Abstract
The Rho family of GTPases are inactivated in a cell context–dependent manner by Rho-GTPase-activating proteins (Rho-GAPs), but their signaling mechanisms are poorly understood. Here we demonstrate that ARHGAP4, one of the Rho-GAPs, forms a complex with SEPT2 and SEPT9 via its Rho-GAP domain and SH3 domain to enable both up- and down-modulation of integrin-mediated focal adhesions (FAs). We show that silencing ARHGAP4 and overexpressing its two mutually independent upstream regulators, SEPT2 and SEPT9, all induce reorganization of FAs to newly express Integrin Beta 1 and also enhance both cell migration and invasion. Interestingly, even if these cell migration/invasion–associated phenotypic changes are induced upon perturbations to the complex, it does not necessarily cause enhanced clustering of FAs. Instead, its extent depends on whether the microenvironment contains ligands suitable for the up-regulated Integrin Beta 1. These results provide novel insights into cell migration, invasion, and microenvironment-dependent phenotypic changes regulated by the newly identified complex.
INTRODUCTION
Focal adhesions (FAs) are sites that connect extracellular matrix (ECM) proteins and intracellular actin bundles to regulate multiple cellular processes such as migration and invasion (Carragher and Frame, 2004; Wozniak et al., 2004; Paluch et al., 2016; Machiyama et al., 2017). Among the proteins that constitute FAs, integrin serves as a transmembrane receptor that exists in a heterodimer composed of different types of α and β subunits. Taking different combinations of α and β subunits, integrin organizes FAs together with other proteins such as focal adhesion kinase (FAK) and paxillin, allowing interacting with the ECM (Hynes, 2002), as well as serving as biomechanical signaling hubs (Sun et al., 2016; Manninen and Varjosalo, 2017). Among the subunits, Integrin Beta 1 is involved in many processes, but signaling pathways that regulate the Integrin Beta 1–mediated FAs remain incompletely understood (Wu et al., 1995; Chen et al., 2016).
The Rho family of GTPases (Rho-family)—a master regulator of a wide range of cellular processes, including migration and invasion—is well known to regulate FAs (Lawson and Burridge, 2014; Blangy, 2017). In fact, the involvement of Rho-family proteins in the clustering and activation of integrin has been reported in many studies (Schwartz and Shattil, 2000). Meanwhile, it remains poorly identified regarding the involvement of Rho-GTPase-activating proteins (Rho-GAPs), a family of proteins comprising 66 different members that regulate the Rho family (Dubash et al., 2009). Based on cell-based screening for all 66 Rho-GAPs, we have recently identified that ARHGAP4, one of the Rho-GAPs, plays a crucial role in maintaining some phenotypic features that epithelial cells possess, and thus its depletion leads to eliciting mesenchymal cell features in a manner reminiscent of the epithelial–mesenchymal transition (EMT; Kang et al., 2020). Specifically, silencing of ARHGAP4 was found to increase the induction of mesenchymal cell morphology, EMT marker expression, cell proliferation, 2D migration, disordered 3D morphogenesis, and traction force generation. We also revealed part of its molecular mechanism, in which ARHGAP4 regulates EMT via binding with SEPT9 and via the activation of FAs. While ARHGAP4 has recently been found to suppress cell motility and axon outgrowth (Vogt et al., 2007) and mediate the Warburg effect (Shen et al., 2019; Shen, Chen et al., 2019), details of how this Rho-GAP controls cell migration/invasion remain to be elucidated.
Here, we aim at revealing how ARHGAP4 impacts FAs and in turn cell migration and invasion. We performed microarray analysis to examine the comprehensive cellular response to ARHGAP4 overexpression and observed a notable change in the expression of integrins. We further identified that ARHGAP4 interacts, via its Rho-GAP domain and SH3 domain, with SEPT2 and SEPT9 to form a complex that allows both up- and down-modulation of FAs and transwell migration/invasion of epithelial cells.
RESULTS
ARHGAP4-induced transcriptional changes lead to cytoskeletal and integrin reorganizations
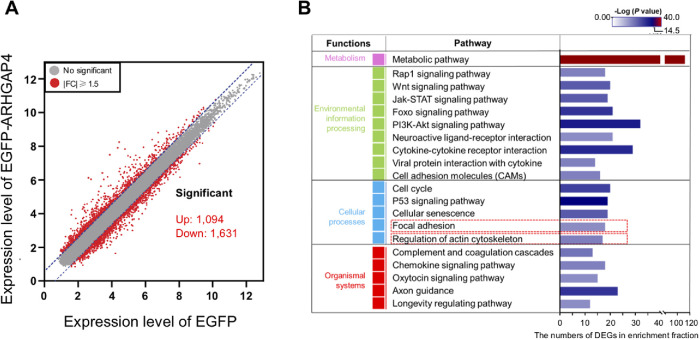
To probe the role of ARHGAP4, microarray analysis was performed on two types of MCF10A cells, one of which stably overexpresses EGFP (EGFP-Ov), and the other EGFP-ARHGAP4 (EGFP-ARHGAP4-Ov). A fold change of ≧1.5 was chosen as the benchmark for extracting differentially expressed genes (DEGs) (Figure 1A). A total of 2725 DEGs were identified between the EGFP-Ov and EGFP-ARHGAP4-Ov cell lines, among which 1094 genes and 1631 genes were found to be up- and down-regulated upon ARHGAP4 overexpression, respectively. All the data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE147988 (DEGs are shown in Supplemental Table S1). Focusing on the categories of “focal adhesion” and “regulation of actin cytoskeleton” (Figure 1B), cellular processes affected by ARHGAP4 overexpression were mapped in comparison with the reference canonical pathways in the KEGG database, from which we detected 17 DEGs in “focal adhesion” and 18 DEGs in “regulation of actin cytoskeleton” (Supplemental Figure S1A; Supplemental Table S2). In this study, we focus on the FA cluster and further characterize the role of ARHGAP4-SEPT2-SEPT9 in FAs. Notably, we found that the reorganization of integrin from an organization with α4 (ITGA4) and/or β1 (ITGB1) to one with α10 (ITGA10) and/or β4 (ITGB4) is induced upon ARHGAP4 overexpression. A similar tendency was observed in HEK293 cells as well (Supplemental Figure S1B).
FIGURE 1:
Microarray analysis to identify the DEGs induced upon ARHGAP4 overexpression in MCF10A cells. (A) Plots to show DEGs with a fold change of ≧1.5 between EGFP-ARHGAP4 and EGFP. (B) Functional classification of the DEGs selected in the KEGG pathway enrichment analysis. The top 20 categories are shown, of which the present study focused on “Focal adhesion.” The horizontal axis represents the number of DEGs in each functional category.
ARHGAP4 forms a complex with SEPT2 and SEPT9 via the Rho-GAP and SH3 domains of ARHGAP4
To excavate the regulatory mechanism of ARHGAP4, we searched by proteomic analysis for proteins that interact with ARHGAP4. Here, we used HEK293 cells, an epithelial cell line, in view of their high transfection efficiency. While we used MCF10A cells for the above microarray analysis as we constructed a stable MCF10A cell line expressing EGFP-ARHGAP4, in a separate analysis we identified similar trends of expression between the two epithelial cell lines regarding the partial DEGs acquired from the microarray (Supplemental Figure S1B).
