Abstract
Previous cross-sectional studies have shown that skin microbiomes in adults are distinct from children. However, the human skin microbiome in individuals as they sexually mature has not been studied as extensively. We performed a prospective, longitudinal study to investigate puberty-associated shifts in skin microbiota. Twelve healthy children were evaluated every 6–18 months for up to 6 years. Using 16S rRNA (V1-V3) and ITS1 amplicon sequencing analyzed with DADA2, we characterized the bacterial and fungal communities of 5 different skin and nares sites. We identified significant alterations in the composition of skin microbial communities, transitioning towards a more ‘adult’ microbiome, during puberty. The microbial shifts were associated with Tanner stages (classification method for degree of sexual maturation) and showed noticeable sex-specific differences. Over time, female children demonstrated predominance of Cutibacterium with decreasing diversity. For fungal communities, Malassezia predominated at most skin sites in more sexually mature subjects, which was more pronounced in female children. The higher relative abundances of these lipophilic taxa, Cutibacterium acnes and Malassezia restricta, were strongly associated with serum sex hormone concentrations with known influence on sebaceous gland activity. Taken together, our results support the relationship between sexual maturation, skin physiology, and skin microbiome.
Introduction
Human skin is an ecosystem that harbors a variety of commensal and pathogenic microbes, including bacteria, fungi, and viruses. Alterations of these microbial communities have been linked to various skin disorders including acne and atopic dermatitis (O’Neill and Gallo, 2018, Paller et al., 2019). Skin microbial communities vary among individuals, which is attributed to several host factors such as age, sex, and body sites (Capone et al., 2011, Costello et al., 2009, Fierer et al., 2008, Grice et al., 2009, Marples, 1982). Age-related shifts in skin microbial communities among different age groups are well-known through several culture-based, targeted PCR-based, and sequencing-based studies (Oh et al., 2012, Somerville, 1969, Sugita et al., 2010). Previous cross-sectional studies measured sexual maturation with Tanner staging (based on secondary sex characteristics from stages 1 to 5) to demonstrate that skin bacterial and fungal communities in postpubertal young adults are distinct from those of younger children (Jo et al., 2016, Oh et al., 2012). Differences in the skin microenvironments in children versus adults have been postulated to contribute to microbiome patterns distinguishing prepubertal children (Tanner1) with higher relative abundances of Firmicutes (Streptococcus), Bacteroidetes and Proteobacteria (β, γ) and higher microbial diversity from young adults (Tanner5) with predominant lipophilic microbes (Corynebacterium, Cutibacterium, and Malassezia). For instance, lipid-rich environments due to mature sebaceous glands in young adults can encourage the growth of lipophilic skin microbial inhabitants (McGinley et al., 1980, Pochi et al., 1979, Ro and Dawson, 2005). While a cross-sectional study of children demonstrated different fungal relative abundances in sebaceous skin of boys as compared to girls (Jo et al., 2016), the effect of sex on skin microbial communities is less studied.
Puberty is characterized by profound physical and sexual development mediated by hormones which likely contribute to skin microbiome shifts. Various microbe-associated skin disorders including acne, hidradenitis suppurativa, and tinea versicolor often begin during puberty when major skin physiological changes occur; however, the skin microbiome in children during sexual maturation is not extensively studied. Herein, we performed a prospective, longitudinal study to investigate the puberty-associated shift in skin microbial communities.
Results
To determine temporal changes in bacterial and fungal communities of healthy children transitioning through puberty, we obtained longitudinal samples from 4 skin sites [antecubital fossa (Ac, inner elbow), volar forearm (Vf, mid-inner forearm), forehead (Fh), retroauricular crease (Ra, behind the ear) ] and nares (N, inside the nostril) of 12 healthy children at different pubertal stages. Nares were included in this study because this site is a reservoir for potential skin pathogens such as Staphylococcus aureus, and previously demonstrated microbial community shifts in young adults versus prepubertal children, similar to other skin sites (Jo et al., 2016, Oh et al., 2012). Characteristics of the study participants are summarized in Supplementary Table S1. A total of 4,475,820 bacterial 16S V1-V3 (27,459 mean reads/sample, 163 samples), and 6,082,563 fungal internal transcribed spacer (ITS)1 (37,089 mean reads/sample, 164 samples) sequenced reads were analyzed (Supplementary Table S2). The taxonomic assignment of bacterial and fungal community members at phylum-genus and species levels of each sample are shown in Supplementary Table S3 and S4.
