Summary
Rice (Oryza sativa) is a staple food crop and serves as a model cereal plant. It contains two biosynthetic gene clusters (BGCs) for the production of labdane‐related diterpenoids (LRDs), which serve important roles in combating biotic and abiotic stress. While plant BGCs have been subject to genetic analyses, these analyses have been largely confined to the investigation of single genes.
CRISPR/Cas9‐mediated genome editing was used to precisely remove each of these BGCs, as well as simultaneously knock out both BGCs.
Deletion of the BGC from chromosome 2 (c2BGC), which is associated with phytocassane biosynthesis, but not that from chromosome 4 (c4BGC), which is associated with momilactone biosynthesis, led to a lesion mimic phenotype. This phenotype is dependent on two closely related genes encoding cytochrome P450 (CYP) mono‐oxygenases, CYP76M7 and CYP76M8, from the c2BGC. However, rather than being redundant, CYP76M7 has been associated with the production of phytocassanes, whereas CYP76M8 is associated with momilactone biosynthesis. Intriguingly, the lesion mimic phenotype is not present in a line with both BGCs deleted.
These results reveal directional cross‐cluster phytotoxicity, presumably arising from the accumulation of LRD intermediates dependent on the c4BGC in the absence of CYP76M7 and CYP76M8, further highlighting their interdependent evolution and the selective pressures driving BGC assembly.
Keywords: CRISPR, disease resistance, diterpenoid, gene cluster, phytoalexin, rice
Introduction
As the staple food for over half of the global population, rice is a critically important crop (Muthayya et al., 2014). In addition, rice serves as a model cereal plant due, in no small part, to the small size of its genome, which led to early sequencing (Goff et al., 2002; Yu et al., 2002). Among the early findings from the rice genome sequence was the presence of two biosynthetic gene clusters (BGCs), associated with the production of antimicrobial phytoalexins, which were among the first defence proteins to be reported (Nutzmann et al., 2016). More specifically, these clusters contained genes encoding enzymes for biosynthesis of labdane‐related diterpenoids (LRDs), which have long been postulated to serve as phytoalexins and allelochemicals in rice (Peters, 2006), with recent results demonstrating roles in resistance to abiotic as well as biotic stress (Lu et al., 2018; Zhang et al., 2021). Indeed, LRDs are now known to serve such roles in cereal crops more generally (Murphy & Zerbe, 2020), further emphasising their importance.
The LRDs are characterised by initial bicyclisation of the general diterpenoid precursor (E,E,E)‐geranylgeranyl diphosphate (GGPP) catalysed by class II diterpene cyclases (Peters, 2010). These cyclases most often produce the eponymous labdadienyl/copalyl diphosphate (CPP), leading to the designation of the relevant enzymes as CPP synthases (CPSs). CPP can be produced in different stereochemical configurations, with the production of ent‐CPP required in all vascular plants (tracheophytes) for gibberellin (GA) phytohormone biosynthesis (Zi et al., 2014). Rice also produces syn‐CPP and LRD phytoalexins derived from this, but also from ent‐CPP (Peters, 2006). Further cyclisation and/or rearrangement of CPP is catalysed by class I diterpene synthases, again with all tracheophytes necessarily containing an ent‐kaurene synthase (KS) for GA biosynthesis, which has given rise to evolutionarily derived enzymes termed KS‐like (KSL) (Zi et al., 2014).
The rice genome was found to contain two BGCs, defined as proximal unrelated genes involved in a common biosynthetic pathway, for LRDs, with one on chromosome (chr.) 2 (c2BGC), and the other on chr. 4 (c4BGC) (Schmelz et al., 2014). These BGCs contain not only sequentially acting CPSs and KSLs, but also cytochrome P450 (CYP) mono‐oxygenases as well as, for the c4BGC, short chain alcohol dehydrogenases (Fig. 1). Notably, the c4BGC is clearly associated with momilactone biosynthesis, while c2BGC is largely associated with phytocassane biosynthesis, representing the two most prevalent LRDs produced by rice (Yamane, 2013), although c2BGC also clearly contains both KSLs and CYPs involved in additional LRD metabolism (Swaminathan et al., 2009; Wu et al., 2011).
Fig. 1.
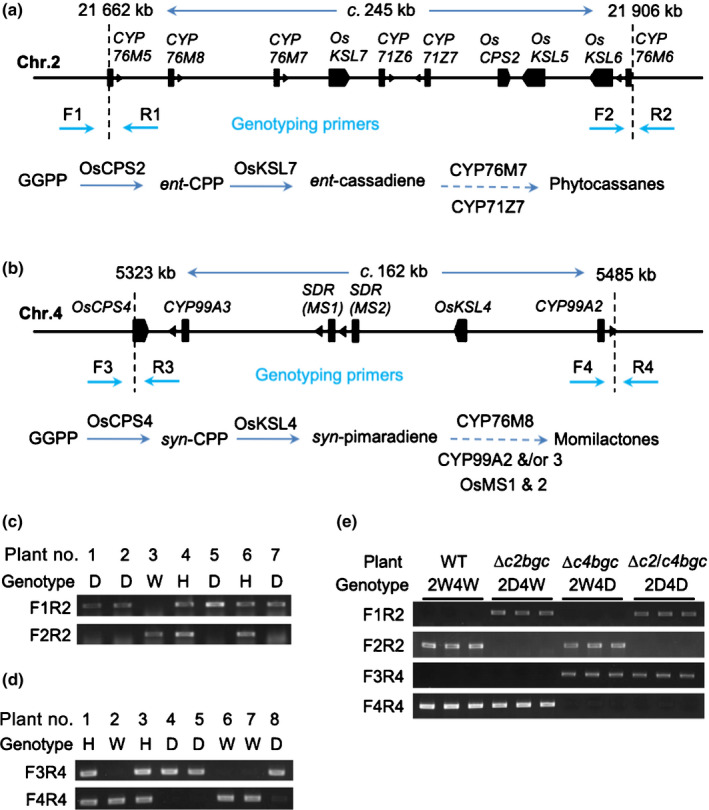
Large deletions of the rice diterpenoid biosynthetic gene clusters (BGCs). (a, b) Gene structure of diterpenoid on chromosomes 2 (a) and 4 (b), and associated biosynthesis pathways. The filled boxes representing genes. Dashed vertical lines represent locations of genomic break sites. Dashed arrows indicate multiple enzymatic reactions leading to production of diterpenoid families (known steps shown in Supporting Information Fig. S1). (c–e) Genotyping of progeny rice plants derived from the fragmental deletion mutants and their crossing using deletion‐specific primers. The numbers 2 and 4 in font of genotype represent chr.2 and chr.4, respectively. CPS, copalyl diphosphate synthase; CYP, cytochrome P450; Genotype D, mutant homozygous for deletion; GGPP, geranylgeranyl diphosphate; H, Heterozygous; KSL, ent‐kaurene synthase‐like; SDR, short‐chain alcohol dehydrogenase/reductas; W, WT.
The occurrence of BGCs, proximal unrelated genes involved in a common biosynthetic pathway, is somewhat unusual in eukaryotic genomes (Nutzmann et al., 2018). These seem to be associated with the production of secondary metabolites not required for normal growth and development. However, not all such biosynthetic pathways are associated with BGCs, leaving the evolutionary pressures underlying their assembly in question. This has been suggested to arise from a combination of positive and negative selection pressures, both for production of bioactive metabolites but also against the accumulation of certain intermediates (Swaminathan et al., 2009; Takos & Rook, 2012). Indeed, investigation of the rice c4BGC provided the first evidence for such negative effects from incomplete pathway inheritance, as a T‐DNA insertion knock‐out mutant of OsKSL4 was found to exhibit significantly reduced seed germination rates (Xu et al., 2012). Notably, this study also utilised a T‐DNA insertion knock‐out mutant of OsCPS4, but found that the momilactones seemed to primarily act as allelochemicals rather than in defence against the fungal blast pathogen Magnaporthe oryzae (Xu et al., 2012), at least in the relevant cultivar (cv) Zhonghua 11, despite the fact that these were the first phytoalexins against M. oryzae found in rice (Cartwright et al., 1981). Conversely, a T‐DNA insertion knock‐down mutant of OsCPS4 found in a different rice cv (Nipponbare) did effect resistance to M. oryzae as well as allelopathic activity (Toyomasu et al., 2014).
