Abstract
NC2 (Dr1-Drap1 or Bur6-Ydr1) has been characterized in vitro as a general negative regulator of RNA polymerase II (Pol II) transcription that interacts with TATA-binding protein (TBP) and inhibits its function. Here, we show that NC2 associates with promoters in vivo in a manner that correlates with transcriptional activity and with occupancy by basal transcription factors. NC2 rapidly associates with promoters in response to transcriptional activation, and it remains associated under conditions in which transcription is blocked after assembly of the Pol II preinitiation complex. NC2 positively and negatively affects approximately 17% of Saccharomyces cerevisiae genes in a pattern that resembles the response to general environmental stress. Relative to TBP, NC2 occupancy is high at promoters where NC2 is positively required for normal levels of transcription. Thus, NC2 is associated with the Pol II preinitiation complex, and it can play a direct and positive role at certain promoters in vivo.
TATA-binding protein (TBP) nucleates the assembly of the RNA polymerase II (Pol II) transcription machinery by specifically recognizing TATA promoter elements and directly interacting with general transcription factors TFIIA and TFIIB (12, 34, 36, 40). TBP is a component of distinct multiprotein complexes that affect Pol II transcription in vitro and in vivo (26, 31). One such TBP complex, TFIID, contains approximately 14 associated factors (TAFs) that contact initiator or downstream promoter elements and that may serve as targets for transcriptional activator proteins (2, 3, 35, 39, 43). It is generally believed that TFIID is the predominant form of TBP that mediates Pol II transcription, although an alternative form(s) of transcriptionally active TBP exists in Saccharomyces cerevisiae cells (22, 29). TBP also associates with Mot1 (1, 7), the multiprotein Not-Ccr4 complex (9, 25, 30), and the NC2 (Dr1-Drap or Bur6-Ydr1) heterodimer (13, 17, 32, 33), and it has been suggested that these proteins function as general negative regulators that inhibit some aspect of TBP function (26).
NC2 (Dr1-Drap) was originally identified in human cells as a biochemical activity that inhibits basal TBP-dependent transcription in vitro (17, 32). NC2 is a heterodimer between two histone fold proteins (13, 33), and a homologous complex (Ydr1-Bur6) is required for growth in yeast cells (10, 19, 37). NC2 directly interacts with TBP and DNA, but the TBP-NC2-TATA complex is transcriptionally inactive in vitro, because it is unable to bind TFIIA or TFIIB and hence the remainder of the basal Pol II machinery (13, 33). In this regard, TBP mutations that inhibit interaction with NC2 are located near surfaces that mediate TFIIA or TFIIB binding (4, 20). NC2 also interacts in vitro with the repression domain of the AREB6 repressor (16) and with the hyperphosphorylated form of Pol II (5).
After this paper was initially submitted, it was shown that NC2 can function in vitro as a positive or negative effector of transcription in a manner that depends on the structure of the core promoter (45). Specifically, in certain kinds of cell extracts, NC2 can stimulate transcription in vitro from Drosophila promoters containing downstream promoter elements (DPEs), whereas it represses transcription from TATA-containing promoters (45). DPEs have a conserved DNA sequence motif that is located a precise distance from the TATA element and mRNA initiation site, and they interact with the TAF60 component of the TFIID complex (2, 24). DPEs appear to be as widely utilized as TATA elements in Drosophila core promoters (24), although they have not been described for yeast promoters. These recent experiments do not address whether the positive role of NC2 reflects a productive association with the preinitiation complex, and in this regard, NC2 blocks the association of TFIIA and TFIIB with promoters in vitro (13, 33). In addition, the positive role of NC2 in these experiments may be due to inhibition of another inhibitory factor in the cell extracts employed. Finally, the physiological significance of these biochemical observations remains to be established.
