Enhancer dememorization resets developmental clock and may unify epigenetic reprogramming modes in mammals and nonmammals.
Abstract
Marked epigenetic reprogramming is essential to convert terminally differentiated gametes to totipotent embryos. It remains puzzling why postfertilization global DNA reprogramming occurs in mammals but not in nonmammalian vertebrates. In zebrafish, global methylome inheritance is however accompanied by extensive enhancer “dememorization” as they become fully methylated. By depleting maternal dnmt1 using oocyte microinjection, we eliminated DNA methylation in early embryos, which died around gastrulation with severe differentiation defects. Notably, methylation deficiency leads to derepression of adult tissue–specific genes and CG-rich enhancers, which acquire ectopic transcription factor binding and, unexpectedly, histone H3 lysine 4 trimethylation (H3K4me3). By contrast, embryonic enhancers are generally CG-poor and evade DNA methylation repression. Hence, global DNA hypermethylation inheritance coupled with enhancer dememorization installs an epigenetic gate that safeguards embryonic programs and ensures temporally ordered gene expression. We propose that “enhancer dememorization” underlies and unifies distinct epigenetic reprogramming modes in early development between mammals and nonmammals.
INTRODUCTION
DNA methylation plays critical roles in embryonic development, genomic imprinting, transposon silencing, and X chromosome inactivation (1). Methylation of DNA at regulatory elements can interact with its binding proteins to exert gene repression (2). In mammals, DNA methytransferase 3A (DNMT3A) and DNMT3B can conduct de novo methylation (3). During DNA replication, DNA methylation is then robustly inherited by DNMT1, facilitated by a critical cofactor ubiquitin-like with PHD and RING finger domains 1 (UHRF1) (4). On the other hand, DNA demethylation is often carried out by members of the ten-eleven translocation (TET) family of enzymes (5). Deficiency in these enzymes often leads to embryonic lethality (2, 3, 5). DNA methylation at regulatory elements is considered to be associated with long-term and stable gene repression (2). While the numbers of promoters that are subjected to dynamic methylation are limited in the genome, DNA methylation at enhancers is highly dynamic during development and cell differentiation (6–8). Enhancer activation is often associated with hypomethylation, and hypermethylation at enhancer is shown to be repressive (9, 10). Enhancers often remain hypomethylated even after decommission, and this state can serve as a developmental memory, as these enhancers can be activated once the corresponding transcription factors (TFs) reappear (11, 12).
In mammals, DNA methylome undergoes marked reprogramming including global demethylation in primordial germ cells (PGCs) and preimplantation embryos, followed by genome-wide remethylation (13, 14). This reprogramming is essential in removing genomic imprints and parental memories (15, 16). Despite locus-specific reprogramming, there is no global DNA methylation reprogramming during early embryonic development in many nonmammalian vertebrates, such as zebrafish (Danio rerio) and Xenopus laevis (17, 18). This is unexpected given that histone marks, by contrast, are shown to undergo global resetting in these animals (19–21). However, the significance of the global methylome inheritance in nonmammalian vertebrates remains elusive. In particular, previous studies showed paradoxically that zebrafish mutants deficient in zygotic dnmt1 and uhrf1 can survive up to a week, a stage well beyond early development as the primordial organ development is largely completed (22–24). While these data argue against a role of DNA methylation in embryonic development in zebrafish, it is also possible that the maternal supplies of dnmt1 mRNAs or proteins may be sufficient to support these mutants beyond early development. Knocking down uhrf1 using morpholino in zebrafish embryos (25) or overexpressing STELLA, a protein that can sequester UHRF1 and induce DNA demethylation, in medaka embryos (26), causes notable mortality by gastrulation. However, a similar knockdown toward dnmt1 yielded no embryonic phenotype (23). It was proposed that this discrepancy may arise from methylation-independent functions of Uhrf1 or specific methylome pattern differences between uhrf1 and dnmt1 mutants (25). Therefore, whether DNA methylation is required for zebrafish embryonic development remains elusive thus far.
Despite the persisting global DNA methylation in zebrafish early embryos, marked local DNA methylation reprogramming occurs at regulatory elements during parental-to-zygotic transition. While promoters undergo “bidirectional” programming including both demethylation and methylation during this process (21, 27, 28), our previous work revealed largely “unidirectional” reprogramming for enhancers which become fully methylated (thus “dememorized”) either before fertilization for sperm or just after fertilization for oocyte (20). Enhancers are not demethylated until the phylotypic stage, when Tet proteins start to be expressed (29, 30). However, the functional significance of this enhancer dememorization and how embryonic enhancers can operate while being unusually hypermethylated (31) remain unanswered.
Here, we interrogated the function of DNA methylome and its inheritance in early zebrafish development, by generating dnmt1 maternal knockdown (mKD) embryos via our recently developed technology OMIS (oocyte microinjection in situ) (32). These embryos showed markedly depleted DNA methylation before zygotic genome activation (ZGA), which then failed to initiate epiboly and died around gastrulation. Careful analyses revealed defective cell differentiation, derepression of transposons, and failed establishment of Polycomb domains. These mutant embryos also showed widespread ectopic activation of adult enhancers and genes. Embryonic and adult enhancers show distinct CG densities and sensitivity to DNA methylation, enabling global DNA methylation as a critical epigenetic gate (“EpiGate”) to separate embryonic and adult programs. The distinct methylation sensitivity between embryonic and adult enhancers is likely sequence coded, as adult enhancers are preferentially CG-rich, while embryonic enhancers are generally CG-poor. Collectively, our study revealed that global DNA methylome inheritance is essential for early development and cell differentiation. Coupled with enhancer dememorization, DNA methylation resets an EpiGate after fertilization that safeguards embryonic programs and prevents premature firing of adult programs, to ensure temporally ordered activation of developmental programs. Furthermore, we propose that enhancer dememorization underlines epigenetic reprogramming of early development in both mammals and nonmammalian vertebrates, despite the distinct methylome reprogramming modes.
RESULTS
Maternal dnmt1 is essential for embryonic DNA methylation and development
To determine which Dnmts are responsible for maintaining embryonic DNA methylomes in zebrafish (Fig. 1A), we performed RNA sequencing (RNA-seq) to oocyte, zygote, pre-ZGA embryos (4-cell and 256-cell), post-ZGA embryos (dome, a stage shortly after ZGA, and shield, a stage when gastrulation occurs), and larva [head and tail, 5 days postfertilization (dpf)]. High expression levels of dnmt1 were found from oocyte to the 256-cell stage, indicating abundant maternal Dnmt1 (fig. S1A). By contrast, the de novo Dnmts (Dnmt3/4/5/3b, the orthologs of mammalian DNMT3B, and Dnmt3aa/3ab, the orthologs of mammalian DNMT3A) show relatively low expression in zebrafish oocytes and early embryos. Our initial attempt to deplete Dnmt1 and DNA methylation by knocking down dnmt1 in early embryos [zygotic knockdown (zKD)] or by knocking out zygotic dnmt1 [zygotic knockout (zKO)] through crossing dnmt1+/− heterozygotes (23) failed to reduce Dnmt1 protein or DNA methylation in zebrafish early embryos (Fig. 1, B and C, and fig. S1, B and C). Consistent with previous studies in zebrafish (22, 23), zKD and zKO embryos can survive more than 1 week after fertilization (fig. S2A). We validated zKO as the methylome analysis showed that the tail and head from zKO mutants exhibited DNA methylation loss at a later stage (larva, 5 dpf) as previously described (Fig. 1C and fig. S1C) (23). We therefore applied OMIS (see Fig. 1B and Materials and Methods), a method we developed recently to target maternal factors (32), to knockdown maternal dnmt1 starting from oocytes rather than from zygotes (termed mKD). Briefly, we injected dnmt1 morpholino (MO) into stage III oocytes [prophase I arrested oocytes, germinal vesicle (GV) stage] in situ while they were still kept in ovary, and the recipient female zebrafish were allowed to recover and mate with wild-type (WT) male to generate embryos naturally about 40 hours after injection. We collected embryos derived from these injected oocytes (rhodamine B traced) at the 256-cell, dome, and shield stages and examined the states of DNA methylation (Fig. 1B). Immunofluorescence (IF) analyses showed nearly absent signals of Dnmt1 protein or 5-methylcytosine (5mC) in mKD embryos from the 256-cell to shield stage (fig. S1B, top). Low-input methylome profiling using STEM-seq (small-scale TELP-enabled methylome sequencing) (33) confirmed marked loss of global DNA methylation in mKD embryos at the 256-cell stage (from 85 to 20%), which is further exacerbated at dome (17%) and shield (7%) stages (Fig. 1C and fig. S1C). Notably, dnmt1 mKD embryos displayed severe defects and died around gastrulation [10 hours postfertilization (hpf)] (Fig. 1, D and E). In WT embryo, cells at the margin region of blastula embryos initiate cell movements, or “epiboly,” to form dorsal-ventral axis at shield stage (34). By contrast, dnmt1 mKD embryos failed to initiate this epiboly and died accompanied with yolk extrusion, likely caused by the ectopic stress from cell movement defects (Fig. 1, D and E, and fig. S2B). dnmt1 mKD embryos contain fewer cells especially after ZGA (fig. S2C), indicating defects in cell proliferation and/or cell apoptosis. Therefore, maternal Dnmt1 is required for both DNA methylome inheritance and early development in zebrafish.
Fig. 1. Maternal dnmt1 is essential for embryonic DNA methylation and development.
(A) Schematic of DNA methylation landscapes in zebrafish (red) and mammals (blue) during early development. mCG, methylated CG. (B) Schematic of dnmt1 mKD via OMIS, zKD, and zKO. Three main developmental stages were examined in this study, including the 256-cell (pre-ZGA), dome, and shield (post-ZGA) stages. ZGA begins around 3 hpf. zKO embryos (dnmt1−/−) could survive until 120 hpf. (C) Violin plots showing average DNA methylation levels across the genome at different developmental stages of control (blue) and dnmt1 mKD embryos (red), zKD embryos (red), and zKO (red) embryos/larvae. (D) Survival curve of control (blue line), dnmt1 mKD embryos (red line), and dnmt1 mKD embryos rescued with either dnmt1 mismatch mRNAs [mKD; WT overexpressed embryos (WT OE), green dashed line] or catalytically mutant dnmt1 mismatch mRNAs (mKD; Mut OE, orange dashed line). Log-rank test was used to calculate P value. (E) Representative images of embryo phenotypes in control, dnmt1 mKD embryos, and dnmt1 mKD embryos rescued with either dnmt1 mismatch mRNAs (WT OE) or catalytically inactive mutant dnmt1 mismatch mRNAs (Mut OE) across different developmental stages. The numbers and ratios of embryos with a particular phenotype in each group are also shown. Scale bar, 250 μm. (F) Immunostaining of 5mC (green) in control and dnmt1 mKD oocytes, 256-cell, and dome embryos, as well as dnmt1 mKD embryos rescued by either dnmt1 mismatch mRNA (WT OE) or catalytically inactive mutant dnmt1 mismatch mRNA (Mut OE). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bars, 50 μm. (G) Violin plot showing the global DNA methylation levels at the 256-cell and dome stages in control (Ctrl, blue), dnmt1 mKD (red), dnmt1 mismatch mRNA rescued (WT OE, green), and catalytically inactive mutant dnmt1 mismatch mRNA rescued embryos (Mut OE, yellow).
The loss of maternal Dnmt1 and embryonic DNA methylomes is responsible for the early lethality
We then asked whether the lethality of dnmt1 mKD embryos is caused by the loss of DNA methylation. First, coinjecting dnmt1 morpholino with dnmt1 mismatch mRNA (with the codons modified to avoid being targeted by MO without changing amino acid coding; see Materials and Methods), but not catalytically inactive mutant dnmt1 mismatch mRNA (with MO target codons modified and catalytic domain mutated; Materials and Methods), during OMIS can efficiently rescue the epiboly delay in dnmt1 mKD embryos (Fig. 1, D and E) and restore 5mC signal at the 256-cell and dome stages, although with final levels moderately lower than the control group (Fig. 1, F and G, and fig. S2D). Second, we examined whether mKD of dnmt1 affects oocyte development. Both IF and low-coverage STEM-seq (due to limited numbers of mutant oocytes) analyses showed that the global DNA methylation in dnmt1 mKD oocytes is grossly retained (Fig. 1F and fig. S3A). In addition, the MOF (maturation, ovulation, and fertilization) (32) rates of oocytes were comparable between control and dnmt1 mKD oocytes (see Materials and Methods and fig. S3B). RNA-seq of oocytes also revealed few dysregulated genes in dnmt1 mKD oocytes compared to control oocytes (fig. S3C). Last, to further rule out a role of oocyte defects in the embryonic lethality caused by dnmt1 mKD, we overexpressed mouse STELLA [also known as DPPA3 (developmental pluripotency associated 3)], a protein that can sequester UHRF1 thus preventing the proper functions of DNMT1 to induce DNA demethylation (35), in zebrafish zygotes (instead of oocytes) (fig. S3, D and E). DNA methylation is severely impaired in Stella overexpressed embryos (Stella OE) (fig. S3E), which showed developmental arrest at 14 hpf (somite stage) (fig. S3F). Evident developmental delay or embryonic lethality was observed as early as shield stage and become prevalent around the somite stage (fig. S3F). By contrast, overexpression of a mutant STELLA deficient in interaction with UHRF1 [KRR mutant (K85E/R86E/R87E); see Materials and Methods] (35) has little impact on 5mC level (fig. S3E) and development (fig. S3F). Hence, we conclude that DNA methylation is essential for early zebrafish development.
