Abstract
Cancer immunotherapy offers substantive benefit to patients with various tumour types, in some cases leading to complete tumour clearance. However, many patients do not respond to immunotherapy, galvanizing the field to define the mechanisms of pre-existing and acquired resistance. Interferon-γ (IFNγ) is a cytokine that has both protumour and antitumour activities, suggesting that it may serve as a nexus for responsiveness to immunotherapy. Many cancer immunotherapies and chemotherapies induce IFNγ production by various cell types, including activated T cells and natural killer cells. Patients resistant to these therapies commonly have molecular aberrations in the IFNγ signalling pathway or express resistance molecules driven by IFNγ. Given that all nucleated cells can respond to IFNγ, the functional consequences of IFNγ production need to be carefully dissected on a cell-by-cell basis. Here, we review the cells that produce IFNγ and the different effects of IFNγ in the tumour microenvironment, highlighting the pleiotropic nature of this multifunctional and abundant cytokine.
In 1965, leukocytes were found to produce an antiviral molecule in response to phytohaemagglutinin that was different to the previously described type I interferons (interferon-α (IFNα) and IFNβ)1. It was not until 1980 that this type II interferon was formally designated IFNγ. In addition to its role in microbial infections, IFNγ has prominent roles in other diseases, such as cancer. Initially, it was thought that IFNγ has only antitumour effects: rejection of a transplanted fibrosarcoma in mice by treatment with a bacterial endotoxin was prevented when the mice were given an IFNγ-neutralizing antibody and IFNγ-neutralized tumours grew faster2. Similarly, studies of mice lacking IFNγ receptor (IFNGR) and signal transducer and activator of transcription 1 (STAT1) showed that endogenous IFNγ prevents the development of carcinogen-induced sarcomas. Additionally, the antitumour effects of the IFNγ-inducing cytokine interleukin-12 (IL-12) were ablated by neutralization of IFNγ3. These early studies uncovered the cytotoxic effects of IFNγ on tumour cells. Since then, protumour effects of IFNγ have emerged. The discovery that IFNγ promotes expression of the inhibitory molecules programmed cell death 1 ligand 1 (PDL1), PDL2, indoleamine 2,3-dioxygenase 1 (IDO1), inducible nitric oxide synthase (iNOS), FAS and FAS ligand (FASL), all of which limit antitumour immunity, has increased caution with the use of IFNγ-modulating cancer immunotherapies.
Programmed cell death 1 ligand 1.
(PDL1). A ligand that binds to programmed cell death 1 (PD1) on T cells to inhibit their activation, proliferation and cytokine production. PDL1 is also known as CD274 and B7-H1, and PDL2 is also known as CD273 or B7-C.
In this Review, we discuss IFNγ in the context of cancer and the challenges of targeting IFNγ therapeutically. Although IFNγ has direct cytotoxic effects on tumour cells, its therapeutic application is currently not possible owing to the broad expression of IFNGR and thus potential cytotoxic effects on antitumour immune cells. Therefore, IFNγ-inducing cancer immunotherapies have the undesirable potential to exacerbate tumour burden. We propose that IFNγ may serve different functions when produced, or responded to, by different immune cells, which thereby act as teammates (with immunostimulating, antitumour functions) or opponents (with immunosuppressing, protumour functions) in the tumour microenvironment (TME). By considering IFNγ production and responses within the TME, it may be possible to develop more mechanistically tailored approaches to bias IFNγ-based cancer immunotherapy towards solely antitumour effects.
Regulation of IFNγ expression
IFNγ expression is tightly regulated by epigenetic, transcriptional, post-transcriptional and post-translational modifications. These mechanisms prevent IFNγ expression by non-immune cells, naive T cells and even some activated immune cells4. Understanding how IFNγ expression is regulated may uncover mechanisms of immune exploitation by tumours to escape immunosurveillance and novel IFNγ-inducing pathways for therapeutic intervention.
Epigenetic and transcriptional regulation of IFNG.
IFNG is actively silenced in naive T cells via methylation and hypoacetylation. Conserved non-coding sequences within the proximal regulatory regions of IFNG allow epigenetic regulation, transcription factor binding and cellspecific expression5. Acetylation of histones H3 and H4 is important for T helper 1 (TH1) cell differentiation, which is abrogated in STAT4-deficient cells, indicating that STAT4 promotes Ifng expression via acetylation and binding to the Ifng promoter6. Interestingly, hyperacetylation of IFNγ-encoding chromatin differs among T cells and can extend outside the IFNG locus, allowing cell-specific epigenetic regulation. For example, epigenetic regulation of IFNG by the long non-coding RNA NeST (also known as Tmevpg1 or Ifng-AS1) and WD repeat-containing protein 5 (WDR5) is critical for TH1 cell expression of IFNγ via the transcription factor T-bet7. Acetylation of H3 and H4 is similar in TH1 cells and CD8+ T cells; however, in CD8+ T cells it occurs independently of T-bet and is greater in memory T cells, in which it requires CD4+ T cell help. Ifng histone modifications induced by T-bet overexpression in CD4+ T cells are sufficient to drive Ifng expression under TH2-polarizing conditions, uncovering epigenetic mechanisms that ensure cell lineage-specific expression of IFNγ8.
Epigenetic regulation.
Control of gene expression through phenotypic changes that do not alter the DNA sequence. Examples include DNA methylation, histone modifications, microRNAs, long non-coding RNAs and nucleosome positioning.
Similarly to the case for other cytokines, IFNG transcription is promoted by various stimuli, including T cell receptor (TCR) engagement, and transcription factors, such as activator protein 1 (AP-1), T-bet, eomesodermin (EOMES), nuclear factor of activated T cells (NFAT) and nuclear factor-κB (NF-κB)9–12. Interestingly, a positive-feedback loop is formed by which activated immune cell products, such as IL-2, hydrogen peroxide and leukotrienes, further induce IFNγ production via second messengers (protein kinase C and cyclic GMP)13–15. In addition, IL-12 and IL-18 promote IFNG transcription16; IL-18 acts as a cofactor by signalling via NF-κB and AP-1, and IL-12 promotes transcription of IL-18 receptor, establishing a positive-feedback loop17. These parallel signalling pathways allow synergistic regulation of IFNγ expression. IL-12 activates STAT4, and IL-18 activates the AP-1 subunit JUN, forming a STAT4–AP-1 complex which enhances STAT4 binding to the IFNG promoter18,19. Consequently, prevention of IL-12 production may result in the equivalent of IFNG deletion by blocking IFNγ production in the TME.
Post-transcriptional regulation of IFNG.
Preformed IFNG mRNA in both the nuclear compartment and the cytoplasmic compartment allows fast translation and secretion upon appropriate stimulation. The mitogen-activated protein kinase p38 induced by IL-12 and IL-18 binds to an AU-rich element in the 3′ untranslated region of IFNG mRNA and stabilizes the mRNA20,21. IFNG mRNA is also negatively regulated indirectly by the microRNAs miR-29, miR-146a and miR-142–3P22–24. However, tumours can hijack these suppressive mechanisms to limit IFNG expression. Various tumour types have been shown to secrete miR-29 and miR-146, which remodel the TME in favour of tumour growth and metastasis25,26.
Secretion of IFNγ.
The protein structure of IFNγ comprises mainly α-helices, allowing two molecules of IFNγ to dimerize in an antiparallel manner through noncovalent bonds27. The functional IFNγ homodimer is immediately secreted and can be detected extracellularly as early as 6 h after TCR activation, and its level peaks at 12–24 h. Once secreted, mouse IFNγ persists in the blood for much longer (0.94 h) than other cytokines such as IL-2 (0.2 h)28,29. This long half-life is thought to be due to expression of IFNGR by platelets, which may bind IFNγ to facilitate systemic transport30. This systemic stabilization of IFNγ may explain the undesired systemic cytotoxic effects that are seen with IFNγ-modulating cancer immunotherapy31.
Producers of IFNγ
Numerous immune cell subsets, including T cells, natural killer (NK) cells, invariant NK T cells (iNKT cells), regulatory T (Treg) cells, γδ T cells and B cells, produce IFNγ in the TME. Although IFNγ has pleiotropic effects, IFNγ production by immune cells is generally antitumorigenic rather than protumorigenic. However, the secretion of other cytotoxic, proinflammatory and anti-inflammatory cytokines together with IFNγ may modulate its activity in the TME. Thus, IFNγ produced by different cell types can have unique and distinct effects on its intended targets and bystanders within the TME. In addition, the way in which IFNγ is secreted by these cells, such as synaptic, leaky synaptic or multidirectional secretion, can affect the outcome of IFNγ production32 (FIG. 1).
Fig. 1 |. Classical IFNγ producers in the tumour microenvironment.

The spatial pattern of interferon-γ (IFNγ) release by T helper 1 (TH1) cells, cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells. TH1 cells release IFNγ, interleukin-2 (IL-2), granulocyte–monocyte colony-stimulating factor (GM-CSF) and lymphotoxin-α (LTα) in a concentrated manner within the antigen-presenting cell (APC)–TH1 cell synapse (synaptic release). Tumour necrosis factor (TNF) is released in many directions, both towards and away from the synapse (multidirectional release). CTLs release IFNγ towards the synapse but the release is not well directed, allowing IFNγ to exert effects on cells beyond, but near, the synapse (leaky synaptic release). The release of TNF and perforin is synaptic. NK cells release IFNγ in a multidirectional manner.
Invariant NK T cells.
(iNKT cells). Innate-like T cells that express a T cell receptor α-chain that recognizes lipid antigens presented by the non-classical MHC molecule CD1d expressed on dendritic cells.
γδ T cells.
T cells that express T cell receptor γ and δ chains and represent 1–4% of the T cell population. They produce interferon-γ (IFNγ) rapidly following activation in a non-MHC-restricted manner by tumour-derived lipids, glycoproteins and phosphorus-containing compounds.
Effector T cells.
CD8+ cytotoxic T lymphocytes (CTLs) are well-known key producers of IFNγ and are crucial for antitumour immunity. Unlike for TH1 cells, expression of IFNγ by CTLs, as well as their production of perforin, granzymes, tumour necrosis factor (TNF) and IL-2, is independent of T-bet and requires the T-bet paralogue EOMES33,34. TCR activation induces EOMES expression, which promotes IFNγ release from CTLs in a leaky synaptic manner; this ensures that the target cell receives a concentrated IFNγ signal but IFNγ also reaches neighbouring cells35 (FIG. 1). This mode of IFNγ secretion differs from that for the cytotoxic molecules TNF and perforin, which are concentrated at the immunological synapse to mediate targeted cell killing. IFNγ+ T cells form a synapse with neighbouring non-antigen-specific CD8+ T cells, which is crucial to promote expansion and differentiation of bystander T cells, which in turn become IFNγ producers36–38. Leaky synaptic release of IFNγ combined with its increased stability and widespread distribution compared with other cytokines allows hours of IFNγ exposure, which is required for the transcriptional effects that mediate effective tumour cell killing. Importantly, IFNγ production by CD8+ T cells is required for a response to therapy using antibody against the immune checkpoint molecule programmed cell death 1 (PD1)39. In addition, the presence of proliferating CD8+IFNγ+ T cells in the TME is a biomarker for therapeutic response to the kinase inhibitor sorafenib in hepatocellular carcinoma40. These findings illustrate the importance of IFNγ production by CD8+ T cells in mediating beneficial widespread and durable cytotoxic tumour killing and proinflammatory effects in the TME.
IFNγ is also the signature cytokine of TH1 cells, which also produce TNF, IL-2, lymphotoxin-α (LTα) and granulocyte–monocyte colony-stimulating factor (GM-CSF). The release of IFNγ, IL-2, LTα and GM-CSF from TH1 cells is synaptic, which directs prosurvival signals to antigen-presenting cells (APCs) (FIG. 1). By contrast, TNF is released from TH1 cells in a multidirectional manner to mediate cytotoxicity in the TME, while promoting dendritic cell (DC) maturation and macrophage activation at the synapse41. The importance of TH1 cells in the TME was shown in a mouse lung carcinoma model in which a switch from a TH2 cell-dominated TME to a TH1 cell-dominated TME promoted tumour clearance with combinatorial immunotherapies, and IFNγ production negatively correlated with tumour size42.
Human TH17 cells, which produce IL-17 and resemble terminally differentiated memory T cells, have been shown to produce other effector cytokines, including IFNγ. Tumour-infiltrating TH17 cells showed a positive correlation with CD4+IFNγ+ T cells and CD8+IFNγ+ T cells, and produced a potent antitumour response in an ovarian cancer model43. Interestingly, IFNγ and IL-17 synergize to induce production of the TH1-type chemokines CXC-chemokine ligand 9 (CXCL9) and CXCL10 by tumour cells to facilitate effector cell recruitment to the TME44.
NK cells.
