Abstract
The function of BAD, a proapoptotic member of the Bcl-2 family, is regulated primarily by rapid changes in phosphorylation that modulate its protein-protein interactions and subcellular localization. We show here that, during interleukin-3 (IL-3) deprivation-induced apoptosis of 32Dcl3 murine myeloid precursor cells, BAD is cleaved by a caspase(s) at its N terminus to generate a 15-kDa truncated protein. The 15-kDa truncated BAD is a more potent inducer of apoptosis than the wild-type protein, whereas a mutant BAD resistant to caspase 3 cleavage is a weak apoptosis inducer. Truncated BAD is detectable only in the mitochondrial fraction, interacts with BCL-XL at least as effectively as the wild-type protein, and is more potent than wild-type BAD in inducing cytochrome c release. Human BAD, which is 43 amino acids shorter than its mouse counterpart, is also cleaved by a caspase(s) upon exposure of Jurkat T cells to anti-FAS antibody, tumor necrosis factor alpha (TNF-α), or TRAIL. Moreover, a truncated form of human BAD lacking the N-terminal 28 amino acids is more potent than wild-type BAD in inducing apoptosis. The generation of truncated BAD was blocked by Bcl-2 in IL-3-deprived 32Dcl3 cells but not in Jurkat T cells exposed to anti-FAS antibody, TNF-α, or TRAIL. Together, these findings point to a novel and important role for BAD in maintaining the apoptotic phenotype in response to various apoptosis inducers.
The Bcl-2 family proteins are key regulators of apoptosis whose function as cell death agonists or antagonists is modulated by transcriptional and posttranscriptional modifications. Posttranscriptional modifications, together with relative levels of expression and subcellular localization of inhibitors (Bcl-2, Bcl-XL) and promoters (BAX, BAD, BID, BIK) determine how cells respond to apoptotic stimuli (27, 37, 16). A study of growth factor-dependent hematopoietic cell lines has shown that BAD, a death-promoting BH3-only member of the Bcl-2 family of proteins, is a key regulator of apoptosis (14). BAD is regulated primarily by phosphorylation (16). Phosphorylation of BAD at serine 112 and 136 residues can be stimulated by interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor, platelet-derived growth factor, nerve growth factor, insulin growth factor, and, under noncytotoxic conditions, even tumor necrosis factor (TNF) (11, 9, 52, 36, 29). This modification is necessary for the association of BAD with the 14-3-3 proteins, which prevents BAD translocation to the mitochondria and its interaction with Bcl-XL or Bcl-2 (52, 19), thus allowing the latter proteins to promote cell survival. Recent studies have also shown BAD phosphorylation at its BH3 domain (serine 155) by survival-promoting kinases (42, 54, 10); this phosphorylation seems necessary for releasing BAD from its association with Bcl-XL (10). Together, these studies suggest that BAD can promote apoptosis when cells have inadequate levels of survival factors. IL-3 deprivation-induced apoptosis of murine 32Dcl3 myeloid precursor cells provides a paradigm of BAD-mediated cell death. Once the cytokine is removed from the culture medium, BAD is rapidly dephosphorylated by the specific serine/threonine phosphatase PP1α (32), dissociates from 14-3-3 proteins, and translocates to the mitochondria, where it interacts with Bcl-XL and Bcl-2 and antagonizes their antiapoptotic functions. A similar pattern of events also occurs after IL-2 deprivation of the murine T-cell line TS1αβ (2). Bcl-2 is also phosphorylated in its loop region during normal cell cycle progression and microtubule-damaging-agent-induced growth arrest and apoptosis (35, 17). Several Bcl-2 family members are also targets for proteolysis. For example, BID, a cytosolic proapoptotic family member (46), can be cleaved by caspase 8 and by granzyme-B; following the cleavage, truncated BID translocates to the mitochondria, where it promotes the release of cytochrome c (25, 24). In some circumstances, Bcl-2 and Bcl-XL are also caspase targets (8, 6). Interestingly, cleavage of these two proteins converts them from prosurvival into proapoptotic molecules able to induce cytochrome c release from the mitochondria (8, 6). The proapoptotic BAX protein is also cleaved in a caspase-dependent manner to a 18-kDa fragment that is reportedly more toxic than its full-length counterpart (47).
In the present study, we show that BAD is cleaved 4 to 6 h after IL-3 deprivation of murine myeloid precursor 32Dcl3 cells. The cleavage is caspase dependent and is abrogated when aspartates 56 and 61 are replaced with glutamate residues. A caspase 3-resistant mutant BAD shows less proapoptotic activity than the wild-type (WT) protein, whereas truncated BAD is a more potent inducer of apoptosis, possibly due to its ability to promote cytochrome c release from the mitochondria. BAD is also cleaved in human Jurkat T cells induced to undergo apoptosis by treatment with anti-Fas antibody, TNF-α, or TRAIL. Once overexpressed, truncated BAD is more potent than the WT form in accelerating Fas-dependent apoptosis.
Together, these data indicate that cleavage of full-length BAD during apoptosis increases its intrinsic cytotoxic properties and enhances the commitment to cell death by maintaining the mitochondrion-dependent activation of the caspase cascade.
MATERIALS AND METHODS
Plasmids. (i) pcDNA3 WT HA-BAD.
Hemagglutinin (HA)-tagged mouse WT BAD cloned in a HindIII- and EcoRI-linearized pcDNA3.1 vector was a kind gift from M. E. Greenberg, Children's Hospital and Department of Neurobiology, Harvard Medical School, Boston, Mass.
(ii) pcDNA tBAD58, pcDNA tBAD68, and pcDNA tBAD84.
Truncated forms of murine BAD were amplified by PCR from pcDNA3 WT HA-BAD using the following primers: 5′BAD58-HindIII (5′-AAGCTTAAGCTTATGAGTGCTACAGATAGGGGC-3′), 5′BAD68-HindIII (5′-AAGCTTAAGCTTATGCTCACTGAGGACCAGCCA-3′), and 5′BAD84-HindIII (5′-AAGCTTAAGCTTATGAGCAACATTCATCAGCAGG-3′), all of which contain two HindIII sites (in bold), and 3′BAD-EcoRI (5′-GAATTCGAATTCTCACTGGGAGGGGGT-GGAG-3′), which contains two EcoRI sites (in boldface). The amplified fragments and the pcDNA3.1 plasmid (Invitrogen, Inc.) were digested with HindIII and EcoRI and ligated.
(iii) pcDNA-D56E, -D61E, and -DM56/61 HA-BAD.
pcDNA3 WT HA BAD was mutagenized by PCR using a mutagenesis kit (Stratagene) and the following primer pairs: 5′-GAGTGAGCAGGAAGAAGCTAGTGCTACAGAT-3′ and 5′-ATCTGTAGCACTAGCTTCTTCCTGCTCACTC-3′ (D56E BAD) and 5′GACGCTAGTGCTACAGAAAGGGGCCTGGGC-3′ and 5′-GCCCAGGCCCCTAACTGTAGCACTAGCGTC-3′ (D61E BAD); DM56/61 BAD was obtained by mutagenesis of pcDNA-D56E BAD at nucleotides specific for codon 61 using the following primers: 5′-GAAGCTAGTGCTACAGAAAGGGGCCTGGGC-3′ and 5′-GCCCAGGCCCCTAACTGTAGCACTAGCTTC-3′. Plasmids were sequenced to verify the presence of mutations.
(iv) pLXSP-HA WT BAD.
The pLXSP-HA WT BAD retroviral construct contains the HA epitope cloned in frame at the 5′ end of WT BAD (32).
(v) pLXSP WT, DM56/61 and tBAD68 BAD-HA.
The pLXSP WT BAD-HA, pLXSP DM56/61 BAD-HA, and pLXSP tBAD68 BAD-HA retroviral constructs were prepared by PCR amplification of BAD sequences from pcDNA WT BAD, DM56/61 BAD, and tBAD68 using the following primers: 5′BAD-HpaI (5′-GTTAACGTTAACATGGGAACCCCAAAGCAGCC-3′) (WT and DM56/61 BAD) and 5′BAD68-HpaI (5′-GTTAACGTTAACATGCTCACTGAGGACCAGCC-3′) (tBAD68), which contain two HpaI sites (in boldface), and 3′BAD-XhoI (5′-CTCGAGCTCGAGATGCTGGGAGGGGGTGGAG-3′) (common to all PCR), which contains two XhoI sites (in boldface). PCR products were digested with HpaI and XhoI and ligated into the HpaI- and XhoI-digested pLXSP-HA vector, which includes an XhoI/SalI fragment containing the HA epitope plus an in-frame stop codon. In-frame cloning of BAD upstream of the 3′-end HA tag was assessed by plasmid sequencing.
(vi) pMIGR1-WT BAD pMIGR1-DM56/61 BAD, and pMIGR1-tBAD68.
pcDNA WT BAD, pcDNA DM56/61 BAD, and pcDNA tBAD68 were HindIII and EcoRI digested, Klenow fragment blunt ended, and cloned upstream of the internal ribosome entry site sequence of the EcoRI-linearized and Klenow fragment-blunt-ended pMIGR1 vector (30).
(vii) pLXSN-Bcl-2.
The pLXSN-Bcl-2 retroviral construct contains the human Bcl-2 cDNA cloned at the EcoRI site of the pLXSN vector (7).
(viii) pTAT-HA WT BAD, pTAT-HA DM56/61 BAD, and pTAT-HA tBAD68.