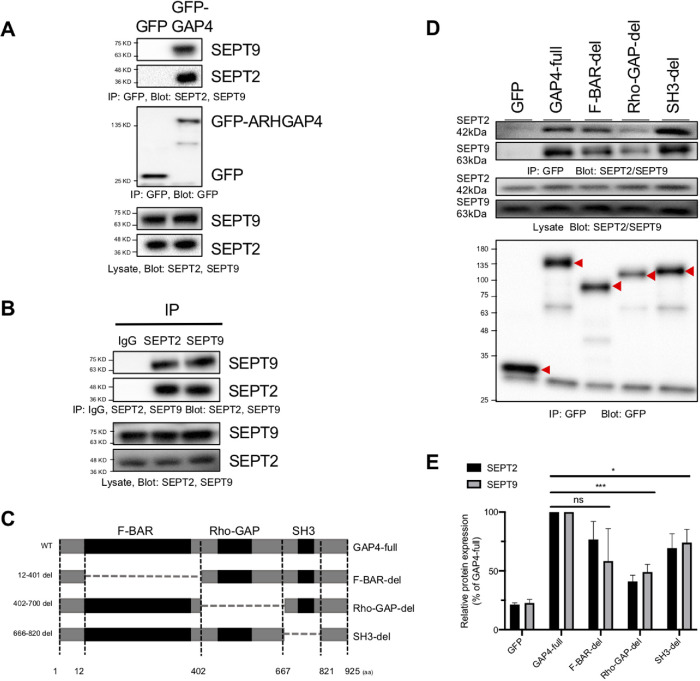
Candidates for interacting proteins of ARHAGP4, including SEPT2 and SEPT9, were obtained in HEK293 cells (Figure 2A), which we are focusing on in the present study; in Bxpc3 cells, in which ARHGAP4 is highly expressed (Supplemental Figure S3A); and in MCF10A cells as well (Supplemental Figure S2A). Here, HEK293 cells transfected with EGFP or EGFP-ARHGAP4 were lysed and subjected to coimmunoprecipitation with anti-GFP antibody. Mass spectrometry on coprecipitated proteins separated by SDS–PAGE suggested that not only SEPT9 (with a molecular weight of 65 kDa) but also SEPT2 (41 kDa) may be associated with ARHGAP4 (Supplemental Figure S2B). We then confirmed by immunoblotting using anti-ARHGAP4, anti-SEPT2, and anti-SEPT9 antibody that the GFP-ARHGAP4-associated complex indeed contains SEPT2 as well as SEPT9 (Figure 2A). To confirm the existence of the ARHGAP4-SEPT complex, coimmunoprecipitation was conducted by pulling down with each of the SEPT2 and SEPT9 antibodies, and the association to ARHGAP4 was confirmed in both cases (Supplemental Figure S2, C and D). We also found that SEPT2 and SEPT9 interact with each other to form a complex in HEK293 cells (Figure 2B). Not only Western blot but also immunofluorescence reveals colocalization of endogenous ARHGAP4 in Bxpc3 cells (Supplemental Figure S3, B and C) and exogenous ARHGAP4 in HEK293 cells (Supplemental Figure S4) with SETP2 and SEPT9. The magnitude of the correlation was modest (Pearson correlation coefficient r ∼ 0.4 for endogenous ARHGAP4 and ∼0.7 for exogenous ARHGAP4), but these results suggest that ARHGAP4 indeed has the potential to bind to the SEPT2–SEPT9 (or SEPT2/9) complex.
FIGURE 2:
Identification of the ARHGAP4-SEPT2/9 complex in HEK293 cells. (A) Coimmunoprecipitation shows that SEPT2 and SEPT9 were pulled down with GFP-tagged ARHGAP4 but not with GFP in HEK293 cells. (B) SEPT2 was pulled down with SEPT9 and vice versa. (C) Schematic structure of ARHGAP4 and its deletion mutants constructed in this study. (D) ARHGAP4 binds to SEPT2/9 via its Rho-GAP domain and SH3 domain. The red arrow heads show GFP-labeled target bands. (E) Quantification of D, with the mean and SD.
ARHGAP4 belongs to Rho-GAPs, which typically contain three primary domains, the F-BAR domain, the Rho-GAP domain, and the SH3 domain. SEPT2 and SEPT9 both belong to the septin protein family, which is a member of a conserved family of GTP-binding proteins. Here, to determine the domain of ARHGAP4 that is indispensable for binding to the SEPT2/9 complex, we transfected each of EGFP-ARHGAP4 and its domain deletion mutants (Figure 2C) into HEK293 cells, and cell lysates were coprecipitated with anti-GFP antibody–conjugated magnetic beads. The precipitates were analyzed by immunoblotting with anti-GFP antibody, SEPT2 antibody, and SEPT9 antibody. The ability of the full-length ARHGAP4 (GAP4-full) and the F-BAR deletion mutant (F-BAR-del) to pull down the SEPT2/9 complex was greater than that of the Rho-GAP deletion (Rho-GAP-del) and SH3 deletion (SH3-del) mutants (Figure 2D). Quantification showed that the affinity of Rho-GAP-del and SH3-del for the SEPT2/9 complex was on the average 55% and 65% of that of the ARHGAP4 full group, respectively (Figure 2E), suggesting that the Rho-GAP and SH3 domains of ARHGAP4 are necessary for its binding to the SEPT2/9 complex.
