Abstract
The core structure of motile cilia and flagella, the axoneme, is built from a stable population of doublet microtubules. This unique stability is brought about, at least in part, by a network of microtubule inner proteins (MIPs) that are bound to the luminal side of the microtubule walls. Rib72A and Rib72B were identified as MIPs in the motile cilia of the protist Tetrahymena thermophila. Loss of these proteins leads to ciliary defects and loss of additional MIPs. We performed mass spectrometry coupled with proteomic analysis and bioinformatics to identify the MIPs lost in RIB72A/B knockout Tetrahymena axonemes. We identified a number of candidate MIPs and pursued one, Fap115, for functional characterization. We find that loss of Fap115 results in disrupted cell swimming and aberrant ciliary beating. Cryo-electron tomography reveals that Fap115 localizes to MIP6a in the A-tubule of the doublet microtubules. Overall, our results highlight the complex relationship between MIPs, ciliary structure, and ciliary function.
INTRODUCTION
Microtubules perform diverse essential functions within the cell, such as acting as tracks for movement of cargoes, forming the mitotic spindle, and serving as the major structural component of cilia/flagella, hereafter referred to as cilia. Functional differentiation of microtubules depends heavily on combinations of different posttranslational modifications as well as microtubule-associated proteins that bind the outside of microtubules (Janke, 2014; Bodakuntla et al., 2019).
Dynamic instability, that is, the ability of most native microtubules to rapidly grow and shrink, is essential for a number of microtubule functions. However, not all populations of microtubules are dynamic. Microtubules found in neuronal axons and the axonemes of cilia do not display highly dynamic properties. In the case of motile cilia axonemes, the microtubules are also resistant to the bending forces that cytoplasmic microtubules are susceptible to (Portran et al., 2017; Xu et al., 2017). This stability is critical for motile cilia function because these microtubules bend during the beating motion that moves cells or extracellular fluid. In humans, defects in ciliary motility result in a wide range of diseases including hydrocephaly, respiratory difficulty, and infertility (Wanner et al., 1996; Lyons et al., 2006; Sawamoto et al., 2006). Gaining an understanding of how motile cilia are assembled and stabilized is critical to understanding the pathology of these disorders.
The structure of axonemal microtubules may be responsible for their unique stability. Unlike cytoplasmic microtubules, the nine peripheral microtubules in axonemes of motile cilia are doublet microtubules that consist of a complete A-tubule with 13 protofilaments and an attached 10 protofilament B-tubule that shares a wall with the A-tubule (Figure 1A). Under physiological conditions, microtubules do not form doublets in vitro, which suggests that other components are necessary to assemble this modified microtubule structure.
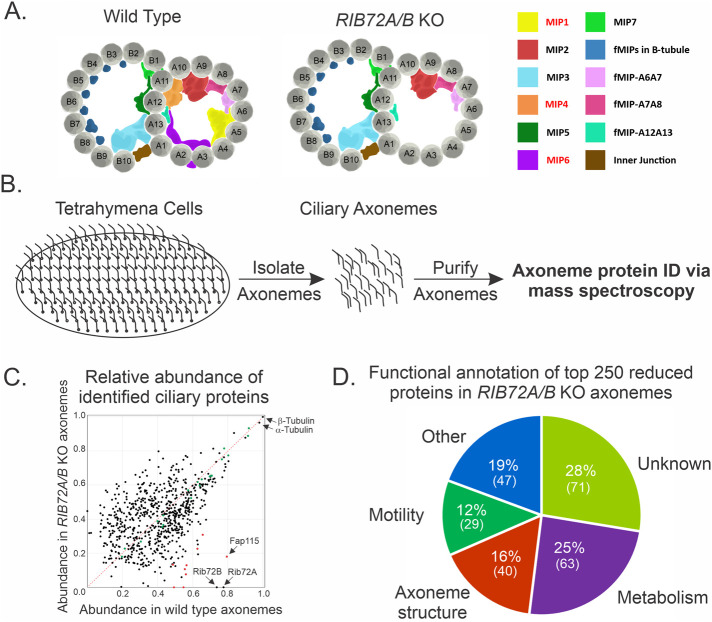
FIGURE 1:
Identification of proteins lost in RIB72A/B knockout Tetrahymena cells. (A) Schematic representation of ciliary doublet microtubules in WT (left) and RIB72A/B KO axonemes showing the loss of A-tubule MIPs. (B) Workflow for the identification of proteins lost in RIB72A/B KO doublet microtubules. Axonemes were isolated from wild-type, RIB72A KO, RIB72B KO, and RIB72A/B KO cells and analyzed by mass spectrometry. (C) Scatterplot showing normalized abundance of doublet microtubule proteins in WT vs. RIB72A/B KO axonemes. Black dots represent proteins and red dots represent candidate MIP proteins. Green dots represent dynein proteins that are not expected to change with the loss of RIB72 proteins. All proteins that fall under the red dashed line are reduced in RIB72A/B KO cells relative to wild-type cells. (D) Functional annotation of top 250 proteins lost in RIB72A/B KO cells based on domain analysis and known homologue characterization.
Cryo-electron tomography (Cryo-ET) studies have led to the discovery of a highly repetitive network of nontubulin densities, termed microtubule inner proteins (MIPs), that attach to the luminal side of doublet microtubule walls (Nicastro et al., 2006, 2011; Sui and Downing, 2006). MIPs have also been visualized in the triplet microtubules of centrioles and basal bodies at the base of ciliary axonemes (Li et al., 2012). Several recent studies have identified MIPs and potential MIP functions in ciliates and flagellates (Ichikawa et al., 2017; Kirima and Oiwa, 2018; Stoddard et al., 2018; Urbanska et al., 2018; Ma et al., 2019; Owa et al., 2019; Khalifa et al., 2020). In addition, recent single-particle cryo-electron microcopy studies have revealed higher resolution structures of doublet microtubules in Chlamydomonas and Tetrahymena cilia, providing a strong basis for identification and characterization of MIPs (Ma et al., 2019; Song et al., 2020). While some studies have provided clues to the functional significance, for example, microtubule lattice spacing angles and intolerance to mechanical stress (Ichikawa et al., 2017, 2019; Owa et al., 2019), there is still a need for additional information about the roles of individual MIPs and MIPs as a group. In addition, some MIP proteins have yet to be identified and characterized, and MIPs have been shown to vary between species and systems.