WT BAD, DM56/61 BAD, and tBAD68 were amplified by PCR from the corresponding pcDNA3.1 plasmids using the following primers: 5′BAD-XhoI (5′-CTCGAGCTCGAGGGAACCCCAAAGCAGCCCT-3′) (WT and DM56/61 BAD) and 5′BAD68-XhoI (5′-CTCGAGCTCGAGCTCACTGAGGACCAGCCAG-3′) (tBAD68), which contain two XhoI sites (in boldface), and 3′BAD-EcoRI (5′-GAATTCGAATTCTCACTGGGAGGGGGTGGAG-3′), which contains two EcoRI sites (in boldface). Amplified fragments and pTAT-HA (kind gift of A. M. Vocero-Akbani, HHMI/Washington University, Medical School, St. Louis, Mo.) were digested with XhoI and EcoRI and ligated overnight. Recircularized plasmids were selected from transformed DH5-α Escherichia coli and sequenced to verify in-frame cloning.
(ix) pCR II human BAD (hBAD).
An MCF-7 cDNA library was obtained by reverse transcription-PCR of total mRNA. BAD cDNA was amplified by PCR using the following primers: 5′hBAD (5′-ATGTTCCAGATCCCAGAGTT-3′) and 3′hBAD (5′-TCACTGGGAGGGGGCGG-3′). The PCR fragments were subcloned in the pCR II vector using a TOPO-TA cloning kit (Invitrogen, Inc.).
(x) pLXSP WT hBAD and pLXSP t-hBAD29.
WT hBAD and tBAD29 BAD (t-hBAD29) were generated by PCR using the following primers: 5′hBAD-HpaI (5′-GTTAACGTTAACATGTTCCAG-ATCCCAGAGTT-3′) and 5′BAD29-HpaI (5′-GTTAACGTTAACATGGGGCCCTCA-GGCTCCG-3′), which contain two HpaI sites (in boldface), and 3′hBAD-XhoI (5′-CTCGAGCTCGAGATGCTGGGAGGGGGCGG-3′) (common to both PCRs), which has two XhoI sites (in boldface). PCR-amplified fragments were digested with HpaI and XhoI and cloned at the 5′ end of the HA tag in an HpaI- and XhoI-digested pLXSP-HA retroviral vector.
Cell lines.
The IL-3-dependent murine 32Dc13 myeloid precursor cell line was maintained at 37°C in IMDM-CM (Iscove's modified Dulbecco medium supplemented with 10% heat-inactivated fetal bovine serum [FBS], 2 mM l-glutamine, penicillin and streptomycin [100 μg/ml each), and 15% WEHI-conditioned medium [WEHI-CM] as a source of IL-3 [50]). Clones of 32Dcl3 cells expressing p210 BCR/ABL were generated in our laboratory and maintained as described previously (39). In assays requiring cell starvation, cells were washed four times in phosphate-buffered saline (PBS) and incubated in IMDM supplemented with 10% FBS or 0.1% bovine serum albumin and 2 mM l-glutamine but lacking WEHI-CM. The 293-derived PHOENIX packaging cells (kind gift of G. P. Nolan, Stanford University) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated FBS, and 2 mM glutamine (GIBCO). The human Jurkat T-cell line (American Type Culture Collection, Rockville, Md.) was cultured in RPMI medium supplemented with 10% heat-inactivated FBS and 2 mM glutamine (GIBCO). Bcl-2-expressing 32Dcl3 and Jurkat T cells were obtained by retrovirus-mediated gene transfer (see below) of pLXSN-Bcl-2.
Retrovirus infection and methylcellulose colony assays.
PHOENIX packaging cells were cultured in DMEM and transfected with the pLXSP retroviral vector (empty or containing HA-WT BAD, HA-DM56/61 BAD, or tBAD68 cDNA), by the calcium phosphate technique (Mammalian Profection Kit; Promega). Immediately before transfection, cells were treated with 25 μM chloroquine to increase transfection efficiency. At 12 h posttransfection, DMEM was replaced with IMDM-CM (for 32Dc13 cells) or RPMI 1640 (for Jurkat T cells), and cells were grown for an additional 24 h. Supernatants were collected, filtered through 0.45-μm-pore-size filters, and supplemented with 4 μg of Polybrene per ml just before the spin-infection procedure. Briefly, 32Dcl3 alls (WT or expressing BCR/ABL) or Jurkat T cells were resuspended (2 × 105 cells/ml) in the virus-containing medium, plated in six-well plates, and centrifuged for 45 min at 600 × g in a 32°C-prewarmed centrifuge. Cells were then incubated for 4 h at 32°C, followed by a second cycle of centrifugation and incubation with fresh virus-containing medium as described above. At the end of the second cycle, the infectious supernatant was replaced with IMDM-CM (32Dcl3 cells) or RPMI 1640 (Jurkat T cells) and cells were incubated overnight at 37°C. The following day, after a third cycle of centrifugation and incubation with fresh infectious supernatant, cells were cultured in regular medium for 16 h and plated in methylcellulose in the presence of puromycin (2.5 μg/ml) and 10% WEHI-CM as source of IL-3 where indicated in Results. Colonies were counted 5 to 7 days later. Individual clones picked from methylcellulose plates of BAD retrovirus-infected cells were transferred to 24-microtiter wells and expanded to generate cell clones stably expressing WT, mutant, or truncated BAD.
Cell viability and apoptosis assays.
The pMCSV-based bicistronic retroviral vector pMIGR1 (30) carrying the green fluorescent protein (GFP) DNA as a reporter gene downstream of the internal ribosome entry site was used to transiently infect 32Dcl3 cells (empty vector or carrying the WT, DM56/61, or tBAD68 cDNA), as described above. Retrovirus-infected cells were assayed for GFP expression by fluorescence-activated cell sorter (FACS) analysis (Coulter Inc., Hialeah, Fla.) and sorted to obtain a pure population of transgene-positive cells. Sorted cells were replated for 2 h in IMDM supplemented with IL-3 to allow recovery from the sorting procedure. To assess viability, cells were stained with ANNEXIN V (Pharmingen, Torrey Pines, Calif.) to measure the exposure of cell surface phosphatidylserine molecules by FACS analysis or with trypan blue to count the number of positive cells at different time points. To distinguish between necrotic and apoptotic cells in the ANNEXIN V assay, only cells not stained with the vital dye 7 amino-actinomycin D (7AAD) were considered positive. Trypan blue exclusion assay was also used for time course experiments of IL-3-deprived 32Dcl3 cells stably expressing WT-BAD, DM56/61 BAD, or tBAD68.
Preparation of subcellular fractions.
Subcellular fractionation was performed as described previously (49). Briefly, cells were lysed in buffer A (20 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA [pH 8.0], 0.1 mM EDTA, 1 mM dithiothreitol, 250 mM sucrose, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 mM benzamidine, 1 mM phenylmethylsulfoxyl fluoride [PMSF], 0.2 mM sodium orthovanadate) and homogenized (20 strokes with a B-pestle Dounce homogenizer). Samples were centrifuged twice (750 × g for 5 min) to remove nuclei and unlysed cells and centrifuged again (10,000 × g for 10 min) to obtain the heavy-membrane (HM) fraction (pellet). The supernatant was centrifuged at 150,000 × g for 1.5 h to obtain the cytosolic fraction. The HM fractions were solubilized in a solution containing 1% Triton X-100, 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 mM benzamidine, 1 mM PMSF, and 0.2 mM sodium orthovanadate.
Western blot, immunoprecipitation, and coimmunoprecipation analyses.
Whole-cell extracts were obtained in a 1% Triton X-100 lysis buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA [pH 8.0], 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 mM benzamidine, 1 mM PMSF, 0.2 M sodium orthovanadate). Western blotting was performed using anti-BAD (C-20, K-17, H-168; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), anti-BCL-XL (Santa Cruz Biotechnology), anti-HA (Babco, Richmond, Calif.), and anti-14-3-3β (Santa Cruz Biotechnology) antibodies. Immunoprecipitations of HA-tagged BAD were carried out using the anti-HA antibody (Babco). Coimmunoprecipitations were carried out in a solution containing 0.2% Triton X-100, 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 mM benzamidine, 1 mM PMSF, and 0.2 M sodium orthovanadate (immunoprecipitation buffer). Precleared extracts were incubated with antibody for 2 h and with protein G-coated agarose beads for an additional 2 h. Beads were washed five times in immunoprecipitation buffer and boiled for 2 min in sample buffer. Samples were loaded on a 4 to 15% gradient polyacrylamide–sodium dodecyl sulfate (SDS) gel for electrophoresis.
In vitro cleavage of 35S-labeled proteins.
pcDNA3.1-WT BAD, -D56E BAD, -D61E BAD, -DM56/61 BAD, -tBAD58, -tBAD68, and -tBAD84 were translated in vitro in the presence of [35S]methionine (TNT coupled reticulocyte lysate systems; Promega). Labeled proteins were incubated with 10 ng of recombinant caspases or with 32Dcl3 apoptotic cell extracts for 1 h at 30°C (in the presence or absence of the broad-range caspase inhibitor z-VAD fmk [50 μM] or the caspase 3-specific inhibitor DEVD fmk [50 nM] where indicated in the figure legends). Apoptotic cell extracts were obtained by lysis of 12-h IL-3-deprived 32Dcl3 cells in buffer A (see above). After homogenization (40 strokes with a B-pestle Dounce homogenizer), lysates were centrifuged at 90,000 × g for 1 h at 4°C and the supernatant was used for assessment of further cleavage of in vitro-translated protein. Reactions were stopped by adding sample buffer, and proteins were resolved on a 4 to 15% gradient polyacrylamide–SDS gel. The gel was subsequently dried and exposed to X-ray film for 2 h.
Purification of TAT-BAD fusion proteins.
