Abstract
Virus infections are huge threats to living organisms and cause many diseases, such as COVID-19 caused by SARS-CoV-2, which has led to millions of deaths. To develop effective strategies to control viral infection, we need to understand its molecular events in host cells. Virus related functional genomic datasets are growing rapidly, however, an integrative platform for systematically investigating host responses to viruses is missing. Here, we developed a user-friendly multi-omics portal of viral infection named as MVIP (https://mvip.whu.edu.cn/). We manually collected available high-throughput sequencing data under viral infection, and unified their detailed metadata including virus, host species, infection time, assay, and target, etc. We processed multi-layered omics data of more than 4900 viral infected samples from 77 viruses and 33 host species with standard pipelines, including RNA-seq, ChIP-seq, and CLIP-seq, etc. In addition, we integrated these genome-wide signals into customized genome browsers, and developed multiple dynamic charts to exhibit the information, such as time-course dynamic and differential gene expression profiles, alternative splicing changes and enriched GO/KEGG terms. Furthermore, we implemented several tools for efficiently mining the virus-host interactions by virus, host and genes. MVIP would help users to retrieve large-scale functional information and promote the understanding of virus-host interactions.
INTRODUCTION
Viruses are everywhere, comprising an enormous proportion of our environment, in both quantity and total mass (1). Many viral infections cause human diseases (2,3). More than 12% new cancer cases were attributable to oncoviruses, such as hepatitis B or C virus (HBV or HCV), Epstein-Barr virus (EBV), Kaposi's sarcoma herpes virus (KSHV), and human papillomavirus (HPV) (4–6). Recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused the COVID-19 disease, and resulted in a global pandemic and millions of deaths (7–9). Viral infections generally cause dysregulated gene expression and abnormal RNA processing (10–13). In mammalians, viral infections can lead to local inflammatory responses and innate immune responses called as ‘cytokine storm’ (2). For example, SARS-CoV-2 broadly alters gene expression programs in human cells and disrupts splicing to suppress host defences (14,15). In addition, SARS-CoV-2 RNAs can bind and repurpose host RNA-binding proteins (RBPs), which is one of the pathogenetic factors (16–18). Moreover, viral infections can also change the epigenetic states and RNA modifications of hosts (19–22). To better understand how viruses affect hosts at molecular level, we need to integrate various types of omics data and systematically analyse the many-to-many virus-host interactions genome-wide.
In recent years, the studies of genome, structure and taxonomy have been rapidly developed for viral species, including ViPR (23), VIPERdb (24,25), IMG/VR v.2.0 (26) and ICTV (27) databases. Moreover, it is found that the molecular network of host in many cancers are perturbated by viral proteins (17). Therefore, the relevant resources of biological pathway and network signatures associated with virus were developed, such as KEGG (28) and PAGER (29,30). In addition, multiple types of raw sequencing data under viral infection are deposited into the NCBI GEO and SRA (31,32) databases. These data were separately generated in different studies to uncover the cellular events in various species with different viral infections. However, an integrative multi-omics database of virus-host interactions for multiple species/viruses, enabling users to mine relevant data jointly, is missing.
Here, we have developed a user-friendly multi-omics portal of viral infections across different species, named MVIP (https://mvip.whu.edu.cn/). We firstly manually collected available high-throughput sequencing data under viral infections, and also the description of these data (metadata). We unified detailed metadata including virus, host species, cell types/tissues, infection time, treatment, assay, target, and publication, etc. We processed >4900 viral infected samples (from 77 virus and 33 host species) with standard pipelines for 22 types of omics data including RNA-seq, ChIP-seq, ATAC-seq, CLIP-seq, small RNA-seq (smRNA-seq), Ribo-seq, RIP-seq etc. Furthermore, we analysed the differentially expressed genes, alternative splicing events, GO and KEGG pathway enrichment, genome-wide binding events, translational states, etc. Then, we integrated these comprehensive data into MVIP, provided customized genome browser with JBrowse2 and UCSC track hub to visualize them simultaneously, and developed dynamic charts to display gene-level information such as differential expression changes in responding to viral infections versus controls. Furthermore, we implemented different search modes and several tools for efficiently mining the virus-host interactions by virus, host and assay type, etc. The database will help users to quickly retrieve and compare different virus-host interactions at multilayers, to efficiently analyse gene dynamic changes, and to visualize large-scale omics data of viral infections with flexible settings.