ARHGAP4 depletion activates and stabilizes FAs
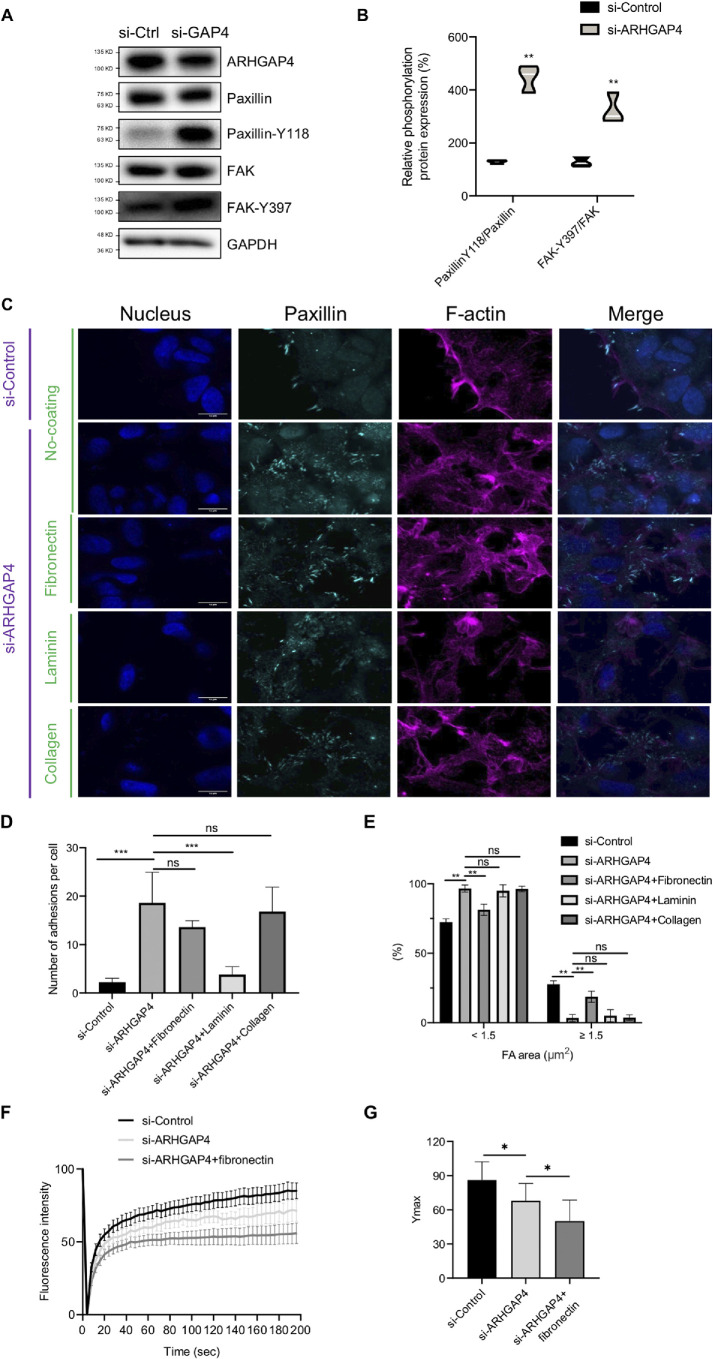
Motivated by the above observations (Figure 1C and Supplemental Figure S1B) that ARHGAP4 induces integrin reorganization, we next analyzed how integrin-mediated FAs are modulated by ARHGAP4 knockdown. Immunoblotting revealed that the expression of paxillin and FAK, major markers of FAs, was significantly increased in ARHGAP4-silenced HEK293 cells compared with controls, while the phosphorylation of FAK at Y397 and that of paxillin at Y118 were both increased (Figure 3, A and B). Immunofluorescence showed that paxillin- or paxillin-Y118-labeled FAs have a tendency to change in spatial organization upon ARHGAP4 knockdown (Figure 3C and Supplemental Figure S5A). To quantify this, the number and size of individual FAs were analyzed. We found that, while ARHGAP4 knockdown augmented the total number, it led to the opposite effect on the size (Figure 3, D and E). Specifically, FAs in control cells are relatively few in total number but large in size; nearly one-fourth of the adhesions are larger than 1.5 μm2 in area. In knockdown cells, in contrast, the total number of FAs increases about threefold, but they almost lose the large-size population while instead acquiring a great number of small patches less than 1.5 μm2 in area that are spread over the peripheral regions of the cells. Consistently, the silencing of ARHGAP4 also caused cell morphological change in HEK293 cells, in which cells were endowed a spread morphology compared with si-Control cells, probably because of the alterations in the FAs (Supplemental Figure S5B). In separate experiments, we analyzed the immunofluorescence of endogenous ARHGAP4 in Bxpc3 cells and found that there is no obvious colocalization between ARHGAP4 and paxillin (Supplemental Figure S6; Pearson correlation coefficient r ∼ 0.1), suggesting that ARHGAP4 is not necessarily localized at FAs but instead may work on their regulation by mediating other FA-related proteins.
FIGURE 3:
FAs are activated upon ARHGAP4 depletion in HEK293 cells. (A) Immunoblot shows that ARHGAP4 depletion increased the expression of paxillin and FAK, and the phosphorylation of FAK and paxillin was both increased. (B) Quantification of Paxillin-Y118/Paxillin and FAK-Y397/FAK levels from blots in A with violin plots, in which the white lines indicate the means. (C) Immunofluorescence of paxillin to observe the effect of ARHGAP4 depletion on FAs in the presence of fibronectin, laminin, or collagen or in the absence of coating. The nuclei (Hoechst 33342) and F-actin (phalloidin) are also shown. Scale, 15 µm. (D, E) Quantification of C in terms of the total numbers per cell, D, and the area distribution of FAs (whether <1.5 or ≧1.5 μm2 in projected area), E. The data are shown by the mean and SD. (F) FRAP curve over time. All the values are normalized between the initial and photobleached values and are shown in percentage. (G) The steady state values of Ymax in F, in which the mean and SD are shown.
Thus, while FAs are activated upon ARHGAP4 depletion in terms of total number, they are fragmented into small pieces. Given that the expression of integrin α4β1—which plays a predominant role in the formation of FAs (Yeh et al., 2017) and acts as a fibronectin receptor (Paloma et al., 1994; Gu, 2007)—is decreased with ARHGAP4 overexpression (Supplemental Figure S1), we assumed that the reduced area of individual FAs following ARHGAP4 knockdown may be attributable to a shortage of fibronectin in the ECM. To test this hypothesis, we examined the effect of adding fibronectin on the area of individual FAs in ARHGAP4-silenced cells. We found that the reduction in area due to knockdown is partly rescued by the coating of fibronectin in the substrate (Figure 3, C–E). We also examined the effect of adding laminin or collagen instead of fibronectin and found that the reduction in area due to ARHGAP4 knockdown is not rescued by these ECM components, consistent with our hypothesis (Figure 3, C–E). These results suggest that the size of individual FAs is not necessarily a straightforward measure to evaluate the intracellular activation of FAs in the study of ARHGAP4 because it is accompanied by replacement of constituent integrin subunits and consequently might be affected by the existing extracellular cues (i.e., the components of the surrounding environment).
To more carefully evaluate the state of FAs, fluorescence recovery after photobleaching (FRAP) experiments, which are commonly used to evaluate the dynamics of proteins in cells, were performed on EGFP-vinculin-labeled FAs in cells treated si-Control or si-ARHGAP4 (Figure 3F and Supplemental Figure S7). We evaluated the effect of si-ARHGAP4 in the absence and presence of additional fibronectin in the substrate. The steady state value of the recovered fluorescence intensity Ymax, which measures the stability or maturity of FAs, was significantly lowered by si-ARHGAP4 and significantly further by the further addition of fibronectin (Figure 3G and Supplemental Figure S7). Taken together, FAs subjected to ARHGAP4 knockdown are actually activated (as suggested by the increase in the expressed number) and hence have a potential to become large in size or mature, but they cannot because of the shortage of suitable ECM proteins (i.e., in this case, fibronectin, a ligand of the up-regulated integrin α4β1), and thus the limited size of FAs is due only to the surrounding unsuitable environment of the cells.
ARHGAP4 suppresses FAs via its Rho-GAP/SH3 domains
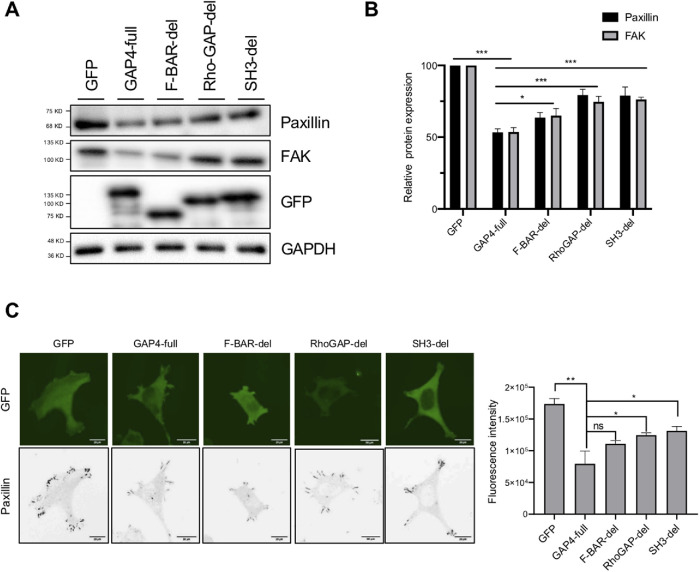
We also found that the overexpression of ARHGAP4 reduces paxillin protein expression compared with controls in immunoblotting (Figure 4A), thus eliciting the opposite effect on FAs from its knockdown. To identify the domain of ARHGAP4 indispensable for this regulation of FAs, we introduced each of the domain deletion mutants (Figure 2C) into cells to examine their effect on paxillin expression on the substrate. Compared with the case with full-length ARHGAP4 overexpression, the expression of paxillin was significantly higher in cells with the F-BAR deletion mutant and was further significantly increased in cells with the Rho-GAP deletion mutant or with the SH3 deletion mutant, as quantified by immunoblotting (Figure 4B). Immunofluorescence also showed a similar tendency (Figure 4C and Supplemental Figure S8). These results suggest that the Rho-GAP and SH3 domains of ARHGAP4 are essential for its ability to down-regulate FAs in terms of the paxillin expression level.