For mitochondrial cytochrome c release assay, BAD proteins fused with the leader sequence of the human immunodeficiency virus-encoded TAT protein (YGRKKRRQRRR) were produced as described previously (43). Briefly, pTAT-6His-HA-WT BAD, pTAT-6His-HA-DM56/61 BAD, and pTAT-6His-HA-tBAD68 plasmids were used to transform the BL-21 E. coli strain and bacteria were subsequently plated in Luria-Bertani (LB) agar plates plus ampicillin. Ten clones from each transformation were picked and cultured overnight at 37°C in 10 ml of ampicillin-LB agar medium plus IPTG (isopropyl-β-d-thiogalactopyranoside) to induce protein expression. The following day, 1 ml from each overnight culture was spun down and resuspended in SDS sample buffer, loaded on a 4 to 15% gradient polyacrylamide–SDS gel, and monitored for BAD protein expression by Coomassie blue staining. High-level-expression clones were cultured overnight at 37°C in 250 ml of ampicillin-LB broth, inoculated in 1 liter of ampicillin-LB broth, and induced with IPTG for 2 h. Cells were spun down, resuspended in buffer Z (8 M urea, 100 mM NaCl, 20 mM HEPES [pH 8.0], 1 mM PMSF), and sonicated in ice. After centrifugation, 10 mM imidazole was added and samples were loaded on buffer Z–imidazole-preequilibrated Ni-nitrilotriacetic acid columns (Qiagen). His-TAT proteins were eluted by stepwise addition of 10 ml each of 100, 250, 500, 1,000 mM NaCl plus 10 mM imidazole. Flow-through, wash, and fraction volumes were checked for BAD protein by SDS-polyacrylamide gel electrophoresis, followed by Coomassie blue staining. Appropriate fractions were pooled and diluted 1:1 (vol/vol) with 20 mM HEPES (pH 8.0)–100 mM NaCl–8 M urea. Samples were applied to 5/5 Mono Q columns attached to a fast protein liquid chromatograph (Amersham Pharmacia Biotech, San Francisco, Calif.) equilibrated in 50 mM NaCl–20 mM HEPES, pH 8.0. His-TAT proteins were eluted stepwise with 150, 250, 350, and 500 mM NaCl and desalted on a PD-10 disposable G-25 Sephadex gravity column (Amersham Pharmacia Biotech) in PBS.
Cytochrome c release assay.
Mitochondria were isolated from 32Dcl3 parental cells as described above. Each TAT-BAD protein (100 mM) was incubated with the mitochondrial suspension (in the presence or absence of 10 ng of caspase 3) for 1 h at 30°C. Samples were centrifuged (10,000 × g for 10 min at 4°C), and supernatants were separated from pellets. Mitochondrial pellet proteins were extracted in lysis buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA [pH 8.0], 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 mM benzamidine, 1 mM PMSF). Mitochondrial extracts and supernatant proteins were resolved on a 15% polyacrylamide–SDS gel. Mitochondrial proteins were blotted with monoclonal anti-COX IV antibody (Molecular Probes, Eugene, Oreg.) as a control for equal loadings and subsequently with a monoclonal anti-cytochrome c antibody to assess the presence of unreleased protein. Supernatant-suspended proteins were blotted with the anti-cytochrome c antibody to assess cytochrome c release from mitochondria.
Anti-CD95 antibody, TNF-α, or TRAIL-induced apoptosis.
Jurkat T cells (2.5 × 105/ml) were treated with the anti-CD95 antibody (0.05 μg/ml; R&D Systems, Minneapolis, Minn.), TNF-α (10 ng/ml), and TRAIL (1 μg/ml) for up to 6 h. Cells were harvested, and protein extracts were blotted with polyclonal anti-BAD serum (H-168; Santa Cruz Biotechnology).
RESULTS
Cleavage of proapoptotic BAD in IL-3 starvation-induced apoptosis of 32Dc13 myeloid precursor cells.
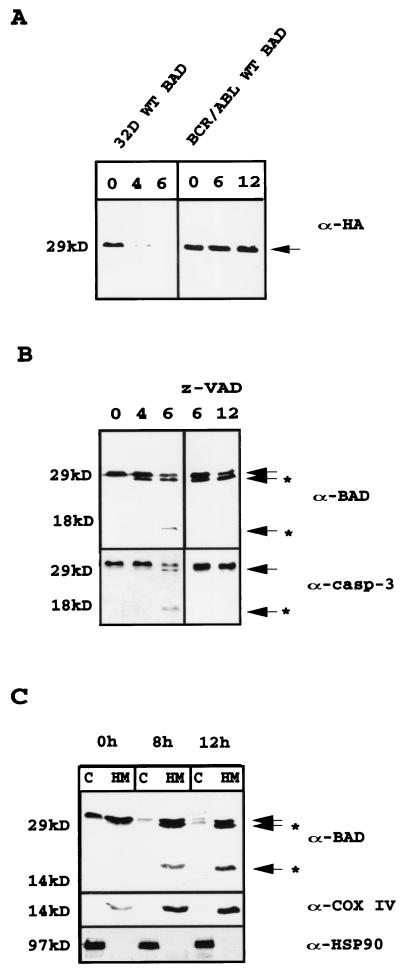
During the process of apoptosis induced by IL-3 starvation of 32Dc13 cells, BAD is dephosphorylated by the serine/threonine phosphatase 1α (32) and translocates from the cytoplasm to the mitochondria. Upon IL-3 deprivation, levels of BAD were also down-modulated (not shown). To investigate in detail this finding, we utilized 32Dc13 cells ectopically expressing an HA-tagged BAD in which the HA epitope is cloned in frame 5′ of BAD. A decrease in BAD protein levels at 4 h after IL-3 withdrawal and almost undetectable levels at 6 h were noted in Western blot experiments with an antibody raised against the HA epitope (Fig. 1A, left gel). Blotting of the same filter with an antibody against the C terminus of BAD (C-20; amino acids [aa] 185 to 204) resolved two fast-migrating BAD-reactive bands, one very close is mass to full-length BAD (∼26 kDa) and another of ∼15 kDa (Fig. 1B, left gel). An antibody specific for the N terminus of BAD (K-17) did not recognize the 15-kDa cleavage product (data not shown), suggesting that the cleavage was downstream of the epitope recognized by this antibody. Only the full-length form of BAD was detected in BCR/ABL-expressing cells (Fig. 1A, right gel), which do not undergo apoptosis after IL-3 withdrawal.
FIG. 1.
(A) Expression of WT HA-BAD in IL-3-starved parental and BCR/ABL-expressing 32Dcl3 cells. Extracts from cells starved for 0, 4, and 6 h (32D) or 0, 6, and 12 h (BCR/ABL) were blotted with anti-HA antibody (α-HA). (B) Effect of z-VAD fmk (50 μM) on levels of WT HA-BAD in IL-3-starved parental 32Dcl3 cells. Western blot analyses were performed with an anti-BAD (C-20, upper gels) antibody. Caspase-3 levels were measured as a control for the inhibitory capacity of z-VAD fmk. Results are representative of three separate experiments. (C) HA-BAD expression in subcellular fractions from retrovirus-infected parental cells. At 0, 8, and 12 h after IL-3 withdrawal, mitochondrial (HM) and cytoplasmic (C) extracts were tested for BAD expression using the anti-BAD (C-20) antibody. Arrows indicate full-length HA-BAD; arrows plus asterisks indicate cleaved products of BAD or caspase 3. Results are representative of three separate experiments.
Role of caspase 3-like activity in BAD cleavage after IL-3 removal.
To assess the possible involvement of a caspase(s) in the cleavage of BAD, expression levels and activation of caspase 3 were analyzed in WT BAD retrovirus-infected parental and BCR/ABL-expressing cells. Cleavage of caspase 3 correlated in part with that of BAD: at 6 h almost 50% of the caspase 3 precursor was cleaved (Fig. 1B, lower left gel). By contrast, caspase 3 was detected as the ∼32-kDa inactive precursor in WT BAD-infected BCR/ABL-expressing cells (data not shown). Treatment of 32Dc13 parental cells with the broad-range caspase inhibitor z-VAD fmk prevented the generation of the 15-kDa cleaved from of BAD but not of the 26-kDa band (Fig. 1B, upper right gel). Activation of caspase 3 was completely suppressed by z-VAD fmk (Fig. 1B, lower left gel). Western blotting of subcellular fractions with the anti-C-terminus BAD antibody (C-20) revealed the 15-kDa cleavage product only in the mitochondrion-enriched protein fraction (Fig. 1C).
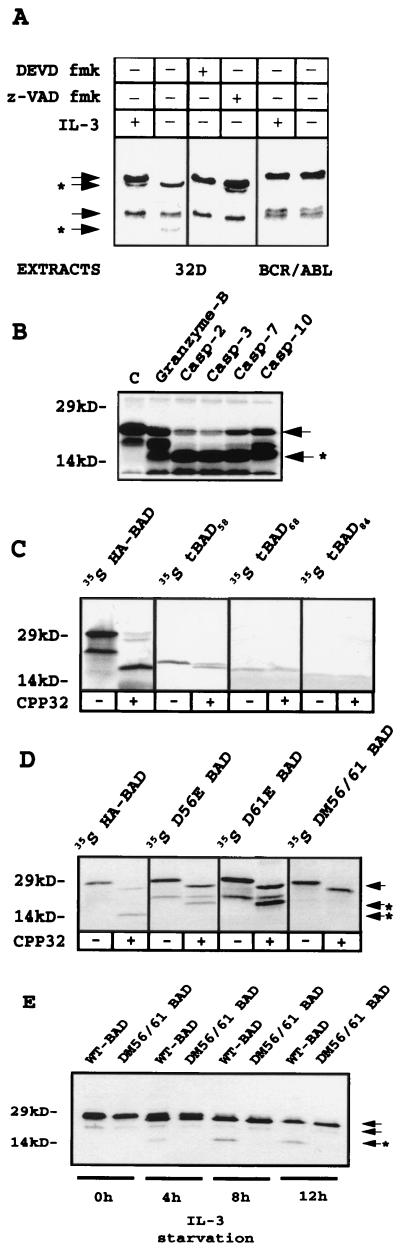
Analysis of the caspase-like activity of apoptotic extracts from 32Dc13 parental cells using in vitro-translated BAD as the substrate demonstrated a pattern of BAD cleavage identical to that seen in the in vivo experiments (Fig. 2A). As expected, cleavage was prevented when apoptotic extracts were incubated with 50 μM z-VAD fmk or with a 50 nM concentration of the caspase-3-specific caspase inhibitor DEVD fmk (Fig. 2A). Extracts from IL-3-deprived BCR/ABL-expressing cells used as a negative control for caspase activity did not cleave BAD (Fig. 2A). Use of recombinant caspases on in vitro-translated BAD showed that BAD was cleaved by caspase 2, 3, 7, and 10 (Fig. 2B) and caspase 8 (not shown); in vitro-translated BAD was also cleaved by granzyme-B (Fig. 2B).