We previously characterized the MIPs Rib72A and Rib72B in the ciliate Tetrahymena thermophila (Stoddard et al., 2018). We found that Rib72 is necessary for the localization of a majority of the MIPs found in the A-tubule of doublet microtubules, and that loss of Rib72 proteins results in reduced ciliary beating function and doublet microtubule stability. Here, we report our proteomics- and bioinformatics-based approach to identify the MIPs lost in RIB72 knockout (KO) Tetrahymena cells. We follow this up with functional characterization of a single MIP candidate, Fap115. Through the use of cryo-ET, we identify that Fap115 is a component of MIP6a, a previously identified structure in the A-tubule of doublet microtubules (Stoddard et al., 2018). Furthermore, we find that the power stroke of motile cilia beating is slowed in FAP115 KO cells. Overall, our work helps us begin to understand the complex network of MIPs within doublet microtubules and importantly, demonstrates an experimental pipeline that will allow for the functional characterization of other candidate MIPs. These steps are essential for understanding the molecular mechanisms of stable cilia assembly.
RESULTS AND DISCUSSION
Rib72 phylogeny
Our previous study identified Tetrahymena Rib72A and Rib72B as MIPs and showed that they are necessary for the localization of a majority of A-tubule MIPs (Figure 1A; Stoddard et al., 2018). To better understand whether Rib72A and Rib72B (and associated A-tubule MIPs) play a role in the formation of motile cilia, we queried a wide panel of eukaryotes (both containing and not containing motile cilia) for the presence of Rib72 homologues (Figure 2). We find that a majority (16 of 21) of species that have motile cilia also have a homologue for Rib72. Almost every species that does not contain motile cilia correspondingly does not have Rib72. One exception to this finding is Caenorhabditis elegans. Interestingly, while C. elegans does not contain motile cilia, they do contain sensory cilia, suggesting that sensory cilia may contain MIPs (Inglis et al., 2018). Given the nonoverlapping roles of Rib72A and Rib72B in Tetrahymena, we also wanted to assess whether other species had two Rib72 proteins. We note that the majority of motile cilia-containing species that have Rib72A (9 of 14) do not have a homologue for Tetrahymena Rib72B. Overall, our findings highlight that Rib72 is highly conserved among species with motile cilia, suggesting an important role in motile cilia formation and/or function, whereas Rib72 was absent in organisms without motile cilia.
FIGURE 2:
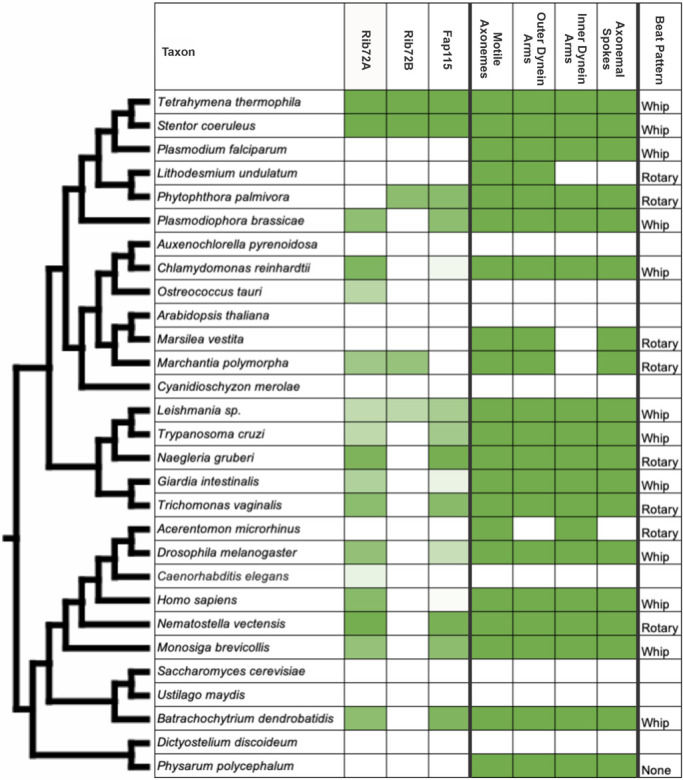
Homology assessment and motile cilia molecular components among a panel of eukaryotes. Homology for Rib72A, Rib72B and Fap115 is based on BLASTp results using Tetrahymena thermophila as the query. The degree of green shading is based on the E-value (log10 scaled) ranging from the lightest green (Caenorhabditis elegans E-value = 5.0 × 10–19) to the darkest green (Stentor coeruleus E-value = 1 × 10–120). The presence of motile cilia structural components for each organism is found on the right and indicated by solid green shading. Beat patterns are briefly described (except where beat pattern was unavailable = blank).
Proteomics/analysis
Determining the molecular identity of MIPs is essential for our understanding of cilia structure. Because the loss of both Rib72A and Rib72B in Tetrahymena leads to loss of a majority of MIP densities within the A-tubule (Figure 1A), we sought to identify these missing MIPs using a proteomics-based approach. To do this, we first isolated ciliary axonemes from wild-type (WT), RIB72A KO, RIB72B KO, and RIB72A/B KO Tetrahymena strains. We then performed mass spectrometry on these axonemes and calculated the relative abundance of each protein identified in each population of ciliary axoneme (Figure 1B). A ratio comparison between RIB72 single and double KO axonemes and WT axonemes reveals which proteins are lost in RIB72 KO cells relative to WT cells (SupplementalTable S1). We next shifted our attention to the double knockout because of the more severe loss of A-tubule MIPs relative to single knockouts (Stoddard et al., 2018). The 1000 most abundant proteins in WT cells were plotted based on their relative abundance in WT axonemes vsversus RIB72A/B KO axonemes (Figure 1C). The top 250 most reduced proteins from this list were then annotated for predicted functional role (Figure 1D). Forty proteins were predicted to associate with microtubules or have homologues that function at the cilia/flagella axonemes. These candidates were considered potential structural proteins of the cilia (Figure 1D). We applied various bioinformatics approaches, including reciprocal best-hit BLAST analysis, domain analysis, comparisons with previously published basal body and ciliary proteomes, and findings from the recent Chlamydomonas MIP map (Keller et al., 2005; Kilburn et al., 2007; Ma et al., 2019) to remove proteins that are unlikely to be Tetrahymena MIPs and to narrow the list to a reasonable number for functional analysis. The identity of the candidates based on available information is shown in Table 1. When comparing our MIP candidate’s protein levels in single RIB72A or RIB72B KO strains versus double RIB72 KO strains, we find that almost every MIP candidate had a greater loss in the double KO than in either single KO alone, suggestive of a combinatorial effect of losing both Rib72 proteins (Table 1).