Several lines of genetic evidence have suggested that NC2 functions as a general negative regulator in yeast cells. First, bur6 mutations were identified by their ability to increase transcription from enhancerless promoters, suggesting that NC2 inhibits basal transcription in vivo (37). Second, overproduction of NC2 is toxic, and this toxicity can be reversed by overproduction of TBP (19). Third, the essential function(s) of NC2 can be overcome by a mutation in TFIIA (46) or the Sin4 component of Pol II holoenzyme (18, 27). Fourth, reduced NC2 function permits cell growth and Pol II transcription in cells with functionally compromised Srb4 (10, 25), a component of Pol II holoenzyme that is universally required for Pol II transcription (15, 41). This functional antagonism between NC2 and Pol II holoenzyme suggests that NC2 is a global negative regulator, although this global effect is observed under a nonphysiological condition where Pol II holoenzyme is functionally compromised. There are a few examples of genes whose transcription decreases upon loss of NC2 function, suggesting that NC2 might play a positive role in transcription in vivo (27, 37). However, there is no evidence addressing whether these positive effects of NC2 on transcription are direct or indirect.
To investigate the mechanism of transcriptional regulation by NC2 in vivo, we directly measure NC2 association with yeast promoters by chromatin immunoprecipitation using an epitope-tagged derivative of Bur6. In addition, we analyze the transcriptional profile of a bur6 mutant strain on a genomic scale using microarrays. Our results indicate that, in contrast to the conventional view, NC2 associates with the Pol II preinitiation complex in vivo. Further, NC2 appears to act directly to increase transcription of certain genes. Thus, NC2 is not simply a general negative regulator that blocks preinitiation complex formation, but rather it selectively affects transcription both positively and negatively.
MATERIALS AND METHODS
Chromatin immunoprecipitations were generally performed in yeast strain FT4 (42) that either did or did not express an epitope-tagged version of Bur6 containing three copies of the HA1 epitope at its amino terminus. The levels of untagged and tagged Bur6 were comparable, as determined by Western blotting using a Bur6 antibody. For the experiment in Fig. 4, isogenic KIN28 and kin28-ts16 strains were used as described previously (8, 23). Cells were grown at 30°C in Casamino Acid medium lacking uracil supplemented with 2% glucose to an optical density at 600 nm of 0.6. Chromatin immunoprecipitations used monoclonal antibodies to the hemagglutinin (HA) epitope (F7 from Santa Cruz) or polyclonal antibodies to TBP or TFIIB on identical samples. Quantitative PCR analyses were performed as described previously (22, 23), except for the experiment in Fig. 2, which was performed in real time using an Applied Biosystems 7700 sequence detector. The NC2/TBP ratios were calculated by dividing background-subtracted NC2 binding by background-subtracted TBP binding. The average of the occupancy ratios for promoters analyzed in Fig. 1 was arbitrarily defined as 1.0. Individual values represent the averages from at least three independent experiments and have an error of approximately ±25%. Therefore, promoters showing values of 2.0 and greater (see Fig. 6) contain relatively high NC2 levels that are clearly beyond experimental error. Detailed information on experimental procedures, genetic reagents, high-density array technology, and data analysis can be found on the World Wide Web at http://www.wi.mit.edu/young/expression/nc2. The bur6 temperature-sensitive strain was generated and kindly provided by Danny Reinberg.
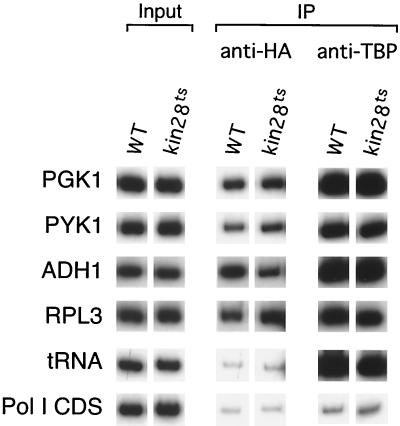
FIG. 4.
Bur6 remains bound at promoters under conditions in which transcription is blocked after assembly of the Pol II machinery. Bur6 and TBP association following thermal inactivation of Kin28, the TFIIH subunit that phosphorylates the Pol II C-terminal tail, is shown. Cells were grown at 24°C and were shifted to 37°C for 75 min to inactivate Kin28.
FIG. 2.
Bur6 association is localized over the promoter. Bur6 and TBP occupancy at the indicated regions of the RPS11B locus (drawing to scale). PCR analysis was performed in real time.
