Abstract
The assembly, disassembly, and functional properties of transcription preinitiation complexes (PICs) of human RNA polymerase I (Pol I) play a crucial role in the regulation of rRNA gene expression. To study the factors and processes involved, an immobilized-promoter template assay has been developed that allows the isolation from nuclear extracts of functional PICs, which support accurate initiation of transcription. Immunoblotting of template-bound factors showed that these complexes contained the factors required to support initiation of transcription, SL1, upstream binding factor (UBF), and Pol I. We have demonstrated that, throughout a single round of transcription, SL1 and UBF remain promoter bound. Moreover, the promoter-bound SL1 and UBF retain the ability to function in transcription initiation. SL1 has a central role in the stable association of the PIC with the promoter DNA. The polymerase component of the PIC is released from the promoter during transcription yet is efficiently recycled and able to reinitiate from “poised” promoters carrying SL1 and UBF, since the PICs captured on the immobilized templates sustained multiple rounds of transcription. Kinetic analyses of initiation of transcription by Pol I revealed that Pol I-dependent transcription is rate limited in a step subsequent to recruitment and assembly of Pol I PICs. The rate of RNA synthesis is primarily determined by the rates at which the polymerase initiates transcription and escapes the promoter, referred to as promoter clearance. This rate-limiting step in Pol I transcription is likely to be a major target in the regulation of rRNA gene expression.
As cells grow, proliferate, and differentiate, there is a varying demand for protein synthesis and, with that, for ribosome biogenesis (14). The rRNAs are precursors and integral components of ribosomes, and as such, their production is controlled coordinately. A dedicated nuclear RNA polymerase I (Pol I) mediates synthesis of the major rRNAs in the nucleolus. A number of studies with both Saccharomyces cerevisiae and higher eukaryotes have suggested that ribosome biogenesis is regulated in a large part at the level of transcription of the rRNA genes by Pol I (reviewed in references 13, 18, 23, 37, and 40). For example, in cancer cells rRNA transcriptional activity and nucleolar size are inversely related to cell doubling time (11), and consequently nucleolar morphology is used by tumor pathologists as a diagnostic and prognostic marker. In order to advance our understanding of this process crucial to the general physiology of the cell, we have investigated the critical steps in the expression of rRNA genes in mammalian cells.
Gene activation begins with the assembly of a transcription preinitiation complex (PIC) at the gene promoter. The early stages of PIC formation involve transcription factors specifically binding to the core promoter of the rRNA genes to allow for the recruitment of Pol I, which itself displays no sequence selectivity. In mammalian cells this entry point for Pol I is provided by at least two transcription factors: selectivity factor SL1 (32), which is composed of the TATA-binding protein (TBP) and three TBP-associated factors (TAFs) of 110, 63, and 48 kDa (8, 9, 48, 49), and upstream binding factor (UBF), the relaxed-specificity DNA binding and multiple-HMG-box-containing factor and activator of Pol I transcription (5, 24, 41). Studies with reconstituted cell-free transcription systems from human cells have suggested a cooperative interaction between SL1 and UBF preceding the recruitment of Pol I and possibly other associated essential factors (5). In cells, the formation of this PIC is likely to be facilitated by or include activities that remodel and derepress chromatin at the gene promoter (31).
Assembly of the PIC is one step in the events that lead to gene activation, and these are conceptually similar for prokaryotic and eukaryotic RNA polymerases. What follows is the isomerization of the PIC from a closed complex to an open complex, initiation of transcription by Pol I, escape or promoter clearance by Pol I, and subsequent elongation through the gene to sequences and factors that signal termination to complete the transcription cycle. Pol I is recycled and may reinitiate transcription from a previously activated and engaged promoter which has prebound transcription factors. Given this multistage event, it is conceivable that every step in the transcription cycle may be subject to tight regulation (for a review, see reference 28), but the control of only those that appear rate limiting will have a major impact on rRNA gene expression.
In this study, we set out to determine whether PIC assembly or subsequent events are the critical rate-limiting steps in a human cell-free transcription system. To this end, we developed an immobilized-ribosomal DNA (rDNA) promoter template assay similar to those which previously have proven instrumental in the analysis of Pol III and Pol II PICs (2, 26, 34, 39). This assay allowed us to capture and purify Pol I PICs from nuclear extracts and study their fate in the transcription cycle. The formation of these complexes on the DNA promoter template is experimentally unbiased; it may follow a strictly sequential or stepwise pathway involving individual components, as reported previously (5, 27, 35, 44), or it may involve partial complexes or perhaps holoenzyme complexes (1, 19, 43, 46). Here we demonstrate that the rate-limiting step in Pol I transcription in vitro is not at the stage of PIC formation but rather is a postassembly event, in which Pol I initiates transcription and clears the promoter.
MATERIALS AND METHODS
Preparation of immobilized templates.
The biotinylated templates were synthesized by PCR using Vent DNA polymerase (New England Biolabs). The plasmid prHu3 (32) was used as template, and the sense primer used had been biotinylated at the 5′ end. The binding sites in the ribosomal promoter of the primers are indicated in Fig. 1 and have the following coordinates: 5′ primers, −193 to −163 (Fr4), −324 to −294 (Fr3), and −515 to −492 (Fr2); 3′ primer, +215 to +239. The DNA fragments were purified by extraction from an agarose gel and by QIAquick spin columns (Qiagen). The 5′-end-biotinylated DNA fragments were immobilized on streptavidin-coated paramagnetic beads (M280 Dynabeads; Dynal) according to the manufacturer's instructions. Typically, 10 to 50 ng of biotinylated DNA was immobilized on 1 μl (10 mg/ml) of beads. Beads were concentrated with a magnetic particle concentrator (Dynal) and washed extensively to remove possible traces of unbound DNA, and they were incubated with bovine serum albumin (5 mg/ml; Merck BDH) to block nonspecific binding sites.
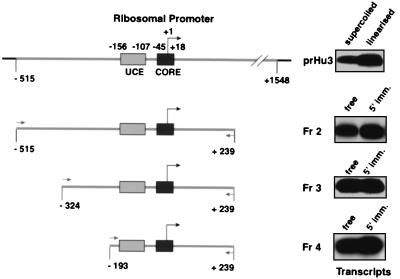
FIG. 1.
Immobilized human rRNA gene promoter fragments containing the UCE and core promoter elements support in vitro transcription. The promoter fragments generated by PCR (Fr2 to Fr4) are schematically outlined. PCR primer binding sites, relative to the transcription start site at +1, and the UCE (−156 to −107) and core region (−45 to +18) in the ribosomal promoter are indicated. prHu3 is a pBR322-derived supercoiled plasmid DNA containing the human ribosomal promoter sequence from −515 to +1548. In vitro transcription assays contained 1 μl of HeLa cell nuclear extract (NE), and a 1.25 pM concentration of the appropriate template. 5′-end-biotinylated DNA was immobilized (5′ imm.) onto 2.5 μl of streptavidin-coated paramagnetic beads (Dynabeads M280; Dynal). Transcripts synthesized in vitro were analyzed in S1 nuclease protection assays with a radiolabeled oligonucleotide overlapping the transcription start site (see Materials and Methods).
