Abstract
Cordycepin or 3′-deoxyadenosine is an interesting anti-cancer drug candidate that is found in abundance in the fungus Cordyceps militaris. It inhibits cellular growth of many cancers including lung carcinoma, melanoma, bladder cancer, and colon cancer by inducing apoptosis, anti-proliferation, anti-metastasis and by arresting the cell cycle. Cordycepin has, however, poor stability and low solubility in water, resulting in loss of its bioactivity. Liposomes can be used to overcome these obstacles. Our aim is to improve cordycepin's anti-colon cancer activity by liposome encapsulation. Cordycepin-encapsulated liposomes were designed and fabricated based on a combination of theoretical and experimental studies. Molecular dynamics (MD) simulations and free energy calculations suggest that phosphatidylcholine (PC) lipid environment is favorable for cordycepin adsorption. Cordycepin passively permeates into PC lipid bilayers without membrane damage and strongly binds to the lipids' polar groups by flipping its deoxyribose sugar toward the bilayer center. Our fabricated liposomes containing 10 : 1 molar ratio of egg yolk PC : cholesterol showed encapsulation efficiency (%EE) of 99% using microfluidic hydrodynamic focusing (MHF) methods. In our in vitro study using the HT-29 colon cancer cell line, cordycepin was able to inhibit growth by induction of apoptosis. Cell viability was significantly decreased below 50% at 125 μg mL−1 dosage after 48 h treatment with non-encapsulated and encapsulated cordycepin. Importantly, encapsulation provided (1) a 2-fold improvement in the inhibition of cancer cell growth at 125 μg mL−1 dosage and (2) 4-fold increase in release time. These in silico and in vitro studies indicate that cordycepin-encapsulated liposomes could be a potent drug candidate for colon cancer therapy.
Cordycepin-encapsulated liposomes could be a potent drug candidate for cancer therapy.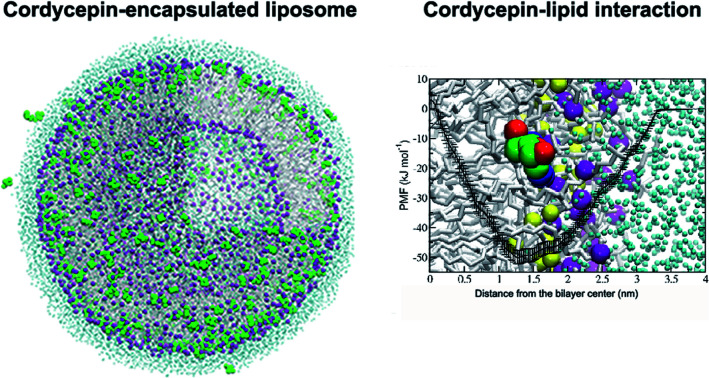
1. Introduction
Cordyceps, a genus of fungi that are well-known and valuable in traditional Chinese medicine, are widespread as active ingredients in food supplements and cosmetic products.1–3 They possess various pharmaceutical properties such as being anti-oxidative, anti-free radical, anti-tumor/cancer, and anti-inflammatory.4–9Cordyceps are mainly composed of polysaccharides, mannitols, ergosterols and nucleosides.10 Their nucleoside compounds have received a lot of attention due to their potent anti-tumor/cancer activities.11–13
Cordycepin, or 3′-deoxyadenosine, is cytotoxic nucleoside analogue mostly found in Cordyceps militaris.10 Many studies have shown that cordycepin is involved in inducing apoptosis,14–16 anti-proliferation,17 anti-metastasis18 and cell cycle arrest19 of various cancers by mediating cell surface receptors, in particular the adenosine receptors. The anti-cancer activities of cordycepin and their inhibition mechanisms against protein targets have been reviewed in ref. 18, 20 and 21. Current understanding of how cordycepin transports into cell membranes is, however, unresolved. This is manifested by the fact that although cordycepin is a potent anti-cancer drug candidate, in vivo studies have reported loss of its activity.22 The half-life of cordycepin in rat blood is low, about 1.6 minutes, but studies have shown that it can be increased by 14-fold after pretreatment by metabolic blockers.22 It has also been shown that the decrease in its in vivo activity is mainly due to biotransformation of adenosine to inosine which is induced by the adenosine deaminase enzyme.23,24
Nanocarriers can be used to improve the stability and solubility of bioactive compounds25–29 and they can passively enter into tumor tissues through leaky vasculature;30–32 tumor vasculature has gaps between them and adjacent endothelium cells ranging from 100–780 nm, while those in healthy vasculature are 5–10 nm.30 Thus, nanocarriers can enhance the tumor targeting ability of drugs by being size-selective. In addition, many studies have shown that nanocarriers also exhibit other advantages over conventional treatment techniques such as providing larger surface area, providing higher absorption rate, increased cellular uptake and drug localization, improved uptake of poorly soluble drugs and decrease in drug resistance.33,34