FIG. 2.
(A) Cleavage of in vitro-translated 35S-labeled murine HA-BAD by extracts of IL-3-starved (12 h) parental cells, untreated or treated with z-VAD fmk (50 μM) or DEVD fmk (50 nM), and nonstarved or starved (12 h) BCR/ABL-expressing 32Dcl3 cells. (B) Cleavage of in vitro-translated 35S-labeled murine BAD by recombinant granzyme-B, caspase 2, caspase 3, caspase 7, and caspase 10. Lane C contains the uncleaved in vitro translation products. (C and D) caspase 3-dependent cleavage of in vitro-translated 35S-labeled full-length, truncated, and mutant forms of murine HA-BAD. Reactions were carried out for 1 h at 30°C and stopped by adding sample buffer. The down-shift of full-length BAD produced by caspase 3 on WT and mutated BAD is probably due to the cleavage of the HA epitope, which contains a caspase 3 canonical cleavage site (DEVD). (E) Kinetics of WT and DM56/61 BAD-HA expression in virus-infected 32Dcl3 cells 0, 4, 8, and 12 h after IL-3 removal. Anti-BAD (C-20) antibody was used for Western blotting. Arrows indicate full-length BAD and a shorter translation product (∼23 kDa), presumably starting from methionine 43 of the full-length coding sequence; arrows plus asterisks indicate the cleavage products.
Determination of BAD cleavage site(s).
To assess the potential role of the 15-kDa BAD fragment in IL-3-deprivation-induced apoptosis, experiments were carried out to localize the caspase 3 cleavage site(s) of BAD. Although the BAD amino acid sequence does not contain canonical caspase 3 recognition sequences, three sites, 53Glu-Gln-Glu-Asp-Ala57(EQEDA), 58Ser-Ala-Thr-Asp-Arg62 (SATDR), and 68Lys-Thr-Glu-Asp-Gln72(KTEDQ), were considered potential caspase-3 targets on the basis of the molecular weight of truncated BAD. Thus, three distinct BAD fragments, one containing both SATDR and KTEDQ sites (aa 58 to 204, tBAD58), a second with KTEDQ only (aa 68 to 204, tBAD68), and a third lacking all three potential sites (aa 84 to 204, tBAD84), were generated by PCR and subcloned under a T7 promoter to obtain in vitro translation products. 35S-labeled proteins were incubated with recombinant caspase 3, and the cleavage pattern of the truncated proteins was compared to that of full-length BAD. Only tBAD58 was cleaved in vitro (Fig. 2C), suggesting that SATDR is a caspase 3 recognition sequence. To confirm that this was indeed a recognition site for caspase 3, Asp 61 was mutated to Glu (D61E BAD) and the in vitro-translated product was tested for susceptibility to caspase 3 cleavage. Since D61E BAD was cleaved as effectively as WT BAD (Fig. 2D), the upstream potential site EQEDA was also mutated individually (D56E BAD) or in combination with D61E (double mutant DM56/61 BAD). Only DM56/61 BAD was resistant to caspase 3 protease activity (Fig. 2D), suggesting that both sites are targets for this caspase. The in vitro data were confirmed by in vivo experiments using 32Dc13 cells stably expressing DM56/61 BAD. Upon induction of apoptosis by IL-3 deprivation, this mutant form of BAD remained intact (Fig. 2E).
Differential effects of WT, DM56/61, and truncated BAD on 32Dc13 cell viability before and after IL-3 starvation.
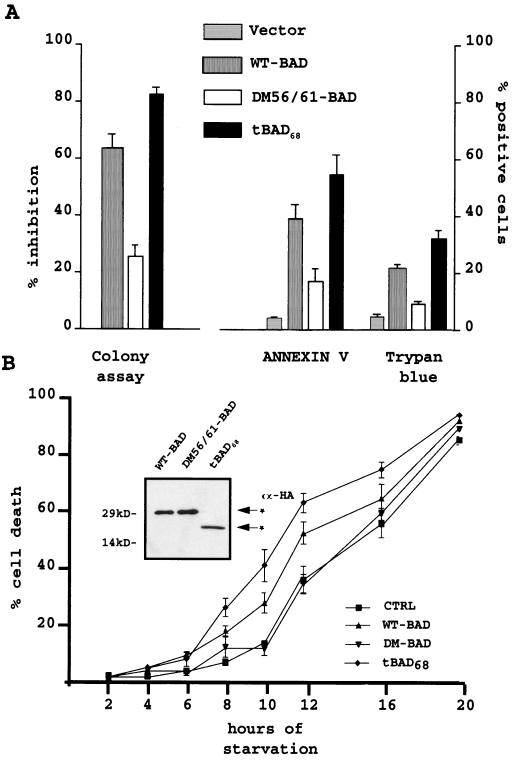
WT BAD induces apoptosis after transient expression in 32Dc13 cells, as measured by the inhibition of clonogenic activity (32). The same approach was used to assess the effect of ectopically expressed DM56/61 and truncated BAD. Parental cells infected with retroviruses encoding the HA-tagged WT BAD, DM56/61 BAD, or tBAD68 and expressing similar levels of HA-tagged proteins (not shown) were plated in semisolid medium in the presence of puromycin used to select for retrovirus-infected cells. In 32Dc13 cells infected with a retrovirus expressing WT BAD, DM56/61 BAD, or tBAD68, clonogenic ability was inhibited to different degrees; compared to what occurred in vector-infected cells, WT BAD induced a 64% decrease in the number of puromycin-resistant colonies, DM56/61 BAD was a less potent inhibitor (33% inhibition), and tBAD68 led to an 82% reduction in colony formation (Fig. 3A). To determine whether the inhibition of colony formation reflected induction of apoptosis, exposure of phosphatidylserine molecules on the outer plasma membrane, an early feature of programmed cell death, and the results of trypan blue exclusion were assessed in cells transduced with WT BAD, DM56/61 BAD, and tBAD68 subcloned in a bicistronic retroviral vector carrying the GFP reporter gene. Twelve hours after infection, GFP-positive cells were sorted by flow cytometry, stained with ANNEXIN V or trypan blue, and counted. In each experiment, tBAD68 was a more potent inducer of cell death (55% ANNEXIN V-positive cells and 33% trypan blue-positive cells) than WT (40 and 25%, respectively) or DM56/61 (18 and 9%, respectively) BAD (Fig. 3A).
FIG. 3.
(A) Colony formation, ANNEXIN V reactivity, and trypan blue staining in 32Dcl3 cells transiently overexpressing WT, double-mutant (DM56/61), or truncated (tBAD68) BAD. For clonogenic assays, parental 32Dcl3 cells were cultured in pLXSP, pLXSP WT BAD-HA pLXSP DM56/61 BAD-HA, or pLXSP tBAD68 BAD-HA retrovirus-containing medium. Twelve hours after infection, cells were plated in methylcellulose in the presence of IL-3 and puromycin (2.5 μg/ml). Colonies were counted 5 days later. The effect of BAD (WT, mutant, and truncated) on the clonogenic ability of infected cells is expressed as the percentage (mean + standard deviation [SD]) of inhibition of the clonogenic ability of vector-infected cells. For ANNEXIN V reactivity or trypan blue uptake, 32Dcl3 cells were cultured with the supernatant from pMIGR1-WT BAD, pMIGR1-DM56/61 HA-BAD, or pMIGR1-tBAD68 HA-BAD retrovirus-infected PHOENIX cells. At 12 h after infection, GFP-positive cells were sorted by flow cytometry. Transiently infected cells were probed with ANNEXIN V 2 h later. Data are expressed as percentages (means + SDs) of ANNEXIN V-positive cells versus total viable cells (7ADD negative). 32Dcl3 GFP-positive cells were also stained with trypan blue and counted. Results are expressed as the percentages (means + SDs) of trypan blue-positive cells among total GFP-positive cells. Five hundred cells were counted in each experiment. Results are representative of four separate experiments. (B) Kinetics of cell death in IL-3-starved parental 32Dcl3 cells (CTRL) and 32Dcl3 cells stably expressing WT, DM56/61, or tBAD68 HA-BAD. Cell death was measured as a percentage (mean + SD) of trypan blue-positive cells among total cells. Five hundred cells were counted in each experiment. Results are representative of three separate experiments. The inset shows a Western blot of WT, BAD, DM56/61 BAD, and tBAD68 expression in the 32Dc13-infected cells used in the cell death assay. Anti-HA antibody (α-HA) was used as the probe.
