Abstract
Skp1p–cullin–F-box protein (SCF) complexes are ubiquitin-ligases composed of a core complex including Skp1p, Cdc53p, Hrt1p, the E2 enzyme Cdc34p, and one of multiple F-box proteins which are thought to provide substrate specificity to the complex. Here we show that the F-box protein Rcy1p is required for recycling of the v-SNARE Snc1p in Saccharomyces cerevisiae. Rcy1p localized to areas of polarized growth, and this polarized localization required its CAAX box and an intact actin cytoskeleton. Rcy1p interacted with Skp1p in vivo in an F-box-dependent manner, and both deletion of its F box and loss of Skp1p function impaired recycling. In contrast, cells deficient in Cdc53p, Hrt1p, or Cdc34p did not exhibit recycling defects. Unlike the case for F-box proteins that are known to participate in SCF complexes, degradation of Rcy1p required neither its F box nor functional 26S proteasomes or other SCF core subunits. Importantly, Skp1p was the only major partner that copurified with Rcy1p. Our results thus suggest that a complex composed of Rcy1p and Skp1p but not other SCF components may play a direct role in recycling of internalized proteins.
Endocytosis is required for a wide range of cellular functions, including transmission of neuronal, metabolic, and proliferative signals, uptake of nutrients, and regulation of cellular homeostasis (31). Internalized proteins travel through two morphologically and biochemically distinct organelles, called early and late endosomes, before reaching the lysosomal-vacuolar compartment, where they are degraded (15). Lysosomal degradation is not the obligatory fate of internalized proteins; many receptors recycle from the early endosome back to the plasma membrane, allowing multiple rounds of ligand binding and internalization. In some specialized cell types the recycling pathway is also used for antigen presentation and transcytosis, as well as recycling of synaptic vesicle components. However, the machinery and molecular mechanisms controlling recycling of plasma membrane proteins are poorly understood.
In the yeast Saccharomyces cerevisiae, the chitin synthase Chs3p and the exocytic v-SNARE Snc1p have been suggested to recycle (23, 27, 51). Chs3p translocates between sites of chitin deposition on the cell surface and an internal structure called the chitosome, which may correspond to an early endosomal compartment (51). Snc1p is involved in fusion of Golgi-derived secretory vesicles with the plasma membrane. Removal of Snc1p from the plasma membrane by endocytosis followed by its targeting back to the Golgi allows reutilization of Snc1p for several rounds of internalization (27). Finally, the Ste3p receptor was recently shown to recycle back to the plasma membrane in response to pheromone (8). Importantly, a quantitative recycling assay based on release of the internalized dye FM4-64 into the extracellular medium was developed (46). Cells defective in the SNAREs Tlg1p and Tlg2p fail to recycle FM4-64 (46), and likewise, Chs3p and Snc1p accumulate in intracellular compartments in tlg1Δ or tlg2Δ cells, suggesting that they may be blocked in endosomal or Golgi structures (23, 27). In wild-type cells, green fluorescent protein (GFP)-Snc1p is localized at the plasma membrane with some punctate staining of internal structures (27). Tlg1p and Tlg2p have been detected in both Golgi and endosomal compartments (22, 41). Snc1p colocalizes with Tlg2p, suggesting that Tlg2p may play a direct role in recycling (27).
Recently, the F-box protein Rcy1p has been shown to be required for both a postinternalization step of endocytosis and recycling of FM4-64 (46). However, the molecular role of Rcy1p in membrane trafficking remains elusive. Rcy1p contains two sequence motifs that may provide clues to its function: an amino-terminal F box (4) and a CAAX box motif at its carboxyl terminus, which may mediate the interaction of Rcy1p with membranes (49). The F box is a degenerate sequence of about 70 amino acids that interacts with Skp1p (11). Skp1p is one of the core components of Skp1p–cullin–F-box protein (SCF) complexes, which comprise a family of E3 ubiquitin-ligases composed of three core subunits (Skp1p, Cdc53p, and Hrt1p [also called Roc1p or Rbx1p]) associated with an F-box protein. SCF complexes also associate with the E2 ubiquitin-conjugating enzyme Cdc34p, which transfers activated ubiquitin onto substrates. SCF complexes were first identified for their essential role during cell cycle progression in promoting ubiquitination and subsequent degradation of the Cdk inhibitors Sic1p and Far1p, as well as the G1 cyclins Cln1p and Cln2p (5, 12, 19, 43). Subsequent studies revealed that SCF complexes control a wide variety of cellular functions, including signal transduction and morphogenesis. For example, SCFGrr1(SCF containing the F-box protein Grr1p), SCFMet30, and SCFCdc4 are required for the degradation of the bud emergence protein Gic2p (24) and the transcriptional regulators Met4p and Gcn4p (30, 36), respectively. F-box proteins have been shown to bind substrates in a phosphorylation-dependent manner and are thus thought to bring specificity to the complex (10). However, among the at least 15 F-box proteins encoded in the yeast genome, only Cdc4p, Grr1p, and Met30p have so far been shown to participate in SCF complexes.
The involvement of the F-box protein Rcy1p raises the possibility that ubiquitination and degradation of unknown substrates may be required for recycling. Here we have investigated the localization and functional properties of Rcy1p during recycling of the plasma membrane protein Snc1p. We found that a complex between Rcy1p and Skp1p was required for recycling, while no other SCF components were associated with Rcy1p or appeared to play a role in recycling. Our data thus imply that Skp1p and F-box proteins may function in both SCF and non-SCF complexes. Similar to Snc1p, Rcy1p accumulated in areas of polarized growth, and this localization required its CAAX motif and an intact actin cytoskeleton, consistent with a direct role of Rcy1p during recycling.
MATERIALS AND METHODS
Yeast strains.
Yeast strains are described in Table 1. All strains are derived from K699 (mata ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+ psi+ ssd1-d2) (W303 background). Standard yeast growth conditions and genetic manipulations were used as described previously (17). Yeast transformations were performed by the lithium acetate procedure (16). Database searches were performed using the SGD (Stanford University) and the National Center for Biotechnology Information BLAST (National Institutes of Health [NIH]) programs. Strains expressing GFP-, myc-, or hemagglutinin (HA)-tagged versions of RCY1 were constructed as described previously (28).
TABLE 1.
Yeast strains
| Strain name | Relevant genotype | Background | Source |
|---|---|---|---|
| K699 | Wild type | W303 | K. Nasmyth |
| RH3622 | erg6::LEU2 | S288c | A. Heese-Peck |
| RDY1251 | skp1::SKP1-9myc | W303 | This study |
| RDY1510 | rcy1::RCY1-TEV-9myc | W303 | This study |
| JMG199 | rcy1::kanMX Matα | W303 | S. Alvaro |
| JMG263 | rcy1::kanMX Mata | W303 | This study |
| JMG192 | rcy1::GAL-HA3-RCY1-HIS3 | W303 | This study |
| JMG201 | rcy1::GAL-GFP-RCY1-HIS3 | W303 | This study |
| JMG283 | rcy1::GAL-HA3-RCY1ΔF-HIS3 | W303 | This study |
| JMG284 | rcy1::GAL-GFP-RCY1ΔF-HIS3 | W303 | This study |
| YMT670 | cdc34-2 | W303 | C. Mann |
| YMT740 | cdc53-1 | W303 | M. Tyers |
| YMT668 | cdc4-1 | W303 | M. Tyers |
| Y552 | skp1-11 | W303 | S. Elledge |
| Y554 | skp1-12 | W303 | S. Elledge |
| rbx1-1 | rbx1::HIS3 × PDK102 (rbx1-1 in pRS314) | W303 | S. Elledge |
| tsyK03 | tlg2::HIS3 | SEY6210 | H. Pelham |
| RH1965 | end4::LEU2 | S288c | H. Riezman |
| RH144 | sec18-1 | S288c | H. Riezman |
| YMB802 | arp2-H330L | S288c | B. Winsor |
| LRB341 | Wild type | W303 | L. Robinson |
| LRB346 | yck1-Δ1 yck2-2ts | W303 | L. Robinson |
DNA manipulations.