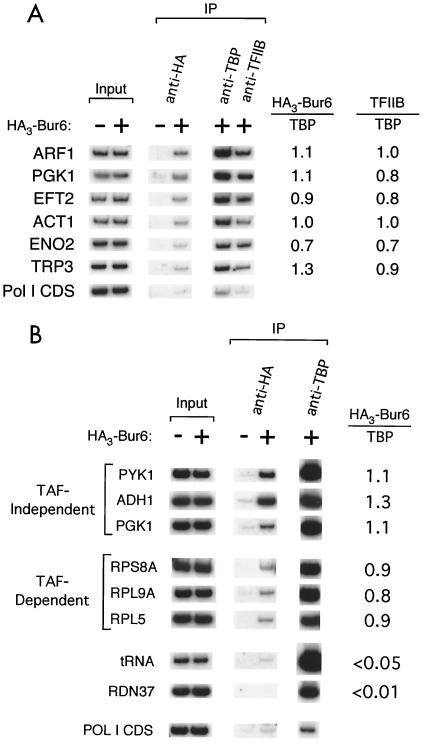
FIG. 1.
Association of NC2 with promoters strongly correlates with association of TBP and TFIIB. (A) Bur6, TBP, and TFIIB occupancy at selected promoters. (B) Bur6 and TBP occupancy at TAF-dependent and TAF-independent promoters. Cross-linked chromatin preparations from HA3-Bur6 and untagged Bur6 were immunoprecipitated with antibodies to the HA epitope, TBP, or TFIIB. Promoter-specific PCR products were generated from input chromatin or immunoprecipitated DNA, and the NC2/TBP or TFIIB/TBP occupancy ratios are indicated.
FIG. 6.
Increased Bur6 occupancy relative to TBP at promoters positively regulated by Bur6. Analysis of genes that are positively or negatively regulated by NC2 as defined by the microarray analysis shown in Fig. 4. (A) Absolute promoter binding (in arbitrary units) by Bur6 (left axis) and TBP (right axis) expressed over background. (B) Plot of background-subtracted Bur6/TBP occupancy ratios. The average occupancy ratios are 1.0 for NC-independent promoters, 1.1 for NC-inhibited promoters, and 3.9 for NC2-stimulated promoters.
RESULTS
NC2 specifically associates with Pol II promoters in a manner that strongly correlates with transcriptional activity and occupancy by TBP and TFIIB. In previous work, we and others demonstrated that the level of transcriptional activity in yeast cells is strongly correlated with the level of TBP association at promoters (23, 28). Moreover, the relative associations of TBP, TFIIB, and TFIIA are very tightly correlated with each other; i.e., the TBP/TFIIA and TBP/TFIIB occupancy ratios are constant at essentially all promoters (22). In contrast, association of the TAFs in the TFIID complex is not strictly correlated with TBP occupancy, and the TAF/TBP occupancy ratio can vary over a 5- to 10-fold range depending on the promoter (22, 29). Given the biochemical properties of NC2 and the genetic evidence that NC2 functions as a global repressor, we expected that NC2 occupancy would be inversely correlated with transcriptional activity and with TFIIA and TFIIB association.
In the initial experiment, we analyzed NC2 occupancy at several Pol II promoters, whose transcriptional activities span a wide range (Fig. 1A). In contrast to our expectation, NC2 associates with promoters in a manner that is strongly correlated with TBP and TFIIB occupancy and hence transcriptional activity. Specifically, the NC2/TBP and NC2/TFIIB occupancy ratios at these promoters are essentially indistinguishable, indicating that NC2 behaves similarly to TFIIA and TFIIB but differently from TAFs. The NC2/TBP ratio is not significantly affected by whether the promoter contains high or low levels of TAFs (and hence TFIID) (Fig. 1B), suggesting that promoter occupancy by TAFs and NC2 is not mutually exclusive. NC2 does not associate with a tRNA promoter, which is transcribed by Pol III, even though this (and most other) tRNA promoter contains canonical TATA elements that specifically bind TBP (14, 44). In addition, NC2 does not associate with the rRNA promoter, which is transcribed by Pol I and shows high TBP occupancy. Finally, mapping experiments on the RPS11B locus indicate that TBP and NC2 colocalize over the promoter (Fig. 2), indicating that NC2 is not associated with the elongating Pol II complex. Thus, NC2 specifically associates with the functional Pol II machinery at promoters.