MATERIAL AND METHODS
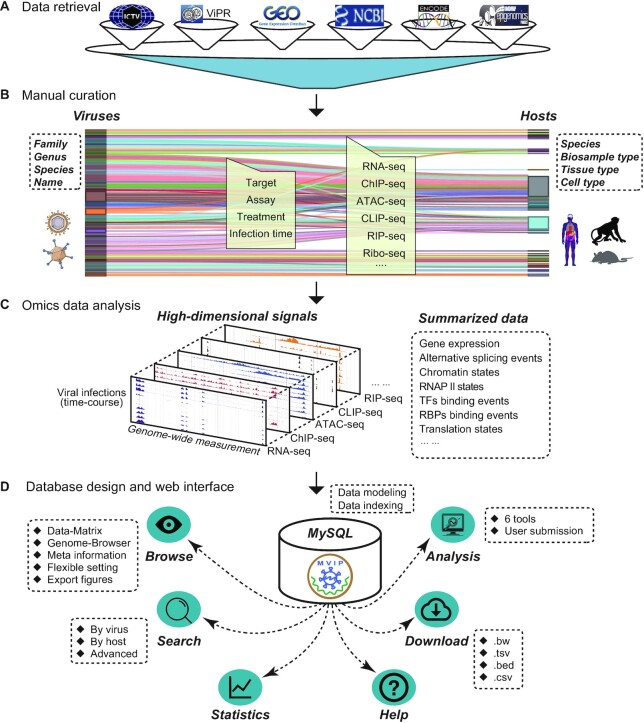
All data sources, metadata information, data processing, and web interface features are briefly summarized in Figure 1. The development of MVIP consists of data collection and curation, omics data processing and analysis, database design and construction, and web interface and tool development. The main steps, used tools and results are briefly illustrated in Supplementary Figure S1 and described in detail as below.
Figure 1.
Schematic view of MVIP. (A) Information retrieval of omics data related to viral infections in various public databases. (B) Manual curation of metadata. (C) Overview of the generated data from the analyses of different types of omics data. (D) Overview of the database design and web interface of MVIP.
Data sources and metadata unification
We firstly searched NCBI GEO DataSets database with keywords ‘virus’ and ‘seq’ up to September 2019 using Entrez E-utility. We further filtered these accessions by requiring the presence of a keyword ‘virus’, ‘viruses’ and ‘viral’ appear in its GEO summary ignoring cases (Figure 1A). Finally, we manually checked the metadata and obtained 4757 samples (291 GEO accessions) with diverse types of high-throughput sequencing data related to viral infection. In addition, due to burst of the pandemic COVID-19 caused by SARS-CoV-2, we retrieved 1547 RNA-seq and 282 scRNA-seq data samples related to SARS-CoV-2 infection from NCBI/GEO database up to December 2020.
Next, we manually collected all the relevant metadata (description of the sequencing data), including virus, host species, cell type or tissue, infection time, treatment, assay, target, publication etc. (Figure 1B). Furthermore, we manually curated and classified these viruses (family, genus, species etc.) using the information from ViPR (23) and ICTV (33) databases. Similarly, according to the classical information of ENCODE (34), Roadmap (35) and NCBI, we manually annotated and unified all hosts on biosample type, tissue type, and cell type (Table 1). All curated metadata of these omics data are summarized in Supplementary Table S1.
Table 1.
Summary of metadata in MVIP
| Class | Count |
|---|---|
| Sample | 6586 |
| Dataset | 4255 |
| Assay | 22 |
| Species | 33 |
| Biosample type | 9 |
| Tissue type | 76 |
| Cell type | 114 |
| Host | 183 |
| Virus family | 34 |
| Virus genus | 53 |
| Virus species | 68 |
| Virus name | 77 |
Omics data processing
The raw Fastq data files of variety of omics data including RNA-seq, ChIP-seq, ATAC-seq, CLIP-seq, smRNA-seq, RIP-seq, Ribo-seq and others, were downloaded from the NCBI GEO and SRA database (36). We aim to ease users to explore these high-dimensional genomic signals and to query the summarized data at different layers of regulation in cells responding to different viruses during the course of infections, thus enabling systematic thinking and the development of biological hypotheses (Figure 1C).
All high-throughput sequencing data that had passed the quality control using fastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) were used in the downstream analysis. The raw reads were filtered to remove the sequencing adaptors and low-quality bases using Trimmomatic (37) or trim_galore (38) programs. For the omics data of RNA-seq, ChIP-seq, ATAC-seq and smRNA-seq, we used the data processing pipelines by following the recommendations in ENCODE project (39). RNA-seq and smRNA-seq (or miRNA-seq) reads were mapped to host and viral genomes using the STAR program (40). The ChIP-seq (or FAIRE-seq), ATAC-seq, CLIP-seq (or irCLIP-seq), RIP-seq (or MeRIP-seq) and GRO-seq reads were mapped using Bowite2 program (41). In addition, the potential chrM and PCR duplicate reads were removed for ATAC-seq data. The read counts of genes and features were computed using the featureCounts program (42). The gene expression quantifications in FPKM (Fragments Per Kb of exon per Million mapped fragments) and TPM (Transcripts Per Kb of exon per Million mapped reads) were computed using the StringTie program (43). The differentially alternative splicing events were identified using the rMATS program (44). The peaks of ChIP-seq, CLIP-seq (or irCLIP-seq), RIP-seq (or MeRIP-seq) and ATAC-seq data were identified using MACS, Clipper (45), Piranha (46) and MACS2 (47), respectively. For Ribo-seq, the potential rRNA reads were filtered before mapping. The cleaned reads were mapped to host and viral genomes using the STAR program in end-to-end mode. Then, the potential chrM and PCR duplicate reads were removed. Next, we calculated the translation efficiency for all ORFs using RiboWave (48).
