Abstract
Chromosomal double-strand breaks (DSBs) stimulate homologous recombination by several orders of magnitude in mammalian cells, including murine embryonic stem (ES) cells, but the efficiency of recombination decreases as the heterology between the repair substrates increases (B. Elliott, C. Richardson, J. Winderbaum, J. A. Nickoloff, and M. Jasin, Mol. Cell. Biol. 18:93–101, 1998). We have now examined homologous recombination in mismatch repair (MMR)-defective ES cells to investigate both the frequency of recombination and the outcome of events. Using cells with a targeted mutation in the msh2 gene, we found that the barrier to recombination between diverged substrates is relaxed for both gene targeting and intrachromosomal recombination. Thus, substrates with 1.5% divergence are 10-fold more likely to undergo DSB-promoted recombination in Msh2−/− cells than in wild-type cells. Although mutant cells can repair DSBs efficiently, examination of gene conversion tracts in recombinants demonstrates that they cannot efficiently correct mismatched heteroduplex DNA (hDNA) that is formed adjacent to the DSB. As a result, >20-fold more of the recombinants derived from mutant cells have uncorrected tracts compared with recombinants from wild-type cells. The results indicate that gene conversion repair of DSBs in mammalian cells frequently involves mismatch correction of hDNA rather than double-strand gap formation. In cells with MMR defects, therefore, aberrant recombinational repair may be an additional mechanism that contributes to genomic instability and possibly tumorigenesis.
Caretaker genes, such as the genes in the mismatch repair (MMR) and nucleotide excision (NER) pathways, are involved in the repair of DNA lesions (6, 12). Loss of function of a caretaker gene can result in a mutator phenotype, thus allowing the numerous mutations necessary for tumorigenesis to accumulate. Such a mutator phenotype is now seen as an important contributor to tumorigenesis (23, 27, 34). Defects in MMR lead to the inherited cancer syndrome hereditary nonpolyposis colorectal cancer (HNPCC) and possibly to a small proportion of sporadic colorectal cancers (references 5 and 6 and references therein), while defects in NER lead to xeroderma pigmentosum (12). In HNPCC, an autosomal dominant disease, one mutated allele of a gene involved in MMR is inherited and a somatic mutation occurs in the remaining wild-type allele, thus promoting colorectal cancer. Mutations in two MMR genes, MSH2 and MLH1, have been typically associated with HNPCC, while mutations in other MMR genes (MSH6, PMS1, and PMS2) are rarer. HNPCC tumors have frequent length alterations of microsatellites, such as contractions or expansions of mononucleotide repeats, which is referred to as microsatellite instability and which is due to the lack of repair of slipped replication intermediates.
The MMR pathway is a conserved pathway which maintains genomic integrity in procaryotes and eucaryotes. The Escherichia coli MutHLS MMR pathway has been well-characterized genetically and biochemically (37) and has served as a paradigm for the yeast and mammalian MMR pathways (reviewed in reference 6). A number of homologs of MutS and MutL have been found in yeast and mammalian cells. The MutS homologs Msh2 and Msh6 form a heterodimer known as MutSα that functions in the repair of single-base mismatches and small (1 bp) insertion-deletion loops (16, 25). Msh2 also pairs with another MutS homolog, Msh3, to form a heterodimer known as MutSβ, which is involved in the repair of larger (2 to 4 bp) insertion-deletion loops (2, 35). Msh2, therefore, plays a central role in eucaryotic MMR since it is responsible for the repair of all mismatches, while Msh6 and Msh3 act to determine the specific types of mismatches that are recognized.
In addition to their role in the repair of replication errors, MMR components have been implicated in a second DNA repair pathway, homologous recombination. In mammalian cells, as in other organisms, homologous recombination is well-established as one of the major pathways for the repair of DNA double-strand breaks (DSBs) (32, 40). As a result of its role in DSB repair, homologous recombination is stimulated 2 to 3 orders of magnitude by a DSB in the genome (33, 48, 50). We have found that the frequency of DSB-induced recombination in mammalian cells can be affected by relatively small degrees of sequence heterology (17). For example, recombination is decreased by approximately sixfold between sequences that are 1.2% divergent. During recombination, a gene conversion tract (GCT) around the DSB is formed, where there is a unidirectional transfer of sequence information from the unbroken donor DNA molecule to the broken DNA molecule. In principal, gene conversion can result from either mismatch correction of heteroduplex DNA (hDNA) or from the processing of a DSB to a gap, such that the only information available is from the donor DNA molecule. In yeast, chromosome ends at DSBs appear to be highly protected (21), such that most DSB-induced gene conversion involves mismatch correction of hDNA (44, 58).
MMR components function in recombination by suppressing recombination between diverged (homologous) sequences, a role that appears to be conserved in bacteria, yeast, and in mammalian cells (reviewed in reference 37). For example, E. coli strains deficient for MutS or MutL have lost the barrier to recombination between diverged sequences on the same chromosome as well as between genera, resulting in chromosomal rearrangements and intergeneric crosses, respectively (43, 45). As a result, these MMR-deficient strains exhibit a “recombinator phenotype” in addition to their well-characterized mutator phenotype, thus contributing an additional level of genetic instability to these mutants. Similarly, in Saccharomyces cerevisiae, loss of MMR proteins significantly increases homeologous recombination between direct repeats (8, 10, 11, 52), as well gene targeting of homeologous sequences (29, 39).
In mammals, Msh2-deficient embryonic stem (ES) murine cells have been shown to be promiscuous for recombination between diverged sequences in gene-targeting experiments (1, 13). This role of MMR proteins in suppressing recombination between homeologous sequences may be particularly important in mammalian cells, since repetitive elements are naturally occurring diverged sequences that make up a large fraction of mammalian genomes. For example, human cells contain approximately 106 copies of Alu elements, which are around 300 bp and are 70 to 98% homologous to the consensus Alu sequence (51). Although rare, Alu-Alu-mediated recombination has been reported for many diseases with deleterious consequences (reviewed in reference 9), such as in acute myeloid leukemia, where recombination between two diverged Alu elements in the ALL1 gene results in a partial gene duplication (54).
The large stimulation of recombination by DSBs led us to investigate the effect of MMR deficiency on DSB-induced recombination in mammalian cells. In this report, we investigate Msh2−/− ES cells for DSB-induced gene-targeting and intrachromosomal recombination between diverged substrates. Both the frequences of recombination and the GCTs are examined, the latter so as to determine whether gene conversion at a DSB occurs primarily by gap repair or MMR of hDNA. Our goal is to begin to ascertain what genomic alterations in addition to unrepaired replication errors could be potentiated in MMR-deficient cells, which may therefore have implications for tumorigenesis in HNPCC.
MATERIALS AND METHODS
DNA and cell line constructions.
Construction of the pneo gene-targeting plasmids, pneo-WT, pneo-5mu, and pneo-8mu, has been described previously (17). The pneo-10mu substrate was made by PCR-induced site-directed mutagenesis as follows: the pneo-8mu plasmid was amplified with primer 1B (5′CAGTCGATGAATCCGGAAAAGCGGCCAT) and primer 4 (5′ACCATGATTACGCCAAGCTT) and with primer 2B (5′CCGCTTTTCCGGATTCATCGACTGTGGC) and primer 3 (5′TGCTCGACGTTGTCACTGAA). The integrity of the neo sequence of the pneo-10mu plasmid was checked in both directions with three primers (Bio Resource Center, Cornell University, Ithaca, N.Y.). To make the intrachromosomal constructs, the 0.8-kb SacI-XbaI fragments derived from the pneo-WT and pneo-8mu plasmids were subcloned into the S2neo/pgkhygo plasmid (33). The pgkhygro gene was replaced by a puro gene (26) from pHA262pur (a kind gift of Hein te Riele, Netherlands Cancer Institute, Amsterdam, The Netherlands). A 1.6-kb PvuII-PvuII fragment of the HPRT gene from pGPD351 (15) and a 2.3-kb XbaI-XbaI fragment of the HPRT gene from the same plasmid were subcloned to flank the S2neo/pgkpuro/pneo repeat, forming 5′ and 3′ targeting arms, respectively.