TABLE 1:
Candidate Tetrahymena MIPs.
| Tetrahymena MIP candidate proteins | |||||
|---|---|---|---|---|---|
| Gene Name | Gene | Protein ID | Ratio of protein levels in RIB72A/BΔ cilia versus wild-type cilia | Ratio of protein levels in RIB72AΔ cilia versus wild-type cilia | Ratio of protein levels in RIB72BΔ cilia versus wild-type cilia |
| Rib72A | TTHERM_00143690 | I7M0S7 | 0.00 | 0.00 | 0.85 |
| Rib72B | TTHERM_00584850 | I7MCU1 | 0.00 | 0.98 | 0.00 |
| Fap107 | TTHERM_00320040 | Q237T1 | 0.00 | 0.70 | 0.00 |
| CAPS | TTHERM_00691900 | I7LT67 | 0.00 | 0.00 | 0.00 |
| Fap129 | TTHERM_00565590 | I7M9I4 | 0.14 | 0.80 | 0.76 |
| Fap143 | TTHERM_00502359 | A4VD56 | 0.19 | 0.77 | 0.54 |
| Fap115 | TTHERM_00193760 | Q23KF9 | 0.23 | 0.86 | 0.68 |
| Fap210a | TTHERM_00643530 | Q23EX8 | 0.23 | 0.90 | 0.52 |
| Rib43A | TTHERM_00624660 | Q240R7 | 0.29 | 0.77 | 0.56 |
| Fap106a | TTHERM_00137550 | I7M279 | 0.36 | 0.81 | 0.76 |
| Fap45a | TTHERM_001164064 | W7XCX2 | 0.47 | 0.89 | 0.67 |
aPredicted B-tubule MIPs.
Loss of Rib72A/B may not only affect other A-tubule MIPs, but also the localization of B-tubule MIPs. In our proteomic analysis, we find a number of B-tubule MIPs to be reduced in RIB72A/B KO Tetrahymena cells. This raises the intriguing possibility that proper assembly of the B-tubule is dependent on the A-tubule MIP network. Some MIPs have been shown to reach through the tubulin lattice and it is possible that projections into the B-tubule could serve as a platform for B-tubule–specific MIP localization (Ichikawa et al., 2017; Ma et al., 2019). A-tubule MIPs also make connections with dynein arms and radial spoke proteins (Ichikawa et al., 2019; Khalifa et al., 2020). A cursory look at whether the abundance of dyneins from our mass spectrometry screen are affected by loss of RIB72 reveals that dynein loss is negligible and none come close to the levels of loss seen in our MIP candidates, suggesting that dyneins may rely on other structural components for localization (Figure 1C). Though dyneins may still localize in the absence of many A-tubule MIPs it is not certain whether they are able to form the types of connections necessary to power the movement of ciliary axoneme as the loss of Rib72 does result in ciliary beating defects (Stoddard et al., 2018). Overall, the intricacies of MIP interactions across the A- and B-tubules, along with connections to dyneins and radial spoke proteins is intriguing and should be further investigated.
One of the MIP candidates, Fap115, was of particular interest for a number of reasons. Fap115 shows a significant reduction (77%) in abundance in RIB72 double KO cilia relative to WT cilia. Fap115 was also previously identified as a component of the Tetrahymena basal body (Kilburn et al., 2007), and a recent study confirmed that Chlamydomonas Fap115 is present inside the A-tubule (Ma et al., 2019). Additionally, we find that 100% of species with motile cilia that contain Rib72 also contain Fap115 (Figure 2), suggesting an essential interaction or Rib72-dependent function. Overall, our proteomics approach was successful in generating a list of candidate Tetrahymena MIPs from which Fap115 was chosen as a protein of most immediate interest and for further analysis.
Fap115 is necessary for ciliary beating
Fap115 in Tetrahymena is a ∼110 kDa, EF-hand domain-containing protein and was one of the original MIP candidates found in earlier 2D gel screens of salt-extracted Chlamydomonas axonemes in the Nicastro lab (unpublished data). Recently, a new and extensive Chlamydomonas mutant library was generated (Li et al., 2016), containing a FAP115 mutant. After PCR verification of the Chlamydomonas mutant, we performed cryo-ET of isolated FAP115 mutant axonemes. The 96-nm axonemal averages revealed that the MIP6a density that is found along protofilament 2 of the A-tubule (PFA2) in WT axonemes was missing in the FAP115 mutant, whereas all other MIP subunits were present (Supplemental Figure S2). This is consistent with the localization of FAP115 in Chlamydomonas by Ma et al. (2019), which used our previous subtomogram averages of RIB72 KOs (Stoddard et al. 2018) for validation of their near-atomic models.
Before analyzing the function of Fap115 in Tetrahymena, we first confirmed its localization in Tetrahymena cilia and basal bodies. For these analyses we generated two strains: a strain expressing an mCherry C-terminal fusion of Fap115 confirmed Fap115 localization to cilia and basal bodies (Figure 3A), and a Tetrahymena knockout strain of FAP115, which was confirmed by RT-PCR, was used for cryo-ET and to analyze the potential role of Fap115 as a MIP in stability or motility function of the ciliary axoneme.
