Abstract
Despite Auger electrons being highly appealing due to their short range and high linear energy transfer to surrounding tissue, the progress in the field has been limited due to the challenge in delivering a therapeutic dose within the close proximity of cancer cells DNA. Here, we demonstrate that the PARP inhibitor 123I-MAPi is a viable agent for systemic administration and treatment of p53 mutant cancers. Significantly, minimal off-site toxicity was observed in mice administered with up to 74 MBq of 127I-PARPi. Taken together, these results lay the foundation for future clinical evaluation and broader pre-clinical investigations. By harnessing the scaffold of the PARP inhibitor Olaparib, we were able to deliver therapeutic levels of Auger radiation to the site of human colorectal cancer xenograft tumors after systemic administration. In-depth toxicity studies analyzed blood chemistry levels and markers associated for specific organs toxicity. Finally, p53+/+ and p53−/− human colorectal cancer cell lines were evaluated for the ability of 123I-MAPi to induce tumor growth delay. Toxicity studies demonstrate both 123I-MAPi and its stable isotopologue, 127I-PARPi, have no significant off-site toxicity when administered systemically. Analysis following 123I-MAPi treatment confirmed its ability to induce DNA damage at the site of xenograft tumors when administered systemically. Finally, we demonstrate that 123I-MAPi generates a therapeutic response in p53−/−, but not p53+/+, subcutaneous xenograft tumors in mouse models. Taken together, these results represent the first example of a PARP Auger theranostic agent capable of delivering a therapeutic dose to xenograft human colorectal cancer tumors upon systemic administration without causing significant toxicity to surrounding mouse organs. Moreover, it suggests that a PARP Auger theranostic can act as a targeted therapeutic for cancers with mutated p53 pathways. This landmark goal paves the way for clinical evaluation of 123I-MAPi for pan cancer therapeutics.
Keywords: PARP, Auger, 123I-MAPi, p53, p21
Introduction
After the discovery of the Auger effect in 1922 by Lise Meitner and by Pierre Auger, scientists have sought to take advantage of this form of radiation for targeted radiotherapy. Targeted radiotherapy itself has seen a rapid expansion over the years with both β- and more recently that of α-particles being utilized to good success [1, 2]. Despite this, the long-distance effective ranges β-particles can induce unwanted off-target effects, irradiating healthy bystander cells and causing damage to vascular and metabolically active structures. This may not be the case when looking at the very short range, high linear energy transfer capabilities of Auger emitting radionuclides [3, 4], making them ideal candidates when seeking to target cancer specific biomarkers in vivo while avoiding off-site radiation induced toxicities [5].
An early example to reach the clinical space is that of 5-[125I]I-iodo-2’-deoxyuridine (125I-UdR), a thymidine analogue reported to target DNA during the S phase of the cell cycle and thus making it highly promising for targeting of rapidly growing cancer cells [6]. However, efficacy in the clinical setting was limited, resulting in heterogeneous response rates in patients [7]. More recent examples include that of 111In-DTPA-octreotide for the treatment in patients with advanced SST-2 positive neuroendocrine malignancies and 111In-DTPA-hEGF for patients with metastatic EGFR-positive breast cancer [8, 9]. In 2020, Kiess and co-workers reported the use of 125I-DClBzL, a PSMA auger radio-therapeutic [10]. In this study, the authors successfully demonstrated that through systemic administration, 125I-DClBzL may serve as a viable option for inducing tumor growth delay in PSMA-positive micro-metastatic models. Other areas of auger research include that of modular nano-transporters and antibodies containing nuclear localization sequences [5].
Given the short range of Auger elections, it is well recognized that successful treatment depends on the delivery of payloads as close as possible to the sub-cellular compartment of interest [11, 12]. Of those sub-cellular compartments available for targeting, it is DNA which is the most attractive for attaining effective treatment with Auger electrons.
Having a vector through which Auger radiation may be shuttled onto the DNA is paramount. One class of compounds to achieve this is that of PARP inhibitors (PARPi). Upon damage to DNA, PARP is recruited to the damage site. PARP inhibitors induce trapping on the DNA, stalling the repair process and inducing further DNA damage. In 2020, Pirovano and co-workers reported the use of an Iodine-123 PARP auger theranostic agent, 123I-MAPi, for the treatment of glioblastoma [13]. Harnessing the PARP1 targeting scaffold of Olaparib, the authors were able to deliver the payload within reach of the cancer cells’ DNA, and therefore within reach of the auger electron. Pirovano and colleagues showed that the effect on cell killing during 123I-MAPi treatment is exclusively due to Auger-induced DNA damage formation at tracer concentrations and that I-123 iodide does not play a role in cell killing at these concentrations. A limitation of this approach, however, was the need to deliver the payload through a convection enhanced delivery (CED) mechanism. While CED is an elegant solution for treating glioblastoma [14], the technique is limited in its availability to the public. Having a pathway to achieving a therapeutic response via systemic administration would consequently be highly appealing. Furthermore, being able to demonstrate a therapeutic response whilst minimizing off-site toxicity would be critical for the long-term development of such a reagent.
To this end, here we discuss the finding of an in-depth toxicity study for 123I-MAPi when systemically administered. Following this, we evaluate 123I-MAPi for its ability to induce DNA damage at the site of the tumor. Aberrant DNA damage checkpoints can be exploited by PARP inhibitors, therefore we evaluated the DNA damage response in the presence and absence of a functional wild-type p53 tumor suppressor which normally works to initiate growth arrest, DNA repair and apoptosis [15]. The p53 checkpoint is often nonfunctional in colorectal cancers [16, 17]. Therefore, we compared the well characterized isogenic human colorectal cancer cell lines HCT116 p53+/+ and HCT116 p53−/− in order to determine the influence of a loss of a functional p53 DNA-damage response [18, 19]. In xenograft mouse models, with these colorectal cancer cell lines, we confirmed that 123I-MAPi treatment translated into a therapeutic response that works better to inhibit the tumors when exploiting the common cancer cell state of loss of tumor suppressor p53 function.
Methods
General.
Unless specified otherwise, all reagents were purchased from Sigma-Aldrich and used as received. 4-(4-fluoro-3-(piperazine-1-carbonyl)benzyl)phthalazine-1(2H)-one was purchased from Synthonix (Wake Forest, NC, USA). 123I-NaI in 0.1N NaOH was purchased from GE Healthcare (Arlington Heights, IL, USA), Nuclear Diagnostic Products (Rockaway, NJ, USA) and BWX Technologies (Vancouver, B.C., Canada). Olaparib (AZD-2281) was purchased from LC Laboratories. Water (18.2 MΩcm−1 at 25 °C) was obtained from an Alpha-Q Ultrapure water system (Millipore) and acetonitrile (MeCN) as well as ethanol (EtOH) were of high-performance liquid chromatography (HPLC) grade purity. HPLC purification and analysis were performed on a Shimadzu UFLC HPLC system equipped with a DGU-20A degasser, an SPD-M20A UV detector, a LC-20AB pump system, and a CBM-20A Communication BUS module. A LabLogic Scan-RAM radio–thin layer chromatography/HPLC detector was used to detect activity.
Cell Culture.
Both human colorectal cell lines, HCT116 p53+/+ and HCT116 p53−/− were kindly provided by the laboratory of Dr. Bert Vogelstein (John Hopkins, Baltimore, MD). Cells were grown in McCoy’s 5A (Iwakate and Grace Modification), 2 mM [+] L-glutamine, 25 mM HEPE (Cat#10–051-Cl, Corning) supplemented with 10% heat inactivated FBS, 50 U/mL penicillin and 50 μg/mL streptomycin. Both cell lines were maintained in a 37 °C environment containing 5% CO2. All media was purchased from the culture media preparation faculty at MSKCC (New York, NY).
Immunohistochemistry.
Tissues were fixed in 4% paraformaldehyde/PBS, permeabilized and stained with anti PARP1 antibody (Invitrogen, PA5–16452).
Immunoblotting.
