Abstract
TAP, the human homologue of the yeast protein Mex67p, has been proposed to serve a role in mRNA export in mammalian cells. We have examined the ability of TAP to mediate export of Rev response element (RRE)-containing human immunodeficiency virus (HIV) RNA, a well-characterized export substrate in mammalian cells. To do this, the TAP gene was fused in frame to either RevM10 or RevΔ78–79. These proteins are nonfunctional Rev mutant proteins that can bind to HIV RNA containing the RRE in vivo but are unable to mediate the export of this RNA to the cytoplasm. However, the fusion of TAP to either of these mutant proteins gave rise to chimeric proteins that were able to complement Rev function. Significantly, cotransfection with a vector expressing NXT1 (p15), an NTF2-related cellular factor that binds to TAP, led to dramatic enhancement of the ability of the chimeric proteins to mediate RNA export. Mutant-protein analysis demonstrated that the domain necessary for nuclear export mapped to the C-terminal region of TAP and required the domain that interacts with NXT1, as well as the region that has been shown to interact with nucleoporins. RevM10-TAP function was leptomycin B insensitive. In contrast, the function of this protein was inhibited by ΔCAN, a protein consisting of part of the FG repeat domain of CAN/Nup214. These results show that TAP can complement Rev nuclear export signal function and redirect the export of intron-containing RNA to a CRM1-independent pathway. These experiments support the role of TAP as an RNA export factor in mammalian cells. In addition, they indicate that NXT1 serves as a crucial cellular cofactor in this process.
During recent years, it has become increasingly clear that regulation of molecular trafficking between the nucleus and cytoplasm of the eukaryotic cell plays an important role in the regulation of cellular gene expression (for reviews, see references 27, 33, and 39). Although the detailed mechanisms for nuclear export and import remain to be elucidated, numerous studies have shed light on these processes. It has been demonstrated that both protein and RNA substrates are recognized by specific import and export receptors. Several of these receptors have been identified, including receptors involved in export of RNA to the cytoplasm.
Different classes of RNA are transported through separate pathways, and export of each of these classes is saturable, indicating the involvement of specific limiting factors (15, 29, 49, 51). The details of the mRNA export pathway have not yet been elucidated. However, in higher eukaryotes, most primary mRNA transcripts contain introns, and as a general rule, these introns are all removed before export from the nucleus (21, 34). Nuclear retention until splicing is completed is believed to ensure that incompletely processed mRNAs do not reach the cytoplasm, where they could give rise to aberrant proteins.
Retroviruses have, for several years, served as important model systems for the analysis of mRNA export (for reviews, see references 9 and 22). For all retroviruses, the primary transcription product expressed from the integrated viral genome is an intron-containing RNA that is subject to splicing within the cellular machinery to generate a singly spliced RNA encoding the envelope proteins. However, the primary transcript also serves as the viral genome that is packaged into particles in the cytoplasm of infected cells. This RNA is also the mRNA for translation of the viral gag-pol gene products. Thus, both intron-containing RNA and completely spliced RNA are exported from the nucleus during viral replication. In the case of complex retroviruses such as human immunodeficiency virus (HIV), the situation is even more complicated, since both singly spliced and multiply spliced RNAs are generated. Thus, several spliced RNAs that contain residual introns reach the cytoplasm as well.
In HIV and several other complex retroviruses, the export of intron-containing RNAs is mediated through the action of specific elements in these RNAs. These elements work in conjunction with virus-encoded proteins that bind directly to these sequences (for a review, see reference 50). In HIV, the cis-acting Rev response element (RRE) is located within the env region of the genome. The RRE forms a stable secondary structure containing several stem-loops and the viral Rev protein binds with high affinity to one of these stem-loops (11, 38). The Rev protein is a small phosphoprotein (116 amino acids [aa] in HIV type 1 [HIV-1]) that contains a nuclear localization signal, as well as a nuclear export signal (NES) (36). The Rev NES, the first such signal to be identified (15, 28, 40), binds to the nuclear export receptor CRM1 (16, 43, 58). The Rev-CRM1 complex interacts with RanGTP, as well as with several nucleoporins, and these interactions are believed to be crucial for Rev-mediated export (1, 6, 17, 65). The function of CRM1 is specifically inhibited by the drug leptomycin B (LMB). This drug has been shown to be a potent inhibitor of HIV replication, as well as of expression of Rev-dependent HIV proteins from subgenomic HIV reporter constructs (16, 47, 63). The Rev-RRE pathway is believed to utilize cellular factors that are important also for the export of U snRNAs and 5S rRNA (15).
The genomes of the simpler retroviruses encode only structural proteins and lack regulatory genes such as rev. In some of these viruses (e.g., Mason-Pfizer monkey virus [MPMV], simian retrovirus type 1 [SRV-1], and Rous sarcoma virus), the export of full-length, intron-containing mRNA is achieved through the action of cis-acting elements in the RNA (8, 13, 14, 44, 66) that interact directly with cellular proteins. The best characterized of these elements is the constitutive transport element (CTE) of type D retroviruses (MPMV and SRV-1) (8). Although the CTE has been demonstrated to be functionally interchangeable with the Rev-RRE system (8, 14, 66), it is clear that these systems utilize at least partially nonoverlapping pathways for the export of their intron-containing mRNAs. In contrast to the Rev-RRE system, CTE-mediated export uses a CRM1-independent export pathway, as CTE function has been reported to be insensitive to LMB (47). In addition, the pathway which the CTE accesses is expected to share factors with the cellular mRNA export pathway, since excess CTE is able to inhibit mRNA export in competition experiments with Xenopus oocytes (49, 51).
Recent experiments have implicated the cellular protein TAP as a potential export receptor for cellular mRNA. TAP is a 619-aa protein that is a mammalian orthologue of Mex67p, a yeast mRNA export factor. TAP has been shown to bind to the CTE in vitro and to enhance the export of various CTE-containing RNA substrates in Xenopus oocyte injection experiments (2, 7, 20). In addition, the expression of human TAP has been reported to be essential for efficient CTE-mediated gene expression in a quail cell line (31). Mex67p has been shown to associate with RNA in vivo and temperature-sensitive Mex67 mutants display a phenotype of rapid nuclear accumulation of poly(A) RNA at the nonpermissive temperature (53).