Single-cell RNA-seq analysis revealed differentiation defects in dnmt1 mKD embryos
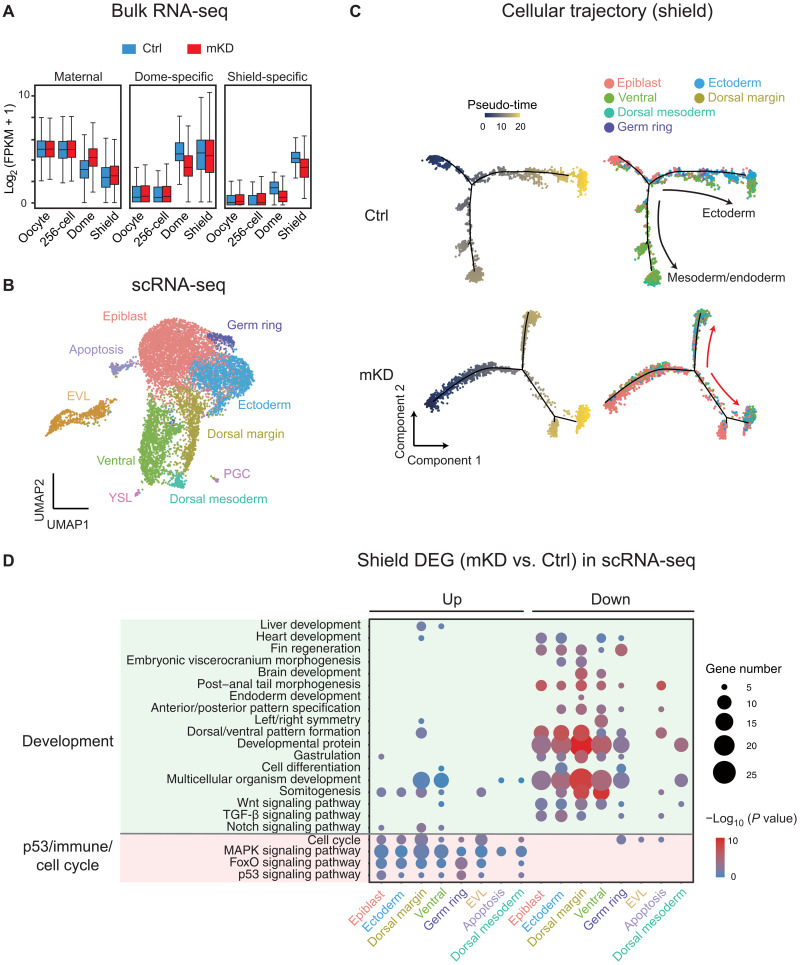
We then asked how transcriptome is altered in these dnmt1-deficient embryos. To do so, we first performed bulk RNA-seq for control and dnmt1 mKD embryos at the 256-cell, dome, and shield stages. Globally, maternal RNA [expressed in oocytes, fragments Per Kilobase of exon model per Million mapped fragments (FPKM) > 10] degradation was delayed in dnmt1 mKD embryos but was nevertheless achieved to a large degree by shield stage (Fig. 2A). Similarly, the activation of dome-specific genes [FPKM >10 in dome embryos and not expressed in oocytes (FPKM <5)] was delayed in mutants at dome stage and was partially recovered at shield stage (Fig. 2A). Activation of shield-specific genes [FPKM >10 in shield embryos and not expressed in oocytes and dome (FPKM <5)] was also partially affected (Fig. 2A). Given the cell heterogeneity of gastrula, we then performed 10× single-cell RNA-seq (scRNA-seq) and profiled 19,563 and 11,852 cells from control and mKD dome embryos, respectively, and 11,288 and 20,595 cells from control and mKD shield embryos, respectively (see Materials and Methods and fig. S4A). We confirmed the scRNA-seq results with a second replicate of lower depth (fig. S4B). Clustering analysis for integrated data of both control and mKD embryos using Seurat (36) identified a total of 10 clusters, including PGCs, enveloping layer cells (EVLs), yolk syncytial layer cells (YSLs), epiblast, ectoderm, germ ring, ventral, dorsal mesoderm, and dorsal margin (Fig. 2B and fig. S4C). Most clusters were also present in dnmt1 mKD embryos, suggesting that these embryos were able to initiate cell differentiation (fig. S5A). However, mutant embryos showed increased epiblast cells (fig. S5A, red arrow) and decreased ectoderm and ventral cells (fig. S5A, blue arrow) at shield stage (fig. S5, A and B), suggesting inefficient lineage differentiation. We then analyzed cellular trajectories of control and mKD embryos in pseudo-time using Monocle 2 (37). Cell differentiation initiates from epiblast along two major trajectories, including ectoderm (mainly located on the animal pole) and mesoderm/endoderm (mainly located in marginal zone, whose precursors are arranged along the dorsal-ventral axis) (34) directions in control embryos (Fig. 2C and fig. S5C), consistent with previous scRNA-seq data in zebrafish early embryos (38, 39). By contrast, dnmt1 mKD shield embryos exhibited multiple branches from epiblast that led to a mixture of differentiated cells and undifferentiated epiblast, suggesting aberrant differentiation programs. Consistently, down-regulated genes of each cell cluster are overwhelmingly enriched for developmental genes and genes related to gastrulation and cell movement (Fig. 2D and fig. S5, D and E). Several lineage markers showed incorrect spatial expression patterns, including tph1b and frzb (for dorsal margin) and eve1 and dld (for ventral margin) (fig. S6). Notably, developmental genes, including some that are typically expressed in adult tissues such as ntsr1, ncaldb, atxn1b, and lrrtm1, were also enriched in up-regulated genes in mKD embryos (Fig. 2D and fig. S6) (discussed in detail later). Hence, scRNA-seq analysis revealed widespread differentiation defects in dnmt1 mKD embryos.
Fig. 2. Bulk and scRNA-seq revealed developmental defects in mKD embryos.
(A) Distribution of RNA levels of maternal genes (left), dome-specific genes (middle), and shield-specific genes (right) across different developmental stages of control (blue) and dnmt1 mKD (red) embryos. (B) Projection of cells with Uniform Manifold Approximation and Projection (UMAP) for control and dnmt1 mKD embryos at dome and shield stages. Cells are colored by clusters. (C) Pseudo-time trajectory of control and dnmt1 mKD embryos at shield stage. Cells were ordered from epiblast to ectoderm or mesoderm and endoderm and colored by pseudo-time (left) or clusters in (B) (right). Red arrows indicate abnormal cell differentiation branches. (D) Bubble plot showing enriched Gene Ontology (GO) terms for differentially expressed genes (DEGs) between control and dnmt1 mKD embryos of each cluster at shield stage. Top enriched terms include development and p53-dependent apoptosis, immune response, and cell cycle–related genes. Size of circle encodes gene number; color of the circle indicates −log10(P value). TGF-β, transforming growth factor–β; MAPK, mitogen-activated protein kinase.
dnmt1 mKD embryo lethality is partially contributed by transposon derepression triggered immune response and p53-mediated cell apoptosis
Notably, up-regulated genes in dnmt1 mKD embryos are also enriched for p53 signaling, cell cycle, and immune response (Fig. 2D and fig. S5E), consistent with the observation in hypomethylated dnmt1 zKO zebrafish larva (>3 dpf) where innate immune response and p53-mediated cell apoptosis are often activated upon the loss of DNA methylation due to the activation of transposons (40). Using WT and p53-null female zebrafish (fig. S7, A and B), we observed increased cell apoptosis in dnmt1 mKD p53+/+ embryos but not in dnmt1 mKD p53−/− embryos (fig. S7C). The dnmt1 mKD p53−/− embryos could now initiate epiboly movements and form dorsal-ventral axis, although with a delayed kinetics. However, these embryos eventually died around 12hpf (fig. S7B), suggesting that p53-dependent cell apoptosis is only partially responsible for embryo lethality in dnmt1 mKD embryos. Furthermore, analysis of the total RNA-seq data revealed derepression of several classes of transposable elements (TEs), in particular, long terminal repeats and their subfamily gypsy, in mKD embryos at dome stage, which is further exacerbated at shield stage (fig. S7, D to F). This is accompanied by increased genome instability, as manifested by increased phosphorylated H2AX signals, a marker of DNA double-strand breaks (DSBs) (fig. S8A) (41). Similar observation was made for STELLA overexpressed zebrafish embryos (fig. S8B). We then asked whether the induced DSBs may be partially responsible for the lethality of DNA methylation–deficient embryos, by treating embryos with Foscarnet (FOS; see Materials and Methods), an inhibitor of reverse transcriptases and polymerases and an effective antiviral agent (42). We used Stella OE embryos in this experiment as dnmt1 mKD using OMIS could not produce large numbers of embryos required for drug treatment and statistical analysis. Although effectively eliminated DSB from the Stella OE embryos, FOS treatment, however, could not rescue their developmental defects (fig. S8, B and C). These data indicate that the transcription of transposon or the resulting transcripts, but not its retrotranscribed DNA or the subsequent DNA damage, may trigger developmental defects in dnmt1 mKD embryo. Phosphorylation of TANK-binding kinase 1 (pTbk1) acts as an indicator of viral sensor signaling activation (43). The treatment of BX795, an inhibitor of pTbk1 that can repress interferon-response genes in zebrafish (40), could rescue a small fraction (9 of 111, 8.1%) of Stella OE embryos (fig. S8C). Therefore, we conclude that p53-dependent cell apoptosis and pTbk1-mediated immune response, likely triggered by transposon derepression, partially contribute to developmental defects in dnmt1 mKD embryos.
The loss of promoter DNA methylation is responsible for part, but not all, of gene derepression in dnmt1 mKD embryos
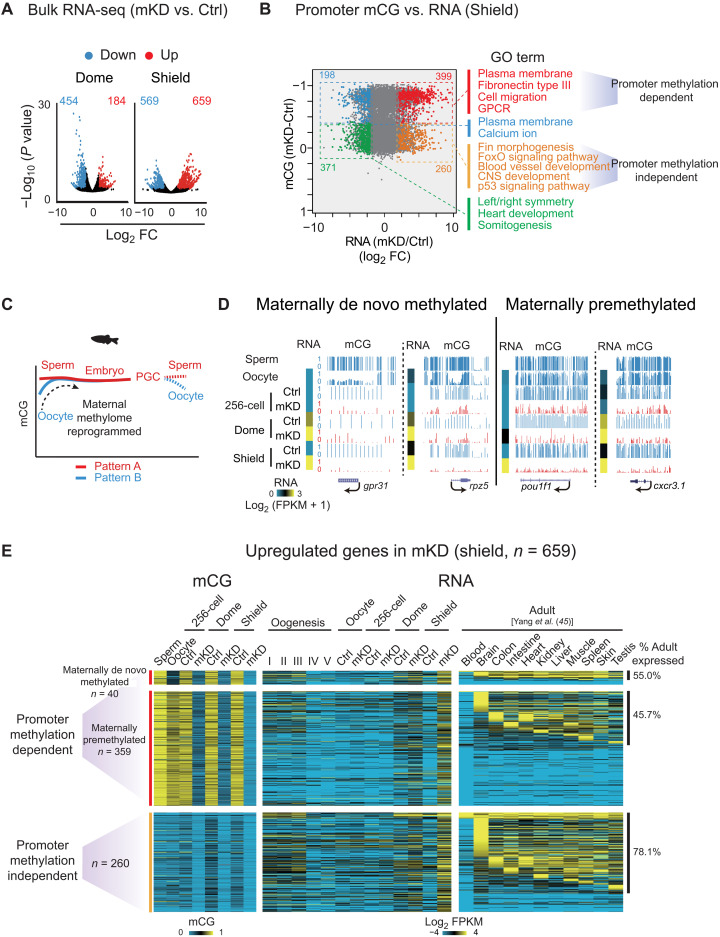
Next, we investigated how the transcription defects in mKD embryos are related to the loss of DNA methylation. As the lingering maternal transcripts may mask embryonic transcription, we excluded “maternal genes” (FPKM > 5 in oocytes) from the analysis. Given the repressive roles of DNA methylation, we focused on up-regulated genes and interrogated their expression and promoter methylation states (Fig. 3, A and B). We first examined shield stage embryos, where gene up-regulation is more evident in mutants (Fig. 3, A and B). About 399 up-regulated genes show the loss of DNA methylation at promoters, which mainly function in plasma membrane and fibronectin (involved in cell movements likely related to epiboly), cell migration, and G protein–coupled receptor (Fig. 3B, red). The rest 260 up-regulated genes showed no significant changes of DNA methylation (Fig. 3B, orange). Ninety-eight percent of these promoters are hypomethylated in both control and mKD embryos. These genes are preferentially enriched for fin morphogenesis, blood vessel development, and CNS (central nervous system) development (Fig. 3B, orange), which appear to be more related to adult tissue development. Examples of adult development–related genes, including ntsr1, atxn1b, ncaldb, and lrrtm1, were also observed in scRNA-seq data (fig. S6). For convenience, we termed these two groups as “promoter methylation–dependent” and “promoter methylation–independent” genes. A similar analysis of dome stage embryos revealed 130 promoter methylation–dependent and 54 promoter methylation–independent genes, with 55.4% of promoter methylation–dependent genes overlapping those at shield stage (fig. S9, A and B). In sum, promoter DNA methylation loss is likely responsible for some, but not all, derepressed genes.
