Abstract
The major distal regulatory sequence for the β-globin gene locus, the locus control region (LCR), is composed of multiple hypersensitive sites (HSs). Different models for LCR function postulate that the HSs act either independently or synergistically. To test these possibilities, we have constructed a series of expression cassettes in which the gene encoding the enhanced green fluorescent protein (EGFP) is under the control of DNA fragments containing single and multiple HSs of the LCR. LCR DNA fragments containing only the minimal region needed for position-independent expression (HS cores) or containing cores plus flanking sequences (HS units) were compared to ascertain whether conserved sequences between the HS cores contributed to enhancement. Expression of these constructs was measured after targeted integration into three defined loci in murine erythroleukemia cells using recombinase-mediated cassette exchange. At all three marked loci, synergistic enhancement of expression was observed in cassettes containing a combination of HS2, HS3, and HS4 units. In contrast, HS2, HS3, and HS4 cores (without flanking sequences) give an activity equivalent to the sum of the activities of the individual HS cores. These data suggest a model in which an HS core plus flanking regions, bound by specific proteins, forms a structure needed for interaction with other HS units to confer strong enhancement by the LCR. The three targeted integration sites differ substantially in their permissivity for expression, but even the largest LCR construct tested could not overcome these position effects to confer equal expression at all three sites.
Genes in the β-globin gene complex (HBBC, containing HBE1, HBG2, HBG1, HBD, and HBB), together with those in the α-globin gene complex (HBZ2, HBA2, and HBA1) encode the developmentally regulated, erythroid-specific family of hemoglobins in vertebrates. Transcription of the mammalian HBBC is regulated both by proximal elements, such as promoters, and by a distal regulatory element known as the locus control region (LCR). The LCR is marked by several hypersensitive sites (HSs) in erythroid chromatin (18, 45) and is required for high-level expression of genes within the HBBC in erythroid cells (reviewed in references 6 and 19). Transfection and transgenic mouse studies show that the LCR confers this high-level expression at many, but perhaps not all, ectopic sites of integration (1, 21, 31). Gain-of-function experiments examining multiple integrated copies of LCR constructs revealed expression that is copy number dependent and independent of the site of integration (e.g., see references 43 and 44), suggesting that the LCR contains a dominant chromatin-opening activity. However, deletion of HS1 to HS6 from the LCR of mouse Hbbc (3, 12), as well as deletion of HS2 to HS5 from the LCR of human HBBC on chromosome 11 (37), leaves the globin genes in an open chromatin domain, albeit expressed at very low to undetectable levels. Thus, the LCR is clearly required for enhancement but it is not necessary for chromatin domain opening at the normal chromosomal position.
The core of each HS can be defined as the minimal DNA fragment capable of conferring high-level expression on a linked globin gene in transgenic mice; these cores tend to be 200- to 400-bp fragments (e.g., see references 35 and 44). Numerous studies have examined the roles of individual HSs in various expression assays (reviewed in references 20 and 22). HS2 contains a strong enhancer that functions both in transient assays and in stably transfected cells. HS3 can also enhance expression of globin genes, with its major function seen after integration. HS4 does not enhance by itself but can contribute increased expression in combination with other HSs (11). HS1 appears to be dispensable, since a naturally occurring deletion encompassing it does not affect β-globin gene expression (28). HS5 is absent from rabbits (7), and no phenotype was observed when HS5 and HS6 were deleted from mouse Hbbc (2). Thus, the bulk of the known function for the LCR maps to the region encompassing HS2, HS3, and HS4.
Despite the substantial effects of HS2 and HS3 in gain-of-function assays, only a small decrease in globin gene expression was observed when HS2 or HS3 (in each case including some flanking DNA) was deleted from the endogenous mouse Hbbc locus (16, 23) or from a YAC with 150 kb encompassing the HBBC in transgenic mice (34). This could be explained by one or more of the remaining HSs substituting for the function of the deleted HS. This, in turn, implies that the remaining HSs function independently of the deleted HS and enhance at a level almost comparable to that of the intact LCR. A distinctly different phenotype was seen when only the HS cores were deleted from the HBBC carried in large YACs in transgenic mice. Deletion of the cores of HS2, HS3, or HS4 (with no flanking DNA) caused a dramatic reduction in the expression of all of the β-like globin genes (8, 9, 32). In these constructs, the remaining HSs of the LCR were unable to form a strong enhancer (despite the fact that the DNase hypersensitivity was retained in several cases), implying that the various HSs have to act together, synergistically, in an LCR holocomplex to enhance globin gene expression (8). Analysis of an extensive set of single-HS deletions in transgenic mice containing the HBBC shows that deletion of any HS makes the transgenic locus susceptible to two different kinds of chromosomal position effects (31), also arguing that the components of the LCR form an interactive holocomplex (47).
Direct evidence for synergistic interactions has been obtained in a few studies. Combinations of three HSs were needed for expression of HBB beyond that obtained with a single HS in transfected murine erythroleukemia (MEL) cells (11). Synergism between HS2 and HS3 was observed for enhanced expression of a rabbit HBE-luciferase reporter gene in stably transfected K562 cells (25, 26). Synergism among HS2, HS3, and HS4 was inferred for long-range activation of an HBG1 reporter gene in stably transfected K562 cells (5). In each of these cases, the LCR constructs contained both the HS cores and flanking DNA, and Jackson et al. (25) showed that the flanking DNA was needed, since only additive increases were observed when combinations of HS cores were used.
Further evidence for the importance of sequences between (or flanking) the HS cores comes from analysis of interspecies sequence alignments, which reveal many conserved blocks both within and between HS cores (22). In a direct test, larger DNA fragments containing single HSs plus flanking DNA showed significantly stronger enhancement of an HBE-luciferase reporter after stable integration into K562 cells (26) than did the cores alone. Indeed, earlier studies had mapped additional functions outside the core of HS2 (10, 29, 44). We refer to a DNA fragment containing the HS core plus flanking DNA as an “HS unit” (25).