NC2 rapidly associates with promoters in response to transcriptional induction.
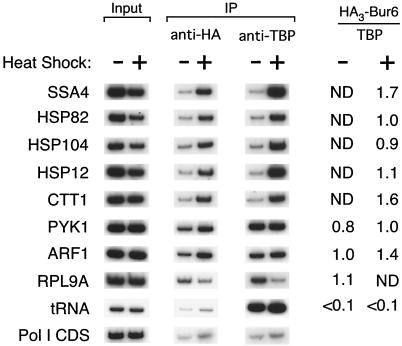
Although the above analysis was performed under steady-state growth conditions, the results suggest that NC2 is recruited to promoters by transcriptional activator proteins. We addressed this issue directly by analyzing NC2 occupancy at heat shock promoters under conditions where transcription was strongly induced upon a rapid heat shock (Fig. 3). Heat shock causes a rapid increase of NC2 occupancy at all heat shock promoters tested, and the NC2/TBP occupancy ratios are comparable to those of the non-heat-shock promoters. Thus, in accord with their abilities to activate transcription, the Hsf1, Msn2, and Msn4 activators cause a rapid association of NC2 with target promoters.
FIG. 3.
Bur6 is rapidly recruited to promoters by transcriptional activators. Bur6 and TBP occupancy at heat shock-inducible and uninducible promoters is shown. Cells were grown at 24°C and were heat shocked for 15 min at 39°C. SSA4 and HSP82 are activated by heat shock factor (Hsf1), HSP12 and CTT1 are activated by the Msn2 and Msn4 activators, and HSP104 is activated by both classes of activator. Heat shock inhibits the RPL9A promoter but does not affect the other promoters tested.
NC2 association correlates with formation of a Pol II preinitiation complex, not transcriptional activity per se.
Phosphorylation of the C-terminal tail of Pol II by the Kin28 subunit of TFIIH is required for transcription at a step after formation of the preinitiation complex such as promoter clearance or elongation. Mutational inactivation of Kin28 results in rapid inhibition of transcription (8), but the Pol II machinery remains stably associated with the promoter in vivo (21, 23). Loss of Kin28 function does not affect NC2 occupancy (Fig. 4), indicating that NC2 associates with promoters even under conditions in which the Pol II machinery is assembled at promoters in an elongation-incompetent state. Thus, NC2 association with promoters correlates with formation of a Pol II preinitiation complex, not transcriptional activity per se.
NC2 positively and negatively affects transcription in a manner that overlaps the response to general environmental stress.
To address the requirement for NC2 at individual promoters, we compared the transcriptional profiles of wild-type and bur6 mutant strains on a genome-wide level using microarrays (Fig. 5). Thermal inactivation of Bur6 resulted in twofold or greater transcriptional effects on approximately 852 genes, which represent 17% of all yeast genes. Of these, 415 genes show decreased transcription, whereas 437 genes display increased transcription. Thus, NC2 can positively or negatively affect transcription of selected genes.
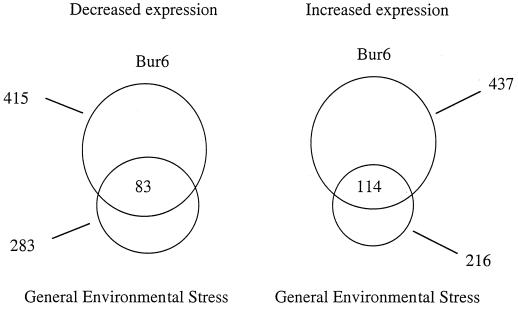
FIG. 5.
Relationship between Bur6 function and the response to environmental stress. Venn diagram indicating the number of genes that are increased or decreased at least twofold in cells in which Bur6 is thermally inactivated (top circles) or in cells subjected to a wide variety of environmental stresses (bottom circles). The number of genes affected by both conditions is indicated at the intersections.