Other genetic studies have been directed at these rice BGCs, albeit also primarily targeted at single genes or at a pair of closely related paralogues to investigate the role of the encoded enzyme(s) in LRD biosynthesis, and occasionally that of the resulting LRDs in microbial disease resistance, rather than probing BGC evolution per se. For example, consistent with their localisation within the c4BGC, RNA‐interference (RNAi)‐mediated knock‐down of the closely related CYP99A2 and CYP99A3 provided the first evidence that these play a role in momilactone biosynthesis (Shimura et al., 2007). Similarly, consistent with their localisation within the c2BGC, RNAi knock‐down of the closely related CYP76M7 and CYP76M8 was used to indicate that these play a role in phytocassane biosynthesis (Wang et al., 2012). Correlation of the c2BGC with phytocassane production was further bolstered by the finding that a T‐DNA insertion knock‐out mutant of the similarly c2BGC localised CYP71Z7 was deficient in a certain transformation (hydroxylation at carbon‐2) in the underlying metabolic network (Ye et al., 2018). Finally, the potential redundancy of ent‐CPP‐derived LRD metabolism in rice was investigated using a T‐DNA insertion knock‐out mutant of OsCPS2, revealing that this is insufficient to completely block production of the phytocassanes or other ent‐CPP‐derived LRDs, presumably due to the presence of OsCPS1 required for GA phytohormone biosynthesis, which enables continued production of ent‐CPP (Lu et al., 2018). This study also further investigated the role of rice LRDs in microbial disease resistance, including the use of the OsCPS4 T‐DNA insertion knock‐out mutant, which uncovered a role for syn‐CPP‐derived (OsCPS4‐dependent) LRDs in nonhost disease resistance to Magnaporthe poae, as well as roles for OsCPS2‐associated (ent‐CPP‐derived) LRDs in defence against bacterial leaf blight pathogen Xanthomonas oryzae pathovar (pv) oryzae (Xoo) as well as M. oryzae (Lu et al., 2018).
Development of the CRISPR/Cas9 technique has offered precise genome editing that has now been applied to rice (Char et al., 2019), and even more specifically to its LRD BGCs in work targeted at the fast‐growing cv Kitaake. In particular, this approach is most often applied to target single genes and has been used in this context to provide a direct comparison of knocking‐out OsCPS2 and/or OsCPS4 in the same genetic background, with crossing of the initially generated cps2 and cps4 single‐mutant lines to generate a cps2x4 double‐mutant line (Zhang et al., 2021). Similarly, the closely related CYP76M7 and CYP76M8 also have been individually knocked out, revealing that CYP76M7 is primarily associated with phytocassane biosynthesis, while CYP76M8 is more important for momilactone production, although these do appear to be partially redundant (Kitaoka et al., 2021). However, the co‐localisation of these two genes in the c2BGC essentially blocks the production of a double‐mutant line by genetic crossing. Fortunately, CRISPR/Cas9 can be applied to simultaneously target multiple genes/loci (Bi & Yang, 2017), enabling the investigation of neighbouring genes such as those found in BGCs, or even removal of large fragments such as entire BGCs (Li et al., 2019). Indeed, deletion of the rice LRD BGCs using CRISPR/Cas9 has previously been reported in protoplasts and, for deletion of the c2BGC, in T0 plants, although this was only present in mono‐allelic form with the c. 245 kb deletion paired with small nucleotide changes at the targeted sites in the other copy of the c2BGC (Zhou et al., 2014).
Here we report that deletion of the c2BGC, as well as the c4BGC, yields lines that are viable and genetically stable in the homozygous form. This enabled a chemotypic and phenotypic investigation of such complete BGC loss, revealing not only the expected effects on rice LRD metabolism and microbial disease resistance, but also the discovery of a lesion mimic phenotype for homozygous c2BGC deletion plants. CRISPR/Cas9 multiplex targeting was then used to demonstrate that the lesion mimic phenotype depends on both CYP76M7 and CYP76M8, as exhibited by cyp76m7/8 double, but not cyp76m7 or cyp76m8 single, mutant lines. Intriguingly, it was further possible to cross the BGC deletion lines to create a viable line missing both BGCs, which no longer exhibited the lesion mimic phenotype. This indicates that the lesions arise from directional phytotoxicity, presumably from c4BGC‐dependent LRD intermediates that accumulate in the absence of the c2BGC, specifically loss of CYP76M7 and CYP76M8.
Materials and Methods
Plant materials and growth conditions
The japonica cultivar (Oryza sativa) Kitaake was used here. The T0 plants with each BGC deleted had been generated previously using CRISPR/Cas9 genome editing (Zhou et al., 2014; Li et al., 2019). Rice seeds were sterilised in 2% sodium hypochlorite and grown aseptically on half‐strength Murashige & Skoog (½MS) medium containing 3% sucrose and 0.5% agar (pH 5.8) in plastic container at 30°C. Here, 2‐wk‐old seedlings were transferred and planted to large plastic boxes containing farm soil and grown in a glasshouse (unless alternatively indicated). Growth conditions were 30°C in daytime, 28°C in nighttime, and with a 13‐h day length.
Phytochemical analyses
The 3‐wk‐old rice leaves were cut into 5‐cm pieces and 0.1 g was induced by floating on a solution of 0.5 mM CuCl2. After 72 h these induced leaves were frozen by submersion into liquid N2 and ground into a fine power, then were extracted by shaking in 3 ml methanol for 72 h in a cold (4°C) room. Each sample had 2.31 µg sclareol added as an internal standard, and filtered through a 0.2‐µm nylon filter (F2513‐2; Thermo Fisher Scientific, Waltham, MA, USA) using a glass syringe, dried under a stream of N2 gas, and resuspended in 1 ml methanol for analysis.
Analyses were carried out via liquid chromatography tandem mass spectrometry (LC‐MS/MS), with 15‐µl samples injected onto a Supelco (Sigma‐Aldrich) Ascentis C18 column (10 cm × 2.1 mm; 3 μm) using an Agilent Technologies 1100 Series high performance liquid chromatography system coupled to both an ultraviolet–visible (UV‐Vis) light diode array detector and an Agilent Technologies Mass Selective Trap SL detector located in the Iowa State University W.M. Keck Metabolomics Research Laboratory. A binary gradient was used, consisting of water with 0.1% acetic acid v/v (buffer A) and acetonitrile with 0.1% acetic acid v/v (buffer B). The solvent gradient elution was programmed as follows: initial, 40% buffer B; 0–13 min, a linear gradient from 40% buffer B to 85% buffer B; 13–14 min, a linear gradient from 85% buffer B to 100% buffer B; 14–14.5 min, a linear gradient from 100% buffer B to 40% buffer B. The known rice LRD phytoalexins were detected by MS/MS analysis, using an isolation selection window of ±0.5 m/z. The isolated masses were re‐ionised and the resulting mass spectra recorded.
The isolated parent ions and quantified secondary ions, along with the corresponding retention time (RT), based on optimisation with the available authentic standards, for the known rice LRD phytoalexins were as follows: momilactone A, m/z 315.2–271.2, RT 11.1 min; momilactone B, m/z 331.2–269.2, RT 9.0 min; oryzalexin A, m/z 303.2–285.2, RT 10.0 min; oryzalexin B, m/z 303.3–286.2, RT 9.7 min; oryzalexin C, m/z 301.2–283.2, RT 9.6 min; oryzalexin D, m/z 287.2–269.2 (147.2), RT 9.6 min; oryzalexin E, m/z 287.2–269.2, RT 11.3 min; oryzalexin S, m/z 287.2–269.2 (241.2), RT 9.7 min; phytocassane A, m/z 317.2–299.2, RT 7.6 min; phytocassane B, m/z 335.2–317.2, RT 7.3 min; phytocassane C, m/z 319.2–301.2, RT 6.8 min; phytocassane D, m/z 317.2–299.2, RT 9.7 min; phytocassane E, m/z 317.2–299.2, RT 8.3 min; phytocassane F, m/z 333.2–315.2, RT 7.3 min; sclareol, m/z 273.2–163.1, RT 13.6 min. The relative amount of each LRD was determined with the 6300 Series Ion Trap LC/MS v.1.8 software package (Thermo Fisher Scientific), by comparison of total ion peak areas to that of the sclareol internal standard.
