Abstract
Candida albicans, the major fungal pathogen in humans, can undergo a reversible transition from ellipsoidal single cells (blastospores) to filaments composed of elongated cells attached end to end. This transition is thought to allow for rapid colonization of host tissues, facilitating the spread of infection. Here, we report the identification of Rfg1, a transcriptional regulator that controls filamentous growth of C. albicans in an environment-dependent manner. Rfg1 is important for virulence of C. albicans in a mouse model and is shown to control a number of genes that have been implicated in this process. The closest relative to Rfg1 in Saccharomyces cerevisiae is Rox1, a key repressor of hypoxic genes. However, Rfg1 does not appear to play a role in the regulation of hypoxic genes in C. albicans. These results demonstrate that a regulatory protein that controls the hypoxic response in S. cerevisiae controls filamentous growth and virulence in C. albicans. The observations described in this paper raise new and intriguing questions about the evolutionary relationship between these processes.
Candida albicans is the most frequently encountered fungal pathogen in humans and is responsible for a wide variety of mucosal and systemic infections (28, 31, 35). With an increase in the number of immunocompromised patients (due primarily to immunosuppressive therapies and to AIDS), the incidence of fungal infections has risen dramatically in recent years (48).
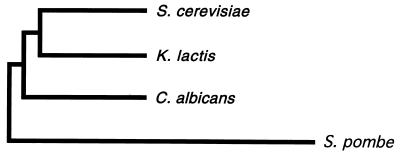
C. albicans and the nonpathogenic yeast Saccharomyces cerevisiae diverged from a common ancestor roughly 200 to 300 million years ago (32) (Fig. 1), and since that time they have been subject to very different selective pressures. C. albicans can survive only in the tissues of warm-blooded animal hosts and became a major human fungal pathogen (28), whereas Saccharomyces species, harmless to humans, are found in a wide variety of environments including soil, fruit, and insects (34, 39). What makes Candida a human pathogen?
FIG. 1.
A phylogenetic tree, constructed by neighbor joining, depicting evolutionary relationships among four fungal species. S. cerevisiae, Kluyveromyces lactis, C. albicans, and S. pombe are positioned on the tree according to the similarity of their small rRNA sequences. Evolutionary dissimilarity corresponds to the horizontal distance joining any two species on the tree (45).
Two general answers can be advanced in response to this question. First, C. albicans may have acquired an additional set of genes that are specifically required for growth in animal hosts. Indeed, now that the C. albicans genome has been completely sequenced, it is known that Candida possesses approximately 2,000 genes that do not contain significant identity to any genes in S. cerevisiae (Stanford DNA Sequencing and Technology Center, http://www-sequence.stanford.edu/group/candida). However, because many of these genes have been identified only recently, the extent to which they contribute to growth in animal hosts is not yet clear. Second, conserved signal transduction and regulatory pathways may have been reconfigured in Candida to respond to the selective pressures of growth in warm-blooded animals. While little direct evidence has been available to support this idea, it is certainly plausible. If some of the same regulatory mechanisms that have been well characterized for S. cerevisiae are used to control the virulence properties of Candida, efforts to develop drugs that combat fungal infections on a molecular level would be accelerated.
One property of C. albicans that has been strongly implicated in virulence is its ability to alter its cell morphology. On rich laboratory media, Candida grows as single, ellipsoidal cells (blastospores). However, under inducing conditions (in the presence of serum, high temperature, neutral pH, or nutrient-poor media) C. albicans can grow in a variety of filamentous forms. The filamentous forms can range from pseudohyphal (cells are attached and elongated but still ellipsoidal) to true hyphal (cells are attached, highly elongated, and cylindrical) growth. The morphological switch is believed to allow C. albicans to rapidly colonize and disseminate in host tissues, facilitating the spread of infection. The reversible blastospore-to-filament transition is likely to be important for virulence: both forms are found in infected tissues, and Candida mutants blocked in the formation of blastospores or filaments are avirulent (3, 21, 28–31). However, certain strains of S. cerevisiae can also undergo the transition to filamentous growth in response to nitrogen starvation; although these strains do not form true hyphae, they can take on a variety of pseudohyphal forms (12). Many of the same gene products appear to function in hyphal growth in Candida and pseudohyphal growth in S. cerevisiae, although the pathways of control do not appear strictly parallel (17, 18, 20). For example, S. cerevisiae does not respond to serum, a major inducer of filamentous growth in Candida.
In this paper, we report the identification of a putative DNA-binding protein, Rfg1, that functions as a transcriptional regulator of hypha-specific genes in Candida. Strains from which the RFG1 gene has been deleted show condition-dependent enhanced filamentous growth and are avirulent in a mouse model. The closest homolog to Rfg1 in S. cerevisiae is the ROX1 gene. In the presence of oxygen, Rox1 is known to bind to the promoters of hypoxic genes and repress transcription via recruitment of the Ssn6-Tup1 corepressor complex (2, 22, 44). In Candida, however, Rfg1 does not appear to regulate hypoxic gene expression but instead appears to be involved exclusively in the control of filamentous growth and virulence. These results provide a direct example of a well-understood, conserved regulatory protein in S. cerevisiae that has apparently been reconfigured in C. albicans to promote virulence. Our findings also raise the intriguing possibility of an evolutionary relationship among filamentous growth, virulence, and hypoxia.
MATERIALS AND METHODS
Strains and media.
The Δrfg1/+ and Δrfg1/Δrfg1 strains were constructed from strain CAI4 (Δura3::imm434/Δura3::imm434) (9) (deletion constructs are described in the next section). In both cases, a URA3-marked disruption cassette was integrated at the genomic locus. Potential deletion strains were screened by PCR to confirm that both ends of the disruption cassette were integrated correctly and that the wild-type copy of RFG1 had been removed. The Δrfg1/Δrfg1::RFG1 strain was generated by integrating a single URA3-marked wild-type copy of the RFG1 gene at the promoter of one of the deleted alleles (the reintegration construct is described in the next section). The Δrfg1/Δrfg1 Δtup1/Δtup1 strain was constructed by deleting both copies of TUP1 (as described previously [4]) in the homozygous rfg1 deletion strain. The Δtup1/Δtup1 strain has been described elsewhere (4). In all experiments, with the exception of those described for Fig. 8, the wild-type strain used was CAF2-1 (Δura3::imm434/URA3) (9). In the virulence experiments (see Fig. 8), the wild-type strain corresponds to RM1 (Δura3::imm434/Δura3::imm434 his1::hisG-URA3-hisG/HIS1) (kindly provided by J. Pla). The S. cerevisiae Δrox1/Δrox1 strain was constructed in a Σ1278b background (a/α ura3-52/ura3-52 leu2Δ1/leu2Δ1) (4) (derived from strains provided by G. Fink and colleagues) using standard two-step gene disruption techniques. C. albicans and S. cerevisiae transformations were carried out as described previously (4, 10). The identity of all strains was confirmed by PCR and/or Southern analysis.