Isolation of Pol I PICs.
Immobilized template DNA (IT-DNA), typically 1 to 20 μl in a 20- to 200-μl total reaction volume, was incubated with gentle agitation in HeLa cell nuclear extract (12) or partially purified transcription factors (8) for 5 to 25 min at 4°C in 50 mM KCl (final concentration)–TM10i buffer (50 mM Tris HCl [pH 7.9], 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM sodium metabisulfite, 1 mM dithiothreitol, 50 ng of bovine serum albumin per μl, 0.03% NP-40) to which an EDTA-free protease inhibitor cocktail (Roche) was added. Under these conditions, we did not detect nonspecific binding of transcription factors to the M280 Dynabeads (data not shown). After separation using a magnetic stand, beads were washed three times with 2 reaction volumes of TM10i–0.05M KCl buffer.
Protein detection.
Proteins were analyzed by immunoblotting. To this end, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred to Immobilon P membranes (Millipore). Primary antibodies used for the detection of proteins were as follows: anti-A190 (Pol I) antibodies (1:250) affinity purified from sheep immunized with a mixture of three peptides derived from A190, the largest subunit of human Pol I (K. I. Panov, G. Miller, and J. C. B. M. Zomerdijk, unpublished results); anti-UBF antibodies (at 1:1,000) from a polyclonal rabbit serum raised against recombinant and purified human UBF (kindly provided by B. McStay); and anti-TAFI110, anti-TAFI63, and anti-TAFI48 (SL1 subunits) (all at 1:1,000) from polyclonal rabbit sera (9, 48). Appropriate secondary antibodies conjugated to horseradish peroxidase were used to detect immunocomplexes on the blot by chemiluminescence according to the manufacturer's instructions (ECL kit; Amersham Pharmacia Biotech).
In vitro transcription.
In vitro transcription reactions were performed as described previously (5, 33) at a final salt concentration of 50 mM KCl. Supercoiled prHu3 DNA (32), pseudo-wild-type rDNA promoter (5), or immobilized linear DNA fragments were used as templates in the transcription reaction. In transcription assays where competitor DNA was used to limit transcription to a single round, we included controls to determine the appropriate ratios of immobilized template, nuclear extracts, and competitor DNA. The last component, when mixed simultaneously with the template and nuclear extract, should totally block transcription. Pol I and SL1 were purified as described previously (8); UBF was purified to near homogeneity from Sf9 cells infected with recombinant UBF baculovirus (K. I. Panov and J. C. B. M. Zomerdijk, unpublished results). Each component alone did not support transcription, and recombinant UBF activated about eightfold reconstituted transcription with SL1 and Pol I (data not shown). Transcription assays were analyzed in an S1 nuclease protection assay after annealing the RNA to a 5′-end-labeled oligonucleotide, which was identical to the region between −20 and +40 of the template strand in the human rRNA gene promoter (5). The pseudo-wild-type-specific oligonucleotide used was identical to the sequences from −20 to +29 of the template strand in the pseudo-wild-type rRNA gene promoter.
Rate constant calculations.
Complete PIC formation is a second-order reaction: template + NE → PIC ⇒ VPIC = k [template] [NE]. For the kinetics experiments, we used nuclear extract/immobilized template ratios such that no significant depletion of transcriptional activity was observed from the NE after the beaded templates had been removed. Thus, all Pol I factors were in vast excess in comparison with template DNA, and the reaction is of the first order, as follows: template + GTFs → PIC; if [GTFs] >> [template], then VPIC ≅ k [GTFs], where GTFs are general transcription factors and Pol I.
A single-round transcription reaction programmed by preformed PICs is of the first order: PIC → Pol I* + template/UBF/SL1 + RNA ⇒ Vsynt = ksynt [PIC], where Pol I* depicts a Pol I that has synthesized a transcript of at least 40 nucleotides (nt) (the length of the oligonucleotide used in S1 nuclease protection) and that upon release from the promoter template was prevented from reinitiation by nonspecific competitor DNA. Thus, analysis of the entire kinetic curves using standard methods for chemical kinetics (as outlined below) yielded the rate constant. To this end, the amounts of RNA produced in in vitro transcription reactions were quantified with a Fuji phosphorimager. After subtracting background, phosphorimager units (PU) from the transcription signal at each time point were divided by the average PU produced at the longest time points (in the plateau region) to obtain the fractional completion at each time point. These values were fed into the following equation: Fc = 1 − e−kt , or −[ln(1 − Fc)] = kt, where Fc is the fractional completion, k is the observed rate constant, and t is time in seconds (see reference 29). The logarithmic plot of the fractional completion, −[ln(1 − Fc)], versus time (t) results in a straight line, whose slope corresponds to k (per second). The values of k and the standard errors were calculated from curve fitting by linear regression using EnzFitter software (Biosoft, Cambridge, United Kingdom). The data from three independent experiments (see Tables 1 and 2) were processed, and rate constants were calculated. From these, a mean rate constant and standard deviation were derived.
TABLE 1.
Time course of transcription from preassembled PICs in three independent experiments
| Time (s) | RNA synthesis (PU) in expt no.:
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| 60 | 7,775 | 9,941 | 6,400 |
| 120 | 10,300 | 11,000 | 8,390 |
| 300 | 11,901 | 13,000 | 12,050 |
| 600 | 15,960 | 15,741 | 15,741 |
| 900 | 17,530 | 18,344 | 18,344 |
| 1,500 | 23,100 | 22,440 | 20,610 |
| 1,800 | 23,981 | 23,110 | 21,855 |
| 2,400 | 24,562 | 23,855 | 22,155 |
TABLE 2.
Transcription from PICs assembled for various periods of time in three independent experiments
| Time (s) | RNA synthesis (PU) in expt no.:
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| 5 | 1,156 | 1,099 | 1,073 |
| 10 | 4,214 | 4,777 | 3,882 |
| 15 | 10,427 | 10,359 | 10,200 |
| 20 | 17,538 | 17,671 | 16,439 |
| 30 | 24,791 | 24,482 | 23,950 |
| 60 | 35,086 | 36,068 | 34,990 |
| 300 | 42,677 | 43,781 | 42,391 |
| 600 | 41,488 | 43,005 | 42,366 |
| 1,200 | 43,171 | 42,991 | 42,998 |
RESULTS
RNA Pol I-mediated transcription in vitro from an immobilized human rRNA gene promoter.