NK cells are innate cytotoxic cells that provide the first line of defence against tumour growth. NK cells recognize non-self targets, such as tumour cells, and mediate cytotoxic effects through the production of IFNγ, which is induced by IL-2 and IL-12, and is potentiated by TNF45,46. IFNγ and TNF are stored in recycled endosomes, which deliver these cytokines to localized areas away from the target cell for multidirectional release47. This makes IFNγ available to promote the activation of inflammatory cells and their recruitment to the TME (FIG. 1). NK cell tumour infiltration positively correlates with better cancer prognosis, and cancer stage negatively correlates with NK cell activity, specifically IFNγ production48–50. A diagnostic test that measures blood NK cell activity for IFNγ production in patients with gastric cancer may be a promising non-invasive test for monitoring disease progression51.
iNKT cells.
When iNKT cells form a synapse with a DC presenting iNKT cell ligand, IL-12 from the DC is released and binds to IL-12 receptor on iNKT cells to induce IFNγ production52. The pattern of IFNγ release by iNKT cells may be synaptic owing to a iNKT cell–DC positive-feedback loop in which IFNγ produced by iNKT cells promotes DC maturation via the upregulation of co-stimulatory molecules53,54. However, iNKT cells also generate IFNγ in response to DC-derived IL-12 in the absence of CD1d-presented antigen55.
Activation of iNKT cells with the synthetic ligand α-galactosylceramide (α-GalCer) promotes antitumour activity in the clinic, which is dependent on an increase in the number of IFNγ+ cells in peripheral blood56,57. Unfortunately, α-GalCer has been shown to produce iNKT cell anergy, contributing to the suboptimal therapeutic effects. Similar to T cells, iNKT cell anergy can be reinvigorated with anti-PD1 or anti-PDL1 therapy, which when combined with use of α-GalCer in mice prolonged the antitumour effect58.
Treg cells.
Treg cells are a canonically immunosuppressive subset of CD4+ T cells characterized by expression of the transcription factor forkhead box P3 (FOXP3) and production of the inhibitory cytokines IL-10, IL-35 and transforming growth factor-β (TGFβ)59,60. FOXP3 maintains Treg cell suppressive identity through AKT inhibition via nuclear sequestration of forkhead box O1 (FOXO1), which represses IFNG transcription61,62. Despite these mechanisms to suppress IFNγ production, FOXP3+IFNγ+ Treg cells are present in various autoimmune diseases and in the TME62. IFNγ+ Treg cells found in autoimmunity and bacterial infections express a TH1-like transcriptional programme that includes expression of CXC-chemokine receptor 3 (CXCR3), which mediates their recruitment to sites of inflammation and suppression of TH1 cells63,64. However, IFNγ+ Treg cells in the TME have impaired suppressive function, which allows greater antitumour immunity and contributes to decreased tumour growth65.
In various mouse models of inflammatory disease, IFNγ and the alarmin IL-33 form a regulatory loop: IL-33 promotes IFNγ production by NK cells and γδ T cells, and IFNγ promotes IL-33 production by keratinocytes and fibroblasts66,67. However, in the TME, IL-33 has a unique role in maintaining stability and function of Treg cells, as IL-33-deficient Treg cells produce IFNγ and exhibit a loss of suppressive function, which potentiates the antitumour effects of immune checkpoint blockade therapy68. Although a direct mechanism of IFNγ repression by IL-33 in Treg cells has not been elucidated, IL-33 supported the induction of a TH2-like environment during chronic inflammation that promoted Treg cell stability and was protumorigenic. Additionally, Treg cell-specific deletion of the IL-33 receptor ST2 blocked tumour development in models of inflammation-induced skin and colon cancer69. Collectively, the impact of this IFNγ–IL-33 axis on Treg cells differs from its impact on other immune cells but supports the antitumorigenic role of IFNγ+ fragile Treg cells in the TME.
γδ T cells.
γδ T cells are another T cell population that produces IFNγ rapidly following activation via T-bet and EOMES. Mice have two subsets of γδ T cells: IL-17+ γδ T cells, which promote tumour growth (owing to increased PDL1 expression and recruitment of immunosuppressive neutrophils and macrophages) and IFNγ+ γδ T cells, which have antitumour effects (owing to increased production of IFNγ, TNF, perforin and granzymes). IL-17+ γδ T cells are rare in humans, whereas IFNγ+ γδ T cells are more common in the TME of human cancers. γδ T cells are recruited to the TME before αβ T cells. Interestingly, γδ T cell-deficient mice have increased incidence of tumour development and growth. Mice with IFNγ-deficient γδ T cells fail to control tumour initiation and growth and also have impaired IFNγ production by αβ T cells70. Similarly to the case for conventional αβ T cells, PD1 blockade in patients with leukaemia reinstated IFNγ production by tumour-infiltrating γδ T cells71. Interestingly, γδ T cell prevalence in the epithelia negatively correlates with epithelial malignancies, whereas γδ T cell prevalence in the TME correlates with good prognosis in various human cancers72,73.
B cells.
In addition to their ability to produce antibodies, B cells mediate antibody-independent functions via cytokine secretion74. A subset of innate CD11ahiCD16/CD32hi B cells produce IFNγ during early stages of bacterial infection, similarly to NK cells75. Interestingly, IFNγ production by CD11ahiCD16/CD32hi B cells requires IFNGR and T-bet expression, as well as Bruton’s tyrosine kinase (BTK; via NF-κB), IL-1β and CD40–CD40L signalling76. However, the role of IFNγ+CD11ahiCD16/CD32hi B cells in the TME and the manner in which IFNγ is released are unknown. It is possible that the antitumour effects induced by immunotherapy with CD40 agonists, which activate NF-κB in DCs and B cells, involve the induction of IFNγ by CD11ahiCD16/CD32hi B cells, but this has yet to be proven77,78.
IFNγ responders in the TME
All nucleated cells constitutively express IFNGR1 and can respond to IFNγ, and therefore its pleiotropic effects in the TME are complex and the overall impact on tumour growth depends on the balance of antitumour IFNγ signalling (tumour cell killing, effector function, cell migration, immune cell proliferation and antigen presentation) acting as a teammate for the immune system and protumour IFNγ signalling (immunosuppression, angiogenesis and tumour cell proliferation) acting as an opponent of the immune system. Details of IFNγ signalling via its receptor are described in BOX 1, and an abbreviated list of notable IFNγ-regulated genes and their known roles in the TME are given in TABLE 1 (see REFS79,80 for comprehensive reviews of IFNγ-induced genes in various diseases). The complex network of IFNγ responders in the TME is delineated here, focusing on T cells, NK cells, APCs, tumour cells, the vasculature and lymphatics (FIG. 2).
Box 1 |. IFNGR and signalling.
Structure and expression of IFNGR
Interferon-γ (IFNγ) receptor (IFNGR) consists of IFNGR1 (α-subunit) and IFNGR2 (β-subunit)173 (see the figure). IFNGR1 is constitutively expressed by all nucleated cells at 200–25,000 molecules per cell. IFNGR1 expression is highest in non-lymphoid tissues such as the skin, nerves, placenta and syncytiotrophoblasts, suggesting a role for IFNγ in embryonic development, tissue homeostasis and tolerance174. the levels of IFNGR1 mrNa expression by immune cells differ, with monocytes expressing the highest levels, then B cells, NK cells and lastly T cells175,176. Conversely, the inducible expression of IFNGR2 by transcription factor SP1, activating protein 2 (AP-2) and nuclear factor-κB (NF-κB) allows regulation of IFNγ-induced signalling177. IFNGR1 has a major role in ligand binding, whereas IFNGR2 has a predominant role in signalling via Janus kinases (JAKs) and signal transducers and activators of transcription (STATs), albeit both receptor subunits are required178. interestingly, only IFNGR1 has a nuclear localization signal for translocation with STAT1 (REF.179).
IFNGR signalling
IFNGR lacks intrinsic kinase activity and requires the adaptors JAK1 and JAK2 and the transcription factor STAT1 to mediate downstream signalling173. IFNγ binding and receptor association triggers JAK2 autophosphorylation, transphosphorylation and activation, which uncovers docking sites for SH2 domain-containing signal transducers180.
Canonical IFNGR signalling.
Each molecule of the STAT1–STAT1 antiparallel homodimer is phosphorylated at Tyr701 by JAK1 and/or JAK2, resulting in immediate receptor dissociation. STAT1 is phosphorylated at Ser727 via a PI3K- and AKT-dependent mechanism that is required for maximum transcriptional activity. Phosphorylated STAT1–STAT1 dimers undergo nuclear translocation via importin α5 (REF.181). STAT1 then binds to IFNγ-activating sites (GASs) containing the consensus sequence TTCN2–4GAA within the promoters of interferon-responsive genes (IRGs)182. IFNγ promotes the transcription of interferon regulatory factor 1 (IRF1) and IRF9, thus further amplifying IFNγ-induced gene transcription182.
Non-canonical IFNGR signalling.
Studies of Stat1-knockout cells reveal STAT1–STAT1-independent IFNγ signalling via STAT3 (REFS183,184). STAT3 and STAT1 are structurally similar and compete for binding to the phosphorylated Tyr419 of IFNGR, although STAT1 has higher affinity185. STAT3 forms either a homodimer (STAT3–STAT3) or a heterodimer (STAT1–STAT3), which regulate GAS3 and GAS2, respectively. As interleukin-6 (IL-6) and IL-10 both signal via STAT3, the expression of cytokine receptors dictates which signalling pathway dominates186. Differing levels of expression of STAT1 and STAT3 by specific cell types results in unique IFNγ-induced transcriptional effects in the tumour microenvironment185,187.
Suppressor of cytokine signalling proteins.
Prolonged IFNγ-induced signalling promotes lethal autoimmune diseases in mice; therefore, intrinsic feedback inhibition mediated by suppressor of cytokine signalling (SOCS) proteins is imperative188. SOCS1 transcription is promoted by SP1 and IRF1, which are induced by T cell receptor activation and IFNγ signalling, respectively189. SOCS1 binds to JAK1 and JAK2 via its kinase inhibitory domain to target them for proteasomal degradation190.
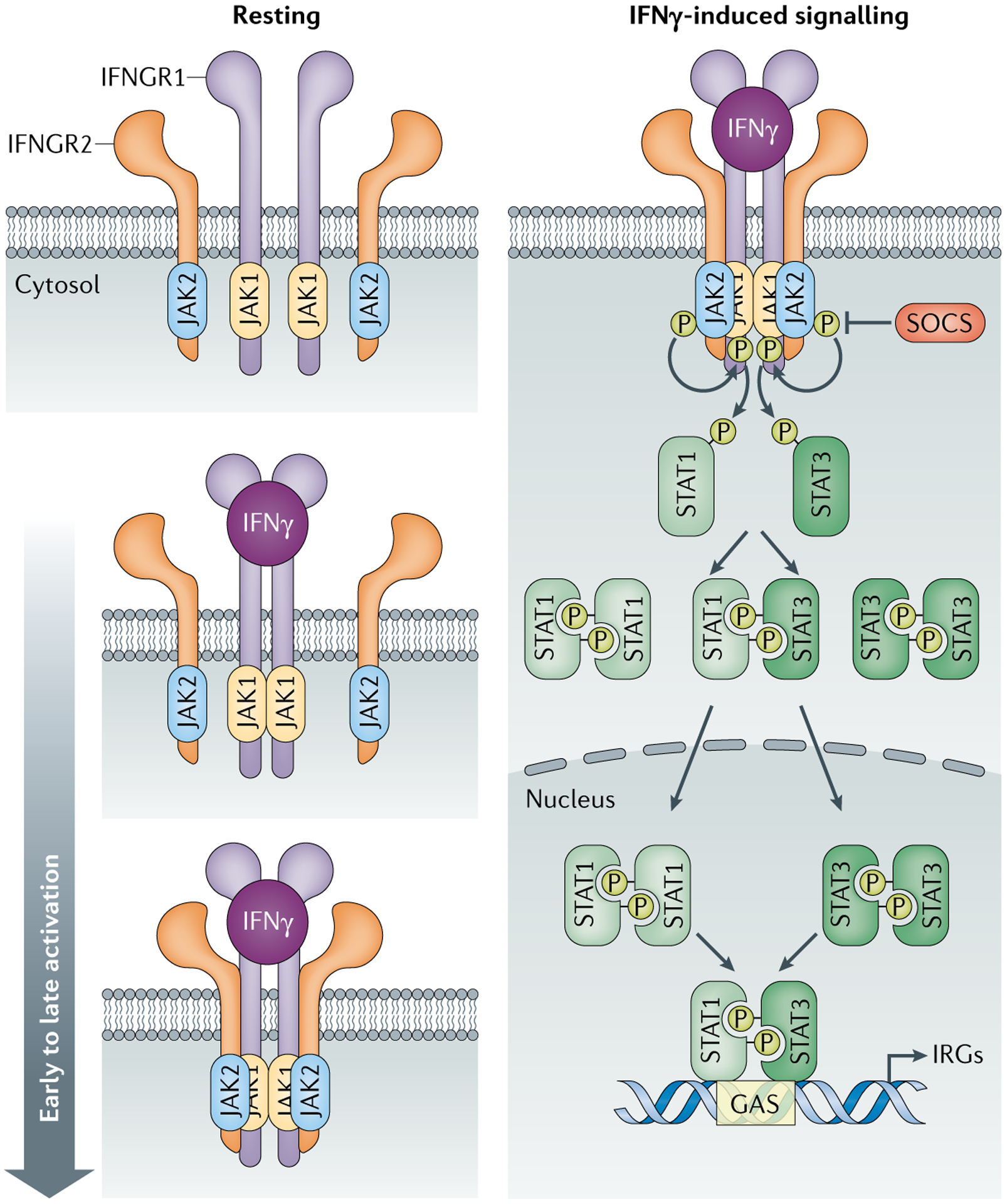
Table 1 |.