Plasmids are listed in Table 2. Standard procedures were used for recombinant DNA manipulations (2). PCRs were performed with the Expand polymerase kit as recommended by the manufacturer (Boehringer Mannheim). Oligonucleotides were synthesized by Genset (Paris, France). Oligonucleotide sequences are available upon request. Site-directed mutagenesis was performed using the method developed by Kunkel et al. (25), and the correct sequences of the mutants were confirmed by sequencing.
TABLE 2.
Plasmids
| Plasmid name | Relevant characteristics | Source or reference |
|---|---|---|
| pYES2-GST-SKP1 | GAL-SKP1-GST URA3 2μm | 14 |
| JMG95 | GAL-GFP-RCY1 URA3 CEN | This study |
| JMG111 | GAL-GFP-RCY1ΔCAAX URA3 CEN | This study |
| JMG125 | GAL-GFP-RCY1ΔFbox URA3 CEN | This study |
| JMG98 | GAL-HA3-RCY1 URA3 CEN | This study |
| JMG51 | HA3-RCY1 TRP1 CEN | This study |
| JMG73 | pEG203 HRT1 | 7 |
| JMG77 | pJG4-6 HRT1 | 7 |
| JMG32 | pJG4-6 GRR1 | 14 |
| pBM110 | pJG4-6 CDC4 | 7 |
| pMT1297 | pEG202 CDC53 | M. Tyers |
| JMG30 | pEG203 SKP1 | This study |
| JMG31 | pJG4-6 SKP1 | 14 |
| pCL5 | pEG203 RCY1 | This study |
| JMG34 | pJG4-6 RCY1 | This study |
| JMG112 | pJG4-6 RCY1-3A | This study |
| JMG113 | pJG4-6 RCY1-ΔF | This study |
| JMG114 | RCY1ΔF TRP1 CEN | This study |
| JMG30 | RCY1ΔCAAX TRP1 CEN | This study |
| JMG118 | GFP-SNC1 URA3 CEN | M. Lewis |
| JMG122 | GFP-SNC1-pem URA3 CEN | M. Lewis |
| JMG121 | GFP-SNC1/SSO1 URA3 CEN | M. Lewis |
| JMG87 | YCK1 URA3 2μm | This study |
Antibodies, Western blotting, phosphatase assays, and microscopy.
Standard procedures were used for yeast cell extract preparation and immunoblotting (14). Immunoblots were quantified using the NIH Image program. Antibodies against glutathione S-transferase (GST) (Qiagen) and the HA epitope (HA11; Babco) were used as recommended by the manufacturers. 9E10 antibodies were produced by the Swiss Institute for Experimental Cancer Research antibody facility, and polyclonal antibodies specific for Gic2p (24) and GFP were kindly provided by M. Jaquenoud (Swiss Institute for Experimental Cancer Research) and P. Silver (Dana Farber, Boston, Mass.), respectively.
For phosphatase assays, extracts (1 optical density unit) prepared from K699 cells transformed with JMG118 (GFP-Snc1p) were resuspended in 100 μl of alkaline phosphatase (calf intestinal phosphatase [CIP]) buffer and split in two parts. One half was incubated with 1 U of CIP for 15 min at 37°C, whereas the other half was incubated under the same conditions but with 1 U of heat-inactivated (10 min at 100°C) CIP. Proteins were precipitated with trichloroacetic acid (10% final concentration), resuspended in gel sample buffer, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
For microscopy, cells were grown to early log phase and photographed on a Zeiss Axiophot fluorescence microscope with a Photometrics CCD camera. GFP-Rcy1p and GFP-Snc1p were visualized using a Chroma GFPII filter (excitation wavelength, 440 to 470 nm) and analyzed with Photoshop 4.0 software (Adobe). Photographs shown are both phase-contrast and fluorescence images of the same cells. Cells expressing GFP-Rcy1p from the inducible GAL promoter were grown to early log phase at 30°C in selective medium containing raffinose (2% final concentration), at which time galactose was added (2% final concentration) and left for 6 h. Temperature-sensitive strains were grown at 25°C and shifted for 1 h to 37°C before analysis of the localization of Rcy1p-GFP. Where indicated, the actin polymerization inhibitor latrunculin A (LAT-A) (200 μM final concentration in dimethyl sulfoxide [DMSO]) or DMSO (as a control) was added. Depolarization of the actin cytoskeleton by LAT-A was monitored by staining the cells with rhodamine-labeled phalloidin (33). Where indicated, α-factor was added to a final concentration of 50 μg/ml.
Determination of half-life.
Cultures were grown to early log phase in rich medium at 30°C (25°C for temperature-sensitive mutants), at which time cycloheximide (CHX) (Sigma) was added to a final concentration of 50 μg/ml (stock solution, 10 mg/ml). Temperature-sensitive strains were shifted to 37°C 3 h before addition of CHX. The proteasome inhibitor MG132 (Sigma) was solubilized in DMSO and added to a final concentration of 50 μM 90 min before addition of CHX. Aliquots were collected at the times indicated, and protein levels were analyzed by immunoblotting with specific antibodies. Immunoblots were quantified using the NIH Image program.
Gel filtration.
Wild-type (K699), cdc4-1 (YMT 668), skp1-11 (Y552), and skp1-12 (Y554) cells harboring pJMG98 (GAL-HA3-RCY1) were grown at 30°C to mid-log phase in selective medium containing raffinose (2% final concentration), and expression of HA3-Rcy1p was induced for 2 h by the addition of galactose (2% final concentration). The cells were pelleted and lysed as described previously (6). The extract was centrifuged first at 4°C for 10 min at 10,000 × g and then for 10 min at 100,000 × g in a TFT80.2 rotor (S100). Approximately 800 μg of the soluble S100 supernatant was loaded on a Superose 6PC 3.2/30 column compatible with the SMART system (Amersham Pharmacia Biotechnology GMBH). Aliquots (50 μl) were collected, concentrated, and analyzed by SDS-PAGE and immunoblotting. Standard molecular mass markers (66 kDa [bovine serum albumin], 234 kDa [catalase], and 660 kDa [thyroglobulin]) were run separately to control the fractionation.
FM4-64 recycling assay.
FM4-64 recycling assays were performed as described previously (46). Yeast cells were allowed to internalize FM4-64 for 12 min at 24°C and then washed three times with ice-cold SD medium. After the last wash, the cells were resuspended in 10 μl of SD medium and kept on ice. Prewarmed SD medium at 24°C (37°C for SCF-deficient mutants) was added to the cells, and the fluorescence was recorded during 600 s.
Two-hybrid assays.
Two-hybrid assays were performed as described previously (2) with the yeast strain K699 containing the lacZ reporter plasmid pSH18.34, using pEG202-based plasmids expressing LexA DNA-binding domain (DBD) fusions and pJG4-5-based plasmids containing fusions to the B42 transcriptional activation domain (AD) (18). Results are reported as Miller units (averages from three independent experiments with standard deviations).
Immunoprecipitations and purification of Rcy1p complex.
Exponentially growing yeast cells were pelleted, washed with water, and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 0.2% Triton X-100) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, and 1 μg of aprotinin per ml). Cells were lysed with glass beads by 10 cycles of vortexing for 40 s followed by a 1-min incubation on ice. After centrifugation (30 min, 100,000 × g), extracts (7 mg) were incubated with 9E10 monoclonal antibodies cross-linked to protein A-Sepharose (9E10 beads, 20 μl) for 90 min at 4°C. The beads were washed four times with 1 ml of lysis buffer lacking protease inhibitors and eluted with 0.1 mg of TEV protease (Gibco BRL) per ml at room temperature for 40 min. The eluted proteins were separated by SDS-PAGE and then visualized by staining with silver or immunoblotting with anti-myc, anti-Skp1, and anti-Cdc53 antibodies.
RESULTS
GFP-Rcy1p is concentrated at sites of polarized growth.