Cell culture and transfections.
The Msh2−/− mutant cell line dMsh2-9 (a kind gift of Hein te Riele) was derived from the E14 ES cell line, which served as a control in our experiments and has both alleles of msh2 disrupted by the hyg marker (13). All cells were cultured on gelatin-coated dishes in standard medium supplemented with 103 U of leukemia inhibitory factor (GIBCO/Life Technologies)/ml as previously described (47). For the construction of cell lines for gene-targeting assays, the Msh2−/− mutant cells were electroporated with 60 μg of the S2neo gene on a plasmid along with 20 μg of a plasmid containing the pgk-puro gene. Doubly resistant hyg and puro colonies were isolated and tested by Southern blot analysis for the presence of a single copy of S2neo. Possibly integrated into the genome of wild-type and Msh2−/− cells adjacent to the S2neo gene are bacterial vector sequences which would have homology to vector sequences in the pneo gene-targeting plasmids. This homology would be interrupted, however, by sequences unique to the S2neo gene, since it extends 0.3 kb 5′ and 0.1 kb 3′ beyond the pneo homology. For the construction of cell lines for the intrachromosomal recombination assays, the H-DR-WT and the H-DR-8mu constructs were integrated into the HPRT locus in murine ES wild-type and Msh2−/− cells by electroporation of 15 to 20 μg of the SacI-XhoI-digested H-DR-WT (or 8mu) construct. Colonies were selected in 6-thioguanine and puromycin, and the resulting cell lines were checked by Southern blot analysis, as shown, for the correct integration of the construct.
For each sample of cells in the gene-targeting and intrachromosomal recombination assays, 2 × 107 cells in 1 ml of phosphate-buffered saline were electroporated with 20 to 25 μg of each uncut plasmid DNA in a 0.4-cm electrode-gap cuvette (250 V, 960 μF). Electroporated cells were aliquoted into four or five 10-cm-diameter dishes. Colonies were selected in media with one or more of the following drugs 20 to 24 h after electroporation and were grown in selection media for 10 to 14 days before colony counts (or after colony expansion): G418 (200 μg/ml), hygromycin (150 μg/ml), puromycin (1.6 μg/ml), or 6-thioguanine (4 μg/ml).
PCR analysis.
A region of the chromosomal neo gene in neo+ intrachromosomal recombinants from wild-type and Msh2−/− cells containing the H-DR-8mu construct was PCR amplified with primers Neol (5′GCCAATATGGATCGGCCATTGAACAA) and Neo3 (5′CCTCAGAAGAACTCGTCAAGA). Amplification was performed by denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and then extension at 72°C for 15 min. Amplified products were digested with I-SceI and appropriate restriction enzymes and were electrophoresed on 2% agarose gels (25% agarose–75% Nusieve agarose).
RESULTS
Gene targeting with diverged repair substrates in wild-type and Msh2−/− ES cells.
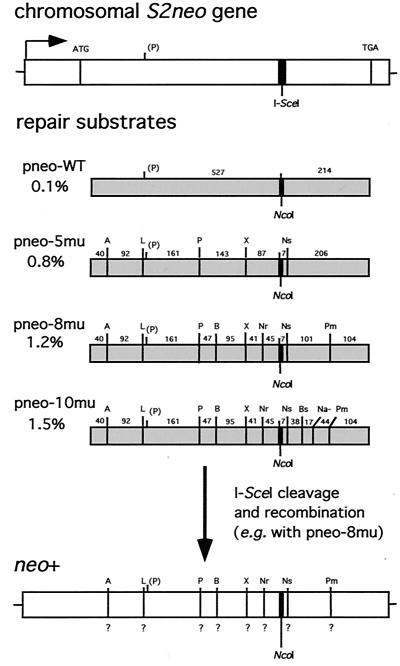
We have previously found in a gene-targeting assay with wild-type ES cells that the efficiency of homologous repair of a DSB decreases as the divergence of the homologous repair substrate increases (17). Using the same assay, we wanted to determine whether the effects of heterology would be overcome in an MMR-deficient genetic background. The gene-targeting assay consists of three components: a chromosomally integrated nonfunctional neo gene (termed S2neo) which contains the 18-bp I-SceI endonuclease cleavage site, diverged neo repair substrates on circular plasmids, and an expression plasmid for the I-SceI endonuclease (Fig. 1). With this design, a DSB introduced into the S2neo gene at the I-SceI site can be repaired from the homologous repair substrates to restore a neo+ gene.
FIG. 1.
DSB-induced gene-targeting strategy. The S2neo gene which is integrated into the genome of wild-type and Msh2−/− murine ES cells contains the 18-bp I-SceI site (thick vertical bar) which can be cleaved in vivo by the I-SceI endonuclease. The S2neo promoter (arrow) is derived from polyomavirus (strain F441) and the herpesvirus thymidine kinase gene (55). The repair substrates are internal neo sequences which are 745 bp in length. The percent divergence of each of these sequences relative to the same region in S2neo is indicated. For simplicity, heterology at the NcoI–I-SceI sites is considered to be one change within the 745-bp segment, although recombination involves loss of the 18-bp cleavage site at this position and a gain of 4 bp at the NcoI site (17). Silent mutations create the following restriction enzyme site polymorphisms in the neo sequence: A, ApaI; L, ApaLI; P, PstI; B, BamHI; X, XbaI; Nr, NruI; Ns, NsiI; Bs, BspEI; Na−, loss of NaeI; Pm, PmlI. Note that there is a naturally occurring PstI site within the neo sequence. Distances (in base pairs) between the 1-bp silent mutations are indicated. For DSB-induced gene targeting, an expression vector for the I-SceI endonuclease and a repair substrate on a plasmid are transfected into wild-type and Msh2−/− cells containing the S2neo gene. One example of the type of analysis after recombination is shown after targeting with pneo-8mu. Since a neo+ gene is selected, the correcting neo gene sequence at the NcoI site has been incorporated. As recombination may occur with or without conversion of adjacent sequences, the presence of the 8 silent mutations (?) was determined by restriction analysis.
The repair substrates, which diverged from S2neo in the range of 0.1 to 1.5%, each consist of an internal neo gene fragment restricting selected recombination events to noncrossover gene conversions. Each substrate had the correcting NcoI site at the position of the I-SceI site in the S2neo gene. Divergence was created in the 745-bp neo fragments by substitutions at third-base codon positions that create phenotypically silent mutations as well as restriction enzyme site polymorphisms (56). Plasmids pneo-5mu and pneo-8mu have neo gene fragments with five and eight silent mutations, respectively, as described previously (17). A new repair substrate with more divergence was created for this study by the addition of two silent mutations downstream of the NcoI and NsiI sites in pneo-8mu. With this new substrate, called pneo-10mu, there were similar densities of heterology both 5′ and 3′ of the correcting NcoI site.