We developed an immobilized-rDNA promoter template assay to isolate and study the properties of Pol I PICs. To examine what would be an appropriate promoter fragment, we used prHu3 DNA (32), which contains a region of the rRNA gene transcription unit from −515 to +1548 relative to the transcription start site at +1, as a template in a PCR. DNA fragments with progressively shortened 5′ ends that contained the previously mapped upstream control element (UCE) and core region of the human rRNA gene promoter (16, 17) were generated (Fig. 1). The 5′-end oligonucleotide used in the PCR was biotinylated to allow for attachment of the PCR fragment to streptavidin-coated paramagnetic beads. In a comparative analysis, we tested equimolar amounts of supercoiled template (prHu3) or linearized prHu3 and promoter fragments as free DNA or as 5′-end-immobilized templates in in vitro transcription assays with HeLa cell nuclear extract. All promoter fragments (as free or immobilized DNA) supported in vitro transcription more efficiently than supercoiled prHu3 DNA (Fig. 1). Remarkably, a bead (at −193) close to the UCE (−156 to −107) in Fr4 did not affect the activity, since this promoter fragment was as efficient as longer fragments in supporting transcription initiation. Under the in vitro transcription conditions used the α-amanitin-resistant and Pol I-mediated transcription was in most cases a little higher when the 5′ end of the promoter fragment was attached to beads, possibly resulting from reduced accessibility of the promoter to repressive activities in the nuclear extract. Attachment of Fr4 at the 3′ end, however, led to reduced levels of transcription (data not shown). This probably resulted from interference with polymerases running off the end of the template and consequently the jamming of polymerases on the template occluding successive rounds of transcription. Taken together, the data indicated that the Fr4 template was a suitable promoter substrate for our subsequent studies of PIC assembly and recycling during initiation of transcription.
Isolation of functional Pol I PICs.
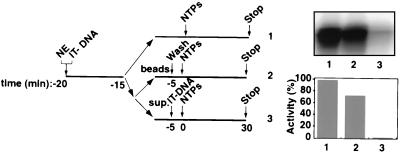
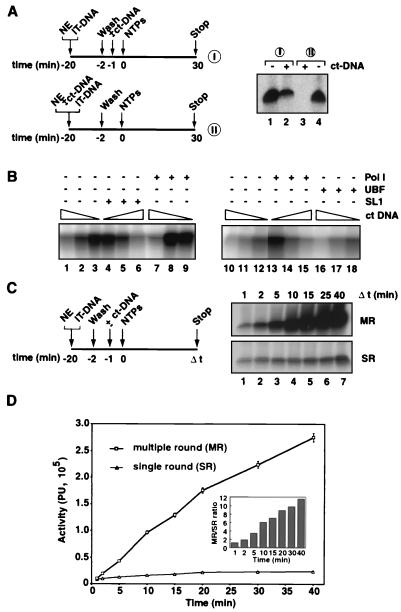
Since the minimal promoter fragment, Fr4 (Fig. 1), supported efficient initiation of transcription in vitro, we wished to determine whether it would allow for the isolation from nuclear extracts of functional PICs containing Pol I. To this end, we incubated the immobilized template with HeLa cell nuclear extract and subsequently washed these templates at various salt concentrations. To ascertain the presence of functional PICs on these templates, we then analyzed their ability to support transcription. Functional PICs were recovered after 50 mM KCl washes (Fig. 2, lane 2), and the recovered transcriptional activity for these PICs on the immobilized templates was 75% of that in the nuclear extract (Fig. 2, compare lane 2 with lane 1). Efficient capture of Pol I factors was evident under conditions where the template was in excess, as the nuclear extract was almost completely depleted of Pol I transcriptional activity by the immobilized template (Fig. 2, lane 3).
FIG. 2.
Functional Pol I transcription PICs captured from nuclear extracts (NEs) onto immobilized promoter templates. The experimental design to test for the isolation of PICs is outlined. Five microliters of immobilized template (50 ng of Fr4 per μl of beads [IT-DNA]) was incubated for 5 min at 4°C with 2 μl of HeLa NE in a 10-μl reaction volume and washed with TM10i–0.05 M KCl buffer. The reaction mixture was split equally into two. In one portion, the immobilized template was left in the NE for the entire time (lane 1), while in the other the beaded template was separated (lanes 2 and 3). The beads were washed in TM10i–0.05 M KCl buffer before initiation of transcription (lane 2), and the supernatant was tested for transcriptional activity by adding back the immobilized promoter template. Transcription in all three reactions was initiated with the addition of ribonucleoside triphosphates NTPs, and the reactions were allowed to proceed for 30 min at 30°C. Transcript synthesis was analyzed by S1 nuclease protection. The autoradiograph shows the transcript levels from in vitro transcription reactions supported by HeLa cell NE and immobilized DNA (lane 1), by isolated PICs on the immobilized DNA (lane 2), and by HeLa cell NE after PIC extraction (lane 3). Phosphorimager quantitation is presented in a bar graph, with the signal in lane 1 set at 100%.
Relaxed DNA binding specificity of Pol I transcription factors during PIC formation.
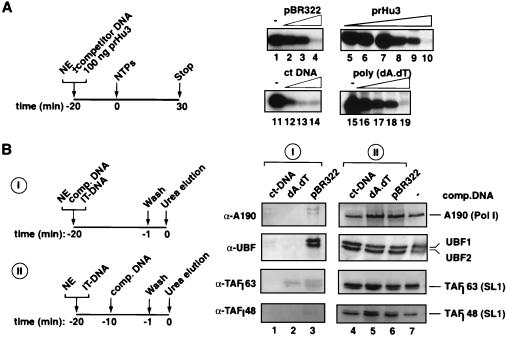
To test for the specificity of the interaction of the Pol I transcription machinery with the immobilized template, we performed competition experiments with increasing concentrations of an assortment of nonspecific DNAs as schematically outlined in Fig. 3A. Supercoiled plasmid DNA (pBR322) and linear DNAs such as sheared calf thymus DNA and poly(dA-dT) all prevented PIC formation, since transcription was completely blocked (Fig. 3A, lanes 4, 13, and 19). Significantly, pBR322 and prHu3 (a pBR322 derivative containing the ribosomal promoter) were equally capable of completely abolishing Pol I-specific transcription at only a 15-fold molar excess (Fig. 3A, lanes 4 and 10). Calf thymus DNA also appeared to be an effective competitor DNA (Fig. 3A, lanes 11 to 14). Taken together, the data imply a relaxed sequence specificity of the Pol I general transcription factors. This is in agreement with DNA binding characteristics observed for individual factors, such as that for UBF (10, 22, 38) and SL1 (3–5, 33; J. K. Friedrich and J. C. B. M. Zomerdijk, unpublished data).
FIG. 3.