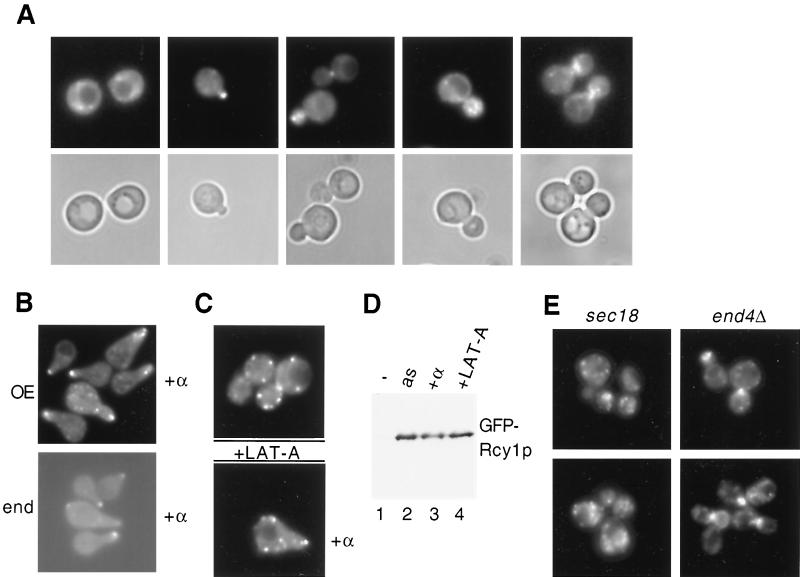
To determine the subcellular localization of Rcy1p, we constructed a GFP-tagged version, which was expressed from its endogenous promoter (Fig. 1B; lower panel) or the galactose-inducible GAL1 promoter (Fig. 1A to E). Cells with RCY1 deleted are cold sensitive (48); GFP-Rcy1p expressed from its own promoter was able to restore growth of rcy1Δ cells at 15°C, suggesting that it is functional in vivo (data not shown). GFP-Rcy1p showed a dynamic localization that changed at different cell cycle stages. In unpolarized G1 cells, GFP-Rcy1p was found in patches in the cytoplasm, while after bud emergence it was concentrated in nascent buds and at the mother-bud neck (Fig. 1A). In addition, GFP-Rcy1p accumulated at the shmoo tips of cells treated with pheromones (Fig. 1B). Thus, GFP-Rcy1p is localized in areas of polarized growth, similar to actin patches (1). To test whether an intact actin cytoskeleton is required for this localization, we treated cells with the potent actin-depolymerizing compound LAT-A (3) and visualized Rcy1p by fluorescence microscopy. Within 5 min after addition of LAT-A, GFP-Rcy1p was found in patches, which were randomly distributed all over the cells (Fig. 1C). As expected, actin was completely depolymerized under these conditions, as determined by staining with rhodamine-phalloidin (data not shown). Western blot analysis showed that neither pheromone nor LAT-A treatments altered Rcy1p levels (Fig. 1D). Interestingly, the polarized distribution of Rcy1p also required functional Sec18p but was not affected in end4Δ and arp2-1 mutant cells, which are defective for endocytosis (Fig. 1E and data not shown) (15). We thus conclude that the polarized localization of Rcy1p depends on an intact actin cytoskeleton and a functional secretion pathway.
FIG. 1.
Rcy1p is localized in areas of polarized growth in an actin-dependent manner. (A) JMG201 cells (rcy1::GAL-GFP-RCY1) were grown to early log phase at 30°C in selective medium. Cells were analyzed by fluorescence (upper panels) and phase-contrast (lower panels) microscopy as described in Materials and Methods. (B) Upper panel, JMG201 cells were grown as described for panel A (overexpression [OE] of GFP-Rcy1p). Four hours after addition of galactose, α-factor (50-μg/ml final concentration) was added and left for 2 h, and GFP-Rcy1p was visualized by fluorescence microscopy. Lower panel, wild-type cells (K699) were transformed with a plasmid expressing GFP-Rcy1p from its own promoter (end) and treated with α-factor for 2 h. (C) JMG201 cells were grown as described above and either treated (lower panel) or not treated (upper panel) with α-factor. After 2 h, LAT-A was added and left for 5 min, the cells were fixed, and the localization of GFP-Rcy1p was visualized. (D) The expression of GFP-Rcy1p in JMG201 cells shown in panels A (lane 2, as), B (lane 3, α-factor), and C (lane 4, LAT-A) was analyzed by immunoblotting with polyclonal antibodies against GFP. Cells with an empty control vector were used as a control (lane 1, −). Note that neither pheromone treatment nor addition of LAT-A alters GFP-Rcy1p levels. (E) The localization of GFP-Rcy1p expressed from the GAL promoter was examined in sec18-1 (RH144) or end4Δ (RH1965) cells by GFP microscopy. sec18-1 cells were incubated at the restrictive temperature (37°C) for 1 h before analysis of the localization of Rcy1p-GFP.
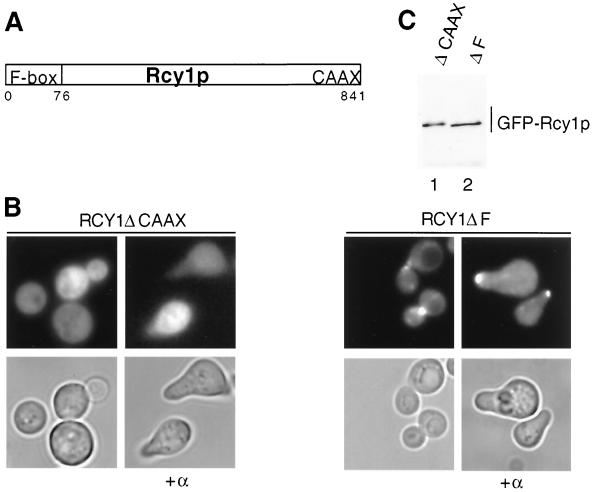
Rcy1p contains two conserved domains (Fig. 2A): an F-box motif at its amino terminus and a CAAX box that is implicated in mediating interactions with membranes. An Rcy1p mutant lacking its last four amino acids (ΔCAAX) was distributed throughout the cytoplasm (Fig. 2B, left panels). In contrast, deletion of the F-box motif (ΔF) had no effect on polarized localization of GFP-Rcy1p in either growing or pheromone-arrested cells (Fig. 2B, right panels). Western blot analysis confirmed that both GFP-Rcy1 proteins were equally expressed and migrated at their expected size on SDS gels (Fig. 2C). Taken together, these results indicate that the polarized localization of Rcy1p is independent of its F box but requires an intact CAAX membrane-binding motif.
FIG. 2.
The polarized localization of Rcy1p requires its CAAX motif but not the F box. (A) Schematic representation of the conserved domains in Rcy1p. Rcy1p contains an F box (amino acids 4 to 76) at its amino terminus and a CAAX box at its carboxyl terminus. Amino acids are numbered at the bottom. (B) Wild-type (K699) cells transformed with either plasmid JMG111 (GAL-GFP-RCY1ΔCAAX) or JMG125 (GAL-GFP-RCY1ΔF) were grown as described for Fig. 1A. Four hours after addition of galactose, cells were treated (right panels) or not treated (left panels) with α-factor. After 2 h, the cells were analyzed by phase-contrast (lower row) and fluorescence (upper row) microscopy. Note that deletion of the CAAX motif alters the localization of GFP-Rcy1p, whereas deletion of the F box has no effect. (C) The expression of GFP-Rcy1p in the cells shown in panel B was analyzed by immunoblotting with polyclonal antibodies against GFP. Lane 1, GFP-Rcy1p-ΔCAAX; lane 2, GFP-Rcy1p-ΔF.
Rcy1p is required for recycling of GFP-Snc1p.