For our assays, we used two wild-type ES cell lines, clones 12 and 5, each of which have a single copy of the S2neo gene randomly integrated into the genome (53; data not shown). To create Msh2−/− ES cell lines containing the S2neo gene, we cotransfected the S2neo gene on a plasmid along with the puromycin N-acetyltransferase gene (pgk-puro) on a separate plasmid into Msh2-deficient ES cells which had both alleles of the msh2 gene disrupted by the hygromycin (hyg) resistance gene (13). Cells were selected in both puromycin and hygromycin, and 28 doubly resistant colonies were isolated. Colonies were screened by Southern blotting for the presence of a single copy of the S2neo gene (data not shown). Four independent Msh2−/− cell lines obtained in this screen (termed clones 14, 18, 28, and 30) were used in subsequent analysis.
Effect of heterology on the frequency of gene targeting.
To examine the effect of heterology on DSB-induced gene targeting, wild-type and Msh2−/− cell lines were electroporated with the I-SceI expression vector pPGK3xnlsI-SceI and each of the pneo repair substrates. As controls, cells were electroporated with these plasmids separately, and cells were also electroporated with pMC1neo, a plasmid containing a functional neo gene. Cells were selected with G418, and neo+ (G418r) recombinant colonies were scored 10 to 14 days later. Control electroporations of the I-SceI expression vector or any of the pneo repair substrates alone yielded very few or no neo+ clones from any of the cell lines (≤10−7), yet neo+ clones were readily obtained from electroporations of the two plasmids together (Table 1; data not shown). As previously seen with the pneo-WT substrate, DSBs resulting from I-SceI expression stimulated gene targeting more than 3 orders of magnitude in the wild-type ES cells (17). This large increase in stimulation was also observed in the Msh2−/− cells. The absolute frequency of targeting was subject to some clonal variation, but overall the range of gene-targeting frequencies with pneo-WT was similar for both wild-type and Msh2 parental cell lines (Table 1).
TABLE 1.
Gene targeting in wild-type cells (Msh2+/+) and Msh2−/− cells with diverged repair substrates
| Cell line and transfectant pneo and pPGK3xnlsI-SceI | Frequency of neo+ colonies (% wild type)a
|
|||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Expt 4 | |
| Clone 12 (WT)b | ||||
| pneo-WT | 1.3 × 10−4 (100) | 1.9 × 10−4 (100) | 2.9 × 10−4 (100) | 2.9 × 10−4 (100) |
| pneo-5mu | 4.7 × 10−5 (37) | 7.9 × 10−5 (41) | 1.3 × 10−4 (44) | 1.4 × 10−4 (47) |
| pneo-8mu | 2.1 × 10−5 (17) | 6.2 × 10−5 (30) | ND | ND |
| pneo-10mu | NDc | ND | 3.4 × 10−5 (12) | 1.5 × 10−5 (5) |
| Clone 14 (Msh2−/−)b | ||||
| pneo-WT | 5.9 × 10−5 (100) | ND | 1.1 × 10−4 (100) | 1.1 × 10−4 (100) |
| pneo-5mu | 3.6 × 10−5 (62) | ND | 1.2 × 10−4 (110) | 9.9 × 10−5 (93) |
| pneo-8mu | 3.4 × 10−5 (58) | ND | ND | ND |
| pneo-10mu | ND | ND | 8.7 × 10−5 (82) | 5.7 × 10−5 (53) |
| Clone 28 (Msh2−/−) | ||||
| pneo-WT | 2.6 × 10−5 (100) | 7.2 × 10−5 (100) | ND | 1.1 × 10−4 (100) |
| pneo-5mu | 1.8 × 10−5 (68) | 6.1 × 10−5 (85) | ND | 7.7 × 10−5 (71) |
| pneo-8mu | 1.5 × 10−5 (56) | 5.6 × 10−5 (78) | ND | ND |
| pneo-10mu | ND | ND | ND | 4.9 × 10−5 (45) |
| Clone 30 (Msh2−/−) | ||||
| pneo-WT | 4.3 × 10−5 (100) | 9.0 × 10−5 (100) | 1.4 × 10−4 (100) | ND |
| pneo-5mu | 4.2 × 10−5 (98) | 7.7 × 10−5 (86) | 1.2 × 10−4 (87) | ND |
| pneo-8mu | 3.8 × 10−5 (85) | 6.8 × 10−5 (76) | ND | ND |
| pneo-10mu | ND | ND | 7.1 × 10−5 (50) | ND |
| pMC1neod | ||||
| Clone 12 (WT) | 4.1 × 10−4 | 1.9 × 10−2 | 1.8 × 10−3 | 2.0 × 10−3 |
| Clone 14 (Msh2−/−) | ND | ND | 9.7 × 10−4 | 1.6 × 10−3 |
| Clone 28 (Msh2−/−) | 4.7 × 10−4 | ND | ND | 1.9 × 10−3 |
| Clone 30 (Msh2−/−) | 4.2 × 10−4 | ND | 2.0 × 10−3 | ND |
The frequency of neo+ colonies was calculated by dividing the number of neo+ cells observed after transfection by the number of electroporated cells (2 × 107), correcting for 50% viability of cells after electroporation. % Wild type, percent freqency of neo+ colonies from transfection with diverged repair substrate and pPGK3xnlsI-SceI divided by frequency of neo+ colonies from transfection with pneo-WT and pPGK3xnlsI-SceI and multiplied by 100.
Not shown are wild type (WT) clone 5 and clone 18 (Msh2−/−), both of which showed a similar decrease in frequency of recombination as wild-type clone 12 and clones 14, 28, and 30 (Msh2−/−), respectively, as the heterology of repair substrates increased. The absolute frequency of recombination for clone 5 was somewhat lower than that for clone 12, although the relative frequencies of the four substrates were similar.
ND, not determined.
The frequency of neo+ colonies was calculated by plating a dilution (1:50) of cells after electroporation of 2 × 107 cells.
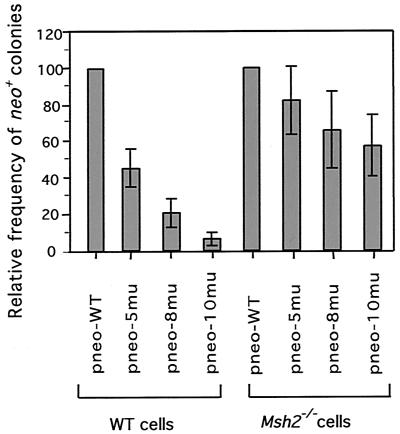
By examining DSB-promoted recombination with the diverged substrates, we found that in wild-type cells, the frequency of neo+ clones decreased as the repair substrates became more diverged from the S2neo gene (Table 1). The mean decrease in frequency from that of pneo-WT was 2.2-fold for pneo-5mu (0.8% divergent) and 4.8-fold for pneo-8mu (1.2% divergent) (Fig. 2), similar to previous results (17). Recombination with pneo-10mu (1.5% divergent) was even more dramatically decreased, i.e., by 16.7-fold. Thus, the addition of two silent mutations in the 101-bp stretch of perfect homology just downstream of the DSB significantly decreased the frequency of recombination.
FIG. 2.
Relative gene-targeting efficiencies of diverged substrates. The frequency of neo+ colonies after DSB-induced recombination was plotted for cell lines transfected with each of the diverged pneo substrates relative to frequencies of cell lines transfected with the pneo-WT substrate. In each case, the transfection included pPGK3xnlsI-SceI to induce a DSB at the S2neo gene. The means of seven independent experiments are shown with error bars indicating the standard deviations. WT, wild type.
