Abstract
Adelgids (Insecta: Hemiptera: Adelgidae) form a small group of insects but harbor a surprisingly diverse set of bacteriocyte-associated endosymbionts, which suggest multiple replacement and acquisition of symbionts over evolutionary time. Specific pairs of symbionts have been associated with adelgid lineages specialized on different secondary host conifers. Using a metagenomic approach, we investigated the symbiosis of the Adelges laricis/Adelges tardus species complex containing betaproteobacterial (“Candidatus Vallotia tarda”) and gammaproteobacterial (“Candidatus Profftia tarda”) symbionts. Genomic characteristics and metabolic pathway reconstructions revealed that Vallotia and Profftia are evolutionary young endosymbionts, which complement each other’s role in essential amino acid production. Phylogenomic analyses and a high level of genomic synteny indicate an origin of the betaproteobacterial symbiont from endosymbionts of Rhizopus fungi. This evolutionary transition was accompanied with substantial loss of functions related to transcription regulation, secondary metabolite production, bacterial defense mechanisms, host infection, and manipulation. The transition from fungus to insect endosymbionts extends our current framework about evolutionary trajectories of host-associated microbes.
Subject terms: Environmental microbiology, Microbial ecology
Introduction
Most plant sap-feeding insects harbor bacterial endosymbionts, which are of great importance in their host ecology and serve as a model for studying microbe–host relationships and genome evolution of host-restricted bacteria [1–3]. However, how these insect–bacteria partnerships arise is still poorly understood, because the precursor of the endosymbionts can rarely be identified [4–6].
Adelgids (Insecta: Hemiptera: Adelgidae) live on Pinaceae conifers and feed on phloem sap or parenchyma cells [7, 8]. The group has nearly 70 species and is sister to the families of phylloxerans (Phylloxeridae) and aphids (Aphididae) within the suborder Sternorrhyncha. Some adelgid species, such as the balsam woolly adelgid (Adelges piceae) and the hemlock woolly adelgid (Adelges tsugae), are well-known forest pests and represent severe threats to firs and hemlocks [9].
Adelgids have a complex multigenerational life cycle, which typically involves sexual generations and an alternation between spruce (Picea), which is the primary host, and another secondary conifer host (Abies, Pinus, Larix, Pseudotsuga, or Tsuga). However, other adelgids reproduce asexually on either of the host genera [8].
Similarly to other plant sap-feeding insects, adelgids harbor maternally inherited bacterial symbionts within specialized cells, so-called bacteriocytes, which form a large bacteriome in the abdomen [10–15]. Although the function of these bacterial partners remains largely unexplored, they are expected to provide essential amino acids and B vitamins scarce in the plant sap diet, similarly to obligate endosymbionts of other plant sap-feeding insects [2, 16]. Besides these obligate nutritional endosymbionts, non-essential facultative symbionts might also occur within the bacteriome (or in other tissues), which can provide selective fitness benefits to insects such as protection against parasites and fungal pathogens, increased heat tolerance, or expansion of host plant range [17–19]. Similarly to obligate mutualists, facultative symbionts are usually maternally inherited but can also spread horizontally within and between insect species via mating [20], parasites [21], and food source, such as plant tissues [22].
Interestingly, most adelgid species contain two phylogenetically distinct bacteriocyte-associated symbionts [10, 11, 13–15]. These symbionts belong to at least six different lineages within the Gammaproteobacteria or the Betaproteobacteria. As an exception, in most A. tsugae populations a single symbiont resides within the bacteriome, but in each population, there is another symbiont free in the hemocoel [10, 11, 23]. Remarkably, specific pairs of symbionts correspond to distinct lineages of adelgids specialized to one of the five secondary host tree genera [11, 15]. A gammaproteobacterial symbiont lineage involving “Candidatus Annandia adelgestsugas” and “Candidatus Annandia pinicola” (hereafter Annandia), is present in both A. tsugae and Pineus species and was likely already associated with ancestral adelgids before diversification into the two major adelgid lineages, Adelges and Pineus, over 87 million years ago [8, 10, 11, 15]. Nevertheless, this putatively ancient symbiont lineage is missing from other adelgids, and the high diversity of symbionts within this small group of insects suggests an evolutionary history involving multiple acquisitions and replacements of bacterial partners [11, 14, 15]. It has been postulated that the frequent turnover of symbionts might be due to fluctuating selective pressure on essential symbiotic functions during evolution coupled to changes in the feeding behavior of adelgids. Adelgids typically feed on nutrient-rich parenchyma versus nutrient-poor phloem on the primary and secondary host trees, respectively, and their host-alternating lifestyles might have emerged repeatedly [11, 23]. The high diversity of symbionts of adelgids is in sharp contrast to the aphid sister group, in which most species have tightly co-evolved with a single obligate symbiont, Buchnera aphidicola, for over 180 million years [2].
To date, whole-genome sequences of adelgid endosymbionts are available for only one species: the hemlock woolly adelgid, A. tsugae [23]. Metabolic potential and genomic characteristics of Annandia resemble those of long-term obligate intracellular symbionts. However, Annandia has lost many genes in essential amino acid synthesis. The accompanying, evolutionary more recent Pseudomonas symbiont of A. tsugae—which is present in the hemocoel—can complement these missing capabilities and thus has a co-obligatory status in the symbiosis [23]. In addition to this obligate dual endosymbiotic system, analysis of a genome fragment of a gammaproteobacterial symbiont (“Candidatus Steffania adelgidicola”) of the Adelges nordmannianae/A. piceae species complex revealed a metabolically versatile, putatively evolutionarily young endosymbiont in this adelgid lineage [13]. Further genomic data on the symbionts would help to infer the history of association of adelgids with distinct bacterial groups.
Here we investigate the bacterial symbionts of the Adelges laricis/Adelges tardus species complex using a metagenomic approach and ask what is the function and putative origin of the dual symbiosis in this lineage of adelgids. A. laricis and A. tardus are morphologically and genetically hardly distinguishable species of adelgids [7, 24]. They contain betaproteobacterial and gammaproteobacterial symbionts, “Candidatus Vallotia tarda” and “Candidatus Profftia tarda” (hereinafter Vallotia and Profftia), respectively. Both symbionts are rod shaped and are co-localized inside the same bacteriocytes. Profftia-related symbionts have only been found in larch-associated lineages of adelgids, while Vallotia symbionts occur in both larch- and Douglas fir-associated lineages of adelgids. Although host–symbiont co-speciation could not be fully resolved with confidence yet, the dual obligatory status of Profftia and Vallotia in the symbiosis seems to be possible given their common occurrence across different populations and life stages of adelgids [11, 14].
Our results demonstrate that both bacteriocyte-associated symbionts are evolutionary recent partners of adelgids complementing each other’s role in essential amino acid biosynthesis. Notably, phylogenomic analyses revealed a close relationship of Vallotia with endosymbionts of Rhizopus fungi. Detailed analysis of genomic synteny and gene content indicated an evolutionary transition from fungus to insect symbiosis accompanied by a substantial loss of functions in the insect symbiont especially in transcription regulation, secondary metabolite production, host infection, and manipulation.
Materials and methods
Sampling
Spruce (Picea) branches with galls of adelgids [7] were collected near Klausen-Leopoldsdorf, Austria (Fig. S1). Galls were stored at −80 °C in the laboratory for subsequent genomic DNA isolation.