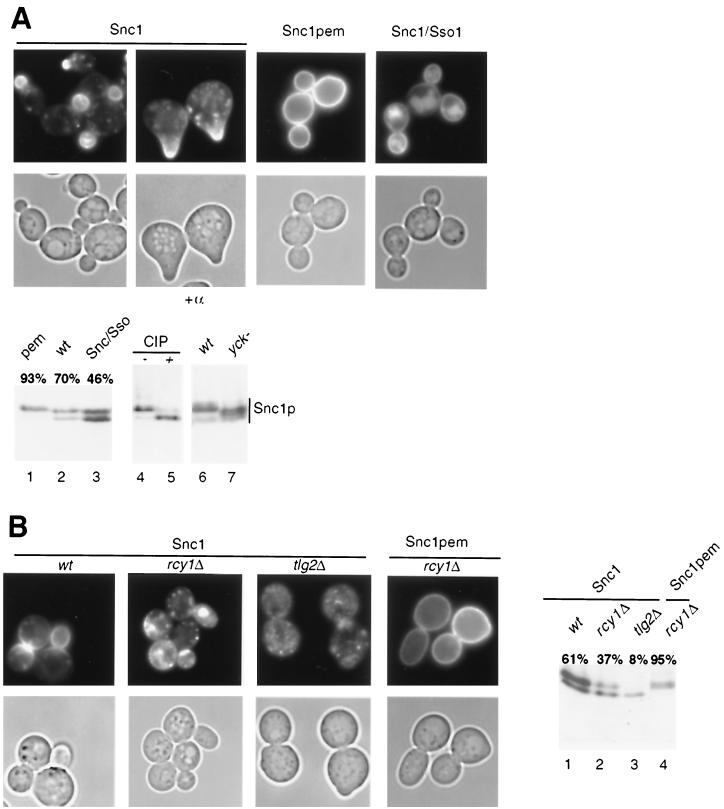
Two assays have been developed to analyze recycling in yeast: first, resecretion of previously internalized FM4-64 can be quantified as a loss of membrane-bound fluorescence (46), and second, changes in localization of GFP-Snc1p can be used as a readout for a functional recycling pathway (27). In wild-type cells, GFP-Snc1p accumulates at the plasma membrane in areas of polarized growth (bud and shmoo), with some intracellular punctate staining which may correspond to an endosomal compartment (Fig. 3A, left panels) (27). In contrast, GFP-Snc1p with mutations in its internalization signal (GFP-Snc1pem) is located exclusively at the plasma membrane (Fig. 3A, middle panels) (27). Conversely, exchange of the transmembrane domain of Snc1p with that of the SNARE Sso1p (GFP-Snc1/Sso1p) abolishes recycling to the plasma membrane, and the chimeric protein is instead targeted to the vacuole (Fig. 3A, right panels) (27). Importantly, immunoblotting of the wild-type and mutant variants of GFP-Snc1p with GFP antibodies revealed that GFP-Snc1p migrated as two distinct bands (Fig. 3A, lane 2). Treatment of the sample with alkaline phosphatase quantitatively converted the slower-migrating form into the faster-migrating one (lanes 4 and 5), demonstrating that the upper band corresponds to a phosphorylated form of GFP-Snc1p. GFP-Snc1pem was hyperphosphorylated (Fig. 3A, lane 1), while GFP-Snc1/Sso1p accumulated as an underphosphorylated species (lane 3). Quantification revealed that approximately 70% of Snc1p is hyperphosphorylated in wild-type cells, compared to over 90% of Snc1pem and 46% of GFP-Snc1/Sso1p. Thus, the cellular localization of GFP-Snc1p correlates with the phosphorylation state of the protein, providing a convenient assay to monitor recycling of GFP-Snc1p. Interestingly, phosphorylation of GFP-Snc1p was partly impaired in mutant cells defective in Yck1p and Yck2p (Fig. 3A, lanes 6 and 7), two yeast casein kinase I isoforms which have been implicated in internalization of plasma membrane proteins (35). In tlg2Δ cells, GFP-Snc1p was predominantly intracellular (Fig. 3B) (27) and accumulated in its underphosphorylated form (lane 3). Likewise, GFP-Snc1p was predominantly intracellular and underphosphorylated in rcy1Δ cells (Fig. 3B, lane 2) (37% in the hyperphosphorylated form), supporting the notion that Rcy1p is involved in recycling of membrane proteins (46). In contrast, GFP-Snc1pem was hyperphosphorylated and localized at the plasma membrane in rcy1Δ cells (Fig. 3B, lane 4) (95% in the hyperphosphorylated form), demonstrating that Rcy1p is not required for phosphorylation or membrane targeting of newly synthesized Snc1p. Finally, the levels of GFP-Snc1p were reduced in rcy1Δ and tlg2Δ cells compared to wild-type cells (Fig. 3B, lanes 1 to 3), possibly because Snc1p is degraded in the vacuole if it cannot recycle back to the plasma membrane.
FIG. 3.
Localization and phosphorylation of the recycling v-SNARE GFP-Snc1p. (A) Wild-type (K699) cells transformed with either plasmid JMG118 (GFP-Snc1p), JMG122 (GFP-Snc1-pem), or JMG121 (GFP-Snc1/Sso1p) were grown in selective SD medium to early log phase and analyzed by phase-contrast (lower row) and fluorescence (upper row) microscopy. Where indicated, cells were treated with α-factor for 2 h. The expression of wild-type protein and the GFP-Snc1p variants was examined by Western blotting using an antibody raised against GFP (lower panel, lanes 1 to 3). The bands were quantified, and the level of the upper band was expressed as a percentage of the total GFP-Snc1 protein. A protein extract prepared from wild-type (K699) cells expressing GFP-Snc1p was incubated with active (+, lane 5) or heat-inactivated (−, lane 4) alkaline phosphatase (CIP). Protein extracts prepared from either wild-type (wt) (LRB341) or yck1-Δ1 yck2-2ts (yck−, LRB346) cells expressing GFP-Snc1p shifted for 2 h to 37°C were analyzed by Western blotting using an antibody raised against GFP (lanes 6 and 7). Note that only part of the GFP-Snc1p shift is dependent on functional Yck kinases and that the phosphorylation state of the protein correlates with its subcellular localization. (B) Wild-type (K699), rcy1Δ (JMG199), and tlg2Δ (tsyK03) cells transformed with the plasmid JMG118 (GFP-Snc1p) or JMG122 (GFP-Snc1pem) were grown and analyzed as described for panel A. Note that GFP-Snc1p, but not GFP-Snc1pem, is mainly intracellular and accumulates in its underphosphorylated form in rcy1Δ and tlg2Δ cells.
The F box and CAAX motif of Rcy1p are both required for its recycling function.
