Abstract
Addition of serum to mitogen-starved cells activates the cellular immediate-early gene (IEG) response. Serum response factor (SRF) contributes to such mitogen-stimulated transcriptional induction of many IEGs during the G0-G1 cell cycle transition. SRF is also believed to be essential for cell cycle progression, as impairment of SRF activity by specific antisera or antisense RNA has previously been shown to block mammalian cell proliferation. In contrast, Srf−/− mouse embryos grow and develop up to E6.0. Using the embryonic stem (ES) cell system, we demonstrate here that wild-type ES cells do not undergo complete cell cycle arrest upon serum withdrawal but that they can mount an efficient IEG response. This IEG response, however, is severely impaired in Srf−/− ES cells, providing the first genetic proof that IEG activation is dependent upon SRF. Also, Srf−/− ES cells display altered cellular morphology, reduced cortical actin expression, and an impaired plating efficiency on gelatin. Yet, despite these defects, the proliferation rates of Srf−/− ES cells are not substantially altered, demonstrating that SRF function is not required for ES cell cycle progression.
Both postmitotic cells and mitotic cells persisting in a resting G0 state can be induced by extracellular stimuli to activate an instantaneous gene expression program called the immediate-early gene (IEG) response (13). Postmitotic cells such as neurons rely on the IEG response to convert, for example, neurotransmitter-mediated activation processes into long-term adaptive responses (19, 46). G0 cells, on the other hand, employ the IEG response to activate transcription factor-encoding genes (like c-fos, junB, or Egr-1), which subsequently regulate gene cascades enabling G1 progression (2, 10, 13, 20). This G0-to-G1 transition is commonly studied by the addition of mitogens (or serum) to serum-deprived cells. The IEG response displays a very rapid and transient gene activation profile that does not require de novo protein biosynthesis but instead utilizes preexisting cellular signaling components and transcription factors. Signaling cascades that mediate the IEG response include the mitogen-activated protein (MAP) kinase network, Ca2+-dependent pathways, NF-κB signaling, and JAK/STAT signaling. The MAP kinase pathway assumes an especially important role in triggering the IEG response (41).
Of the many transcription factors that are targeted by MAP kinase signaling, the serum response factor (SRF) has been characterized in significant detail. SRF (28), a ubiquitously expressed MADS box protein (29, 35, 36), mediates both the signal-stimulated transcriptional induction of IEG and the activation of cell type-specific genes. The latter include muscle-specific genes or neuronal genes. SRF binds to serum response elements (SREs) in the promoters of these genes by recognizing the CArG-box sequence (CC[A/T]6GG) (24, 29, 40). SRF can recruit additional proteins to SREs, e.g., the Ets family ternary complex factors (TCFs), muscle-specific accessory factors, or others (for review, see reference 42; Nordheim, submitted). The Ets/TCFs are direct targets of MAP kinase signaling cascades (8, 41), instrumental for rapid and transient IEG induction of, e.g., c-fos and Egr-1. SRF can also activate genes in a TCF-independent fashion (9, 17, 18), primarily through the involvement of Rho signaling in response to, e.g., lysophosphatidic acid (LPA) stimulation (15, 16).
The indicated contribution of SRF to mitogen-stimulated transcriptional induction of IEGs during the G0-G1 transition (13) and the requirement of the SRF homologue MCM1 at the G2-M transition in Saccharomyces cerevisiae (3, 22) suggested an essential regulatory function for SRF in both G0 exit and the ensuing cell cycle progression. This notion was strengthened by experiments in which SRF activity was impaired by using specific antisera or antisense RNAs blocking the proliferation of rat embryo fibroblasts (7) or myoblasts (39), respectively. In stark contrast to these cell-based assays, we have demonstrated that cells of Srf−/− mutant embryos are viable and support early embryonic growth in utero up to E6.0 (4). To explore SRF function in cells of the early embryo, we have used the embryonic stem (ES) cell as an in vitro model system for cellular proliferation, differentiation, and early embryonic development (37, 44, 45).
We demonstrate here that wild-type murine ES cells cannot be rendered quiescent upon withdrawal of serum yet still contain a vigorous IEG response upon serum readdition. SRF-deficient ES cells were found to have a severe defect in both IEG activation and actin gene expression, but the accompanying alterations in cellular morphology did not prevent normal rates of ES cell proliferation.
MATERIALS AND METHODS
ES cells.
Germ line competent D3 and E14 ES cells were provided by R. Jaenisch (Whitehead Institute, Cambridge, Mass.) and K. Rajewsky (Genetics Institute, Cologne, Germany), respectively. Srf−/+ ES cells, derived from the E14 line, were described previously (4, 44). Selection for enhanced G418 resistance and characterization of Srf−/− ES cells, as well as the generation of Srf−/−rescue ES cells, are described in reference 44.
ES cell culture conditions. (i) General growth conditions.
ES cells were kept without feeders on gelatin-coated dishes in Dulbecco modified Eagle medium, containing 4.5 g of glucose/liter and 3.7 g of NaHCO3/liter supplemented with 2mM l-glutamine, 100 U of penicillin/ml, 100 g of streptomycin/ml, 0.1 mM mercaptoethanol, 15% fetal calf serum (FCS), and 1,000 U of leukemia inhibitory factor/ml (referred to as complete medium). Cultivation was at 37°C in a humidified atmosphere at 5% CO2, and cells were passaged every 48 h.
(ii) ES cell culture conditions for IEG induction.
For serum-withdrawal experiments, cells were seeded and grown for 12 to 24 h in complete medium for optimal attachment. The medium was then exchanged and cells were kept for 24 to 38 h in complete medium lacking FCS. The expression of IEGs was assayed before and 10, 30, 60, or 180 min after the addition of 15% FCS, 100 ng of phorbol ester tetradecanoyl phorbol acetate (TPA)/ml, or 20 μM LPA (final concentrations). LPA stimulations were done for 30 min only. The kinetic profiles of IEG induction proved somewhat variable in ES cells, explaining the differences in the expression kinetics of Fig. 5A and B and 5C and D, respectively, as well as the magnitude of the deviations shown in Fig. 6, which represent the mean of five separate experiments, each sampled at the 30-min time point.