IFNγ-responsive genes and their role in the tumour microenvironment
| Function in the TME | Gene symbol | Protein | Effect on tumour | Mechanism | Transcription factors |
|---|---|---|---|---|---|
| Antigen presentation | HLA-A, HLA-B, HHL-C | MHC class I molecules | Antitumorigenic | Antigen presentation to T cells | STAT1, IRF1, NF-κB |
| Protumorigenic | Inhibitory receptor engagement on NK cells | ||||
| CD80 | CD80 (also known as B7–1) | Antitumorigenic | Co-stimulatory molecule for CD28 on T cells | STAT1, STAT3, IRF1 | |
| CD86 | CD86 (also known as B7–2) | Antitumorigenic | Co-stimulatory molecule for CD28 on T cells | STAT3, IRF1 | |
| CTSS | Cathepsin S | Antitumorigenic | Processing of tumour-specific peptides for antigen presentation | STAT1, STAT3, IRF1 | |
| Proliferation | CDKN1A | Cyclin-dependent kinase inhibitor 1A (also known as p21 or CIP1) | Antitumorigenic | Cell cycle arrest of tumour cells (G1 → S phase) | STAT1, STAT3, IRF1 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (also known as p27 or KIP1) | Antitumorigenic | Cell cycle arrest of tumour cells (G1 → S phase) | STAT1, IRF1 | |
| RB1 | Retinoblastoma-associated protein | Protumorigenic | Cell cycle progression in tumour cells (G1 → S phase) | STAT1, STAT3, IRF1, NF-κB | |
| MYC | MYC | Protumorigenic | Cell cycle progression in tumour cells (G1 → S phase); promotes cellular metabolism | STAT1, STAT3, IRF1, NF-κB | |
| Apoptosis | FAS | FAS | Antitumorigenic | Apoptosis of tumour cells | STAT1, IRF1, |
| Protumorigenic | Apoptosis of B cells, T cells and endothelial cells | NF-κB | |||
| FASL | FAS ligand | Antitumorigenic | Apoptosis of tumour cells | STAT1, IRF1, | |
| Protumorigenic | Apoptosis of B cells, T cells and endothelial cells | NF-κB | |||
| CTSD | Cathepsin D | Antitumorigenic | Apoptosis of tumour cells | STAT3 | |
| Immunosuppression | CD274 | Programmed cell death 1 ligand 1 | Protumorigenic | T cell exhaustion; Treg cell, M2 macrophage and TH17 cell differentiation; inhibition of phagocytosis | STAT3, NF-κB |
| PDCD1LG2 | Programmed cell death 1 ligand 2 | Protumorigenic | T cell exhaustion and cell death | STAT1, STAT3, IRF1, NF-κB | |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | Protumorigenic | T cell anergy and death; Treg cell expansion; angiogenesis; TGFp production | STAT1, STAT3, IRF1, NF-κB | |
| Effector | TBX21 | T-bet | Antitumorigenic | TH1 cell differentiation; IFNγ production | STAT3, NF-κB |
| SERPINB9 | Serpin B9 | Antitumorigenic | Inactivation of granzyme B to protect lymphocytes | IRF1 | |
| Protumorigenic | Inactivation of granzyme B to protect tumours | ||||
| ITK | IL-2-inducible T cell kinase | Antitumorigenic | T cell activation | STAT1, STAT3, | |
| Protumorigenic | TH2 cell differentiation | IRF1, NF-κB | |||
| Migration | CXCR3 | CXC-chemokine receptor 3 | Antitumorigenic | Migration of TH1 cells, CTLs, NK cells and NKT cells | STAT1, STAT3, IRF1, NF-κB |
| CXCL9 | CXC-chemokine ligand 9 | Antitumorigenic | Lymphocyte migration; TH1 and TH17 cell differentiation; inhibition of angiogenesis | STAT1, STAT3, IRF1, NF-κB | |
| CXCL10 | CXC-chemokine ligand 10 | Antitumorigenic | Lymphocyte migration; TH1 and TH17 cell differentiation; inhibition of angiogenesis | STAT1, STAT3, NF-κB | |
| CXCL11 | CXC-chemokine ligand 11 | Antitumorigenic | Lymphocyte migration | STAT3 | |
| Protumorigenic | TH2 cell differentiation; angiogenesis; IL-10 production | ||||
| CCL2 | CC-chemokine ligand 2 (also known as MCP1) | Protumorigenic | Monocyte migration; tumour-associated macrophage and TH2 cell differentiation; IL-4 production | STAT1, STAT3 | |
| CCL5 | CC-chemokine ligand 5 (also known as RANTES) | Antitumorigenic | T cell migration | STAT3, IRF1, NF-κB | |
| Protumorigenic | Monocyte and Treg cell migration; angiogenesis | ||||
| ICAM1 | Intracellular adhesion molecule 1 | Antitumorigenic | Transendothelial leukocyte migration; inhibition of M2 macrophage differentiation | STAT1, NF-κB | |
| Transcription | IRF1 | Interferon regulatory factor 1 | Antitumorigenic | Apoptosis; inhibition of proliferation; antigen presentation | STAT1, IRF1, NF-κB |
| BCL6 | B cell lymphoma 6 | Antitumorigenic | Germinal centre formation; T follicular helper cell formation | STAT1 | |
| Protumorigenic | DNA damage; proliferation; tumour cell migration and invasion | ||||
| NFKB1 | Nuclear factor-κB | Antitumorigenic | Lymphocyte survival and proliferation; inflammatory cytokine production | STAT1, STAT3, NF-κB | |
| Protumorigenic | Angiogenesis; tumour proliferation and invasion | ||||
| CREB1 | cAMP-responsive element-binding protein 1 | Antitumorigenic | Leukocyte survival and proliferation; B and T cell activation; TH1 cell differentiation; antibody production; IFNγ production | STAT3, IRF1, NF-κB | |
| Protumorigenic | Tumour cell proliferation, migration and invasion; TH2 cell differentiation; Treg cell stability; IL-10 production | ||||
| JUN | Activator protein 1 subunit JUN | Antitumorigenic | T cell activation, proliferation and differentiation; production of proinflammatory cytokines; inhibition of T cell exhaustion | STAT1, STAT3, NF-κB | |
| Protumorigenic | Neoplastic transformation; tumour cell proliferation | ||||
| FOS | Cellular oncogene FOS | Antitumorigenic | Tumour cell apoptosis; T cell activation and differentiation | STAT1, STAT3, IRF1, NF-κB | |
| Protumorigenic | Apoptosis; neoplastic transformation; tumour cell proliferation; IL-10 production |
cAMP, cyclic AMP; CTL, cytotoxic T lymphocyte; IFNγ, interferon-γ; IL, interleukin; NF-κB, nuclear factor-κB; NK cell, natural killer cell; NKT cell, natural killer T cell; STAT, signal transducer and activator of transcription; TGFβ, transforming growth factor-β; TH, T helper; TME, tumour microenvironment; Treg cell, regulatory T cell.
Fig. 2 |. IFNγ responders in the tumour microenvironment.
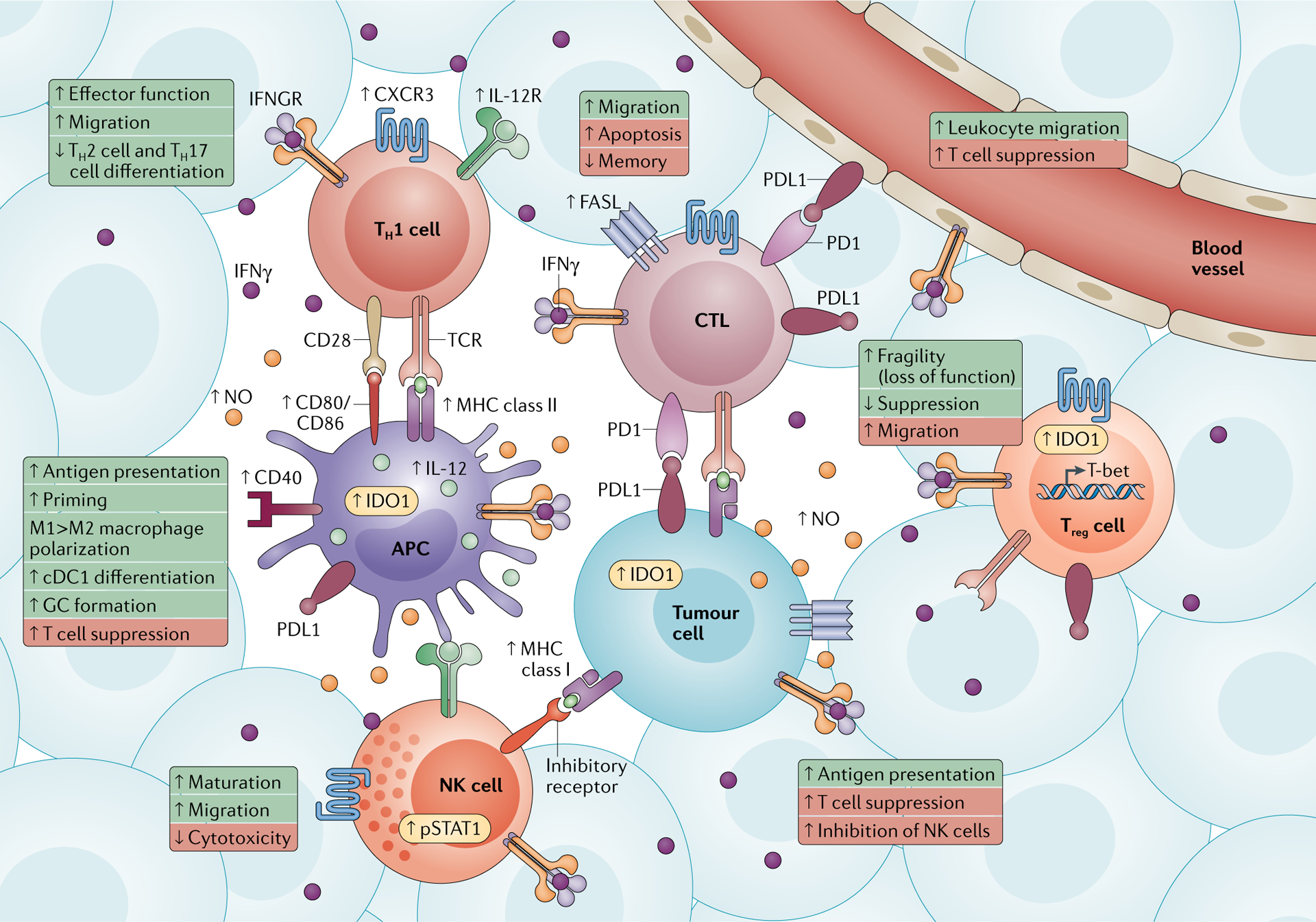
Interferon-γ (IFNγ)-induced signalling occurs in many types of immune cells (such as T helper 1 (TH1) cells, cytotoxic T lymphocytes (CTLs), antigen-presenting cells (APCs), regulatory T (Treg) cells and natural killer (NK) cells) and non-immune cells (vasculature and tumour cells) within the tumour microenvironment. Key proteins upregulated by IFNγ and the interacting ligands and receptors are shown. The biological consequences of IFNγ-induced signalling in each cell type are summarized in boxes. IFNγ responses that make these cell types teammates are in green, whereas responses that make them opponents are in red. CXCR3, CXC-chemokine receptor 3; cDC1, conventional type 1 dendritic cell; FASL, FAS ligand; GC, germinal centre; IDO1, indoleamine 2,3-dioxygenase 1; IL-12R, interleukin-12 receptor; IFNGR, interferon-γ receptor; NO, nitric oxide; PD1, programmed cell death 1; PDL1, programmed cell death 1 ligand 1; pSTAT1, phosphorylated signal transducer and activator of transcription 1; TCR, T cell receptor.
Cytotoxic T lymphocytes.