Fig. 3. Promoter DNA methylation–dependent and –independent gene derepression in mKD embryos.
(A) Volcano plots showing gene expression changes between control and dnmt1 mKD embryos at dome and shield stages. Red and blue dots indicate up-regulated and down-regulated genes, respectively. The numbers indicate counts of dysregulated genes. FC, fold change. (B) Scatter plots comparing alteration of gene expression and promoter mCG between control and dnmt1 mKD embryos at shield stage. Red and orange dots indicate promoter DNA methylation–dependent and –independent up-regulated genes, respectively; blue and green dots indicate down-regulated genes with decreased and constant promoter DNA methylation, respectively. The numbers of dysregulated genes and enriched GO terms in corresponding group (color coded) are also shown. GPCR, G protein–coupled receptor. (C) Schematic of DNA methylation reprogramming from gametes to the next generation in zebrafish. Sperm, early embryo, and PGC exhibit highly similar methylomes (pattern A) (27, 28). Oocyte has a distinct methylome (pattern B) that will be reprogrammed to pattern A after fertilization. (D) UCSC Genome Browser snapshots showing promoter mCG in sperm, oocyte, 256-cell, dome, and shield stage embryos for maternally de novo methylated [mCG(dome or shield − oocyte) > 0.4; mCG(oocyte < 0.4)] and maternally premethylated genes [mCG(dome or shield – oocyte) ≤ 0.4; mCG(oocyte ≥ 0.4)]. RNA of the 256-cell, dome, and shield stage embryos is also shown (heatmap). (E) Heatmaps showing promoter mCG and RNA expression of promoter DNA methylation–dependent or –independent genes in (B) of oogenesis, early embryos, and adult tissues (45). Promoter DNA methylation–dependent genes were further classified into maternally de novo methylated and maternally premethylated genes groups based on the mCG levels in oocyte and early embryos. The ratios of tissue expressed genes (FPKM > 5) in each group are also shown, and statistical significance for the enrichment was assessed with one-sided Fisher’s exact test.
Notably, despite the inheritance of global DNA methylome after fertilization, promoter-specific DNA methylation reprogramming does occur in zebrafish early embryos (27, 28). In particular, promoter methylation on the maternal genome is reconfigured (including both methylation and demethylation) to a pattern that is similar to that of the paternal genome (Fig. 3C) (27, 28). This “sperm-like” methylome also persists to PGCs (44). The significance of this reprogramming, however, remains elusive. We focused on promoters that are hypermethylated in dome and shield stage embryos, among which 842 promoters are hypomethylated in oocytes, hence indicating de novo methylation after fertilization (“maternally de novo methylated” promoters) (Fig. 3D and fig. S9C). Those that remain hypermethylated in oocyte and dome/shield stage embryos are thus “maternally premethylated” promoters (n = 7318) (Fig. 3D). About 41.0% of maternally de novo methylated genes are expressed during oogenesis or PGCs, and 32.5% are expressed during embryonic development or in adult tissues (fig. S9, C and D). However, only 4.8% (n = 40 of 842) of these maternally de novo methylated genes are derepressed in mKD embryos by shield stage. Therefore, it appears that the function of this oocyte-to-embryo reprogramming is not just restricted to gene repression during the imminent ZGA. Rather, these data raise an interesting possibility that this reprogramming may restore promoter methylation to a “ground” state to facilitate both ZGA and future development.
We then asked when these derepressed genes normally express in WT zebrafish. Using a collected RNA-seq data from a total of 11 adult tissues (45), we found that 46.6% of promoter methylation–dependent group are expressed in at least one adult tissue [55.0% for maternally de novo methylated (n = 40) and 45.7% for maternally premethylated (n = 359), Fig. 3E]. The ratio of tissue expressed genes is, however, substantially higher for promoter methylation–independent group (78.1%; Fig. 3E; also compared to 53.3% of random genes). This also echoes Gene Ontology (GO) term analysis, which showed that promoter methylation–independent group genes are preferentially involved in later development (such as “fin morphogenesis,” “blood vessel development,” and “CNS development”) (Fig. 3B). Overall, 389 (59.0%) up-regulated genes are expressed in adult tissues. To confirm that these genes are preferentially expressed at late stages during development, we examined their expression using scRNA-seq data from 4 to 24 hpf of WT embryos (39). We found that only 19.3% (75 of 389) are expressed before 24 hpf (phylotypic stage), while the rest 80.7% (311) are activated at least after 24 hpf (fig. S9E). Therefore, these data indicate that adult programs are aberrantly activated in dnmt1 mKD embryos.
DNA methylation is required for the proper establishment of Polycomb domains
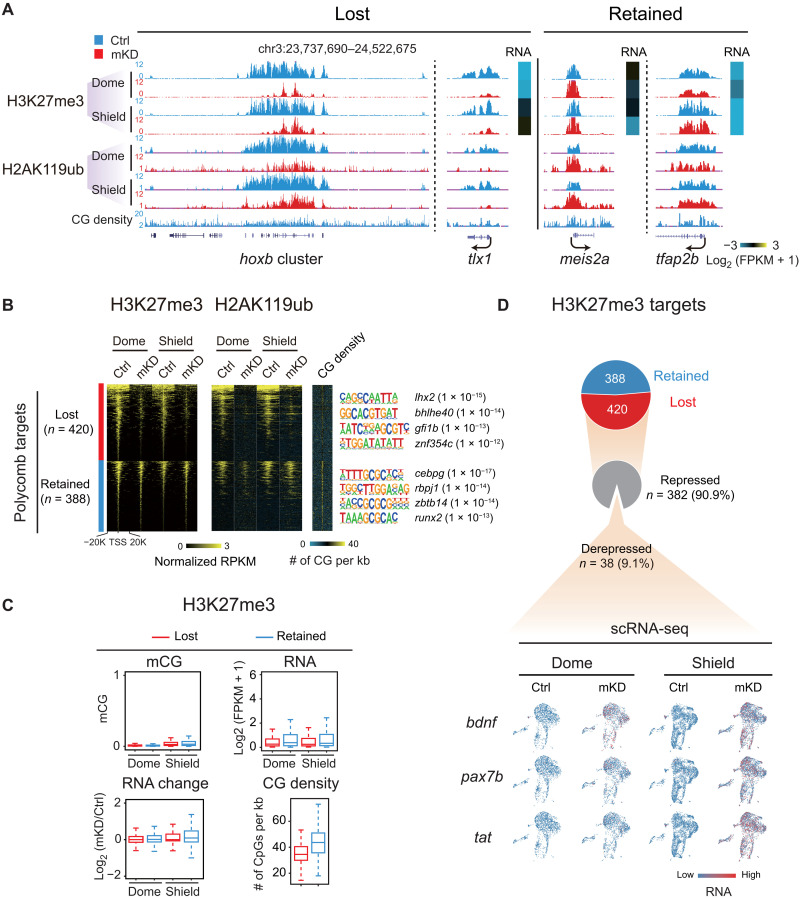
We then investigated what may contribute to promoter methylation–independent gene derepression in dnmt1 mKD embryos. Histone modifications undergo extensive global loss and reestablishment during early zebrafish development (20, 21, 46). In particular, Polycomb domains, marked by the repressive marks trimethylated histone H3 at lysine 27 (H3K27me3) and mono-ubiquitinated histone H2A at lysine 119 (H2AK119ub), are established around ZGA (47, 48). These marks, deposited by Polycomb repressive complexes 2 (PRC2) and PRC1, respectively, are critical repressors for key developmental genes (49). Thus, we collected dnmt1 mKD embryos at dome and shield stages and performed H3K27me3 and H2AK119ub CUT&RUN (50). Notably, H3K27me3 and H2AK119ub reestablishment are severely impaired in dnmt1 mKD embryos (Fig. 4, A and B, and fig. S10A). About 51.7% (420 of 808) promoter H3K27me3 peaks are lost or strongly reduced in dnmt1 mKD embryos. The rest (48.3%, 388 of 808) appear to be largely intact (Fig. 4B). These two groups show similar enrichment for developmental genes (fig. S10B), although their promoters appear to enrich for distinct TF motifs (Fig. 4B). The “retained” group shows higher CG levels at promoters (Fig. 4, B and C), consistent with the notion that CG-rich sequences can recruit Polycomb (51). The fact that these marks at some, but not other, genes are affected suggests that this is not simply due to developmental delay. IF analysis also excluded a possibility of global H3K27me3 decrease in mKD embryos (fig. S10C). A similar trend was observed for H2AK119ub (Fig. 4, A and B). This result is reminiscent of the observation in mouse embryonic stem cells (mESCs), where the loss of global DNA methylation leads to decrease in promoter H3K27me3 (51), and supports the model that the absence of DNA methylation in the genome elsewhere may allow the spreading and/or recruitment of Polycomb, which, in turn, dilutes Polycomb and H3K27me3 away from promoters (51). Nevertheless, the decreased H3K27me3 and H2AK119ub did not cause apparent widespread gene derepression (Fig. 4, C and D), as only 38 (9.1%) genes that lost promoter H3K27me3 showed moderate derepression, including bdnf, pax7b, and tat (Fig. 4D). Therefore, these data demonstrate that DNA methylation is crucial for proper establishment of Polycomb domains at developmental gene promoters in early zebrafish embryos. However, the loss of repressive marks, such as H3K27me3 and H2AK119ub, cannot fully explain promoter methylation–independent gene derepression in dnmt1 mKD embryos.
Fig. 4. dnmt1 mKD results in defective Polycomb domain establishment.
(A) UCSC Genome Browser snapshot showing H3K27me3 and H2AK119ub of control (blue) and dnmt1 mKD (red) embryos at dome and shield stages. Heatmap showing RNA expression of related genes. (B) Heatmaps showing the normalized H3K27me3 and H2AK119ub enrichment around promoters (TSS ± 20 kb) of Polycomb target in control (blue) and dnmt1 mKD (red) embryos at dome and shield stages. CG density and enriched TF motifs are shown too (right). (C) Box plots showing promoter mCG, RNA, and CG density levels for two clusters of Polycomb targets [lost (red) and retained (blue), defined in (B)] at dome and shield stages of control (top). RNA changes between control and dnmt1 mKD embryos (RNA change) are also shown (bottom left). (D) Pie chart showing the percentages of Polycomb target genes with lost or retained H3K27me3 in dnmt1 mKD embryos at shield stage and, among “lost” group genes, the percentages of genes that are derepressed [log2FC (mKD/Ctrl) > 2] and remain repressed (gray) (top). The feature plots of scRNA-seq show RNA expression of example derepressed genes in control and dnmt1 mKD embryos at dome and shield stages (bottom). Each dot indicates one cell. Red, high expression; blue, low expression.