DNA isolation
Before DNA isolation, adelgids were collected from the galls using teasing needles. For high-throughput sequencing, the symbionts were enriched by sequential filtration. The insects were washed twice in buffer A + EDTA solution (35 mM Tris-HCl, 250 mM sucrose, 25 mM KCl, 10 mM MgCl2, 250 mM EDTA; pH 7.5) and were subsequently homogenized in fresh solution with a plastic pestle. The suspension was then sequentially filtered through 53 and 30 μm pore size meshes and 5 μm membrane syringe filters. Samples were centrifuged at 7000 rpm for 5 min at 4 °C, and the supernatants were discarded. Pellets were resuspended in buffer A and centrifuged again at 7000 rpm for 5 min at 4 °C. This washing step was repeated once and pellets were next re-suspended in 1×TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.5). High-molecular-weight (HMW) DNA was isolated by an sodium dodecyl sulfate-based DNA extraction method using 1% cetyltrimethylammonium bromide and 1.5% polyvinylpyrrolidone in the extraction buffer [25].
For nanopore sequencing, DNA was isolated from whole insects. Adelgids were rinsed in 1× phosphate-buffered saline three times. DNA was extracted using the Promega Wizard HMW DNA Extraction Kit by homogenizing the adelgids directly in the HMW Lysis Buffer A. The Short Read Eliminator XS Kit from Circulomics was used to remove short DNA fragments.
DNA samples were stored at −20 °C.
Sequencing and genome assembly
From the symbiont-enriched samples, a paired-end library was sequenced on a HiSeq 2000 sequencer (Illumina). Sequencing reads of 2 × 100 bp were quality filtered and trimmed with PRINSEQ [26] and were assembled with SPAdes v3.1 [27]. Using a subset of 30 million read pairs, a single contig representing the circular Profftia chromosome was obtained with 52-fold coverage, while the assembly of the Vallotia genome remained fragmented. To improve this assembly, reads were mapped on the Profftia genome using the Burrows–Wheeler Alignment (BWA) tool and the BWA-MEM algorithm [28], and matching sequences were removed from further analysis. Contigs >1000 bp obtained in a novel assembly with the remaining reads were further analyzed against a custom protein database containing single-copy markers found in 99% of prokaryote genomes using blastx [29]. Phylogenetic information of the best hits was assessed in Megan v4.70.4 [30]. Ribosomal RNAs (rRNAs) were inferred by RNAmmer [31]. Based on these, seven contigs representing the chromosome and a single contig corresponding to a putative circular plasmid of Vallotia have been identified.
In order to close the Vallotia genome, we performed nanopore long-read sequencing on an ONT MinION flow cell. Porechop (https://github.com/rrwick/Porechop) and Canu v2.1 [32] were used to remove adapters and for read-error correction, respectively. Nanopore and short reads were mapped against the previously assembled symbiont sequences using Minimap2 v2.17 [33] and Bowtie2 v2.3.5.1 [34], respectively. We pseudo-randomly subsampled 3% of those mapping back to the Vallotia contigs, and this read set was used for a targeted reassembly using Unicycler v0.4.9b [35] with k-mer sizes of 33, 55, and 77 and in “bold” mode. The novel assembly resulted in two circular molecules corresponding to the chromosome and the previously assembled plasmid of Vallotia, with a coverage of 323× and 258×, respectively.
Genome analysis
The putative origin of replication was identified with GenSkew (http://genskew.csb.univie.ac.at/). We used the ConsPred genome annotation pipeline for gene prediction and annotation [36]. Genome annotations were curated with the help of the UniPro UGENE software [37]. We identified pseudogenes by using the intergenic and hypothetical protein regions as queries in blastx searches against nr and the UniProt Swiss-Prot database with an e-value <1e−3 confidence threshold. Pseudogenes were identified as remains of genes, which were either truncated (having a length <80% of the reference) or were interrupted by internal stop codons and/or frameshift mutations. Pseudogene coordinates were set according to the best blast hit in the UniProt Swiss-Prot database, if applicable.
The presence of mobile genetic elements was inferred with blastn and blastx searches against the ISfinder database [38]. Metabolic pathways were explored with the help of the Ecocyc, Biocyc, and Metacyc databases [39] and the Pathway Tools software [40]. Orthologous proteins shared by the relevant genomes or unique to either of the symbionts were identified by using OrthoMCL with a 1e−5 e-value threshold [41]. Distribution of the predicted proteins among the main functional categories was explored by using eggNOG-mapper v2 with the DIAMOND sequence comparison option and a 1e−3 e-value threshold [42]. Genome alignments were performed by Mauve [43]. Synteny between the chromosomes and plasmids of Vallotia and the closely related fungus-associated endosymbiont, Mycetohabitans rhizoxinica (accession numbers: FR687359, FR687360) were visualized by Circos based on single-copy shared genes [44]. A close up of collinear regions was created by using the Easyfig tool [45]. The assembled genomes have been submitted to the European Nucleotide Archive under the project accession number PRJEB40648.
Phylogenetic analyses
A phylogenomic approach was used to infer the phylogenetic positions of the endosymbionts. Protein sequences of closely related species within the Burkholderiales and Enterobacteriales were collected from the Assembly database of NCBI. Single-copy marker genes were identified by Phyla-AMPHORA [46] using the Brandon Seah (2014) Phylogenomics tools (online: https://github.com/kbseah/phylogenomics-tools). Individual sets of genes were aligned with Muscle 3.8.31 [47]. Poorly aligned positions were removed with Gblocks 0.91b [48] using default parameters apart from the following settings: allowed gap positions with half, the minimum length of a block was 5. Alignments of 108 and 45 proteins were concatenated and used for the calculation of phylogenetic trees for Vallotia and Profftia, respectively.
For both endosymbionts, we generated maximum likelihood trees with IQ-TREE after selecting the best-fit substitution models with ModelFinder as available at http://iqtree.cibiv.univie.ac.at [49–51]. SH-like approximate likelihood ratio test (SH-aLRT) and ultrafast bootstrap support values, both based on 1000 iterations, were calculated [52]. The best-fit models were LG + F + R5 and LG + R5 for the Vallotia and Profftia tree, respectively. Besides, Bayesian phylogenetic analyses were performed by MrBayes 3.2.7a [53] with the LG + I + G model and 4 gamma categories on the CIPRES Science Gateway v.3.3. web interface [54]. Two runs, each with 4 chains were performed until convergence diagnostics fell below 0.01. A 50% majority consensus tree was created with a relative burn-in of 25%.
Results and discussion
Profftia and Vallotia are related to free-living bacteria and fungus-associated endosymbionts
Previous 16S rRNA-based phylogenetic analyses suggested an affiliation of Profftia with free-living gammaproteobacteria and a close phylogenetic relationship between Vallotia and betaproteobacterial endosymbionts of Rhizopus fungi [14]. Biased nucleotide composition and accelerated sequence evolution of endosymbiont genomes [2, 3] often result in inconsistent phylogenies and may cause grouping of unrelated taxa [55, 56]. Thus, to further investigate the phylogenetic relationships of the A. laricis/tardus symbionts, we used conserved marker genes for maximum likelihood and Bayesian phylogenetic analyses.
Phylogenetic analysis of 45 single-copy proteins demonstrated that Profftia opens up a novel insect symbiont lineage most similar to Hafnia species and an isolate from the human gastrointestinal tract within the Hafniaceae, which has been recently designated as a distinct family within the Enterobacteriales [57] (Fig. S2). Hafnia strains are frequently identified in the gastrointestinal tract of humans and animals and were also found in insects [58, 59]. The phylogenomic placement of Profftia in our analysis is in agreement with previous 16S rRNA-based analyses [14].