The Pol I transcription PIC displays stable DNA binding with a relaxed sequence specificity. (A) PIC formation from nuclear extracts (NEs) was allowed to proceed in the presence of various competitor DNAs as outlined. The effects on transcription of 20-min preincubation at 4°C of 100 ng of prHu3 DNA (template), various amounts of competitor DNA, and 2 μl of NE at 50 mM KCl in a 12.5-μl total reaction volume were analyzed by S1 nuclease protection. Competitor DNAs used are indicated above the lanes: pBR322 (0.5, 1, and 2 μg for lanes 2, 3, and 4, respectively), sheared calf thymus DNAs (ct-DNAs) (0.1, 0.5, and 1 μg for lanes 12, 13, and 14, respectively), and poly (dA-dT) (0.1, 0.2, 0.5, and 1 μg for lanes 16, 17, 18, and 19, respectively). Increasing amounts of prHu3 template itself were titrated in transcription reactions (0.05, 0.1, 0.2, 0.5, 1, and 2 μg for lanes 5 through 10, respectively). Control transcriptions for each of the competitor DNA experiments were performed (lanes 1, 11, and 15). (B) The experimental setup to analyze the immobilized templates for the presence of components of the Pol I transcription machinery upon challenge with nonspecific DNA at different stages during the assembly is outlined. Twenty microliters of IT-DNA (50 ng Fr4 DNA per μl of M280 Dynabeads) was incubated at 4°C for 20 min with 40 μl of HeLa cell NE in a total reaction volume of 160 μl, in the presence of nonspecific competitor DNA (I). Subsequently, the beads were washed in TM10i–0.05 M KCl buffer, and bound proteins were eluted with 5 M urea. The setup for panels II was the same as for panels I except that competitor DNA was added 10 min after NE and IT-DNA were mixed. Urea-eluted proteins were analyzed by immunoblotting with antibodies raised against the largest subunit of human Pol I, A190, against UBF, and against two subunits of SL1, TAPI63 and TAPI48. In lanes 1 and 4, 20 μg of ct-DNA was used; in lanes 2 and 5, 20 μg of poly(dA-dT) was used; and in lanes 3 and 6, 20 μg of pBR322 DNA was used. The control lane, lane 7, contains no competitor DNA in the reaction mixture.
We next analyzed by immunoblotting the binding of factors to immobilized templates and the influence of nonspecific competitor DNAs (Fig. 3B). Competitor DNAs effectively blocked the association of SL1 and Pol I with the immobilized template when added simultaneously (Fig. 3B, lanes 1 to 3) but did not displace these factors after PIC formation (Fig. 3B, lanes 4 to 6). Sequence-specific promoter DNA binding of UBF1 and UBF2 was also affected by competitor DNA, with the exception of supercoiled plasmid DNA, for which perhaps higher concentrations of nonspecific DNA are required in order to effectively compete. Note that on the minimal ribosomal promoter both splice variants of UBF, UBF1 and UBF2 (21, 36), which are present in the nuclear extracts in about equimolar amounts, appeared to bind stoichiometrically (Fig. 3B, lanes 4 to 7). This suggests that perhaps UBF in the PIC is a heterodimer, despite the reported reduced DNA affinity and a much reduced transcriptional activation function of UBF2 (30). Taken together, the data illustrate the relaxed specificity of DNA binding of Pol I factors during PIC formation. They further indicate that the PIC, once formed, is relatively stable, since no appreciable decrease in factor binding could be observed upon addition of competitor DNA (Fig. 3B, compare lanes 4 to 6 with lane 7). Moreover, the data suggest that the inhibition of transcription by competitor DNAs occurred by blocking promoter binding and hence assembly of functional PICs.
The binding of Pol I factors to the immobilized template correlated with their transcriptional activities.
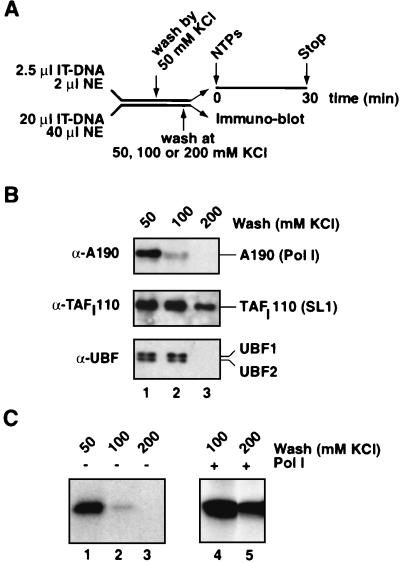
We next determined the stability of assembled PICs at increasing salt concentrations. To this end, PICs were assembled onto immobilized promoter fragments at a 50 mM salt concentration and were subsequently washed at various salt concentrations (Fig. 4A). The complexes, or partial complexes, that remained on the template were analyzed for their protein composition in immunoblots and in transcription assays. The Pol I enzyme is the least salt-stable component of the PIC, and the vast majority of the largest subunit of Pol I, and therefore presumably the enzyme complex, dissociated from the template at 100 mM KCl, under conditions where SL1 and UBF remained bound (Fig. 4B, lane 2). Indeed, the transcriptional activity from this engaged template was drastically reduced (Fig. 4C, compare lanes 1 and 2) but could be recovered by adding back purified Pol I (Fig. 4C, lane 4).
FIG. 4.
Stability of Pol I PICs at the ribosomal promoter. (A) In parallel, for transcription and immunoblotting assays, PICs were assembled for 20 min at 4°C in nuclear extracts (NEs) (2 and 40 μl, respectively, and final volumes of 20 and 160 μl, respectively) with immobilized ribosomal promoter templates (IT-DNA, 2.5 and 20 μl of 50 ng of Fr4 DNA per μl of beads, respectively) as outlined. The immobilized DNA-Pol I transcription complexes were then washed at 50 mM KCl in TM10i buffer before being subjected to washes in the same buffer but with increased salt concentrations (50 to 200 mM KCl). (B) For immunobloting, proteins were eluted from the IT-DNA with 5 M urea, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and blotted onto polyvinylidene difluoride membranes which were probed with antibodies against A190, TAFI110, and UBF (lanes 1 to 3). (C) The parallel reactions were assayed for transcriptional activity, and transcripts were detected by S1 nuclease protection (lanes 1 to 3). In addition, to test for transcriptional recovery of the templates that had been washed at 100 and 200 mM KCl, 1 μl of purified Pol I was added back to the transcription reaction mixtures (lanes 4 and 5, respectively).
The two UBF splice variants, UBF1 and UBF2, dissociated at a higher (200 mM KCl) salt concentration. Remarkably, SL1 remained bound to the promoter under those conditions (Fig. 4B, lane 3) and this SL1 was functional, as it supported transcription upon addition of Pol I (Fig. 4C, lane 5). In fact, it took over 700 mM KCl to elute a fraction of SL1 from the template (data not shown). This was in agreement with the salt concentrations required to elute SL1 from heparin columns during fractionation of HeLa nuclear extracts (5, 8, 32). The binding of factors to the immobilized templates paralleled the transcriptional activities from these templates, and hence the binding reflected primarily functional complexes rather than nonspecifically bound factors.
Efficient reinitiation of transcription by Pol I from isolated PICs.
