Abstract
Iron is an indispensable metabolic cofactor in both pro- and eukaryotes, which engenders a natural competition for the metal between bacterial pathogens and their human or animal hosts. Bacteria secrete siderophores that extract Fe3+ from tissues, fluids, cells, and proteins; the ligand gated porins of the Gram-negative bacterial outer membrane actively acquire the resulting ferric siderophores, as well as other iron-containing molecules like heme. Conversely, eukaryotic hosts combat bacterial iron scavenging by sequestering Fe3+ in binding proteins and ferritin. The variety of iron uptake systems in Gram-negative bacterial pathogens illustrates a range of chemical and biochemical mechanisms that facilitate microbial pathogenesis. This document attempts to summarize and understand these processes, to guide discovery of immunological or chemical interventions that may thwart infectious disease.
Graphical Abstract
INTRODUCTION
Since 1947, when Pappenheimer saw the regulation of diphtheria toxin production by iron availability,1 the link between iron acquisition and bacterial pathogenesis seemed logical. Twenty years later, Bullen and Rogers2 noted the impact of excess iron on innate immune defense to infection, which began a series of their articles describing the antagonism between prokaryotic iron requirements and iron sequestration by human hosts.2-10 The research that validated those ideas exponentially expanded over the ensuing 50 years to create an immense body of data. This paper will review those findings to the present day, especially as they relate to iron uptake by Gram (−) bacterial pathogens that acquire different forms of Fe3+ through TonB-dependent transport systems in their cell envelopes. One goal is to explain the transport strategies that carbapenem-resistant Enterobacterales11,12 (formerly Enterobacteriaceae13-15) (CRE; see Abbreviations for a list of all abbreviations and acronyms) and the notorious group of Enterococcus, Staphylococcus, Klebsiella, Acinetobacter, Pseudomonas, and Enterobacter (ESKAPE) pathogens16,17 and other dangerous or multiply drug resistant organisms, utilize to circumvent the innate immune defenses of eukaryotic hosts. In the process, we will consider the nature and relationships of dozens of Gram (−) bacterial outer membrane (OM) receptor proteins that bind and transport organic iron complexes. These summaries consider genetic, microbiological, biochemical, and structural biological data with both clinical and mechanistic relevance. Our discourse focuses on the iron uptake systems of pathogenic organisms of current worldwide concern as a result of their unrelenting development of antibiotic resistance. Our overview is not all-inclusive of Gram (−) bacterial iron uptake systems, nor comprehensive with regard to clinical remedies that may arise against such phenomena. Instead, we address the possibility of antibiotic discovery against TonB-dependent iron uptake in the target bacteria. TonB is a ubiquitous, essential protein component of Gram (−) bacterial iron uptake pathways, so inhibition of TonB action is potentially effective to limit bacterial growth, and thereby stem the severity of CRE/ESKAPE pathogenesis. To simplify designation of the many small molecules and proteins under consideration, we adopted the convention of abbreviating prokaryotic molecules with a capitalized first-letter (e.g., enterobactin, Ent) and eukaryotic proteins with all capitals (e.g., siderocalin, SCN).
1. NOVEL THERAPEUTICS AGAINST GRAM (−) BACTERIAL PATHOGENS
Today’s world faces a long-standing threat that intensified over the past several decades: the uncertain outcomes of bacterial infection. In 2009, for example, Gram (−) bacteria caused two-thirds of the mortality among ~100 000 bacteria-associated deaths in U.S. hospitals; 20% were resistant to all known antibiotics.18 At that time, the WHO identified Gram (−) CRE/ESKAPE pathogens as critical priorities for antibiotic discovery.19 Ten years later, in 2019, the CDC reported more than 35,000 US deaths,20 as a result of more than 2.8 million infections with ESKAPE and other antibiotic-resistant bacteria. During the same time, pharmaceutical companies lessened efforts to combat microbial pathogens.21-25 Antimicrobial treatments are typically either inhibitors of essential biochemical pathways in the pathogen (antibiotics) or molecular constructs (vaccines) that stimulate adaptive immunity in the host. Both approaches have a history of clinical applications that saved millions of lives. Unfortunately, natural selection of variations in the pathogens that lead to resistance undermines both approaches. Antibiotic resistance often arises from mutations that alter cell envelope permeability or decrease the susceptibility of target enzymes to inhibition or other mechanisms.26-28 Vaccine inefficacy stems from changes in the antigenic determinants of the pathogen that supersede the epitopes of the vaccine construct. Hence, one challenge is to identify new pathways, proteins, or other molecules that are vulnerable targets for drug or vaccine development.
Gram (−) bacterial antibiotic resistance largely derives from the selective permeability of the OM and inner membrane (IM) of the cell envelope. The former excludes large or hydrophobic antibiotics but internalizes solutes and nutrients,27,28 whereas the latter contains pumps that expel antibiotics.29,30 Without new antibiotics,31,32 soon no therapeutic options will exist for an expanding number of bacterial pathogens. Many multidrug resistant bacteria became problematic in the past decade, including members of the CRE/ESKAPE pathogen group.33,34 Plus, in 2019, the CDC added other Gram (−) species as urgent or serious threats: Campylobacter, Neisseria, Salmonella, and Shigella.20 The high rate of antibiotic resistance in such strains, that produce the majority of nosocomial infections in the U.S., makes them potentially lethal. These bacteria also often contain uniquely adapted systems for “iron piracy”35 from humans and animals. The clinical options against CRE are so limited that physicians must resort to abandoned toxic drugs like colistin,36 an old antibiotic that was kept in reserve as a last resort against bacterial infections. If CRE acquire colistin resistance, then they become predicted “superbugs”31,32 that are untreatable by all known antibiotics. Colistin resistant Escherichia coli already appeared in the U.S.,37 underscoring the urgent need for new antibacterial agents.
2. IRON ACQUISITION AND BACTERIAL PATHOGENESIS
From a biochemical or metabolic perspective, iron is the most valuable metal in biological systems. Over 80 enzymes require iron-containing heme (Hn) or non-Hn cofactors that help catalyze the metabolic biochemistry of bacteria, fungi, and animals. Examples include aconitase and succinate dehydrogenase in the Krebs cycle, proton-pumping oxidoreductases in the electron transport chain, class Ia ribonucleotide reductases in de novo DNA synthesis, monooxygenases like cytochrome P450, and catalases and superoxide dismutases that detoxify reactive oxygen species. This central role of iron in aerobic biochemistry makes it a determinant of bacterial pathogenesis, invasiveness, and molecular competition at the host–pathogen interface: the eukaryotic innate immune system sequesters iron, but successful pathogens overcome this defense mechanism and capture the metal.38-51 The eukaryotic components of cellular iron trafficking include the Fe3+-binding proteins transferrin (TF), lactoferrin (LF) and ferritin (FTN), an intricate intracellular network of regulatory and delivery proteins hepcidin, hepphaestin, hemoglobin-haptoglobin, Hn-hemopexin, ferroportin, ceruloplasmin, serum albumin, and lipocalins (LCN),52-54 including LCN2, that is now called siderocalin55 (SCN). Their prokaryotic counterparts are components of diverse, omnipresent iron uptake systems that bacteria employ to obtain iron in the host. In Gram (−) cells, iron acquisition usually begins with the elaboration of siderophores (Gr. iron carrier), low molecular weight organic chelators56,57 that complex adventitious, or sequestered iron with unparalleled high affinity: Ent has a binding affinity constant of 1052 M−1.58,59 Over 500 different siderophores are known and characterized.60 The second part of Gram (−) bacterial iron uptake is an equally large group of discriminating, high affinity cell surface receptors that bind ferric siderophores and other iron complexes (KD ~ 10−10 M61,62). Since their discovery,63,64 these ~80 kDa proteins were recognized as the OM components of multiprotein, energy- and TonB-dependent cell envelope transport systems.65 Their nomenclature has evolved with the understanding of their properties, as iron-regulated membrane proteins (IRMP66), iron-regulated OM proteins (IROMP67), ligand-gated porins (LGP68), or TonB-dependent transporters (TBDT69). None of these acronyms is perfect (see following), but LGP perhaps best describes their mechanistic attributes. Like ligand-gated ion channels,70-72 the binding of a ferric siderophore, other metal complex, or eukaryotic iron-containing protein activates LGP to conformational motion35,73,74 that signals their occupancy and stimulates interactions with TonB. The ensuing actions of TonB, as energized by electrochemical proton motive force (PMF), enable uptake of the iron complex or free iron through the OM into the periplasm.
Since about 1970,6 the biochemical connections between pro- and eukaryotic iron homeostasis were apparent, and many researchers, but especially J. J. Bullen,4,8,75,76 noted the relationship between bacterial iron acquisition and infection. Fifty years of research on these systems confirmed that bacteria need iron for metabolism, they produce biosynthetic and transport systems to obtain it, and their success toward this end influences the outcome of their infections. Conversely, iron deprivation, or disruption of iron uptake processes, retards bacterial growth,66,77-81 reducing or eliminating virulence.35,82-89 Although none of the prokaryotic uptake systems are yet fully understood,69,90,91 Gram (−) bacterial Fe3+ transport begins when LGP adsorb iron complexes, and facilitated by TonB, internalize them through the OM bilayer. TonB-dependent iron acquisition systems contribute to colonization of eukaryotic hosts.48,92-99 Overall, an assortment of experimental approaches accumulated comprehensive evidence that iron acquisition is a determinant of pathogenesis:
Iron deprivation slows bacterial growth;100,66,77,79-82,101 bacteria secrete siderophores to combat low-iron stress.60,66,81
Gram (−) bacterial pathogens, including species of Escherichia, Salmonella, Neisseria, Vibrio, Acinetobacter, Klebsiella, Yersinia, Pseudomonas, Hemophilus, and more, acquire iron with TonB-dependent transporters.73,88,102-120
Microbial iron scavenging and host iron sequestration are antagonistic processes that influence infection.4,7,75,121
Iron sequestration reduces or eliminates bacterial virulence.35,82-89,122-131
Successful pathogens capture iron from their hosts.35,38,39,41-51,83,96,132-147
Vaccination with bacterial iron transporters creates protective humoral and/or cellular immunity.148-157
“Trojan Horse” siderophore antibiotics, that enter bacteria through iron transporters, show broad-spectrum activity against Gram (−) bacteria.158-163
Despite the many connections between iron and infectious disease, and the variety of studies that repeatedly verified the relationship between iron acquisition and bacterial colonization164 or virulence,92,94,99,165-167 some findings challenged the idea that iron uptake promoted bacterial pathogenesis.168 The explanation of this discrepancy is that bacterial pathogens often elaborate multiple aposiderophores and acquire even more ferric siderophores. So, single mutations that abrogate a particular iron uptake pathway may not impair host colonization or virulence168 because other iron uptake pathways compensate for the deficiency. Laboratory E. coli K-12 strains, for example, produce at least seven TonB-dependent transport systems for ferric iron,51,104,169-171 and wild E. coli clinical isolates encode even more172-174 that either internalize ferric siderophores57,175 or extract iron from eukaryotic proteins.35,74 Other bacterial pathogens, like Acinetobacter baumannii, produce as many as 10 different siderophores in iron deficient environments.176
Once this knowledge of iron uptake multiplicity and redundancy was known, it raised doubts that siderophore pathways are appropriate targets for antibiotic development. However, all Gram (−) bacteria acquire ferric iron with TonB-dependent LGP, so TonB itself is a conserved common component of all these OM uptake reactions. The actions of TonB are ostensibly susceptible chemical inhibition, which will reduce iron acquisition and therefore also reduce bacterial proliferation in humans and animals. Furthermore, mutant bacteria lacking TonB, or producing mutant TonB proteins, will not obtain iron in the host environment and therefore fail to thrive or colonize.88,164 So, antibiotics that target TonB may suffer less from resistance. These points suggest that TonB-dependent iron uptake pathways are viable candidates for antibiotic discovery.
A large percentage of existing antibiotics target bacterial cell envelope biochemistry.177-180 Compounds that block OM iron transport will similarly focus on a process that is uniquely prokaryotic: eukaryotes acquire iron by different mechanisms.181-184 Iron potentiates the activity of the quinone antibiotic streptonigrin against E. coli,185 Neisseria gonorrheae,186,187 and Haemophilus influenzae,188 but not a single natural antibiotic is known to antagonize bacterial iron transport systems, a fact that questions the likelihood of finding new antibiotics against them. Nevertheless, the recent licensing of cefiderocol,189-191 that utilizes a TonB-dependent LGP to introduce a bacteriocidal antibiotic into the bacterial periplasm, illustrates the clinical potential of such pathways. Finally, it is perhaps most pertinent that the innate immune system encodes numerous proteins that reduce iron availability to invading microbes, underscoring the potential of seeking chemical or immunological interventions that similarly interfere with prokaryotic iron uptake.
3. OVERVIEW OF TONB-DEPENDENT IRON TRANSPORT SYSTEMS
Gram (−) bacterial LGP are surface receptors that recognize and bind metal complexes. Then, activated by TonB, they internalize the ferric siderophore or porphyrin through the OM bilayer. Virtually all Gram (−) bacterial pathogens obtain iron with TonB-dependent systems, which explains the interest in blocking TonB action, but the incomplete information about LGP transport mechanisms74,90,192,193 complicates the use of iron deprivation against pathogenesis. Furthermore, complex multiprotein arrays (in E. coli, 13 cell envelope proteins) collaborate in the uptake of each iron atom. Their functions include high affinity ligand recognition, transmembrane signal transduction, internal conformational motion, catalytic protein–protein interactions driven by energy transmission between membranes, and active transport in two distinct membranes energized by both the electrochemical gradient and ATP hydrolysis. Most notably, active iron OM transport occurs through a closed membrane channel, across a bilayer that is unable to sustain an ion gradient, necessitating a novel means of energization. Hence, besides its clinical potential, the topic has theoretical importance.
LGP are omnipresent in Gram (−) bacterial cell envelopes to varying degrees of representation. Members of Enterobacterales encode many (7–20) that act in iron or other metal uptake, but Proteobacteria in other Families may contain many more (Pseudomonadaceae, 35–38; Caulobacteriacae, 63; Xanthamonadaceae, 42–7069) that are predicted to span other substrate specificities. Most of these functions were assigned by bioinformatic analyses and are not yet experimentally verified. In the case of Caulobacter crescentus, five iron-regulated Omps were identified, but only one transport function was identified as the receptor for hemin.194 The most mechanistically well characterized LGP catalyze iron195-199 or cobalt195-199 uptake, but many other transport specificities are proposed in the LGP superfamily.200 LGP often also act as receptors for bacteriocins and phage. E. coli FepA (EcoFepA) (we abbreviate bacterial proteins to also designate the genus and species of their origin: e.g., Klebsiella peumoniae FepA, KpnFepA; A. baumannii BauA, AbaBauA; Pseudomonas aeruginosa FpvA, PaeFpvA, etc.), for example, is the cognate receptor for the TonB-dependent colicins B and D64,201,202 and bacteriophage H8.203 The architecture of EcoFepA196 typifies the tertiary structure of all LGP: a 150-residue N-terminal globular domain situated within a 22-stranded C-terminal porin β-barrel (Figure 1). The eight LGP of E. coli K-12 acquire different types of metal complexes: ferric catecholates (FepA,196 Fiu,204 Cir195), ferric hydroxamates (FhuA,198,199 FhuE,205 IutA206), ferric citrate (FecA197), and cyanocobalamin (vitamin B12; BtuB207). FptA208 and FpvA209 of P. aeruginosa, which show the same overall fold, transport iron complexes of pyochelin210,211 (Pch) and pyoverdine212,213 (Pvd), respectively. The discriminating specificity of these receptors for their ligands,61,202,214 whose binding potentiates the active transport mechanism, is what led to the designation LGP.61,202,214 They are unlike diffusive porins,27 in that they bind ligands with high affinity and require energy and TonB action to accomplish ligand internalization. The common designation TBDT69 is intuitively accurate but potentially confuses these OM uptake systems with completely different active transporters in the IM. Both classes of membrane proteins perform active transport, but they are structurally different, function by different mechanisms, utilize different energy sources, and inhabit different membranes. Hence, we reserve the term “transporter” for ATP-binding cassette (ABC) transporters and electrochemical gradient-coupled (e.g., PMF-dependent) major facilitator transporters in the bacterial IM.
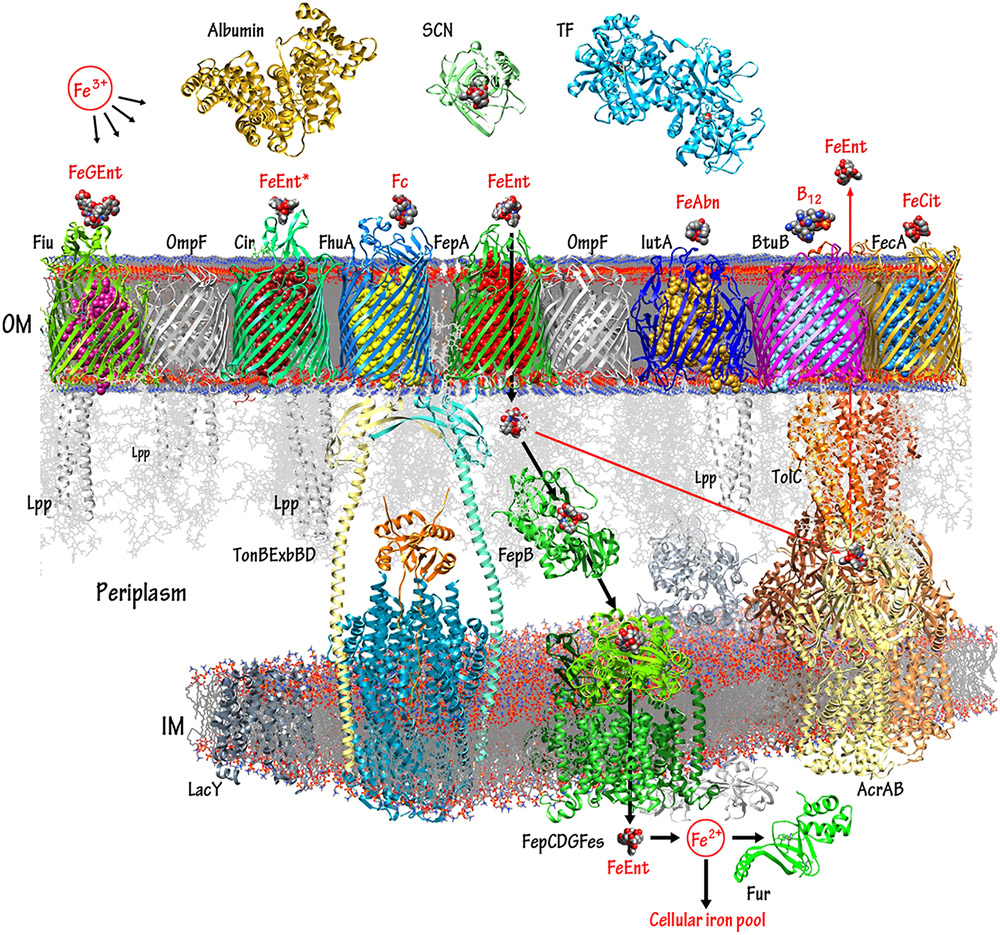
Figure 1.
TonB-dependent iron and B12 transport pathways in Gram (−) bacteria. The diagram displays selected components of the E. coli OM, periplasm, and IM, rendered by CHIMERA (UCSF) from their RCSB crystallographic coordinates. Proteins that participate in metal flux are portrayed in colors; other cell envelope components are shown in shades of gray. Bacteria and fungi secrete siderophores that chelate extracellular iron. In human and animal hosts, the innate immune system proteins albumin, SCN, and TF antagonize bacterial iron acquisition, by adsorbing siderophores, ferric siderophores, or free iron from blood, serum, lymph, and other fluids. Nevertheless, high affinity bacterial OM LGP bind specific ferric siderophores (or vitamin B12) and actively transport them into the periplasm. The bacterial TonB/ExbBD complex spans the cell envelope and utilizes IM PMF to energize the OM active transport reactions.91,237 TonB/ExbBD is modeled from the crystallographic coordinates of the TonB C-terminus,241 the ExbBD proteins,232,233,707 and other data;236,644,708 the full complex was not yet structurally delineated. The import (black arrows) and export (red arrows) pathways of FeEnt typify those of other metal complexes: after binding and TonB-dependent internalization by FepA, FeEnt binds to the periplasmic protein FepB that delivers it to the IM ABC-transporter FepCDG, which hydrolyzes ATP as it transports the ferric siderophore to the cytoplasm. During or after the IM uptake process, Fes hydrolyzes the lactone backbone of FeEnt, which effectively releases Fe3+ for reduction to Fe2+.709 Ferrous iron enters cellular iron pools, and equilibrium with the global regulator, Fur.710-712 Alternatively, if surplus FeEnt exists in the periplasm, then the AcrABTolC export complex expels the excess to the exterior.62 The depiction of FepCDGFes was modeled from the crystal structure of BtuCD.
3.1. OM Iron Transport: LGP Crystal Structures
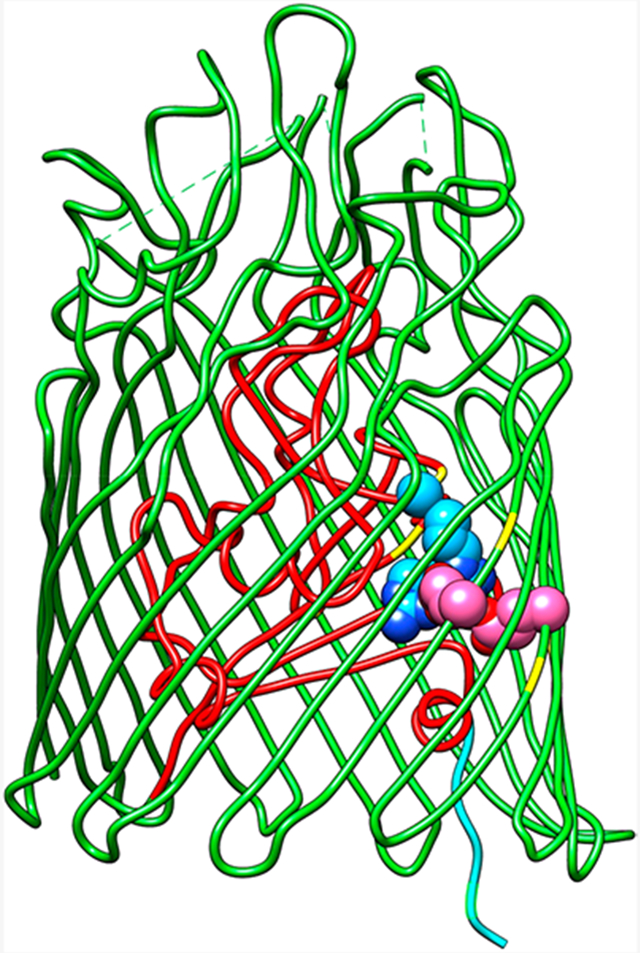
In 1990, Weiss et al.215 determined the first detailed crystal structure of a porin from Rhodobacter capsulatus. It was followed by the crystal structure of E. coli OmpF (EcoOmpF).216 Buchanan et al.196 submitted a description of the crystal structure of EcoFepA in September of 1998, a few weeks before that of EcoFhuA.198,199 Since then, 18 more LGP structures were resolved (Table 1). The transmembrane β-barrels of these OM proteins are central to the understanding LGP functionality, because they classify them in the porin superfamily.27,200 The 22-stranded β-sheets surround a structurally distinct, N-terminal, ~150 amino acid globule that regulates the movement of molecules through the pore.
Table 1.
LGP of CRE/ESKAPE and Other Pathogensa
| LGPb | strainc | metal complexd | protein ligandse | aa | massf | pIg | E. coli K-12 orthologueh | NCBI refi | PDB |
|---|---|---|---|---|---|---|---|---|---|
| Commensal E. coli | |||||||||
| FecA | MG1655 | FeCit | ND | 741 | 81 707 | 5.61 | NSI | NP_418711.1 | 1PNZ |
| FepA | MG1655 | FeEnt | colB, D; H8, mE495 | 724 | 79 771 | 5.4 | 53% IroN | NP_415116.1 | 1FEP |
| FhuA | MG1655 | Fc | colM, T1, T5, φ80 | 714 | 78 742 | 5.3 | NSI | NP_414692.1 | 1BY5 |
| Fiu | MG1655 | FeDHBS | ND | 727 | 78 432 | 5.75 | NSI | NP_415326.1 | 6BPN |
| FhuE | MG1655 | FeRta | ND | 693 | 77 411 | 4.89 | NSI | NP_415620.1 | 6E4V |
| Cir | MG1655 | FeDHBS | colIa, Ib, | 638 | 71 149 | 5.2 | 35% FepA | NP_416660.1 | 2HDF |
| BtuB | MG1655 | B12 | ColE1, E3, BF23 | 594 | 66 325 | 5.35 | NSI | NP_418401.1 | 1NQE |
| Pathogenic E. coli | |||||||||
| YddB | UPEC 042 | ND | ND | 771 | 87 206 | 6.06 | NSI | CBG34449.1 | 6OFR |
| IutA | 083:H1j | FeAbn | cloacin DF13, colV | 708 | 78 061 | 5.23 | NSI | WP_000973516.1 | ND |
| IroN | 083:H1j | FeGEnt | ND | 701 | 76 525 | 5.79 | 52% FepA | ADR29866.1 | ND |
| YncD | UPEC C15 | ND | ND | 677 | 74 900 | 5.32 | NSI | AKC11926.1 | 6V81 |
| FyuA | 0157:H7 | FeYbt | pesticin | 551 | 71 387 | 5.52 | NSI | EFB2704300.1 | ND |
| ChuA | 0157:H7 | Hn | ND | 632 | 69 436 | 5.27 | NSI | NP_312407.1 | ND |
| LGP1 | 0157:H7 | ND | ND | 687 | 76 150 | 5.48 | NSI | QGF16871.1 | ND |
| LGP2 | 0157:H7 | ND | ND | 634 | 71 005 | 5.69 | NSI | QGF15879.1 | ND |
| K. pneumoniae | |||||||||
| FepA4 | Kp52.145 | FeEnt | ND | 728 | 80 070 | 5.67 | 72% FepA | WP_004179434.1 | ND |
| FepA1 | Kp52.145 | FeEnt | ND | 717 | 79 665 | 5.41 | 81% FepA | CDO13414.1 | ND |
| FhuA | Kp52145 | Fc | ND | 715 | 79 054 | 5.34 | 89% FhuA | WP_048972727.1 | ND |
| IutA | hvKP1 | FeAbn | cloacin DF13 | 708 | 78 043 | 5.23 | NSI | CDO11693.1 | ND |
| Fiu | KP52145 | FeDHBS | ND | 727 | 78 023 | 5.71 | 77% Fiu | WP_171841556.1 | ND |
| FepA2 | KP52145 | ND | ND | 701 | 77 382 | 5.31 | 51% FepA | CDO16709.1 | ND |
| FhuE | KP52145 | FeRTAl | ND | 695 | 76 897 | 5.25 | 50% FhuE | AYK02175.1 | ND |
| IroN | KP52145k | FeGEnt | ND | 700 | 76 760 | 6.41 | 51% FepA | WP_042940746.1 | ND |
| FcuA | KP52145 | ND | ND | 703 | 76 166 | 5.57 | NSI | EMB11413.1 | ND |
| YncD | hvKP1 | ND | ND | 677 | 74 569 | 5.75 | NSI | EMB10697.1 | ND |
| FyuA | hvKP1 | FeYbt | pesticin | 652 | 71 400 | 5.52 | NSI | CDO15344.1 | ND |
| Cir | Kp52.145 | FeDHBS | 632 | 70 367 | 5.35 | 82% Cir | EMB11539.1 | ND | |
| ChuA | hvKP1 | Hn | ND | 613 | 67 571 | 5.27 | 100% ChuA | WP_001322816.1 | ND |
| BtuB | KP52145 | B12 | ND | 592 | 66 035 | 5.32 | 57% BtuB | CDO16333.1 | ND |
| LGP1 | KP52145 | ND | ND | 737 | 81 135 | 5.62 | NSI | QDA45483.1 | ND |
| LGP2 | KP52145 | ND | ND | 680 | 74 658 | 5.64 | 33% FhuA | EMB11926.1 | ND |
| A. baumannii | |||||||||
| Fiu | 17978 | FeDHBS | ND | 771 | 84 285 | 6.89 | 34% Fiu | AZM39353.1 | ND |
| BfnH | 17978 | FeBfn | ND | 728 | 80 491 | 5.84 | NSI | ABO12082.2 | ND |
| FepA | 17978 | FeEnt | ND | 730 | 80 248 | 5.67 | 45% FepA | WP_005135700.1 | ND |
| PiuA | 17978 | FeDHBS | ND | 736 | 80 054 | 6.23 | 31% Fiu | ABO10929.1 | 5FP1 |
| PirA | 17978 | FeDHBS | ND | 714 | 78 022 | 5.43 | NSI | SCX98474.1 | 5FR8 |
| BauA | 17978 | FeAcn | ND | 712 | 77 497 | 7.6 | NSI | ABO12804.2 | ND |
| BauA | 19606 | FeAcn | ND | 703 | 76 016 | 5.62 | NSI | WP_001073039.1 | 6H7V |
| FhuA | 19606 | Fc | ND | 679 | 75 739 | 5.59 | 25% FhuA | ABO12348.2 | ND |
| FbsN | 19606 | FeFbn | ND | 629 | 68 763 | 6.82 | 25% FhuA | ABO12983.2 | ND |
| BtuB | 19606 | B12 | ND | 598 | 65 781 | 5.71 | 25% BtuB | ABO13283.2 | ND |
| LGP1 | 19606 | ND | ND | 862 | 93 996 | 5.08 | NSI | CAA0247590.1 | ND |
| LGP2 | 19606 | ND | ND | 781 | 88 811 | 6.63 | 34% FecA | ABO13864.1 | ND |
| LGP3 | 19606 | ND | ND | 697 | 78 847 | 5,8 | NSI | ABO13728.2 | ND |
| LGP4 | 19606 | ND | ND | 674 | 77 325 | 5.52 | NSI | EEX02122.1 | ND |
| LGP5 | 19606 | ND | ND | 681 | 75757 | 5.48 | 25% FhuA | ABO11495.2 | ND |
| LGP6 | 19606 | ND | ND | 669 | 74 690 | 5.18 | 42% FepA | ABO13298.2 | ND |
| P. aeruginosa | |||||||||
| HxuA | PAO1 | Hn | ND | 965 | 95 071 | 7.33 | NSI | CRQ69633.1 | ND |
| HasR | PAO1 | Hn | ND | 855 | 94 205 | 5.85 | NSI | NP_252098.1 | ND |
| FpvA | PAO1 | FePvd | pyocins S2, S3, S4 | 772 | 86 469 | 5.27 | NSI | NP_251088.1 | 2IAH |
| PupA | PAO1 | FePch | ND | 777 | 86 005 | 5.28 | NSI | AMU01031.1 | ND |
| FoxA | PAO1 | FxB | ND | 773 | 85 273 | 5.05 | 32% FhuA | NP_251156.1 | 6196 |
| ChuA | PAO1 | Hn | ND | 737 | 81 892 | 5.99 | NSI | AAC13289.1 | ND |
| FepA1 | PAO1 | FeEnt? | ND | 735 | 80 919 | 5.85 | 71% FepA | MXH37568.1 | ND |
| PiuD | PAO1 | FeDHBS | ND | 731 | 80 149 | 5.68 | NSI | WP_132667204.1 | 5NEC |
| FepA2 | PAO1 | FeEnt? | ND | 717 | 79 687 | 5.19 | 81% FepA | MXH36562.1 | 5NEC |
| P. aeruginosa | |||||||||
| PfeA | PAO1 | FeEnt | ND | 721 | 78 503 | 5.8 | 60% IroN | NP_251378.1 | 6Q5E |
| PiuA | PAO1 | ND | ND | 729 | 78 313 | 5.7 | 77% Fiu | MXH35875.1 | 5FOK |
| ChtA | PAO1 | FeRTAl | ND | 714 | 78 166 | 5.45 | NSI | PTC33848.1 | ND |
| IutA | PAO1 | FeAbn | ND | 708 | 78 161 | 4.96 | 72% IutA | MXH36073.1 | ND |
| PirA | PAO1 | FeDHBS | ND | 714 | 77 992 | 5.43 | 56% FepA | AAG04320.1 | 5FP2 |
| FhuA | PAO1 | Fc | ND | 702 | 77 881 | 5.37 | 61% FhuA | MXH37021.1 | ND |
| IroN | PAO1 | FeGEnt | ND | 714 | 77 866 | 5.38 | 60% IroN | WP_058129121.1 | ND |
| FptA | PAO1 | FePch | pyocin E5 | 682 | 75 597 | 5.58 | NSI | NP_252911.1 | 1XKW |
| FhuE | PAO1 | FeRTAl | ND | 689 | 75 467 | 5.14 | 46% FhuE | CRQ23141.1 | ND |
| FvbA | PAO1 | FeVbn | ND | 665 | 73 731 | 5.46 | NSI | WP_003093526.1 | ND |
| Cir | PAO1 | FeDHBS | ND | 632 | 70 398 | 5.31 | 80% Cir | MXH34319.1 | ND |
| BtuB | PAO1 | B12 | ND | 598 | 66 521 | 5.38 | 57% BtuB | MXH38591.1 | ND |
| LGP1 | PAO1 | ND | ND | 821 | 92 251 | 6.39 | NSI | BAQ41081.1 | ND |
| Y. pestis | |||||||||
| HasR | KIM6+ | Hn | ND | 795 | 89 654 | 7.96 | 56% HasR | WP_002209485.1 | ND |
| IutA | KIM6+ | FeAbn | ND | 745 | 82 439 | 5.36 | 67% IutA | WP_087813403.1 | ND |
| FhuE | KIM6+ | FeRTAl | ND | 717 | 79 827 | 7.8 | 27% FhuE | WP_002211883.1 | ND |
| FhuA | KIM6+ | Fc | ND | 716 | 78 382 | 5.99 | 26% FhuA | AAS63640.1 | ND |
| huA | KIM6+ | Hn | ND | 690 | 75 802 | 5.24 | 68% ChuA | WP_002209062.1 | ND |
| Cir | KIM6+ | FeDHBS | ND | 679 | 75 555 | 6.0 | 38% Cir | WP_071526008.1 | ND |
| BtuB | KIM6+ | B12 | ND | 672 | 72 172 | 5.45 | 65% BtuB | WP_058987704.1 | ND |
| Psn | KIM6+ | FeYbt | pesticin | 651 | 71 442 | 5.62 | NSI | AAC69592.1 | 4EPA |
| LGP1 | KIM6+ | Cu++? | ND | 698 | 76 423 | 8.82 | 28% BtuB | WP_002208882.1 | ND |
| LGP2 | KIM6+ | Hn? | ND | 667 | 76 274 | 5.34 | 45% YoeA | WP_002211632.1 | ND |
| LGP3 | KIM6+ | FeDHBS | ND | 678 | 74 129 | 6.2 | 31% Cir | AAM84435.1 | ND |
| Other Gram (−) Pathogens | |||||||||
| HasR | S. marscescens m | Hn | ND | 865 | 94 847 | 6.2 | NSI | CAE46936.1 | 3CSN |
| FauA | B. pertussis | alcaligin | ND | 699 | 77 593 | 6.75 | NSI | WP_014905926.1 | 3EFM |
| FrpB | N. meningitidis n | FeEnt? | ND | 692 | 76 823 | 9.42 | NSI | AAF42315.1 | 4AIP |
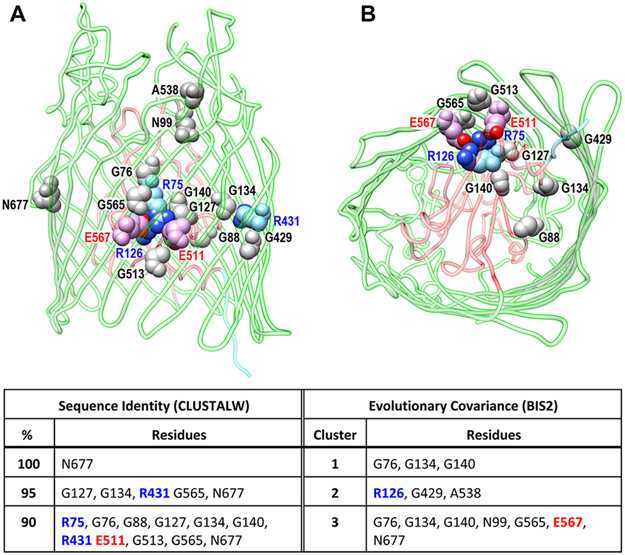
We identified 79 LGP that participate in the uptake of ferric siderophores, heme or other metal complexes. This list, that is not fully comprehensive, illustrates the breadth of metal chelate recognition in Gram (−) bacterial pathogens. We used sequences of the mature proteins for analysis by CLUSTALΩ, the results of which appear in Figures S1 and 5.
LGP reflect standard nomenclature; if function is unknown, the protein is enumerated: e.g., EcoLGP1, EcoLGP2, etc.
Bacterial strain from which the genomic information originated.
Ferric siderophore or metal porphyrin; see text for abbreviations.
Abbreviations: col, colicin; m, microcin.
Mass (Da) of the mature protein.
Isoelectric point of the mature protein.
Extent of identity to the closest homologue in E. coli K-12 strain MG1655; NSI; NSI, no significant identity (i.e., <25%).
Entries originated from the NCBI PROTEIN database.
Structural gene resides on pNRG857c in strain O83:H1.
Structural gene resides on pLVPK in strain Kp52.145, also called pII.
LGP that recognize RTA often also bind FxB or coprogen.
S. marcescens strain SM365.
N. meningitidis strain MC58.
3.1.1. N-Terminal Globular Domain (N-Domain).
The N-terminal portion of LGP contain structural features that enable its biochemical functions. A four-stranded β-sheet obstructs LGP channels. The N-terminal region contains the “TonB-box”,217,218 a short sequence (7–11 residues) that mediates signal transduction to TonB. When ferrichrome (Fc) binds to FhuA, or B12 binds to BtuB, their loops undergo changes that propagate through the N-domain, altering the disposition of the TonB-box at the periplasmic interface.197,207 The exact sequence and molecular mechanics of these conformational changes are unknown, but they occur in response to high affinity binding of a metal complex to the surface loops of the LGP that coalesce around its ligand by induced fit.197,219,220 Two large loops sit atop the N-domain globule, and as many as 11 more surface loops ranging from 2 to 40 residues bridge adjacent β-strands in the C-domain β-barrel. Loop motion during ligand binding is the basis for concomitant or ensuing movement of the TonB-box at the internal surface of the receptor, creating a trans-OM signaling pathway that activates the actions of TonB in the periplasm.