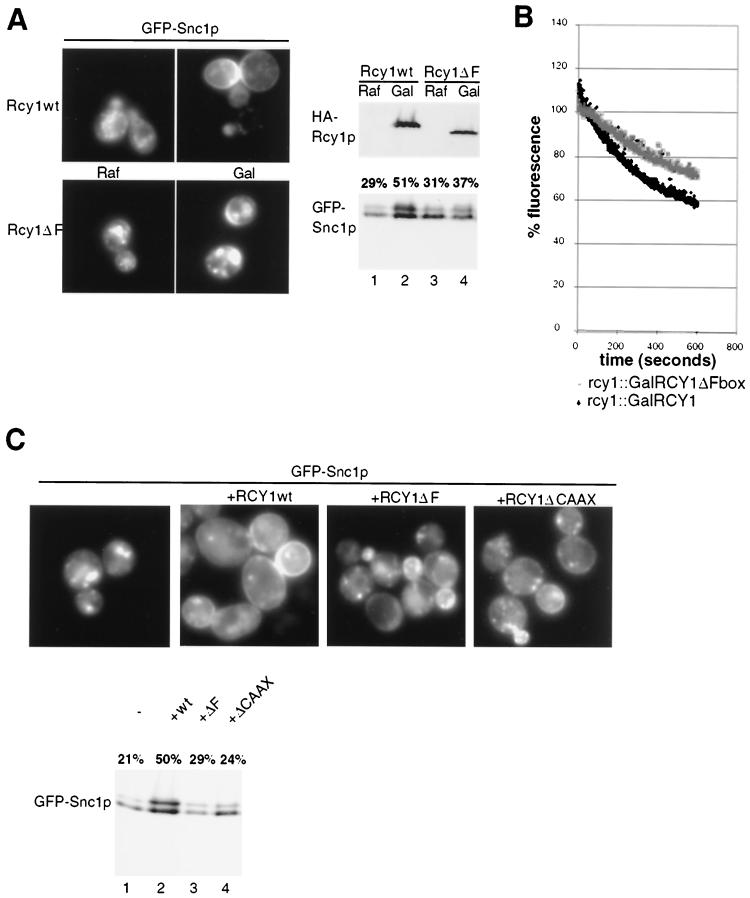
To determine whether the F-box motif of Rcy1p is required for its recycling function, we constructed strains in which the RCY1 locus was replaced by either HA3-tagged wild-type RCY1 or the ΔF-box mutant of RCY1 under the control of the galactose-inducible promoter. As expected, both strains showed intracellular localization and underphosphorylation of GFP-Snc1p when grown on medium containing raffinose (GAL promoter off) (Fig. 4A, lanes 1 to 3). Two hours after addition of galactose, both the wild-type and ΔF mutant forms of HA3-Rcy1p were expressed at comparable levels (upper panels). However, in contrast to wild-type HA3-Rcy1p, the HA3-Rcy1p-ΔF mutant failed to restore plasma membrane localization and phosphorylation of GFP-Snc1p (lower panels), suggesting that an intact F box is required for the recycling function of Rcy1p. Consistent with this result, cells expressing Rcy1p-ΔF exhibited a recycling defect of the fluorescent dye FM4-64, which is comparable to the defect measured in rcy1Δ cells (Fig. 4B) (46). Identical results were obtained with rcy1Δ cells transformed with low-copy-number plasmids expressing either wild-type Rcy1p or the ΔF mutant protein from the endogenous promoter (Fig. 4C). Recycling also required an intact CAAX motif; Rcy1p-ΔCAAX expressed from the endogenous promoter failed to restore membrane localization and phosphorylation of GFP-Snc1p in rcy1Δ cells (Fig. 4C), suggesting that Rcy1p needs to associate with membranes. Taking the results together, we conclude that both the F box and the CAAX motif of Rcy1p are important for its recycling function in vivo.
FIG. 4.
The F and CAAX boxes of Rcy1p are required for its recycling function. (A) JMG192 (rcy1::GAL-3HA-RCY1) and JMG283 (rcy1:: GAL-3HA-RCY1ΔF) cells transformed with the plasmid JMG118 (GFP-Snc1p) were grown at 30°C to early log phase in selective medium containing 2% raffinose (Raf). Cultures were divided, and 2% galactose (Gal) was added to one half. After 2 h, the subcellular localization (left panels) and phosphorylation state (lower right panel) of GFP-Snc1p were analyzed as described in the legend to Fig. 3. The expression of HA3-Rcy1p was controlled by Western blotting with HA11 antibodies (upper right panel). (B) JMG192 (rcy1::GAL-HA3-RCY1) and JMG283 (rcy1::GAL-HA3-RCY1ΔF) cells were grown as described for panel A. FM4-64 recycling assays were carried out as described in Materials and Methods. (C) JMG199 (rcy1Δ) cells cotransformed with plasmid JMG118 (GFP-Snc1p) and either an empty control vector (−) or plasmid JMG51 (HA3-Rcy1p), JMG114 (Rcy1p-ΔF), or JMG130 (Rcy1p-ΔCAAX) were grown to early log phase and examined for localization (left panels) and phosphorylation state (right panels) of GFP-Snc1p as described in the legend to Fig. 3.
Skp1p, but not the SCF pathway, is required for recycling.
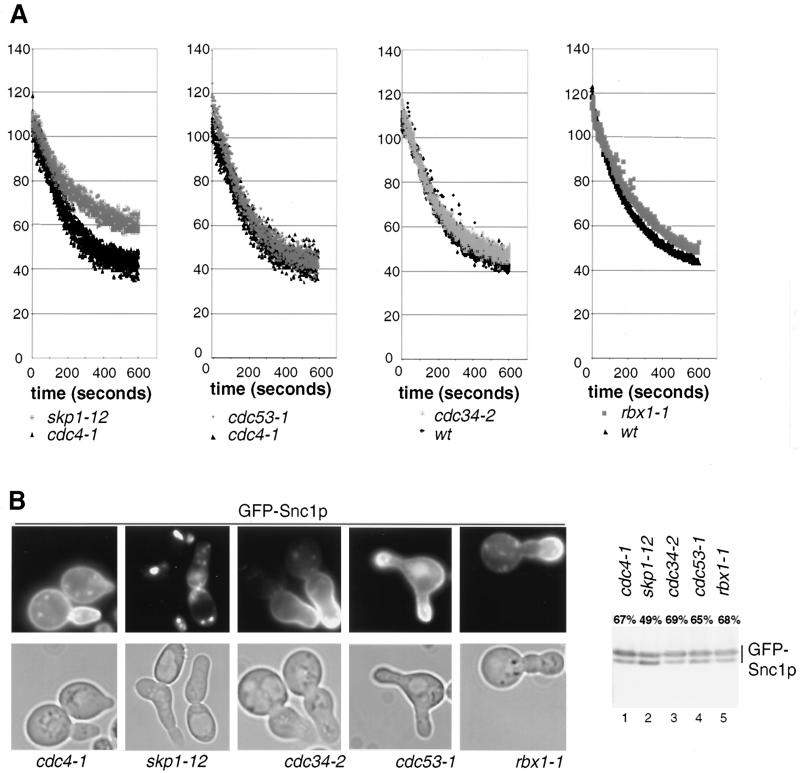
To investigate whether Rcy1p functions as a subunit of an SCF ubiquitin-ligase complex, we tested whether SCF-deficient cells are defective for recycling of FM4-64 and GFP-Snc1p (Fig. 5). The skp1-12 and skp1-11 mutant cells showed a partial but reproducible defect in recycling of FM4-64 (Fig. 5A and data not shown). In contrast, cells defective in Cdc53p, Cdc34p, and Hrt1p showed no recycling defect at nonpermissive temperature, indicating that these SCF core components may not be involved in recycling. As a control we used either wild-type or cdc4-1 cells, which arrest at the same cell cycle stage as skp1-11, cdc34-2, cdc53-1, and rbx1-1 mutants. To corroborate these results, we examined the localization and phosphorylation state of GFP-Snc1p. Cells were shifted to the restrictive temperature for 3 h, and GFP-Snc1p was analyzed by fluorescence microscopy (Fig. 5B, left panels) or immunoblotting (right panel). Clearly, GFP-Snc1p was located predominantly at the plasma membrane and accumulated in its hyperphosphorylated state in all SCF mutants except the skp1-12 mutant, implying that recycling occurs efficiently in these cells. In 40% of skp1-12 cells, GFP-Snc1p was clearly intracellular (as shown in Fig. 5B), while in the rest of the cells the phenotype was less severe (20%) or hardly detectable (40%). Quantitation of the immunoblots revealed that 49% of GFP-Snc1p was in its hyperphosphorylated form in skp1-12 cells (Fig. 5B, lane 2), compared to approximately 70% in wild-type or SCF mutants (lanes 1, 3, 4, and 5). A weak vacuolar staining of GFP-Snc1p was visible in cdc53-1 cells; these cells grow poorly compared to other SCF mutants, and we observed that a growth defect is often associated with vacuolar mislocalization of GFP-Snc1p. Taken together, these results suggest that recycling requires Skp1p but may not involve the function of the core SCF components Cdc34p, Cdc53p, and Hrt1p, although we cannot exclude the possibility that the mutant alleles used are specifically defective for cell cycle progression but not recycling.
FIG. 5.