Sarkosyl has been used widely to limit transcription from templates to a single round, yet the mechanism of this action is ill defined. As an alternative, competitor DNA has been successfully used previously (29), with the advantage that it has predictable effects by preventing factors released from templates from rebinding (Fig. 3). We used it here to analyze transcription by Pol I in single and multiple rounds. Competitor DNA, which completely blocked transcription when added simultaneously with the nuclear extract to the immobilized template (Fig. 5A, lane 3), only partially inhibited transcription when it was added after PICs were allowed to assemble on the template (Fig. 5A, lane 2). Most likely reinitiation by Pol I was prevented under those conditions. In agreement with this interpretation, at intermediate concentrations of competitor DNA where partial inhibition of transcription was observed (Fig. 5B, lanes 1 to 3 and 10 to 12), addition of Pol I (Fig. 5B, lanes 7 to 9 and 13 to 15), but not of SL1 (Fig. 5B, lanes 4 to 6) or UBF (Fig. 5B, lanes 16 to 18), restored transcription. In time course experiments, as expected for a single round, transcription occurred primarily in the first few minutes, whereas transcript synthesis continued for over 40 min in the absence of competitor DNA, consistent with multiple rounds of transcription (Fig. 5C). Remarkably, factors initially part of PICs on the immobilized templates, that is, SL1, UBF, and Pol I, were recycled without significant loss of activity through many transcription cycles (Fig. 5D).
FIG. 5.
Pol I PICs assembled onto immobilized templates support specific initiation in single and multiple rounds of transcription. (A) The experimental design was as follows. A total of 2.5 μl of immobilized template (50 ng of Fr4 per μl of beads [IT-DNA]) was incubated with 2 μl of HeLa cell nuclear extract (NE) in a 20-μl reaction volume at 4°C for 20 min and washed with TM10i–0.05 M KCl buffer. Calf thymus DNA (ct-DNA) (1.25 μg) was added, after (I) or during (II) the assembly of PICs. Transcription was initiated with the addition of ribonucleoside triphosphates (NTPs), and the reaction was allowed to proceed for 30 min at 30°C. Transcript synthesis was analyzed by S1 nuclease protection. The autoradiograph shows the transcriptional activity of isolated PICs in the absence (lane 1) and the presence (lane 2) of ct-DNA, added after PIC assembly, and transcriptional activity of templates to which ct-DNA was or was not added during the assembly (lanes 3 and 4, respectively). (B) The most sensitive component of the PIC to competitor DNA is Pol I. One hundred nanograms of prHu3 promoter DNA, various amounts of ct-DNA (0, 0.1, and 1 μg; see triangles above the lanes), and 1 μl of NE at 50 mM KCl in a 15-μl total reaction volume were preincubated at 4°C for 10 min. In two separate experiments (lanes 1 to 9 and 10 to 18) either nothing (lanes 1 to 3 and 10 to 12), highly purified SL1 (1 μl, lanes 4 to 6), UBF (100 ng, lanes 16 to 18), or Pol I (5 μl, lanes 7 to 9 and 13 to 15) was added. The mixtures were incubated for a further 10 min and then a 30-min transcription reaction was initiated with NTPs. Transcription in the two experiments was analyzed by S1 nuclease protection and autoradiography. (C) Experimental design for the analysis of time-dependent RNA synthesis from preassembled Pol I PICs in single and multiple rounds of transcription is outlined. Immobilized templates (IT-DNA), 50 μl of 50 ng of Fr4 per μl of beads, were incubated for 18 min at 4°C with 400 μl of HeLa cell NE in a 1,600-μl total reaction volume. Templates were washed in TM10i–0.05 M KCl buffer and split into two equal portions. One portion was left as it was, and to the other, 12.5 μg of ct-DNA was added to limit transcription to a single round. Transcription was initiated by the addition of NTPs. Transcription was allowed to proceed at 30°C (there is no detectable transcription at 0°C) and time points (t) were established by transfer of 25-μl aliquots (2.5 μl of IT-DNA) into a transcription stop solution. The transcript levels in the transcription time course assay were determined by S1 nuclease protection, and a single representative experiment is shown. The top autoradiograph shows the levels of initiation in multiple rounds of transcription (MR) and the bottom autoradiograph shows the levels of initiation in single-round transcription reactions (SR). (D) Transcript levels of three independent time course transcription experiments were quantified with the aid of a phosphorimager, and transcriptional activities (in arbitrary PU) for single rounds and multiple rounds were plotted against time. The inset represents the ratios of multiple- to single-round transcriptional activities over the 40-min period.
SL1 and UBF remained promoter bound in a single round and in multiple rounds of transcription.
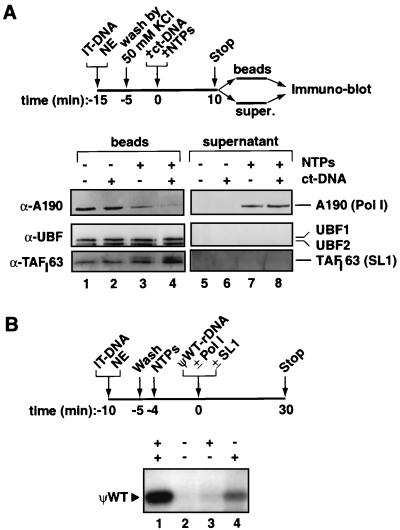
We next determined the fate of factors during transcription in which Pol I had cleared the promoter. Previous in vitro transcription experiments had suggested that SL1 and UBF remained bound to the promoter to support multiple rounds of transcription (7, 20, 27, 44). Here we demonstrated this directly by analyzing the promoter-bound and released-factor fractions in single and multiple rounds of transcription. PICs were assembled from nuclear extracts on the immobilized promoter, and these were subsequently washed to remove unbound factors. Transcription was then initiated, and promoter-bound and released factors were analyzed by immunoblotting (Fig. 6). As shown in the control reactions, the assembled PICs were stable at the competitor DNA concentration used (Fig. 6A, lanes 1 and 2), in agreement with the analyses presented in Fig. 3B. SL1 (as represented by TAFI63) and UBF1 and UBF2 remained on the template throughout the transcription reaction (Fig. 6A, lanes 7 and 3, respectively). This suggested that both SL1 and UBF were bound at the promoter all the time or were reloaded during every round of transcription. To distinguish between these possibilities, promoter occupancy by these factors in a single round of transcription was analyzed by blocking reassociation with competitor DNA. The levels of promoter-bound factors SL1 and UBF remained unaffected, and indeed no release was detectable (Fig. 6A, lanes 4 and 8, respectively). Hence, SL1 and UBF remained on the promoter template during initiation and transcript elongation by Pol I in a single round of transcription. Release of Pol I was not observed in the absence of ribonucleotides (Fig. 6A, lanes 2 and 6), or with ATP alone (data not shown), but a fraction of Pol I was released from the template in the transcription reaction as the enzymes ran off the DNA end (Fig. 6A, lanes 3 and 7). This became more apparent when reassociation of Pol I with the template during reinitiation of transcription was blocked with competitor DNA (Fig. 6A, lanes 4 and 8).
FIG. 6.