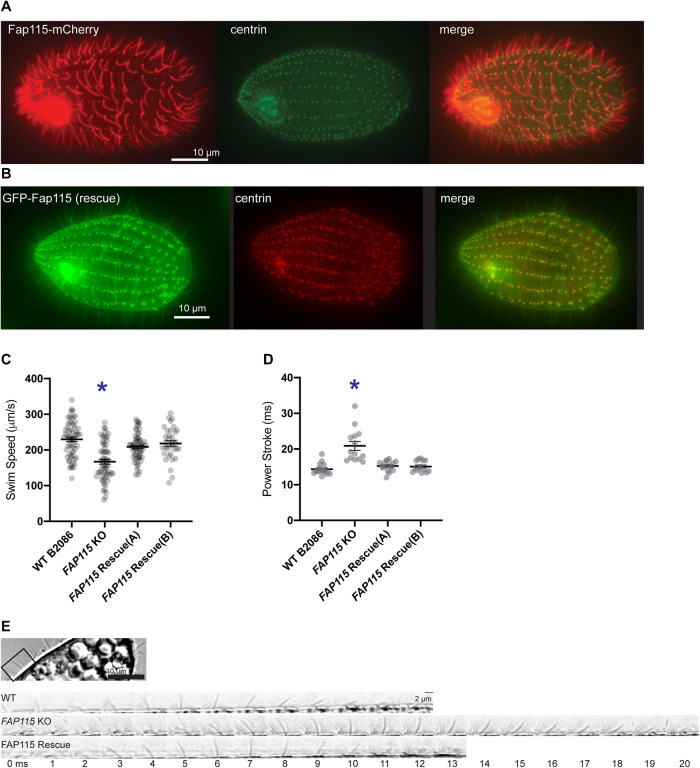
FIGURE 3:
Fap115 is a ciliary protein. (A) Fap115-mCherry localizes to cilia and colocalizes with the basal body protein centrin (antibody staining). (B) GFP-Fap115 localizes to cilia and basal bodies in the rescue of FAP115 KO. (C) Swim speed measurements in wild-type (WT), FAP115 KO, and FAP115 rescue. (D) Duration of individual cilia power strokes measured in wild-type (WT), FAP115 KO, and FAP115 rescue strains based on images such as those shown in E. (E) Sample image of a Tetrahymena cell, showing identification of a single cilium (top) and single frames from time-lapse images of cilia power strokes measured in WT, FAP115 KO, and FAP115 rescue strains (bottom). *, p < 0.01; error bars = SEM.
Ciliary microtubules must be built to resist mechanical strain during ciliary beating (Portran et al., 2017; Xu et al., 2017). It has been shown that MIPs play a role in providing stability to the axoneme, because loss of Rib72 renders axonemes unusually flexible during the typically rigid force generating power stroke (Stoddard et al., 2018). Additionally, loss of MIPs in Chlamydomonas axonemes caused a stark decrease in structural stability of doublet microtubules (Owa et al., 2019).
Tetrahymena FAP115 KO cells showed a decreased swim speed when compared with WT cells (Figure 3C). Swim speed was restored to WT level with the reintroduction of Fap115 to the FAP115 KO Tetrahymena strain (GFP-Fap115 rescue; Figure 3, B and C, and Supplemental Figure S4). To assess the possible cause of the decreased swim speed, we analyzed the ciliary density of the FAP115 KO. However, ciliary density was not altered (Supplemental Figure S3A), so we performed high-speed imaging to evaluate the ciliary beat pattern, because a defect in the shape, frequency, or coordination of ciliary beating could affect swim speed. High-speed time-lapse images of free-swimming cells were recorded, and movement of individual cilia was analyzed. There was no clear abnormal deformation of the cilia during the power stroke, as was previously seen in RIB72 KO cilia (Stoddard et al. 2018). However, the duration of the power stroke was elongated in the FAP115 KO (Figure 3, D and E), albeit the swim speed and power stroke defects in FAP115 KO were not as pronounced as the defects previously seen in RIB72A, RIB72B, and RIB72A/B double KO cells (Stoddard et al., 2018). Mass spectrometry of FAP115 KO axonemes shows that MIP proteins reduced or missing in RIB72 KOs are not greatly reduced in FAP115 KO axonemes (Supplemental Figure S3D), so it is not surprising that we do not see a more severe defect. FAP115 KO Tetrahymena cells also did not differ from wild type in growth rate or phagocytosis (Supplemental Figure S3, B and C).
Because the cilia in the FAP115 KO are not severely disrupted, it is likely that either A-tubule MIPs besides Fap115 are individually necessary for stabilization of the axoneme or that the collective connection of A-tubule MIPs works together to stabilize the axoneme. Much like we see when analyzing ciliary beating defects, it would be useful in the future to determine whether there are specific axoneme-stabilizing MIPs or whether the stability breaks down suddenly after loss of a number of MIPs. This would suggest a concerted function for the stability of cilia. Our findings do show that there are gradations of defects caused by losing individual MIPs and the entire system is not broken by the loss of a single MIP. In future studies, it will be important to determine the exact contributions of each MIP or subset of MIPs to ciliary beating efficiency.
Fap115 localizes to the A-tubule in Tetrahymena cilia
We determined the location of Tetrahymena Fap115 using cryo-ET and subtomogram averaging of isolated FAP115 KO axonemes. Our averages showed complete loss of a single bilobed MIP density, MIP6c, that in WT axonemes has 8 nm longitudinal periodicity and connects A-tubule protofilaments A2 and A3 (Figure 4, A–E). The loss of a single density in the FAP115 KO, as compared with loss of multiple MIP densities in the RIB72A or RIB72B KO axonemes (Stoddard et al., 2018), is consistent with the more subtle ciliary phenotypes seen in the FAP115 KO. These experiments demonstrate that Fap115 localizes to PFA2-3 in the A-tubule, presumably interacting with Rib72A/B. The loss of the Fap115 density was also observed previously in the RIB72A or RIB72B KO (Stoddard et al., 2018), suggesting the dependence of Rib72A/B for the recruitment of Fap115 at PFA2 and PFA3 during the cilia assembly.