Stably expressing mixed-cell populations (Fig. 3B, inset) were selected from parental cells infected with HA-tagged WT BAD, DM56/61 BAD, or tBAD68; deprived of IL-3; and counted at various time points to monitor the viability by trypan blue exclusion assay. Cells ectopically expressing tBAD68 were more sensitive to IL-3 starvation (40% trypan blue-positive cells after 10 h of IL-3 deprivation) than cells expressing WT (28% at the same time point) or DM56/61 (10%) BAD (Fig. 3B).
Subcellular localization and phosphorylation of WT BAD DM56/61 BAD, and tBAD68.
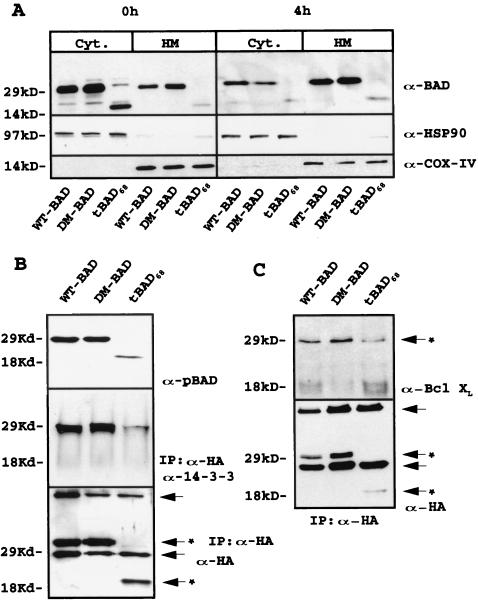
Clones resulting from infection of wild-type 32Dc13 cells with WT BAD, DM56/61 BAD, or tBAD68 retrovirus were analyzed for ectopic BAD expression, subcellular localization, and phosphorylation. After the initial harvest, a few clones expressed high levels of tBAD68; however, these clones rapidly lost expression of the tBAD68 transgene and only a few low-level-expression clones could be maintained in culture. Cells infected with WT or DM56/61 BAD maintained higher levels of transgene expression. To investigate subcellular distribution and serine phosphorylation of wild-type and mutant BAD, extracts of retrovirus-infected 32Dc13 cells were fractionated and blotted with anti-BAD antibody or the anti-phosphorylated BAD antibody (the anti-pSer112 and pSer136 antibody mix). Like WT BAD, DM56/61 BAD and tBAD68 were mostly cytoplasmic in the presence of IL-3 (Fig. 4A, upper left gel); after IL-3 deprivation, the localization of the three forms was mainly mitochondrial (Fig. 4A, upper right gel). While phosphorylation of WT an DM56/61 BAD was readily detectable, tBAD68 was barely detectable in its phosphorylated form when cell extracts from retrovirus-infected cells were blotted with the anti-pSer112 and anti-pSer136 BAD antibody mix (Fig. 4B, upper gel).
FIG. 4.
(A) BAD (WT, mutant, and truncated) expression in subcellular fractions of retrovirus-infected parental 32Dc13 cells. Western blots show BAD levels in mitochondrion (HM)- and cytoplasm (Cyt)-enriched fractions from parental cells in the presence of IL-3 or 4 h after its removal (upper gels). Anti-BAD (C-20) antibody (α-BAD) was used as the probe. Levels of the cytoplasmic marker HSP90 (middle gels) and mitochondrial marker subunit IV of cytochrome oxidase (COX-IV, lower gels) were measured as a control for equal loadings and the purity of subcellular fractions. Results are representative of three separate experiments. (B) Serine phosphorylation and 14-3-3 interaction of WT, DM56/61, and tBAD68 HA-BAD in retrovirus-infected parental cells. Western blot analyses were carried out with a mix of anti-pSer112 and -pSer136 antibodies to monitor phosphorylated BAD (upper gel). To study the interaction between 14-3-3 protein and WT, DM56/61, and tBAD68 HA-BAD, cell extracts were immunoprecipitated with the anti-HA antibody and blotted with anti-14-3-3 antibody (middle gel). Levels of WT, DM56/61, and tBAD68 HA-BAD were measured as a control for equal loadings (lower gel). Results are representative of three separate experiments. Arrows indicate heavy- and light-chain immunoglobulins; arrows with asterisks indicate antibody-specific reactive bands. IP, immunoprecipitate. (C) Interaction of HA-BAD (WT, mutant, and truncated) with BCL-XL in retrovirus-infected 32Dc13 cells. Anti-HA immunoprecipitates from WT, DM56/61, and tBAD68 HA-BAD retrovirus-infected 32Dc13 cells were blotted with anti-BCL-XL antibody (upper gel). Levels of HA-tagged protein were measured as a control for immunoprecipitation (lower gel). Results are representative of three separate experiments. Arrows indicate heavy- and light-chain immunoglobulins; arrows with asterisks indicate antibody-specific reactive bands.
Phosphorylation of BAD correlates with its ability to interact with 14-3-3β, an association necessary for sequestration of BAD in the cytoplasm (10). Coimmunoprecipitation experiments to determine the extent of this association in BAD-expressing 32Dc13 cells cultured in the presence of IL-3 revealed a strong interaction of WT and DM56/61 BAD with 14-3-3β, while the interaction with tBAD68 (Fig. 4B, middle gel) was very faint even though the amounts of immunoprecipitated HA-BAD were comparable (Fig. 4B, lower gel).
DM56/61 BAD and tBAD68 interact with Bcl-XL in 32Dc13 cells.
Because the interaction of BAD with Bcl-2 (48) and Bcl-XL (52) is crucial for BAD proapoptotic activity (21, 28), we tested whether the diminished sensitivity to IL-3 deprivation of 32Dc13 cells expressing the mutant form of BAD resistant to caspase cleavage compared to the sensitivity of cells expressing the truncated form (Fig. 3) might rest in an altered ability of ectopically expressed BAD mutants to interact with Bcl-XL. Thus, whole extracts from IL-3-starved 32Dc13 cells retrovirally infected with DM56/61 BAD-HA, tBAD68-HA or WT BAD-HA (positive control) were immunoprecipitated using anti-HA antibody followed by blotting with anti-Bcl-XL antibody. Levels of immunoprecipitated BCL-XL were similar in cells infected with a retrovirus carrying WT BAD, DM56/61 BAD, or truncated tBAD68, but expression of the last was less abundant (Fig. 4C), suggesting that truncated BAD interacts with Bcl-XL at least as effectively as WT or DM56/61 BAD.
WT BAD, DM56/61 BAD and tBAD68 differentially induce cytochrome c release from isolated mitochondria.
Mitochondria play a crucial role in apoptosis by releasing apoptogenic factors such as cytochrome c (15, 1, 23) and apoptosis-inducing factor (40) from the intermembrane space into the cytoplasm. Proapoptotic BAX (20), BID (38), and BIK (38) induce the release of cytochrome c from mitochondria both in vivo and in vitro. To determine whether BAD induces cytochrome c release and whether its truncated form, like truncated BID, is more potent in promoting this effect, partially purified mitochondria from 32Dc13 cells were incubated with WT BAD, DM56/61 BAD, or tBAD68 protein in a cell-free system and examined for cytochrome c release. WT, mutated, and truncated forms of BAD all induced the release of cytochrome c from isolated mitochondria (Fig. 5A, upper gel); however, tBAD68 was more effective than DM56/61 or WT BAD. Interestingly, cytochrome c release induced by WT BAD was enhanced upon incubation with recombinant caspase 3, which generated the truncated 15-kDa product (not shown). Consistent with the hypothesis that cleavage of BAD enhances its proapoptotic function, the cytochrome c-inducing activity of cleavage-resistant DM56/61 BAD was not affected by incubation with active caspase-3 (Fig. 5A). To normalize the cytochrome c release activity of BAD to the amount of mitochondria used in the assay, pellet extracts were blotted with anti-COX IV antibody used as a mitochondrial marker of equal loadings (Fig. 5A, middle gel) and the release of cytochrome c was plotted in arbitrary units. As shown in Fig. 5B, this analysis indicated that truncated BAD was two to threefold more effective than wild-type BAD in inducing cytochrome c release. Western blotting of pellet-extracted proteins with an anti-cytochrome c antibody confirmed the release of cytochrome c from the mitochondrion samples incubated with tBAD or with WT BAD and caspase 3 (Fig. 5A, lower gel).
FIG. 5.
(A) Mitochondrial cytochrome c release induced by recombinant WT, DM56/61, and tBAD68 BAD proteins. Recombinant WT, double-mutant, and truncated BAD proteins fused to the TAT leader sequence were generated and purified as described in Materials and Methods. Partially purified mitochondria (15 μl) from parental 32Dc13 cells were incubated with a vehicle (lane 1; 250 mM sucrose buffer) or 100 nM WT BAD (lanes 2 and 3), double-mutant BAD (lanes 4 and 5), or truncated BAD (lane 6) in the absence (lanes 2, 4, and 6) or presence (lanes 1, 3, and 5) of 1 μl of recombinant caspase 3 (10 ng) in 30 μl. After 1 h at 30°C, samples were centrifuged at 12,000 × g for 10 min at 4°C. Supernatants (Sup) were subjected to an SDS–4 to 15% gradient polyacrylamide gel and transferred to a nitrocellulose filter. The filter was probed with a monoclonal anti-cytochrome c antibody (α-Cytochrome C) (upper gel). Mitochondrial pellets were lysed, and protein extracts were blotted with an antibody to subunit IV of cytochrome oxidase COX-IV) as a control for equal loadings (middle gel). The same filter was blotted with anti-cytochrome c antibody to monitor the presence of residual protein in the mitochondrial fraction (lower gel). (B) Densitometric analysis of cytochrome c release. The intensities of the cytochrome c band in the supernatants were measured and normalized against that of COX-IV detected in the pellet extracts, used as control (CTRL) of mitochondrial loading. Data are the ratios of the value of each cytochrome c band to that of its COX-IV counterpart. Results are representative of two separate experiments.