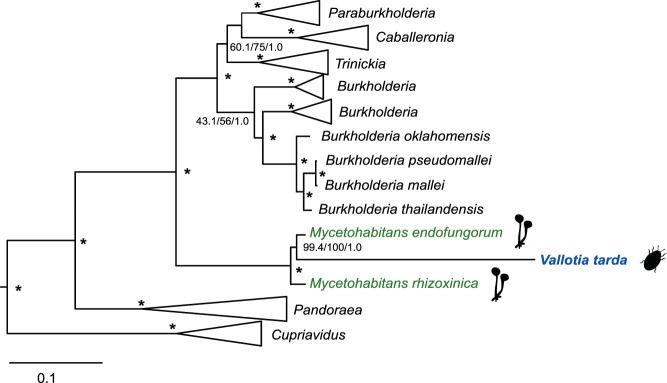
Vallotia formed a monophyletic group with Mycetohabitans endofungorum and M. rhizoxinica, endosymbionts of Rhizopus fungi within the Burkholderiaceae [60, 61] with strong support in phylogenetic analyses based on a concatenated set of 108 proteins (Figs. 1 and S3; previous taxonomic assignments of the fungus-associated symbionts were as Burkholderia/Paraburkholderia endofungorum and rhizoxinica, respectively). Interestingly, Vallotia and M. endofungorum appeared as well-supported sister taxa within this clade. This implies a closer phylogenetic relationship between Vallotia and M. endofungorum and a common origin of adelgid endosymbionts from within a clade of fungus-associated bacterial symbionts. Lengths of branches leading to the fungus-associated endosymbionts were similar to those of free-living bacteria in the data set; however, Vallotia had a remarkably longer branch marking a rapid rate of sequence evolution characteristic of obligate intracellular bacteria [2, 3]. M. endofungorum and M. rhizoxinica have been identified in the cytosol of the zygomycete Rhizopus microsporus, best known as the causative agent of rice seedling blight [61, 62]. The necrotrophic fungus secretes potent toxins, rhizoxin and rhizonin, which are produced by the endosymbionts. The bacterial partners are obligatory for their host as they tightly control its sporulation, while they benefit from host nutrients and spread with the fungal spores [63, 64]. Additionally, related bacterial strains have also been found in association with Rhizopus fungi worldwide in a diverse set of environments, including other plant species, soil, food, and even human tissues [65, 66].
Fig. 1. Phylogenomic analysis showing the affiliation of the adelgid endosymbiont “Candidatus Vallotia tarda” and its closest relatives, the fungus-associated endosymbionts M. rhizoxinica and M. endofungorum within the Burkholderiaceae.
Selected members of Oxalobacteraceae (Janthinobacterium agaricidamnosum [HG322949], Collimonas pratensis [CP013234], and Herbaspirillum seropedicae [CP011930]) were used as outgroup. Maximum likelihood and Bayesian analyses were performed based on a concatenated alignment of 108 proteins. Maximum likelihood tree is shown. SH-aLRT support (%) and ultrafast bootstrap support (%) values based on 1000 replicates, and Bayesian posterior probabilities are indicated on the internal nodes. Asterisks stand for a maximal support in each analysis (100%/1).
Taken together, phylogenomic analyses support that Profftia and Vallotia open up novel insect symbionts lineages most closely related to free-living bacteria within the Hafniaceae and a clade of fungus-associated endosymbionts within the Burkholderiaceae, respectively. Given the well-supported phylogenetic positioning of “Candidatus Vallotia tarda” nested within a clade formed by Mycetohabitans species, we propose the transfer of “Candidatus Vallotia tarda” to the Mycetohabitans genus, as “Candidatus Mycetohabitans vallotii” (a detailed proposal for the re-classification is given in the Supplementary Material).
Vallotia and Profftia are evolutionary young symbionts of adelgids
The complete sequence of the Profftia chromosome had a length of 1,225,795 bp and a G + C content of 31.9% (Table 1). It encoded for 645 proteins, one copy of each rRNA, 35 transfer RNAs (tRNAs), and 10 non-coding RNAs (ncRNAs). It had tRNAs and amino acid charging potential for all 20 standard amino acids. However, protein-coding sequences (CDSs) made up only 52.4% of the genome, and 21 pseudogenes indicated an ongoing gene inactivation.
Table 1.
Genomic features of Profftia and Vallotia.
| Organism | Replicon | Genome size (bp) | GC (%) | Coding density (%) | CDS | rRNA gene | tRNA gene | Pseudogene |
|---|---|---|---|---|---|---|---|---|
| Profftia | Chromosome | 1,225,795 | 31.9 | 52.4 | 645 | 3 | 35 | 21 |
| Vallotia | Chromosome | 1,123,864 | 42.9 | 64.9 | 751 | 9 | 41 | 47 |
| Plasmid | 72,431 | 36.1 | 46.2 | 29 | 0 | 0 | 5 |
The Vallotia chromosome had a length of 1,123,864 bp. It had a G + C content and a coding density of 42.9 and 64.9%, respectively. However, a 72,431-bp-long contig showed a characteristically lower G + C content (36.1%) and contained only 46.2% putative CDSs. This contig had identical repeats at its ends, and genome annotation revealed neighboring genes for a plasmid replication initiation protein, and ParA/ParB partitioning proteins, which function in plasmid and chromosome segregation between daughter cells before cell division [67]. We thus assume that this contig corresponds to a circular plasmid of Vallotia. Vallotia has three rRNA operons, similarly to its close relative, M. rhizoxinica [68]. In total, the Vallotia genome encoded 780 proteins (29 on the putative plasmid), 41 tRNAs, and 52 predicted pseudogenes (5 on the putative plasmid).
The host-restricted lifestyle has a profound influence on bacterial genomes. Relaxed purifying selection on many redundant functions and increased genetic drift can lead to the accumulation of slightly deleterious mutations and the proliferation of mobile genetic elements [69–72]. Disruption of DNA repair genes can increase mutation rates, which promote gene inactivation [73]. Non-functional genomic regions get subsequently lost, and ancient obligate endosymbionts typically have tiny (≪0.8 Mb), gene-dense genomes with AT-biased nucleotide composition [2, 74, 75]. Facultative symbionts also possess accelerated rates of sequence evolution but have larger genomes (>2 Mb) with variable coding densities following the age of their host-restricted lifestyle [76]. The number of pseudogenes in Vallotia and Profftia is higher than in ancient intracellular symbionts, which suggests an intermediate state of genomic reduction [2]. The only moderately reduced size and AT bias together with the low protein-coding density of the Vallotia and Profftia genomes was most similar to those of evolutionary young co-obligate partners of insects [76], for instance, “Ca. Pseudomonas adelgestsugas” in A. tsugae [23], Serratia symbiotica in Cinara cedri [77, 78], and the Sodalis-like symbiont of Philaenus spumarius, the meadow spittlebug [79].
The evolutionary link between Vallotia and fungus-associated endosymbionts
High level of genomic synteny between Vallotia and M. rhizoxinica
Intracellular symbionts usually show a low level of genomic similarity to related bacteria. Rare examples of newly emerged bacteriocyte-associated symbionts of herbivorous insects pinpoint their source from plant-associated bacteria [4], gut bacteria [5], and other free-living bacteria [6].
Genome alignments showed a low level of collinearity between the genomes of Profftia and its closest relatives. Among the relatives of Vallotia, a closed genome is available for M. rhizoxinica [68]. We therefore mostly focused on this fungus-associated symbiont as a reference for comparison with Vallotia.