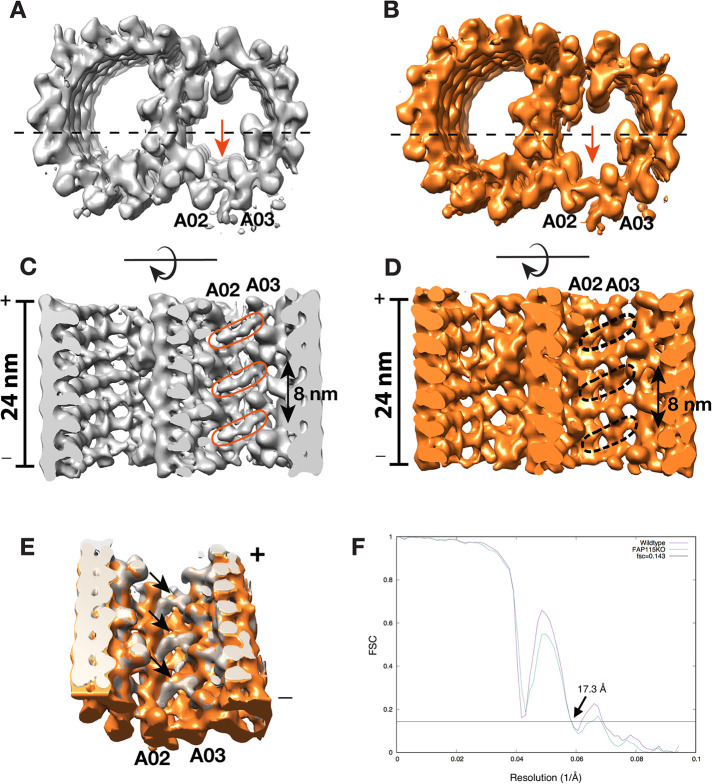
FIGURE 4:
Cryo-ET and subtomogram average of the axonemal doublet microtubule from Tetrahymena wild-type and FAP115 KO. (A, B) Longitudinal view of the averages. Wild-type (gray) and FAP115 KO (orange) are viewed from the minus ends of the doublet. The red arrows indicate the missing density in the KO axoneme that is present in the wild type. The dashed lines indicate the cross-section direction in C and D. In C, the red elliptical circles highlight the density likely attributed to Fap115, the equivalent positions in the mutant doublet in D, where the density is missing, are indicated by a black dashed circle. (E) Overlay of two averages. The black arrows indicate the likely positions of Fap115 in the wild-type structure. They show 8 nm periodicity. (F) Both structures are at 17.3 Å, determined by the FSC at 0.143 cutoff. The dips in certain frequency in the FSC curves are due to a narrow defocus range, 2.3–3.0 µm, used during the data collection. In both wild-type and FAP115 KO averages, the longitudinal segment length is 24 nm, including one 16-nm repeat of Rib72A/B. Doublet microtubules (DMTs) were averaged based on 16-nm repeat of Rib72A/B.
Across phylogeny, the presence of MIP homologues in motile cilia-containing organisms suggests high conservation of MIP structural architecture. In the two systems with the most extensively characterized MIPs, Chlamydomonas and Tetrahymena, the structures and organization of MIPs appear to be highly similar (Ichikawa et al., 2017; Ma et al., 2019). Dependence of Fap115 localization on Rib72A/B is consistent with high-resolution structures in Chlamydomonas showing interactions between Rib72 and Fap115 (Ma et al., 2019). It should be noted that Fap115 in Tetrahymena is four times the size of Fap115 in Chlamydomonas. This larger size is similar to Fap115 homologues in Giardia and Paramecium. Because Fap115 is made up of multiple EF-hand domains combined by linkers, it is conceivable that the EF-hands in these proteins function and localize in a similar manner. It is also possible that cilia in other organisms have multiple proteins that share the role of a single MIP in Tetrahymena.
Much of the work on stability of the axoneme will be tied to how axonemes are assembled and the timing of MIP integration into the axoneme. Axonemal microtubules assemble an order of magnitude slower than cytoplasmic microtubules and it is probable that MIPs are being incorporated into doublet microtubules coincident with tubulin addition to the axoneme (Sánchez and Dynlacht, 2016). The absence of Fap115 in the RIB72 KO cells suggests a sequential and hierarchical assembly of MIPs. Rib72A/B are still present in the FAP115 KO, but Fap115, along with most other A-tubule MIPs, is absent in RIB72A/B KOs. There is evidence that local curvature of the doublet microtubules is affected by MIP structures (Ichikawa et al., 2017, 2019; Khalifa et al., 2020), but which proteins create the curvature and how it may affect recruitment and binding of additional MIPs is yet to be understood. Structural specificity of microtubules and direct binding to tubulin or other MIPs are likely mechanisms for early assembly of the MIP architectures.
Many questions about MIPs remain, and future research in the field is necessary to reveal the identities, structures, and functions of the network of MIPs.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Tetrahymena strains
The WT (B2086.2) Tetrahymena strain was obtained from the Tetrahymena Stock Center (Cornell University, Ithaca, NY). All strains were grown in the standard medium 2% SPP (2% proteose peptone, 0.2% glucose, 0.1% yeast extract, and 33 µM FeCl3) at 30°C. RIB72A/B single and double KO strains are described in Stoddard et al. (2018).
Endogenous macronuclear FAP115 (Ttherm_00193760) in B2086.2 cells was tagged with a C-terminal mCherry from the 4T2-1 vector containing C-terminal FAP115 sequence and downstream sequence as homology for recombination.
The FAP115 macronuclear knockout was generated by replacing the macronuclear FAP115 gene in B2086.2 with the fragment of 4T2-1, containing mCherry and paromomycin resistance, using 0.5-1 kb homology from upstream and downstream regions of the gene. Tetrahymena were transformed with particle bombardment (Cassidy-Hanley et al., 1997; Hai and Gorovsky, 1997), then transformants were identified by resistance to paromomycin. For the FAP115 KO, paromomycin concentrations were increased until the cells were fully assorted as knockouts, confirmed with RT-PCR. We used two independently assorted FAP115 KO strains in our analyses.