Immunoprecipitation (IP) experiments were used to identify several potential TAP cofactors (32). One of these, p15, has been shown to be a novel protein with significant homology to NTF2, a RanGDP-binding protein involved in nuclear import (41, 48). A functional relationship between TAP and p15 was indicated by the fact the two proteins could function together to partially complement a mex67-mtr2 defect in yeast (32). p15 was also independently identified in a database search for NTF2-related proteins and given the name NXT1 (5). Biochemical characterization of NXT1 showed it to be a RanGTP-binding protein that stimulates nuclear export in permeabilized-cell assays (5, 46).
In this report, we show that TAP fused to export-incompetent Rev mutant proteins can substitute for Rev and achieve nucleocytoplasmic export of intron-containing RNA. This export function is dramatically enhanced by coexpression of NXT1. The region of TAP that is essential to the achievement of RNA export maps to the C-terminal domains of TAP that interact with NXT1 and nucleoporins, respectively.
MATERIALS AND METHODS
Plasmid constructs.
The subgenomic HIV-1 reporter constructs pCMVgagpol-RRE and pCMVgagpol-CTE have been previously described (57). The Rev protein-expressing plasmids pCMV-Rev (55), pCMV-RevM10 (36), and pCMVRev Δ78–79 (12) have also been previously described. Secreted alkaline phosphatase (SEAP) (4) was expressed from pCMV-SEAP (14).
A TAP cDNA clone encoding aa 61 to 619 of TAP was a gift from J. U. Jung (64). This form of TAP is called full-length TAP in this paper. TAP was PCR amplified using oligonucleotide primers which generated BamHI sites flanking the open reading frame (ORF). The PCR product was purified and ligated into the BamHI site of pCMV (35). Constructs encoding Rev-TAP fusion proteins were generated by use of the splicing by overlap extension (SOE)-PCR method (26). The upstream external oligonucleotide primer generated an NcoI site at the start of the Rev ORF, and the downstream primer after the end of the TAP ORF contained a BamHI site. The Rev-TAP hybrid fragment that was generated was cleaved with NcoI and cloned between the NcoI and T4 polymerase-repaired XhoI sites of a variant of pCMV (35) in which a promoter-proximal NcoI site was generated (pCMV-NX). The resulting constructs encode chimeric proteins where Rev residues 1 to 116 are fused to TAP residues 61 to 619. These constructs were made with three different Rev proteins (wild-type Rev, RevM10, and RevΔ78–79). A similar SOE-PCR strategy was utilized to create plasmids expressing RevM10 fusion proteins containing different fragments of the TAP gene and to create internal deletions in TAP. The NcoI-digested fragments encoding the various fusion protiens were ligated into pcDNA-FLAG (5) to generate the FLAG-RevM10-TAP plasmids. Details of these plasmids will be furnished upon request. The sequences of all regions amplified by PCR were confirmed by DNA sequencing. This analysis was performed in the University of Virginia automated sequencing facility on an Applied Biosystems 377 Prism DNA Sequencer using dye terminator chemistry with Taq polymerase.
The pCDNA-FLAG-NXT1 expression plasmid has been previously described (5). pCMV-EGFP TAP was constructed by inserting the BamHI fragment from pCMV-TAP into the BamHI site of pEGFP-C2 (Clontech).
Cell lines and transient transfections.
293T/17 and CMT3-COS cells were maintained in Iscove's minimal essential medium supplemented with 10% bovine calf serum. Transient transfections of CMT3-COS cells were performed using a modification of the DEAE-dextran method as previously described (25). 293T/17 cells were transfected using a calcium phosphate transfection protocol (19, 62). Transfections were performed using 0.25 μg of pCMV-SEAP, 5 μg of pCMVRev or pCMV-Rev-TAP, and 5 μg of the pCMVgagpol reporter constructs, unless otherwise indicated.
The plasmids pBC12 and pBC12-ΔCAN were gifts from Bryan Cullen (Duke University). pBC12-ΔCAN expresses aa 1864 to 2090 of CAN/Nup214 (6). In the titration experiments using pBC12-ΔCAN, the total amount of DNA was kept constant with the pBC12 plasmid.
SEAP and p24 antigen quantitation.
Supernatants were collected at 72 h posttransfection, centrifuged in a microcentrifuge to remove residual cells and debris, and stored at −20°C until assayed. p24 (HIV capsid protein) expression levels were determined using a commercial enzyme-linked immunosorbent assay kit (NEN). SEAP activity in the supernatants was measured using the Tropix Phospha-Light Chemiluminescent Reporter kit (Tropix cat. no. BP100).
RNA fractionation and Northern blot analysis.
The methods used for nuclear and cytoplasmic RNA extraction, poly(A) RNA selection, and Northern blot analysis were previously described (23, 24). Cells were harvested 65 h posttransfection. [32P]CTP-labeled DNA probes were generated using the T7 Quickprime Kit (Pharmacia). The gag-pol-specific probe was generated using the SacI-BglII (nucleotides 682 to 2093) fragment of the BH10 HIV-1 proviral clone. The SEAP-specific probe was generated using the BamHI fragment of the human SEAP-encoding gene (nucleotides 213 to 1698). Visualization and quantitation of Northern blots were performed with a Molecular Dynamics PhosphorImager and ImageQuant analysis software.
In vitro translation and binding reactions.
The interactions between NXT1 and the RevM10-TAP fusion protein and its deletion derivatives were analyzed by immunoprecipitation using 35S-labeled proteins. FLAG epitope-tagged RevM10-TAP was synthesized in the transcription-translation rabbit reticulocyte lysate system (TNT; Promega) using Tran35S-label (31.5 μCi/50-μl reaction mixture; Amersham Life Sciences). NXT1 was synthesized in a similar manner without the epitope tag. Lysates containing FLAG-RevM10-TAP (2 μl) and NXT1 (2 μl) were combined in a total volume of 20 μl in phosphate-buffered saline (PBS) supplemented with bovine serum albumin (0.2 mg/ml), protease inhibitors (aprotinin, leupeptin, and pepstatin at 10 μg/ml each, phenylmethylsulfonyl fluoride at 1 mM), and dithiothreitol (2 mM). After 1 h of incubation at 25°C, the binding reaction mixture was combined with 20 μl of protein G beads containing 2 μg of anti-FLAG antibody M2 (Sigma). The binding reaction mixture was mixed end over end overnight at 4°C. The beads were then collected by centrifugation and washed twice with PBS–0.1% NP-40, twice with PBS–0.5 M NaCl, and twice again with PBS–0.1% NP-40. The beads were resuspended in Laemmli gel sample buffer, boiled, and resolved by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE). 35S-labeled proteins were visualized by standard autoradiography methods.
Immunolocalization and fluorescence microscopy.