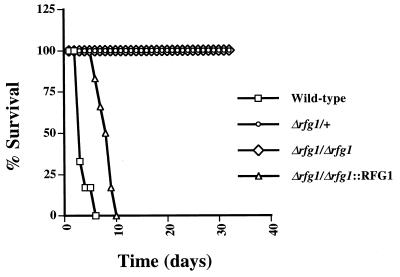
FIG. 8.
Virulence of different Candida strains. Cells (106) of wild-type, Δrfg1/+, Δrfg1/Δrfg1, and Δrfg1/Δrfg1::RFG1 (a reintegrant bearing a single copy of wild-type RFG1 reintegrated at the original locus) strains were injected separately by tail vein into six female BALB/c mice. Survival of the mice was monitored over a period of 32 days.
Standard, uninducing conditions for C. albicans growth were yeast extract-peptone-dextrose (YEPD) medium at 30°C (13). YEPD media (4× and 1/16×) were made by altering the concentrations of yeast extract and peptone accordingly (glucose concentration was kept constant). Serum medium consisted of YEPD plus 10% fetal calf serum. Spider, Lee's, minimal synthetic dextrose (SD), and cornmeal agar media were prepared as described previously (13, 19, 20, 46). SD plates containing 5-fluoroorotic acid and uridine were used to counterselect against the URA3 marker (9). SLAD plates were prepared as described by Gimeno et al. (12) and contained 195 pM ammonium sulfate as the sole nitrogen source.
DNA constructions.
The C. albicans high-copy-number library was constructed as follows. C. albicans genomic DNA was partially digested with Tsp509I to generate inserts with an average length of ∼4.3 kb (kindly provided by B. Braun). These inserts were then ligated into S. cerevisiae vector pRS426 (2μm URA3) (37) cut with EcoRI and phosphatase treated. The library contains ∼50,000 clones, of which >97% contain inserts, and covers the haploid C. albicans genome approximately 14-fold. The 1L49C clone contains a 3.4-kb insert that was sequenced in its entirety (Biomolecular Resource Center, DNA Sequencing Facility, University of California, San Francisco, San Francisco, Calif.).
The S. cerevisiae ROX1 overexpression plasmid was constructed by cloning a PCR fragment containing the ROX1 open reading frame (ORF) (generated from the Σ1278b strain) into the BamHI and HindIII sites of the p426 vector (2μm URA3) (26), just downstream of the ADH1 promoter. A PCR-generated fragment containing the RFG1 ORF was digested with BamHI, filled in, cut with HindIII, and cloned into phosphatase-treated p426 digested with HindIII and SalI (filled in). Both PCR fragments were sequenced in their entirety to verify that they contained no errors. The rox1 deletion plasmid was made by cloning a 546-bp SphI-PstI-digested PCR fragment containing the ROX1 promoter into the SphI and PstI sites of YIplac211 (11). The resulting construct was then digested with BamHI and Asp718, phosphatase treated, and used as a vector to clone a ∼0.5-kb BglII-Asp718-digested PCR fragment containing the last 6 amino acids (aa) of ROX1 and the ROX1 3′ untranslated region. The final construct, YIplac211 Δrox1, was linearized by cutting with ClaI and used for two rounds of two-step gene disruption in the S. cerevisiae Σ1278b strain.
The rfg1 deletion construct was generated by cloning a ∼0.4-kb SphI-PstI-digested PCR fragment, containing part of the RFG1 promoter and the first 107 aa of Rfg1, and a ∼0.4-kb BglII-Asp718-digested PCR fragment, containing the last 7 aa of Rfg1 and the RFG1 3′ untranslated region, into plasmid pBB510 (marked with hisG-URA3-hisG) (5) digested with HindIII-Asp718-BamHI-NsiI or HindIII-Asp718-BglII-PstI by four-way ligation. The resulting rfg1 deletion constructs contain the hisG-URA3-hisG cassette cloned in both orientations to allow for sequential deletion of both RFG1 alleles followed by rapid PCR identification of the four unique ends; both constructs were linearized for transformation by digestion with HindIII and Asp718. The RFG1 reintegration plasmid was made as follows: a 1.7-kb SalI-PstI-digested PCR fragment containing the RFG1 promoter and the first 107 aa of Rfg1 and a 1.9-kb PstI-HindIII fragment from 1L49C containing the remainder of the Rfg1 coding sequence and the RFG1 3′ untranslated region were cloned into pAU71 (a rahB-URA3-rahB-marked vector kindly provided by A. Uhl) cut with SalI and HindIII by three-way ligation. The coding region of the PCR fragment was sequenced to verify that there were no errors. The reintegration construct was linearized for transformation by cleavage with SwaI, a site unique in the RFG1 promoter.
RNA preparation and analysis.
Overnight cultures were diluted to an optical density at 600 nm (OD600) of ∼1.0 (OD600 of ∼0.8 for S. cerevisiae strains), and after 2 h of growth under the appropriate conditions, cells were harvested and chilled on ice. RNA was prepared using hot acid phenol, as described previously (1), and quantitated by spectrophotometer. Approximately 5 μg of each sample was loaded onto a denaturing gel and subjected to Northern analysis. Uniform loading was confirmed by visualization of ethidium bromide-stained rRNA subunits and by comparison of ACT1 expression levels. PCR was used to generate probes to the appropriate genes. Probes were purified on a Qiaquick column and labeled with [α-32P]dATP using a random priming kit (Amersham Pharmacia). All blots were prehybridized and hybridized at 70°C and washed twice for 10 min at 42°C. Quantitation was performed by phosphorimager, and autoradiographs were scanned and digitized (Adobe Photoshop 5.0) for presentation.
For hypoxia experiments, overnight cultures were diluted as described previously. Aerobic cultures were grown normally for 2 h at 30°C in air. In anaerobic cultures, hypoxic conditions were generated by bubbling pure nitrogen through the medium for 2 h at 30°C with gentle agitation, as described elsewhere (49). RNA was prepared in an identical manner for both S. cerevisiae and C. albicans strains.