A complication in interpreting the results of the above-described studies with transgenic mice and stably transfected cells is the effect of the integration site on expression. For instance, a set of clones containing the same DNA construct but with a different number of copies at different integration sites shows a range of expression levels, even after correction for copy number (e.g., see references 25 and 26). Although significant differences were observed between constructs, one cannot rule out some contribution from position effects, even when examining pools of clones. Moreover, lines of transgenic mice carrying one or two copies of 150-kb YACs or large BACs encompassing the HBBC (with an intact LCR) are subject to position effects (1, 36). Thus, interpretation of studies comparing different LCR mutations within the context of a large region containing the HBBC in transgenic mice is still complicated by the influence of particular integration sites.
Therefore, techniques have been developed that target integration of a single copy of a test construct to specified chromosomal locations (4, 33, 40, 41). These techniques allow expression of a series of test constructs to be compared at the same site. The integrated constructs are subject to exactly the same effects of flanking DNA, and complications arising from site-to-site variation in position effects are eliminated. At the same time, only single copies of the integrant are obtained, thus removing copy number as a variable in the experiment. In this study, we used the technique of recombinase-mediated cassette exchange, or RMCE (4, 15), to compare the effects of LCR HS cores and HS units, singly and in combination, on expression of an HBB-enhanced green fluorescent protein (EGFP) reporter integrated into MEL cells. The constructs were examined at three different chromosomal locations, RL4, RL5, and RL6, in both orientations (14). We found that HS2, HS3, and HS4 units (but not cores) can interact to produce a level of enhancement beyond the sum of the effects observed with the individual HS cores. Thus, DNA fragments containing the cores plus flanking DNA are needed for synergism among the HSs of the LCR. Despite this synergistic activation, even large DNA fragments containing all of the major functional sequences in the LCR cannot overcome the position and orientation effects seen at these integration sites.
MATERIALS AND METHODS
Plasmids for RMCE.
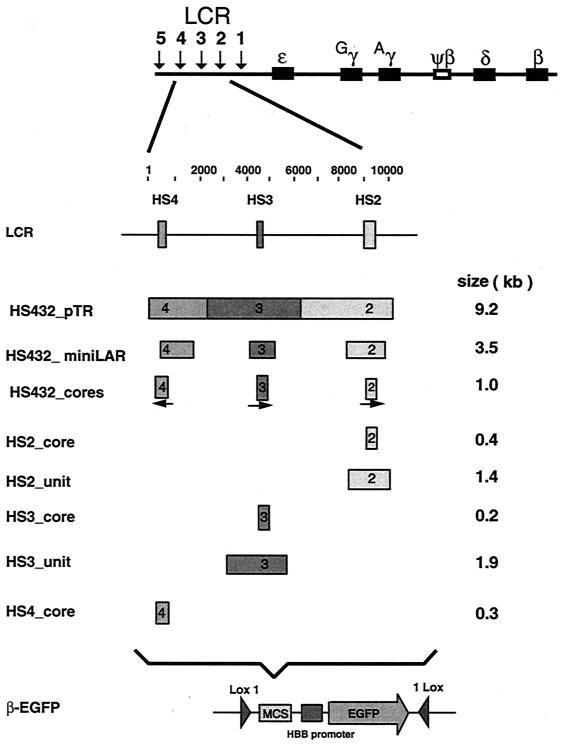
The expression plasmid L1-β-EGFP-1L, into which all of the LCR fragments were inserted, was described by Feng et al. (15). Briefly, the promoter of the human β-globin gene (HBB), on a DNA fragment extending from −374 to +44 relative to the cap site, replaced the cytomegalovirus (CMV) promoter in plasmid pEGFP-N1 (Clontech, Palo Alto, Calif.), and inverted Lox sites L1 (Lox1) and 1L (1Lox) were placed at the 5′ and 3′ ends of the cassettes, respectively (15). An oligonucleotide containing restriction endonuclease cleavage sites designed for insertion of LCR DNA fragments (MCS in Fig. 1) was added 5′ to the HBB promoter, and a series of DNA fragments from the β-globin LCR were inserted (Fig. 1). The restriction endonucleases used and the positions of the DNA fragments containing HSs are listed in Table 1. Plasmid L1-HS432_pTR-1L contains a 9.2-kb LCR segment encompassing HS2, HS3, and HS4 obtained from pTR68 (38). Plasmid L1-HS432_mLAR-1L contains linked DNA fragments containing units of HS2, HS3, and HS4 (a total of 3.5 kb) excised from plasmid mLAR (17). Plasmid L1-HS432_cores-1L contains a linked set of cores for HS2, HS3, and HS4 (a total of 1.0 kb, with HS4 in the opposite orientation to HS3 and HS2) as described by Sadelain et al. (39). Cre expression plasmid pBS185(CMV-CRE) was obtained from Clontech.
FIG. 1.
Maps of the human LCR fragments and EGFP expression cassettes used in this study. The top line shows the HBB complex, and the next line shows the positions of the HS2, HS3, and HS4 cores, using the coordinates in GenBank locus HUMHBB (accession number U01317). The next eight lines show the segments of the LCR inserted into the expression cassette, which is diagrammed on the last line. It contains the HBB promoter linked to an EGFP reporter gene, preceded by multiple cloning sites (MCS) and flanked by Lox1 and 1Lox. The names of the cassettes are on the left, and the sizes of the LCR inserts are on the right.
TABLE 1.
LCR constructs tested in transfected cells
| LCR construct | Total size (kb) | Restriction enzyme sites | HS(s) in construct | Size of fragment (kb) | Position in HUMHBB |
|---|---|---|---|---|---|
| 432_pTR | 9.22 | EcoRI-BglII | HS4, HS3, HS2 | 9.22 | 1–9218 |
| 432_mLAR | 3.55 | SacI-SacI | HS4 | 1.25 | 951–2199 |
| SacI-PvuII | HS3 | 0.85 | 4274–5122 | ||
| KpnI-StuI | HS2 | 1.42 | 7764–9186 | ||
| 432_cores | 1.00 | SacI-AvaI | HS4 | 0.29 | 951–1239 |
| Oligonucleotide-Fnu4HI | HS3 | 0.27 | 4509–4775 | ||
| HindIII-SnaBI | HS2 | 0.42 | 8486–8909 | ||
| HS2_core | 0.37 | HindIII-XbaI | HS2 | 0.37 | 8486–8860 |
| HS2_unit | 1.46 | KpnI-BglII | HS2 | 1.46 | 7764–9218 |
| HS3_core | 0.23 | HphI-Fnu4HI | HS3 | 0.23 | 4550–4775 |
| HS3_unit | 1.91 | HindIII-HindIII | HS3 | 1.91 | 3266–5172 |
| HS4_core | 0.29 | SacI-AvaI | HS4 | 0.29 | 951–1239 |
Transfection, selection, and screening.