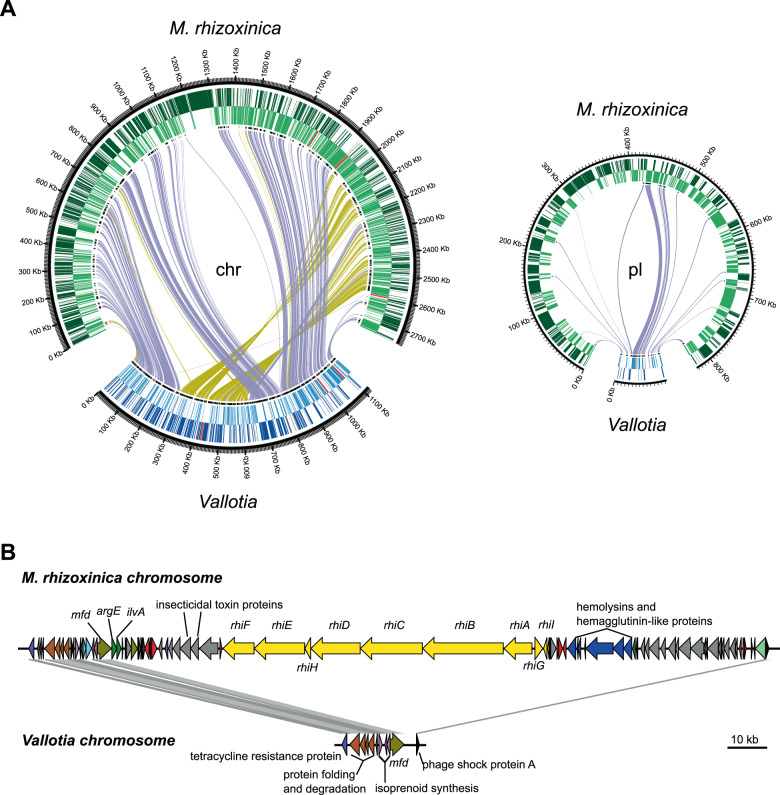
The Vallotia chromosome showed a surprisingly high level of synteny with the chromosome of M. rhizoxinica (Fig. 2A). However, its size was only ~40% of the fungus-associated symbiont chromosome. The putative plasmid of Vallotia was perfectly syntenic with the larger of the two plasmids of M. rhizoxinica (pBRH01), although the Vallotia plasmid was >90% smaller in size (72,431 bp versus 822,304 bp) [68]. Thus, the Vallotia plasmid showed a much higher level of reduction than the chromosome, which together with its lower G + C content and gene density suggests differential evolutionary constraints on these replicons.
Fig. 2. High level of collinearity between the genomes of Vallotia and M. rhizoxinica.
A Circos plot showing the synteny between the chromosome and plasmid of Vallotia and M. rhizoxinica, an endosymbiont of Rhizopus fungi. The outermost and the middle rings show genes in forward and reverse strand orientation, respectively. These include rRNA genes in red and tRNA genes in dark orange. The innermost ring indicates single-copy genes shared by M. rhizoxinica and Vallotia in black. Purple and dark yellow lines connect forward and reverse matches between the genomes, respectively. B Close up of the largest deletion on the chromosome of M. rhizoxinica and the syntenic region on the Vallotia chromosome. Genes are colored according to COG categories. Yellow: secondary metabolite biosynthesis; red: transposase; gray: unknown function; khaki: replication, recombination and repair; pink: lipid transport and metabolism; brown: protein turnover and chaperones; dark green: amino acid transport and metabolism; light green: cell envelope biogenesis; black: transcription. The figure was generated by Easyfig.
The conservation of genome structure contrasts with the elevated number of transposases and inactive derivatives making up ~6% of the fungus-associated symbiont genome [68]. Transition to a host-restricted lifestyle is usually followed by a sharp proliferation of mobile genetic elements coupled with many genomic rearrangements [80–82]. However, mobile genetic elements get subsequently purged out of the genomes of strictly vertically transmitted symbionts via a mutational bias toward deletion and because of lack of opportunity for horizontal acquisition of novel genetic elements [71, 74]. Independent origins of the fungus and adelgid symbioses from free-living precursors would have likely resulted in extensive genome rearrangements due to the accumulation of mobile genetic elements, as seen, for instance, between different S. symbiotica strains in aphids [81]. In contrast to the fungus-associated symbiont, mobile elements are notably absent from the Vallotia genome, suggesting that they might have been lost early after the establishment of the adelgid symbiosis conserving high collinearity between the fungus- and adelgid-associated symbiont genomes. M. rhizoxinica is transmitted also horizontally among fungi and might have more exposure to foreign DNA, therefore at least part of the mobile elements could possibly be inserted into its genome after the host switch of the Vallotia precursor [61, 62].
The observed high level of genome synteny between Vallotia and M. rhizoxinica genomes is consistent with the phylogenetic position of Vallotia interleaved within the clade of Rhizopus endosymbionts. This points toward a direct evolutionary link between these symbioses and a symbiont transition between the fungus and insect hosts.
Shrinkage of the insect symbiont genome
Deletion of large genomic fragments—spanning many functionally unrelated genes—represents an important driving force of genome erosion especially at early stages of symbioses when selection on many functions is weak [3, 83]. Besides, gene loss also occurs individually and is ongoing, albeit at a much lower rate, even in ancient symbionts [75, 84, 85]. Both small and large deletions could be seen when comparing the Vallotia and M. rhizoxinica genomes. Several small deletions as small as one gene were observed sparsely in the entire length of the Vallotia genome within otherwise collinear regions. The largest genomic region missing from Vallotia encompassed 165 kbp on the M. rhizoxinica chromosome (Fig. 2B). The corresponding intergenic spacer was only 3843-bp long on the Vallotia genome between a phage shock protein and the Mfd transcription-repair-coupling factor, present both in Vallotia and M. rhizoxinica. Interestingly, this large genomic fragment included the large rhizoxin biosynthesis gene cluster (rhiIGBCDHEF), which is responsible for the production of rhizoxin, a potent antimitotic macrolide serving as a virulence factor for R. microsporus, the host of M. rhizoxinica [86]. A homologous gene cluster was also found in Pseudomonas fluorescens, and it has been suggested that it has been horizontally acquired by M. rhizoxinica [68, 86]. The rhi cluster is also present in M. endofungorum, therefore it was most likely already present in the genome of the common ancestor of the fungus- and adelgid-associated symbionts and got subsequently lost in Vallotia. Rhizoxin blocks microtubule formation in various types of eukaryotic cells [86, 87], thus the loss of this gene cluster in ancestral Vallotia could have contributed to the establishment of the adelgid symbiosis. However, this large deleted genomic region also contained several transposases and many other genes, such as argE and ilvA, coding for the final enzymes for ornithine and 2-oxobutanoate productions, which were located adjacent to each other at the beginning of this fragment. The largest deletion between the plasmids encompassed nearly 137 kbp of the megaplasmid of M. rhizoxinica and involved several non-ribosomal peptide synthetases (NRPS), insecticidal toxin complex (Tc) proteins, and a high number of transposases among others. M. rhizoxinica harbors 15 NRPS gene clusters [68] in total, all of which are absent in Vallotia. NRPSs are large multienzyme machineries that assemble various peptides, which might function as antibiotics, signal molecules, or virulence factors [88]. Insecticidal toxin complexes are bacterial protein toxins, which exhibit powerful insecticidal activity [89]. Two of such proteins are also present in the large deleted chromosomal region in close proximity to the rhizoxin biosynthesis gene cluster (Fig. 2B); however, their role in M. rhizoxinica remains elusive.