BAD cleavage in human cell lines after induction of apoptosis.
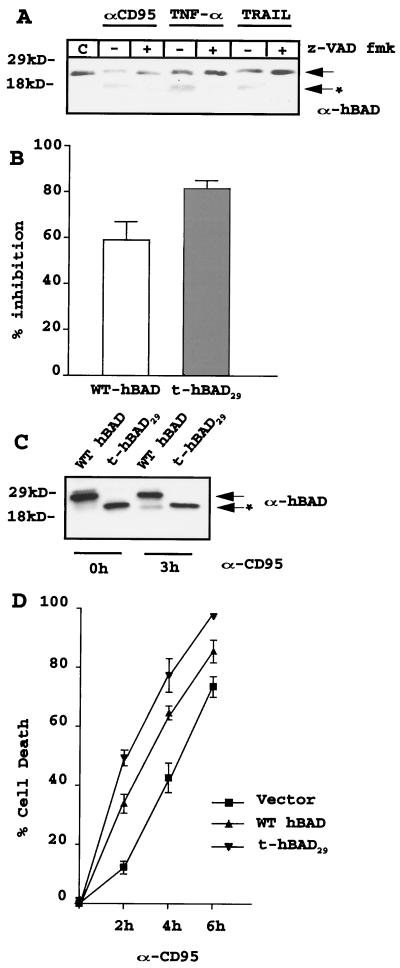
Comparison of the amino acid sequences of mouse and human BAD revealed that the latter lacks the initial 43 residues and the SATDR caspase 3 consensus sequence (which includes aspartate 61 of the mouse sequence) but that the EQEDA cleavage site (which includes aspartate 56 of the mouse sequence) is conserved, except for the replacement of alanine with serine. To determine whether BAD cleavage occurs only in mouse cells and only in the course of growth factor deprivation-induced apoptosis, we monitored the possible presence of BAD-cleaved forms in human Jurkat T cells induced to apoptosis by the death ligand anti-CD95 antibody, TNF-α, or TRAIL. Once incubated with Jurkat T cells, the three agents led to cleavage of the endogenous BAD detected as an ∼16-kDa band after blotting of apoptotic cell extracts with a polyclonal anti-BAD antibody. This cleavage was caspase dependent since it was abolished by cotreatment of Jurkat T cells with the caspase inhibitor z-VAD fmk (Fig. 6A). Identical results were obtained with the human T-lymphoma line HUT-78 (data not shown).
FIG. 6.
(A) Caspase-dependent cleavage of endogenous BAD in human Jurkat T cells after treatment with anti-Fas antibody (αCD95; 0.05 ng/ml), TNF-α (10 ng/ml), or TRAIL (1 μg/ml). Cells (2 × 105/ml) were exposed to anti-CD95 antibody, TNF-α, or TRAIL for 6 h in the absence or presence of the caspase inhibitor z-VAD fmk (50 μM) and harvested. Cell extracts were subjected to an SDS–4 to 15% gradient polyacrylamide gel, transferred to a nitrocellulose membrane, and blotted with anti-BAD polyclonal rabbit antibody. Untreated cell extracts were used as a negative control for caspase activity. Results are representative of two separate experiments. The arrow indicates full-length hBAD; the arrow plus the asterisk indicates cleavage products. (B) Inhibition of colony formation by transient overexpression of WT or truncated hBAD. Parental Jurkat T cells were incubated with infectious supernatant from pLXSP, pLXSP WT hBAD, or pLXSP t-hBAD29 retrovirus-infected cells. After three cycles of infection (see Materials and Methods), cells were plated in methylcellulose in the presence of puromycin (2.5 μg/ml). Colonies were counted 7 days later. The effect of BAD on the clonogenic capacity of infected cells is expressed as a percentage (mean + SD) of inhibition of the clonogenic ability of vector-infected cells. Results are representative of three separate experiments. (C) Expression of exogenous WT and t-hBAD29 in infected-Jurkat T cells. A Western blot shows expression of WT and t-hBAD29 in a representative clone of Jurkat T cells, untreated or treated (3 h) with 0.05 ng of anti-CD95 antibody per ml. Protein extracts were blotted with a polyclonal anti-hBAD serum (H-168). Results are representative of two separate experiments. The arrow indicates full-length hBAD; the arrow with the asterisk indicates the cleavage product. (D) Kinetics of apoptosis in Jurkat T cells (empty vector or BAD transduced) treated with the anti-CD95 antibody for 0, 2, 4, and 6 h. Cell death was measured as a percentage (mean + SD) of trypan blue-positive cells among total cells. Results are representative of three separate experiments.
After cloning of the full-length hBAD cDNA, a truncated form of BAD lacking the EQEDS recognition sequence (aa 29 to 168, t-hBAD29) was generated by PCR and inserted in a retroviral expression vector. Jurkat T cells infected with the retrovirus carrying the HA-tagged full-length hBAD or h-BAD29 were plated in semisolid medium in the presence of puromycin to select retrovirus-infected cells. Compared to what occured with empty vector-infected cells, expression of either full-length or truncated BAD decreased the clonogenic efficiency of Jurkat T cells, although the truncated form of BAD was more toxic (∼84% reduction in colony number) than its full-length counterpart (∼58% reduction) (Fig. 6B).
To confirm that h-tBAD29 cannot be further cleaved during apoptosis, Jurkat cells expressing full-length BAD or hBAD29 were treated with the anti-CD95 antibody and monitored by Western blotting for fast-migrating BAD or HA cross-reactive bands. While full-length WT hBAD was cleaved into a smaller product, there was no further cleavage of the t-hBAD29 form (Fig. 6C), indicating that the only caspase cleavage site of hBAD was at aspartate 28. Full-length hBAD in which aspartic acid at codon 28 is replaced by glutamic acid was resistant to caspase cleavage in vitro and in retrovirus-transduced cells (not shown). Consistent with the effects of truncated BAD in murine 32Dc13 cells (Fig. 3), expression of t-hBAD29 accelerated Fas-dependent apoptosis of Jurkat T cells (Fig. 6D).
BAD cleavage in 32Dc13 and Jurkat cells ectopically expressing Bcl-2.
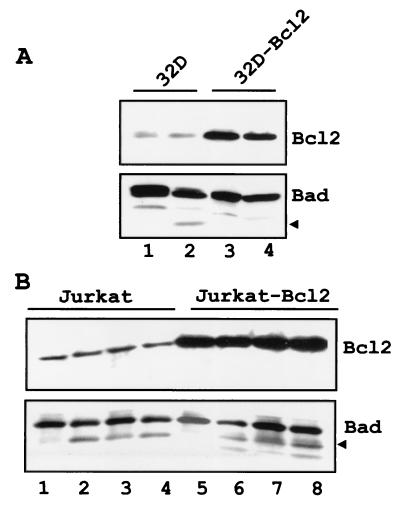
The enhanced cytochrome c release by truncated BAD suggests the existence of an autoregulatory loop by which caspase-3 cleavage of BAD may promote a second wave of caspase activation. To further assess whether BAD cleavage is downstream of caspase activation, the generation of truncated BAD was monitored in Bcl-2-expressing 32Dc13 cells and Jurkat cells after exposure to apoptotic stimuli. Upon IL-3 deprivation, truncated BAD was detected in parental but not in Bcl-2-expressing 32Dc13 cells (Fig. 7A). By contrast, ectopically expressed Bcl-2 did not prevent the generation of truncated BAD in Jurkat cells treated with anti-CD95 antibody, TNF-α, or TRAIL (Fig. 7B). Of note, BAD was not cleaved in serum-deprived Bcl-2-expressing Jurkat cells (not shown). Thus, the pattern of BAD cleavage reflects the fact that caspase activation in growth factor 32Dc13 cells is downstream of the point of action of Bcl-2 but that it is direct in Jurkat cells treated with anti-CD95 antibody, TNF-α, or TRAIL.
FIG. 7.
(A) Ectopic Bcl-2 expression blocks BAD cleavage in IL-3-starved 32Dc13 cells. Parental (lanes 1 and 2) and Bcl-2-expressing (lanes 3 and 4) 32Dc13 cells were infected with pLXSP-HA WT BAD; after selection in puromycin (2.5 μg/ml), transduced cells (2 × 105/ml) expressing high levels of murine BAD were deprived of IL-3 for 8 h (lanes 2 and 4) and harvested. Cell extracts were subjected to an SDS–4 to 15% gradient polyacrylamide gel, transferred to a nitrocellulose membrane, and blotted with the anti-C-terminus (C-20) BAD antibody. (B) Ectopic Bcl-2 expression does not block BAD cleavage in death ligand-treated Jurkat T cells. Parental (lanes 1 to 4) and Bcl-2-expressing (lanes 5 to 8) Jurkat cells were infected with pLXSP WT hBAD; after selection in puromycin (2.5 μg/ml), transduced cells (2 × 105/ml) expressing high levels of hBAD were treated for 6 h with anti-FAS antibody (lanes 2 and 6), TNF-α (lanes 3 and 7), and TRAIL (lanes 4 and 8) and harvested. Cell extracts were subjected to an SDS–4 to 15% gradient polyacrylamide gel, transferred to a nitrocellulose membrane, and blotted with polyclonal anti-BAD serum (H-168). Arrows indicate cleaved BAD.