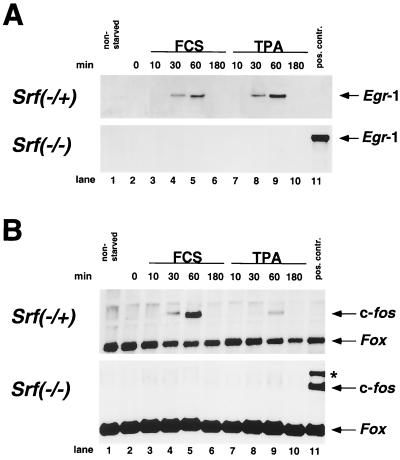
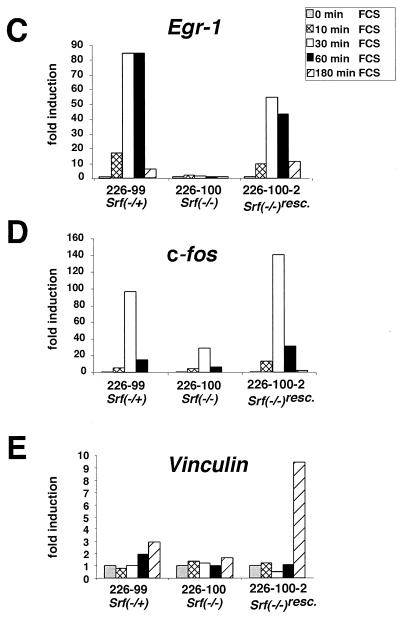
FIG. 5.
Severe impairment in Srf−/− ES cells of IEG induction by serum or TPA. (A and B) Northern blot analysis of Egr-1 (A) and c-fos. (B) RNA expression in the Srf−/+ ES line 44 (top panels) or the Srf−/− ES line 226-100 (bottom panels) under normal growth conditions (lane 1), after serum withdrawal for 24 h (lane 2), or after serum withdrawal and subsequent induction with either 15% serum (lanes 3 to 6) or 100 ng of TPA/ml (lanes 7 to 10) for the indicated times. Lane 11 represents a positive hybridization control included in the Srf−/− blot. The Egr-1 blot (A) was reprobed for c-fos (B). In addition, a Fox probe was used to verify equivalent RNA loading in each lane (33). The asterisk (B) indicates a residual Egr-1 signal still present from prior probing of the blot. (C to E) Quantitative RT-PCR measuring expression of Egr-1 (C), c-fos (D), and Vinculin (E) in ES cell lines of the indicated Srf genotype, as well as in Srf−/− ES cells that ectopically express human Srf cDNA (Srf−/−rescue). mRNA induction factors relative to prestimulation levels are given. Cells were either serum starved for 36 h or serum starved and subsequently restimulated with 15% serum for the indicated times. mRNA levels were normalized to mRNA levels of the Hprt gene, and values are given as fold induction over nonstimulated cells. Each kinetic measurement was performed in duplicate.
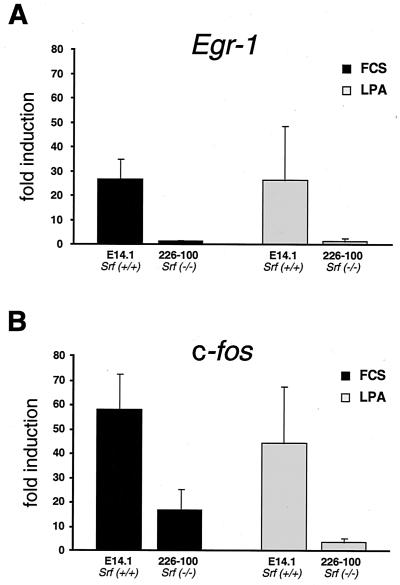
FIG. 6.
Severe impairment in Srf−/− ES cells of IEG induction by LPA. Quantitative RT-PCR measuring expression of Egr-1 (A) and c-fos (B) in ES cell lines of the indicated Srf genotypes. mRNA induction factors relative to prestimulation levels are given for the 30-min time points subsequent to LPA addition. Standard deviations representing five independent experiments are provided (see Materials and Methods).
Northern blotting.
RNA was isolated by the guanidinium isothiocyanate method (5). RNA (20 μg) was electrophoresed and transferred to Gene Screen nylon membrane (NEN, Boston, Mass.). Filters were successively hybridized with [32P]dCTP-labeled human Egr-1, v-fos, and, for standardization, mouse fox cDNA probes (33), under standard conditions.
Western blotting.
Preparation of cell extracts was as described previously (11), and Western blotting using a pan-actin antibody (Acurate, Westbury, N.Y.) was performed according to reference 44.
Quantitative RT-PCR.
Total RNA preparation (RNeasy kit; QIAGEN) and first-strand cDNA synthesis (Superscript II; Gibco) were done according to the manufacturers' protocols. One microgram of total RNA treated with DNase I was used for reverse transcription (RT). One-twentieth of the RT reaction was included in a 25-μl PCR reaction. For quantitative analysis, the SYBR Green PCR technology was used (Perkin-Elmer). Real-time detection of the PCR product was monitored by measuring the increase in fluorescence caused by the binding of SYBR Green to double-stranded DNA with the ABI PRISM 7700 sequence detector. In order to obtain relative quantification, a threshold cycle (Ct), the cycle at which a statistically significant increase in fluorescence occurs, was derived from the resulting PCR profiles of each sample. Ct is a measure for the amount of template present in the starting reaction. To correct for different amounts of total cDNA in the starting reaction, Ct values of an endogenous control (Hprt) were subtracted from those of the corresponding sample, resulting in ΔCt. The relative quantitation value is expressed as 2−ΔCt. Induction of specific mRNAs by IEG stimulation is then plotted as the induction factor relative to uninduced mRNA levels in starved cells.
Primers used were as follows, where F is the forward primer and R is the reverse primer: β-Actin, F(GGCGCTTTTGACTCAGGAT) and R(GGGATGTTTGCTCCAACCAA); c-fos, F(AAGGGAACGGAATAAGATGGC) and R(CAACGCAGACTTCTCATCTTCAA); Egr-1, F(GCCGAGCGAACAACCCTA) and R(TCCACCATCGCCTTCTCATT); Hprt, F(GCCTAAGATGAGCGCAAGTTG) and R(TACTAGGCAGATGGCCACAGG); and Vinculin, F(CCAAGGTCAGAGAAGCCTTCC) and R(CGTAGCTGTTCAAGGTCTGGT).
Cell cycle analysis.