MEL cells marked by integration of a plasmid containing L1-CMV-HYTK-1L at one of three different chromosomal locations, RL4, RL5, or RL6 (14), were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% bovine calf serum, 2% penicillin and amphotericin B (Fungizone), and hygromycin at 1 mg/ml for at least 3 days prior to electroporation. RMCE was performed as described by Bouhassira et al. (4), using 200 μg of the test plasmid, 50 μg of the CRE expression plasmid, and electroporation at 450 V and 500 μF. Cells in which the HYTK cassette is replaced with the test construct are resistant to gancyclovir. Thus, transfected cells were plated in soft agar containing DMEM, 10% bovine calf serum, 2% penicillin and amphotericin B, and 12 μM gancyclovir. Individual colonies were picked after 2 weeks and expanded in liquid culture containing gancyclovir.
Colonies containing a single integrant at RL4, RL5, and RL6 were identified by analysis of genomic DNA. Southern blots containing 20 μg of genomic DNA were digested with BglII and hybridized to a DNA probe from the gene for EGFP. In cases where orientation could not be identified from a single digest, a double digestion with BglII and HindIII was performed and a DNA fragment containing HS4 was used as a probe.
EGFP measurements.
To measure the level of EGFP, a sample of each cell culture containing 106 cells was resuspended in phosphate-buffered saline and 2 μM propidium iodide and analyzed on a flow cytometer (Beckman-Coulter XL-MCL). Rainbow calibration particles (RCP-30-5; Spherotech Inc.) coated with defined amounts of fluorescein were analyzed under the same conditions as the cells and used to construct a standard curve. This standard curve was used to convert the median of the distribution of EGFP levels to relative molecules of equivalent fluorescein (MEFL). The means of these median values for multiple clones (in most cases, three clones) and the standard deviations were calculated for both orientations of each cassette. The significance of differences in the means was calculated using Students t test. Cells were assayed for EGFP within 6 weeks after isolation.
The amount of EGFP fluorescence expected for additive effects of HS cores was calculated by first subtracting the fluorescence level for the β-EGFP cassette from that for each HS core cassette. These adjusted levels were then added to the fluorescence level for the β-EGFP cassette, effectively adding in the signal from the HBB promoter only once. Fluorescence substantially above this level was interpreted as a synergistic effect.
Induction.
To test the inducibility of various LCR-containing cassettes, 5 × 104 cells from selected clones were incubated in DMEM containing 4 mM N,N′-hexamethylene-bis-acetamide (HMBA) at 37°C for 6 days. Increased production of hemoglobin was evident by the red color of the induced cell pellet. The EGFP levels in the induced cells and uninduced cells were compared by flow cytometry.
RESULTS
Synergism among HS units after integration at the permissive RL5 locus.
The expression cassette used in these studies contained the gene (EGFP) encoding EGFP and the human HBB promoter flanked by Lox sites in the inverted orientation (Lox1 and 1Lox, Fig. 1). Various fragments of the β-globin LCR were ligated into the multiple cloning sites upstream of the promoter (Fig. 1). Each plasmid was electroporated into marked MEL cells along with a Cre expression plasmid so that the entire cassette replaced a HYTK cassette at the desired integration site (15), rendering the targeted cells resistant to gancyclovir. Individual clones were then selected and screened for single-copy integrants at the desired sites. Levels of EGFP were measured by flow cytometry.
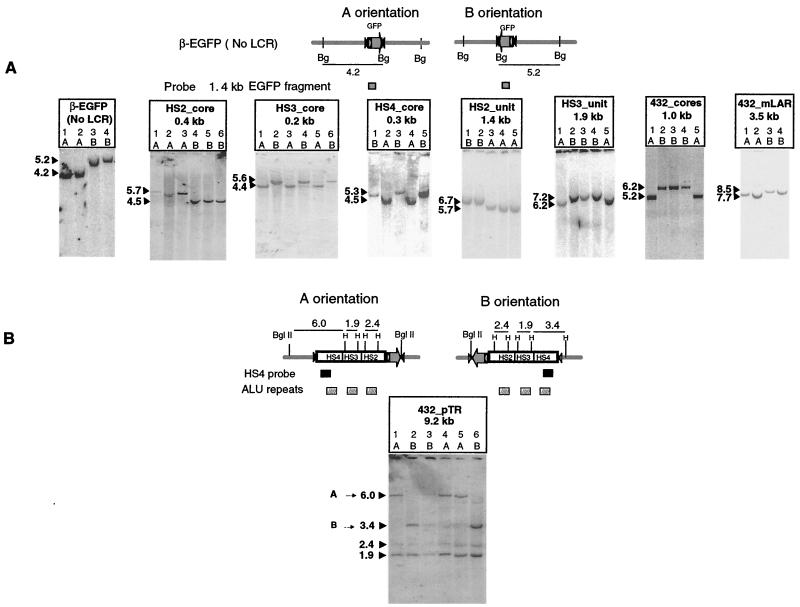
Figure 2 shows the results of genomic Southern blot assays of clones selected after cassette exchange at RL5 in MEL cells. After integration of the reporter construct β-EGFP (i.e., no LCR), digestion of the genomic DNA with BglII, followed by hybridization with an EGFP probe, yielded a 4.2-kb fragment in orientation A and a 5.2-kb fragment in orientation B (Fig. 2A). The sizes expected after replacement with the LCR-containing cassettes were calculated by adding the size of the LCR fragments to the size of the BglII fragments seen for β-EGFP. For all constructs, clones were found for which the sizes of the observed bands closely matched the expected sizes for both orientations. At least three individual clones for each orientation were obtained from this screen. Given the large size of the BglII fragments containing the HS432_pTR cassette, a single enzyme digest could not distinguish the orientation. By utilizing a double digest with HindIII and BglII and hybridizing the blot with an HS4 probe (Fig. 2B), fragments of 6.0 or 3.4 kb were obtained, which are distinctive for orientations A and B, respectively. Common bands of 1.9 and 2.4 kb result from a cross hybridization between Alu repeat sequences in the probe and those in the fragments containing HS2 and HS3.