As well as being important producers of IFNγ, CTLs express IFNGR and respond to IFNγ in the TME (FIG. 2). In the context of an infection, IFNγ regulates the contraction phase of CTL responses via FAS–FASL-mediated and BIM-mediated apoptosis, both of which are induced by STAT1 signalling81. High levels of IFNγ during the CTL expansion phase limit the size of the memory population by inhibiting IL-7Rα expression, thus reducing signalling by the prosurvival cytokine IL-7 (REFS82,83). The mechanism by which IFNγ regulates IL-7Rα expression remains elusive, but it may involve an AKT–FOXO1 pathway84,85. Treatment of patients with IFNγ-inducing immunotherapies induces effector and memory CD8+ T cell expansion, but whether these expanding cells have low IFNGR expression protecting them from apoptosis and promoting IL-7Rα expression is unclear86,87. However, in vitro, activated IFNγ+ CTLs do not express IFNGR2 nor do they upregulate IFNγ-inducible genes in response to IFNγ88. Conversely, CTLs in mouse models of low tumour burden express more IFNGR than naive T cells do, and IFNγ induction by treatment with antibodies against cytotoxic T lymphocyte antigen 4 (CTLA4) or PD1 resulted in activation-induced cell death, which limited effector memory formation and resulted in tumour growth89. Due to this dichotomy, assessment of tumour burden, CTL infiltration, IFNGR expression and IFNγ levels before checkpoint blockade may help to identify treatment-responsive patients.
IFNγ promotes the recruitment of immune cells to the TME through transcriptional regulation of CXCL9, CXCL10 and CXCL11, and their cognate receptor CXCR3 on T cells, NK cells, monocytes, DCs and cancer cells90. Increased chemotaxis of activated CTLs to the TME enhances cytotoxic effects and limits tumour growth. A tumour-selective oncolytic vaccinia virus engineered to express CXCL11 induced CXCR3+ CTL recruitment into the TME of a mouse mesothelioma model and elicited profound antitumour effects91. IFNγ also promotes CTL motility via chemokine-independent mechanisms92.
IFNγ has other protumorigenic effects on CTLs besides apoptosis induction. Overexpression of Ifngr2 on CTLs did not affect their development or proliferation but limited their cytotoxic activity in response to antigenic stimulation by an unknown mechanism88. IFNγ also upregulates the expression of PDL1 and/or PDL2 on many cell types. Owing to the expression of both PD1 and PDL1 by T cells, self-inhibition may occur in trans in the TME93 (FIG. 2).
CD4+ effector T cells.
Similarly to IFNγ+ CTLs, IFNγ-producing TH1 cells decrease IFNGR2 expression following differentiation, enhancing their survival and thus antitumour effects in the TME94. TH1 cells actively repress TH17 cell polarization via T-bet and inhibition of RUNX1, which provides reinforced TH1 cell commitment and expression of a gene transcriptional programme that is most effective for tumour clearance95,96 (FIG. 2). Additionally, IFNγ prevents TH2 cell polarization via suppressor of cytokine signalling 1 (SOCS1) and T-bet, which inhibit IL-4 receptor (IL-4R) signalling and GATA3 expression and function, respectively97,98. Interestingly, upon TCR stimulation in CD4+ TH cell precursors, IFNGR1 becomes localized at the immunological synapse along with STAT1, a process that is inhibited by IL-4R expression in TH2 cells. This co-recruitment of IFNGR1 and STAT1 to the immunological synapse creates a ‘TH1 cell readiness’ upon TCR stimulation and ‘primes’ the cells to quickly polarize and mediate TH1 cell signalling. Ultimately, IFNGR2 expression is downregulated in TH1 cells; therefore, the continuation of dominant TH2 cell/TH17 cell antagonism may be reinforced by T-bet, rather than STAT1.
IFNγ also has protumorigenic effects on TH1 cells. PDL1 expression on tumour-infiltrating effector T cells prevents TH1 cell differentiation, providing an additional negative-feedback loop in the TME to limit IFNγ production99. However, it is unclear which PD1+ cells interact with PDL1+CD4+ effector T cells in the TME and/or whether there is self-inhibition among TH1 cells. IFNγ also promotes apoptosis via reduction of BCL-2 expression, upregulation of Fas and Fasl, and production of a detrimental oxidative environment100–102.
Treg cells.
The effects of IFNγ on Treg cells have been studied in various disease states and are a topic of ongoing debate. As mentioned earlier, Treg cells have been shown to adopt a TH-like (T-bet+IFNγ+) effector phenotype to better suppress the appropriate effector responses in models of autoimmune disease and bacterial infection63 (FIG. 2). In the TME however, IFNγ drives a ‘fragile’ Treg cell phenotype, in which Treg cells lose suppressive activity yet maintain FOXP3 expression, to undermine their protumour activity65,103. Strikingly, mice with Ifngr1-knockout Treg cells are resistant to anti-PD1 therapy in tumour models. A potential mechanism is that IFNγ-resistant Treg cells change from a TH1-like state to a TH2-like state, the latter of which exhibits the highest viability and activation potential of Treg cells (protumorigenic) and is enriched in melanoma and colorectal cancer104. These findings suggest that in the TME, IFNγ-induced Treg cell dysfunction allows full reinvigoration of CTL-mediated antitumour effects unleashed by anti-PD1 therapy. IFNγ also induces PDL1 expression on Treg cells, and high numbers of PDL1+ Treg cells in non-small-cell lung cancer correlate with better responses to PD1 and/or PDL1 blockade105. These data suggest that PDL1+ Treg cells create a barrier to antitumour immunity that can be disrupted only with PD1 and/or PDL1 blockade. Supporting these seemingly paradoxical findings, it has been shown that an IFNγ–STAT1-induced TH1-like Treg cell programme promotes suppression of TH1 cells (protumour) but maintenance of Treg cell stability through the delayed induction of IL-12 receptor, thereby protecting Treg cells from STAT4-dependent dysfunction106.
IFNγ also promotes antitumorigenic effects by inducing IDO1 expression, which catalyses the breakdown of tryptophan into kynurenines, which induce T cell apoptosis via caspase 8 activation and mitochondrial cytochrome c release107 (FIG. 2). Interestingly, expression of IDO1, PDL1 and CTLA4 on Treg cells is interconnected in peripheral blood of patients with melanoma, and strongly correlates with advanced disease and negative outcome108. Several small-molecule IDO1 inhibitors are being investigated in the clinic as ‘immunometabolic adjuvants’ to widen the therapeutic window and limit autoimmune side effects of current cancer therapies109. However, a recent phase I/II study with pembrolizumab (anti-PD1) plus the IDO1 inhibitor epacadostat did not show clinical benefit in patients with solid tumours110.
NK cells.
Data suggest that antitumorigenic functions of NK cells are activated by IFNγ. Phosphorylation of STAT1 on Tyr701 in NK cells occurs following transactivation by IFNγ+ iNKT cells in response to IL-12 from DCs111 (FIG. 2). Studies using Stat1Y701F-knock-in mice revealed that Tyr701-phosphorylated STAT1 is required for NK cell maturation, suggesting that STAT1 activation promotes antitumour immunity112. Indeed, NK cell tumour infiltration is largely dependent on IFNγ-induced CXCR3 expression, as Ifngr1-knockout mice and Cxcr3-knockout mice have fewer tumourinfiltrating NK cells113 (FIG. 2). Additionally, IFNγ produced by bystander T cells acts on NK cells to promote maturation and tumour killing via TNF-related apoptosis-inducing ligand (TRAIL), expression of which is enhanced by IFNγ-induced interferon regulatory factor 1 (IRF1)114.
Conversely, phosphorylation of Ser727 on STAT1 in resting NK cells by cyclin-dependent kinase 8 results in decreased production of granzyme B and perforin, thus decreasing NK cell cytotoxicity (protumorigenic). Stat1S727A-knock-in mice are more resistant to leukaemia and melanoma than controls, and are completely resistant to breast cancer metastasis115.
Antigen-presenting cells.
A key antitumorigenic function of IFNγ is the induced expression of MHC class I and class II molecules by APCs, such as DCs, macrophages and B cells, for presentation of tumour antigens to T cells116 (FIG. 2). IFNγ induces STAT1 and IRF1 binding to promoter IV of MHC class II transactivator (CIITA), which is the non-DNA-binding master regulator of MHC class II transcription117. IFNγ induces MHC class I expression via IRF1 binding to the promoter of NLRC5, which is a transcriptional regulator of MHC class I118. IFNγ also induces expression of the co-stimulatory molecules CD80 and CD86 by APCs, which promote T cell activation via CD28 engagement (FIG. 2). The induction of CD80 and CD86 expression offsets the immunosuppression induced by competitive binding of CTLA4 to CD28 (REFS119,120).
IFNγ also drives antitumorigenic effects via DC differentiation into conventional type 1 DCs (cDC1s) through the expression of CD80, CD86, MHC class I, CD40, CD54 and CC-chemokine receptor 7 (CCR7), and the production of IL-1β and IL-12, which promote TH1 cell differentiation and activation of CD8+ T cells121. Indeed, for full therapeutic efficacy, cDC1s are required to respond to IFNγ produced by CD8+ T cells during anti-PD1 therapy122.
The impact of IFNγ on B cells has only recently been described in models of autoimmunity and implies that IFNγ–STAT1 signalling is required for spontaneous development of germinal centres and T follicular helper cells, suggesting a potential antitumour effect. Specifically, IFNγ, in combination with B cell receptor and CD40 activating signals, induces expression of the germinal centre master transcription factor B cell lymphoma 6 (BCL-6)123. IFNγ also works in concert with IL-12 to promote antibody class switching from IgM to IgG2a, therefore generating higher-affinity and specialized antibodies with antibody-dependent cytotoxicity, thus potentially promoting tumour antigen processing and presentation124. Although a direct role for IFNγ in B cells and germinal centre formation in cancer is not known, recent studies described a patient survival advantage with the presence of B cells and tertiary lymphoid structures in the TME125,126.
IFNγ was initially named ‘macrophage activation factor’ due to its role in driving classical activation of macrophages, commonly referred to as ‘IFNγ priming’. IFNγ signalling prepares macrophages for activation by Toll-like receptor (TLR)-induced inflammatory responses. IFNγ priming drives macrophages towards a proinflammatory and antitumorigenic, M1-like phenotype via the downregulation of miR-3473b, thereby suppressing an M2-like phenotype127. In addition, IFNγ blocks sterol regulatory element-binding protein 1-dependent fatty acid synthesis in M2-like tumour-associated macrophages (TAMs), an effect that is required for responses to anti-PD1 therapy in a mouse model of melanoma39. M1-like macrophages have increased phagocytic and tumoricidal activity compared with M2-like macrophages that is important for tumour surveillance128. TAMs produce CXCL9 and CXCL10 in response to IFNγ, which not only promotes immune cell infiltration of the TME but may also inhibit angiogenesis129.
In terms of protumorigenic activities, APCs are the predominant PDL1-expressing immune cell population in the TME. In addition to cancer cells and cancer-associated stroma, DCs and TAMs express IDO1 in response to IFNγ, which promotes immunosuppression through metabolic disruption and angiogenesis130 (FIG. 2). Kynurenines produced by IDO1 induce TGFβ production in DCs, which promotes Treg cell differentiation and immunosuppression131.
IFNγ also promotes iNOS expression in myeloid cells, which catabolizes the essential amino acid l-arginine to the free radical nitric oxide (NO). The role of NO in the TME is paradoxical and complex due to its effect on stromal cells, immune cells and tumour cells. The overall impact of NO largely depends on its expression level, the duration of exposure and the genetic makeup of the tumour132. NO promotes antitumour effects by inducing apoptosis, chromosome condensation and DNA fragmentation of immune cells, but it has protumour effects by inducing genomic instability of tumour cells via p53 and promoting angiogenesis. The TH2-type cytokine-inducible enzyme arginase antagonizes NO production via competition with iNOS for l-arginine. Arginase has immunosuppressive effects through inhibition of immune cell proliferation, cytokine production, TCR activation and promotion of apoptosis133. The overall balance of TH1-type and TH2-type cytokines dictates the impact of l-arginine catabolism on tumour growth.
Tumour cells.
Cancer cells are key responders to IFNγ in the TME and, like for immune cells, IFNγ drives both immunoactivating (teammate) and immunosuppressing (opponent) effects. The immunoactivating activity of IFNγ on tumour cells is largely attributed to induced tumour cell expression of MHC class I and secretion of CXCL9, CXCL10 and CXCL11 by tumour cells, monocytes, endothelial cells and fibroblasts to promote lymphocyte migration and inhibit angiogenesis (antitumorigenic)134. Conversely, CXCL11 has pleiotropic activities due to binding to CXCR7, which promotes angiogenesis and tumour growth. CXCL9 and CXCL10 promote TH1 and TH17 effector cell function, whereas CXCL11 promotes a TH2 cell response and regulatory function via IL-10 (REFS135,136). Pharmacological approaches are in development to create biased synthetic ligands that favour CXCL9 or CXCL10 T cell signalling via CXCR3, rather than CXCL11-induced signalling, to promote antitumour immunity.