To generate an exogenous expression rescue strain (with GFP-tagged Fap115), FAP115 cDNA amplified from WT Tetrahymena cells was cloned into a plasmid containing RPL29 homology arms (pBS-MTT-GFP-gtw; see Stoddard et al., 2018). The construct was linearized and introduced to FAP115 KO Tetrahymena cells by biolistic bombardment. Two independently assorted cycloheximide-resistant strains were saved and used for analysis. Induction using CdCl2 (0.5 µg/ml) was between 1 and 24 h, as noted.
Proteomics
Tetrahymena ciliary axonemes were isolated from WT (B2086.2), single knockout (RIB72A or RIB72B), and double knockout (RIB72A/RIB72B) strains via calcium shock (Rosenbaum and Carlson, 1969). These axonemes were pelleted by centrifugation, then solubilized, reduced, and alkylated with 4% (wt/vol) sodium dodecyl sulfate, 10 mM Tris (2-carboxyethyl) phosphine hydrochloride, 40 mM chloroacetamide, 100 mM Tris, pH 8.5, and boiled at 95°C for 10 min. Axonemes were then sonicated with a probe sonicator (Branson) for 1 s on, then 1 s off for a total of 1 min. Approximately 100 µg of total protein were processed using the filter-aided sample preparation method (Wis´niewski, 2016). Briefly, samples were diluted 10-fold with 8 M urea, 0.1 M Tris, pH 8.5, and applied to the top of an Amicon Ultra 0.5 ml, 30 kD NMWL cutoff (Millipore) ultrafiltration device. Samples were washed in the devices three times with 8 M urea, 0.1 M Tris, pH 8.5, and another three times with 2 M urea, 0.1 M Tris, pH 8.5. Endoproteinase Lys-C (Wako) was added first at a 1:100 protease to protein ratio, then incubated ∼2 h on a nutator at room temperature. Trypsin (Pierce) was added next also at 1:100, then incubated overnight again on a nutator at room temperature. Tryptic peptides were eluted via centrifugation and desalted using a C-18 spin column (Pierce) according to the manufacturer’s instructions. Samples were suspended in 3% (vol/vol) acetonitrile/0.1% (vol/vol) trifluoroacetic acid for 0.5 µg/µl peptide by A280 nm, and 1 µl was directly injected onto a C18 1.7 µm, 130 Å, 75 µm × 250 mm M-class column (Waters), using a Waters M-class UPLC. Peptides were gradient eluted at 300 nl/min from 3% to 20% acetonitrile over 100 min into an Orbitrap Fusion mass spectrometer (Thermo Scientific). Precursor mass spectra (MS1) were acquired at a resolution of 120,000 from 380 to 1500 m/z with an AGC target of 2E5 and a maximum injection time of 50 ms. Dynamic exclusion was set for 20 s with a mass tolerance of ± 10 ppm. Precursor peptide ion isolation width for MS2 fragment scans was set at 1.6 Da using the quadrupole, and the most intense ions were sequenced by Top Speed with a 3-s cycle time. All MS2 sequencing was performed using higher energy collision dissociation at 35% collision energy and scanned out in the linear ion trap. An AGC target of 1E4 and 35 s maximum injection time was used. Orbitrap data raw files were searched against the Uniprot T. thermophila database (05-04-2020) using Maxquant with cysteine carbamidomethylation as a fixed modification and methionine oxidation and protein N-terminal acetylation as variable modifications. All peptides and proteins were thresholded at a 1% false discovery rate. The ratio of reduction in Rib72 KO axonemes versus WT axonemes was determined by first calculating the relative protein abundance in each dataset (WT, RIB72A KO, RIB72B KO, RIB72A/B KO). Then, the ratio of the abundance in the RIB72 KO axonemes to WT abundance was calculated for each individual protein.
Homology assessment of Rib72A/B and Fap115
Analysis of MIP conservation was performed using existing genetic data from a diverse taxonomic sample composed of 29 different model organisms (21 with motile cilia and 8 without). Taxon sampling was adapted from previous phylogenetic analysis of microtubule-based organelles (Carvalho-Santos et al., 2010, 2011). Our phylogenetic tree represented in Figure 2 was updated by using Genbank’s Common Tree and resolved using available literature and Open Tree of Life (opentreeoflife.org). Individual branches were confirmed by PhyloT (https://phylot.biobyte.de). Only model organisms with accessible whole genome sequences were considered for analysis to be confident about homology assessment. Amino acid sequences for Rib72A/B and Fap115 were taken from the Tetrahymena Genome Database (Ciliate.org). Protein homology was determined by BLASTp search across the nonredundant protein sequence database (nr), including the respective model organism identifier in the search set using the BLASTp algorithm with default parameters. A reciprocal best-hit BLAST search was done for the top search result in each species to determine whether the protein of interest is also the best hit in the T. thermophila genome and to discriminate between orthologues and paralogues (Tatusov et al., 1997; Bork et al., 1998). To do this, the highest scoring accession was then searched in BLAST using the same settings but modifying the search set to only include T. thermophila as the organism. If the protein of interest was the best hit in the reciprocal search, then it was considered homologous. Proteins in the taxon sampling were confirmed to be homologous if the protein BLAST yielded a result with an E-value less than or equal to 0.001 and had a positive reciprocal best-hit BLAST to the protein of interest in T. thermophila.
Fluorescence microscopy
Cells were fixed and imaged as described previously (Stoddard et al., 2018). Briefly, log-phase cells were pelleted, then resuspended in a 3% formaldehyde solution. Cells were incubated in primary α-TtCen1 (Stemm-Wolf et al., 2005) antibody at 4°C overnight and secondary antibody (Alexa 488 or 594 anti-rabbit) for 1 h at room temperature. Antibodies were diluted 1:1000 in phosphate-buffered saline (PBS) + 1% bovine serum albumen (BSA). Cells were washed three times with PBS + 0.1% BSA after each antibody incubation. Cells were mounted in Citifluor (EMS, Hatfield, PA) for viewing. Projection images were generated using ImageJ software.