In addition, we have processed Bisulfite-seq, and GRO-seq data using gemBS (49) and Homer (http://homer.ucsd.edu/homer/), respectively. There are six raw datasets associated with five rare species such as Myotis daubentoniid, Chlorocebus aethiops and Beta macrocarpa, were not processed, because their genomes and annotations are not well defined. For scRNA-seq data, we directly retrieved and used their processed data in GEO database. The main steps in different pipelines are described in Supplementary Figure S1.
We managed the data analyses with Snakemake program, a reproducible workflow management system (50), and executed the pipelines on Linux servers. All used programs and packages with their version information are listed in Supplementary Table S2, and the statistics of the processed files are summarized in Table 2.
Table 2.
Summary of processed files during analysis
| File type | Count | Description |
|---|---|---|
| FeatureCounts (.tsv) | 4615 | The read counts for annotated genes |
| Stringtie (.tsv) | 4378 | The FPKM and TPM for annotated genes |
| Differential expression (.csv) | 1950 | Differentially expression analysis results of viral infection vs. controls |
| GO/KEGG (.tsv) | 1846 | GO/KEGGs analysis results of DEGs |
| Alternative splicing (.csv) | 5465 | Alternative splicing events analysis of viral infected datasets |
| Peaks (.bed, .tsv, .txt, .png) | 2470 | Peaks calling and annotation results of protein binding data |
| Translation efficiency (.txt) | 80 | Translation efficiency of ORFs |
| Methylation (.bed.gz) | 64 | Three types of methylation value (CHG, CHH, CPG) |
| Mapping (.bw) | 10,361 | Genomic signals from mapped reads |
DEGs analysis and peak annotation
The differentially expressed genes (DEGs) are identified using DESeq2 (51) or edgeR (52) and under the cut-offs of P-value ≤0.05 and at least 2-fold change. Users can adjust the cut-offs for customized sensitivity and specificity. Currently, MVIP provides 1950 results from differential expression analysis (Table 2), and enables the visualization of these DEGs by volcano plot in the result page. Furthermore, the GO-term and KEGG enrichment analysis were performed using the R package clusterProfiler (53) for identified DEGs. The peaks were annotated by the R package ChIPseeker (54). MVIP supports visualization of the peaks in multiple ways, including displaying peak coverage signals over chromosomes and showing profiles of peaks relative to the transcription start site (TSS). We used pie charts to show the genomic features of peaks such as promoter, 5′ UTR, 3′ UTR, exon, and intron, using the ‘annotatePeak’ function. The peak profiles around the TSS region (±3 kb) were visualized using the ‘peakHeatmap’ function.
Database design and construction
We designed a set of data models with suitable indexes in MVIP MySQL database to efficiently store, update, query, view, and analyse the metadata and processed data (Figure 1D). Due to the complex virus-host interactions (Figure 2A) and diverse types of omics data (Figure 2B, C), we organized the metadata of all multi-omics sequencing data in a hierarchical structure following the principles developed for ENCODE project (55).
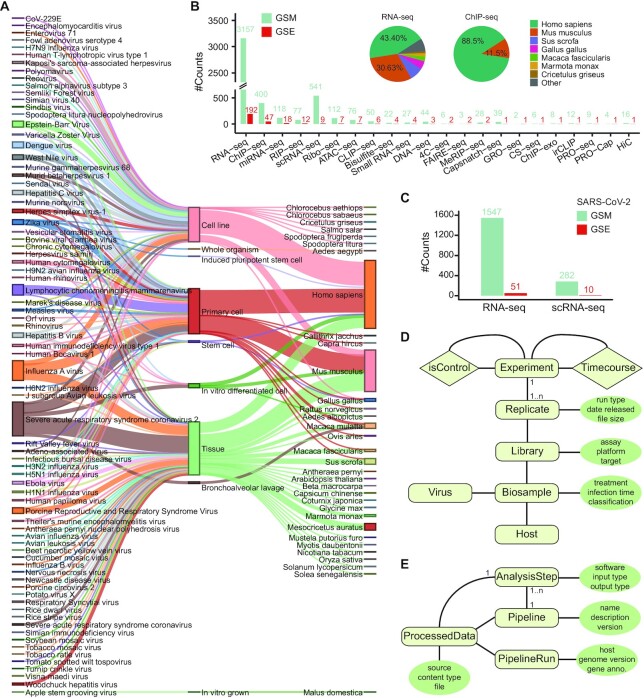
Figure 2.