Recycling of FM4-64 and GFP-Snc1p in SCF-deficient cells. (A) Wild-type (K699), cdc4-1 (YMT668), cdc34-2 (YMT670), cdc53-1 (YMT740), skp1-12 (Y554), and rbx1-1 (rbx1-1) cells were analyzed for their ability to recycle FM4-64 as described in Materials and Methods. (B) cdc4-1 (YMT668), cdc34-2 (YMT670), skp1-12 (Y554), cdc53-1 (YMT730), and rbx1-1 (rbx1-1) cells transformed with the plasmid JMG118 (GFP-Snc1p) were grown in selective medium at 25°C to early log phase, shifted for 3 h to 37°C, and examined for both localization (left panel) and phosphorylation (right panel) of GFP-Snc1p as described in the legend to Fig. 3. Note that SCF-deficient cells accumulate with the typical hyperpolarized morphology.
Degradation of Rcy1p is not mediated by an SCF-dependent ubiquitination pathway.
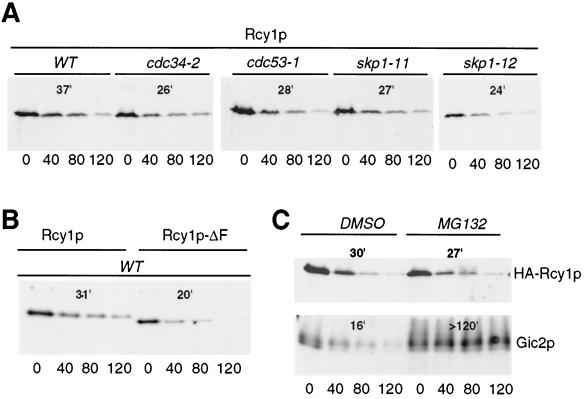
Several F-box proteins that are subunits of SCF E3-ligase complexes are constitutively turned over by ubiquitin-dependent degradation (14, 50). For example, degradation of the F-box proteins Cdc4p and Grr1p requires their F-box motif, the SCF core components Cdc53p and Skp1p, and the 26S proteasome. In addition, Met30p and the mammalian F-box protein Skp2 are also degraded in an SCF- and/or proteasome-dependent manner (36, 47). To examine whether the F-box protein Rcy1p may be degraded by a similar mechanism, we measured its turnover by CHX chase experiments (Fig. 6). Rcy1p was degraded in wild-type cells with a half-life of approximately 35 min. However, in contrast to Grr1p, Cdc4p, and Met30p (14, 36), Rcy1p was not stabilized in cdc34-2, cdc53-1, skp1-11, or skp1-12 mutants (Fig. 6A). Moreover, deletion of the F box of Rcy1p did not stabilize the protein (Fig. 6B), indicating that assembly into an SCF complex does not mediate its degradation. Finally, Rcy1p was not stabilized in cells treated with the proteasome inhibitor MG132 (Fig. 6C), suggesting that Rcy1p may not be degraded by 26S proteasomes. In contrast, the SCFGrr1 target Gic2p was fully stabilized in MG132-treated cells, confirming that 26S proteasomes were efficiently inhibited under these conditions. Finally, Rcy1p was not stabilized in pep4Δ cells (data not shown), implying that it may not be degraded in lysosomes. Taken together, these results suggest that degradation of Rcy1p does not require SCF core subunits or functional 26S proteasomes, supporting the notion that Rcy1p may not assemble into a functional SCF complex.
FIG. 6.
Rcy1p is rapidly degraded by a proteasome- and SCF-independent pathway. (A) The half-life of HA3-Rcy1p was determined by CHX chase in either wild-type (K699), cdc34-2 (YMT670), cdc53-1 (YMT730), skp1-11 (Y552), or skp1-12 (Y554) cells transformed with JMG98 (GAL-HA3-RCY1). Cells were grown in medium containing 2% galactose to mid-log phase, at which time CHX was added (time zero) to a 50-μg/ml final concentration. Aliquots were removed at the times indicated (in minutes) and analyzed by immunoblotting with HA11 antibodies. The blots were quantified as described in Materials and Methods, and the half-lives are shown in minutes. (B) The half-lives of Rcy1p and Rcy1p-ΔF were determined by CHX chase experiments. JMG192 (rcy1::GAL-HA3-RCY1) and JMG283 (rcy1::GAL-HA3-RCY1ΔF) cells were grown to mid-log phase, and HA3-Rcy1p levels were analyzed as described above. (C) The half-life of Rcy1p was measured by CHX chase in erg6Δ (RH3622) cells in the presence of the proteasome inhibitor MG132. RH3622 cells transformed with JMG98 (GAL-HA3-RCY1) were grown to mid-log phase, at which time either the solvent DMSO or MG132 was added. After 2 h, CHX was added, and aliquots were analyzed for the presence of HA3-Rcy1p by immunoblotting with HA11 antibodies (upper panel). For a control, degradation of Gic2p was monitored in the same cells by immunoblotting with Gic2p antibodies (lower panel).
Rcy1p binds to Skp1p through its F-box motif but may not be a component of an SCF complex.
To determine whether Rcy1p interacts with Skp1p, we performed two-hybrid and coimmunoprecipitation experiments (Table 3 and Fig. 7). Like the F-box proteins Cdc4p and Grr1p (7, 14), full-length Rcy1p strongly interacted with Skp1p, irrespective of whether it was fused to an AD or a DBD (Table 3). Mutating three conserved residues in the F box of Rcy1p to alanine residues (Rcy1p-3A) or deletion of this motif (Rcy1p-ΔF) impaired the interaction with Skp1p (Table 3), although the mutant proteins were efficiently expressed (data not shown). Thus, these results suggest that Rcy1p interacts with Skp1p through its amino-terminal F-box motif.
TABLE 3.
Two-hybrid analysis of the interaction between Rcy1p and Skp1pa
| AD | DBD | Miller units (mean ± SD) |
|---|---|---|
| Skp1 | None | 59 ± 11 |
| None | Skp1 | 6 ± 4 |
| Rcy1 | None | 9 ± 5 |
| None | Rcy1 | 3 ± 4 |
| Skp1 | Rcy1 | 1,818 ± 134 |
| Rcy1 | Skp1 | 2,187 ± 156 |
| Rcy1-3A | Skp1 | 631 ± 73 |
| Rcy1-ΔF | Skp1 | 91 ± 12 |
Skp1 and Rcy1p were fused to either the LexA DBD or the B42 transcriptional AD. Expression of the β-galactosidase reporter was quantified as described in Materials and Methods. Note that Rcy1p interacts with Skp1p in an F-box-dependent manner.
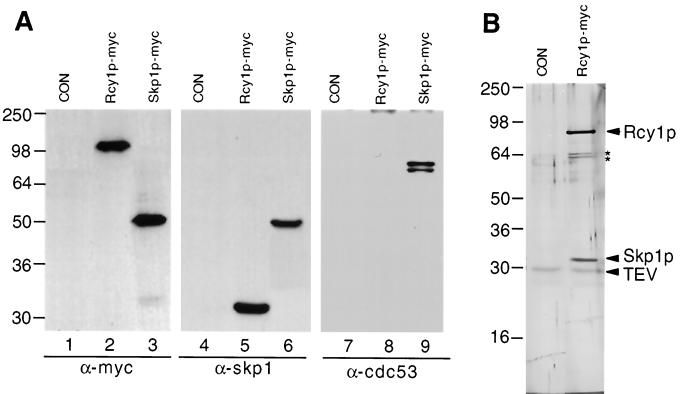
FIG. 7.
Rcy1p coimmunoprecipitates with Skp1p but not other components of the SCF pathway. (A) Lysates prepared from either wild-type (K699) (CON), RDY1510 (Rcy1p-myc), or RDY1251 (Skp1p-myc) cells were immunoprecipitated with 9E10 anti-myc antibodies and were analyzed by Western blotting for the presence of associated proteins using antibodies raised against the myc epitope (lanes 1 to 3), Skp1p (lanes 4 to 6), or Cdc53p (lanes 7 to 9). The positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) Lysate prepared from either control cells (CON, K699) or RDY1510 cells (RCY1-TEV-9myc) was immunoprecipitated with 9E10 anti-myc antibodies, and bound proteins were eluted by incubation with TEV protease as described in Materials and Methods. The eluate was analyzed by SDS-PAGE followed by silver staining. The positions of Rcy1p, Skp1p, and TEV protease are marked by arrowheads; two unspecifically bound proteins are indicated by asterisks. Molecular mass markers (in kilodaltons) are indicated on the left.