Skin and nares bacterial and fungal communities comparisons across pubertal stages
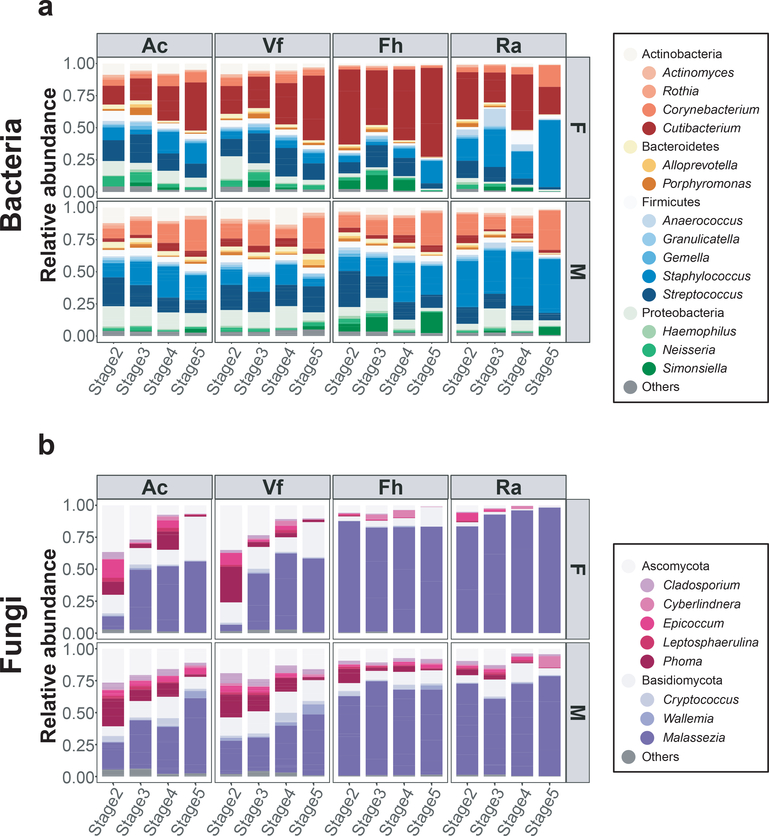
To investigate the microbial compositions of children during pubertal development, we examined relative abundances of major taxa (>1% average relative abundance across all subjects) of different pubertal stages at the genus level. Based on prior skin microbiome studies demonstrating differences among body sites and between male and female children, relative abundances of bacteria and fungi were visualized by Tanner stage as averages for the four sites and sex (Figure 1). Generally, female children had higher relative abundances of Cutibacterium and less Corynebacterium at all Tanner stages in all skin sites as compared to male children. For fungi, Malassezia predominated at most skin sites. Sebaceous skin (Fh, Ra) had higher Malassezia relative abundances than nonsebaceous skin (Ac, Vf) in both sexes across all Tanner stages. The nares bacterial and fungal communities were similar to the skin sex- and Tanner stage-associated patterns (Supplementary Figure S1).
Figure 1. Major skin bacterial and fungal taxa in healthy children.
(a, b) Average relative abundances of major bacterial and fungal genera by Tanner stage and by sex from 4 skin sites, antecubital fossa (Ac), volar forearm (Vf), forehead (Fh), retroauricular crease (Ra). Colors represent distinct bacterial and fungal genera (less common genera are represented by the phyla color).
Although sexual maturation during puberty is classically measured using Tanner stage, most studies use chronological age in studying skin microbiomes in children. To determine whether Tanner stage versus chronological age was more strongly associated with relative abundances of microbial taxa, linear regression models were used, adjusting for sex and sites. R2 values for Tanner stage were higher than those for age for most bacterial and fungal taxa, although the differences were not statistically significant (Supplementary Table S5). Collectively, the results suggest that skin bacterial and fungal communities of healthy children change with Tanner stage in a sex- and site-dependent manner.