FIG. 2.
Southern blot hybridization data showing integration of expression cassettes at RL5 of MEL cells by RMCE. (A) Analysis with BglII (Bg). The maps at the top show the sizes of the inserts obtained from integration of the β-EGFP (no LCR) cassette into RL5 MEL cells in each orientation and the BglII fragments detected with an EGFP probe. Below the maps are blot hybridization data for genomic DNA purified from gancyclovir-resistant clones, digested with BglII, and hybridized with labeled DNA containing the EGFP coding sequence. Representative clones are showed for each construct in each orientation. (B) Analysis with BglII and HindIII (H). To distinguish the two orientations of the HS432_pTR cassette at RL5, genomic DNA was digested with BglII and HindIII and hybridized to an HS4 probe. Orientation A was identified by the presence of a 6.0-kb band, while orientation B has the 3.4-kb band. Positions of the hybridization probe and the Alu repeats are indicated below the maps.
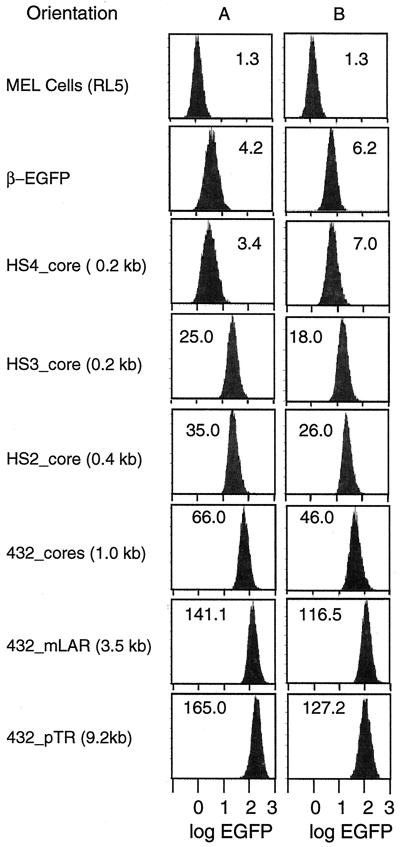
Analysis of EGFP fluorescence by flow cytometry showed that locus RL5 is permissive for expression in both orientations. Even in the absence of any LCR enhancers, EGFP was produced in all of the cells (Fig. 3, β-EGFP construct). Addition of HS3 or HS2 caused an increase in the median level of fluorescence, indicating a higher level of expression of the EGFP gene (Fig. 3), whereas HS4 had no effect. All of the cells in the population increased their EGFP signal when an enhancer was added; the fraction of cells expressing EGFP did not change. The mean fluorescence for at least three individual clones is plotted in Fig. 4 for comparisons among the cassettes. HS2 has a stronger effect than HS3 (4.4- to 6.5-fold enhancement for orientations B and A of the HS2_unit cassette, compared to 2.6- to 4.0-fold for the HS3_unit cassette). No significant difference was seen between the HS cores and HS units (P > 0.9 by Student's t test) for both HS3_unit versus core and HS2_unit versus core. No difference in EGFP fluorescence was observed between the two orientations with the individual HSs; however, orientation A with a combination of HSs enhanced 1.5- to 2.0-fold more than orientation B. This orientation difference is significant for HS432_cores, HS432_mLAR, and HS432_pTR cassettes (P < 0.001).
FIG. 3.
Flow cytometric analysis of clones containing β-EGFP cassettes (with and without the LCR) in MEL cells at locus RL5. Ten thousand cells were counted, and the results were plotted as the number of cells (y axis) versus the intensity of EGFP fluorescence (x axis). The MEL cells are RL5 cells before transfection; this shows the background fluorescence in the absence of EGFP expression. A and B are the orientations in which the cassette is integrated into the RL5 locus. The median of the EGFP peak is given in each graph.
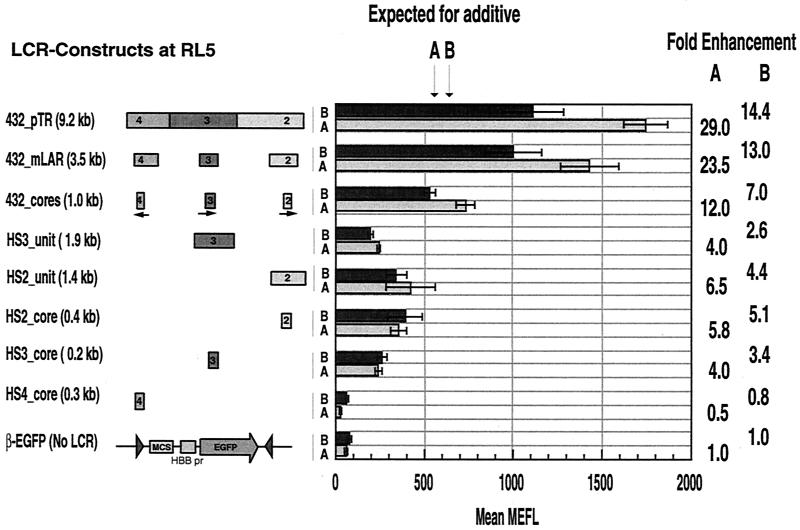
FIG. 4.
Expression of EGFP in MEL cell clones containing β-EGFP (with and without the LCR) cassettes at RL5. EGFP levels were measured in multiple clones containing a single integrant of the indicated constructs in each orientation (A or B). EGFP levels are reported as MEFL. The median values for at least three clones were averaged and plotted as bars with standard deviations. The EGFP levels expected for addition of the effects of individual HS4, HS3, and HS2 cores are indicated at the top. The ratio of the mean MEFL for each cassette to the mean MEFL for the β-EGFP cassette is listed as the fold enhancement on the right. pr, promoter; MCS, multiple cloning sites.