FIGURE 4:
ARHGAP4 down-regulates FAs in HEK293 cells via its Rho-GAP domain and SH3 domain. (A) Immunoblot shows that ARHGAP4 lowers the paxillin and FAK expression, to which the F-BAR domain partly contributes and the Rho-GAP/SH3 domains contribute more significantly. (B) Quantification of A with the mean and SD. (C) Immunofluorescence of paxillin in cells overexpressed with EGFP, EGFP-labeled paxillin, and its mutants, and its quantification with the mean and SD. Scale, 20 µm.
SEPT2 and SEPT9 both activate FAs in a manner similar to ARHGAP4 depletion
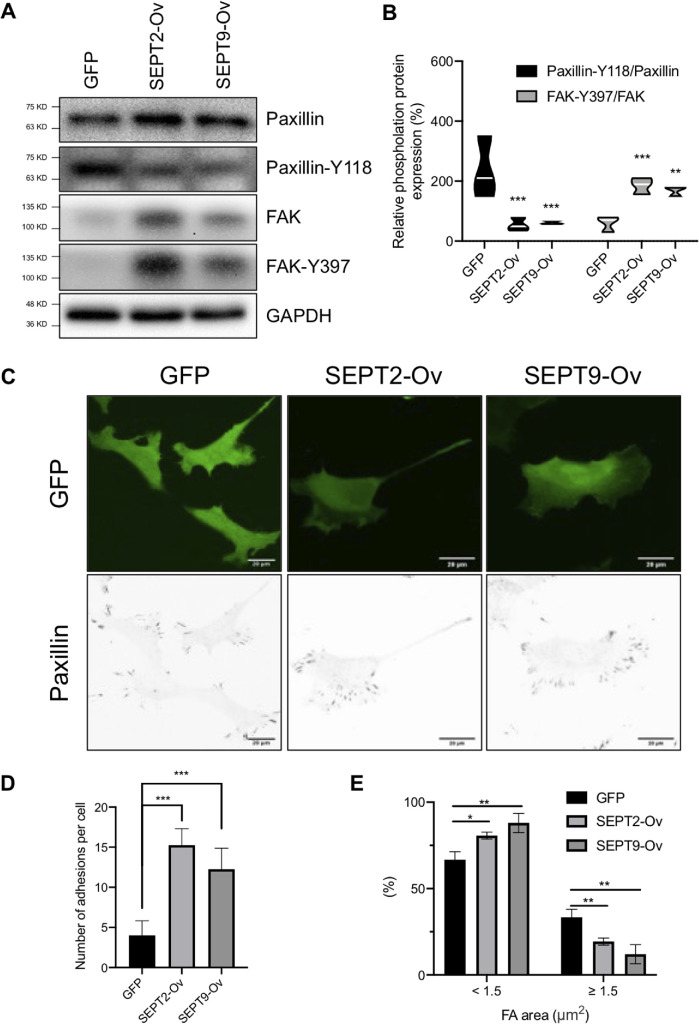
These results show that the domain of ARHGAP4 critical to regulating FAs is consistent with the one critical to forming a complex with SEPT2/9 (Figures 2 and 4). These results led us to consider the effect of SEPT2/9 on the regulation of FAs. We then investigated the effect of overexpressing SEPT2 and SEPT9 on FAs. We found that SEPT2 and SEPT9 caused a significant increase in paxillin, FAK, and FAK (Y397) protein expression, but a decrease in paxillin-Y118 (Figure 5A). Quantification showed that the overexpression of SEPT2 and SEPT9 resulted in a significant decrease and increase of, on average, 5.2-fold and 2.1-fold in paxillin-Y118/paxillin and FAK-Y397/FAK expression, respectively (Figure 5B). Immunofluorescence also showed a remarkable increase in the total number of paxillin-labeled FAs for both the overexpression of SEPT2 and that of SEPT9, but a similar downward trend of paxillin-Y118 with immunoblot (Figure 5C and S9). Quantification of the images confirmed the significant increase in the number of FAs upon the overexpression of SEPT2 and SEPT9 (Figure 5D) and further showed a significant decrease in the large-size population more than 1.5 μm2 in area, which is instead compensated for by a significant increase in the small-size population of less than 1.5 μm2 (Figure 5E). These responses of FAs to SEPT2 and SEPT9 overexpression were both similar to the case of ARHGAP4 depletion, allowing FAs to be activated.
FIGURE 5:
SEPT2 and SEPT9 both activate FAs in HEK293 cells. (A) Immunoblot shows that SEPT2 and SEPT9 increase the expression of paxillin and FAK and the phosphorylation of FAK, but decrease the phosphorylation of paxillin. (B) Violin plots depicting the phosphorylation of paxillin and FAK, with the white lines representing the means. (C) Immunofluorescence of paxillin in cells overexpressed with EGFP, EGFP-labeled SEPT2, and EGFP-labeled SEPT9. Scale, 20 µm. (D, E) Quantification (mean and SD) of C in terms of the total number per cell, D, and the area distribution of FAs with two categories of <1.5 or ≧1.5 μm2 in projected area, E.