Sex-dependent differences in bacterial and fungal communities of skin and nares
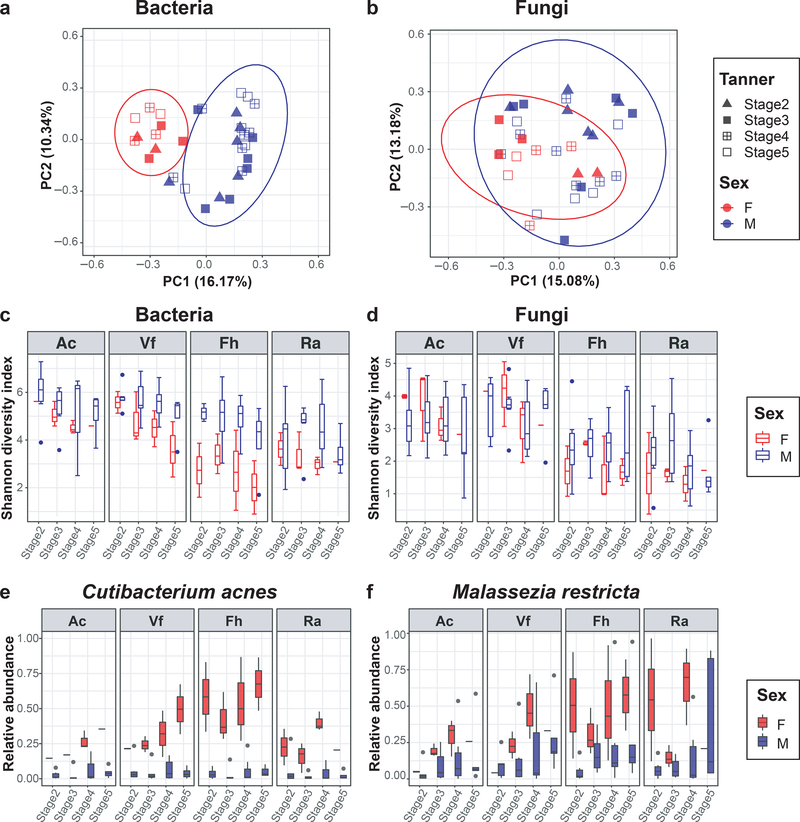
Based on known distinct physiological changes in boys and girls during puberty, principal coordinates analysis (Bray-Curtis dissimilarity) was used to investigate how the skin and nares microbiome compositions in the two sexes differed across samples. Skin bacterial communities in male children were statistically significantly different from female children at the amplicon sequence variants (ASV) level (Figure 2a and b) (P<0.001, AMOVA). In addition, pairwise comparison of individuals in each sex demonstrated that microbial communities in female children were more similar to each other than male children (Supplementary Figure S2a and b).
Figure 2. Comparisons of skin bacterial and fungal communities in female and male children during puberty.
(a, b) Principal coordinates analysis of Bray-Curtis dissimilarity. All skin samples from different skin sites belonging to an individual were averaged. Colors and shapes of the symbols represent different sex and different Tanner stage, respectively. Ellipses indicate the 95% confidence intervals based on standard error. (c, d) Alpha (Shannon) diversity of bacterial and fungal communities, based on sex, Tanner stages, and skin sites. (e, f) Temporal changes of the mean relative abundances of Cutibacterium acnes and Malassezia restricta across Tanner stage at 4 skin sites. Each box represents the lower quartile, median, and upper quartile of the value, and whiskers represent value up to 1.5 times the interquartile range. The dots at the end of box represent outliers. Skin sites abbreviated as antecubital fossa (Ac), volar forearm (Vf), forehead (Fh), retroauricular crease (Ra).
In further analyzing these microbiome changes across pubertal stages, the microbial communities in female children converged to be more similar to one another with higher Tanner stages, but not in male children. Based on a cross-sectional study showing differences between Tanner1–3 versus Tanner4–5 individuals (Oh et al., 2012), community differences were further studied between early (Tanner2–3) and late (Tanner4–5) stages. The interpersonal similarity within Tanner4–5 (versus Tanner2–3) female children was statistically significantly higher for bacteria at Ac and Ra and fungi at Ac (P<0.001) (Supplementary Table S6). Furthermore, female children showed significant reduction in skin microbial diversity (Shannon index) at most skin sites with increasing Tanner stage (bacteria, P<0.001 for Ac and P<0.05 for Fh and Ra; fungi, P<0.001 for Vf and P<0.05 for Ac and Fh) (Figure 2c and d, Supplementary Table S7). Male children showed a trend toward decreased skin microbial diversity over time, with statistically significant decreased bacterial diversity at Fh (P=0.044) and fungal diversity at Ra (P<0.001). The nares microbial diversities were significantly decreased with increasing Tanner stages only for fungi in males (P<0.001) (Supplementary Figure S2c and d, Supplementary Table S7).