Consistent with this finding, HA3-Rcy1p fractionated on a Superose 6 column with peak of approximately 200 kDa, which was dependent on functional Skp1p and its F box (data not shown). To examine the Skp1p-Rcy1p interaction by coimmunoprecipitation, we constructed strains in which the chromosomal loci of RCY1 and SKP1 were replaced by 9myc epitope-tagged alleles. Rcy1p-myc and Skp1p-myc were efficiently immunoprecipitated with anti-myc antibodies (Fig. 7A, lanes 2 and 3). Importantly, Skp1p efficiently coimmunoprecipitated with Rcy1p-myc (lane 5), confirming that Skp1p is indeed present in a complex with Rcy1p. Several F-box proteins are part of large SCF complexes, which besides Skp1p also contain Cdc53p and Hrt1p. As expected, therefore, Cdc53p readily coimmunoprecipitated with Skp1p-myc (Fig. 7A, lane 9). However, we were unable to detect Cdc53p in Rcy1p-myc immunoprecipitates, although the F-box protein YBR280 precipitated with Cdc53p in parallel experiments (40). Consistent with these results, we were unable to detect an interaction between Cdc53p and Rcy1p by two-hybrid analysis, whereas Grr1p and Cdc53p bound efficiently (data not shown). Taken together, these results support the possibility that Rcy1p may not be a component of an SCF E3-ubiquitin-ligase complex that contains Cdc53p. To further investigate the subunit composition of Rcy1p-containing complexes, we purified the complex to near homogeneity and visualized its components by SDS-PAGE followed by silver staining (Fig. 7B). To this end, lysates prepared from cells expressing Rcy1-TEV-9myc were immunoprecipitated with anti-myc antibodies, and bound proteins were eluted by cleavage with TEV protease. Two main bands were evident at 90 and 32 kDa, and these proteins were identified by immunoblotting as Rcy1p and Skp1p, respectively (data not shown). Importantly, no proteins with the size of Cdc53p or Hrt1p were detectable, although these proteins are major components in SCFCdc4 and SCFGrr1 complexes (39). Thus, these results strongly suggest that Skp1p is the only major partner of Rcy1p, implying that Rcy1p may not be part of a classical SCF complex.
DISCUSSION
Here we show that Rcy1p is required for recycling of the plasma membrane protein Snc1p. Internalization and phosphorylation of Snc1p occurred efficiently in rcy1Δ cells, but the protein failed to recycle back to the plasma membrane. Rcy1p localized in cytoplasmic patches which concentrate in areas of polarized growth, consistent with a direct role in recycling. The recycling function of Rcy1p depends on its CAAX motif and an amino-terminal F box, which was required to bind to Skp1p. Biochemical characterization revealed that Rcy1p was bound to Skp1p but did not associate with the SCF pathway components Cdc53p, Hrt1p, and Cdc34p. Taken together, our results suggest that the F-box protein Rcy1p may directly regulate recycling in a complex with Skp1p but may not function as a substrate-specific adapter in SCF complexes.
Localization of Rcy1p.
We found that Rcy1p localizes in cytoplasmic patches, which concentrate in areas of polarized growth (bud, bud neck, and shmoo tip). Several recycled proteins also concentrate in these zones: Snc1p accumulates near the plasma membrane of nascent buds or tips of mating projections and at the mother-bud neck (27) (Fig. 3A), while Chs3p localizes to the bud neck region (23). Together these data suggest that Rcy1p may play a direct role in recycling, a conclusion which is supported by the observed homology of its carboxyl-terminal domain with hSec10, a protein involved in membrane trafficking (48).
The localization of Rcy1p resembles actin patches and was sensitive to LAT-A, suggesting that Rcy1p may be associated with actin. However, Rcy1p was still found in patches in cells treated with LAT-A, although actin patches were completely disrupted under these conditions. We thus favor a model in which Rcy1p is not associated with actin patches but needs an intact actin cytoskeleton to reach its polarized localization at the plasma membrane.
The polarized localization and function of Rcy1p also depend on its CAAX motif, suggesting that membrane association of Rcy1p is important in vivo. CAAX boxes are found in a number of eukaryotic proteins, including many small GTPases and the nonclassically secreted S. cerevisiae mating pheromone a-factor (49). Proteins terminating in a CAAX motif are modified at their carboxyl termini in a sequential three-step process consisting of isoprenylation, proteolysis, and carboxyl methylation (49). This processing is performed by enzymes associated with the endoplasmic reticulum (38), but it is not known how CAAX box proteins are transported from the endoplasmic reticulum membrane to their final location. They could either diffuse passively across the cytoplasm or travel along the cytoplasmic face of organelles such as those of the secretory pathway. Supporting the latter possibility, we found that Rcy1p was mislocalized in sec18-1 mutant cells, which are defective for polarized secretion.
rcy1Δ cells contain an enlarged compartment that is often located in the region of the bud (46), suggesting that recycling proteins travel through this organelle. Consistent with this hypothesis, Tlg1p localized to this compartment (46), and similarly, we observed that GFP-Snc1p often accumulates in large structures near the bud in rcy1Δ cells (Fig. 3B and data not shown). However, colocalization experiments between Rcy1p, Snc1p, and Tlg1p will be required to confirm whether these proteins indeed travel through a common compartment.
A complex between Rcy1p and Skp1p is required for recycling.
We found that Rcy1p interacts in vivo with Skp1p in an F-box-dependent manner. Supporting these results, skp1 mutants or rcy1Δ cells expressing Rcy1p-ΔF showed a defect in recycling of FM4-64 and GFP-Scn1p. The recycling defect of cells expressing Rcy1p-ΔF is unlikely to be due to misfolding of the mutant protein, because Rcy1p-ΔF is normally expressed and degraded (Fig. 4A and 6B) and localized like the wild-type protein to sites of polarized growth (Fig. 2B). We thus conclude that a complex between Skp1p and Rcy1p is specifically involved in recycling.
F-box proteins are thought to function as substrate-specific adapters for SCF complexes (32), raising the possibility that ubiquitin-dependent degradation may be involved in recycling of plasma membrane proteins. However, several lines of evidence indicate that the SCF components Cdc53p, Cdc34p, and Hrt1p are not part of the Skp1p-Rcy1p recycling complex: (i) we were unable to detect an interaction between Rcy1p and either Hrt1p or Cdc53p by coimmunoprecipitation and two-hybrid assays; (ii) in contrast to F-box proteins associated with SCF complexes, degradation of Rcy1p required neither 26S proteasomes, SCF core subunits, nor its F box; (iii) mutant cells defective in Cdc34p, Cdc53p, or Hrt1p did not exhibit defects in recycling of either Snc1p or FM4-64; and (iv) Skp1p was the only component that copurified in stoichiometric amounts with Rcy1p. In mammalian cells, Hrt1p interacts with at least five different cullins, including Cul1 and Cul2, which may constitute a subfamily of complexes which are based on adapters termed SOCS box proteins (45). Cdc53p has two yeast homologs, Rtt101p and Ygr003w, but cells with both of these open reading frames deleted did not exhibit a recycling defect (C. Lafourcade and M. Peter, unpublished results). Thus, the Skp1p-Rcy1p complex may function in recycling without any cullin requirement, implying that Rcy1p is not a substrate-specific adapter in a novel SCF complex. Rcy1p is not the first F-box protein that is implicated in functions other then ubiquitin-dependent degradation. The F-box protein Ctf13p interacts with Skp1p and is a component of the CBF3 kinetochore complex (37). It has been proposed that excess amounts of Ctf13p may be specifically eliminated by ubiquitin-dependent degradation, thereby providing a counting mechanism to assemble one and only one complex at every kinetochore. Like Ctf13p, Rcy1p is an unstable protein, but it is not degraded by a proteasome-dependent mechanism. Recently, the F-box proteins Rav1p and Rav2p have been shown to form a novel protein complex with Skp1p and the vacuolar ATPase, suggesting that they regulate its activity (40). Taken together, these results provide evidence that association of Skp1p with some F-box proteins could fulfill functions other than targeting of substrates for degradation.