ARHGAP4 down-regulates Integrin Beta 1-linked FAs under negative regulation by SEPT2/9
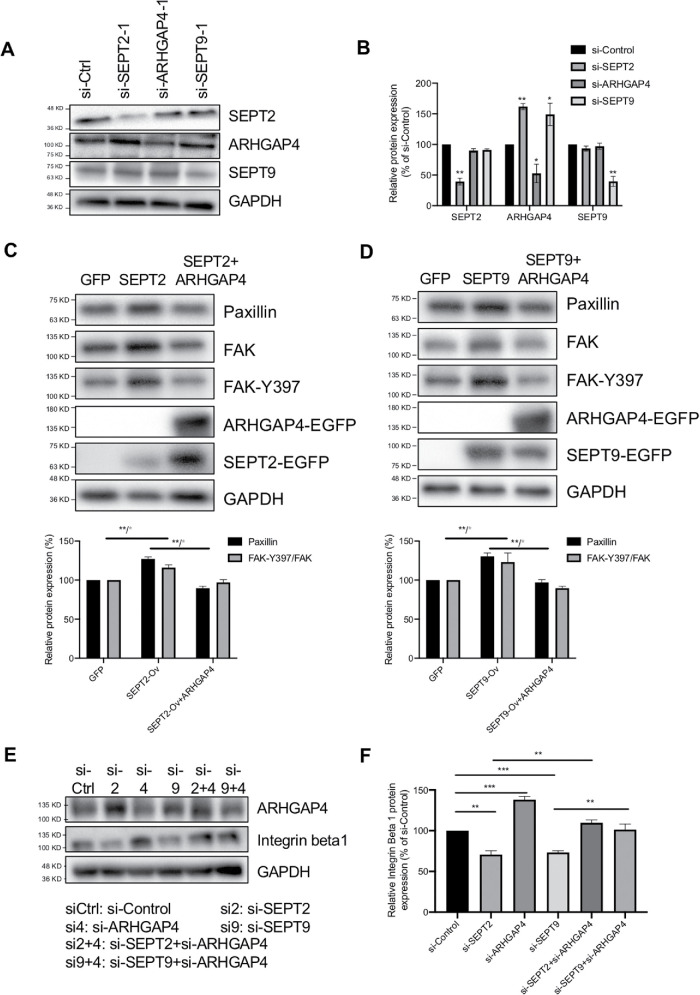
Motivated by the similar cell responses between ARHGAP4 depletion and SEPT2/9 overexpression, we next investigated the mutual relationship among those three proteins. HEK293 cells were transfected with one of the control siRNA and ARHGAP4-, SEPT2-, and SEPT9-targeting siRNAs for immunoblot analysis. By the depletion of SEPT2 and SEPT9, a significant increase in expression was induced for ARHGAP4 but not to its family members, SEPT9 and SEPT2 (Figure 6, A and B). In contrast, no significant alteration in SEPT2 and SEPT9 expression was induced by the depletion of ARHGAP4. To rule out experimental errors, two groups of siRNA were used for each gene (shown in Supplemental Figure S10). These results suggest that SEPT2 and SEPT9 both work upstream of ARHGAP4 to inhibit its activity, while SEPT2 and SEPT9 do that independent of each other.
FIGURE 6:
The ARHGAP4-SEPT2/9 complex regulates FAs by mediating Integrin Beta 1 expression in HEK293 cells. (A) Immunoblots show that SEPT2 and SEPT9 depletion up-regulate ARHGAP4 expression; meanwhile, ARHGAP4 depletion does not affect SEPT2 and SEPT9. (B) Quantification of A with the mean and SD. (C) Immunoblots show that FAK and paxillin expression is increased with SEPT2 overexpression, and is reversed by ARHGAP4 overexpression. (D) Immunoblots show that FAK and paxillin expression is increased with SEPT9 overexpression, and is reversed by ARHGAP4 overexpression. (E) Immunoblots show that the expression of Integrin Beta 1 is decreased by SEPT2 depletion but reversed by ARHGAP4 depletion, is increased by ARHGAP4 depletion, and is decreased by SEPT9 depletion but reversed by ARHGAP4 depletion. (F) Quantification of E. Data are shown by the mean and SD.
To investigate the relevance to the regulation of FAs, we overexpressed ARHGAP4 together with either SEPT2 or SEPT9. While paxillin was increased in expression with the overexpression of SEPT2 (Figure 6C) as well as that of SEPT9 (Figure 6D), the additional overexpression of ARHGAP4 remarkably rescued the paxillin response. We addressed the same issue using siRNAs, with a focus on Integrin Beta 1. We found that the expression of Integrin Beta 1 is decreased by the silencing of SEPT2 and SEPT9 compared with controls but is increased by the silencing of ARHGAP4 (Figure 6, E and F). On the other hand, the decrease in Integrin Beta 1 expression due to the silencing of SEPT2 and of SEPT9 is reversed by the additional ARHGAP4 silencing. These results suggest that ARHGAP4 suppresses the activation of Integrin Beta 1–associated FAs under negative regulation by SEPT2/9.
ARHGAP4-SEPT2/9 complex regulates cell migration and invasion in transwell cultures
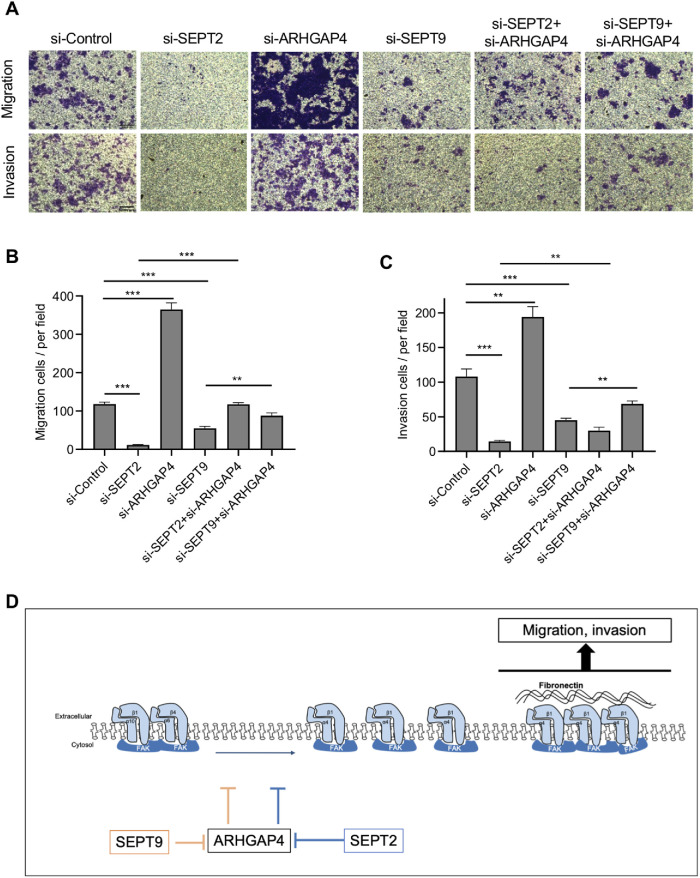
Integrin-linked FAs associated with the actin cytoskeleton provide cells with the main sites for adhering to the ECM, which in turn control cell motility (Carragher and Frame, 2004; Paluch et al., 2016). Motivated by this view and our above results, we hypothesized that the alteration in FAs mediated by the ARHGAP4-SEPT2/9 complex changes cell motility. Here, we performed a transwell assay, in which the migration and invasion of HEK293 cells into media without and with Matrigel, respectively, was analyzed. We observed a significant decrease in the number of migrating cells in both cases of SEPT2 depletion and SEPT9 depletion (Figure 7, A and B). Meanwhile, migrating cells were increased in number upon ARHGAP4 depletion compared with controls, thus exhibiting the opposite effect to SEPT2/9. We also observed that the down-regulation of migration ability due to SEPT2/9 depletion was partly rescued by the simultaneous depletion of ARHGAP4. A similar tendency was observed in the three-dimensional cell invasion into Matrigel (Figure 7, A and C). These results suggest that ARHGAP4 suppresses cell migration and invasion under the negative regulation by SEPT2/9.
FIGURE 7:
The ARHGAP4-SEPT2/9 complex regulates the migration and invasion of HEK293 cells. (A) Photographs of cells (purple) undergoing migration (upper) and invasion (lower) at the conditions described. Scale, 5 mm. (B) Quantification with the mean and SD of A showing migrating cells per the field of view. (C) Quantification with the mean and SD of A showing invading cells per the field of view. (D) Model of the signaling pathway.