Tumors were removed and snap frozen. Proteins were extracted by homogenization of the tissue with RIPA buffer (0.1% SDS, 1% IGEPAL NP-40, 0.5% Deoxycholate, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM Tris-Cl pH 8.0, 1mM PMSF, 8.5 μg/ml Aprotinin, 2 μg/ml Leupeptin and phosphatase inhibitor cocktail). RIPA buffer was added at a ratio of 0.4 ml per 0.1 g tissue (1:4 w:v). Incubation was carried out on ice for 10 min to maximize protein solubilization and the sample was vortexed every 2 minutes. Additional sonication of the lysate (for 3 times) for 30 sec pulses/ 30 sec rest on ice at 98% amplitude was carried out after the incubation. Samples were centrifuged at 13,000 rpm for 15 min at 4°C. 50 ug of cell extracts were run on 10% or 12% SDS-PAGE to separate samples followed by electrotransfer onto Polyvinylidene fluoride membrane (GE). The membrane was blocked with 5% non-fat milk (Biorad) or 5% Bovine Serum Albumin (BSA, Sigma) in 1X TBS-0.1% Tween-20 followed by incubation with primary antibody overnight at 4°C. The membrane was washed with 1X TBS-0.1% Tween-20 3 times and incubated with secondary antibody for 1hr at room temperature. The signal was detected by chemiluminescence with Super Signal Kit (Pierce) and autoradiography with Hyblot CL films (Denville Scientific) or Typhoon FLA 7000 laser scanner (GE Healthcare). Densitometric quantification was carried out with ImageJ software (NIH). Values for the proteins of interest were normalized for values of loading controls. Statistical analyses were conducted in Graphpad Prism 9. Results are expressed as mean + SEM. Statistical significance for hypothesis testing was performed by multiple t tests (and nonparametric tests).
Synthesis of 127I-PARPi.
127I-PARPi was synthesized according to a previously published procedure [20]. Briefly, 4-(4-fluoro-3-(piperazine-1-carbonyl)benzyl)phthalazin-1(2H)-one (100 mg, 0.27 mmol) was dissolved in 5 mL of DCM and added to 50 mg (0.21 mmol) of 3-iodobenzoic acid, followed by 125 mg (0.33 mmol) of N,N,N’,N’-tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate (HBTU) and 100 μL (0.73 mmol) of Et3N. The reaction mixture was then stirred overnight at room temperature. The excess solvent was removed in vacuo and the crude product purified by flash column chromatography (DCM:MeOH 1% - 5%) to yield the title compound (69 mg, 0.12 mmol, 55%) as a white foamy solid. 1H-NMR (CDCl3) δ = 10.48 (s, 1H), 8.40 – 8.39 (m, 1H), 7.74 – 7.66 (m, 5H), 7.27 – 7.26 (d, 2H), 7.09 – 7.07 (d, 2H), 4.22 (s, 2H), 3.73 – 3.14 (m, 8H).
Synthesis of 4-(4-fluoro-3-(4-(3-(tributylstannyl)benzoyl) piperazine-1-carbonyl)benzyl) phthalazin-1(2H)-one (Sn-MAPi).
Sn-MAPi was synthesized according to a previously published procedure [21]. Briefly, to a flame dried round bottom flask under an atmosphere of argon was added, anhydrous DCM (10 mL), 4-(4-fluoro-3-(piperazine-1-carbonyl)benzyl)phthalazin-1 (2H) -one (64 mg, 0.17 μmol), HBTU (88 mg, 0.24 mmol), DIPEA (42 μL, 0.12 mmol) and N-succinimidyl-4-(tributylstannyl)benzoate (100 mg, 0.19 mmol). The reaction mixture was stirred overnight before removing the excess solvent in vacuo. The reaction was extracted with EtOAc (30 mL) and washed with Brine (3 × 30 mL). The organic phase was collected, and the excess solvent removed in vacuo. The crude mixture was purified by flash column chromatography (DCM:MeOH 0–10%) affording 4-(4-fluoro-3-(4-(3-(tributylstannyl)benzoyl) piperazine-1-carbonyl)benzyl) phthalazin-1(2H)-oneas a white solid (37.1 mg, 0.049 mmol, 29%); 1H NMR (500 MHz, Chloroform-d) δ 10.20 (bs, 1H), 8.46 (dt, J = 6.9, 3.8 Hz, 1H), 7.79 – 7.67 (m, 3H), 7.48 (d, J = 31.8 Hz, 2H), 7.79 – 7.67 (m, 4H), 7.03 (s, 1H), 4.28 (s, 2H), 3.94 – 3.23 (m, 8H), 1.50 (h, J = 9.1, 8.3 Hz, 6H), 1.31 (h, J = 7.3 Hz, 6H), 1.14 – 0.97 (m, 6H), 0.86 (t, J = 7.3 Hz, 9H).
Synthesis of 123I-MAPi.
123I-MAPi was synthesized according to a previously published procedure.17 Briefly, to a 1.5 mL microcentrifuge tube was added 4-(4-fluoro-3- (4-(3-(tributylstannyl)benzoyl)piperazine-1-carbonyl)benzyl) phthalazin-1(2H)-one (100 μg, 0.132 μmol) in anhydrous MeCN (10 μL), N-Chlorosuccinimide (NCS) (50 μg, 0.37 μmol) in a solution of anhydrous methanol (25 mμL), acetic acid (10 μL), anhydrous acetonitrile (20 μL) and 123I-NaI in NaOH 0.1 M (0.34–0.41 GBq, 9.29–11.04 mCi). After addition, the reaction was stirred at 650 rpm for 15 min at 25 °C. The crude reaction was purified by RP-HPLC (C18 Waters Atlantis T3) column (C18-RP, 5 mm, 6 × 250 mm) Solvent A: H2O, Solvent B: MeCN; flow rate: 1 mL/min; gradient: 0 – 5 min 5% B; 5 – 15 min 5 – 95% B; 15 – 18 min 95% B; 18 – 20 min 95 – 5% B). Under these conditions, 123I-MAPi has retention time of about 15 min (activity yield (AY): 53 ± 5% (n = 6)), which was collected and concentrated to dryness in vacuum. The compound has radiochemical purity > 99% and molar activity (MA) of greater than 10 GBq μmol−1 (EOS).
123I-MAPi Stability Study.
123I-MAPi (reconstituted in 30% PEG/PBS) was added to a solution of 10% fetal-bovine serum and incubated at 37 ºC for pre-determined times. Once elapsed, an aliquot was taken and injected directly onto RP-HPLC for analysis.
Uptake Assay.
Uptake of 123I-MAPi was tested in vitro (three replicates) as previously described [13]. 2.5 × 105 cells (HCT116 p53+/+, HCT116 p53−/−) were plated in a 6-well plate 24 h prior to the experiment (n = 3). Media was changed and 1 h later 3.7 kBq of 123I-MAPi were added to the cells. For blocking, cells were incubated with a 100-fold molar excess of Olaparib 1 h before adding 123I-MAPi. Media was removed and cells were washed with PBS and lysed (1M NaOH) at 1 h. The lysates were collected, and uptake was determined by radioactivity on a γ-counter.
Viability Assay.
8 × 103 cells (HCT116 p53+/+, HCT116 p53−/−) were plated in a 96-well plate in 200 μL of media 24 hours prior to the experiment (n = 3). Each cell line was incubated with 0 – 1,700 kBq of 123I-MAPi (50 μL per well (250 μL total), reconstituted in 10% PEG/PBS) or an equivalent dose of Olaparib (50 μL per well (250 μL total), reconstituted in 10% PEG/PBS) for 4 days at 37 ºC (three replicates, n = 3 each). Viability was determined by Alamar Blue assay as indicated by the manufacturer. Specifically, Alamar blue reagent, as provided by the manufacturer, was diluted 1 in 10 in media. Subsequently, the remaining media in each well of the 96-well plate was removed and the diluted Alamar blue reagent in media was added (200 μL) directly to each well. The 96 well plate was incubated at 37 ºC for four hours before analysis was performed. Analysis was performed using an iD5 microplate reader.
Animal Work.
All in vivo experiments were performed with female athymic nude CrTac:NCr-Fo mice purchased from Envigo RMS, Inc. at age 6–8 weeks. During subcutaneous injections, mice were anesthetized using 2% isoflurane gas in 2 L/minute medical air. Each cell line was injected in the right shoulder subcutaneously (HCT116 p53+/+: 1 × 106 in 100 μL 1:1 media/matrigel, HCT116 p53−/−: 1 × 106 in 100 μL 1:1 media/matrigel). For bio-distribution studies and DNA damage studies, all tumors were grown for 3–4 weeks such that average tumor volumes were 300 – 400 mm3. For intravenous injections, the lateral tail vein was used. Mice were warmed with a heat lamp, placed in a restrainer, and the tail was sterilized with alcohol pads before injection.
123I-MAPi Toxicity study in Healthy Mice.