Combinations of HSs gave EGFP fluorescence levels higher than those seen for the individual HSs. Linking of the HS4, HS3, and HS2 cores produced a fluorescence equivalent to that expected for addition of the effects of the individual HS cores (Fig. 4). In particular, the linked cores in orientation A produced EGFP fluorescence only slightly above that expected for addition of the effects of the cores, and in orientation B, the effect was slightly below that expected for an additive effect. In contrast, combinations of the units for HS4, HS3, and HS2 enhanced at markedly greater levels (up to 29.0-fold, compared to 12-fold for the combination of cores). This level of fluorescence is substantially greater than that expected for addition of the cores (Fig. 4). This difference is statistically significant for both the HS432_pTR cassette versus the HS432_cores cassette and the HS432_mLAR cassette versus the HS432_cores cassette (P < 0.001 for each comparison). Hence, HSs of the LCR can interact to obtain greater-than-additive (i.e., synergistic) increases in enhancement. This effect requires DNA fragments containing cores plus flanking DNA (HS units), since combinations of the cores do not show synergism.
The EGFP levels measured by fluorescence assay correlate well with the amount of EGFP RNA in the cells. The amount of EGFP RNA in the total RNA from clones carrying cassettes at RL5 was measured by an RNA protection assay. The fold increases in EGFP RNA corresponded well to the fold increases in EGFP fluorescence (data not shown). Thus, the effects of the LCR fragments are exerted on the amount of stable RNA, which is consistent with an effect on the level of transcription of the cassette.
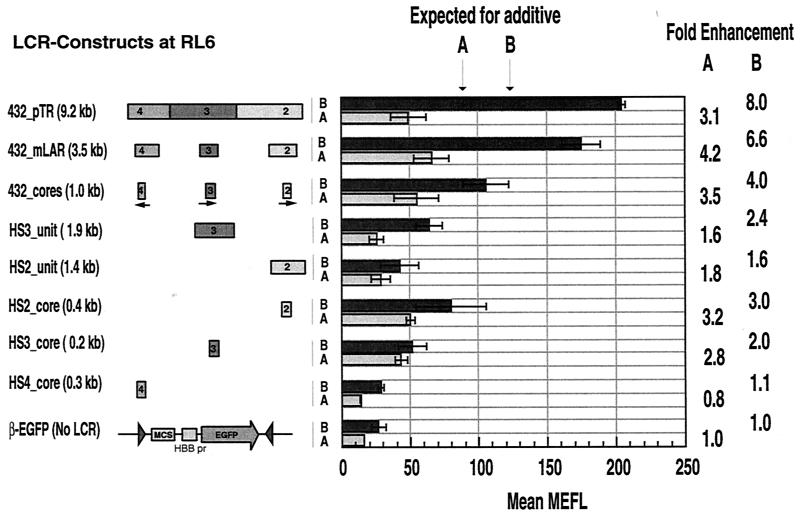
Synergism among HS units in one orientation at the less permissive locus, RL6.
The set of expression cassettes was also integrated at locus RL6, which is not as permissive for expression as is RL5 (14). Screening of multiple clones from each transfection allowed the selection of several clones containing each cassette in each orientation. In the absence of LCR fragments, the β-EGFP cassette at RL6 is not expressed at a detectable level, since the flow profiles are virtually indistinguishable from those of the parental cells (data not shown). Addition of LCR fragments stimulated expression (Fig. 5), and this effect was seen in all of the cells in the population. However, the level of expression for each construct was considerably lower than that seen at RL5 (compare Fig. 5 to Fig. 4). Examination of multiple clones shows that the HS2 and HS3 cores enhance significantly (P < 0.01) but HS4 does not (Fig. 5). Combining the HS4, HS3, and HS2 cores does not increase the level of enhancement; expression of the HS432_cores cassette is not significantly different from that of the HS2_core cassette in either orientation. In contrast, for orientation B, inclusion of multiple HS units in the cassette increased enhancement significantly, from 4.0-fold for the HS432_cores cassette to 8.0-fold for the HS432_pTR cassette (P < 0.001). The increase in enhancement observed for the two cassettes with HS4, HS3, and HS2 units is substantially beyond that expected from addition of the effects of the HS cores. Hence, like the situation at RL5, sequences outside the core are needed to obtain synergistic enhancement among the HSs.
FIG. 5.
Expression of EGFP in MEL cell clones containing β-EGFP (with or without the LCR) cassettes at RL6. EGFP levels were measured in multiple clones containing a single integrant of the indicated constructs in each orientation (A or B). In most cases, the median values for at least three clones were averaged and plotted as bars with standard deviations. pr, promote; MCS, multiple cloning sites.
Surprisingly, inclusion of additional HSs in cassettes in orientation A did not produce any increase beyond that of the HS2 core (Fig. 5). The HS432_pTR construct gave only a 3.1-fold enhancement, which is comparable to the 3.2-fold obtained with the HS2 core alone. Thus, orientation A at RL6 has a limited capacity to respond to enhancers. Feng et al. (14) observed no differences between orientations A and B at RL6 for expression of a construct comparable to the HS432_mLAR cassette. This may reflect a stable alteration at this locus, perhaps epigenetic, in the line of RL6-marked cells used in these experiments.
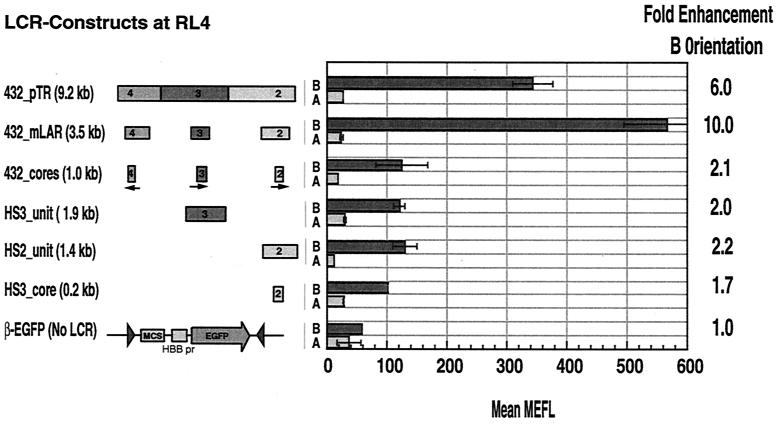
Synergism among HS units in one orientation at RL4.
