Abstract
Proteins of the kinesin superfamily define a class of microtubule-dependent motors that play crucial roles in cell division and intracellular transport. In the mouse, several kinesin motors have been characterized and are suggested to play roles in axonal and/or dendritic transport. One such kinesin is KifC2. Sequence and secondary structure analysis revealed that KifC2 is a member of the C-terminal motor family. Northern and Western blot analyses indicated that KifC2 is specifically expressed in both the central and peripheral nervous systems. The cellular locations of the KifC2 proteins were found to be mainly in neural cell bodies and dendrites but also in axons. To understand the in vivo function of the KifC2 gene, we used homologous recombination in embryonic stem cells to construct knockout mouse strains for the KifC2 gene. Homozygous KifC2 mutants were viable and reproduced normally, and their development was apparently normal. These results suggest that KifC2 is dispensable for normal neural development and behavior in the mouse.
Microtubule-dependent motors of the kinesin superfamily have undergone structural and functional diversification during evolution and play crucial roles in cell division and intracellular transport (1, 5, 8). Members of this superfamily share extensive sequence similarity within the motor domain but display diversification in their tail domains. The motor domain is composed of an ∼330-amino-acid catalytic domain that hydrolyzes ATP and interacts with the microtubule track and of a short ∼40-amino-acid neck domain that is important for processive movement and control of direction (2, 13). The tail domains have been suggested to provide different cargo-binding or regulatory partners and to confer the ability to form different types of oligomers. As a group, kinesins can be categorized by their motility as either plus-end- or minus-end-directed motors (1, 5). Most kinesins, such as true kinesin (conventional kinesin or kinesin I), have an N-terminal catalytic motor domain fused to one of many different neck and tail domains and are plus-end-directed motors. Members of another group of kinesins, called C-terminal motors, have their catalytic motor domain at the C terminus and variable tail domains at the N terminus. So far, all tested members of the C-terminal kinesins are minus-end-directed motors. Because of their motility polarization, most C-terminal kinesin motors are believed to play roles in mitotic and meiotic spindle assembly or in driving or maintaining spindle pole separation (1, 3).
Neurons are highly polarized cells that contain long axons and dendrites. Because the cell body is the primary site of biosynthesis, a continuous flow of material must be transported long distances from the cell body to the peripheral regions of the neuron. Biochemical and intracellular localization studies of kinesin superfamily proteins suggest that several kinesin motors may power these transport events in neurons (6, 8). One such mouse kinesin motor is KifC2 (7, 12), which is a C-terminal motor originally isolated from a mouse brain cDNA library using a PCR-based cloning technique. Unlike most C-terminal motors, KifC2 is specifically expressed in neural tissues such as the brain, spinal cord, and sciatic nerve. The cellular location of the KifC2 proteins is mainly in neural cell bodies and dendrites but also in axons, suggesting that KifC2 has a role in dendritic and axonal transport (7, 12). Electron microscopic analysis of immunoisolated KifC2-bound organelles using anti-KifC2 revealed that KifC2 associates with multivesicular body-like organelles, suggesting that KifC2 functions as the motor for the transport of the multivesicular body-like organelles in axons or dendrites (12). However, the precise role that KifC2 plays is not clear.
In this paper, we report our results on the generation and analysis of a knockout mouse strain for the KifC2 gene. To understand the in vivo function of the KifC2 gene, we used homologous recombination in embryonic stem cells to construct a mouse strain lacking the KifC2 gene. Homozygous KIFC2 mutants were viable, reproduced normally, and apparently developed normally. These results suggest that KIFC2 is dispensable for normal development and behavior in the mouse.
MATERIALS AND METHODS
Cloning and mapping of the KifC2 gene.
Full-length KifC2 cDNA was used for isolating KifC2 genomic clones from a mouse 129/SvJ genomic phage library (a gift from the laboratory of J. Marth). The genomic clones we isolated from the library were then cloned into the vector Bluescript (Stratagene, La Jolla, Calif.). The map of the KifC2 gene was obtained by digestion of the genomic clones using different restriction enzymes and probing with different regions of the KifC2 cDNA. Southern blotting and other molecular biological techniques were performed according to standard methods.
Generation of targeting vector and ES cells.