123I-MAPi (74 MBq reconstituted in 30% PEG/PBS) or vehicle (30% PEG/PBS) was injected intravenously into healthy mice (n = 3/cohort). At pre-determined time points (1 day, 7 days, 14 days), cohorts were euthanized, and the associated organs, tissue and blood harvested for analysis. Organs/tissues included were: liver, gallbladder, spleen, stifle joint, sternum, vertebral column, kidneys, head-forebrain (optic chiasm, thalamus/hypothalamus), head-hindbrain (pons, midcerebellum/medulla oblongata), blood plasma.
127I-PARPi Toxicity study in Healthy Mice.
For the investigation into 127I-PARPi related toxicity, the study was carried out under GMP guidelines. 127I-PARPi (2.05 mg/kg reconstituted in 30% PEG/PBS) or vehicle (30% PEG/PBS) was injected intravenously into healthy mice (n = 10/cohort (5 male, 5 female)). At pre-determined time points (2 days, 15 days), cohorts were euthanized, and the associated organs, tissue and blood harvested for analysis. Organs/tissues included were: artery (aorta), body cavity (nasal), bone marrow (sternum, smear), bone (femur, sternum), brain, epididymis, esophagus, eye, gallbladder, ganglion, dorsal root, lumbar, gland (adrenal, clitoral, lacrimal, harderian, mammary, parathyroid, pituitary, preputial, preputial, prostate, salivary (submandibular, sublingual, parotid, seminal vesical, thyroid, Zymbal’s), gut-associated lymphoid tissue, heart, joint (femorotibial), kidney, large intestine (cecum, colon, rectum), larynx, liver, lung, lymph node (lumbar), muscle (skeletal), nerve (optic, sciatic, tibial), ovary oviduct, pancreas, skin, small intestine (duodenum, ileum, jejunum), spinal cord, spleen, stomach, testis, thymus, tongue, trachea, ureter, urinary bladder, uterus, vagina.
Survival Studies.
Mice were inoculated subcutaneously with HCT116 p53+/+ cells (106 cells in 100 μL 1:1 growth media/matrigel) or HCT116 p53−/− cells (106 cells in 100 μL 1:1 growth media/matrigel). The tumors were grown until an average of 100 ± 10 mm3 was achieved throughout the group. At this point, the mice were randomized into separate cohorts and injected intravenously with 123I-MAPi (approximately 74 MBq in 100 μL 30% PEG300 in 0.9% sterile saline) or 30% PEG300 in 0.9% sterile saline vehicle (100 μL). For the HCT116 p53−/− cell line, an additional cohort was injected with 127I-PARPi (approx. 80 μg/kg ± 10%). Mice were monitored daily thereafter. Study endpoints was determined on the basis of animals’ sign of discomfort, pain, significant weight loss, tumor size greater than 800 mm3 or ulceration greater than 5 mm in diameter.
Organ Level Dosimetry.
For dosimetry studies, healthy athymic nude mice were injected intravenously with 123I-MAPi at pre-determined time points and organs were collected for biodistribution. The 123I-MAPi time-dependent biodistribution data (from 1 to 48 h post-intravenous injection), initially expressed as the percent injected dose per gram (%ID/g) decay-corrected to the time of injection, were converted to organ standard uptake values (SUVs) for translation of these data from mouse to man, that is, to calculate the fraction of the injected dose in each human organ. The time-activity data for each organ were fit to either a mono-exponential or bi-exponential time-activity function, as appropriate, using EXCEL (Microsoft, Redmond, WA). The time-integrated activity coefficient (formerly known as the cumulated activity or residence time) in human organ i, τi, in μCi-h/μCi or simply h, was then calculated by analytically integrating the time-activity function in organ i, replacing the biological clearance constant, (λb)j. for each component j of the fitted exponential function with the corresponding effective clearance constant, , where is the physical decay constant of 123I. Assuming, in first order, that SUVs are independent of body mass and thus the same among species, the mean SUV in mouse organ i, SUVOrgan i|Mouse, was converted to the fraction of the injected dose in each human organ i, FIDOrgan i|Human, using the following formula:
and the organ and total-body masses of the 70-kg Standard Man anatomic model [22].
The resulting organ 123I time-integrated activity coefficient were entered into the OLINDA computer program [23] to yields the 123I mean organ absorbed doses and effective dose in rad/mCi and rem/mCi, respectively, for the 70-kg Standard Man anatomic model.
The mouse organ absorbed doses were also calculated by multiplying the time-integrated activity coefficient concentration in each mouse organ, in μCi-h/μCi/g, by the 123I energy per decay (formerly the equilibrium dose constant) for particulate radiations (i.e., conversion and Auger electrons), 0.0601 g-rad/μCi-h [24], assuming complete local of the particulate radiations and ignoring the small dose contribution of the 123I x- and gamma-rays.
Statistical Significance.
Data are expressed as mean ± SD. Statistical analyses were performed with GraphPad Prism. Non-parametric two-tailed Student’s t-tests with assumption of unequal standard deviations were used to calculate statistics. P-values < 0.05 were considered significant.
Data and Material Availability.
All data necessary for interpreting the manuscript have been included in the main manuscript or in the supplementary materials. Additional information may be requested from the authors.
Results
Synthesis of 123I-MAPi and in vitro evaluation.
For this work, 123I-MAPi was synthesized according to our previously reported single step 123I-iododestannylation of the corresponding precursor with good molar activities (≥ 10 GBq/μmol), radiochemical yields (53% ± 5% (n = 6)) and high radiochemical purities (> 99%) achieved throughout the study. 123I-MAPi demonstrated high serum stability up to 24 hours when incubated at 37 ºC (Supporting Information, Fig. S1A) and blood half-life consistent to what has been observed in the past (Supporting Information, Fig. S1B).
Toxicity study of 127I-PARPi in healthy athymic nude mice.
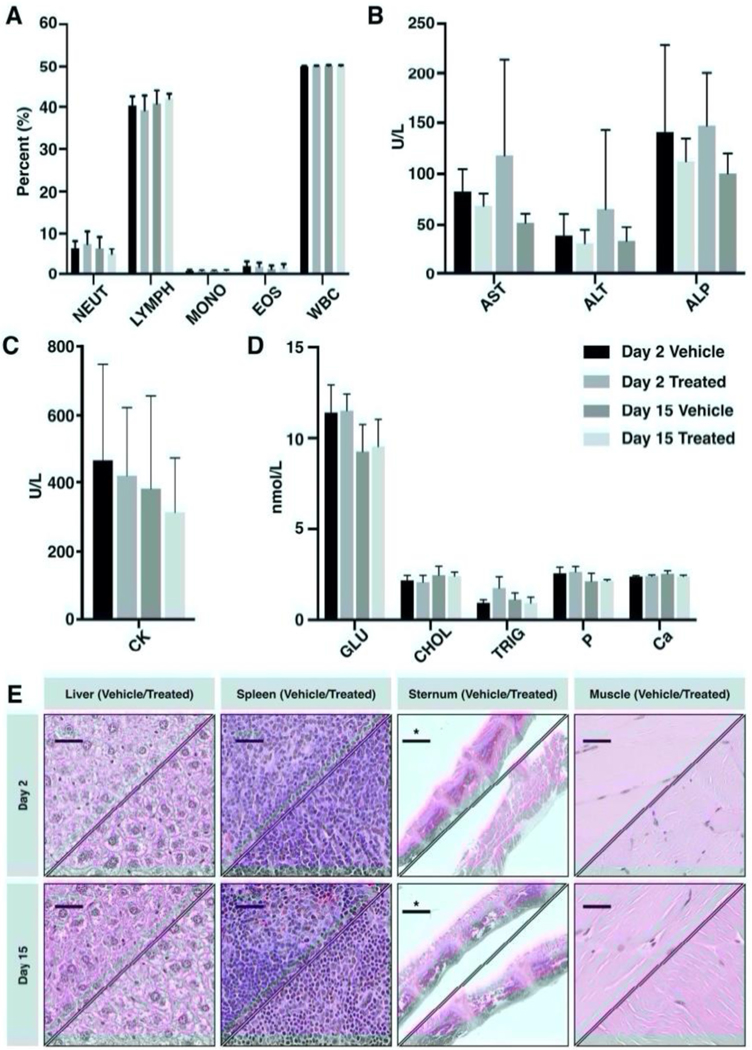
To investigate wider toxicity and influence of 127I-PARPi upon other organs and blood chemistry, healthy athymic nude mice (n = 10/cohort), were administered with 127I-PARPi (2.05 mg/kg) or vehicle intravenously and the animals sacrificed at 24 hours or 2 weeks post compound administration (Table S1). Following this, blood draws were taken along with selected organs for pathology, immunohistochemistry and blood chemistry analysis. For all markers evaluated, analysis may be found in the supporting information. Blood analysis did not show any significant difference over the course of 15 days for treated and vehicle (Fig. 1A). Markers associated with liver toxicity including ALP, ALT and AST levels demonstrated no significant variation compared to vehicle cohorts (Fig. 1B), similarly to CK (Fig 1C) and markers of organ toxicity (Fig. 1D), a result which was confirmed through H&E analysis of the corresponding tissues (Fig. 1E).