Immunolocalization was performed essentially as previously described (56). For RevTAP immunolocalization, transfected CMT3-COS cells were washed in PBS and fixed in 3% formaldehyde in PBS at 25°C for 15 min. Fixed cells were washed in PBS and permeabilized with 0.2% Triton X-100 for 15 min on ice. Cells were then washed and stained with a rabbit anti-Rev polyclonal antibody diluted 1/100 (45). After extensive washing, secondary staining was performed with goat anti-rabbit Alexafluor 488 (Molecular Probes). Cells expressing enhanced green fluorescent protein (GFP)-TAP were processed in the same way, except for the addition of antibodies. Samples were visualized on a Diaphot 300 fluorescence microscope (Nikon).
IP and Western blot analysis.
Transfected 293T cells (106) were lysed in radioimmunoprecipitation assay (RIPA) buffer (45) 65 h posttransfection and spun at 12,000 rpm in an Eppendorf tabletop centrifuge. A 1-μl sample of rabbit anti-Rev serum (45) was added to the cleared lysate. Samples were gently mixed for 1 h at 4°C. The immune complexes were collected on 2 mg of swollen protein A-Sepharose for 1 h at 4°C. Beads were then washed sequentially three times in cold RIPA buffer, once in cold RIPA buffer plus 0.5 M NaCl, and once in TNE (45). Proteins were resolved by SDS–15% PAGE (acrylamide/bisacrylamide ratio, 30/0.14). Western blot analysis was performed essentially as previously described (23). Briefly, proteins were transferred to an Immobilon-P membrane (Millipore) and the membrane was blocked in 5% milk and probed with an anti-Rev mouse monoclonal antibody (3H6) (45). After washing, the blot was incubated with a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Amersham) and the proteins were visualized using ECL (Amersham).
RESULTS
Development of an assay for TAP export function in mammalian cells.
To define regions that are important for TAP nuclear export function in mammalian cells, we utilized a previously well-characterized HIV-based reporter system (8, 14, 54). This system is based on the nuclear export of intron-containing RNA encompassing the gag-pol region of the HIV genome. In the absence of a specific transport element, this RNA is not exported from the nucleus. However, in the presence of either the MPMV CTE or Rev-RRE, the RNA reaches the cytoplasm, where it is translated into the HIV Gag and Pol proteins, giving rise to pseudoviral particles that bud into the supernatant medium. These particles can be quantitated using a commercial enzyme-linked immunosorbent assay that measures p24, the major HIV capsid protein.
We initially analyzed the effects of TAP on MPMV CTE function in transfected cells using this assay system. However, we were unable to see any effects of overexpression of TAP on the levels of p24 expressed from an HIV gag-pol reporter construct containing this element in any of the mammalian cell lines tested (CMT3/COS, HeLa, and 293T cells; data not shown). We thus decided to map regions of TAP important for export by fusing the TAP-encoding gene in frame to Rev-encoding genes containing mutated NESs. To this end, two different mutant forms of Rev (RevM10 and RevΔ78-79) were fused in frame to TAP (aa 61 to 619). In addition, the TAP gene was also fused in frame to the wild-type Rev gene. The Rev mutant proteins used for these fusions contain functional nuclear localization signal- and RRE-binding domains but contain mutations within the NES. Both of these mutant proteins have been shown to be nonfunctional and to display a transdominant negative phenotype. It has been previously shown that NES function can be complemented by fusion of other proteins containing leucine-rich NESs (e.g., human T-cell leukemia virus Rex and c-ABL) to NES-deficient Rev proteins (e.g., RevM10) (28, 61).
To analyze whether the Rev mutant-TAP fusion proteins were able to function in conjunction with the HIV RRE, the fusion constructs were transfected into 293T cells together with pCMVgagpol-RRE. The cells were also transfected with a plasmid that expresses SEAP, which is secreted into the supernatant medium (10). The SEAP protein is expressed from a spliced RNA that would be expected to reach the cytoplasm through the regular cellular mRNA export pathway. Thus, this construct provides the means to correct for differences in transfection efficiency and potential general effects of the expressed proteins on cellular RNA export pathways. As controls, cells were also transfected with the pCMVgagpol-RRE plasmid alone or with this plasmid in cotransfections with constructs expressing either the wild-type Rev or the RevM10 protein. Supernatant medium was harvested from the cells at 72 h posttransfection and analyzed for p24 and SEAP expression.
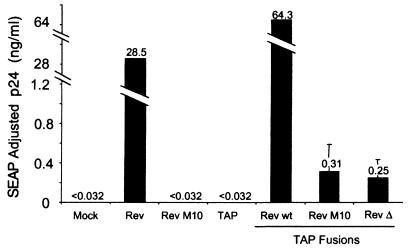
The results of this experiment are shown in Fig. 1. The graph displays the amount of p24 expressed in the various transfections adjusted for differences in SEAP activity. The SEAP values showed a less-than-twofold difference between the different supernatants (data not shown). As expected, no significant p24 activity was observed in the transfection with pCMVgagpol-RRE alone (Mock). In contrast, a high level of p24 expression was obtained in the supernatants of cells cotransfected with plasmids expressing either the wild-type Rev or the wild-type Rev–TAP fusion protein. This confirms previous results showing an absolute requirement for Rev to obtain p24 activity in this system (14, 54). Cotransfection of the pCMVgagpol-RRE plasmid with a plasmid expressing RevM10 or TAP did not give rise to any p24 activity above the background levels obtained with pCMVgagpol-RRE alone. However, a small amount of p24 activity was observed when the pCMVgagpol-RRE plasmid was cotransfected with the plasmid expressing either RevM10-TAP or RevΔ78–79–TAP. This was observed in several independent transfection experiments (data not shown). These results suggested that TAP was able to complement the NES defect in Rev, albeit only to a small extent.
FIG. 1.
Fusion to TAP functionally complements export-deficient Rev proteins. 293T cells were transfected with plasmids expressing the intron-containing gag-pol–RRE reporter mRNA, the indicated transactivator constructs, and a plasmid expressing SEAP from a completely spliced mRNA. At 72 h posttransfection, supernatants were collected and analyzed for p24 levels and SEAP activity. The values shown are averages of duplicate SEAP-normalized supernatant p24 levels. wt, wild type.
NXT1 dramatically increases the function of the Rev-TAP fusion proteins on the HIV RRE.