For analysis of DNA content, cells were harvested, incubated for 15 min in staining solution (0.5 mg of propidium iodide [PI]/ml, 4 mM sodium citrate, 0.03% Triton X-100), treated for 30 min with RNase A, and diluted with phosphate-buffered saline (PBS). Fluorescence intensity was determined by flow cytometry on a Becton Dickinson FACScan, and the percentages of G1-, S-, and G2-phase cells were calculated with the LYSIS (Becton Dickinson) or MODFIT software program. To measure DNA synthesis, serum-starved cells were cultured in the presence of 30 μm of bromodeoxyuridine (BrdU) (Roche) for 30 min, trypsinized, and stored in 70% ethanol for >2 h. Cells were rehydrated in PBS containing 1% heat-inactivated fetal serum (PBS/IFS) treated with 0.3 ml of 2 M HCl at room temperature for 20 min, washed in PBS/IFS, neutralized with 0.1 M sodium borate (pH 8.0) for 2 min, and washed again in PBS/IFS. BrdU was identified by labeling the cells with anti-BrdU monoclonal antibodies at a 1:10 dilution (Roche) and fluorescein isothiocyanate-conjugated goat anti-mouse antibody. Cells were then resuspended in modified PI staining solution (0.4 mg of PI/ml, 4 mM sodium citrate, 25 μg of RNase A/ml) for 15 min and analyzed by flow cytometry as described above.
SEM.
ES cells were grown on gelatin-coated coverslips for up to 72 h. Samples were fixed with 1.6% glutaraldehyde in 20 mM HEPES–120 mM NaCl for 5 min at room temperature and for 1 h at 4°C, postfixed with 1% osmium tetroxide in PBS for 1 h on ice, washed with H2O, and treated with 1% aqueous uranyl acetate for 1 h at 4°C. For scanning electron microscopy (SEM) ES cell colonies grown on coverslips were dehydrated in ethanol and critical-point dried from CO2. The samples were sputter coated with 8 nm of gold-palladium and examined at 20 kV accelerating voltage in a Hitachi S-800 field emission scanning electron microscope.
Indirect immunofluorescence.
Cells were grown on gelatin-coated coverslips in complete growth medium for 48 h. The samples were then fixed in 4% formaldehyde and permeabilized in 0.2% Triton X-100. Nonspecific binding was blocked by incubating the cells for 1 h in 1% bovine serum albumin in PBS at 37°C before staining with a rabbit anti-E-cadherin antibody (1:300 in PBS; gift of H. Beug) for 1 h at 37°C. Incubation with Oregon green-conjugated secondary antibody (Molecular Probes; 1:200 in 0.2% bovine serum albumin) was performed for 30 min at 37°C. To visualize filamentous actin, 1 U of Texas red phalloidin (Molecular Probes) was included in the mixture. Coverslips were washed four times with PBS and once in water, air dried, and mounted in Moviol. Image acquisition was done with LSM 510 (Zeiss).
RESULTS
To investigate the contributions of SRF to cellular growth control, we generated and characterized Srf−/− ES cells (44) derived from the Srf−/+ ES cells that were originally used for blastocyst injections and the generation of Srf−/+ mice (4). The homozygous Srf−/− ES cells were obtained from the heterozygous Srf−/+ ES cells upon selection for elevated cellular resistance levels to the drug G418. In the Srf−/− ES cells obtained, no protein was observed that might be antigenically related to SRF and no SRF-derived DNA binding activity could be seen (44). Rescued cells (Srf−/−rescue) expressed SRF at levels three to four times higher than wild-type ES cells (44).
Normal proliferation but impaired colony formation of Srf−/− ES cells.
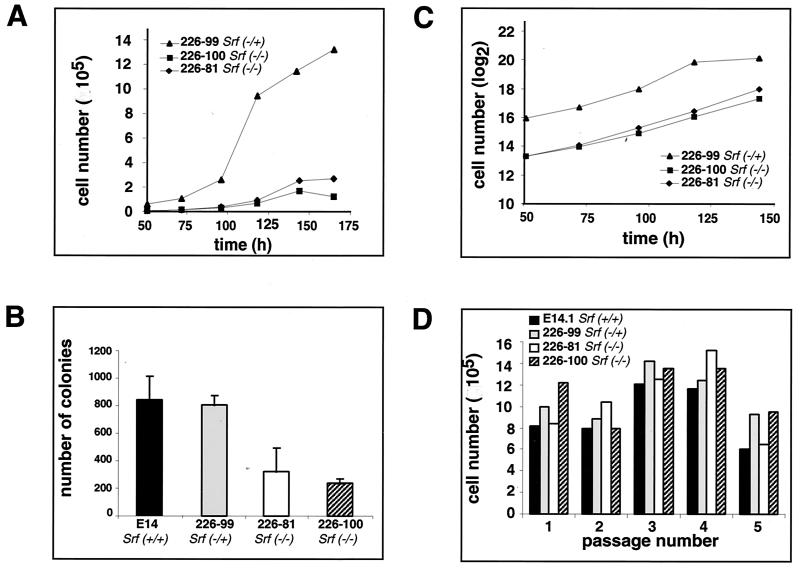
Since SRF activity appears to be essential for the growth of in vitro cultured somatic cells (7, 39), we sought to determine if SRF deficiency altered the proliferation characteristics of ES cells. We first examined the growth rates of one Srf−/+ and two Srf−/− ES clones. Equal numbers of cells were seeded at low densities in leukemia inhibitory factor-containing medium, and the proliferation kinetics of colonized cells were monitored by counting total cell numbers for a period of 7 days. Proliferation started after a lag period of 1 to 2 days in all clones. The growth curves revealed that the colonies of the Srf−/+ clone increased more rapidly in cell number than did those of each of the two Srf−/− clones (Fig. 1A). Additionally, after 150 h of culture, the total cell numbers of SRF-deficient cells plateaued (E81 Srf−/−) or even declined (E100 Srf−/−), whereas Srf−/+ cell numbers increased, reaching a higher saturation cell count than SRF-deficient cells. At this late phase of the growth curve, ES cell colonies had become rather big and cells began to display signs of differentiation (data not shown). In the case of Srf−/− cells this was accompanied by an enhanced tendency to undergo apoptosis (unpublished observation). During the early phase of the growth curve, i.e., up to 125 h, the observed lower increase in the Srf−/− ES cell number might have been caused by either an impaired plating efficiency or an altered proliferation capacity. We therefore first investigated the efficiencies of colony formation subsequent to plating. We seeded identical numbers of ES cells of different Srf genotypes and counted the number of colonies formed after 6 days of culturing. Both Srf−/− ES clones displayed significantly reduced efficiencies in colony formation compared to the wild type and Srf−/+ clones (Fig. 1B). We believe that this difference is the prime cause for the differences in the cumulative cell numbers over time, as observed in Fig. 1A with cultures of ES cells either containing or lacking SRF.