3.1.2. C-Terminal Transmembrane β-Barrel (C-Domain).
Like other OM proteins (Omp),215,216,221-223 LGP contain an antiparallel, amphiphilic β-sheet that circumscribes an aqueous channel. The β-strands in the sheet are linked on the periplasmic side by short reverse turns, and on the outer surface by usually expansive loops that are populated with residues involved in ligand recognition and high-affinity binding. The diameter of the 22-stranded C-terminal β-barrel approximates 50 Å, a size that potentially compromises the permeability barrier of the OM bilayer.28 However, the N-domain restricts passage of molecules through this large hydrophilic pore to ligands that specifically bind and activate the energy- and TonB-dependent uptake mechanism.68
3.2. TonB/ExbBD Physiology
Genetic studies on iron uptake by E. coli, as well as its susceptibility to bacteriophage or colicins,64,202,224-227 identified the tonB locus as a crucial component of the transport system. tonB mutants were unable to thrive in iron-deficient media and showed pleiotropic transport deficiencies. exbBD strains were similarly implicated,217,228 but the impact of exbBD-deficiency was less dramatic, likely because they may be substituted by TolQR.229,230 Subsequent research revealed that TonB, ExbB, and ExbD form a multimeric protein complex in the IM,231 with TonB presumably at the center of this assembly.232,233 Genetic, biochemical, bioinformatic, and biophysical data65,233-238 show that TonB is an IM-anchored protein that spans the periplasm (Figure 1). When an LGP binds a metal complex, the TonB-box of its N-domain repositions at the periplasmic interface,198,199 allowing protein–protein interactions with the TonB C-terminal domain (TonB CTD).239,240 Binding of the TonB-box of LGP to the TonB C-terminal domain (CTD)239,240 facilitates the movement of iron through the LGP channel. Models postulate238,241,242 and evidence exists237 that TonB transmits energy from the IM to the OM by rotary motion, driven by the electrochemical gradient across the IM [for review see ref 91]. ExbB and ExbD231 participate in this reaction. TonB-dependent, PMF-driven activity of LGP62,227,243,244 allows their accumulation of iron against its concentration gradient. Thus, Gram (−) bacterial pathogens obtain iron from human and animal hosts by TonB-dependent uptake systems that are virulence determinants.4,7,62,121,227,243,244 The ubiquity of TonB in Gram (−) bacterial metal transport makes it a potential target for drug discovery.
3.3. Periplasmic and IM Iron Transport
After traversing the OM through LGP, iron complexes adsorb to periplasmic binding proteins245,246 (like EcoFepB,247,248 Figure 2) and then to IM ABC-transporters249-251 that intake iron into the cytoplasm. For the prototypic FeEnt acquisition system, FepB247,252,253 (Figures 1 and 2) transfers FeEnt to the IM ABC-transporter complex FepCDG254 (Figure 1). During or after entry into the cytoplasm, Fes hydrolyzes the lactone scaffold of FeEnt, concomitantly reducing and releasing ferric iron into intracellular pools as Fe2+.255-261
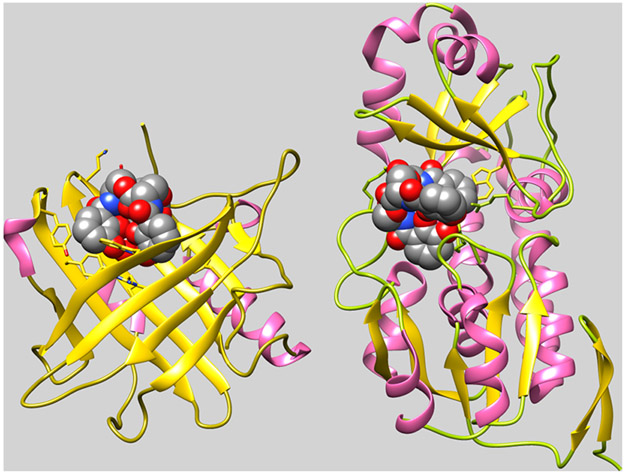
Figure 2.
Binding of FeEnt by HsaSCN and EcoFepB. Comparison of the crystallographic structures of human SCN (3CMP) and E. coli FepB (3TLK), with bound FeEnt, shows two different structural folds for FeEnt binding. Both contain α- (pink) and β- (gold) structures, but the former human serum protein binds FeEnt in the mouth of a seven-stranded β-barrel, whereas the latter periplasmic protein binds it in the central cleft of a bilobed globule. In both cases, however, affinity for the aromatic, triply negatively charged ferric siderophore derives from interactions with cationic (SCN: R81, R130, R134; FepB: R78, R242, R301) and aromatic (SCN: Y52, W79, Y100, Y106, F123, Y132; FepB: F300, W209) side chains in the binding protein. Adsorption of FeEnt to EcoFepA involves similar contributions of charge713 and aromaticity714,715 to the overall affinity.
4. SIDEROPHORES
After the isolation of ferrichrome from the smut fungus Ustilago sphaerogena,56 and mycobactin from the acid-fast bacterium Mycobacterium johnei,262 more than 500 other siderophores were discovered. We will not review the basic chemistry of siderophores because numerous other descriptions57,59,60,263 already exist. Suffice it to say that based on their complexation of Fe3+, siderophores fall into three main groups: catecholate, hydroxamate, and mixed chemistry chelators (Figure 2). Compounds in the latter category contain a variety of chemical groups that may share electrons with the iron nucleus: carboxylates, imidazoles, oxazolines, quinones, thiazolidines, and more. Although siderophores in different categories have characteristic properties that affect their affinities for iron(III), the ferric ion favors complexation by oxygen, rather than nitrogen or sulfer, and siderophores reflect this preference.
4.1. Complexation of Fe3+ by Siderophores
Many siderophores are virulence factors of the bacteria that produce them.264 They capture Fe3+ in the host environment because they generally possess higher affinity for iron than host proteins.264,265 Besides the prototypic tricatecholate compound Ent that many Gram (−) bacteria in the Family Enterobacterales produce, and the prototypic trihydroxamate ferrichromes (Fc) that many fungi produce, other siderophores of interest are the monocatecholates dihydroxybenzoic acid (DHBA) and dihydroxybenzoyl serine (DHBS), citrate (Cit), the citrate-based hydroxamate aerobactin (Abn), and the mixed chemistry chelates acinetobactins (Acn), baumannoferrins (Bfn), fimsbactins (Fbn), yersiniabactin (Ybt), pyochelin (Pch), and pyoverdine (Pvd). In nature, the monocatecholates are relevant degradation products of Ent, vibriobactin266 (Vbn), and corynebactin267 (Crn; also called bacillibactin657) that are all cyclic trimers of 2,3-DHBS. The individual units are joined by ester linkages between the alpha carboxyls and side chain hydroxyl groups of Ser (Figure 3). Additionally, Ent may be derivatized by the addition of glucose to two of its three catechol groups to create glucosylated Ent (GEnt). Both Ent and GEnt are potentially labile compounds because their cyclic lactone backbone is susceptible to acid or base hydrolysis, and their catecholate groups may oxidize to quinones. These chemical processes produce a series of natural hexadentate, tetradentate, and bidentate catecholate compounds in the prokaryotic microenvironment, each with different metal complexation activities and affinities. Several attributes of trimeric, hexadentate Ent/GEnt contribute to their immense affinity for Fe3+, that is, 30 logs higher than that of a bidentate ligand like DHBA or DHBS. First, the architecture of Ent/GEnt locates their catecholate hydroxyls in perfectly symmetrical geometry around the metal center, without strain.263,268 Second, Fe3+ prefers a hard acidic ligand, like oxygen, rather than soft basic ligands like nitrogen or sulfur. Lastly, and most importantly, trimeric Ent/GEnt exemplify the chelate effect: metal complexes of polydentate ligands (in this case, hexadentate) are much more stable than complexes of chemically similar mono- or bidentate ligands. Complexation by three isolated bidentate ligands (i.e., DHBA/DHBS) requires three individual productive collisions between the metal and the ligands, whereas chelation by a single hexadentate ligand (i.e., Ent/GEnt) occurs by an initial collision that attaches the first oxygen, followed by binding of the second oxygen by rotation, and the remaining oxygens by motion of the aposiderophore that enables them to surround the iron center. A hexadentate chelate also better resists dissociation. When a mono- or bidentate group is displaced, it is lost into the bulk solution. But, if any of the oxygens of the hexadentate Ent/GEnt are displaced, other oxygens still remain attached, and it is only a matter of time until the displaced oxygen(s) find(s) the metal again and reattach. All of these conditions stabilize the complex with hexadentate Ent/GEnt, relative to an iron chelate formed by bidentate monocatecholate groups.
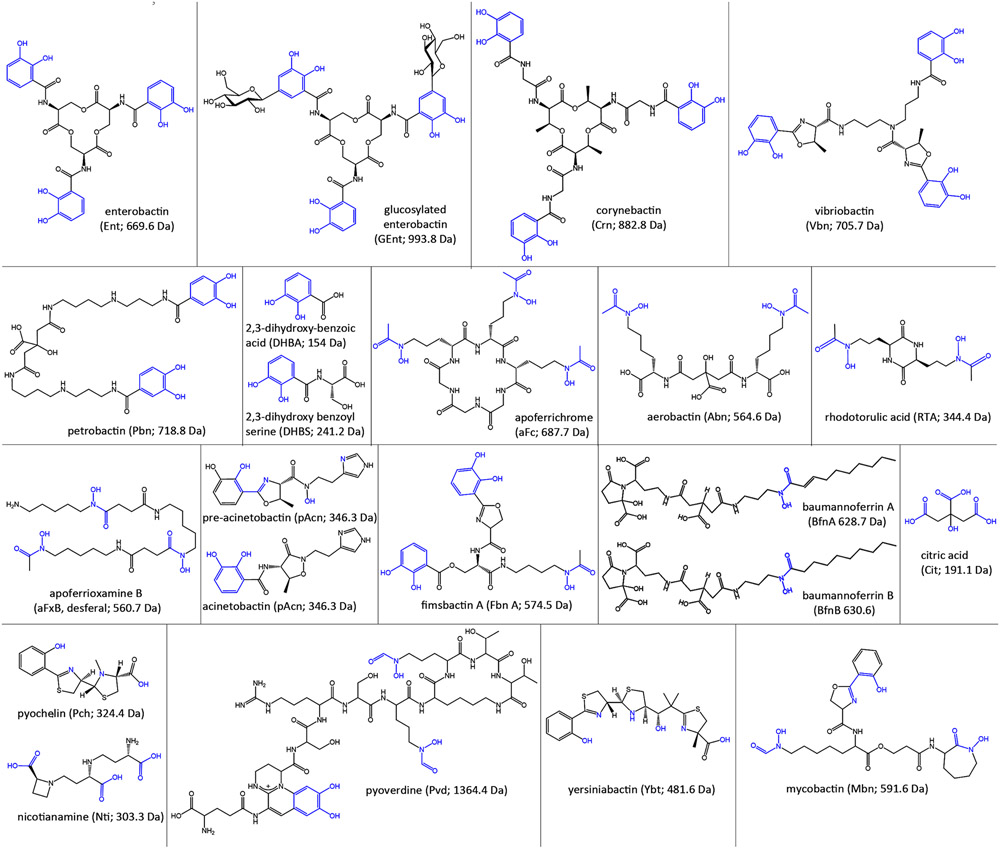
Figure 3.
Siderophores. Pathogenic bacteria secrete and/or utilize a variety of catecholate, hydroxamate, and mixed chelation siderophores, usually less than 1000 Da in mass. The illustrations show the structures, abbreviations, and masses of the aposiderophores, with their iron chelation moieties colored blue.
4.2. Siderophore Biosythesis in Bacterial Pathogenesis
An average man contains about 5 g of iron, creating an overall concentration of ~3 mM. Most of that iron was acquired in the oxidized ferric [Fe3+ or Fe(III)] form and then reduced to the ferrous [Fe2+ or Fe(II)] state within cells. Human proteins complex both ferrous and ferric iron, for use as a biochemical or redox cofactor, as part of intracellular iron homeostasis, and as a means of blocking microbial iron acquisition. As an initial response to bacterial infection mammalian hosts increase the production of LF and TF, two iron binding proteins of the innate immune system. This upregulation minimizes the concentration of adventitious extracellular iron,269 confronting invading bacteria with an iron-depleted environment.270 Iron sequestration renders bacteria iron-deficient, which retards their metabolism and propagation.271,272 Within cells, proteins or small molecules complex iron for metabolic purposes, but also because free iron promotes the formation of reactive oxygen species by the Fenton reaction.264,273 Faced by iron unavailability, bacteria upregulate their iron acquisition systems, many of which are implicated in bacterial pathogenesis.5,35,49,274 They usually comprise two components: (i) siderophores, that chelate iron(III) with high affinity, and (ii) OM LGP, that avidly bind ferric siderophore complexes and actively transport them into bacterial cells. Microbial siderophores surmount the low solubility of free Fe3+ in aqueous solutions (10−18 M)4 and antagonize innate immune proteins that adsorb free iron in blood, serum, and cellular secretions. They capture the extra- and intracellular iron of humans and animals for utilization by invading bacteria that may also directly extract and transport35,275,276 iron from eukaryotic iron-binding proteins. Ent, the native catecholate siderophore of the family Enterobacterales, has the highest affinity for Fe3+ (Ka = 1052 M−1),58 which allows it to remove iron from proteins277,278 that have lower affinity (e.g., transferrin; Ka = 1020 M−1).279 Other microbial siderophores also remove iron from TF, LF, or FTN.211,212 It is often stated that the host environment is iron deficient, but the siderophores of pathogens invade and scavenge iron from eukaryotic metabolic and storage pools, effectively raising the concentration of available iron from submicromolar to much higher, potentially millimolar levels. In this way, siderophores confer an advantage to bacteria during infection of host tissues,124,134,264,280-282 so it is not surprising that heightened production of one or more siderophores is a virulence factor283,284 in Gram (−) bacterial pathogenesis. Siderophore production by invading pathogens overcomes host-imposed iron restriction,
4.3. Utilization of Xenosiderophores
Ent and FepA, the receptor for FeEnt, are prototypic components of Gram (−) bacterial TonB-dependent iron uptake systems. FeEnt is recognized and transported by both commensal and pathogenic Gram (−) species in the same and other Families.120 This natural ferric catecholate uptake system illustrates a common attribute of microbial habitats: utilization of xenosiderophores. The term refers to siderophores that are recognized and acquired by a different organism than the one that produced them. So, although A. baumannii, Yersinia enterocolitica, Neisseria gonorrheae, and P. aeruginosa lack the ability to synthesize Ent, all four species encode transport systems to assimilate FeEnt.120,272,285 This ability to utilize gratuitous ferric siderophores is an asset during colonization and pathogenesis.286 The gut, for example, is populated by thousands of different bacterial species287-289 that may simultaneously produce dozens or hundreds of different siderophores,60 so access to this conglomeration of iron chelates is potentially valuable to microbes that do not elaborate their own siderophores (e.g., Listeria monocytogenes290). In response, host epithelial cells and neutrophils produce SCN55 that preferentially binds apo- and ferric catecholates (Figure 2). SCN binds FeEnt with about 10-fold less affinity than EcoFepA and about the same affinity as other orthologues of EcoFepA (Table 1), so depending on its concentration, SCN has the ability to compete for ferric catecholates and thereby inhibit bacterial growth.264,291 Nevertheless, members of Enterobacterales adapted to evade SCN by glucosylating two of the three the catecholate rings of Ent. SCN binds the glucosylated form of the siderophore (GEnt/FeGEnt; also called ferric salmochelin) with much lower affinity.291 Species of Klebsiella, Salmonella,292 Escherichia,293,294 and Enterobacter295 encode the iroA gene cluster to glucosylate Ent.44 The iroA system contains the biosynthetic iroBCDE genes: IroB glucosylates Ent to form GEnt; IroC mediates GEnt export out of the cell; IroE cleaves the trilactone backbone of GEnt to a linear form that may traverse the IM, while IroD cleaves the linearized aposiderophore to generate a monomer and dimer.296-299 The iroA region also encodes IroN, the cognate receptor for FeGEnt. The ability of numerous pathogenic bacterial genera to glucosylate Ent and transport FeGEnt, even in the presence of SCN, allows proliferation in places that are inhospitable to other Gram (−) bacteria. Surprisingly, because it produces a number of SCN-resistant siderophores,300 including GEnt, the E. coli Nissle 1917 strain301 is employed as a probiotic treatment.302-304 It is thought to protect from diarrheal infections (for example, by S. enterica) by outcompeting the other pathogens for iron.305,306,264 These observations, combined with widespread Ent biosynthesis and FeEnt transport by Gram-negative bacteria, highlight the importance of catecholate siderophores in bacterial colonization and/or pathogenesis.164
4.4. Exchange of Iron among Siderophore Ligands
The myriad of chemically distinct siderophores264 warrants the question: why are so many different molecules needed to scavenge iron in the microenvironent? Especially given their prodigious affinity for Fe3+, why do not the catecholate siderophores monopolize bacterial iron uptake processes to the exclusion of other less avid microbial chelators? A clue to the answer may reside in the fact that the multitude of siderophores translates into another multitude of unique ferric siderophore receptors in the OM. The variety of ferric siderophore structures requires a variety of LGP recognition specificities that allows individual organisms to preferentially bind and transport particular iron complexes. As explained below, this selectivity for certain ferric siderophores has advantages to proliferation in certain environments. Second, in considering how siderophores of lower affinity compete against Ent and its derivatives for complexation of iron, it is important to note that iron chelation reactions are equilibria: in a solution of multiple aposiderophores, the distribution of iron among them reflects the affinities of the different organic ligands, their concentrations, and the pH because exchange of ferric iron between organic ligands occurs more rapidly at acidic pH. For example, Ent (KA = 1052 M−1), acquires iron from FcA (KA = 1034 M−1),59 but even at millimolar concentrations of both compounds this exchange reaction takes hours to occur (t1/2 = 4.5 h at neutrality307). In native, even iron-deficient habitats, the concentrations of microbial aposiderophores usually do not exceed micromolar levels, so the rates of ligand exchange around Fe 3+ will be slower. The upshot is that ferric siderophore complexes are relatively kinetically stable regardless of their chelation chemistry. Once formed, a lower affinity iron complex like FeAbn has a sufficient lifespan to provide iron to cells expressing its surface receptor, IutA,308 even in the presence of Ent or other potent catecholate siderophores. Furthermore, living bacterial cells act as a thermodynamic sink, driving iron–siderophore chelation equilibria toward the ferric complexes that they bind and transport.
4.5. Redundant Iron Acquisition Systems
The advantage of redundancy during iron acquisition becomes apparent when considering K. pneumoniae.309 Hypervirulent K. pneumoniae, that causes pyogenic liver abscesses310,311 (predominantly in Asia312-315), elaborates copious amounts of multiple siderophores (≥30 μg/mL)283,316 that are virulence determinants. In this sense, hypervirulent K. pneumoniae differs from classical K. pneumoniae. Hypervirulent K. pneumoniae strains harbor plasmids that encode synthesis of Ent and GEnt, as well as uptake systems for their ferric complexes.317 Humans and animals respond with serum SCN, that tightly binds Ent and FeEnt, reducing their ability to supply iron to invading bacteria. Serum albumin also adsorbs Ent, albeit with lower affinity.318 However, glycosylation of Ent by hypervirulent K. pneumoniae and other pathogens impedes its recognition by SCN.55,291,319 Besides catecholates, hypervirulent K. pneumoniae secrete the hydroxamate Abn and the mixed chelator Ybt.320 Deletion of the Abn biosynthetic locus iucA reduced the virulence of hypervirulent K. pneumoniae; loss of other siderophores did not affect its pathogenesis in a murine infection model.283 The citrate-based siderophore Abn is well-known to confer bacterial invasiveness,155,321,322 and its production is a virulence determinant of hypervirulent K. pneumoniae.323 Like GEnt and FeGEnt, neither Abn nor FeAbn bind to SCN or albumin. Thus, despite its much lower affinity for Fe3+ (KA = 1024 M−1),59 unlike Ent, Abn remains active and available in host fluids and tissues, where it may remove iron from TF and LF (Ka = 10 20 M−1).277 Biosynthesis of all four siderophores, Ent, GEnt, Abn, and Ybt, is common in hypervirulent K. pneumoniae. However, studies on the relationships of these four siderophores to the pathogenesis of K. pneumoniae284 reiterated the influence of Abn, that consistently associated with virulence.284,309,324,325 For instance, the majority of K. pneumoniae isolated from pyogenic liver abscesses produced Abn, while only 2% isolated from other sites (respiratory tract, urine, blood, or stool) secreted it.326 The other siderophores are less involved in the virulence of hypervirulent K. pneumoniae. As noted, K. pneumoniae glucosylates Ent and produces Ybt, but neither GEnt nor Ybt correlate with its systemic infections. One potential explanation for the persistent presence of GEnt in hypervirulent K. pneumoniae is that it may act in concert with the toxin microcin E492 during colonization.323 In classical K. pneumoniae, on the other hand, GEnt production does correlate with invasive colonization of specific lung tissues.280 Additionally, in extraintestinal pathogenic E. coli (ExPEC), the expression of IroN, the cognate receptor for FeGEnt, promotes biofilm formation.327 Thus overall, the apparent redundancy in the siderophores of bacterial iron acquisition systems is a misconception. They all complex Fe3+, but each one has chemical nuances that define their chelation properties, their ability to extract iron from eukaryotic proteins, and their interaction with or persistence in animal fluids or tissues. Together, these properties may produce unique, unexpected contributions to bacterial pathogenesis.
4.6. Siderophores and Tissue Tropism
The production of Ybt by K. pneumoniae illustrates other aspects of microbial iron acquisition during pathogenesis. Although named for its discovery in Yersinia pestis,328,329 several other infectious bacteria produce Ybt,330,331 that is, an atypical siderophore with mixed chelation groups. The genes encoding Ybt reside in the pigmentation (pgm) locus of the high pathogenicity island (HPI) of Y. pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica. The HPI also occurs throughout Enterobacterales in Citrobacter, Enterobacter, Klebsiella, Salmonella, Serratia, and all known pathotypes of Escherichia.332 Murine infection studies with Y. pestis revealed that the siderophore facilitates establishment of the pathogen at peripheral sites.330 While Ybt is essential for bubonic plague, it is dispensable and has varying degrees of involvement for septicemic and pneumonic plague.332-334 With regard to K. pneumoniae, classical strains that produce Ybt cause pneumonia in the murine infection model, whereas Ybt nonproducers that only produce Ent are at best opportunistic and only establish an infection in SCN-deficient mice. Despite the fact that classical K. pneumoniae causes septicemia, wound, and urinary tract infections (UTI), Ybt+ K. pneumoniae strains are predominantly found in the respiratory tract over blood, urine, or stool samples.326 Hypervirulent K. pneumoniae abundantly produces Ybt, but it was not found to enhance its pathogenesis, perhaps because copious Abn production masked the impact of Ybt.323 For uropathogenic E. coli, the noncatecholate siderophores Abn and Ybt were advantageous to colonization, and their receptors IutA and FyuA, respectively, correlated with bacterial invasion of the bladder and kidney.335 Vaccination of mice with FyuA, furthermore, protected the animals against ascending UTI to the bladder and kidney.336,337 Next, the mixed chelation chemistry of Ybt, that includes three electron pairs from nitrogen and three pairs from oxygen, imparts multifunctionality: besides Fe3+, Ybt may bind Cu 2+, which protects Y. pestis against reactive oxygen by mimicking a superoxide dismutase that converts oxygen to less harmful forms.338 In uropathogenic E. coli, this property confers a higher intracellular survival rate than that observed for nonpathogenic strains. Lastly, Ybt production illustrates that a particular siderophore may influence the site of an infection and allow the siderophore-producer to capture a replicative niche within the host.264 Whereas Ybt+ classical K. pneumoniae strains caused bronchopneumonia in normal mice, resulting in moderate bacterial load in the lungs and spleen, otherwise isogenic Ent+ K. pneumoniae caused inflammation and bacterial density in the airways.280,283,284,326 In SCN-deficient mice, on the contrary, introduction of Ent+ or Ent+, GEnt+ classical K. pneumoniae strains caused perivascular invasion, higher bacterial load, greater involvement of the spleen, and lower survival. These differences likely arise because of the better ability of Ent, relative to Ybt, to strip iron from transferrin, which is rich in the perivascular space. Consequently, in the absence of SCN, that neutralizes Ent but not Ybt, Ent-producers outcompete Ybt-producers for the available iron.280,326 These data also show the antagonism of bacterial dissemination by SCN. In summary, ferric siderophore uptake systems have multiple attributes that contribute to the infection of humans and animals. When viewed from the perspective of the diversity of their chemistry and iron chelation properties, their individual characteristics allow bacteria to adapt to the different conditions and iron sources in specific tissues.
4.7. Utilization of Hn
Erythrocyte hemoglobin constitutes the biggest source of iron in the mammalian body. Bacteria acquire iron from hemoglobin by hemolysis or cell death that releases Hn into the plasma. Alternatively, Hn may adsorb to cell surfaces, such as the intestinal lumen. Hn utilization is a factor in the virulence and pathogenesis of A. baumannii, E. coli, Haemophilus influenzae, Neisseriameningitidis, P. aeruginosa, Shigella dysenteriae, Vibrio cholerae and Y. pestis, that all utilize iron from hemoglobin.339-345 The iron in Hn is usually in the ferrous state, but once removed from the porphyrin by Hn oxidases, and especially in the presence of siderophores, the equilibrium shifts toward ferric iron, that is readily complexed by both siderophores and TF.346 A. baumannii LAC-4, P. aeruginosa, and N. meningitidis oxidize Hn to biliverdin,347 concomitantly converting Fe2+ to Fe3+, which is accessible to siderophores for iron supply to the pathogens.
5. FERRIC SIDEROPHORE TRANSPORT BY LGP OF BACTERIAL PATHOGENS
Infectious Gram (−) bacteria evolved a variety of molecular strategies to initiate the iron uptake process, that involve many different ferric siderophore or Hn receptors on the cell surface. However, all these specific systems of diverse Gram (−) bacteria share an underlying mechanistic component: the TonB/ExbBD complex that converts PMF-driven bioenergetics in the IM into biochemical processes that drive active transport through the OM (see also sections 3.2 and 8.1). The Gram (−) CRE/ESKAPE bacteria encompass various examples and paradigms with regard to iron acquisition during pathogenesis. These include K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli, as well as other Gram (−) pathogens (S. enterica, Y. pestis, Serratia marscescens) that collectively provide unambiguous evidence of the close relationship between iron acquisition and bacterial virulence.
Just a glance through the activities of the iron-transporting LGP (Table 1) emphasizes the significance of generic and specialized iron uptake mechanisms to bacterial pathogenesis. Starting with pathogenic variants of E. coli (UPEC and EHEC), and in pathogens of other genera, uptake of FeAbn, FeGEnt, and Hn consistently associates with invasiveness, tissue tropism, or infectivity. Hn scavengers (P. aeruginosa, Y. pestis), furthermore, produce both hemophore-dependent and -independent transport systems to obtain it. Utilization of other ferric siderophores is often highly specialized: besides Y. pestis, the CRE pathogens E. coli and K. pneumoniae both acquire FeYbt via FyuA, whereas P. aeruginosa does not encode such a receptor in its genome. A. baumannii is a general exception to these commonalities, in that most strains transport neither FeAbn nor Hn. However, analyses of genomic sequences suggest that A. baumannii encodes several unique iron uptake systems. A. baumannii produces three unique siderophores (Acn, Bfn, Fbn), it utilizes FeEnt,272 and its chromosome encodes at least six other LGP of currently unknown functions. Exclusively genomic inferences are sometimes incorrect,194,348 so the full understanding of the iron transport capabilities of A. baumannii await experimental characterization. After TonB-dependent OM uptake, iron translocation into the cytoplasm involves multiple binding and transport reactions in the periplasm and the IM. Consequently, the identification of a homologous LGP orthologue in the OM does not necessarily guarantee the ability of A. baumannii to use a particular ferric siderophore as an iron source.
5.1. E. coli
Most E. coli strains are harmless to animals and humans and live in the gut as commensal microbes. Strains like the probiotic Nissle 1917 are beneficial to human physiology, alleviating symptoms of colitis and inflammatory bowel disease.349 However, E. coli also acquires pathogenesis determinants, including siderophore biosynthetic genes, toxins, or other molecules that promote tissue invasion or tropism. Pathogenic E. coli fall into numerous categories: ETEC (entero-toxigenic), EIEC (entero-invasive), EHEC (enterohemorrhagic (including the widespread O157:H7), EPEC (entero-pathogenic), EAEC (entero-aggregative), and AIEC [adherent-invasive, which includes uropathogenic E. coli (UPEC), that causes ~90% of urinary tract infections (UTI)]. These pathogenic isolates often rely on iron acquisition mechanisms that are not found in laboratory strains. For instance, the EHEC pathogen 0157:H7 acquires iron from Hn or hemoglobin through the outer membrane receptor ChuA.118
Tang and Saier167 compared the laboratory E. coli strain MG1655350 to serovars in five of the pathogenic categories. Not only are certain LGP exclusively found in the pathogens, but some strains have multiple receptors for a single ferric siderophore or Hn. MG1655 expresses six receptors for iron uptake, whereas UPEC strains produce 10–15, again illustrating the direct connection between iron uptake versatility and virulence. It is noteworthy that 4 of 7 pathogenic strains had two chromosomal tonB homologues,167 although their functional differences are not known.
Prototypic laboratory E. coli K-12 strains, like the sequenced paradigm MG1655, do not chromosomally encode the FeAbn receptor IutA, but it is present in the genome of pathogenic E. coli, like the AIEC strain O83:H7.351 As first discovered for EcoIutA,155 LGP are often encoded and mobilized on plasmids that transfer among bacteria in natural habitats. IutA (NRG857_30235) and IroN (NRG857_30015) are encoded on the E. coli plasmids pAPECO103-ColBM, pAPEC-O1-ColBM, and pVM01 (from the APEC strain E3), on the S. enterica serovar Kentucky plasmid pCVM29188_146, and on the K. pneumoniae CG43 plasmid pVLK. The chromosome of K. pneumoniae CG43 also contains another FepA paralogue (NRG857_02640). Furthermore, in ExPEC the FeGEnt receptor, IroN, contributes to biofilm formation, independently of GEnt production.327 E. coli O157:H7 encodes several novel LGP (Table 1), including ChuA, that recognizes and transports Hn.
5.2. K. pneumoniae
Classical K. pneumoniae is a nonmotile Gram (−) bacillus in the Family Enterobacterales. Most K. pneumoniae isolates are encapsulated, nontransformable, nontransducible, and probably virulent. However, mutations in LPS and capsule biosynthesis,352-356 DNA methylation,357 and iron acquisition358 may attenuate K. pneumoniae. Besides its ubiquity in surface water and soil, it is a commensal bacterium in the gastrointestinal tract and a common opportunistic nosocomial pathogen. It may infiltrate the urinary tract, bloodstream, or lungs, and it may contaminate surgeries, resulting in wound and urinary tract infections, pneumonia, bacteremia, and sepsis. Infections with classical K. pneumoniae may progress to pyogenic liver abscesses, meningitis, endophthalmitis, and sepsis. Such “community-acquired infections” are public health threats, and the increasing propensity of this organism to acquire antibiotic resistance augments its threat to human and animal health. Especially, carbapenem-resistant strains of classical K. pneumoniae, that are resistant to nearly all known antibiotics,359 cause 40–50% mortality from bloodstream infections.360 Classical K. pneumoniae strains that express extended spectrum β-lactamases (ESBLs) are resistant to cephalosporins and monobactams.253,265 Besides the classical K. pneumoniae pathotype, a hypervirulent variant emerged that causes hepatic abscesses, endopthalmitis, meningitis, osteomyelitis, and necrotizing fasciitis, even in otherwise healthy individuals.361-366 Acquired drug resistance makes classical K. pneumoniae difficult to eliminate but does not enhance its virulence. Hypervirulent K. pneumoniae, on the other hand, may acquire both antibiotic resistance genes and novel virulence genes together on a large plasmid309 and, in some cases, additional chromosomal elements as well. The biomarkers on the virulence plasmid differentiate hypervirulent from classical K. pneumoniae. Hypervirulent K. pneumoniae has the ability to infect healthy individuals and frequently causes invasive infections that further distinguish it from classical K. pneumoniae. Thus, clinical isolates of this superbug show a worrisome confluence of drug resistance and virulence determinants that threaten a medical crisis, including hypermucoviscous capsule, lipopolysaccharide (LPS), siderophores, and fimbriae.324,367 Other factors also play a role in the virulence of K. pneumoniae: the OM permeability properties of its porins, IM efflux pumps, and systems involved in allantoin metabolism. In many cases, the contributions of these factors to pathogenesis are not yet fully understood.367
5.2.1. Overview of TonB-Dependent Iron Uptake by K. pneumoniae.
Relative to wild-type strains, TonB-deficient K. pneumoniae are attenuated in murine infection models.95 K. pneumoniae chromosomally encodes biosynthesis of four different siderophores: Ent, GEnt, Abn, and Ybt. The production of multiple iron acquisition systems counteracts host neutralization of any individual one of them, and different siderophores may promote colonization of different tissues in the host,264,368 in both cases increasing the survival of the pathogen. Among four siderophores secreted by classical K. pneumoniae, Ent has the highest affinity for Fe3+ (KA = 1052 M−1)58 and Abn has the lowest (KA = 1023 M−1),59 but this Ent/GEnt/Abn example illustrates, as discussed above (section 4.2), that avidity for iron is not always the factor that determines the contributions of a particular iron uptake process to virulence, invasiveness, or pathogenesis.369 Ent production is ubiquitous among both classical and hypervirulent K. pneumoniae that utilize ferric catecholates in both the wild and host environments. As in E. coli, the genes encoding the Ent biosynthetic enzymes of K. pneumoniae reside in the chromosomal entABCDEF gene cluster, while genes encoding FeEnt transport are in the chromosomal fepABCDEG gene cluster. Both Ent production and FepA expression are upregulated during infection by K. pneumoniae, which enhances colonization of the lungs.370,371
5.2.2. GEnt.
Production of SCN by neutrophils and on mucosal surfaces opposes the actions of Ent/FeEnt by competing with KpnFepA for binding of the apo- and ferric siderophore.326,367 Consequently, SCN minimizes iron uptake, which retards growth of bacterial pathogens in host fluids and tissues.122,326,367,372 Increased production of SCN also causes acute inflammatory effects, resulting in secretion of IL-8 that recruits neutrophils to the infection site.122 Host production of SCN illustrates the active role of innate immunity in combating bacterial iron uptake, but like other bacterial pathogens, K. pneumoniae responds by glucosylating Ent. The chromosomal- or plasmid-encoded iroA gene cluster (iroBCDEN) contains the genes for enzymes involved in GEnt biosynthesis319 as well as for the OM FeGEnt receptor, IroN.367 KpnIroN is only 53.2% identical to EcoFepA, considerably less than normally seen between LGP orthologues (the identity between KpnFepA and EcoFepA is ~80%). This lower extent of identity still infers the same structural fold, and may rationalize the different ligand selectivities of the two proteins: KpnIronN recognizes both FeEnt and FeGEnt, but EcoFepA only binds FeEnt and not FeGEnt.285 Because SCN does not adsorb GEnt/FeGEnt, neither does the glucosylated siderophore induce inflammation at the infection site. Thus, concomitant glucosylation of Ent and expression of IroN combine as a virulence determinant in both classical and hypervirulent K. pneumoniae that supersedes the host innate immune response. GEnt producers are more virulent that Ent producers in an SCN-sufficient host. For instance, GEnt expression enhances colonization of the nasophyrynx by classical K. pneumoniae.122 Although only 2–4% of the hospital-acquired classical K. pneumoniae strains carry the iroA gene cluster, more than 90% of hypervirulent strains isolated from pyogenic liver abscesses carry the genes and produce GEnt.122
5.2.3. Ybt.
SCN does not recognize the mixed chelation siderophore Ybt that was originally identified in a pathogenicity island of Yersinia enterocolitica.329,373 18% of classical K. pneumoniae and 90% of hypervirulent clinical isolates produce Ybt.122,367,372 Ybt has robust affinity for Fe3+ (KA = 1036.6 M−1)374 but significantly lower than Ent/GEnt. FyuA binds and transports FeYbt; it is also a receptor for pesticin.375,376 The IM ABC-transporter YbtPQ conveys FeYbt into the cytoplasm.367 Because SCN does not bind FeYbt,280,326 K. pneumoniae strains that produce it create increased bacterial loads during lung infections. TF antagonizes Ybt in plasma, but as a result of its lower affinity for Fe3+, at equivalent concentrations the equilibrium favors the ferric siderophore. K. pneumoniae strains that only produce Ybt are unable to infect immunocompetent individuals,326 but elaboration of Ybt is a virulence determinant for classical K. pneumoniae strains in mouse infection models.122,367
5.2.4. Abn.
The citrate-based hydroxamate siderophore Abn was originally isolated from Aerobacter aerogenes.377 It has the lowest affinity for iron59 of the siderophores secreted by K. pneumoniae, but Abn rapidly removes iron from TF,277 and unlike the catecholate chelators, neither SCN nor serum albumin remove it from circulation.318 Hence, Abn-mediated iron acquisition is unexpectedly efficacious in the host. Furthermore, 90% of hypervirulent K. pneumoniae strains excrete Abn, compared to only 6% of classical strains,378,379 directly linking Abn to bacterial pathogenesis. In K. pneumoniae, as in virulent E. coli strains that make Abn and transport FeAbn,49,380 the biosynthetic iucABCD enzyme system utilizes L-lysine and citrate to produce the hydroxamate siderophore. The sequential activities include a hydroxylase (IucD), an acetyltransferase (IucB) and the Abn synthetase (IucA), that stereospecifically adds N6-acetyl-N6-hydroxylysine to the primary carboxylate of citrate.381 Transfer of a plasmid carrying the IucABCD genes of hypervirulent K. pneumoniae to laboratory E. coli led to production of Abn,324 confirming this pathway. The structural gene of the FeAbn receptor, iutA,308,382 usually resides on the same plasmid as the Abn biosynthetic loci. Furthermore, in hypervirulent K. pneumoniae, the rmpA gene, that increases capsule production, is on the same plasmid.264,367 Consequently, Abn-mediated iron acquisition and hypermucoviscous capsule are often linked in hypervirulent strains. Elevated levels of both Abn production (6–10-fold) and hypermucoviscous capsule are defining characteristics of these strains, relative to classical K. pneumoniae. Among the four siderophores that hypervirulent K. pneumoniae secretes, Abn accounts for more than 90% of iron transport activity,383 is a critical factor for growth and survival in human ascites or ex vivo serum and confers virulence in murine systemic or pulmonary infection models.284 This role as a primary virulence factor likely derives from a combination of Abn’s indifference to SCN and its enhanced production by hypervirulent K. pneumoniae. Although hypervirulent K. pneumoniae normally also produces Ent/GEnt, the contributions of the glucosylated catecholate to systemic infections are not fully defined.284,323 In the absence of Abn, otherwise wild-type classical K. pneumoniae, that still excrete Ent, GEnt, and Ybt, showed higher bacterial load in the lungs of mice, relative to an isogenic entB ybtS strain that did not produce either catecholate siderophore. All three siderophores were required for the dissemination of classical K. pneumoniae to the spleen and induction of proinflammatory cytokines.265 These data support both the primary importance of Abn during pathogenesis by K. pneumoniae, as well as the ability of particular siderophore iron acquisition pathways to allow bacterial proliferation in specific host tissues.