Liposomes are one of the most popular nanocarriers due to their biocompatibility, biodegradability, high-loading capacity, controlled release, easy production and low cost.25,27,28,35,36 Liposomes with their sizes in the range of 100–200 nm have been broadly used to deliver anti-cancer agents.25,36 They can transport through the leaky tumor vasculature and also increase anti-cancer drugs half-lives.32,37 Cordycepin-encapsulated liposomes have been shown to reduce the growth of hepatoma tumors in mice whereas treatment using free cordycepin and blank liposomes has been shown to have no effect.38 Cordycepin-encapsulated liposomes and cordycepin-encapsulated transferrin-conjugated liposomes have also been shown to increase reactive oxygen species (ROS) and alter mitochondrial function in liver cancer cells associated with the induction of apoptosis;39 although elevated levels of ROS are typically associated with adverse health effects, increase in ROS has also been shown to induce apoptosis.40 This encapsulation also enhanced the release time of cordycepin by 4-fold.39
Computer simulations have been extensively used to study many biomolecular interactions such as protein–ligand interactions, protein–protein interactions and membrane transport, including drug delivery systems.41–46 MD simulations have also been used to investigate the binding mechanisms of cordycepin and many protein targets, for example uric acid transporter 1 (URAT1) associated with anti-hyperuricemic effect47 and Cdk-2 in cervical cancer.48 Atomistic insight into binding properties can be used as guidance for the design of new potent proteins inhibitors.49 In addition, the loading mechanism(s) of cordycepin into nanodrug carriers such as cyclodextrins (CDs) can be explored by MD simulations;50 simulations have suggested that cordycepin forms highly stable inclusion complex with CDs in agreement with experiments.50 In particular, the binding energies showed that the inclusion complex of cordycepin and α-CD was the strongest in comparison to β-CD and γ-CD (which have larger cavities).50 Translocation free energy of adenine DNA base, which is a part of cordycepin, indicated that adenine could permeate into palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer and be stably located around the lipid carbonyl groups (the adsorption free energy was about −5 kJ mol−1).51
In the current work, computational and experimental studies are applied to study the encapsulation of cordycepin into liposomes and its anti-colon cancer activity. The simulation results were used as a guideline for liposome design. Our thus designed cordycepin-encapsulated liposomes were experimentally formulated and evaluated by in vitro testing of their anti-colon cancer activity. The liposomes, mainly composed of PC lipids, were fabricated by microfluidic hydrodynamic focusing (MHF) methods52 and they can enhance encapsulation efficiency by 1.3–2.3 fold, compared to the previous related studies.38,39,53 Results regarding cytotoxicity and induction of apoptosis show that our fabricated cordycepin-encapsulated liposomes could improve anti-HT-29 colon cancer activity.
2. Materials and methods
2.1. Computational methods
Force field testing
The initial structure of cordycepin was created using the Automated Topology Builder (ATB).54 The united atom Gromacs53a6 force field55,56 was used for cordycepin and solvents. The octanol/water partitioning of cordycepin was estimated and compared with experiments.57 The force field details are described in ESI (Table S1†). The partition coefficient can be obtained from the difference of the free energies of cordycepin dissolved in the octanol and water, ΔGo and ΔGw, respectively, as
| ΔΔGo/w = Go − Gw = −RT ln Po/w, | 1 |
where R is the gas constant (0.008314 kJ mol−1 K−1) and T is temperature in Kelvins.
The free energies were estimated using the thermodynamic integration (TI) technique.58,59 A cordycepin molecule was fully dissolved in single point charge (SPC) water60 and octanol in simulation boxes of 3.0 × 3.0 × 3.0 nm3 and 4.7 × 4.7 × 4.7 nm3, respectively. The simulations were performed under the NVT ensemble (constant number of atoms, volume and temperature) for 5 ns and the last 1 ns was used to analysis. The temperature was held constant at 298 K by the Parrinello–Donadio–Bussi velocity rescale thermostat.61,62 The coupling parameter (λ) was varied from 0 to 1 using intervals of 0.05. The Bennett Acceptance Ratio (BAR) was used to calculated the free energy differences.63 The free energy difference at each λ with the error bar for the last 1 ns are plotted in Fig. S1.† The computed octanol/water partition coefficients are shown in Table S2† and are in good agreement with experimental data57 suggesting the present octanol force field is reliable. The errors in octanol solvation free energy in water and in octanol with respect to the experiments were 0.1% and 2.8% (Table S2†). The solvation free energies of cordycepin indicated that cordycepin is preferably dissolved in octanol as compared to water. The calculation of Po/w gave −1.30 kJ mol−1, which is close to the experimental value of −0.91 kJ mol−1.57 The partitioning coefficient of cordycepin suggests that cordycepin favors being encapsulated inside the liposomes.