Figure 1. In vivo toxicity of 127I-PARPi in healthy female athymic nude mice after injection with 2.05 mg/kg of 127I-PARPi.
(A) Immune cell population levels; (B) Blood chemistry levels of biomarkers associated with liver toxicity; (C) Blood chemistry levels of biomarkers associated with muscle toxicity; (D) Selected markers associated with other key organs including liver (GLU), heart (CHOL), pancreas (TRIG), bone (P and Ca); (E) Immunohistochemical analysis of key organs: top left section shows a representative vehicle mouse and bottom right section shows a representative 127I-PARPi treated mouse example. Scale bar represents 100 μm, scale bar with asterisk represents 1 mm.
Biodistribution of 123I-MAPi and Dosimetry Studies.
Human colorectal cancer HCT116 p53+/+ and HCT116 p53−/− cell lines were used to grow xenograft tumors in athymic nude mice. Cells were implanted on the right shoulder (n = 3/group) of each animal and were allowed to form tumors. Animals were treated when implanted tumors were ~100 mm3 size. For blocking, Olaparib was administered intravenously 1 hour prior to intravenously injection of 123I-MAPi. At 1 hour (Fig. S2A, S2B, and Table S2A,B) and 2 hours (Fig. S3A, S3B, and Table S3A,B) time points post administration of 123I-MAPi, mice were sacrificed and acute biodistribution studies carried out. 123I-MAPi demonstrated hepatobiliary clearance, a result in line with other previously reported agents. Inverse to those results observed in vitro, HCT116 p53−/− tumors demonstrated higher specific uptake compared to HCT116 p53+/+ tumors at 1-hour post compound administration.
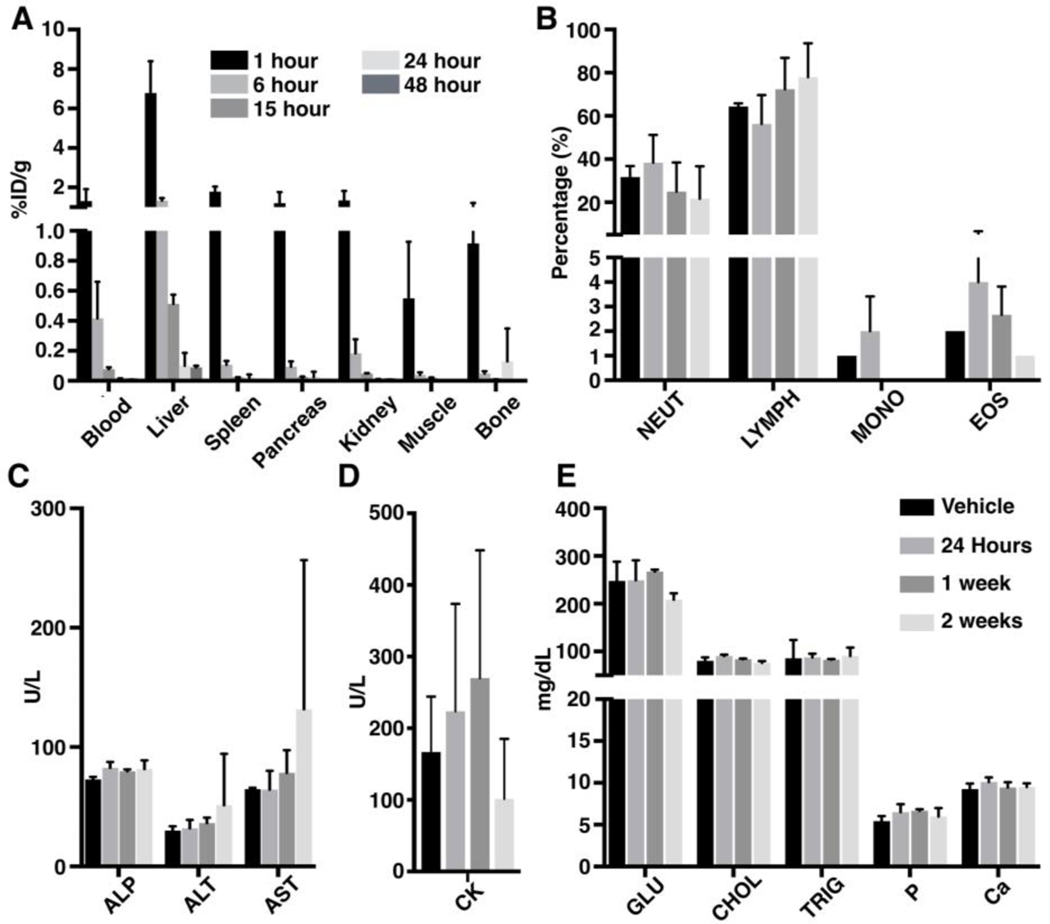
For dosimetry studies, healthy athymic nude mice were injected intravenously with 123I-MAPi at pre-determined time points. As observed for tumor bearing mice, 123I-MAPi was hepatobiliary cleared. Clearance from all major organs had occurred by 15 hours post i.v. injection (Fig. 2A, Table S4). Uptake in the thyroid was observed at all time-points, a result likely due to the in vivo de-halogenation of 123I-MAPi. Following bio-distribution studies, dosimetry analysis was carried out (Fig. S2A, S2B), and human dose projections were calculated (Table S5).
Figure 2. In vivo toxicity of 123I-MAPi in healthy female athymic nude mice after injection with 74 MBq of 123I-MAPi.
(A) Biodistribution of 123I-MAPi in key organs at selected time points; (B) Immune cell population levels; (C) Blood chemistry levels of biomarkers associated with liver toxicity; (D) Blood chemistry levels of biomarkers associated with muscle toxicity; (E) Selected markers associated with other key organs including liver (GLU), heart (CHOL), pancreas (TRIG), bone (P and Ca).
Toxicity study of 123I-MAPi in healthy athymic nude mice.
To interrogate off-target effects of 123I-MAPi, healthy athymic nude mice (n = 3/cohort) were administered with 123I-MAPi (74 MBq) intravenously and the animals sacrificed at 24 hours, 1 week or 2 weeks post compound administration. Following this, blood draws were taken along with selected organs for pathology, immunohistochemistry and blood chemistry analysis. Blood chemistry revealed slightly increased levels of lymphocytes at the two-week time point while other cell populations demonstrated minimum variation compared to vehicle control (Table S6). Immune cell populations were analyzed and did not show any significant change compared to the controls (Fig. 2B). Markers associated with liver toxicity including ALP, ALT and AST levels demonstrated increased levels in a single treated mouse at the two-week time point (Fig. 2C). CK levels, a marker associated with muscle damage were shown to be raised at one-week post treatment (Fig. 2D), however, this increase was reversed by the two-week time point. Finally, a range of blood-based markers were evaluated including glucose, cholesterol, triglyceride, phosphorus and calcium levels. In all cases, minimal to no variations were observed (Fig. 2E).
123I-MAPi and in vitro evaluation.