Fungal resistance assays
Magnaporthe oryzae strains CA89 and O254 were used, and infection assays were performed as previously described (Sesma & Osbourn, 2004). Briefly, rice seedlings at the third leaf stage were used for infection. Four pots of 20 plants were used per line and per experiment. Each pot was sprayed with 5 ml of a suspension of 105 conidia per millilitre. The sprayed plants were then incubated in the growth chamber with humid conditions (85% relative humidity, 25°C, and a 16 h : 8 h, light : dark photoperiod) for 6 d to score disease symptoms. Leaf lesions were scanned and analysed with Assess 2.0 software (American Phytopathological Society). Blast lesions were evaluated based on lesion number and lesion extension of disease symptoms as previously reported (Tucker et al., 2010). One‐way ANOVA statistical analyses were performed to compare wild‐type and ∆c2bgc and ∆c4bgc plants. The Dunnett's significant difference test was used for post‐ANOVA pairwise analysis of significance, set at 5% (P < 0.05).
Bacterial resistance assays
Xanthomonas oryzae pv oryzae (Xoo) strain PXO99A and its hrpC, a gene encoding a type III secretion system, mutant PXO99AME7 (ME7, for short) were used for leaf blight disease assays. Xoo was grown for 2–3 d on TSA (1% (w/v) tryptone, 1% sucrose, 0.1% glutamic acid, and 1.5% agar, pH 6.8) plates at 28°C, scraped off and resuspended in sterile deionised water. The suspension was then adjusted to an optical density of 0.5 at 600 nm before infection. Here, 2‐wk‐old seedlings were inoculated using a needleless syringe infiltration method (Zhu et al., 1998), with five infiltrated spots on each leaf. The leaves were then harvested 2 d after injecting and ground sufficiently in a sterile mortar by adding a specified volume of sterile water. The subsequent solution was serially diluted and spread on TSA plates to count colonies, then converted into colony formation units per infiltration spot. Bacterial counting was performed three times in parallel for each rice line. One‐way ANOVA statistical analyses were applied to compare wild‐type and ∆c2bgc and ∆c4bgc plants after bacterial inoculation. Dunnett's significant difference test was used for post‐ANOVA pairwise analysis of significance, set at 5% (P < 0.05) (Fig. 4a–c; as described later).
Fig. 4.
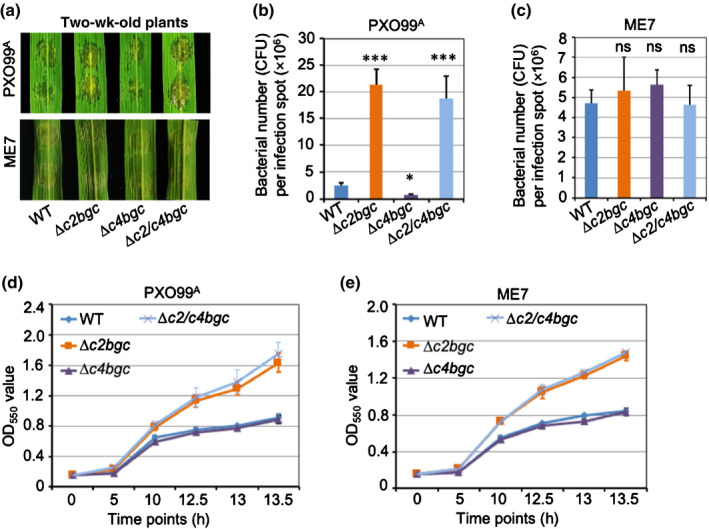
In vivo and in vitro plant defence of ∆c2bgc, ∆c4bgc and ∆c2/c4bgc rice plants against Xanthomonas oryzae pv. oryzae (Xoo). (a) Representative leaves from 2‐wk‐old plants of the wild‐type (WT), ∆c2bgc, ∆c4bgc and ∆c2/c4bgc lines after inoculation with Xoo strain PXO99A (PXO99A) (upper panel) or the type III secretion system, mutant PXO99AME7 (ME7) strain (lower panel) via needless syringe infiltration. ME7 is incapable of type III effector secretion, is nonpathogenic and does not induce the microbial defence response in rice. (b, c) Susceptibility of WT, ∆c2bgc, ∆c4bgc and ∆c2/c4bgc lines to infection with Xoo strain PXO99A (b) or ME7 (c) measured by bacterial colony counting 2 d post‐inoculation. *, P < 0.05; ***, P < 0.001; ns, no significant difference, calculated using post‐ANOVA pairwise analysis of significance with Dunnett's significant difference test, for ∆c2bgc, ∆c4bgc and ∆c2/c4bgc plants relative to WT. Error bars represent ± SD. (d, e) Growth curves of PXO99A (d) and its mutant ME7 (e) in medium containing root exudates from WT (control), ∆c2bgc, ∆c4bgc and ∆c2/c4bgc mutant plants, as indicated. Error bars represent ± SD.
Next, 8‐wk‐old rice plants were inoculated using the leaf‐tip‐clipping method (Kauffman et al., 1973). Symptoms were scored by measuring lesion lengths after 14 d for the leaf‐clipping inoculations. Lesion length measurements are the averages of 20 leaves. Lesion length of ∆c2bgc mutant plants was compared with wild‐type (WT) plants after PXO99A inoculation. ***, P < 0.001, calculated using post‐ANOVA pairwise analysis of significance with the Dunnett's test (Fig. 5g; as described later).
Fig. 5.
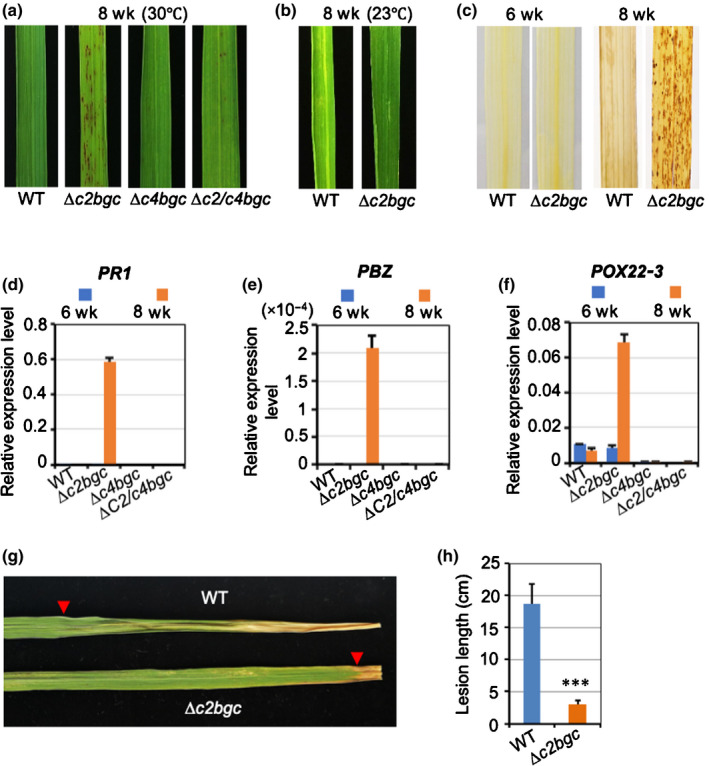
Deletion of c2BGC leads to a lesion mimic (hypersensitive response) phenotype in rice. (a) Representative leaves from wild‐type (WT), ∆c2bgc, ∆c4bgc and ∆c2/c4bgc plants grown in a glasshouse (30°C) for 2 months after germination. (b) Representative leaves from WT and ∆c2bgc plants at the heading stage grown in a growth chamber at 23°C. (c) 3,3′‐Diaminobenzidine (DAB) staining of the ∆c2bgc mutant and WT leaves before and after appearance of lesions (i.e. the indicated time after germination). (d–f) Relative expression level of defence‐related genes PBZ, PR1 and POX22‐3 in leaves from ∆c2bgc, ∆c4bgc and ∆c2/c4bgc plants compared with WT. The leaf tissues were sampled before (6 wk) and after (8 wk) formation of lesions. Error bars represent ± SD. (g) Representative leaves from WT and ∆c2bgc 8‐wk‐old plants infected with Xanthomonas oryzae pathovar oryzae (Xoo) (PXO99A) via the leaf‐tip clipping method to distinguish lesion length from lesion mimic spots. Arrowheads point to the edge of the infection progression. (h) Lesion length analysis for ∆c2bgc and WT plants after PXO99A inoculation. ***, P < 0.001; calculated using post‐ANOVA pairwise analysis of significance with Dunnett's significant difference test for ∆c2bgc plants relative to WT. Error bars represent ± SD.