DISCUSSION
It is well known that the Rho family plays essential roles in controlling the organization of FAs (Rottner et al., 1999; Guo, 2006). However, individual roles of their upstream regulators—Rho-GAPs—are still incompletely characterized (Haga and Ridley, 2016; Hodge and Ridley, 2016; Muller et al., 2020). We previously analyzed the whole Rho-GAP genes and successfully identified that ARHGAP4 depletion promotes cellular EMT under negative regulation by SEPT9 (Kang et al., 2020, 2021). In the present study, aiming at further exploring the signaling mechanism and its consequences for cell functions, we performed microarray analysis and detected ARHGAP4-induced transcriptional changes related to the activity of FAs (Figure 1). Motivated by this result, we focused on FAK and paxillin protein expression as markers of FAs and demonstrated that ARHGAP4 forms a complex with SEPT2 as well as SEPT9 via the Rho-GAP and SH3 domains of ARHGAP4 in HEK293 cells (Figure 2). This finding on the interaction between SEPT2 and SEPT9 is consistent with a previous report on retinal pigmented epithelial cells (Ghossoub et al., 2013). We revealed that ARHGAP4 down-regulates FAs in a manner mediated by its Rho-GAP domain and SH3 domain (Figures 3 and 4), thus implying the involvement of SEPT2/9 given the critical domains described above. Indeed, we found that SEPT2 and SEPT9 negatively and independently regulate ARHGAP4 (Figures 5 and 6). We also showed that SEPT2 and SEPT9 stabilize FAs in HEK293 cells (Figure 5), which is consistent with a previous report in which septins were shown to be required for the stabilization and maturation of nascent adhesions (Dolat et al., 2014). Furthermore, we demonstrated that the activation of FAs induced by ARHGAP4 depletion is accompanied by the up-regulation of Integrin Beta 1, a fibronectin receptor (Figure 6), which is consistent with the enhanced stabilization of FAs upon ligation to fibronectin (Figure 3). Another intriguing finding in this regard is that, even when the increased cell migration and invasion take place due to perturbations to oncogenes, proto-oncogenes, or tumor suppressor genes, they may not necessarily lead to prompt enlargement of the FAs; instead, their extent depends on environmental cues, including whether the surrounding ECM contains ligands suitable for the newly up-regulated integrin subsets (Figure 7D). We also found that the roles of the ARHGAP4-SEPT2/9 complex in the activation of FAs are in line with those in the increase in cell migration and invasion in transwell cultures (Figure 7). These findings provide new insights into the molecular mechanism of tumorigenesis, metastasis, and embryonic development.
While cell biological studies on the function of ARHGAP4 have been rather limited in number, there has been a significant increase in them in recent years. To date, almost all fundamental research on ARHGAP4 has reported prominent functions in inhibiting cell migration and invasion (Vogt et al., 2007; Shen et al., 2019; Shen et al., 2020). Our study is the first to our knowledge that has revealed its molecular mechanism dominated by the SEPT2/9-driven modulation of FAs via its Rho-GAP and SH3 domains. Here, the ARHGAP4-SEPT2/9 complex was found to enable both up- and down-modulation of FAs to eventually regulate the extent of migration and invasion of the cells. Another finding, particularly interesting from a physiological viewpoint, is that the perturbations to the complex are accompanied by reorganization of integrin components. The consequence of this is that the level of clustering of individual FAs becomes dependent on whether the microenvironment contains ECM ligands suitable for the reorganized integrin components. These results provide novel molecular insights into understanding how the microenvironment affects cells with mutations in the ARHGAP4-SEPT2/9-associated pathway.
Integrin family proteins are transmembrane heterodimers composed of an α- and a β-subunit. There are 18 α and 8 β subunits, making 24 different combinations whose composition determines their potential to bind to specific extracellular ligands (Takada et al., 2007; Barczyk et al., 2010). FAs are such integrin-mediated cell–ECM contact sites that are clustered according to the intracellular and environmental cues (Sun et al., 2016; Manninen and Varjosalo, 2017) to consequently regulate many cell functions, which include cell migration and invasion (Fortea and Belmonte, 2018; Miller, Hu et al., 2020). Our microarray results revealed ARHGAP4-associated changes in the integrin subunit. Specifically, integrins α10 and β4 are more expressed in ARHGAP4-overexpressed cells, whereas integrin α4β1 is more expressed in control cells (Supplemental Figure S1). Accumulated studies have shown that the only subunits that form heterodimers with β4 and α10 are α6 (as a laminin receptor) and β1 (as a collagen receptor), respectively, to be expressed in epithelial cells (Gilcrease, 2007; Manninen, 2015; Matlin et al., 2017). On the other hand, integrin α4β1 works as a leukocyte and/or fibronectin receptor and has been implicated in pro-inflammation and cancer progression (Danen and Yamada, 2001). Thus, along with our previous findings on its involvement in EMT (Kang et al., 2020), the ARHGAP4-elicited replacement of the integrin components suggests the role of ARHGAP4 as a tumor-suppressor gene. Consistent with this, the DEGs that decreased with ARHGAP4 expression (such as KRAS, NRAS, and PKCα) are related to cancer, proliferation, and migration (Supplemental Table S2). With a view to revealing the role of ARHGAP4 in cell migration and invasion, we have focused on the categories of “focal adhesion” and “regulation of actin cytoskeleton” to interpret the microarray results; thus, the DEGs in the other categories will be the subject of future investigation.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture and transfection
HEK293 cells (JCRB Cell Bank, Ibaraki, Japan) were cultured with E-MEM, including L-glutamine and phenol red (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, USA), 1% nonessential amino acid (Wako), and 1% penicillin–streptomycin solution (Wako) in a 5% CO2 incubator at 37°C. MCF10A cells (ATCC, Manassas, USA) were cultured in DMEM/Ham’s F12 medium (Wako) supplemented with 5% horse serum (Thermo Fischer Scientific, Waltham, USA), 0.25 IU/ml insulin (Wako), 0.5 mg/ml hydrocortisone (Wako), 100 ng/ml cholera toxin (Wako), 20 ng/ml EGF (Wako), and 1% penicillin–streptomycin solution (Wako). Cells were transfected with plasmid and siRNA using ScreenFect A plus (Wako) and RNAiMAX (Thermo Fisher Scientific), respectively, according to the manufacturer’s instructions. Briefly, seeded cells reaching 75% of the confluent state were used for transfection. A concentration of 20 μM silencer and 1250 ng plasmids were used in culture dishes with diameter 35 mm, resulting in ∼100% and ≥90% transfection efficiency, respectively. Cells were then used for the experiments 48 h after the transfection. In separate analyses, to generate MCF10A cells stably expressing EGFP or EGFP-ARHGAP4, cells were selected in growth media containing G418 (final concentration, 60 μg/ml, Wako), in which cloning rings were used for picking up the cluster of monoclonal cells after the transfection until the final stable cell lines were obtained.
Plasmid and siRNA
Plasmids encoding human ARHGAP4 gene combined with EGFP were constructed as described previously (Kang et al., 2020). Plasmids encoding F-BAR-deletion, Rho-GAP-deletion, and SH3-deletion mutants of ARHGAP4 were constructed using a KOD-Plus-Mutagenesis Kit (Toyobo, Osaka, Japan). The cDNAs encoding human SEPT2 and SEPT9 were amplified by PCR using the Q5 High-Fidelity 2X Master Mix (NEB, Ipswich, USA) with primers listed in Supplemental Table S3 and inserted into the pEGFP vector. The siRNAs targeting ARHGAP4, SEPT2, and SEPT9 and the negative control siRNA (Thermo Fisher Scientific; Supplemental Table S4) were transfected into cells as described above.