FIG. 1.
Proliferation kinetics, colony-forming capabilities, and growth rates of Srf−/− ES cells. (A) Proliferation kinetics at low cell density. Growth curves of ES cell lines with the indicated Srf genotypes were generated by seeding 2 × 103 cells at day 0 and counting total cell numbers at the indicated time points. One representative experiment of three independent ones is shown. (B) Efficiencies of colony formation upon plating at low cell density. A total of 2 × 103 cells of each of the indicated ES cell lines was plated on gelatin-coated dishes, and the resulting colonies were counted after 6 days of cultivation. Each value represents the mean of duplicate measurements, except for ES cell line 226-100, where four measurements are averaged. (C) Rates of proliferation at low cell density. Shown are semilogarithmic plots of cell numbers counted in panel A during the growth periods of exponential growth.The slopes of the graphs indicate proliferation rates. (D) ES cell proliferation at high cell density. Cells (4 × 105) of the indicated ES cell lines were plated on gelatin-coated dishes (10 cm). At each passage (24 h after plating), ES cells were trypsinized and counted.
Given the observed differences in plating efficiency, the proliferation rates of heterozygous and homozygous mutant cell lines shown in Fig. 1A nevertheless appeared to be similar, as judged by the slopes of the semilogarithmic plots of cell numbers over time (Fig. 1C). Also, no significant differences in proliferation rates were observed between wild-type and Srf−/+ ES cells (data not shown). To investigate more closely the rates of proliferation of SRF-deficient ES cells, we also monitored growth under high cell-density conditions over several passages. Cells (4 × 105) were seeded on a gelatin-coated 10-cm dish, trypsinized every 24 h, counted, and replated at the above-mentioned density. Under these conditions, differences in plating efficiency or spontaneous differentiation between SRF-containing and SRF-deficient cells were not apparent (unpublished data) and similar total cell counts were reached after each 24-h cycle (Fig. 1D). Therefore, also under high-density plating conditions, no significant SRF-dependent differences in ES cell proliferation were observed. Collectively we infer from these data that SRF does not contribute in an important way to the rates at which ES cells proliferate.
Altered morphology of Srf−/− ES colonies is accompanied by impaired cortical actin expression.
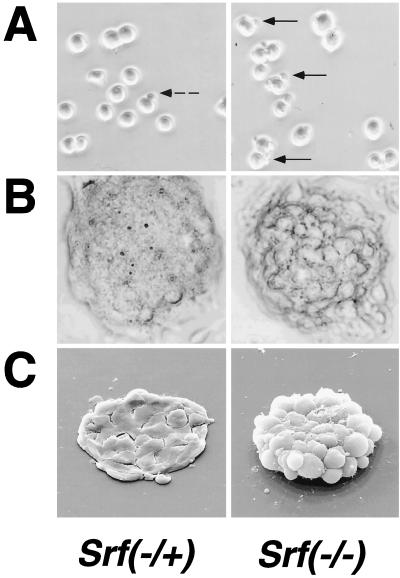
The observed differences in plating efficiency of Srf−/− ES cell clones (Fig. 1B) warranted a closer inspection of their morphologies. Light microscopic analysis of ES cells revealed protrusions on many of the SRF-deficient cells (Fig. 2A, right panel), features rarely seen with Srf−/+ ES cells (Fig. 2A, left panel) or with wild-type ES cells (data not shown). After 72 h of culture, colonies of cells deficient in SRF were less compact and appeared to display less tight cell-cell interaction (Fig. 2B). This was confirmed by SEM, which demonstrated that individual cells within an Srf−/− ES colony established fewer contacts with neighboring cells than those within an Srf−/+ ES colony (Fig. 2C). We speculate that the decreased plating efficiency is a direct consequence of these altered structural characteristics.
FIG. 2.
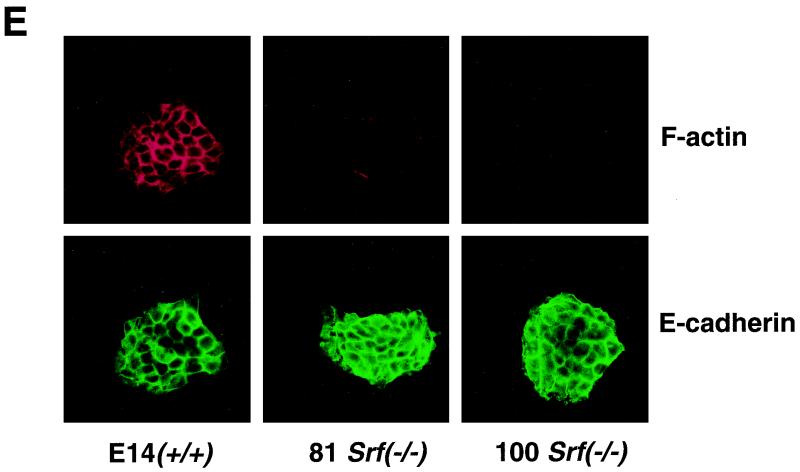
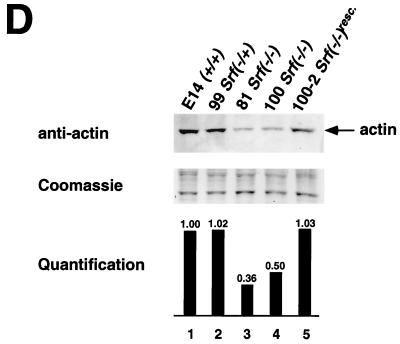
Altered cellular morphologies and actin expression levels of Srf−/− ES cells and their colonies. (A) Light microscopy of individual ES cells. (B) Light microscopy of ES cell colonies. (C) SEM of ES cell colonies. (A to C) Cell and colony morphologies of Srf−/+ (left panels) or Srf−/− (right panels) ES cells were determined after initial cultivation for 48 h in complete growth medium, trypsinization, and subsequent plating for either 3 h (A) or 72 h (B and C). Cell protrusions (marked by arrows in panel A) were observed very frequently in the homozygous mutant ES cells but were found rarely in Srf−/+ cells. (D) Western blot analysis of total cellular actin protein in ES cells. Whole cell extracts from ES cells of the indicated Srf genotypes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted, and probed with a pan-actin antiserum (upper panel). Coomassie staining of the gel after blotting confirms equal loading (middle panel). Densitometric quantification of the actin signals is shown in the lower panel, where the signal from the E14.1 Srf+/+ wild-type cells was arbitrarily set at one. (E) Histological distributions of F-actin and E-cadherin in cell colonies derived from ES cells of different Srf genotypes. Wild-type cells (left panel) and the two Srf−/− cell lines 226-81 (middle panel) and 226-100 (right panel) were grown on coverslips for 48 h. Expression of F-actin (upper row) and E-cadherin (lower row) was detected with Texas red phalloidin and an E-cadherin-specific antiserum, respectively.