Loss of DNA methylation results in aberrant activation of adult enhancers with ectopic H3K4me3 and TF binding
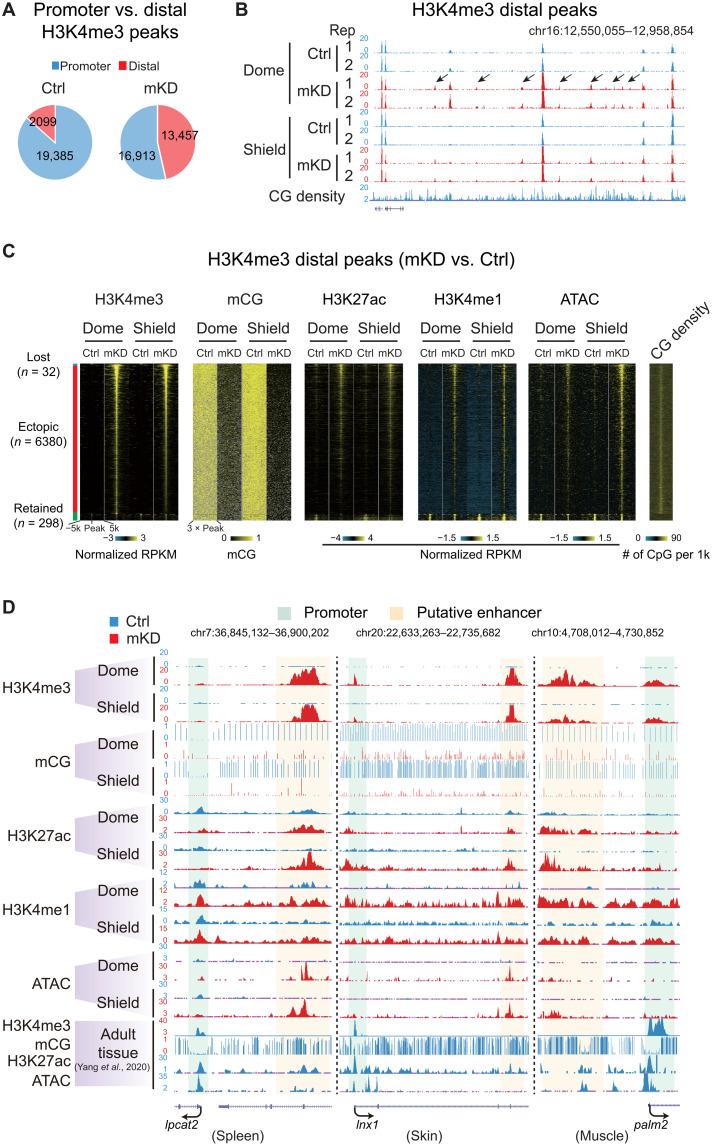
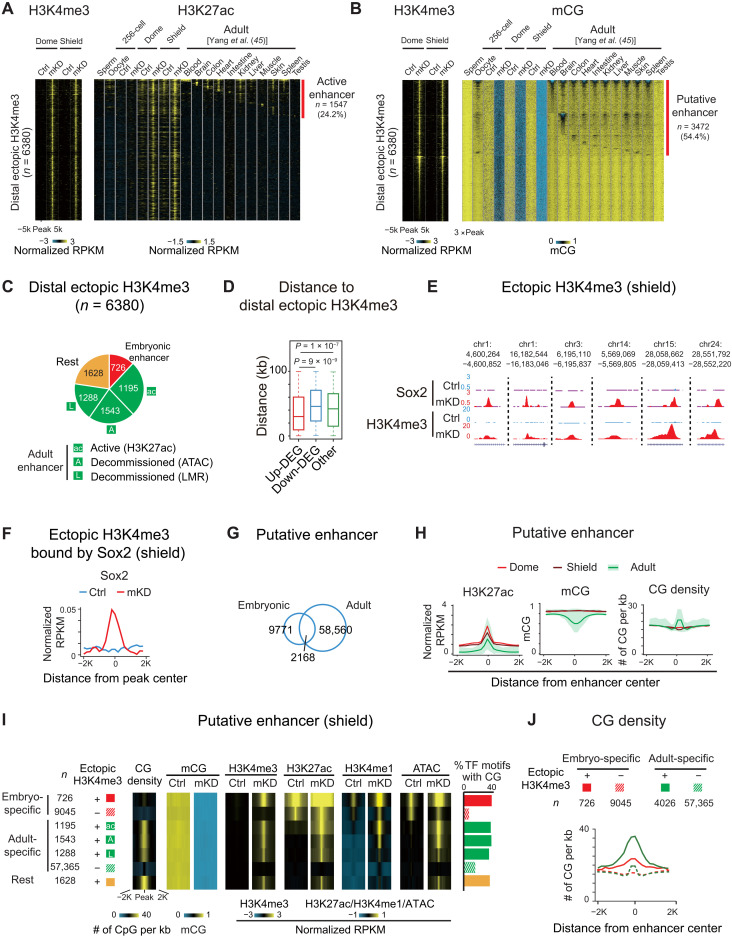
Besides promoters, distal regulatory elements, such as enhancers, play crucial roles in gene regulation. To ask whether they may play a role in gene derepression in dnmt1 mKD mutants, we profiled histone marks H3K4me3, H3K4me1, acetylation of histone H3 at lysine 27 (H3K27ac) (see Materials and Methods), and chromatin accessibility using assay for transposase-accessible chromatin using sequencing (ATAC-seq). Notably, we found that H3K4me3, a typical permissive promoter mark, is highly dynamic upon the loss of DNA methylation especially in distal regions (Fig. 5, A and B). While the overall numbers of promoter H3K4me3 peaks showed a moderate decrease from 19,385 to 16,913, the distal H3K4me3 peaks increase markedly from 2099 to 13,457 (Fig. 5A). As a result, the ratio of distal H3K4me3 peaks among all H3K4me3 peaks increases substantially from 9.8% in control to 44.3% in mKD embryos. Even after excluding weak distal H3K4me3 peaks [normalized reads per kilobase of bin per million of reads sequenced (RPKM) < 0.5 in both control and mKD samples], there are still 7580 distal H3K4me3 peaks left. To rule out the possibility that these regions may be unannotated promoters, we further excluded distal H3K4me3 peaks that overlap with any H3K4me3 peaks (promoter mark) in adult tissues (45) and unmethylated regions in the 256-cell embryos (when all enhancers are presumably methylated, leaving only promoters unmethylated) (see Materials and Methods) (20). This still yielded a total of 6380 distal H3K4me3 peaks. This widespread distal H3K4me3 is unique to mutant embryos, as we only identified 32 distal H3K4me3 peaks specifically in control embryos and 298 peaks present in both control and mKD embryos (Fig. 5C). To understand the nature of these ectopic distal H3K4me3 sites, we mapped the states of DNA methylation, H3K27ac, H3K4me1, open chromatin (ATAC-seq), and CG density in different groups based on whether a distal H3K4me3 peak is lost, ectopically acquired, or retained in mutant (Fig. 5C). Ectopic distal H3K4me3 sites also showed substantially increased H3K27ac, H3K4me1, and chromatin accessibility, as manifested globally (Fig. 5C) and also at individual genes (Fig. 5D), indicating enhanced regulatory activities. Notably, a fraction of these regions (11.4%) also showed H3K27ac in control embryos (dome and shield), suggesting that these elements are likely active in WT embryos and their activities further increased in mutants (discussed later).
Fig. 5. Loss of DNA methylation causes ectopic activation of putative adult enhancers and aberrant acquisition of distal H3K4me3 in early embryos.
(A) Pie charts showing distributions of promoter (blue) and distal (red) H3K4me3 sites in control and mKD embryos at shield stage. (B) UCSC Genome Browser snapshot showing H3K4me3 at dome and shield stages of control (blue) and dnmt1 mKD (red) embryos, and CG density. Arrows indicate ectopic H3K4me3 sites. (C) Heatmaps showing distal H3K4me3, mCG, H3K27ac, H3K4me1, open chromatin (ATAC-seq), and CG density in either control or dnmt1 mKD embryos. Peaks were classified into three clusters based on the dynamics of distal H3K4me3: lost (H3K4me3 lost in dnmt1 mKD embryos, blue), ectopic (H3K4me3 acquired in dnmt1 mKD embryos, red), and retained (H3K4me3 present in both control and mKD, green). (D) UCSC Genome Browser snapshots showing H3K4me3, mCG, H3K27ac, H3K4me1, and open chromatin (ATAC-seq) at dome and shield stages of control (blue) and dnmt1 mKD (red) embryos and adult tissues (spleen, left; skin, middle; muscle, right) (45). Green shadow indicates promoter, and orange shadow indicates putative enhancer.
Given H3K27ac, accessible chromatin, and, in particular, H3K4me1, are considered as hallmarks for enhancers (52), we then asked whether these elements are possibly enhancers. As most of them are not active in control early embryos, we examined their chromatin states in 11 adult tissues (45). Encouragingly, 24.2% (1547 of 6380) are marked by H3K27ac, which marks active enhancers (53), in at least one adult tissue (Fig. 6A). Enhancers can also stay in poised or decommissioned states, which no longer bear H3K27ac, but are still marked by accessible chromatin or DNA hypomethylation (11, 12). Notably, more than half (54.4%, 3472 of 6380) of these ectopic distal H3K4me3 overlap with lowly methylated regions (LMRs) identified in tissues (Fig. 6B). The ectopic H3K4me3 peaks also precisely align with the center of LMRs. Overall, we found that among all 6380 ectopic distal H3K4me3 sites, the majority (63.1%, 4026 of 6380) overlap with putative enhancers that are either active (marked by H3K27ac) or poised/decommissioned (marked by LMRs or accessible chromatin) in at least one of 11 adult tissue lineages (Fig. 6C). By contrast, only a small fraction (11.4%) overlaps with early embryonic putative enhancers (dome and shield stages). The rest 1628 (25.5%) peaks did not overlap with adult enhancers or embryonic enhancers (Fig. 6C, rest). We expect that this number would further decrease when adding additional tissue types and developmental stages. Last, we asked whether the presumably derepressed enhancers are within the proximity of derepressed genes. These putative enhancers are closer to up-regulated genes but not to down-regulated genes or non–DEGs (differentially expressed genes) (Fig. 6D). Hence, these data suggest that these ectopic distal H3K4me3 sites preferentially occupy putative adult enhancers, which are linked to derepression of adult genes.
Fig. 6. DNA methylation separates embryonic and adult programs through blocking high CG density adult enhancers.
(A and B) Heatmaps showing H3K4me3, H3K27ac (A), and mCG (B) around distal ectopic H3K4me3 in embryos and adult tissues (45). Peaks with H3K27ac signals are defined as active enhancers (A) (red). LMRs are defined as putative enhancers (B) (red). (C) Pie chart showing distribution of distal ectopic H3K4me3 overlapping with embryonic (red), active adult (green; “ac”, distal H3K27ac), decommissioned adult (green; “A”, ATAC-seq; “L”, LMRs; without H3K27ac) enhancers, and the rest ectopic H3K4me3 sites (orange). (D) Box plot showing distance from center of distal ectopic H3K4me3 peaks to up-regulated (red), down-regulated (blue), and non-DEGs (green) at shield stage. (E and F) UCSC Genome Browser snapshots (E) and line chart (F) showing Sox2 binding and H3K4me3 (E) and Sox2 binding distribution around ectopic H3K4me3 acquiring Sox2 (F) in control (blue) and dnmt1 mKD (red) embryos. (G) Venn diagram showing overlap between embryonic (dome and shield) and adult tissues (45) enhancers. (H) Line charts showing H3K27ac, mCG, and CG densities in dome (red), shield (dark red) embryos, and adult tissues (45) (green; line, average value; shade, range of all tissues) around putative enhancers defined at each stage. (I) Heatmaps showing average CG density, mCG, H3K4me3, H3K27ac, H3K4me1, and ATAC-seq of putative enhancers. Classification of enhancer (left): red/crossed red, embryonic-specific enhancer with/without distal ectopic H3K4me3; green/crossed green, adult enhancers with [defined in (C)]/without distal ectopic H3K4me3; orange, the rest distal ectopic H3K4me3. Bar chart shows the ratios of enriched TF motifs (P < 1 × 10−20) in putative enhancers containing CGs (right). (J) Line chart showing CG densities around embryonic specific enhancers with (red line)/without (red dashed line) distal ectopic H3K4me3, as well as adult enhancer with (green line)/without (green dashed line) distal ectopic H3K4me3.
Last, we asked whether ectopically activated enhancers can recruit TFs. The pluripotency factor SoxB1 is critical for ZGA and early development in zebrafish (54). SoxB1 includes six Sox genes sox1a, sox1b, sox2, sox3, sox19a, and sox19b. We chose Sox2, one of the earliest zygotic genes activated (4.3 hpf) (54) for which an antibody is conveniently available and performed CUT&RUN (50) in control and dnmt1 mKD embryos at shield stage (fig. S11A). Reassuringly, Sox2 binding in both control and mKD mutants enriches for Sox2 motif (fig. S11B). Ectopic Sox2 binding occurs in at least 570 ectopic H3K4me3 sites, preferentially aligning at centers of these H3K4me3 peaks (Fig. 6, E and F). About 12.7% (186 of 1463) of ectopic H3K4me3 sites with sox2 motif acquire Sox2 binding, while the number decreased to 7.8% (384 of 4917) for ectopic H3K4me3 without sox2 motif (P = 3 × 10−8) (fig. S11C). These Sox2 ectopic binding sites are also closer to derepressed genes in mKD mutants, although it did not reach statistical significance due to the limited numbers of genes (fig. S11D). Hence, ectopically activated adult enhancers recruit TFs and are correlated with gene derepression.
