Abstract
Saccharomyces cerevisiae mutants lacking the structure-specific nuclease Rad27 display an enhancement in recombination that increases as sequence length decreases, suggesting that Rad27 preferentially restricts recombination between short sequences. Since wild-type alleles of both RAD27 and its human homologue FEN1 complement the elevated short-sequence recombination (SSR) phenotype of a rad27-null mutant, this function may be conserved from yeast to humans. Furthermore, mutant Rad27 and FEN-1 enzymes with partial flap endonuclease activity but without nick-specific exonuclease activity partially complement the SSR phenotype of the rad27-null mutant. This suggests that the endonuclease activity of Rad27 (FEN-1) plays a role in limiting recombination between short sequences in eukaryotic cells.
Dispersed, short (less than 300 bp), repetitive DNA sequences are a dominant feature of all eukaryotic genomes (8). Recombination between these sequences creates a variety of genome rearrangements (45, 61) and can cause disease in humans (12). Discouraging recombination between short sequences enhances genome stability by reducing the incidence of these rearrangements.
In Saccharomyces cerevisiae and mammalian cells, short repeats recombine less frequently per unit length than longer sequences (25, 46, 57), while sequences below 30 bp recombine extremely poorly (35). Recent discoveries suggest a genetic basis for the limitation of short-sequence recombination (SSR) in yeast. Novel mutations in the RAD3, SSL1, and SSL2 genes, which encode components of the nucleotide excision repair (NER) apparatus and transcription factor TFIIH (14, 69), disrupt the control of SSR and increase genome rearrangement (2, 3, 29, 34). Genome instability may be due to slow removal of sequences at the ends of broken DNA molecules by an unidentified nuclease or nucleases. The persistence of these sequences increases the likelihood that they will recombine with other sequences in the genome. Interestingly, these factors also influence Ty1 retrotransposition, which gives rise to distinct genome rearrangements (28, 29).
A search for nucleases that affect SSR led us to examine the effects of null alleles of several structurally related nuclease genes in yeast. Previously, it was shown that Rad2, a NER endonuclease (13, 20), does not play a role in SSR (2). Similarly, in this report we describe evidence that Exo1, a 5′-3′ double-stranded exonuclease that plays a role in homologous recombination, mismatch repair, and the repair of UV-damaged DNA (16, 41, 54, 63, 66), and Din7, a mitochondrial protein that affects mitochondrial DNA stability (15), do not significantly affect SSR.
Rad27, a fourth, related protein, is a structure-specific endo- and exonuclease homologous to the human FEN-1 enzyme (5, 21, 31, 42, 51, 55, 71). Yeast cells carrying a null allele of the RAD27 gene display a broad array of defects in genome stability, including an elevated spontaneous mutation rate (64), expansion and contraction of micro- and minisatellite sequences (19, 26, 27, 48, 56), telomeric repeat fluctuation (40), increased spontaneous recombination (19, 32, 55, 64, 67), and increased chromosome breakage (19, 67). These are attributed to a failure in the removal of RNA residues from the 5′ ends of Okazaki fragments during lagging-strand DNA synthesis (19, 64, 67). Importantly, this biochemical defect also affects the repair of DNA double-strand breaks (DSBs) by homologous recombination (22).
Interestingly, a variety of genome rearrangements involving very short (3 to 12 bp) repeats have been observed in rad27-null mutant cells but not in wild-type cells (10, 64). These rearrangements could result from aberrant repair of DSBs by homologous recombination. The lethal effect of blocking homologous recombination in rad27 mutant cells indicates that DSB repair is critical for their survival (59, 64).
In this report we show that Rad27 inhibits the insertion of DNA fragments into the genome, a model for the repair of broken chromosomes (36, 43). Similar to recombination in the rad3, ssl1, and ssl2 mutants described previously (2, 3, 29, 34), SSR was stimulated severalfold in the rad27-null mutant, while recombination between long sequences was unaffected.
Complementation of the elevated SSR phenotype by FEN1 suggests that this function may be conserved from yeast to humans. Interestingly, mutant alleles of FEN1 and RAD27 that encode proteins with partial endonuclease activity but no exonuclease activity partially complement the elevated SSR of the rad27 mutant, indicating that endonuclease activity is required to limit SSR. Further, we show that Rad27 is required to insert DNA fragments with nonhomologous sequences at their ends into genomic sequences by SSR, suggesting that Rad27 plays a role in processing recombining molecules. These results suggest that Rad27 may play a role in maintaining genome stability by cleaving and destabilizing intermediates that are important determinants of the efficiency of SSR.
MATERIALS AND METHODS
Yeast strains and plasmids.
Plasmids used in this study were constructed using established methods (47) and are listed in Table 1. Plasmids and DNA fragments were introduced into yeast by electroporation. All yeast strains used in this study were isogenic with W303-1A (62) and are listed in Table 2. The rad5-535 allele was found to have no effect on the recombination assays performed in this study (unpublished data). Strain construction and maintenance followed established procedures (52). The construction of the rad27::LEU2 allele was described previously (18). The LEU2 marker in this allele was replaced (44) with the hisG::URA3::hisG universal disruptor (1) by transforming the strain IC2-1 (18) with a 4.9-kb XhoI/SalI leu2::hisG::URA3::hisG fragment from pLAY266. Ura+ Leu− transformants were plated on 5-fluoroorotic acid medium (7) to create the rad27::leu2::hisG allele, which was confirmed on Southern blots (unpublished data). The exol::hisG::URA3::hisG allele was constructed by transforming W961-5A (34) with a 4.6-kb XhoI/XbaI exo1::hisG::URA3::hisG fragment from pLAY281. The exo1::hisG allele was obtained by 5-fluoroorotic acid selection and confirmed by Southern blot (unpublished data). The din7::KAN-MX allele was constructed by transforming W1011-3B to G418 resistance (200 μg/ml) with a PCR-generated DNA fragment carrying the KAN-MX gene (68) bordered by DIN7 sequences. The primers 