5.2.5. FepB.
In K. pneumoniae, FepB participates in the uptake of FeEnt/FeGEnt; a ΔfepB strain was attenuated in lung colonization and tissue dissemination in vivo. However, the reductions in virulence engendered by ΔfepB were distinct from the FeEnt/FeGEnt uptake defects alone and unrelated to acquisition of FeYbt.253 Studies of the transcriptional regulator RamA in another member of Enterobacterales, S. enterica, coincidentally revealed that the periplasmic FeEnt/FeGEnt binding protein FepB contributes to the survival of S. enterica in RAW 264.7 macrophages and to its virulence in BALB/c mice.253
5.3. A. baumannii
A. baumannii is a short, nonmotile, rod-shaped, oxidase-negative, Gram (−) coccobacillus in the Family Moraxcellaceae. In 2019, A. baumannii constituted over 20% of all hospital-acquired infections; nearly half of these isolates were carbapenem-resistant Acinetobacter384 that are recognized as an urgent threat to human health from their propensity to cause pneumonia, wound, bloodstream, and urinary tract infections. The rapidly increasing resistance of this bacterium to most antibiotics is a concern for healthcare systems around the globe.20 Additionally, A. baumannii has an abnormally high mortality rate compared to other Gram (−) pathogens (up to 70% from extreme drug resistant (XDR) strains).385 A. baumannii further illustrates the impact of iron acquisition on pathogenesis, in that it overcomes the iron-limiting conditions of the host by secreting atypical siderophores and altering its OM protein composition to optimize the uptake of its own and other ferric siderophores.386-390
5.3.1. Fur-Mediated Regulation of Iron Acquisition.
Iron deficiency affects the virulence of A. baumannii, which responds by upregulating genes involved in iron acquisition and other processes like respiration, biofilm formation, and motility.391-393 As in all Gram (−) bacteria, the ferric uptake regulator (Fur) controls expression of the iron transport systems of A. baumannii. Fur negatively controls expression by binding to the “Fur box,” a conserved DNA sequence upstream of iron-related biosynthetic and transport genes. The primary structure of Fur from A. baumannii strain BM2580 is 63% identical to that of Fur from E. coli K-12.387 Fur boxes were also identified in the genomes of other A. baumannii strains.391,394 The iron uptake systems of A. baumannii are also upregulated during growth at 28 °C compared to 37 °C,395 and during growth in serum, supporting their role the organism’s virulence in vivo.396 Additionally, at lower temperatures the BLUF-type photoreceptor BlsA interacts with Fur to photoregulate genes involved in Acn biosynthesis and FeAcn uptake.397 In the dark at 23 °C, BlsA antagonizes the actions of Fur to upregulate the production of both biosynthetic and transport genes; growth in blue light or at 37 °C eliminates this effect.
5.3.2. Siderophores: Acn, Bfn, Fbn.
The native, chromosomally encoded siderophores of A. baumannii are the mixed chelate Acn,390 the hydroxymates BfnA and BfnB,398 and the mixed chelation compounds Fbn.399 Other putative siderophores biosynthetic gene clusters exist among particular isolates of A. baumannii that are as yet uncharacterized.388,400-403
5.3.2.1. Acn.
The most-studied A. baumannii siderophore gene cluster encodes Acn390, that contains catecholate, hydroxamate, and imidazole groups. The Acn gene cluster occurs in the majority of clinical isolates and sequenced genomes, with the exception of A. baumannii SDF.391,400,404 Fur boxes control transcription of the structural genes for the enzymes involved in Acn biosynthesis and for FeAcn transport, in response to extracellular iron levels.394 Bioinformatic analyses identified three putative systems within the Acn cluster: basA-J for acinetobactin synthesis, bauA-F for A. baumannii acinetobactin utilization, and barAB for A. baumannii acinetobactin release to the environment.394,405 Acn belongs to the nonribosomal peptide synthetase (NRPS) class of siderophores406 and consists of 2,3-DHBA, threonine, and N-hydroxyhistamine.406 The entA gene, that encodes production of DHBA, resides elsewhere in the genome, away from the acinetobactin gene cluster.394,407 Acn exists in two forms: preacinetobactin (pAcn) contains an isooxazolidinone ring system that undergoes a pH-dependent isomerization to an oxazoline ring in Acn408 (Figure 3). This response to the acidic conditions typically found at sites of acute infection makes pAcn/Acn virulence factors.409 Both siderophore isomers bind iron as a 2:1 complex, and both enable A. baumannii growth in low iron conditions.408,410-412 BauA, the pertinent OM receptor, was crystallized in complex with FepAcn.413 However, the exact selectivity of BauA for iron complexes of pAcn and Acn is uncertain because both compounds promote growth. Furthermore, the next entity in the transport pathway, the periplasmic binding protein BauB, was crystallized in complex with FeAcn. BauB binds both FepAcn and FeAcn with nanomolar affinity.410,411 The fact that the crystallized BauA protein originates from A. baumannii ATCC 19606,413 whereas bauA was originally annotated in strain ATCC 17978, within the classic Acn biosynthesis and transport gene cluster,391 further confuses the issue. The 19606 and 17978 BauA primary structures are only 56.6% identical, suggesting that they are functionally different receptors. Although it lacks the ability to produce Ent or Fc, A. baumannii assimilates both ferric siderophores through LGP encoded in its genome.264,272,285,414
Similar to hypervirulent K. pneumoniae, the presence or absence of genes for biosynthesis and uptake of various siderophores in the A. baumannii genome often affects the extent of its virulence. Acn synthesis is required for A. baumannii pathogenesis: the biosynthetic and transport proteins BasD and BauA, respectively, are virulence determinants for A. baumannii ATCC 19606T in the Galleria mellonella larvae infection model415,416 and in a murine model of systemic infection.415 BasD was required for full virulence of A. baumannii ATCC 19606T in a murine model of wound infections,417 and Acn biosynthesis enabled persistence of A. baumannii ATCC 19606T in human alveolar epithelial cells, ultimately resulting in apoptosis.415,418 Siderophores, especially Acn, promote survival of A. baumannii in serum, where they strip iron from TF and LF.176,419
5.3.2.2. Bfn.
A second siderophore biosynthetic cluster exists in A. baumannii AYE and other strains, that encodes the hydroxamates Bfn A and B.398 The genetic and biochemical details of Bfn biosynthesis are less well-defined. A. baumannii AYE does not have a functional entA locus and therefore does not produce Acn.398,407 The Bfn biosynthetic and transport gene cluster consists of bfnA-L and exists in the majority of sequenced strains.398 Bfn compounds are nonribosomal peptide synthetase-independent (NIS) siderophores, based on their production by BfnA and BfnD synthetases.398 BfnH is the LGP that recognizes Bfn.398 Bfn A and B are chemically similar to acinetoferrin, another hydroxamate from Acinetobacter hemolyticus ATCC 17906T.420 The role of Bfn A and B in pathogenesis are not yet known.
5.3.2.3. Fbn.
A. baumannii produces a third unique iron acquisition system, based on the mixed chelation (catecholhydroxamate) siderophores Fbn A-F, that were found in Acinetobacter baylyi ADP1 and four other sequenced strains including A. baumannii ATCC 17978.391,399,400,411 FbnA is the predominant siderophore in this group (~85% of total mass); FbnB-F are likely biosynthetic intermediates or shunt byproducts.399 Like Acn, Fbn classify as NRPS siderophores, but unlike Acn, they contain a single hydroxamate and two catecholate groups that together create a 1:1 complex with Fe3+.411 A. baumannii ATCC 17978 excretes less Fbn than Acn,411 and the role of Fbn in pathogenesis is currently undefined. Regarding their transport, FeFbnA initially binds to FbsN,399 then to BauB (the FeAcn periplasmic binding protein) with nanomolar affinity. The fbn gene cluster does not encode a periplasmic binding protein; broad recognition specificity is common in periplasmic siderophore binding proteins, as occurs in FhuD421 and FepB248,421,422 of E. coli. Consequently, FeFbnA transport competitively antagonizes FeAcn uptake.411
5.3.3. Hn Utilization.
Like many bacterial pathogens, A. baumannii displays hemolytic activity when grown in iron-deficient conditions423 that releases Hn from erythrocyte hemoblobin as an iron source. The genomes of numerous sequenced strains and clinical isolates of A. baumannii contain plc1 and plc2 that encode phospholipase C enzymes with the capability of lysing host red blood cells.400,404,423,424 Plc1 and Plc2 are upregulated and hemolytic to horse and human erythrocytes in low-iron conditions, suggesting a role in iron acquisition from Hn found in erythrocytes.423 The former enzyme, but not the latter, is critical for the virulence of A. baumannii ATCC 19606 in the G. mellonella infection model.423 Three different phospholipase D enzymes in A. baumannii also impact its virulence,404,425,426 but their role in iron acquisition is not yet defined.
Once inserted into protoporphyrin IX by ferrochelatase, iron is stable in Hn unless the porphyrin ring system is compromised. Bacterial pathogens, including A. baumannii, liberate iron from Hn by the actions of oxidases. Although A. baumannii generally uses Hn as an iron source, different strains show variability in Hn utilization.427-430 Separate gene clusters encode Hn uptake systems and a Hn oxygenase (HemO) that cleaves the porphyrin and releases iron.400,428,430 For example, the highly virulent A. baumannii LAC-4 contains a cell envelope Hn uptake system, that acquires Hn at sites of infection, and it produces HemO, that degrades Hn to release iron.428,429 On the other hand, A. baumannii ATCC 17978 encodes the Hn uptake loci but lacks hemO. Consequently, it does not grow well with Hn as a sole iron source, underscoring the importance of Hn degradation to its pathogenesis.428,429
5.3.4. LGP.
TonB-dependent ferric siderophore transport into A. baumannii encompasses as many as 21 predicted LGP, although at present only a few are biochemically characterized.400 As noted above, AbaBauA recognizes FeAcn, while AbaBfnH398 and AbaFbsN399 are the predicted receptors for FeBfn and FeFbn, respectively. The latter two designations are not yet experimentally verified. Besides the three siderophores of its own creation, A. baumannii utilizes other xenosiderophores, including FeEnt,272,431 via the orthologue AbaFepA. Accordingly, A. baumannii apparently also acquires iron from monocatecholate compounds, catalyzed by AbaPiuA and AbaPirA, that confer sensitivity of A. baumannii ATCC 19606T to siderophore–monobactam antibiotic conjugates.432 Despite the solution of crystal structures for PiuA and PirA from both A. baumannii and P. aeruginosa,432 their biochemical roles in siderophore uptake, bacterial physiology and infection are currently undefined. A. baumannii utilizes iron from ferrichrome but not from ferrichrome A.272 Other proteins were implicated in iron transport by A. baumannii, including homologues of OmpW and OprD.433 Lastly, AbaFepA is a virulence determinant of A. baumannii ATCC 17978 in a mouse sepsis infection model, showing the importance of xenosiderophore utilization in vivo.431
5.3.5. TonB/ExbBD.
All sequenced A. baumannii genomes contain three separate tonB gene clusters that were likely horizontally acquired from different sources.434 In A. baumannii ATCC 19606T, the three systems are tonB1/exbB1/exbD1.1/exbD1, tonB2, and tonB3/exbB3/exbD3. The genes in these loci are variably expressed, but only tonB3 contains an upstream Fur box, and iron deprivation only upregulates tonB3, implicating it in iron homeostasis.393,434 The analyses of individual mutants in these loci were inconclusive,393,434 but both AbaTonB2 and AbaTonB3 functionally complement the absence of EcoTonB during iron-limited growth of E. coli.434 AbaTonB1 and AbaTonB2 are both required, but neither are individually sufficient, for full virulence of A. baumannii ATCC 19606T in the G. mellonella infection model, nor in a mouse sepsis model. In the latter case, Runci et al.393 inoculated mice with wild-type A. baumannii ATCC 19606T, and a second group with a ΔtonB3 derivative of the same strain. After 24 h, none of the mice infected with the wild-type survived, whereas all the mice inoculated with the tonB3-deficient strain survived. These data indicated that TonB3 is essential to the virulence of A. baumannii ATCC 19606T. Overall, AbaTonB3 was both necessary and sufficient for virulence in either experimental model.393,434 Together these studies suggest that tonB3/exbB3/exbD3 is the major system that facilitates iron acquisition by A. baumannii.
5.3.6. Uptake of Fe2+.
Like E. coli, S. typhimurium and other Gram (−) bacteria, A. baumannii utilizes the ferrous iron transport system, FeoAB, regulated by FeoC, during growth in reducing environments where ferrous iron may predominate.400 The eukaryotic intracellular environment is one such situation. An feoA mutant of A. baumannii ATCC 17978 was deficient in iron-limiting conditions with regard to growth, biofilm formation, adhesion, and virulence.435 Two feoB homologues exist in some A. baumannii strains, including ATCC 17978, that contains both chromosomal and plasmid (pAB3) loci.435 In A. baumannii ATCC 19606T, feoAB is controlled by Fur, via an upstream Fur box.393 The FeoAB system is a virulence determinant in both Gram (–)88 and (+)436 bacteria. However, a ΔfeoB derivative of A. baumannii ATCC 19606T was not impaired in low-iron media, nor in the G. mellonella nor mouse infection models. Conversely, its growth was restricted in human serum,393 and feoB was required for full virulence in a mouse sepsis model.414 On balance, despite some conflicting data, the FeoAB system is an important contributor to iron acquisition by many prokaryotes, including A. baumannii, in the host environment.
5.4. P. aeruginosa
P. aeruginosa is an encapsulated, catalase- and oxidase-positive Gram (−) bacillus in the Family Pseudomonadaceae that forms biofilms and causes disease in plants, animals, and humans. It is an opportunistic pathogen that thrives in numerous host environments, infects a variety of tissues, and causes particularly serious disease in patients with cystic fibrosis. Their lungs and digestive pathways accumulate copious sticky mucus, creating an environment that P. aeruginosa readily colonizes with biofilms that correlate with its virulence.437 P. aeruginosa also infects the urinary tract and burn wounds, often resulting in septicemia. Like other pathogens it faces the reduced accessibility of iron in animal tissues, as a result of TF, LF, and FTN.44 However, depending on the infection (acute vs chronic), P. aeruginosa adjusts its preferred iron source to minimize the metabolic input needed to obtain it,438 and it employs diverse mechanisms of iron acquisition.
5.4.1. Elaboration of Siderophores.
P. aeruginosa synthesizes Pch211 and Pvd,212 that are atypical siderophores (Figure 3). The former is similar to Ybt, in that it contains a hydroxyphenyl and two thiazolidine rings (Ybt contains three thiazolidines), but Pch (324 Da) is smaller than Ybt (482 Da). Pch has considerably lower affinity for Fe3+ (KA = 105 M −1)439 than most siderophores, it binds iron with a 2:1 stoichiometry and may form mixed iron complexes with other ligands.209 Its biosynthesis requires expression of fewer genes than Pvd, so when environmental iron levels drop below micromolar levels, P. aeruginosa produces Pch first.440 The larger, mixed chelation siderophore Pvd (1365 Da) has an uncommon structure, that includes a dihydroxyquinoline complexation moiety and 6–14 l- or d-amino acids, in some cases cyclized, that provide additional hydroxamate or carboxylate ligands to the hexadentate iron center. Its more typical high affinity for Fe3+ (KA = 1032 M−1)441,442 allows Pvd to remove iron from TF/LF,443,444 and it is not recognized and removed from circulation by SCN (see below). Pseudomonads in the biosphere produce over 60 chemical variants of pyoverdine (also called pseudobactins445,446). Strains that do not produce pyoverdine are less virulent in murine443 and in rabbit447 infection models, comparable to the reduction in virulence seen for TonB-deficient mutants.448 The role of Pvd in pathogenesis goes beyond its ability to capture iron, because it regulates its own synthesis, as well as the production of two extracellular virulence factors, the protease Prp and exotoxin A.73,449 Besides Pch and Pvd, studies on the effect of airway mucus secretions on the growth of P. aeruginosa revealed another siderophore biosynthesis and transport system.450 A mutant strain devoid of Pch and Pvd survived iron deficiency by obtaining iron with a prevously unknown, alternative mechanism. Using transposon-mutagenesis, Gi et al.450 identified a multigene locus encoding the synthesis and uptake of nicotianamine, a tricarboxylate iron chelator formed from S-adenosyl methionine, that was originally described in plants.451,452 Nicotianamine may play a role in the survival and fitness of P. aeruginosa in human lungs.450
5.4.2. Uptake of Xenosiderophores.
The genome of P. aeruginosa encodes more than 30 putative LGP, many of which bind and transport ferric siderophores that originated from other bacteria and fungi. PfeA and PirA bind and transport FeEnt, produced by Enterobacterales;453-455 FoxA transports ferrioxamine B,456 a siderophore from Streptomyces; FiuA transports the fungal siderophore ferrichrome;457,458 FemA utilizes iron complexes of mycobactin and carboxymycobactin;459 FecA transports FeCit;460 ChtA acts as receptor for iron complexes of rhizobactin, Abn, and schizokinen;461 FvbA for the uptake of FeVbn.462 The role of these ferric xenosiderophores in the mammalian pathogenesis of P. aeruginosa is not yet fully known, but P. aeruginosa has an ability to sense its environment, as illustrated by the fact that some strains stop making their own siderophores when grown together with Streptomyces ambofaciens and instead up-regulate the expression of FoxA to profit from the presence of ferrioxamine B.463 Similar accommodations may occur during infection of mammalian hosts, depending on other microorganism that may be present.
5.4.3. Hn Utilization.
P. aeruginosa uses two distinct TonB-dependent systems for acquisition of Hn from proteins like hemoglobin or hemopexin:464 Pseudomonas haem uptake (Phu) and haem assimilation system (Has). The Phu system involves an LGP, designated PhuR, that recognizes and directly extracts Hn from the target proteins, while the Has system encompasses an additional, secreted hemophore (HasAp) that binds Hn and then adsorbs to the LGP HasR.465-467 Single mutants in either systems still utilize Hn as sole iron source, but a double mutant is unable to do so.464 Experiments suggest that PhuR is the primary Hn receptor, whereas the Has system is centered on sensing Hn in the environment.468,469 A third system, Hxu, was identified by proteomics.470 Like Has, it may play a prominent role in sensing Hn but only a minor role in Hn uptake. However, in the absence of HasR, P. aeruginosa upregulates the OM receptor HxuA, suggesting its participation in Hn uptake.
5.4.4. Extraction of Iron from FTN.
Roughly 25% of all the iron in the human body resides in cytoplasmic FTN. Each molecule holds as many as 4000 iron atoms, which makes it a potentially useful source of iron for invading pathogens. However, the iron within FTN exists as a ferrihydrite that has only marginal solubility. To further complicate utilization of its iron, FTN is present in miniscule amounts in circulation: the plasma FTN concentration is 1.5–30 ng/mL; TF, by comparison, is present at 100-fold higher concentrations.471 However, the lungs of CF patients contain a 70-fold higher extracellular concentration of FTN,472 suggesting that it may act as an iron source for growth of strains adapted to thrive in that environment. It is relevant that both catecholate and hydroxamate siderophores readily remove iron from FTN.278 Careful measurements with wild-type and siderophore-deficient strains implied that Pch and Pvd acquired iron from FTN to support growth of P. aeruginosa, even without any proteolytic degradation of the multimeric protein cage. FTN also supported growth of siderophore-deficient P. aeruginosa strains, but only in the presence of extraneous proteases.473 In this case, after degradation of the protein framework, the actions of extracellular reducing agents (e.g., phenazines like PCA) presumably produce Fe2+, allowing iron uptake by the FeoAB system.
5.4.5. Cell Surface Signaling during Iron Uptake.
As originally described for E. coli FecA,474 certain LGP accomplish signal transduction through the cell envelope to the cytoplasm in response to recognition and binding of a cognate ligand on the cell surface. Binding of FeCit by EcoFecA positively upregulates transcription of its structural gene, ultimately increasing the rate of FeCit uptake. In P. aeruginosa, numerous LGP have the ability to regulate gene expression.475 This phenomenon, now called cell-surface signaling (CSS), was first shown in P. aeruginosa for the self-regulated uptake of FePvd by FpvA, but it was later observed for receptors of Hn and citrate, that act as xenosiderophores. CSS LGP like FecA and FpvA contain a 70–80 residue N-terminal extension that contains two α-helices sandwiched between two β-sheets.476,477 In the well-studied case of FpvA, binding of FePvd activates the alternate sigma factors PvdS and FpvI, leading to enhanced expression of the Pvd biosynthetic genes and fpvA itself.478,479 In the absence of FePvd the activities of PvdS and FpvI are inhibited by an antisigma factor, FpvR, that spans the cytoplasmic membrane.480,449,481 When Pvd captures iron and associates with FpvA, the binding reaction stimulates proteolytic degradation of FpvR, releasing the alternate sigma factors to act on RNA polymerase, which improves recognition of promoters and up-regulates both the synthesis of pyoverdine, FpvA, and virulence factors.
5.4.6. Iron Uptake in Biofilms.
The biofilms of P. aeruginosa are complex structures that may contain additional different microorganisms, all of which are surrounded by extracellular polysaccharides, DNA, and polypeptides.482-484 Biofilm formation facilitates evasion of host immune recognition, phagocytosis, and bacteriocidal actions by the host.485 Although Pch is not needed for biofilm production, Pvd biosynthesis is required,486,487 indicating a need for iron uptake during biofilm formation. In the absence of Pvd, supplementation of media with FeCit restores biofilm formation, supporting this conclusion. High-throughput genetic screens using Pvd fluorescence as an assay of its production revealed 55 genes that affect Pvd production: their absence decreased Pvd biosynthesis.484 The loci fell into a few classes; several genes related to biofilm production affected Pvd biosynthesis, including genes for flagellin biosynthesis, chemotaxis, type IV pili assembly, cup fimbriae biogenesis, and exopolysaccharide synthesis.484 A direct correlation exists between Pvd production and biofilm formation, leading to the conclusion that FePvd provides the iron needed for biofilm biosynthesis in iron-deficient conditions. Nevertheless, Pvd biosynthesis is not needed for biofilm formation in iron-replete conditions.
5.4.7. Extracellular Reduction of Iron.
Production of siderophores combats the low solubility of ferric iron in aerobic aqueous environments but also create a dilemma for Gram (−) bacteria because ferric siderophore complexes are often too large and bulky to penetrate the 10 Å channels of general porins. LGP solve this OM transport problem, but at an energetic cost (in the case of EcoFepA, ~4 ATP per molecule of FeEnt internalized62). Ferrous iron, on the other hand, is soluble and predominates in partially anaerobic, acidic environments.488 This setting may exist in the lungs of CF patients, due to the disproportionate amount of mucus that accumulates on their epithelial surfaces, where the bacterium forms biofilms. In that context, P. aeruginosa secretes PCA, a secondary metabolite with reducing potential. PCA is present in large amounts in the mucus of patients with advanced P. aeruginosa infections,489,490 portending a role for this uptake system in virulence. Once reduced to Fe2+, iron may diffuse through OM general porin491 channels to the periplasm, where the FeoAB transporter passes it to the cytoplasm. The human protein calprotectin targets PCA-mediated iron uptake by P. aeruginosa. Calprotectin sequesters Mn(II), Zn(II), and Fe(II),492 and it inhibits phenazine biosynthesis in P. aeruginosa, resulting in iron-deprivation. Hence, its activity resembles the sequestration of ferric catecholates by SCN, as a nutritional immunity defense mechanism against the pathogen493,494
5.4.8. Periplasmic Transport of Pvd.
FePvd is bound and transported to the periplasm by the TonB-dependent OM receptors FpvAI and FpvB.442,495 In Gram (−) bacteria, ferric siderophores generally remain intact, even when bound to periplasmic binding proteins, until their internalization through the IM into the cytoplasm. In P. aeruginosa, however, it was suggested that reduction of iron occurs in the periplasm and that both Fe2+ and the aposiderophore traverse the IM into the bacterial cytoplasm. For example, after OM transport through TonB-dependent LGP FpvAI or FpvB, FePvd is bound by periplasmic FpvC or FpvF,496 after which the IM FpvG reduces the iron complex, releasing Fe 2+ for binding by periplasmic FpvC. According to this mechanism, the ABC-type transporter FpvDE ultimately transports ferrous iron into the cytoplasm. An overview of experiments concerning the fascinating path of pyoverdines from outside to inside the bacterial cell is found in Vigoroux et al.497
5.4.9. Mitochondrial Toxicity of Pvd.
Besides the well-known the role of Pvd in the virulence of P. aeruginosa, it directly contributes to cytotoxicity. When Pvd enters Caenorhabditis elegans498 or mammalian cells,483 it fatally disrupts mitochondrial homeostasis. Cells that suffer this type of damage destroy and recycle their mitochondria, in a process known as autophagy, which may be viewed as an arm of innate immunity against pathogens like P. aeruginosa.
5.5. Y. pestis
Y. pestis is a nonmotile, facultative anaerobic, rod-shaped, Gram (−) coccobacillus in the Family Yersiniaceae, that does not produce spores. It causes potentially fatal diseases: bubonic, pneumonic, and septicemic plague.499-502 In Y. pestis, as in many pathogens, the ability to infect humans and animals hinges on the activation of virulence determinants, some of which acquire nutrients in the host. Iron uptake systems are required for intracellular growth of Y. pestis so they are tightly regulated during its colonization of humans and animals.503-506 The Y. pestis genome encodes multiple iron or Hn transporters, including TonB-dependent OM systems, IM ABC- and non-ABC transporters. “Ironomic” studies of Y. pestis identified 16 iron uptake systems, but only two of them, the LGP FyuA, that internalizes FeYbt, and the IM ABC-transporter Yfe, that transports iron or manganese, correlate with its virulence.505,507-515
5.5.1. The High Pathogenicity Island.
The TonB-dependent uptake system for FeYbt is encoded in a high pathogenicity island termed pgm, that includes the biosynthetic ybtPQXS operon, the irp12ybtUTE loci, and the OM transport locus, psn.516 The entire pgm region may be deleted by recombination, creating a strain with attenuated virulence.517-520 In light of its high affinity for Fe3+ (KA = 4 × 1036 M−1),374 Ybt readily extracts iron from LF and TF.502,521 Ybt biosynthesis requires so-called “high molecular weight proteins” (HMWP), including the iron-regulated proteins (Irp), YbtU, YbtS, YbtT, and YbtE.516 After secretion of Ybt to the extracellular host environment, where it complexes iron, FeYbt encounters the LGP YpePsn (99.4% identical to KpnFyuA), that binds and transports it to the periplasm. The YbtPQ ABC-transporter passes FeYbt to the cytoplasm.522-524 ybtX, in the same operon, is not required for iron uptake; the growth of a YbtX-deficient strain was not impaired in iron-deficient conditions. YbtX was essentials, however, for Zn uptake,524 suggesting that the same system transports two different metals. This conclusion faces some conceptual problems because the chemical attributes of FeYbt and uncomplexed Zn2+ are significantly different.
5.5.2. Iron Acquisition As a Determinant of Plague.
The Ybt system is a virulence factor for Y. pestis in the early progression of bubonic plague. Loss of any gene that compromises the overall iron uptake system (i.e., siderophore biosynthesis or ferric siderophore transport) renders Y. pestis avirulent in mice after subcutaneous inoculation, although virulence of the same strains was not reduced upon intravenous inoculation.502,525 In a pneumonic plague model, the biosynthesis-deficient irp strain showed greater loss of virulence than a transport-deficient psn strain. These results suggest either redundancy in the FeYbt uptake process, or secondary functions of Ybt, distinct from its role in iron acquisition, during the progression of pneumonic plague.502,525 For example, the presence of Ybt may activate transcription of relevant genes or other virulence factors in Y. pestis.502,525 In avian pathogenic E. coli, that also synthesize Ybt and transport FeYbt, irp2 and fyuA are virulence determinants: inactivation of either irp2 or fyuA on its high-pathogenicity island impaired cell adhesion, inhibited transcription of other virulence genes and reduced pathogenicity.526
5.5.3. OM LGP and IM Transporters.
The OM receptor for FeYbt is a 651 residue protein, termed Psn in Y. pestis and FyuA in K. pneumoniae. YpePsn and KpnFyuA are 99.4% identical, with only four amino acid differences between the two proteins. Like all LGP, they contain a 22-stranded β-barrel that wraps around an N-terminal globular domain.527 The two proteins function equivalently in Y. pestis and K. pneumoniae, as evidenced by their recognition of both FeYbt and pesticin in the two organisms.527-529 Furthermore, an engineered hybrid toxin that contains the receptor binding domain of pesticin, and the N-terminus of T4 lysozyme, that degrades peptidoglycan (PG), killed both Y. pestis and pathogenic E. coli. The hybrid toxin crossed the OM and was unaffected by Pim, a protein that inhibits degradation of PG in some Yersinia pathogens. Such hybrid toxins or siderophores may target bacterial pathogens expressing particular receptor proteins that correlate with virulence.527
The cryo-EM structure of a predicted IM ABC transporter of FeYbt, YbtPQ from uropathogenic E. coli, unexpectedly revealed the conformation of a type IV exporter.530 Furthermore, a transmembrane helix within YbtP unwinds upon release of substrate, while the nucleotide binding domain remains tightly packed even in the absence of a bound nucleotide. These findings suggest a different mechanism of ferric siderophore uptake by YbtPQ,530 relative to other ABC-transporters, like the BtuCD complex that acquires B12.531,532 The YbtPQ IM ABC-transporter, that contributes to the virulence of Y. pestis, spans a 5.6 kb region of the Y. pestis genome and contains the yfeABCD operon and the yfeE locus.509,511,521 yfeABCD is regulated by iron availability, mediated by Fur; yfeE is tightly linked to yfeABCD, transcribed in the opposite direction, and expressed independently of Fur.281 Deletion of yfeE inhibited growth in iron-deficient conditions, indicating that YfeE participates in iron transport despite the fact that its expression is not iron-regulated. Although the exact function of the 184 residue Yfe protein is unknown, it contributes to the pathogenesis of bubonic plague:509,533 a ybt, yfe double mutant was avirulent in mice after intravenous inoculation, suggesting that yfe is essential for its virulence during the later stage of the disease. By the subcutaneous route, a ybt+yfe strain had reduced virulence relative to its ybt+yfe+ parent. These data suggested that Yfe participates early in the progression of bubonic plague but may not be absolutely essential.509 On the other hand, the TonB-dependent Ybt system was essential for the virulence in the early stage of bubonic stage but not in the later stage. Hence, Ybt and Yfe system may act together to drive the progression of bubonic plague through different stages.509,533,534 Regarding other components, YfeA resembles a periplasmic substrate binding protein. Its crystal structures535 classified YfeA as a cluster A-1 substrate binding protein, whose other members directly bind metal ions, including zinc, manganese, and iron. YfeA contains two metal binding sites: site 1 shows polyspecificity for Zn2+, Mn2+, and Fe2+ ions and alters its substrate binding specificity in response to environmental conditions. Binding site 1 in YfeA tightly binds metal ions because incubation with EDTA does not remove metals ions from it. Site 2 binds Zn2+ and Mn2+, but not Fe2+, and its biological contributions are undefined.536
6. ADSORPTION OF APO- AND FERRIC SIDEROPHORES BY SCN
The innate immune system produces TF, LF, and FTN, that antagonize bacteria by scavenging, sequestering, and storing iron,537-540 whereas SCN adsorbs both apo- and ferric siderophores in body fluids, eliminating them from circulation and thereby reducing their availability to the bacteria that produced them. Before the discovery of this activity,55 SCN was known as neutrophil gelatinase-associated lipocalin (NGAL), a 25 kDa protein that was found covalently bound to matrix metalloproteinase 9 from human neutrophils.541 The same protein was also discovered by the increase of its mRNA in mouse kidney cells infected by SV40 virus and named lipocalin 2 (LCN2).542 Lipocalins were known as acute phase proteins from myelocytes that were stored in neutrophil granules and overexpressed in epithelial cells during inflammation.543 Experiments suggested that LCN2 was an alternative means of delivering iron to epithelial cells during development, especially in the absence of TF,55 although the form of delivered iron was unknown.544 A so-called “mammalian siderophore” (2,5-DHB) was found bound to LCN2, implicating it in the delivery of iron to cells.545 Consistent with these postulates, animals responded to infection by suppressing production of 2,5-DHB and upregulating LCN2.546 Another member of the lipocalin superfamily, 24p3/NGAL, delivers iron to the cytoplasm of cells by endocytosis.544 Goetz et al.55 subsequently observed that FeEnt adsorbed to NGAL/LCN2, creating a red-colored complex and appropriately renamed the protein as SCN.
6.1. Specificity
Characterization of the SCN-FeEnt binding reaction by tryptophan fluorescence quenching analysis showed high affinity binding of the ferric siderophore to purified SCN (KD = 0.4 nM), which led to the conclusion that SCN competed with the bacterial receptor FepA to capture FeEnt during bacterial infections. SCN also bound the apo-siderophore with about 10-fold lower affinity (KD = 3.5 uM). Structural delineation of the SCN–FeEnt complex found a binding interaction mediated by ionic and cation–π interactions between anionic FeEnt (3−) and the cationic side chains of residues R81, K125, and K134 in the SCN calyx.547 Additional experiments revealed broad recognition of siderophores (Figure 2) by SCN: it adsorbs tricatecholates, carboxylates, hydroxamates,547 the monocatecholate breakdown products of Ent (DHBA, DHBS),548 and carboxymycobactins.549 Epithelial cells secrete another lipocalin, LCN1, in tears and respiratory secretions,550 that binds an array of siderophores, including hydroxamates, but with lesser affinity, so its role in innate immunity may be secondary to the more efficient SCN.547 Transcriptional microarrays that monitor host gene expression demonstrated that colonization of nasal passages by Streptococcus pneumoniae or H. influenzae induced expression of host SCN to higher levels.551 Neither of these pathogens produces their own siderophores, but they utilize xenosiderophores of other microorganisms, so enhanced production of SCN naturally counteracts their infectivity. Similarly, the presence of bacteria in bronchial epithelium induces the influx of myeloid cells, resulting in increased SCN production. Consistent with these inferences, SCN-deficient mice were more susceptible to intraperitoneal or intratracheal infection with E. coli,552 and more bacteria were found in their lungs. The protective effects of SCN were counteracted by administration of high doses of Fc.553 These data all supported a major role for SCN in counteracting bacterial pathogenesis.
6.2. Glycosylated Catecholates: GEnt/FeGEnt
Despite the efficacy of SCN in neutralizing microbial siderophores, bacterial pathogens adapt to escape SCN-mediated defenses. As noted above, they synthesize and/or utilize GEnt/FeGEnt (see also below, section 5.2.2), that SCN does not efficiently bind.124 The micromolar affinities of SCN for GEnt/FeGEnt are a thousand-fold lower than for Ent/FeEnt,124 so the protein does not clear the glucosylated catecholates from blood, serum, lymph, or other fluids.554-556
6.3. Mixed Chelation Siderophores
Other noncatecholate siderophores also evade inhibition by SCN, including Abn, Ybt, and Pvd and Pch from P. aeruginosa. In part because of their SCN-resistance, Abn enhances the hypervirulent phenotype of K. pneumoniae in lung infections,324 and Ybt increases the virulence of Y. pestis in the manifestations of bubonic plague.332 Molecular modeling of the binding interactions between Ent or Pvd to SCN concluded that SCN readily adsorbs the former but not the latter.438 Although Pvd docked to SCN in nine potential positions, none of them occurred in the binding cleft that adsorbs other siderophores. These data rationalize the finding that Pvd promotes colonization of P. aeruginosa in patients with cystic fibrotic lungs.438 SCN also acts in the microbial ecology of the gut. Nonpathogenic E. coli that produce GEnt (e.g., Nissle 1917) outcompete and reduce the numbers of pathogenic S. typhimurium in the intestines of normal mice that were used as a model of acute colitis and chronically persistent infections. In SCN-deficient mice, however, Nissle 1917 did not outcompete S. typhimurium.305 Bacillus anthracis similarly evolved mechanisms to escape SCN-mediated iron deprivation. It produces two siderophores, Crn and petrobactin (Pbn); SCN adsorbs the former but not the latter. However, a combinatorial, genetically engineered version of SCN selectively bound Pbn instead of Cbn, with even higher affinity (KD = 20 pM). The novel binding protein, called “petrocalin” was crystallographically solved, and when administered together with SCN it suppressed growth of Bacillus cereus under iron-limiting conditions. The reprogrammed SCN, petrocalin, may offer new treatment options for serious infections caused by B. anthracis.557 Similar reshaping of SCN improved its binding of siderophores from P. aeruginosa.558
7. SIDEROPHORE–ANTIBIOTIC CONJUGATES (TROJAN HORSE ANTIBIOTICS)
7.1. Uptake of Trojan Horse Antibiotics by E. coli
The potential clinical applications of siderophores were hypothesized and realized soon after their discovery.56 The first example was the ability of apoferrioxamine B (Figure 3, also called desferrioxamine B, desferal),559-561 to combat hemochromatosis and reduce iron overload by excretion of ferrioxamine B in the urine.559 Coulton et al.562 subsequently discovered that even when conjugated to a large polymer, Fc was capable of TonB-dependent iron supply to E. coli, and Rogers et al.563,564 showed that transition metal complexes of Ent had bacteriostatic effects on pathogenic bacteria. Soon thereafter, β-lactams conjugated to catecholates and related moieties were found active against E. coli, other members of Enterobacterales, and P. aeruginosa.565-570 Furthermore, uptake of the catechol-containing cephalosporin E0702 (Figure 4) was TonB-dependent,571 and it produced spontaneously resistant, 100- to 1000-fold less susceptible mutants of E. coli that mapped to the tonB locus.571 Lastly, the antibacterial potency of E0702 was enhanced in iron-deficient conditions but lost in iron-replete and reduced in anaerobic conditions,119 where TonB-independent pathways facilitate uptake of soluble Fe2+, leading to downregulation of LGP expression.572 These studies provided the key evidence that Gram (−) bacteria actively transport catechol-containing antibiotic compounds with iron-regulated LGP. Miller et al.573 designated siderophore–antibiotic conjugates as Trojan Horse antibiotics.
Figure 4.
Structures of siderophore-β-lactam Trojan Horse antibiotics. These monocatecholate and hydroxypyridinone siderophore–antibiotic conjugates target ferric catecholate uptake pathways in Gram (−) cells. The iron chelation moieties are colored blue. Cefiderocol (FDC) contains a monocatecholate siderophore moiety and is the only FDA-approved siderophore-conjugated antibacterial drug.