Embryonic and adult enhancers exhibit distinct DNA methylation sensitivity and CG densities
While DNA methylation appears to repress adult enhancers, it was reported that embryonic enhancers are hypermethylated and hence are insensitive to DNA methylation in zebrafish early embryos (29, 31). This is attributed to the absence of TET proteins, the key regulatory enzymes of DNA demethylation, as its expression is not detectable in zebrafish embryos until 24 hpf (the phylotypic stage) (fig. S11E) (29, 30). It remains elusive why DNA methylation can repress adult enhancers but not embryonic enhancers. Furthermore, given the large numbers of adult enhancers present in the genome (n = 60,728 across 11 adult tissues) (Fig. 6G), clearly not all adult enhancers are derepressed and acquire ectopic H3K4me3 in dnmt1 mutant embryos. To understand why certain enhancers are selectively sensitive to DNA methylation and are prone to ectopic H3K4me3 acquisition, we identified early embryonic enhancers (dome and shield stages) and adult enhancers (across 11 adult tissues) using distal H3K27ac and removed those overlapping with annotated promoters or H3K4me3 in adult tissues (promoter mark) (see Materials and Methods). We confirmed that while both carry H3K27ac (as defined), embryonic enhancers and adult enhancers are hypermethylated and hypomethylated, respectively (Fig. 6H and fig. S11F). Adult enhancers, but not embryonic enhancers, showed elevated CG densities compared to the background (Fig. 6H). This high CG density of adult enhancers is much more evident for those that acquire ectopic H3K4me3 upon the loss of DNA methylation (H3K4me3+), but less so for those that did not gain ectopic H3K4me3 (H3K4me3−) (Fig. 6, I and J). This is consistent with the notion that CG-rich sequences can attract histone methyltransferases such as MLL1/2, which contain the CXXC domain that recognizes unmethylated CpG regions (55). Furthermore, adult enhancers that show ectopic H3K4me3 are generally inaccessible in control early embryos but become accessible in dnmt1 mKD mutants (Fig. 6I). By contrast, H3K4me3− adult enhancers are inaccessible in both control and mKD embryos. This difference appears to be also true for embryonic enhancers. A small portion of embryonic enhancers (7.4%, n = 726) also acquired H3K4me3 upon the loss of DNA methylation (Fig. 6I). Despite the overall low CG density of embryonic enhancers, these H3K4me3+ embryonic enhancers also showed a slightly higher CG density compared to H3K4me3− enhancers (Fig. 6, I and J). Only H3K4me3+, but not H3K4me3−, embryonic enhancers showed increased chromatin accessibility in dnmt1 mKD embryos (Fig. 6I). Notably, the CG densities of H3K4me3+ embryonic enhancers are still substantially lower than those of H3K4me3+ adult enhancers (Fig. 6I and fig. S11G). In sum, these data revealed that adult enhancers are preferentially CG-rich, more sensitive to DNA methylation, and are prone to acquire ectopic H3K4me3 and increased chromatin accessibility upon the loss of DNA methylation.
Enhancers are activated by interacting TFs (52, 56). Therefore, we asked whether the differential activation of H3K4me3+ and H3K4me3− adult enhancers may be related to different sets of TFs and whether these TFs are present in early embryos. By searching for TF motif in these enhancers, we found distinct motifs present between H3K4me3+ and H3K4me3− adult enhancers, as well as between adult active enhancers (H3K27ac+) and adult decommissioned enhancers (H3K27ac− but ATAC+/LMR+) (fig. S11G). Most TFs of which the motifs are found in H3K4me3+ adult enhancers are expressed in both embryos and adult tissues. By contrast, many TFs of which the motifs are enriched in H3K4me3− adult enhancers are highly expressed in adult tissues but not in early embryos (fig. S11G, red arrow). In this analysis, we averaged gene expression for TFs from the same family but with almost identical motifs (such as gata and fox). Hence, the activation of H3K4me3+ adult enhancers in dnmt1 mKD embryos may be due to both their sensitivity to DNA methylation and the presence of corresponding TFs in early embryos. Meanwhile, the GREAT (Genomic Regions Enrichment of Annotations Tool) analysis (57) revealed that genes near H3K4me3+ adult enhancers are enriched for those functioning in fibroblast growth factor and Wnt signaling pathway; on the other hand, H3K4me3− adult enhancers are more enriched for kinase signaling pathway, regulation of cell cycle, etc. (fig. S11H). Last, we reasoned that if DNA methylation interferes TF binding at CG-rich enhancers, then these TFs may be more likely to contain CG in their motifs. TF motifs identified from embryonic enhancers are less likely to contain CGs than those from adult enhancers (Fig. 6I, far right, bar chart). However, exceptions are TF motifs identified in H3K4me3+ embryonic and adult enhancers, which preferentially have CGs. In sum, these data suggest that adult enhancers are preferentially CG-rich and interact with CG-containing TFs. By contrast, embryonic enhancers tend to be CG-poor and interact with CG-less TFs. Given the absence of Tet proteins in early embryos (fig. S11E) (29), these data suggest that the inherited DNA methylation, coupled by enhancer dememorization, present an epigenetic gate that prevents premature firing of adult enhancers and transcription programs without interfering embryonic programs (Fig. 7A).
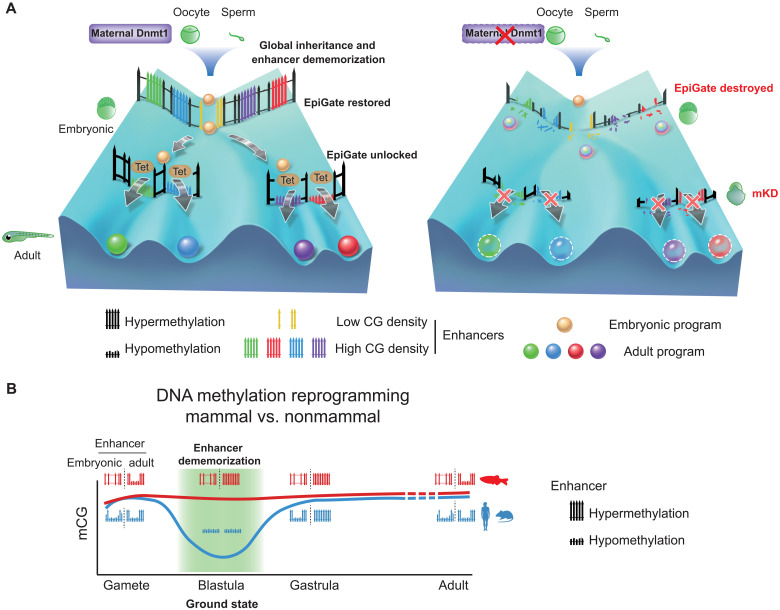
Fig. 7. Inherited methylome coupled by enhancer dememorization resets an epigenetic gate that safeguards embryonic programs.
(A) Maternal Dnmt1–mediated inherited global DNA methylome coupled with enhancer dememorization plays an instrumental role in restoring a full methylome to ultimately safeguard embryonic development against premature activation of adult programs. In WT embryos, inherited methylome sets up an EpiGate after fertilization and effectively repress adult enhancers, which are preferentially CG-rich. Embryonic enhancers, which are CG-poor, and insensitive to DNA methylation, can function while hypermethylated, and instruct embryonic transcription program. At the phylotypic stage, expression of tet genes demethylates CG-rich adult enhancers, allowing their activation and cell lineage differentiation. While in maternal dnmt1 mKD embryos, inherited DNA methylome failed to be maintained after fertilization, hence destroying the EpiGate. This leads to aberrant activation of adult programs in early embryos, accompanied by developmental failure and embryonic lethality around gastrulation. (B) Enhancer dememorization resets the developmental clock by restoring a ground-state (green shade) free of parental epigenetic memories in both mammals (blue, human and mouse) and nonmammalian vertebrate (red, zebrafish) around blastula stage. This epigenetic resetting is achieved through global DNA remethylation and demethylation in mammals and through enhancer hypermethylation in zebrafish. Embryonic enhancers are then demethylated in gastrula only in mammals, where TETs are expressed, but not in zebrafish, where TETs are still silenced. Zebrafish embryonic enhancers are nevertheless functioning presumably due to their low CG densities and insensitivity to DNA methylation. Adult enhancers are subsequently demethylated by TETs in both mammals and zebrafish. In mammals, embryonic enhancers also remain hypomethylated in adult tissues despite being decommissioned (11, 12).
DISCUSSION
The reprogramming of DNA methylation in mammals is critical for successful parental-to-embryonic transition and epigenetic memory resetting between generations. However, many nonmammalian vertebrates appear to lack such global reprogramming (17, 18, 27, 28). To date, why DNA methylation undergoes these distinct reprogramming modes between mammals and nonmammals remains elusive. This is particular intriguing given histone marks undergo global resetting in both mammals and nonmammalian vertebrates (19–21, 58). Here, we sought to decipher this mystery by depleting maternal dnmt1 in zebrafish early embryos, which revealed an essential role for inherited DNA methylation in early embryonic development. Moreover, this methylome, when coupling with enhancer dememorization, restores a full methylome to guard against premature activation of adult programs through repressing adult enhancers (Fig. 7A). Hence, enhancer dememorization resets the developmental clock by restoring a “ground-state” free of parental epigenetic memories. This epigenetic resetting is similarly achieved in mammals but through distinct paths, as global demethylation and remethylation essentially also remove parental epigenetic memories at enhancers (Fig. 7B). Therefore, enhancer dememorization may underlie and potentially unify distinct epigenetic reprogramming modes between mammals and nonmammalian vertebrates.
Inherited DNA methylation is essential for early development and proper cell differentiation
scRNA-seq analysis revealed that a subset of developmental genes showed down-regulation in dnmt1 mKD embryos, which are likely indirectly caused by failed differentiation. Differentiation defects are partially attributed to TE derepression–induced immune response and p53-mediated apoptosis and cell cycle arrest, as inhibiting p53 or immune signaling pathways partially rescued the differentiation defects. However, most of these animals still experience embryonic lethality, suggesting that these developmental defects likely stem beyond transposon derepression. We recently showed that, in mESCs that are deficient for all regulatory enzymes of DNA methylation (DNMT3A/3B/3C and TET1/2/3) except for DNMT1, the global DNA methylome is well maintained but becomes static (59). While the silencing of transposons is expected to be not affected, this mESC line still failed to differentiate, suggesting that TE silencing–independent function of DNA methylation may also contribute to differentiation defects.
Promoter DNA methylation reprogramming during the oocyte-to-embryo transition
Despite the global inheritance of methylome, dynamic methylation reprogramming does occur at regulatory elements such as promoters and enhancers in early zebrafish embryos (20, 27, 28). In particular, the maternal methylome is conformed to a state that highly resembles that of sperm, including both gain and loss of DNA methylation at specific promoters. Notably, this reprogramming does not depend on sperm, as it can occur even when sperm DNA was disrupted (28). Therefore, we previously proposed that both oocyte and sperm are perhaps reprogrammed to an “embryonic state” (20). While this reprogramming occurs after fertilization for the maternal genome, it may occur even before fertilization for sperm, as supported by the full methylation of enhancers in sperm. The significance of this intriguing phenomenon, however, remained unclear. One plausible possibility is that this transformation may help prepare gene activation and silencing during the forthcoming ZGA. However, our data showed that among genes that are hypomethylated in oocytes but become hypermethylated in embryos, only a small subset of genes is derepressed in dnmt1 mKD embryos. Most of these genes remain silenced by shield stage (fig. S9, C and D), suggesting that this conversion do not seem to solely serve the immediate gene repression after fertilization. Alternatively, this reprogramming may be important not only for ZGA but also for future development. By resetting adult and gametic epigenetic memories to a ground state, DNA methylation reprogramming may facilitate future gene regulation when stage-specific activators and repressors of promoters and enhancers appear in a spatiotemporally controlled manner.
Reprogramming of enhancers resets an epigenetic gate that prevents precocious activation of adult programs
The idea that DNA methylome reprogramming may have a larger impact beyond ZGA and create a ground state is strongly supported by further analyses of enhancers. Nearly all enhancers in zebrafish gametes are dememorized through DNA hypermethylation in early development (20). This essentially creates a methylome free of past enhancer memories and thus represents a likely ground epigenetic state. Moreover, hypermethylation of enhancers also prevents precocious activation of adult enhancers in early embryos.
Mechanistically, this is probably due to the absence of Tet proteins that are not expressed until the phylotypic stage. By contrast, mammalian TET proteins are expressed throughout pre- and postimplantation stages and play critical roles at enhancers during gastrulation (60). The motivation underlying Tet’s absence in early zebrafish embryos remains unknown. One possibility may lie in the different cell cycle speed in early embryos between mammals (such as 24 hours per cell cycle for cleavage-stage mouse embryos) and cold-blooded vertebrate animals (such as 15 min per cell cycle for zebrafish pre-ZGA embryos). It is tempting to speculate that TET proteins, even if expressed, may not properly function at enhancers in these rapidly dividing cells. After 24 hpf, the cell cycle prolongs to more than 3 hours (61), which is perhaps more accessible for the epigenome editing enzymes such as Tets.
The absence of TET proteins presumably also creates a potential challenge for embryonic enhancers to function given their hypermethylated states. Our analyses revealed that embryonic enhancers tend to be CG-poor and are thus less likely to be affected by DNA methylation. In addition, TFs that potentially bind these enhancers tend to contain fewer CGs in their recognition motifs (fig. S11H). We propose that these enhancers may have adopted these sequence features during evolution to survive without TET proteins. By contrast, adult enhancers tend to be relatively CG-rich and are bound by TFs that contain CGs in their motifs. Upon the loss of DNA methylation, many putative CG-rich adult enhancers become aberrantly activated, as indicated by their acquisition of active marks such as H3K4me1, H3K27ac, and H3K4me3, increased chromatin accessibility, and precocious activation of their neighbor genes. Therefore, these data demonstrate that DNA methylome may play a critical role in ensuring temporally ordered enhancer activation during development. Collectively, our data showed that the inherited global DNA methylome, coupled by enhancer dememorization, plays an essential role in embryonic development by repressing transposons and restoring an epigenetic gate that guards against premature activation of adult programs (Fig. 7A). Future studies are warranted to determine whether similar mechanisms (such as enhancer dememorization) can be applied to other reprogramming processes to reset the epigenetic clock and restore cells from differentiated or aged states back to a ground state of totipotency.