5′-CAATTAAAGAGAATTCAAAAACAGGTG TCCCTGAAAAAATACATGTATCAAACACGTACGCTGCAGGTCGA C-3′ and 5′-CTCCCTCTCCGATAACACGTCCTGCGTATCCACTAGCGGTTGCTCCACTTTCTTATCGATCAATTCGAGCTCG-3′ were used to amplify KAN-MX sequences from pFA6a-kanMX4 (68). To construct the his3::ura3::LEU2 allele, a 2.0-kb HpaI/SmaI LEU2 clone was inserted into StuI- and ApaI-digested pLAY144 (2), bisecting the URA3 gene in the his3::URA3 segment and creating pLAY315. A 3.2-kb HindIII fragment from pLAY315 containing the his3::ura3::LEU2 segment was used to transform W961-5A to leucine prototrophy. Leu+ His− transformants were confirmed to have replaced the wild-type HIS3 allele with the his3::ura3::LEU2 allele by Southern blotting (unpublished data). The sam2::his3::TRP1 allele was generated by transforming W961-5A to tryptophan prototrophy with a PCR-generated DNA fragment carrying the TRP1 gene flanked by 91 and 36 bp of HIS3 coding sequence bordered on both sides by 45 bp of the SAM2 coding sequence. The oligonucleotides 5′-GGTCCTCAAGGTGACGCTGGTTTGACCGTAGAAAGATTATTGTCAAGCTTTTAAA ATAGGCCC-3′ and 5′-GGAGAAGGCACCACCACCGACGGATGAGGCACCACCGTAAGCGTCAAGCTTTCAAGAAAATGC-3′ were used to amplify the his3::TRP1 sequence from pLAY365. Trp+ transformants were confirmed to have the sam2::his3::TRP1 allele by Southern blotting (unpublished data).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pRS414 | Yeast centromere plasmid containing TRP1 selectable marker | 53 |
| pFA6a-kanMX4 | Plasmid containing KAN-MX selectable marker | 68 |
| pET-FEN-1 | 1.2-kb FEN1 cDNA in pET28-b for overexpression of FEN-1 | 18 |
| pET-fen-1-E160D | 1.2-kb fen1-E160D cDNA in pET28-b for overexpression of FEN-1-E160D | 18 |
| pBS-RAD27 | 1.2-kb RAD27 PCR clone in pBluescript | This study |
| pET-RAD27 | 1.2-kb RAD27 sequence replaces FEN1 sequence in pET-FEN-1 for overexpression of Rad27 | This study |
| pET-rad27-E158D | 1.2-kb rad27-E158D sequence from in vitro-mutagenized pBS-RAD27 in pET28-b for overexpression of Rad27-E158D | This study |
| pLAY144 | 1.1-kb BamHI URA3 sequence replacing 60-bp BglII segment of 1.3-kb HIS3 sequence | 2 |
| pLAY266 | 3.8-kb BglII/BamHI hisG::URA3::hisG fragment replacing 1.2-kb HpaI/EcoRV fragment of LEU2 sequence in pUC18-LEU2 | Glenn Manthey |
| pLAY281 | 3.8-kb BglII/BamHI hisG::URA3::hisG fragment replaces 1.2-kb DraI segment of 2.0-kb EX01 sequence | This study |
| pLAY298 | 3.2-kb SacI fragment carrying the wild-type FEN1 coding sequence under the control of the yeast ADH1 promoter and terminator inserted into the polylinker of pRS414 | This study |
| pLAY303 | 1.5-kb ApaI fragment containing the fen1-E160D mutation swapped for wild-type FEN1 sequence in pLAY298 | This study |
| pLAY315 | 2.0-kb HpaI/SmaI LEU2 sequence replaces 60 bp of URA3 coding sequence between ApaI and StuI sites in pLAY144 | This study |
| pLAY327 | 1.2-kb PCR-generated RAD27 coding sequence with NotI ends swapped with the 1.2-kb NotI fragment carrying the FEN1 coding sequence in pLAY298 | This study |
| pLAY362 | 720-bp ClaI/BstXI fragment carrying rad27-E158D mutation swapped for wild-type sequence in pLAY327 | This study |
| pLAY365 | 1.5-kb BamHI TRP1 sequence replacing 60-bp BglII segment of 1.3-kb HIS3 sequence | This study |
TABLE 2.
Yeast strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| W961-5A | MATaHIS3 | 34 |
| W1011-3B | MATα HIS3 | John McDonald |
| IC2-1 | MATaHIS3 rad27::LEU2 | 18 |
| ABX367 | MATa/α HIS3/his3::ura3::LEU2 | This study |
| ABX382 | MATa/α HIS3/his3::ura3::LEU2 rad27::leu2::hisG/rad27::leu2::hisG | This study |
| ABX451 | MATa/α HIS3/his3::ura3::LEU2 din7::KAN-MX/din7::KAN-MX | This study |
| ABX456 | MATa/α HIS3/his3::ura3::LEU2 exo1::hisG/exo1::hisG | This study |
| ABX457-2A | MATα his3::ura3::LEU2 sam2::his3::TRP1 | This study |
| ABX457-1B | MATα his3::ura3::LEU2 sam2::his3::TRP1 rad27::leu2::hisG | This study |
| ABT301 | Same as ABX382, with addition of pLAY298 (FEN1) | This study |
| ABT302 | Same as ABX382, with addition of pLAY327 (RAD27) | This study |
| ABT303 | Same as ABX382, with addition of pLAY303 (fen1-E160D) | This study |
| ABT306 | Same as ABX382, with addition of pRS414 (empty plasmid) | This study |
| ABT344 | Same as ABX382, with addition of pLAY362 (rad27-E158D) | This study |
All strains are isogenic to W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 rad5-535) (61). Only deviations from this genotype are listed.
Cloning and in vitro mutagenesis of RAD27 and FEN1.
The RAD27 coding sequence was amplified by PCR from yeast genomic DNA using the oligonucleotides 5′-CCCCGGGCCCGCGGCCGCATGGGTATTAAAGGTTTG-3′ and 5′-CCCCGGGCCCGCGGCCGCTCATCTTCTTCCCTTTGT-3′. These primers include NotI (underlined) sequences for cloning. The 1.2-kb RAD27 PCR product was digested with NotI and inserted in place of the FEN1 gene in pLAY298 to create pLAY327 for expression in yeast. This plasmid puts RAD27 under the control of the ADH1 promoter and terminator and possesses a centromere and a TRP1 selectable marker. RAD27 was also amplified using the oligonucleotides 5′-ACAGCACGCGGCCGCCATGGGTATTAAAGGTTTGA-3′ and 5′-ACCTAGGAGCGGCCGCTTACTCGAGTCTTCTTCCCTTTGTGACT-3′, which include NotI (underlined) and NcoI or XhoI (in boldface) sequences. NcoI- and XhoI-digested RAD27 PCR product was cloned into NcoI- and XhoI-digested pET28b and pBluescript to create pET-RAD27 for overexpression in Escherichia coli and pBS-RAD27 for in vitro mutagenesis. pBS-RAD27 was mutagenized using the RAPID PCR site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and the oligonucleotides 5′-CCAACGGAAGCTGATGCTCAATGTGC-3′ and 5′-GCACATTGAGCATCAGCTTCCGTTGG-3′ (substituted codon in bold). The rad27-E158D mutation was verified by DNA sequencing. A 1.2-kb NcoI/XhoI rad27-E158D fragment was cloned into pET28b to make pET-rad27-E158D and to express Rad27-E158D in E. coli. A 710-bp ClaI/BstXI rad27-E158D fragment was swapped with wild-type sequences in pLAY327 to make pLAY362 and to express Rad27-E158D in yeast. Wild-type FEN-1 was overexpressed in E. coli cells from the plasmid pET-FEN-1 (18). pET-fen-1-E160D (18) was used for the overexpression of FEN-1-E160D. Wild-type and mutant FEN1 genes under the control of the ADH1 promoter and terminator from pDB20-FEN-1 and pDB20-FEN-1-E160D (18) were inserted into pRS414 to create pLAY298 and pLAY303 and to express the proteins in yeast.
Measurement of flap endonuclease and nick exonuclease activities, enzyme kinetics, and substrate flap-length effects of wild-type and mutant proteins.