Recycling of Pol I and transcription factors SL1 and UBF during a transcription cycle. (A) Schematic representation of PIC isolation and analyses by immunoblotting of template-bound and released factors in single and multiple rounds of transcription reactions. One hundred twenty-five microliters of IT-DNA (70 ng of Fr4 per μl of beads) was incubated for 20 min at 4°C with 250 μl of nuclear extract (NE) in a total reaction volume of 1,000 μl (adjusted by TM10i buffer). Beads were washed extensively in TM10i buffer containing 50 mM KCl and afterwards were split into four samples (25 μl of beads per sample). To the reactions nonspecific DNA (2 μg of calf thymus DNA [ct-DNA]) and 0.5 mM concentrations of the ribonucleoside triphosphates (NTPs) were added as indicated. TM10i buffer was added up to a final volume of 25 μl. Tubes were incubated at 30°C for 10 min and subsequently beads were separated from supernatants with a magnet. Beads (1 M KCl extraction of IT-DNA [lanes 1 to 4]) and supernatants (lanes 5 to 8) were analyzed for Pol I factors by immunoblotting with antibodies against the largest human Pol I subunit (A190), UBF, and TAFI63 (a subunit of SL1). Samples in lanes 3 and 7 were derived from reactions that allowed for multiple rounds of transcription, whereas those in lanes 4 and 8 were from reactions that had been restricted to a single round by the addition of competitor DNA. (B) A pseudo-wild-type template (ΨWT-rDNA) (5) was used to analyze the ability of Pol I released upon transcription from one template to support initiation of transcription from another, as outlined schematically. Immobilized templates (2 μl [IT-DNA]) were incubated at 4°C for 10 min with 0.5 μl of HeLa cell NE. Templates were washed in TM10i–0.05 M KCl buffer to remove excess and unbound proteins, and transcription was initiated with NTPs. Transcription was allowed to proceed for 10 min at 30°C, and the released Pol I was assayed for the ability to support specific transcription initiation from a distinct second template, the pseudo-wild-type template which had or had not been supplemented with purified SL1 and/or Pol I. Transcription from this second template (100 ng) was allowed to proceed for 30 min at 30°C, and RNA synthesis was assayed in an S1 nuclease protection assay with an oligonucleotide specific for this pseudo-wild-type template. The pseudo-wild-type template supports accurate transcription initiation with purified factors SL1 and Pol I (lane 1). Pol I is released under transcription conditions from IT-DNA and is able to support accurate transcription initiation from the ΨWT-rDNA template supplemented with, but not without, SL1 (lanes 4 and 2, respectively). There is little transcription detectable from the ΨWT-rDNA template due to release of SL1 from IT-DNA (lane 3).
Furthermore, Pol I released from one template (wild-type rDNA promoter) and incubated with another template (pseudo-wild-type rDNA promoter), which had prebound highly purified SL1 (lacking Pol I), supported transcription initiation (Fig. 6B, lane 4). Since Pol I was able to freely partition between wild-type and pseudo-wild-type templates in these reactions, a reduced level of transcription was observed (Fig. 6B, compare lanes 4 and 1). Moreover, a basal level of transcription with just SL1 and Pol I, in the absence of UBF, was observed (Fig. 6B) (J. K. Friedrich and J. C. B. M. Zomerdijk, unpublished). The purified Pol I and SL1 fractions individually did not support transcription initiation (data not shown). The results indicated that the released Pol I complex from the first template had retained the ability to initiate transcription and suggested that the Pol I complex did not leave behind at the first template factors that it required for initiation on the second promoter.
The rate-limiting step in Pol I initiation of transcription is subsequent to PIC formation.
We next asked whether PIC formation itself or RNA synthesis is rate limiting for initiation of transcription mediated by Pol I. Note that during multiple rounds of transcription several reactions occur (for example, besides RNA synthesis, new PICs are assembled on engaged promoters and factors are recycled), and therefore under those conditions no single rate constant can be calculated using the approach described in Materials and Methods. Therefore, the rate constant of RNA synthesis was calculated from preassembled PICs in single-round transcription experiments of the kind outlined in Fig. 5. PICs were allowed to assemble for a fixed period of time, after which the transcription reactions were initiated by the addition of ribonucleotide triphosphates and competitor DNA to prevent reinitiation by Pol I. These reactions were then terminated after various periods of time. The transcript signals for three independent time course experiments were quantified (Table 1) and a rate constant for RNA synthesis of 1.94 × 10−3 ± 0.04 × 10−3 s−1 was derived.
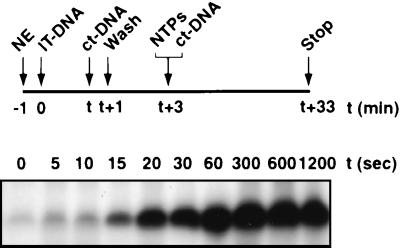
Next, we wished to determine how this rate constant compared to that for stable and functional PIC formation. In this experiment, PICs were allowed to assemble from nuclear extract on the immobilized templates for various lengths of time, after which further assembly was blocked with an excess of nonspecific DNA (Fig. 7). The immobilized template was washed to remove unbound factors and was subsequently tested for activity in a single round of transcription. Some PICs had already formed within 5 s. Quantification of the transcript signals from three independent experiments (Table 2) gave a rate constant for PIC assembly of 3.16 × 10−2 ± 0.14 × 10−2 s−1. Thus, the assembly of PICs was relatively fast, with a >1 order of magnitude (16-fold) difference in rate constant compared with that for RNA synthesis, and these data imply that in vitro a postassembly step, not PIC formation, is rate limiting in initiation of transcription by Pol I.
FIG. 7.
Analysis of the rate of Pol I PIC formation. A schematic outline of the experiments to analyze time-dependent Pol I PIC formation is presented. Immobilized template (2.5 μl of 50 ng of Fr4 per μl of beads [IT-DNA]) was incubated at 4°C with 20 μl of HeLa cell nuclear extract (NE) in an 80-μl reaction volume. The rate of assembly of PIC was not significantly different at 20°C from that of assembly at 4°C. Four micrograms of calf thymus DNA (ct-DNA) was added after various periods of time (t) to stop PIC assembly, and beads were washed in TM10i buffer containing 50 mM KCl. Transcription was initiated by the addition of ribonucleoside triphosphates (NTPs), and ct-DNA (1.25 μg) was included to limit transcription to a single round. Transcript levels from these PICs assembled in a time-dependent manner were determined by S1 nuclease protection.
DISCUSSION
Functional Pol I PICs assemble from nuclear extracts on the immobilized rDNA promoter.