Since TAP has been shown to interact with the NXT1 protein and the TAP and NXT1 genes were shown to complement a mex67-mtr2 mutant of yeast (32), we next decided to determine whether overexpression of NXT1 would have any effect on Rev-TAP fusion protein function. To this end, we cotransfected a pCDNA3-based plasmid expressing the NXT1 protein into cells together with the HIV pCMVgagpol-RRE construct alone or with pCMVgagpol-RRE and a plasmid expressing RevM10-TAP or the wild-type Rev protein. As a control, cells were also transfected with these plasmids and the empty vector pCDNA3. As in the previous experiments, in each case, the cells were also cotransfected with the SEAP plasmid. Supernatant medium was harvested at 72 h posttransfection and analyzed for p24 and SEAP activity. The results of this experiment are shown in Fig. 2.
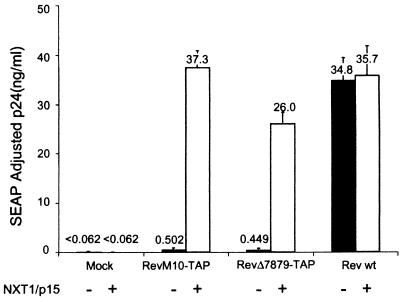
FIG. 2.
NXT1 dramatically enhances the export function of Rev-TAP fusion proteins. Transfections were performed as described in the legend to Fig. 1, except that 2 μg of pCDNA-NXT1 (empty bars) or pCDNA (black bars) was included, as indicated. wt, wild type.
Whereas NXT1 had no effect on expression from pCMVgagpol-RRE alone (Mock), a very dramatic increase in p24 activity was seen in the cotransfections of this plasmid with NXT1 and RevM10-TAP or RevΔ78-79–TAP. In fact, the levels of p24 activity were equivalent to those obtained with the gag-pol-RRE plasmid in conjunction with the wild-type Rev protein. This effect was specific for the Rev-TAP chimeric proteins, since NXT1 did not increase the p24 levels in the case of the unfused wild-type Rev protein. The SEAP levels in transfected cells were not affected by cotransfection of NXT1 (data not shown). Thus, the NXT1 stimulation in this assay was likely to reflect specific interactions between TAP and this protein.
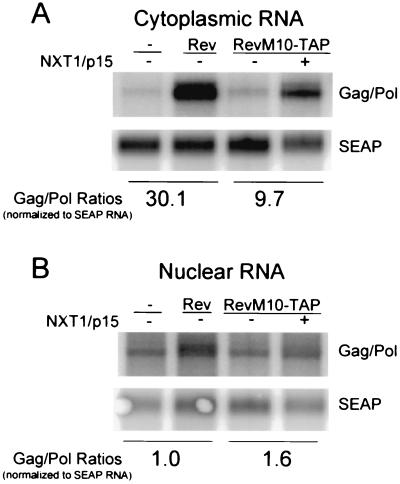
To determine if the NXT1-mediated enhancement of RevM10-TAP function occurred at the level of RNA export, Northern blot assays were performed on nuclear and cytoplasmic poly(A)-selected RNAs from transfected cells. The blots were analyzed with a probe specific for the HIV gag-pol message, as well as with a probe specific for the control SEAP mRNA. As can be seen in Fig. 3A, virtually no gag-pol–RRE RNA was detected in the cytoplasmic fraction in the absence of Rev protein. The wild-type Rev protein increased the relative levels of gag-pol–RRE RNA in the cytoplasm 30-fold. This confirms our previous experiments that have shown a strict requirement for Rev to obtain RNA export in this system (8). Expression of RevM10-TAP alone resulted in the accumulation of only a small amount of cytoplasmic gag-pol–RRE RNA in the cytoplasm. However, a nearly 10-fold relative increase in the cytoplasmic levels of gag-pol–RRE mRNA was seen with NXT1 and RevM10-TAP. In contrast to what was observed in the cytoplasmic RNA fractions, the relative levels of gag-pol RNA in the nucleus were not dramatically different (Fig. 3 B). This has been observed previously and suggests that the RNA that is retained in the nucleus in the absence of a functional Rev protein is unstable and rapidly degraded (37, 52). Taken together, these experiments show that coexpression of NXT1 is essential for the ability of the TAP fusion construct to provide an efficient export function. They also show that NXT1 enhances expression at the level of RNA export.
FIG. 3.
NXT1-mediated enhancement of RevM10-TAP function occurs at the level of RNA export. Northern blot analysis of cytoplasmic (A) and nuclear (B) RNAs is shown. 293T cells were transfected with the indicated constructs. Nuclear or cytoplasmic poly(A)-selected RNA was isolated and analyzed. Blots were hybridized with probes complementary to HIV Gag (Gag/Pol) or SEAP (SEAP) coding sequences.
NXT1 coexpression does not affect RevM10-TAP function by stabilization or by changing the steady-state localization of the fusion protein.
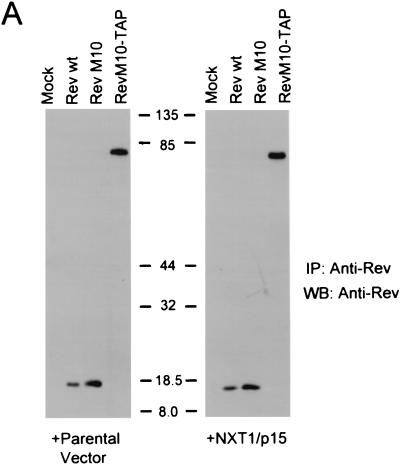
We next decided to analyze the expression of Rev, RevM10, and RevM10-TAP in transfected cells in the presence or absence of NXT1 using IP Western blot analysis. This analysis was performed to compare the levels of wild-type Rev and RevM10 protein expression to that of RevM10-TAP and to exclude the possibility that NXT1 affected RevM10-TAP function simply by stabilizing this protein. The results of these experiments (Fig. 4A) showed that all of the Rev proteins were efficiently expressed in transfected cells and that coexpression of NXT1 did not have any observable effects on the level of expression of either of these proteins.
FIG. 4.
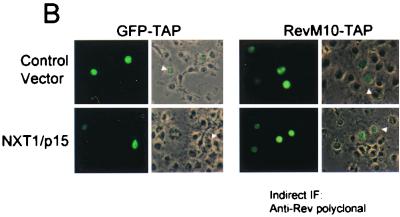
Expression of NXT1 does not alter the steady-state localization of M10-TAP or dramatically affect RevM10-TAP expression levels. (A) IP-Western blot (WB) analysis of proteins from transfected cells. Rev and RevM10-TAP fusion proteins were immunoprecipitated from lysates of transfected cells using an anti-Rev polyclonal antibody and separated using SDS-PAGE. Western blot analysis was performed with an anti-Rev monoclonal antibody (3H6) (45). Blots were visualized with ECL. Positions of commercial molecular weight standards (103 Bio-Rad) are indicated. (B) Fluorescence microscope analysis of GFP-TAP and RevM10-TAP proteins. Transfected CMT-3/COS cells were fixed and permeabilized. RevM10-TAP-expressing cells were stained using a primary anti-Rev rabbit polyclonal antibody and an Alexafluor-488-conjugated secondary antibody (Molecular Probes). Cells were visualized by fluorescence microscopy, and representative fields were photographed. wt, wild type. IF, immunofluorescence.