Protein overexpression and purification from E. coli extracts were carried out as described previously (18, 49, 50). Purity of the preparations was assessed on Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gradient gels. 5′ flap and nicked double-strand substrates were prepared as described in Hosfield et al. (23). Flap endonuclease assays were performed in a volume of 13 μl with 0.5 to 25.0 pmol of 32P-labeled flap substrate, TM buffer (10 mM Tris [pH 8.0], 10 mM MgCl2), and 5 to 40 nmol of wild-type or mutant enzyme. The substrates for the flap-length dependence assay are described in Fig. 3. Substrates were synthesized by the DNA synthesis center of the City of Hope National Medical Center and purified on 15% acrylamide gels as described by Frank et al. (18). Seventy-five nanomoles of FEN-1 was used for all of the flap-length dependence assays. All reactions were incubated at 30°C for 10 min and then quenched with an equal volume of stop solution (U.S. Biochemical Corp., Cleveland, Ohio). The products were resolved on denaturing 15% acrylamide gels and visualized by autoradiography. The percentage of substrate cleaved was determined using the IPLabGel program and was converted to substrate concentration cleaved per unit time as described previously (18). Vmax and Km values were determined from double-reciprocal plots of product formed versus substrate concentration (data not shown).
FIG. 3.
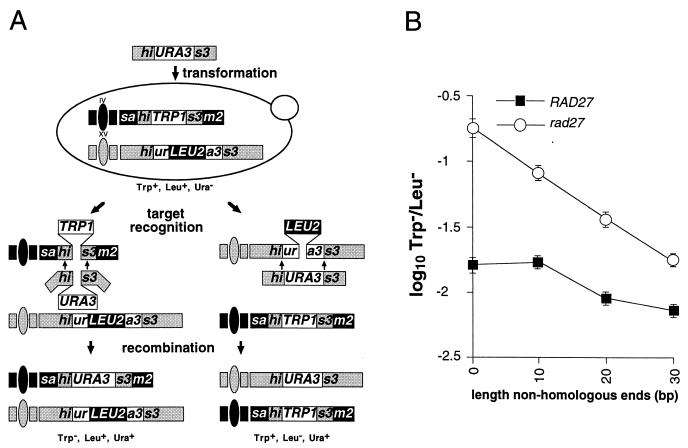
Relationship between length of nonhomologous DNA sequences at ends of substrate DNA fragments and their integration into genomes of wild-type and rad27-null mutant cells. (A) DNA fragment insertion assay involving nonhomologous terminal sequences. DNA fragments comprised of the URA3 gene bordered by various lengths of HIS3 sequence were introduced into haploid cells by electroporation. The two relevant recombination targets are the previously described his3::ura3::LEU2 allele at the HIS3 locus on chromosome XV and the sam2::his3::TRP1 allele at the SAM2 locus on chromosome IV. All of the sequences on the fragments can align with the his3::ura3::LEU2 allele, whereas only the HIS3 sequences can align with the sam2::his3::TRP1 allele. All of the HIS3 sequences on the smallest his3::URA3 fragment (127 bp) can align with the HIS3 sequences in the sam2::his3::TRP1 allele (not pictured), while the +10, +20, and +30 fragments have an additional 10, 20, or 30 bp of HIS3 sequence at both ends that cannot. These nonhomologous ends must be removed for insertion into the sam2::his3::TRP1 allele to occur. Insertion into the his3::ura3::LEU2 allele deletes the 2.0-kb LEU2 sequence, which results in a Trp+ Leu− Ura+ cell, while insertion into the sam2::his3::TRP1 allele deletes the 1.5-kb TRP1 sequence and creates a Trp− Leu+ Ura+ cell. (B) Plot of the ratio of DNA fragment insertions into the sam2::his3::TRP1 and his3::ura3::LEU2 targets versus length of terminal HIS3 sequence on the fragment that is not homologous to sam2::his3::TRP1 in wild-type and rad27 mutant cells. The four different his3::URA3 fragments were separately introduced into wild-type (ABX457-2A) and rad27 mutant (ABX457-1B) cells by electroporation. Ura+ transformants were replica plated to media lacking tryptophan or leucine to determine whether the fragments had inserted into the sam2::his3::TRP1 allele or the his3::ura3::LEU2 allele. The mean ratios of Trp− Leu+ Ura+ to Trp+ Leu− Ura+ transformants (T−/L−) obtained with the four DNA fragments were determined from a minimum of five independent trials. Log10 values of the mean ratios ± two standard errors were plotted against the length of the nonhomologous sequences at the ends of the fragment.
DNA fragment insertion assay.
DNA fragments with different lengths of HIS3 sequence flanking the URA3 gene were prepared by restriction endonuclease digestion of pLAY144 (2) or by PCR. Phenol-extracted and gel-purified DNA fragments were used to transform diploid cells by electroporation, selecting for expression of URA3. Transformation efficiency differed by less than 3.5-fold in wild-type and rad27 mutant cells.
Recombination between the his3::URA3 fragments and several different genomic sequences (Fig. 1) was distinguished by replica plating to appropriate drop-out media and confirmed by Southern blotting DNA from selected transformants (unpublished data). Recombination at the HIS3 allele replaces 60 bp of HIS3 coding sequence with the 1.1-kb URA3 marker and results in His− Leu+ Ura+ transformants. Recombination at the his3::ura3::LEU2 allele replaces a 2-kb LEU2 marker with 60 bp of URA3 coding sequence and results in His+ Leu− Ura+ transformants. Recombination at either of the ura3-1 alleles replaces a point mutation in the URA3 coding sequence by gene conversion and results in His+ Leu+ Ura+ transformants. The fraction of transformants that were due to gene conversion at the ura3-1 alleles remained between 50 and 60% in both the wild type and the rad27 mutant, regardless of the DNA fragment used. However, the ratio of recombination events at the HIS3 allele versus the his3::ura3::LEU2 allele varied significantly with changes in the length of the HIS3 sequences on the DNA fragments. The mean ratio of insertions at HIS3 versus his3::ura3::LEU2 (H−/L−) for each strain was determined from at least five trials. Each trial consisted of a minimum of 400 Ura+ transformants.
FIG. 1.