Molecular dynamics (MD) simulations
Atomistic MD simulations were performed to explore the interactions of cordycepin and lipids associated with the early stages of cordycepin encapsulation and delivery process. The system preparations and simulations were done using the Gromacs 5.1.1 software.64 The 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) lipid bilayer was chosen since PC lipids are the main component of the present liposomes and they also show significantly elevated levels in colon cancer cells with respect to normal cells.65,66
The initial structure of the 128 DPPC lipid bilayer and lipid parameters were taken from a previous study.67 Cordycepin molecules were initially placed in the water phase 3.5 nm away from the bilayer center; the number of cordycepins was either 1 or 18, corresponding to 1 : 374 and 1 : 20 cordycepin : lipid molar ratios, respectively. All systems were solvated with 10 367 SPC water molecules.60 To eliminate possible overlaps and voids, energy minimization was first performed using the method of steepest descents. The simulations were performed under the NPT ensemble (constant number of atoms, pressure and temperature) for 1 μs. The last 400 ns were used for analysis. The integration time step was 2 fs and data were collected every 2 ps. Periodic boundary conditions were applied in all directions. Cut-off of 1.0 nm was used for both the Lennard-Jones and the real-space part of the electrostatic interactions. The long-range electrostatic interactions were treated by the particle-mesh Ewald (PME) method68–70 with the reciprocal-space interactions evaluated on a 0.12 nm grid with cubic interpolation of order four. All bond lengths were constrained by the P-LINCS algorithm.71 Cordycepin molecules, lipids and water were thermostated separately at 298 K using the Parrinello–Donadio–Bussi velocity rescale algorithm.61,62 The Parrinello–Rahman barostat72 was applied separately in the xy-plane and the bilayer normal (semi-isotropic barostat). Pressure was held constant at 1 bar with a time constant of 1 ps and compressibility of 4.5 × 10−5 bar−1. The optimized simulation protocols had been intensively tested73,74 and extensively used.75–78 Molecular visualizations were done using the Visual Molecular Dynamics (VMD) software.79
Translocation free energy
The free energy of cordycepin translocating through a DPPC lipid bilayer was calculated by umbrella sampling80 with the weighted histogram analysis method (WHAM).81 The cordycepin molecule was restrained with respect to the center of mass (COM) of the lipid bilayer, using a harmonic restraint with a force constant of 1000 kJ mol−1 nm−2. The distance along the z-axis between the COMs of the cordycepin molecule and the bilayer's center was varied from 0 to 4 nm with 0.1 nm interval (a total 41 simulation windows). All systems were simulated under the NPT ensemble at temperature of 298 K. Each window was run for 100 ns. The total simulation time was about 4.1 μs and the trajectories of the last 20 ns were extracted for free energy calculation. Statistical errors were estimated by the bootstrap analysis.82
2.2. Experimental methods
Reagents
Cordycepin from Cordyceps militaris and phosphatidylcholine (PC) from egg yolk were obtained from Sigma-Aldrich®. Their purity was more than 98%, verified by high performance liquid chromatography (HPLC). The Dulbecco's Modified Eagle Medium (DMEM), antibiotics and fetal bovine serum (FBS) were purchased from Gibco. LIVE/DEAD®. All other chemicals and reagents used in these experiments were of analytical grade.
Fabrication of cordycepin-encapsulated liposomes
Cordycepin-encapsulated liposomes were prepared by the MHF method. The lipid phase of the liposomes was prepared by mixing cholesterol and egg yolk PC at molar ratio of 1 : 10, dissolved in ethanol with didecyldimethylammonium (DDAB) surfactant. Cordycepin was dissolved in water and then mixed to the lipid phase at molar ratio of 1 : 20 using turbulent micromixing technique. The flow rate ratio (FRR) of water and lipid was 2 : 1. Ethanol was evaporated under vacuum at pressure of 100 torr.
Particle size distribution and zeta potential measurements
Particle sizes and the zeta potential of liposome-encapsulated cordycepin were determined by the dynamic light scattering (DLS) technique (Malvern Instruments Zetasizer Nano ZX). DLS measurements were carried out using He–Ne laser (λ0 = 633 nm, θ = 173°). The cordycepin-encapsulated liposomes were in solution and diluted 100 times in deionized (DI) water before measurements.
Entrapment efficiency measurements
The cordycepin-encapsulated liposomes were added to Amicon® ultra-15 centrifugal filter unit with Ultracel® regenerated cellulose membrane (Merch Millipore Ltd., Darmstadt, Germany) and centrifuged at 15 000 × g for 1 hour. Consequently, non-encapsulated cordycepin was separated and then dissolved in water. The cordycepin concentration was measured by HPLC (Agilent®) with UV light (λ = 254 nm), column C18 at 40 °C, solute by 15%MeOH. The percentage of entrapment efficiency (%EE) can be calculated by the following equation:
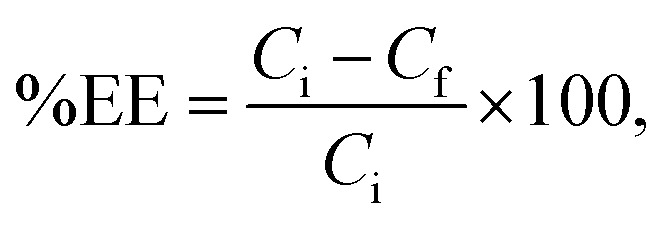 |
2 |
where Ci is the initial concentration of cordycepin added to the liposomes and Cf is the concentration of free cordycepin.