In previous studies, it has been shown that the TAP protein localizes mainly to the nucleus at steady state (2, 32). This has been shown to be true also for GFP-TAP fusion proteins (2, 3). When the localization of a GFP-TAP fusion protein was compared to the localization of RevM10-TAP in transfected CMT3-COS cells, both proteins were found mainly in the nucleus at steady state (Fig. 4B). This localization was not noticeably altered by coexpression of NXT1.
TAP-mediated export requires both NXT1 and nucleoporin interaction domains.
We were next interested in determining which regions within the TAP protein are essential for export activity in the context of the RevM10-TAP fusion protein. To address this issue, constructs were made with various fragments of the TAP ORF fused to RevM10. The resulting plasmids were then assayed for the ability to induce p24 expression in conjunction with the HIV RRE in transient-transfection experiments. These experiments were done in the presence or absence of cotransfected NXT1 as described above.
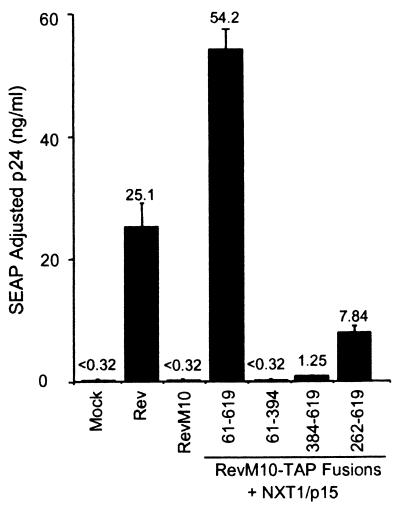
The diagram presented in Fig. 5 shows all of the different constructs that were tested. The results show that fusion of RevM10 to TAP with either of two N-terminal deletions resulted in proteins that were still able to induce some p24 expression (262-619 and 384-619 in Fig. 6). The levels of p24 were much lower with these mutant proteins compared to that obtained with the fusion protein containing TAP aa 61 to 619, the protein analyzed in the experiments described above; however, even the more extensive deletion gave some p24 activity over the background. These results thus showed that the N-terminal portion of the TAP protein is not essential for its ability to provide an RNA export function in the context of a RevM10-TAP fusion protein.
FIG. 5.
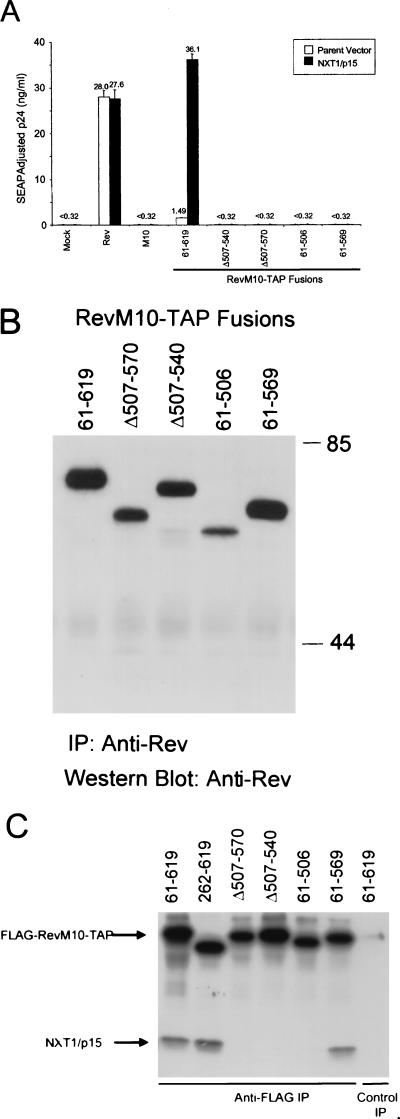
Schematic diagram of RevM10-TAP deletion mutant proteins. The indicated fragments of TAP-encoding DNA were fused to the end of the ORF encoding RevM10 using SOE-PCR. Solid lines indicate ORFs, and dotted lines represent deleted regions. These mutant DNAs were cloned into the mammalian expression vector pCMV (35). Functional data generated with these mutant constructs are shown in Fig. 6 and 7. NUP, nucleoporin.
FIG. 6.
The C-terminal region of TAP is essential for export activity. Plasmids expressing the indicated portions of TAP (Fig. 5) fused to RevM10 were transfected into 293T cells and tested for activity as described in the legend to Fig. 1. A plasmid expressing NXT1 was cotransfected as indicated. Plasmids expressing Rev or RevM10 were also tested as controls. The values shown are averages of duplicate SEAP-normalized p24 levels.
In contrast to what was observed with the N-terminal TAP mutant proteins, deletions of the C-terminal portion of TAP (aa 395 to 619) resulted in a fusion protein that was unable to induce any p24 expression from HIV gag-pol–RRE (61–394 in Fig. 6). The analysis of additional deletion mutants showed that deletion of as little as 50 aa from the C-terminal end of TAP resulted in fusion proteins that were completely inactive both in the presence and in the absence of cotransfected NXT1 (61-569 in Fig. 7A). The C-terminal portion of TAP has been shown to contain domains that interact with NXT1 and nucleoporins, respectively (2, 32). Based on these findings, we went on to make two internal deletion mutant proteins (Δ507-540 and Δ507-570) that were designed to specifically disrupt the NXT1-binding domain (32). Both of these deletion constructs failed to induce any p24 activity in transfected cells (with or without NXT1) (Fig. 7A).
FIG. 7.