The Vallotia genome encodes a subset of functions of the fungus-associated endosymbionts
The number of protein-coding genes of Vallotia is less than one-third of those of M. rhizoxinica and M. endofungorum, although metabolic functions are already reduced in the fungus-associated endosymbionts compared to free-living Burkholderia species [68] (Figs. S4 and S5). When compared to the two genomes of the fungus-associated endosymbionts, only 53 proteins were specific to Vallotia (Fig. S6). All of these were short (on average 68 amino acid long) hypothetical proteins and most of them showed no significant similarity to other proteins in public databases. Whether these Vallotia-specific hypothetical proteins might be over-annotated/non-functional open reading frames or orphan genes with a yet unknown function [90, 91] needs further investigation. Four genes were present in Vallotia and M. rhizoxinica but were missing in M. endofungorum. These encoded for BioA and BioD in biotin biosynthesis, NagZ in cell wall recycling, and an MFS transporter. Fifteen genes, including, for instance, the MreB rod-shape-determining protein, glycosyltransferase and hit family proteins, genes in lipopolysaccharide, lipoate synthesis, and the oxidative pentose phosphate pathway, were shared between Vallotia and M. endofungorum only. The rest of the Vallotia genes, coding for 91% of all of its proteins, were shared among the fungus- and insect-associated endosymbionts.
Comparing the genes present in both endosymbionts to those shared only by the fungus-associated endosymbionts (Fig. S7), we can infer selective functions maintained or lost during transition to insect endosymbiosis. Translation-related functions have been retained in the greatest measure in the group shared by all endosymbionts. Functions, where higher proportion of genes were specific to the fungus endosymbioses, were related to transcription, inorganic ion transport and metabolism, secondary metabolite biosynthesis, signal transduction, intracellular trafficking, secretion, vesicular transport, and defense mechanisms. Most of the proteins specific to either of the fungus-associated symbionts were homologous to transposases and integrases, transcriptional regulators, or had an unknown function.
Fungus-associated endosymbionts encode a high number of transcriptional regulators (~5% of all genes in M. rhizoxinica) [68], but Vallotia has retained only a handful of such genes, which is a feature similar to other insect symbionts and might facilitate the overproduction of essential amino acids [75, 92].
M. rhizoxinica is resistant against various β-lactams and has an arsenal of efflux pumps that might provide defense against antibacterial fungal molecules, the latter might also excrete virulence factors to the fungus cytosol (type I secretion) [68]. Besides, M. rhizoxinica encodes several genes for pilus formation; adhesion proteins; and type II, type III, and type IV secretion systems, which likely play a central role in host infection and manipulation in the bacteria–fungus symbiosis [68, 93, 94]. However, all of the corresponding genes are missing in Vallotia. Thus, neither of these mechanisms likely play a role in the adelgid symbiosis. Indeed, we could not even detect remnants of these genes in the Vallotia genome, except for a type II secretion system protein as a pseudogene. Loss of these functions is consistent with a strictly vertical transmission of Vallotia between host generations. Transovarial transmission likely does not require active infection mechanisms, and the endosymbionts rather move between the insect cells in a passive manner via an endocytic/exocytic vesicular route [12, 95]. In contrast, M. rhizoxinca is also able to spread horizontally among fungi and re-infect cured Rhizopus strains under laboratory conditions [61, 62].
Differential reduction of metabolic pathways in Vallotia and Profftia
Although compared to their closest free-living relatives both Vallotia and Profftia have lost many genes in all functional categories, both retained the highest number of genes in translation-related functions (Fig. S4). Besides, functions related to cell division, nucleotide and coenzyme transport and metabolism, DNA replication and repair, posttranslational modification, and cell envelope biogenesis are reduced to a lesser extent in both endosymbionts. As a consequence, most of the genes of Vallotia and Profftia are devoted to translation and cell envelope biogenesis, which make up higher proportions of their genomes than in related bacteria (Fig. S5). Retention of a minimal set of genes involved in central cellular functions such as translation, transcription, and replication is a typical feature of reduced genomes, even extremely tiny ones of long-term symbionts [75]. However, ancient intracellular symbionts usually miss a substantial number of genes for the production of the cell envelope and might rely on host-derived membrane compounds [96–98].
Based on pathway reconstructions, both Vallotia (Fig. S8) and Profftia (Fig. S9) have a complete gene set for peptidoglycan, fatty acid, and phospholipid biosynthesis and retained most of the genes for the production of lipid A, LPS core, and the Lpt LPS transport machinery. Besides, we found a partial set of genes for O antigen biosynthesis in the Vallotia genome. Regarding the membrane protein transport and assembly, both adelgid endosymbionts have the necessary genes for Sec and signal recognition particle translocation and the BAM outer membrane protein assembly complex. Profftia also has a complete Lol lipoprotein trafficking machinery (lolABCDE), which can deliver newly matured lipoproteins from the inner membrane to the outer membrane [99]. In addition, Profftia has a near-complete gene set for the Tol-Pal system; however, tolA has been pseudogenized suggesting an ongoing reduction of this complex. Further, both adelgid endosymbionts have retained mrdAB and mreBCD having a role in the maintenance of cell wall integrity and morphology [100, 101]. The observed well-preserved cellular functions for cell envelope biogenesis and integrity are consistent with the rod-shaped cell morphology of Profftia and Vallotia [14], contrasting the spherical/pleomorphic cell shape of ancient endosymbionts, such as Annandia in A. tsugae and Pineus species [10, 11, 15].
Regarding the central metabolism, Vallotia lacks 6-phosphofructokinase but has a complete gene set for gluconeogenesis and the tricarboxylic acid (TCA) cycle. TCA cycle genes are typically lost in long-term symbionts but are present in facultative and evolutionarily recent obligate endosymbionts [79, 82, 102]. Interestingly, Vallotia does not have a recognized sugar transporter. Similarly to M. rhizoxinica [68], a glycerol kinase gene next to a putative glycerol uptake facilitator protein is present on its plasmid. However, the latter gene has a frameshift mutation and a premature stop codon in the first 40% of the sequence and whether it can still produce a functional protein remains unknown.
Profftia can convert acetyl-CoA to acetate for energy but lacks TCA cycle genes, a feature characteristic to more reduced genomes, such as, for instance, Annandia in A. tsugae [23]. Profftia has import systems for a variety of organic compounds, such as murein tripeptides, phospholipids, thiamine, spermidine and putrescine, 3-phenylpropionate, and a complete phosphotransferase system for the uptake of sugars.
NADH dehydrogenase, ATP synthase, and cytochrome oxidases (bo/bd-1) are encoded on both adelgid symbiont genomes. However, Vallotia is not able to produce ubiquinone and six pseudogenes in its genome indicate a recent inactivation of this pathway (Fig. S10).
Profftia retained more functions in inorganic ion transport and metabolism, while Vallotia had a characteristically higher number of genes related to amino acid biosynthesis (see its function below) and nucleotide transport and metabolism (Fig. S4). For instance, Profftia can take up sulfate and use it for assimilatory sulfate reduction and cysteine production, and it has also retained many genes for heme biosynthesis (Fig. S9). However, it cannot produce inosine-5-phosphate and uridine 5’-monophosphate precursors for the de novo synthesis of purine and pyrimidine nucleotides and thus would need to import these compounds.