The set of cassettes with different portions of the LCR was also tested at a third locus, RL4. Clones in each orientation for all cassettes were obtained for analysis. Feng et al. (14) have shown that orientation A of RL4 is not permissive for expression of cassettes regulated by the CMV promoter-enhancer or an enhancer containing HS4, HS3, and HS2 of the LCR (comparable to the HS432_mLAR cassette). The data in Fig. 6 confirm this observation and show, in addition, that for all of the LCR fragments tested, including the 9.2-kb fragment encompassing all of HS4, HS3, and HS2 (HS432_pTR cassette), only a low level of EGFP is produced in orientation A. In contrast, orientation B is permissive for expression. For this orientation, all of the cells in the population express EGFP and, hence, there is no change in the fraction of cells expressing EGFP with different LCR fragments (data not shown).
FIG. 6.
Expression of EGFP in MEL cell clones containing β-EGFP (with or without the LCR) cassettes at RL4. EGFP levels were measured in multiple clones containing a single integrant of the indicated constructs in each orientation (A or B). Averages for clones carrying the same cassettes in a given orientation are plotted as bars with standard deviations. pr, promoter; MCS, multiple cloning sites.
In permissive orientation B, combinations of the HS units give substantially higher levels of enhancement (6- to 10-fold) than does the combination of HS cores (2.1-fold). This is consistent with the need for flanking sequences for synergistic interactions observed at RL5 and RL6.
LCR fragments with HS4, HS3, and HS2 cannot overcome position effects.
Figure 7 compares the levels of expression and enhancement by cassettes containing multiple HS sites at the three different chromosomal locations, using the most highly expressing orientation for each site. Data for cassettes with no LCR confirm that RL5 is the most permissive locus, RL4 (orientation B) allows an intermediate level of expression, while RL6 is the least active. This has been inferred by analysis of LCR-containing cassettes (14) and is directly demonstrated in the comparison in Fig. 7. If any of the DNA fragments from the LCR used in this study were capable of overcoming all position effects, they should be able to enhance to comparable levels of expression at all three loci. This was not seen. Instead, each LCR construct was able to enhance expression but the levels of expression and fold enhancement were highest at RL5, intermediate at RL4, and lowest at RL6. Even the largest DNA fragment tested, encompassing the most active portions of the LCR, did not increase expression to the same level at all three integration sites.
FIG. 7.
Expression of EGFP in MEL cell clones obtained through RMCE at three different loci. Expression levels for cassettes containing combinations of HSs in the more highly expressing orientation at RL4, RL5, and RL6 are presented. The mean values are plotted as bars with standard deviations. pr, promoter; MCS, multiple cloning sites.
Effects of induction.
The effects of different LCR fragments on inducibility were tested with clones from each cassette in the higher-expressing orientation at integration sites RL5 and RL4. Cultures of clones grown in the absence or presence of the inducer HMBA were assayed for EGFP fluorescence by flow cytometry. All of the cassettes at RL5 induced 3.5- to 6.5-fold (Fig. 8A). At RL4, several LCR-containing cassettes induced four- to fivefold, including combinations of units, combination of cores, and the HS3 unit alone (Fig. 8B). Induction of the HS2 unit was only twofold, whereas the HS3 core and the cassette with no LCR showed no response to induction. At both RL4 and RL5, the amount of induction for cassettes containing the HS3 unit exceeded that of those containing the HS3 core (P < 0.05 for clones at RL5).
FIG. 8.
Induction of EGFP expression in RMCE clones. Expression levels for uninduced clones (U) or clones induced for 6 days with 4 mM HMBA (I) are plotted. (A) Clones with cassettes integrated at RL5. (B) Clones with cassettes integrated at RL4. The fold increase upon induction is given at the right.
DISCUSSION
We have examined the expression of a series of single-copy cassettes containing different combinations of functional elements from the LCR at identical integration sites in MEL cells. In contrast to experiments using randomly integrated constructs, data from this technique are not complicated by variations in position effects or copy numbers. We show that the components of the LCR do not work strictly independently, but rather they can interact to produce synergistic enhancement of the HBB promoter. These data support a model in which the HSs of the LCR interact in a holocomplex to provide a high level of enhancement (5, 8, 9, 24, 31, 47).
The criterion for synergism in the current study is the stimulation of expression of an HBB-EGFP reporter to a level greater than the sum of that stimulated by the individual HS cores. Combining the HS4, HS3, and HS2 cores enhanced expression to a level comparable to the sum of the enhancements by the cores. In contrast, combinations of larger DNA fragments containing the HSs (HS units) enhanced to a significantly higher level, indicating synergistic interactions. A contrasting interpretation is that the HS units individually are capable of enhancing to a greater extent than are the HS cores, thereby producing the larger enhancements in combination. Although stronger enhancement by HS units than HS cores has been seen in some studies (e.g., see reference 26), this result was not obtained with single-copy integrants at the sites examined in this study; in uninduced cells, the enhancements by cores and units are the same for HS3 and HS2. No HS4 unit was tested in the current series of experiments, since no difference was observed in previous experiments comparing the HS4 core and HS4 units from the rabbit LCR (26). This leaves open the possibility that the human HS4 unit included in the HS432_miniLAR cassette has an enhancement activity greater than that of the HS4 unit, thereby accounting for the greater activity of the combinations of units. However, this seems unlikely, given the absence of an enhancement effect for HS4 seen in several studies (20, 22). Thus, for the loci examined here, the enhancement expected for the sum of the HS units is the same as that expected for the sum of the HS cores in this system. The data in the current study can be interpreted as reflecting synergistic interactions among the HSs.
The observation of synergism among larger DNA fragments of the LCR, but not among the HS cores, shows that additional sequences outside the cores are needed for synergistic enhancement. The ability of HS units (i.e., cores plus flanking regions) to synergize in enhancement was seen at all three of the loci examined. A similar result was obtained in our earlier study of randomly integrated LCR-containing reporter constructs, but synergism was observed between rabbit but not human LCR HS units (25). This earlier study used a different promoter (from the rabbit HBE gene) and a different cell line, K562; it was limited to combinations of HS2 and HS3; and it was subject to the complications of different position effects. Any or all of these factors could contribute to the lack of observable synergism between human HS units in that study. Our current approach, using RMCE to examine expression of cassettes integrated at defined chromosomal locations, is more sensitive and robust. These data clearly show that the HS4, HS3, and HS2 units of the human LCR can interact synergistically to enhance expression from the HBB promoter. In support of this observation, recent studies utilizing LCR segments in gene therapy vectors also show that a combination of larger LCR fragments, comparable to our HS units, produce higher levels of expression for a longer time than do combinations of HS cores (30).