The NXT1- and nucleoporin-binding domains of TAP are required for export activity. (A) Plasmids expressing the indicated portions of TAP (Fig. 5) fused to RevM10 were transfected into 293T cells and tested for activity as described in the legend to Fig. 1. A plasmid expressing NXT1 was cotransfected as indicated. Plasmids expressing Rev or RevM10 were also tested as controls. The values shown are averages of duplicate SEAP-normalized p24 levels. (B) Lysates of 293T cells transfected with plasmids expressing C-terminal deletion mutant RevM10-TAP were subjected to IP-Western blot analysis as described in the legend to Fig. 4A. The values on the right are molecular weights in thousands. (C) FLAG-RevM10-TAP and NXT1 binding in vitro. Full-length RevM10-Tap (61-619) and the indicated RevM10-TAP deletion mutant proteins were assayed for in vitro NXT1-binding activity. 35S-labeled FLAG-RevM10-TAP fusion proteins and NXT1 protein were synthesized in vitro in rabbit reticulocyte lysates. FLAG-RevM10-TAP lysates were mixed with NXT1 lysates and allowed to bind. FLAG-RevM10-TAP proteins were immunoprecipitated with M2 anti-FLAG monoclonal antibody or a control irrelevant antibody, resolved by SDS-PAGE, and visualized by autoradiography. The bands corresponding to the proteins are indicated.
To verify that all of the fusion proteins with C-terminal mutations in TAP were expressed efficiently in transfected cells, we performed an IP-Western blot analysis on lysates from transfected cells (Fig. 7B). This experiment showed that all of the mutant proteins were expressed and that most of the mutant proteins were expressed at levels that did not vary significantly from the expression of the original RevM10-TAP fusion protein, 61-619. In addition, all of these mutant proteins showed a nuclear localization similar to that of the full-length RevM10-TAP fusion protein (data not shown).
The proteins expressed from the various deletion constructs were also tested for the ability to associate with NXT1 in an in vitro binding assay using FLAG-tagged proteins translated in a reticulocyte lysate system. The full-length FLAG-RevM10-TAP fusion protein, 61-619, and the N-terminal deletion mutant protein, 262-619, bound significant amounts of NXT1 in this assay (Fig. 7C). Although, the 262-619 mutant protein did not function very efficiently in the export assay (Fig. 6), it appeared to bind NXT1 as well as the 61-619 protein. In contrast, the fusion protein lacking aa 507 to 540 or aa 507 to 570 of TAP, as well as the deletion mutant protein lacking aa 507 to 619, failed to bind detectable amounts of NXT1. Importantly, the 61-569 protein that lacks the C-terminal 50 aa of TAP and failed to induce p24 in transfected cells (Fig. 7A) retained the ability to bind to NXT1 in vitro. This shows that the NXT1- binding domain maps between aa 262 and 569 of TAP. The C-terminal portion of TAP has previously been shown to be essential for nuclear-rim association and interaction with nucleoporins. Taken together, the results of these experiments indicate that both the NXT1- and C-terminal nucleoporin-binding domains of TAP are of critical importance for the ability of the RevM10-TAP fusion protein to mediate export of intron-containing RNA.
RevM10-TAP-mediated RNA export utilizes a CRM1-independent export pathway.
Although Rev-RRE and CTE are functionally interchangeable in the export of intron-containing RNA, they have been shown to utilize, at least partially, nonoverlapping pathways (6, 47, 49, 65). Rev export is dependent on a leucine-rich NES, which has been shown to interact with the export receptor CRM1 (16, 58). CRM1-mediated export has been shown to be specifically inhibited by LMB, and Rev-mediated export is inhibited by this drug (63). In contrast, CTE-mediated export has been reported to be LMB insensitive, indicating that it gains access to a CRM1-independent export pathway (47).
In order to examine the effects of LMB on export mediated by the RevM10-TAP fusion protein, we treated cells cotransfected with RevM10-TAP, NXT1, and pCMVgagpol-RRE with LMB from 24 to 72 h posttransfection. As controls in these experiments, we used cells transfected with pCMVgagpol-RRE and Rev, as well as cells transfected with pCMVgagpol-CTE. The results of these experiments are shown in Fig. 8.
FIG. 8.
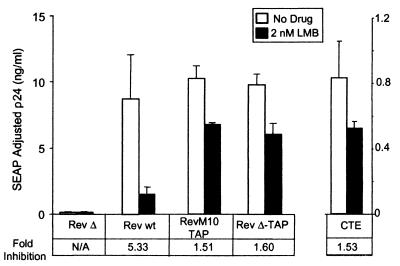
TAP-mediated export uses a CRM1-independent pathway. Duplicate transfections of 293T cells were performed with plasmids expressing the indicated Rev or Rev-TAP fusion proteins. pcDNA-NXT1 and pCMV-SEAP were included in all transfections. At 24 h posttransfection, the medium was replaced with a medium containing 2 nM LMB (black bars) or an equivalent amount of ethanol solvent (white bars). Samples were collected 12 h later. Fold inhibition was calculated taking the ratio of the SEAP-adjusted p24 value obtained with no drug to the SEAP-adjusted p24 value of the LMB-treated sample. N/A, not applicable. wt, wild type.
Confirming previously published results, Rev function was inhibited more than fivefold by LMB in these experiments (47, 63). In contrast, only a very small drop in p24 activity was observed with either of the Rev-TAP fusion proteins. This was comparable to the reduction in p24 activity that was observed with the pCMVgagpol-CTE plasmid. These results thus demonstrate that the TAP fusion proteins promote export of HIV gag-pol-RRE RNA through a CRM1-independent pathway.
RevM10-TAP activity is inhibited by an FG repeat fragment from CAN/Nup214(ΔCAN).
It has previously been demonstrated that both CRM1 and Rev function can be inhibited by ΔCAN, a fragment of CAN/Nup214 that contains some of the FG repeats. ΔCAN binds to CRM1, and overexpression of ΔCAN blocks the ability of CRM1 to interact with the nuclear pore complex (17). In contrast, CTE function has been reported to be unaffected by ΔCAN (6, 31, 65). However, recent publications have shown that CAN also binds to the nuclear porecomplex-binding domain of TAP and FG repeat-containing fragments of CAN, similar to ΔCAN, have been shown to interact with the TAP protein (2, 32). We thus decided to determine the effect of ΔCAN expression on RevM10-TAP function. To this end, we transfected cells with pCMVgagpol-RRE and either Rev or RevM10-TAP together with increasing amounts of a plasmid expressing ΔCAN. In parallel, we performed a similar experiment using pCMVgagpol-CTE. pCMVSEAP was included in each transfection as a control for potential nonspecific effects of the addition of ΔCAN.