For in vitro antimicrobial activity assays, 150 germinated seeds from each rice line were grown in a sterile beaker with 10 ml of ½MS medium for 15 d in 30°C growth chamber. The liquid solution containing root exudates from each line was collected separately. Liquid TSA medium containing 50% (v/v) of the root exudates was used for Xoo culture. The initial Xoo suspension was adjusted to an optical density (OD) of 0.15 at 600 nm and then grown with shaking in a 30°C incubator. OD600 values were collected at the specified time points for the growth curves for bacteria.
3,3′‐Diaminobenzidine staining
For measurement of H2O2 accumulation, leaves were collected at 45 d after germination and immediately submerged in a solution containing 1 mg ml−1 DAB, and then incubated at room temperature in the dark for 8 h. The stained leaves were then decoloured in 95% boiling ethanol for 10 min and soaked for 2 d in 95% ethanol until all chlorophyll had been removed.
RNA isolation and gene expression analysis
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. First‐strand cDNA synthesis for RT‐PCR was carried out using iScript™ cDNA Synthesis kit (Bio‐Rad) and random primers. A real‐time PCR reaction was conducted in the CFX96 Touch Real‐Time PCR Detection System (Bio‐Rad) using SYBR Green Supermix (Bio‐Rad) according to the manufacturer’s instructions. The primer pairs used to specifically amplify target genes and Actin1 are listed in Supporting Information Table S3.
Genome editing and mutant creation
CRISPR editing was used to mutate CYP76M7 and CYP76M8 genes. The CRISPR/Cas9 protocol has been described previously (Zhou et al., 2014). Two guide RNA sequences targeting both CYP76M7 and CYP76M8 genes were designed, synthesised and used for the CRISPR/Cas9 construct, in order to mutate both genes simultaneously. Immature embryo‐derived callus cells of rice (cv Kitaake) were transformed using the Agrobacterium‐mediated rice transformation method. Transformed calli and regenerated plantlets were selected on medium containing hygromycin B. T0 lines were used for mutation screening, and T‐DNA free homozygous mutant lines were selected in the corresponding T1 and T2 generation plants. The sequences of guide RNA and the corresponding primers used for mutation detection and plant genotyping are listed in Table S1.
Results
Hereditary stability of BGC deletions
To further investigate genetic stability of the BGC deletions the previously reported T0 lines (Zhou et al., 2014) were selfed to generate T1 and T2 generations, with single plants homozygous for deletions of c2BGC or c4BGC and free from CRISPR constructs selected by PCR analysis. Specifically, for analysis of the c2BGC loci, one pair of primers, F1 and R1 flanking the 5′ site (located at 21 662 kb of chr. 2), and another pair of primers, F2 and R2 flanking the 3′ site (located at 21 906 kb of chr. 2), which were the locations of the sequences targeted for construction of this deletion, were designed (Fig. 1a; Table S1). PCR amplicons are only expected with the F1 and R1 or F2 and R2 primers from WT genomic DNA (gDNA), while amplicons with F1 and R2 were only expected from gDNA in which this region has been deleted (Figs 1a, S2a). Similarly, for analysis of the c4BGC, primer pairs F3 and R3, flanking the 5′ site (located at 5323 kb of chr. 4) or F4 and R4, flanking the 3′ site (located at 5485 kb of chr. 4) were used to detect the target sites used for this deletion, while F3 and R4 would only amplify gDNA with this c. 160 kb region deleted (Figs 1b, S2b; Table S1). Using these primers, individual plants were analysed during this selection process. The observed 1 : 2 : 1 segregation of WT, heterozygous and homozygous deletions (Fig. 1c,d; Table S2) indicated the hereditary stability of the large deletion mutations of both the c2BGC and c4BGC. For each BGC, a line homozygous for the deletion that was also CRISPR/Cas9 construct free (as verified by the lack of targeted PCR amplicons) was selected for further investigation; these are from this point forwards referred to as ∆c2bgc and ∆c4bgc. Further analysis of these lines by RT‐PCR indicated that the genes from the relevant (deleted) BGC were no longer expressed in these plants, consistent with their loss from the genome (Fig. S3a,b). The sequences of gene‐specific oligonucleotides for expression are provided in Table S3.
A mutant line homozygous for deletion of both c2GBC and c4GBC was generated by crossing ∆c2bgc and ∆c4bgc mutant lines, with selection for homozygous loss of both BGCs via the same PCR‐based approach as described previously (Fig. 1e). This line, with deletions of both c2GBC and c4GBC, is from this point forwards referred to as ∆c2/c4bgc.
Effect of BGC deletions on LRD production
As it has been shown that rice production of LRDs can be induced by the application of copper chloride, enabling the detection of most of these natural products (Lu et al., 2018), investigation of such elicited leaf extracts was utilised here. In particular, to investigate how BGC deletion affected LRD production, leaves from the ∆c2bgc, ∆c4bgc and ∆c2/c4bgc lines were induced and extracted, with analysis via an LC‐MS method developed for this purpose (Lu et al., 2018). Consistent with the multiple genes from the c2BGC associated with phytocassane biosynthesis (Fig. 1), these LRDs were no longer detectable from ∆c2bgc seedlings (Fig. 2a). By contrast, consistent with the remaining presence of the relevant OsKSL10, as well as OsCPS1 (Fig. S1), production of oryzalexins A–F remained essentially unaffected (Fig. 2d). Conversely, consistent with the previously demonstrated importance of CYP76M8 to momilactone biosynthesis (Kitaoka et al., 2021), production of these LRDs was significantly reduced (Fig. 2b). Perhaps reflecting the increased availability of syn‐CPP, production of the derived oryzalexin S was significantly increased (Fig. 2c). In ∆c4bgc plants, consistent with the multiple genes from this BGC associated with momilactone biosynthesis (Fig. 1), these LRDs were no longer detectable (Fig. 2b). Similarly, the OsCPS4‐dependent (syn‐CPP‐derived) oryzalexin S also is essentially not produced (Fig. 2c). By contrast, production of phytocassanes was significantly increased (Fig. 2a), perhaps due to the increased availability of GGPP as has been speculated for cps mutants (Zhang et al., 2021). Conversely, production of oryzalexins A–F was reduced (Fig. 2d). As expected, phytocassanes, momilactones and oryzalexin S were no longer detectable in ∆c2/c4bgc plants, and production of oryzalexins A–F was significantly reduced (Fig. 2a–d).
Fig. 2.
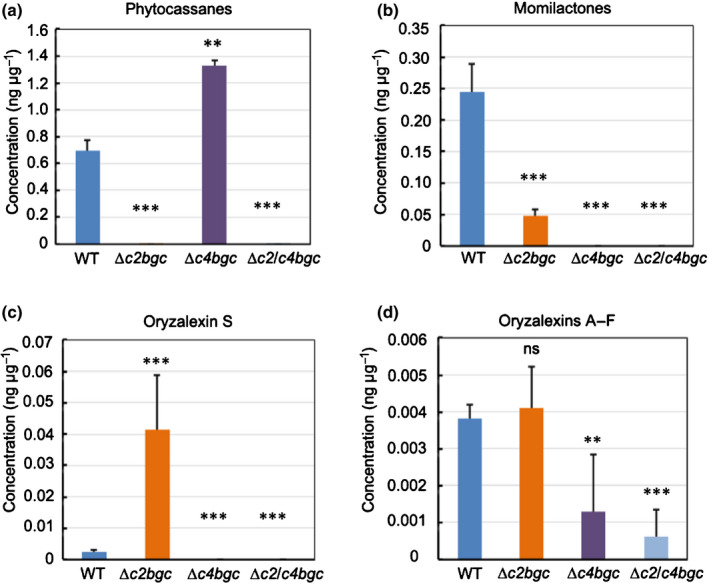
Phytochemical profiling of ∆c2bgc and ∆c4bgc rice plants. (a–d) Effect of deleting the biosynthetic gene cluster from chromosome 2 (c2BGC) or the BGC from chromosome 4 (c4BGC) on labdane‐related diterpenoid metabolism in rice relative to wild‐type (WT) plants (average from three plants with error bars indicating standard deviation), specifically from analysis of leaves from 3‐wk‐old seedlings that were induced with CuCl2. **, P < 0.01; ***, P < 0.001; ns, no significant difference; calculated using post‐ANOVA pairwise analysis of significance with the Dunnett's significant difference test for ∆c2bgc, ∆c4bgc and ∆c2/c4bgc plants relative to the relevant WT line. Error bars represent ± SD.