In examining which bacteria and fungi contributed to the observed sex-specific microbiome variances, clustering of samples from female children was strongly driven by Cutibacterium, whereas the differences in male children were driven by several bacterial genera including Corynebacterium, Staphylococcus, Streptococcus, and Haemophilus (Supplementary Figure S2e and f). Although fungal communities showed no distinguishing taxa between sexes, sebaceous skin (Fh, Ra) displayed much tighter clustering in female children than in male children, mostly driven by Malassezia. For nares, bacterial and fungal communities demonstrated no distinguishing genera by sex. Taken together, the skin microbiomes in female children become less diverse and more similar over time as compared to male children in our cohort, primarily by increasing Cutibacterium.
Based on the contributions of several taxa in differentiating the skin microbiomes between male and female children, changes of relative abundances of major taxa were analyzed at the genus and species levels. Similar to culture-based studies (Somerville, 1969), Streptococcus relative abundances decreased with increased Tanner stages in both female and male children (Supplementary Figure S3a). The relative abundances of the two lipophilic species, Cutibacterium acnes and Malassezia restricta, in female children were consistently higher than those of male children, and were increased at most skin sites with increasing Tanner stage (Figure 2e and f, Supplementary Figure S3b and c).
To systematically examine the changes in relative abundances of major taxa between two groups, Tanner2–3 and Tanner4–5, a linear regression model was used (Supplementary Table S8). In females, the relative abundances of most bacterial genera including Alloprevotella, Granulicatella, Gemella, Streptococcus, and Neisseria were statistically significantly decreased at certain skin sites. Streptococcus was significantly less abundant in Tanner4–5 at Ra (P<0.001). Correspondingly, the relative abundances of Corynebacterium and Cutibacterium were increased in more mature females, statistically significantly at Ac (P<0.001). Males exhibited similar trends of decreased relative abundances in many skin bacterial genera in Tanner4–5 with a statistically significant decrease in Streptococcus at Fh (P=0.001). At the species level, the relative abundances of C. acnes were significantly increased with increasing Tanner stages in female children at Ac (P<0.001). The skin commensal Streptococcus epidermidis was more abundant in Tanner4–5 in both sexes, statistically significantly at Fh in females (P=0.001) and Ac in males (P<0.001). Conversely, Streptococcus mitis showed decreasing trends with increased Tanner stages (female, P=0.003 and P<0.001 for Ac and Ra; male, P<0.001 for Fh).
For skin fungal communities, both female and male children demonstrated lower Ascomycota fungi and higher Malassezia relative abundances in late Tanner4–5, especially on nonsebaceous sites; the changes were statistically significant in female children only (other Ascomycota, P<0.001 for Ac and Vf; Malassezia, P<0.001 for Vf). Species-level analyses of Malassezia showed that the relative abundances of M. restricta were significantly increased with increasing Tanner stages in female children at nonsebaceous skin (P=0.003 for Ac and Vf), whereas M. globosa was less abundant in late Tanner stages of both sexes at sebaceous skin (Fh, Ra), although the changes were not statistically significant.
Nares communities showed similar trends to skin sites for many of these bacterial and fungal genera (relative abundances decreased for Streptococcus and increased for Cutibacterium, Corynebacterium, Malassezia) when comparing late versus early Tanner stages, although the differences were not statistically significant. At the species level, an important pathobiont S. aureus was less abundant in both sexes in Tanner4–5 versus Tanner2–3, though not statistically significant. These results indicate that skin and nares microbial communities of female children shift towards a more ‘adult’ microbiome predominated by lipophilic microbes, during puberty.