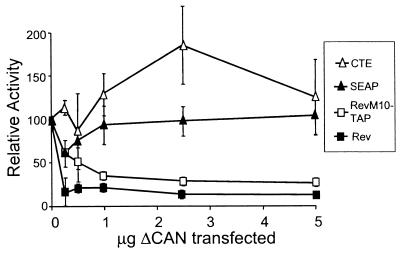
As shown in Fig. 9, ΔCAN inhibited both Rev and RevM10-TAP function in a dose-dependent manner. In contrast, SEAP activity was not inhibited by ΔCAN, indicating that expression of this protein had no overall toxic effects on the cell. The dose-response curves were similar for Rev and RevM10-TAP, clearly indicating that ΔCAN was able to specifically inhibit the function of both of these proteins. As previously reported, ΔCAN showed no inhibitory effect on CTE function (6, 31, 65). In fact, a small increase in CTE activity was observed in several of the transfections. This was observed in several independent transfection experiments (data not shown). Thus, ΔCAN efficiently inhibited the TAP export function but had no effect on the CTE function in 293T cells.
FIG. 9.
RevM10-TAP-mediated export is sensitive to the transdominant negative nucleoporin ΔCAN. Duplicate transfections of 293T cells were performed with increasing amounts of a plasmid expressing ΔCAN, and the total amount of plasmid DNA was held constant with the parental pBC12 plasmid. This was done in combination with either pCMVgagpol-CTE or pCMVgagpol-RRE and the pCMV-Rev or pCMV-RevM10-TAP and pcDNA-NXT1 plasmids, as indicated pCMV-SEAP was included in all transfections. Samples were collected and analyzed as described in the legend to the Fig. 1. The CTE, Rev, and RevM10-TAP values shown are SEAP-normalized averages of duplicate transfections expressed as percentages of the value obtained in the absence of ΔCAN. The SEAP curve was generated by using the average of all of the SEAP values from the CTE, Rev, and RevM10-TAP transfections.
DISCUSSION
Several recent studies have suggested that the mammalian protein TAP plays an important role in mRNA export. TAP and NXT1 have been shown to partially complement a defect in mex67-mtr2 in yeast (32). In addition, TAP binds to the CTE of type D retroviruses and is able to enhance the CTE function in Xenopus oocytes (7, 20) and quail cells (31). However, direct evidence that TAP plays a role in RNA export in mammalian cells has been lacking. Our results, as well as those of previous experiments using other assay systems, have failed to demonstrate any effect of overexpression of TAP on the CTE function in human or monkey cells (31). The experiments presented in this paper show that TAP can mediate RNA export in mammalian cells and map the regions of TAP that are required for this export. To show this, we used an assay system that uses intron-containing HIV gag-pol RNA as an export substrate in conjunction with a RevM10-TAP fusion protein. The RNA substrate contains the HIV RRE instead of the CTE but is otherwise identical to the reporter that was initially used to demonstrate the CTE function in mammalian cells (8, 14).
It has previously been shown that the export function of NES-defective Rev proteins can be complemented by using fusions with other leucine-rich NESs. This strategy was initially utilized to demonstrate that the human T-cell leukemia virus type 1 Rex protein contains a functional NES (28). Subsequently, similar Rev fusion constructs were used to demonstrate that c-Abl (61) and transcription factor IIIa (18) contain Rev-like NES domains that can complement the Rev function. A similar approach was also used to analyze the ability of the M9 NES in hnRNPA1 to complement the Rev NES function. However, the Rev-M9 chimeras were unable to induce detectable expression from a Rev-dependent reporter containing the RRE (36).
In a previous publication, only the amino-terminal 300 aa of TAP were shown to be required for enhancement of export of an intron lariat RNA containing the CTE in Xenopus oocytes (7, 20). In contrast, the nucleoporin-binding domain was shown to be essential when a U6 RNA containing the CTE was analyzed in the same system (2). However, even with this substrate, NXT1 did not appear to be an essential cofactor, since deletion of the domain of TAP that binds to this protein had no effect on the ability of TAP to enhance CTE function (2). Experiments in which the nucleoporin-binding domain of TAP was fused to heterologous proteins have indicated that this region contains a functional NES for protein export in mammalian cells (3, 31). TAP enhancement of the CTE function in quail cells was also shown to require this domain (31). However, the NXT1-binding domain does not appear to be essential for either protein export or CTE-mediated RNA export in the quail cell system (31).
In contrast to the results obtained with the oocyte and quail cell systems, deletions within the domain of TAP that binds to NXT1 were shown to completely abolish TAP-mediated RNA export in our mammalian expression system. The reason for these seemingly contradictory results are not clear. One potential explanation is that, as previously suggested, TAP requires different interactions with cellular cofactors, depending on the nature of the cargo (2, 60).
The fact that expression of NXT1 causes such a dramatic increase in the ability of the RevM10-TAP chimeric protein to promote RNA export activity was perhaps the most surprising finding of this study. NXT1 expression increased cytoplasmic RNA accumulation almost 10-fold. This effect was specific for the RevM10-TAP fusion protein, as we observed no effect of NXT1 expression on Rev activity. In addition, this factor had no effect on expression from the control SEAP plasmid nor did it enhance expression from reporters containing the MPMV CTE. Since the 293T cells with which the transfections were performed already express NXT1 (unpublished observations), it is not clear why overexpression of this protein was required for efficient export. One potential explanation might be that the endogenous protein is limited and may already be in a complex with TAP and/or other proteins, making it unavailable for binding to the fusion protein.
Although previous studies have shown that NXT1 binds to TAP (30, 32) and that the combination of these two proteins can serve to complement a mex67-mtr2 defect in yeast (32), it is not clear how this protein promotes the RevM10-TAP function. However, since in vitro binding studies have indicated that NXT1 is able to specifically bind RanGTP (5), it seems possible that this protein functions to recruit RanGTP to the RevM10-TAP–RNA export complex. A mutational analysis of NXT1 should reveal if the RanGTP-binding activity of this protein correlates with its ability to enhance the RevM10-TAP function.
The amino-terminal half of TAP is not essential for export activity in our system. This portion of TAP has been shown to be involved in RNA binding and specific binding to the CTE (20, 31). In addition, it interacts with several RNA-binding proteins (59, 60). In the RevM10-TAP fusion protein, the RNA-binding domain of Rev is intact and would therefore be expected to target TAP specifically to the RRE RNA, eliminating the need for TAP to provide RNA targeting. The amino-terminal domain of TAP also contains a leucine-rich repeat and has been reported to contain a functional NES (3, 7). However, our results clearly indicate that this domain is not sufficient to provide RevM10-TAP-mediated RNA export, since no export activity was observed with the fusion protein that contained this domain alone.