In contrast to results obtained with wild-type ES cells, in Msh−/− cells, the frequencies of neo+ colonies obtained with each of the three diverged repair substrates were only slightly reduced from the frequency obtained with the pneo-WT control (Table 1). Substrates pneo-5mu and pneo-8mu had mean decreases of 1.2- and 1.5-fold in the frequency of recombination, respectively, while pneo-10mu had only a 1.7-fold decrease. Thus, Msh2−/− cells had 10-fold more recombination with pneo-10mu relative to recombination with pneo-WT than did wild-type cells, threefold more recombination with pneo-8mu, and twofold more recombination with pneo-5mu (Fig. 2). Clearly, the barrier to recombination imposed by heterology is significantly overcome in Msh2-deficient cells.
Intrachromosomal recombination with diverged repair substrates in wild-type and Msh2−/− ES cells.
Gene targeting provides a rapid assay to evaluate the effect of sequence divergence, but a limitation is that one of the recombining partners is on a plasmid. We therefore decided to examine the effect of sequence divergence in a more physiological setting in which both neo partners would be integrated in chromosomal DNA. For this, we constructed intrachromosomal recombination substrates and examined both the frequency and products of recombination. So as to eliminate possible position effects on recombination, we integrated the neo genes at the same chromosomal locus in wild-type and Msh2−/− cells. The locus we chose for integration is the X-linked hypoxanthine phosphoribosyltransferase (HPRT) locus since integrations which disrupt the HPRT gene can be selected in XY murine ES cells by using the base analog 6-thioguanine.
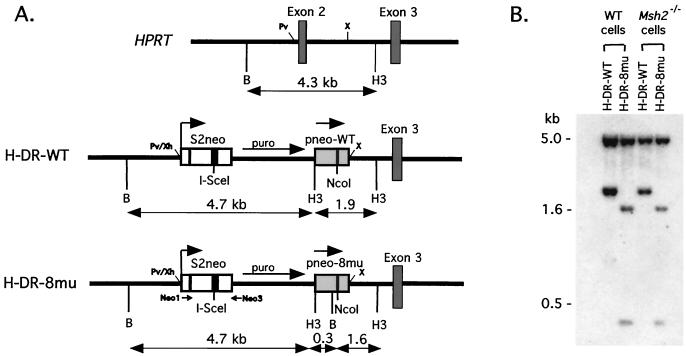
The site of integration at the HPRT locus and the intrachromosomal recombination substrates are shown in Fig. 3A. The recombination substrates contained the S2neo gene and a repair template derived from a pneo plasmid, which were oriented as direct repeats and separated by the pgk-puro gene. Two recombination substrates were constructed which differed by the pneo-derived repair substrate. In the control, H-DR-WT (HPRT–direct repeat–pneo-WT), the internal neo fragment was derived from pneo-WT, whereas in the test construct H-DR-8mu (HPRT–direct repeat–pneo-8mu), it was derived from pneo-8mu. As in the gene-targeting experiments, DSB-promoted recombination between an I-SceI-cleaved S2neo gene and the internal neo fragment resulted in a neo+ gene. Because crossover in these intrachromosomal recombination substrates would have resulted in a truncated neo gene, only recombinants which had undergone a noncrossover gene conversion were selected, thus simplifying the interpretation of our results to one repair pathway.
FIG. 3.
Intrachromosomal recombination substrates integrated at the HPRT locus. (A) The HPRT locus at exons 2 and 3 is shown along with gene-targeted HPRT loci containing intrachromosomal repair substrates. The H-DR-WT substrate contains a direct neo repeat of S2neo and pneo-WT, and the H-DR-8mu substrate contains a direct neo repeat of S2neo and pneo-8mu (Fig. 1). Separating the neo repeat in both substrates is a pgk-puro gene for selection. Repair substrates were cloned into the PvuII (Pv) and XbaI (X) sites of the HPRT locus, as indicated, using XhoI (Xh) and XbaI sites of the substrates, and this deleted exon 2. Distances between the BamHI (B) and HindIII (H3) sites are shown in kilobases. Neo1 and Neo3 indicate the primers used for PCR amplification. Neo1 has no overlap with pneo-8mu, and Neo3 has an overlap of 6 nucleotides. (B) Southern blot analysis of BamHI-HindII-digested genomic DNA indicated that the intrachromosomal substrates were correctly targeted and had integrated as a single copy in the wild-type and Msh2−/− cell lines. The probe was a 1.1-kb XhoI-HindIII fragment containing the entire neo+ gene. Genomic DNA from parental wild-type and Msh2−/− cells did not hybridize with this probe, as expected (data not shown).
The intrachromosomal recombination substrates were electroporated into wild-type and Msh2−/− ES cells. Cells were selected in puromycin and 6-thioguanine, and doubly resistant colonies were picked 10 to 14 days later. Clones were examined by Southern blotting, and the majority (83%) had correctly integrated the substrates into the HPRT locus (Fig. 3B). For each of the wild-type and Msh2−/− ES cell lines, two or three clones containing the H-DR-WT or H-DR-8mu constructs were further characterized.
Effect of heterology on the frequency of intrachromosomal recombination.
To examine homologous recombination between the two neo repeats, cells were electroporated with the I-SceI expression vector or a control vector, pPGKlacZ. Selection was in medium containing G418, and G418r (neo+) colonies were isolated 10 to 14 days later. Results from these experiments are shown in Table 2. As with gene targeting, DSBs induced intrachromosomal recombination in both wild-type and Msh2−/− ES cells, resulting in a similar frequency of recombination for both cell lines. The absolute frequencies of DSB-induced intrachromosomal recombination and gene targeting with the pneo-WT repair substrate were similar (approximately 2 × 10−4). However, since the spontaneous level of intrachromosomal recombination was readily detectable, unlike in our gene-targeting assay, DSBs stimulated intrachromosomal recombination only about 2 orders of magnitude, rather than the >3 orders of magnitude seen in the gene-targeting experiments.
TABLE 2.
Intrachromosomal recombination in wild-type (Msh2+/+) and Msh2−/− cells
| Cell line | Frequency of neo+ recombinants (10−4)a
|
|
|---|---|---|
| DSB inducedb | Spontaneousc | |
| Wild-type clones | ||
| H-DR-WT | 1.8 ± 0.9 | 0.05 ± 0.05 |
| H-DR-8mu | 0.4 ± 0.1 | 0.004 ± 0.005 |
| Msh2−/− clones | ||
| H-DR-WT | 1.9 ± 0.5 | 0.08 ± 0.06 |
| H-DR-8mu | 1.6 ± 0.3 | 0.05 ± 0.02 |
Data are averages of four experiments with standard deviations, using two or three independently derived clones for each cell line.
Cells were transfected with the I-SceI expression vector, pPGK3xnlsI-SceI.
Cells were transfected with a control plasmid, pPGKlacZ.
In wild-type cells, sequence divergence led to a significant decrease in both break-induced and spontaneous recombination. For break-induced recombination, the frequency of neo+ colonies obtained from clones containing the H-DR-8mu substrate (1.2% divergent) relative to those containing the control H-DR-WT substrate (0.1% divergent) decreased by 4.5-fold (Table 2). The frequency of spontaneous recombination was also decreased, approximately 12-fold, although the decrease in the rate of recombination has not been measured. In Msh2−/− cells, the frequency of neo+ colonies arising from transfection of the I-Sce I expression vector was not substantially different between cell lines containing the H-DR-WT and H-DR-8mu substrates, nor did the spontaneous recombination frequency differ. Thus, there is little barrier to both DSB-promoted and spontaneous recombination from sequence divergence in the absence of the MMR pathway.