Virulence experiments.
Growth curves for all strains to be used in virulence studies were determined by growing cells in YEPD media at 30°C. OD600 was determined for each strain at 1-h intervals, and doubling times were calculated as described by Rieg et al. (33). All strains showed a doubling time of ∼1.0 h, with the exception of the Δrfg1/Δrfg1 strain (∼1.2 h). C. albicans strains were then grown overnight, and their cell density was determined by counting individual cells on the hemocytometer. Strains were diluted in saline to a concentration of 2 × 106 cells/ml, and this concentration was confirmed, again, by hemocytometer. A 0.5-ml amount of each strain (106 cells) was then injected intravenously by tail vein into six female BALB/c mice (8 to 10 weeks old; Charles River Co., Cambridge, Mass.). Survival was monitored over a period of 32 days.
RESULTS
Identification of Rfg1.
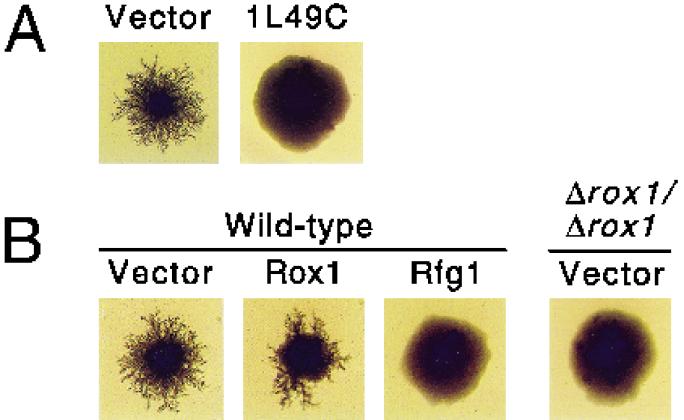
Because C. albicans is a diploid organism that lacks a well-characterized sexual cycle, the rapid identification of genes by standard genetic techniques has been problematic. S. cerevisiae, however, undergoes the transition to pseudohyphal growth upon nitrogen starvation and, in the past, has proven to be a useful tool for the identification of Candida genes that affect filamentous growth (20, 42). Previous work had demonstrated that filamentous growth in Candida is under both positive and negative transcriptional control (4, 21). In order to identify potential negative regulators of filamentous growth, a library bearing C. albicans genomic fragments was constructed in an S. cerevisiae high-copy-number vector. This library was used to transform the Σ1278b strain of S. cerevisiae, and transformants were plated on SLAD plates containing 195 pM ammonium sulfate as the sole nitrogen source. Under these nitrogen-poor conditions, the majority of colonies grow in the pseudohyphal form (12). Transformants were then screened for reduced filamentous growth (RFG). Out of approximately 9,400 transformants (covering the Candida haploid genome about 2.6-fold), a single clone, 1L49C, showed a reduced filamentous growth phenotype that was reproducible upon retransformation. Colonies expressing this clone exhibited little, if any, filament formation even when grown over 12 days on SLAD plates (Fig. 2A).
FIG. 2.
Growth of diploid S. cerevisiae strains under nitrogen starvation conditions. All strains were grown on SLAD medium (12) containing 195 pM ammonium sulfate for 12 days at 30°C and photographed at approximately 9× magnification. (A) A Σ1278b strain contained YCplac111 (a LEU2-marked CEN plasmid) (11) in addition to the indicated constructs. Vector, pRS426 (URA3 2μm) (37); 1L49C, pRS426 containing the 3.4-kb insert that confers reduced filamentous growth. (B) Wild-type and homozygous ROX1 deletion strains (Δrox1/Δrox1) contained YCplac111 in addition to the indicated constructs. Vector, p426 (URA3 2μm ADH1 promoter) (26); Rox1, p426 expressing ROX1 under ADH1 control; Rfg1, p426 expressing RFG1 under ADH1 control.
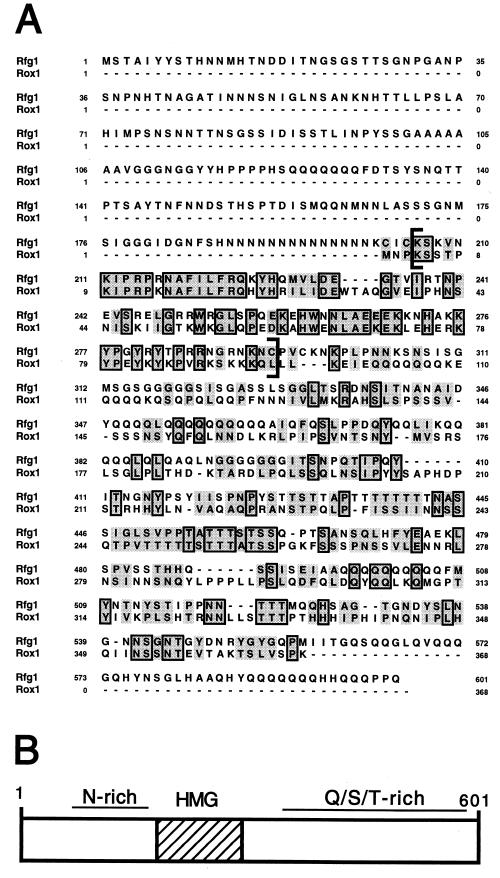
Sequence analysis of the 1L49C clone revealed a 3.4-kb insert containing a 1.8-kb ORF which we have termed the RFG1 gene (GenBank accession no. AF330198). Expression of this ORF alone under control of an S. cerevisiae ADH1 promoter generated a phenotype nearly identical to that of the original 1L49C clone (Fig. 2B). RFG1 encodes a 601-aa, 65-kDa protein with an N terminus rich in asparagine and a C terminus rich in glutamine, serine, and threonine (Fig. 3). Perhaps the most striking feature of Rfg1 is an 89-aa high-mobility group (HMG) DNA-binding domain (aa 203 to 291) that is 52% identical to that encoded by the S. cerevisiae ROX1 gene (Fig. 3A). Rox1 is a key transcriptional repressor of hypoxic genes in S. cerevisiae, and the HMG domain of this protein has been shown to bind directly to hypoxic operator sites in vitro (2, 23, 50). The Rfg1 HMG domain also showed weaker identity to a number of non-S. cerevisiae fungal mating-type proteins including mating-type M-specific polypeptide MC of Schizosaccharomyces pombe, pheromone response factor 1 of Ustilago maydis, and MAT-1-3 of Gibberella fujikuroi (14, 16).