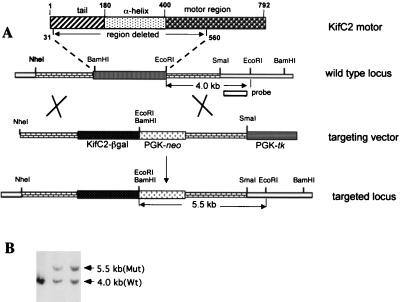
We made a targeting vector to delete a 4.0-kb DNA fragment between BamHI and EcoRI of the KifC2 gene (Fig. 1A), which encodes amino acid residues 31 to 560 of the KifC2 protein. In the targeting vector, the deleted region was replaced with a 4.8-kb β-galactosidase (β-Gal)- and phosphoglycerate kinase (PGK)-neo cassette. This cassette contains a promoterless β-Gal, which should use the KifC2 endogenous promoter to drive expression, and a complete PGK-driven neo gene (4). The vector was made so that the first 30 amino acid residues of the KifC2 motor were fused to the third amino acid of the β-Gal; thus, the β-Gal activity could be used to track the expression pattern of the KifC2 gene. To increase the frequency of homologous recombination in embryonic stem (ES) cells, the targeting vector also included a negative selection marker, PGK-tk (Fig. 1A). The vector was linearized with NheI (Fig. 1A) and introduced into R1 ES cells by electroporation as previously described (9). ES clones resistant to both G418 and ganciclovir were analyzed by Southern blotting. The wild-type and targeted KifC2 alleles were detected as 4.0-kb and 5.5-kb bands, respectively, by Southern blotting of the ES cell genomic DNA digested with EcoRI and probed with a 1.5-kb SmaI and EcoRI DNA fragment (Fig. 1A). The targeted KifC2 allele was confirmed by Southern blotting with different restriction enzymes and probes.
FIG. 1.
Generation and analysis of KifC2 mutants in ES cells. (A) Strategy for generating KifC2 knockout mice. One 4.0-kb DNA fragment between BamHI and EcoRI was replaced with a β-Gal-PGK-neo-pA cassette. This cassette contains a promoterless β-Gal, which should use the KifC2 endogenous promoter to drive expression, and a complete PGK-driven neo gene. The N-terminal 30 amino acid residues of KifC2 are fused to the third amino acid of β-Gal. The targeting vector was linearized with NheI and introduced into RI ES cells as described in Materials and Methods. (B) Southern analysis of ES cells. ES cell genomic DNA was cut with EcoRI and probed with a 1.5-kb SmaI-EcoRI DNA fragment shown in Fig. 1A. The wild-type and targeted KifC2 alleles were detected as 4.0- and 5.5-kb bands, respectively.
Generation and genotypes of KifC2 knockout mice.
Chimeric mice (129/SvJ-derived ES cells in blastocysts of C57BL/6J mice) were generated as previously described (9). Heterozygous mice were used for interbreeding to produce homozygous (KifC2−/−) and heterozygous (KifC2+/−) deletion animals as well as wild-type (KifC2+/+) animals. A set of three primers was used for genotyping the KifC2 mice by PCR. One forward primer based on the KifC2 sequence (CTCTCTGCTCATCTACATCTTC) is common for both wild-type and targeted alleles. Two reverse primers based on the KifC2 sequence (GAGTCGTCCCGCAGCTCTCTTCTGCCCCCAA) and the β-Gal sequence (GGGGATGTGCTGCAAGGCGA) were used for PCR to amplify 800- and 300-bp DNA fragments specifically for the wild-type and targeted alleles, respectively. The genotypes were confirmed by Southern blotting.
Analysis of KifC2 knockout mice.
Gross and histopathological analyses employed standard techniques. Littermate KifC2+/+ and KifC2−/− mice were examined for appearance, posture, circadian activity, home cage assessment, rotarod task performance, balance, and fear conditioning. These behavioral tests were carried out using standard protocols.
Antibody production and Western blot analysis.
Affinity purified rabbit antibodies against mouse KifC2 protein (Affinity Bioregents, Inc.) recognize the C-terminal 14-amino-acid residues (CSGLTLEPPGDPPP) of the KifC2 protein. Western blot analysis of mouse brain tissue was performed as previously described (14).
RESULTS AND DISCUSSION
Deletion of the KifC2 gene in mice.