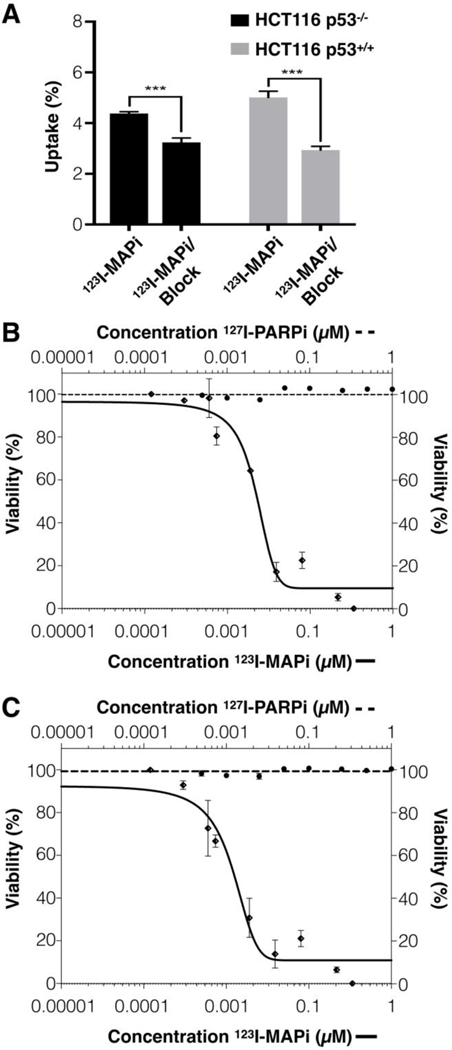
In line with our previous studies, the in vitro performance of 123I-MAPi was evaluated in both HCT116 p53+/+ and HCT116 p53−/− cell lines. In vitro internalization assays were performed with 123I-MAPi (37 kBq/well) being added to an adherent monolayer of each respective cell line and uptake being measure through gamma counting at 1-hour post incubation. In vitro, HCT116 p53+/+ demonstrated higher levels of specific uptake when compared to HCT116 p53−/− (Fig. 3A). To show target specificity, a 100-fold excess of Olaparib was used for blocking studies.
Figure 3. In vitro cytotoxicity of 123I-MAPi and p53 status.
(A) In vitro cellular uptake of 123I-MAPi 1 hour post compound administration. For blocking doses, a 100-fold excess of 127I-PARPi was administered intravenously 1 hour prior to administration of 123I-MAPi; Alamar Blue toxicity assay comparing the efficacy of 123I-MAPi and olaparib. Nonlinear fit four parameters variable slope (B) 123I-MAPi HCT116 p53−/− EC50 22 nmol/L. (C) 123I-MAPi HCT116 p53+/+ EC50 11 nmol/L.
Following in vitro validation results, we tested the cancer specificity of 123I-MAPi in HCT116 p53+/+ and HCT116 p53−/− cell lines via incubation with 123I-MAPi at predetermined concentrations. In all cases, 123I-MAPi demonstrated a significantly increased potency when compared to Olaparib. 123I-MAPi showed the highest potency against that HCT116 p53+/+ (EC50 = 11 nmol/L, Fig. 3B) followed by HCT116 p53−/− (EC50 = 22 nmol/L, Fig. 3C.
Ex vivo evaluation of 123I-MAPi to induce DNA damage response.
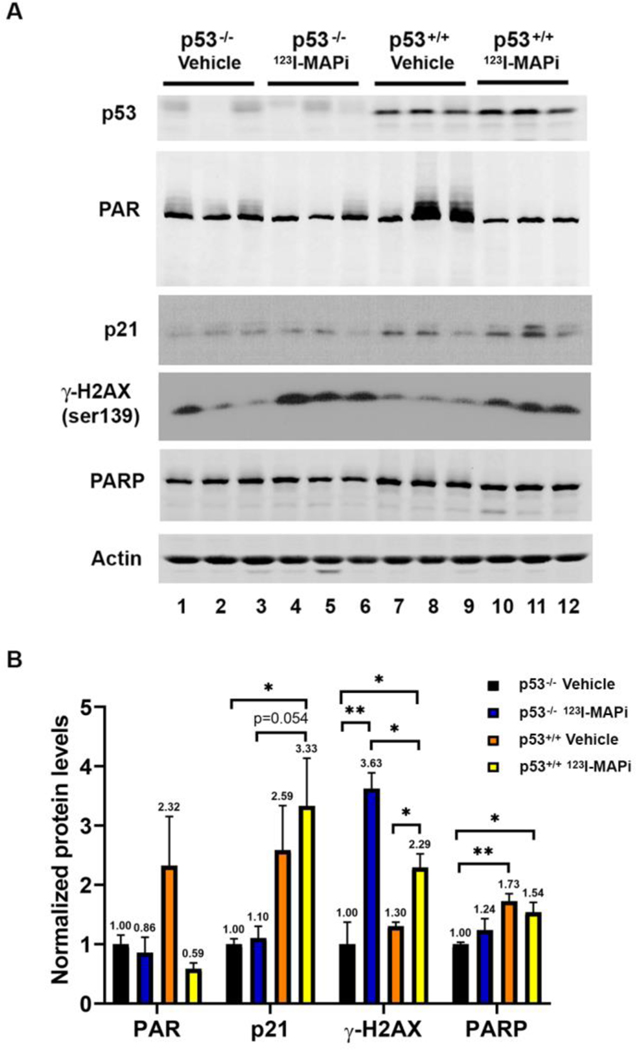
To evaluate the ability for 123I-MAPi to induce DNA damage, and associated responses, at the site of the tumor, athymic nude mice were implanted with the well described human colorectal cancer model HCT116 p53+/+ and HCT116p53−/− cell lines [18, 19]. The tumors were allowed to grow to 300 – 400 mm3 (n = 12/cohort). At this point, mice were administered either vehicle or 123I-MAPi (74 MBq) intravenously before harvesting tumors at the 24-hour time point. Tumors were stored in 4% PFA for embedding and subsequent immunohistochemistry (n = 3/cohort), or snap frozen and stored in liquid N2 (n = 3/cohort) for subsequent immunoblotting. Western blot analysis demonstrated that while tumors from mice with HCT116 p53−/− cells had no increase in the cyclin dependent kinase inhibitor p21, the tumors from mice with HCT116 p53+/+ cells demonstrated wild-type p53 protein activity with concomitant increased p21 cyclin dependent kinase inhibitor (Fig. 4A, compare lanes 1–6 with 7–12). Moreover, the tumors from treated mice with HCT116 p53−/− displayed more DNA damage than the tumors from treated mice with HCT116 p53+/+ when compared by increased levels of γH2AX (Fig. 4A, compare lanes 4–6 to lanes 10–12). Quantitation of the western blot results demonstrated that 123I-MAPi treatment caused statistically significant increased p21 in HCT116 p53+/+ as compared to HCT116 p53−/− tumors (Fig. 4B). The quantitation also showed that treated mice with HCT116 p53−/− tumors had a statistically significant increase in DNA damage in their tumors compared to that observed in HCT116 p53+/+ tumors (Fig. 4B).
Figure 4. Loss of wild type p53 (p53−/−) reduced p21 response, and increased γH2AX, after 123I-MAPi -mediated double strand breaks (DSBs).
(A) Western blot analysis of p53, PAR, p21, γH2AX and PARP protein levels in HCT116 p53−/− or HCT116 p53+/+ tumors 24 hours post administration with either vehicle control or 123I-MAPi (n = 3/cohort). (B) Quantification of western blot in panel A. ImageJ analysis was performed for PAR, p21, γH2AX, and PARP protein levels normalized to actin. The graph represents an average of three mice/group. The p-value was determined by multiple t tests and nonparametric tests. *P < 0.05, **P < 0.01.
Therapeutic efficacy of 123I-MAPi in subcutaneous colorectal cancer models.
Having established the ability for 123I-MAPi (74 MBq) to induced DNA damage at the site of the tumor but not in off-target organs when administered systemically, we turned our attention to investigating the therapeutic efficacy of 123I-MAPi in athymic nude mice with either HCT116 p53+/+ or HCT116 p53−/− cell lines when implanted subcutaneously. Both respective cell lines were implanted in the right shoulder and allowed to grow until the average tumor size across all tumors was 100 mm3. At this stage, mice were randomized into cohorts and therapy studies initiated. Study endpoints were determined on the basis of animals’ sign of discomfort, pain, significant weight loss, tumor size greater than 800 mm3 or ulceration greater than 5 mm in diameter (Fig. S4A, S4B).