DISCUSSION
Upon IL-3 deprivation of 32Dc13 myeloid precursor cells, proapoptotic BAD is cleaved in the N terminus to generate two smaller products: one very similar in size to the full-length protein (26 kDa) and the second of ∼15 kDa. The nature of the high-molecular-weight form of cleaved BAD is unclear; by contrast, the block in generation of the 15-kDa truncated form by treatment of 32Dc13 cells with a pancaspase inhibitor suggests that it is the product of a caspase-like activity. Cleavage of in vitro-translated BAD by apoptotic cell extracts was also abolished by a very low concentration of a caspase 3 inhibitor, consistent with the potential role of this caspase as a BAD protease. Indeed, recombinant caspase 3 was able to cleave in vitro-translated BAD, although similar cleavage was also achieved by caspase 2, 7, and 10 and granzyme-B. In vivo and in vitro experiments identified aspartate 56 and 61 as the caspase cleavage sites since their replacement with glutamate completely abolished BAD cleavage. The 15-kDa cleavage product of BAD was detected only in the mitochondrial fraction, possibly due to highly efficient translocation to the mitochondria after cleavage in the cytoplasm or to cleavage of BAD at the mitochondrial outer membrane. This latter mechanism is consistent with reports indicating the localization and activation of caspase(s) in the mitochondria (26, 4, 22, 41).
In 32Dc13 cells, truncated BAD was more potent than the full-length protein in inhibiting colony formation by retrovirus-transduced cells whereas the mutant form was a weak death inducer. The reduced death-inducing effect of an ectopically expressed caspase-resistant mutant BAD is probably caused by its apparent ability to interfere with the cleavage of endogenous BAD (not shown). The cytotoxic effect of truncated BAD was due to induction of apoptosis, as indicated by exposure of phosphatidylserine on the outer plasma membrane and trypan blue positivity, early and late markers, respectively, of apoptosis. Moreover, cleavage of BAD and enhanced apoptotic activity of its truncated form were not limited to growth factor deprivation-induced apoptosis in mouse cells; apoptosis induced by death ligands in human Jurkat T cells was also accompanied by generation of the cleaved product of BAD and occurred more rapidly in cells overexpressing truncated BAD than in cells overexpressing wild-type BAD. Cleavage of BAD was suppressed by Bcl-2 overexpression in growth factor-deprived 32Dc13 cells but not in death ligand-treated Jurkat T cells.
The activity of BAD is regulated by changes in phosphorylation and subcellular localization. Upon calcium influx increase or growth factor deprivation, phosphorylated BAD is rapidly dephosphorylated by the specific serine-phosphatase calcineurin (45) or PP1α (32, 2) and translocates to the mitochondrion outer membrane, where, through its BH3 domain, it interacts with anti-apoptotic Bcl-2 and Bcl-XL antibodies (53, 21, 28). The subcellular distribution of overexpressed mutated and truncated BAD was similar to that of full-length BAD both in exponentially growing cells and after induction of apoptosis by IL-3 deprivation. However, truncated BAD was barely phosphorylated at serine residues 112 and 136 and interacted inefficiently with 14-3-3β. The low-level phosphorylation of truncated BAD might rest in an enhanced susceptibility to protein phosphatases. Alternatively, truncated BAD may serve as a poor substrate for some BAD kinases; this may interfere with some of the steps regulating the shuttling of BAD between different subcellular compartments. For example, mitochondrial BAD can be released into the cytoplasm once phosphorylated, directly or indirectly, by mitochondrion-localized protein kinases such as protein kinase and Raf-1 (44, 18, 31). If truncated BAD is no longer a target for these protein kinases, the “escape” mechanism of BAD inactivation at the mitochondria might be compromised. Mitochondrial BAD can be phosphorylated at serine 155 (42, 54, 10), which is localized inside the BH3 domain and, upon phosphorylation, renders BAD incapable of interacting with Bcl-XL or Bcl-2. Since phosphorylation of serine 136 seems to be required for serine 155 phosphorylation (10), it is possible that truncated BAD is also poorly phosphorylated at serine 155. This poor phosphorylation would enhance the interaction of truncated BAD with Bcl-XL, consistent with results of the coimmunoprecipitation experiments, which suggest that truncated BAD has a similar or even higher affinity for Bcl-XL than WT BAD. Enhanced interaction with Bcl-XL is important for the apoptosis-inducing effects of the buried BH3 domain members of the Bcl-2 family, whose three-dimensional structure indicates that, in resting conditions, their BH3 pocket is masked (16). The BH3 domain becomes available for interaction with Bcl-2 and Bcl-XL, after caspase 8 cleavage of BID or homodimerization of BAX (16). Based on a threading analysis and a comparison of secondary structure, BAD belongs to the subgroup of exposed BH3 domain proteins; however, there are no crystallographic data to exclude the possibility of an enhanced interaction of truncated BAD with Bcl-XL.
Common to all the proapoptotic members of the Bcl-2 family is the pertubation of mitochondrial membrane permeability, often accompanied by cytochrome c release in the cytosol with activation of downstream caspases (16, 15, 51, 12, 13). In the case of BID, its caspase 8 cleavage product is a more potent inducer of cytochrome c release than is the uncleaved molecule (25, 24). Similarly, truncated BAD is a more potent inducer of cytochrome c release than full-length WT or mutated BAD. Interestingly, the differences in activity between truncated and full-length WT BAD were abolished by treating the latter with active caspase-3, which cleaves in the N terminus and generates truncated BAD. The enhanced cytochrome c release by truncated BAD might rest in its persistence in the mitochondria, where it may antagonize antiapoptotic Bcl-XL or enhance the cytochrome c-releasing activity of proapoptotic molecules.
Compared to its murine counterpart, human BAD lacks the first 43 amino acids, raising the possibility that caspase-dependent cleavage is not necessary in further enhancing its apoptosis-inducing effects. However, BAD cleavage was evidenced in human Jurkat T cells induced to undergo apoptosis by treatment with the death ligands anti-Fas antibody, TNF-α, and TRAIL. Caspase cleavage occurred after residue aspartate 28, and, as in mouse cells, the truncated form of hBAD lacking the first 28 amino acids was more active than full-length BAD as a proapoptotic molecule. Overexpressed truncated BAD also accelerated FAS-dependent apoptosis of Jurkat T cells. Since caspase 3 can be activated directly by caspase 8 in Fas-mediated apoptosis (33, 34), the cleavage of BAD by caspase 3 might serve as a mechanism to amplify the mitochondrial phase of the apoptotic response initiated by truncated-BID-induced cytochrome c release (25, 24). Since murine BAD is also cleaved by caspase 8, it remains possible that BAD cleavage in Jurkat cells is consequent to death ligand activation of caspase 8. A feedback amplification loop linking caspase 3 activation to mitochondrial dysfunction might also occur in genotoxic-stress-induced apoptosis (5). In that model of apoptosis, ionizing radiation and etoposide induced the release of cytochrome c. The early release of low levels of cytochrome c into the cytosol preceded the activation of caspase 9 and 3 with no effects on ATP levels or mitochondrial transmembrane potential, whereas late-stage cytochrome c release was accompanied by reductions in ATP levels and transmembrane potential (5). In that system, activation of caspase 3 might lead to BAD cleavage, contributing to the late-stage massive release of cytochrome c. A peculiar model of apoptosis is that of natural killer (NK) cell-dependent killing of target cells, wherein NK cells are the effectors of nonspecific immunity and eliminate nonself cells by release of porine and granzyme-B. Porines are involved in the entry of granzyme-B into target cells, where the proteolytic activity of granzyme-B activates the caspase cascade, leading to apoptosis. In such a model of apoptosis, BID is activated and cleaved directly by granzyme-B, even in cells lacking caspase 3 or 8 (3). Since granzyme-B cleaves BAD in vitro in a manner similar to that of caspase 3, it is possible that this cleavage also occurs in vivo in the NK cell model of apoptosis. In conclusion, caspase 3-dependent cleavage of BAD might serve as a general mechanism to enhance BAD proapoptotic activity, perhaps through the ability of truncated BAD to escape mitochondrial phosphorylation and its release into the cytoplasm. This, in turn, would lead, directly or indirectly, to an increase in cytochrome c release and the maintenance of caspase activation.
ACKNOWLEDGMENTS
We thank W. S. Pear for the kind gift of the pMIGR1 vector.
F.C. was supported, in part, by a doctoral fellowship of the University of Catania Medical School. This work was supported, in part, by NIH grants to B.C. and E.S.A. (PO1 CA78890).