When yeast cells are subjected to a broad range of environmental stress conditions, there is a common response involving approximately 500 genes (6, 11). Depending on the specific gene, these various environmental stress conditions result in positive or negative regulation of transcription. Strikingly, the set of NC2-affected genes significantly overlaps the set of genes that are coregulated in response to a broad range of environmental stress conditions. Approximately 40% of the 500 genes that are positively or negatively affected by environmental stress are affected in the same manner by loss of NC2 function. This relationship between NC2 regulation and environmental stress is specific and not due to thermal inactivation per se, because the NC2 pattern of expression has never been observed in comparable analyses of numerous temperature-sensitive mutants in other components of the Pol II machinery (15). One model to explain this relationship is that loss of NC2 affects the transcription of a gene(s) that results in the generation of a stress signal. Alternatively, environmental stress could result in the transient inactivation of NC2.
Increased NC2 association at promoters positively affected by NC2.
Formally, the transcriptional profile of the bur6 mutant strain indicates that NC2 behaves selectively in vivo as both a positive and negative factor, although it does not establish whether NC2 acts directly at the affected promoters. To address this issue, we examined NC2 occupancy at promoters at which NC2 appears to act positively or negatively (Fig. 6). For all five promoters in which NC2 appears to act positively (i.e., gene expression is reduced upon loss of NC2 function), the NC2/TBP occupancy ratio is 2.5- to 5-fold higher than observed on NC2-independent promoters. Four of these NC2-stimulated promoters (the exception being CDC31) are also stimulated in response to general stress (6, 11). In contrast, the six promoters whose activity appears to be negatively regulated by NC2 show NC2/TBP occupancy ratios comparable to those of NC2-independent promoters. Thus, high NC2 levels are specifically observed on promoters that require NC2 for normal levels of expression, indicating that NC2 can perform a direct and positive role in transcription.
DISCUSSION
NC2 associates with the Pol II preinitiation complex in vivo.
Several lines of evidence indicate that NC2 associates with the Pol II preinitiation complex in yeast cells. First, the Pol II preinitiation complex is defined in vivo by the constant TBP/TFIIA/TFIIB occupancy ratio (22) and its very strong correlation with Pol II occupancy and transcriptional activity (23, 28). In terms of promoter occupancy, NC2 behaves like a general Pol II transcription factor, indicating that it associates with the preinitiation complex. Second, NC2 association is not simply due to its ability to interact with TBP and form TBP-NC2-TATA complexes, because NC2 associates specifically with active Pol II promoters. High TBP occupancy (Pol I and Pol III promoters) or canonical TATA elements (most Pol III promoters and many inactive Pol II promoters) are clearly insufficient for NC2 association in vivo. Third, the hypothesis that the observed NC2 occupancy represents an association with TBP (or TFIID) alone or with a partially assembled preinitiation complex (e.g., lacking TFIIB or Pol II holoenzyme) is inconsistent with the strong correlation of NC2 occupancy with general factors. Furthermore, loss of TFIIB or the Srb4 component of Pol II holoenzyme significantly reduces TBP occupancy, indicating that partial Pol II preinitiation complexes are unstable in vivo (23, 28).
NC2 functions directly to increase transcription of certain genes in vivo.
In virtually all microarray (or more limited) experiments involving yeast strains with mutations in specific transcriptional regulatory proteins, some genes show increased transcription whereas other genes show decreased transcription. By themselves, however, such experiments do not allow the determination of whether the observed positive or negative effects on gene expression are due to the direct action of the transcriptional regulatory protein at the affected promoter. The possibility of indirect effects is particularly relevant for proteins that do not exhibit sequence-specific binding to DNA, such as general factors, TBP-associated proteins, or components of chromatin-modifying activities. Hence, it is essential to develop independent criteria for distinguishing direct from indirect effects on transcription.