SRF directs the expression of Actin genes (26), and the actin cytoskeleton profoundly influences cell surface activities, like cell-cell and cell-substrate interactions (34). We therefore investigated cytoskeletal actin expression (β-actin) in Srf−/− ES cells. Figure 2D reveals a 50 to 65% drop in actin protein expression levels in SRF-deficient cells, accompanied by a strongly reduced intensity of cortical actin networks (Fig. 2E). While reduced in intensity, the cortical actin arrangement—at this level of resolution—does not appear severely disturbed in structural terms. At the same time, expression and cellular localization of E-cadherin appear unaffected by the lack of SRF (Fig. 2E).
Cell cycle progression of Srf−/− ES cells.
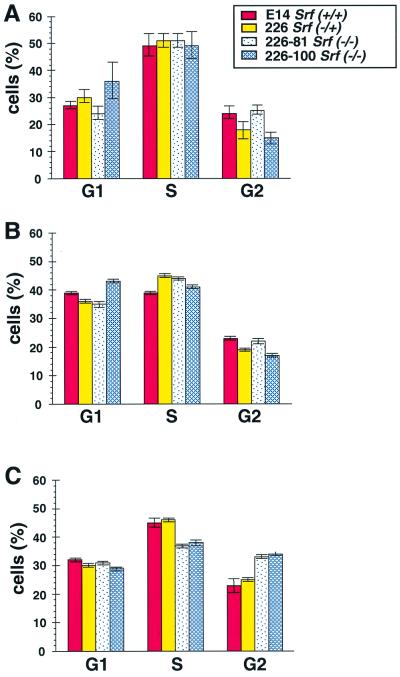
The apparently normal proliferation rate of SRF-deficient ES cells contrasted with earlier demonstrations of SRF being required for the proliferation of somatic cells. SRF is known to regulate gene activation at specific stages of the cellular growth cycle (i.e., the G0-G1 transition as quiescent cells enter into the active growth cycle, progression of cycling cells through G1, and the G2-M transition). This cell cycle-specific nature of SRF function suggested that ES cells lacking SRF may display specific alterations in the progression through the cell cycle, despite having a growth rate similar to wild-type cells. The parental ES cell line E14 (Srf+/+), the heterozygous line 226 (Srf−/+), and the two homozygous lines 226–81 and 226–100 (each Srf−/−) were studied regarding their cell cycle progression. In nonstarved, normally growing cultures (Fig. 3A), the majority of cells were found in the S phase and lower but comparable amounts of cells were found in each of the G1 and G2 phases. No obvious differences in these distributions were seen between wild-type, heterozygote, or homozygote mutant ES cells.
FIG. 3.
Progression through the cell cycle of Srf−/− ES cells. Shown is the distribution within the cell cycle of different ES cell lines. ES cells of the indicated genotypes were cultivated under three different conditions (A to C). (A) Cell cultivation under continuous, normal growth conditions for 3 days. (B) Cells after serum starvation for 38 h. (C) Cells after serum starvation for 38 h and subsequent readdition of serum for 24 h. (A to C) Cells were stained with PI for determinations of DNA content and cell cycle distribution. The percentages of cells found in the G1, S, and G2 phases are shown.
Despite the normal proliferation rate and cell cycle distribution of Srf−/− ES cells cultured in the presence of serum, we speculated that serum restimulation of quiescent Srf−/− ES cells may be altered in some manner. Thus, we analyzed the cell cycle distribution profiles of the various ES cell lines after serum withdrawal and upon subsequent stimulation with serum. In none of the ES clones tested did serum withdrawal lead to efficient growth arrest. After a 38-h period of starvation, the percentage of G1 cells was slightly but significantly increased at the expense of S-phase cells, but there was no difference between the cells of different Srf genotypes (Fig. 3B). When wild-type and heterozygote ES cells were reexposed to serum for 24 h (Fig. 3C), the cell cycle distribution again resembled that of asynchronously growing cells. Restimulation of starved mutant cells with serum, however, led to the accumulation of 10% more of the cells in the G2-M phase of the cell cycle, a small but significant alteration in the cell cycle distribution of the cells (Fig. 3C). Thus, while SRF function does not appear to be essential for the cell cycle progression of ES cells, it may nonetheless contribute to the G2-M checkpoint function, as has been previously suggested (21).
Cell cycle progression of wild-type ES cells.
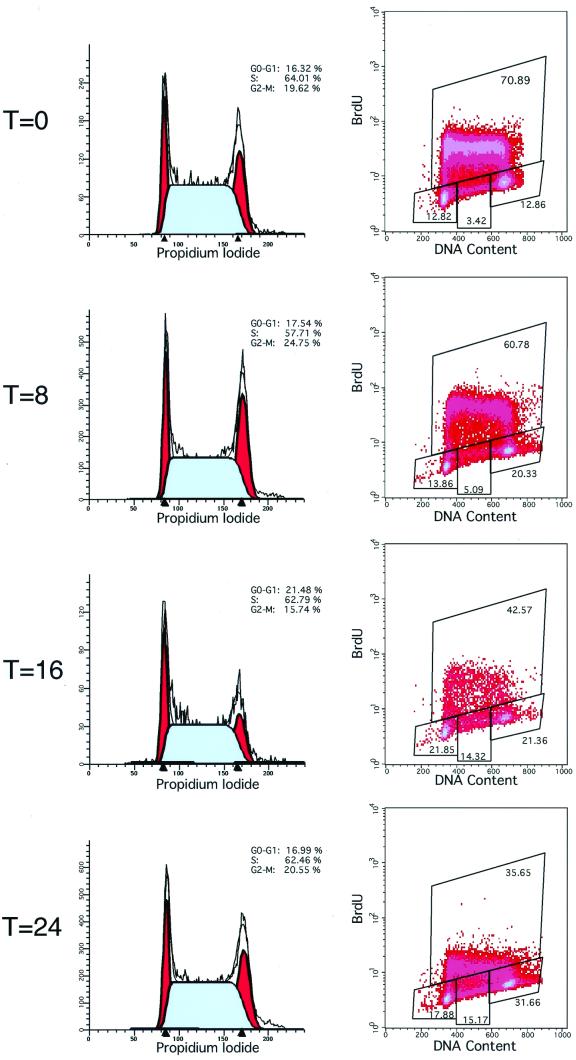
We were surprised that the E14 ES cells failed to accumulate in G1 in the absence of serum. We wished to test whether this was a characteristic of ES cells in general and whether the ES cells are altered in any way in the absence of serum. To do so, we examined a second independently derived ES cell line, termed D3, which is also capable of germ line transmission. As noted for E14 ES cells above, the cell cycle profile of D3 cells was not substantially altered up to 24 h after withdrawal of serum (Fig. 4). In this experiment we also examined DNA synthesis by the uptake of the thymidine analog BrdU. Although serum-starved cells retained normal cell cycle distribution, their rate of DNA synthesis gradually slowed, as evidenced by a progressive decrease in the rate of uptake of BrdU. Thus, ES cells appear to be sensitive to serum withdrawal, as evidenced by a slowing in the rate of DNA synthesis, yet nonetheless lack the ability to undergo G0 exit.