Effect on susceptibility to the fungal blast pathogen
Given the previously demonstrated differential effect of rice LRD phytoalexins on resistance to various species from the phytopathogenic fungal genus Magnaporthe and even strains of M. oryzae (Lu et al., 2018), two M. oryzae strains (O254 and CA89) were examined here with ∆c2bgc and ∆c4bgc mutant plants as well as the parental/WT cv Kitaake. The O254 strain was from India, while CA89 was from California in the USA. Susceptibility was assayed by spray‐inoculating rice seedlings at the three‐leaf stage and analysing the diseased (discoloured) leaf area (Fig. 3). Consistent with the previous finding that the inducible ent‐CPP‐derived LRDs, which are largely dependent on OsCPS2, are effective phytoalexins against this pathogen (Lu et al., 2018), ∆c2bgc and ∆c2/c4bgc mutant plants were significantly more susceptible to both strains, although this effect was much more pronounced with strain O254 than with CA89. By contrast, ∆c4bgc mutant plants did not exhibit significantly increased susceptibility to either strain of M. oryzae (Fig. 3a–d).
Fig. 3.
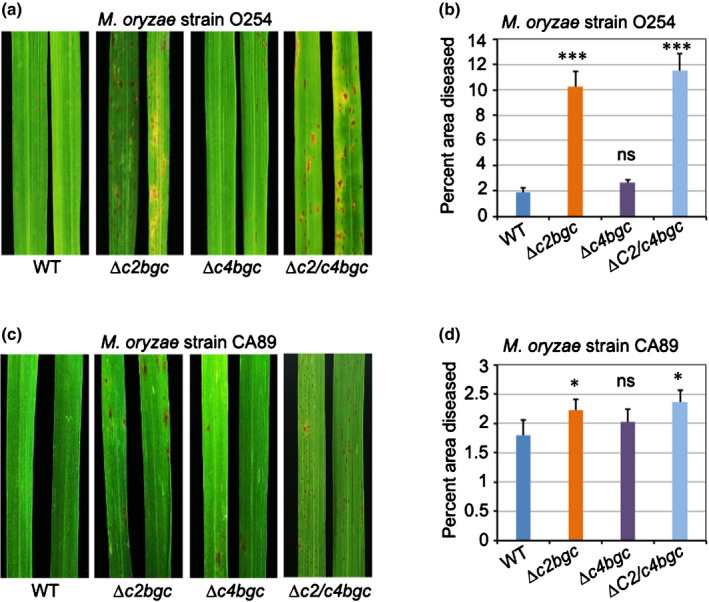
Susceptibility of ∆c2bgc, ∆c4bgc and ∆c2/c4bgc plants to Magnaporthe oryzae. (a, c) Representative leaves after infection with M. oryzae strains O254 (a) and CA89 (c) of rice plants at the three‐leaf stage from the wild‐type (WT), ∆c2bgc, ∆c4bgc and ∆c2/c4bgc lines as indicated. (b, d) Percentage of diseased (discoloured) area vs total leaf area for ∆c2bgc, ∆c4bgc, ∆c2/c4bgc and WT plants after infection with M. oryzae strains O254 (b) and CA89 (d). *, P < 0.05; ***, P < 0.001; ns, no significant difference; calculated using post‐ANOVA pairwise analysis of significance with Dunnett's significant difference test, for ∆c2bgc and ∆c4bgc plants relative to the relevant WT line. Error bars represent ± SD.
Effect on susceptibility to the bacterial leaf blight pathogen
It has previously been suggested that OsCPS2‐dependent, ent‐CPP‐derived, but not OsCPS4‐dependent (syn‐CPP‐derived), LRDs are effective against the rice bacterial leaf blight pathogen Xoo (Lu et al., 2018; Zhang et al., 2021). This was further examined here with ∆c2bgc and ∆c4bgc mutant plants compared with WT, using Xoo strain PXO99A to infect 2‐wk‐old seedlings via infiltration and measuring bacterial numbers at 2 d post‐infection (Fig. 4). As expected, ∆c2bgc and ∆c2/c4bgc plants were significantly more susceptible to Xoo (Fig. 4a), with almost a 10‐fold increase in colony forming units (CFU) relative to WT (Fig. 4b), while ∆c4bgc plants instead exhibited a slight but significant decrease in CFU (Fig. 4a,b).
c2BGC‐dependent LRDs as phytoalexins and phytoanticipins against Xoo
The vast majority of rice LRDs are considered to be phytoalexins whose production is inducible (Peters, 2006), with accumulation of momilactones and phytocassanes specifically observed in Xoo‐infected leaf tissues (Klein et al., 2015). However, only the oryzalides and related (ent‐isokaurene‐derived) LRDs, whose production is not strongly inducible (Watanabe et al., 1996), have been shown to exhibit antibiotic activity against Xoo and, hence, serve as phytoanticipins. The effectiveness of ent‐CPP‐derived (OsCPS2‐dependent) LRDs has been demonstrated by the increased susceptibility of cps2 knock‐out mutant plants to infection by Xoo (Lu et al., 2018; Zhang et al., 2021). However, it remains unclear if this effect was due to the activity of the relevant phytoanticipins (oryzalides and related LRDs) or phytoalexins (phytocassanes) against this bacterial phytopathogen. This was examined here with the Xoo mutant strain PXO99AME7 (from this point forwards ME7), which is incapable of type III effector secretion and considered to be nonpathogenic, as it does not induce the plant defence response (Sugio et al., 2007). Notably, ME7 grew at the same rate in the ∆c2bgc, ∆c4bgc and ∆c2/c4bgc mutant plants relative to WT (Fig. 4a,c). To verify that the c2BGC‐dependent LRDs still exhibited antimicrobial activity against the Xoo mutant strain ME7, as well as the parental pathogenic strain PXO99A, bioassays were performed using root exudates, which are known to contain some LRDs (Toyomasu et al., 2008). In particular, root exudates from WT, ∆c2bgc, ∆c4bgc, and ∆c2/c4bgc seedlings were collected (separately) and was added to liquid culture medium, then used to grow PXO99A and ME7 (again separately), with the growth rate observed by OD measurements (Fig. 4d,e). Consistent with general antibiotic activity of the c2BGC‐dependent LRDs against Xoo, both PXO99A and ME7 grew faster in medium with root exudates from ∆c2bgc and ∆c2/c4bgc mutant plants than with those from WT or ∆c4bgc mutant plants (Fig. 4d,e). The contrast between the unaffected growth of ME7 on ∆c2bgc mutant plants relative to the inhibitory effect of ∆c2bgc root exudates on ME7 growth in liquid culture indicated that rice produced c2BGC‐dependent (ent‐CPP‐derived) LRDs as phytoalexins against Xoo in their leaves. In addition, given the effectiveness of such LRDs from uninduced root exudates against Xoo growth, it appears that these natural products also served as phytoanticipins in the rhizosphere.
A lesion mimic phenotype with ∆c2bgc plants
Consistent with the effect recently reported for cps2 and cps4, as well as cps2x4, knock‐out mutants in cv Kitaake (Zhang et al., 2021), the ∆c2bgc and ∆c4bgc mutant plants exhibited a drought‐sensitive phenotype (Fig. S4). Notably, the ∆c2bgc, but not ∆c4bgc or ∆c2/c4bgc, mutant plants were found also to exhibit a lesion mimic phenotype c. 7 wk after germination, in the booting stage (Fig. 5a). This lesion mimic phenotype was temperature sensitive as, under low‐temperature growth conditions (23°C), lesions were only rarely observed even in the heading stage of ∆c2bgc mutant plants (Fig. 5b).