FIG. 3.
Sequence and characteristics of Rfg1. (A) Comparison of the amino acid sequences of C. albicans Rfg1 and S. cerevisiae Rox1 proteins. Regions of identity are boxed, and regions of similarity are shaded. The sequence in brackets corresponds to the HMG DNA-binding domain. Dashes indicate gaps in the alignment. Rfg1 contains ∼200 more aa at its N terminus than does Rox1. Sequences were aligned using the Pileup program (GCG, Inc.). (B) Schematic representation of Rfg1. The HMG domain is indicated by the hatched box.
Rfg1 regulates C. albicans filamentous growth in a nutrient-dependent manner.
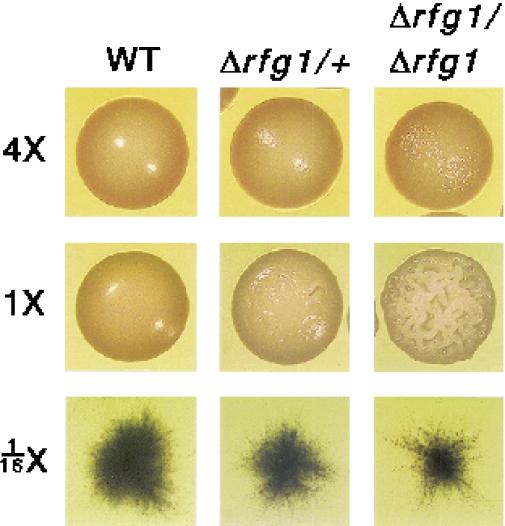
In order to determine whether Rfg1 functions as a regulator of filamentous growth in C. albicans, we generated both heterozygous (Δrfg1/+) and homozygous (Δrfg1/Δrfg1) deletion strains. These strains showed a general increase in filamentous growth relative to that for the parent strain on YEPD medium; however, this phenotype was most pronounced after several days of growth, suggesting that the strains were sensitive to depletion of nutrients on the plates. In order to test this possibility directly, wild-type, Δrfg1/+, and Δrfg1/Δrfg1 strains were plated on solid media containing 4×, 1×, and 1/16× concentrations of YEP with the glucose concentration kept constant. As shown in Fig. 4, at 1× YEPD, Δrfg1/Δrfg1 mutants exhibit a highly crinkled (or “lacy”) colony morphology compared to that of the “smooth” wild-type strain. This crinkled morphology is diagnostic of a high proportion of filamentous cells in the colony, and this surmise was verified by microscopic examination of cells taken from colonies. In fact, Δrfg1/Δrfg1 colonies have a rough skin which is composed almost exclusively of filaments and a smooth interior which is composed primarily of blastospores (data not shown). A plate assay has also indicated that filamentous growth observed for rfg1 deletion strains is invasive (data not shown). The Δrfg1/+ heterozygous strains show a phenotype intermediate between that of the wild-type and the homozygous deletion strains. At 4× YEPD, however, the enhanced filamentation of the rfg1 mutant strains appears to be almost completely abolished. Wild-type C. albicans is known to form filaments on nutrient-poor media (1/16× YEPD, Fig. 4) (28). Under these conditions, rfg1 homozygous mutants may show a slight reduction in filamentous growth. These results show that on 1× YEPD Rfg1 formally acts as a repressor of filament formation whereas on 1/16× YEPD it may slightly activate filamentous growth.
FIG. 4.
Growth of wild-type and rfg1 C. albicans strains at different nutrient concentrations. Wild-type (WT), Δrfg1/+, and Δrfg1/Δrfg1 strains were grown overnight in 1× YEPD, diluted, and plated out on YEPD plates containing 4, 1, and 1/16 times the normal concentrations of yeast extract and peptone (glucose concentration was kept constant). Strains were grown for 3 days at 30°C, and individual colonies were photographed at approximately 3× magnification.
Wild-type, Δrfg1/+, and Δrfg1/Δrfg1 strains were also grown under a variety of different conditions which are known to induce filamentous growth. The mutants did not appear significantly different from wild-type strains when grown on cornmeal agar plates. However, on Spider medium, serum, Lee's pH 4.5 mannitol, and Lee's pH 6.8 mannitol plates, rfg1 strains were severely defective for filament formation but may also have been reduced in growth rate, complicating the assessment of the role of RFG1 on filamentous growth per se on these media.
Rfg1 functions as a condition-specific transcriptional regulator of hypha-specific genes.
We next sought to determine whether the effects of rfg1 mutations on filamentous growth correlated with changes in the expression of known hypha-specific genes. RNA was prepared from wild-type and Δrfg1/Δrfg1 strains grown in liquid media under a variety of different inducing conditions, and transcript levels of specific target genes were determined by Northern analysis (Fig. 5).
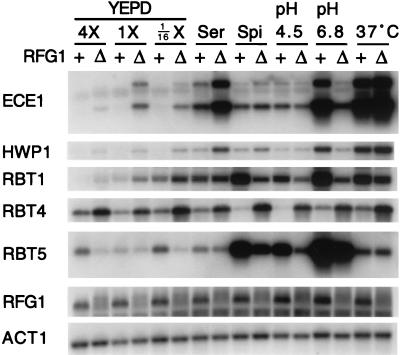
FIG. 5.
Expression of various filament-specific genes under different environmental conditions in wild-type and rfg1 strains. Wild-type (+) and Δrfg1/Δrfg1 (Δ) strains were grown overnight in 1× YEPD, diluted to an OD600 of ∼1.0, and grown for 2 h at 30°C in the indicated media (YEPD medium was prepared as described in the legend to Fig. 3; Ser, 1× YEPD + 10% fetal calf serum; Spi, Spider; pH 4.5, Lee's pH 4.5 mannitol; pH 6.8, Lee's pH 6.8 mannitol; 37°C, 1× YEPD at 37°C). RNA was prepared from each strain, and Northern analysis was carried out using probes to the indicated genes.
Two general trends appear to emerge from these data. First, under standard noninducing conditions (1× YEPD) Rfg1 functions as a transcriptional repressor. For example, the ECE1, HWP1, RBT1, and RBT4 transcripts are all derepressed in the Δrfg1/Δrfg1 strain when cells are grown in 1× YEPD (Fig. 5, lanes 3 and 4). Second, under specific inducing conditions, Rfg1 functions as a strong transcriptional activator. For example, induction of the ECE1, HWP1, and RBT1 transcripts is significantly reduced in the rfg1 deletion strain when cells are grown in Lee's pH 6.8 mannitol medium (Fig. 5, lanes 13 and 14). We do note, however, that there are some exceptions to these trends (e.g., RBT4).