Overview of metadata and data models. (A) Sankey diagram of virus, cell types, and host species in metadata. (B) Data counts of different assays by GEO series (GSE) and sample (GSM) for curated data up to Sep. 2019. The insert pie charts represent the distribution of RNA-seq and ChIP-seq samples by species. (C) Data counts of RNA-seq and scRNA-seq by GSE and GSM for SARS-CoV-2 related data up to December 2020. (D) Modeling of metadata. (E) Modeling of data analysis pipeline and processed data.
As shown in Figure 2D, each experiment, the unit of a sequencing study, has one or more replicates. Each replicate has corresponding sequencing data for the library constructed from specific assay (e.g. RNA-seq) and for specific target (e.g. ChIP-seq antibody). The sequencing library has its biosample information including the virus, host (e.g. tissue and cell type), the infection time, and specific treatment. Here, we took special efforts to curate the control experiment, such as mock control without virus infection for an experiment, and input control for a ChIP-seq or CLIP-seq assay. In addition, we annotated the time-course studies composing of a series of experiments to investigate the dynamics after virus infection. We also integrated the metadata for the processed files, which are generated from an analysis step in running a pipeline with specific software, genome, gene annotation, and input files (Figure 2E).
For querying data across multiple experiments, such as gene expression in multiple human samples by a specific virus, we stored the data in MySQL tables by organism, and concatenated the expression values together with comma symbol to be saved as a text field, for speeding up the response to queries.
Web interface and genome browser
MVIP is developed using MySQL MariaDB and running in a Docker container deployed on a Linux-based Apache Web server. We used Python 3.7 and Django 3.1.7 for server-side scripting to provide query and computation supports in the backend of the database, and used Typescript 4.3 (https://www.typescriptlang.org/) and React.js 17.0.4 (https://reactjs.org/) framework for developing a user-friendly interactive web interface (Figure 1D). We applied Material-UI 4.12.3 (https://material-ui.com), and ant-design charts 1.2.7 (https://charts.ant.design/) as graphical visualization frameworks. We recommend to visit MVIP using a modern web browser that supports the HTML5 standard such as Google Chrome, Firefox, Safari, or Microsoft Edge.
All mapping results to both host and viral genomes were converted to bigWig format and peak files were converted to bigBed files. We embedded a customized JBrowse2-based browser to visualize those genomic signals (56,57). Meanwhile, we constructed a MVIP Track Hub, allowing visualization of MVIP data for hosts in UCSC genome browser (58). The tracks are organized by organism with super-tracks and composite-tracks. The URL for MVIP track hub is https://mvip.whu.edu.cn/db/mvip/hub.txt, with which users can connect using the URL (http://genome.ucsc.edu/cgi-bin/hgHubConnect#unlistedHubs) to add a hub. Then, users can explore the MVIP data simultaneously with other existing UCSC data tracks. The genes presented in MVIP web pages have links to UCSC genome browser with MVIP track hub automatically connected.
RESULTS
Currently, MVIP contains 6586 sample including 4980 viral infected samples and 1606 control samples, involving 77 viruses, 33 host species, 114 cell types and 76 tissues (Figure 2A and Table 1). The samples related to SARS-CoV-2 and influenza A virus (IAV) account for one-third of the infection samples (Figure 2A), which are derived from RNA-seq, scRNA-seq, ChIP-seq, miRNA-seq, Ribo-seq etc.
There are 22 types of high-throughput sequencing data related to viral infection, however the data counts of those types, by either GEO series (GSE) or GEO sample (GSM), are not evenly distributed (Figure 2B). The RNA-seq data account for about 66%, which is partially due to that RNA-seq is the easiest and most widely used technique, currently. These samples are enriched in Homo sapiens and Mus musculus, which take proportions 43.4% and 30.63% in RNA-seq, 88.5% and 11.5% in ChIP-seq, respectively (Figure 2B, insert). Due to burst of the pandemic COVID-19 caused by SARS-CoV-2, We further curated 1547 RNA-seq and 282 scRNA-seq data samples (corresponding to 51 and 10 GEO series, respectively) related to SARS-CoV-2 infection from NCBI/GEO database up to December 2020 (Figure 2C). These metadata and processed data are saved in the well-designed database (Figure 2D-E).
In MVIP web page, users can access the data through different modules, including Data-Matrix, Search, Genome-Browser, Analysis and Download (Figure 3A).