CTE-mediated export has previously been reported to be insensitive to LMB, a drug believed to specifically inhibit the activity of CRM1 (42, 47). In contrast, Rev-RRE-mediated export is efficiently inhibited by this drug (63). The results presented here show that RevM10-TAP-mediated RNA export is also insensitive to LMB, indicating that TAP-mediated export of RNA in this system does not use CRM1. This is the first demonstration that RRE-mediated export of intron-containing RNA can be redirected to a pathway that does not use this receptor.
The nucleoporin fragment ΔCAN has previously been demonstrated to inhibit the function of CRM1 and Rev-mediated RNA export (6, 65). In contrast, it has been reported that ΔCAN expression fails to inhibit RNA export mediated by the CTE (6, 31, 65). Our findings confirm these results and, in addition, show that RevM10-TAP-mediated export is inhibited by ΔCAN. This was unexpected, since it has been proposed that ΔCAN, like LMB, is a selective inhibitor of export mediated through Rev-like NESs (6, 31). However, it should be noted that the C-terminal region of TAP that is essential for export activity in our system has been demonstrated to bind specifically to FG repeat-containing fragments of CAN/Nup214 in vitro (2, 32), as well as in the yeast two-hybrid binding assay (30, 32). This provides a potential explanation for the ability of ΔCAN to inhibit RevM10-TAP-mediated export. Independently of the mechanism of inhibition, these results suggest clear differences between the CTE export pathway and the pathway utilized by RevM10-TAP in conjunction with the RRE. This is also suggested by the fact that the NXT1-binding domain of TAP has been shown to be dispensable for TAP enhancement of the CTE function in Xenopus oocytes, as well as in quail cells (2, 7, 20, 31). In contrast, NXT1 appears to be an essential TAP cofactor in general mRNA export (32). Thus, the system presented here may be a better model for such export than assays that utilize the CTE.
ACKNOWLEDGMENTS
We thank Bryan Cullen and J. U. Jung for the gift of plasmids and Barbara Wolff (Novartis) for providing LMB.
This work was supported by NIH grants AI34721 to M.-L.H. and AI47008 to D.R. and American Cancer Society grant RPG-98-048-01-CSM to B.M.P. Salary support for M.-L.H. and D.R. was provided by the Charles H. Ross Jr. and Myles H. Thaler Endowments at the University of Virginia.
REFERENCES
- 1.Askjaer P, Bachi A, Wilm M, Bischoff F R, Weeks D L, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj I W, Fornerod M. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachi A, Braun I C, Rodrigues J P, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bear J, Tan W, Zolotukhin A S, Tabernero C, Hudson E A, Felber B K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 5.Black B E, Lévesque L, Holaska J M, Wood T C, Paschal B M. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol Cell Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R, Malim M H. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 1992;216:362–368. doi: 10.1016/0076-6879(92)16033-g. [DOI] [PubMed] [Google Scholar]
- 11.Daly T J, Cook K S, Gray G S, Maione T E, Rusche J R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 12.Dundr M, Leno G H, Lewis N, Rekosh D, Hammarskjoid M L, Olson M O. Location of the HIV-1 Rev protein during mitosis: inactivation of the nuclear export signal alters the pathway for postmitotic reentry into nucleoli. J Cell Sci. 1996;109:2239–2251. doi: 10.1242/jcs.109.9.2239. [DOI] [PubMed] [Google Scholar]
- 13.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 17.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component, Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridell R A, Fischer U, Luhrmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham F L, van der Eb A J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973;54:536–539. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- 20.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 21.Hamm J, Mattaj I W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 22.Hammarskjöld M-L. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 23.Hammarskjöld M-L, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammarskjöld M-L, Li H, Rekosh D, Prasad S. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J Virol. 1994;68:951–958. doi: 10.1128/jvi.68.2.951-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammarskjöld M-L, Wang S C, Klein G. High-level expression of the Epstein-Barr virus EBNA1 protein in CV1 cells and human lymphoid cells using a SV40 late replacement vector. Gene. 1986;43:41–50. doi: 10.1016/0378-1119(86)90006-5. [DOI] [PubMed] [Google Scholar]
- 26.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 27.Hood J K, Silver P A. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 28.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang Y, Bogerd H P, Cullen B R. Analysis of cellular factors that mediate nuclear export of RNAs bearing the Mason-Pfizer monkey virus constitutive transport element. J Virol. 2000;74:5863–5871. doi: 10.1128/jvi.74.13.5863-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung J U, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komeili A, O'Shea E K. Nuclear transport and transcription. Curr Opin Cell Biol. 2000;12:355–360. doi: 10.1016/s0955-0674(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 34.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 35.Lewis N, Williams J, Rekosh D, Hammarskjöld M-L. Identification of a cis-acting element in human immunodeficiency virus type 2 (HIV-2) that is responsive to the HIV-1 rev and human T-cell leukemia virus types I and II rex proteins. J Virol. 1990;64:1690–1697. doi: 10.1128/jvi.64.4.1690-1697.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malim M H, Bohnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 37.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 39.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 40.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 41.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 44.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orsini M J, Thakur A N, Andrews W W, Hammarskjold M L, Rekosh D. Expression and purification of the HIV type 1 Rev protein produced in Escherichia coli and its use in the generation of monoclonal antibodies. AIDS Res Hum Retrovir. 1995;11:945–953. doi: 10.1089/aid.1995.11.945. [DOI] [PubMed] [Google Scholar]
- 46.Ossareh-Nazari B, Maison C, Black B E, Lévesque L, Paschal B M, Dargemont C. RanGTP-binding protein NXT1 facilitates nuclear export of different classes of RNA in vitro. Mol Cell Biol. 2000;20:4562–4571. doi: 10.1128/mcb.20.13.4562-4571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paschal B M, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 51.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith A J, Cho M-I, Hammarskjöld M-L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith A J, Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spector D L, Goldman R D, Leinwand L A. Cells: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 57.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjöld M-L, Rekosh D. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 59.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stutz F, Bachi A, Doerks T, Braun I C, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y, Hope T J. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wigler M, Pellicer A, Silverstein S, Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978;14:725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- 63.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 64.Yoon D W, Lee H, Seol W, DeMaria M, Rosenzweig M, Jung J U. Tap: a novel cellular protein that interacts with tip of herpesvirus saimiri and induces lymphocyte aggregation. Immunity. 1997;6:571–582. doi: 10.1016/s1074-7613(00)80345-3. [DOI] [PubMed] [Google Scholar]
- 65.Zolotukhin A S, Felber B K. Nucleoporins Nup98 and Nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]