In order to determine whether RFG1 itself is regulated at the transcriptional level, we examined RFG1 expression under the various conditions (Fig. 5). Little or no change was observed, suggesting that the condition-specific Rfg1 transcriptional activity is regulated at a posttranslational level.
tup1 mutation is epistatic to rfg1 mutation.
A major pathway required for the negative regulation of pseudohyphal and hyphal growth is defined by the C. albicans homolog of the S. cerevisiae TUP1 global transcriptional repressor. Deletion of both copies of the C. albicans TUP1 gene results in constitutive filamentous growth that is not responsive to serum or other inducers; tup1 null mutants are also completely defective for virulence in a mouse model (3). In S. cerevisiae, Tup1 forms a complex with Ssn6 which, in turn, is recruited to the promoters of target genes via interaction with promoter-specific DNA-binding proteins (15, 44, 47). This complex is responsible for transcriptional repression of a-specific mating genes, haploid-specific genes, glucose-repressed genes, oxygen-repressed genes, and DNA damage-inducible genes (8, 15, 25, 36, 43, 50). Since C. albicans TUP1 complements an S. cerevisiae Δtup1 mutant, the Candida protein has been proposed to function by a similar mechanism to repress genes important for filamentous growth and virulence (4). Consistent with this notion, several previously characterized hypha-specific transcripts (HWP1, ECE1, HYR1, and ALS1) show significant derepression in Δtup1/Δtup1 strains (5).
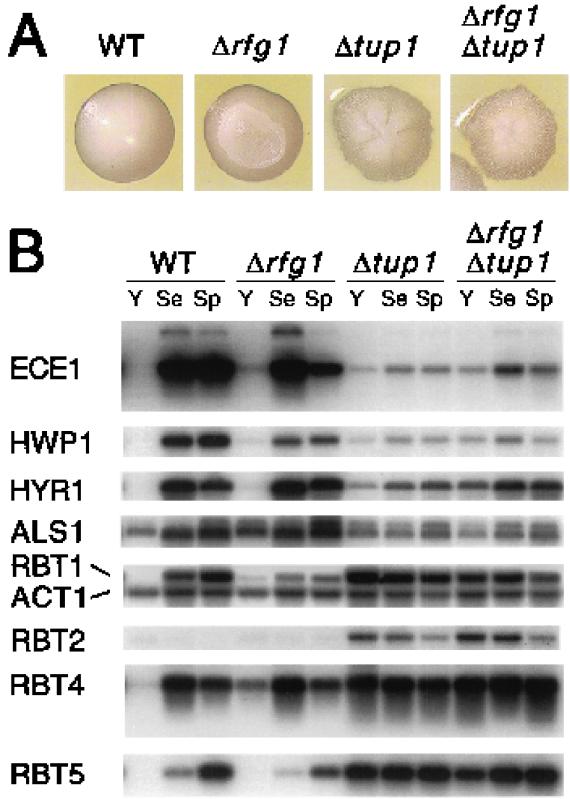
In C. albicans, Rfg1 regulates a number of genes that are also regulated by Tup1 (Fig. 5), and in S. cerevisiae, the Rfg1 homolog (Rox1) is known to direct repression of hypoxic transcripts via recruitment of the Ssn6-Tup1 corepressor complex. We therefore sought to test whether Candida Rfg1 acts through Tup1 by determining whether a tup1 deletion was epistatic to an rfg1 deletion. As shown in Fig. 6A, Δrfg1/Δrfg1 colonies have a crinkled phenotype whereas those of the Δtup1/Δtup1 strain have a more “mountainous” appearance; the tup1 deletion strain also adheres much more strongly to the agar than does the Δrfg1/Δrfg1 strain (data not shown). With respect to both colony morphology and adherence, the Δtup1/Δtup1 Δrfg1/Δrfg1 mutant appears nearly identical to the tup1 deletion strain (Fig. 6A).
FIG. 6.
Epistasis analysis of tup1 and rfg1 mutants. (A) A wild-type (WT) strain and strains bearing homozygous deletions of rfg1, tup1, and rfg1 tup1 were streaked on SD plates lacking uracil and grown for 3 days at 30°C. Individual colonies were photographed at approximately 3× magnification. (B) The strains described for panel A were grown overnight in 1× YEPD, diluted to an OD600 of ∼1.0, and grown for 2 h at 30°C in the indicated media (Y, 1× YEPD at 30°C; Se, 1× YEPD + 10% fetal calf serum at 37°C; Sp, Spider medium at 37°C). RNA was prepared from each strain, and Northern analysis was carried out using probes to the indicated genes.
To assess the relationship between Tup1 and Rfg1 at the transcriptional level, we examined serum and Spider medium induction (at 37°C) of a variety of Tup1-regulated transcripts in wild-type, Δrfg1/Δrfg1, Δtup1/Δtup1, and Δrfg1/Δrfg1 Δtup1/Δtup1 strains (Fig. 6B). In general, the transcriptional profile of the double mutant appeared very similar, if not identical, to that of the tup1 deletion strain, suggesting that TUP1 and RFG1 function in the same pathway. In the case of a few genes, however, there are some differences in the transcriptional effects observed for Δtup1/Δtup1 and Δtup1/Δtup1 Δrfg1/Δrfg1 strains, raising the possibility that RFG1 may be capable of regulating transcription in a TUP1-independent manner.
One TUP1-repressed gene, RBT2, is not induced when cells are grown in serum or Spider medium. RBT2 transcript levels appear nearly identical in Δtup1/Δtup1 and Δtup1/Δtup1 Δrfg1/Δrfg1 strains and are not significantly affected in the rfg1 deletion strain. These findings, combined with the previous results, are consistent with the notion that Rfg1 plays a role in the regulation of a subclass of Tup1 target genes specifically involved in filamentous growth and virulence.