We developed an immobilized-rDNA promoter template assay to analyze Pol I preinitiation complex assembly from nuclear extracts, the transcriptional activity of the PIC, its stability and disassembly during the transcription cycle, and its recycling in reinitiation of Pol I transcription. A minimal rDNA promoter encompassing the UCE and core elements (−193 to +239) is sufficient to efficiently capture functional PICs from nuclear extracts. Immunoblotting demonstrated the presence of SL1, both UBF splice variants, and Pol I. Moreover, these isolated and washed PICs supported both single and multiple rounds of transcription. The degree of binding paralleled the level of transcription from these templates, despite the relaxed sequence specificity of UBF and SL1, suggesting that the bound factors were functional and that these reflected authentic PICs. We have shown that the same amount of nonspecific DNA that prevented PIC formation at the promoter did not disrupt a preformed PIC, suggesting that cooperative interactions between factors within the PIC and between factors and DNA stabilize the complex at the promoter. For example, interactions between SL1 and UBF (5), between UBF and Pol I (45), and between SL1 and hRRN3 in Pol I (35a) have been reported. We note that the DNA binding specificity of the PICs assembled from purified SL1, UBF, and Pol I is comparable to the moderate specificity displayed by PICs isolated from nuclear extract (J. K. Friedrich, K. I. Panov, and J. C. B. M. Zomerdijk, unpublished results). Furthermore, we have demonstrated directly that UBF and SL1 remain promoter associated in single and multiple rounds of Pol I-dependent transcription. Pol I is transiently associated with the PIC and escapes the promoter to support pre-rRNA elongation and subsequent reinitiation. Pol I was the most sensitive of the components in the PIC to elevated salt concentrations and was most readily competed away with nonspecific DNA in experiments in which we limited transcription to a single round.
The rate-limiting step in rRNA gene activation in this cell-free system has been delineated. By limiting transcription to a single round, we measured rates and derived rate constants for the assembly of PICs and for those complexes to productively synthesize RNA. We found that the rate-limiting step in transcription by Pol I is postrecruitment and postassembly of the general transcription factors and Pol I.
General transcription factor behavior during the transcription cycle.
In agreement with earlier suggestions concerning the fate of transcription factors during the transcription cycle based on experiments using template commitment and competition assays with partially purified transcription factor fractions (20, 27, 44, 47), we provide direct evidence here that SL1 and UBF remain bound to the promoter in single and multiple rounds of transcription. Intriguingly, lower-resolution studies on the colocalization of Pol I transcription factors in vivo indicated that SL1, UBF, and Pol I remained associated with rDNA throughout the cell cycle, even during mitosis, when Pol I transcription was repressed (25, 42). Thus, the rRNA gene promoters appear to be “bookmarked” for reactivation upon exit from mitosis. In light of these observations and our results on the recycling of factors in a transcription cycle, the regulation of the efficiency of reinitiation by Pol I could significantly contribute to the control of rRNA gene expression.
Interestingly, SL1 remained associated with the ribosomal promoter under (elevated salt) conditions under which both forms of UBF dissociated. In the absence of UBF and Pol I, the stable SL1 and promoter DNA complex was still functional, since upon adding Pol I back initiation of transcription was restored. These results point to a key role for human SL1 in the formation of productive Pol I PICs and in the recruitment of Pol I (35a).
Reinitiation of transcription by Pol I.
Importantly, the isolated and rinsed human Pol I PICs on the immobilized templates used in this study supported very efficient reinitiation. Thus, the reinitiation intermediates at the Pol I promoter, which we have shown included SL1 and UBF, acted to recruit Pol I to form functional reinitiation complexes. This may be unique to the system under study, as, for example, prewashed Pol II PICs captured from yeast extracts on beaded-promoter templates supported transcription to levels reminiscent of only a single round of transcription (39). We could not detect a specific dissociation of reinitiation activity upon more extensive washing (at 50 mM KCl) of the immobilized-template PICs. In fact, under those conditions progressively more Pol I dissociated from the PICs and an accompanying decrease in transcription, both in single and in multiple rounds, was observed (data not shown). The efficient reinitiation of transcription without apparent loss over a 40-min period suggests that the Pol I components are efficiently recycled and reactivated by as yet unknown factors that associate with and survive the conditions of PIC isolation on these immobilized templates. However, we have observed a dramatically reduced efficiency of reinitiation of transcription supported by PICs assembled from purified factors rather than from a nuclear extract (K. I. Panov, G. Miller, and J. C. B. M. Zomerdijk, unpublished results). This underscores the presence in the nuclear extract of distinct activities which support reinitiation, and these are currently under investigation (T. Kasciukovic, G. Miller, K. I. Panov, and J. C. B. M. Zomerdijk, unpublished results). Reinitiation is likely to present a pivotal point of control in rRNA gene expression during cell growth, proliferation, and differentiation.
A postrecruitment step, promoter clearance, limits the rate of Pol I-dependent transcription.
The transcription cycle comprises multiple steps. We have determined the kinetic parameters of PIC formation and RNA synthesis. The rate of RNA synthesis from preformed PICs is determined by the rate of promoter clearance and the rate of elongation, promoter clearance being a multistage event comprising promoter opening, initiation, and promoter escape. However, this is correct only under conditions where no reinitiations occur. Therefore, for these kinetic studies we limited transcription to a single round. We added competitor DNA after PIC assembly to limit transcription to a single round, as it prevented reassociation of Pol I with SL1 and UBF at the promoter. Indeed, we have demonstrated that a precisely titrated amount of competitor DNA reduced and limited transcription principally to the first few minutes, with little RNA synthesis thereafter. Moreover, an intermediate concentration of competitor DNA primarily inhibited Pol I activity, since transcription was rescued by additional Pol I and not by SL1 or UBF.
Under our experimental conditions, functional PICs assembled with an observed rate constant of about 0.032 s−1, which was only threefold below that for Pol II and purified Pol II general transcription factors (29). This difference may be innate to the particular transcription machineries. Importantly, the rate constant for RNA synthesis from preassembled Pol I PICs (0.002 s−1) was over 1 order of magnitude lower than that for PIC formation. Intriguingly, this rate constant for RNA synthesis is almost identical to that determined for Pol II (29). Elongation of Pol I transcription is relatively fast. In rodent extracts, the rate of transcript elongation by Pol I was about 2 nt/s (15), and it was 30 nt/s for highly purified mouse Pol I (6). The latter converts to a rate constant of >0.03 s−1, again very similar to that determined for Pol II (29). Thus, since RNA synthesis and not PIC assembly is rate limiting in Pol I transcription and elongation is apparently fast, we conclude that Pol I-dependent transcription, like Pol II-mediated transcription, is rate limited in promoter clearance. We speculate that this step is likely to be a target for Pol I transcriptional regulators. Future research will be directed to understanding detailed mechanisms and identifying factors that may modulate this evidently important and rate-limiting step in the control of rRNA gene expression.
ACKNOWLEDGMENTS
We thank Struan Wilkie for technical assistance and Stefan Roberts for advice during the development of the immobilized-template assay. We thank Brian McStay for providing us with antibodies against human UBF. We thank the National Cell Culture Center (Minneapolis, Minn.) for growing HeLa cells. We thank our colleagues in the Zomerdijk laboratory and Tom Owen-Hughes, Neil Perkins, Stefan Roberts, and Jackie Russell for advice and critical reading of the manuscript.