GCTs from intrachromosomal neo+ recombinants.
During DSB repair, gene conversion can occur by mismatch correction of hDNA, by repair of a gap formed around the break site, or by a combination of both mechanisms. In wild-type cells, mismatches in hDNA would be expected to be corrected rapidly, resulting in daughter cells with an identical genotype. In Msh2−/− cells, however, mismatches in hDNA would be expected to remain uncorrected. Segregation of mismatches after replication and cell division would produce daughter cells with two different genotypes (Fig. 4A), similar to postmeiotic and postmitotic segregation found in yeast (36, 42, 44). Since cells were plated immediately after electroporation of the I-SceI expression vector, both daughter cells from a recombinant cell should have been represented in a neo+ colony. Unlike mismatch correction of hDNA, however, repair of a double-stranded gap is entirely dependent on sequence information from the homologous repair template. Therefore, gap repair involving a particular silent mutation would be expected to lead to complete conversion in both Msh2−/− and wild-type cells, resulting in neo+ colonies with a single genotype.
FIG. 4.
Analysis of DSB-induced intrachromosomal recombination in wild-type and Msh2−/− cell lines. (A) After in vivo expression of the I-SceI endonuclease, a DSB was introduced in the S2neo gene (top bar) of the H-DR-8mu substrate. Recombinational repair was initiated using the pneo-8mu template (bottom bar), restoring the NcoI site at the former position of the I-SceI site (thick hatch marks) to form a neo+ gene and, in this case, creating hDNA at the downstream NsiI and PmlI restriction site polymorphisms (thin hatch marks). Correction of the mismatched hDNA in a wild-type cell, followed by replication and division, results in both daughter cells containing the same genotype. However, in Msh2−/− cells, MMR correction does not occur, and the uncorrected strands are segregated to daughter cells. (B) PCR analysis of two neo+ clones after DSB-induced recombination in the H-DR-8mu substrate. The neo+ gene coding region was amplified by PCR as an 810-bp fragment from genomic DNA with primers Neo1 and Neo3 and electrophoresed on agarose gels following digestion with the indicated restriction enzymes (Fig. 3). The amplified product from each of the neo+ clones was not cleaved by I-SceI (data not shown) but was cleaved by NcoI (Nco) and other restriction enzymes, as indicated by arrows. In the wild-type recombinant 3A-10, the NsiI (Ns) and PmlI (Pm) sites were fully cleaved, indicating a population of cells with one genotype (as in lane A). By contrast, in the Msh2−/− recombinant 17A-9, both the NsiI and the PmlI sites were partially cleaved, indicating a population of cells with two genotypes (as in lane A). Note that due to the naturally occurring PstI (P) site, the amplified fragment in each of the clones was shifted when cleaved with PstI. M, mixed digest; MW, 1-kb ladder molecular weight marker; A, ApaI; L, ApaLI; B, BamHI; X, XbaI; Nr, NruI.
To gain insight into mechanisms of gene conversion, we analyzed (GCTs) of neo+ recombinants obtained from our intrachromosomal recombination assays. The recombinants were derived from transfection of the I-SceI expression vector into clones containing the H-DR-8mu construct. Genomic DNA was isolated from each of the recombinants, and PCR was performed to amplify the converted S2neo (now neo+) gene. The neo primers used in this analysis flanked the position of the DSB site and were mainly outside the region of homology of pneo-8mu so as not to amplify the donor repair substrate (Fig. 3). PCR products were digested with restriction enzymes corresponding to the silent mutations in the repair substrate to determine which of the silent mutations had been incorporated into the neo+ gene after gene conversion.
PCR analysis for two recombinants is shown in Fig. 4B. Products from both recombinants were completely cleaved by NcoI, as was expected since these clones are G418 resistant, indicating that neo gene function was restored. The incorporation of the NcoI site required the loss of heterology from the I-SceI site, 5 bp from the 5′ side and 9 bp from the 3′ side plus the 4-base overhang on each side. In addition to the NcoI site, the PCR product from the recombinant derived from wild-type ES cells was also completely cleaved by NsiI and PmlI, enzymes which have restriction sites 3′ of the NcoI site. Thus, this clone had an observed GCT of 114 bp. This is a unidirectional GCT, as none of the six polymorphisms 5′ of the NcoI site was incorporated. (The PCR product was also cleaved by PstI at the naturally occurring site 5′ of the NcoI site which is present in both the S2neo gene and the pneo-8mu repair substrate but was not cleaved at the polymorphic PstI site present only in pneo-8mu.) The recombinant derived from the Msh2−/− cell line had a PCR product which was partially cleaved by NsiI and PmlI. This implies that a small gap of less than 8 bp formed on the 3′ side of the I-SceI cleavage site (i.e., 7 bp plus the 1-bp silent mutation at the NsiI site; Fig. 1). In addition, it implies that hDNA was present at the position of the NsiI site (8 bp from the I-SceI site) as well as that of the PmlI site (102 bp from the I-SceI site) but that as a result of the MMR deficiency the hDNA remained uncorrected and segregated to daughter cells.
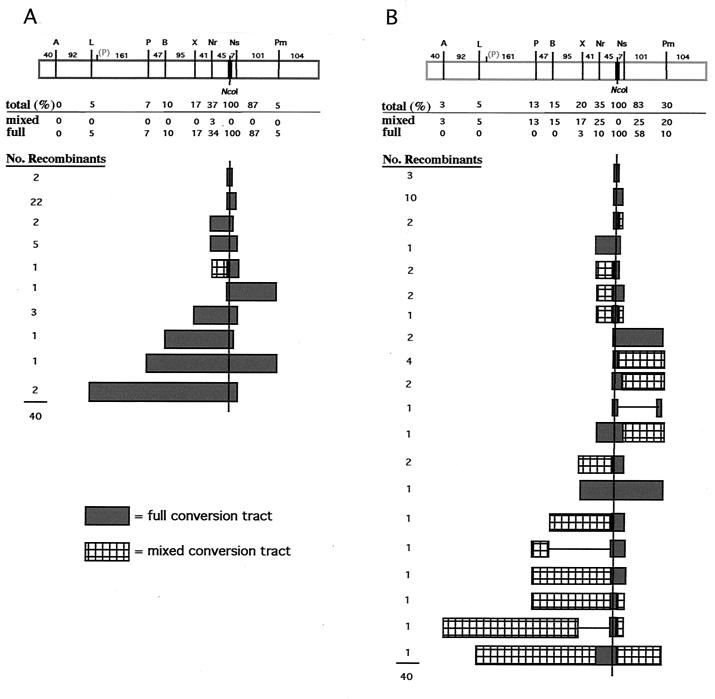
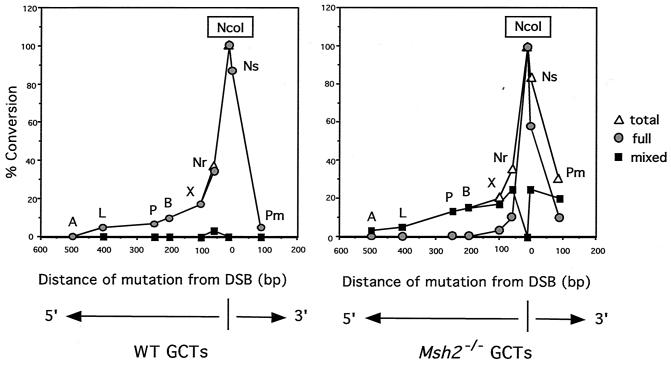
A summary of GCTs from 80 neo+ recombinants that were analyzed is presented in Fig. 5. Each of the recombinants from both cell lines had lost the I-SceI site and had acquired the NcoI site. Flanking restriction site polymorphisms in recombinants from the wild-type cells, with one exception, were either completely cleaved or completely uncleaved by the diagnostic restriction enzymes (Fig. 5A). This indicated that daughter cells derived from the initial recombinant cell contained the same genotype. In the one exception, one of the two polymorphic restriction sites incorporated into the recombinant (NruI) was partially cleaved. The GCTs were generally short. The majority (80%) of the observed GCTs in the recombinants were 58 bp or less, maximally incorporating the two mutations nearest the NcoI site (NsiI and NruI), although two tracts were observed to be as long as 406 bp. (Note: This is a minimum length of conversion, as it is unknown how far conversion extended between the polymorphic markers.) The distribution of silent mutations incorporated in the GCTs inversely correlated with distance from the DSB site, with only the nearby NsiI site incorporated in the majority of clones. The GCTs were exclusively continuous, such that each of the mutations between the outermost converted restriction site and the NcoI site was converted. Overall, the GCTs derived from intrachromosomal DSB-promoted recombination were very similar to those we observed previously in gene-targeting experiments (17).