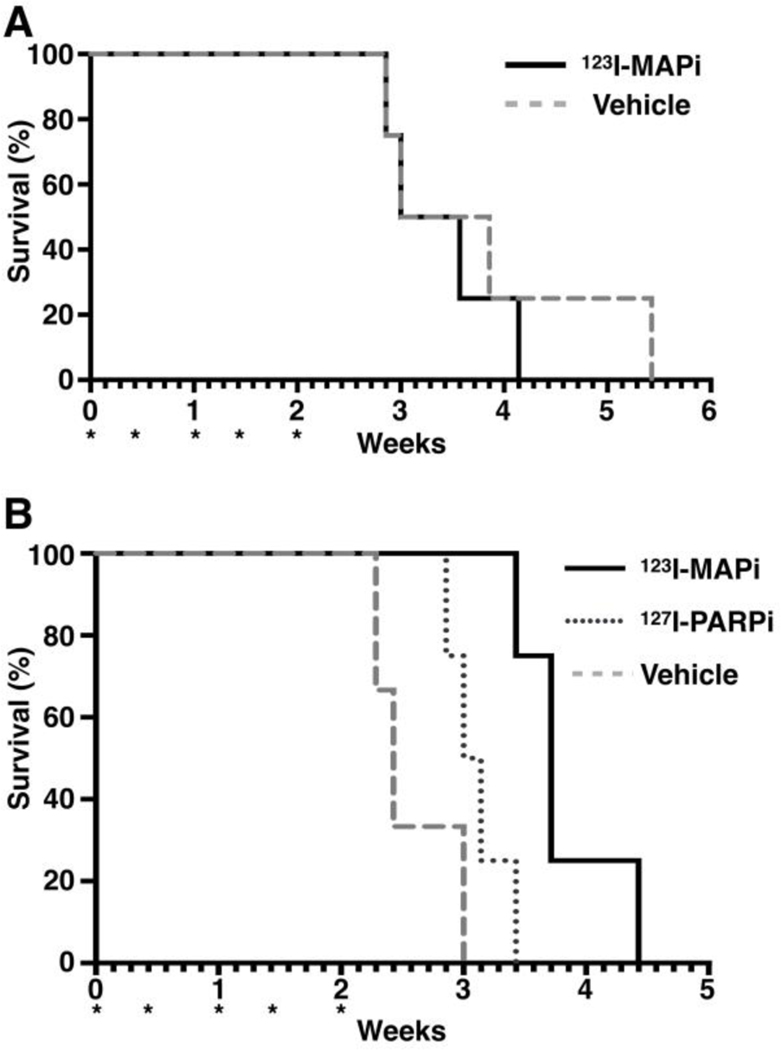
Mice were treated every 3 – 4 days with either vehicle (30% PEG/PBS) or 123I-MAPi (74 MBq) for 5 cycles of treatment with daily weight measurements taken (Fig. S5A, S6B). Within HCT116 p53+/+ cohorts, 123I-MAPi (median survival = 3.286 weeks, Table 1) was shown to have no statistically significant efficacy upon tumor growth delay compared to the vehicle control cohort (median survival = 3.429 weeks, p = 0.5924, Table 1). For HCT116 p53−/−, survival data confirmed 123I-MAPi (median survival = 3.174 weeks Table 1) to be statistically significant for overall survival compared to the vehicle control group (median survival = 2.429 weeks, (+53%), p = 0.0044, Table 1). Tumor uptake was slightly lower to what has been previously observed with the same molecule probably due to the chosen cell line model.
Table 1.
Summary of survival data for selected cell lines.
| Cohort | n | Median survival (weeks) | P |
|---|---|---|---|
| HCT116 p53+/+ Vehicle | 4 | 3.429 | N/A |
| HCT116 p53+/+ 123I-MAPi | 4 | 3.286 | 0.5924 (ns) |
| HCT116 p53−/− Vehicle | 6 | 2.429 | N/A |
| HCT116 p53−/− 127I-MAPi | 4 | 3.071 | 0.0538 (ns) |
| HCT116 p53−/− 123I-MAPi | 4 | 3.714 | 0.0169*, 0.0044** |
Significance when comparing 123I-MAPi (2 mCi) treatment cohort vs. 127I-PARPi treatment cohort;
Significance when comparing 123I-MAPi (2 mCi) treatment cohort vs. vehicle treatment cohort.
To determine if this efficacy was due to the Auger effect or that of PARP inhibition, a cold dose of 127I-PARPi at an equivalent concentration (approx. 80 μg/kg ± 10%) to that of 123I-MAPi was administered. Mice were sacrificed when tumors reached 800 mm3. Although an improved median survival compared to control group was observed (+26%), this was not statistically significant (Fig. 5).
Figure 5. Kaplan Meier curves for selected cell lines.
(A) HCT116 p53+/+; (B) HCT116 p53−/−; *Represents day of treatment. For each mouse either vehicle or 123I-MAPi (2 mCi) was administered intravenously; Study endpoints was determined on the basis of animals’ sign of discomfort, pain, significant weight loss, tumor size greater than 800 mm3 or ulceration greater than 5 mm in diameter.
Correlating p53 status, response and PARP1 expression.
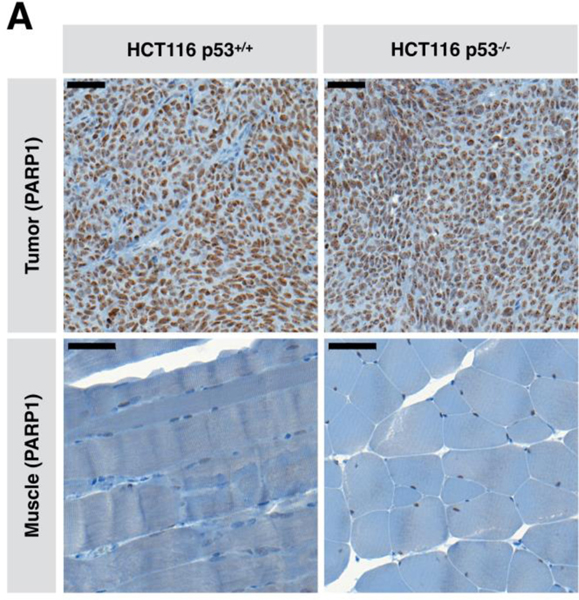
To evaluate if p53 status effects levels of expression of PARP1, both HCT116 p53+/+ and HCT116 p53−/− cell lines – as confirmed by western-blot analysis – were implanted subcutaneously in athymic nude mice (n = 3/cohort) and allowed to grow to approximately 350 – 450 mm3 before removal, PFA fixation and sectioning to examine PARP1 status. IHC staining for PARP1 demonstrated both tumors to express PARP1. This confirmed the data shown in Fig 4 that the colorectal cancer cells with or without wild-type p53 expression had expression of PARP protein and PARPi inhibited poly-ADP-ribose (PAR) activity (Fig. 6).
Figure 6. Immunohistochemistry of PARP in tumor and muscle.
(A) Immunohistochemistry to determine the PARP1 status in HCT116 p53+/+ and HCT116 p53−/− colorectal cell lines and muscle; Scale bar: 50 μm.
Discussion
The use of Auger containing theranostic agents is a field that has seen a renaissance in recent years with several promising candidates being disclosed [5]. In 2020, our lab reported the use of 123I-MAPi, a PARP Auger theranostic agent for the treatment of glioblastoma through a convection enhanced delivery (CED) process [13, 21]. The success of this work was built on the hypothesis that although large fractions 123I-MAPi pass through the hepatobiliary system during excretion, metabolism confines the agent’s intracellular localization to the peri-nuclear region of the cell and therefore puts it outside the range of an Auger emitter to achieve significant damage upon cellular DNA [25]. In comparison, PARP1 in cancer cells is expressed at a higher level in the nuclear region of the cell, and within the range of the DNA – suggesting that the likelihood of inducing DNA damage by 123I-MAPi is increased. In our previous work, a modest toxicity study was carried out with the results demonstrating 123I-MAPi to be non-toxic when administered through a CED pathway. With the aim of this work to achieve a therapeutic response via systemic administration, it was likely that a higher initial dose would be required and therefore, further investigations into off-site toxicity were necessary.
As highlighted, several organs are well described for expressing high levels of PARP1 including – but not limited to – liver, spleen, pancreas and bone marrow [26]. Despite this, no toxicity was observed when127I-PARPi, the stable isotopologue of 123I-MAPi, was administered intravenously at 2.05 mg/kg, as confirmed through immunohistochemical and blood chemistry analysis. Following this, we turned our attention to 123I-MAPi. Biodistribution studies were carried out at multiple pre-determined time points, thus allowing for dosimetry studies to be performed. In a single treated mouse, raised ALT and AST levels suggest minor levels of liver toxicity. 5 g body weight was occasionally observed in an animal but was not considered a major sign of toxicity after consultation with a veterinary. To further probe the toxicity, the respective tissues were subject to pathology studies. Pathology showed no significant differences between treated animals and controls. All other markers considered in this study demonstrated no toxicity when treated with 74 MBq of 123I-MAPi. Having shown that 123I-MAPi led to no significant off-site toxicity when administered at 74 MBq/mouse, we moved to confirm if 123I-MAPi induced DNA damage at the site of tumors after intravenous administration.