Swim speed assay
Tetrahymena cells were grown at 30°C in SPP media (2% protease peptone, 0.1% yeast extract, 0.2% glucose, 0.003% FeCl3) to midlog phase, 2 × 105 cells/ml. Cells were then placed on a slide and imaged using differential interference contrast (DIC) imaging with a 20× objective at 10 frames/s on a Nikon Ti microscope (Nikon, Japan) using NIS Elements software (Nikon). ImageJ with the ImageJ software plugin MTrackJ (E. Meijering, Biomedical Imaging Group of Rotterdam, Netherlands) was used to track and quantify the movement of Tetrahymena cells. Twenty cells were quantified per condition, and the experiment was performed in triplicate.
Cilia wave beating analysis
Log-phase Tetrahymena cells were place in a drop on a cover glass, then DIC images of free-swimming cells (above the plane of the cover glass) were captured at 996 frames/s with a 100× NA, 1.45 oil immersion objective lens on a Nikon Ti-E microscope (Nikon, Japan) with a Hamamatsu Flash4.0 V3 camera fitted with a V3 Firebird Camlink Board, using Nikon Elements software. Images were analyzed using ImageJ. Multiple cilia from four to five cells per strain were analyzed in each experiment. This experiment was completed in triplicate.
Cell growth assay
WT and FAP115 KO Tetrahymena cells were grown at 30°C in 2% SPP medium to a concentration of 2 × 104 cells/ml, as measured by a hemocytometer. Cell density was quantified by hemocytometer every 2 h for 8 h, then at 24 h. This experiment was completed in duplicate.
Phagocytosis assay
Tetrahymena cells were incubated with 0.5% India ink in 1% SPP media at 25°C. At 30 min after the India ink was added, cells were washed in 2% SPP and fixed with 3% formaldehyde. Cells were mounted on slides and the average number of food vacuoles per cell was quantified (n = 20 cells) using phase contrast imaging with a 10× objective on an upright light microscope (Nikon Ti, Japan). Twenty cells per condition were quantified for each experiment. This experiment was completed in duplicate.
Cryo sample preparation and cryo-ET
Cryo-ET and image processing of isolated axonemes from Chlamydomonas WT and fap115 mutant were performed as reported previously (Stoddard et al., 2018; Gui et al., 2019, respectively). The fap115 mutant strain was obtained from the CLiP collection (CLiP ID: LMJ.RY0402.105592; Li et al., 2016).
WT data were recorded on a Tecnai F30 transmission electron microscope (TEM) using a charge-coupled device camera, whereas the fap115 KO mutant data were acquired on a Titan Krios TEM using a K2 direct electron detector. Chlamydomonas data shown are averages of the 96 nm axonemal repeat.
For cryo-ET on Tetrahymena axonemes, 10 nm colloid gold (Sigma) was mixed with ciliary axonemes isolated from the WT or FAP115 KO. A 4 µl sample was applied onto Quantifoil grid Rh/Cu 200 R2/2 (Quantifoil), and was plunge frozen in liquid ethane using a Vitrobot (FEI).
Tomography tilt series were collected on a field emission gun microscope (Titan Krios, FEI) operated at 300 kV. The microscope was equipped with a postcolumn energy filter Bio-Quantum GIF (Gatan). The slit width was set at 25 eV. SerialEM (Mastronarde, 2005) was used for the tomography tilt series collection. Images were recorded on a K3 direct electron detector (Gatan) in the superresolution and dose-fractionation mode. The nominal magnification was 33,000×, resulting in a pixel size of 2.65 Å. The specimen was tilted in 2° increments in a bidirectional scheme, that is, starting from 0° to −60° and then from 0° to +60°. The total accumulative dose on the sample was limited to 80 electron/Å2.
For tomogram reconstruction and subtomogram averaging, the dose-fractionated movies at each tilt were motion-corrected and summed using MotionCor2 (Zheng et al., 2017). The tilt series images were aligned based on the gold fiducials using IMOD (Kremer et al., 1996) and TomoAlign (Fernandez et al., 2018). The contrast transfer function for each tilt series was determined and corrected by TomoCTF (Fernandez et al., 2006). The tomograms were reconstructed by TomoRec (Fernandez et al., 2019) that took into account the beam-induced sample motion during data collection. Respectively, for the WT, 5235 subtomograms from 15 datasets, for the FAP115 KO construct, 5926 subtomograms from 18 datasets were used for the subtomogram averaging. The subtomogram alignment and average were carried out, without using any external reference, by the program MLTOMO implemented in the Xmipp software package (Scheres et al., 2009). Extensive classification of subtomogram was carried out in order to find the previously described 16 nm periodicity of Rib72A/B. The subtomograms were shifted accordingly in the longitudinal direction of the axoneme so that these MIPs are in-register. This was followed by additional alignment in a “gold-standard” scheme in 2×-binned format (effective pixel size 5.30 Å) (Scheres and Chen, 2012). The final resolution, based on the Fourier Shell Correlation (FSC) cutoff at 0.143, is reported as 17.3 Å for both the WT and the FAP115 KO structure (Figure 4F). The structures have been deposited in Electron Microscopy Data Bank with access codes EMD-22712 (B2086, WT) and EMD-22713 (FAP115 KO).
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health (NIH) Grant no. R01GM-127571 (PI—M.W.), Grant no. R01GM-083122 (PI—D.N.), and Grant no. R35GM-118099 (PI—D.A.), and the Cancer Prevention and Research Institute of Texas RR140082 (PI—D.N.). Cryo-ET sample preparation was performed by Fei Guo at the University of California, Davis BioEM facility, which is supported by the user fees, the Department of Molecular and Cellular Biology (MCB), the College of Biological Sciences, the Office of Research, and the Provost’s Office. Technical Director, Fei Guo, is supported by discretionary funds provided by Jodi Nunnari (MCB). Cryo-ET data were collected at University of California, San Francisco (UCSF), the Louise Mashal Gabbay Cellular Visualization Facility at Brandeis University (Waltham, MA) and the UT Southwestern Cryo-Electron Microscopy Facility that is supported in part by Cancer Prevention and Research Institute of Texas Core Facility Support Award RP170644. We thank David Bulkley and Glenn Gilbert in the Bay Area CryoEM Center at UCSF for technical support. This center is supported in part by NIH grants including Grant no. 1S10OD-020054 and Grant no. 1S10OD-021741. We also thank Daryn Baker and Steve Hinds for maintaining computational infrastructure at Santa Clara University to allow for assessment of homology. We thank Nhan Phan (UT Southwestern) for biochemical assistance with the Chlamydomonas fap115 mutant. We thank Rachel Howard-Till and Usha Kar for critical review.