Coimmunoprecipitation assay and mass spectrometry
HEK293 cells transfected with EGFP-ARHGAP4 or its mutants were lysed with nondenatured lysis buffer (20 mM Tris-HCl, 137 mM NaCl, 1% NP-40, 2 mM EDTA, 1 mM DTT, 10 μg/ml leupeptin,10 μg/ml pepstatin, and 1 mM PMSF; pH 8.0). Equal amounts of the cell lysate transfected with different plasmids were reacted overnight with GFP antibody together with magnetic beads at 4°C. Immunoprecipitates and interacting proteins were stained by a silver stain MS kit (Wako). The silver staining bands with significant differential expression were sequenced by mass spectrometry at the Center for Medical Innovation and Translational Research of Osaka University Medical School. Q-Exactive (Thermo Fisher Scientific) was used for mass spectrum analysis by an UltiMate 3000 Nano LC system (Thermo Fisher Scientific). Proteins were identified by immunoblotting with anti-ARHGAP4, anti-SEPT2, and anti-SEPT9 antibody.
Transcriptomic analysis
The total RNA was collected by a PureLink RNA Mini Kit (Thermo Fischer Scientific) from MCF10A cells stably expressing EGFP or EGFP-ARHGAP4. The data were summarized and normalized by the robust multiaverage (RMA) method implemented in Affymetrix Power Tools (Thermo Fischer Scientific) to obtain differentially expressed genes (DEGs). Gene enrichment and functional annotation analysis for the significant probe list were performed using the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database (Kanehisa and Goto, 2000). All data analysis and visualization of DEGs were conducted using R 3.3.3 (Azevedo et al., 2019).
Western blotting
Cells transfected with plasmids or siRNAs were lysed in RIPA buffer (50 mM Tris-HCl, 100 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1 mM Na3VO4, 1 mM NaF, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 1 µg/ml leupeptin; pH 7.4) and subjected to centrifugation at 15,000 × g for 30 min to collect the supernatants. A Pierce BCA protein assay kit (Thermo Fisher Scientific) was used to calculate the protein concentration. Proteins were fractionated on 5–10% gradient acrylamide gels (Bio-rad, Berkeley, USA). Following SDS–PAGE, samples in gels were transferred onto polyvinylidene fluoride membranes (Wako). After blocking in 5% BSA, primary antibodies against target proteins (Supplemental Table S5) were used and detected using HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (Bio-rad). The target bands were obtained by Immobilon Western Kit (Millipore, Burlington, USA) and analyzed by ImageLab software (Bio-rad).
Immunofluorescence staining and fluorescence imaging
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (Wako) at 37°C for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, blocked with 5% normal goat serum, and incubated with the primary antibody (Supplemental Table S5) for 1.5 h and then with fluorescent dye–labeled anti-rabbit or anti-mouse IgG second antibody (Thermo Fischer Scientific) for 1 h at room temperature. For the staining of the nucleus and F-actin, cells were incubated with Hoechst 33342 and phalloidin, respectively, and then with SlowFade (Thermo Fischer Scientific). Zenon Tricolor rabbit IgG labeling kit (Thermo Fischer Scientific) was used for labeling the three antibodies, ARHGAP4, SEPT2, and SEPT9, to show the colocalization. Fluorescence images were acquired using a fluorescence microscope (IX73, Olympus, Tokyo, Japan) or a confocal laser scanning microscope (FV1000, Olympus) equipped with a UPlan Apo 60× oil objective lens (NA = 1.42) and analyzed by ImageJ software.
Image analysis of individual FAs
To analyze the response of individual FAs to various perturbations such as ARHGAP4 depletion and overexpression of SEPT2/9, immunofluorescence staining of paxillin was performed to label FAs in HEK293 cells according to the method described above. The outlines of all the individual FAs that were visible to the naked eye were carefully traced in ImageJ software using an electric pen device (Cintiq 13HD, DTK-1301, Wacom) to determine if their size is larger than a specific threshold (set here as 1.5 µm2) or not. At least 500 FAs were analyzed for each group. We then analyzed how the ratio of the two groups changes in response to the perturbations to the cells. This evaluation on the response of FAs is achieved with higher reliability compared with a case based on the cumulative distribution function because it is hard to accurately determine the absolute values of the size of such small individual FAs. The choice of the threshold of 1.5 µm2 is to some extent arbitrary, but this value is within the characteristic size range of FAs; indeed, similar approaches have been taken in previous studies (e.g., Angely et al., 2020).
Fluorescence recovery after photobleaching
U2OS cells (ATCC) expressing EGFP-vinculin were cultured on a glass-bottomed dish in a stage incubator (Tokai Hit, Fujinomiya, Japan). FRAP experiments were performed using a confocal microscope (FV1000) with a 60× oil immersion objective lens on cells treated, 48 h before the experiments, with si-Control or si-ARHGAP4. To evaluate the effect of fibronectin in the substrate, cells were plated on a glass-bottomed dish coated in advance with fibronectin (10 µl/mg, Sigma). Photobleaching was induced using a 405- and a 440-nm-wavelength laser on individual vinculin-labeled FAs, and images were taken every 4 s. The recovery curve was fitted to a single exponential function by the least-squares method, by which the steady state value of the fluorescence intensity Ymax was computed to measure the stability of FAs.
Cell migration and invasion assay
For migration and invasion assay, 2 × 105 cells were seeded onto the upper chambers of a 24-well transwell plate with an 8-μm pore size (Corning, NY, USA). For the invasion assay, the upper chamber was coated in advance with Matrigel (Corning) diluted in no-serum medium at a ratio of 1:8. In both assays, cells were seeded on the surface of the upper chamber in the serum-deprived medium supplied with 0.1% BSA. The lower room was filled with a medium containing 10% FBS. After incubation for 24 h for migration assay and 36 h for invasion assay, cells were fixed with 4% paraformaldehyde for 30 min at 37°C. Cells that remained on the apical side of the chamber were gently scraped off using wetted cotton swabs, stained with 0.1% crystal violet, and then imaged using a bright-field microscope (CKX41, Olympus).
Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad Software, San Diego, USA), in which p-values were calculated using a one-way analysis of variance followed by Tukey’s test or a two-tailed paired t-test for multiple comparisons. Data were shown as mean ± SD from more than three independent experiments. Statistical significance was set, compared with respective controls, as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Supplementary Material
Acknowledgments
N.K. and S.L. are supported by a Chinese Scholarship Council (CSC) Scholarship. This work was supported in part by JSPS KAKENHI grants (15H03004 and 18H03518).
Abbreviations used:
- DEGs
differentially expressed genes
- ECM
extracellular matrix
- EMT
epithelial-mesenchymal transition
- FAK
focal adhesion kinase
- FAs
focal adhesions
- FRAP
fluorescence recovery after photobleaching
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Rho-family
Rho family of GTPases
- Rho-GAPs
Rho-GTPase-activating proteins.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-01-0010) on September 15, 2021.