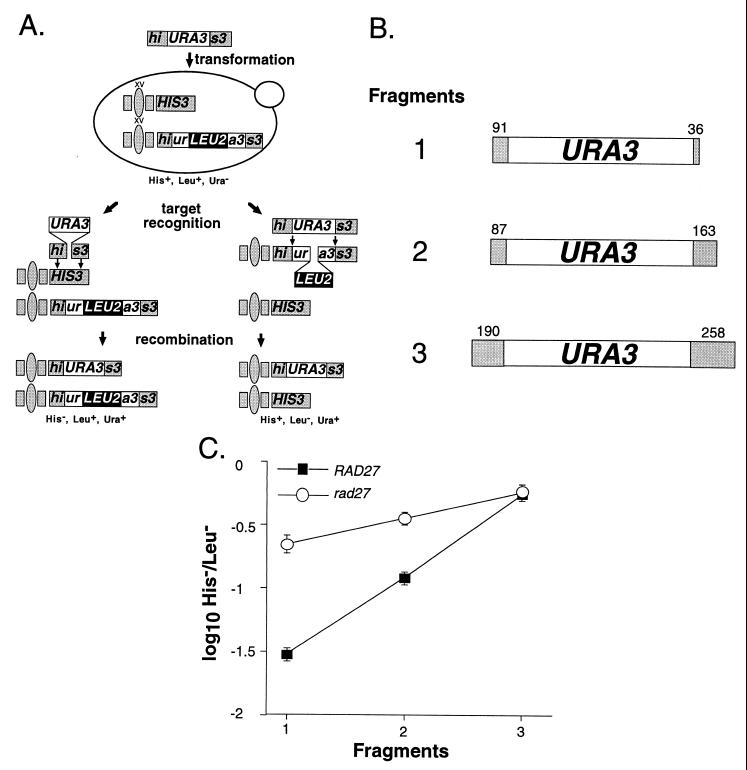
DNA fragment insertion into the genomes of wild-type and rad27-null mutant cells. (A) DNA fragment insertion assay. DNA fragments containing a 1.1-kb URA3 clone flanked by various lengths of HIS3 sequence (his3::URA3) were electroporated into diploid yeast cells. The two relevant recombination targets are a wild-type HIS3 allele at the HIS3 locus on one copy of chromosome XV and a his3::ura3::LEU2 allele at the HIS3 locus on the homologue. Alignment of the his3::URA3 fragment with the wild-type HIS3 allele involves only the terminal HIS3 sequences, whereas alignment with the his3::ura3::LEU2 allele involves all of the fragment. Insertion of the fragment into the HIS3 allele by recombination replaces 60 bp of the HIS3 coding sequence with the 1.1-kb URA3 marker, resulting in a cell that is His− Leu+ Ura+, whereas insertion into the his3::ura3::LEU2 allele replaces a 2.0-kb LEU2 marker with 60 bp of URA3 sequence that results in a His+ Leu− Ura+ cell. (B) DNA fragments used in the insertion assay. The three his3::URA3 DNA fragments used in the experiments are depicted. The HIS3 sequences at the ends of the fragments are depicted as shaded boxes with sequence lengths marked above in base pairs. The URA3 sequence in each fragment is 1,071 bp long. (C) A plot of the ratio of DNA fragment insertions of each his3::URA3 fragment into the HIS3 and his3::ura3::LEU2 targets in wild-type and rad27 mutant diploids. The three different his3::URA3 fragments were separately electroporated into wild-type (ABX367) and rad27 homozygous (ABX382) diploids. Ura+ transformants were replica plated to medium lacking His or Leu to determine which transformants were due to insertion into the HIS3 or his3::ura3::LEU2 alleles. The mean ratios of His− Leu+ Ura+ to His+ Leu− Ura+ transformants (H−/L−) obtained with the three DNA fragments were determined from a minimum of 10 independent trials with each fragment in each strain. Log10 values of the mean ratios ± two standard errors were plotted for each of the fragments.
The ratios of DNA fragment insertion into the HIS3 and his3::ura3::LEU2 alleles were not significantly influenced by the lengths of DNA being deleted from or transplaced into the genome during recombination. Very similar ratios were obtained when the smallest his3::URA3 fragment was inserted into the HIS3 allele, which requires deletion of 60 bp (Fig. 1C), as when it was inserted into the sam2::his3::TRP1 allele (see Fig. 3B), which requires deletion of 1.5 kb. We also found that inserting a 1.5-kb TRP1 marker into the URA3 sequence of the smallest his3::URA3 fragment did not significantly affect the relative frequencies of recombination with the HIS3 and his3::ura3::LEU2 genomic targets (unpublished data), indicating that transplacing 60 bp of DNA into the his3::ura3::LEU2 allele was equivalent to 1.5 kb.
Ectopic recombination between the ura3-1 alleles at the URA3 loci and the his3::ura3::LEU2 allele at the HIS3 locus occured at a frequency of ∼10−7 (4), which is 10- to 100-fold lower in frequency than fragment insertion into the his3::ura3::LEU2 allele and indicates that it does not interfere with the assay. Frequencies of ectopic recombination between the leu2-3,112 alleles at the LEU2 loci and the his3::ura3::LEU2 allele should occur at a similarly low frequency and should not interfere with the assay.
Nonhomologous ends assay.
his3::URA3 fragments with different lengths of HIS3 sequence were obtained by PCR using pLAY144 as template. The four primer pairs 5′-AAGCTTTTAAAGAGGCCC-3′ and 5′-AAGCTTTCAAGAAAATGC-3′, 5′-CTCTCGGTCAAGCTTTTA-3′ and 5′-CTAGCCTCTGCAAAGCTT-3′, 5′-GCGGGATTGCTCTCGGTC-3′ and 5′-GGTAATTCTGCTAGCCTC-3′, and 5′-ACTGAAGACTGCGGGATT-3′ and 5′-AACGTGGAGGGTAATTCT-3′ were used to generate his3::URA3 fragments with 91 and 36 bp, 101 and 46 bp, 111 and 56 bp, and 121 and 66 bp of HIS3 sequence flanking the 1.1-kb URA3 marker, respectively. PCR products were used to transform haploid wild-type and rad27 mutant cells by electroporation, selecting for Ura+ recombinants.
As described above, recombination events at the different genomic targets (see Fig. 3) are distinguishable by replica plating to appropriate drop-out media and were confirmed by Southern blot (unpublished data). Just like the previous assay, recombination with the his3::ura3::LEU2 allele at the HIS3 locus replaces a 2-kb LEU2 marker with 60 bp of URA3 coding sequence and results in Trp+ Leu− Ura+ transformants. Recombination with the sam2::his3::TRP1 allele at the SAM2 locus replaces 1.5 kb of TRP1 coding sequence with the 1.1-kb URA3 marker and results in Trp− Leu+ Ura+ transformants. The fragments can also recombine with the ura3-1 allele at the URA3 locus. Fragment recombination at the URA3 locus remained between 25 and 30% of the total, regardless of fragment length or cell type. However, the ratio of insertions at the sam2::his3::TRP1 and his3::ura3::LEU2 targets changed significantly depending on the length of the HIS3 sequences on the fragment. The mean ratio of insertions into the sam2::his3::TRP1 and his3::ura3::LEU2 targets (T−/L−) was determined from at least five trials. Each trial involved a minimum of 400 Ura+ transformants.
RESULTS
DNA fragment insertion assay.
We used the insertion of DNA fragments into the genome to study the effect of DNA sequence length on homologous recombination in wild-type and mutant yeast. This approach was chosen because DNA fragment length can be readily changed and because DNA fragment insertion is similar to the repair of broken chromosomes by homologous recombination (30, 36, 38). In this assay, homozygous wild-type and rad27 mutant diploids were transformed with three different DNA fragments consisting of a URA3 marker flanked by varying lengths of HIS3 sequence. The two relevant genomic targets for the his3::URA3 fragments were a wild-type HIS3 allele on one copy of chromosome XV and the his3::ura3::LEU2 allele on the other (Fig. 1A). Insertion of a fragment into the HIS3 allele is governed only by the short HIS3 sequences at its ends, while insertion into the his3::ura3::LEU2 allele utilizes its entire length. The length of the terminal HIS3 sequences on the three fragments, and therefore homology to the HIS3 allele, varies over 3.5-fold (Fig. 1B). However, overall fragment length and homology to the his3::ura3::LEU2 allele vary less than 21% (Fig. 1B). Since significant changes in terminal HIS3 sequence length have little effect on overall fragment length, the three fragments should have a significantly different propensity to recombine with the HIS3 allele but a similar propensity to recombine with the his3::ura3::LEU2 allele. Therefore, the ratio of DNA fragment insertions into the HIS3 allele versus the his3::ura3::LEU2 allele (H−/L−) indicates how well the cell can use the terminal HIS3 sequences for recombination.