Abbreviation used:
- MIP
microtubule inner protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-12-0786).
REFERENCES
- Bodakuntla S, Jijumon AS, Villablanca C, Gonzalez-Billault C, Janke C (2019). Microtubule-associated proteins: structuring the cytoskeleton. Trends Cell Biol. 29, 804–819. [DOI] [PubMed] [Google Scholar]
- Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y (1998). Predicting function: from genes to genomes and back. J Mol Biol 283, 707–725. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M (2011). Evolution: tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M (2010). Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123, 1414–1426. [DOI] [PubMed] [Google Scholar]
- Fernandez J-J, Li S, Agard DA (2019). Consideration of sample motion in cryo-tomography based on alignment residual interpolation. J Struct Biol 205, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J-J, Li S, Crowther RA (2006). CTF determination and correction in electron cryotomography. Ultramicroscopy 106, 587–596. [DOI] [PubMed] [Google Scholar]
- Fernandez J-J, Li S, Bharat TAM, Agard DA (2018). Cryo-tomography tilt-series alignment with consideration of the beam-induced sample motion. J Struct Biol 202, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Song K, Tritschler D, Bower R, Yan S, Dai A, Augspurger K, Sakizadeh J, Grzemska M, Ni T, et al. (2019). Scaffold subunits support associated subunit assembly in the Chlamydomonas ciliary nexin-dynein regulatory complex. Proc Natl Acad Sci USA 116, 23152–23162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Gorovsky MA (1997). Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc Natl Acad Sci USA 94, 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Khalifa AAZ, Kubo S, Dai D, Basu K, Maghrebi MAF, Vargas J, Bui KH (2019). Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins. Proc Natl Acad Sci USA 116, 19930–19938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Liu D, Kastritis PL, Basu K, Hsu TC, Yang S, Bui KH (2017). Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat Commun 8, 15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Ou G, Leroux MR, Scholey JM (2018). The sensory cilia of Caenorhabditis elegans (WormBook). [DOI] [PMC free article] [PubMed]
- Janke C (2014). The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol 206, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF (2005). Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol CB 15, 1090–1098. [DOI] [PubMed] [Google Scholar]
- Khalifa AAZ, Ichikawa M, Dai D, Kubo S, Black CS, Peri K, McAlear TS, Veyron S, Yang SK, Vargas J, et al. (2020). The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications. ELife 9, e52760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Culver BP, Yates JR, Winey M (2007). New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol 178, 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirima J, Oiwa K (2018). Flagellar-associated protein FAP85 is a microtubule inner protein that stabilizes microtubules. Cell Struct Funct 43, 1–14. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez J-J, Marshall WF, Agard DA (2012). Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J 31, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. The Plant Cell 28, 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons RA, Saridogan E, Djahanbakhch O (2006). The reproductive significance of human fallopian tube cilia. Hum Reprod Update 12, 363–372. [DOI] [PubMed] [Google Scholar]
- Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, Zhang R (2019). Structure of the decorated ciliary doublet microtubule. Cell 179, 909–922.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW (2011). Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci USA 108, E845–E853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948. [DOI] [PubMed] [Google Scholar]
- Owa M, Uchihashi T, Yanagisawa H-A, Yamano T, Iguchi H, Fukuzawa H, Wakabayashi K-I, Ando T, Kikkawa M (2019). Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun 10, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Schaedel L, Xu Z, Théry M, Nachury MV (2017). Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol 19, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Carlson K (1969). Cilia regeneration in Tetrahymena and its inhibition by colchicine. J Cell Biol 40, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez I, Dynlacht BD (2016). Cilium assembly and disassembly. Nat Cell Biol 18, 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, Spassky N, Murcia NS, Garcia-Verdugo JM, Marin O, Rubenstein JLR, et al. (2006). New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 311, 629–632. [DOI] [PubMed] [Google Scholar]
- Scheres SHW, Chen S (2012). Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW, Melero R, Valle M, Carazo J-M (2009). Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17, 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Shang Z, Fu X, Lou X, Grigorieff N, Nicastro D (2020). In situ structure determination at nanometer resolution using TYGRESS. Nat Methods 17, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf AJ, Morgan G, Giddings TH Jr, White EA, Marchione R, McDonald HB, Winey M (2005). Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell 16, 3606–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard D, Zhao Y, Bayless BA, Gui L, Louka P, Dave D, Suryawanshi S, Tomasi RF-X, Dupuis-Williams P, Baroud CN, et al. (2018). Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol Biol Cell 29, 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH (2006). Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature 442, 475–478. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ (1997). A genomic perspective on protein families. Science 278, 631–637. [DOI] [PubMed] [Google Scholar]
- Urbanska P, Joachimiak E, Bazan R, Fu G, Poprzeczko M, Fabczak H, Nicastro D, Wloga D (2018). Ciliary proteins Fap43 and Fap44 interact with each other and are essential for proper cilia and flagella beating. Cell Mol Life Sci 75, 4479–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner A, Salathé M, O’Riordan TG (1996). Mucociliary clearance in the airways. Am J Respir Crit Care Med 154, 1868–1902. [DOI] [PubMed] [Google Scholar]
- Wis´niewski JR (2016). Quantitative evaluation of filter aided sample preparation (FASP) and multienzyme digestion FASP protocols. Anal Chem 88, 5438–5443. [DOI] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, Nachury MV (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.