The DNA sequences between the HS cores contain clusters of consereved sequence blocks, or phylogenetic footprints (22), but they do not have enhancement activity themselves (42). Thus, they appear to be modulators of enhancement by the cores of the HSs. Ongoing studies show that specific proteins can bind to conserved sites between the HS cores. Proteins bound to the DNA flanking the cores may play a structural role, placing the activator proteins bound to the HS cores in an optimal orientation, perhaps by facilitating interactions with other HS units.
Surprisingly, even large LCR constructs were unable to overcome position effects at RL4, RL5, and RL6. The largest segment tested, a 9.2-kb fragment with HS4, HS3, and HS2 and all of the intercore sequences, contains all of the major regulatory elements mapped in the LCR. These results indicate either that important LCR functions needed to overcome these position effects lie outside of this fragment or that the LCR cannot overcome them. The recent demonstrations that even large YACs and BACs containing the human HBBC (1, 36) or mouse Hbbc (27) are sensitive to position effects argue that the LCR cannot overcome all position effects. This raises the possibility that strategies based on random integration are not effective long-term approaches to genetic therapy of disorders of the β-globin genes. However, more studies are needed to ascertain whether the ability to overcome all position effects (which may not be possible) is required or whether strong enhancers providing robust expression at a subset of integration sites will suffice.
The technique of RMCE overcomes limitations due to differences in position effects and copy number, but it also restricts the number of loci examined. We have measured expression at three loci that differ markedly in permissivity for expression; a cassette containing the HBB promoter but not the LCR is not expressed at a detectable level at RL6, whereas this cassette is readily expressed at RL5 and RL4 (orientation B). Expression is enhanced by appropriate DNA fragments from the LCR at all three loci, and in all cases, flow cytometry showed that all of the cells in the population contained EGFP. Thus, enhancement occurring by an increase in the fraction of cells expressing the reporter gene (46) was not observed in the current experiments. Further studies are required to determine whether the enhancement seen here results exclusively from an increased rate of expression, an increase in the fraction of time that a cell is expressing (13), or some combination of effects (4). EGFP is a stable protein, and cells that have ceased expression of the EGFP gene will continue to show EGFP fluorescence for some time, thus making it difficult to see transiently nonexpressing cells. Although we have studied three loci with strikingly different expression properties, it is possible that other sites are subject to stronger negative regulation and our current studies would not reveal regulatory functions needed to overcome these stronger negative effects. For instance, clones containing each of the expression cassettes at RL5 were monitored for 6 months but no evidence of silencing was observed (data not shown). Thus, the ability of enhancers to prevent silencing cannot be assayed at the RL5 locus.
Two types of orientation effect were observed in these studies. Cassettes at RL4 are expressed in only one orientation, and cassettes at RL6 are less responsive to enhancers in one orientation. Recent studies (14) show that silencing correlated with methylation of regulatory elements in the cassette but not with changes in chromatin structure (DNase-hypersensitive sites still formed at HS2 and HS3). Presumably, some sequences flanking the RL4 and RL6 integration sites influence methylation and expression. Two possibilities include effects of the direction of replication and interfering transcription from an adjacent gene. Studies of the sequences flanking the integration sites should be informative.
In contrast to previous results (26), we see no difference between the enhancement by individual HS cores and individual HS units for the HBB-EGFP hybrid reporter gene at these loci in MEL cells. Several possibilities could explain the difference. For instance, the collection of random integration sites sampled in the earlier studies with K562 cells may have been less permissive for expression than the sites in MEL cells examined in this study and, hence, were more dependent on the functions provided by the sequences flanking individual HS cores. Also, the HBE-luciferase reporter used in previous studies may be more sensitive to the effects of the sequences flanking the cores. Alternatively, it is possible that the set of clones examined previously for HS cores happened to be subject to more negative position effects than the set of clones examined for HS units and, hence, the result could reflect the limitations of the assay available at that time. Comparison of cassettes containing HS units and cores at even less permissive loci by RMCE may provide a better evaluation of whether individual units have stronger activity than single cores. Indeed, some of the data obtained by RMCE are suggestive of a role of the sequences flanking the cores even for single HSs. For example, treatment of clones containing the HS3_unit cassette with HMBA produced a stronger induction than did treatment of clones containing the HS3_core cassette, at both RL4 and RL5. Thus, the sequences flanking the cores do play a role in LCR function. Multiple lines of evidence now support the conclusion that they are needed for synergistic interactions between the HS cores, and in some (but not all) assays, they are needed for optimal function of a single HS.
ACKNOWLEDGMENTS
Plasmids containing mLAR, a fragment spanning the region including HS5 through HS2, and the linked cores of HS2, HS3, and HS4 were generously provided by Mark Groudine, Tim Townes, and Michel Sadelain, respectively.
This work was supported by NIH grants DK27635 (to R.H.), LM05110/HG02238 (to W.M.), and HL38655 and HL554350 (to E.B.). J.M. is supported partly by the South Africa National Research Foundation.