Rfg1 fails to regulate hypoxic transcripts in response to oxygen starvation in C. albicans
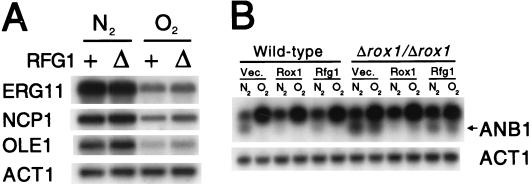
As discussed above, the gene with the greatest sequence similarity to RFG1 is ROX1, a major regulator of the hypoxic response in S. cerevisiae. This similarity raised the intriguing possibility that Rfg1, a regulator of filamentous growth in Candida, might respond to hypoxic conditions. To test this idea, RNA was prepared from liquid cultures of wild-type and Δrfg1/Δrfg1 strains of Candida grown in the presence or absence of oxygen and mRNA levels of three Candida genes suspected (from homology with S. cerevisiae) to be regulated by hypoxia were determined. As shown in Fig. 7A, all three putative hypoxic genes from Candida (ERG11, NCP1, and OLE1) were indeed derepressed upon oxygen starvation. Unlike deletion of ROX1 in S. cerevisiae (6, 22), however, deletion of RFG1 in C. albicans fails to cause a derepression of these transcripts in the presence of oxygen. These results indicate that Rfg1 does not play a central role in the C. albicans hypoxic response (with respect to at least three major hypoxic transcripts) and suggest that this response is mediated by a different regulator(s). However, we cannot formally exclude the possibility that Rfg1 regulates other hypoxic genes that have not yet been examined. Thus far, no genes closely related to RFG1 have been reported by the C. albicans sequencing project at the Stanford DNA Sequencing and Technology Center (http://www-sequence.stanford.edu/group/candida).
FIG. 7.
Expression of C. albicans and S. cerevisiae transcripts under hypoxic conditions. (A) Wild-type (+) and Δrfg1/Δrfg1 (Δ) C. albicans strains were grown overnight in 1× YEPD, diluted to an OD600 of ∼1.0, and grown for 2 h in YEPD at 30°C in the presence (O2) or absence (N2) of oxygen. Hypoxic conditions were generated by bubbling pure nitrogen (N2) through the liquid cultures (49). RNA was prepared from each strain, and Northern analysis was carried out using probes to the indicated genes. (B) Wild-type and Δrox1/Δrox1 S. cerevisiae strains bearing the indicated plasmids (as described for Fig. 1B) were grown overnight in SD medium with selection, diluted to an OD600 of ∼0.8, and grown for 2 h in the same medium at 30°C in the presence (O2) or absence (N2) of oxygen (as described above). RNA was prepared from each strain, and Northern analysis was carried out using probes to the indicated genes. Note that the ANB1 probe also hybridizes to a second, closely related gene (upper band) (22). As indicated by the arrow, the bottom band corresponds to ANB1.
To determine whether Rfg1 can function as a repressor of hypoxic genes when ectopically expressed in S. cerevisiae, we transformed wild-type and Δrox1/Δrox1 strains with high-copy plasmids expressing ROX1 and RFG1 and examined expression of ANB1 (a Rox1 target gene) in the presence and absence of oxygen (Fig. 7B). As previously observed, ANB1 is induced in the absence of oxygen and repressed in its presence, and deletion of ROX1 relieves this repression (22). Repression is restored by expression of ROX1 but only slightly affected by RFG1 expression. Thus, RFG1 appears to poorly repress transcription of the hypoxic genes in S. cerevisiae.
Both Rfg1 and Rox1 can regulate filamentous growth in S. cerevisiae.
Our finding that the Candida RFG1 gene, when overexpressed, could repress filamentous growth in S. cerevisiae (Fig. 2B) led us to question whether Rox1, which contains a homologous DNA-binding domain, could also play a role in S. cerevisiae filament formation. We transformed the Σ1278b strain with a high-copy plasmid expressing ROX1 and grew transformants on SLAD plates. Overexpression of ROX1 in S. cerevisiae led to at most a minor decrease in filament formation compared to the dramatic reduction observed upon overexpression of RFG1 (Fig. 2B). Pseudohyphal growth was significantly impaired, however, in Σ1278b strains having both copies of ROX1 deleted.
The Δrfg1/Δrfg1 strain is avirulent in a mouse model.
As a final experiment, we determined whether RFG1 was important for virulence of C. albicans in vivo. Twenty-four mice (six per strain) were injected by tail vein with 106 cells of wild-type, Δrfg1/+, Δrfg1/Δrfg1, and Δrfg1/Δrfg1::RFG1 strains. The final strain contains a single wild-type copy of RFG1 reintegrated at its original locus in the rfg1 homozygous deletion strain. As shown in Fig. 8, mice injected with wild-type C. albicans died over a period of 2 to 6 days. Mice carrying the Δrfg1/Δrfg1::RFG1 reintegrant strain were slightly reduced for virulence and died between day 6 and day 10. In marked contrast, however, all mice injected with the Δrfg1/+ and Δrfg1/Δrfg1 strains were alive after 32 days. In general, animals carrying the Δrfg1/+ strain had lost weight and were visibly sick but still active. Mice injected with the Δrfg1/Δrfg1 strain were mostly in good health throughout the course of the experiment.
On the surface, the difference in virulence between the Δrfg1/+ and Δrfg1/Δrfg1::RFG1 strains appears puzzling, since both strains contain a single copy of RFG1. However, several factors could account for this difference: for example, slight differences in the expression levels or activities of the two RFG1 alleles could exist, or expression of the reintegrated copy of RFG1 could be elevated because it is in proximity to an actively transcribed URA3 gene. Consistent with these hypotheses, Δrfg1/+ and Δrfg1/Δrfg1::RFG1 strains showed small differences at the phenotypic level, with the Δrfg1/Δrfg1::RFG1 strain appearing closer to the wild type (data not shown). Despite these differences, our results clearly suggest that RFG1 is important for virulence and may be a regulator of virulence-specific genes in C. albicans.
DISCUSSION
In this paper, we report the identification of Rfg1, an HMG domain protein that plays an important role in controlling C. albicans morphogenesis and virulence. The nearest relative of Rfg1 in S. cerevisiae, Rox1, regulates the response to hypoxia (22, 23, 50). Because Rfg1 does not regulate the hypoxic response in Candida, these results raise questions about the divergence of C. albicans and S. cerevisiae since they shared a common ancestor, an issue which is discussed in detail below.
Rfg1 functions as a regulator of filamentous growth and virulence in C. albicans
Rfg1 appears to function in both the positive and negative regulation of filamentous growth in C. albicans, depending upon the environmental conditions. For example, on 1× YEPD medium rfg1 mutants are hyperfilamentous compared to the wild type, whereas on some types of media that normally induce filamentous growth, the rfg1 mutants show a defect in filamentation.