Release study
Cordycepin release was determined using in vitro release test (IVRT) method, which has been described by Dana et al.83 The cordycepin-encapsulated liposomes were mixed with phosphate-buffered saline (PBS) buffer pH 7 and incubated at 37 °C. Free cordycepin was separated from the encapsulated cordycepin using Amicon® ultrafiltration membrane with a molecular weight cut-off (MWCO) of 50 kDa and collected at various time points. The amount of released cordycepin was analyzed by HPLC (Agilent, USA). Methanol/distilled water (15 : 85, v/v) was used as the mobile phase. Elution was performed at a flow rate of 0.6 mL min−1 on HiQSil C18HS column (250 mm × 4.6 mm, 5 μm) at 40 °C using UV detector with the wavelength of 260 nm.
Cell culture
The HT-29 human colon cancer cell line was provided by Nano Delivery System laboratory, Nanotechnology center (NSTDA, Thailand). The cells were grown in DMEM medium with 10% FBS, penicillin (100 units per mL), streptomycin (100 units per mL), and l-glutamine (2 mM) at 37 °C of 5% CO2. The spheroidal HT-29 cells were cultured in low-attachment 96-well plate under the same growth conditions as the HT-29 cell line. After 24 hours, spheroids were ready to be used for treatment.
Cell viability tests
Inhibition of cell viability of the HT-29 human colon cancer cell was evaluated by an MTT (3,4,5-dimethylthiazol 2,5 diphenyltetrazolium bromide) assay. Cells were seeded in 96-well plate at 37 °C with 5% CO2. After 72 hours, the two-fold serial dilutions (initial concentration of 500 μg mL−1 diluted to 0.49 μg mL−1) of free cordycepin, encapsulated cordycepin and free liposomes in fresh culture medium were prepared. The old culture medium was removed and was then replaced by the dilute solutions. After 48 hours of incubation, the cells were removed from the drug-containing medium which was replaced by a fresh medium. MTT dye was added to the wells and the cells were incubated for further 3 hours followed by the MTT assay procedure. Then, medium was replaced again by MTT reagents. The results were measured at the optical density (OD) of 590 nm. For observation of spheroidal cells, the HT-29 cell line was seeded to low-attachment 96-well plate. After 24 hours, the tumors were treated by drug-containing medium for 48 hours. Spheroid cell viability was evaluated using the LIVE/DEAD™ Viability/Cytotoxicity Kit (Invitrogent). Three-dimensional spheroidal cell assemblies present a better model for tumors than two-dimensional cell lines closely mimicking features of solid tumors.84
Apoptosis tests
The HT-29 cell line was grown and treated with free cordycepin and encapsulated cordycepin for 48 hours following the same conditions as in the cell viability tests. Then, cells were stained with the reagents in the Apoptosis detection kit I (BD Biosciences). Caspases 3/7 activity and Annexin V-FITC/propidium iodide (PI) staining as indicator of apoptosis/necrosis were examined.85
3. Results and discussion
3.1. Insight into interaction of cordycepin and lipid
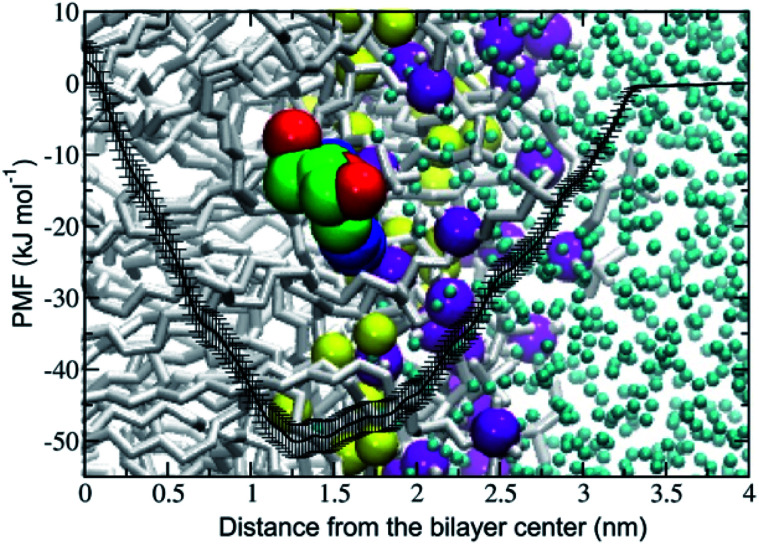
The potential of mean force (PMF) of transferring a cordycepin molecule into a DPPC lipid bilayer, Fig. 1, shows that cordycepin prefers to permeate inside the membrane interior. The absorption free energy, the free energy difference between cordycepin in water and cordycepin at the minimum position of 1.3 nm from the bilayer center was found to be −50 kJ mol−1. Translocation free energy of 53 kJ mol−1 was required to move cordycepin from the minimum position further to the bilayer center. The free energy calculation indicates that cordycepin can easily enter the lipid bilayer by strongly binding to the lipids' polar group.
Fig. 1. Potential of mean force (PMF) of moving a cordycepin molecule into a DPPC lipid bilayer as a function of distance in the z-direction from the bilayer center (ξ = 0 nm). The free energy in the water phase at ξ = 4 nm was defined to be equal to zero. Cordycepin preferably located around the DPPC carbonyl groups and was tilted with respect to the bilayer normal by turning its deoxyribose sugar ring toward the bilayer's center. A snapshot at the end of the simulation at 1 μs and at 1.3 nm distance from the bilayer center is shown. The yellow and purple van der Waals (vdW) spheres represent the carbonyl and phosphate groups, respectively.