MATERIALS AND METHODS
Zebrafish strain and fertilized egg microinjection
The WT AB (female) strains, p53 M214K line, and dnmt1s872 lines were used in most experiments. Embryos derived from dnmt1s872 heterozygous intercrosses were identified by polymerase chain reaction (PCR) genotyping at desired stages. Ethical approval was obtained from the Animal Care and Use Committee of Tsinghua University. All experimental animal procedures were performed under anesthesia, and all efforts were made to minimize suffering.
For 1-cell microinjection, mouse Stella mRNAs were injected into naturally fertilized 1-cell stage embryos, according to a commonly used zebrafish microinjection protocol (62). After injection, embryos were grown in fresh Holtfreter solution [KCl (0.05 g/liter), CaCl2 (0.1 g/liter), NaHCO3 (0.025 g/liter), and NaCl (3.5 g/liter) (pH 7.0)] at 28.5°C and were staged according to standard morphological criteria (61). The dose of mouse Stella mRNA was 550 pg per embryo.
dnmt1 mKD with OMIS
Briefly, on the first day of OMIS (32), adult females at 5 to 12 months old were anesthetized in tricaine (550 μg/ml) (Sigma-Aldrich, catalog no. A5040) in a petri dish. Then, the fish was placed on a damp sponge with specific buffer [5.4 mM KCl, 136.8 mM NaCl, 4.2 mM NaHCO3, 0.44 mM KH2PO4, 0.25 mM Na2HPO4, and 0.5% (w/v) bovine serum albumin (BSA)]. A cut was made on one side of belly to expose the ovary. The diluted MOs were microinjected into each oocyte. Rhodamine B (Sigma-Aldrich, catalog no. R8881) was coinjected with MOs as a dye. After injection, the wound on the belly was sewed with a surgical sewing needle carefully and quickly. Once the operation was done, the female was transferred into fish water supplemented with tricaine (32 μg/ml), penicillin (10 U/ml), and streptomycin (10 μg/ml) (HyClone, catalog no. SV30010). Then, the fish was transferred to fish water containing gradually reduced concentrations of tricaine. In the evening of the second day, the female was paired with a WT male. In the morning of the third day, the pair started to chase and lay fertilized eggs naturally. The injected oocyte-derived embryos were identified by coinjected dye (rhodamine B) at very early developmental stages. The injection doses of dnmt1 MO and standard control MO (cMO) were both 5 ng per oocyte in the same assay. The sequences of MOs are 5′-ACAATGAGGTCTTGGTAGGCATTTC-3′ (dnmt1 MO) and 5′-CCTCTTACCTCAGTTACAATTTATA-3′ (cMO). MOs were dissolved in ribonuclease (RNase)–free water and heated to 65°C for 10 min before microinjection.
Tissue collection
Embryos at 5 dpf were euthanized with tricaine, and head and tail were dissected carefully by tweezers. After brief grinding, tissues were frozen at −80°C for later usage.
IF and imaging
The embryos at the defined time points after fertilization were fixed by 4% polyformaldehyde overnight at 4°C. Then, they were dechorionated manually and dehydrated with methanol. The whole-mount IFs with Dnmt1 antibody (Santa Cruz Biotechnology, catalog no. sc-20701), 5mC antibody (Abcam, catalog no. ab10805), and pH2AX antibody [Cell Signaling Technology, catalog no. 2577S], were done with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, catalog no. D1306) staining and performed as previously described (20). The secondary antibodies were Alexa Fluor 488–conjugated anti-rabbit and Alexa Fluor 488–conjugated anti-mouse (Jackson ImmunoResearch; 1:200 dilution). After staining, embryos were deyolked by tweezers and mounted on glass slides in mounting medium (Sigma-Aldrich, catalog no. P3130) at animal polar upturned position. Images were acquired on 710 or 880 META laser scanning confocal microscope and manipulated by ZEN software. Treated or untreated embryos were anesthetized at desired stages with 0.02% tricaine and mounted in 5% methyl cellulose (Sigma-Aldrich, catalog no. M-6385) for observation, and phenotype pictures were taken under Nikon SMZ1500 microscope.
dnmt1 rescue
The full-length dnmt1 coding sequence from zebrafish was cloned into pXT7 vector and linearized by Sma I digestion. mRNA was synthesized in vitro using mMESSAGE mMACHINE kit (Ambion, catalog no. AM1344) and purified using RNeasy Mini kit (QIAGEN, catalog no. 74104). To avoid targeting dnmt1 mRNA by MO, the target sequence of MO was mutated without affecting amino acid sequence (dnmt1 mismatch mRNA). Catalytically inactive mutant dnmt1 mismatch mRNA was designed according to s872 mutant (23), which contains a stop codon in catalytic domain resulting the loss of methyltransferase activity (mutant dnmt1 mismatch mRNA). These mRNAs were coinjected with MO into GV oocytes by OMIS (32) for rescue experiment. Embryos at desired stages were then collected for further analysis.
Mouse Stella overexpression
The construct containing full-length Stella coding sequence from mouse was generated and linearized by Not I digestion. The mutant Stella (KRR, K85E/R86E/R87E) contains three amino acids mutations within the nuclear export signal as previously described (35). mRNA was synthesized in vitro using mMESSAGE mMACHINE kit (Ambion, catalog no. AM1344) and purified using RNeasy Mini kit (QIAGEN, catalog no. 74104). These mRNAs were injected into zygotes, and embryos at desired stages were collected for further analysis.
Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling
ApopTag Red In Situ Apoptosis Detection Kit (Millipore, catalog no. S7165) was used to probe cell apoptosis in zebrafish embryo. Collected embryos at desired stages were fixed by 4% polyformaldehyde overnight at 4°C, then dechorionated manually, and dehydrated with methanol. After being stored at −20°C for 1 hour, embryos were rehydrated with 0.1% Triton X-100 in phosphate-buffered saline (PBS), then refixed with 4% polyformaldehyde for 20 min at room temperature, and put into precooled ethanol-ethyl acetate mixture (volume ratio, 2:1). Next, 50 μl of equilibration buffer was added into tubes contained embryos at room temperature for 1 hour and changed with 55 μl of terminal deoxynucleotidyl transferase (TdT) reaction system (38.5 μl of reaction buffer and 16.5 μl of TdT enzyme) at 37°C for more than 1 hour to add digoxin-labeled deoxyuridine triphosphate in DNA breaks. To stop reaction and visualize digoxin signals, embryos were washed with stop/wash buffer and incubated with anti-digoxin antibody coupled with rhodamine buffer (34 μl of blocking buffer and 31 μl of antibody) at 37°C for 30 min or overnight. DNA was stained with DAPI. After staining, embryos were mounted in the same way as IF and imaged on 710 or 880 META laser scanning confocal microscope.
Inhibitor treatment
To inhibit the reverse transcription, embryos were treated with 50 μM FOS (Selleck, catalog no. S3076). To inhibit the immune response, embryos were treated with 0.1 μM or 0.01 μM BX795 (Selleck, catalog no. S1274). Embryos treated with dimethyl sulfoxide were used as control. All these embryos were examined for phenotypes and fixed for immunostaining at desired stages. Phenotype at 24 hpf was quantified and summarized.
STEM-seq library preparation
STEM-seq was carried out as described previously (33). The deyolked embryos and tissues were lysed with 20 μl of lysis buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA (pH 8.0), and 0.5% NP-40] and 2 μl of protease K (Roche, catalog no. 10910000) for at least 3 hours at 55°C. After heat inactivation, spike-in λ-DNA (Promega, catalog no. D150A) was added at a mass ratio of 1:200. Bisulfite conversion was performed with the EpiTect Fast Bisulfite Conversion Kit (QIAGEN, catalog no. 59824). The converted DNA was subjected to column purification and desulfonation on MinElute DNA spin columns (QIAGEN, catalog no. 59824) with carrier RNA (QIAGEN, catalog no. 59824) according to the manufacturer’s instructions. The purified DNA was eluted in 30 μl of elution buffer and ready for TELP library preparation (63).
Total RNA-seq library preparation and sequencing
The embryos were dechorionated manually by tweezers and transferred into 750 μl of TRIzol (Invitrogen, catalog no. 15596018). About 10 fresh embryos were transferred into TRIzol and vortexed until no visible particles. Chloroform (150 μl) (Amresco, catalog no. 0757) was added and mixed thoroughly. The mixture was then transferred into Phasemaker tube (Invitrogen, catalog no. A33248) and spun at 14,000 rpm for 15 min. Next, the top phase was taken out from the tube, and 1 μl of lysophosphatidic acid was added (Sigma-Aldrich, catalog no. 56575) and mixed well using pipettes. Then, RNA was precipitated by adding 750 μl of isopropanol (Sigma-Aldrich, catalog no. 59304) at −20°C overnight. At the next day, the tube was spun at 14,000 rpm for 30 min, and supernatant was removed. The pellet was washed with fresh 70% ethanol, resuspended in 20 μl of RNase-free water and stored at −80°C for later usage.
NEBNext rRNA Depletion Kit (NEB, catalog no. E6310S) was used to deplete ribosomal RNA (rRNA) according to the manufacturer’s instructions. Briefly, rRNA was hybridized with probes and digested with RNase H, and then excess probes were digested with deoxyribonuclease I. After that, NEBNext RNA sample purification beads were used to purify rRNA-depleted RNA. Purified RNA was fragmented before cDNA synthesis at 95°C for 8 min. Double-stranded cDNA was synthesized with NEBNext first-strand (NEB, catalog no. E7771S) and second-strand synthesis modules (NEB, catalog no. E7550S) and then purified with AMPure XP beads (Beckman Coulter, catalog no. A63882). Synthesized cDNA was subjected to library preparation with NEBNext Ultr II DNA Library Prep Kit (NEB, catalog no. E7645S). DNA was end-repaired, adenylated, and ligated to TruSeq sequencing adaptors. DNA was amplified using KAPA HiFi HotStart ReadyMix (KAPA Biosystems, catalog no. RR2602). The amplified DNA was size-selected using AMPure XP beads for 200– to 500–base pair (bp) DNA fragments. All libraries were sequenced by Illumina HiSeq 1500 or 2500 or XTen platform according to the manufacturer’s instructions.
scRNA-seq library preparation and sequencing
Cell dissociation protocol was based on a previously described method (38) with modifications to adapt it for 10× Genomics platform. Briefly, dnmt1 mKD and control embryos were collected 20 min after fertilization and cultured as mentioned before. Then, 15 embryos at dome or shield stages for each sample were transferred into plastic petri dishes that had previously been coated with 2% agarose and soaked with Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium (Gibco/Life Technologies, catalog no. 11330032) at least 2 hours. Next, embryos at desired stages were dechorionated and deyolked manually by forceps and transferred to a 1.5-ml Eppendorf tube with 50 μl of DMEM/F12 medium. Dissections were performed for up 15 min. The volume of DMEM/F12 medium containing embryos was adjusted to 200 μl, and then cells were mechanically dissociated by flicking the tube 30 times and pipetting mixture 10 times through a 200-μl tip. The volume was adjusted to 1 ml with PBS containing 1.0% BSA and spun to pellet cells at 300g for 30s. The supernatant was removed, and cells were resuspended in 80 μl of PBS containing 0.1% BSA and 20% OptiPrep (STEMCELL Technologies, catalog no. 07820), aiming for a concentration above 300 cells/μl. Cells were then passed through a cell sieve (100 μm for dome stage and 70 μm for shield stage).
scRNA-seq library was performed with Chromium Next GEM Single-cell kit (10× Genomics, catalog no. PN-1000121) based on the standard protocol. All libraries were sequenced by Illumina HiSeq 1500 or 2500 or XTen platform according to the manufacturer’s instructions.
CUT&RUN library preparation and sequencing
CUT&RUN was conducted as previously described (50, 64) with modifications in cell permeation to adapt it for zebrafish embryos. Embryos were deyolked by tweezers manually and transferred into a 1.5-ml conventional, non–low binding tube (Axygen). Then, tubes were flicked several times to disperse cells in embryos and resuspended by 60 μl of washing buffer [20 mM Hepes-KOH (pH 7.5), 150 mM NaCl, and 0.5 mM Spermidine and Roche complete protease inhibitor]. Ten microliters of concanavalin-coated magnetic beads (PolyScience, catalog no. 86057) for each sample were gently washed twice, resuspended by binding buffer [20 mM Hepes-KOH (pH 7.5), 10 mM KCl, 1 mM CaCl2, and 1 mM MnCl2], and added carefully to the cells. The cells with beads were incubated at 23°C for 30 min on ThermoMixer (Eppendorf) at 400 rpm, then held at magnetic stand to exclude buffer, and resuspended by 75 μl of antibody buffer (washing buffer supplied with 0.02% digitonin and 2 mM EDTA, freshly made) with antibodies against H3K4me3 (in-house) (65), H3K27me3 (Active Motif, catalog no. 61017), H2AK119ub (Cell Signaling Technology, catalog no. 8240 s), or Sox2 (Active Motif, catalog no. 39843) diluted at ratio of 1:100. Then, the samples were incubated at 4°C on ThermoMixer overnight at 400 rpm. On the second day, the samples were washed by digitonin-washing buffer several times on magnetic stand, resuspended with 50 μl of digitonin-washing buffer supplied with pA–micrococcal nuclease (pA-MNase) (700 ng/ml), and incubated at 4°C on ThermoMixer for 3 hours at 400 rpm. After that, the cells were washed by digitonin-washing buffer on magnetic stand and resuspended by 100 μl of digitonin-washing buffer on ice for at least 2 min. Targeted region digestion was activated by adding 2 μl of 100 mM CaCl2 for 30 min in ice, then stopped by 100 μl of 2× stop buffer [340 mM NaCl, 20 mM EDTA (pH 8.0), 4 mM EGTA (pH 8.0), RNase (50 μg/ml), glycogen (100 μg/ml), and 0.02% digitonin suppled with spike-in DNA), and fully vortexed. To release fragments, the samples were incubated at 37°C on ThermoMixer at 400 rpm for 20 min. Then, the supernatants were purified by phenol chloroform and ethanol purification and subjected to TruSeq library construction using NEBNext Ultra II DNA Library Prep kit (NEB, catalog no. E7645S) as standard protocols. The amplified DNA was size-selected using AMPure Beads for 200- to 800-bp DNA fragments. All libraries were sequenced by Illumina HiSeq 1500 or 2500 or XTen platform according to the manufacturer’s instructions.