Notably, although core genes in DNA replication and repair [70] are well preserved, multiple pseudogenes may indicate an ongoing erosion of DNA repair functions in the genomes. These include genes for the UvrABC nucleotide excision repair complex in both adelgid symbionts, helicases (recG, recQ), mismatch repair genes (mutL, mutS; the MutHLS complex is also missing in Profftia), and alkA encoding a DNA glycosylase in Vallotia.
Taken together, their moderately reduced, gene-sparse genomes but still versatile metabolic capabilities support that Vallotia and Profftia are evolutionarily recently acquired endosymbionts. This is following their occurrence in lineages of adelgids, which likely diversified relatively recently, ~60 and ~47 million years ago, respectively, from the remaining clades of Adelgidae [8].
Vallotia and Profftia are both obligatory nutritional symbionts
Complementary functions in essential amino acid provision
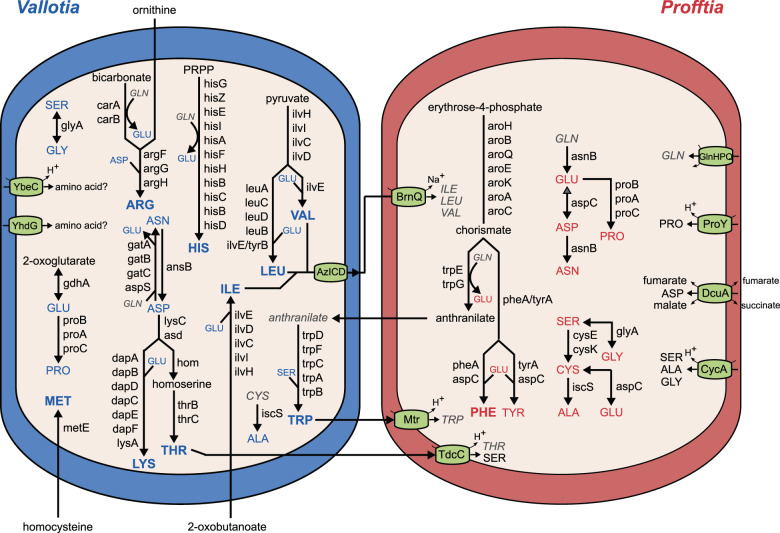
Vallotia and Profftia complement each other’s role in the essential amino acid synthesis, thus have a co-obligatory status in the A. laricis/A. tardus symbiosis (Fig. 3). Although Vallotia likely generates most essential amino acids, solely Profftia can produce chorismate, a key precursor for the synthesis of phenylalanine and tryptophan. Profftia is likely responsible for the complete biosynthesis of phenylalanine as it has a full set of genes for this pathway. It can also convert chorismate to anthranilate; however, further genes for tryptophan biosynthesis are only present in the Vallotia genome. Thus, Vallotia likely takes up anthranilate for tryptophan biosynthesis. Anthranilate synthase (trpEG), is subject to negative feedback regulation by tryptophan [103], thus partition of this rate-limiting step between the co-symbionts can enhance overproduction of the amino acid and might stabilize dual symbiotic partnerships at an early stage of coexistence. The production of tryptophan is partitioned between Vallotia and Profftia similarly as seen in other insect symbioses [77, 78, 104], and it is also shared but is more redundant between the Annandia and Pseudomonas symbionts of A. tsugae [23]. The Vallotia–Profftia system generally shows a lower level of functional overlap between the symbionts and is more unbalanced than the Annandia–Pseudomonas association. In the latter, redundant genes are present also in the synthesis of phenylalanine, threonine, lysine, and arginine, and Annandia can produce seven and the Pseudomonas partner five essential amino acids with the contribution of host genes [23].
Fig. 3. Division of labor in amino acid biosynthesis and transport between Vallotia and Profftia showing co-obligatory status of endosymbionts of A. laricis/tardus.
Amino acids produced by Vallotia and Profftia are shown in blue and red, respectively. Bolded texts indicate essential amino acids. The insect host likely supplies ornithine, homocysteine, 2-oxobutanoate, and glutamine. Other compounds that cannot be synthesized by the symbionts are shown in gray italics.
The Vallotia genome encodes for all the enzymes for the synthesis of five essential amino acids (histidine, leucine, valine, lysine, threonine). ArgG and tyrB among the essential amino acid synthesis-related genes are only present on the plasmid of Vallotia, which might be a reason that the plasmid is still part of its genome. However, neither of the endosymbionts can produce ornithine, 2-oxobutanoate, and homocysteine de novo, which are key for the biosynthesis of arginine, isoleucine, and methionine, respectively. The corresponding functions are also missing from the Annandia–Pseudomonas system [23]. These compounds are thus likely supplied by the insect host, as seen for instance in aphids, mealybugs, and psyllids, where the respective genes are present in the insect genomes and are typically overexpressed within the bacteriome [97, 105, 106]. The metC and argA genes are still present as pseudogenes in Vallotia suggesting a recent loss of these functions in methionine and arginine biosynthesis, respectively.
In most plant sap-feeding insects harboring a dual symbiotic system, typically the more ancient symbiont provides most of the essential amino acids [77, 107]. Given its prominent role in nutrient provision and its presence in both larch- and Douglas fir-associated adelgids, Vallotia might be the older symbiont. Loss of functions in chorismate and anthranilate biosynthesis might have led to the fixation of Profftia in the system.
Vallotia and Profftia have more redundant functions in non-essential amino acid production (Fig. 3). Only Profftia can produce cysteine and tyrosine, while none of the symbionts can build up glutamine, thus this latter amino acid is likely supplied by the insect bacteriocytes.
The presence of relevant transporters can complement missing functions in amino acid synthesis (Fig. 3). For instance, Profftia has a high-affinity glutamine ABC transporter and three symporters (BrnQ, Mtr, TdcC), which can import five among the essential amino acids that can be produced by Vallotia. Vallotia might excrete isoleucine, valine, and leucine via AzICD, a putative branched-chain amino acid efflux pump [108], and these amino acids could be taken up by Profftia via BrnQ and would be readily available also for the insect host.
B vitamin provision by Vallotia
Regarding the B vitamin synthesis, Vallotia is likely able to produce thiamine (B1), riboflavin (B2), pantothenate (B5), pyridoxine (B6), biotin (B7), and folic acid (B9) (Fig. S11). Although Vallotia misses some genes of the canonical pathways, alternative enzymes and host-derived compounds might bypass these reactions, as detailed in the Supplementary Material. Profftia has only a few genes related to B vitamin biosynthesis. Three pseudogenes (ribAEC) in the riboflavin synthesis pathway indicate that these functions might have been lost recently in this symbiont (Fig. S11).
Conclusions
Our results demonstrated that both endosymbionts of A. laricis/A. tardus are evolutionary recent bacterial partners of adelgids with moderately reduced genomes. Profftia and Vallotia open up novel insect symbiont lineages within the Hafniaceae and Burkholderiaceae, respectively. Given its phylogenetic position within a clade of Mycetohabitans species, we propose the reclassification of the betaproteobacterial symbiont as “Candidatus Mycetohabitans vallotii”.