Cordycepin is preferably located around the lipids' carbonyl groups and it tilts its ribose sugar ring toward the bilayer center by 72.3 ± 9.5° with respect to the bilayer normal. The tilt angle (θ) is given in Fig. S2.† The density profiles of cordycepin, and the lipids' phosphates and carbonyls for biased and unbiased simulations are shown in Fig. S3.† Permeation into lipid bilayer interior has been shown for several molecules analogous to cordycepin.51,86 As already discussed earlier, adenine has been shown to permeate into the POPC lipid tail region51 reaching its free energy minimum of approximately −5 kJ mol−1 at about 1.2 nm from the bilayer center.51 In comparison, the anti-cancer drug ellipticine, which is composed of purine (adenine analog) and isoquinoline has been reported to permeate into POPC lipids' tail region and have absorption energy of −30 kJ mol−1 at the minimum position of about 1.0 nm from the bilayer's center.68 The experimental emission spectra of ellipticine in DPPC liposome shows that it is incorporated in DPPC liposomes by partition coefficient (Kp) of 1.1 × 104.87
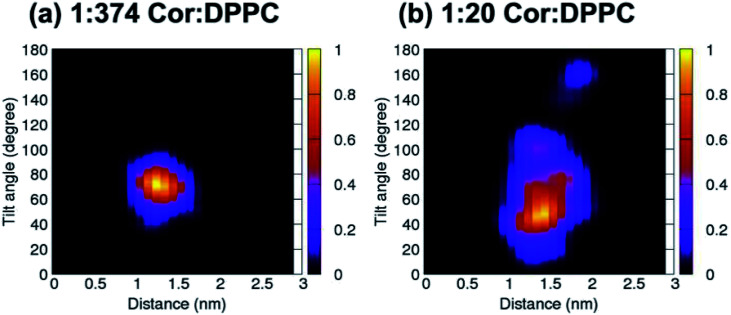
Unbiased simulations in Fig. 2a show that cordycepin preferably adsorbs into the bilayer at the position and orientation in agreement with Fig. 1. At 1 : 374 Cor : DPPC molar ratio (Fig. 2a), the most probable distance from the bilayer center was 1.30 nm and cordycepin had the tilt angle of 72.4°. The distance from the bilayer center increased with increasing cordycepin concentration. As a result, cordycepin molecules were shifted close to the lipids' carbonyl groups at high concentrations (Fig. 2b and Fig. S3b†). Most of the cordycepin molecules (89% or 16 molecules out of total of 18) still preferably turned their deoxyribose rings toward the bilayer center, while the remaining cordycepin molecules flipped their deoxyribose rings facing outward of the bilayer center. At 1 : 20 Cor : DPPC molar ratio (consistent with the experimental conditions), the presence of cordycepin clusters caused a slight increase in bilayer thickness and area per lipid (Fig. S4†), but there was no noticeable membrane deformation (Fig. S4†).
Fig. 2. 2D contour plot showing cordycepin's distance from the bilayer center and its tilt angle for the systems with (a) 1 : 374 and (b) 1 : 20 Cor : DPPC molar ratio. All cordycepin molecules were located stably at DPPC's carbonyl groups and had mostly turned their sugar ring toward the bilayer's center (θ < 90°) at 1 : 20 Cor : DPPC molar ratio. Some cordycepins preferably flipped their adenine rings close to the bilayer center instead (θ > 90°).
Strong hydrogen bonding (Hbond) between cordycepin and DPPC's phosphate and carbonyl groups was observed. The total number of Hbonds between each cordycepin molecule and lipids' polar groups was in the range of 1–4 (Fig. S5†). The number of Hbonds to carbonyls was slightly higher than to the phosphate groups suggesting that binding to carbonyls is preferable. For the phosphate and carbonyl groups at 1 : 374 Cor : DPPC molar ratio, the maxima in the Hbond distributions were at 1 and 3 bonds, respectively (Fig. S5a†). At the higher concentration 1 : 20 Cor : DPPC molar ratio, the cordycepin molecules preferably formed Hbonds to the phosphate and carbonyl, 1–2 bond(s) and 2–3 bonds, respectively (Fig. S5b†). These observations are consistent with high encapsulation efficiency observed in experiment. Permeation of cordycepin into the lipid membrane without membrane damage was observed.