FIG. 4.
Cell cycle progression of wild-type ES cells upon serum withdrawal. Wild-type ES cells were labeled with BrdU for 30 min and harvested prior to or following 8, 16, or 24 h of serum starvation as indicated. Cell cycle distribution (left panels) was determined by staining cells for DNA content with PI, and the rates of DNA synthesis (right panels) were determined by immunostaining cells for BrdU incorporation (y axis) and plotting against DNA content (x axis). The percentages of cells in the G1, S, and G2 phases are indicated.
SRF-mediated induction of the IEGs Egr-1 and c-fos is dispensable in the ES cell.
Given the relatively inefficient exit of ES cells from the active cell cycle into G0 and the lack of a requirement for SRF for ES cell proliferation, we first wondered whether wild-type ES cells were able to mount an efficient IEG activation response. The activation of SRF complexes by the MAP kinase cascade has been implicated in the transcriptional up-regulation of IEGs such as c-fos and Egr-1 in the G0-G1 transition. This step is believed to be essential for resting cells to reenter the cell cycle (13). We therefore investigated the pattern of Egr-1 and c-fos induction following stimulation of ES cells with intact SRF function, either wild-type (data not shown) or Srf−/+ ES cells (Fig. 5). Serum-withdrawn ES cells were stimulated for up to 3 h with either serum or the phorbol ester TPA, and the resulting mRNA levels were measured over time by Northern blot analysis. ES cells containing at least one intact Srf allele were able to mount a robust IEG response, as judged by c-fos and Egr-1 gene activation profiles.
A strongly contrasting result was seen with the Srf−/− cells. They exhibited a drastically impaired induction of both Egr-1 and c-fos following serum stimulation. Whereas no induction at all could be seen for Egr-1 in the Srf−/− ES cells, a residual response was observed with c-fos. This data set was fully confirmed by separate experiments which investigated independently derived RNA preparations by quantitative RT-PCR analysis (Fig. 5C and D and Fig. 6). While the induction of other IEGs was not examined, we suggest that it is similarly compromised in these cells.
To confirm that the lack of IEG induction in the mutant ES cells was due to a deficiency of SRF, we reintroduced the SRF protein back into these cells by ectopic expression of the cDNA from the cytomegalovirus promoter. In these Srf−/−rescue ES cells the levels of SRF expression are three- to fivefold higher than those of the endogenous protein (44). Indeed, the Srf−/−rescue ES clone reacquired a very efficient serum-dependent induction of both c-fos and Egr-1 (Fig. 5C and D). This induction was both rapid and transient, in a manner similar to that seen in wild-type ES cells expressing endogenous SRF. These data provide for the first time direct genetic evidence that SRF is an important regulator of a component of the cellular IEG activation response.
We also investigated the expression profiles of the β-Actin and Vinculin genes, both of which are known to be SRE regulated yet do not fall into the class of transiently induced IEGs (25, 27). Indeed, in SRF-containing heterozygous ES cells both β-Actin (data not shown) and Vinculin (Fig. 5E) are stimulated 1.5 to 3-fold by serum addition. This activation, however, did not follow the rapid and transient IEG activation kinetics but rather mRNAs accumulated gradually over a 3-h time period. The basal RNA levels of β-Actin as well as the serum-stimulated induction of both β-Actin and Vinculin were reduced in Srf−/− ES cells. Again, reintroduction of SRF protein into the Srf−/−rescue ES cells restored the serum-stimulated expression of both genes (Fig. 5E and data not shown). From these data we conclude that in ES cells SRF controls the expression levels of SRE-containing genes. In doing so, it regulates rapidly responding, transiently activated IEGs, such as Egr-1 and c-fos, as well as β-Actin and Vinculin, other SRE-containing genes that do not display the same transient activation profiles.
LPA-induced signaling targets endogenous genes via SRF.
TCF-independent activation of SRF target genes was reported to occur upon stimulation with serum and LPA (9, 15, 17, 18). These results were obtained with transiently transfected or microinjected reporter constructs, and the chromosomal state of the reporters was found to matter (1). The availability of SRF-deficient cells provided us with the tool to examine directly a potential involvement of SRF in the stimulation of endogenous IEGs by LPA. Fig. 6 demonstrates that SRF is indeed required for efficient activation of both the c-fos and Egr-1 genes by LPA. As seen before with serum induction (Fig. 5D), LPA-stimulated induction of ES cells lacking SRF still permits significant residual activation of c-fos (Fig. 6B) but not of Egr-1 (Fig. 6A). This analysis provides the first genetic proof for a direct involvement of SRF in LPA-stimulated induction of cellular target genes.
DISCUSSION
Mouse embryos lacking the transcription factor SRF develop until the onset of gastrulation at around E6.0 (4). At that point an essential requirement for SRF exists since the Srf−/− embryos fail to develop detectable mesodermal cells and die in utero. Up until E6.0, however, it is apparent that many cell division cycles occur in a fashion that is apparently little affected by the absence of SRF. Prior to this unexpected finding, SRF was believed to be essential for the proliferation of all mammalian cells (7, 39). In the present work we made use of the ES cell system to explore the mechanisms that permit the cells of the early Srf−/− mouse embryo to proliferate. To that end, we have used Srf−/− ES cells to investigate the requirement for SRF during ES cell proliferation, cell cycle progression, and IEG activation.