Here, we utilize relative promoter association in vivo as such an independent criterion. Specifically, we show that the NC2/TBP occupancy ratios at all five NC2-stimulated promoters tested are significantly higher (average, 3.9-fold; range, 2.5- to 5-fold) than the ratios observed for NC2-independent or NC2-inhibited promoters. This observation provides strong evidence that NC2 performs a direct transcriptional role at the NC2-stimulated promoters tested and presumably at most other NC2-stimulated promoters. Indeed, it is very difficult to formulate a plausible hypothesis in which the positive effects of NC2 are indirect, given that analysis of more than 25 genes reveals a strict relationship between increased NC2/TBP occupancy ratios and positive NC2 effects on transcription. Our results do not distinguish whether NC2 is directly or indirectly responsible for the NC2-dependent repression of selected genes.
It is important to note that NC2-TBP occupancy ratios are arbitrarily defined in absolute terms and hence do not provide any information about the stoichiometry of NC2 and TBP molecules on promoters. Although we presume that a Pol II preinitiation complex contains one molecule each of TBP, TFIIB, and TFIIA, we have no experimental information on how many molecules of NC2 associate with a preinitiation complex in vivo. However, we suspect that that the high NC2/TBP occupancy ratios observed at NC2-stimulated promoters do not reflect multiple NC2 molecules associated with an individual preinitiation complex but rather reflect increased association of NC2 with preinitiation complexes assembled at these promoters. For this reason, we believe that NC2 associates with, but is not a stoichiometric component of, the preinitiation complex at the vast majority of promoters (i.e., those with NC2/TBP occupancy ratios of 1.0).
Molecular implications.
Our conclusion that NC2 associates with functional Pol II preinitiation complexes and can perform a direct and positive role in transcription is in apparent conflict with the ability of NC2 to inhibit TBP-TFIIB-TATA and TBP-TFIIA-TATA complex formation and basal transcription in vitro. However, these biochemical experiments were performed with purified proteins at nonphysiological concentrations in the absence of Pol II holoenzyme. We suspect that NC2 interactions (direct or indirect) with Pol II holoenzyme alleviate or override the inhibitory effects observed with purified general factors, perhaps by competitive binding or conformational alteration of the relevant protein surfaces. In support of this idea, NC2 interacts genetically with the Srb4 (10) and Sin4 (18, 27) components of yeast Pol II holoenzyme. NC2 also interacts in vitro with the hyperphosphorylated form of Pol II (5), although NC2 remains associated with promoters in the absence of Kin28 function (Fig. 4), conditions that block phosphorylation of the Pol II C-terminal domain (21). Thus, under physiological conditions, our results are inconsistent with the model that NC2 globally represses Pol II transcription by inhibiting TBP function and assembly of the preinitiation complex, although we cannot exclude the possibility that this model operates at certain promoters.
Though unexpected, the ability of yeast NC2 to selectively and directly increase transcription in vivo is in broad accord with the concurrent and unexpected observation that NC2 is selectively required for activity of promoters containing DPEs in vitro (45). Furthermore, our observation that NC2 and TAFs (and hence TFIID) can cooccupy promoters in vivo is consistent with the requirement for TFIID (i.e., not TBP) to mediate NC2-dependent activation of TATA-less promoters in vitro (45). However, TAFs are not required for NC2 to associate with promoters in vivo, and it is possible that a TBP-NC2 complex represents a non-TFIID form of transcriptionally active TBP inferred from previous studies (22, 29). Finally, the results from the microarray analysis that NC2 can have both positive and negative effects on transcription in yeast cells are in broad accord with recent results obtained with Drosophila promoters in vitro (45).
At present, we do not know whether the selective positive effects of NC2 in yeast cells directly correspond to DPE-dependent transcription in vitro. DPEs have yet to be described in yeast promoters, and it is unclear whether this reflects the true absence of DPEs or complications due to the atypical structure of yeast core promoters. In Drosophila melanogaster and most eukaryotes, DPEs are located a precise distance downstream of both the TATA and initiator elements (24). In yeast, the distance between TATA and initiator elements is considerably larger and highly variable and promoter regions are AT rich (38), thereby making it difficult to define a TATA-less promoter and to know where a yeast DPE should be located. Nevertheless, it is interesting that NC2 positively affects his3 transcription that depends on a weak TATA element but not on a canonical TATA element (27).