Similarly to APCs, tumours present antigens on their surface to T cells via MHC class I (antitumour); however, MHC class I molecules can also serve as a marker of ‘self ‘ which engages inhibitory receptors on NK cells to prevent killing (protumour)137 (FIG. 2). MHC class I expression on tumour cells is variable, with immunosuppressive tumours often downregulating MHC class I expression, thus escaping immunosurveillance.
IFNγ regulates many survival and apoptotic pathways in tumour cells. For example, IFNγ induces apoptosis through IFNGR on tumour cells (antitumour). Knockdown of Ifngr1 in B16 melanoma cells results in impaired tumour rejection with anti-CTLA4 therapy, suggesting that IFNγ produced by the revigorated effector response must act directly on the tumour cells to elicit antitumour effects138. Conversely, protumour effects of IFNγ on tumours are mediated through induction of PDL1, IDO1, iNOS, FAS and FASL expression (FIG. 2). Prevention of IFNγ signalling decreased PDL1 expression by tumour cells and increased IFNγ-responsive gene expression by immune cells, including exhausted T cells139. Tumour cells are the main source of IDO1 in the TME and a major source of NO; however, expression can differ among tumour types140. Tumour-derived iNOS promotes angiogenesis, which allows increased vascularization and tumour growth141. Tumour cells express FAS and FASL, with the former mediating antitumorigenic effects (tumour cell apoptosis by cytolytic effector cells) and the latter mediating protumorigenic effects (apoptosis of immune effector cells).
Vasculature and lymphatics.
The vasculature and lymphatics within the TME are underappreciated IFNγ responders, with both protumorigenic and antitumorigenic effects. Angiogenesis within the TME has been targeted by therapeutics for years but has produced mixed clinical results142. The lymphatics have recently been shown to serve not only as passive conduits for immune cell exchange in the TME but also as important regulators of inflammation and immunity.
IFNγ directly promotes protumorigenic effects on the lymphatics; T cell-derived IFNγ inhibits lymphangiogenesis via downregulation of lymphatic vessel endothelial hyaluronan receptor 1, podoplanin and prospero homeobox protein 1 on lymphatic endothelial cells, the last of which is a key transcription factor required for the growth, proliferation and invasion of lymphatics143. IFNγ does not affect the initiation of lymphangiogenesis but instead inhibits the continuation of lymphatic vessel formation, resulting in reduced lymphatic density144. Similarly to its effects on other cells, IFNγ induces PDL1 expression on lymphatics, which limits CD8+ T cell accumulation in the TME and prevents tumour control145. IFNγ also promotes neovascularization in the TME indirectly through the induction of CXCL9, CXCL10 and IDO1 expression146.
IFNγ also has indirect antitumorigenic effects to limit angiogenesis via the polarization of TAMs to an M1-like phenotype, which limits the amount of vascular endothelial growth factor that will be secreted by M2-like TAMs147. Additionally, IFNγ production induced by IL-12 or pulse IL-2 therapy led to endothelial cell apoptosis via FAS–FASL, promoting tumour regression.
IFNγ and cancer immunotherapy
Almost all cancer immunotherapies, such as recombinant cytokines, vaccines, checkpoint inhibitors, chimeric antigen receptor T cell therapy and TLR agonists, modulate IFNγ148–151 (BOX 2). These therapies aim to induce inflammation to aid tumour clearance; however, IFNγ-driven adaptive immune resistance can precipitate therapeutic resistance or disease exacerbation. Many preclinical studies of IFNγ-modulating immunotherapies over the past decade have aimed to exploit the antitumour effects and block the protumour effects of IFNγ in the TME.
Box 2 |. Promising IFNγ-modulating cancer immunotherapies in clinical trials.
Checkpoint blockade
Interferon-γ (IFNγ)-induced expression of programmed cell death 1 ligand 1 (PDL1) creates an anergic immunological state. Various antibodies blocking programmed cell death 1 (PD1; nivolumab, pembrolizumab, toripalimab and tislelizumab), PDL1 (atezolizumab, durvalumab and avelumab) and cytotoxic T lymphocyte antigen 4 (CTLA4; ipilimumab) are approved by the US Food and Drug Administration, with many more in clinical trials. interestingly, IFNG expression in the tumour microenvironment is a strong predictor of therapeutic response138,164.
CAR T cell therapy
Autologous CD8+ T cells from patients with cancer are engineered to express a chimeric antigen receptor (CAR) specific for a tumour antigen; they are then expanded and transferred back to the patient191. First-generation CD19-targeted CAR T cells led to complete cures in patients with B cell acute lymphoblastic leukaemia; however, antigen loss and T cell exhaustion precipitated resistance192. second-, third- and fourth-generation CAR T cell therapies have been developed targeting various different antigens (including CD22, CD30, CD147, carcinoembryonic antigen, mesothelin, NKG2D, prostate-specific membrane antigen, tumour-associated mucin 1, B cell maturation antigen and disialoganglioside) and have been modified to increase antitumour efficacy and overcome immunosuppression in the tumour microenvironment.
IFNγ1b
Intratumoural injection of non-glycosylated recombinant IFNγ (IFNγ1b) was performed on the basis that it would increase intratumoural concentrations of interferon-inducible chemokines and increase immune cell infiltration of melanoma. However, the results were disappointing owing to upregulation of the immunosuppressive IFNγ-induced molecules indoleamine 2,3-dioxygenase 1 (IDO1) and PDL1 (REF.193). IFNγ1b in conjunction with the anti-PD1 antibody pembrolizumab is currently in a phase ii clinical trial.
Recombinant IL-12
Human recombinant interleukin-12 (IL-12; edodekin alfa and NM-IL12) is currently being tested in phase I/II clinical trials with or without chemotherapy. it has been shown to result in stimulation of natural killer cells, T cells and natural killer T cells and in production of IFNγ and chemokines194,195.
Cancer vaccines
Cancer vaccines present tumour-associated neoantigens, and sometimes patient-specific tumour antigens, to T cells and B cells, resulting in activation, maturation, proliferation and antibody production196. Current vaccines in development use antigen-loaded antigen-presenting cells, adenoviruses and DNA or RNA approaches197.
TLR agonists
Toll-like receptors (TLRs), expressed by innate immune cells, are activated by microbial products during infection and induce cytokine production and co-stimulatory molecules for T cell activation198. the TLR7 and TLR8 agonist resiquimod (R848) promotes a strong T helper 1-type antitumour response via IFNγ and IL-12 production by natural killer cells and dendritic cells, respectively199–201. However, resiquimod has poor pharmacokinetic profiles, and when administered systemically it promotes widespread immune activation, leading to autoimmune responses. Novel delivery methods for resiquimod using nanoparticles or thermosensitive liposomes are being tested to limit these toxic effects, and have produced promising results in combination with anti-PD1 in preclinical models202.
Adaptive immune resistance.
The upregulation of immunosuppressive mechanisms in response to chronic proinflammatory stimuli.
Therapeutic approaches to deliver IFNγ to the TME.
The first clinical use of recombinant IFNγ was more than 30 years ago for the treatment of cancer and viral infections. Treatment with modified recombinant human IFNγ1b however generated disappointing results in the clinic, production was costly and the protein had a short half-life152,153. IFNγ fusion proteins have since been engineered with longer half-lives and tissue-specific homing to enhance therapeutic effects and limit adverse effects154. However, the toxicity of these IFNγ fusion proteins remains a challenge owing to widespread expression of IFNGR and receptor trapping of IFNγ that prevents effective tumour targeting155.
The induction of IFNγ expression in the TME through alternative, gene-based approaches, such as viral transduction, also has technical limitations, such as transgene size, selective integration and expression efficiency156. Clinical use of a replication-defective adenovirus encoding human IFNγ showed beneficial responses in most patients with cutaneous T cell lymphoma in a phase II clinical trial31. Oncolytic viruses encoding IFNγ allow concentrated cytokine release in the TME, which activates DCs and enhances T cell-mediated antitumour effects, prolonging survival of tumour-bearing mice157. Delivery of an oncolytic adenovirus encoding the IFNγ-inducing cytokine IL-12, in conjunction with the collagen-associated extracellular matrix proteoglycan decorin to limit Treg cell expansion, produced a potent antitumour response in a mouse model of breast cancer158.
Non-viral genetic approaches include the delivery of IFNG gene therapy via plasmids, vectors and liposomes. The best route of delivery has been shown to be a promoter and plasmid backbone that results in constant and steady IFNγ production and lacks an initial burst, which is responsible for adverse effects159. Recently, an Ifng-loaded lipoplex and an antigen-loaded liposome had synergistic effects of targeting DCs to present antigens and produce IFNγ in mice. This lipoplex–liposome combination resulted in tumour clearance and enhanced mouse survival that was dependent on CTL activation160. Despite early optimism, approaches targeting IFNγ to the TME have largely failed to provide any clinical benefit. IFNγ-induced adaptive immune resistance highlights the importance of improving the delivery of and increasing the specificity and duration of IFNγ-induced immunotherapies and simultaneously limiting or blocking the expression or activity of IDO1, PDL1, NO, FAS and FASL.
Antitumour-biased IFNγ agonists.
A recent novel receptor engineering approach resulted in increased affinity of IFNGR2 for the IFNγ–IFNGR1 complex, and crystallization of this hexameric complex (in a 2:2:2 ratio) has revealed numerous targets for biochemical intervention to decouple the protumour and antitumour effects of IFNγ signalling161. Recently developed antitumour-biased IFNγ agonists for IFNGR retain IFNγ-induced upregulation of MHC class I expression but have impaired upregulation of PDL1 expression161. These biased agonists dimerize with one molecule of endogenous IFNγ to prevent full assembly of the hexameric ligand–receptor complex. These findings illustrate that the second IFNGR2 molecule of the hexameric complex may be redundant, and loss of this single IFNGR2 molecule maintains MHC class I induction, yet limits PDL1 induction. The therapeutic potential of these antitumour-biased agonists is promising, but lessons from IFNγ1b suggest that the addition of fusion proteins to induce favourable pharmacokinetics may be required.
Immune checkpoint blockade.
As mentioned earlier, IFNγ has a prominent role in immune checkpoint blockade with anti-PD1 or anti-PDL1. IFNγ was found to be localized to regions of high PDL1 expression on the surface of melanomas, implying that CTLs may trigger autoinhibition through IFNγ-driven PDL1 expression162. This mechanism of adaptive immune resistance may explain tumour escape from immunosurveillance. As well as driving upregulation of PDL1 expression, IFNγ production by CTLs is required to mediate their therapeutic effects39,139,163. The requirement for CD8+ T cells and CD4+ T cells in mediating the response to anti-PD1 therapy in mouse models is now well established. Anti-PD1-induced IFNγ production by CTLs acts on cDC1s to produce IL-12, all of which are required to elicit a therapeutic response122. Although the cellular target (or targets) of IL-12 remain unclear, it is possible that IL-12-mediated downregulation of IFNGR2 may protect CTLs from IFNγ-induced apoptosis.
Resistance to cancer immunotherapy
Despite the potential for immunotherapy to transform the cell context and cytokine milieu of the TME, only a subset of patients have a complete response. Various clinical studies have been conducted at the genetic, epigenetic and metabolic levels to better understand adaptive immune resistance mechanisms of IFNγ-modulating cancer immunotherapies.
Genetic mutations.
Genomic and transcriptomic studies of responders and non-responders to checkpoint blockade therapy identified IFNγ-stimulated genes as key mediators of the therapeutic response164. Specifically, non-responders to anti-CTLA4 therapy had defects in IFNγ signalling within the tumour, showing downregulation of ten genes (IFNG, STAT1, CCR5, CXCL9, CXCL10, CXCL11, IDO1, PRF1, GZMA and HLA-DRA) that constitute an IFNγ signature138,. Additionally, JAK1 or JAK2 loss-of-function tumour mutations resulted in the lack of response to IFNγ and anti-PD1 therapy165. Interestingly, JAK1 or JAK2 mutations promoted both adaptive and primary resistance to anti-PD1 therapy, with primary resistance evident in JAK1-mutated or JAK2-mutated, PDL1-negative tumours166.
Epigenetic modulations.
Immune evasion through epigenetic silencing of CXCL9 and CXCL10 in tumour cells was associated with poor patient outcome, and pharmacological removal of these epigenetic marks increased effector T cell infiltration and efficacy of anti-PDL1 therapy167. CXCL9 and CXCL10 are also silenced by enhancer of zeste homologue 2 (EZH2), which itself is antagonized by binding of AT-rich interaction domain-containing protein 1A (ARID1A). Interestingly, ARID1A mutations are highly prevalent in various cancers, especially ovarian cancer (50%), resulting in a decreased TH1 cell signature and poor clinical benefit following checkpoint blockade168.
Metabolic disruptions.