To explore the in vivo function of the KifC2 gene, we generated a mouse strain lacking a 4.0-kb DNA fragment in the KifC2 gene. The strategy for targeting the KifC2 gene in ES cells is shown in Fig. 1A. The 4.0-kb DNA fragment encodes amino acid residues 31 to 560 of the KifC2 protein, which has a total of 792 amino acids (7, 12). This deletion includes both tail and α coiled-coil domains as well as half of the motor domain. Since the deleted motor domain contains functional ATP binding and ATPase activity sites, it is likely that the deletion will completely abolish the function of KifC2. When the targeting vector was introduced into R1 ES cells by electroporation, both G418- and ganciclovir-resistant clones were selected and analyzed by Southern analysis. Several independent clones with a deletion in the KifC2 gene were obtained (Fig. 1B). In Southern blot analysis, when the ES cell genomic DNA was digested with EcoRI and probed with a 1.5-kb SmaI and EcoRI fragment, the wild-type and targeted KifC2 alleles were detected as 4.0-kb and 5.5-kb bands, respectively (Fig. 1B). The deletion in these ES cells was confirmed with different restriction enzymes and probes in Southern analysis.
When the 129/SvJ-derived ES cells with a deletion in the KifC2 gene were injected into the blastocysts of C57BL/6J mice, several chimeras were generated. When the chimeras were backcrossed to C57BL/6J mice, heterozygous mice were obtained. The heterozygous mice were used for interbreeding to produce knockout (KifC2−/−) and wild-type (KifC2+/+) mice. The deletion was confirmed by Southern blot (Fig. 2A), Northern blot (Fig. 2B), and finally by Western blot (Fig. 2C) analyses. In Southern blot analysis, as in ES cells, when the mouse tail genomic DNA was digested with EcoRI and probed with the 1.5-kb SmaI and EcoRI fragment, the wild-type and targeted KifC2 alleles were detected as 4.0- and 5.5-kb bands, respectively (Fig. 2A). In Northern blot analysis of total RNA obtained from the brains of the knockout mice and wild-type littermates, no KifC2-specific mRNA was observed in KifC2 homozygous mutants with probes corresponding either to the deleted region (probe A) or to a region adjacent to the deleted region (probe B) (Fig. 2B). In Western blot analysis of the brain lysates from the knockout mice and wild-type littermates, no KifC2-specific protein was detected in KifC2 homozygous mutants by using antibodies against the C-terminal 14 amino acids of the KifC2 protein. These results clearly indicate that the mice we generated are truly null for the KifC2 gene.
FIG. 2.
Analysis of KifC2 knockout mice. (A) Southern analysis of the KifC2 knockout mice. Genomic DNA was isolated from tails of the KifC2 mice and digested with EcoRI. Southern blots were probed as described for Fig. 1B. (B) Northern analysis of KifC2 knockout mice. Total RNA was isolated from the brains of the KifC2 mice (+/+, +/−, and −/−) and the duplicate Northern blots were probed with probe A (a cDNA fragment corresponding to the deleted region of the KifC2 gene) and probe B (a cDNA fragment corresponding to the 3′ nondeleted region of the KifC2 gene). (C) Western analysis of KifC2 knockout mice. Brain lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The blot was probed with antibodies against KifC2, which were made using the C-terminal 14 amino acids of the KifC2 protein. (D) Diagram of the structural domains of the KifC2 protein, the probes used for Northern blots, and the antibodies used for Western blots.
Analysis of KifC2 knockout mice.
Mice that carried one copy of the deleted gene were interbred to generate litters that were +/+, +/−, and −/− for KifC2, as determined using genomic PCR and Southern blot analysis. Of 276 offspring from heterozygous parents, there were 68 wild-type (+/+), 136 heterozygous (+/−), and 72 homozygous (−/−) animals. The ratios of +/+, +/−, and −/− mice from the heterozygous parents yielded the predicted Mendelian ratios of 1:2:1 [0.95 < P(χ2 = 0.17) > 0.90, as determined by chi-square test] as expected for nonlethal alleles. Thus, the KifC2 knockout pups were no less viable than their wild-type and heterozygous littermates. Mice heterozygous or homozygous for the KifC2 deletion appeared healthy (up to 2 years of age), developed normally, and did not display any impairment of reproductive capacity and neonatal survival. The breeding pairs with either heterozygous × heterozygous or homozygous × homozygous animals produced litters similar in size to those produced by wild-type breeding pairs, demonstrating that the absence of the KifC2 protein does not hinder the fertility of male or female mice.
Since KifC2 is specifically expressed in the nervous system, we examined whether the deletion of the KifC2 gene affects mouse behaviors, using a variety of tests. Mice were tested in a rotarod apparatus to assess their motor coordination, balance, and ataxia; fear conditioning was used to examine long-term memory. No differences between the KifC2 knockout mice and their wild-type littermates were observed.