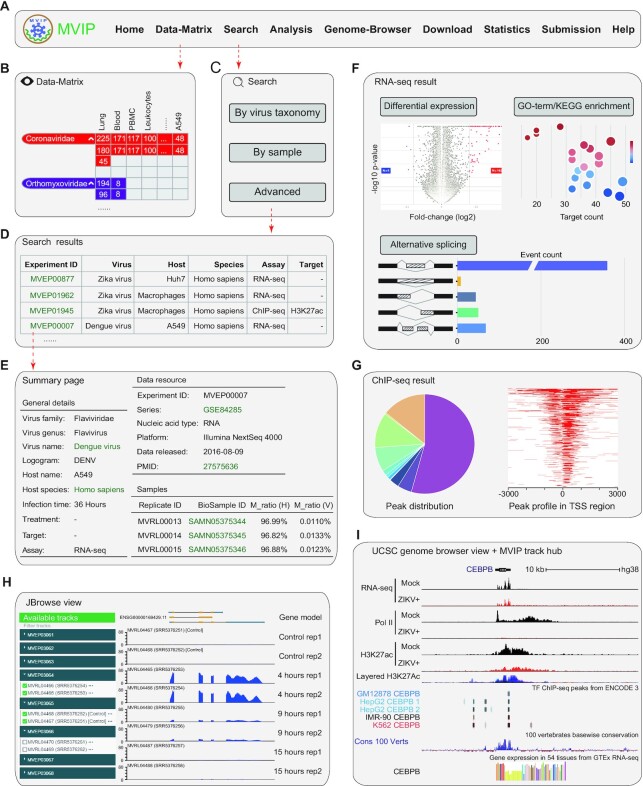
Figure 3.
Main modules and usage of MVIP. (A) Navigation bar of the main modules in MVIP web page. (B) Matrix-like viewer of available multi-omics data. (C) Three search modes for filtering MVIP records. (D) An example of search results including experiment ID, virus, host, species, assay, and target. (E) An example of the summary page describing an experiment. (F) An example of the analysis results for RNA-seq data. (G) An example of ChIP-seq analysis results. (H) An example of JBrowse2 view of omics data signals. (I) An example of UCSC genome browser view via MVIP track hub along with ENCODE ChIP-seq, UCSC conservation data and GTEx RNA-seq data.
A data-matrix interface for browsing the omics data
The ‘Data-Matrix’ page is an interactive and digitized table that allows users to quickly search and browse omics data. The matrix is organized by row for ordered viruses and column for cell types or tissues (Figure 3B), which can be filtered from the left panel. To view the details of omics data, users can click the URL over the numbers (Figure 3B), linking to the records displayed by page.
Search interface for retrieving omics data
MVIP provides user-friendly search options supporting auto-completion to retrieve various omics data under viral infection. Users can query the omics data of interest through three ways: ‘By virus taxonomy’, ‘By sample’ and ‘Advanced’ (Figure 3C). Based on the virus taxonomy query, users can select a virus according to ‘Virus Family’ and ‘Virus Genus’ of interest. Clicking the ‘Search’ button will present users the omics data associated with the virus. In sample-based query mode, users can select a host of interest according to ‘BioSample Type’ and ‘Tissue Type’ and clicking ‘Search’ button will give users the omics data associated with host that under various viral infections. In advanced mode, users can query related omics data by selecting more search options, including ‘Virus Family’, ‘Virus Genus’, ‘Virus Name’, ‘Host Name’, ‘Assay’ and ‘Cell Type’.
The brief information of searched results is displayed in a table supporting sorting and filtering (Figure 3D). The interactive table describes the omics data including MVIP ID, virus name, logogram, host, assay, target, species, GEO ID and Pubmed ID. Users can click the link on MVIP ID to view the details, such as data summary, sample information, and analysis results (Figure 3E). For RNA-seq data, MVIP provides 4 classes of analysis results including differential expression, GO-term enrichments, KEGG pathway enrichment, and alternative splicing (Figure 3F). In addition, MVIP also enables ‘Threshold’ options supporting users to set custom thresholds to select DEGs with different stringencies. For each gene listed in the table, the gene ID and gene symbol have links out to the Ensembl and GeneCard databases, respectively. The corresponding genomic signals of the genes can be viewed in our local JBrowse2-based genome browser directly, or in UCSC genome browser via MVIP track hub. Moreover, users can export the results of interest in the current page, or download the complete results via ‘Download complete table’ button or from the ‘File Details’ panel. Meanwhile, MVIP provides four analysis results associated with peaks, including the annotation information, visualization of peak coverage signals over chromosomes, peak profiles around the TSS region, and the distribution of peaks in genome (Figure 3G).
Customized genome browsers and data visualization
To help user view and compare various omics data under viral infections, we developed a customized genome browser using JBrowse2 (Figure 3H). By entering the genomic location or gene ID, users can conveniently explore the available track data related to the gene of interest. All tracks are classified based on host species, viruses, and assays, and similar tracks are organized into track groups, in which the tracks can be shown by toggling the checkboxes.
For genomes available in UCSC genome browser, users can also view our MVIP data with many other data in UCSC simultaneously, which are in the same genomic coordinates and enable users to distill hypotheses from jointly exploring them. For example, as shown in Figure 3I, we observe that the CEBPB gene is repressed upon ZIKA infection (ZIKA+ versus mock control) in RNA, Pol II, and H3K27ac levels. The results are consistent with the original report (59), indicating the correctness of our processing. Interestingly, in combining with CEBPB ChIP-seq from ENCODE data in UCSC genome browser, we see that CEBPB protein has multiple binding sites around its own gene locus, and two sites are very conserved from UCSC’s 100 vertebrate basewise conservation track (Figure 3I bottom). Integration of these existing data suggests a CEBPB auto-regulatory loop functioning during ZIKA infection.