IFNγ can disrupt tumour cell metabolism and promote antitumour effects. IFNγ-induced downregulation of cystine–glutamate antiporter (Xc−) on tumour cells impairs cystine uptake and promotes lipid peroxidation and ferroptosis. Accordingly, a favourable outcome with anti-PD1 therapy was seen in patients with reduced expression of Xc−; however, the mechanism of IFNγ-mediated suppression of Xc− expression is unclear169. As mentioned, IFNγ-mediated induction of IDO1 expression with checkpoint blockade therapy has been postulated as a mechanism of adaptive immune resistance. Although monotherapy with IDO1 modulators was disappointing, they are safe and well tolerated. Preclinical studies have shown synergy with IDO1 inhibition and CTLA4 or PD1 blockade, and clinical studies have produced a wide range of response rates170. The complex role of IDO1 in various tumours needs further investigation to ensure optimal therapeutic targeting.
Finally, by profiling IFNγ-regulated genes within the TME of patients before and after therapy, it may be possible to predict the proinflammatory and anti-inflammatory influence of IFNγ with specific treatments to better predict therapeutic responses164.
Concluding remarks
Although IFNγ was discovered more than 50 years ago, the complex nature of this pleotropic cytokine in the TME is continuously being unravelled. The principles learned in infectious disease, graft-versus-host disease and autoimmune disease have provided insight into the role of IFNγ in the TME. However, the immune context of tumours can differ greatly from that of other disease states, and IFNγ induced with novel immunotherapies creates conflicting IFNγ-induced antitumour or protumour signalling events.
The categorization of IFNγ-induced signalling as antitumour or protumour is proposed to depend largely on the duration (acute versus chronic) and magnitude of IFNγ signalling. Interestingly, IFNγ is captured by phosphatidylserine residues on the surface of cells and slowly released to mediate autocrine and paracrine signalling (‘catch and release’) and contributes to preserving or delaying IFNγ signalling171. In the TME, the duration and magnitude of IFNγ signalling are also largely dictated by tumour burden and the state of immune cell infiltrate, respectively.
Initial IFNγ exposure recruits teammates (via CXCL9, CXCL10, CXCL11 and CXCR3) to promote antigen presentation (MHC class I and class II), T cell priming and activation (CD80, CD86 and CD40) and tumour cell killing (FAS and FASL). However, prolonged IFNγ exposure converts teammates into opponents, promoting protumorigenic effects via immunosuppression (PDL1, IDO1, FAS and FASL), angiogenesis (CXCL9, CXCL10, CXCL11, IDO1 and iNOS) and tumour cell proliferation172. In addition, opponents are likely to be present in the TME initially; however, teammates may dominate to promote overall antitumorigenic effects. Thus, studies that clearly measure and map IFNγ production and response over time in the TME are warranted.
There are several other key questions that warrant further investigation, including whether there are as-yet-unidentified IFNγ-induced genes, which IFNγ-producing cells are most important for antitumour effects, which IFNγ-expressing cells in the TME mediate resistance to immunotherapy and how IFNγ could be introduced to turn ‘cold’ tumours ‘hot’ (BOX 3). Future studies must use novel approaches to tease apart the proinflammatory effects from the anti-inflammatory effects of IFNγ to design better therapeutics to bias its antitumour capabilities and prevent immune escape.
Box 3 |. Outstanding questions on role of IFNγ in tumours.
Are there novel genes induced by interferon-γ (IFNγ) yet to be identified and characterized? With the recent advances in single-cell transcriptomics, additional proinflammatory and anti-inflammatory genes induced by IFNγ may be identified. Novel IFNγ-induced proinflammatory molecules may be induced therapeutically, whereas novel IFNγ-induced anti-inflammatory molecules could serve as druggable targets to combat resistance to IFNγ-inducing cancer immunotherapies.
Are all immune cells that can produce IFNγ required to do so to elicit an antitumour response? It is possible that a specific subset of IFNγ+ cells in the tumour microenvironment (TME) switch from acting as teammates to acting as opponents during disease or therapy. Alternatively, do different cell populations drive protumour effects versus antitumour effects? IFNγ production by cytotoxic T lymphocytes, T helper 1 cells and natural killer cells in the TME has been extensively studied, and they all appear to produce profound antitumour effects. However, the role of IFNγ+ regulatory T cells and B cells in the TME remains unclear, and they may also need to produce IFNγ to promote protumour or antitumour effects. For instance, tumour-infiltrating IFNγ+ regulatory T cells may have other effector functions besides suppressive ability, a phenomenon that could be required to unleash the antitumour effects of T helper 1 cells and cytotoxic T lymphocytes. Furthermore, IFNγ+CD11ahiCD16/CD32hi B cells in the TME may be an understudied population mediating the therapeutic effects of CD40 ligands in the clinic. Preclinical studies that selectively delete IFNγ expression from individual cell populations would aid in addressing these key questions.
Do the location and identity of IFNγ+ and IFNGR2+ cells in the TME explain why some patients respond to therapy whereas others are resistant? Furthermore, is it possible to determine which IFNγ+ cells are located close to cells expressing indoleamine 2,3-dioxygenase 1 (IDO1) and programmed cell death 1 ligand 1 (PDL1), thus inducing protumour effects? PDL1 is highly expressed by antigen-presenting cells and tumour cells, which are most commonly surveyed by CD4+ effector T cells, cytotoxic T lymphocytes and natural killer cells. Through identification of these protumour IFNGR2+ cell types, bispecific antibodies composed of an IFNγ receptor 2 (IFNGR2) blocking antibody conjugated to a cell-specific homing antibody could prove advantageous.
How can IFNγ be introduced into the TME of ‘cold’ immune-excluded solid tumours? Intratumoural injection of recombinant IFNγ is possible for easily accessible tumours such as melanoma, but not all tumours. This is an important challenge for many cell-based immunotherapies, and therefore direct IFNγ delivery through chimeric antigen receptor T cells, adenovirus vectors and vaccines may be viable and may negate the pharmacological and toxicity issues associated with systemic administration of IFNγ agents. Alternatively, indirect IFNγ-inducing therapeutic strategies such as pattern recognition receptor agonists could be used and have been shown to overcome resistance to checkpoint blockade203,204.
Acknowledgements
The authors thank everyone in the Vignali laboratory for all their constructive comments and advice. This work was supported by the US National Institutes of Health (F32 CA247004-01 and T32 CA082084 to A.M.G.; P01 AI108545, R01 CA203689 and P30 CA047904 to D.A.A.V.).
Competing interests
D.A.A.V. is a co-founder and shareholder of Novasenta and Tizona, a shareholder of Oncorus and Werewolf, has patents licensed and receives royalties from Astellas and Bristol Myers Squibb, is a scientific advisory board member for Tizona, Werewolf, F-Star and Bicara, is a consultant for Astellas, Bristol Myers Squibb, Almirall and Incyte, and receives research funding from Bristol Myers Squibb, Astellas and Novasenta. The other authors declare no competing interests.
References
- 1.Wheelock Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science 149, 310–311 (1965). [PubMed] [Google Scholar]
- 2.Dighe, Richards, Old & Schreiber Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity 1, 447–456 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Nastala et al. Recombinant IL-12 administration induces tumor regression in association with IFN-gamma production. J. Immunol 153, 1697–1706 (1994). [PubMed] [Google Scholar]
- 4.Young, Dray & Farrar Expression of transfected human interferon-gamma DNA: evidence for cell-specific regulation. J. Immunol 136, 4700–4703 (1986). [PubMed] [Google Scholar]
- 5.Soutto, Zhou & Aune Cutting edge: distal regulatory elements are required to achieve selective expression of IFN-gamma in Th1/Tc1 effector cells. J. Immunol 169, 6664–6667 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Fields, Kim & Flavell Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J. Immunol 169, 647–650 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Gomez et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152, 743–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shnyreva et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc. Natl Acad. Sci. USA 101, 12622–12627 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiani et al. Regulation of interferon-gamma gene expression by nuclear factor of activated T cells. Blood 98, 1480–1488 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Beals, Sheridan, Turck, Gardner & Crabtree Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275, 1930–1934 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Dong, Davis & Flavell MAP kinases in the immune response. Annu. Rev. Immunol 20, 55–72 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Park et al. A mechanism underlying STAT4-mediated up-regulation of IFN-gamma induction in TCR-triggered T cells. Int. Immunol 16, 295–302 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Johnson & Torres Leukotrienes: positive signals for regulation of gamma-interferon production. J. Immunol 132, 413–416 (1984). [PubMed] [Google Scholar]
- 14.Kasahara, Hooks, Dougherty & Oppenheim Interleukin 2-mediated immune interferon (IFN-gamma) production by human T cells and T cell subsets. J. Immunol 130, 1784–1789 (1983). [PubMed] [Google Scholar]
- 15.Johnson, Russell & Torres Second messenger role of arachidonic acid and its metabolites in interferon-gamma production. J. Immunol 137, 3053–3056 (1986). [PubMed] [Google Scholar]
- 16.Vignali & Kuchroo IL-12 family cytokines: immunological playmakers. Nat. Immunol 13, 722–728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rex et al. A comprehensive pathway map of IL-18-mediated signalling. J. Cell Commun. Signal 14, 257–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamura et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature 378, 88–91 (1995). [DOI] [PubMed] [Google Scholar]; The authors cloned IL-18 (also known as IGIF) and showed in vitro induction of IFNγ with recombinant IL-18 via an IL-12-independent pathway.
- 19.Nakahira et al. Synergy of IL-12 and IL-18 for IFN-gamma gene expression: IL-12-induced STAT4 contributes to IFN-gamma promoter activation by up-regulating the binding activity of IL-18-induced activator protein 1. J. Immunol 168, 1146–1153 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Mavropoulos, Sully, Cope & Clark Stabilization of IFN-gamma mRNA by MAPK p38 in IL-12- and IL-18-stimulated human NK cells. Blood 105, 282–288 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Hodge, Martinez, Julias, Taylor & Young Regulation of nuclear gamma interferon gene expression by interleukin 12 (IL-12) and IL-2 represents a novel form of posttranscriptional control. Mol. Cell. Biol 22, 1742–1753 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiner et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang et al. Regulation of human natural killer cell IFN-γ production by microRNA-146a via targeting the NF-κB signaling pathway. Front. Immunol 9, 293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma et al. MicroRNA-142–3p inhibits IFN-γ production via targeting of RICTOR in Aspergillus fumigatus activated CD4+ T cells. Ann. Transl. Med 7, 649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, Zhang, Wu & Jiang Diverse roles of miR-29 in cancer (review). Oncol. Rep 31, 1509–1516 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Shahriar et al. The dual role of mir-146a in metastasis and disease progression. Biomed. Pharmacother 126, 110099 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Ealick et al. Three-dimensional structure of recombinant human interferon-gamma. Science 252, 698–702 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Miyakawa et al. Prolonged circulation half-life of interferon γ activity by gene delivery of interferon γ-serum albumin fusion protein in mice. J. Pharm. Sci 100, 2350–2357 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Charych et al. Modeling the receptor pharmacology, pharmacokinetics, and pharmacodynamics of NKTR-214, a kinetically-controlled interleukin-2 (IL2) receptor agonist for cancer immunotherapy. PLoS ONE 12, e0179431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molinas, Wietzerbin & Falcoff Human platelets possess receptors for a lymphokine: demonstration of high specific receptors for HuIFN-gamma. J. Immunol 138, 802–806 (1987). [PubMed] [Google Scholar]
- 31.Dummer et al. Phase II clinical trial of intratumoral application of TG1042 (adenovirus-interferon-gamma) in patients with advanced cutaneous T-cell lymphomas and multilesional cutaneous B-cell lymphomas. Mol. Ther 18, 1244–1247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altan-Bonnet & Mukherjee Cytokine-mediated communication: a quantitative appraisal of immune complexity. Nat. Rev. Immunol 19, 205–217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearce et al. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science 302, 1041–1043 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Szabo et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 295, 338–342 (2002). [DOI] [PubMed] [Google Scholar]; This study shows the requirement of T-bet expression for IFNγ production in CD4+ TH cells and NK cells but not in CD8+ T cells, indicating that distinct transcription factors control IFNγ in immune cell subsets.
- 35.Sanderson et al. Cytotoxic immunological synapses do not restrict the action of interferon-γ to antigenic target cells. Proc. Natl Acad. Sci. USA 109, 7835–7840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gérard et al. Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8+ T cells. Nat. Immunol 14, 356–363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thibaut et al. Bystander IFN-γ activity promotes widespread and sustained cytokine signaling altering the tumor microenvironment. Nat. Cancer 1, 302–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that IFNγ diffuses extensively throughout the TME to produce a widespread field of IFNγ. Sustained IFNγ signalling via STAT1 and IRF8 was required to modulate tumour cell expression of MHC class I and PDL1.
- 38.Hoekstra et al. Long-distance modulation of bystander tumor cells by CD8+ T cell-secreted IFNγ. Nat. Cancer 1, 291–301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study visualizes the long-range diffusion of IFNγ produced by low-frequency activated CD8+ T cells in the TME.