To further characterize the uptake pathways, Curtis et al.574 studied catechol–cephalosporins and evaluated their antibacterial activity against mutants with disruptions in one or more TonB-related genes. Single mutants in tonB, exbB, exbD, and cir had significantly elevated minimal inhibitory concentrations (MIC) for catechol-conjugated cephalosporins, but only mutant strains lacking both fiu and cir had elevated MIC values (as much as 1000-fold), which was comparable to tonB mutants.574 The double fiu-cir mutant also exhibited dramatic MIC shifts for the catechol–cephalosporin E0702 and the hydroxypyridinone–monobactam, pirazmonam (Figure 4).204 Uptake of the 55Fe-chelate of a catechol–cephalosporin confirmed that both TonB and Fiu/Cir participate in transport of the ferric catechol–cephalosporin complex, consistent with the MIC shifts observed for the mutants.574 Wild-type cells transported both the unliganded and ferric E0702 at equivalent rates, but uptake of both forms was lost in the fiu-cir double mutant.204
Trojan Horse antibiotics involve, mimic, or capitalize on the native iron acquisition systems and siderophores of the target bacteria. Consequently, the chemistry and biology of the iron acquisition pathways are essential to the design of antibiotic compounds against them. A priori, it is difficult to evaluate the contributions of the various potential catecholate iron complexes in the microenvironment of a bacterium that is producing and excreting Ent/GEnt. The efficacy of the degradation products (such as mono- and dicatecholates) in complexing and supplying iron may depend on whether other, potentially more avid siderophores are present. However, members of Enterobacterales and other Gram (−) Families produce TonB-dependent uptake systems for ferric monocatecholates. Nikaido et al.204 suggested that the natural ligands of Fiu and Cir are the monocatechol hydrolytic products of Ent: DHBS and/or DHBA. The latter is most relevant as a biosynthetic byproduct, rather than a degradation byproduct. Besides the prokaryotic monocatechols, the eukaryotic catecholamine stress hormones epinephrine, norepinephrine, and dopamine are relevant to this phenomenon.575-577 They are proposed to release iron from TF/LF, making it available to support bacterial growth, mediated by the scavenging actions of bacterial siderophores or the catecholamine iron complexes themselves.577 Even in the absence of TF/LF, dopamine promoted S. enterica growth and increased iron uptake from the medium.575 Bordetella bronchiseptica utilizes ferric-norepinephrine to support growth of a siderophore-deficient mutant due to the presence of three TonB-dependent catecholamine transporters.576 These receptors can also recognize the Ent component DHBA.576 It is noteworthy that in the presence of equivalent concentrations of tricatecholate siderophores like Ent, GEnt or Vbn, monocatecholates are not thought to significantly contribute to extracellular iron scavenging. The intact tricatecholates will monopolize Fe3+ because of their much higher affinity. Nevertheless, as discussed above (section 4.4), the affinity of a chelate for iron is not the only consideration that determines its overall importance to iron utilization. Siderophores complex Fe3+ over a broad range of affinities;59,60 lower affinity siderophores like Pch439 and Abn59 still effectively bind iron, and their cognate LGP internalize the iron complexes via TonB-dependent reactions. Gram (−) bacteria also efficiently transport monocatecholate ferric complexes supplied at appropriate external concentrations.578-580 Analogously, LGP (e.g., EcoFiu and EcoCir) actively transport catecholate siderophore–antibiotics (such as those in Figure 4) by virtue of their chemical similarity to native monocatecholate compounds. In contrast to most natural siderophores, and like the degradation products of Ent/GEnt, synthetic siderophore β-lactam conjugates (SβLC) have a single bidentate Fe3+ chelation ligand (Figure 4) and show orders of magnitude lower affinity for Fe3+581 than Ent/GEnt58 or other relevant siderophores (e.g., Pvd582). Iron-regulated LGP are expressed at higher levels in iron-deficient conditions,119,204,574,583 so the antibacterial activity of Trojan Horse conjugates increases in low-iron media119,204,574,583 despite the fact that Fe3+ levels are below the KD of FeSβLC binding to their OM receptors and further decrease as bacteria secrete high-affinity siderophores.583 This situation suggests the possibility that the relevant LGP also recognize and transport apo-SβLC. Some ferric siderophore receptors bind the corresponding aposiderophore (e.g., Ent/FepA; Pvd/FpvA584), but this association has minimal biological significance because LGP optimally recognize the metal center of ferric siderophores, often stereospecifically.585-589 Ferric siderophores always adsorb to their receptors with higher affinity than the corresponding aposiderophores. Consequently, in a binding equilibrium involving both forms, the ferric siderophore predominates. It is plausible that both apo- and ferric complexes of SβLC may be recognized and transported by LGP, but uptake of an aposiderophore is intuitively counterproductive and has not been demonstrated. Besides the apo- and ferric complexes of SβLC,204 SβLC may form complexes with alternative divalent cations (Zn2+, Ca2+, or Mg2+) that are present in culture media or host tissues, and may also form mixed, “piggyback” complexes with ferric siderophores (e.g., pAcn or Pvd) to gain entry through the cell envelope.590
7.2. Spectrum of Trojan Horse Antibiotic Activity
Besides the susceptibility of E. coli to siderophore–antibiotics, SβLC are potent and effective against other Gram (−) bacteria, including P. aeruginosa and A. baumannii.119,591-593 The repertoire of iron-regulated LGP varies among Gram (−) pathogens, but orthologues of E. coli Fiu, FepA, and Cir exist across the Family Enterobacterales432 and other Families as well, although these relationships are mostly bioinformatically defined and not yet experimentally validated. In P. aeruginosa432,592,594-597 and A. baumannii,432 for example, the OM proteins PiuA/D and PirA were implicated in SβLC uptake. Disruption of one or both of the genes encoding these proteins, in both organisms, reduced susceptibility to SβLC.432,592,594-596 PiuA is most homologous to Fiu; PirA is most homogous to FepA.432 Expression of both proteins is regulated by Fur598,599 in response to extracellular iron availability and potentially by iron uptake through other pathways. The P. aeruginosa genome encodes 34 different LGP, whose specificities and natural ligands are mostly undefined. Iron deprivation leads to >2-fold upregulation for nearly half of the P. aeruginosa LGP; expression of FptA, the receptor for FePch,594 increases as much as 120-fold. The expression levels of these proteins during infection is not known, but in a P. aeruginosa piuA mutant as many as 7 LGP significantly affected susceptibility to SβLC,594 suggesting that multiple OM proteins may actively transport SβLC via TonB-dependent pathways. In each target organism, SβLC potency is a function of the relevant LGP expression level, its affinity for the siderophore–antibiotic, potential competition with other natural iron-binding ligands, and overall uptake efficiency.
7.3. Cefiderocol, The First FDA-Approved Trojan Horse Antibiotic
The first and only FDA-approved Trojan Horse antibiotic is the catecholate–cephalosporin conjugate cefiderocol (Fetroja; FDC, Figure 4), that was authorized for treatment of complicated urinary tract infections, including pyelonephritis.592,600-602 FDC is potent against critical Gram (−) pathogens including carbapenem-resistant P. aeruginosa and A. baumannii, due to the combination of uptake via LGP, good stability against all classes of carbapenemases, and its covalent inhibition of target PBPs.581,592,594,602 The branded name of FDC, Fetroja, reflects its transport through iron (Fe) siderophore uptake like a Trojan Horse (troja). Because of the enhanced uptake of FDC by LGP and in vivo pharmacokinetic/pharmacodynamics (PK/PD) correlations to MIC, the standard medium for FDC susceptibility testing is iron-depleted, cation-adjusted Mueller–Hinton broth (final [Fe] < 0.10 mg/L, [Ca] = 22.5 mg/L, [Mg] = 11.25 mg/L, [Zn] = 0.65 mg/L).583 FDC utilizes the typical pathogenspecific LGP identified for other SβLC (e.g., Fiu-Cir or PiuA-PirA),592,594,596 but an alternative uptake pathway for FDC uptake through FptA may exist in a pyochelin-dependent manner.590
Earlier SβLC, such as MB-1 (Figure 4) and SMC-3176, failed to show efficacy in animal infection models and suffered from “adaptive resistance” in vitro.596,603,604 FDC avoids the adaptive resistance liability compared to MC-1, MB-1, and SMC-3176.604,605 In the case of P. aeruginosa, increased levels of Pvd were implicated in adaptive resistance to MB-1.603,606 Higher Pvd levels would be expected to result in more efficient Fe3+ uptake, higher cytosolic Fe levels, and downregulation of the preferred SβLC LGP. Although FDC seems avoids the adaptive resistance liability of other SβLC, in vivo efficacy studies with humanized exposures of FDC identified clinical isolates of both P. aeruginosa and A. baumannii against which FDC underperformed or did not demonstrate expected efficacy.605 A significant 10–20% of these isolates did not achieve bacterial stasis or ≥1-log10-CFU reduction despite having MIC values that predicted susceptibility. The results with many isolates also showed high variability due to inconsistent responses to FDC among the replicates.605 Whether these issues relate to adaptive resistance is not known. While FDC has potent antibacterial activity against Gram (−) pathogens, as a cephalosporin, it is still hydrolyzed by clinically relevant β-lactamases.602 It is worth noting that SβLC that show adaptive resistance liability (e.g., MC-1, MB-1, and SMC-3176) all contain hydroxypyridinone chelation groups, whereas FDC is based on chelation by a catecholate (Figure 4).596 Another catechol-containing β-lactamase inhibitor LN-1-255 (a substituted penicillin sulfone) was reported, though nothing was known about whether it promotes adaptive resistance or is transported by an LGP.607 The only other SβLC to enter clinical trials, BAL30072, was also a hydroxypyridinone siderophore (Figure 4). It did not show adaptive resistance,432,594,595 but its development was suspended in phase 1.608
7.4. Non-β-Lactam Siderophore Conjugates
Decades before FDA approval of FDC, a diverse group of non-β-lactam siderophore–antibiotic conjugates were described and studied. The research began with natural sideromycins, albomycins,609-613 and salmycins;614 the former showed broad spectrum antibacterial activity against Gram (+) and Gram (−) bacteria in a murine infection model,615 but the latter were less effective, likely as a result of its chemical lability. Synthetic siderophore–antibiotic conjugates were subsequently developed that required some form of cleavage for full activity.616-619 While the targets of β-lactam antibiotics, penicillin-binding proteins, reside in the periplasm, many other systems or pathways that are susceptible to antibiotic action reside in the cytoplasm. So although LGP may deliver siderophore–antibiotics into the periplasm, the IM poses a second permeability barrier, especially to charged/polar compounds.620 However, appropriate IM ABC transporters may recognize and internalize the ferric siderophore moiety of Trojan Horse compounds, with concomitant uptake of the attached antibiotic. Nolan and co-workers created Ent-antibiotic conjugates, beginning with Ent-β-lactam conjugates that are active in the periplasm.618 The uptake of the conjugate depended on FepA and provided 1000-fold lower MIC than the β-lactam alone.618 They later conjugated Ent to ciprofloxacin (CIP), whose targets (DNA gyrase and topoisomerase IV) reside in the cytoplasm.617 In the latter case, the Ent-CIP conjugate crossed the OM via FepA-TonB/ExbBD and then crossed the IM by FepCDG.617 Ent-CIP was inactive unless hydrolyzed by the salmochelin esterase IroD in the cytoplasm to release the DHBS-CIP monomer.617 An alternative strategy for cytoplasmic release of CIP employs a disulfide linker in the Ent-CIP conjugate that may be cleaved by the cytoplasmic low-molecular-weight thiols like glutathione.617
Miller and co-workers used other siderophores and cleavage strategies for cytoplasmic antibiotic delivery and release.616,619 They first conjugated desferrioxamine B to CIP with potential esterase- and phosphatase-susceptible linkers.621 The esterase-triggered conjugate had weaker activity than that of the parent CIP, while the phosphatase-triggered conjugate was inactive.619 These results revealed the extent of optimization needed when considering all the uptake and implementation variables: OM and IM transport, enzymatic cleavage, and target engagement. The Gram (−) OM creates a potentially insurmountable permeability barrier27,28 to many antibiotics that are active against Gram (+) cells. For example, oxazolidinones target the ribosome but are limited to Gram-positive pathogens, even though their ribosomal target is conserved in Gram (−) bacteria like E. coli. Consequently, Miller and co-workers designed a clever siderophore–cephalosporin–oxazolidinone conjugate,616 whose cleavage depended on periplasmic hydrolysis by cephalosporinases, releasing the free oxazolidinone.616 This conjugate, that contains a bis-catechol siderophore, boosted cephalosporin activity against periplasmic PBP. Although expression of the cephalosporinase impacted the potency of the cephalosporin core, the consequent release of the oxazolidinone provided a significant boost in antibacterial potency.622 Another study conjugated bis-catechol or bis-catechol-monohydroxamate to teicoplanin, which normally targets the PG d-Ala-d-Ala termini of only Gram-positive bacteria because it cannot penetrate the OM of Gram (−) bacteria.623 Interestingly, the siderophore–teichoplanin conjugates demonstrated potent activity in A. baumannii but not E. coli or P. aeruginosa, despite the fact that their PG targets are identical.623 These data suggest differences in the LGP-mediated uptake pathways among the different bacteria. Compared to SβLC, the challenges to synthesize, characterize, and develop synthetic sideromycins are daunting. Nevertheless, antimicrobial resistance is inevitable for every new drug, so the development of all types of Trojan Horse antibacterials remains a desirable long-term goal.
8. MECHANISTIC INSIGHT FROM BIOINFORMATIC ANALYSES OF TONB-DEPENDENT SYSTEMS
As a consequence of their high rate of antibiotic resistance, the Gram (−) CRE/ESKAPE pathogens33,34 cause a large fraction of nosocomial infections, and clinical options against them are limited.31,32,36 Both strategic design of antibiotics and large scale screening of chemical libraries for compounds that may block iron acquisition in these organisms hinge on the understanding of LGP transport biochemistry. The bacteria under discussion in this analysis represent four phylogenetic Families: Enterobacterales, Moraxellaceae, Pseudomonadaceae, and Yersiniaceae. Together, they inhabit different natural environments, but each one has adapted to infect humans and animals, in part from OM permeability properties that differ from those of E. coli, the prototype of Enterobacterales. For example, clinical isolates of K. pneumoniae, in the same Family, have much lower overall OM permeability from the absence of certain porins624-627 and higher serum resistance from enhanced capsule formation.628,629 The latter trait was maximized in the highly virulent, hypermucoviscous form that also manifests more efficient iron acquisition.630,631 Both P. aeruginosa632-635 and A. baumannii,636-638 in the Families Pseudomonadaceae and Moraxellaceae, respectively, have similarly low OM permeability, and their iron-regulated formation of biofilms639-641 constitutes an additional virulence determinant. Lastly, Ybt, the primary siderophore of Y. pestis, was co-opted by members of the other families in ways that augment their virulence.326,358,376 Such adaptations illustrate the connections between cell envelope architecture, the mechanisms of TonB-dependent iron acquisition, and pathogenic virulence.
8.1. Sequence Diversity in TonB
TonB action encompasses a number of biochemical activities that are potential targets for chemical inhibition: binding to the TonB-box of LGP,239,240 physical interactions with ExbBD,232,233 and associations with PG238 that may involve monomer–dimer conversions91,241 and other currently un-delineated aspects of the transport process. Besides their microbiological and ecological diversity, each ESKAPE bacterium acquires multiple ferric siderophores, some of which correlate with their invasiveness, tissue tropism, or overall virulence (K. pneumoniae, FeAbn; P. aeruginosa, FePvd; A. baumannii, FeAcn; Y. pestis, FeYbt). Consequently, their TonB proteins must physically interact with multiple iron-transporting LGP. The primary structures of these TonB orthologues are unusually variable. The extent of EcoTonB (NCBI: NP_415768.1) sequence identity to KpnTonB (CAA48498.1), PaeTonB (Q51368.2), and AbaTonB (AHB92731.1) is 75%, 37%, and 25%, respectively; a CLUSTALW2 alignment of the four TonB proteins showed only 11% identity (26 of 239 residues). This divergence among TonB proteins indicates that the component proteins of the individual OM iron transport systems are uniquely adapted to one another in each species and explains why the LGP from one species do not necessarily function in other species. Structural data is only available for the C-terminal domain of EcoTonB, but both bioinformatic predictions and biochemical data suggest that the 239 aa EcoTonB encompasses a hydrophobic N-terminal helix in the IM,238,642 a central rigid region that spans the periplasm,236,643,644 and a globular, periplasmic C-terminal domain (CTD) that associates with the LGP in the OM.239-241,645,646 Most of the conserved residues among the four noted TonB proteins reside in the central, ~75-residue rigid region (9 Pro, 4 Lys and 2 Glu), whereas the ~75-residue CTD, that recruits TonB-box peptides of LGP into a four-stranded β-sheet, contains only six conserved identical residues (5 Val, 1 Phe). This structural variability suggests that it is unlikely to find a generic, broad-spectrum inhibitor of TonB activity that functions across distantly related bacterial pathogens. Yet, an HTS screen against EcoTonB discovered numerous compounds that also inhibited the activity of AbaTonB,272 so despite a priori skepticism, a broad-spectrum anti-TonB antibiotic is conceivable.
The conserved identical residues in the CTD of ESKAPE TonB proteins localize to internal regions of both the dimeric241 and monomeric239,240,647 forms. The hydrophobic nature of the conserved residues and their internal localization infer that they stabilize the domain’s tertiary structure. Since the completion of the crystal structures of the TonB CTD in different forms, including its association with the TonB-box of LGP,241,645-647 few experiments reflected on the functional relationships between its monomeric and dimeric forms. Yet, the crystallographic demonstration of interactions between the monomeric TonB CTD and LGP N-termini validated the relevance of the monomer, and the interactions of the dimer with PG in the bacterial periplasm had implications on the potential mechanism of TonB action.91,238
8.2. Commonality of FeEnt Transport by Prototypic FepA Proteins
Each of the four CRE/ESKAPE organisms secretes different siderophores and utilizes different ferric xenosiderophores by the actions of unique LGP, but they all also efficiently transport FeEnt120,285 with orthologues of EcoFepA. The E. coli protein is an accessible prototype of both active OM transport and biochemical interactions with TonB. The comparative bioinformatic analysis of EcoFepA orthologues (see below) reveals unexpected aspects about the capture of ferric catecholates by Gram (−) bacterial pathogens.
8.2.1. EcoFepA.
Laboratory E. coli K-12 strains produce Ent and transport FeEnt through EcoFepA, but pathogenic E. coli (UPEC, EHEC; Table 1) also glucosylate the siderophore and transport FeGEnt through orthologues of IroN. The primary structures of EcoFepA and EcoIroN are only 52% identical, intimating a potentially significant divergence of specificity and function.
8.2.2. KpnFepA.
K. pneumoniae encodes four apparent orthologues of EcoFepA in its genome: three in the chromosome [loci 1658 (KpnFepA1), 2380 (KpnFepA2), 4984 (KpnFepA4)] and one (locus 0027: KpnIroN) from an endogenous plasmid. The resulting four FeEnt receptors are 82%, 53%, 73%, and 53% identical to EcoFepA, respectively.
8.2.3. AbaFepA.
A. baumannii produces a single LGP (AbaFepA) that catalyzes FeEnt uptake, and has 46% sequence identity with EcoFepA. Despite being the most distant orthologue to EcoFepA in the CRE/ESKAPE group, AbaFepA still retains sufficient identity (i.e., >30%648,649) to predict a conserved tertiary structure.648
8.2.4. PaeFepA.
Like K. pneumoniae, P. aeruginosa strain PAO1 contains the structural genes for 3 FepA orthologues, as well as IroN, all in its chromosome. PaeFepA1, PaeFepA2, and PaePfeA share 71%, 81%, and 61% identity with EcoFepA; the sequence of PaeIroN is 60% identical to that of EcoIroN.
When grown to low-iron stress in iron-deficient MOPS media,650 the four CRE/ESKAPE pathogens comparably transport FeEnt like E. coli K-12.285 Despite their sequence divergence, the ESKAPE FepA orthologues all have sufficient identity to predict a nearly identical overall structural fold as EcoFepA (1FEP196), but like TonB, their primary structures have evolved in K. pneumoniae, A. baumannii, and P. aeruginosa such that they are not generally interchangeable among the four species. Closely related KpnFepA transports FeEnt when expressed in E. coli, but more distant PaeFepA and AbaFepA do not partner with EcoTonB to catalyze FeEnt uptake in E. coli (Nairn, Newton, Kumar and Chakravorty, unpublished data). This situation reinforces the notion that TonB and LGP concomitantly evolved in the different species. Therefore, compounds that inhibit TonB-dependent Fe3+ uptake in E. coli may not similarly block iron uptake in K. pneumoniae, and even less so in P. aeruginosa or A. baumannii. Hence, each CRE/ESKAPE pathogen will likely exhibit different susceptibilities to compounds in chemical libraries, and potential inhibitors of TonB-dependent processes may require species-specific evaluation and/or optimization to attain appropriate efficacy.
8.3. Other Ferric Catecholate Transporters
At least six LGP participate in uptake of ferric siderophores in E. coli K-12: Fiu, FecA, FepA, FhuE, FhuA, and Cir. Expanding the scope to pathogenic E. coli adds IutA, IroN, ChuA, and FyuA;651 consideration of vitamin B12 (cyanocobalamin) includes BtuB.652 The fact that among 11 E. coli LGP, five (FepA, IroN, Fiu, FecA, Cir) function in the uptake of ferric catecholates underscores the importance of this class of siderophore to bacterial iron acquisition. With the exception of IroN, the structures of these ferric catecholate transporters were independently crystallographically determined. However, the scope of their recognition specificities and binding preferences are only now becoming fully known.285 Orthologues of EcoFepA are broadly distributed among members of Enterobacterales and other Families, to selectively recognize and transport FeEnt, but the chemical lability of the catecholate trilactone siderophore makes its degradation products, that include mono-catecholates (Figure 3), also relevant to iron acquisition by the spectrum of Gram (−) bacteria. Within a few days of forming and purifying FeEnt,66 its visible absorbance spectrum begins to change, even if the ferric siderophore is stored on ice or frozen. Chromatography over Sephadex LH20 reveals a rapidly mobile, purple peak of oxidized FeEnt (FeEnt*) that separates from the crimsoncolored authentic FeEnt.214 These changes primarily derive from oxidation of the catechol groups at the metal center to quinones. Furthermore, the lactone backbone is susceptible to cleavage by acid, base, and esterases. Consequently, the monomeric iron–catecholate complexes, formed by the degradation products of FeEnt, inhabit the environments that bacteria experience in the host. Cir574 and Fiu204 participate in the transport of monocatecholate iron complexes [e.g., Fe(DHBS)3] and catechol-containing antibiotics.653,654 GEnt is secreted by uropathogenic E. coli, S. enterica, and K. pneumoniae, whereas Crn267,655,656 is a similar but distinct catecholate from Gram (+) bacteria. Collectively, the catecholate siderophores create a myriad of possible iron complexes that bacterial pathogens may utilize to different extents and priorities.
The approximately one dozen iron-transporting LGP in E. coli expand into a plethora of LGP in some other organisms. The genomes of C. crescentus and P. aeruginosa encode 66 and 34 LGP, respectively. Besides a similar cadre of iron-regulated LGP,194 one of the C. crescentus receptors performs TonB-dependent transport of maltodextrins,658,659 and others are predicted to transport a variety of substrates besides metal complexes. The primary, secondary, and tertiary structures of proteins in the LGP superfamily create a consistent structure/function paradigm. The external hydrophobic surfaces of their amphiphilic transmembrane β-barrels interact with OM lipids, while their internal hydrophilic surfaces circumscribe an aqueous channel and envelop the N-terminal, 150-residue globular domain that interacts with TonB/ExbBD to regulate ligand movement through the pore. Large surface loops that selectively bind ligands and short reverse β-turns between the β-strands of the barrel complete LGP architecture. Despite this conserved format, variability of their surface loops results in a relatively low overall sequence identity in the superfamily. The E. coli LGP only average about 20% overall identity between any two individual proteins despite much higher of levels of identity in the strands of their β-barrels. The highest degree of conservation of primary structure among E. coli LGP is the 52% identity that occurs between FepA and IroN, the receptors for FeEnt and FeGEnt. The low overall sequence identity among EcoLGP is somewhat unexpected because they all physically interact with EcoTonB and likely function by the same general mechanism. However, sequence divergence in the surface loops confers unique ligand recognition specificity to each individual LGP. Furthermore, this ligand selectivity occurs in the context of antigenic variation in the same external loops to evade the vertebrate immune response. The biochemical selectivity created by the external loops impacts the actions of Trojan Horse antibiotics. For instance, both mono- or di- glucosylated Ent (GEnt) derivatized with ampicillin or amoxicillin showed improved antibacterial activity and evaded scavenging by SCN from the host. Both siderophore–antibiotics had a narrow application range that selectively killed pathogenic E. coli (expressing IroN) but not nonpathogenic E. coli (lacking IroN).660 These findings suggest that the recognition of particular iron complexes by LGP may be exploited with Trojan Horse compounds to only target pathogens that produce those siderophores and/or utilize their iron complexes. These are significant advantages over wide-spectrum antibiotics and potentially superior for clinical applications.
8.4. Biphasic Ligand Adsorption
The initial stage of ligand adsorption is a mechanistically well-defined aspect of LGP-mediated iron transport. Payne et al.661 demonstrated biphasic binding kinetics for the interactions of both FeEnt and ColB with EcoFepA. Subsequent studies refined this conclusion for EcoFepA219 and EcoFecA,197 in the former case with chemical cross-linking studies and in the latter case by crystallographic depictions that showed conspicuous motion of surface loop 7 (L7) in the transition from the ligand-free to the FeCit-bound form of the receptor. Crystallographic characterizations and simulations of PaePfeA found potential FeEnt binding sites in both its the surface loops and within its external vestibule, supporting the two-stage nature of FeEnt binding.662 Furthermore, fluoresceination of individual Cys substitutions in EcoFepA220 allowed descriptions of the loop motion that LGP undergo during ligand adsorption: stopped-flow measurements of fluorescence quenching showed that they move at different rates, individually and independently, as they capture FeEnt from the environment. The two-stage kinetic process thus resolves into rapid initial interactions of the ligand with surface loop residues that engender a slower second stage of conformational motion as loops coalesce around the metal complex by induced fit, creating a high-affinity form at equilibrium. The process of ligand acquisition by the surface loops of FepA is analogous to a hand catching a ball from the air: the ball collides with the open hand, and then the fingers individually move to close around it.220 In that sense, in iron-deficient conditions the Gram (−) bacterial cell surface becomes infused with thousands of molecular hands, each adapted to catch a particular type of iron complex for subsequent translocation into the cell.
As noted for FepA, biphasic binding kinetics correlate with a structural transition from a form with open, extended loops to a form with contracted loops that surround the bound ligand.219 Crystal structures of EcoFiu reiterated this conclusion.663 Like other TonB-dependent transporters, Fiu contains a 22-stranded β-barrel, covered by extracellular loops. Crystallography revealed two distinct structural states of Fiu: a conformation with disordered extracellular loops that form an open cavity to the extracellular environment, and a conformation with ordered, closed loops.663 By opening and closing in this manner during ligand transport the dynamic actions of LGP maintain the natural permeability barrier of the OM. They allow ligand recognition and uptake while still excluding deleterious compounds like bile salts and antibiotics that may compromise the integrity of the IM bilayer, PG biosynthesis, or other processes in the periplasm.663 As Dick van der Helm described it, the FepA channel functions like an air-lock: the external loops close before the internal domain opens.
8.5. Evolutionary Covariance and Conserved Sites of Mechanistic Importance
Despite 50 years of research on TonB-dependent membrane transport, the underlying molecular mechanism of metal internalization remains incompletely defined. Nevertheless, the mountain of available genomic information, mined by bioinformatic algorithms, yields insight into this conundrum. We aligned and analyzed 79 LGP sequences (Table 1) by CLUSTALΩ664 (Figure S1) and BIS2,665 which identified sequence conservation that reflects on the biochemical transport mechanisms of ferric siderophore and Hn receptors. The analyses also described the phylogenetic relationships of the proteins (Supporting Information, Figure S2). The collection of 79 proteins, that transport at least 16 different metal complexes, exposed an unexpected characteristic of LGP N-domains: the most significantly conserved and simultaneously covariant amino acids in the LGP N-termini are glycines (Figure 5). Among seven highly conserved residues (>90% identity) in the N-domains of 79 LGPs of bacterial pathogens, five were Gly. The other two conserved amino acids have basic side chains (R75, R126 in EcoFepA), that map adjacent to and pair with conserved acidic amino acids on the interior of the C-terminal β-barrel (E511, E567 in EcoFepA). In the full length primary structures of the 79 proteins, a total of 16 residues exhibited extensive identity, or evolutionary covariance, or both. Together they defined a charge cluster in LGP infrastructure,90 situated among a group of conserved glycines (Figure 5). These data suggest two attributes of LGP ligand transport: an electrostatic channel-gating mechanism and conformational flexibility that promotes uptake functionality. The association of R75-E511 and R126-E567 in an ionic cluster on the channel wall, directly above the TonB-box and across the pore from the N-domain–β-barrel junction, strongly suggests a protonation-based trigger to the ligand internalization process. Protonation of E511 and E567 is a key to unlocking this electrostatically closed channel because it will free the N-domain to movement. As in the case of LacY, the protein environment surrounding the R75-E511 and R126-E567 pairings may significantly change the pKa values of the acidic side chains.666,667 This inference concurs with the PMF-dependence of LGP-mediated transport,62,668 but in an unexpected way that raises the question, does this biochemistry precede, coincide with, or follow the interaction of the TonB-box of LGP with the C-terminus of TonB? Second, five conserved Gly surrounding the charge cluster in the N-domain minimizes φ/ψ steric hindrance to conformational motion, which reinforces conclusions from site-directed disulfide bonds within EcoFepA: disulfide links in the N-domain that precluded conformational motion also prevented FeEnt transport.669 Thus, both bioinformatic and experimental results point to structural rearrangements within the N-terminal globule, while resident in the transmembrane channel, allowing passage of FeEnt into the periplasm.
Figure 5.
Conserved mechanistic charge cluster in the LGP interior. After aligning the primary structures of 79 LGP from commensal and pathogenic Gram (−) bacteria (Table 1) by CLUSTALΩ664 and analyzing the aligned files for evolutionary covariance by BIS2,665 we mapped 16 conserved (>90%) or coevolved amino acids to the tertiary structure of EcoFepA (PDB 1FEP) using CHIMERA (UCSF716). (A) Side view of EcoFepA: the N-domain (residues 1–150) is depicted in ribbon format and colored red; the C-domain β-barrel (residues 151–7240 is depicted in ribbon format and colored green. Among the 16 residues of interest (shown in space-filling fomat), one (N677, colored gray) was conserved in all the LGP. (B) −90° X-axis rotation of the view in (A) creates a perspective inside the β-barrel, from the periplasm. Four polar charged side chains (R75, R126, E511, E567; colored sky blue and red, respectively; heteroatoms N and O colored blue and red, respectively) create an electrostatic lock that, in the absence of protonation, holds the N-domain to the β-barrel directly above the TonB-box region (in ribbon format, colored cyan). A group of eight glycines (colored gray), located in either the interior of the N-domain (G76, G88, G127, G134, G140) or in the strands of the β-barrel (G429, G513, G565), surround the charge cluster, potentially maximizing the flexibility of the protein structure in this region.
9. INTERVENTION AGAINST GRAM (−) BACTERIAL PATHOGENS
The indispensability of iron in aerobic metabolism, combined with the uniqueness of prokaryotic cell envelope iron acquisition systems, makes TonB-dependent transport activity a potentially susceptible target in the Gram (−) cell envelope. It is conceivable to either block uptake of iron complexes by immunochemical inhibition, or to chemically target the mechanisms of LGP biochemistry.
9.1. Immunological Approaches
Certain anti-LGP antibodies prevent the recognition and binding of ferric siderophores by adsorbing to loops that participate in the recognition process,670-674 and these immunogenic epitopes are the basis of vaccines.675-677 However, rough E. coli K-12 strains were used for many of the immunochemical analyses of these phenomena, which raises questions about their application to unattenuated, wild bacterial pathogens that are usually encapsulated and produce complete lipopolysaccharide (LPS) O-antigens. Full-length LPS shields Omp surface epitopes from antibody binding,678,679 capsule accentuates this effect, and the cell envelopes of CRE/ESKAPE pathogens encompass both of these traits. Consequently, efficacious human vaccines against CRE/ESKAPE organisms from Gram (−) bacterial LGP are an uncertain prospect, and existing data substantiate these concerns.680 Therapeutic monoclonal antibodies, that react with single or multiple epitopes of specific iron transporters, face similar obstacles to recognition of surface proteins in pathogenic bacteria and are costly to produce for clinical use. It is fair to say that the conceptual promise of immunological intervention against TonB-dependent iron uptake systems faces practical problems that will be difficult to circumvent or supersede.
9.2. Biochemical Targets of Antibiotic Action
The overall biochemistry of Gram (−) bacterial iron acquisition offers both specific and general molecular targets that are vulnerable to chemical inhibition. The former, specific category includes cell surface ligand binding reactions, intrinsic LGP mechanisms, and the activities of periplasmic binding proteins, ABC transporters, and ferric reductases that function during iron uptake. The latter, general category focuses on TonB: physical interactions between LGP and TonB, intrinsic TonB/ExbBD mechanisms, and interactions between TonB and ExbBD. The latter category is more desirable, but the notion of compounds that broadly inhibit TonB/ExbBD in a group of diverse pathogens is undercut by the known sequence diversity in the target proteins of these bacteria. The diversity originates from coevolution of LGP and TonB together in the unique cell envelopes of the ESKAPE organisms, as they propagate in different wild and host environments (see following). From the current understanding of TonB/ExbBD physiology,91,237 inhibitors may block iron uptake by adsorbing to TonB’s C-terminal domain (CTD), that interacts with PG238 and with ligand-bound iron transporters,239,240,681 or to the regions of TonB that interact with ExbBD.232,233 The sequence diversity that was noted in the TonB proteins of the ESKAPE organisms (see section 8.1, above) also occurs in their FepA orthologues: the extent of EcoFepA (NP_415116.1) sequence identity to KpnFepA (EYB77073.1), PaeFepA (NP_251378.1), and AbaFepA (KFG14278.1) is 81%, 61%, and 46%, respectively. Hence, in each individual bacterium, the LGP is adapted to the properties and components of its own cell envelope, including the nuances of the TonB/ExbBD complex. Hypothetically, chemicals that block TonB-dependent iron uptake in E. coli may not inhibit, or may have less efficacy against, K. pneumoniae, P. aeruginosa, and A. baumannii. Thus a search for broad-spectrum inhibitors of TonB action that are efficacious against all Gram (−) cells may not succeed. Specific inhibitors of TonB systems in each individual CRE/ESKAPE organism are a potentially realistic goal for HTS of chemical libraries. Because the activities of TonB/ExbBD occur in the periplasm or IM, potential antibiotics against TonB-dependent activity must also overcome the size and hydrophobicity barriers of the Gram (−) bacterial OM.28 Efficacy depends in part on the cell envelope permeability properties of the individual target organisms, so it is most apt to directly screen chemical libraries against the pathogen of interest and subsequently assess whether specific hits that inhibit TonB-dependent processes may have generic activity. Alternatively, one may seek compounds that target and block specific iron transporters instead of TonB/ExbBD, for example, specific inhibitors of FeEnt uptake in E. coli, K. pneumoniae, or A. baumannii, or of FePvd uptake in P. aeruginosa. However, this approach is potentially compromised by the numerous, redundant iron uptake systems that exist in bacterial pathogens.
9.3. Fluorescent High-Throughput Screening (FLHTS)
A search for therapeutic chemicals requires an assay that identifies them. Kaback and colleagues extensively explored Cys scanning mutagenesis of the lactose permease of the E. coli cell envelope682-684 and extended the approach to alkylation of single Cys residues685-688 with fluorescent686,689-692 or paramagnetic693-696 probes. These biophysical modifications allowed determinations of internal distances, conformational change, sugar binding, and other parameters. Their studies with the lactose permease required preliminary mutagenesis to eliminate seven native Cys residues, followed by site-directed introduction of Cys at positions of interest into the so-called “Cys-less” LacY. However, Cys is often conveniently absent from Gram (−) bacterial OM proteins or involved in stable disulfide bonds when it is present. EcoFepA, for example, contains a single pair of Cys residues in L7 that are unreactive unless reduced.697 Many porins, including most LGP, are devoid of Cys. Cao et al.698 took advantage of this fact and employed site-directed fluoresceination to create a spectro-scopic assay of FeEnt uptake by EcoFepA in living bacterial cells.698 This methodology observes TonB-dependent FeEnt uptake by monitoring fluorescence quenching as bacterial transport the ferric siderophore. The in vivo approach surmounts our current inability to reconstitute TonB-dependent systems in vitro and confers the advantages of living cell-based assays: convenience, predictability, miniaturization, automation, and multiplexing.699
9.3.1. Specific Fluoresceination of Heterologous Proteins.
Once modified with a fluorophore in its surface loops, an LGP becomes a sensitive biophysical sensor for the detection, quantification, and flux of a particular metal complex in the environment. Fluoresceination usually does not impair either the specificity or affinity of an LGP for its ligand.220,285 Hence, it is feasible to design and create fluorescent LGP sensors in the individual pathogens of interest. Each CRE/ESKAPE bacterium poses challenges from their particular cell envelope architecture and biochemistry, which complicates the interpretation of HTS data and makes it advantageous to directly screen chemical libraries against the individual pathogens of interest. Lastly, each bacterium may transport specific ferric siderophores that correlate with their virulence (K. pneumoniae, FeAbn; P. aeruginosa, FePvd, FePch; A. baumannii, FeAcn); FLHTS methods have the ability to modify and analyze virtually any LGP in a living bacterium. The structural folds of approximately 20 LGP are crystallographically solved (Table 1), which simplifies the identification of optimal sites for localization of fluorescent probes.220,285,700 Unsolved proteins may be modeled from the structures of solved orthologues. For example, AbaFepA in A. baumannii ATCC 17978 shares 46% sequence identity with EcoFepA in E. coli MG1655, resulting in a nearly conserved predicted tertiary structure196 that accurately suggested good locations for fluoresceination.272 However, caution is advised, because modeling of an orthologue/parologue with lower identity (e.g., EcoFiu, 20% identical to EcoFepA) may significantly err in the delineation and disposition of LGP secondary structures and surface loop regions.
9.3.2. Universal Fluorescent Sensors.
Further development of the FLHTS concept revealed another approach that obviates the need to genetically engineer the LGP of individual pathogens for site-directed fluoresceination. Production of EcoFepA-FM in a ΔtonB E. coli host creates a “sensor strain” that detects [FeEnt] and sensitively reports FeEnt depletion from solution. The E. coli sensor strain binds but cannot transport FeEnt because of its TonB-deficiency. By monitoring FeEnt-mediated quenching, the sensor strain observes FeEnt uptake by other bacteria in the same solution. This method creates a “universal” fluorescence assay of FeEnt uptake by any organism and readily adapts to uptake of any iron complex by any bacterium.285 Both species-specific272,701 and universal285 FLHTS assays effectively function in microtiter plate format.