A direct evolutionary link between Vallotia and endosymbionts of Rhizopus fungi was supported by a high level of genomic synteny. The phylogenetic position of Vallotia interleaved within the clade of Rhizopus endosymbionts and the lack of functions specific to the adelgid symbiont point toward a transition of endosymbiosis from fungi to insect hosts. The evolutionary origin of insect symbionts from fungus-associated endosymbionts is, according to our knowledge, unprecedented. Rhizopus endosymbionts are equipped with many functions for infection and overcoming host defense. Chitinase, chitosanase, and a putative chitin-binding protein have also been found among the putatively Sec exported proteins of M. rhizoxinica [68], which besides the infection of fungi could have had a role in the transmission into an insect. Their host, R. microsporus, is a plant pathogen fungus with a broad environmental distribution. Thus, a potential route for acquisition of the symbiont by insects could have been via plant tissues, the food source of adelgids, similar to plant-mediated symbiont transmission observed for intracellular insect symbionts [22].
Vallotia has a pivotal role in essential nutrient provision, but both endosymbionts are obligatory to their insect host. It has been suggested that repeated replacements of symbionts among adelgids might be a consequence of periods with relaxed selection on symbiont functions due to different feeding behavior of adelgids on primary and secondary host trees and multiple origins of their host-alternating lifestyles [11]. Annandia, the ancient symbiont of adelgids, has lost many functions in essential amino acid biosynthesis, which could support this hypothesis [23], although the Vallotia–Profftia system does not show this pattern.
Taken together, our comprehensive genomic analysis of co-obligate endosymbionts of adelgids revealed a novel path for the evolution of bacteria–insect symbioses from a clade of fungus-associated ancestors.
Supplementary information
Acknowledgements
We would like to acknowledge Alexander Siegl and Thomas Penz for their help in collecting adelgids and Irene Lichtscheidl-Schultz (Core Facility Cell Imaging and Ultrastructure Research, Cell Imaging Lab, University of Vienna) for the close-up photos of adelgids. Genome sequencing was performed by the Next Generation Sequencing Facility at Vienna BioCenter Core Facilities (VBCF), member of the Vienna BioCenter (VBC), Austria. This study was funded by the Austrian Science Fund (FWF) project P22533-B17. Work in the laboratory of MH is supported by Austrian Science Fund project DOC 69-B. AM-M was supported by a Marie Skłodowska-Curie Individual Fellowship (840270, LEECHSYMBIO) of the European Union.
Author contributions
ERT and MH initiated the study. GS, ERT, and AM-M carried out lab experiments. GS, FS, and AM-M performed genome reconstructions and genomic analyses. GS made phylogenetic analyses. GS, FS, AM-M, and MH interpreted the results. All authors took part in preparing the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01056-z.
References
- 1.Douglas AE. Phloem-sap feeding by animals: problems and solutions. J Exp Bot. 2006;57:747–54. doi: 10.1093/jxb/erj067. [DOI] [PubMed] [Google Scholar]
- 2.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–90. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 3.Wernegreen JJ. Endosymbiont evolution: predictions from theory and surprises from genomes. Ann NY Acad Sci. 2015;1360:16–35. doi: 10.1111/nyas.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manzano-Marín A, Coeur D’acier A, Clamens A-L, Orvain C, Cruaud C, Barbe V, et al. Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids’ di-symbiotic systems. ISME J. 2020;14:259–73. doi: 10.1038/s41396-019-0533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pons I, Renoz F, Noël C, Hance T. Circulation of the cultivable symbiont Serratia symbiotica in aphids is mediated by plants. Front Microbiol. 2019;10:764–77. doi: 10.3389/fmicb.2019.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, von Niederhausern AC, et al. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genet. 2012;8:e1002990. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havill NP, Foottit RG. Biology and evolution of Adelgidae. Annu Rev Entomol. 2007;52:325–49. doi: 10.1146/annurev.ento.52.110405.091303. [DOI] [PubMed] [Google Scholar]
- 8.Havill NP, Foottit RG, von Dohlen CD. Evolution of host specialization in the Adelgidae (Insecta: Hemiptera) inferred from molecular phylogenetics. Mol Phylogenet Evol. 2007;44:357–70. doi: 10.1016/j.ympev.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Favret C, Havill NP, Miller GL, Sano M, Victor B. Catalog of the adelgids of the world (Hemiptera, Adelgidae) Zookeys. 2015;534:35–54. doi: 10.3897/zookeys.534.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Dohlen CD, Spaulding U, Shields K, Havill NP, Rosa C, Hoover K. Diversity of proteobacterial endosymbionts in hemlock woolly adelgid (Adelges tsugae) (Hemiptera: Adelgidae) from its native and introduced range. Environ Microbiol. 2013;15:2043–62. doi: 10.1111/1462-2920.12102. [DOI] [PubMed] [Google Scholar]
- 11.von Dohlen CD, Spaulding U, Patch KB, Weglarz KM, Foottit RG, Havill NP, et al. Dynamic acquisition and loss of dual-obligate symbionts in the plant-sap-feeding Adelgidae (Hemiptera: Sternorrhyncha: Aphidoidea) Front Microbiol. 2017;8:1037–52. doi: 10.3389/fmicb.2017.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalik A, Gołas A, Kot M, Wieczorek K, Szklarzewicz T. Endosymbiotic microorganisms in Adelges (Sacchiphantes) viridis (Insecta, Hemiptera, Adelgoidea: Adelgidae): molecular characterization, ultrastructure and transovarial transmission. Arthropod Struct Dev. 2013;42:531–8. doi: 10.1016/j.asd.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Toenshoff ER, Penz T, Narzt T, Collingro A, Schmitz-Esser S, Pfeiffer S, et al. Bacteriocyte-associated gammaproteobacterial symbionts of the Adelges nordmannianae/piceae complex (Hemiptera: Adelgidae) ISME J. 2012;6:384–96. doi: 10.1038/ismej.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toenshoff ER, Gruber D, Horn M. Co-evolution and symbiont replacement shaped the symbiosis between adelgids (Hemiptera: Adelgidae) and their bacterial symbionts. Environ Microbiol. 2012;14:1284–95. doi: 10.1111/j.1462-2920.2012.02712.x. [DOI] [PubMed] [Google Scholar]
- 15.Toenshoff ER, Szabó G, Gruber D, Horn M. The pine bark adelgid, Pineus strobi, contains two novel bacteriocyte-associated gammaproteobacterial symbionts. Appl Environ Microbiol. 2014;80:878–85. doi: 10.1128/AEM.03310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gündüz EA, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 2009;276:987–91. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duron O, Hurst GD. Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol. 2013;11:45–9. doi: 10.1186/1741-7007-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldhaar H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol. 2011;36:533–43. [Google Scholar]
- 19.Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–66. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 20.Moran NA, Dunbar HE. Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci USA. 2006;103:12803–6. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehrer L, Vorburger C. Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol Lett. 2012;8:613–5. doi: 10.1098/rsbl.2012.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chrostek E, Pelz-Stelinski K, Hurst GDD, Hughes GL. Horizontal transmission of intracellular insect symbionts via plants. Front Microbiol. 2017;8:2237–45. doi: 10.3389/fmicb.2017.02237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weglarz KM, Havill NP, Burke GR, von Dohlen CD. Partnering with a pest: Genomes of hemlock woolly adelgid symbionts reveal atypical nutritional provisioning patterns in dual-obligate bacteria. Genome Biol Evol. 2018;10:1607–21. doi: 10.1093/gbe/evy114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zurovcova M, Havelka J, Stary P, Vechtova P, Chundelova D, Jarosova A, et al. ‘DNA barcoding’ is of limited value for identifying adelgids (Hemiptera: Adelgidae) but supports traditional morphological taxonomy. Eur J Entomol. 2010;107:147–56. [Google Scholar]
- 25.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–22. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421–30. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–60. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–36. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinmaier T, Platzer A, Frank J, Hellinger H-J, Tischler P, Rattei T. ConsPred: a rule-based (re-)annotation framework for prokaryotic genomes. Bioinformatics. 2016;32:3327–9. doi: 10.1093/bioinformatics/btw393. [DOI] [PubMed] [Google Scholar]
- 37.Okonechnikov K, Golosova O, Fursov M. the UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–7. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 38.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–9. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 2017;34:2115–22. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Wu M. A phylum-level bacterial phylogenetic marker database. Mol Biol Evol. 2013;30:1258–62. doi: 10.1093/molbev/mst059. [DOI] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 49.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–5. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–95. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE). New Orleans, LA, USA, 2010; pp 1–8.