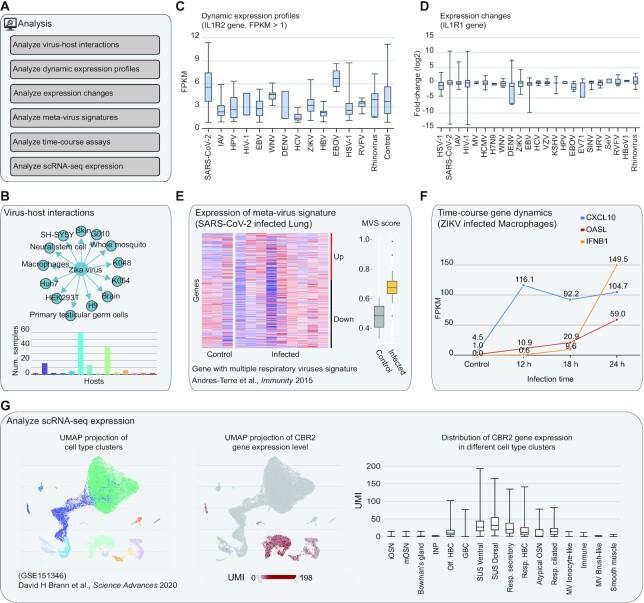
Online analysis tools and user cases
In the Analysis page, MVIP provides six practical analysis tools to directly answer a set of common biological questions (Figure 4A). With ‘Analyze virus-host interactions’ tool, users can submit a virus or a host to analyse the virus–host interactions with omics data (Figure 4B). With ‘Analyze dynamic expression profiles by gene’ tool, users can submit one gene or a gene list of interest, then MVIP will show the dynamic expression profiles (FPKM or TPM) of genes under various viral infections in comparison with control group (Figure 4C). With ‘Analyze expression changes by gene’ tool, users can submit one gene or a gene list of interest, then MVIP will show the fold-changes between infection versus corresponding controls (Figure 4D).
Figure 4.
Analysis tools in MVIP web server. (A) List of six analysis tools in MVIP. (B–G) Interactive figure examples generated from these six tools.
With ‘Analyze meta-virus signature’ tool, for a given virus and host, users submit a list of genes with or without defined changes (up- or down-regulation), MVIP will show the heatmap of gene expression in virus infected and control samples (Figure 4E). If changes are defined, such as the common viral transcriptional signature in (60), the MVS scores will be computed for all samples and presented as boxplots for the viral infection and control groups, respectively. MVIP also provides several gene lists with known signatures from literature (30). With ‘Analyze gene dynamics in time-course assay’ tool, users can view the expression dynamics at different time-points after viral infection for a list of submitted genes (Figure 4F). Using ‘Analyze scRNA-seq expression’ tool, users can search scRNA-seq dataset with processed expression data, and users can submit a gene of interest to view the cell type UMAP, its expression distribution on the UMAP and that in different cell types or conditions (Figure 4G).
Data download and statistics
MVIP provides a list-like tool for downloading the omics data, gene expression and analysis results associated with various viral infections, in ‘.bw’, ‘.tsv’, ’.bed’ and ‘.csv’ formats. Users can download these data through clicking the links to the corresponding filenames. In the ‘Statistics’ page, MVIP provides users with digital and graphical displays about assays, cell types, and tissue types information.
Data submission and update
Because the analysis of any omics data takes enormous time and space of computation, MVIP does not support online analysis of user data currently. We will routinely and continuously update MVIP with new data and tools. Meanwhile, we have created a Submission page for users to notify us new omics data related to viral infection. We recommend users to submit the GEO or SRA accessions with optional metadata. We will collect and curate the data, analyze them using our pipeline and resource, and then integrate the results into the database for all users in a timely manner.
DISCUSSION
To the best of our knowledge, MVIP is the first database providing comprehensive and multi-dimensional large-scale data for multiple species responding to various virus infections. Currently available virus-related databases mainly focus on viral sequence information, including OpenFluDB (61), RVDB (62), MMRdb (63) and the three NAR databases referred in the introduction section. The Viruses.STRING (64) database only provides the virus–host protein–protein interactions.
MVIP fills the gap for various genomic data under viral infection, integrates the largest number (>6500 samples) and most diverse types of omics data, and provided a global network of broad virus-host interactions. Moreover, MVIP provides several user-friendly custom dynamic charts and useful tools to help users better investigate molecular events under viral infections. In addition, MVIP provides analysis results for multiple types of sequencing data. However, we have not processed the raw data of Hi-C, Capsnatch-seq and scRNA-seq data currently, due to the complexity of the analysis or sample heterogeneity. We plan to construct pipelines to process these omics data in future updates. Meanwhile, we will upgrade the post-mapping analyses such as peak calling when better programs are available.