STAR chromatin immunoprecipitation sequencing library preparation and sequencing
The STAR chromatin immunoprecipitation sequencing (ChIP-seq) was performed as previously described (20). Embryos were deyolked by repeatedly blowing with a 200-μl pipette, and cell pellets were collected by spinning down at 5000 rpm for 5 min at 4°C. After centrifuge, cell pellet was lysed in 40 μl of lysis buffer (0.5% NP-40, 0.5% Tween 20, 0.1% SDS, and proteinase inhibitor) with pipetting up and down several times. MNase (0.1 U) (Sigma-Aldrich, catalog no. N3755) was added for chromatin digestion at 37°C for 5 min. The reaction was then terminated by adding 1 μl of 0.5 M EGTA. Immunoprecipitation (IP) sample was incubated with 1 μg of H3K4me3 antibody (in-house) (65) and 2 μg of H3K27me3 antibody (Active Motif, catalog no. 61017) overnight with rotation at 4°C. In the next day, the sample was incubated with 300 μg of Dynabeads Protein A or Protein G (Life Technologies, catalog no. 10001D or 10003D) for 2 hours with rotation at 4°C. The beads were washed five times in 150 μl of radioimmunoprecipitation assay buffer and once in 150 μl of LiCl buffer. After washing, tubes were spun briefly, and the supernatant was removed. For each IP sample, beads were resuspended with 27 μl of double-distilled H2O (ddH2O) and 1 μl of 10× Ex-Taq buffer (Takara). One microliter of proteinase K (Roche, catalog no. 10910000) was then added, and the mix was incubated at 55°C for 90 min to elute DNA from beads. The supernatant was then transferred to a new tube, and the proteinase K was inactivated at 72°C for 40 min. One microliter of recombinant Shrimp Alkaline Phosphatase (rSAP) (NEB, catalog no. M0371) was then added to dephosphorylate 3′ end of DNA at 37°C for 1 hour. rSAP was inactivated at 65°C for 10 min. The resulting sample was subjected to TELP library preparation as previously described (63). The amplified DNA was size-selected using AMPure Beads for 200- to 800-bp DNA fragments. All libraries were sequenced by Illumina HiSeq 1500 or 2500 or XTen platform according to the manufacturer’s instructions.
ATAC-seq library preparation and sequencing
The miniATAC-seq procedure was performed as previously described (66) with modifications to adapt it for zebrafish embryos. Briefly, dispersed cells from deyolked zebrafish embryos were transferred into 6 μl of lysis buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.02% digitonin) in ice for 10 min. The ATAC reaction was performed by adding 4 μl of ddH2O, 4 μl of 5× TTBL, and 5 μl TTE mix V5 (Vazyme Biotech Co.,Ltd, catalog no. TD502) at 37°C for 30 min and then stopped by adding 5 μl of 5× TS stop buffer at room temperature for 5 min. DNA was extracted by phenol chloroform and ethanol purification after adding 40 ng of carrier RNA and 103 μl of tris-EDTA. Then, DNA was PCR-amplified with 10 μl of index (Vazyme Biotech Co.,Ltd, catalog no. TD202), 10 μl of 5× TAB and 1 μl tris-acetate-EDTA (Vazyme Biotech Co.,Ltd, catalog no. TD502) with the program of 72°C for 3 min, 98°C for 30 s, (98°C for 15 s, 60°C for 30 s, and 72°C for 3 min) with 18 cycles, and 72°C for 5 min. The amplified DNA was size-selected using AMPure Beads for 200- to 800-bp DNA fragments. All libraries were sequenced by Illumina HiSeq 1500 or 2500 or XTen platform according to the manufacturer’s instructions.
STEM-seq data processing
All STEM-seq datasets were mapped to the danRer7 reference genome by Bismark (67). Reads were trimmed with cutadaptor (68) using parameters: --minimum-length 20 --pair-filter = any. Alignments were performed with the following parameters: -N 1 -X 600 --score_min L,0,-0.6. Multimapped reads and PCR duplicates were removed. The function bismark_methylation_extractor was used to calculate the DNA methylation level.
Total RNA-seq data processing
The total RNA-seq data were first processed using Trim Galore! with default parameters to trim the adapter-containing and low-quality reads. The filtered data were then mapped to the zebrafish reference genome (danRer7) by STAR (version: STAR_2.5.3a_modified) (69) with the following parameters: --outSAMstrandField intronMotif --outSAMattributes All --outSAMunmapped Within --outSAMattrIHstart 0 --outWigStrand Stranded --outFilterMultimapNmax 20 --twopassMode Basic. The gene expression level was normalized to FPKM values using Cufflinks (version 2.2.1) (70).
scRNA-seq data processing
The scRNA-seq data were processed with Cell Ranger 3.1 for genome alignment (danRer10) and transcript counting. Next, the raw count matrices were imported to Seurat 3.0 for identification of cell subtypes. To filter low-quality cells, cells with unique detected genes lower than 2000 and higher than 6000 genes were discarded, and cells with high percentages of mitochondrial genes (>5%) were also removed. To make data comparable between cells and samples, we performed SCTransform (36) normalization for each dataset independently. To further eliminate the effects caused by sequencing depths and percentages of mitochondrial genes, unique molecular identifier (UMI) variance and percent.mt were regressed out from SCTransform normalized data with regularized negative binomial model and linear model, respectively. Samples of dome and shield stages were integrated by identifying anchors with the top 3000 gene features. During the integration, the control cells were set as reference. Dimensional reduction was performed on the integrated data with principal components analysis, and then the first 30 PCs were selected for cluster identification and Uniform Manifold Approximation and Projection (UMAP) visualization. Positive marker genes for each single-cell subtype were identified as following: The expression of a gene is detected in at least 25% of cells of current cell subtype, and its expression is higher than the average expression levels of the rest cells [average log(fold change) > 0.25 and false discovery rate (FDR) < 0.01].
Cellular trajectory analysis
To better study the major differentiation lineages, we removed identified apoptotic cells, EVLs, YSLs, and PGCs that are more distally related lineages. Raw count matrices were imported to Monocle 2 (37), and the lists of DEGs in each cluster identified through Seurat were used for the construction of cell trajectory. The root of each trajectory was defined by epiblast cells.
CUT&RUN, ChIP-seq, and ATAC-seq data processing
All reads were aligned to the zebrafish reference genome (danRer7) using Bowtie2 (version 2.2.2) (71) with the parameters –t –q –N 1 –L 25. All unmapped reads, nonuniquely mapped reads, and PCR duplicates were removed. For downstream analysis, the read counts were normalized by computing the numbers of RPKM. To minimize the batch and cell type variation, RPKM values across whole genome were further z-score–normalized. To visualize the signals in the UCSC Genome Browser, each read was extended by 250 bp, and the coverage for each base was counted.
Differential gene expression analysis
Read counts of genes were summarized with featureCounts (72). Next, the read count matrices were imported to DESeq2 (73) to perform differential expression analysis for genes in control and dnmt1 mKD embryos. Genes with an FDR of <0.01 and a log2(fold change) of >2 were considered as significantly DEGs.
Analysis of maternal, dome-specific, and shield-specific genes
The maternal genes were identified by high expression level in oocyte (FPKM > 10), but low at post-ZGA stages (fold change of dome/oocyte < 0.5). Dome-specific expressed genes were defined by low expression in oocyte and the 256-cell stage (FPKM < 5) and high expression at dome stage (FPKM > 10). Similarly, shield-specific genes were defined with low expression levels in oocyte, embryos at the 256-cell and dome stages (FPKM < 5), and high expression at shield stage (FPKM > 10).
Analysis of nonmaternal ZGA genes
Because of the possible confounding effects from maternally inherited RNA transcripts, nonmaternal ZGA genes were defined as low expression level in oocyte and the 256-cell stage (FPKM < 5) but high expression level at dome stage (FPKM of dome > 5 and fold change of dome/256-cell >2).
GO analysis
Functional annotation was performed using the Database for Annotation, Visualization and Integrated Discovery bioinformatics resource (74). GO terms for each functional cluster were summarized to a representative term, and P values were plotted to show the significance.
Identification of CUT&RUN, ChIP-seq, and ATAC-seq peaks
Peaks were called using MACS2 (75) with the parameters -p 1e-5 --nomodel -g 1.3e10. The called peaks with weak signals were filtered in the further analysis. Where appropriate, a random set of peaks with identical lengths was used as controls.
Identification of putative enhancers
For adult enhancers, H3K27ac data were used to identify active enhancers, and ATAC-seq and DNA methylation data were used to identify decommissioned enhancers (12). To ensure the consistency of data processing, the H3K27ac, ATAC-seq, and DNA methylation data collected from previous study (45) were reanalyzed with our pipelines as described above. The LMRs were obtained from the same study (45).
To define active enhancers, H3K27ac peaks at least ±2 kb away from annotated promoters were firstly selected as candidate enhancers. To exclude possible unannotated promoters, any H3K27ac sites that overlap with H3K4me3 peaks of adult tissues (45) were also excluded. Furthermore, to exclude possible unannotated promoters in embryos, we took advantage of the notion that hypomethylated regions often mark promoters in zebrafish (76) and enhancers are hypermethylated in zebrafish early embryos, leaving only promoters unmethylated (20). Therefore, we chose the 256-cell stage as a representative stage when enhancer dememorization is completed. All unmethylated regions (UMRs)/LMRs (20) at the 256-cell stage of WT embryos are merged together and identified as possible promoters and were further excluded from the enhancer lists to generate the final putative enhancer lists.
TF motif identification at enhancers
The findMotifsGenome.pl script from HOMER program (77) was used to identify the enriched motifs at enhancers. TFs expressed at least at one stage with motif enrichment P < 1 × 10−20 were included.
Identification of lost, ectopic, and retained histone mark peaks
We clustered H3K4me3, H3K27me3, or H2AK119ub peaks and categorized them into lost, ectopic, and retained groups based on the following cutoffs. The lost peaks were identified with a log2(fold change) of <−2 and a normalized RPKM of <−1 in mKD embryonic cells; the ectopic peaks were defined by a log2(fold change) of >2 and a normalized RPKM of >1 in mKD embryonic cells; the retained peaks were defined by a log2(fold change) of <2 and a normalized RPKM of >0.5 in both control and mKD samples.
Distance calculation for ectopic distal H3K4me3 peaks and DEGs
We calculated the distances from the transcription start sites (TSSs) of up-regulated, down-regulated, or non-DEGs to the center of distal ectopic H3K4me3 peaks. Statistical significance between groups was estimated with Mann Whitney U Test.
Quantification and statistical analysis
At least two biological replicates were used for RNA-seq, scRNA-seq, CUT&RUN, STAR-ChIP, and ATAC-seq experiments. The reproducibility between replicates was estimated with Pearson correlation. All box plots were plotted using R and Python. In box plots, the horizontal line shows the median, the box encompasses the interquartile range, and whiskers extend to 5th and 95th percentiles. Statistical significance for the enrichment of dnmt1 mKD embryo’s up-regulated genes in adult tissue–expressed genes was assessed with one-sided Fisher’s exact test.
Acknowledgments
We appreciate comments from members of the Xie laboratory during preparation of the manuscript. We would like to express our gratitude to S. Bultmann and H. Leonhardt from Ludwig Maximilians University of Munich for sharing mouse Stella plasmid and discussion, S. Henikoff from Fred Hutchinson Cancer Research Center for sharing pA-MNase, and Q. Tao from Tsinghua University for providing the DNMT1 antibody. We are also grateful to the cell facility at the Tsinghua Center of Biomedical Analysis for assistance with imaging and the biocomputing facility at Tsinghua University. We would like to thank Mr. Jinyang Li for assistance with artistic drawing.
Funding: This work was funded by the National Natural Science Foundation of China 31725018 (W. Xie) and 31988101 (W. Xie and A.M.), National Key R&D program of China 2019YFA0508901 (W. Xie), and Beijing Municipal Science & Technology Commission (Z181100001318006) (W. Xie), and the THU-PKU Center for Life Sciences (W. Xie and A.M.). W. Xie is a Howard Hughes Medical Institute international research scholar.