- 55.Husník F, Chrudimský T, Hypša V. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 2011;9:87–103. doi: 10.1186/1741-7007-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, et al. Phylogeny of Gammaproteobacteria. J Bacteriol. 2010;192:2305–14. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adeolu M, Alnajar S, Naushad S, Gupta RS. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:5575–99. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 58.Janda JM, Abbott SL. The genus Hafnia: from soup to nuts. Clin Microbiol Rev. 2006;19:12–28. doi: 10.1128/CMR.19.1.12-28.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lundgren JG, Lehman RM, Chee-sanford J. Bacterial communities within digestive tracts of ground beetles (Coleoptera: Carabidae) Ann Entomol Soc Am. 2007;100:275–82. [Google Scholar]
- 60.Estrada-de los Santos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp ET, Briscoe L, et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes. 2018;9:389–412. doi: 10.3390/genes9080389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Partida-Martinez LP, Groth I, Schmitt I, Richter W, Roth M, Hertweck C. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int J Syst Evol Microbiol. 2007;57:2583–90. doi: 10.1099/ijs.0.64660-0. [DOI] [PubMed] [Google Scholar]
- 62.Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–8. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 63.Moebius N, Üzüm Z, Dijksterhuis J, Lackner G, Hertweck C. Active invasion of bacteria into living fungal cells. Elife. 2014;3:e03007. doi: 10.7554/eLife.03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Partida-Martinez LP, Monajembashi S, Greulich K-O, Hertweck C. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr Biol. 2007;17:773–7. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 65.Dolatabadi S, Walther G, Gerrits van den Ende AHG, de Hoog GS. Diversity and delimitation of Rhizopus microsporus. Fungal Divers. 2014;64:145–63. [Google Scholar]
- 66.Lackner G, Möbius N, Scherlach K, Partida-Martinez LP, Winkler R, Schmitt I, et al. Global distribution and evolution of a toxinogenic Burkholderia-Rhizopus symbiosis. Appl Environ Microbiol. 2009;75:2982–6. doi: 10.1128/AEM.01765-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dubarry N, Pasta F, Lane D. ParABS Systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J Bacteriol. 2006;188:1489–96. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011;12:210–23. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–30. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 71.Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–33. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 72.Wernegreen JJ. For better or worse: genomic consequences of intracellular mutualism and parasitism. Curr Opin Genet Dev. 2005;15:572–83. doi: 10.1016/j.gde.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 73.Bourguignon T, Kinjo Y, Villa-Martín P, Coleman NV, Tang Q, Arab DA, et al. Increased mutation rate is linked to genome reduction in prokaryotes. Curr Biol. 2020;30:3848.e4–55.e4. doi: 10.1016/j.cub.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 74.Kuo C-H, Ochman H. Deletional bias across the three domains of life. Genome Biol Evol. 2009;1:145–52. doi: 10.1093/gbe/evp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 76.Lo W-S, Huang Y-Y, Kuo C-H. Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol Rev. 2016;40:855–74. doi: 10.1093/femsre/fuw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamelas A, Gosalbes MJ, Manzano-Marín A, Peretó J, Moya A, Latorre A. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011;7:e1002357. doi: 10.1371/journal.pgen.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manzano-Marín A, Latorre A. Snapshots of a shrinking partner: genome reduction in Serratia symbiotica. Sci Rep. 2016;6:1–11. doi: 10.1038/srep32590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koga R, Moran NA. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 2014;8:1237–46. doi: 10.1038/ismej.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plague GR, Dunbar HE, Tran PL, Moran NA. Extensive proliferation of transposable elements in heritable bacterial symbionts. J Bacteriol. 2008;190:777–9. doi: 10.1128/JB.01082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manzano-Marín A, Latorre A. Settling down: The genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol. 2014;6:1683–98. doi: 10.1093/gbe/evu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Degnan PH, Leonardo TE, Cass BN, Hurwitz B, Stern D, Gibbs RA, et al. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 2010;12:2060–9. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:research0054.1–0054.12. [DOI] [PMC free article] [PubMed]
- 84.Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol. 2003;6:512–8. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 85.Silva FJ, Latorre A, Moya A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 2001;17:615–8. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- 86.Partida-Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem. 2007;8:41–5. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- 87.Hamel E. Natural products which interact with tubulin in the vinca domain: maytansine, rhizoxin, phomopsin a, dolastatins 10 and 15 and halichondrin B. Pharm Ther. 1992;55:31–51. doi: 10.1016/0163-7258(92)90028-x. [DOI] [PubMed] [Google Scholar]
- 88.Süssmuth RD, Mainz A. Nonribosomal peptide synthesis—principles and prospects. Angew Chem Int Ed Engl. 2017;56:3770–821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 89.Landsberg MJ, Jones SA, Rothnagel R, Busby JN, Marshall SDG, Simpson RM, et al. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc Natl Acad Sci USA. 2011;108:20544–9. doi: 10.1073/pnas.1111155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ochman H. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet. 2002;18:335–7. doi: 10.1016/s0168-9525(02)02668-9. [DOI] [PubMed] [Google Scholar]
- 91.Satoshi F, Nishikawa K. Estimation of the number of authentic orphan genes in bacterial genomes. DNA Res. 2004;11:219–31. [PubMed] [Google Scholar]
- 92.Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 2005;187:4229–37. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lackner G, Moebius N, Hertweck C. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 2011;5:252–61. doi: 10.1038/ismej.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nivaskumar M, Francetic O. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta. 2014;1843:1568–77. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 95.Szklarzewicz T, Michalik A. Transovarial transmission of symbionts in insects. In: Kloc M, editor. Oocytes. Cham, Switzerland: Springer International Publishing; 2017. [DOI] [PubMed] [Google Scholar]
- 96.Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio. 2014;5:e01697–14. doi: 10.1128/mBio.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–78. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 98.Shigenobu S, Wilson ACC. Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont. Cell Mol Life Sci. 2011;68:1297–309. doi: 10.1007/s00018-011-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaplan E, Greene NP, Crow A, Koronakis V. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain. Proc Natl Acad Sci USA. 2018;115:E7389–97. doi: 10.1073/pnas.1806822115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 101.Tamaki S, Matsuzawa H, Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980;141:52–7. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pabst MJ, Kuhn JC, Somerville RL. Feedback regulation in the anthranilate aggregate from wild type and mutant strains of Escherichia coli. J Biol Chem. 1973;248:901–14. [PubMed] [Google Scholar]
- 104.Sloan DB, Moran NA. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 2012;29:3781–92. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hansen AK, Moran NA. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108:2849–54. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, et al. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 2014;31:857–71. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–18. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Belitsky BR, Gustafsson MC, Sonenshein AL, Von Wachenfeldt C. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J Bacteriol. 1997;179:5448–57. doi: 10.1128/jb.179.17.5448-5457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.