3.2. Fabrication of cordycepin-encapsulated liposome and its physicochemical properties
The nanoliposomes, composed of 10 : 1 molar ratio of egg yolk PC and cholesterol, were formed with high cordycepin encapsulation efficiency (EE) of 99% using the MHF technique.52,88 Notably, the %EE of the cordycepin-encapsulated liposomes, previously reported in ref. 38, 39 and 53 were 43%, 65% and 75%, respectively. In those studies, different lipid components and preparation techniques had been used in which encapsulations were prepared at 4 : 1 molar ratio of soy PC : cholesterol, 3 : 1 mass ratio of soy PC : cholesterol, and 3 : 1 mass ratio of soy lecithin : cholesterol by using the ammonium sulfate gradient loading method, ethanal dilution-sonication method and supercritical gas anti-solvent (SC-GAS) method, respectively. The average particle diameter size of the cordycepin-encapsulated liposome was 138.0 ± 1.2 nm. Based on size, it is plausible that these liposomes are able to enter into the tumor tissues via the leaky tumor vasculature, but they are too large to enter into the healthy tissue. The zeta potential of cordycepin-encapsulated liposomes was −6.51 ± 0.79 mV. The value of the zeta potential has been reported to depend on the lipid species that compose the liposomes: for DPPC and DOPC liposomes the zeta potential, determined in phosphate buffer at 20 °C, has been reported to be −10.2 and +3.2 mV, respectively.89 The zeta potential of cordycepin-encapsulated in liposome in the study of Bi et al.,39 which used mainly hydrogenated soy phosphatidylcholine and cholesterol, was +9.2 ± 3.5 mV.
3.3. In vitro release from cordycepin-encapsulated liposomes
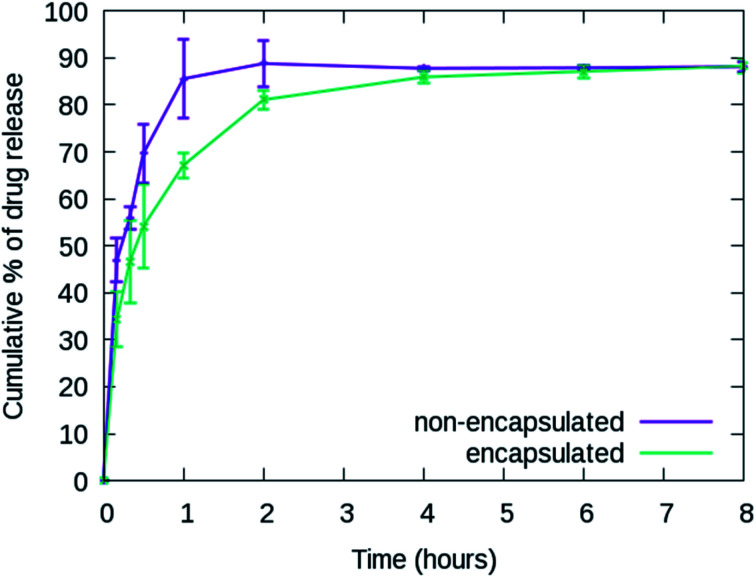
The cumulative release percentages of cordycepin at 37 °C in PBS buffer are shown in Fig. 3. The cumulative percentage increased with time and reached ∼88% within the first 2 h and it extended to more than 8 h for the cordycepin-encapsulated liposome. Encapsulation led to a 4-fold enhancement in release time compared to free cordycepin. This improvement is in the range observed in a previous study of Bi et al. who observed that the cumulative percentage reached around 90% at 6 h for free cordycepin while it took over 24 h for encapsulated-cordycepin.39 Note that, the filtration membrane in our study has about 4-times higher MWCO than Bi et al. had.39 The high MWCO refers to large membrane pore size. The rate constant (K) is determined by fitting to A × (1 − e(−Kx)) + B, the haft-time (t1/2) is 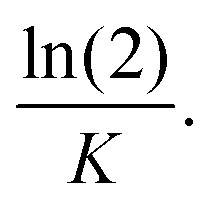 The results in Table S3† show that the release rate constant (K) of non-encapsulated cordycepin was significantly higher than that of the encapsulated cordycepin by 2.9-fold. Consequently, the half-time of non-encapsulated cordycepin was decreased by 2.5-fold compared to encapsulated cordycepin.
The results in Table S3† show that the release rate constant (K) of non-encapsulated cordycepin was significantly higher than that of the encapsulated cordycepin by 2.9-fold. Consequently, the half-time of non-encapsulated cordycepin was decreased by 2.5-fold compared to encapsulated cordycepin.
Fig. 3. In vitro release profiles of non-encapsulated and encapsulated cordycepin. The cumulative percentage reached ∼88% within 2 and 8 hours for free cordycepin and cordycepin-encapsulated liposomes, respectively.