Sex hormone concentrations and BMI are associated with lipophilic taxa
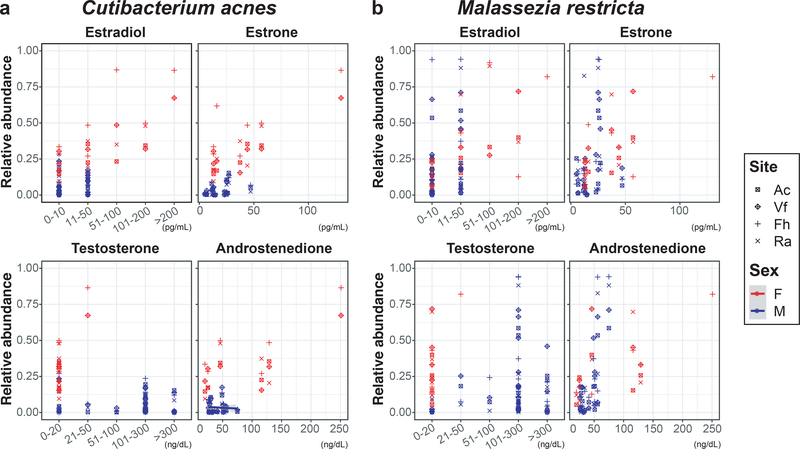
To investigate pubertal skin microbial shifts with physiological changes, the relative abundances of lipophilic taxa were evaluated in comparison to serum sex hormone concentrations and body mass index (BMI) (Supplementary Table S9 and S10). A strong association was found between the relative abundances of the lipophilic species and several serum sex hormones in female children when adjusting for site and Tanner stage, but not in male children. In female children, the relative abundances of C. acnes and M. restricta were positively associated with higher concentrations of estradiol, estrone, and testosterone (P<0.001) (Figure 3). However, inverse associations were observed for the relative abundances of C. acnes with DHEA-Sulfate and progesterone and M. restricta with sex hormone binding globulin (P<0.001) (Supplementary Figure S4). There were no associations between the relative abundances of the two taxa and 11-deoxycortisol. Male children exhibited a statistically significant positive association between M. restricta relative abundances and androstenedione concentrations (P<0.001), unlike female children. In addition, the relative abundances of C. acnes and M. restricta were positively associated with BMI in female children (P<0.001). Altogether, these results suggest that the shifts in skin microbial communities toward a more ‘adult’ microbiome may be mediated by the changes of sex hormone concentrations with subsequent increased sebaceous gland activity during pubertal development.
Figure 3. The association of the two lipophilic species, (a) Cutibacterium acnes and (b) Malassezia restricta, with serum estrogen and androgen concentrations.
Colors and shapes of the symbols represent different sex and different skin sites, respectively. Antecubital fossa (Ac), volar forearm (Vf), forehead (Fh), retroauricular crease (Ra).
Discussion
Although progressive alterations in vaginal and gut microbiome with respect to pubertal development have been reported (Hickey et al., 2015, Yuan et al., 2020), the influence of sexual maturation on skin and nares microbiomes during puberty has not been studied as systematically. In this longitudinal study, we identified significant alterations in the composition and diversity of skin microbial communities during puberty based on Tanner staging with shifts approaching an adult-like microbiome. Due to the interpersonal variability in the age of onset and progression of puberty, Tanner stage is a widely accepted measure of the degree of sexual maturation based on primary and secondary sexual characteristics (Marshall and Tanner, 1969, 1970). Concordantly, we found that skin microbial shifts more closely associated with Tanner stages as compared to chronological age.
With increasing Tanner stages, Streptococcus and diverse Ascomycota fungi of the skin and S. aureus of the nares showed decreasing trends in both sexes. In contrast, more sexually mature individuals demonstrated increasingly predominant lipophilic Cutibacterium, Corynebacterium, and Malassezia, which are known predominant members of the adult skin microbiome, consistent with previous observations comparing postpubertal adults versus younger children.