MVIP currently focuses on the host responses, and we plan to investigate and integrate the molecular events of viruses, such as viral subgenomic RNA dynamics we recently found for SARS-CoV-2 (65). With the enhanced functionalities on data visualization and analysis, MVIP would provide new convenient resources for a wide variety of biologists including virologists, microbiologists, immunologists, cancer and molecular biologists, physicians, and bioinformaticians, etc.
DATA AVAILABILITY
The MVIP database is freely available for the research community at https://mvip.whu.edu.cn/. Users are not required to register or login to use the database, and to download the curated and processed data.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members in Zhou lab for insightful discussions during the process of this investigation. We are grateful to Drs Dong Wang and Jinsong Qiu for critical reading of the manuscript. Part of the computation in this work was done on the supercomputing system in the Supercomputing Center of Wuhan University.
Contributor Information
Zhidong Tang, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Weiliang Fan, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Qiming Li, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Dehe Wang, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Miaomiao Wen, Institute for Advanced Studies, Wuhan University, Wuhan 430072, China.
Junhao Wang, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Xingqiao Li, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Yu Zhou, State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China; Institute for Advanced Studies, Wuhan University, Wuhan 430072, China; RNA Institute, Wuhan University, Wuhan 430072, China; Frontier Science Center for Immunology and Metabolism, Wuhan University, Wuhan 430072, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31922039, 31871316]; Hubei Provincial Natural Science Foundation of China [2020CFA057, 2020CFA017]; National Key R&D Program of China [2017YFA0504400]; Fundamental Research Funds for the Central Universities [2042020kf1069]. Funding for open access charge: National Natural Science Foundation of China [31922039].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wobus C.E., Nguyen T.H.. Viruses are everywhere—what do we do. Curr. Opin. Virol. 2012; 2:60–62. [DOI] [PubMed] [Google Scholar]
- 2. Koyuncu O.O., Hogue I.B., Enquist L.W.. Virus infections in the nervous system. Cell Host Microbe. 2013; 13:379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peng R., Wu L.-A., Wang Q., Qi J., Gao G.F.. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021; 46:848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mesri E.A., Feitelson M.A., Munger K.. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014; 15:266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Martel C., Ferlay J., Franceschi S., Vignat J., Bray F., Forman D., Plummer M.. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012; 13:607–615. [DOI] [PubMed] [Google Scholar]
- 6. Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L.et al.. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009; 10:321–322. [DOI] [PubMed] [Google Scholar]
- 7. Wu A., Wang L., Zhou H.-Y., Ji C.-Y., Xia S.Z., Cao Y., Meng J., Ding X., Gold S., Jiang T.et al.. One year of SARS-CoV-2 evolution. Cell Host Microbe. 2021; 29:503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F.. The proximal origin of SARS-CoV-2. Nat. Med. 2020; 26:450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J.et al.. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020; 382:1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suryawanshi R.K., Koganti R., Agelidis A., Patil C.D., Shukla D. Dysregulation of Cell Signaling by SARS-CoV-2. Trends Microbiol. 2021; 29:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mahalingam S., Peter J., Xu Z., Bordoloi D., Ho M., Kalyanaraman V.S., Srinivasan A., Muthumani K.. Landscape of humoral immune responses against SARS-CoV-2 in patients with COVID-19 disease and the value of antibody testing. Heliyon. 2021; 7:e06836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watanabe T., Watanabe S., Kawaoka Y.. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010; 7:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Speck S.H., Ganem D. Viral latency and its regulation: lessons from the γ-herpesviruses. Cell Host Microbe. 2010; 8:100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Shen Q., Wu S., Chen S., Lomakin I.B.et al.. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020; 80:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C.et al.. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020; 183:1325–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S., Lee Y., Choi Y., Son A., Park Y., Lee K.-M., Kim J., Kim J.-S., Kim V.N.. The SARS-CoV-2 RNA interactome. Mol. Cell. 2021; 81:2838–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M.et al.. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012; 487:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison A.G., Lin T., Wang P.. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020; 41:1100–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Isaacson M.K., Ploegh H.L.. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009; 5:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisfeld A.J., Halfmann P.J., Wendler J.P., Kyle J.E., Burnum-Johnson K.E., Peralta Z., Maemura T., Walters K.B., Watanabe T., Fukuyama S.et al.. Multi-platform ’omics analysis of human ebola virus disease pathogenesis. Cell Host Microbe. 2017; 22:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A.et al.. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021; 594:246–252. [DOI] [PubMed] [Google Scholar]
- 22. Lieberman P.M. Epigenetics and genetics of viral latency. Cell Host Microbe. 2016; 19:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z.et al.. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012; 40:D593–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho P.T., Montiel-Garcia D.J., Wong Jonathan.J., Carrillo-Tripp M., Brooks C.L., Johnson J.E., Reddy V.S.. VIPERdb: a tool for virus research. Annu. Rev. Virol. 2018; 5:477–488. [DOI] [PubMed] [Google Scholar]