SSR is increased in rad27 but not exo1 or din7 mutant cells.
It was previously shown that Rad2 does not play a role in the control of SSR (2). Here we show that SSR was unaffected by the loss of the structurally similar proteins Din7 and Exo1, as the H−/L− ratios obtained with fragment 1 (Fig. 1B), which has the shortest HIS3 ends (127 bp total), were not significantly different from that of the wild type in exo1- and din7-null mutant cells (Table 3). In contrast, we found that the H−/L− ratio in rad27-null mutant cells was sevenfold higher than that of the wild type, indicating that Rad27 suppresses recombination between short sequences (Table 3). However, the stimulation was only threefold with fragment 2, which has 250 bp of HIS3 sequence, and nonexistent with fragment 3, which has 448 bp of HIS3 sequence (Fig. 1C). Therefore, in this assay rad27 stimulates recombination between short sequences but not long ones.
TABLE 3.
SSR phenotypes of wild-type and nuclease-deficient cells
| Genotype | H−/L− ratioa |
|---|---|
| Wild-type diploid | 0.03 ± 0.01 |
| din7::KANMX/din7::KANMX | 0.03 ± 0.01 |
| exo1::hisG/exo1::hisG | 0.048 ± 0.01 |
| rad27::leu2::hisG/rad27::leu2::hisG | 0.22 ± 0.04 |
Diploid strains ABX367, ABX382, ABX451, and ABX456 were transformed with the shortest DNA fragment, which contains 91 and 36 bp of DNA flanking the URA3 gene. The ratio of transformants that were due to fragment insertion into the HIS3 allele (His− Leu+ Ura+) versus those that were due to insertion into the his3::ura3::LEU2 allele (His+ Leu− Ura+) was determined as described in the text. The means from a minimum of five trials ± two standard errors are reported. Means that differ by greater than two standard errors are different at the 95% confidence level.
The flap endonuclease activities of Rad27 and FEN-1 are implicated in the control of SSR.
Previous studies have shown that mutating a conserved glutamate at residue 160 in the active site of human FEN-1 leads to marked changes in DNA binding and/or 5′ flap cleavage (18, 50). Substituting an aspartate for the glutamate (E160D) has no effect on substrate binding but partially reduces 5′ flap cleavage efficiency (18). Here we show that FEN-1-E160D and the analogous Rad27 mutant enzyme (E158D) possess similar partial flap endonuclease activities (Table 4). The Kms of FEN-1-E160D and Rad27-E158D for the 5′ flap substrate were nearly equivalent to that of the wild type, but the Vmaxs of FEN-1-E160D and Rad27-E158D were only 59 and 69%, respectively, of that of the wild type. Neither mutant protein possessed any measurable capacity for exonucleolytic cleavage (Table 4). These results demonstrate that the enzymatic functions defined by these assays are separable.
TABLE 4.
5′-Flap endonuclease and 5′-nick-specific exonuclease activities of wild-type and mutant FEN-1 and Rad27 enzymes
| Enzymea | Flap endonucleaseb
|
Nick exonucleasec
|
||
|---|---|---|---|---|
| Km (nM) | Vmax (nM/min) | Km (nM) | Vmax (nM/min) | |
| Rad27 | 48.0 | 3.37 | 47.5 | 2.9 |
| Rad27-E158D | 48.9 | 2.33 | >150 | <0.1 |
| FEN-1 | 48.1 | 3.07 | 47.2 | 2.3 |
| FEN1-E160D | 48.6 | 1.82 | >150 | <0.1 |
From 5 to 40 nmol of enzyme was reacted with 0.1 to 25 pmol of 32P-labeled flap substrate prepared as described previously (23). Double-reciprocal plots of reaction velocities, determined as described previously (18), versus substrate concentrations were used to determine the values for Km and Vmax.
From 5 to 40 nmol of enzyme was reacted with 0.1 to 25 pmol of 32P-labeled nicked duplex DNA substrate prepared as described previously (23). The values for Km and Vmax were determined as described above.
Recently, the human FEN1 gene was shown to fully complement the temperature-sensitive growth and methyl methanesulfonate sensitivity of the rad27-null mutant, while the fen1-E160D allele only partially complemented it (18). These results suggest that the cellular functions of these proteins may be conserved and are linked to flap endonuclease activity. We tested whether FEN1 also complements the SSR phenotype of the rad27 mutant by comparing the H−/L− ratio from rad27 cells containing FEN1 on a plasmid to the wild-type ratio. We found that the ratios were not significantly different, demonstrating that FEN1 complements the SSR phenotype of rad27-null mutant cells (Table 5). Interestingly, the ratios obtained with the fen1-E160D and rad27-E158D mutant alleles were significantly above those from cells containing wild-type FEN1 and RAD27 but significantly below the ratio obtained from cells containing an empty plasmid (Table 5). Since the intermediate level of complementation by the fen1-E160D and rad27-E158D alleles matches the flap endonuclease activity data described above (Table 4) and since the levels of Rad27 or FEN-1 protein are similar in cells containing either the wild-type or mutant alleles (data not shown), we suggest that the effects of Rad27 and FEN-1 on SSR are a function of their flap endonuclease activities but not their nick exonuclease activities. In support of these results we found that exonucleolytic degradation at DSBs in rad27 cells was not significantly different from that in wild-type cells (data not shown).
TABLE 5.
Complementation of the rad27 SSR phenotype by alleles of FEN1 and RAD27
| Allele on plasmida | H−/L− ratiob |
|---|---|
| None | 0.18 ± 0.04 |
| RAD27 | 0.05 ± 0.006 |
| rad27-E158D | 0.08 ± 0.004 |
| FEN1 | 0.04 ± 0.003 |
| fen1-E160D | 0.10 ± 0.01 |
All strains are rad27::leu2::hisG/rad27::leu2::hisG homozygotes. All plasmids are derived from pRS414, which carries a yeast centromere and a TRP1 selectable marker (52). Expression of the FEN1 and RAD27 alleles is under the control of the yeast ADH1 promoter and terminator.
Strains ABT301, ABT302, ABT303, ABT306, and ABT344 were transformed with a his3::URA3 fragment that contains 91 and 36 bp of HIS3 DNA flanking the URA3 gene. H−/L− ratios were determined as described in the text. The means from a minimum of 10 trials ± two standard errors are reported. Means that differ by greater than two standard errors are different at the 95% confidence level.
Length dependence of 5′ flap cleavage.