We observed significant sex-specific differences in skin microbial communities during puberty, particularly among bacteria. Microbial communities in female children were less diverse and converged to be more similar to one another over time as compared to male children, driven by higher Cutibacterium relative abundances. These findings may be due to earlier physiological change in equivalent pubertal stage in females relative to males and potentially related to higher adolescent acne prevalence in female children at younger ages (Lynn et al., 2016). Despite sex-specific differences in skin physiology including skin pH, thickness, hydration, and sebum level in adults and prior findings of higher bacterial loads in adult male versus female skin in culture-based studies (Giacomoni et al., 2009, Marples, 1982), most prior skin microbiome studies have not shown significant sex-based differences when considering the entire microbial community. The relative abundances of certain bacterial taxa such as Corynebacterium and Cutibacterium were different between sexes at different skin sites in a few studies of young adults, postulated to be from intrinsic (skin lipid, hydration, pH) and extrinsic factors (washing behavior, cosmetic use). (Fierer et al., 2008, Staudinger et al., 2011). Since extrinsic factors (e.g. skin care products, topical and systemic medications, bathing) that primarily influence the skin microbiome were controlled in this study, sex-specific differences potentially result from asynchronous progression of pubertal development between male and female children. The maturation-associated microbial shifts in male children may be delayed when compared to females in response to later physiological changes by sexual maturation despite controlling for Tanner stage. A cross-sectional microbiome study of volar forearms in healthy individuals ages 5 to 59 years showed that relative abundances of major skin commensal (C. acnes, Corynebacterium spp., S. epidermidis) continue to change with age extending into adulthood (Shi et al., 2016). Concordantly, culture-based studies showed that C. acnes increased after puberty, more so in late adolescence and peaking in young adulthood after age 20 (Leyden et al., 1975). Although our cohort included Tanner5, the children were 16 years and younger. Thus, larger, longitudinal studies into late adolescence and young adulthood are needed to confirm the sex-based differences in skin microbial community and to understand the influence of dynamic maturation-associated skin microbial community on long-term skin health.
In addition, the higher relative abundances of the lipophilic species, C. acnes and M. restricta, were strongly associated with androgens (testosterone, androstenedione) and estrogens (estradiol, estrone), suggesting sex hormones may shape skin microbial communities. Both females and males experience surges in serum androgens and estrogens during puberty. Androgens play a role in sebaceous and sweat gland activity and hair growth at puberty, while estrogens are associated with skin thickness, water content, and barrier function (Zouboulis et al., 2007). Because increased sebaceous gland activity is primarily dependent upon androgen (Strauss et al., 1962), microbial shifts during puberty may be mediated by androgen-driven increased sebaceous gland secretion and higher lipid contents on skin. Skin lipid quantity and composition have been shown to be correlated to total populations of lipophilic C. acnes (McGinley et al., 1980). Since pubertal variations in skin lipid compositions (e.g. wax esters) have been reported (Ramasastry et al., 1970), additional investigations into the influence of skin lipid compositions on the growth of various skin lipophilic microbes would be of great interest. Other androgen-independent skin physiological (lower pH, increased skin thickness) or immunological changes during pubertal development may also account for the maturation-associated skin microbial shifts (Oh et al., 2012).
Whether maturation-associated microbial shifts may contribute to the predilection of microbe-related skin disorders in certain age groups remains unclear. C. acnes-associated acne vulgaris and Malassezia-associated tinea versicolor are more common in adolescents and young adults than in younger children, whereas cases of S. aureus-associated atopic dermatitis tend to decrease in the majority of children during puberty. The limitations of the current study include small sample size and uneven number of samples between sexes, including fewer samples in Tanner2 and Tanner5 females. However, microbial communities were analyzed by sex and by early versus late Tanner stages consistent with prior results. Based on the skin physiological changes during the menstrual cycle and the premenstrual exacerbation of microbe-associated dermatoses, e.g., acne and rosacea, in-depth skin microbiome investigation throughout the menstrual cycle may be informative.
In summary, this study explores puberty-associated skin microbial shifts in conjunction with hormonal analysis, considering the influence of sex hormones on the host-microbiome interactions. Skin microbial communities shifted during puberty, transitioning toward a more ‘adult’ microbiome with predominance of lipophilic microbes, potentially from lipid-rich skin environments driven by androgen-dependent sebaceous gland activity. These findings support the relationship between sexual maturation, skin physiology, and skin microbiome.
Materials and Methods
Subjects
Twelve healthy children (4 girls and 8 boys) were recruited from the Washington, DC metropolitan region, USA, from 2012 to 2018. Sample collection was approved by the Institutional Review Boards of the NHGRI (http://www.clinicaltrials.gov/ct2/show/NCT00605878) and the NCI (https://clinicaltrials.gov/ct2/show/NCT00001505). Written informed consent was obtained from parents or guardians of the participating subjects, and, when age appropriate, children also provided assent. Subjects were evaluated every 6–18 months for up to 6 years at NIH Clinical Center. During the visits, subjects provided medical history and underwent swab sampling, physical examination, and blood draws for serum sex hormone concentrations. Pubertal staging, BMI calculations, and exclusion criteria are detailed in the Supplementary Methods. The study was performed in accordance with and following the Declaration of Helsinki Principles.