REFERENCES
- Angely C, Ladant D, Planus E, Louis B, Filoche M, Chenal A, Isabey D (2020). Functional and structural consequences of epithelial cell invasion by Bordetella pertussis adenylate cyclase toxin. PLoS One 15, e0228606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo LS, Pestana IA, Nery A, Bastos WR, Souza CMM (2019). Influence of the flood pulse on mercury accumulation in detritivorous, herbivorous and omnivorous fish in Brazilian Amazonia. Ecotoxicology 28, 478–485. [DOI] [PubMed] [Google Scholar]
- Barczyk M, Carracedo S, Gullberg D (2010). Integrins. Cell Tissue Res 339, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A (2017). Tensins are versatile regulators of Rho GTPase signalling and cell adhesion. Biol Cell 109, 115–126. [DOI] [PubMed] [Google Scholar]
- Carragher NO, Frame MC (2004). Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol 14, 241–249. [DOI] [PubMed] [Google Scholar]
- Chen MB, Lamar JM, Li R, Hynes RO, Kamm RD (2016). Elucidation of the roles of tumor Integrin beta1 in the extravasation stage of the metastasis cascade. Cancer Res 76, 2513–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen EHJ, Yamada KM (2001). Fibronectin, integrins, and growth control. J Cell Physiol 189, 1–13. [DOI] [PubMed] [Google Scholar]
- Dolat L, Hunyara JL, Bowen JR, Karasmanis EP, Elgawly M, Galkin VE, Spiliotis ET (2014). Septins promote stress fiber–mediated maturation of focal adhesions and renal epithelial motility. J Cell Biol 207, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubash AD, Menold MM, Samson T, Boulter E, García-Mata R, Doughman R, Burridge K (2009). Chapter 1. Focal adhesions: new angles on an od structure. Int Rev Cell Mol Biol 277, 1–65. [DOI] [PubMed] [Google Scholar]
- Fortea MB, Belmonte FMN (2018). Mechanosensitive adhesion complexes in epithelial architecture and cancer onset. Curr Opin Cell Biol 50, 42–49. [DOI] [PubMed] [Google Scholar]
- Ghossoub R, Hu Q, Failler M, Rouyez M-C, Spitzbarth B, Mostowy S, Wolfrum U, Saunier S, Cossart P, Jamesnelson W, et al. (2013). Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. J Cell Sci 126, 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilcrease MZ (2007). Integrin signaling in epithelial cells. Cancer Lett 247, 1–25. [DOI] [PubMed] [Google Scholar]
- Gu J (2007). Regulation of integrinfunctions by N-glycans. Yakugaku Zasshi 127, 571–578. [DOI] [PubMed] [Google Scholar]
- Guo F, Debidda M, Yang L, Williams DA, Zheng Y (2006). Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J Biol Chem 281, 18652–18659. [DOI] [PubMed] [Google Scholar]
- Haga RB, Ridley AJ (2016). Rho GTPases: regulation and roles in cancer cell biology. Small GTPases 7, 207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RG, Ridley AJ (2016). Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol 17, 496–510. [DOI] [PubMed] [Google Scholar]
- Hynes RO (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang N, Matsui TS, Deguchi S (2021). Statistical profiling reveals correlations between the cell response to and the primary structure of Rho-GAPs. Cytoskeleton (Hoboken) doi: 10.1002/cm.21659 [DOI] [PubMed] [Google Scholar]
- Kang N, Matsui TS, Liu S, Fujiwara S, Deguchi S (2020). Comprehensive analysis on the whole Rho-GAP family reveals that ARHGAP4 suppresses EMT in epithelial cells under negative regulation by Septin9. FASEB J 34, 8326–8340. [DOI] [PubMed] [Google Scholar]
- Lawson CD, Burridge K (2014). The on–off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases 5 e27958-27951-e27958-27912. doi: 10.4161/sgtp.27958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiyama H, Yamaguchi T, Watanabe TM, Fujita H (2017). A novel c-Src recruitment pathway from the cytosol to focal adhesions. FEBS Lett 591, 1940–1946. [DOI] [PubMed] [Google Scholar]
- Manninen A (2015). Epithelial polarity—generating and integrating signals from the ECM with integrins. Exp Cell Res 334, 337–349. [DOI] [PubMed] [Google Scholar]
- Manninen A, Varjosalo M (2017). A proteomics view on integrin-mediated adhesions. Proteomics 17, 1–16. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Myllyma¨ki SM, Manninen A (2017). Laminins in epithelial cell polarization: old questions in search of new answers. Cold Spring Harb Perspect Biol 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AE, Hu P, Barker TH (2020). Feeling things out: bidirectional signaling of the cell–ECM interface, implications in the mechanobiology of cell spreading, migration, proliferation, and differentiation. Adv Healthc Mater 1901445, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PM, Rademacher J, Bagshaw RD, Wortmann C, Barth C, van Unen J, Alp KM, Giudice G, Eccles RL, Heinrich LE, et al. (2020). Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat Cell Biol 22, 498–511. [DOI] [PubMed] [Google Scholar]
- Paloma SI-A, Carmen D-JN, Angeles G-P (1994). Activation of the Alpha 4 Beta 1 integrin through the Beta 1 subunit induces recognition of the RGDS sequence in fibronectin. J Cell Biol 126, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch EK, Aspalter IM, Sixt M (2016). Focal adhesion–independent cell migration. Annu Rev Cell Dev Biol 32, 469–490. [DOI] [PubMed] [Google Scholar]
- Rottner K, Hall A, Smal JV (1999). Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol 9, 640–648. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Shattil SJ (2000). Signaling networks linking integrins and Rho family GTPases. Trends Biochem Sci 25, 388–391. [DOI] [PubMed] [Google Scholar]
- Shen Y, Chen G, Gao H, Li Y, Zhuang L, Meng Z, Liu L (2020). miR-939-5p contributes to the migration and invasion of pancreatic cancer by targeting ARHGAP4. Onco Targets Ther 13, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Chen G, Zhuang L, Xu L, Lin J, Liu L (2019). ARHGAP4 mediates the Warburg effect in pancreatic cancer through the mTOR and HIF-1alpha signaling pathways. Onco Targets Ther 12, 5003–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Xu L, Ning Z, Liu L, Lin J, Chen H, Meng Z (2019). ARHGAP4 regulates the cell migration and invasion of pancreatic cancer by the HDAC2/beta-catenin signaling pathway. Carcinogenesis 40, 1405–1414. [DOI] [PubMed] [Google Scholar]
- Sun Z, Guo SS, Fassler R (2016). Integrin-mediated mechanotransduction. J Cell Biol 215, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Ye X, Simon S (2007). The integrins. Genome Biol 8, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt DL, Gray CD, Young WS, Orellana SA, Malouf AT (2007). ARHGAP4 Is a novel RhoGAP that mediates inhibition of cell motility and axon outgrowth. Mol Cell Neurosci 36, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ (2004). Focal adhesion regulation of cell behavior. Biochim Biophys Acta 1692, 103–119. [DOI] [PubMed] [Google Scholar]
- Wu C, Fields AJ, Kapteyn BA, McDonald JA (1995). The role of α4β1 integrin in cell motility and fibronectin matrix assembly. J Cell Sci 108, 821–829. [DOI] [PubMed] [Google Scholar]
- Yeh YC, Ling JY, Chen WC, Lin HH, Tang MJ (2017). Mechanotransduction of matrix stiffness in regulation of focal adhesion size and number: reciprocal regulation of caveolin-1 and beta1 integrin. Sci Rep 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.