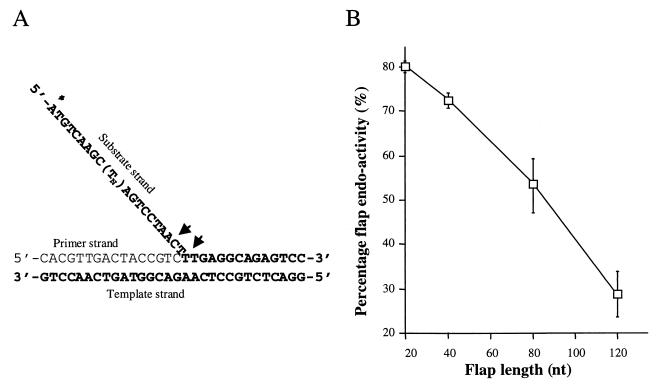
Bambara and colleagues showed that 5′ flap cleavage by FEN-1 requires that the flap remain single stranded until its junction with duplex DNA (37). This implies that the enzyme must slide unimpeded from the 5′ end of the flap to the junction with duplex DNA. We examined the capacity of FEN-1 to cleave flaps of various lengths (Fig. 2). The flaps were comprised of different lengths of oligodeoxythymidine in order to minimize secondary structure formation (11). We found that flap cleavage efficiency decreased markedly with increasing flap length, from 80% cleavage with a 20-bp flap to 28% with a 120-bp flap. Given the very similar in vitro activities of FEN-1 and Rad27 (see Fig. 4), we expect that Rad27 would display similar characteristics. As discussed below, the diminished capacity of FEN-1 and Rad27 to cleave long flaps might in part explain why losing Rad27 preferentially affects SSR.
FIG. 2.
Effect of DNA flap length on FEN-1 activity. (A) DNA substrates. The substrate consists of three DNA strands. (Tn) indicates the number of T residues (0, 20, 60, or 100) in the substrate strand. This corresponds to total flap lengths of 20, 40, 80, and 120 nucleotides. The asterisk denotes the location of the 32P label. (B) Plot of flap endonuclease activities with different flap lengths. Five picomoles of 32P-labeled flap substrate was incubated with 75 nmol of FEN-1, and reaction products were run on analytical gels and autoradiographed. Substrate and product levels were quantitated by densitometry and converted to cleavage efficiencies by dividing product signals by the total of the substrate and product signals and multiplying by 100. nt, nucleotides.
FIG. 4.
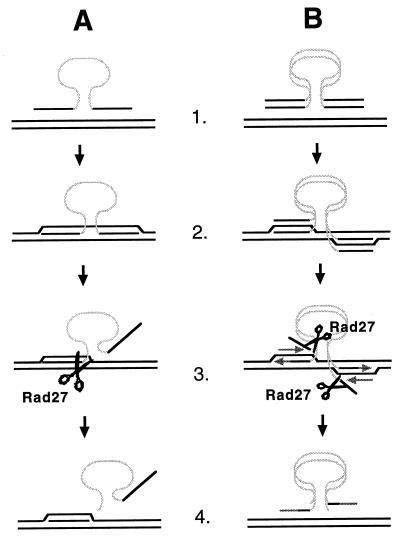
Models of how Rad27 and FEN-1 may limit recombination between short sequences. (A) Single-strand assimilation. 1. A single strand of the his3::URA3 fragment is pictured with HIS3 genomic target sequences. HIS3 sequences are represented by black lines and URA3 sequences are represented by a gray line. 2. A single strand of the his3::URA3 fragment invades homologous genomic sequences, creating a heteroduplex. 3. Short heteroduplexes may be unwound by a helicase(s). The unannealed 5′ single strand is a substrate for endonucleolytic cleavage by Rad27. 4. Recombination is aborted. (B) Break-induced repair. 1. A double-stranded his3::URA3 fragment is pictured with HIS3 genomic target sequences. HIS3 sequences are represented by black lines and URA3 sequences are represented by light gray lines. 2. The ends of the double-stranded his3::URA3 fragment invade HIS3 genomic sequences, forming four-stranded heteroduplex intermediates. 3. The short heteroduplexes may be unwound by the advancing lagging-strand polymerase or by a helicase(s), creating unannealed 5′ single strands that are cleaved by Rad27. 4. Recombination is aborted because the unannealed 5′ strands of the DNA fragment have been cleaved away, leaving the newly synthesized Okazaki fragments unable to ligate onto the ends of the fragment.
Rad27 is required for removal of nonhomologous ends during SSR.
Previously, Fishman-Lobell and Haber (17) found that a null mutation in the RAD1 gene blocks recombination when sequences at the ends of DSBs obscure homology with a recombination partner. Subsequently, it was shown that Rad1- Rad10 is a structure-specific endonuclease (58, 65) that can cleave terminal 3′ single strands displaced when adjacent sequences pair with homologous sequences on another DNA molecule (6). Rad1 may cleave other intermediates during several different types of recombination events (2, 60).
Since Rad27 cleaves 5′ single strands at the junction with double-stranded DNA (31), a second DNA fragment insertion assay was designed to test whether it also plays a role in removing nonhomologous sequences from the ends of DNA molecules undergoing homologous recombination (Fig. 3A), perhaps implying a role in the processing of other recombination intermediates. Four different DNA fragments consisting of a URA3 marker bordered by varying lengths of HIS3 sequence were used to transform haploid wild-type and rad27-null mutant cells. One recombination target is the his3::ura3::LEU2 allele described above (Fig. 1A). All of the fragments are homologous to this target along their entire length. The other relevant target, sam2::his3::TRP1, consists of a TRP1 marker bordered by 36- and 91-bp HIS3 segments at the SAM2 locus on chromosome IV. The smallest his3::URA3 fragment has the same HIS3 sequences as the sam2::his3::TRP1 target, while the other fragments have 10, 20, or 30 bp of additional HIS3 sequences at both ends. Recombination between the smallest fragment and sam2::his3::TRP1 is unblocked, whereas insertion of the other fragments into this target requires that 10, 20, or 30 bp of terminal HIS3 sequences be removed from both ends. The different his3::URA3 fragments have the same length of homology with the sam2::his3::TRP1 target, and their homology with the his3::ura3::LEU2 target varies very little (less than 5%). Since there is little difference in the lengths of homology shared by the fragments and the targets, this should have little effect on the ratios of fragment insertion into the two targets (T−/L−). Therefore, changes in these ratios should indicate how well the cell can remove any excess HIS3 sequences from the ends of a fragment during recombination with the sam2::his3::TRP1 target (Fig. 4B).
The results of these assays indicate that Rad27 plays an important role in the removal of nonhomologous sequences from the ends of recombining DNA molecules. Each addition of 10 bp of nonhomologous sequences onto the ends of the DNA fragments decreased insertion into the sam2::his3::TRP1 target relative to the his3::ura3::LEU2 target approximately twofold in rad27 mutant cells (Fig. 3B). The nonhomologous sequences had much less of an impact on the ratio of insertion into the two targets in wild-type cells, as the addition of 10 bp of nonhomologous sequences had no effect and even of 30 bp had little more than a twofold effect (Fig. 3B). Therefore, the presence of Rad27 facilitates the removal of sequences that can otherwise inhibit recombination. Since the exonuclease activity of Rad27 appears to play little, if any, role in the control of SSR (Table 4; data not shown), we conclude that the 5′ flap endonuclease of Rad27 can cleave flaps that form at the ends of recombining molecules.