ES cell morphology and cortical actin expression are influenced by SRF.
Srf−/− ES cells display morphological characteristics indicative of altered cell surface activities, including cell-substrate and cell-cell interactions. The reduction in plating efficiency on gelatin substrates, as observed when cells were seeded at low density, may be the direct cause of the smaller increases in the total number of Srf−/− ES cells over time seen in our growth curves. We do not yet know the fate of those individual ES cells that are unable to give rise to colony formation on the growth plates. However, preliminary data indicate that Srf−/− ES cells display reduced adhesion to various matrices (unpublished data). The nonadherent cells are unable to form colonies and can be expected to die by anoikis, i.e., the apoptosis that occurs in certain cell types when cell matrix contact is lost. We did not observe signs of apoptosis in continuously growing Srf−/− ES cells (unpublished). However, it is possible that detached cells might have been lost during the experiment.
Differences in colony morphology and altered cell surface-directed cellular activities may, in part, be due to the observed reduced expression levels of the cytoskeletal proteins actin and vinculin in continuously proliferating Srf−/− ES cells (unpublished). Disturbance of the cortical actin network has also been implicated in the formation of cytoplasmic extrusions (31), structures also displayed by the Srf−/− ES cells. Similar alterations in cell morphology were also seen with embryoid bodies derived from Srf−/− ES cells (44). This supports the notion that SRF fulfills an important role in directing the expression of structural proteins of the cytoskeleton, thereby facilitating the buildup of structures crucial for cell shape and cell adhesion. More detailed investigations of cytoskeletal structures and associated adhesion properties of Srf−/− ES cells are currently under way.
SRF is not required for ES cell proliferation.
Although a comparison of Srf−/+ and Srf−/− ES cell growth curves revealed smaller increases in the total number of Srf−/− ES cells over time, the rates of proliferation of both types of ES cell were found to be comparable. This was true for both low and high cell-density growth conditions. This is in distinct contrast to the block in proliferation observed when SRF function was inhibited in rat embryo fibroblasts (7) and myoblasts (39). Therefore, in contrast to the somatic cells investigated, ES cells lacking SRF function do not have a severe impairment in their proliferative capacity, and this correlates well with the unaffected early development of Srf−/− mouse embryos up to E6.0. Similarly, the product of the SRF target gene c-fos is not required for proliferation of ES cells (6). However, we cannot formally rule out the possibility that our Srf−/− ES cells acquired unidentified mutations during neomycin selection that allowed them to grow in the absence of SRF.
In addition, Srf−/− cells displayed a cell cycle distribution identical to wild-type cells, whether asynchronously growing or deprived of serum. However, slightly more cells accumulated in G2-M following restimulation of serum-starved Srf−/− cells than wild-type cells. SRF function in the G2-M phase of the cell cycle may be associated with effects on microtubular reorganization (21). Since yeast cells lacking the SRF homologue Mcm1 are completely arrested at the G2-M transition (3), our data may suggest that murine ES cells may also activate critical SRF-dependent genes in a cell cycle-specific fashion at the G2-M transition.
SRF is required for IEG activation.
In SRF-negative ES cells, Egr-1 and c-fos were found to be drastically impaired in their immediate-early response upon induction with serum or TPA. This provides genetic proof that SRF contributes in an essential way to the cellular immediate-early response of SRE-containing genes, a concept previously suggested by the use of transiently expressed SRF effector and reporter systems of altered binding specificity (14, 18).
Even in Srf−/− ES cells, however, we observed a modest induction of c-fos following serum stimulation. This may be due to other transcription factors binding to the v-sis-inducible element (SIE) present in the c-fos promoter, which is lacking in the Egr-1 promoter. Indeed, serum stimulation-mediated activation of the c-fos promoter independently of SRF, by factors that bind to the SIE, has previously been demonstrated (23). Robertson et al. argued for functional interdependence of the c-fos SIE and SRE elements, based on the assessment of cis-element mutagenesis in transgenic mice and explanted cells (32). The drastic reduction in c-fos IEG activation that we see in ES cells lacking only SRF is consistent with this notion. It will be interesting to investigate by genomic footprinting (12) whether the c-fos SRE in Srf−/− ES cells is occupied by SRE binding proteins other than SRF.
Our analysis also provides genetic evidence for LPA-induced activation of c-fos and Egr-1 to depend upon functional SRF, thereby strengthening previous findings (15). Rho family small GTPases are suggested to be involved (16), a signaling scenario that is likely to be influenced by SRF target genes like β-Actin (see above and reference 38). We note that treatment of serum-withdrawn ES cells with LPA was sufficient to elicit a significant transcriptional response of endogenous genes, indicating that in these cells the required signaling events can all be activated by LPA alone (1). The expression of β-Actin and Vinculin, known to be regulated by SRF (25, 27), was similarly affected by SRF deficiency in both asynchronously growing and serum-restimulated cells (data not shown). More than 60 genes have been identified with functional SREs in their promoters (A. Nordheim, unpublished data), and we speculate that transcriptional regulation of most of these genes will be impaired in the absence of SRF. This altered regulation of SRF-dependent genes is likely to affect many physiological processes in organisms that lack SRF function.
ES cells do not assume a quiescent G0 state upon serum withdrawal.
One striking difference between ES and somatic cells is the response to serum withdrawal. Serum-deprived somatic cells undergo G0 exit (43), while similarly treated ES cells fail to do so. ES cells continue to proliferate in the absence of serum, even with substantially reduced c-fos and Egr-1 expression, and they appear to proliferate normally in the absence of SRF. These data suggest that the IEG program and SRF function are dispensable in the ES cell, in part because ES cells lack the capacity to undergo G0 exit and cell cycle reentry. Furthermore, the relatively normal growth in vitro of SRF-deficient ES cells is reminiscent of the normal growth of cells of the early Srf−/− embryo, which are able to support embryonic development until E6.0. We speculate that, like ES cells, the cells of the early mouse embryo also undergo continuous cell division cycles without G0 exit and therefore do not require the mechanisms to undergo cell cycle reentry. Thus, rather than containing cellular factors that complement SRF deficiency and maintain the IEG program, the cells of the early embryo simply may not require SRF function and IEG activation. Recently, Poser et al. described SRF as contributing to a novel phosphatidylinositol 3-kinase-mediated mechanism of cell cycle reentry seen with HeLa and 3T3-L1 cells (30). It will be interesting to determine whether such an SRF-dependent mechanism is also operative in ES cells.