9.4. Summary of Antibiotic Discovery
Several small-scale HTS studies conceived assays that targeted TonB-dependent uptake systems. Yep et al.273 employed a whole-cell growth-based high throughput screen of 149 243 compounds against UPEC under iron-limiting conditions and found 16 compounds that arrested bacterial growth only under iron-limiting conditions, that were all bacteriostatic, and that did not inhibit proton motive force. Two of the compounds lost inhibitory activity against a TonB-deficient strain. Nairn et al.272 used FLHTS to identify inhibitors of TonB function in E. coli K-12 and A. baumannii. In a screen of 17 441 compounds, 165 primary hits inhibited TonB-dependent FeEnt uptake at a level of at least 30%. Among 20 of the primary hits that were further analyzed with respect to TonB-dependent ferrichrome uptake, colicin killing, and proton-motive force-dependent lactose transport, six of the compounds blocked TonB activity in all tests without affecting lactose uptake. Lastly, Bailey et al.702 conducted HTS of 110 000 compounds for inhibitors of Abn biosynthesis324 in hypervirulent K. pneumoniae. As noted above, Abn is a virulence factor for HvKpn. The HTS system utilized a sensitive malachite green-based assay, in which an inorganic pyrophosphatase cleaved the byproduct pyrophosphate to produce inorganic phosphate. The screening assay identified potent inhibitors of IucA, but these compounds also showed undesirable attributes, especially inhibition of unrelated enzymes.
10. CONCLUSIONS AND FUTURE DIRECTIONS
As this overview illustrates, the correlations between prokaryotic iron acquisition and the pathogenesis of humans and animals are diverse, numerous, and well supported by extensive data. Bacterial pathogens secrete siderophores that capture iron from host cells or proteins and facilitate bacterial tropism or invasiveness to particular tissues. Additionally, a multitude of unique LGP of different specificities populate the outer membranes of Gram (−) bacteria, allowing recognition and transport the many ferric xenosiderophores that they may encounter in wild or host environments. The various examples of these phenomena spotlight the potential for chemical therapeutics that block prokaryotic iron uptake. The redundancy and complexity of these systems, and the relative inaccessibility of TonB in the periplasm, created skepticism about LGP systems as targets for antibiotic discovery. However, opportunity exists for the identification of efficacious compounds against TonB-dependent uptake systems. Random high-throughput screening of chemical libraries, and rational design of novel compounds that target iron transport biochemistry or related cell envelope processes are viable approaches. Both will profit from additional findings that better explain the mechanisms of TonB-dependent transport.
10.1. Antibiotic Development
Although cefiderocol is now available for clinical applications against Gram (−) bacterial infections, numerous questions remain about it and related Trojan Horse antibiotics. For example, which among the four or five ferric catecholate receptors in each ESKAPE pathogen act to receive and transport ferric monocatecholate–antibiotic conjugates? This query highlights the fact that for many LGP, the full scope of their ligand recognition attributes are not well-defined. What are their natural ligands (human or bacterial), do they include catecholamine stress hormones, and what are the affinities of their binding interactions and the rates of their iron transport reactions? Little experimental data exists to illuminate the uptake of monocatecholate iron complexes. Presumably the Trojan Horse catecholate conjugates follow the entry same routes as iron complexes of the hydrolytic and/or oxidized degradation products of Ent, but which LGP participate in these transport events in the various CRE/ESKAPE organisms? Once through the OM into the bacterial periplasm, FeSβLC complexes enter a biochemical no-man’s land that is difficult to experimentally characterize or observe. What periplasmic binding proteins recognize these complexes, does the AcrAB-TolC export pathway counteract their OM uptake,62 and how do such large molecules gain entry into bacterial cells through IM membrane permeases? The answers to these questions may explain the molecular mechanisms of adaptive resistance that are major obstacles to continuing use of antibiotics like Fetroja.
10.2. HTS of Chemical Libraries
Once an HTS assay method, like FLHTS,703 is developed and optimized for the primary screening of a chemical library, the main impediments to antibiotic discovery within the library derive from the secondary screening process. For example, primary hits against EcoTonB by FLHTS were subject to a funnel of criteria that attempted to exclude nonrelevant inhibition. In the FeEnt uptake FLHTS assay, besides authentic inhibitors of TonB action, primary hits may include: (i) specific antagonists of FeEnt uptake, (ii) nonspecific fluorescence quenchers, (iii) metabolic poisons that interfere with active transport, (iv) membrane disruptors that compromise cellular integrity, and (v) pan assay interference compounds (PAINS704-706). Secondary screens on candidate compounds are laborious and time-consuming. Whereas primary HTS of a chemical library only requires a few weeks of experiments, the ensuing secondary screens are much slower because they usually involve biochemical characterizations of each individual candidate inhibitor. Analysis of only 20 or 30 compounds may involve months of work. Nairn et al.272 screened a small chemical scaffold library of 17 500 compounds and found 165 primary hits. Applications of the same method, that yielded a 1% hit rate to a more typical library of 500 000 chemicals will produce ~5000 primary hits that are unmanageable except by HTS methods. Hence, the expansion and optimization of secondary assays to HTS formats is an important aspect of antibiotic discovery in chemical libraries. It is likely that the novel antibiotics we seek are present in those chemical collections, but it will take cleverness and technical innovations to find them.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R21AI115187 to P. E. Klebba and S. M. Newton. D.A. Six was supported in whole or in part with Federal funds from the Defense Threat Reduction Agency (DTRA) under Contract No. HDTRA117C0070 and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) under Contract No. 75N93020C00016.
ABBREVIATIONS
Siderophore or Porphyrin
- Ent*
degraded enterobactin
- GEnt
glucosylated enterobactin
- DHBA
2,3-dihydroxybenzoic acid
- DHBS
2,3-dihydroxybenzoyl serine
- Crn
corynebactin (= bacillibactin)
- Pbn
petrobactin
- Vbn
vibriobactin
- Fc
ferrichrome
- FcA
ferrichrome A
- FxB
ferrioxamine B (apoFxB: desferal)
- Abn
aerobactin
- Acn
acinetobactin
- Fbn
fimsbactin
- Bfn
baumannoferrin
- Ybt
yersiniabactin
- Pvd
pyoverdin
- Pch
pyochelin
- Nti
nicotianamine
- Mbn
mycobactin
- Hn
heme
- B12
cyanocobalamin (vitamin B12)
- Cit
citrate
- TF
transferrin
- LF
lactoferrin
- FTN
ferritin
- NGAL
neutrophil gelatinase-associated lipocalin
- LCN2
lipocalin 2
- SCN
siderocalin (= NGAL, LCN2)
- FDC
Fetroja (= cefidericol)
- CIP
ciprofloxacin
Phrase or Series of Words
- CRE
carbapenem-resistant Enterobacterales
- ESKAPE
Enterococcus, Staphylococcus, Klebsiella, Acinetobacter, Pseudomonas, Enterobacter
- OM
outer membrane
- IM
inner membrane
- PG
peptidoglycan
- IRMP
iron-related or -regulated membrane proteins
- IROMP
iron-regulated outer membrane proteins
- LGP
ligand-gated porin
- TBDT
TonB-dependent transporter
- PMF
proton-motive force
- CTD
C-terminal domain
- ABC
ATP-binding cassette
- HPI
high pathogenicity island
- UTI
urinary tract infection
- ExPEC
extra-intestinal pathogenic E. coli
- ETEC
entero-toxigenic E. coli
- EHEC
entero-hemorrhagic E. coli
- EPEC
entero-pathogenic E. coli
- EAEC
entero-aggregative E. coli
- AIEC
adherent-invasive E. coli
- UPEC
uropathogenic E. coli
- ESBL
extended spectrum β-lactamase
- XDR
extreme drug resistant
- CSS
cell-surface signaling
- HMWP
high molecular weight protein
- PK/PD
pharmacokinetic/pharmacodynamic
- MIC
mimimum inhibitory concentration
- PAINS
pan assay interference compounds
Biography
Phillip E. Klebba is a Distinguished Professor of Biochemistry and Molecular Biophysics at Kansas State University. He received his doctorate in biochemistry with J.B. Neilands at UC Berkeley, and completed postdoctoral study in microbiology and immunology with L.T. Rosenberg at Stanford Medical School, and with Hiroshi Nikaido at UC Berkeley, studying the immunology and transport biochemistry of bacterial porins. Relevant to this review, he was a Professor of Medical Microbiology at the Medical College of Wisconsin, a visiting scientist with M. Hofnung at Institut Pasteur, and with A. Charbit at Institut Necker Enfant Malades, and a visiting professor with H.R. Kaback at the UCLA David Geffen School of Medicine. His current research involves the development of fluorescent sensors to monitor membrane transport, toward the understanding of TonB-dependent iron acquisition and development of new antibiotics.
Salete M. Newton is a Research Professor of Biochemistry and Molecular Biophysics at Kansas State University. She received her doctorate in biochemistry with Sergio Olavo Pinto da Costa at Universidade de Sao Paulo and performed research with B.A.D. Stocker at Stanford University, studying the biotechnology of vaccines. She was a visiting scientist with M. Hofnung at Institut Pasteur and with A. Charbit at Institut Necker Enfant Malades and a visiting professor with H.R. Kaback at the UCLA David Geffen School of Medicine. Her current research focuses on the biochemistry of bacterial iron acquisition.
David A. Six is a Principal Scientist in Biology at Venatorx Pharmaceuticals. He obtained his M.S. and doctorate in Chemistry with E.A. Dennis at UC San Diego, working on the enzymology of cytosolic phospholipase A2. After postdoctoral work on lipopolysaccharide with C.R.H. Raetz at Duke University, he led antibacterial drug discovery programs at Novartis Institutes for BioMedical Research in Infectious Diseases, where he developed assays to measure bacterial compound accumulation. His work at Venatorx supports the clinical-stage cefepime–taniborbactam combination and novel non-β-lactam inhibitors of penicillin-binding proteins.
Ashish Kumar is a predoctoral researcher in Biochemistry and Molecular Biophysics with P.E. Klebba and S.M. Newton at Kansas State University. His research addresses the development of fluorescent sensors to detect infectious bacteria in blood, tissue, and food samples.
Taihao Yang is a predoctoral researcher in Biochemistry and Molecular Biophysics with P.E. Klebba and S.M. Newton at Kansas State University. His current research focuses on iron transporters of Gram-negative bacteria, with the objective of developing next generation antibiotics against bacterial pathogens.
Brittany L. Nairn (formerly Mortensen) is an Assistant Professor of Biology at Bethel University in St. Paul, MN. She completed her doctorate in microbiology and immunology with T. Kawula at UNC Chapel Hill and performed postdoctoral study with E.P. Skaar at Vanderbilt and with P.E. Klebba and S.M. Newton at Kansas State University and with M.C. Herzberg at the University of Minnesota. Her research interests include biofilm formation, metal acquisition, and pathogenesis of bacteria, focusing on Acinetobacter baumannii and Streptococcus gordonii.
Colton Munger is an undergraduate researcher in Biochemistry and Molecular Biophysics with P.E. Klebba and S.M. Newton at Kansas State University.
Somnath Chakravorty is a postdoctoral scientist at the University of Buffalo Medical School. He received his doctorate in Microbiology with R. Gachhui at Jadavpur University, India, and performed postdoctoral research with P.E. Klebba and S.M. Newton at Kansas State University, working on iron uptake by ESKAPE pathogens. His current work with T.A. Russo at UB includes includes study of virulence factors in Hypervirulent Klebsiella pneumoniae and XDR Acinetobacter baumannii, and the role of iron acquisition in pathogenesis of K. pneumoniae.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrev.0c01005.
Phylogenetic relationships of Gram (−) bacterial LGP, derived from pairwise comparisons of the mature protein sequences listed below the cladogram/phylogram (PDF)
The authors declare the following competing financial interest(s): D.A.S. is employed and compensated by Venatorx, and may own stock or stock options in Venatorx as part of his remuneration for employment.
Contributor Information
Phillip E. Klebba, Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, Kansas 66506, United States.
Salete M. C. Newton, Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, Kansas 66506, United States
David A. Six, Venatorx Pharmaceuticals, Inc., Malvern, Pennsylvania 19355, United States.
Ashish Kumar, Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, Kansas 66506, United States.
Taihao Yang, Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, Kansas 66506, United States.
Brittany L. Nairn, Department of Biological Sciences, Bethel University, St. Paul, Minnesota 55112, United States
Colton Munger, Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, Kansas 66506, United States.
Somnath Chakravorty, Jacobs School of Medicine and Biomedical Sciences, SUNY Buffalo, Buffalo, New York 14203, United States.
REFERENCES
- (1).Pappenheimer AM Jr Diphtheria toxin; a reinvestigation of the effect of iron on toxin and porphyrin production. J. Biol. Chem 1947, 167, 251–9. [PubMed] [Google Scholar]
- (2).Bullen JJ; Rogers HJ; Cushnie GH Abolition of passive immunity to bacterial infections by iron. Nature 1967, 214, 515–6. [DOI] [PubMed] [Google Scholar]
- (3).Bullen JJ Iron-binding proteins in milk and resistance to Escherichia coli infection in infants. Postgrad Med. J 1975, 51, 67–70. [PubMed] [Google Scholar]
- (4).Bullen JJ The significance of iron in infection. Clin. Infect. Dis 1981, 3, 1127–38. [DOI] [PubMed] [Google Scholar]
- (5).Bullen JJ Iron and infection. Eur. J. Clin. Microbiol 1985, 4, 537–9. [DOI] [PubMed] [Google Scholar]
- (6).Bullen JJ; Rogers HJ Bacterial iron metabolism and resistance to infection. J. Med. Microbiol 1970, 3, P8–P9. [PubMed] [Google Scholar]
- (7).Bullen JJ; Rogers HJ; Griffiths E Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol 1978, 80, 1–35. [DOI] [PubMed] [Google Scholar]
- (8).Bullen JJ; Rogers HJ; Spalding PB; Ward CG Iron and infection: the heart of the matter. FEMS Immunol. Med. Microbiol 2005, 43, 325–30. [DOI] [PubMed] [Google Scholar]
- (9).Bullen JJ; Ward CG; Rogers HJ Iron, infection, and the role of bicarbonate. FEMS Microbiol. Lett 1990, 71, 27–29. [DOI] [PubMed] [Google Scholar]
- (10).Bullen JJ; Ward CG; Rogers HJ The critical role of iron in some clinical infections. Eur. J. Clin. Microbiol. Infect. Dis 1991, 10, 613–7. [DOI] [PubMed] [Google Scholar]
- (11).Perez F; Van Duin D Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable patients. Cleve Clin J. Med 2013, 80, 225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Smith HZ; Kendall B Carbapenem Resistant Enterobacteriacea. In Stat Pearls; StatPearls: Treasure Island, FL, 2020. [Google Scholar]
- (13).Adeolu M; Alnajar S; Naushad S; Gupta RS Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol 2016, 66, 5575–5599. [DOI] [PubMed] [Google Scholar]
- (14).Lasko MJ; Nicolau DP Carbapenem-Resistant Enterobacterales: Considerations for Treatment in the Era of New Antimicrobials and Evolving Enzymology. Curr. Infect Dis Rep 2020, 22, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).McAdam AJ Enterobacteriaceae? Enterobacterales? What Should We Call Enteric Gram-Negative Bacilli? A Micro-Comic Strip. J. Clin. Microbiol 2020, 58, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Boucher HW; Talbot GH; Bradley JS; Edwards JE; Gilbert D; Rice LB; Scheld M; Spellberg B; Bartlett J Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis 2009, 48, 1–12. [DOI] [PubMed] [Google Scholar]
- (17).Rice LB Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis 2008, 197, 1079–81. [DOI] [PubMed] [Google Scholar]
- (18).Elemam A; Rahimian J; Mandell W Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clin. Infect. Dis 2009, 49, 271–4. [DOI] [PubMed] [Google Scholar]
- (19).Tacconelli E; Magrini N Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics World Health Organization; 2017. [Google Scholar]
- (20).Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2019; https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf, DOI; DOI: 10.15620/cdc:82532. [DOI] [Google Scholar]
- (21).Pollack A Rising Threat of Infections Unfazed by Antibiotics. New York Times; February 26, 2010. [Google Scholar]
- (22).Pollack A Deadly Germs Largely Ignored by Drug Firms. New York Times, February 27 2010; p 1. [Google Scholar]
- (23).Ventola CL The antibiotic resistance crisis: part 1: causes and threats. P & T 2015, 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- (24).Hu C Pharmaceutical Companies Are Backing Away from a Growing Threat That Could Kill 10 Million People a Year by 2050. Business Insider, 2018. [Google Scholar]
- (25).Jacobs A Deadly Germs, Lost Cures: Crisis Looms in Antibiotics as Drug Makers Go Bankrupt. New York Times, December 26, 2019. [Google Scholar]
- (26).Nikaido H Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol 1998, 1, 516–523. [DOI] [PubMed] [Google Scholar]
- (27).Nikaido H Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev 2003, 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nikaido H; Vaara M Molecular basis of bacterial outer membrane permeability. Microbiol. Rev 1985, 49, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yu EW; McDermott G; Zgurskaya HI; Nikaido H; Koshland DE Jr Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science (Washington, DC, U. S.) 2003, 300, 976–80. [DOI] [PubMed] [Google Scholar]
- (30).Zgurskaya HI; Nikaido H Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 7190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Khan SN; Khan AU Breaking the Spell: Combating Multidrug Resistant ‘Superbugs’. Front. Microbiol 2016, 7, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yaneja N; Kaur H Insights into Newer Antimicrobial Agents Against Gram-negative Bacteria. Microbiol. Insights 2016, 9, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bassetti M; Righi E Development of novel antibacterial drugs to combat multiple resistant organisms. Langenbecks Arch Surg 2015, 400, 153–65. [DOI] [PubMed] [Google Scholar]
- (34).Pogue JM; Kaye KS; Cohen DA; Marchaim D Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin. Microbiol. Infect 2015, 21, 302–12. [DOI] [PubMed] [Google Scholar]
- (35).Cornelissen CN; Sparling PF Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol. Microbiol 1994, 14, 843–50. [DOI] [PubMed] [Google Scholar]
- (36).Li J; Nation RL; Milne RW; Turnidge JD; Coulthard K Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 25, 11–25. [DOI] [PubMed] [Google Scholar]
- (37).McGann P; Snesrud E; Maybank R; Corey B; Ong AC; Clifford R; Hinkle M; Whitman T; Lesho E; Schaecher KE Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First report of mcr-1 in the USA. Antimicrob. Agents Chemother 2016, 60, 4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Buckling A; Harrison F; Vos M; Brockhurst MA; Gardner A; West SA; Griffin A Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa. FEMS Microbiol. Ecol 2007, 62, 135–141. [DOI] [PubMed] [Google Scholar]
- (39).Dhople AM; Ibanez MA; Poirier TC Role of iron in the pathogenesis of Mycobacterium avium infection in mice. Microbios 1996, 87, 77–87. [PubMed] [Google Scholar]
- (40).Payne SM; Neilands IB Iron and virulence in the family Enterobacteriaceae. Critical Rev. Microbiol 1988, 16, 81–111. [DOI] [PubMed] [Google Scholar]
- (41).Payne SM Iron and virulence in Shigella. Mol. Microbiol 1989, 3, 1301–6. [DOI] [PubMed] [Google Scholar]
- (42).Payne SM Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993, 1, 66–9. [DOI] [PubMed] [Google Scholar]
- (43).Schoolnik GK Microarray analysis of bacterial pathogenicity. Adv. Microb. Physiol 2002, 46, 1–45. [DOI] [PubMed] [Google Scholar]
- (44).Smith KD Iron metabolism at the host pathogen interface: lipocalin 2 and the pathogen-associated iroA gene cluster. Int. J. Biochem. Cell Biol 2007, 39, 1776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Snyder JA; Haugen BJ; Buckles EL; Lockatell CV; Johnson DE; Donnenberg MS; Welch RA; Mobley HL Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun 2004, 72, 6373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Stojijkovic I; Hwa V; Saint Martin L; O'Gaora P; Nassif X; Heffron F; So M The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol 1995, 15, 531–541. [DOI] [PubMed] [Google Scholar]
- (47).Sturm AW Iron and virulence of Haemophilus ducreyi in a primate model. Sex. Transm. Dis 1997, 24, 64–8. [DOI] [PubMed] [Google Scholar]
- (48).Torres AG; Redford P; Welch RA; Payne SM TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun 2001, 69, 6179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Williams PH Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect. Immun 1979, 26, 925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Wong HC; Liu CC; Yu CM; Lee YS Utilization of iron sources and its possible roles in the pathogenesis of Vibrio parahaemolyticus. Microbiol. Immunol 1996, 40, 791–8. [DOI] [PubMed] [Google Scholar]
- (51).Zhou D; Hardt WD; Galan JE Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun 1999, 67, 1974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Anderson CP; Shen M; Eisenstein RS; Leibold EA Mammalian iron metabolism and its control by iron regulatory proteins. Biochim. Biophys. Acta, Mol. Cell Res 2012, 1823, 1468–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Anderson GJ; Frazer DM Current understanding of iron homeostasis. Am. J. Clin. Nutr 2017, 106, 1559s–1566s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Cronin SJF; Woolf CJ; Weiss G; Penninger JM The Role of Iron Regulation in Immunometabolism and Immune-Related Disease. Front Mol. Biosci 2019, 6, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Goetz DH; Holmes MA; Borregaard N; Bluhm ME; Raymond KN; Strong RK The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–43. [DOI] [PubMed] [Google Scholar]
- (56).Neilands JB A Crystalline Organo-iron Pigment from a Rust Fungus (Ustilago sphaerogena). J. Am. Chem. Soc 1952, 74, 4846–7. [Google Scholar]
- (57).Neilands JB Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem 1995, 270, 26723–6. [DOI] [PubMed] [Google Scholar]
- (58).Harris WR; Carrano CJ; Raymond KN Spectrophotometric determination of the proton-dependent stability constant of ferric enterobactin. J. Am. Chem. Soc 1979, 101, 2213–2214. [Google Scholar]
- (59).Neilands JB Microbial iron compounds. Annu. Rev. Biochem 1981, 50, 715–31. [DOI] [PubMed] [Google Scholar]
- (60).Hider RC; Kong X Chemistry and biology of siderophores. Nat. Prod. Rep 2010, 27, 637–57. [DOI] [PubMed] [Google Scholar]
- (61).Di Masi DR; White JC; Schnaitman CA; Bradbeer C Transport of vitamin B12 in Escherichia coli: common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J. Bacteriol 1973, 115, 506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Newton SM; Trinh V; Pi H; Klebba PE Direct measurements of the outer membrane stage of ferric enterobactin transport: postuptake binding. J. Biol. Chem 2010, 285, 17488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).McIntosh MA; Earhart CF Effect of iron of the relative abundance of two large polypeptides of the Escherichia coli outer membrane. Biochem. Biophys. Res. Commun 1976, 70, 315–22. [DOI] [PubMed] [Google Scholar]
- (64).Pugsley AP; Reeves P The role of colicin receptors in the uptake of ferrienterochelin by Escherichia coli K-12. Biochem. Biophys. Res. Commun 1977, 74, 903–11. [DOI] [PubMed] [Google Scholar]
- (65).Noinaj N; Guillier M; Barnard TJ; Buchanan SK TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol 2010, 64, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Klebba PE; McIntosh MA; Neilands JB Kinetics of biosynthesis of iron-regulated membrane proteins in Escherichia coli. J. Bacteriol 1982, 149, 880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Morton DJ; Williams P Characterization of the outer-membrane proteins of Haemophilus parainfluenzae expressed under iron-sufficient and iron-restricted conditions. Microbiology 1989, 135, 445–51. [DOI] [PubMed] [Google Scholar]
- (68).Rutz JM; Liu J; Lyons JA; Goranson J; Armstrong SK; McIntosh MA; Feix JB; Klebba PE Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science (Washington, DC, U. S.) 1992, 258, 471–5. [DOI] [PubMed] [Google Scholar]
- (69).Schauer K; Rodionov DA; de Reuse H New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci 2008, 33, 330. [DOI] [PubMed] [Google Scholar]
- (70).Abbott GW KCNQs: Ligand- and Voltage-Gated Potassium Channels. Front. Physiol 2020, 11, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Moreira C; Calixto AR; Richard JP; Kamerlin SCL The role of ligand-gated conformational changes in enzyme catalysis. Biochem. Soc. Trans 2019, 47, 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Vetter I; Carter D; Bassett J; Deuis JR; Tay B; Jami S; Robinson SD High-Throughput Fluorescence Assays for Ion Channels and GPCRs. Adv. Exp. Med. Biol 2020, 1131, 27–72. [DOI] [PubMed] [Google Scholar]
- (73).Cornelis P Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol 2010, 86, 1637–45. [DOI] [PubMed] [Google Scholar]
- (74).Guerinot ML Microbial iron transport. Annu. Rev. Microbiol 1994, 48, 743–72. [DOI] [PubMed] [Google Scholar]
- (75).Bullen J; Griffiths E; Rogers H; Ward G Sepsis: the critical role of iron. Microbes Infect. 2000, 2, 409–15. [DOI] [PubMed] [Google Scholar]
- (76).Griffiths E; Rogers HJ; Bullen JJ Iron, plasmids and infection. Nature 1980, 284, 508–9. [DOI] [PubMed] [Google Scholar]
- (77).Adams TJ; Vartivarian S; Cowart RE Iron acquisition systems of Listeria monocytogenes. Infect. Immun 1990, 58, 2715–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Conte MP; Longhi C; Buonfiglio V; Polidoro M; Seganti L; Valenti P The effect of iron on the invasiveness of Escherichia coli carrying the inv gene of Yersinia pseudotuberculosis. J. Med. Microbiol 1994, 40, 236–40. [DOI] [PubMed] [Google Scholar]
- (79).Coulanges V; Andre P; Vidon DJ Effect of siderophores, catecholamines, and catechol compounds on Listeria spp. Growth in iron-complexed medium. Biochem. Biophys. Res. Commun 1998, 249, 526–30. [DOI] [PubMed] [Google Scholar]
- (80).Cowart RE; Foster BG Differential effects of iron on the growth of Listeria monocytogenes: minimum requirements and mechanism of acquisition. J. Infect. Dis 1985, 151, 721–30. [DOI] [PubMed] [Google Scholar]
- (81).Neilands JB Iron absorption and transport in microorganisms. Annu. Rev. Nutr 1981, 1, 27–46. [DOI] [PubMed] [Google Scholar]
- (82).Conte MP; Longhi C; Polidoro M; Petrone G; Buonfiglio V; Di Santo S; Papi E; Seganti L; Visca P; Valenti P Iron availability affects entry of Listeria monocytogenes into the enter-ocytelike cell line Caco-2. Infection and immunity 1996, 64, 3925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Furman M; Fica A; Saxena M; Di Fabio JL; Cabello FC Salmonella typhi iron uptake mutants are attenuated in mice. Infect. Immun 1994, 62, 4091–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Gregory EM; Yost FJ Jr; Fridovich I Superoxide dismutases of Escherichia coli: intracellular localization and functions. J. Bacteriol 1973, 115, 987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Rouquette C; Bolla JM; Berche P An iron-dependent mutant of Listeria monocytogenes of attenuated virulence. FEMS Microbiol. Lett 1995, 133, 77–83. [DOI] [PubMed] [Google Scholar]
- (86).Rozalska B; Lisiecki P; Sadowska B; Mikucki J; Rudnicka W The virulence of Staphylococcus aureus isolates differing by siderophore production. Acta Microbiol. Pol 1998, 47, 185–194. [PubMed] [Google Scholar]
- (87).Tai SS; Lee CJ; Winter RE Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect. Immun 1993, 61, 5401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Tsolis RM; Baumler AJ; Heffron F; Stojiljkovic I Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infection and immunity 1996, 64, 4549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Ward CG; Bullen JJ; Rogers HJ Iron and infection: new developments and their implications. J. Trauma 1996, 41, 356–64. [DOI] [PubMed] [Google Scholar]
- (90).Klebba PE Three paradoxes of ferric enterobactin uptake. Front. Biosci., Landmark Ed 2003, 8, s1422–s1436. [DOI] [PubMed] [Google Scholar]
- (91).Klebba PE ROSET Model of TonB Action in Gram-Negative Bacterial Iron Acquisition. J. Bacteriol 2016, 198, 1013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Abdelhamed H; Lawrence ML; Karsi A The Role of TonB Gene in Edwardsiella ictaluri Virulence. Front. Physiol 2017, 8, 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Beddek AJ; Sheehan BJ; Bosse JT; Rycroft AN; Kroll JS; Langford PR Two TonB systems in Actinobacillus pleuropneumoniae: their roles in iron acquisition and virulence. Infect. Immun 2004, 72, 701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Holden KM; Browning GF; Noormohammadi AH; Markham PF; Marenda MS TonB is essential for virulence in avian pathogenic Escherichia coli. Comp Immunol Microbiol Infect Dis 2012, 35, 129–38. [DOI] [PubMed] [Google Scholar]
- (95).Hsieh PF; Lin TL; Lee CZ; Tsai SF; Wang JT Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis 2008, 197, 1717–27. [DOI] [PubMed] [Google Scholar]
- (96).Lu F; Miao S; Tu J; Ni X; Xing L; Yu H; Pan L; Hu Q The role of TonB-dependent receptor TbdR1 in Riemerella anatipestifer in iron acquisition and virulence. Vet. Microbiol 2013, 167, 713–718. [DOI] [PubMed] [Google Scholar]
- (97).Morton DJ; Hempel RJ; Seale TW; Whitby PW; Stull TL A functional tonB gene is required for both virulence and competitive fitness in a chinchilla model of Haemophilus influenzae otitis media. BMC Res. Notes 2012, 5, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Stork M; Di Lorenzo M; Mourino S; Osorio CR; Lemos ML; Crosa JH Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun 2004, 72, 7326–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Zhang SR; Zhang L; Sun L Identification and analysis of three virulence-associated TonB-dependent outer membrane receptors of Pseudomonas fluorescens. Dis. Aquat. Org 2014, 110, 181–91. [DOI] [PubMed] [Google Scholar]
- (100).Carver PL Metal ions and infectious diseases. An overview from the clinic. Met. Ions Life Sci 2013, 13, 1–28. [DOI] [PubMed] [Google Scholar]
- (101).Cowart RE Reduction of iron by extracellular iron reductases: implications for microbial iron acquisition. Arch. Biochem. Biophys 2002, 400, 273–81. [DOI] [PubMed] [Google Scholar]
- (102).Elkins C; Chen CJ; Thomas CE Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infection and immunity 1995, 63, 2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Henderson DP; Payne SM Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun 1994, 62, 5120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Seliger SS; Mey AR; Valle AM; Payne SM The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol 2001, 39, 801–12. [DOI] [PubMed] [Google Scholar]
- (105).Bjarnason J; Southward CM; Surette MG Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J. Bacteriol 2003, 185, 4973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Bruske AK; Anton M; Heller KJ Cloning and sequencing of the Klebsiella pneumoniae tonB gene and characterization of Escherichia coli-K. pneumoniae TonB hybrid proteins. Gene 1993, 131, 9–16. [DOI] [PubMed] [Google Scholar]
- (107).Kingsley RA; Reissbrodt R; Rabsch W; Ketley JM; Tsolis RM; Everest P; Dougan G; Baumler AJ; Roberts M; Williams PH Ferrioxamine-mediated Iron(III) utilization by Salmonella enterica. Appl. Environ. Microbiol 1999, 65, 1610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Angerer A; Klupp B; Braun V Iron transport systems of Serratia marcescens. J. Bacteriol 1992, 174, 1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Benevides-Matos N; Biville F The Hem and Has haem uptake systems in Serratia marcescens. Microbiology 2010, 156, 1749–1757. [DOI] [PubMed] [Google Scholar]
- (110).Cobessi D; Meksem A; Brillet K Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: heme binding by an induced fit mechanism. Proteins: Struct., Funct., Genet 2010, 78, 286–94. [DOI] [PubMed] [Google Scholar]
- (111).Paquelin A; Ghigo JM; Bertin S; Wandersman C Characterization of HasB, a Serratia marcescens TonB-like protein specifically involved in the haemophore-dependent haem acquisition system. Mol. Microbiol 2001, 42, 995–1005. [DOI] [PubMed] [Google Scholar]
- (112).Turner PC; Thomas CE; Stojiljkovic I; Elkins C; Kizel G; Ala’Aldeen DA; Sparling PF Neisserial TonB-dependent outer-membrane proteins: detection, regulation and distribution of three putative candidates identified from the genome sequences. Microbiology 2001, 147, 1277–90. [DOI] [PubMed] [Google Scholar]
- (113).Bitter W; Marugg JD; de Weger LA; Tommassen J; Weisbeek PJ The ferric-pseudobactin receptor PupA of Pseudomonas putida WCS358: homology to TonB-dependent Escherichia coli receptors and specificity of the protein. Mol. Microbiol 1991, 5, 647–655. [DOI] [PubMed] [Google Scholar]
- (114).Poole K; Zhao Q; Neshat S; Heinrichs DE; Dean CR The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology 1996, 142, 1449–58. [DOI] [PubMed] [Google Scholar]
- (115).Perry RD; Shah J; Bearden SW; Thompson JM; Fetherston JD Yersinia pestis TonB: role in iron, heme, and hemoprotein utilization. Infect. Immun 2003, 71, 4159–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Stojiljkovic I; Hantke K Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992, 11, 4359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Thompson JM; Jones HA; Perry RD Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun 1999, 67, 3879–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Torres AG; Payne SM Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol 1997, 23, 825–33. [DOI] [PubMed] [Google Scholar]
- (119).Fung-Tomc J; Bush K; Minassian B; Kolek B; Flamm R; Gradelski E; Bonner D Antibacterial activity of BMS-180680, a new catechol-containing monobactam. Antimicrob. Agents Chemother 1997, 41, 1010–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Rutz JM; Abdullah T; Singh SP; Kalve VI; Klebba PE Evolution of the ferric enterobactin receptor in gram-negative bacteria. J. Bacteriol 1991, 173, 5964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Bullen JJ Proceedings: Iron and infection. Br. J. Haematol 1974, 28, 139–140. [PubMed] [Google Scholar]
- (122).Bachman MA; Miller VL; Weiser JN Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009, 5, No. e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Collins HL Withholding iron as a cellular defence mechanism–friend or foe? Eur. J. Immunol 2008, 38, 1803–6. [DOI] [PubMed] [Google Scholar]
- (124).Fischbach MA; Lin H; Liu DR; Walsh CT How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol 2006, 2, 132–8. [DOI] [PubMed] [Google Scholar]
- (125).Haley KP; Skaar EP A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes Infect. 2012, 14, 217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Kim H; Sandaruwan Elvitigala DA; Lee Y; Lee S; Whang I; Lee J Ferritin H-like subunit from Manila clam (Ruditapes philippinarum): molecular insights as a potent player in host antibacterial defense. Fish Shellfish Immunol. 2012, 33, 926–936. [DOI] [PubMed] [Google Scholar]
- (127).McLaughlin HP; Hill C; Gahan CG The impact of iron on Listeria monocytogenes; inside and outside the host. Curr. Opin. Biotechnol 2011, 22, 194–199. [DOI] [PubMed] [Google Scholar]
- (128).Parrow NL; Fleming RE; Minnick MF Sequestration and scavenging of iron in infection. Infect. Immun 2013, 81, 3503–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Pieracci FM; Barie PS Iron and the risk of infection. Surg Infect (Larchmt). 2005, 6, S41–S46. [DOI] [PubMed] [Google Scholar]
- (130).Radtke AL; O’Riordan MX Intracellular innate resistance to bacterial pathogens. Cell. Microbiol 2006, 8, 1720–9. [DOI] [PubMed] [Google Scholar]
- (131).Wakeman CA; Skaar EP Metalloregulation of Gram-positive pathogen physiology. Curr. Opin. Microbiol 2012, 15, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Allen CE; Burgos JM; Schmitt MP Analysis of novel iron-regulated, surface-anchored hemin-binding proteins in Corynebacterium diphtheriae. J. Bacteriol 2013, 195, 2852–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Cadieux B; Lian T; Hu G; Wang J; Biondo C; Teti G; Liu V; Murphy ME; Creagh AL; Kronstad JW The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J. Infect. Dis 2013, 207, 1339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Caza M; Kronstad JW Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol 2013, 3, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Cornelis P; Dingemans J Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol 2013, 3, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Erac B; Yilmaz FF; Hosgor Limoncu M; Ozturk I; Aydemir S [Investigation of the virulence factors of multidrug-resistant Acinetobacter baumannii isolates]. Mikrobiyol Bul. 2014, 48, 70–81. [PubMed] [Google Scholar]
- (137).Jung WH; Do E Iron acquisition in the human fungal pathogen Cryptococcus neoformans. Curr. Opin. Microbiol 2013, 16, 686–91. [DOI] [PubMed] [Google Scholar]
- (138).Konings AF; Martin LW; Sharples KJ; Roddam LF; Latham R; Reid DW; Lamont IL Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect. Immun 2013, 81, 2697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Mortensen BL; Skaar EP The contribution of nutrient metal acquisition and metabolism to Acinetobacter baumannii survival within the host. Front. Cell. Infect. Microbiol 2013, 3, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Nagy TA; Moreland SM; Andrews-Polymenis H; Detweiler CS The ferric enterobactin transporter Fep is required for persistent Salmonella enterica serovar typhimurium infection. Infect. Immun 2013, 81, 4063–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Pandey SD; Choudhury M; Yousuf S; Wheeler PR; Gordon SV; Ranjan A; Sritharan M Iron-regulated protein HupB of Mycobacterium tuberculosis positively regulates siderophore biosynthesis and is essential for growth in macrophages. J. Bacteriol 2014, 196, 1853–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Alonzo F 3rd; Benson MA; Chen J; Novick RP; Shopsin B; Torres VJ Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol 2012, 83, 423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Pich OQ; Merrell DS The ferric uptake regulator of Helicobacter pylori: a critical player in the battle for iron and colonization of the stomach. Future Microbiol. 2013, 8, 725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Runyen-Janecky LJ Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol 2013, 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Wells RM; Jones CM; Xi Z; Speer A; Danilchanka O; Doornbos KS; Sun P; Wu F; Tian C; Niederweis M Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 2013, 9, No. e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Wilks A; Ikeda-Saito M Heme Utilization by Pathogenic Bacteria: Not All Pathways Lead to Biliverdin. Acc. Chem. Res 2014, 47, 2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Zapotoczna M; Jevnikar Z; Miajlovic H; Kos J; Foster TJ Iron-regulated surface determinant B (IsdB) promotes Staphylococcus aureus adherence to and internalization by non-phagocytic human cells. Cell. Microbiol 2013, 15, 1026–41. [DOI] [PubMed] [Google Scholar]