DISCUSSION
Previous work with several mutants demonstrated a correlation between increased DSB stability and enhanced frequencies of SSR (2, 3, 29, 34). This led to the investigation of how null alleles of several nuclease genes affect SSR. A novel DNA fragment insertion assay was used to show that the exo1 and din7 alleles have negligible effects on SSR (Table 3), while the rad27 allele confers a significant increase in SSR (Fig. 1). Combined with results previously obtained with a rad2 mutant (34), these data support the conclusion that RAD27 is the only member of this group of genes that plays a significant role in the control of SSR.
Our assay for his3::URA3 fragment insertion into genomic HIS3 sequences does not require that the recombination events perfectly conserve the homology between the fragment and genomic sequences. Since rad27 mutants accumulate mutations that appear to be due to imprecise recombination (64), we examined the precision of fragment insertion events in rad27 mutant cells. Analysis of the DNA sequences of 40 independent insertions of the smallest his3::URA3 fragment into the HIS3 allele of rad27 cells revealed no alterations of the HIS3 sequences undergoing recombination (data not shown). In combination with the observation that no fragment insertion is obtained in cells lacking Rad52 (G. Manthey and A. Bailis, unpublished observations), a central factor in homologous recombination (39), we suggest that the loss of Rad27 stimulates precise recombination between short sequences by a Rad52-dependent mechanism.
The biological functions of FEN-1 and Rad27 may be conserved, since the expression of human FEN1 in rad27 mutant yeast cells complements their temperature-sensitive growth, methyl methanesulfonate sensitivity, and elevated SSR (18) (Table 5). This apparent conservation is both structural and functional, as changing conserved glutamate residues to aspartates in both proteins leads to similar changes in enzymatic activity (Table 4). Since the mutant alleles partially complement the rad27 SSR phenotype (Table 5), we conclude that the endonuclease activities of Rad27 and FEN-1 are important for SSR control in yeast and, perhaps, human cells. While the effect of mutations in genes that encode TFIIH-NER core proteins on SSR correlates with defects in DSB processing (2, 3, 29, 34), the loss of Rad27 has no impact on the exonucleolytic degradation of DSBs (data not shown). This suggests that Rad27 may restrict SSR by a mechanism that is different from that of the TFIIH-NER apparatus. We are currently testing this by epistasis analysis.
We found that Rad27 is needed for efficient SSR when the DNA fragment has sequences at both ends that are not homologous to the genomic target (Fig. 3), implying that Rad27 cleaves 5′ overhangs that block SSR. This is similar to the observation by Wu et al. (70) that 5′ overhang removal by Rad27 appears to be important in the recovery of certain nonhomologous end-joining (NHEJ) products. Both results imply that basepairing, or heteroduplex formation, precedes trimming of 5′ tails from recombining molecules. Therefore, unlike recombination between long, homologous sequences, SSR does not seem to require prior degradation of the 5′ ends of DNA molecules (39). Interestingly, nonhomologous sequences at the ends of DNA fragments that shared hundreds of base pairs of homology with their genomic targets had no effect on fragment insertion in wild-type or rad27 mutant cells (M. Navarro and A. Bailis, unpublished observations). This implies that nonhomologous 5′ tails can be removed by a Rad27-independent mechanism during recombination between longer sequences. We speculate that this process may remove RNA primer residues from the 5′ ends of newly synthesized DNA molecules in the absence of Rad27.
Why is recombination between short sequences, but not long sequences, increased in the rad27 mutant? Genomic DSB formation is enhanced in rad27 mutants, which increases the incidence of many recombination events (10, 19, 64). In our assays, an increase in the frequency of DSBs at the genomic targets is just as likely to increase recombination with the longer DNA fragments as with the shorter DNA fragments. However, the genome-wide increase in DSBs in rad27 mutant cells may deplete the factors that normally suppress the insertion of DNA fragments into the genome by SSR. Alternatively, the failure by rad27 cells to remove nonhomologous sequences from the ends of recombining DNA molecules (Fig. 4) suggests that Rad27 might restrict recombination at a stage following the formation of DSBs. For example, following heteroduplex formation, Rad27 might remove unannealed 5′ single strands generated by an advancing lagging-strand polymerase or helicase unwinding (33). The TFIIH-NER helicases Rad3 and Ssl2, which play a role in SSR control (2, 3, 29, 34), and Dna2, a helicase that works in concert with Rad27 during DNA replication (9), are potential candidates for this activity. If unwinding is extensive, Rad27 could remove enough DNA to terminate recombination as discussed below. Decreasing the length of the sequences shared by the recombination partners would increase the likelihood of complete heteroduplex unwinding, 5′ flap cleavage (Fig. 2), or both.
Several existing models for DNA fragment insertion into the genome (30, 36, 39, 43) were used to construct models for how Rad27 may restrict SSR. The first model (Fig. 4A) is based on the single-strand assimilation model of Leung et al. (30). Following formation of a heteroduplex between a single strand of the DNA fragment and genomic sequences, helicase unwinding creates a 5′ flap that includes the selectable marker. Flap cleavage by Rad27 would terminate recombination because the fragment containing the marker would not have homology to the genomic target on both sides. An important caveat limiting the attractiveness of this model is that the 5′ flap would exceed 1 kb and be a poor substrate for Rad27 (Fig. 2). We favor another model (Fig. 4B) based on the break-copy model of Morrow et al. (36), which is similar to the break-induced repair model for the repair of genomic DSBs (39). Following heteroduplex formation between the ends of the DNA fragment and the genomic target, the ends of the fragment simulate the ends of broken chromosomes by copying genomic sequences. DNA fragment insertion is concluded when the replication forks reach the centromere or the end of the chromosome or by Holliday junction resolution at nicks or gaps. Recombination could be impeded by unwinding the heteroduplex and cleaving the resulting 5′ flap before an Okazaki fragment can be ligated onto the 5′ strand. This would terminate fragment insertion unless the lagging-strand polymerase switches from a chromosomal template to the DNA fragment, which would be unlikely to occur if the remaining heteroduplex between the 3′ strand and the target is also unstable.
Kolodner and colleagues observed genome rearrangements involving very short repeats in rad27 mutant cells that were not observed in wild-type cells (10, 64). We have observed that deletions between 100-bp repeats are fourfold more stimulated in rad27 mutant cells than deletions between 400-bp repeats (unpublished data). These results suggest that Rad27 plays a role in limiting recombination between an abundant class of genomic sequences which could contribute to the etiology of cancer. Previously, it was suggested that Rad27 limits recombination by limiting DSB formation (19, 59, 64). In addition, the work described here suggests that Rad27 and FEN-1 restrict SSR by processing recombination intermediates. Armed with the evolving understanding of the relationship between FEN-1 and its substrates at the atomic level (23, 24), our future studies will continue to focus on the molecular details of how Rad27 and FEN-1 maintain genome stability.
ACKNOWLEDGMENTS
This work was supported by U.S. Public Health Service grants GM57484 to A.M.B. and CA73764 to B.S. and by an APRC suppplemental grant (CA85344) to B.S. and A.M.B. from the National Institutes of Health, as well as by funds from the Beckman Research Institute of the City of Hope and the City of Hope National Medical Center.