PARP inhibitors are often used as treatment for BRCA1 mutant cancers [27, 28]. PARP inhibitors exploit cancer cells DNA damage defects and cause synthetic lethality under such conditions. Little focus has been placed on exploiting p53 loss-of-function (LOF) but a great deal has focused on reactivating wild-type activity [15]. Dysfunctional p53 has lost the ability to induce signals for cell cycle arrest, apoptosis and DNA repair [29, 30]. As such, the loss of p53 function has the potential to increase PARP inhibitor to efficacy [31, 32]. PARP inhibitors induce the strongest death responses when they trap PARP on damaged DNA [27].
Colorectal cancers are well known to have mutations in the TP53 gene [16, 17]. We examined a well characterized colorectal cancer cell model with, and without, wild-type p53 function (HCT116 p53+/+ compared to HCT116 p53−/−) [18, 19]. This allowed us to compare how p53 function influenced 123I-MAPi Auger associated DNA damage. When we examined ex vivo tumor tissue we determined that 123I-MAPi treatment caused more DNA damage in HCT116 p53−/− than in their HCT116 p53+/+ tumor counterparts. This was measured by the amount of γH2AX. Moreover, the increased DNA damage in HCT116 p53−/− tumors correlated with reduced xenograft tumor volume, and increased survival. Mice with HCT116 p53+/+ tumors did not have a statistically significant increase in 123I-MAPi induced survival. These data suggest that colorectal cancers with LOF p53 pathways respond with the classic synthetic lethal paradigm. This specificity for killing LOF HCT116 p53−/− tumors with 123I-MAPi treatment, while leaving the surrounding tissue free from damage, is a breakthrough that has long been desired for targeting the numerous cancers types that express loss-of-function mutant p53. The observed discrepancy of cytotoxicity observed in vitro vs in vivo could be explained by the choice of the experimental assay. Further data are required in order to characterize this finding and more investigations in this sense are ongoing. A larger more thorough study might be needed in order to fully characterize the behavior of 123I-MAPi treatment in different in vitro p53 settings. This could clarify the discrepancy observed between in vivo and in vitro findings in the present study. Nevertheless, our data suggest that there actually is a correlation between treatment potency and p53 status in mice models, opening the way to broader hypothesis on the efficacy and applications of Auger radioligand therapy.
Due to its prevalence within cancer patients, mutations in p53 have been pursued extensively as a potential target for the treatment of cancers [15]. Tumor protein p53 (TP53/p53) – a tumor suppression protein – has been well documented for its ability to respond to oncogenic stress [30]. If the cancer does not have a p53 mutation, this allows for the elimination of incipient tumor cells prior to tumor formation and disease progression. In the case of mutated or null p53 however, evasion to p53 mediated cell death can be taken thus prompting tumor growth. In the United States, colorectal cancer is the third most common with over 100,000 new cases of colon cancer and over 40,000 new cases of rectal cancer being reported in 2020 [33]. Of those patients to be diagnosed, approximately 50% have a p53 mutation (mtp53), a trend which is common throughout many forms of cancer and one which is associated with poor outcomes. To this end, having a targeted radiotherapy that can exploit the loss of function found in mtp53 would be highly desirable. A growing body of literature has suggested that PARP inhibitors, in combination with external beam radiation, may offer a solution for patients bearing a p53 mutation [34, 35]. With this knowledge in hand, our results with 123I-MAPi as an all-in-one reagent that initiates high levels of DNA damage through the shuttling of Auger radiation to the site of the DNA are very compelling as a mechanism to induce synthetic lethality in p53 loss of function colorectal cancer.
In vitro toxicity studies confirmed 123I-MAPi to have low nanomolar potency against both HCT116 p53+/+ and HCT116 p53−/− cell lines compared to the stable isotopologue, 127I-PARPi. Furthermore, when administered intravenously to tumor bearing mice, ex vivo western blot analysis showed increased levels of DNA damage at 24 hours post compound administration when compared to vehicle control. Crucially however, for p53+/+ cell lines, in response to treatment with 123I-MAPi, activation of p21 was observed. Taken together, we hypothesize that in p53+/+ cell lines, the transient activation of p21 and associated cell cycle arrest allows for DNA damage to undergo repair and therefore, tumorigeneses to proceed. In p53−/− cells, the lack of p21 activation allows for DNA damage to persist and subsequent cell death to be induced, thus leading to tumor growth delay. A recent study has shown that the position of cells in the division cycle during which they sustain DNA damage can lead to different activation of p21 that can then act as a survival factor by inducing growth arrest [36]. Affirming this finding, the survival study in mice comparing p53−/− and p53+/+ tumors treated with 123I-MAPi showed similar outcomes. Cohorts were treated every 3–4 days in order to maximize the efficacy of 123I-MAPi and prevent intermittent tumor growth and recovery. Pleasingly, after 5 cycles of treatment, a clear distinction between p53−/− and p53+/+ tumor response rates was observed as confirmed with vehicle cohorts as control. The difference between cell line only studies and the evaluations of the tumors in mice may have a lot to do with the available time given in between doses for DNA repair in addition to outcomes driven by the immune response [37]. For p53−/− bearing mouse models, a 127I-PARPi cohort was added to determine if the efficacy observed in the 123I-MAPi cohorts was due to Auger associated damage. No significant increase in median survival was observed for 127I-PARPi treated mice.
To conclude, here we demonstrate for the first time, the ability for Auger containing PARP theranostics agents to be viable for the treatment of tumor bearing mice after systemic injection. The success of this work was built upon the identification of p53 mutant as a viable target for inducing synthetic lethality during 123I-MAPi treatment. Furthermore, minimal to no systemic toxicity was observed in non-targeting organs. In so doing, this forms the foundation for broader pre-clinical and clinical evaluation of 123I-MAPi in p53 mutant models.
Supplementary Material
Figure S1. In vitro evaluation of 123I-MAPi serum stability and blood half-life.
Table S1. Complete Toxicity Report of 127I-PARPi in healthy mice at 2 days and 15 days post compound administration.
Figure S2. Biodistribution of 123I-MAPi in human colorectal cancer HCT116 p53+/+ (A) and HCT116 p53−/− (B) mouse models 1 hour post administration.
Table S2. Bio-distribution tables for 123I-MAPi in selected cell lines expressed as %ID/g.
Figure S3. Biodistribution of 123I-MAPi in human colorectal cancer (A) HCT116 p53+/+ and (B) HCT116 p53−/− mouse models 2 hours post administration.
Table S3. Bio-distribution tables for 123I-MAPi in selected cell lines expressed as %ID/g.
Table S4. Bio-distribution of 123I-MAPi in healthy athymic nude mice. All numbers expressed as %ID/g.
Table S5. Dosimetry Data for 123I-MAPi in healthy athymic nude mice and the equivalent dose in humans.
Table S6. Complete Toxicity Report of 123I-MAPi in healthy mice at 24 hours, 7 days, 14 days.
Figure S4. Tumor growth curves of either HCT116 p53+/+ or HCT116 p53−/− mouse models when treated with vehicle, 127I-PARPi or 123I-MAPi.
Figure S5. Weights of mice of either HCT116 p53+/+ or HCT116 p53−/− mouse models when treated with vehicle, 127I-PARPi or 123I-MAPi.
Acknowledgements.
This work was supported by National Institutes of Health grants P30 CA008748, R01 CA204441 (TR) and R01 CA239603 (J.B.), the Breast Cancer Research Foundation (JB, BCRF-20-011), the Memorial Sloan Kettering Imaging and Radiation Sciences Program and a research grant from Theragnostics, Inc. The funding sources were not involved in study design, data collection and analysis, writing of the report or the decision to submit this article for publication.
Abbreviations.
- PARP
Poly (ADP-ribose) polymerase
- MAPi
Meitner-Auger PARP inhibitor
- DNA
Deoxyribonucleic acid
- PFA
paraformadheide
- IHC
immunohistochemistry
- H&E
Hematoxylin and eosin
Footnotes
Conflicts of Interest.
T.R. is shareholder of Summit Biomedical Imaging, LLC. T.R. is a coinventor on filed U.S. patent (WO2016164771). T.R. is a co-inventor on U.S. patent (WO2012074840) held by the General Hospital Corporation. T.R. is a co-inventor on U.S. patent (WO2016033293) held by MSK. T.R. is a paid consultant for Theragnostics, Inc.