FIG. 5.
GCTs after DSB-induced intrachromosomal recombination in cells containing the H-DR-8mu substrate. GCTs were derived from four independent experiments, and 40 recombinants of each of the wild-type (A) and Msh2−/− (B) cell lines were analyzed. The pneo-8mu repair template is shown at the top (rectangle), with the positions of silent mutations and the distances (in base pairs) between the mutations. When GCTs were being calculated, an additional base pair was added for the conversion of the silent mutation and 4 bp was added for the incorporation of the NcoI site. The percent of recombinants incorporating each silent mutation is shown. This number is broken down to the percent of recombinants which had partial restriction cleavage at the site (i.e., mixed) and the percent of recombinants which had complete cleavage (i.e., fully converted). The correcting NcoI site occurred in 100% of the neo+ recombinants as expected, since it restores a functional gene. The number of recombinants with each of the indicated GCTs is shown.
In contrast to recombinants from the wild-type cells, the majority of recombinants derived from Msh2−/− cells frequently had GCTs in which the restriction site polymorphisms were incompletely converted (Fig. 5B). Many of the restriction sites in the amplified PCR products were only partially cleaved by one or more restriction enzymes, indicating that segregation of unrepaired hDNA had occurred in these recombinants. This strongly suggests that in wild-type cells, hDNA repair is a mechanism of gene conversion during DSB repair. Other differences were also seen between the mutant and wild-type cells. Taking into account the partially converted GCTs, only 52% of the GCTs were 58 bp or less, compared with 80% in the wild-type cells. The longest observed GCT was 508 bp, and 1 of the 40 recombinants was found to have incorporated the most distant polymorphism, ApaI, located 487 bp 5′ of the NcoI site. We have not observed the ApaI polymorphism in any of the 120 GCTs we analyzed from wild-type recombinants derived from either intrachromosomal recombination (this report) or gene targeting (17). The most distant 3′ polymorphism at the PmlI site was also found to be more frequently incorporated into the Msh2−/− recombinants. We also for the first time observed discontinuous GCTs in three of the Msh2−/− recombinants. The GCT of one of these clones, for example, was converted at the NcoI and PmlI sites, but not at the NsiI site.
Although restriction site polymorphisms most distant from the DSB site were more frequently converted in the Msh2−/− cells, the two closest sites were converted at a frequency similar to that in the wild-type cells. When examining these sites to see if they were fully converted or partially converted, we found that many of the more distant sites were partially converted in the Msh2−/− cells, whereas the sites closest to the DSB site were often fully converted. That is, the NsiI site was fully converted in the majority of recombinants containing this site (23 of 33 recombinants); however, the next closest site, NruI, was present in 14 recombinants but was fully converted in only four of them. The exceptional long GCT that was fully converted in the Msh2−/− cells was 202 bp. The presence of fully converted restriction site polymorphisms suggests that gap repair also contributes to gene conversion, although less frequently than hDNA repair.
Segregation of gene converstion tracts from Msh2−/− recombinants.
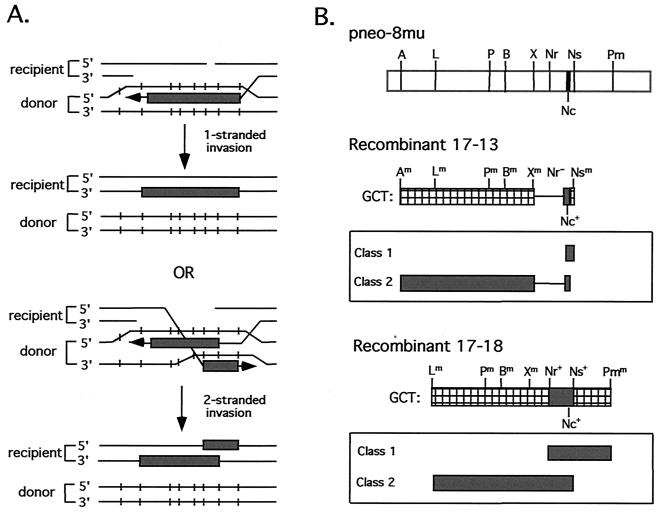
The bidirectional GCTs from Msh2−/− cells raised the question as to how the partially converted silent mutations would segregate. That is, would mutations 5′ of the DSB site segregate as a group, and if so, would this group of mutations segregate away from mutations 3′ of the DSB site or segregate together with 3′ mutations? If sequence information incorporated into hDNA was derived from only one strand of the repair substrate and this strand spanned the break site, then the 5′ and 3′ mutations would be expected to segregate together (Fig. 6A). Alternatively, if sequence information was derived from both DNA strands of the repair substrate, one strand 5′ to the break site and the other 3′ to the break site, the two groups of mutations would be expected to segregate independently.
FIG. 6.
(A) Two mechanisms for the derivation of bidirectional tracts. (B) Segregation of GCTs from two Msh2−/− intrachromosomal recombinants. Both recombinants have a bidirectional GCT (i.e., a detectable GCT on both sides of the NcoI site) that is mixed. After subcloning, each recombinant yielded two classes of subclones, as shown.
To address this, we subcloned two neo+ recombinants, 17-13 and 17-18 (Fig. 6B). Cells from each recombinant were diluted and plated into a 96-well plate, and wells were then examined after a few days for the growth of a single colony. Subclones were expanded for genomic DNA preparation and PCR analysis. The bidirectional GCT of recombinant 17-13 can be designated Am Lm Pm Bm Xm Nr− Nc+ Nsm, where “m” indicates mixed (partial) cleavage by the indicated restriction enzyme, “+” indicates complete cleavage, and “−” indicates no cleavage. Two classes of subclones were identified from this recombinant. In one class, the genotype was A+ L+ P+ B+ X+ Nr− Nc+, and in the other class it was Nc+ Ns+. The GCT of recombinant 17-18 is the longest of all of the neo+ recombinants and is designated Lm Pm Bm Xm Nr+ Nc+ Ns+ Pmm. This recombinant also gave rise to two classes of subclones. In this case, the genotype of one class was L+ P+ B+ X+ Nr+ Nc+ Ns+ and the genotype of the other class was Nr+ Nc+ Ns+ Pm+. Since in both recombinants the mutations segregated independently, silent polymorphisms appear to have been incorporated on both strands of the hDNA. Presumably, one strand was 5′ to the break site and the other strand was 3′ to the break site.