REFERENCES
- 1.Adams J M, Cory S. The Bc1–2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 2.Ayllon V, Martinez-A C, Garcia A, Cayla X, Rebollo A. Protein phosphatase 1alpha is a Ras-activated BAD phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J. 2000;19:2237–2246. doi: 10.1093/emboj/19.10.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry M, Heibein J A, Pinkoski M J, Lee S-F, Moyer R W, Green D R, Bleackley R C. Granzyme B short-circuits the need for caspase-8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly clearing Bid. Mol Cell Biol. 2000;20:3781–3794. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandler J M, Cohen G M, MacFarlane M. Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. J Biol Chem. 1998;273:10815–10818. doi: 10.1074/jbc.273.18.10815. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q, Gong B, Almasan A. Distinct stages of cytochrome c release from mitochondria: evidence for a feedback amplification loop linking caspase activation to mitochondrial dysfunction in genotoxic stress-induced apoptosis. Cell Death Differ. 2000;7:227–233. doi: 10.1038/sj.cdd.4400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng E H, Kirsch D G, Clem R J, Rovi R, Kastan M B, Bedi A, Ueno K, Hardwick J M. Conversion of Bc1–2 to a Bax-like death effector by caspases. Science. 1997;278:1966–1978. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 7.Cirinnà M, Trotta R, Salomoni P, Kossev P, Wasik M, Perrotti D, Calabretta B. Bcl-2 expression restores the leukemogenic potential of a BCR/ABL mutant defective in transformation. Blood. 2000;96:3915–3921. [PubMed] [Google Scholar]
- 8.Clem R J, Cheng E H, Karp C L, Kirsch D G, Ueno K, Takahashi A, Kastan M B, Griffin D E, Earnshaw W C, Velivona M A, Hardwick J M. Modulation of cell death by BCL-XL through caspase interaction. Proc Natl Acad Sci USA. 1998;95:554–559. doi: 10.1073/pnas.95.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 10.Datta S R, Katsov A, Hu L, Petros A, Fesik S W, Yaffe M B, Greenberg M E. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 11.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 12.Eskes R, Antonsson B, Osen-Sand A, Montessiut S, Richter C, Sadoul R, Mazzei G, Nichols A, Martinou J C. Bax-induced cytochrome c release from mitochondria is independent of the permeability transition pore but highly dependent on Mg2+ ions. J Cell Biol. 1998;143:217–224. doi: 10.1083/jcb.143.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finucane D M, Bossy-Wetzel E, Waterhouse N J, Cotter T G, Green D R. Bax-induced caspase activation and apoptosis via cytochrome release from mitochondria is inhibitable by Bcl-XL. J Biol Chem. 1999;274:2225–2233. doi: 10.1074/jbc.274.4.2225. [DOI] [PubMed] [Google Scholar]
- 14.Franke T F, Cantley L C. Apoptosis: a bad kinase makes good. Nature. 1997;390:116–117. doi: 10.1038/36442. [DOI] [PubMed] [Google Scholar]
- 15.Green D R, Reed J C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 16.Gross A, McDonnell J M, Korsmeyer S J. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 17.Haldar S, Basu A, Croce C M. Serine-70 is one of the critical sites for drug-induced Bcl-2 phosphorylation in cancer cells. Cancer Res. 1998;58:1609–1615. [PubMed] [Google Scholar]
- 18.Harada H, Becknell B, Wilm M, Mann M, Huang L J, Taylor S S, Scott J D, Korsmeyer S J. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 19.Hsu S Y, Karpia A, Zhu L, Hsueh A J. Interference of BAD (Bcl-XL/Bcl-2-associated death promoter)-induced apoptosis in mammalian cells by 14-3-3 isoforms and P11. Mol Endocrinol. 1997;12:1858–1867. doi: 10.1210/mend.11.12.0023. [DOI] [PubMed] [Google Scholar]
- 20.Jürgensmeier J M, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed J C. Bax directly induces release of cytochrome c from isolated mitochondria. Proc Natl Acad Sci USA. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelekar A, Chang B S, Harlan J E, Fesik S W, Thompson C B. Bad is a BH3 domain-containing protein that forms an inactive dimer with Bcl-XL. Mol Cell Biol. 1997;17:7040–7046. doi: 10.1128/mcb.17.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krebs J F, Armstrong R C, Srinivasan A, Aja T, Wong A M, Aboy A, Sayers R, Pham B, Vu T, Hoang K, Karanewsky D S, Leist C, Schmitz A, Wu J C, Tomaselli K J, Fritz L C. Activation of membrane-associated procaspase-3 is regulated by Bcl-2. J Cell Biol. 1999;144:915–926. doi: 10.1083/jcb.144.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroemer G, Dallaporta B, Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol. 1998;60:619–642. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Zhu H, Xu C J, Yuan J. Cleavage of BID by caspase-8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 25.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. BID, a Bcl-2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 26.Mancini M, Nicholson D W, Roy S, Thornberry N A, Peterson E P, Casciola-Rosen L A, Rosen A. The caspase-3 precursor has a cytosolic and mitochondrial distribution: implications for apoptotic signaling. J Cell Biol. 1998;140:1485–1495. doi: 10.1083/jcb.140.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oltvai Z N, Korsmeyer S. Checkpoints of dueling dimers foil death wishes. Cell. 1994;79:189–192. doi: 10.1016/0092-8674(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 28.Ottilie S, Diaz J L, Horne W, Chang J, Wang Y, Wilson G, Chang S, Fritz L C, Oltersdorf T. Dimerization properties of human BAD. Identification of a B domain and analysis of its binding to mutant BCL-2 and BCL proteins. J Biol Chem. 1997;272:30866–30872. doi: 10.1074/jbc.272.49.30866. [DOI] [PubMed] [Google Scholar]
- 29.Pastorino J G, Tafani M, Farber J L. Tumor necrosis factor induces phosphorylation and translocation of BAD through a phosphatidylinositide-3-OH kinase-dependent pathway. J Biol Chem. 1998;274:19411–19416. doi: 10.1074/jbc.274.27.19411. [DOI] [PubMed] [Google Scholar]
- 30.Pear W S, Miller J P, Xu L, Pui J C, Soffer B, Quackenbush R C, Pender A M, Bronson R, Aster J C, Scott M L, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 31.Salomoni P, Wasik M A, Riedel R F, Reiss K, Choi J K, Skorski T, Calabretta B. Expression of constitutively active Raf-1 in the mitochondria restores anti-apoptotic and leukemogenic potential of a transformation-deficient BCR/ABL mutant. J Exp Med. 1998;187:1995–2007. doi: 10.1084/jem.187.12.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomoni P, Condorelli F, Sweeney S M, Calabretta B. Versatility of BCR/ABL-expressing leukemic cells in circumventing pro-apoptotic BAD effects. Blood. 2000;96:676–684. [PubMed] [Google Scholar]
- 33.Salvesen G S, Dixit V M. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 34.Scaffidi C, Fulda S, Srinivasan A, Li C, Friesen F, Tomaselli K J, Debatin K M, Krammer P H, Peter M E. Two CD45 (APO-1/FAS) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scatena C D, Stewart Z A, Mays D, Tang L J, Keefer C J, Leach S D, Pieteupal J A. Mitotic phosphorylation of Bcl-2 during normal cell cycle progression and taxol-induced growth arrest. J Biol Chem. 1998;273:30777–30784. doi: 10.1074/jbc.273.46.30777. [DOI] [PubMed] [Google Scholar]
- 36.Scheid M P, Duronio V. Dissociation of cytokine-induced phosphorylation of BAD and activation of PKB/Akt: involvement of BAD upstream of BAD phosphorylation. Proc Natl Acad Sci USA. 1998;95:7439–7444. doi: 10.1073/pnas.95.13.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedlak T W, Oltvai Z N, Yang E, Wang K, Boise L H, Thompson C B, Korsmeyer S J. Multiple Bcl-2 family members demonstrate selective dimerizations with Bax. Proc Natl Acad Sci USA. 1995;92:7834–7838. doi: 10.1073/pnas.92.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu S, Tsujimoto Y. Pro-apoptotic BH3-only Bcl-2 family members induce cytochrome c release, but not mitochondrial membrane potential loss, and directly modulate voltage-dependent anion channel activity. Proc Natl Acad Sci USA. 2000;97:577–582. doi: 10.1073/pnas.97.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi J K, Trotta R, Wlodarski P, Perrotti D, Chan T O, Wasik M A, Tsichlis P N, Calabretta B. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Susin S A, Lorenzo H K, Zamzani N, Marzo I, Snow B E, Brothers G M, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Prevost M C. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 41.Susin S A, Lorenzo H K, Zamzani N, Marzo I, Brenner C, Larochette-Prevost L, Alzari P M, Kroemer G. Mitochondrial release of caspase-2 and -9 during the apoptotic process. J Exp Med. 1999;189:381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan Y, Demeter M R, Ruan H, Comb M J. BAD Ser155 phosphorylation regulates BAD: Bcl-XL interaction and cell survival. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 43.Vocero-Akbani A M, Heyden N V, Lissy N A, Ratner L, Dowdy S F. Killing HIV-infected cells by transduction with an HIV protease-activated caspase-3 protein. Nat Med. 1999;5:29–33. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 44.Wang H-G, Rapp U R, Reed J C. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87:629. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 45.Wang H-G, Pathan N, Ethell I M, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke T F, Reed J C. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 46.Wang K, Yian X-M, Chao D T, Milliman C L, Korsmeyer S J. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 47.Wood D E, Newcomb E W. Cleavage of Bax enhances its cell death function. Exp Cell Res. 2000;256:375–382. doi: 10.1006/excr.2000.4859. [DOI] [PubMed] [Google Scholar]
- 48.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. BAD, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Car J, Peng T-I, Jones D P, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 50.Ymer S, Tucker W Q, Sanderson C J, Hapel A J, Campbell H D, Young I G. Constitutive synthesis of interleukin-3 by leukemia cell line WEHI-3B is due to retroviral insertion near the gene. Nature. 1985;317:255–258. doi: 10.1038/317255a0. [DOI] [PubMed] [Google Scholar]
- 51.Zamzami N, Brenner C, Marzo I, Susin S A, Kroemer G. Subcellular and submitochondrial mode of action of Bcl-2-like oncoproteins. Oncogene. 1998;16:2265–2282. doi: 10.1038/sj.onc.1201989. [DOI] [PubMed] [Google Scholar]
- 52.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 53.Zha J, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer S J. BH3 domain of BAD is required for heterodimerization with BCL-XL and pro-apoptotic activity. J Biol Chem. 1997;272:24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 54.Zhou X-M, Lui Y, Payne G, Lutz R J, Chittenden T. Growth factors inactivate the cell death, BAD, by phosphorylation of its BH3 domain on serine 155. J Biol Chem. 2000;275:25046–25051. doi: 10.1074/jbc.M002526200. [DOI] [PubMed] [Google Scholar]