J.K.F. received an MRC Ph. D. studentship. J.C.B.M.Z. is a Wellcome Trust Senior Research Fellow in the Basic Biomedical Sciences.
REFERENCES
- 1.Albert A-C, Denton M, Kermekchiev M, Pikaard C S. Histone acetyltransferase and protein kinase activities copurify with a putative Xenopus RNA polymerase I holoenzyme self-sufficient for promoter-dependent transcription. Mol Cell Biol. 1999;19:796–806. doi: 10.1128/mcb.19.1.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias J A, Dynan W S. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J Biol Chem. 1989;264:3223–3229. [PubMed] [Google Scholar]
- 3.Beckmann H, Chen J L, O'Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Jantzen H M, Tjian R. Assembly of alternative multiprotein complexes directs rRNA promoter selectivity. Genes Dev. 1990;4:943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- 5.Bell S P, Learned R M, Jantzen H M, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 6.Cavanaugh A H, Thompson E A., Jr Hormonal regulation of transcription of rDNA: glucocorticoid effects upon initiation and elongation in vitro. Nucleic Acids Res. 1985;13:3357–3369. doi: 10.1093/nar/13.9.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cizewski V, Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983;11:7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 9.Comai L, Zomerdijk J C, Beckmann H, Zhou S, Admon A, Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994;266:1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 10.Copenhaver G P, Putnam C D, Denton M L, Pikaard C S. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994;22:2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs R L. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 13.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 14.Hadjiolov A A. The nucleolus and ribosome biogenesis. Cell Biol Monogr. 1985;12:1–290. [Google Scholar]
- 15.Haglund R E, Rothblum L I. Isolation, fractionation and reconstitution of a nuclear extract capable of transcribing ribosomal DNA. Mol Cell Biochem. 1987;73:11–20. doi: 10.1007/BF00229371. [DOI] [PubMed] [Google Scholar]
- 16.Haltiner M M, Smale S T, Tjian R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol. 1986;6:227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haltiner-Jones M, Learned R M, Tjian R. Analysis of clustered point mutations in the human ribosomal RNA gene promoter by transient expression in vivo. Proc Natl Acad Sci USA. 1988;85:669–673. doi: 10.1073/pnas.85.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannan K M, Hannan R D, Rothblum L I. Transcription by RNA polymerase I. Front Biosci. 1998;3:d376–d398. doi: 10.2741/a282. http://www.bioscience.org/1998/v3/d/hannan/list.htm . [Online.] http://www.bioscience.org/1998/v3/d/hannan/list.htm. . [DOI] [PubMed] [Google Scholar]
- 19.Hannan R D, Cavanaugh A, Hempel W M, Moss T, Rothblum L. Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res. 1999;27:3720–3727. doi: 10.1093/nar/27.18.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson S L, Ryan K, Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989;3:212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- 21.Hisatake K, Nishimura T, Maeda Y, Hanada K, Song C Z, Muramatsu M. Cloning and structural analysis of cDNA and the gene for mouse transcription factor UBF. Nucleic Acids Res. 1991;19:4631–4637. doi: 10.1093/nar/19.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu C H, McStay B, Jeong S W, Reeder R H. xUBF, an RNA polymerase I transcription factor, binds crossover DNA with low sequence specificity. Mol Cell Biol. 1994;14:2871–2882. doi: 10.1128/mcb.14.5.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob S T, Ghosh A K. Control of RNA polymerase I-directed transcription: recent trends. J Cell Biochem. 1999;32–33(Suppl.):41–50. doi: 10.1002/(sici)1097-4644(1999)75:32+<41::aid-jcb6>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 24.Jantzen H M, Admon A, Bell S P, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 25.Jordan P, Mannervik M, Tora L, Carmo Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasher M S, Pintel D, Ward D C. Rapid enrichment of HeLa transcription factors IIIB and IIIC by using affinity chromatography based on avidin-biotin interactions. Mol Cell Biol. 1986;6:3117–3127. doi: 10.1128/mcb.6.9.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato H, Nagamine M, Kominami R, Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986;6:3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingston R E, Green M R. Modeling eukaryotic transcriptional activation. Curr Biol. 1994;4:325–332. doi: 10.1016/s0960-9822(00)00071-3. [DOI] [PubMed] [Google Scholar]
- 29.Kugel J F, Goodrich J A. Promoter escape limits the rate of RNA polymerase II transcription and is enhanced by TFIIE, TFIIH, and ATP on negatively supercoiled DNA. Proc Natl Acad Sci USA. 1998;95:9232–9237. doi: 10.1073/pnas.95.16.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn A, Voit R, Stefanovsky V, Evers R, Bianchi M, Grummt I. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 1994;13:416–424. doi: 10.1002/j.1460-2075.1994.tb06276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langst G, Blank T A, Becker P B, Grummt I. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Learned R M, Cordes S, Tjian R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol. 1985;5:1358–1369. doi: 10.1128/mcb.5.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Learned R M, Learned T K, Haltiner M M, Tjian R T. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 35.McStay B, Hu C H, Pikaard C S, Reeder R H. xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X. laevis. EMBO J. 1991;10:2297–2303. doi: 10.1002/j.1460-2075.1991.tb07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. B. M. Zomerdijk. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 36.O'Mahony D J, Smith S D, Xie W, Rothblum L I. Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paule M R. Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Berlin, Germany: Springer-Verlag; 1998. [Google Scholar]
- 38.Putnam C D, Copenhaver G P, Denton M L, Pikaard C S. The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol Cell Biol. 1994;14:6476–6488. doi: 10.1128/mcb.14.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranish J A, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeder R H. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 41.Reeder R H, Pikaard C S, McStay B. UBF, an architectural element for RNA polymerase I promoters. Nucleic Acids Mol Biol. 1995;9:251–263. [Google Scholar]
- 42.Roussel P, Andre C, Comai L, Hernandez Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SaezVasquez J, Pikaard C S. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc Natl Acad Sci USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnapp A, Grummt I. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem. 1991;266:24588–24595. [PubMed] [Google Scholar]
- 45.Schnapp G, Santori F, Carles C, Riva M, Grummt I. The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J. 1994;13:190–199. doi: 10.1002/j.1460-2075.1994.tb06248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seither P, Iben S, Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J Mol Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- 47.Wandelt C, Grummt I. Formation of stable preinitiation complexes is a prerequisite for ribosomal DNA transcription in vitro. Nucleic Acids Res. 1983;11:3795–3809. doi: 10.1093/nar/11.11.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zomerdijk J C B M, Beckmann H, Comai L, Tjian R. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science. 1994;266:2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]
- 49.Zomerdijk J C B M, Tjian R. Structure and assembly of human selectivity factor SL1. In: Paule M R, editor. Transcription of eukaryotic ribosomal RNA genes by RNA polymerase I. Berlin, Germany: Springer-Verlag; 1998. pp. 67–73. [Google Scholar]