DISCUSSION
We have examined the effect of heterology on DSB-induced homologous recombination in mammalian cells, comparing wild-type and Msh2−/− ES cells. We found that in wild-type cells, the frequency of recombination between diverged sequences in a gene-targeting assay was reduced as the sequence divergence increased, consistent with what was previously observed (17), but that the effect of heterology was significantly overcome in Msh2−/− cells. Thus, for a plasmid substrate with 1.5% heterology relative to the chromosomal sequence, DSB-induced recombination is reduced 17-fold in wild-type cells when normalized to recombination with a nearly identical substrate, but only 1.7-fold in Msh2−/− cells. Similar to gene targeting, DSB-induced intrachromosomal recombination between diverged sequences was also reduced to a greater extent in wild-type cells than in Msh2−/− cells. The products of recombination in the Msh2 mutant were also significantly altered. Most notably, the majority of GCTs derived from intrachromosomal recombination had mixed polymorphic markers present in the homologous repair substrate, implying that hDNA formation followed by MMR correction is a major mechanism for the conversion of markers near a DSB in mammalian cells.
These results are in agreement with those previously reported for mammalian cells in which the barrier to spontaneous recombination between diverged sequences was found to be relaxed in MMR-deficient cells (1, 13, 14). The fold reduction of recombination in wild-type cells in these experiments was similar to or even greater than what we saw in the wild-type cells with our most diverged substrate. However, in our experiments we found that recombination was not fully restored in the Msh2−/− ES cells, whereas in the previous reports the barrier to recombination was found to be almost completely abrogated by Msh2 mutation. This difference may be attributable to the design of the gene-targeting experiments. Our experiments examined DSB-induced recombination of small DNA fragments containing single nucleotide substitutions, whereas the previous experiments investigated recombination in the absence of induced chromosomal damage using much larger fragments that contained different types of polymorphisms.
In addition to the frequency of recombination, we have also examined GCTs in the recombinants derived from the Msh2 mutant cell line. This is the first time, to our knowledge, that GCTs in mammalian cells in an MMR-defective background have been studied. For this study, we examined phenotypically silent mutations spaced every 50 to 100 bp or less in the repair substrate. DSB repair by gene conversion has been proposed to occur by two possible mechanisms: (i) hDNA formation followed by mismatch correction and (ii) gap formation after both strands adjacent to the DSB are processed. Examination of the GCTs in cells which lacked MMR activity allowed us to infer which mechanism of gene conversion was occurring during DSB repair in wild-type cells. We observed that 55% of Msh2−/− neo+ recombinants had mixed GCTs, which were predicted by hDNA correction, whereas only 2.5% of the wild-type recombinants had mixed GCTs. Considering the silent mutations individually, most mutations when converted were partially converted in the recombinants (i.e., mixed; Fig. 7), suggesting that the conversion involved hDNA correction. The mutation that was the one exception was the one nearest the DSB (the NsiI site), which is frequently fully converted, even in Msh2−/− cells. Thus, with the exception of the mutation at the NsiI site, 39 of 48 (81%) conversion events involved hDNA formation. The frequency for each mutation depends on the distance from the DSB site, since mutations at some distance from the site (≥183 bp) were found to be incorporated exclusively by hDNA correction (Fig. 7).
FIG. 7.
Summary of gene conversion frequencies for each silent mutation in wild-type and Msh2−/− cells. The conversion frequency for each mutation (Fig. 5) was plotted as a function of distance from the DSB.
Gap formation was expected to account for the remaining conversion events. Fully converted mutations were anticipated to occur in the Msh2 mutant only if both strands of the DNA adjacent to the DSB were degraded by nucleases. Information for the repair would then have come solely from the diverged donor substrate. In three conversion events, gap formation may have extended 100 bp or more from the DSB. Gap formation, however, was especially evident at the NsiI site located 8 bp from the I-SceI site, since it was fully converted in 70% (58 out of 83; Fig. 5) of the conversion events (Fig. 7). In the remaining events at this site, processing at the DNA ends led to the loss of the I-SceI site but also to the retention of nucleotides only 8 bp further away, although it is not clear if the sequence heterology of the I-SceI site affected the incorporation of this mutation. In summary, therefore, we infer that gene conversion occurs primarily by hDNA correction, but that sequences near the DSB can be incorporated by gap formation, the frequency depending on the distance from the DSB. These results are generally in good agreement with those obtained with yeast, in which it has been found that sequences close to the end of a DSB are preserved during gene conversion (22, 44, 58), although the results here suggest that there may be somewhat more nibbling of ends in mammalian cells. Evidence for hDNA correction during DSB-promoted and spontaneous gene conversion in wild-type mammalian cells has previously been obtained using palindromic markers which are not efficiently repaired by MMR (15, 30, 31). However, some other aspects of the GCTs (i.e., length and discontinuity) differ from results obtained here with single polymorphisms.
Current models for mitotic gene conversion favor mechanisms in which recombination is coupled to replication (3, 19, 24, 38, 40, 46). In these models, a 3′ end invades the unbroken homologous template and initiates repair synthesis. The newly synthesized strand is then incorporated into the broken molecule, which can result in hDNA formation. In principle, invasion of either one or both 3′ ends flanking the DSB could initiate repair synthesis. Our bidirectional GCTs are consistent with invasion of the template from both 3′ ends, since markers on each side of the DSB segregate at the next cell division (Fig. 6), as would be expected from hDNA formation involving opposite strands. The majority of recombinants (27 out of 40; 68%) did not show bidirectional tracts, however. In these cases, two-ended invasion may have occurred but may not have incorporated the polymorphic markers. Alternatively, one-ended invasion may have occurred, as has been proposed for other mammalian gene conversion events (24, 46).
Our results also indicate a trend toward longer GCTs in the Msh2 mutant. In 80% of the wild-type recombinants, the observed GCTs were 58 bp or less, consistent with previous results (17), whereas in the Msh2−/− recombinants, only 52% of GCTs were as short. We saw a sixfold increase in the Msh2−/− cells in GCTs extending to the most distant 3′ polymorphism from the break site, PmlI. We also saw for the first time the incorporation of the most distant 5′ polymorphism, ApaI, in one of the mutant recombinants. The mean observed GCT was also found to be somewhat longer (by 20%) in Msh2−/− cells than in wild-type cells. Two mechanisms could account for the longer GCTs: (i) MMR proteins may regulate the length of hDNA formation, as suggested previously (7), or (ii) the length of hDNA is the same, but in wild-type cells, correction of hDNA at a distance from the DSB is in favor of the recipient DNA molecule rather than the donor. Support for the latter mechanism is that the strand copied from the donor molecule would be expected to be broken at the end of the hDNA tract, and strand breaks are known to influence the direction of mismatch correction. Nevertheless, in bacteria, MMR proteins have been shown to inhibit RecA-catalyzed strand transfer (59), consistent with a direct effect on hDNA length.