3.4. Effects of cordycepin-encapsulated liposomes on HT-29 cell viability and apoptosis induction
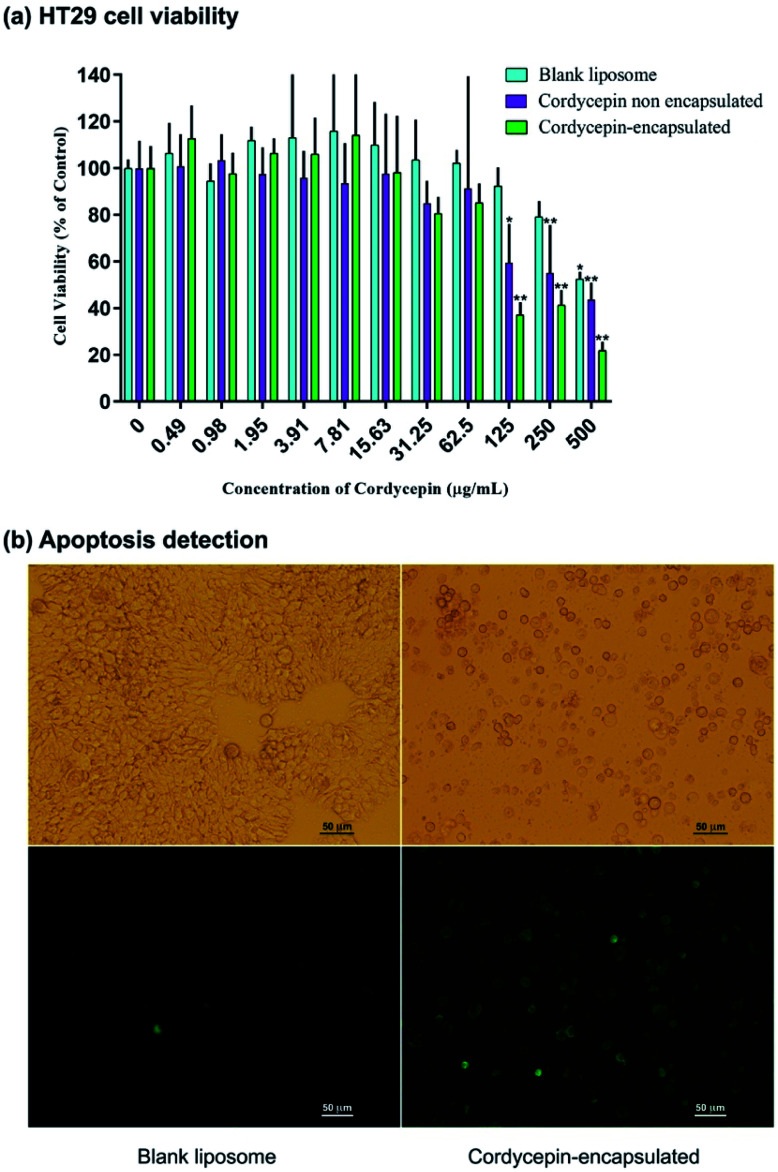
The effects of cordycepin-encapsulated liposomes on HT-29 cell viability after 48 h treatment were tested by MTT assay. The results revealed that both cordycepin-encapsulated liposomes and free cordycepin significantly restricted the growth of HT-29 cancer cells in a dose-dependent manner (Fig. 4a). After cordycepin-encapsulated liposome exposure with doses of 125 μg mol−1 for 48 h, cell viability dramatically decreased below 50%. Cordycepin-encapsulated liposomes induced HT-29 cell death and they were more effective than free cordycepin and blank liposomes at doses higher than 125 μg mol−1. At 125 μg mL−1 dosage the percentage of cell viability after being treated by blank liposomes, free cordycepin and encapsulated cordycepin were 92%, 59% and 37%, respectively. The cordycepin-encapsulated liposomes are 1.5 times more efficient in killing cancer cells at 125 μg mL−1 dosage than free cordycepin. Next, the induction of apoptosis by cordycepin-encapsulated and blank liposomes was checked (Fig. 4b). Cells were shrunk and detached after treatment with cordycepin-encapsulated liposomes for 24 hours, while after treatment with blank liposomes the cells were still alive (Fig. 4b (top)). Apoptotic cell death was detected by Annexin V assay, marked in green in Fig. 4b (bottom).
Fig. 4. (a) Dose-dependence of HT-29 cell viability after 48 h treatment by blank liposomes (cyan), non-encapsulated cordycepin (purple) and encapsulated cordycepin (green). For statistical analysis, One-way ANOVA with post hoc Dunnett's test was performed. *P < 0.001 and **P < 0.0001 were considered as statistically significant. (b) HT-29 cells under microscope after treatment by blank liposomes and cordycepin-encapsulated liposomes. (Top) The cells' morphology changed after cordycepin-encapsulated treatment. (Bottom) Apoptotic cell death was detected after cordycepin-encapsulated treatment. The dead cells are shown in green.
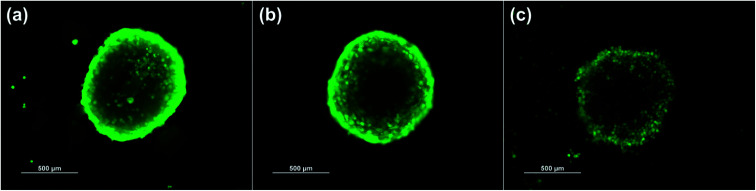
The HT-29 (spheroidal) cell viability after 48 h treatment was also investigated as shown in Fig. S6.† The results are in agreement with the HT-29 cell line in that the percentage of cell viability decreased below 50% at 125 μg mL−1 dosage. Images of HT-29 cells are shown in Fig. 5a–c before treatment, blank liposomes and after treatment with cordycepin-encapsulated liposomes at 125 μg mL−1 dosage, respectively. After treatment by cordycepin-encapsulated liposomes at 125 μg mL−1 dosage, the spheroidal cell was damaged. In contrast, the HT-29 cells appeared as regular spheroids after treatment with blank liposomes at the same dosage.