Ubiquitination and recycling.
Ubiquitination of plasma membrane proteins has been established as a general signal for their internalization in both yeast and mammalian cells, but the machinery mediating these processes is still poorly understood. It is not clear, for example, how cells can distinguish between ubiquitin as an internalization or targeting signal to proteasomes. A single ubiquitin is sufficient to internalize Ste2p (20), but monoubiquitinated proteins are not recognized by proteasomes. Formation of the ubiquitin chains on the permeases Fur4p and Gap1p takes place via ubiquitin-Lys63, not -Lys48, which is commonly used for ubiquitin attachment (35). Thus, the number and specific linkage of ubiquitin to substrates could influence its intracellular fate.
Could the Skp1-Rcy1p complex function during recycling by ubiquitinating substrates at the plasma membrane, thereby targeting them for internalization? Although we cannot exclude this possibility, we consider it unlikely, because mutants defective in Rcy1p or Skp1p efficiently internalize GFP-Snc1p as well as the α-factor receptor Ste2p and the uracil permease Fur4p (46; C. Marchal, personal communication). Thus, the Rcy1p-Skp1p complex functions at a step after internalization. Moreover, Rsp5p, a HECT domain-containing E3 unrelated to SCF ligases, is required for ubiquitination of most internalized proteins at the plasma membrane (35). In mammals, it has been shown that ubiquitination of the growth hormone receptor is blocked in cells deficient for its internalization, suggesting that ubiquitination of the growth hormone receptor occurs in an internal compartment (44). Furthermore, it was recently proposed that ubiquitination of the epidermal growth factor receptor occurs in endosomal compartments and regulates lysosomal targeting of the receptor (26).
Mechanism of recycling.
Recycled proteins travel from an early, sorting endosome compartment back to plasma membrane, but it is not known whether they are first targeted to the Golgi and/or whether they can directly reach the plasma membrane. In yeast, Snc1p has been shown to recycle from endosomes to the Golgi, and FM4-64 efficiently labels late Golgi cisternae after short incubations, indicating that recycling through the Golgi may be a general process (27). In mammalian cells, the major part of transferrin receptors recycle directly from endosomal compartments to the plasma membrane without passing through the Golgi (42). Better characterization of the Rcy1p-positive compartment together with the identification of additional targets may clarify whether such a direct recycling mechanism also exists in yeast.
The molecular mechanism of recycling remains unknown but may be regulated by phosphorylation. The phosphorylation state of Snc1p correlates with its localization: Snc1p is hyperphosphorylated when located at the plasma membrane but becomes dephosphorylated after its internalization. Interestingly, we found that Snc1p is phosphorylated in a Yck1-2p-dependent manner, and at least Yck2p is concentrated at the plasma membrane in areas of polarized growth (34). Moreover, we identified Yck1p as a multicopy suppressor of the cold sensitivity of rcy1Δ cells (data not shown), consistent with a role for phosphorylation during recycling. Members of the casein kinase I family have previously been implicated in membrane trafficking; Yck1-2p-dependent phosphorylation of pheromone receptors and permeases at the plasma membrane triggers their ubiquitination, which is a signal for internalization (9, 13, 21, 29). Thus, phosphorylation of Snc1p at the plasma membrane may trigger its internalization, whereas dephosphorylation in an endosomal or Golgi compartment may be needed for recycling of the protein.
ACKNOWLEDGMENTS
We thank M. Lewis, H. Pelham, S. Elledge, and P. Silver for providing antibodies, plasmids, and strains. We are grateful to M. Blondel and W. Krek for helpful suggestions, to N. Perrinjaquet and P. Pagé for excellent technical assistance, to members of our laboratories for discussion, and to R. Iggo for critical reading of the manuscript. Special thanks go to S. Avaro and C. Lafourcade for help and advice during the early stages of this work.
J.-M.G. is supported by an EMBO postdoctoral fellowship; M.P. is supported by the Swiss National Science Foundation, the Swiss Cancer League, and a Helmut Horten Incentive Award.
REFERENCES
- 1.Amberg D C. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol Biol Cell. 1998;9:3259–3262. doi: 10.1091/mbc.9.12.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1991. [Google Scholar]
- 3.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 6.Blondel M, Alepuz P M, Huang L S, Shaham S, Ammerer G, Peter M. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 1999;13:2284–2300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondel M, Galan J M, Peter M. Isolation and characterization of HRT1 using a genetic screen for mutants unable to degrade Gic2p in Saccharomyces cerevisiae. Genetics. 2000;155:1033–1044. doi: 10.1093/genetics/155.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Davis N G. Recycling of the yeast a-factor receptor. J Cell Biol. 2000;151:731–738. doi: 10.1083/jcb.151.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decottignies A, Owsianik G, Ghislain M. Casein kinase I-dependent phosphorylation and stability of the yeast multidrug transporter Pdr5p. J Biol Chem. 1999;274:37139–37146. doi: 10.1074/jbc.274.52.37139. [DOI] [PubMed] [Google Scholar]
- 10.Deshaies R J. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies R J. SCF and cullin/ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 12.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Davis N G. Feedback phosphorylation of the yeast a-factor receptor requires activation of the downstream signaling pathway from G protein through mitogen-activated protein kinase. Mol Cell Biol. 2000;20:563–574. doi: 10.1128/mcb.20.2.563-574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galan J M, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geli I M, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press Inc.; 1991. [PubMed] [Google Scholar]
- 18.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 19.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicke L. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 21.Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holthuis J C, Nichols B J, Dhruvakumar S, Pelham H R. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holthuis J C M, Nichols B J, Pelham H R B. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol Biol Cell. 1998;9:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaquenoud M, Gulli M P, Peter K, Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W Y, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis J M, Nichols B J, Prescianotto-Baschong C, Riezman H, Pelham H R. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine S M, McKenzie III A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. Casein kinase I-dependent phosphorylation within a PEST sequence, and ubiquitination at nearby lysines, signal endocytosis of yeast uracil permease. J Biol Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- 30.Meimoun A, Holtzman T, Weissman Z, McBride H J, Stillman D J, Fink G R, Kornitzer D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 32.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peter M, Neiman A M, Park H O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson L C, Bradley C, Bryan J D, Jerome A, Kweon Y, Panek H R. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol Biol Cell. 1999;10:1077–1092. doi: 10.1091/mbc.10.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotin D, Staub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- 36.Rouillon A, Barbey R, Patton E E, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell I D, Grancell A S, Sorger P K. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt W K, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seol, J. H., A. Shevchenko, A. Shevchenko, and R. J. Deshaies. Prospecting the Skp1-anchored proteome by mass spectrometry identifies RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol., in press. [DOI] [PubMed]
- 41.Seron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel M O, Guillaud P, Devilliers G, Rossanese O W, Glick B S, Riezman H, Keranen S, Haguenauer-Tsapis R. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheff D R, Daro E A, Hull M, Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 44.Strous G J, van Kerkhof P, Govers R, Rotwein P, Schwartz A L. Growth hormone-induced signal transduction depends on an intact ubiquitin system. J Biol Chem. 1997;272:40–43. doi: 10.1074/jbc.272.1.40. [DOI] [PubMed] [Google Scholar]
- 45.Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 46.Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirbelauer C, Sutterlüty H, Blondel M, Gstaiger M, Peter M, Reymond F, Krek W. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 2000;19:5362–5375. doi: 10.1093/emboj/19.20.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wysocki R, Roganti T, Van Dyck E, de Kerchove D'Exaerde A, Foury F. Disruption and basic phenotypic analysis of 18 novel genes from the yeast Saccharomyces cerevisiae. Yeast. 1999;15:165–171. doi: 10.1002/(SICI)1097-0061(19990130)15:2<165::AID-YEA351>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 49.Zhang F L, Casey P J. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 50.Zhou P, Howley P M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 51.Ziman M, Chuang J S, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]