc. Natl. Acad. Sci. U.S.A 117, 20926–20931 (2020). doi: 10.1073/pnas.2005496117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leypunskiy E et al. , The cyanobacterial circadian clock follows midday in vivo and in vitro. eLife 6, e23539 (2017). doi: 10.7554/eLife.23539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow GK et al. , Monitoring protein-protein interactions in the cyanobacterial circadian clock in real time via electron paramagnetic resonance spectroscopy. Biochemistry 59, 2387–2400 (2020). doi: 10.1021/acs.biochem.0c00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heisler J, Chavan A, Chang Y-G, LiWang A, Real-time in vitro fluorescence anisotropy of the cyanobacterial circadian clock. Methods Protoc. 2, 42 (2019). doi: 10.3390/mps2020042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theile CS et al. , Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc 8, 1800–1807 (2013). doi: 10.1038/nprot.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guimaraes CP et al. , Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc 8,1787–1799 (2013). doi: 10.1038/nprot.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo B et al. , Monitoring ATP hydrolysis and ATPase inhibitor screening using 1H NMR. Chem. Commun 50, 12037–12039 (2014). doi: 10.1039/C4CC04399E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terauchi K et al. , ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 104, 16377–16381 (2007). doi: 10.1073/pnas.0706292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith RM, Williams SB, Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A 103, 8564–8569 (2006). doi: 10.1073/pnas.0508696103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SE, Golden SS, Circadian rhythms in cyanobacteria. Microbiol Mol. Biol. Rev 79, 373–385 (2015). doi: 10.1128/MMBR.00036-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kageyama H, Kondo T, Iwasaki H, Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J. Biol. Chem 278, 2388–2395 (2003). doi: 10.1074/jbc.M208899200 [DOI] [PubMed] [Google Scholar]
- 33.Tseng R et al. , Cooperative KaiA-KaiB-KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J. Mol. Biol 426, 389–402 (2014). doi: 10.1016/j.jmb.2013.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Straughn PB et al. , Modulation of response regulator CheY reaction kinetics by two variable residues that affect conformation. J. Bacteriol 202, e00089–20 (2020). doi: 10.1128/JB.00089-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West AH, Stock AM, Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci 26, 369–376 (2001). doi: 10.1016/S0968-0004(01)01852-7 [DOI] [PubMed] [Google Scholar]
- 36.Tseng R et al. , Structural basis of the day-night transition in a bacterial circadian clock. Science 355, 1174–1180 (2017). doi: 10.1126/science.aag2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y-G et al. , A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349, 324–328 (2015). doi: 10.1126/science.1260031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS, LdpA: A component of the circadian clock senses redox state of the cell. EMBO J. 24, 1202–1210 (2005). doi: 10.1038/sj.emboj.7600606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T, Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A 107, 3263–3268 (2010). doi: 10.1073/pnas.0909924107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rust MJ, Golden SS, O’Shea EK, Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331, 220–223 (2011). doi: 10.1126/science.1197243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim Y-I, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS, Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc. Natl. Acad. Sci. U.S.A 109, 17765–17769 (2012). doi: 10.1073/pnas.1216401109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paddock ML, Boyd JS, Adin DM, Golden SS, Active output state of the Synechococcus Kai circadian oscillator. Proc. Natl. Acad. Sci. U.S.A 110, E3849–E3857 (2013). doi: 10.1073/pnas.1315170110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyd JS, Bordowitz JR, Bree AC, Golden SS, An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc. Natl. Acad. Sci. U.S.A 110, 13950–13955 (2013). doi: 10.1073/pnas.1312793110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ungerer J, Pakrasi HB, Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci. Rep 6, 39681 (2016). doi: 10.1038/srep39681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakajima M, Ito H, Kondo T, In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett. 584, 898–902 (2010). doi: 10.1016/j.febslet.2010.01.016 [DOI] [PubMed] [Google Scholar]
- 46.Chew J, Leypunskiy E, Lin J, Murugan A, Rust MJ, High protein copy number is required to suppress stochasticity in the cyanobacterial circadian clock. Nat. Commun 9, 3004 (2018). doi: 10.1038/s41467-018-05109-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J, Chew J, Chockanathan U, Rust MJ, Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A 111, E3937–E3945 (2014). doi: 10.1073/pnas.1408692111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaur M, Ng A, Kim P, Diekman C, Kim Y-I, CikA modulates the effect of KaiA on the period of the circadian oscillation in KaiC phosphorylation. J. Biol. Rhythms 34, 218–223 (2019). doi: 10.1177/0748730419828068 [DOI] [PubMed] [Google Scholar]
- 49.Murakami R et al. , The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J. Biol. Chem 287, 29506–29515 (2012). doi: 10.1074/jbc.M112.349092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valencia S J. et al. , Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells 17, 398–419 (2012). doi: 10.1111/j.1365-2443.2012.01597.x [DOI] [PubMed] [Google Scholar]
- 51.Snijder J et al. , Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science 355, 1181–1184 (2017). doi: 10.1126/science.aag3218 [DOI] [PubMed] [Google Scholar]
- 52.Murakami R et al. , Cooperative binding of KaiB to the KaiC hexamer ensures accurate circadian clock oscillation in cyanobacteria. Int. J. Mol. Sci 20, 4550 (2019). doi: 10.3390/ijms20184550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snijder J et al. , Insight into cyanobacterial circadian timing from structural details of the KaiB-KaiC interaction. Proc. Natl. Acad. Sci. U.S.A 111, 1379–1384 (2014). doi: 10.1073/pnas.1314326111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garces RG, Wu N, Gillon W, Pai EF, Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 23, 1688–1698 (2004). doi: 10.1038/sj.emboj.7600190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Mori T, Johnson CH, Circadian clock-protein expression in cyanobacteria: Rhythms and phase setting. EMBO J. 19, 3349–3357 (2000). doi: 10.1093/emboj/19.13.3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holtman CK et al. , High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 12, 103–115 (2005). doi: 10.1093/dnares/12.2.103 [DOI] [PubMed] [Google Scholar]
- 57.Imai K, Kitayama Y, Kondo T, Elucidation of the role of Clp protease components in circadian rhythm by genetic deletion and overexpression in cyanobacteria. J. Bacteriol 195, 4517–4526 (2013). doi: 10.1128/JB.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