- (148).Cassat JE; Skaar EP Iron in infection and immunity. Cell Host Microbe 2013, 13, 509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Silva-Gomes S; Vale-Costa S; Appelberg R; Gomes MS Iron in intracellular infection: to provide or to deprive? Front. Cell. Infect. Microbiol 2013, 3, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Larrie-Bagha SM; Rasooli I; Mousavi-Gargari SL; Rasooli Z; Nazarian S Passive immunization by recombinant ferric enterobactin protein (FepA) from Escherichia coli O157. Iran J. Microbiol 2013, 5, 113–119. [PMC free article] [PubMed] [Google Scholar]
- (151).Abergel C; Rigal A; Chenivesse S; Lazdunski C; Claverie JM; Bouveret E; Benedetti H Crystallization and preliminary crystallographic study of a component of the Escherichia coli tol system: TolB. Acta Crystallogr., Sect. D: Biol. Crystallogr 1998, 54, 102–4. [DOI] [PubMed] [Google Scholar]
- (152).Ons E; Bleyen N; Tuntufye HN; Vandemaele F; Goddeeris BM High prevalence iron receptor genes of avian pathogenic Escherichia coli. Avian Pathol. 2007, 36, 411–414. [DOI] [PubMed] [Google Scholar]
- (153).Abergel RJ; Clifton MC; Pizarro JC; Warner JA; Shuh DK; Strong RK; Raymond KN The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport. J. Am. Chem. Soc 2008, 130, 11524–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Baghal SM; Gargari SL; Rasooli I Production and immunogenicity of recombinant ferric enterobactin protein (FepA). Int. J. Infect. Dis 2010, 14, e166–e170. [DOI] [PubMed] [Google Scholar]
- (155).Warner PJ; Williams PH; Bindereif A; Neilands JB ColV plasmid-specific aerobactin synthesis by invasive strains of Escherichia coli. Infect. Immun 1981, 33, 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Abbott MB; Gaponenko V; Abusamhadneh E; Finley N; Li G; Dvoretsky A; Rance M; Solaro RJ; Rosevear PR Regulatory domain conformational exchange and linker region flexibility in cardiac troponin C bound to cardiac troponin I. J. Biol. Chem 2000, 275, 20610–7. [DOI] [PubMed] [Google Scholar]
- (157).Wolf SL; Hogan JS; Smith KL Iron uptake by Escherichia coli cultured with antibodies from cows immunized with high-affinity ferric receptors. J. Dairy Sci 2004, 87, 2103–7. [DOI] [PubMed] [Google Scholar]
- (158).Budzikiewicz H Siderophore-antibiotic conjugates used as trojan horses against Pseudomonas aeruginosa. Curr. Top. Med. Chem 2001, 1, 73–82. [DOI] [PubMed] [Google Scholar]
- (159).Kline T; Fromhold M; McKennon TE; Cai S; Treiberg J; Ihle N; Sherman D; Schwan W; Hickey MJ; Warrener P; Witte PR; Brody LL; Goltry L; Barker LM; Anderson SU; Tanaka SK; Shawar RM; Nguyen LY; Langhorne M; Bigelow A; Embuscado L; Naeemi E Antimicrobial effects of novel siderophores linked to beta-lactam antibiotics. Bioorg. Med. Chem 2000, 8, 73–93. [DOI] [PubMed] [Google Scholar]
- (160).Miethke M; Marahiel MA Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev 2007, 71, 413–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Dolence EK; Lin CE; Miller MJ; Payne SM Synthesis and siderophore activity of albomycin-like peptides derived from N5-acetyl-N5-hydroxy-L-ornithine. J. Med. Chem 1991, 34, 956–68. [DOI] [PubMed] [Google Scholar]
- (162).Mislin GL; Schalk IJ Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics. 2014, 6, 408–420. [DOI] [PubMed] [Google Scholar]
- (163).Wencewicz TA; Long TE; Mollmann U; Miller MJ Trihydroxamate siderophore-fluoroquinolone conjugates are selective sideromycin antibiotics that target Staphylococcus aureus. Bioconjugate Chem. 2013, 24, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Pi H; Jones SA; Mercer LE; Meador JP; Caughron JE; Jordan L; Newton SM; Conway T; Klebba PE Role of catecholate siderophores in gram-negative bacterial colonization of the mouse gut. PLoS One 2012, 7, No. e50020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Zughaier SM; Cornelis P Editorial: Role of Iron in Bacterial Pathogenesis. Front. Cell. Infect. Microbiol 2018, 8, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Ali MK; Kim RY; Karim R; Mayall JR; Martin KL; Shahandeh A; Abbasian F; Starkey MR; Loustaud-Ratti V; Johnstone D; Milward EA; Hansbro PM; Horvat JC Role of iron in the pathogenesis of respiratory disease. Int. J. Biochem. Cell Biol 2017, 88, 181–195. [DOI] [PubMed] [Google Scholar]
- (167).Tang F; Saier MH Jr Transport proteins promoting Escherichia coli pathogenesis. Microb. Pathog 2014, 71–72, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Benjamin WH Jr; Turnbough CL Jr; Posey BS; Briles DE The ability of Salmonella typhimurium to produce the siderophore enterobactin is not a virulence factor in mouse typhoid. Infect. Immun 1985, 50, 392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Desai PJ; Garges E; Genco CA Pathogenic neisseriae can use hemoglobin, transferrin, and lactoferrin independently of the tonB locus. J. Bacteriol 2000, 182, 5586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Najimi M; Lemos ML; Osorio CR Identification of iron regulated genes in the fish pathogen Aeromonas salmonicida subsp. salmonicida: genetic diversity and evidence of conserved iron uptake systems. Vet. Microbiol 2009, 133, 377–382. [DOI] [PubMed] [Google Scholar]
- (171).Perry RD; Mier I Jr.; Fetherston JD Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. BioMetals 2007, 20, 699–703. [DOI] [PubMed] [Google Scholar]
- (172).Braun V Iron uptake by Escherichia coli. Front. Biosci., Landmark Ed 2003, 8, s1409–a1421. [DOI] [PubMed] [Google Scholar]
- (173).Miller CE; Williams PH; Ketley JM Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology 2009, 155, 3157–4165. [DOI] [PubMed] [Google Scholar]
- (174).Nielubowicz GR; Mobley HL Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol 2010, 7, 430–441. [DOI] [PubMed] [Google Scholar]
- (175).Neilands JB; Peterson T; Leong SA High affinity iron transport in microorganisms. In Inorganic Chemistry in Biology and Medicine; ACS Symposium Series 140; Martell AE, Ed.; American Chemical Society, 1980; Vol. 140, pp 263–278. [Google Scholar]
- (176).Sheldon JR; Skaar EP Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020, 16, No. e1008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Baquero F; Levin BR Proximate and ultimate causes of the bactericidal action of antibiotics. Nat. Rev. Microbiol 2021, 19, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Brown AR; Gordon RA; Hyland SN; Siegrist MS; Grimes CL Chemical Biology Tools for Examining the Bacterial Cell Wall. Cell Chem. Biol 2020, 27, 1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Geisinger E; Huo W; Hernandez-Bird J; Isberg RR Acinetobacter baumannii: Envelope Determinants That Control Drug Resistance, Virulence, and Surface Variability. Annu. Rev. Microbiol 2019, 73, 481–506. [DOI] [PubMed] [Google Scholar]
- (180).Kuhn A The Bacterial Cell Wall and Membrane-A Treasure Chest for Antibiotic Targets. Subcell. Biochem 2019, 92, 1–5. [DOI] [PubMed] [Google Scholar]
- (181).Agarwal AK; Yee J Hepcidin. Adv. Chronic Kidney Dis 2019, 26, 298–305. [DOI] [PubMed] [Google Scholar]
- (182).Gao G; Li J; Zhang Y; Chang YZ Cellular Iron Metabolism and Regulation. Adv. Exp. Med. Biol 2019, 1173, 21–32. [DOI] [PubMed] [Google Scholar]
- (183).Testi C; Boffi A; Montemiglio LC Structural analysis of the transferrin receptor multifaceted ligand(s) interface. Biophys. Chem 2019, 254, 106242. [DOI] [PubMed] [Google Scholar]
- (184).Yanatori I; Kishi F DMT1 and iron transport. Free Radical Biol. Med 2019, 133, 55–63. [DOI] [PubMed] [Google Scholar]
- (185).Yeowell HN; White JR Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother 1982, 22, 961–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Cohen MS; Chai Y; Britigan BE; McKenna W; Adams J; Svendsen T; Bean K; Hassett DJ; Sparling PF Role of extracellular iron in the action of the quinone antibiotic streptonigrin: mechanisms of killing and resistance of Neisseria gonorrhoeae. Antimicrob. Agents Chemother 1987, 31, 1507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Dyer DW; McKenna W; Woods JP; Sparling PF Isolation by streptonigrin enrichment and characterization of a transferrin-specific iron uptake mutant of Neisseria meningitidis. Microb. Pathog 1987, 3, 351–63. [DOI] [PubMed] [Google Scholar]
- (188).Holland J; Towner KJ; Williams P Isolation and characterisation of Haemophilus influenzae type b mutants defective in transferrin-binding and iron assimilation. FEMS Microbiol. Lett 1991, 77, 283–288. [DOI] [PubMed] [Google Scholar]
- (189).Sato T; Yamawaki K Cefiderocol: Discovery, Chemistry, and In Vivo Profiles of a Novel Siderophore Cephalosporin. Clin. Infect. Dis 2019, 69, S538–s543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (190).Simner PJ; Patel R Cefiderocol Antimicrobial Susceptibility Testing Considerations: the Achilles’ Heel of the Trojan Horse? J. Clin. Microbiol 2020, 59, e951–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Zhanel GG; Golden AR; Zelenitsky S; Wiebe K; Lawrence CK; Adam HJ; Idowu T; Domalaon R; Schweizer F; Zhanel MA; Lagacé-Wiens PRS; Walkty AJ; Noreddin A; Lynch JP Iii; Karlowsky JA Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289. [DOI] [PubMed] [Google Scholar]
- (192).Anderson JE; Sparling PF; Cornelissen CN Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol 1994, 176, 3162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Klebba PE Transport Biochemistry of FepA; ASM Press, 2004; pp 147–157. [Google Scholar]
- (194).Balhesteros H; Shipelskiy Y; Long NJ; Majumdar A; Katz BB; Santos NM; Leaden L; Newton SM; Marques MV; Klebba PE TonB-Dependent Heme/Hemoglobin Utilization by Caulobacter crescentus HutA. J. Bacteriol 2017, 199, e00723–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Buchanan SK; Lukacik P; Grizot S; Ghirlando R; Ali MM; Barnard TJ; Jakes KS; Kienker PK; Esser L Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J. 2007, 26, 2594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Buchanan SK; Smith BS; Venkatramani L; Xia D; Esser L; Palnitkar M; Chakraborty R; van der Helm D; Deisenhofer J Crystal structure of the outer membrane active transporter FepA from Escherichia coli [see comments]. Nat. Struct. Biol 1999, 6, 56–63. [DOI] [PubMed] [Google Scholar]
- (197).Ferguson AD; Chakraborty R; Smith BS; Esser L; van der Helm D; Deisenhofer J Structural basis of gating by the outer membrane transporter FecA. Science (Washington, DC, U. S.) 2002, 295, 1715–1719. [DOI] [PubMed] [Google Scholar]
- (198).Ferguson AD; Hofmann E; Coulton JW; Diederichs K; Welte W Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide [see comments]. Science (Washington, DC, U. S.) 1998, 282, 2215–2220. [DOI] [PubMed] [Google Scholar]
- (199).Locher KP; Rees B; Koebnik R; Mitschler A; Moulinier L; Rosenbusch JP; Moras D Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 1998, 95, 771–778. [DOI] [PubMed] [Google Scholar]
- (200).Saier MH Jr Families of proteins forming transmembrane channels. J. Membr. Biol 2000, 175, 165–180. [DOI] [PubMed] [Google Scholar]
- (201).Pugsley AP; Reeves P Iron uptake in colicin B-resistant mutants of Escherichia coli K-12. J. Bacteriol 1976, 126, 1052–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Wayne R; Frick K; Neilands JB Siderophore protection against colicins M, B, V, and Ia in Escherichia coli. J. Bacteriol 1976, 126, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Rabsch W; Ma L; Wiley G; Najar FZ; Kaserer W; Schuerch DW; Klebba JE; Roe BA; Laverde Gomez JA; Schallmey M; Newton SM; Klebba PE FepA- and TonB-dependent Bacteriophage H8: Receptor Binding and Genomic Sequence. J. Bacteriol 2007, 189, 5568–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Nikaido H; Rosenberg EY Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J. Bacteriol 1990, 172, 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Hantke K Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet 1983, 191, 301–6. [DOI] [PubMed] [Google Scholar]
- (206).Bindereif A; Neilands JB Cloning of the aerobactin-mediated iron assimilation system of plasmid ColV. J. Bacteriol 1983, 153, 1111–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Chimento DP; Mohanty AK; Kadner RJ; Wiener MC Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Mol. Biol 2003, 10, 394–401. [DOI] [PubMed] [Google Scholar]
- (208).Cobessi D; Celia H; Folschweiller N; Schalk IJ; Abdallah MA; Pattus F The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J. Mol. Biol 2005, 347, 121–134. [DOI] [PubMed] [Google Scholar]
- (209).Cobessi D; Celia H; Pattus F Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol 2005, 352, 893–904. [DOI] [PubMed] [Google Scholar]
- (210).Cox CD Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J. Bacteriol 1980, 142, 581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Liu PV; Shokrani F Biological activities of pyochelins: iron-chelating agents of Pseudomonas aeruginosa. Infect. Immun 1978, 22, 878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Elliott RP Some properties of pyoverdine, the water-soluble fluorescent pigment of the pseudomonads. Appl. Microbiol. 1958, 6, 241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Philson SB; Llinás M Siderochromes from Pseudomonas fluorescens. I. Isolation and characterization. J. Biol. Chem 1982, 257, 8081–8085. [PubMed] [Google Scholar]
- (214).Wayne R; Neilands JB Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J. Bacteriol 1975, 121, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Weiss MS; Wacker T; Weckesser J; Weite W; Schulz GE The three-dimensional structure of porin from Rhodobacter capsulatus at 3 A resolution. FEBS Lett. 1990, 267, 268–272. [DOI] [PubMed] [Google Scholar]
- (216).Cowan SW; Schirmer T; Rummel G; Steiert M; Ghosh R; Pauptit RA; Jansonius JN; Rosenbusch JP Crystal structures explain functional properties of two E. coli porins. Nature 1992, 358, 727–733. [DOI] [PubMed] [Google Scholar]
- (217).Guterman SK; Dann L Excretion of enterochelin by exbA and exbB mutants of Escherichia coli. J. Bacteriol 1973, 114, 1225–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Kadner RJ Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol. Microbiol 1990, 4, 2027–33. [DOI] [PubMed] [Google Scholar]
- (219).Scott DC; Newton SM; Klebba PE Surface loop motion in FepA. J. Bacteriol 2002, 184, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Smallwood CR; Jordan L; Trinh V; Schuerch DW; Gala A; Hanson M; Shipelskiy Y; Majumdar A; Newton SM; Klebba PE Concerted loop motion triggers induced fit of FepA to ferric enterobactin. J. Gen. Physiol 2014, 144, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Kreusch A; Neubuser A; Schiltz E; Weckesser J; Schulz GE Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 A resolution. Protein Sci. 1994, 3, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Meyer JE; Hofnung M; Schulz GE Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenylmaltotrioside. J. Mol. Biol 1997, 266, 761–75. [DOI] [PubMed] [Google Scholar]
- (223).Schirmer T; Keller TA; Wang YF; Rosenbusch JP Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution [see comments]. Science (Washington, DC, U. S.) 1995, 267, 512–514. [DOI] [PubMed] [Google Scholar]
- (224).Davies JK; Reeves P Genetics of resistance to colicins in Escherichia coli K-12: cross- resistance among colicins of group B. J. Bacteriol 1975, 123, 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Guterman SK Colicin B: mode of action and inhibition by enterochelin. J. Bacteriol 1973, 114, 1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Pugsley AP; Reeves P Characterization of group B colicin-resistant mutants of Escherichia coli K-12: colicin resistance and the role of enterochelin. J. Bacteriol 1976, 127, 218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Wang CC; Newton A An additional step in the transport of iron defined by the tonB locus of Escherichia coli. J. Biol. Chem 1971, 246, 2147–51. [PubMed] [Google Scholar]
- (228).Fischer E; Gunter K; Braun V Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol 1989, 171, 5127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Braun V The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol 1989, 171, 6387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Braun V; Herrmann C Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol 1993, 8, 261–8. [DOI] [PubMed] [Google Scholar]
- (231).Ollis AA; Manning M; Held KG; Postle K Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol 2009, 73, 466–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Celia H; Botos I; Ni X; Fox T; De Val N; Lloubes R; Jiang J; Buchanan SK Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Commun. Biol 2019, 2, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Celia H; Noinaj N; Zakharov SD; Bordignon E; Botos I; Santamaria M; Barnard TJ; Cramer WA; Lloubes R; Buchanan SK Structural insight into the role of the Ton complex in energy transduction. Nature 2016, 538, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Bassford PJ Jr; Bradbeer C; Kadner RJ; Schnaitman CA Transport of vitamin B12 in tonB mutants of Escherichia coli. J. Bacteriol 1976, 128, 242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Cadieux N; Phan PG; Cafiso DS; Kadner RJ Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 10688–10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Evans JS; Levine BA; Trayer IP; Dorman CJ; Higgins CF Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 1986, 208, 211–6. [DOI] [PubMed] [Google Scholar]
- (237).Jordan LD; Zhou Y; Smallwood CR; Lill Y; Ritchie K; Yip WT; Newton SM; Klebba PE Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 11553–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Kaserer WA; Jiang X; Xiao Q; Scott DC; Bauler M; Copeland D; Newton SM; Klebba PE Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J. Bacteriol 2008, 190, 4001–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (239).Pawelek PD; Croteau N; Ng-Thow-Hing C; Khursigara CM; Moiseeva N; Allaire M; Coulton JW Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 2006, 312, 1399–402. [DOI] [PubMed] [Google Scholar]
- (240).Shultis DD; Purdy MD; Banchs CN; Wiener MC Outer membrane active transport: structure of the BtuB:TonB complex. Science 2006, 312, 1396–9. [DOI] [PubMed] [Google Scholar]
- (241).Chang C; Mooser A; Pluckthun A; Wlodawer A Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem 2001, 276, 27535–40. [DOI] [PubMed] [Google Scholar]
- (242).Zhai YF; Heijne W; Saier MH Jr Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta, Biomembr 2003, 1614, 201–10. [DOI] [PubMed] [Google Scholar]
- (243).Bradbeer C; Woodrow ML Transport of vitamin B12 in Escherichia coli: energy dependence. J. Bacteriol 1976, 128, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (244).Wang CC; Newton A Iron transport in Escherichia coli: relationship between chromium sensitivity and high iron requirement in mutants of Escherichia coli. J. Bacteriol 1969, 98, 1135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Chu BCH; Vogel HJ A structural and functional analysis of type III periplasmic and substrate binding proteins: their role in bacterial siderophore and heme transport. Biol. Chem 2011, 392, 39–52. [DOI] [PubMed] [Google Scholar]
- (246).Fukamizo T; Kitaoku Y; Suginta W Periplasmic solute-binding proteins: Structure classification and chitooligosaccharide recognition. Int. J. Biol. Macromol 2019, 128, 985–993. [DOI] [PubMed] [Google Scholar]
- (247).Chu BC; Otten R; Krewulak KD; Mulder FA; Vogel HJ The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB. J. Biol. Chem 2014, 289, 29219–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Sprencel C; Cao Z; Qi Z; Scott DC; Montague MA; Ivanoff N; Xu J; Raymond KM; Newton SM; Klebba PE Binding of ferric enterobactin by the escherichia coli periplasmic protein fepB. J. Bacteriol 2000, 182, 5359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Delepelaire P Bacterial ABC transporters of iron containing compounds. Res. Microbiol 2019, 170, 345–357. [DOI] [PubMed] [Google Scholar]
- (250).Ford RC; Beis K Learning the ABCs one at a time: structure and mechanism of ABC transporters. Biochem. Soc. Trans 2019, 47, 23–36. [DOI] [PubMed] [Google Scholar]
- (251).Holland IB Rise and rise of the ABC transporter families. Res. Microbiol 2019, 170, 304–320. [DOI] [PubMed] [Google Scholar]
- (252).Li B; Li N; Yue Y; Liu X; Huang Y; Gu L; Xu S An unusual crystal structure of ferric-enterobactin bound FepB suggests novel functions of FepB in microbial iron uptake. Biochem. Biophys. Res. Commun 2016, 478, 1049–53. [DOI] [PubMed] [Google Scholar]
- (253).Palacios M; Broberg CA; Walker KA; Miller VL A Serendipitous Mutation Reveals the Severe Virulence Defect of a Klebsiella pneumoniae fepB Mutant. mSphere 2017, 2, e00341–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Shea CM; McIntosh MA Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein- dependent systems in Escherichia coli. Mol. Microbiol 1991, 5, 1415–28. [DOI] [PubMed] [Google Scholar]
- (255).Abergel RJ; Warner JA; Shuh DK; Raymond KN Enterobactin protonation and iron release: structural characterization of the salicylate coordination shift in ferric enterobactin. J. Am. Chem. Soc 2006, 128, 8920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (256).Cianciotto NP An update on iron acquisition by Legionella pneumophila: new pathways for siderophore uptake and ferric iron reduction. Future Microbiol 2015, 10, 841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (257).Ecker DJ; Lancaster JR Jr; Emery T Siderophore iron transport followed by electron paramagnetic resonance spectroscopy. J. Biol. Chem 1982, 257, 8623–6. [PubMed] [Google Scholar]
- (258).Kazmi SA; Shorter AL; McArdle JV; Ashiq U; Jamal RA Studies on the redox characteristics of ferrioxamine E. Chem. Biodiversity 2010, 7, 656–65. [DOI] [PubMed] [Google Scholar]
- (259).Leong J; Neilands JB Mechanisms of siderophore iron transport in enteric bacteria. J. Bacteriol 1976, 126, 823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (260).Mies KA; Wirgau JI; Crumbliss AL Ternary complex formation facilitates a redox mechanism for iron release from a siderophore. BioMetals 2006, 19, 115–26. [DOI] [PubMed] [Google Scholar]
- (261).Schalk IJ; Guillon L Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 2013, 44, 1267–77. [DOI] [PubMed] [Google Scholar]
- (262).Francis J; Macturk HM; Madinaveitia J; Snow GA Mycobactin, a growth factor for Mycobacterium johnei. I. Isolation from Mycobacterium phlei. Biochem. J 1953, 55, 596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Raymond KN; Dertz EA; Kim SS Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (264).Holden VI; Bachman MA Diverging roles of bacterial siderophores during infection. Metallomics: integrated biometal science 2015, 7, 986–95. [DOI] [PubMed] [Google Scholar]
- (265).Holden VI; Breen P; Houle S; Dozois CM; Bachman MA Klebsiella pneumoniae Siderophores Induce Inflammation, Bacterial Dissemination, and HIF-1α Stabilization during Pneumonia. mBio 2016, 7, e01397–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (266).Griffiths GL; Sigel SP; Payne SM; Neilands JB Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem 1984, 259, 383–5. [PubMed] [Google Scholar]
- (267).Budzikiewicz H; Bössenkamp A; Taraz K; Pandey A; Meyer JM Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC14067 (Brevibacterium sp. DSM 20411). Z. Naturforsch., C: J. Biosci 1997, 52, 551–554. [Google Scholar]
- (268).Raymond KN; Isied SS; Brown LD; Fronczek FR; Nibert JH Coordination isomers of biological iron transport compounds. VI. Models of the enterobactin coordination site. A crystal field effect in the structure of potassium tris(catecholato)-chromate(III) and -ferrate(III) sesquihydrates, K3(M(O2C6H4)3)-1.5H2O, M = Cr, Fe1. J. Am. Chem. Soc 1976, 98, 1767–74. [DOI] [PubMed] [Google Scholar]
- (269).Nairz M; Haschka D; Demetz E; Weiss G Iron at the interface of immunity and infection. Front. Pharmacol 2014, 5, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (270).Nairz M; Dichtl S; Schroll A; Haschka D; Tymoszuk P; Theurl I; Weiss G Iron and innate antimicrobial immunity-Depriving the pathogen, defending the host. J. Trace Elem. Med. Biol 2018, 48, 118–133. [DOI] [PubMed] [Google Scholar]
- (271).Silva-Gomes S; Vale-Costa S; Appelberg R; Gomes MS Iron in intracellular infection: to provide or to deprive? Front. Cell. Infect. Microbiol 2013, 3, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Nairn BL; Eliasson OS; Hyder DR; Long NJ; Majumdar A; Chakravorty S; McDonald P; Roy A; Newton SM; Klebba PE Fluorescence high-throughput screening for inhibitors of TonB action. J. Bacteriol 2017, 199, e00889–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Yep A; McQuade T; Kirchhoff P; Larsen M; Mobley HL Inhibitors of TonB function identified by a high-throughput screen for inhibitors of iron acquisition in uropathogenic Escherichia coli CFT073. mBio 2014, 5, No. e01089–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (274).Braun V Bacterial iron transport related to virulence. Contrib Microbiol 2004, 12, 210–33. [DOI] [PubMed] [Google Scholar]
- (275).Cornelissen CN Subversion of nutritional immunity by the pathogenic Neisseriae. Pathog. Dis 2018, 76, ftx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Cornelissen CN; Hollander A TonB-Dependent Transporters Expressed by Neisseria gonorrhoeae. Front. Microbiol 2011, 2, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (277).Konopka K; Bindereif A; Neilands JB Aerobactin-mediated utilization of transferrin iron. Biochemistry 1982, 21, 6503–8. [DOI] [PubMed] [Google Scholar]
- (278).Tidmarsh GF; Klebba PE; Rosenberg LT Rapid release of iron from ferritin by siderophores. J. Inorg. Biochem 1983, 18, 161–8. [DOI] [PubMed] [Google Scholar]
- (279).Aisen P; Leibman A; Zweier J Stoichiometric and site characteristics of the binding of iron to human transferrin. J. Biol. Chem 1978, 253, 1930–7. [PubMed] [Google Scholar]
- (280).Bachman MA; Lenio S; Schmidt L; Oyler JE; Weiser JN Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 2012, 3, e00224–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).Bearden SW; Staggs TM; Perry RD An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol 1998, 180, 1135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).Caza M; Lepine F; Dozois CM Secretion, but not overall synthesis, of catecholate siderophores contributes to virulence of extraintestinal pathogenic Escherichia coli. Mol. Microbiol 2011, 80, 266–82. [DOI] [PubMed] [Google Scholar]
- (283).Russo TA; Olson R; Fang CT; Stoesser N; Miller M; MacDonald U; Hutson A; Barker JH; La Hoz RM; Johnson JR; et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol 2018, 56, 776–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (284).Russo TA; Olson R; MacDonald U; Beanan J; Davidson BA Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun 2015, 83, 3325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (285).Chakravorty S; Shipelskiy Y; Kumar A; Majumdar A; Yang T; Nairn BL; Newton SM; Klebba PE Universal fluorescent sensors of high-affinity iron transport, applied to ESKAPE pathogens. J. Biol. Chem 2019, 294, 4682–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (286).Zhang Y; Sen S; Giedroc DP Iron Acquisition by Bacterial Pathogens: Beyond Tris-Catecholate Complexes. ChemBioChem 2020, 21, 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (287).Guarner F; Malagelada JR Gut flora in health and disease. Lancet 2003, 361, 512–9. [DOI] [PubMed] [Google Scholar]
- (288).Lynch SV; Pedersen O The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med 2016, 375, 2369–2379. [DOI] [PubMed] [Google Scholar]
- (289).Shreiner AB; Kao JY; Young VB The gut microbiome in health and in disease. Curr. Opin. Gastroenterol 2015, 31, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (290).Klebba PE; Charbit A; Xiao Q; Jiang X; Newton SM Mechanisms of iron and haem transport by Listeria monocytogenes. Mol. Membr. Biol 2012, 29, 69–86. [DOI] [PubMed] [Google Scholar]
- (291).Abergel RJ; Moore EG; Strong RK; Raymond KN Microbial evasion of the immune system: structural modifications of enterobactin impair siderocalin recognition. J. Am. Chem. Soc 2006, 128, 10998–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (292).Karlinsey JE; Stepien TA; Mayho M; Singletary LA; Bingham-Ramos LK; Brehm MA; Greiner DL; Shultz LD; Gallagher LA; Bawn M; Kingsley RA; Libby SJ; Fang FC Genome-wide Analysis of Salmonella enterica serovar Typhi in Humanized Mice Reveals Key Virulence Features. Cell Host Microbe 2019, 26, 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (293).Peigne C; Bidet P; Mahjoub-Messai F; Plainvert C; Barbe V; Médigue C; Frapy E; Nassif X; Denamur E; Bingen E; Bonacorsi S The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun 2009, 77, 2272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (294).Sobieszczańska BM Distribution of genes encoding iron uptake systems among enteroaggregative Escherichia coli strains isolated from adults with irritable bowel syndrome. Clin. Microbiol. Infect 2008, 14, 1083–6. [DOI] [PubMed] [Google Scholar]
- (295).Matteoli FP; Passarelli-Araujo H; Pedrosa-Silva F; Olivares FL; Venancio TM Population structure and pangenome analysis of Enterobacter bugandensis uncover the presence of bla(CTX-M-55), bla(NDM-5) and bla(IMI-1), along with sophisticated iron acquisition strategies. Genomics 2020, 112, 1182–1191. [DOI] [PubMed] [Google Scholar]
- (296).Bäumler AJ; Tsolis RM; Van Der Velden AW; Stojiljkovic I; Anic S; Heffron F Identification of a new iron regulated locus of Salmonella typhi. Gene 1996, 183, 207–213. [DOI] [PubMed] [Google Scholar]
- (297).Hantke K; Nicholson G; Rabsch W; Winkelmann G Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 3677–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (298).Bister B; Bischoff D; Nicholson GJ; Valdebenito M; Schneider K; Winkelmann G; Hantke K; Süssmuth RD The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica §. BioMetals 2004, 17, 471–481. [DOI] [PubMed] [Google Scholar]
- (299).Zhu M; Valdebenito M; Winkelmann G; Hantke K Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 2005, 151, 2363–2372. [DOI] [PubMed] [Google Scholar]
- (300).Valax P; Georgiou G Molecular characterization of beta-lactamase inclusion bodies produced in Escherichia coli. 1. Composition. Biotechnol. Prog 1993, 9, 539–47. [DOI] [PubMed] [Google Scholar]
- (301).Sonnenborn U Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS microbiology letters 2016, 363, fnw212. [DOI] [PubMed] [Google Scholar]
- (302).Goerg KJ; Schlörer E [Probiotic therapy of pseudomembranous colitis. Combination of intestinal lavage and oral administration of Escherichia coli]. Dtsch. Med. Wochenschr 1998, 123, 1274–8. [DOI] [PubMed] [Google Scholar]
- (303).Kruis W; Schütz E; Fric P; Fixa B; Judmaier G; Stolte M Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther 1997, 11, 853–8. [DOI] [PubMed] [Google Scholar]
- (304).Rembacken BJ; Snelling AM; Hawkey PM; Chalmers DM; Axon AT Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 1999, 354, 635–9. [DOI] [PubMed] [Google Scholar]
- (305).Deriu E; Liu JZ; Pezeshki M; Edwards RA; Ochoa RJ; Contreras H; Libby SJ; Fang FC; Raffatellu M Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 2013, 14, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (306).Massip C; Branchu P; Bossuet-Greif N; Chagneau CV; Gaillard D; Martin P; Boury M; Sécher T; Dubois D; Nougayrède JP; Oswald E Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 2019, 15, No. e1008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (307).Klebba PE Regulation of the biosynthesis of the iron-related membrane proteins in Escherichia coli, 1981. [Google Scholar]
- (308).Van Tiel-Menkveld GJ; Mentjox-Vervuurt JM; Oudega B; de Graaf FK Siderophore production by Enterobacter cloacae and a common receptor protein for the uptake of aerobactin and cloacin DF13. J. Bacteriol 1982, 150, 490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (309).Struve C; Roe CC; Stegger M; Stahlhut SG; Hansen DS; Engelthaler DM; Andersen PS; Driebe EM; Keim P; Krogfelt KA Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio 2015, 6, No. e00630–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (310).Breurec S; Melot B; Hoen B; Passet V; Schepers K; Bastian S; Brisse S Liver Abscess Caused by Infection with Community-Acquired Klebsiella quasipneumoniae subsp. quasipneumoniae. Emerging Infect. Dis 2016, 22, 529–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (311).Lee IR; Molton JS; Wyres KL; Gorrie C; Wong J; Hoh CH; Teo J; Kalimuddin S; Lye DC; Archuleta S; Holt KE; Gan YH Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci. Rep 2016, 6, 29316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Cheng DL; Liu YC; Yen MY; Liu CY; Wang RS Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch. Intern. Med 1991, 151, 1557–1559. [PubMed] [Google Scholar]
- (313).Liu YC; Cheng DL; Lin CL Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med 1986, 146, 1913–1916. [PubMed] [Google Scholar]
- (314).Wang JH; Liu YC; Lee SS; Yen MY; Chen YS; Wang JH; Wann SR; Lin HH Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis 1998, 26, 1434–1438. [DOI] [PubMed] [Google Scholar]
- (315).Wang YY; Lee FY; Chang FY; Lee SD; Fung CP A vanishing liver abscess complicated with Klebsiella pneumoniae chest wall abscess: a case report. J. Microbiol Immunol Infect 1998, 31, 249–252. [PubMed] [Google Scholar]
- (316).Russo TA; Shon AS; Beanan JM; Olson R; MacDonald U; Pomakov AO; Visitacion MP Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One 2011, 6, No. e26734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Lam MMC; Wyres KL; Judd LM; Wick RR; Jenney A; Brisse S; Holt KE Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018, 10, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (318).Konopka K; Neilands JB Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 1984, 23, 2122–7. [DOI] [PubMed] [Google Scholar]
- (319).Fischbach MA; Lin H; Zhou L; Yu Y; Abergel RJ; Liu DR; Raymond KN; Wanner BL; Strong RK; Walsh CT; Aderem A; Smith KD The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 16502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (320).Searle LJ; Meric G; Porcelli I; Sheppard SK; Lucchini S Variation in siderophore biosynthetic gene distribution and production across environmental and faecal populations of Escherichia coli. PLoS One 2015, 10, No. e0117906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (321).Der Vartanian M Differences in excretion and efficiency of the aerobactin and enterochelin siderophores in a bovine pathogenic strain of Escherichia coli. Infect. Immun 1988, 56, 413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (322).Valvano MA; Crosa JH Aerobactin iron transport genes commonly encoded by certain ColV plasmids occur in the chromosome of a human invasive strain of Escherichia coli K1. Infect. Immun 1984, 46, 159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (323).Russo TA; Marr CM Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev 2019, 32, e00001–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (324).Bailey DC; Alexander E; Rice MR; Drake EJ; Mydy LS; Aldrich CC; Gulick AM Structural and functional delineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae. J. Biol. Chem 2018, 293, 7841–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (325).Russo TA; Olson R; Macdonald U; Metzger D; Maltese LM; Drake EJ; Gulick AM Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun 2014, 82, 2356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (326).Bachman MA; Oyler JE; Burns SH; Caza M; Lepine F; Dozois CM; Weiser JN Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun 2011, 79, 3309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (327).Magistro G; Hoffmann C; Schubert S The salmochelin receptor IroN itself, but not salmochelin-mediated iron uptake promotes biofilm formation in extraintestinal pathogenic Escherichia coli (ExPEC). Int. J. Med. Microbiol 2015, 305, 435–445. [DOI] [PubMed] [Google Scholar]
- (328).Gehring AM; DeMoll E; Fetherston JD; Mori I; Mayhew GF; Blattner FR; Walsh CT; Perry RD Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol 1998, 5, 573–86. [DOI] [PubMed] [Google Scholar]
- (329).Haag H; Hantke K; Drechsel H; Stojiljkovic I; Jung G; Zahner H Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J. Gen. Microbiol 1993, 139, 2159–65. [DOI] [PubMed] [Google Scholar]
- (330).Forman S; Paulley JT; Fetherston JD; Cheng YQ; Perry RD Yersinia ironomics: comparison of iron transporters among Yersinia pestis biotypes and its nearest neighbor, Yersinia pseudotuberculosis. BioMetals 2010, 23, 275–94. [DOI] [PubMed] [Google Scholar]
- (331).O’Connor L; Fetherston JD; Perry RD The feoABC Locus of Yersinia pestis Likely Has Two Promoters Causing Unique Iron Regulation. Front. Cell. Infect. Microbiol 2017, 7, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (332).Perry RD; Fetherston JD Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (333).Bobrov AG; Kirillina O; Fetherston JD; Miller MC; Burlison JA; Perry RD The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol. Microbiol 2014, 93, 759–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (334).Fetherston JD; Kirillina O; Bobrov AG; Paulley JT; Perry RD The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun 2010, 78, 2045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (335).Garcia EC; Brumbaugh AR; Mobley HL Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun 2011, 79, 1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (336).Brumbaugh AR; Smith SN; Mobley HL Immunization with the yersiniabactin receptor, FyuA, protects against pyelonephritis in a murine model of urinary tract infection. Infect. Immun 2013, 81, 3309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (337).Vigil PD; Stapleton AE; Johnson JR; Hooton TM; Hodges AP; He Y; Mobley HL Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio 2011, 2, No. e00066–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (338).Chaturvedi KS; Hung CS; Giblin DE; Urushidani S; Austin AM; Dinauer MC; Henderson JP Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem. Biol 2014, 9, 551–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (339).Cornelis P; Dingemans J Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol 2013, 3, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (340).Fang Z; Sampson SL; Warren RM; Gey van Pittius NC; Newton-Foot M Iron acquisition strategies in mycobacteria. Tuberculosis (Oxford, U. K.) 2015, 95, 123–30. [DOI] [PubMed] [Google Scholar]
- (341).Lemos ML; Osorio CR Heme, an iron supply for vibrios pathogenic for fish. BioMetals 2007, 20, 615–26. [DOI] [PubMed] [Google Scholar]
- (342).Lyles KV; Eichenbaum Z From Host Heme To Iron: The Expanding Spectrum of Heme Degrading Enzymes Used by Pathogenic Bacteria. Front. Cell. Infect. Microbiol 2018, 8, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (343).Richard KL; Kelley BR; Johnson JG Heme Uptake and Utilization by Gram-Negative Bacterial Pathogens. Front. Cell. Infect. Microbiol 2019, 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (344).Siudeja K; Olczak T [Mechanisms and regulation of iron and heme utilization in Gram-negative bacteria]. Postepy Biochem. 2005, 51, 198–208. [PubMed] [Google Scholar]