ACKNOWLEDGMENTS
G. Schratt and B. Weinhold contributed equally to this work.
We thank Marianne Petry for expert technical assistance. We acknowledge the gift of anti-E-cadherin antiserum from H. Beug (IMP, Vienna, Austria). We appreciate the comments on this manuscript by O. Heidenreich.
This work was supported through grants of the VolkswagenStiftung (I/74039 to U.R. and I/74043 to A.N.) and the DFG (NO 120/10-1 and NO 120/7-4 to A.N.). A.S.L. and R.A.W. acknowledge financial support through NIH grant R35CA39826.
REFERENCES
- 1.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 2.Almendral J M, Sommer D, Macdonald-Bravo H, Burckhardt J, Perera J, Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988;8:2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arsenian S, Weinhold B, Oelgeschlager M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Field S J, Johnson R S, Mortensen R M, Papaioannou V E, Spiegelman B M, Greenberg M E. Growth and differentiation of embryonic stem cells that lack an intact c-fos gene. Proc Natl Acad Sci USA. 1992;89:9306–9310. doi: 10.1073/pnas.89.19.9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gauthier-Rouviere C, Cavadore J C, Blanchard J M, Lamb N J, Fernandez A. p67SRF is a constitutive nuclear protein implicated in the modulation of genes required throughout the G1 period. Cell Regul. 1991;7:575–588. doi: 10.1091/mbc.2.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gille H, Sharrocks A D, Shaw P E. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 9.Graham R, Gilman M. Distinct protein targets for signals acting at the c-fos serum response element. Science. 1991;251:189–192. doi: 10.1126/science.1898992. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 11.Heidenreich O, Neininger A, Schratt G, Zinck R, Cahill M A, Engel K, Kotlyarov A, Kraft R, Kostka S, Gaestel M, Nordheim A. MAPKAP kinase 2 phosphorylates serum response factor in vitro and in vivo. J Biol Chem. 1999;274:14434–14443. doi: 10.1074/jbc.274.20.14434. [DOI] [PubMed] [Google Scholar]
- 12.Herrera R E, Shaw P E, Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- 13.Herschman H R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 14.Hill C S, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- 15.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatadic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 17.Hill C S, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansen F-E, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson C M, Hill C S, Chawla S, Treisman R, Bading H. Calcium controls gene expression via three distinct pathways that can function independently of the Ras/mitogen-activated protein kinases (ERKs) signaling cascade. J Neurosci. 1997;17:6189–6202. doi: 10.1523/JNEUROSCI.17-16-06189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau L F, Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985;4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S H, Lee H H, Chen J J, Chuang C F, Ng S Y. Serum response element-regulated transcription in the cell cycle: possible correlation with microtubule reorganization. Cell Growth Differ. 1994;5:447–455. [PubMed] [Google Scholar]
- 22.Maher M, Cong F, Kindelberger D, Nasmyth K, Dalton S. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol Cell Biol. 1995;15:3129–3137. doi: 10.1128/mcb.15.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer D J, Campbell G S, Cochran B H, Argetsinger L S, Larner A C, Finbloom D S, Carter-Su C, Schwartz J. Growth hormone induces a DNA binding factor related to the interferon-stimulated 91-kDa transcription factor. J Biol Chem. 1994;269:4701–4704. [PubMed] [Google Scholar]
- 24.Minty A, Kedes L. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol Cell Biol. 1986;6:2125–2136. doi: 10.1128/mcb.6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moiseyeva E P, Weller P A, Zhidkova N I, Corben E B, Patel B, Jasinska I, Koteliansky V E, Critchley D R. Organization of the human gene encoding the cytoskeletal protein vinculin and the sequence of the vinculin promoter. J Biol Chem. 1993;268:4318–4325. [PubMed] [Google Scholar]
- 26.Muscat G E O, Gustafson T A, Kedes L. A common factor regulates skeletal and cardiac α-actin gene transcription in muscle. Mol Cell Biol. 1988;8:4120–4133. doi: 10.1128/mcb.8.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng S Y, Gunning P, Liu S H, Leavitt J, Kedes L. Regulation of the human beta-actin promoter by upstream and intron domains. Nucleic Acids Res. 1989;17:601–615. doi: 10.1093/nar/17.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 29.Pellegrini L, Tan S, Richmond T J. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 30.Poser S, Impey S, Trinh K, Xia Z, Storm D R. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J. 2000;19:4955–4966. doi: 10.1093/emboj/19.18.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priddle H, Hemmings L, Monkley S, Woods A, Patel B, Sutton D, Dunn G A, Zicha D, Critchley D R. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J Cell Biol. 1998;142:1121–1133. doi: 10.1083/jcb.142.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson L M, Kerppola T K, Vendrell M, Luk D, Smeyne R J, Bocchiaro C, Morgan J I, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 33.Rüther U, Komitowski D, Schubert F R, Wagner E F. c-fos expression induces bone tumors in transgenic mice. Oncogene. 1989;4:861–865. [PubMed] [Google Scholar]
- 34.Schmidt A, Hall M N. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz-Sommer Z, Hue I, Huijser P, Flor P J, Hansen R, Tetens F, Lonnig W E, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shore P, Sharrocks A D. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 37.Smith A G, Heath J K, Donaldson D D, Wong G G, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 38.Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 39.Soulez M, Rouviere C G, Chafey P, Hentzen D, Vandromme M, Lautredou N, Lamb N, Kahn A, Tuil D. Growth and differentiation of C2 myogenic cells are dependent on serum response factor. Mol Cell Biol. 1996;16:6065–6074. doi: 10.1128/mcb.16.11.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 41.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 42.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z M, Yang H, Livingston D M. Endogenous E2F-1 promotes timely G0 exit of resting mouse embryo fibroblasts. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinhold B, Schratt G, Arsenian S, Berger J, Kamino K, Schwarz H, Rüther U, Nordheim A. Srf(−/−) ES cells display non-cell autonomous impairment in mesodermal differentiation. EMBO J. 2000;19:5835–5844. doi: 10.1093/emboj/19.21.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams R L, Hilton D J, Pease S, Willson TA, Stewart C L, Gearing D P, Wagner E F, Metcalf D, Nicola N A, Gough N M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 46.Xia Z, Dudek H, Miranti C K, Greenberg M E. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]