REFERENCES
- 1.Alami R, Greally J M, Tanimoto K, Hwang S, Feng Y Q, Engel J D, Fiering S, Bouhassira E E. Beta-globin YAC transgenes exhibit uniform expression levels but position effect variegation in mice. Hum Mol Genet. 2000;9:631–636. doi: 10.1093/hmg/9.4.631. [DOI] [PubMed] [Google Scholar]
- 2.Bender M, Reik A, Close J, Telling A, Epner E, Fiering S, Hardison R, Groudine M. Description and targeted deletion of 5′ HS5 and 6 of the mouse β-globin locus control region. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 3.Bender M A, Bulger M, Close J, Groudine M. β-globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 4.Bouhassira E, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 5.Bresnick E, Tze L. Synergism between hypersensitive sites confers long-range gene activation by the β-globin locus control region. Proc Natl Acad Sci, USA. 1997;94:4566–4571. doi: 10.1073/pnas.94.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 7.Bulger M, von Doorninck J H, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bungert J, Dave U, Lim K-C, Kieuw K H, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 9.Bungert J, Tanimoto K, Patel S, Liu Q, Fear M, Engel J D. Hypersensitive site 2 specifies a unique function within the human β-globin locus control region to stimulate globin gene transcription. Mol Cell Biol. 1999;19:3062–3072. doi: 10.1128/mcb.19.4.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caterina J J, Ciavatta D J, Donze D, Behringer R R, Townes T M. Multiple elements in human β-globin locus control region 5′ HS2 are involved in enhancer activity and position-independent transgene expression. Nucleic Acids Res. 1994;22:1006–1011. doi: 10.1093/nar/22.6.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis P, Antoniou M, Grosveld F. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epner E, Reik A, Cimbora D, Telling A, Bender M, Fiering S, Enver T, Martin D, Kennedy M, Keller G, Groudine M. The β-globin LCR is not necessary for an open chromatin structure or transcription of the mouse β-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y Q, Alami R, Bouhassira E E. Enhancer-dependent transcriptional oscillations in mouse erythroleukemia cells. Mol Cell Biol. 1999;19:4907–4917. doi: 10.1128/mcb.19.7.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y Q, Lorincz M, Fiering S, Greally J M, Bouhassira E. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol Cell Biol. 2001;21:298–309. doi: 10.1128/MCB.21.1.298-309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y Q, Seibler J, Alami R, Eisen A, Westerman K A, Leboulch P, Fiering S, Bouhassira E E. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 16.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 17.Forrester W C, Novak U, Gelinas R, Groudine M. Molecular analysis of the human β-globin locus activation region. Proc Natl Acad Sci USA. 1989;86:5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser P, Grosveld F. Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- 20.Grosveld F, Antoniou M, Berry M, de Boer E, Dillon N, Ellis J, Fraser P, Hanscombe O, Hurst J, Imam A, Lindenbaum M, Philipsen S, Pruzina S, Strouboulis J, Raguz-Bolognesi S, Talbot D. The regulation of human globin gene switching. Philos Trans R Soc Lond B Biol Sci. 1993;339:183–191. doi: 10.1098/rstb.1993.0015. [DOI] [PubMed] [Google Scholar]
- 21.Grosveld F, van Assendelft G B, Greaves D, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 22.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 23.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Analysis of mice containing a targeted deletion of β-globin locus control region hypersensitive site 3. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H, Yamamoto M. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J Biol Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 25.Jackson J D, Miller W, Hardison R C. Sequences within and flanking hypersensitive sites 3 and 2 of the β-globin locus control region required for synergistic versus additive interaction with the ɛ-globin gene promoter. Nucleic Acids Res. 1996;24:4327–4335. doi: 10.1093/nar/24.21.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson J D, Petrykowska H, Philipsen S, Miller W, Hardison R. Role of DNA sequences outside the cores of DNase hypersensitive sites (HSs) in functions of the β-globin locus control region: domain opening and synergism between HS2 and HS3. J Biol Chem. 1996;271:11871–11878. doi: 10.1074/jbc.271.20.11871. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman R M, Pham C T, Ley T J. Transgenic analysis of a 100-kb human beta-globin cluster-containing DNA fragment propagated as a bacterial artificial chromosome. Blood. 1999;94:3178–3184. [PubMed] [Google Scholar]
- 28.Kulozik A E, Sail S, Bellan-Koch A, Bartram C R, Kohne E, Kleihauer E. The proximal element of the β-globin locus control region is not functionally required in vivo. J Clin Investig. 1991;87:2142–2146. doi: 10.1172/JCI115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D, Chang J C, Moi P, Jiu W, Kan Y W, Curtin P T. Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc Natl Acad Sci USA. 1992;89:3899–3903. doi: 10.1073/pnas.89.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May C, Rivella S, Callegari J, Heller G, Gaensler K M, Luzzatto L, Sadelain M. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 31.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 32.Navas P A, Peterson K R, Li Q, Skarpidi E, Rohde A, Shaw S E, Clegg C H, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol. 1998;18:4188–4196. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Gorman S, Fox D T, Wahl G M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 34.Peterson K, Clegg C, Navas P, Norton E, Kimbrough T, Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human β-globin LCR on the developmental regulation of globin gene expression in β-YAC transgenic mice. Proc Natl Acad Sci USA. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipsen S, Talbot D, Fraser P, Grosveld F. The β-globin dominant control region: hypersensitive site 2. EMBO J. 1990;9:2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porcu S, Kitamura M, Witkowska E, Zhang Z, Mutero A, Lin C, Chang J, Gaensler K M. The human beta globin locus introduced by YAC transfer exhibits a specific and reproducible pattern of developmental regulation in transgenic mice. Blood. 1997;90:4602–4609. [PubMed] [Google Scholar]
- 37.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan T M, Behringer R R, Martin N C, Townes T M, Palmiter R D, Brinster R L. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human β-globin gene expression in transgenic mice. Genes Dev. 1989;3:314–323. doi: 10.1101/gad.3.3.314. [DOI] [PubMed] [Google Scholar]
- 39.Sadelain M, Wang C H, Antoniou M, Grosveld F, Mulligan R C. Generation of a high-titer retroviral vector capable of expressing high levels of the human β-globin gene. Proc Natl Acad Sci USA. 1995;92:6728–6732. doi: 10.1073/pnas.92.15.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer B. Multiplex Cre/lox recombination permits selective site-specific DNA targeting to both a natural and an engineered site in the yeast genome. Nucleic Acids Res. 1996;24:4608–4613. doi: 10.1093/nar/24.23.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seibler J, Schubeler D, Fiering S, Groudine M, Bode J. DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- 42.Slightom J, Bock J, Tagle D, Gumucio D, Goodman M, Stojanovic N, Jackson J, Miller W, Hardison R. The complete sequences of the galago and rabbit β-globin locus control regions: extended sequence and functional conservation outside the cores of DNase hypersensitive sites. Genomics. 1997;39:90–94. doi: 10.1006/geno.1996.4458. [DOI] [PubMed] [Google Scholar]
- 43.Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves D R. A dominant control region from the human β-globin locus conferring integration site-independent gene expression. Nature. 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 44.Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 1990;9:2169–2178. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuan D, Solomon W, Li Q, London I M. The β-like globin gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin D I K. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]