- 25. Montiel-Garcia D., Santoyo-Rivera N., Ho P., Carrillo-Tripp M., Iii C.L.B., Johnson J.E., Reddy V.S.. VIPERdb v3.0: a structure-based data analytics platform for viral capsids. Nucleic Acids Res. 2021; 49:D809–D816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paez-Espino D., Roux S., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Huntemann M., Reddy T.B.K., Pons J.C., Llabrés M.et al.. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2019; 47:D678–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B.. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018; 46:D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M.. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021; 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yue Z., Zheng Q., Neylon M.T., Yoo M., Shin J., Zhao Z., Tan A.C., Chen J.Y.. PAGER 2.0: an update to the pathway, annotated-list and gene-signature electronic repository for Human Network Biology. Nucleic Acids Res. 2018; 46:D668–D676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yue Z., Zhang E., Xu C., Khurana S., Batra N., Dang S.D.H., Cimino J.J., Chen J.Y.. PAGER-CoV: a comprehensive collection of pathways, annotated gene-lists and gene signatures for coronavirus disease studies. Nucleic Acids Res. 2021; 49:D589–D599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M.et al.. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011; 39:D1005–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kodama Y., Shumway M., Leinonen R.. on behalf of the International Nucleotide Sequence Database Collaboration The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B.. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018; 46:D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R.et al.. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010; 28:1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leinonen R., Sugawara H., Shumway M.International Nucleotide Sequence Database Collaboration . The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Utturkar S., Dassanayake A., Nagaraju S., Brown S.D.. Bacterial differential expression analysis methods. Methods Mol. Biol. Clifton NJ. 2020; 2096:89–112. [DOI] [PubMed] [Google Scholar]
- 39. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K.et al.. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liao Y., Smyth G.K., Shi W.. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L.. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015; 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen S., Park J.W., Lu Z., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lovci M.T., Ghanem D., Marr H., Arnold J., Gee S., Parra M., Liang T.Y., Stark T.J., Gehman L.T., Hoon S.et al.. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013; 20:1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uren P.J., Bahrami-Samani E., Burns S.C., Qiao M., Karginov F.V., Hodges E., Hannon G.J., Sanford J.R., Penalva L.O.F., Smith A.D.. Site identification in high-throughput RNA-protein interaction data. Bioinforma. Oxf. Engl. 2012; 28:3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu Z., Hu L., Shi B., Geng S., Xu L., Wang D., Lu Z.J.. Ribosome elongating footprints denoised by wavelet transform comprehensively characterize dynamic cellular translation events. Nucleic Acids Res. 2018; 46:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Merkel A., Fernández-Callejo M., Casals E., Marco-Sola S., Schuyler R., Gut I.G., Heath S.C.. gemBS: high throughput processing for DNA methylation data from bisulfite sequencing. Bioinforma. Oxf. Engl. 2019; 35:737–742. [DOI] [PubMed] [Google Scholar]
- 50. Mölder F., Jablonski K.P., Letcher B., Hall M.B., Tomkins-Tinch C.H., Sochat V., Forster J., Lee S., Twardziok S.O., Kanitz A.et al.. Sustainable data analysis with Snakemake. F1000Research. 2021; 10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu G., Wang L.-G., Han Y., He Q.-Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu G., Wang L.-G., He Q.-Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinforma. Oxf. Engl. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 55. Hong E.L., Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M.et al.. Principles of metadata organization at the ENCODE data coordination center. Database J. Biol. Databases Curation. 2016; 2016:baw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L.et al.. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D.et al.. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinforma. Oxf. Engl. 2014; 30:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carlin A.F., Vizcarra E.A., Branche E., Viramontes K.M., Suarez-Amaran L., Ley K., Heinz S., Benner C., Shresta S., Glass C.K.. Deconvolution of pro- and antiviral genomic responses in Zika virus-infected and bystander macrophages. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E9172–E9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Andres-Terre M., McGuire H.M., Pouliot Y., Bongen E., Sweeney T.E., Tato C.M., Khatri P.. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity. 2015; 43:1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liechti R., Gleizes A., Kuznetsov D., Bougueleret L., Le Mercier P., Bairoch A., Xenarios I.. OpenFluDB, a database for human and animal influenza virus. Database J. Biol. Databases Curation. 2010; 2010:baq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goodacre N., Aljanahi A., Nandakumar S., Mikailov M., Khan A.S.. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. mSphere. 2018; 3:e00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Almansour I., Alhagri M.. MMRdb: measles, mumps, and rubella viruses database and analysis resource. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019; 75:103982. [DOI] [PubMed] [Google Scholar]
- 64. Cook H.V., Doncheva N.T., Szklarczyk D., von Mering C., Jensen L.J.. Viruses.STRING: a virus-host protein-protein interaction database. Viruses. 2018; 10:E519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang D., Jiang A., Feng J., Li G., Guo D., Sajid M., Wu K., Zhang Q., Ponty Y., Will S.et al.. The SARS-CoV-2 subgenome landscape and its novel regulatory features. Mol. Cell. 2021; 81:2135–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MVIP database is freely available for the research community at https://mvip.whu.edu.cn/. Users are not required to register or login to use the database, and to download the curated and processed data.