- (345).Tong Y; Guo M Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys 2009, 481, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (346).de Léséleuc L; Harris G; KuoLee R; Xu HH; Chen W Serum resistance, gallium nitrate tolerance and extrapulmonary dissemination are linked to heme consumption in a bacteremic strain of Acinetobacter baumannii. Int. J. Med. Microbiol 2014, 304, 360–9. [DOI] [PubMed] [Google Scholar]
- (347).Wilks A; Ikeda-Saito M Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc. Chem. Res 2014, 47, 2291–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (348).Jin B; Newton SM; Shao Y; Jiang X; Charbit A; Klebba PE Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol. Microbiol 2006, 59, 1185–1198. [DOI] [PubMed] [Google Scholar]
- (349).Kokesová A; Frolová L; Kverka M; Sokol D; Rossmann P; Bártová J; Tlaskalová-Hogenová H Oral administration of probiotic bacteria (E. coli Nissle, E. coli O83, Lactobacillus casei) influences the severity of dextran sodium sulfate-induced colitis in BALB/c mice. Folia Microbiol. (Dordrecht, Neth.) 2006, 51, 478–84. [DOI] [PubMed] [Google Scholar]
- (350).Blattner FR; Plunkett G 3rd; Bloch CA; Perna NT; Burland V; Riley M; Collado-Vides J; Glasner JD; Rode CK; Mayhew GF; Gregor J; Davis NW; Kirkpatrick HA; Goeden MA; Rose DJ; Mau B; Shao Y The complete genome sequence of Escherichia coli K-12. Science (Washington, DC, U. S.) 1997, 277, 1453–62. [DOI] [PubMed] [Google Scholar]
- (351).Nash JH; Villegas A; Kropinski AM; Aguilar-Valenzuela R; Konczy P; Mascarenhas M; Ziebell K; Torres AG; Karmali MA; Coombes BK Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics 2010, 11, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (352).Chhibber S; Aggarwal S; Yadav V Contribution of capsular and lipopolysaccharide antigens to the pathogenesis of Klebsiella pneumoniae respiratory tract infection. Folia Microbiol. (Dordrecht, Neth.) 2003, 48, 699–702. [DOI] [PubMed] [Google Scholar]
- (353).Evrard B; Balestrino D; Dosgilbert A; Bouya-Gachancard JL; Charbonnel N; Forestier C; Tridon A Roles of capsule and lipopolysaccharide O antigen in interactions of human monocyte-derived dendritic cells and Klebsiella pneumoniae. Infect. Immun 2010, 78, 210–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (354).Fresno S; Jimenez N; Canals R; Merino S; Corsaro MM; Lanzetta R; Parrilli M; Pieretti G; Regue M; Tomas JM A second galacturonic acid transferase is required for core lipopolysaccharide biosynthesis and complete capsule association with the cell surface in Klebsiella pneumoniae. J. Bacteriol 2007, 189, 1128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (355).Hetem DJ; Bootsma MCJ; Troelstra A; Bonten MJM Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus. Emerging Infect. Dis 2013, 19, 1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (356).Hsieh PF; Lin TL; Yang FL; Wu MC; Pan YJ; Wu SH; Wang JT Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 2012, 7, No. e33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (357).Mehling JS; Lavender H; Clegg S A Dam methylation mutant of Klebsiella pneumoniae is partially attenuated. FEMS Microbiol. Lett 2007, 268, 187–93. [DOI] [PubMed] [Google Scholar]
- (358).Lawlor MS; O’Connor C; Miller VL Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun 2007, 75, 1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (359).Munoz-Price LS; Poirel L; Bonomo RA; Schwaber MJ; Daikos GL; Cormican M; Cornaglia G; Garau J; Gniadkowski M; Hayden MK; Kumarasamy K; Livermore DM; Maya JJ; Nordmann P; Patel JB; Paterson DL; Pitout J; Villegas MV; Wang H; Woodford N; Quinn JP Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis 2013, 13, 785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (360).Tumbarello M; Viale P; Viscoli C; Trecarichi EM; Tumietto F; Marchese A; Spanu T; Ambretti S; Ginocchio F; Cristini F; Losito AR; Tedeschi S; Cauda R; Bassetti M Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis 2012, 55, 943–50. [DOI] [PubMed] [Google Scholar]
- (361).Iwasaki Y; Inokuchi R; Harada S; Aoki K; Ishii Y; Shinohara K Bacterial Meningitis Caused by Hypervirulent Klebsiella pneumoniae Capsular Genotype K54 with Development of Granuloma-like Nodal Enhancement in the Brain during the Subacute Phase. Intern. Med 2017, 56, 373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (362).Kwon JM; Jung HL; Shim JW; Kim DS; Shim JY; Park MS Klebsiella pneumoniae liver abscess in an immunocompetent child. Korean J. Pediatr 2013, 56, 407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (363).Ng D; Frazee B Necrotizing fasciitis caused by hypermucoviscous Klebsiella pneumoniae in a Filipino female in North America. West J. Emerg Med 2015, 16, 165–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (364).Prokesch BC; TeKippe M; Kim J; Raj P; TeKippe EM; Greenberg DE Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis 2016, 16, e190–e195. [DOI] [PubMed] [Google Scholar]
- (365).Takahashi K; Miura A; Yamaguchi T; Kanematsu M Novel cord-like structures on MRI in a case of hypervirulent Klebsiella pneumoniae. Intern. Med 2015, 54, 355–6. [DOI] [PubMed] [Google Scholar]
- (366).Vandevelde A; Stepanovic B On a Boat: A Case in Australia of Endophthalmitis and Pyogenic Liver, Prostatic, and Lung Abscesses in a Previously Well Patient due to Klebsiella pneumoniae. Case Rep. Infect Dis 2014, 2014, 137248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (367).Paczosa MK; Mecsas J Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev 2016, 80, 629–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (368).Miethke M; Marahiel MA Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev 2007, 71, 413–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (369).Brock JH; Williams PH; Licéaga J; Wooldridge KG Relative availability of transferrin-bound iron and cell-derived iron to aerobactin-producing and enterochelin-producing strains of Escherichia coli and to other microorganisms. Infect. Immun 1991, 59, 3185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (370).Lai YC; Peng HL; Chang HY Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun 2001, 69, 7140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (371).Lawlor MS; O’Connor C; Miller VL Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun 2007, 75, 1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (372).Wasserman SI; Soter NA; Center DM; Austen KF Cold urticaria. Recognition and characterization of a neutrophil chemotactic factor which appears in serum during experimental cold challenge. J. Clin. Invest 1977, 60, 189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (373).Heesemann J; Hantke K; Vocke T; Saken E; Rakin A; Stojiljkovic I; Berner R Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol. Microbiol 1993, 8, 397–408. [DOI] [PubMed] [Google Scholar]
- (374).Perry RD; Balbo PB; Jones HA; Fetherston JD; DeMoll E Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 1999, 145, 1181–1190. [DOI] [PubMed] [Google Scholar]
- (375).Fetherston JD; Bertolino VJ; Perry RD YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol 1999, 32, 289–99. [DOI] [PubMed] [Google Scholar]
- (376).Koh EI; Hung CS; Henderson JP The Yersiniabactin-Associated ATP Binding Cassette Proteins YbtP and YbtQ Enhance Escherichia coli Fitness during High-Titer Cystitis. Infect. Immun 2016, 84, 1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (377).Gibson F; Magrath DI The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim. Biophys. Acta, Gen. Subj 1969, 192, 175–84. [DOI] [PubMed] [Google Scholar]
- (378).Vernet V; Philippon A; Madoulet C; Vistelle R; Jaussaud R; Chippaux C Virulence factors (aerobactin and mucoid phenotype) in Klebsiella pneumoniae and Escherichia coli blood culture isolates. FEMS Microbiol. Lett 1995, 130, 51–57. [DOI] [PubMed] [Google Scholar]
- (379).Koczura R; Kaznowski A Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog 2003, 35, 197–202. [DOI] [PubMed] [Google Scholar]
- (380).Warner PJ; Williams PH; Bindereif A; Neilands JB ColV plasmid-specific aerobactin synthesis by invasive strains of Escherichia coli. Infect. Immun 1981, 33, 540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (381).Russo TA; Gulick AM Aerobactin Synthesis Proteins as Antivirulence Targets in Hypervirulent Klebsiella pneumoniae. ACS Infect. Dis 2019, 5, 1052–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (382).Oudega B; van der Molen J; de Graaf FK In vitro binding of cloacin DF13 to its purified outer membrane receptor protein and effect of peptidoglycan on bacteriocin-receptor interaction. J. Bacteriol 1979, 140, 964–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (383).Russo TA; Olson R; Macdonald U; Metzger D; Maltese LM; Drake EJ; Gulick AM Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun 2014, 82, 2356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (384).Ayobami O; Willrich N; Harder T; Okeke IN; Eckmanns T; Markwart R The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerging Microbes Infect. 2019, 8, 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (385).Wong D; Nielsen TB; Bonomo RA; Pantapalangkoor P; Luna B; Spellberg B Clinical and Pathophysiological Overview of Acinetobacter Infections: a Century of Challenges. Clin. Microbiol. Rev 2017, 30, 409–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (386).Actis LA; Tolmasky ME; Crosa LM; Crosa JH Effect of iron-limiting conditions on growth of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol 1993, 31, 2812–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (387).Daniel C; Haentjens S; Bissinger MC; Courcol RJ Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol. Lett 1999, 170, 199–209. [DOI] [PubMed] [Google Scholar]
- (388).Dorsey CW; Beglin MS; Actis LA Detection and analysis of iron uptake components expressed by Acinetobacter baumannii clinical isolates. Journal of clinical microbiology 2003, 41, 4188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (389).Goel VK; Kapil A; Das B; Rao DN Influence of iron on growth and extracellular products of Acinetobacter baumannii. Jpn. J. Med. Sci. Biol 1998, 51, 25–33. [DOI] [PubMed] [Google Scholar]
- (390).Yamamoto S; Okujo N; Sakakibara Y Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch. Microbiol 1994, 162, 249–54. [DOI] [PubMed] [Google Scholar]
- (391).Eijkelkamp BA; Hassan KA; Paulsen IT; Brown MH Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 2011, 12, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (392).Nwugo CC; Gaddy JA; Zimbler DL; Actis LA Deciphering the iron response in Acinetobacter baumannii: A proteomics approach. J. Proteomics 2011, 74, 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (393).Runci F; Gentile V; Frangipani E; Rampioni G; Leoni L; Lucidi M; Visaggio D; Harris G; Chen W; Stahl J; Averhoff B; Visca P Contribution of Active Iron Uptake to Acinetobacter baumannii Pathogenicity. Infect. Immun 2019, 87, e00755–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (394).Mihara K; Tanabe T; Yamakawa Y; Funahashi T; Nakao H; Narimatsu S; Yamamoto S Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 2004, 150, 2587–97. [DOI] [PubMed] [Google Scholar]
- (395).De Silva PM; Chong P; Fernando DM; Westmacott G; Kumar A Effect of Incubation Temperature on Antibiotic Resistance and Virulence Factors of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother 2018, 62, e01514–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (396).Jacobs AC; Sayood K; Olmsted SB; Blanchard CE; Hinrichs S; Russell D; Dunman PM Characterization of the Acinetobacter baumannii growth phase-dependent and serum responsive transcriptomes. FEMS Immunol. Med. Microbiol 2012, 64, 403–12. [DOI] [PubMed] [Google Scholar]
- (397).Tuttobene MR; Cribb P; Mussi MA BlsA integrates light and temperature signals into iron metabolism through Fur in the human pathogen Acinetobacter baumannii. Sci. Rep 2018, 8, 7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (398).Penwell WF; DeGrace N; Tentarelli S; Gauthier L; Gilbert CM; Arivett BA; Miller AA; Durand-Reville TF; Joubran C; Actis LA Discovery and Characterization of New Hydroxamate Siderophores, Baumannoferrin A and B, produced by Acinetobacter baumannii. ChemBioChem 2015, 16, 1896. [DOI] [PubMed] [Google Scholar]
- (399).Proschak A; Lubuta P; Grun P; Lohr F; Wilharm G; De Berardinis V; Bode HB Structure and biosynthesis of fimsbactins A-F, siderophores from Acinetobacter baumannii and Acinetobacter baylyi. ChemBioChem 2013, 14, 633–8. [DOI] [PubMed] [Google Scholar]
- (400).Antunes LC; Imperi F; Towner KJ; Visca P Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol 2011, 162, 279–84. [DOI] [PubMed] [Google Scholar]
- (401).Dorsey CW; Tolmasky ME; Crosa JH; Actis LA Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology 2003, 149, 1227–38. [DOI] [PubMed] [Google Scholar]
- (402).Echenique JR; Arienti H; Tolmasky ME; Read RR; Staneloni RJ; Crosa JH; Actis LA Characterization of a high-affinity iron transport system in Acinetobacter baumannii. J. Bacteriol 1992, 174, 7670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (403).Yakkala H; Samantarrai D; Gribskov M; Siddavattam D Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLoS One 2019, 14, No. e0218204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (404).Vallenet D; Nordmann P; Barbe V; Poirel L; Mangenot S; Bataille E; Dossat C; Gas S; Kreimeyer A; Lenoble P; Oztas S; Poulain J; Segurens B; Robert C; Abergel C; Claverie JM; Raoult D; Médigue C; Weissenbach J; Cruveiller S Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 2008, 3, No. e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (405).Dorsey CW; Tomaras AP; Connerly PL; Tolmasky ME; Crosa JH; Actis LA The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology 2004, 150, 3657–67. [DOI] [PubMed] [Google Scholar]
- (406).Wuest WM; Sattely ES; Walsh CT Three siderophores from one bacterial enzymatic assembly line. J. Am. Chem. Soc 2009, 131, 5056–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (407).Penwell WF; Arivett BA; Actis LA The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS One 2012, 7, No. e36493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (408).Shapiro JA; Wencewicz TA Acinetobactin Isomerization Enables Adaptive Iron Acquisition in Acinetobacter baumannii through pH-Triggered Siderophore Swapping. ACS Infect. Dis 2016, 2, 157–68. [DOI] [PubMed] [Google Scholar]
- (409).Harding CM; Hennon SW; Feldman MF Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol 2018, 16, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (410).Bailey DC; Bohac TJ; Shapiro JA; Giblin DE; Wencewicz TA; Gulick AM Crystal Structure of the Siderophore Binding Protein BauB Bound to an Unusual 2:1 Complex Between Acinetobactin and Ferric Iron. Biochemistry 2018, 57, 6653–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (411).Bohac TJ; Fang L; Giblin DE; Wencewicz TA Fimsbactin and Acinetobactin Compete for the Periplasmic Siderophore Binding Protein BauB in Pathogenic Acinetobacter baumannii. ACS Chem. Biol 2019, 14, 674–687. [DOI] [PubMed] [Google Scholar]
- (412).Shapiro JA; Wencewicz TA Structure-function studies of acinetobactin analogs. Metallomics: integrated biometal science 2017, 9, 463–470. [DOI] [PubMed] [Google Scholar]
- (413).Moynié L; Serra I; Scorciapino MA; Oueis E; Page MG; Ceccarelli M; Naismith JH Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogen Acinetobacter baumannii. eLife 2018, 7, e42270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (414).Subashchandrabose S; Mobley HL Back to the metal age: battle for metals at the host-pathogen interface during urinary tract infection. Metallomics: integrated biometal science 2015, 7, 935–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (415).Gaddy JA; Arivett BA; McConnell MJ; Lopez-Rojas R; Pachon J; Actis LA Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun 2012, 80, 1015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (416).Russo TA; MacDonald U The Galleria mellonella Infection Model Does Not Accurately Differentiate between Hypervirulent and Classical Klebsiella pneumoniae. mSphere 2020, 5, e00850–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (417).Fleming ID; Krezalek MA; Belogortseva N; Zaborin A; Defazio J; Chandrasekar L; Actis LA; Zaborina O; Alverdy JC Modeling Acinetobacter baumannii wound infections: The critical role of iron. J. Trauma Acute Care Surg 2017, 82, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (418).Wilson BR; Bogdan AR; Miyazawa M; Hashimoto K; Tsuji Y Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med 2016, 22, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (419).de Léséleuc L; Harris G; KuoLee R; Chen W In vitro and in vivo biological activities of iron chelators and gallium nitrate against Acinetobacter baumannii. Antimicrob. Agents Chemother 2012, 56, 5397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (420).Okujo N; Sakakibara Y; Yoshida T; Yamamoto S Structure of acinetoferrin, a new citrate-based dihydroxamate siderophore from Acinetobacter haemolyticus. BioMetals 1994, 7, 170–176. [DOI] [PubMed] [Google Scholar]
- (421).Rohrbach MR; Braun V; Koster W Ferrichrome transport in Escherichia coli K-12: altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. Journal of bacteriology 1995, 177, 7186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (422).Thulasiraman P; Newton SM; Xu J; Raymond KN; Mai C; Hall A; Montague MA; Klebba PE Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J. Bacteriol 1998, 180, 6689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (423).Fiester SE; Arivett BA; Schmidt RE; Beckett AC; Ticak T; Carrier MV; Ghosh R; Ohneck EJ; Metz ML; Sellin Jeffries MK; Actis LA Iron-Regulated Phospholipase C Activity Contributes to the Cytolytic Activity and Virulence of Acinetobacter baumannii. PLoS One 2016, 11, No. e0167068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (424).Camarena L; Bruno V; Euskirchen G; Poggio S; Snyder M Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010, 6, No. e1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (425).Jacobs AC; Hood I; Boyd KL; Olson PD; Morrison JM; Carson S; Sayood K; Iwen PC; Skaar EP; Dunman PM Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun 2010, 78, 1952–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (426).Stahl J; Bergmann H; Göttig S; Ebersberger I; Averhoff B Acinetobacter baumannii Virulence Is Mediated by the Concerted Action of Three Phospholipases D. PLoS One 2015, 10, No. e0138360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (427).Choi SR; Britigan BE; Narayanasamy P Iron/Heme Metabolism-Targeted Gallium(III) Nanoparticles Are Active against Extracellular and Intracellular Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2019, 63, e02643–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (428).de Leseleuc L; Harris G; KuoLee R; Xu HH; Chen W Serum resistance, gallium nitrate tolerance and extrapulmonary dissemination are linked to heme consumption in a bacteremic strain of Acinetobacter baumannii. Int. J. Med. Microbiol 2014, 304, 360–9. [DOI] [PubMed] [Google Scholar]
- (429).Giardina BJ; Shahzad S; Huang W; Wilks A Heme uptake and utilization by hypervirulent Acinetobacter baumannii LAC-4 is dependent on a canonical heme oxygenase (abHemO). Arch. Biochem. Biophys 2019, 672, 108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (430).Zimbler DL; Penwell WF; Gaddy JA; Menke SM; Tomaras AP; Connerly PL; Actis LA Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. BioMetals 2009, 22, 23–32. [DOI] [PubMed] [Google Scholar]
- (431).Subashchandrabose S; Smith S; DeOrnellas V; Crepin S; Kole M; Zahdeh C; Mobley HL Acinetobacter baumannii Genes Required for Bacterial Survival during Bloodstream Infection. mSphere 2016, 1, e00013–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (432).Moynié L; Luscher A; Rolo D; Pletzer D; Tortajada A; Weingart H; Braun Y; Page MG; Naismith JH; Köhler T Structure and Function of the PiuA and PirA Siderophore-Drug Receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother 2017, 61, e02531–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (433).Catel-Ferreira M; Marti S; Guillon L; Jara L; Coadou G; Molle V; Bouffartigues E; Bou G; Shalk I; Jouenne T; Vila-Farrés X; Dé E The outer membrane porin OmpW of Acinetobacter baumannii is involved in iron uptake and colistin binding. FEBS Lett. 2016, 590, 224–31. [DOI] [PubMed] [Google Scholar]
- (434).Zimbler DL; Arivett BA; Beckett AC; Menke SM; Actis LA Functional features of TonB energy transduction systems of Acinetobacter baumannii. Infect. Immun 2013, 81, 3382–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (435).Álvarez-Fraga L; Vázquez-Ucha JC; Martínez-Guitián M; Vallejo JA; Bou G; Beceiro A; Poza M Pneumonia infection in mice reveals the involvement of the feoA gene in the pathogenesis of Acinetobacter baumannii. Virulence 2018, 9, 496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (436).Newton SM; Klebba PE; Raynaud C; Shao Y; Jiang X; Dubail I; Archer C; Frehel C; Charbit A The svpA-srtB locus of Listeria monocytogenes: Fur-mediated iron regulation and effect on virulence. Mol. Microbiol 2005, 55, 927–940. [DOI] [PubMed] [Google Scholar]
- (437).Lamont IL; Konings AF; Reid DW Iron acquisition by Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. BioMetals 2009, 22, 53–60. [DOI] [PubMed] [Google Scholar]
- (438).Peek ME; Bhatnagar A; McCarty NA; Zughaier SM Pyoverdine, the Major Siderophore in Pseudomonas aeruginosa, Evades NGAL Recognition. Interdiscip. Perspect. Infect. Dis 2012, 2012, 843509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (439).Brandel J; Humbert N; Elhabiri M; Schalk IJ; Mislin GL; Albrecht-Gary AM Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans 2012, 41, 2820–34. [DOI] [PubMed] [Google Scholar]
- (440).Dumas Z; Ross-Gillespie A; Kummerli R Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc. R. Soc. London, Ser. B 2013, 280, 20131055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (441).Albrecht-Gary AM; Blanc S; Rochel N; Ocaktan AZ; Abdallah MA Bacterial iron transport: coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem 1994, 33, 6391–6402. [Google Scholar]
- (442).Cezard C; Farvacques N; Sonnet P Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr. Med. Chem 2014, 22, 165–86. [DOI] [PubMed] [Google Scholar]
- (443).Meyer JM; Neely A; Stintzi A; Georges C; Holder IA Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infection and immunity 1996, 64, 518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (444).Xiao R; Kisaalita WS Iron acquisition from transferrin and lactoferrin by Pseudomonas aeruginosa pyoverdin. Microbiology 1997, 143, 2509–15. [DOI] [PubMed] [Google Scholar]
- (445).Teintze M; Hossain MB; Barnes CL; Leong J; van der Helm D Structure of ferric pseudobactin, a siderophore from a plant growth promoting Pseudomonas. Biochemistry 1981, 20, 6446–57. [DOI] [PubMed] [Google Scholar]
- (446).Teintze M; Leong J Structure of pseudobactin A, a second siderophore from plant growth promoting Pseudomonas B10. Biochemistry 1981, 20, 6457–62. [DOI] [PubMed] [Google Scholar]
- (447).Xiong YQ; Vasil ML; Johnson Z; Ochsner UA; Bayer AS The oxygen- and iron-dependent sigma factor pvdS of Pseudomonas aeruginosa is an important virulence factor in experimental infective endocarditis. J. Infect. Dis 2000, 181, 1020–6. [DOI] [PubMed] [Google Scholar]
- (448).Minandri F; Imperi F; Frangipani E; Bonchi C; Visaggio D; Facchini M; Pasquali P; Bragonzi A; Visca P Role of Iron Uptake Systems in Pseudomonas aeruginosa Virulence and Airway Infection. Infect. Immun 2016, 84, 2324–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (449).Lamont IL; Beare PA; Ochsner U; Vasil AI; Vasil ML Siderophore-mediated signaling regulates virulence factor production in Pseudomonasaeruginosa. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 7072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (450).Gi M; Lee KM; Kim SC; Yoon JH; Yoon SS; Choi JY A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci. Rep 2015, 5, 14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (451).Becker R; Grun M; Scholz G Nicotianamine and the distribution of iron into the apoplasm and symplasm of tomato (Lycopersicon esculentum Mill.): I. Determination of the apoplasmic and symplasmic iron pools in roots and leaves of the cultivar Bonner Beste and its nicotianamine-less mutant chloronerva. Planta 1992, 187, 48–52. [DOI] [PubMed] [Google Scholar]
- (452).Takahashi M; Terada Y; Nakai I; Nakanishi H; Yoshimura E; Mori S; Nishizawa NK Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 2003, 15, 1263–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (453).Dean CR; Poole K Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol 1993, 8, 1095–103. [DOI] [PubMed] [Google Scholar]
- (454).Dean CR; Poole K Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol 1993, 175, 317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (455).Cornelis P; Bodilis J A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Rep 2009, 1, 256–62. [DOI] [PubMed] [Google Scholar]
- (456).Llamas MA; Sparrius M; Kloet R; Jimenez CR; Vandenbroucke-Grauls C; Bitter W The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol 2006, 188, 1882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (457).Ó Cuiv P; Keogh D; Clarke P; O’Connell M FoxB of Pseudomonas aeruginosa functions in the utilization of the xenosiderophores ferrichrome, ferrioxamine B, and schizokinen: evidence for transport redundancy at the inner membrane. J. Bacteriol 2007, 189, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (458).Banin E; Lozinski A; Brady KM; Berenshtein E; Butterfield PW; Moshe M; Chevion M; Greenberg EP; Banin E The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 16761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (459).Llamas MA; Mooij MJ; Sparrius M; Vandenbroucke-Grauls CM; Ratledge C; Bitter W Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol. Microbiol 2008, 67, 458–72. [DOI] [PubMed] [Google Scholar]
- (460).Marshall B; Stintzi A; Gilmour C; Meyer JM; Poole K Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 2009, 155, 305–315. [DOI] [PubMed] [Google Scholar]
- (461).Cuiv PO; Clarke P; O’Connell M Identification and characterization of an iron-regulated gene, chtA, required for the utilization of the xenosiderophores aerobactin, rhizobactin 1021 and schizokinen by Pseudomonas aeruginosa. Microbiology 2006, 152, 945–54. [DOI] [PubMed] [Google Scholar]
- (462).Elias S; Degtyar E; Banin E FvbA is required for vibriobactin utilization in Pseudomonas aeruginosa. Microbiology 2011, 157, 2172–2180. [DOI] [PubMed] [Google Scholar]
- (463).Galet J; Deveau A; Hotel L; Frey-Klett P; Leblond P; Aigle B Pseudomonas fluorescens pirates both ferrioxamine and ferricoelichelin siderophores from Streptomyces ambofaciens. Appl. Environ. Microbiol 2015, 81, 3132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (464).Ochsner UA; Johnson Z; Vasil ML Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 2000, 146, 185–198. [DOI] [PubMed] [Google Scholar]
- (465).Letoffe S; Redeker V; Wandersman C Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol. Microbiol 1998, 28, 1223–34. [DOI] [PubMed] [Google Scholar]
- (466).Wandersman C; Delepelaire P Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol 2004, 58, 611–47. [DOI] [PubMed] [Google Scholar]
- (467).Wandersman C; Delepelaire P Haemophore functions revisited. Mol. Microbiol 2012, 85, 618–31. [DOI] [PubMed] [Google Scholar]
- (468).Smith AD; Modi AR; Sun S; Dawson JH; Wilks A Spectroscopic Determination of Distinct Heme Ligands in Outer- Membrane Receptors PhuR and HasR of Pseudomonas aeruginosa. Biochemistry 2015, 54, 2601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (469).Smith AD; Wilks A Differential contributions of the outer membrane receptors PhuR and HasR to heme acquisition in Pseudomonas aeruginosa. J. Biol. Chem 2015, 290, 7756–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (470).Otero-Asman JR; Garcia-Garcia AI; Civantos C; Quesada JM; Llamas MA Pseudomonas aeruginosa possesses three distinct systems for sensing and using the host molecule haem. Environ. Microbiol 2019, 21, 4629–4647. [DOI] [PubMed] [Google Scholar]
- (471).Macedo MF; Cruz E; Lacerda R; Porto G; de Sousa M Low serum transferrin levels in HFE C282Y homozygous subjects are associated with low CD8(+) T lymphocyte numbers. Blood Cells, Mol., Dis 2005, 35, 319–25. [DOI] [PubMed] [Google Scholar]
- (472).Stites SW; Plautz MW; Bailey K; O’Brien-Ladner AR; Wesselius LJ Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med 1999, 160, 796–801. [DOI] [PubMed] [Google Scholar]
- (473).Dehner C; Morales-Soto N; Behera RK; Shrout J; Theil EC; Maurice PA; Dubois JL Ferritin and ferrihydrite nanoparticles as iron sources for Pseudomonas aeruginosa. JBIC, J. Biol. Inorg. Chem 2013, 18, 371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (474).Zimmermann L; Hantke K; Braun V Exogenous induction of the iron dicitrate transport system of Escherichia coli K-12. J. Bacteriol 1984, 159, 271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (475).Harle C; Kim I; Angerer A; Braun V Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995, 14, 1430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (476).Brillet K; Journet L; Celia H; Paulus L; Stahl A; Pattus F; Cobessi D A beta strand lock exchange for signal transduction in TonB-dependent transducers on the basis of a common structural motif. Structure (Oxford, U. K.) 2007, 15, 1383–91. [DOI] [PubMed] [Google Scholar]
- (477).Wirth C; Meyer-Klaucke W; Pattus F; Cobessi D From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor. J. Mol. Biol 2007, 368, 398–406. [DOI] [PubMed] [Google Scholar]
- (478).Cunliffe HE; Merriman TR; Lamont IL Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. Journal of bacteriology 1995, 177, 2744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (479).Redly GA; Poole K Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol 2003, 185, 1261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (480).Beare PA; For RJ; Martin LW; Lamont IL Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol 2003, 47, 195–207. [DOI] [PubMed] [Google Scholar]
- (481).Llamas MA; Imperi F; Visca P; Lamont IL Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev. 2014, 38, 569–97. [DOI] [PubMed] [Google Scholar]
- (482).Flemming HC; Wingender J The biofilm matrix. Nat. Rev. Microbiol 2010, 8, 623–33. [DOI] [PubMed] [Google Scholar]
- (483).Kang D; Kirienko DR; Webster P; Fisher AL; Kirienko NV Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (484).Kang D; Kirienko NV High-Throughput Genetic Screen Reveals that Early Attachment and Biofilm Formation Are Necessary for Full Pyoverdine Production by Pseudomonas aeruginosa. Front. Microbiol 2017, 8, 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (485).Alhede M; Bjarnsholt T; Givskov M; Alhede M Pseudomonas aeruginosa biofilms: mechanisms of immune evasion. Adv. Appl. Microbiol 2014, 86, 1–40. [DOI] [PubMed] [Google Scholar]
- (486).Banin E; Vasil ML; Greenberg EP Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 11076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (487).Banin E; Brady KM; Greenberg EP Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol 2006, 72, 2064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (488).Andrews SC; Robinson AK; Rodriguez-Quinones F Bacterial iron homeostasis. FEMS Microbiol Rev. 2003, 27, 215–37. [DOI] [PubMed] [Google Scholar]
- (489).Hunter RC; Klepac-Ceraj V; Lorenzi MM; Grotzinger H; Martin TR; Newman DK Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am. J. Respir. Cell Mol. Biol 2012, 47, 738–45. [DOI] [PubMed] [Google Scholar]
- (490).Hunter RC; Asfour F; Dingemans J; Osuna BL; Samad T; Malfroot A; Cornelis P; Newman DK Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. mBio 2013, 4, e00557–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (491).Cowan SW; Garavito RM; Jansonius JN; Jenkins JA; Karlsson R; Konig N; Pai EF; Pauptit RA; Rizkallah PJ; Rosenbusch JP; et al. The structure of OmpF porin in a tetragonal crystal form. Structure (Oxford, U. K.) 1995, 3, 1041–50. [DOI] [PubMed] [Google Scholar]
- (492).Nakashige TG; Zhang B; Krebs C; Nolan EM Human calprotectin is an iron-sequestering host-defense protein. Nat. Chem. Biol 2015, 11, 765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (493).Zygiel EM; Nelson CE; Brewer LK; Oglesby-Sherrouse AG; Nolan EM The human innate immune protein calprotectin induces iron starvation responses in Pseudomonas aeruginosa. J. Biol. Chem 2019, 294, 3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (494).Zygiel EM; Nolan EM Exploring Iron Withholding by the Innate Immune Protein Human Calprotectin. Acc. Chem. Res 2019, 52, 2301–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (495).Bonneau A; Roche B; Schalk IJ Iron acquisition in Pseudomonas aeruginosa by the siderophore pyoverdine: an intricate interacting network including periplasmic and membrane proteins. Sci. Rep 2020, 10, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (496).Brillet K; Ruffenach F; Adams H; Journet L; Gasser V; Hoegy F; Guillon L; Hannauer M; Page A; Schalk IJ An ABC transporter with two periplasmic binding proteins involved in iron acquisition in Pseudomonas aeruginosa. ACS Chem. Biol 2012, 7, 2036–45. [DOI] [PubMed] [Google Scholar]
- (497).Vigouroux A; Aumont-Nicaise M; Boussac A; Marty L; Lo Bello L; Legrand P; Brillet K; Schalk IJ; Morera S A unique ferrous iron binding mode is associated with large conformational changes for the transport protein FpvC of Pseudomonas aeruginosa. FEBS J. 2020, 287, 295–309. [DOI] [PubMed] [Google Scholar]
- (498).Kirienko NV; Ausubel FM; Ruvkun G Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 1821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (499).Pollitzer R Plague; WHO, Geneva, 1954; pp 409–482. [Google Scholar]
- (500).Perry RD; Fetherston JD Yersinia pestis–etiologic agent of plague. Clin. Microbiol. Rev 1997, 10, 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (501).Hinnebusch B; Erickson D Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. In Bacterial Biofilms; Springer, 2008; pp 229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (502).Fetherston JD; Kirillina O; Bobrov AG; Paulley JT; Perry RD The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun 2010, 78, 2045–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (503).Guerinot ML Microbial iron transport. Annu. Rev. Microbiol 1994, 48, 743–772. [DOI] [PubMed] [Google Scholar]
- (504).Mietzner TA; Morse SA The role of iron-binding proteins in the survival of pathogenic bacteria. Annu. Rev. Nutr 1994, 14, 471–493. [DOI] [PubMed] [Google Scholar]
- (505).Crosa JH; Mey AR; Payne SM Iron Transport in Bacteria; ASM Press: Washington, DC, 2004; Vol. 410. [Google Scholar]
- (506).Payne SM; Mey AR Pathogenic Escherichia coli, Shigella, and Salmonella. Iron transport in bacteria 2014, 197–218. [Google Scholar]
- (507).Bearden SW; Fetherston JD; Perry RD Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infection and immunity 1997, 65, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (508).Bearden SW; Staggs TM; Perry RD An ABC Transporter System of Yersinia pestis Allows Utilization of Chelated Iron by Escherichia coliSAB11. J. Bacteriol 1998, 180, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (509).Bearden SW; Perry RD The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol 1999, 32, 403–414. [DOI] [PubMed] [Google Scholar]
- (510).Gong S; Bearden SW; Geoffroy VA; Fetherston JD; Perry RD Characterization of the Yersinia pestisYfu ABC Inorganic Iron Transport System. Infect. Immun 2001, 69, 2829–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (511).Perry RD; Abney J; Mier I; Lee Y; Bearden SW; Fetherston JD Regulation of the Yersinia pestis Yfe and Ybt iron transport systems. In The Genus Yersinia; Springer, 2004; pp 275–283. [DOI] [PubMed] [Google Scholar]
- (512).Kirillina O; Bobrov AG; Fetherston JD; Perry RD Hierarchy of iron uptake systems: Yfu and Yiu are functional in Yersinia pestis. Infect. Immun 2006, 74, 6171–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (513).Perry RD; Mier I; Fetherston JD Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. BioMetals 2007, 20, 699. [DOI] [PubMed] [Google Scholar]
- (514).Forman S; Paulley JT; Fetherston JD; Cheng Y-Q; Perry RD Yersinia ironomics: comparison of iron transporters among Yersiniapestis biotypes and its nearest neighbor, Yersinia pseudotuberculosis. BioMetals 2010, 23, 275–294. [DOI] [PubMed] [Google Scholar]
- (515).Pieper R; Huang S-T; Parmar PP; Clark DJ; Alami H; Fleischmann RD; Perry RD; Peterson SN Proteomic analysis of iron acquisition, metabolic and regulatory responses of Yersinia pestis to iron starvation. BMC Microbiol. 2010, 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (516).Perry R; Fetherston J Iron and heme uptake systems. In Yersinia Molecular and Cellular Biology; Horizon Bioscience: Norfolk, UK, 2004; pp 257–283 [Google Scholar]
- (517).Iteman I; Guiyoule A; de Almeida A; Guilvout I; Baranton G; Carniel E Relationship between loss of pigmentation and deletion of the chromosomal iron-regulated irp2 gene in Yersinia pestis: evidence for separate but related events. Infect. Immun 1993, 61, 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (518).Carniel E; Guilvout I; Prentice M Characterization of a large chromosomal” high-pathogenicity island” in biotype 1B Yersinia enterocolitica. Journal of bacteriology 1996, 178, 6743–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (519).Rakin A; Noelting C; Schubert S; Heesemann J Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun 1999, 67, 5265–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (520).Lesic B; Carniel E The high pathogenicity island: a broad-host-range pathogenicity island. In Yersinia Molecular and Cellular Biology; Horizon Bioscience: Norfolk, UK, 2004; pp 285–306. [Google Scholar]
- (521).Perry RD; Balbo PB; Jones HA; Fetherston JD; DeMoll E Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 1999, 145, 1181–1190. [DOI] [PubMed] [Google Scholar]
- (522).Rakin A; Saken E; Harmsen D; Heesemann J The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol 1994, 13, 253–263. [DOI] [PubMed] [Google Scholar]
- (523).Fetherston JD; Lillard J; Perry RD Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J. Bacteriol 1995, 177, 1824–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (524).Fetherston JD; Bertolino VJ; Perry RD YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol 1999, 32, 289–299. [DOI] [PubMed] [Google Scholar]
- (525).Sebbane F; Jarrett C; Gardner D; Long D; Hinnebusch BJ Role of the Yersinia pestis yersiniabactin iron acquisition system in the incidence of flea-borne plague. PLoS One 2010, 5, No. e14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (526).Tu J; Xue T; Qi K; Shao Y; Huang B; Wang X; Zhou X The irp2 and fyuA genes in High Pathogenicity Islands are involved in the pathogenesis of infections caused by avian pathogenic Escherichia coli (APEC). Pol. J. Vet. Sci 2016, 19, 19–21. [DOI] [PubMed] [Google Scholar]
- (527).Lukacik P; Barnard TJ; Keller PW; Chaturvedi KS; Seddiki N; Fairman JW; Noinaj N; Kirby TL; Henderson JP; Steven AC; Hinnebusch BJ; Buchanan SK Structural engineering of a phage lysin that targets gram-negative pathogens. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 9857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (528).Fetherston JD; Lillard JW Jr; Perry RD Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. Journal of bacteriology 1995, 177, 1824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (529).Xu H; Tie K; Zhang Y; Feng X; Cao Y; Han W Design, expression, and characterization of the hybrid antimicrobial peptide T-catesbeianin-1 based on FyuA. J. Pept. Sci 2018, 24, e3059. [DOI] [PubMed] [Google Scholar]