Although yeast NC2 associates with promoters in a manner analogous to general transcription factors, it selectively stimulates or inhibits transcription of particular genes. This property is broadly consistent with the observations in vitro that NC2 acts during assembly of the preinitiation complex and functions positively or negatively depending on the structure of the core promoter (45). This functional dichotomy and the relatively high NC2 occupancy at NC2-stimulated promoters might reflect preferential NC2 interactions with certain DNA sequences or NC2-dependent conformational changes of TBP and/or TFIID that alter promoter recognition. Alternatively, as promoter specificity is affected by multiple forms of transcriptionally active TBP (22, 29) and perhaps by multiple forms of Pol II holoenzyme, NC2 might stimulate or inhibit transcription depending on which isoform of the Pol II machinery is present at a particular promoter.
ACKNOWLEDGMENTS
We are particularly grateful to Danny Reinberg for providing the bur6 temperature-sensitive strain and for fruitful discussions on NC2 function. We also thank Jim Kadonaga for communicating unpublished information about positive and negative functions of NC2 in vitro and Laurent Kuras and Mario Mencia for advice and commentary throughout the course of the work.
This work was supported by research grants from the National Institutes of Health to K.S. (GM 30186 and GM 53720) and R.A.Y. (GM 34365).
REFERENCES
- 1.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 2.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 4.Cang V, Auble D T, Prelich G. A new regulatory domain on the TATA-binding protein. EMBO J. 1999;18:6662–6671. doi: 10.1093/emboj/18.23.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castano E, Gross P, Wang Z X, Roeder R G, Oelgeschlager T. The C-terminal domain-phosphorylated II0 form of RNA polymerase II is associated with the transcription repressor NC2 (Dr1/DRAP1) and is required for transcription activation in human nuclear extracts. Proc Natl Acad Sci USA. 2000;97:7184–7189. doi: 10.1073/pnas.140202297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Causton H C, Ren B, Koh S S, Harbison C T, Kanin E, Jennings E G, Lee T I, True H L, Lander E S, Young R A. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chicca J J, Auble D T, Pugh B F. Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol Cell Biol. 1998;18:1701–1710. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cismowski M, Laff G, Soloman M, Reed S. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart M A, Struhl K. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 10.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiger J H, Hahn S, Lee S, Sigler P B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 13.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 14.Heard D J, Kiss T, Filipowicz W. Both Arabidopsis TATA-binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: evidence for gene-specific changes in DNA binding specificity of TBP. EMBO J. 1993;12:3519–3528. doi: 10.1002/j.1460-2075.1993.tb06026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda K, Halle J P, Stelzer G, Meisterernst M, Kawakami K. Involvement of negative cofactor NC2 in active repression by zinc finger-homeodomain transcription factor AREB6. Mol Cell Biol. 1998;18:10–18. doi: 10.1128/mcb.18.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Cabane K, Hampsey M, Reinberg D. Genetic analysis of the Ydr1-Bur6 repressor complex reveals an intricate balance among transcriptional regulatory proteins in yeast. Mol Cell Biol. 2000;20:2455–2465. doi: 10.1128/mcb.20.7.2455-2465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Na J G, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T K, Zhao Y, Ge H, Bernstein R, Roeder R G. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 21.Komarnitsky P, Cho E-J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 23.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;389:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 24.Kutach A K, Kadonaga J T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T I, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 27.Lemaire M, Xie J, Meisterernst M, Collart M A. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol Microbiol. 2000;36:163–173. doi: 10.1046/j.1365-2958.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- 28.Li X-L, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;389:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 29.Li X-Y, Bhaumik S R, Green M R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 30.Liu H-Y, Badarinarayana V, Audino D C, Rappsilber J, Mann M, Denis C L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 32.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 33.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 34.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 35.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 36.Orphanides G, Lagrange T, Reinberg D. The general initiation factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 37.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- 39.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 40.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 41.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 43.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 44.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 45.Willy P J, Kobayashi R, Kadonaga J T. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 46.Xie J, Collart M, Lemaire M, Stelzer G, Meisterernst M. A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 2000;19:672–682. doi: 10.1093/emboj/19.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]