The transcriptional profile of genes whose transcription is induced during the switch to filamentous growth (discussed in detail below) mirrors these phenotypic effects: under certain conditions, some genes are derepressed in the rfg1 deletion strain compared to the parent strain; under other conditions, some of these same genes are underexpressed when RFG1 is deleted. At this point, we do not know which of these effects result from the direct action of Rfg1 on target genes and which are indirect effects; however, Rfg1 acts formally as both a repressor and an activator of gene expression. Finally, the fact that Rfg1 mutants are avirulent in a mouse model suggests that the regulatory circuit defined by Rfg1 plays an important role in pathogenesis.
Genes controlled by Rfg1.
The results in Fig. 5 and 6 indicate that Rfg1 functions as a transcriptional regulator of at least eight genes, including cell wall or (in some cases) putative cell wall components that are specifically expressed when Candida grows in the filamentous forms. We briefly summarize what is known about each of these genes before returning to the role of Rfg1 in their regulation. One regulated gene, HWP1, is expressed on the surface of hyphal cells and has been shown to form covalent links with host tissues. Mouse studies indicate that C. albicans strains having HWP1 deleted are severely defective for virulence (40). A second gene, RBT1, is closely related to HWP1 and is thought to be a component of the cell wall, although its function is not known (3). HYR1 encodes a nonessential putative glycosylated cell wall protein. Expression of another Rfg1-regulated gene, ECE1, is known to correlate with the extent of cell elongation during filament formation. A fifth gene, ALS1, has been shown to induce adherence to endothelial and epithelial cells when expressed in S. cerevisiae and may belong to a family of Candida adhesins. RBT4, another regulated gene, is highly similar to the PR, or pathogenesis-related, family of proteins in plants. PR proteins are secreted, have a very stable three-dimensional structure, and are known to possess antifungal activity (27, 41). Recent experiments have shown that RBT4 is strongly required for virulence in both mouse and rabbit models. This protein has been hypothesized to play a role in the ability of C. albicans to damage host cells and/or competing microbes (3). The last gene, RBT5, bears identity to the cell surface proline-rich antigen protein of Coccidioides immitis, a pathogenic fungus (7). Both proteins contain a conserved CRoW motif that is thought to form a disulfide bond-linked structure in the extracellular environment (3). Although we have not yet examined the effect of RFG1 on a wide variety of genes, we can say that RFG1 specifically regulates several genes that themselves are important for filamentous growth and virulence.
In general, Rfg1 functions as a transcriptional repressor of these genes when cells are grown in noninducing conditions, such as 1× YEPD. rfg1Δ cells are hyperfilamentous under these same conditions, and it is likely that this morphological phenotype results from the derepression of a set of genes, some of which we have examined directly. RFG1 is also required for full transcriptional activation of certain genes under specific inducing conditions. For example, RFG1 is essential for the full activation of three genes (RBT1, HWP1, and ECE1) in neutral pH (Lee's pH 6.8 mannitol medium).
Mechanism of Rfg1 action.
Several lines of evidence suggest that Rfg1 directs transcriptional repression at hypha-specific promoters via recruitment of the Ssn6-Tup1 complex. First, the Rfg1 protein sequence contains significant identity to that of Rox1, a repressor of hypoxic genes in S. cerevisiae known to function through Ssn6-Tup1 (2, 22, 44). Second, a number of the same hypha-specific genes are derepressed in both tup1 and rfg1 deletion strains (5). Third, with regard to colony morphology, tup1 mutations are epistatic to rfg1 mutations, which is to be expected if Rfg1 is one of several DNA-binding proteins that regulate hyphal transcripts via Ssn6-Tup1 repression. Finally, for several hypha-specific genes the extent of transcriptional derepression observed with Δrfg1/Δrfg1 Δtup1/Δtup1 double mutants is the same as that in the single mutants.
An S. cerevisiae hypoxic regulatory pathway controls filamentous growth and virulence in C. albicans
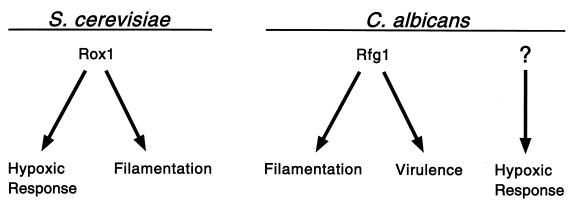
Our identification of Rfg1, an HMG protein related to the Rox1 repressor of hypoxic genes, was based on its ability to repress pseudohyphal growth in S. cerevisiae. Although these two proteins share a DNA-binding domain that is 52% identical and both can function in S. cerevisiae, they are not interchangeable in this organism. For example, overexpression of RFG1, but not ROX1, causes a large reduction in pseudohyphal growth in S. cerevisiae. Rox1, but not Rfg1, is capable of directing high levels of repression at the promoter of a hypoxic gene, ANB1. On the other hand, both proteins are capable of functioning as strong repressors of an S. cerevisiae filament-specific gene, FLO11 (data not shown). The differences could be due to differences in DNA-binding specificities of the two proteins or to differences in their associations with other proteins. In any case, Rfg1 in Candida specifically controls genes important for filamentous growth and virulence but not the major hypoxic genes; a second regulator, not yet identified, must direct repression of hypoxic genes in C. albicans. In S. cerevisiae, Rox1 is well established as a central regulator of hypoxic genes (22, 23), and in this paper, we show that it also appears important for filamentous growth, at least in the Σ1278b strain (Fig. 9).
FIG. 9.
Relationships among filamentous growth, virulence, and hypoxia in S. cerevisiae and C. albicans. In S. cerevisiae, Rox1 (the Rfg1 homolog) functions as a major regulator of hypoxic genes and is also important for pseudohyphal growth. In C. albicans, Rfg1 regulates both virulence and invasive filamentous growth. Another pathway, not yet identified, probably controls the hypoxic response in C. albicans.
These observations suggest that, in an evolutionary sense, filamentous growth and the response to oxygen starvation may be closely related to each other. Previous studies of Candida have implicated O2 levels in the control of filamentous growth (24, 38); a requirement for oxygen in rapid filamentous growth, almost certainly an ATP-driven process, seems logical considering that a majority of the cell's energy needs are met by oxidative phosphorylation.