Author contributions: Xiaotong W. collected zebrafish samples and conducted most zebrafish experiments with help from W.S. Xiaotong W. performed total RNA-seq, scRNA-seq, ATAC-seq, and CUT&RUN experiments with the help from L.L. and W. Xia and performed data analysis with the help from B.Z., H.Z., and B.L. B.Z. performed STAR ChIP-seq and STEM-seq experiments with the help from Y.Z. and data analysis. H.Z. performed bulk RNA-seq and scRNA-seq data analyses. Q.W. helped with mouse Stella overexpression experiments. Xi Wu helped with data analysis. W. Xie supervised the project or related experiments. X.W., B.Z., H.Z., and W. Xie wrote the manuscript with the help from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data have been deposited to the Gene Expression Omnibus with the accession number GSE175951.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
REFERENCES AND NOTES
- 1.Cedar H., Bergman Y., Programming of DNA methylation patterns. Annu. Rev. Biochem. 81, 97–117 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Jones P. A., Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Goll M. G., Bestor T. H., Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Wu X., Zhang Y., TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Stadler M. B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., Wirbelauer C., Oakeley E. J., Gaidatzis D., Tiwari V. K., Schübeler D., DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Xie W., Schultz M. D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J. W., Tian S., Hawkins R. D., Leung D., Yang H., Wang T., Lee A. Y., Swanson S. A., Zhang J., Zhu Y., Kim A., Nery J. R., Urich M. A., Kuan S., Yen C. A., Klugman S., Yu P., Suknuntha K., Propson N. E., Chen H., Edsall L. E., Wagner U., Li Y., Ye Z., Kulkarni A., Xuan Z., Chung W. Y., Chi N. C., Antosiewicz-Bourget J. E., Slukvin I., Stewart R., Zhang M. Q., Wang W., Thomson J. A., Ecker J. R., Ren B., Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gifford C. A., Ziller M. J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A. K., Kelley D. R., Shishkin A. A., Issner R., Zhang X., Coyne M., Fostel J. L., Holmes L., Meldrim J., Guttman M., Epstein C., Park H., Kohlbacher O., Rinn J., Gnirke A., Lander E. S., Bernstein B. E., Meissner A., Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith Z. D., Meissner A., DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Song Y., van den Berg P. R., Markoulaki S., Soldner F., Dall’Agnese A., Henninger J. E., Drotar J., Rosenau N., Cohen M. A., Young R. A., Semrau S., Stelzer Y., Jaenisch R., Dynamic enhancer DNA methylation as basis for transcriptional and cellular heterogeneity of ESCs. Mol. Cell 75, 905–920.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hon G. C., Rajagopal N., Shen Y., McCleary D. F., Yue F., Dang M. D., Ren B., Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 45, 1198–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jadhav U., Cavazza A., Banerjee K. K., Xie H., O’Neill N. K., Saenz-Vash V., Herbert Z., Madha S., Orkin S. H., Zhai H., Shivdasani R. A., Extensive recovery of embryonic enhancer and gene memory stored in hypomethylated enhancer DNA. Mol. Cell 74, 542–554.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckersley-Maslin M. A., Alda-Catalinas C., Reik W., Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wu H., Zhang Y., Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skvortsova K., Iovino N., Bogdanović O., Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 19, 774–790 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Cleaton M. A. M., Edwards C. A., Ferguson-Smith A. C., Phenotypic outcomes of imprinted gene models in mice: Elucidation of pre- and postnatal functions of imprinted genes. Annu. Rev. Genomics Hum. Genet. 15, 93–126 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Macleod D., Clark V. H., Bird A., Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat. Genet. 23, 139–140 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Veenstra G. J. C., Wolffe A. P., Constitutive genomic methylation during embryonic development of Xenopus. Biochim. Biophys. Acta 1, 39–44 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Akkers R. C., van Heeringen S. J., Jacobi U. G., Janssen-Megens E. M., Françoijs K. J., Stunnenberg H. G., Veenstra G. J. C., A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev. Cell 17, 425–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B., Wu X., Zhang W., Shen W., Sun Q., Liu K., Zhang Y., Wang Q., Li Y., Meng A., Xie W., Widespread enhancer dememorization and promoter priming during parental-to-zygotic transition. Mol. Cell 72, 673–686.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Murphy P. J., Wu S. F., James C. R., Wike C. L., Cairns B. R., Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Cell 172, 993–1006.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Sadler K. C., Krahn K. N., Gaur N. A., Ukomadu C., Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad. Sci. U.S.A. 104, 1570–1575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson R. M., Bosch J. A., Goll M. G., Hesselson D., Dong P. D. S., Shin D., Chi N. C., Shin C. H., Schlegel A., Halpern M., Stainier D. Y. R., Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev. Biol. 334, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tittle R. K., Sze R., Ng A., Nuckels R. J., Swartz M. E., Anderson R. M., Bosch J., Stainier D. Y. R., Eberhart J. K., Gross J. M., Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev. Biol. 350, 50–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu J., Loughlin E. A., Gaur N. A., SenBanerjee S., Jacob V., Monson C., Kent B., Oranu A., Ding Y., Ukomadu C., Sadler K. C., UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is required for zebrafish embryogenesis. Mol. Biol. Cell 23, 59–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulholland C. B., Nishiyama A., Ryan J., Nakamura R., Yiğit M., Glück I. M., Trummer C., Qin W., Bartoschek M. D., Traube F. R., Parsa E., Ugur E., Modic M., Acharya A., Stolz P., Ziegenhain C., Wierer M., Enard W., Carell T., Lamb D. C., Takeda H., Nakanashi M., Bultmann S., Leonhardt H., Recent evolution of a TET-controlled and DPPA3/STELLA-driven pathway of passive DNA demethylation in mammals. Nat. Commun. 11, 5972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang L., Zhang J., Wang J. J., Wang L., Zhang L., Li G., Yang X., Ma X., Sun X., Cai J., Zhang J., Huang X., Yu M., Wang X., Liu F., Wu C. I., He C., Zhang B., Ci W., Liu J., Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 153, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potok M. E., Nix D. A., Parnell T. J., Cairns B. R., Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdanović O., Smits A. H., De La Calle Mustienes E., Tena J. J., Ford E., Williams R., Senanayake U., Schultz M. D., Hontelez S., Van Kruijsbergen I., Rayon T., Gnerlich F., Carell T., Veenstra G. J. C., Manzanares M., Sauka-Spengler T., Ecker J. R., Vermeulen M., Gómez-Skarmeta J. L., Lister R., Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 48, 417–426 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida R. D., Loose M., Sottile V., Matsa E., Denning C., Young L., Johnson A. D., Gering M., Ruzov A., 5-Hydroxymethyl-cytosine enrichment of non-committed cells is not a universal feature of vertebrate development. Epigenetics 7, 383–389 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Kaaij L. J. T., Mokry M., Zhou M., Musheev M., Geeven G., Melquiond A. S. J., de Jesus Domingues A. M., de Laat W., Niehrs C., Smith A. D., Ketting R. F., Enhancers reside in a unique epigenetic environment during early zebrafish development. Genome Biol. 17, 146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X., Shen W., Zhang B., Meng A., The genetic program of oocytes can be modified in vivo in the zebrafish ovary. J. Mol. Cell Biol. 10, 479–493 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Xiang Y., Yin Q., Du Z., Peng X., Wang Q., Fidalgo M., Xia W., Li Y., Zhao Z. A., Zhang W., Ma J., Xu F., Wang J., Li L., Xie W., Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 50, 96–105 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Langdon Y. G., Mullins M. C., Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu. Rev. Genet. 45, 357–377 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Zhang Z., Chen J., Liu W., Lai W., Liu B., Li X., Liu L., Xu S., Dong Q., Wang M., Duan X., Tan J., Zheng Y., Zhang P., Fan G., Wong J., Xu G. L., Wang Z., Wang H., Gao S., Zhu B., Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564, 136–140 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrell J. A., Wang Y., Riesenfeld S. J., Shekhar K., Regev A., Schier A. F., Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner D. E., Weinreb C., Collins Z. M., Briggs J. A., Megason S. G., Klein A. M., Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chernyavskaya Y., Mudbhary R., Zhang C., Tokarz D., Jacob V., Gopinath S., Sun X., Wang S., Magnani E., Madakashira B. P., Yoder J. A., Hoshida Y., Sadler K. C., Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development 144, 2925–2939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X., Darzynkiewicz Z., Cytometric assessment of histone H2AX phosphorylation: A reporter of DNA damage. Methods Mol. Biol. 314, 73–80 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vashishtha A. K., Kuchta R. D., Effects of acyclovir, Foscarnet, and ribonucleotides on herpes simplex virus-1 DNA polymerase: Mechanistic insights and a novel mechanism for preventing stable incorporation of ribonucleotides into DNA. Biochemistry 55, 1168–1177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma X., Helgason E., Phung Q. T., Quan C. L., Iyer R. S., Lee M. W., Bowman K. K., Starovasnik M. A., Dueber E. C., Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. U.S.A. 109, 9378–9383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skvortsova K., Tarbashevich K., Stehling M., Lister R., Irimia M., Raz E., Bogdanovic O., Retention of paternal DNA methylome in the developing zebrafish germline. Nat. Commun. 10, 3054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H., Luan Y., Liu T., Lee H. J., Fang L., Wang Y., Wang X., Zhang B., Jin Q., Ang K. C., Xing X., Wang J., Xu J., Song F., Sriranga I., Khunsriraksakul C., Salameh T., Li D., Choudhary M. N. K., Topczewski J., Wang K., Gerhard G. S., Hardison R. C., Wang T., Cheng K. C., Yue F., A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature 588, 337–343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vastenhouw N. L., Schier A. F., Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 24, 374–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vastenhouw N. L., Zhang Y., Woods I. G., Imam F., Regev A., Liu X. S., Rinn J., Schier A. F., Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.G. J. M. Hickey, C. Wike, X. Nie, Y. Guo, M. Tan, P. Murphy, B. R. Cairns, Establishment of developmental gene silencing by ordered polycomb complex recruitment in early zebrafish embryos. bioRxiv 2021.03.16.435592 [Preprint]. 16 March 2021. 10.1101/2021.03.16.435592. [DOI] [PMC free article] [PubMed]
- 49.Margueron R., Reinberg D., The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skene P. J., Henikoff S., An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janssen S. M., Lorincz M. C., Interplay between chromatin marks in development and disease. Nat. Rev. Genet. 10.1038/s41576-021-00416-x , (2021). [DOI] [PubMed] [Google Scholar]
- 52.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., Jaenisch R., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., Divito K. R., Fleming E. S., Giraldez A. J., Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes A. L., Kelley J. R., Klose R. J., Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation. Biochim. Biophys. Acta 1863, 194567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spitz F., Furlong E. E. M., Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 57.McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia W., Xie W., Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Reports 15, 1158–1175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q., Yu G., Ming X., Xia W., Xu X., Zhang Y., Zhang W., Li Y., Huang C., Xie H., Zhu B., Xie W., Imprecise DNMT1 activity coupled with neighbor-guided correction enables robust yet flexible epigenetic inheritance. Nat. Genet. 52, 828–839 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Parry A., Rulands S., Reik W., Active turnover of DNA methylation during cell fate decisions. Nat. Rev. Genet. 22, 59–66 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Yuan S., Sun Z., Microinjection of mRNA and morpholino antisense oligonucleotides in zebrafish embryos. J. Vis. Exp. 27, 1113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng X., Wu J., Brunmeir R., Kim S. Y., Zhang Q., Ding C., Han W., Xie W., Xu F., TELP, a sensitive and versatile library construction method for next-generation sequencing. Nucleic Acids Res. 43, e35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia W., Xu J., Yu G., Yao G., Xu K., Ma X., Zhang N., Liu B., Li T., Lin Z., Chen X., Li L., Wang Q., Shi D., Shi S., Zhang Y., Song W., Jin H., Hu L., Bu Z., Wang Y., Na J., Xie W., Sun Y. P., Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q., Liu W., Kou X., Zhao Y., He W., Li C., Chen B., Li Y., Wang Q., Ma J., Yin Q., Kee K., Meng A., Gao S., Xu F., Na J., Xie W., Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Wu J., Xu J., Liu B., Yao G., Wang P., Lin Z., Huang B., Wang X., Li T., Shi S., Zhang N., Duan F., Ming J., Zhang X., Niu W., Song W., Jin H., Guo Y., Dai S., Hu L., Fang L., Wang Q., Li Y., Li W., Na J., Xie W., Sun Y., Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557, 256–260 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 24, 1138–1143 (2011). [Google Scholar]
- 69.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dennis G., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A., DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, R60 (2003). [PubMed] [Google Scholar]
- 75.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Shirley X. S., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long H. K., Sims D., Heger A., Blackledge N. P., Kutter C., Wright M. L., Grützner F., Odom D. T., Patient R., Ponting C. P., Klose R. J., Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. eLife 2, e00348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11