To investigate the basis for lesion formation, leaves of the ∆c2bgc mutant plants were stained with DAB, with the observed formation of reddish‐brown formazan precipitates indicative of H2O2 accumulation (Fig. 5c). Such reactive oxygen species can trigger either necrosis or programmed cell death (Van Breusegem & Dat, 2006), with the more specific hypersensitive response further coupled to defence responses (Balint‐Kurti, 2019), which is often associated with lesion mimic mutants (Bruggeman et al., 2015). Therefore, it was hypothesised that defence against pathogens in ∆c2bgc mutant plants might be enhanced. This was first investigated by analysis of the expression level of rice defence‐related genes, including PBZ (Kim et al., 2004), PR1 (Mitsuhara et al., 2008) and inducible peroxidase POX22‐3 (Chittoor et al., 1997), in ∆c2bgc leaves before and after lesion appearance. All the defence‐related genes were significantly upregulated in ∆c2bgc mutant leaves with the appearance of the lesions, with no change in expression levels in ∆c4bgc and ∆c2/c4bgc mutant or WT plants during the same growth stage (Fig. 5d–f). Indeed, following lesion formation, the ∆c2bgc mutant plants exhibited enhanced resistance against Xoo PXO99A relative to WT at the same growth stage (Fig. 5g,h). These results indicated that the lesion mimic phenotype exhibited by loss of the c2BGC arose from a hypersensitive response that appeared later in rice plant development.
CYP76M7 and CYP76M8 are responsible for lesion mimic phenotype
To determine the gene(s) from the c2BGC responsible for the lesion mimic phenotype the relevant previously reported cv Kitaake mutant lines were examined – that is, cps2 (Zhang et al., 2021), as well as cyp76m7 and cyp76m8 (Kitaoka et al., 2021). However, none of these single gene knock‐outs exhibited this phenotype. Fortuitously, it was recently noted that RNAi knock‐down of the closely related paralogues CYP76M7 and CYP76M8 from the c2BGC led to a lesion mimic phenotype (Ye et al., 2018). Therefore, a multiplex CRISPR/Cas9 approach was used to knock out both genes simultaneously (Fig. S5), with the resulting cyp76m7/8 double‐mutant plants found to exhibit the lesion mimic phenotype (Fig. 6a), as well as drought sensitivity (Fig. S6). As found for ∆c2bgc mutant plants, this lesion mimic phenotype was temperature sensitive, as under low‐temperature growth conditions (23°C) lesions were similarly only rarely observed even in the heading stage (Fig. 6b). DAB staining of cyp76m7/8 double‐mutant leaves was observed to form reddish‐brown formazan, indicating the accumulation of H2O2 (Fig. 6c). The expression level of rice defence‐related genes, including PBZ, PR1 and POX22‐3, also was found to be markedly increased (Fig. 6d–f). Accordingly, the cyp76m7/8 double mutant then seemed to be sufficient to induce the same hypersensitive response seen with ∆c2bgc plants.
Fig. 6.
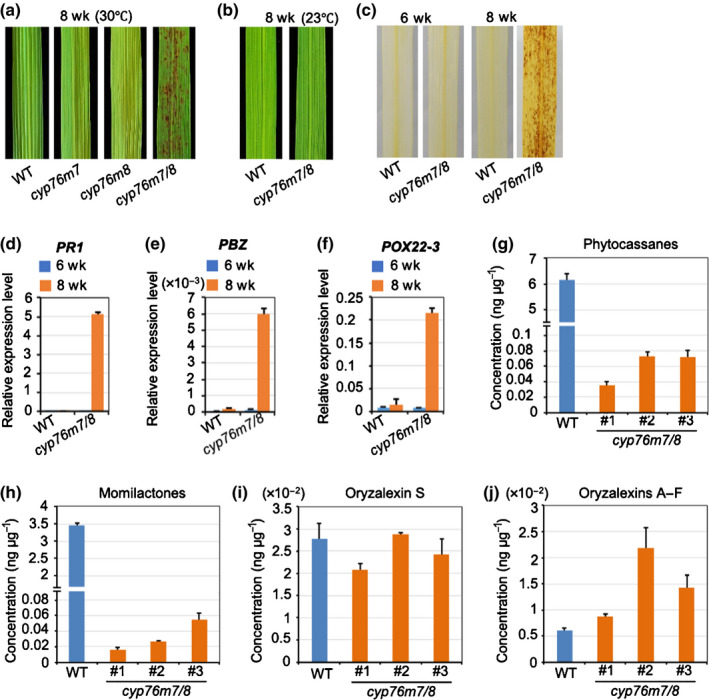
Lesion mimic phenotypes and phytochemical profiling of rice double knock‐out mutants of cyp76m7 and cyp76m8. (a) Representative leaves of wild‐type (WT), cyp76m7, cyp76m8 and cyp76m7/8 plants grown in a glasshouse (30°C) for 2 months after germination. (b) Representative leaves from the heading stage of WT and cyp76m7/8 plants grown in a growth chamber at 23°C. (c) Diaminobenzidine (DAB) staining of cyp76m7/8 and WT leaves before and after appearance of lesions. (d–f) Relative expression level of defence‐related genes PBZ, PR1 and POX22‐3 in leaves from cyp76m7/8 plants compared with WT. The leaf tissues were sampled before (6 wk) and after (8 wk) formation of lesions. Error bars represent ± SD. (g–j) Effect of cyp76m7/8 double knock‐out mutant on the labdane‐related diterpenoid metabolism relative to WT plants. Error bars represent ± SD.
Investigation of secreted LRDs demonstrated that the cyp76m7/8 double mutant exhibited significant reductions of both phytocassanes or momilactones, with no consistent effect on either oryzalexin S or oryzalexins A–F (Fig. 6g–j). Although both phytocassanes and momilactones were significantly reduced in cyp76m7/8 double‐mutant plants, the lesion mimic phenotype did not appear to be due to the reduction in LRD accumulation. In particular, despite similar reductions in these prevalent LRDs, the cps2x4 (Zhang et al., 2021) and ∆c2/c4bgc (Figs 5a, 7) double mutants did not display a lesion mimic phenotype. This contrast indicates that the LRDs are not simply acting as antioxidants, that is, to the block accumulation of reactive oxygen species.
Fig. 7.
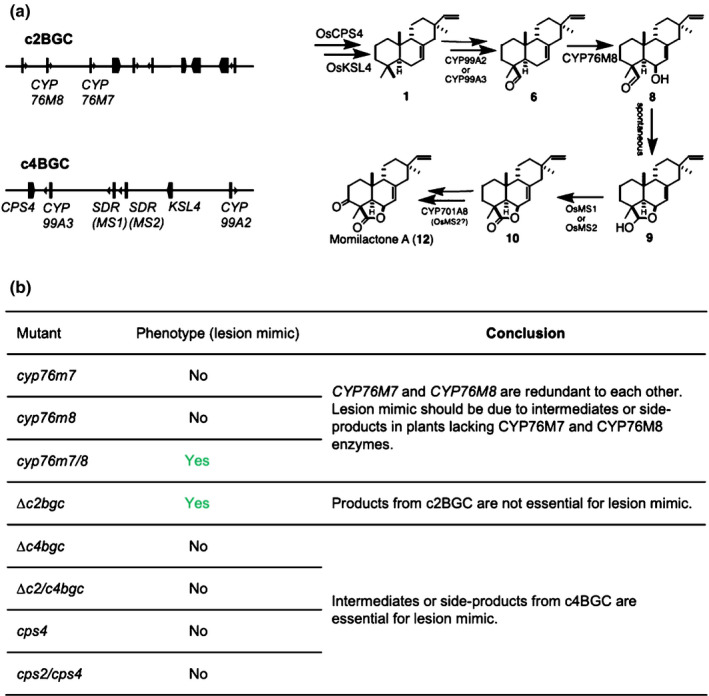
Intermediates or side‐products from the biosynthetic gene cluster from chromosome 4 (c4BGC) are genetically suggested to be reasons for lesion mimic in edited rice. (a) Genes from the BGC from chromosome 2 (c2BGC) (CYP76M8, plus CYP76M7) and c4BGC collaborate in the biosynthesis of momilactones. (b) Lesion mimic phenotype of single, double mutants and BGC deletion mutants, which suggests that intermediates or side‐products from c4BGC are essential for lesion mimic. CYP, cytochrome P450; KSL, ent‐kaurene synthase‐like.