Sample collection, DNA extraction, PCR amplification, and sequencing
Given the topographical diversity of the bacterial and fungal communities resulting from spatial physiological attributes in human skin, samples were obtained from 4 different skin sites; Ac (“moist”), Vf (“dry”), and Fh and Ra (“sebaceous”) (Grice et al., 2009). Nares, a reservoir for colonization of potential pathogens was also included.
DNA was extracted and amplified from the skin and control swab samples, and the bacterial 16S ribosomal RNA (rRNA) V1-V3 region and the fungal ITS1 region were sequenced as described previously (Fadrosh et al., 2014, Oh et al., 2012). All amplicon products were pooled in equal concentrations for sequencing. Next generation sequencing using the Illumina MiSeq (Illumina, San Diego, CA) platform was performed at NIH Intramural Sequencing Center. Reagents and collection procedure controls were tested and demonstrated no significant background contamination. Details of sample collection, DNA extraction, PCR, and amplicon library construction are described in Supplementary Methods.
Sequence analysis pipeline
For 16S rRNA V1-V3 and ITS1 sequence analysis, a Divisive Amplicon Denoising Algorithm 2 (DADA2) pipeline (version 1.16) was used to infer exact ASVs as described previously (Callahan et al., 2016). Taxonomic classifications were assigned down to the genus level using DADA2 with references of Ribosomal Database Project (RDP) database (RDP trainset 16/release 11.5) for 16S reads (Cole et al., 2014) and database from Findley et al. (2013) for ITS1 reads, respectively. Species-level identification was performed with the NCBI Reference Sequence database and RDP database (version 3) (O’Leary et al., 2016) and UNITE ITS database (version 04.02.20) (Nilsson et al., 2019). Unmapped or ambiguously assigned ASV reads were manually interrogated against the NCBI nucleotide database with Basic Local Alignment Search Tool based on top hit at identity threshold ≥99.0% and alignment coverage ≥80%. The cured species information was merged into DADA2-compatible taxonomy file.
16S rRNA V1-V3 reads assigned to chloroplasts and mitochondria were removed prior to downstream analyses. Samples with reads of less than 1,500 were excluded. Details of sequence analysis and taxonomic classifications are described in Supplementary Methods.
Community analysis
Bacterial and fungal community analyses were performed with genus-level taxonomic classifications. Alpha diversity was quantified using the Shannon index, and beta diversity was analyzed using the Bray-Curtis dissimilarity, both based on ASV level-composition. Given the distinct physiological niches of skin sites versus nares (Jo et al., 2016, Oh et al., 2012), skin sites and nares were analyzed separately. All analyses were carried out in R software version 3.6.1.
Statistics
Linear regression models were used to analyze changes in Shannon index, Bray-Curtis dissimilarity, and taxonomic relative abundances per Tanner stage, and association with sex hormones and BMI adjusting for sex and sampling site. Models were estimated using generalized estimating equations with a working independence structure, and associated inferences were based on sandwich variance estimates which accounted for non-normality, heteroscedasticity, and possible correlations within clusters of observations (Liang and Zeger, 1986). Analysis of molecular variance (AMOVA) was used with Bray-Curtis dissimilarity to test differences of microbial communities between groups by sex and Tanner stage (Excoffier et al., 1992). The statistical analyses were done in a sex-specific fashion. Each research hypothesis was tested at a significance level of 0.05. Where necessary, a Bonferroni adjustment was made to control the familywise error rate at the nominal level.
Supplementary Material
Acknowledgements
We thank Diana M. Proctor and other members of the Kong and Segre labs for discussion and underlying efforts. This work was supported by NIH Intramural Research Programs of NHGRI, NCI, NICHD, and NIAMS, and 2019 Research Funds of Jeonbuk National University (J.P.) and National Research Foundation of Korea (NRF) (J.P., No.2019R1C1C1007049). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Abbreviations
- Ac
Antecubital fossa
- AMOVA
analysis of molecular variance
- ASV
amplicon sequence variants
- BMI
body mass index
- DADA2
Divisive Amplicon Denoising Algorithm 2
- Fh
Forehead
- ITS
internal transcribed spacer
- N
nares
- Ra
Retroauricular crease
- RDP
Ribosomal Database Project
- rRNA
ribosomal RNA
- Vf
Volar forearm
Footnotes
Conflict of Interest
The authors state no conflict of interest.