We thank J. McDonald for yeast strain W1011-3B and D. Garfinkel, J. Wilson, and several anonymous reviewers for comments on the manuscript. We also thank J. Termini, T. Krontiris, R. J. Lin, J. Haber, D. Gordenin, S. Rosenberg, G. Smith, R. Rothstein, D. Botstein, and the members of the Bailis and Shen laboratories for stimulating discussions.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailis A M, Maines S. Nucleotide excision repair gene function in short-sequence recombination. J Bacteriol. 1996;173:2136–2140. doi: 10.1128/jb.178.7.2136-2140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailis A M, Maines S, Negritto M C. The essential helicase gene RAD3 suppresses short-sequence recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3998–4008. doi: 10.1128/mcb.15.8.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailis A M, Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homologous genes by an excision repair dependent process. Genetics. 1990;126:535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bambara R A, Murante R S, Henricksen R A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 6.Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2062–2065. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 7.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 8.Britten R J, Kohne D E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 9.Budd M E, Campbell J L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Kolodner R D. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination deficient mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 11.Cheng D M, Dhingra M M, Sarma R H. Spatial configuration of deoxyribotrinucleoside diphosphate in aqueous solution. Nucleic Acids Res. 1978;5:4399–4416. doi: 10.1093/nar/5.11.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deininger P L, Batzer M A. Alu repeats and human disease. Mol Genet Metab. 1999;68:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 13.de Laat W L, Jaspers N G J, Hoeijmakers J H J. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 14.Feaver W J, Svejstrup J Q, Bardwell L, Bardwell A J, Buratowski S, Gulyas K D, Donahue T F, Friedberg E C, Kornberg R D. Dual roles of a multiprotein complex from Saccharomyces cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 15.Fikus M U, Mieczkowski P A, Koprowski P, Rytka J, Sledziewska-Gojska E, Ciesla Z. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 18.Frank G, Qiu J, Somsouk M, Weng Y, Somsouk L, Nolan J P, Shen B. Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J Biol Chem. 1998;273:33064–33072. doi: 10.1074/jbc.273.49.33064. [DOI] [PubMed] [Google Scholar]
- 19.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 20.Habraken Y, Sung P, Prakash L, Prakash S. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature. 1993;366:365–368. doi: 10.1038/366365a0. [DOI] [PubMed] [Google Scholar]
- 21.Harrington J J, Lieber M R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes A, Haber J E. Double-strand break repair requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 23.Hosfield D J, Mol C D, Shen B, Tainer J A. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 24.Hwang Y H, Baek K, Kim H-Y, Cho Y. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat Struct Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- 25.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast Rth1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1998;269:238–239. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 27.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B-S, Lichtenstein C P, Faiola B, Rinckel L A, Wysock W, Curcio M J, Garfinkel D J. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics. 1998;148:1743–1761. doi: 10.1093/genetics/148.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee B-S, Bi L, Garfinkel D J, Bailis A M. Nucleotide excision repair/TFIIH helicases Rad3 and Ssl2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol Cell Biol. 2000;20:2436–2445. doi: 10.1128/mcb.20.7.2436-2445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung K Y, Malkova A, Haber J E. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc Natl Acad Sci USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieber M. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 32.Lobachev K S, Stenger J E, Kozyreva O G, Jurka J, Gordenin D A, Resnick M A. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovett S T, Sutera V A., Jr Suppression of RecJ exonuclease mutant of Escherichia coli by alterations in DNA helicases II (uvrD) and IV (helD) Genetics. 1995;140:27–45. doi: 10.1093/genetics/140.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maines S, Negritto M C, Wu X, Manthey G M, Bailis A M. Novel mutations in the RAD3 and SSL1 genes perturb genome stability by stimulating recombination between short repeats in Saccharomyces cerevisiae. Genetics. 1998;150:963–976. doi: 10.1093/genetics/150.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manivasakam P, Weber S C, McElver J, Schiestl R H. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:2799–2800. doi: 10.1093/nar/23.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrow D M, Connelly C, Hieter P. “Break copy” duplication a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murante R S, Rust L, Bambara R A. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 38.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parenteau J, Wellinger R J. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol Cell Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu J, Guan M X, Bailis A M, Shen B. Saccharomyces cerevisiae exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair. Nucleic Acids Res. 1998;26:3077–3083. doi: 10.1093/nar/26.13.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reagan M S, Pittenger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothstein R. Double-strand break repair, gene conversion, and postdivision segregation. Cold Spring Harbor Symp Quant Biol. 1984;49:629–638. doi: 10.1101/sqb.1984.049.01.071. [DOI] [PubMed] [Google Scholar]
- 44.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 45.Rothstein R, Helms C, Rosenberg N. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1198–1207. doi: 10.1128/mcb.7.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubnitz J, Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 49.Shen B, Nolan J P, Sklar L A, Park M S. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J Biol Chem. 1996;271:9173–9176. doi: 10.1074/jbc.271.16.9173. [DOI] [PubMed] [Google Scholar]
- 50.Shen B, Nolan J P, Sklar L A, Park M S. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen B, Qiu J, Hosfield D, Tainer J A. Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem Sci. 1998;23:171–173. doi: 10.1016/s0968-0004(98)01199-2. [DOI] [PubMed] [Google Scholar]
- 52.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- 53.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′ to 3′ exonuclease required for lagging strand synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 56.Spiro C, Pelletier R, Rolfsmeier M L, Dixon M J, Lahue R S, Gupta G, Park M S, Chen X, Mariappan S V S, McMurray C T. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 57.Sugawara N, Haber J E. Characterization of double-strand-break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sung P, Reynolds P, Prakash L, Prakash S. Purification and characterization of the Saccharomyces cerevisiae Rad1/Rad10 endonuclease. J Biol Chem. 1993;26:23691–23699. [PubMed] [Google Scholar]
- 59.Symington L S. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 1998;26:5589–5595. doi: 10.1093/nar/26.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Symington L S, Kang L E, Moreau S. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:4649–4656. doi: 10.1093/nar/28.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szankasi P, Gysler C, Zehntner U, Leupold U, Kohli J, Munz P. Mitotic recombination between dispersed but related tRNA genes of S. pombe generates a reciprocal translocation. Mol Gen Genet. 1986;202:394–402. [Google Scholar]
- 62.Thomas B J, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination of a GAL10 transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 65.Tomkinson A E, Bardwell A J, Bardwell L, Tappe N J, Friedberg E C. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature. 1993;362:860–862. doi: 10.1038/362860a0. [DOI] [PubMed] [Google Scholar]
- 66.Tran H T, Gordenin D A, Resnick M A. The 3′→5′ exonucleases of DNA polymerases δ and ɛ and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vallen E A, Cross F R. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Friedberg E C. Transcription factor b (TFIIH) is required during nucleotide excision repair in yeast. Nature. 1994;368:74–76. doi: 10.1038/368074a0. [DOI] [PubMed] [Google Scholar]
- 70.Wu X, Wilson T E, Lieber M R. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci USA. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu F X, Biswas E E, Biswas S B. Purification and characterization of the DNA polymerase α associated exonuclease: the RTH1 gene product. Biochemistry. 1997;36:5947–5954. doi: 10.1021/bi962889v. [DOI] [PubMed] [Google Scholar]