Discussion
While deletion of the two BGCs for LRD production in rice has been previously reported in heterozygous form (Zhou et al., 2014), the results reported here demonstrated that this removal of substantial portions of the genome, and loss of associated biosynthetic capacity, is genetically stable. The effects of these deletion mutants on LRD production are consistent with the previously reported role of the CYP76M subfamily members (especially CYP76M8) from c2BGC in momilactone biosynthesis, which is alternatively associated with the c4BGC (Kitaoka et al., 2021). The ability of Δc2bgc mutant plants to produce some momilactones presumably is due to the previously reported ability of CYP76M14, located elsewhere in the genome, to catalyse the equivalent reaction (Kitaoka et al., 2021). This contrasts with the essentially complete loss of momilactone production exhibited by the cyp76m7/8 double‐mutant lines, which may be explained by the inefficiency of the other CYP76M subfamily members in catalysing the relevant reaction. In particular, when only CYP76M7 and CYP76M8 are knocked out, rather than deletion of the whole c2BGC, other substrates with which the remaining CYP76M subfamily members exhibit greater catalytic efficiency may then be present, preventing these from acting in momilactone biosynthesis. Alternatively, this effect may be due to altered expression of these subfamily members in Δc2bgc mutant plants relative to the cyp76m7/8 double‐mutant lines.
The effects of the two BGC deletion mutants on microbial disease resistance also was consistent with previously reported results (Lu et al., 2018; Zhang et al., 2021). However, the results from the Xoo bioassays reported here demonstrated that the c2BGC‐dependent (ent‐CPP‐derived) LRDs exhibited direct antibiotic activity against this bacterial pathogen. Moreover, it is now also evident that these natural products serve as phytoalexins in the leaves as well as phytoanticipins in the rhizosphere. In addition, the c2BGC‐dependent (ent‐CPP‐derived) LRDs exhibited differential effects on fungal blast resistance with the two strains of M. oryzae used here. Given that the LRD phytoalexins accumulated significantly faster in resistant, relative to susceptible, rice cultivars (Hasegawa et al., 2010), it appears that the relevance of the LRD phytoalexins to the defence response depends on timely detection of the microbial pathogen by the rice host. Therefore, the differential results are presumably due to partial genetic (in)compatibility between cv Kitaake rice and the M. oryzae strains used here, just as seen in other known plant–pathogen interactions (Delplace et al., 2021).
The effect of the BGC deletion mutants on drought resistance again is consistent with previously reported results (Zhang et al., 2021). In particular, the complete loss of either phytocassanes or momilactones, the most prevalent LRDs in rice, in Δc2bgc and Δc4bgc mutant plants (respectively), resembles the effect of the cps2 and cps4 knock‐out mutants in which this phenotype was first observed. This supports the hypothesis that LRDs act as a regulatory switch, with accumulation above a certain threshold promoting stomatal closure (Zhang et al., 2021). The cyp76m7/8 double knock‐out mutant lines, with significant reductions in both phytocassanes and momilactones, also exhibited this phenotype, again consistent with this hypothesis.
Perhaps most interesting is the observation of a lesion mimic phenotype with Δc2bgc mutant plants. The results reported here indicated that this results from a hypersensitive response, which includes induction of the rice microbial defence response. Fortuitously, it was possible to dissect the c2BGC and identify the relevant genes for this phenotype as the closely related CYP76M7 and CYP76M8. Intriguingly, despite exhibiting similar catalytic activity in recombinant settings, these are not redundant, with CYP76M7 serving a more important role in phytocassane biosynthesis, while CYP76M8 is more important for momilactone production (Kitaoka et al., 2021). Nevertheless, only the cyp76m7/8 double‐mutant plants exhibited the lesion mimic phenotype.
It has been postulated that phytoalexins exhibit antioxidant activity, at least in vitro (Ahuja et al., 2012), and the loss of such antioxidants would be consistent with the observed accumulation of H2O2 during the appearance of the lesion mimic phenotype. However, the lack of this phenotype in other mutant lines that exhibit analogous loss of LRD biosynthesis as Δc2bgc, most markedly Δc2/c4bgc, demonstrated that this is not simply due to antioxidant loss. Instead, the results suggested that phytotoxicity occurred in the absence of the c2BGC, particularly CYP76M7 and CYP76M8, but required the presence of the c4BGC, leading to a hypersensitive response. Intriguingly, it has recently been shown that in Nicotiana attenuata knocking down the expression of CYPs involved in diterpenoid biosynthesis can lead to accumulation of phytotoxic intermediates (Li et al., 2021), and it seems likely that similar phytotoxic accumulation of LRD intermediates dependent on the c4BGC may occur in rice in the absence of CYP76M7 and CYP76M8 from the c2BGC. This is most likely to be due to intermediates from the c4BGC‐associated momilactone biosynthesis, which has recently been elucidated (De La Pena & Sattely, 2021; Kitaoka et al., 2021), particularly the syn‐pimaradien‐19‐al substrate of CYP76M8. However, given the more general role of OsCPS4 for all syn‐CPP‐derived LRDs in rice (not all of which are known; see Fig. S1), this requires further investigation.
Regardless of the underlying mechanism, the observed directional phytotoxicity between these two BGCs provides further insight into their evolution. While the role for such negative selection pressure in BGC assembly might suggest that CYP76M7 and CYP76M8 should have been ‘recruited’ from the c2BGC to c4BGC, this ignores their largely distinct biosynthetic roles and evolutionary history. In particular, an extensive phylogenomic comparison indicated that phytocassane biosynthesis and assembly of the associated c2BGC arose first, with momilactone biosynthesis with assembly of the associated c4BGC occurring later (Miyamoto et al., 2016). Therefore, the CYP76M subfamily member originally recruited to the c2BGC must have been involved in phytocassane production, with subsequent gene duplication giving rise to a copy that became specialised for momilactone biosynthesis (CYP76M8). The interdependent evolution of these two BGCs has been previously noted (Kitaoka et al., 2021). Nevertheless, the negative selection pressure (i.e. phytotoxicity) demonstrated here provides further insight into the constraints this has imposed. In particular, this evolutionary process seems to have been sufficient to alleviate the usual recruitment that might have otherwise been expected to move CYP76M8 to the c4BGC. In any case, it is expected that further dissection of these intriguing genetic loci will provide additional insights into the underlying mechanisms, as well as evolutionary pressures for such BGC assembly.
Author contributions
RL and JZ designed and performed experiments. ZL, RJP and BY conceived the project and obtained financial support. RL and JZ wrote the initial draft of the manuscript, RJP, ZL and BY revised the manuscript. RL and JZ contributed equally to this work.
Supporting information
Fig. S1 Biosynthesis pathways of ent‐CPP and syn‐CPP‐derived diterpenes and phytochemical analysis of oryzalexins.
Fig. S2 Genomic DNA sequence of ∆c 2 bgc and ∆c 2 bgc mutations.
Fig. S3 Relative expression level of biosynthetic genes from chromosome 2 and 4 gene clusters.
Fig. S4 Effect of ∆c2bgc and ∆c4bgc mutants on drought resistance.
Fig. S5 Mutations of CYP76M7 and CYP76M8 loci in cyp76m7/8 double‐mutant lines.
Fig. S6 Effect of cyp76m7, cyp76m8 and cyp76m7/8 mutations on drought resistance.
Table S1 Primers for mutation detection.
Table S2 Segregation of M2 and F2 populations.
Table S3 Primers for RT‐PCR.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was partially supported by the US Department of Agriculture – National Institute of Food and Agriculture (grant no. 2020‐67013‐32557 to RJP and BY) and the National Institutes of Health (grant no. GM131885 to RJB). The authors are grateful to the National Small Grain Collection for providing rice seed, to Bo Liu and Huanbin Zhou for initially generating the gene cluster deletion lines, and to Guo‐Liang Wang for kindly providing the two strains of M. oryzae.
Contributor Information
Zhaohu Li, Email: lizhaohu@cau.edu.cn.
Reuben J. Peters, Email: rjpeters@iastate.edu.
Bing Yang, Email: yangbi@missouri.edu.
Data availability
Data are available in the Supporting Information.