Data Availability
The sequence data from this study have been submitted to NCBI BioProject under accession number PRJNA704382.
References
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nature methods 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol 2011;131(10):2026–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic acids research 2014;42(Database issue):D633–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009;326(5960):1694–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1992;131(2):479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A 2008;105(46):17994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013;498(7454):367–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomoni PU, Mammone T, Teri M. Gender-linked differences in human skin. J Dermatol Sci 2009;55(3):144–9. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science 2009;324(5931):1190–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey RJ, Zhou X, Settles ML, Erb J, Malone K, Hansmann MA, et al. Vaginal microbiota of adolescent girls prior to the onset of menarche resemble those of reproductive-age women. mBio 2015;6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI, et al. Diverse Human Skin Fungal Communities in Children Converge in Adulthood. J Invest Dermatol 2016;136(12):2356–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden JJ, McGinley KJ, Mills OH, Kligman AM. Age-related changes in the resident bacterial flora of the human face. J Invest Dermatol 1975;65(4):379–81. [DOI] [PubMed] [Google Scholar]
- Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika 1986;73(1):13–22. [Google Scholar]
- Lynn DD, Umari T, Dunnick CA, Dellavalle RP. The epidemiology of acne vulgaris in late adolescence. Adolesc Health Med Ther 2016;7:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marples RR. Sex, constancy, and skin bacteria. Arch Dermatol Res 1982;272(3–4):317–20. [DOI] [PubMed] [Google Scholar]
- Marshall WA, Tanner JM. Variations in pattern of pubertal changes in girls. Arch Dis Child 1969;44(235):291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child 1970;45(239):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinley KJ, Webster GF, Ruggieri MR, Leyden JJ. Regional variations in density of cutaneous propionibacteria: correlation of Propionibacterium acnes populations with sebaceous secretion. J Clin Microbiol 1980;12(5):672–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 2019;47(D1):D259–D64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill AM, Gallo RL. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 2018;6(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med 2012;4(10):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol 2019;143(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochi PE, Strauss JS, Downing DT. Age-related changes in sebaceous gland activity. J Invest Dermatol 1979;73(1):108–11. [DOI] [PubMed] [Google Scholar]
- Ramasastry P, Downing DT, Pochi PE, Strauss JS. Chemical composition of human skin surface lipids from birth to puberty. J Invest Dermatol 1970;54(2):139–44. [DOI] [PubMed] [Google Scholar]
- Ro BI, Dawson TL. The role of sebaceous gland activity and scalp microfloral metabolism in the etiology of seborrheic dermatitis and dandruff. J Investig Dermatol Symp Proc 2005;10(3):194–7. [DOI] [PubMed] [Google Scholar]
- Shi B, Bangayan NJ, Curd E, Taylor PA, Gallo RL, Leung DYM, et al. The skin microbiome is different in pediatric versus adult atopic dermatitis. J Allergy Clin Immunol 2016;138(4):1233–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville DA. The normal flora of the skin in different age groups. Br J Dermatol 1969;81(4):248–58. [DOI] [PubMed] [Google Scholar]
- Staudinger T, Pipal A, Redl B. Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make-up. J Appl Microbiol 2011;110(6):1381–9. [DOI] [PubMed] [Google Scholar]
- Strauss JS, Kligman AM, Pochi PE. The effect of androgens and estrogens on human sebaceous glands. J Invest Dermatol 1962;39:139–55. [DOI] [PubMed] [Google Scholar]
- Sugita T, Suzuki M, Goto S, Nishikawa A, Hiruma M, Yamazaki T, et al. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med Mycol 2010;48(2):229–33. [DOI] [PubMed] [Google Scholar]
- Yuan X, Chen R, Zhang Y, Lin X, Yang X. Gut microbiota: effect of pubertal status. BMC Microbiol 2020;20(1):334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouboulis CC, Chen WC, Thornton MJ, Qin K, Rosenfield R. Sexual hormones in human skin. Horm Metab Res 2007;39(2):85–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data from this study have been submitted to NCBI BioProject under accession number PRJNA704382.