In MMR-deficient bacteria, gross chromosomal rearrangements involving diverged DNA are increased as a result of the recombinator phenotype of these mutants (43). Results in this report, as well as those previously published, demonstrate that MMR-deficient mammalian cells also have increased recombination as a result of a relaxation of recombination between diverged sequences. These results would seem to predict that MMR-deficient mammalian cells, like bacteria, would exhibit frequent gross chromosomal rearrangements. Surprisingly, tumors derived from HNPCC patients are generally euploid and do not display gross chromosomal structure defects, even while cell lines from sporadic tumors show continuous chromosomal instability (reference 4 and references therein; 18, 28). Possibly, more subtle chromosomal rearrangements than those that would be apparent from chromosome number analyses are present in MMR-deficient cells but have yet to be detected.
Nevertheless, several factors may contribute to the maintenance of a normal genotype in cells with MMR deficiency. First, the barrier to recombination between diverged elements is not completely overcome. Although DSBs induce the highest levels of recombination detected thus far (32), DSB-induced recombination between sequences that are only 1.5% diverged are still not fully restored, even with MMR mutation. Considering that repetitive elements in mammalian genomes are often significantly more diverged (e.g., an average of 15% for Alu elements; 51), recombination between highly diverged elements is predicted to be suppressed at least partially in MMR-deficient cells. Second, the outcome of DSB-induced recombination in mammalian cells has a strong bias towards noncrossover events (24, 46). Therefore, recombination between repetitive elements would not be predicted to have a discernible outcome in most instances because there would not be an exchange of flanking markers. Third, in addition to having a role in recombination between diverged sequences, MMR proteins in yeast are known to have a second role in recombination, such that their mutation would decrease recombination instead of leading to its enhancement. This role involves the removal of nonhomologous tails which are 30 nucleotides or longer and which are formed during some types of recombination (41, 49, 55). Because dispersed repetitive elements are flanked by DNA which is not homologous, initiation of recombination outside two dispersed elements, unlike the events assayed in this report, is likely to lead to the formation of nonhomologous tails involving the flanking DNA. Although this role for MMR proteins has yet to be demonstrated in mammalian cells, efficient removal of these tails in yeast requires Msh2 (41, 49, 55), suggesting that recombination between dispersed elements could be reduced by Msh2 mutation.
Despite these multiple levels of control that maintain chromosome integrity in MMR-deficient cells, the experiments presented here nevertheless demonstrate that these cells have a recombinator phenotype for DSB-induced events, raising the possibility that these cells undergo promiscuous recombination at some level. Although tumorigenesis in HNPCC has been clearly linked to the mutator phenotype arising from an inability to repair replication errors (20), recombination between diverged sequences in MMR-deficient cells may contribute to the genomic alterations important for the development of the disease. Additional experiments will be necessary to further delineate the effect of MMR mutation on homologous recombination in mammalian cells.
ACKNOWLEDGMENTS
We thank Hein te Riele (Amsterdam, The Netherlands) for the Msh2 mutant cell line and Roger Johnson and other members of the Jasin laboratory.
B.E. was supported by an NRSA fellowship (GM18831). This work was supported by NIH (GM54688) and NSF (MCB-9728333) grants to M.J.
REFERENCES
- 1.Abuin A, Zhang H, Bradley A. Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol Cell Biol. 2000;20:149–157. doi: 10.1128/mcb.20.1.149-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 4.Bocker T, Schlegel J, Kullmann F, Stumm G, Zirngibl H, Epplen J T, Ruschoff J. Genomic instability in colorectal carcinomas: comparison of different evaluation methods and their biological significance. J Pathol. 1996;179:15–19. doi: 10.1002/(SICI)1096-9896(199605)179:1<15::AID-PATH553>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Boland C R. Hereditary nonpolyposis colorectal cancer. In: Vogelstein B, Kinzler K W, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill; 1998. pp. 333–346. [Google Scholar]
- 6.Buermeyer A B, Deschenes S M, Baker S M, Liskay R M. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Jinks-Robertson S. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol Cell Biol. 1998;18:6525–6537. doi: 10.1128/mcb.18.11.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper D N, Krawczak M, Antonarkis S E. The nature and mechanisms of human gene mutation. In: Vogelstein B, Kinzler K W, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill; 1998. pp. 65–94. [Google Scholar]
- 10.Datta A, Adjiri A, New L, Crouse G F, Jinks Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Laat W L, Jaspers N G, Hoeijmakers J H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 13.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 14.de Wind N, Dekker M, Claij N, Jansen L, van Klink Y, Radman M, Riggins G, van der Valk M, van't Wout K, te Riele H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 15.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond J T, Li G M, Longley M J, Modrich P. Isolation of an hMSH2–p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 17.Elliott B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eshleman J R, Casey G, Kochera M E, Sedwick W D, Swinler S E, Veigl M L, Willson J K, Schwartz S, Markowitz S D. Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53. Oncogene. 1998;17:719–725. doi: 10.1038/sj.onc.1201986. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurin C C, Federici M G, Kang L, Boyd J. Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res. 1999;59:462–466. [PubMed] [Google Scholar]
- 21.Haber J E. DNA repair. Gatekeepers of recombination. Nature. 1999;398:665. doi: 10.1038/19423. , 667. [DOI] [PubMed] [Google Scholar]
- 22.Haber J E, Ray B L, Kolb J M, White C I. Rapid kinetics of mismatch repair of heteroduplex DNA that is formed during recombination in yeast. Proc Natl Acad Sci USA. 1993;90:3363–3367. doi: 10.1073/pnas.90.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg R A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 24.Johnson R D, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 26.Lacalle R A, Pulido D, Vara J, Zalacain M, Jimenez A. Molecular analysis of the pac gene encoding a puromycin N-acetyl transferase from Streptomyces alboniger. Gene. 1989;79:375–380. doi: 10.1016/0378-1119(89)90220-5. [DOI] [PubMed] [Google Scholar]
- 27.Lengauer C, Kinzler K W, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 28.Lengauer C, Kinzler K W, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 29.Leung W, Malkova A, Haber J E. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc Natl Acad Sci USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Baker M D. Use of a small palindrome genetic marker to investigate mechanisms of double-strand-break repair in mammalian cells. Genetics. 2000;154:1281–1289. doi: 10.1093/genetics/154.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Read L R, Baker M D. The mechanism of mammalian gene replacement is consistent with the formation of long regions of heteroduplex DNA associated with two crossing-over events. Mol Cell Biol. 2001;21:501–510. doi: 10.1128/MCB.21.2.501-510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang F, Han M, Romanienko P J, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loeb L A. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 35.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 36.McGill C, Shafer B, Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57:459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- 37.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 38.Nassif N, Penney J, Pal S, Engels W R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pâques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pâques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 43.Petit M A, Dimpfl J, Radman M, Echols H. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray B L, White C I, Haber J E. Heteroduplex formation and mismatch repair of the “stuck” mutation during mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5372–5380. doi: 10.1128/mcb.11.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 46.Richardson C, Moynahan M E, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Washington, D.C.: IRL Press; 1987. pp. 71–112. [Google Scholar]
- 48.Rouet P, Smith F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sargent R G, Brenneman M A, Wilson J H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmid C W. Alu: structure, origin, evolution, significance and function of one-tenth of human DNA. Prog Nucleic Acid Res Mol Biol. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- 52.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smih F, Rouet P, Romanienko P J, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strout M P, Marcucci G, Bloomfield C D, Caligiuri M A. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc Natl Acad Sci USA. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugawara N, Pâques F, Colaiacovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9921. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taghian D G, Nickoloff J A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 58.Weng Y S, Nickoloff J A. Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics. 1998;148:59–70. doi: 10.1093/genetics/148.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]