References
- Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends in Plant Science 17: 73–90. [DOI] [PubMed] [Google Scholar]
- Balint‐Kurti P. 2019. The plant hypersensitive response: concepts, control and consequences. Molecular Plant Pathology 20: 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Yang B. 2017. Gene editing with TALEN and CRISPR/Cas in rice. Progress in Molecular Biology and Translational Science 149: 81–98. [DOI] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, Delarue M. 2015. To die or not to die? Lessons from lesion mimic mutants. Frontiers in Plant Science 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright DW, Langcake P, Pryce RJ, Leworthy DP, Ride JP. 1981. Isolation and characterization of two phytoalexins from rice as momilactones A and B. Phytochemistry 20: 535–537. [Google Scholar]
- Char SN, Li R, Yang B. 2019. CRISPR/Cas9 for mutagenesis in rice. Methods in Molecular Biology 1864: 279–293. [DOI] [PubMed] [Google Scholar]
- Chittoor JM, Leach JE, White FF. 1997. Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae . Molecular Plant–Microbe Interactions 10: 861–871. [DOI] [PubMed] [Google Scholar]
- De La Pena R, Sattely ES. 2021. Rerouting plant terpene biosynthesis enables momilactone pathway elucidation. Nature Chemical Biology 17: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delplace F, Huard‐Chauveau C, Berthomé R, Roby D. 2021. Network organization of the plant immune system: from pathogen perception to robust defense induction. The Plant Journal. doi: 10.1111/tpj.15462. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Mitsuhara I, Seo S, Imai T, Koga J, Okada K, Yamane H, Ohashi Y. 2010. Phytoalexin accumulation in the interaction between rice and the blast fungus. Molecular Plant–Microbe Interactions 23: 1000–1011. [DOI] [PubMed] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. 1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae . Plant Disease Reporter 56: 537–541. [Google Scholar]
- Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY. 2004. Proteomic analysis of pathogen‐responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea . Proteomics 4: 3569–3578. [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Zhang J, Oyagbenro RK, Brown B, Wu Y, Yang B, Li Z, Peters RJ. 2021. Interdependent evolution of biosynthetic gene clusters for momilactone production in rice. Plant Cell 33: 290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AT, Yagnik GB, Hohenstein JD, Ji Z, Zi J, Reichert MD, MacIntosh GC, Yang B, Peters RJ, Vela J et al. 2015. Investigation of the chemical interface in the soybean‐aphid and rice‐bacteria interactions using MALDI‐Mass spectrometry imaging. Analytical Chemistry 87: 5294–5301. [DOI] [PubMed] [Google Scholar]
- Li J, Halitschke R, Li D, Paetz C, Su H, Heiling S, Xu S, Baldwin IT. 2021. Controlled hydroxylations of diterpenoids allow for plant chemical defense without autotoxicity. Science 371: 255–260. [DOI] [PubMed] [Google Scholar]
- Li R, Char SN, Yang B. 2019. Creating large chromosomal deletions in rice using CRISPR/Cas9. Methods in Molecular Biology 1917: 47–61. [DOI] [PubMed] [Google Scholar]
- Lu X, Zhang J, Brown B, Li R, Rodriguez‐Romero J, Berasategui A, Liu B, Xu M, Luo D, Pan Z et al. 2018. Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell 30: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I, Iwai T, Seo S, Yanagawa Y, Kawahigasi H, Hirose S, Ohkawa Y, Ohashi Y. 2008. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense‐related signal compounds (121/180). Molecular Genetics and Genomics 279: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Fujita M, Shenton MR, Akashi S, Sugawara C, Sakai A, Horie K, Hasegawa M, Kawaide H, Mitsuhashi W et al. 2016. Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. The Plant Journal 87: 293–304. [DOI] [PubMed] [Google Scholar]
- Murphy K, Zerbe P. 2020. Specialized diterpenoid metabolism in monocot crops: biosynthesis and chemical diversity. Phytochemistry 172: 112289. [DOI] [PubMed] [Google Scholar]
- Muthayya S, Sugimoto JD, Montgomery S, Maberly GF. 2014. An overview of global rice production, supply, trade, and consumption. Annals of the New York Academy of Sciences 1324: 7–14. [DOI] [PubMed] [Google Scholar]
- Nutzmann HW, Huang A, Osbourn A. 2016. Plant metabolic clusters – from genetics to genomics. New Phytologist 211: 771–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutzmann HW, Scazzocchio C, Osbourn A. 2018. Metabolic gene clusters in eukaryotes. Annual Review of Genetics 52: 159–183. [DOI] [PubMed] [Google Scholar]
- Peters RJ. 2006. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317. [DOI] [PubMed] [Google Scholar]
- Peters RJ. 2010. Two rings in them all: the labdane‐related diterpenoids. Natural Products Reports 27: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ. 2014. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. The Plant Journal 79: 659–678. [DOI] [PubMed] [Google Scholar]
- Sesma A, Osbourn AE. 2004. The rice leaf blast pathogen undergoes developmental processes typical of root‐infecting fungi. Nature 431: 582–586. [DOI] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko K‐W, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N et al. 2007. Identification of a biosynthetic gene cluster in rice for momilactones. Journal of Biological Chemistry 282: 34013–34018. [DOI] [PubMed] [Google Scholar]
- Sugio A, Yang B, Zhu T, White FF. 2007. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice. Proceedings of the National Academy of Sciences, USA 104: 10720–10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. 2009. CYP76M7 is an ent‐cassadiene C11α‐hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Rook F. 2012. Why biosynthetic genes for chemical defense compounds cluster. Trends in Plant Science 17: 383–388. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kagahara T, Okada K, Koga J, Hasegawa M, Mitsuhashi W, Sassa T, Yamane H. 2008. Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Bioscience, Biotechnology, and Biochemistry 72: 562–567. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Usui M, Sugawara C, Otomo K, Hirose Y, Miyao A, Hirochika H, Okada K, Shimizu T, Koga J et al. 2014. Reverse‐genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiologia Plantarum 150: 55–62. [DOI] [PubMed] [Google Scholar]
- Tucker SL, Besi MI, Galhano R, Franceschetti M, Goetz S, Lenhert S, Osbourn A, Sesma A. 2010. Common genetic pathways regulate organ‐specific infection‐related development in the rice blast fungus. Plant Cell 22: 953–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. 2006. Reactive oxygen species in plant cell death. Plant Physiology 141: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ. 2012. Characterization of CYP76M5‐8 indicates metabolic plasticity within a plant biosynthetic gene cluster. Journal of Biological Chemistry 287: 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kono Y, Esumi Y, Teraoka T, Hosokawa D, Suzuki Y, Sakurai A, Watanabe M. 1996. Studies on a quantitative analysis of oryzalides and oryzalic acids in rice plants by GC‐SIM. Bioscience, Biotechnology, and Biochemistry 60: 1460–1463. [Google Scholar]
- Wu Y, Hillwig ML, Wang Q, Peters RJ. 2011. Parsing a multifunctional biosynthetic gene cluster from rice: biochemical characterization of CYP71Z6 & 7. FEBS Letters 585: 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Galhano R, Wiemann P, Bueno E, Tiernan M, Wu W, Chung IM, Gershenzon J, Tudzynski B, Sesma A et al. 2012. Genetic evidence for natural product‐mediated plant–plant allelopathy in rice (Oryza sativa). New Phytologist 193: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H. 2013. Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Bioscience, Biotechnology, and Biochemistry 77: 1141–1148. [DOI] [PubMed] [Google Scholar]
- Ye Z, Yamazaki K, Minoda H, Miyamoto K, Miyazaki S, Kawaide H, Yajima A, Nojiri H, Yamane H, Okada K. 2018. In planta functions of cytochrome P450 monooxygenase genes in the phytocassane biosynthetic gene cluster on rice chromosome 2. Bioscience, Biotechnology, and Biochemistry 82: 1021–1030. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li R, Xu M, Hoffmann RI, Zhang Y, Liu B, Zhang M, Yang B, Li Z, Peters RJ. 2021. A (conditional) role for labdane‐related diterpenoid natural products in rice stomatal closure. New Phytologist 230: 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. 2014. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Research 42: 10903–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Yang B, Chittoor JM, Johnson LB, White FF. 1998. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Molecular Plant–Microbe Interactions 11: 824–832. [DOI] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ. 2014. To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annual Review of Plant Biology 65: 259–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Biosynthesis pathways of ent‐CPP and syn‐CPP‐derived diterpenes and phytochemical analysis of oryzalexins.
Fig. S2 Genomic DNA sequence of ∆c 2 bgc and ∆c 2 bgc mutations.
Fig. S3 Relative expression level of biosynthetic genes from chromosome 2 and 4 gene clusters.
Fig. S4 Effect of ∆c2bgc and ∆c4bgc mutants on drought resistance.
Fig. S5 Mutations of CYP76M7 and CYP76M8 loci in cyp76m7/8 double‐mutant lines.
Fig. S6 Effect of cyp76m7, cyp76m8 and cyp76m7/8 mutations on drought resistance.
Table S1 Primers for mutation detection.
Table S2 Segregation of M2 and F2 populations.
Table S3 Primers for RT‐PCR.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Data are available in the Supporting Information.