- 39.Liu et al. Treg cells promote the SREBP1-dependent metabolic fitness of tumor-promoting macrophages via repression of CD8+ T cell-derived interferon-γ. Immunity 51, 381–397.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalathil, Hutson, Barbi, Iyer & Thanavala Augmentation of IFN-γ+CD8+ T cell responses correlates with survival of HCC patients on sorafenib therapy. JCI Insight 4, e130116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huse, Lillemeier, Kuhns, Chen & Davis T cells use two directionally distinct pathways for cytokine secretion. Nat. Immunol 7, 247–255 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Dai, Hellstrom, Yip, Sjögren & Hellstrom Tumor regression and cure depends on sustained th1 responses. J. Immunother 41, 369–378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kryczek et al. Human TH17 cells are long-lived effector memory cells. Sci. Transl Med 3, 104ra100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kryczek et al. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood 114, 1141–1149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubota, Lian, Lohwasser, Salcedo & Takei IFN-gamma production and cytotoxicity of IL-2-activated murine NK cells are differentially regulated by MHC class I molecules. J. Immunol 163, 6488–6493 (1999). [PubMed] [Google Scholar]
- 46.Almishri et al. TNFα augments cytokine-induced NK cell IFNγ production through TNFR2. J. Innate Immun 8, 617–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reefman et al. Cytokine secretion is distinct from secretion of cytotoxic granules in NK cells. J. Immunol 184, 4852–4862 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Villegas et al. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer 35, 23–28 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Lee et al. A high-throughput assay of NK cell activity in whole blood and its clinical application. Biochem. Biophys. Res. Commun 445, 584–590 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Henriksen et al. Favorable prognostic impact of natural killer cells and T cells in high-grade serous ovarian carcinoma. Acta Oncol. 59, 652–659 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Lee et al. Natural killer cell activity for IFN-gamma production as a supportive diagnostic marker for gastric cancer. Oncotarget 8, 70431–70440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno et al. IFN-gamma-producing human invariant NKT cells promote tumor-associated antigen-specific cytotoxic T cell responses. J. Immunol 181, 2446–2454 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Das et al. The adaptor molecule SAP plays essential roles during invariant NKT cell cytotoxicity and lytic synapse formation. Blood 121, 3386–3395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf, Choi & Exley Novel approaches to exploiting invariant NKT cells in cancer immunotherapy. Front. Immunol 9, 384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brigl, Bry, Kent, Gumperz & Brenner Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol 4, 1230–1237 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Zhang et al. α-GalCer and iNKT cell-based cancer immunotherapy: realizing the therapeutic potentials. Front. Immunol 10, 1126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takami, Ihara & Motohashi Clinical application of iNKT cell-mediated anti-tumor activity against lung cancer and head and neck cancer. Front. Immunol 9, 2021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parekh et al. PD-1/PD-L blockade prevents anergy induction and enhances the anti-tumor activities of glycolipid-activated invariant NKT cells. J. Immunol 182, 2816–2826 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawant, Hamilton & Vignali Interleukin-35: expanding its job profile. J. Interferon Cytokine Res 35, 499–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vignali, Collison & Workman How regulatory T cells work. Nat. Rev. Immunol 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 491, 554–559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lucca et al. TIGIT signaling restores suppressor function of Th1 Tregs. JCI Insight 4, e124427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koch et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol 10, 595–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levine et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Overacre-Delgoffe et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell 169, 1130–1141.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a novel fragile state of intratumoural Treg cells characterized by the gain of IFNγ production and loss of suppressive function. A Treg cell response to IFNγ was required for benefit from anti-PD1 therapy.
- 66.Seltmann, Werfel & Wittmann Evidence for a regulatory loop between IFN-γ and IL-33 in skin inflammation. Exp. Dermatol 22, 102–107 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Liang et al. IL-33 promotes innate IFN-γ production and modulates dendritic cell response in LCMV-induced hepatitis in mice. Eur. J. Immunol 45, 3052–3063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hatzioannou et al. An intrinsic role of IL-33 in Treg cell-mediated tumor immunoevasion. Nat. Immunol 21, 75–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ameri et al. IL-33/regulatory T cell axis triggers the development of a tumor-promoting immune environment in chronic inflammation. Proc. Natl Acad. Sci. USA 116, 2646–2651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao et al. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J. Exp. Med 198, 433–442 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoeres, Holzmann, Smetak, Birkmann & Wilhelm PD-1 signaling modulates interferon-γ production by gamma delta (γδ) T-cells in response to leukemia. Oncoimmunology 8, 1550618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gentles et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Girardi et al. Regulation of cutaneous malignancy by gammadelta T cells. Science 294, 605–609 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Fillatreau B cells and their cytokine activities implications in human diseases. Clin. Immunol 186, 26–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ballesteros-Tato, Stone & Lund Innate IFNγ-producing B cells. Cell Res. 24, 135–136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris, Goodrich, Gerth, Peng & Lund Regulation of IFN-gamma production by B effector 1 cells: essential roles for T-bet and the IFN-gamma receptor. J. Immunol 174, 6781–6790 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Vitale et al. Development of CDX-1140, an agonist CD40 antibody for cancer immunotherapy. Cancer Immunol. Immunother 68, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piechutta & Berghoff New emerging targets in cancer immunotherapy: the role of cluster of differentiation 40 (CD40/TNFR5). ESMO Open 4, e000510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhat et al. Comprehensive network map of interferon gamma signaling. J. Cell Commun. Signal 12, 745–751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study develops a comprehensive IFNγ signalling network curated from the literature and organized into a publicly available browser.
- 80.Samarajiwa, Forster, Auchettl & Hertzog INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37, D852–D857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Refaeli, Van Parijs, Alexander & Abbas Interferon gamma is required for activation-induced death of T lymphocytes. J. Exp. Med 196, 999–1005 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prabhu et al. Gamma interferon regulates contraction of the influenza virus-specific CD8 T cell response and limits the size of the memory population. J. Virol 87, 12510–12522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Badovinac, Porter & Harty CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol 5, 809–817 (2004). [DOI] [PubMed] [Google Scholar]
- 84.Ouyang, Beckett, Flavell & Li An essential role of the forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiang, Banks & Accili Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J. Biol. Chem 285, 27396–27401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stoycheva et al. IFN-γ regulates CD8+memory T cell differentiation and survival in response to weak, but not strong, TCR signals. J. Immunol 194, 553–559 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Pedicord, Montalvo, Leiner & Allison Single dose of anti-CTLA-4 enhances CD8 + T-cell memory formation, function, and maintenance. Proc. Natl Acad. Sci. USA 108, 266–271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tau, Cowan, Weisburg, Braunstein & Rothman Regulation of IFN-gamma signaling is essential for the cytotoxic activity of CD8(+) T cells. J. Immunol 167, 5574–5582 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pai et al. Clonal deletion of tumor-specific T cells by interferon-γ confers therapeutic resistance to combination immune checkpoint blockade. Immunity 50, 477–492.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that tumour-specific CD8+ T cells express higher levels of IFNGR than T cells, thus inducing apoptosis, an immune-intrinsic mechanism of resistance to checkpoint blockade.
- 90.Colvin, Campanella, Sun & Luster Intracellular domains of CXCR3 that mediate CXCL9, CXCL10, and CXCL11 function. J. Biol. Chem 279, 30219–30227 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Liu et al. CXCL11-armed oncolytic poxvirus elicits potent antitumor immunity and shows enhanced therapeutic efficacy. Oncoimmunology 5, e1091554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhat, Leggatt, Waterhouse & Frazer Interferon-γ derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 8, e2836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bonaventura et al. Cold tumors: a therapeutic challenge for immunotherapy. Front. Immunol 10, 168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pernis et al. Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science 269, 245–247 (1995). [DOI] [PubMed] [Google Scholar]
- 95.Holzer, Reinhardt, Lang, Handgretinger & Fischer Influence of a mutation in IFN-γ receptor 2 (IFNGR2) in human cells on the generation of Th17 cells in memory T cells. Hum. Immunol 74, 693–700 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Lazarevic et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol 12, 96–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Naka et al. SOCS-1/SSI-1-deficient NKT cells participate in severe hepatitis through dysregulated cross-talk inhibition of IFN-gamma and IL-4 signaling in vivo. Immunity 14, 535–545 (2001). [DOI] [PubMed] [Google Scholar]
- 98.Hwang, Szabo, Schwartzberg & Glimcher T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307, 430–433 (2005). [DOI] [PubMed] [Google Scholar]
- 99.Diskin et al. PD-L1 engagement on T cells promotes self-tolerance and suppression of neighboring macrophages and effector T cells in cancer. Nat. Immunol 21, 442–454 (2020). [DOI] [PubMed] [Google Scholar]; This study shows that PDL1 ‘back-signalling’ in CD4+ T cells decreased activation and TH1 cell polarization and induced anergy in CD8+ T cells. PDL1+ T cells restrained PD1+ effector T cells and macrophages and induced a protumorigenic microenvironment.
- 100.Zhou, Weyman, Liu, Almasan & Zhou IFN-gamma induces apoptosis in HL-60 cells through decreased Bcl-2 and increased Bak expression. J. Interferon Cytokine Res 28, 65–72 (2008). [DOI] [PubMed] [Google Scholar]
- 101.Xu, Fu, Plate & Chong IFN-gamma induces cell growth inhibition by Fas-mediated apoptosis: requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 58, 2832–2837 (1998). [PubMed] [Google Scholar]
- 102.Rakshit et al. Interferon-gamma induced cell death: regulation and contributions of nitric oxide, cJun N-terminal kinase, reactive oxygen species and peroxynitrite. Biochim. Biophys. Acta 1843, 2645–2661 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Overacre & Vignali Treg stability: to be or not to be. Curr. Opin. Immunol 39, 39–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Halim et al. An atlas of human regulatory T helper-like cells reveals features of Th2-like Tregs that support a tumorigenic environment. Cell Rep. 20, 757–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu et al. Stromal PD-L1-positive regulatory T cells and PD-1-positive CD8-positive T cells define the response of different subsets of non-small cell lung cancer to PD-1/PD-L1 blockade immunotherapy. J. Thorac. Oncol 13, 521–532 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Koch et al. T-bet+ Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity 37, 501–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fallarino et al. T cell apoptosis by tryptophan catabolism. Cell Death Differ. 9, 1069–1077 (2002). [DOI] [PubMed] [Google Scholar]
- 108.Chevolet et al. Characterization of the in vivo immune network of IDO, tryptophan metabolism, PD-L1, and CTLA-4 in circulating immune cells in melanoma. Oncoimmunology 4, e982382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prendergast, Malachowski, DuHadaway & Muller Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. 77, 6795–6811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mitchell et al. Epacadostat plus pembrolizumab in patients with advanced solid tumors: phase I results from a multicenter, open-label phase I/II trial (ECHO-202/KEYNOTE-037). J. Clin. Oncol 36, 3223–3230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carnaud et al. Cutting edge: cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J. Immunol 163, 4647–4650 (1999). [PubMed] [Google Scholar]
- 112.Putz et al. Novel non-canonical role of STAT1 in natural killer cell cytotoxicity. Oncoimmunology 5, e1186314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wendel, Galani, Suri-Payer & Cerwenka Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 68, 8437–8445 (2008). [DOI] [PubMed] [Google Scholar]
- 114.Park et al. IFN-gamma enhances TRAIL-induced apoptosis through IRF-1. Eur. J. Biochem 271, 4222–4228 (2004). [DOI] [PubMed] [Google Scholar]
- 115.Putz, Gotthardt & Sexl STAT1-S727 - the license to kill. Oncoimmunology 3, e955441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Früh & Yang Antigen presentation by MHC class I and its regulation by interferon gamma. Curr. Opin. Immunol 11, 76–81 (1999). [DOI] [PubMed] [Google Scholar]
- 117.Steimle, Siegrist, Mottet, Lisowska-Grospierre & Mach Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science 265, 106–109 (1994). [DOI] [PubMed] [Google Scholar]
- 118.Meissner et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl Acad. Sci. USA 107, 13794–13799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li, Yang, Inoue, Mori & Akiyoshi The expression of costimulatory molecules CD80 and CD86 in human carcinoma cell lines: its regulation by interferon gamma and interleukin-10. Cancer Immunol. Immunother 43, 213–219 (1996). [DOI] [PubMed] [Google Scholar]
- 120.Collins et al. The interaction properties of costimulatory molecules revisited. Immunity 17, 201–210 (2002). [DOI] [PubMed] [Google Scholar]
- 121.Pan et al. Interferon-gamma is an autocrine mediator for dendritic cell maturation. Immunol. Lett 94, 141–151 (2004). [DOI] [PubMed] [Google Scholar]
- 122.Garris et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 49, 1148–1161.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jackson et al. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J. Exp. Med 213, 733–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Metzger et al. Interleukin-12 acts as an adjuvant for humoral immunity through interferon-gamma-dependent and -independent mechanisms. Eur. J. Immunol 27, 1958–1965 (1997). [DOI] [PubMed] [Google Scholar]
- 125.Cillo et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 52, 183–199.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bruno New predictors for immunotherapy responses sharpen our view of the tumour microenvironment. Nature 577, 474–476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu et al. IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b. J. Immunol 193, 3036–3044 (2014). [DOI] [PubMed] [Google Scholar]
- 128.Sadlik et al. Lymphocyte supernatant-induced human monocyte tumoricidal activity: dependence on the presence of gamma-interferon. Cancer Res. 45, 1940–1945 (1985). [PubMed] [Google Scholar]
- 129.Haabeth et al. Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat. Commun 2, 240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu et al. Targeting the IDO1 pathway in cancer: from bench to bedside. J. Hematol. Oncol 11, 100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yan et al. IDO upregulates regulatory T cells via tryptophan catabolite and suppresses encephalitogenic T cell responses in experimental autoimmune encephalomyelitis. J. Immunol 185, 5953–5961 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vannini, Kashfi & Nath The dual role of iNOS in cancer. Redox Biol. 6, 334–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grzywa et al. Myeloid cell-derived arginase in cancer immune response. Front. Immunol 11, 938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Strieter et al. The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J. Biol. Chem 270, 27348–27357 (1995). [DOI] [PubMed] [Google Scholar]
- 135.Tokunaga et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - a target for novel cancer therapy. Cancer Treat. Rev 63, 40–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Karin, Wildbaum & Thelen Biased signaling pathways via CXCR3 control the development and function of CD4+ T cell subsets. J. Leukoc. Biol 99, 857–862 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Kärre, Ljunggren, Piontek & Kiessling Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319, 675–678 (1986). [DOI] [PubMed] [Google Scholar]
- 138.Gao et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that copy-number loss of IFNγ pathway activating genes and amplification of IFNγ pathway inhibitors are associated with clinical resistance to anti-CTLA4 therapy.