- (530).Wang Z; Hu W; Zheng H Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter. Science advances 2020, 6, No. eaay7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (531).Borths EL; Locher KP; Lee AT; Rees DC The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 16642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (532).Pinkett HW; Lee AT; Lum P; Locher KP; Rees DC An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science (Washington, DC, U. S.) 2007, 315, 373–7. [DOI] [PubMed] [Google Scholar]
- (533).Fetherston JD; Mier I; Truszczynska H; Perry RD The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect. Immun 2012, 80, 3880–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (534).Une T; Brubaker RR In vivo comparison of avirulent Vwa- and Pgm-or Pstr phenotypes of yersiniae. Infect. Immun 1984, 43, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (535).Skare JT; Ahmer B; Seachord CL; Darveau RP; Postle K Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem 1993, 268, 16302–16308. [PubMed] [Google Scholar]
- (536).Radka CD; DeLucas LJ; Wilson LS; Lawrenz MB; Perry RD; Aller SG Crystal structure of Yersinia pestis virulence factor YfeA reveals two polyspecific metal-binding sites. Acta Crystallographica Section D: Structural Biology 2017, 73, 557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (537).Bezkorovainy A Antimicrobial properties of iron-binding proteins. Advances in experimental medicine and biology 1981, 135, 139–54. [DOI] [PubMed] [Google Scholar]
- (538).Crichton RR; Charloteaux-Wauters M Iron transport and storage. Eur. J. Biochem 1987, 164, 485–506. [DOI] [PubMed] [Google Scholar]
- (539).Orsi N The antimicrobial activity of lactoferrin: current status and perspectives. BioMetals 2004, 17, 189–96. [DOI] [PubMed] [Google Scholar]
- (540).Otto BR; Verweij-Van Vught AM; MacLaren DM Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol 1992, 18, 217–33. [DOI] [PubMed] [Google Scholar]
- (541).Triebel S; Bläser J; Reinke H; Tschesche HA 25 kDa alpha 2-microglobulin-related protein is a component of the 125 kDa form of human gelatinase. FEBS Lett. 1992, 314, 386–8. [DOI] [PubMed] [Google Scholar]
- (542).Hraba-Renevey S; Türler H; Kress M; Salomon C; Weil R SV40-induced expression of mouse gene 24p3 involves a post-transcriptional mechanism. Oncogene 1989, 4, 601–608. [PubMed] [Google Scholar]
- (543).Xiao X; Yeoh BS; Vijay-Kumar M Lipocalin 2: An Emerging Player in Iron Homeostasis and Inflammation. Annu. Rev. Nutr 2017, 37, 103–130. [DOI] [PubMed] [Google Scholar]
- (544).Yang J; Goetz D; Li JY; Wang W; Mori K; Setlik D; Du T; Erdjument-Bromage H; Tempst P; Strong R; Barasch J An iron delivery pathway mediated by a lipocalin. Mol. Cell 2002, 10, 1045–56. [DOI] [PubMed] [Google Scholar]
- (545).Devireddy LR; Hart DO; Goetz DH; Green MR A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell 2010, 141, 1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (546).Liu Z; Reba S; Chen WD; Porwal SK; Boom WH; Petersen RB; Rojas R; Viswanathan R; Devireddy L Regulation of mammalian siderophore 2,5-DHBA in the innate immune response to infection. J. Exp. Med 2014, 211, 1197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (547).Fluckinger M; Haas H; Merschak P; Glasgow BJ; Redl B Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob. Agents Chemother 2004, 48, 3367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (548).Doneanu CE; Strong RK; Howald WN Characterization of a noncovalent lipocalin complex by liquid chromatography/electrospray ionization mass spectrometry. J. Biomol. Tech 2004, 15, 208–212. [PMC free article] [PubMed] [Google Scholar]
- (549).Holmes MA; Paulsene W; Jide X; Ratledge C; Strong RK Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure (Oxford, U. K.) 2005, 13, 29–41. [DOI] [PubMed] [Google Scholar]
- (550).Lassagne H; Gachon AMF; Nakashima Y; Yanagita T; Ozawa M; Muramatsu T Cloning of a human lacrimal lipocalin secreted in tears. Exp. Eye Res 1993, 56, 605–609. [DOI] [PubMed] [Google Scholar]
- (551).Nelson AL; Barasch JM; Bunte RM; Weiser JN Bacterial colonization of nasal mucosa induces expression of siderocalin, an iron-sequestering component of innate immunity. Cell. Microbiol 2005, 7, 1404–17. [DOI] [PubMed] [Google Scholar]
- (552).Borregaard N; Cowland JB Neutrophil gelatinase-associated lipocalin, a siderophore-binding eukaryotic protein. BioMetals 2006, 19, 211–5. [DOI] [PubMed] [Google Scholar]
- (553).Wu H; Santoni-Rugiu E; Ralfkiaer E; Porse BT; Moser C; Hoiby N; Borregaard N; Cowland JB Lipocalin 2 is protective against E. coli pneumonia. Respir. Res 2010, 11, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (554).Goetz DH; Holmes MA; Borregaard N; Bluhm ME; Raymond KN; Strong RK The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–1043. [DOI] [PubMed] [Google Scholar]
- (555).Flo TH; Smith KD; Sato S; Rodriguez DJ; Holmes MA; Strong RK; Akira S; Aderem A Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004, 432, 917–921. [DOI] [PubMed] [Google Scholar]
- (556).Fischbach MA; Lin H; Zhou L; Yu Y; Abergel RJ; Liu DR; Raymond KN; Wanner BL; Strong RK; Walsh CT; et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 16502–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (557).Dauner M; Eichinger A; Lücking G; Scherer S; Skerra A Reprogramming Human Siderocalin To Neutralize Petrobactin, the Essential Iron Scavenger of Anthrax Bacillus. Angew. Chem., Int. Ed 2018, 57, 14619–14623. [DOI] [PubMed] [Google Scholar]
- (558).Dauner M; Skerra A Scavenging Bacterial Siderophores with Engineered Lipocalin Proteins as an Alternative Antimicrobial Strategy. ChemBioChem 2020, 21, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (559).Heilmeyer L; Woehler F [Modern problems of hemochromatosis with special reference to desferrioxamine treatment]. Dtsch. Med. Wochenschr 1962, 87, 2661–7. [DOI] [PubMed] [Google Scholar]
- (560).Risell E; Schnack H [Experiences with a new iron compound, desferrioxamine]. Wien Klin Wochenschr 1962, 74, 577–580. [PubMed] [Google Scholar]
- (561).Tripod J; Keberle H [Biological assay with desferrioxamine]. Helv. Physiol. Pharmacol. Acta 1962, 20, 291–293. [PubMed] [Google Scholar]
- (562).Coulton JW; Naegeli HU; Braun V Iron supply of Escherichia coli with polymer-bound ferricrocin. Eur. J. Biochem 1979, 99, 39–47. [DOI] [PubMed] [Google Scholar]
- (563).Plaha DS; Rogers HJ; Williams GW Studies of the antibacterial effect of the scandium complex of enterochelin. J. Antibiot 1984, 37, 588–95. [DOI] [PubMed] [Google Scholar]
- (564).Rogers HJ; Synge C; Woods VE Antibacterial effect of scandium and indium complexes of enterochelin on Klebsiella pneumoniae. Antimicrob. Agents Chemother 1980, 18, 63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (565).Ohi N; Aoki B; Shinozaki T; Moro K; Noto T; Nehashi T; Okazaki H; Matsunaga I Semisynthetic beta-lactam antibiotics. I. Synthesis and antibacterial activity of new ureidopenicillin derivatives having catechol moieties. J. Antibiot 1986, 39, 230–41. [DOI] [PubMed] [Google Scholar]
- (566).Basker MJ; Branch CL; Finch SC; Guest AW; Milner PH; Pearson MJ; Ponsford RJ; Smale TC Studies on semi-synthetic 7 alpha-formamidocephalosporins. I. Structure-activity relationships in some semi-synthetic 7 alpha-formamidocephalosporins. J. Antibiot 1986, 39, 1788–91. [DOI] [PubMed] [Google Scholar]
- (567).Burton G; Best DJ; Dixon RA; Kenyon RF; Lashford AG Studies on 6 alpha-substituted penicillins. II. Synthesis and structure-activity relationships of 6 beta-(2-aryl-2-sulfoacetamido)-6 alpha-methoxy penicillanic acids. J. Antibiot 1986, 39, 1419–29. [DOI] [PubMed] [Google Scholar]
- (568).Mochida K; Ono Y; Yamasaki M; Shiraki C; Hirata T; Sato K; Okachi R Aminothiazolylglycyl derivatives of carbacephem antibiotics. II. Synthesis and antibacterial activity of novel aminothiazolyl cephem compounds with hydroxypyridone moiety. J. Antibiot 1987, 40, 182–9. [DOI] [PubMed] [Google Scholar]
- (569).Nakagawa S; Sanada M; Matsuda K; Hazumi N; Tanaka N Biological activity of BO-1236, a new antipseudomonal cephalosporin. Antimicrob. Agents Chemother 1987, 31, 1100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (570).Labott SM; Martin RB The stress-moderating effects of weeping and humor. J. Human Stress 1987, 13, 159–64. [DOI] [PubMed] [Google Scholar]
- (571).Watanabe NA; Nagasu T; Katsu K; Kitoh K E-0702, a new cephalosporin, is incorporated into Escherichia coli cells via the tonB-dependent iron transport system. Antimicrob. Agents Chemother 1987, 31, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (572).Carpenter C; Payne SM Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem 2014, 133, 110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (573).Miller MJ; McKee JA; Minnick AA; Dolence EK The design, synthesis and study of siderophore-antibiotic conjugates. Siderophore mediated drug transport. Biol. Met 1991, 4, 62–9. [DOI] [PubMed] [Google Scholar]
- (574).Curtis NA; Eisenstadt RL; East SJ; Cornford RJ; Walker LA; White AJ Iron-regulated outer membrane proteins of Escherichia coli K-12 and mechanism of action of catechol-substituted cephalosporins. Antimicrob. Agents Chemother 1988, 32, 1879–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (575).Dichtl S; Demetz E; Haschka D; Tymoszuk P; Petzer V; Nairz M; Seifert M; Hoffmann A; Brigo N; Wurzner R; Theurl I; Karlinsey JE; Fang FC; Weiss G Dopamine Is a Siderophore-Like Iron Chelator That Promotes Salmonella enterica Serovar Typhimurium Virulence in Mice. mBio 2019, 10, e02624–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (576).Armstrong SK; Brickman TJ; Suhadolc RJ Involvement of multiple distinct Bordetella receptor proteins in the utilization of iron liberated from transferrin by host catecholamine stress hormones. Mol. Microbiol 2012, 84, 446–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (577).Sandrini SM; Shergill R; Woodward J; Muralikuttan R; Haigh RD; Lyte M; Freestone PP Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J. Bacteriol 2010, 192, 587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (578).Hantke K Dihydroxybenzoylserine–a siderophore for E. coli. FEMS Microbiol. Lett 1990, 67, 5–8. [DOI] [PubMed] [Google Scholar]
- (579).Lopez-Goni I; Moriyon I; Neilands JB Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect. Immun 1992, 60, 4496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (580).Young IG; Cox GB; Gibson F 2,3-Dihydroxybenzoate as a bacterial growth factor and its route of biosynthesis. Biochim. Biophys. Acta, Gen. Subj 1967, 141, 319–31. [DOI] [PubMed] [Google Scholar]
- (581).Ito A; Nishikawa T; Matsumoto S; Yoshizawa H; Sato T; Nakamura R; Tsuji M; Yamano Y Siderophore Cephalosporin Cefiderocol Utilizes Ferric Iron Transporter Systems for Antibacterial Activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother 2016, 60, 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (582).Cody YS; Gross DC Characterization of Pyoverdin(pss), the Fluorescent Siderophore Produced by Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol 1987, 53, 928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (583).Huband MD; Ito A; Tsuji M; Sader HS; Fedler KA; Flamm RK Cefiderocol MIC quality control ranges in iron-depleted cation-adjusted Mueller-Hinton broth using a CLSI M23-A4 multi-laboratory study design. Diagn. Microbiol. Infect. Dis 2017, 88, 198–200. [DOI] [PubMed] [Google Scholar]
- (584).Schalk IJ; Yue WW; Buchanan SK Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol 2004, 54, 14–22. [DOI] [PubMed] [Google Scholar]
- (585).Brillet K; Reimmann C; Mislin GL; Noäl S; Rognan D; Schalk IJ; Cobessi D Pyochelin enantiomers and their outer-membrane siderophore transporters in fluorescent pseudomonads: structural bases for unique enantiospecific recognition. J. Am. Chem. Soc 2011, 133, 16503–9. [DOI] [PubMed] [Google Scholar]
- (586).Hoegy F; Lee X; Noel S; Rognan D; Mislin GL; Reimmann C; Schalk IJ Stereospecificity of the siderophore pyochelin outer membrane transporters in fluorescent pseudomonads. J. Biol. Chem 2009, 284, 14949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (587).Matzanke BF; Muller GI; Raymond KN Hydroxamate siderophore mediated iron uptake in E. coli: stereospecific recognition of ferric rhodotorulic acid. Biochem. Biophys. Res. Commun 1984, 121, 922–30. [DOI] [PubMed] [Google Scholar]
- (588).Neilands JB; Erickson TJ; Rastetter WH Stereospecificity of the ferric enterobactin receptor of Escherichia coli K-12. J. Biol. Chem 1981, 256, 3831–2. [PubMed] [Google Scholar]
- (589).Youard ZA; Reimmann C Stereospecific recognition of pyochelin and enantio-pyochelin by the PchR proteins in fluorescent pseudomonads. Microbiology 2010, 156, 1772–1782. [DOI] [PubMed] [Google Scholar]
- (590).Ito AIN; Ishii R; Tsuji M; Maki H; Sato T; Yamano Y Changes of Responsible Iron-Transporters for the Activity of Cefiderocol against Pseudomonas aeruginosa Depending on the Culture Conditions. In ASM Microbe; ASM: San Francisco, CA, 2019. [Google Scholar]
- (591).Ito A; Nishikawa T; Ota M; Ito-Horiyama T; Ishibashi N; Sato T; Tsuji M; Yamano Y Stability and low induction propensity of cefiderocol against chromosomal AmpC β-lactamases of Pseudomonas aeruginosa and Enterobacter cloacae. J. Antimicrob. Chemother 2018, 73, 3049–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (592).Ito A; Sato T; Ota M; Takemura M; Nishikawa T; Toba S; Kohira N; Miyagawa S; Ishibashi N; Matsumoto S; Nakamura R; Tsuji M; Yamano Y In Vitro Antibacterial Properties of Cefiderocol, a Novel Siderophore Cephalosporin, against Gram-Negative Bacteria. Antimicrob. Agents Chemother 2018, 62, e01454–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (593).Moynie L; Milenkovic S; Mislin GLA; Gasser V; Malloci G; Baco E; McCaughan RP; Page MGP; Schalk IJ; Ceccarelli M; Naismith JH The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model. Nat. Commun 2019, 10, 3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (594).Luscher A; Moynie L; Auguste PS; Bumann D; Mazza L; Pletzer D; Naismith JH; Kohler T TonB-Dependent Receptor Repertoire of Pseudomonas aeruginosa for Uptake of Siderophore-Drug Conjugates. Antimicrob. Agents Chemother 2018, 62, e00097–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (595).van Delden C; Page MG; Kohler T Involvement of Fe uptake systems and AmpC beta-lactamase in susceptibility to the siderophore monosulfactam BAL30072 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 2013, 57, 2095–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (596).Kim A; Kutschke A; Ehmann DE; Patey SA; Crandon JL; Gorseth E; Miller AA; McLaughlin RE; Blinn CM; Chen A; Nayar AS; Dangel B; Tsai AS; Rooney MT; Murphy-Benenato KE; Eakin AE; Nicolau DP Pharmacodynamic Profiling of a Siderophore-Conjugated Monocarbam in Pseudomonas aeruginosa: Assessing the Risk for Resistance and Attenuated Efficacy. Antimicrob. Agents Chemother 2015, 59, 7743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (597).McPherson CJ; Aschenbrenner LM; Lacey BM; Fahnoe KC; Lemmon MM; Finegan SM; Tadakamalla B; O’Donnell JP; Mueller JP; Tomaras AP Clinically relevant Gram-negative resistance mechanisms have no effect on the efficacy of MC-1, a novel siderophore-conjugated monocarbam. Antimicrob. Agents Chemother 2012, 56, 6334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (598).de Lorenzo V; Giovannini F; Herrero M; Neilands JB Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J. Mol. Biol 1988, 203, 875–884. [DOI] [PubMed] [Google Scholar]
- (599).Escolar L; Perez-Martin J; de Lorenzo V Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol 1999, 181, 6223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (600).Shields RK Case Commentary: the Need for Cefiderocol Is Clear, but Are the Supporting Clinical Data? Antimicrob. Agents Chemother 2020, 64, e00059–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (601).Wu JY; Srinivas P; Pogue JM Cefiderocol: A Novel Agent for the Management of Multidrug-Resistant Gram-Negative Organisms. Infect Dis Ther 2020, 9, 17–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (602).Yamano Y In Vitro Activity of Cefiderocol Against a Broad Range of Clinically Important Gram-negative Bacteria. Clin. Infect. Dis 2019, 69, S544–S551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (603).Tomaras AP; Crandon JL; McPherson CJ; Banevicius MA; Finegan SM; Irvine RL; Brown MF; O’Donnell JP; Nicolau DP Adaptation-based resistance to siderophore-conjugated antibacterial agents by Pseudomonas aeruginosa. Antimicrob. Agents Chemother 2013, 57, 4197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (604).Ghazi IM; Monogue ML; Tsuji M; Nicolau DP Humanized Exposures of Cefiderocol, a Siderophore Cephalosporin, Display Sustained in vivo Activity against Siderophore-Resistant Pseudomonas aeruginosa. Pharmacology 2018, 101, 278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (605).Monogue ML; Tsuji M; Yamano Y; Echols R; Nicolau DP Efficacy of Humanized Exposures of Cefiderocol (S-649266) against a Diverse Population of Gram-Negative Bacteria in a Murine Thigh Infection Model. Antimicrob. Agents Chemother 2017, 61, e01022–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (606).Tomaras AP; Crandon JL; McPherson CJ; Nicolau DP Potentiation of antibacterial activity of the MB-1 siderophore-monobactam conjugate using an efflux pump inhibitor. Antimicrob. Agents Chemother 2015, 59, 2439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (607).Vázquez-Ucha JC; Martínez-Guitián M; Maneiro M; Conde-Pérez K; Álvarez-Fraga L; Torrens G; Oliver A; Buynak JD; Bonomo RA; Bou G; González-Bello C; Poza M; Beceiro A Therapeutic Efficacy of LN-1–255 in Combination with Imipenem in Severe Infection Caused by Carbapenem-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother 2019, 63, e01092–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (608).Page MGP The Role of Iron and Siderophores in Infection, and the Development of Siderophore Antibiotics. Clin. Infect. Dis 2019, 69, S529–S537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (609).Albomycin and grisein. Br. Med. J 1958, 1, 391. [PubMed] [Google Scholar]
- (610).Gamburg RL [Use of albomycin in pneumónia in children]. Pediatriia 1951, 5, 37–44. [PubMed] [Google Scholar]
- (611).Luckey M; Pollack JR; Wayne R; Ames BN; Neilands JB Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J. Bacteriol 1972, 111, 731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (612).Rafalowicz A [New antibiotic albomycin]. Pol Tyg Lek (Wars) 1953, 8, 957–958. [PubMed] [Google Scholar]
- (613).Shorin VA; Luo S [Effect of aerobic and anaerobic conditions of growth of bacteria on antibacterial action of albomycin and of other antibiotics]. Dokl. Akad. Nauk SSSR 1954, 96, 645–647. [PubMed] [Google Scholar]
- (614).Roosenberg JM 2nd; Miller MJ Total synthesis of the siderophore danoxamine. J. Org. Chem 2000, 65, 4833–8. [DOI] [PubMed] [Google Scholar]
- (615).Braun V; Pramanik A; Gwinner T; Köberle M; Bohn E Sideromycins: tools and antibiotics. BioMetals 2009, 22, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (616).Liu R; Miller PA; Vakulenko SB; Stewart NK; Boggess WC; Miller MJ A Synthetic Dual Drug Sideromycin Induces Gram-Negative Bacteria To Commit Suicide with a Gram-Positive Antibiotic. J. Med. Chem 2018, 61, 3845–3854. [DOI] [PubMed] [Google Scholar]
- (617).Neumann W; Nolan EM Evaluation of a reducible disulfide linker for siderophore-mediated delivery of antibiotics. JBIC, J. Biol. Inorg. Chem 2018, 23, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (618).Zheng T; Nolan EM Enterobactin-Mediated Delivery of beta-Lactam Antibiotics Enhances Antibacterial Activity against Pathogenic Escherichia coli. J. Am. Chem. Soc 2014, 136, 9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (619).Ji C; Miller MJ Chemical syntheses and in vitro antibacterial activity of two desferrioxamine B-ciprofloxacin conjugates with potential esterase and phosphatase triggered drug release linkers. Bioorg. Med. Chem 2012, 20, 3828–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (620).Silver LL A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem 2016, 24, 6379–6389. [DOI] [PubMed] [Google Scholar]
- (621).Chen J; Yang J; Ren P; Zhou J Prokaryotic expression and function analysis of Lateolabrax japonica hepcidin. Shuisheng Shengwu Xuebao 2012, 34, 554–561. [Google Scholar]
- (622).Liu M; Tanaka WN; Zhu H; Xie G; Dooley DM; Lei B Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem 2008, 283, 6668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (623).Ghosh M; Miller PA; Miller MJ Antibiotic repurposing: bis-catechol- and mixed ligand (bis-catechol-mono-hydroxamate)-teicoplanin conjugates are active against multidrug resistant Acinetobacter baumannii. J. Antibiot 2020, 73, 152–157. [DOI] [PubMed] [Google Scholar]
- (624).Dahmen S; Mansour W; Charfi K; Boujaafar N; Arlet G; Bouallegue O Imipenem resistance in Klebsiella pneumoniae is associated to the combination of plasmid-mediated CMY-4 AmpC beta-lactamase and loss of an outer membrane protein. Microb. Drug Resist 2012, 18, 479–83. [DOI] [PubMed] [Google Scholar]
- (625).Martinez-Martinez L Extended-spectrum beta-lactamases and the permeability barrier. Clin. Microbiol. Infect 2008, 14, 82–9. [DOI] [PubMed] [Google Scholar]
- (626).Nguyen Van JC; Gutmann L [Resistance to antibiotics caused by decrease of the permeability in gram-negative bacteria]. Presse Med. 1994, 23 (522), 527–531. [PubMed] [Google Scholar]
- (627).Nordmann P [Gram-negative bacteriae with resistance to carbapenems]. Med. Sci. (Paris) 2010, 26, 950–9. [DOI] [PubMed] [Google Scholar]
- (628).Broberg CA; Palacios M; Miller VL Klebsiella: a long way to go towards understanding this enigmatic jet-setter. F1000Prime Rep. 2014, 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (629).Hsieh PF; Liu JY; Pan YJ; Wu MC; Lin TL; Huang YT; Wang JT Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J. Infect. Dis 2013, 208, 1580–9. [DOI] [PubMed] [Google Scholar]
- (630).Shon AS; Bajwa RP; Russo TA Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013, 4, 107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (631).Shon AS; Russo TA Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol. 2012, 7, 669–71. [DOI] [PubMed] [Google Scholar]
- (632).Hancock RE; Speert DP Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updates 2000, 3, 247–255. [DOI] [PubMed] [Google Scholar]
- (633).Nakae T Role of membrane permeability in determining antibiotic resistance in Pseudomonas aeruginosa. Microbiol. Immunol 1995, 39, 221–9. [DOI] [PubMed] [Google Scholar]
- (634).Poole K Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 2005, 49, 479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (635).Sugawara E; Nagano K; Nikaido H Alternative folding pathways of the major porin OprF of Pseudomonas aeruginosa. FEBS J. 2012, 279, 910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (636).Dong R; Guan C; Hu D; Xin TT; Qu Y The correlation study on antimicrobial resistance and biofilm related genes in clinical isolates of Acinetobacter baumannii. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2013, 25, 493–494. [PubMed] [Google Scholar]
- (637).Smani Y; Pachon J Loss of the OprD homologue protein in Acinetobacter baumannii: impact on carbapenem susceptibility. Antimicrob. Agents Chemother 2013, 57, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (638).Sugawara E; Nikaido H OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J. Bacteriol 2012, 194, 4089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (639).Oglesby-Sherrouse AG; Djapgne L; Nguyen AT; Vasil AI; Vasil ML The complex interplay of iron, biofilm formation, and mucoidy affecting antimicrobial resistance of Pseudomonas aeruginosa. Pathog. Dis 2014, 70, 307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (640).Smith DJ; Lamont IL; Anderson GJ; Reid DW Targeting iron uptake to control Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir. J 2013, 42, 1723–36. [DOI] [PubMed] [Google Scholar]
- (641).Wiens JR; Vasil AI; Schurr MJ; Vasil ML Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio 2014, 5, No. e01010–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (642).Chu BC; Peacock RS; Vogel HJ Bioinformatic analysis of the TonB protein family. Biometals 2007, 16, 467–483. [DOI] [PubMed] [Google Scholar]
- (643).Hannavy K; Barr GC; Dorman CJ; Adamson J; Mazengera LR; Gallagher MP; Evans JS; Levine BA; Trayer IP; Higgins CF TonB protein of Salmonella typhimurium. A model for signal transduction between membranes. J. Mol. Biol 1990, 216, 897–910. [DOI] [PubMed] [Google Scholar]
- (644).Hannavy K; Higgins CF TonB; a model for signal transduction between membranes. Biochem. Soc. Trans 1991, 19, 530–2. [DOI] [PubMed] [Google Scholar]
- (645).Kodding J; Killig F; Polzer P; Howard SP; Diederichs K; Welte W Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem 2005, 280, 3022–8. [DOI] [PubMed] [Google Scholar]
- (646).Peacock RS; Andrushchenko VV; Demcoe AR; Gehmlich M; Lu LS; Herrero AG; Vogel HJ Characterization of TonB interactions with the FepA cork domain and FecA N-terminal signaling domain. BioMetals 2006, 19, 127–42. [DOI] [PubMed] [Google Scholar]
- (647).Peacock RS; Weljie AM; Peter Howard S; Price FD; Vogel HJ The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol 2005, 345, 1185–1197. [DOI] [PubMed] [Google Scholar]
- (648).Ginalski K Comparative modeling for protein structure prediction. Curr. Opin. Struct. Biol 2006, 16, 172–7. [DOI] [PubMed] [Google Scholar]
- (649).Kryshtafovych A; Venclovas C; Fidelis K; Moult J Progress over the first decade of CASP experiments. Proteins: Struct., Funct., Genet 2005, 61 (7), 225–36. [DOI] [PubMed] [Google Scholar]
- (650).Neidhardt FC; Bloch PL; Smith DF Culture medium for enterobacteria. J. Bacteriol 1974, 119, 736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (651).Do J; Zafar H; Saier MH, Jr Comparative genomics of transport proteins in probiotic and pathogenic Escherichia coli and Salmonella enterica strains. Microb. Pathog 2017, 107, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (652).Bassford PJ, Jr; Kadner, R. J. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J. Bacteriol 1977, 132, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (653).Hantke K Dihydroxybenzolyserine—a siderophore for E. coli. FEMS Microbiol. Lett 1990, 67, 5–8. [DOI] [PubMed] [Google Scholar]
- (654).Nikaido H; Rosenberg EY Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J. Bacteriol 1990, 172, 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (655).Bluhm ME; Hay BP; Kim SS; Dertz EA; Raymond KN Corynebactin and a serine trilactone based analogue: chirality and molecular modeling of ferric complexes. Inorg. Chem 2002, 41, 5475–8. [DOI] [PubMed] [Google Scholar]
- (656).Bluhm ME; Kim SS; Dertz EA; Raymond KN Corynebactin and enterobactin: related siderophores of opposite chirality. J. Am. Chem. Soc 2002, 124, 2436–7. [DOI] [PubMed] [Google Scholar]
- (657).May JJ; Wendrich TM; Marahiel MA The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem 2001, 276, 7209–17. [DOI] [PubMed] [Google Scholar]
- (658).Lohmiller S; Hantke K; Patzer SI; Braun V TonB-dependent maltose transport by Caulobacter crescentus. Microbiology 2008, 154, 1748–54. [DOI] [PubMed] [Google Scholar]
- (659).Neugebauer H; Herrmann C; Kammer W; Schwarz G; Nordheim A; Braun V ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol 2005, 187, 8300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (660).Chairatana P; Zheng T; Nolan EM Targeting virulence: salmochelin modification tunes the antibacterial activity spectrum of β-lactams for pathogen-selective killing of Escherichia coli. Chemical science 2015, 6, 4458–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (661).Payne MA; Igo JD; Cao Z; Foster SB; Newton SM; Klebba PE Biphasic binding kinetics between FepA and its ligands. J. Biol. Chem 1997, 272, 21950–5. [DOI] [PubMed] [Google Scholar]
- (662).Moynié L; Milenkovic S; Mislin GL; Gasser V; Malloci G; Baco E; McCaughan RP; Page MG; Schalk IJ; Ceccarelli M The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model. Nat. Commun 2019, 10, 3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (663).Grinter R; Lithgow T The structure of the bacterial iron-catecholate transporter Fiu suggests that it imports substrates via a two-step mechanism. J. Biol. Chem 2019, 294, 19523–19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (664).Larkin MA; Blackshields G; Brown NP; Chenna R; McGettigan PA; McWilliam H; Valentin F; Wallace IM; Wilm A; Lopez R; Thompson JD; Gibson TJ; Higgins DG Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–8. [DOI] [PubMed] [Google Scholar]
- (665).Dib L; Carbone A Protein fragments: functional and structural roles of their coevolution networks. PLoS One 2012, 7, No. e48124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (666).Grytsyk N; Sugihara J; Kaback HR; Hellwig P pK(a) of Glu325 in LacY. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (667).Kaback HR; Guan L It takes two to tango: The dance of the permease. J. Gen. Physiol 2019, 151, 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (668).Bradbeer C The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol 1993, 175, 3146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (669).Majumdar A; Trinh V; Moore KJ; Smallwood CR; Kumar A; Yang T; Scott DC; Long NJ; Newton SM; Klebba PE Conformational rearrangements in the N-domain of Escherichia coli FepA during ferric enterobactin transport. J. Biol. Chem 2020, 295, 4974–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (670).Esmaeilkhani H; Rasooli I; Hashemi M; Nazarian S; Sefid F Immunogenicity of Cork and Loop Domains of Recombinant Baumannii acinetobactin Utilization Protein in Murine Model. Avicenna J. Med. Biotechnol 2019, 11, 180–186. [PMC free article] [PubMed] [Google Scholar]
- (671).Lin J; Hogan JS; Smith KL Inhibition of in vitro growth of coliform bacteria by a monoclonal antibody directed against ferric enterobactin receptor FepA. J. Dairy Sci 1998, 81, 1267–74. [DOI] [PubMed] [Google Scholar]
- (672).Lin J; Hogan JS; Smith KL Growth responses of coliform bacteria to purified immunoglobulin G from cows immunized with ferric enterobactin receptor FepA. J. Dairy Sci 1999, 82, 86–92. [DOI] [PubMed] [Google Scholar]
- (673).Murphy CK; Kalve VI; Klebba PE Surface topology of the Escherichia coli K-12 ferric enterobactin receptor. J. Bacteriol 1990, 172, 2736–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (674).Wolf SL; Hogan JS; Smith KL Iron uptake by Escherichia coli cultured with antibodies from cows immunized with high-affinity ferric receptors. J. Dairy Sci 2004, 87, 2103–7. [DOI] [PubMed] [Google Scholar]
- (675).Kurupati P; Teh BK; Kumarasinghe G; Poh CL Identification of vaccine candidate antigens of an ESBL producing Klebsiella pneumoniae clinical strain by immunoproteome analysis. Proteomics 2006, 6, 836–44. [DOI] [PubMed] [Google Scholar]
- (676).Larrie-Bagha SM; Rasooli I; Mousavi-Gargari SL; Rasooli Z; Nazarian S Passive immunization by recombinant ferric enterobactin protein (FepA) from Escherichia coli O157. Iran J. Microbiol 2013, 5, 113–119. [PMC free article] [PubMed] [Google Scholar]
- (677).Lin J; Hogan JS; Aslam M; Smith KL Immunization of cows with ferric enterobactin receptor from coliform bacteria. J. Dairy Sci 1998, 81, 2151–8. [DOI] [PubMed] [Google Scholar]
- (678).Bentley AT; Klebba PE Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. J. Bacteriol 1988, 170, 1063–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (679).Klebba PE; Benson SA; Bala S; Abdullah T; Reid J; Singh SP; Nikaido H Determinants of OmpF porin antigenicity and structure. J. Biol. Chem 1990, 265, 6800–10. [PubMed] [Google Scholar]
- (680).Tuntufye HN; Ons E; Pham AD; Luyten T; Van Gerven N; Bleyen N; Goddeeris BM Escherichia coli ghosts or live E. coli expressing the ferri-siderophore receptors FepA, FhuE, IroN and IutA do not protect broiler chickens against avian pathogenic E. coli (APEC). Vet. Microbiol 2012, 159, 470–478. [DOI] [PubMed] [Google Scholar]
- (681).Josts I; Veith K; Tidow H Ternary structure of the outer membrane transporter FoxA with resolved signalling domain provides insights into TonB-mediated siderophore uptake. eLife 2019, 8, e48528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (682).Frillingos S; Ujwal ML; Sun J; Kaback HR The role of helix VIII in the lactose permease of Escherichia coli: I. Cys-scanning mutagenesis. Protein Sci. 1997, 6, 431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (683).Frillingos S; Sahin-Toth M; Wu J; Kaback HR Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998, 12, 1281–99. [DOI] [PubMed] [Google Scholar]
- (684).Frillingos S; Gonzalez A; Kaback HR Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. off. Biochemistry 1997, 36, 14284–90. [DOI] [PubMed] [Google Scholar]
- (685).Nie Y; Ermolova N; Kaback HR Site-directed alkylation of LacY: effect of the proton electrochemical gradient. J. Mol. Biol 2007, 374, 356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (686).Jiang X; Nie Y; Kaback HR Site-directed alkylation studies with LacY provide evidence for the alternating access model of transport. Biochemistry 2011, 50, 1634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (687).Frillingos S; Kaback HR Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry 1996, 35, 3950–6. [DOI] [PubMed] [Google Scholar]
- (688).Ermolova N; Madhvani RV; Kaback HR Site-directed alkylation of cysteine replacements in the lactose permease of Escherichia coli: helices I, III, VI, and XI. Biochemistry 2006, 45, 4182–9. [DOI] [PubMed] [Google Scholar]
- (689).Smirnova IN; Kasho VN; Kaback HR Direct sugar binding to LacY measured by resonance energy transfer. Biochemistry 2006, 45, 15279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (690).Smirnova I; Kasho V; Sugihara J; Kaback HR Probing of the rates of alternating access in LacY with Trp fluorescence. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 21561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (691).Majumdar DS; Smirnova I; Kasho V; Nir E; Kong X; Weiss S; Kaback HR Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 12640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (692).Jung K; Jung H; Kaback HR Dynamics of lactose permease of Escherichia coli determined by site-directed fluorescence labeling. Biochemistry 1994, 33, 3980–5. [DOI] [PubMed] [Google Scholar]
- (693).Voss J; Hubbell WL; Kaback HR Distance determination in proteins using designed metal ion binding sites and site-directed spin labeling: application to the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 12300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (694).Voss J; He MM; Hubbell WL; Kaback HR Site-directed spin labeling demonstrates that transmembrane domain XII in the lactose permease of Escherichia coli is an alpha-helix. Biochemistry 1996, 35, 12915–8. [DOI] [PubMed] [Google Scholar]
- (695).Wu J; Voss J; Hubbell WL; Kaback HR Site-directed spin labeling and chemical crosslinking demonstrate that helix V is close to helices VII and VIII in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 10123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (696).Smirnova I; Kasho V; Choe JY; Altenbach C; Hubbell WL; Kaback HR Sugar binding induces an outward facing conformation of LacY. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16504–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (697).Liu J; Rutz JM; Klebba PE; Feix JB A site-directed spin-labeling study of ligand-induced conformational change in the ferric enterobactin receptor, FepA. Biochemistry 1994, 33, 13274–83. [DOI] [PubMed] [Google Scholar]
- (698).Cao Z; Warfel P; Newton SM; Klebba PE Spectroscopic observations of ferric enterobactin transport. J. Biol. Chem 2003, 278, 1022–8. [DOI] [PubMed] [Google Scholar]
- (699).Michelini E; Cevenini L; Mezzanotte L; Coppa A; Roda A Cell-based assays: fuelling drug discovery. Anal. Bioanal. Chem 2010, 398, 227–38. [DOI] [PubMed] [Google Scholar]
- (700).Cao Y; Bazemore-Walker CR Proteomic profiling of the surface-exposed cell envelope proteins of Caulobacter crescentus. J. Proteomics 2014, 97, 187–94. [DOI] [PubMed] [Google Scholar]
- (701).Nakae T; Nikaido H Multiple molecular forms of uridine diphosphate glucose pyrophosphorylase from Salmonella typhimurium. II. Genetic determination of multiple forms. J. Biol. Chem 1971, 246, 4397–403. [PubMed] [Google Scholar]
- (702).Bailey DC; Buckley BP; Chernov MV; Gulick AM Development of a High-Throughput Biochemical Assay to Screen for Inhibitors of Aerobactin Synthetase IucA. SLAS Discov 2018, 23, 1070–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (703).Hanson M; Jordan LD; Shipelskiy Y; Newton SM; Klebba PE High-Throughput Screening Assay for Inhibitors of TonB-Dependent Iron Transport. J. Biomol. Screening 2016, 21, 316–22. [DOI] [PubMed] [Google Scholar]
- (704).Baell JB; Nissink JWM Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017-Utility and Limitations. ACS Chem. Biol 2018, 13, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (705).Lagorce D; Oliveira N; Miteva MA; Villoutreix BO Pan-assay interference compounds (PAINS) that may not be too painful for chemical biology projects. Drug Discovery Today 2017, 22, 1131–1133. [DOI] [PubMed] [Google Scholar]
- (706).Pouliot M; Jeanmart S Pan Assay Interference Compounds (PAINS) and Other Promiscuous Compounds in Antifungal Research. J. Med. Chem 2016, 59, 497–503. [DOI] [PubMed] [Google Scholar]
- (707).Garcia-Herrero A; Peacock RS; Howard SP; Vogel HJ The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol. Microbiol 2007, 66, 872–89. [DOI] [PubMed] [Google Scholar]
- (708).Brewer S; Tolley M; Trayer IP; Barr GC; Dorman CJ; Hannavy K; Higgins CF; Evans JS; Levine BA; Wormald MR Structure and function of X-Pro dipeptide repeats in the TonB proteins of Salmonella typhimurium and Escherichia coli. J. Mol. Biol 1990, 216, 883–95. [DOI] [PubMed] [Google Scholar]
- (709).Brickman TJ; McIntosh MA Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem 1992, 267, 12350–5. [PubMed] [Google Scholar]
- (710).Escolar L; de Lorenzo V; Perez-Martin J Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol 1997, 26, 799–808. [DOI] [PubMed] [Google Scholar]
- (711).Escolar L; Perez-Martin J; de Lorenzo V Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J. Bacteriol 1998, 180, 2579–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (712).Escolar L; Perez-Martin J; de Lorenzo V Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol 1998, 283, 537–47. [DOI] [PubMed] [Google Scholar]
- (713).Newton SM; Allen JS; Cao Z; Qi Z; Jiang X; Sprencel C; Igo JD; Foster SB; Payne MA; Klebba PE Double mutagenesis of a positive charge cluster in the ligand-binding site of the ferric enterobactin receptor, FepA. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 4560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (714).Annamalai R; Jin B; Cao Z; Newton SM; Klebba PE Recognition of ferric catecholates by FepA. J. Bacteriol 2004, 186, 3578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (715).Cao Z; Qi Z; Sprencel C; Newton SM; Klebba PE Aromatic components of two ferric enterobactin binding sites in escherichia coli fepA. Mol. Microbiol 2000, 37, 1306–17. [DOI] [PubMed] [Google Scholar]
- (716).Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 2004, 25, 1605–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.