Rox1 is a key regulator of the hypoxic response in a yeast (S. cerevisiae) harmless to humans. Here, we show that the most closely related protein in the major human fungal pathogen C. albicans directs invasive filamentous growth and virulence. After many years of selective pressures in warm-blooded mammalian hosts, the C. albicans Rfg1 protein appears to have lost its ability to function as a major hypoxic regulator, expanded its ability to direct strong, invasive filamentous growth, and taken on a new role as an important regulator of virulence. It will be interesting to see how many other routine regulatory pathways have been redirected toward virulence in pathogenic fungi.
ACKNOWLEDGMENTS
We thank J. Pla, G. Fink, B. Braun, A. Uhl, D. Inglis, and C. Hull for plasmids, strains, primers, and probes and R. Khalaf and R. Zitomer for communicating results prior to publication. We are especially grateful to D. Inglis for assistance in carrying out the virulence experiments. We thank members of the Johnson laboratory for fruitful discussions during the course of the experiments. Sequence data for C. albicans were obtained from the Stanford DNA Sequencing and Technology Center website at http://www-sequence.stanford.edu/group/candida.
Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. This work was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG-1512, to D.K. and by National Institutes of Health grant GM-37049 to A.D.J.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- 2.Balasubramanian B, Lowry C V, Zitomer R S. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol Cell Biol. 1993;13:6071–6078. doi: 10.1128/mcb.13.10.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun B R, Head W S, Wang M X, Johnson A D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 5.Braun B R, Johnson A D. TUP1, CPH1, and EFG1 make independent contributions to filamentation in Candida albicans. Genetics. 2000;155:57–67. doi: 10.1093/genetics/155.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deckert J, Perini R, Balasubramanian B, Zitomer R S. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics. 1995;139:1149–1158. doi: 10.1093/genetics/139.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dugger K O, Villareal K M, Ngyuen A, Zimmermann C R, Law J H, Galgiani J N. Cloning and sequence analysis of the cDNA for a protein from Coccidioides immitis with immunogenic potential. Biochem Biophys Res Commun. 1996;218:485–489. doi: 10.1006/bbrc.1996.0086. [DOI] [PubMed] [Google Scholar]
- 8.Elledge S J, Zhou Z, Allen J B, Navas T A. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 9.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 11.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro-mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 12.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 14.Hartmann H A, Kahmann R, Bolker M. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 1996;15:1632–1641. [PMC free article] [PubMed] [Google Scholar]
- 15.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 16.Kelly M, Burke J, Smith M, Klar A, Beach D. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988;7:1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologues of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Kohler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 21.Lo H J, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 22.Lowry C V, Zitomer R S. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc Natl Acad Sci USA. 1984;81:6129–6133. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowry C V, Zitomer R S. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4651–4658. doi: 10.1128/mcb.8.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mardon D, Balish E, Phillips A W. Control of dimorphism in a biochemical variant of Candida albicans. J Bacteriol. 1969;100:701–707. doi: 10.1128/jb.100.2.701-707.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukai Y, Harashima S, Oshima Y. AAR1/TUP1 protein, with a structure similar to that of the β subunit of G proteins, is required for a1-α2 and α2 repression in cell type control of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3773–3779. doi: 10.1128/mcb.11.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 27.Niderman T, Genetet I, Bruyere T, Gees R, Stintzi A, Legrand M, Fritig B, Mosinger E. Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odds F C. Candida and candidosis. 2nd ed. London, United Kingdom: Baillière Tindall; 1988. [Google Scholar]
- 29.Odds F C. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- 30.Odds F C. Morphogenesis in Candida albicans. Crit Rev Microbiol. 1985;12:45–93. doi: 10.3109/10408418509104425. [DOI] [PubMed] [Google Scholar]
- 31.Odds F C. Pathogenesis of Candida infections. J Am Acad Dermatol. 1994;31:S2–S5. doi: 10.1016/s0190-9622(08)81257-1. [DOI] [PubMed] [Google Scholar]
- 32.Pesole G, Lotti M, Alberghina L, Saccone C. Evolutionary origin of nonuniversal CUG (Ser) codon in some Candida species as inferred from a molecular phylogeny. Genetics. 1995;141:903–907. doi: 10.1093/genetics/141.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rieg G, Fu Y, Ibrahim A S, Zhou X, Filler S G, Edwards J E. Unanticipated heterogeneity in growth rate and virulence among Candida albicans AAF1 null mutants. Infect Immun. 1999;67:3193–3198. doi: 10.1128/iai.67.7.3193-3198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose A H, Harrison J S, editors. The yeasts. New York, N.Y: Academic Press, Inc.; 1987. [Google Scholar]
- 35.Rubin R H. Fungal and bacterial infections in the immunocompromised host. Eur J Clin Microbiol Infect Dis. 1993;12(Suppl. 1):S42–S48. doi: 10.1007/BF02389877. [DOI] [PubMed] [Google Scholar]
- 36.Schultz J, Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims W. Effect of carbon dioxide on the growth and form of Candida albicans. J Med Microbiol. 1986;22:203–208. doi: 10.1099/00222615-22-3-203. [DOI] [PubMed] [Google Scholar]
- 39.Skinner F A, Passmore S M, Davenport R R, editors. Biology and activities of yeast. London, United Kingdom: Academic Press; 1980. [Google Scholar]
- 40.Staab J F, Bradway S D, Fidel P L, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 41.Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- 42.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trumbly R J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 44.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 45.Van de Peer Y, Hendriks L, Goris A, Neefs J-M, Vancanneyt M, Kersters K, Berny J-F, Hennebert G L, de Wachter R. Evolution of basidiomycetous yeasts as deduced from small ribosomal subunit RNA sequences. Syst Appl Microbiol. 1992;15:250–258. [Google Scholar]
- 46.Vidotto V, Caramello S, Gallo M G. A new medium for the production of chlamydoconidia by Candida albicans. Mycopathologia. 1986;95:73–75. doi: 10.1007/BF00437163. . (Erratum, 97:135–136, 1997.) [DOI] [PubMed] [Google Scholar]
- 47.Williams F E, Varanasi U, Trumbly R J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinner S H, Scheld W M. Brief review of fungal infections. Eur J Clin Microbiol Infect Dis. 1993;12(Suppl. 1):S146–S149. [Google Scholar]
- 49.Zitomer R S, Limbach M P, Rodriguez-Torres A M, Balasubramanian B, Deckert J, Snow P M. Approaches to the study of Rox1 repression of the hypoxic genes in the yeast Saccharomyces cerevisiae. Methods. 1997;11:279–288. doi: 10.1006/meth.1996.0422. [DOI] [PubMed] [Google Scholar]
- 50.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]