At a gross phenotypic level, the absence of KifC2 had no discernible impact. KifC2−/− mice were indistinguishable from their wild-type littermates with respect to body weight and body length. In addition, they exhibited no macroscopic or microscopic alterations in all organs examined (retina, brain, and kidney). Figure 3 shows the morphology of brain (Fig. 3A and B) and spinal cord (Fig. 3C and D) from wild-type (Fig. 3A and C) and KifC2 knockout (Fig. 3B and D) mice. No differences in the morphology of brain and spinal cord between the KifC2 knockout mice and their littermates were observed. We found no differences in white or red blood cell counts or in levels of hemoglobin between wild-type and knockout mice. These results suggest that KIFC2 is dispensable for normal development and many behaviors.
FIG. 3.
Morphology of brain (A, B) and spinal cord (C, D) from wild-type (A, C) and knockout (B, D) mice. Brains and spinal cords were isolated from adult animals, perfused with paraformaldehyde, postfixed, and embedded in paraffin. Sections were stained with cresyl violet.
Functions of KifC2 motor.
The neural expression and association with multivesicular bodies of KifC2 previously reported had suggested that KifC2 would play an important role in the nervous system. However, when we disrupted the KifC2 gene in mice, the nervous system apparently functioned normally in the homozygous mutants. This conclusion is supported by mouse behavioral tests and histological analyses in which we found no difference between the KifC2 knockout mice and their wild-type littermates.
As a C-terminal kinesin motor, the loss of function of KifC2 in the mutants might be provided by other mouse C-terminal kinesin motors such KifC1(12), KifC3 (10, 15, 16), KifC4 (15), and KifC5 (11). Since KifC1, KifC4, and KifC5 have a different expression pattern than KifC2, we think it is unlikely that KifC1, KifC4, or KifC5 can provide the KifC2 function. In contrast, considering the ubiquitous expression of the KifC3 gene (16), it is conceivable that KifC3 may be able to complement the function of the KifC2 gene. However, when mice with both KifC2 and KifC3 deleted were generated, they turned out to be viable, reproduce normally, and develop apparently normally, similar to mice with either single knockout. Thus, the function of the KifC2 protein will have to be explored further. The experiments on mice that we report in this paper may facilitate exploration of the function of the KifC2 gene in the future.
ACKNOWLEDGMENTS
We thank David Hanlon for KifC2 genomic clones and for performing some preliminary experiments on mapping the KifC2 gene.
L.S.B.G. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bloom G S, Endow S A. Motor proteins 1: kinesins. Protein Profile. 1995;2:1105–1171. [PubMed] [Google Scholar]
- 2.Endow S A. Determinants of molecular motor directionality. Nat Cell Biol. 1999;1:E163–E167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- 3.Endow S A. Microtubule motors in spindle and chromosome motility. Eur J Biochem. 1999;262:12–18. doi: 10.1046/j.1432-1327.1999.00339.x. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein L S, Philp A V. The road less traveled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein L S, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 7.Hanlon D W, Yang Z, Goldstein L S. Characterization of KIFC2, a neuronal kinesin superfamily member in mouse. Neuron. 1997;18:439–451. doi: 10.1016/s0896-6273(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 8.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 9.Joyner A L. Gene targeting: a practical approach. Oxford, England: IRL Press; 1993. [Google Scholar]
- 10.Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc Natl Acad Sci USA. 1997;94:9654–9659. doi: 10.1073/pnas.94.18.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navolanic P M, Sperry A O. Identification of isoforms of a mitotic motor in mammalian spermatogenesis. Biol Reprod. 2000;62:1360–1369. doi: 10.1095/biolreprod62.5.1360. [DOI] [PubMed] [Google Scholar]
- 12.Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N. KIFC2 is a novel neuron-specific C-terminal type kinesin superfamily motor for dendritic transport of multivesicular body-like organelles. Neuron. 1997;18:425–438. doi: 10.1016/s0896-6273(00)81243-x. [DOI] [PubMed] [Google Scholar]
- 13.Vale R D, Fletterick R J. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Goldstein L S. Characterization of the KIF3C neural kinesin-like motor from mouse. Mol Biol Cell. 1998;9:249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Hanlon D W, Marszalek J R, Goldstein L S. Identification, partial characterization, and genetic mapping of kinesin-like protein genes in mouse. Genomics. 1997;45:123–131. doi: 10.1006/geno.1997.4901. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Xia C-H, Roberts E A, Bush K, Nigam S K, Goldstein L S B. Molecular cloning and functional analysis of mouse C-terminal kinesin motor KifC3. Mol Cell Biol. 2001;21:765–770. doi: 10.1128/MCB.21.3.765-770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]