- 139.Benci et al. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell 178, 933–948.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows adaptive immune resistance within the TME by which protumorigenic IFNγ signalling in tumour cells opposes antitumorigenic IFNγ signalling in immune cells to limit tumour killing by adaptive and innate immune cells.
- 140.Watcharanurak et al. Effects of upregulated indoleamine 2, 3-dioxygenase 1 by interferon γ gene transfer on interferon γ-mediated antitumor activity. Gene Ther. 21, 794–801 (2014). [DOI] [PubMed] [Google Scholar]
- 141.Kostourou et al. The role of tumour-derived iNOS in tumour progression and angiogenesis. Br. J. Cancer 104, 83–90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Teleanu, Chircov, Grumezescu & Teleanu Tumor angiogenesis and anti-angiogenic strategies for cancer treatment. J. Clin. Med 9, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kataru et al. T lymphocytes negatively regulate lymph node lymphatic vessel formation. Immunity 34, 96–107 (2011). [DOI] [PubMed] [Google Scholar]
- 144.Zampell et al. Lymphatic function is regulated by a coordinated expression of lymphangiogenic and anti-lymphangiogenic cytokines. Am. J. Physiol. Cell Physiol 302, C392–C404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lane et al. IFNγ-activated dermal lymphatic vessels inhibit cytotoxic T cells in melanoma and inflamed skin. J. Exp. Med 215, 3057–3074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mondal et al. IDO1 is an integral mediator of inflammatory neovascularization. EBioMedicine 14, 74–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun et al. Inhibition of tumor angiogenesis by interferon-γ by suppression of tumor-associated macrophage differentiation. Oncol. Res 21, 227–235 (2014). [DOI] [PubMed] [Google Scholar]
- 148.Xu Th1 cytokine-based immunotherapy for cancer. Hepatobiliary Pancreat. Dis. Int 13, 482–494 (2014). [DOI] [PubMed] [Google Scholar]
- 149.Benmebarek et al. Killing mechanisms of chimeric antigen receptor (CAR) T cells. Int. J. Mol. Sci 20, 1283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Müller et al. Toll-like receptor ligands and interferon-γ synergize for induction of antitumor M1 macrophages. Front. Immunol 8, 1383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ni & Lu Interferon gamma in cancer immunotherapy. Cancer Med. 7, 4509–4516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Razaghi, Owens & Heimann Review of the recombinant human interferon gamma as an immunotherapeutic: Impacts of production platforms and glycosylation. J. Biotechnol 240, 48–60 (2016). [DOI] [PubMed] [Google Scholar]
- 153.Gleave et al. Interferon gamma-1b compared with placebo in metastatic renal-cell carcinoma. N. Engl. J. Med 338, 1265–1271 (1998). [DOI] [PubMed] [Google Scholar]
- 154.Ando et al. Prevention of adverse events of interferon γ gene therapy by gene delivery of interferon γ-heparin-binding domain fusion protein in mice. Mol. Ther. Methods Clin. Dev 1, 14023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hemmerle & Neri The dose-dependent tumor targeting of antibody-IFNγ fusion proteins reveals an unexpected receptor-trapping mechanism in vivo. Cancer Immunol. Res 2, 559–567 (2014). [DOI] [PubMed] [Google Scholar]
- 156.Lee et al. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 4, 43–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bourgeois-Daigneault et al. Oncolytic vesicular stomatitis virus expressing interferon-γ has enhanced therapeutic activity. Mol. Ther. Oncolytics 3, 16001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oh, Choi, Hong & Yun Oncolytic adenovirus coexpressing interleukin-12 and decorin overcomes Treg-mediated immunosuppression inducing potent antitumor effects in a weakly immunogenic tumor model. Oncotarget 8, 4730–4746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ando, Takahashi, Nishikawa, Watanabe & Takakura Constant and steady transgene expression of interferon-γ by optimization of plasmid construct for safe and effective interferon-γ gene therapy. J. Gene Med 14, 288–295 (2012). [DOI] [PubMed] [Google Scholar]
- 160.Yuba et al. pH-sensitive polymer-liposome-based antigen delivery systems potentiated with interferon-γ gene lipoplex for efficient cancer immunotherapy. Biomaterials 67, 214–224 (2015). [DOI] [PubMed] [Google Scholar]
- 161.Mendoza et al. Structure of the IFNγ receptor complex guides design of biased agonists. Nature 567, 56–60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors solve the crystal structure of the hexameric IFNγ–IFNGR1–IFNGR2 signalling complex, which is then used as a blueprint to develop antitumorigenic biased ligands that induce MHC class I expression but not PDL1 expression.
- 162.Taube et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl Med 4, 127ra37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gubin et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 175, 1014–1030.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ayers et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study analyses RNA expression profiles of patients treated with anti-PD1 (pembrolizumab) and finds that IFNγ-responsive genes involved in antigen presentation, chemokine expression, cytotoxic activity and adaptive immune resistance were necessary for a clinical benefit from therapy.
- 165.Zaretsky et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shin et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 7, 188–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Peng et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li et al. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J. Clin. Invest 130, 2712–2726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhai et al. IDO1 in cancer: a Gemini of immune checkpoints. Cell Mol. Immunol 15, 447–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Oyler-Yaniv et al. Catch and release of cytokines mediated by tumor phosphatidylserine converts transient exposure into long-lived inflammation. Mol. Cell 66, 635–647.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study discovers that IFNγ is captured by phosphatidylserine on the surface of cells to mediate slow IFNγ release driving long-term transcriptional effects.
- 172.Benci et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Farrar & Schreiber The molecular cell biology of interferon-gamma and its receptor. Annu. Rev. Immunol 11, 571–611 (1993). [DOI] [PubMed] [Google Scholar]
- 174.Valente et al. Distribution of interferon-gamma receptor in human tissues. Eur. J. Immunol 22, 2403–2412 (1992). [DOI] [PubMed] [Google Scholar]
- 175.Schmiedel et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Monaco et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 26, 1627–1640.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ebensperger et al. Genomic organization and promoter analysis of the gene Ifngr2 encoding the second chain of the mouse interferon-gamma receptor. Scand. J. Immunol 44, 599–606 (1996). [DOI] [PubMed] [Google Scholar]
- 178.Green, Young & Valencia Current prospects of type II interferon γ signaling and autoimmunity. J. Biol. Chem 292, 13925–13933 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Larkin, Johnson & Subramaniam Differential nuclear localization of the IFNGR-1 and IFNGR-2 subunits of the IFN-gamma receptor complex following activation by IFN-gamma. J. Interferon Cytokine Res 20, 565–576 (2000). [DOI] [PubMed] [Google Scholar]
- 180.Gough, Levy, Johnstone & Clarke IFNgamma signaling — does it mean JAK-STAT? Cytokine Growth Factor Rev. 19, 383–394 (2008). [DOI] [PubMed] [Google Scholar]
- 181.Melen et al. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem 278, 28193–28200 (2003). [DOI] [PubMed] [Google Scholar]
- 182.Decker, Kovarik & Meinke GAS elements: a few nucleotides with a major impact on cytokineinduced gene expression. J. Interferon Cytokine Res 17, 121–134 (1997). [DOI] [PubMed] [Google Scholar]
- 183.Ramana et al. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc. Natl Acad. Sci. USA 98, 6674–6679 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study determines that the IFNγ-induced immediate-early genes Myc, Jun and Jund, the transcription factor C/EBPβ and the chemokines CCL3 and CCL7 are induced via a non-canonical STAT1-independent mechanism.
- 184.Stephens, Lumpkin & Fishman Activation of signal transducers and activators of transcription 1 and 3 by leukemia inhibitory factor, oncostatin-M, and interferon-gamma in adipocytes. J. Biol. Chem 273, 31408–31416 (1998). [DOI] [PubMed] [Google Scholar]
- 185.Greenlund et al. STAT recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity 2, 677–687 (1995). [DOI] [PubMed] [Google Scholar]
- 186.Qi et al. Elucidating the crosstalk mechanism between IFN-gamma and IL-6 via mathematical modelling. BMC Bioinformatics 14, 41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Qing, Costa-Pereira, Watling & Stark Role of tyrosine 441 of interferon-gamma receptor subunit 1 in SOCS-1-mediated attenuation of STAT1 activation. J. Biol. Chem 280, 1849–1853 (2005). [DOI] [PubMed] [Google Scholar]
- 188.Gresser, Tovey, Maury & Chouroulinkov Lethality of interferon preparations for newborn mice. Nature 258, 76–78 (1975). [DOI] [PubMed] [Google Scholar]
- 189.Madonna et al. The IFN-gamma-dependent suppressor of cytokine signaling 1 promoter activity is positively regulated by IFN regulatory factor-1 and Sp1 but repressed by growth factor independence-1b and Krüppel-like factor-4, and it is dysregulated in psoriatic keratinocytes. J. Immunol 185, 2467–2481 (2010). [DOI] [PubMed] [Google Scholar]
- 190.Liau et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun 9, 1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Eshhar, Waks, Gross & Schindler Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl Acad. Sci. USA 90, 720–724 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Maude, Teachey, Porter & Grupp CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood 125, 4017–4023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mauldin et al. Intratumoral interferon-gamma increases chemokine production but fails to increase T cell infiltration of human melanoma metastases. Cancer Immunol. Immunother 65, 1189–1199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Strauss et al. First-in-human phase i trial of a tumor-targeted cytokine (NHS-IL12) in subjects with metastatic solid tumors. Clin. Cancer Res 25, 99–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lyerly, Osada & Hartman Right time and place for IL12: targeted delivery stimulates immune therapy. Clin. Cancer Res 25, 9–11 (2019). [DOI] [PubMed] [Google Scholar]
- 196.Chen et al. Neoantigen identification strategies enable personalized immunotherapy in refractory solid tumors. J. Clin. Invest 129, 2056–2070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hollingsworth & Jansen Turning the corner on therapeutic cancer vaccines. NPJ Vaccines 4, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kawai & Akira TLR signaling. Cell Death Differ. 13, 816–825 (2006). [DOI] [PubMed] [Google Scholar]
- 199.Hart, Athie-Morales, O’Connor & Gardiner TLR7/8-mediated activation of human NK cells results in accessory cell-dependent IFN-gamma production. J. Immunol 175, 1636–1642 (2005). [DOI] [PubMed] [Google Scholar]
- 200.Zobywalski et al. Generation of clinical grade dendritic cells with capacity to produce biologically active IL-12p70. J. Transl. Med 5, 18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wagner et al. Modulation of TH1 and TH2 cytokine production with the immune response modifiers, R-848 and imiquimod. Cell Immunol. 191, 10–19 (1999). [DOI] [PubMed] [Google Scholar]
- 202.Rodell et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng 2, 578–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chuang et al. Adjuvant effect of Toll-like receptor 9 activation on cancer immunotherapy using checkpoint blockade. Front. Immunol 11, 1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang et al. Development of thermosensitive resiquimod-loaded liposomes for enhanced cancer immunotherapy. J. Control. Release 330, 1080–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