References
- 1.Sgouros G, Bodei L, McDevitt MR, Nedrow JR. Radiopharmaceutical therapy in cancer: clinical advances and challenges. Nat Rev Drug Discov. 2020; 19: 589–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrmann K, Schwaiger M, Lewis JS, Solomon SB, McNeil BJ, Baumann M, et al. Radiotheranostics: a roadmap for future development. Lancet Oncol. 2020; 21: e146–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poty S, Francesconi LC, McDevitt MR, Morris MJ, Lewis JS. alpha-Emitters for Radiotherapy: From Basic Radiochemistry to Clinical Studies-Part 2. J Nucl Med. 2018; 59: 1020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poty S, Francesconi LC, McDevitt MR, Morris MJ, Lewis JS. alpha-Emitters for Radiotherapy: From Basic Radiochemistry to Clinical Studies-Part 1. J Nucl Med. 2018; 59: 878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ku A, Facca VJ, Cai Z, Reilly RM. Auger electrons for cancer therapy - a review. EJNMMI Radiopharm Chem. 2019; 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macapinlac HA, Kemeny N, Daghighian F, Finn R, Zhang J, Humm J, et al. Pilot clinical trial of 5-[125I]iodo-2’-deoxyuridine in the treatment of colorectal cancer metastatic to the liver. J Nucl Med. 1996; 37: 25S–9S. [PubMed] [Google Scholar]
- 7.Rebischung C, Hoffmann D, Stefani L, Desruet MD, Wang K, Adelstein SJ, et al. First human treatment of resistant neoplastic meningitis by intrathecal administration of MTX plus (125)IUdR. Int J Radiat Biol. 2008; 84: 1123–9. [DOI] [PubMed] [Google Scholar]
- 8.Capello A, Krenning E, Bernard B, Reubi JC, Breeman W, de Jong M. 111In-labelled somatostatin analogues in a rat tumour model: somatostatin receptor status and effects of peptide receptor radionuclide therapy. Eur J Nucl Med Mol Imaging. 2005; 32: 1288–95. [DOI] [PubMed] [Google Scholar]
- 9.Hu M, Scollard D, Chan C, Chen P, Vallis K, Reilly RM. Effect of the EGFR density of breast cancer cells on nuclear importation, in vitro cytotoxicity, and tumor and normal-tissue uptake of [111In]DTPA-hEGF. Nucl Med Biol. 2007; 34: 887–96. [DOI] [PubMed] [Google Scholar]
- 10.Shen CJ, Minn I, Hobbs RF, Chen Y, Josefsson A, Brummet M, et al. Auger radiopharmaceutical therapy targeting prostate-specific membrane antigen in a micrometastatic model of prostate cancer. Theranostics. 2020; 10: 2888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillyar CR, Cornelissen B, Vallis KA. Uptake, internalization and nuclear translocation of radioimmunotherapeutic agents. Ther Deliv. 2014; 5: 319–35. [DOI] [PubMed] [Google Scholar]
- 12.Cornelissen B, Vallis KA. Targeting the nucleus: an overview of Auger-electron radionuclide therapy. Curr Drug Discov Technol. 2010; 7: 263–79. [DOI] [PubMed] [Google Scholar]
- 13.Pirovano G, Jannetti SA, Carter LM, Sadique A, Kossatz S, Guru N, et al. Targeted Brain Tumor Radiotherapy Using an Auger Emitter. Clin Cancer Res. 2020; 26: 2871–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta AM, Sonabend AM, Bruce JN. Convection-Enhanced Delivery. Neurotherapeutics. 2017; 14: 358–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017; 170: 1062–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker SJ, Fearon ER, Nigro JM, Himilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinoma. Science. 1989; 244: 217–21. [DOI] [PubMed] [Google Scholar]
- 17.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007; 318: 1108–13. [DOI] [PubMed] [Google Scholar]
- 18.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998; 282: 1497–501. [DOI] [PubMed] [Google Scholar]
- 19.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999; 104: 263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jannetti SA, Carlucci G, Carney B, Kossatz S, Shenker L, Carter LM, et al. PARP-1-Targeted Radiotherapy in Mouse Models of Glioblastoma. J Nucl Med. 2018; 59: 1225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson TC, Jannetti SA, Guru N, Pillarsetty N, Reiner T, Pirovano G. Improved radiosynthesis of (123)I-MAPi, an auger theranostic agent. Int J Radiat Biol. 2020: 1–7. [DOI] [PMC free article] [PubMed]
- 22.Cristy M, Eckerman KF. Specific absorbed fractions of energy at various ages from internal photon sources: 7, Adult male. United States; 1987. p. 46.
- 23.Stabin MG, Sparks RB, Crowe E. OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med. 2005; 46: 1023–7. [PubMed] [Google Scholar]
- 24.Wong FC. MIRD: Radionuclide Data and Decay Schemes. Journal of Nuclear Medicine. 2009; 50: 2091-. [Google Scholar]
- 25.Carney B, Kossatz S, Reiner T. Molecular Imaging of PARP. J Nucl Med. 2017; 58: 1025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carney B, Kossatz S, Lok BH, Schneeberger V, Gangangari KK, Pillarsetty NVK, et al. Target engagement imaging of PARP inhibitors in small-cell lung cancer. Nat Commun. 2018; 9: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017; 355: 1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papadimitriou M, Mountzios G, Papadimitriou CA. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat Rev. 2018; 67: 34–44. [DOI] [PubMed] [Google Scholar]
- 29.Bargonetti J, Prives C. Gain-of-function mutant p53: history and speculation. J Mol Cell Biol. 2019; 11: 605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019; 26: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu WG, Polotskaia A, Xiao G, Di L, Zhao Y, Hu W, et al. Identification, validation, and targeting of the mutant p53-PARP-MCM chromatin axis in triple negative breast cancer. NPJ breast cancer. 2017; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ireno IC, Wiehe RS, Stahl AI, Hampp S, Aydin S, Troester MA, et al. Modulation of the poly (ADP-ribose) polymerase inhibitor response and DNA recombination in breast cancer cells by drugs affecting endogenous wild-type p53. Carcinogenesis. 2014; 35: 2273–82. [DOI] [PubMed] [Google Scholar]
- 33.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020; 70: 145–64. [DOI] [PubMed] [Google Scholar]
- 34.Jannetti SA, Zeglis BM, Zalutsky MR, Reiner T. Poly(ADP-Ribose)Polymerase (PARP) Inhibitors and Radiation Therapy. Front Pharmacol. 2020; 11: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel M, Nowsheen S, Maraboyina S, Xia F. The role of poly(ADP-ribose) polymerase inhibitors in the treatment of cancer and methods to overcome resistance: a review. Cell Biosci. 2020; 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nathans JF, Cornwell JA, Afifi MM, Paul D, Cappell SD. Cell cycle inertia underlies a bifurcation in cell fates after DNA damage. Sci Adv. 2021; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh M, Saha S, Bettke J, Nagar R, Parrales A, Iwakuma T, et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell. 2021; 39: 494–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. In vitro evaluation of 123I-MAPi serum stability and blood half-life.
Table S1. Complete Toxicity Report of 127I-PARPi in healthy mice at 2 days and 15 days post compound administration.
Figure S2. Biodistribution of 123I-MAPi in human colorectal cancer HCT116 p53+/+ (A) and HCT116 p53−/− (B) mouse models 1 hour post administration.
Table S2. Bio-distribution tables for 123I-MAPi in selected cell lines expressed as %ID/g.
Figure S3. Biodistribution of 123I-MAPi in human colorectal cancer (A) HCT116 p53+/+ and (B) HCT116 p53−/− mouse models 2 hours post administration.
Table S3. Bio-distribution tables for 123I-MAPi in selected cell lines expressed as %ID/g.
Table S4. Bio-distribution of 123I-MAPi in healthy athymic nude mice. All numbers expressed as %ID/g.
Table S5. Dosimetry Data for 123I-MAPi in healthy athymic nude mice and the equivalent dose in humans.
Table S6. Complete Toxicity Report of 123I-MAPi in healthy mice at 24 hours, 7 days, 14 days.
Figure S4. Tumor growth curves of either HCT116 p53+/+ or HCT116 p53−/− mouse models when treated with vehicle, 127I-PARPi or 123I-MAPi.
Figure S5. Weights of mice of either HCT116 p53+/+ or HCT116 p53−/− mouse models when treated with vehicle, 127I-PARPi or 123I-MAPi.