Fig. 5. Images of spheroidal HT-29 live cells in the cases of (a) before treatment, (b) after treatment with blank liposomes and (c) after treatment with cordycepin-encapsulated liposomes. The interfaces become more diffuse and intensity lessens after treatment with cordycepin-encapsulated liposomes.
4. Conclusions
The present in silico and in vitro studies suggest that liposome encapsulation enhances anti colon cancer activity of cordycepin. MD simulations and free energy calculations provided the atomistic details of interactions between cordycepin and lipids, which served as a guideline for the design of cordycepin-encapsulated liposomes; the phosphatidylcholine (PC) lipid is appropriate for cordycepin adsorption. Cordycepin strongly binds to the lipids' polar groups via hydrogen bonding and consequently stably adsorbs around the lipids' carbonyl groups by tilting its deoxyribose sugar ring toward the bilayer's center. Cordycepin was observed to permeate into the PC lipid bilayer without membrane damage at 1 : 20 cordycepin : lipid molar ratio. These findings support high encapsulation efficiency and non-toxicity of cordycepin treatment.
Cordycepin-encapsulated liposomes mainly comprising PC lipids (containing 10 : 1 molar ratio of egg yolk PC : cholesterol) were fabricated using microfluidic hydrodynamic focusing (MHF), and they showed very high percentage of encapsulation efficiency (%EE) of 99%. The in vitro study of HT-29 colon cancer cell line showed that cordycepin could inhibit the HT-29 colon cancer cell growth by inducing apoptosis. Cell viability was dramatically decreased, to be lower than 50% at 125 μg mL−1 dosage after 48 treatment by free cordycepin and cordycepin-encapsulated liposome. Importantly, however, encapsulation by liposomes appears to significantly improve the inhibition of cancer cell growth: compared to treatment with free cordycepin, cell viability showed almost 2-fold decrease after treatment with cordycepin-encapsulated liposomes at 125 μg mL−1 dosage. In addition, encapsulation lead to a 4-fold increase in release time. Finally, the nanosized (138.0 ± 1.2 nm) cordycepin-encapsulated liposome are also size-selective in that they are able to enter into the tumor tissues through the leaky tumor vasculature which is involved in the enhancement of colon cancer cell growth inhibition – tumor vasculature has gaps of size 100–780 nm while healthy vasculature has gaps in much smaller range of 5–10 nm.30
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
This research was supported by National Research Council of Thailand and Thailand Research Fund (TRF) Research Scholar (Grant No. RSA6180021). JW and WS acknowledge support from Postdoctoral Fellowship from Kasetsart University. MK thanks the Natural Sciences and Engineering Research Council of Canada (NSERC) the Canada Research Chairs Program for financial support. Computing facilities were provided by the Department of Physics, Faculty of Science, Kasetsart University.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00038a
References
- Chang Y. Jeng K.-C. Huang K.-F. Lee Y.-C. Hou C.-W. Chen K.-H. Cheng F.-Y. Liao J.-W. Chen Y.-S. Effect of Cordyceps militaris supplementation on sperm production, sperm motility and hormones in Sprague-Dawley rats. Am. J. Chin. Med. 2008;36(5):849–859. doi: 10.1142/S0192415X08006296. [DOI] [PubMed] [Google Scholar]
- Shashidhar M. Giridhar P. Sankar K. U. Manohar B. Bioactive principles from Cordyceps sinensis: a potent food supplement–a review. J. Funct. Foods. 2013;5(3):1013–1030. doi: 10.1016/j.jff.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde K. Bahkali A. Moslem M. Fungi—an unusual source for cosmetics. Fungal Divers. 2010;43(1):1–9. doi: 10.1007/s13225-010-0043-3. [DOI] [Google Scholar]
- Li S. Li P. Dong T. Tsim K. Anti-oxidation activity of different types of natural Cordyceps sinensis and cultured Cordyceps mycelia. Phytomedicine. 2001;8(3):207–212. doi: 10.1078/0944-7113-00030. [DOI] [PubMed] [Google Scholar]
- Liu J.-y. Feng C.-p. Li X. Chang M.-c. Meng J.-l. Xu L.-j. Immunomodulatory and antioxidative activity of Cordyceps militaris polysaccharides in mice. Int. J. Biol. Macromol. 2016;86:594–598. doi: 10.1016/j.ijbiomac.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Tuli H. S. Sandhu S. S. Sharma A. K. Pharmacological and therapeutic potential of Cordyceps with special reference to Cordycepin. 3 Biotech. 2014;4(1):1–12. doi: 10.1007/s13205-013-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Wang J. Wang W. Zhang H. Zhang X. Han C. The Chemical Constituents and Pharmacological Actions of Cordyceps sinensis. J. Evidence-Based Complementary Altern. Med. 2015:575063. doi: 10.1155/2015/575063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y. Jung S.-J. Ha K.-C. Sin H.-S. Jang S.-H. Chae H.-J. Chae S.-W. Anti-inflammatory effects of Cordyceps mycelium (Paecilomyces hepiali, CBG-CS-2) in Raw264.7 murine macrophages. Orient. Pharm. Exp. Med. 2015;15(1):7–12. doi: 10.1007/s13596-014-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiderle F. R. Baggio C. H. Borato D. G. Santana-Filho A. P. Sassaki G. L. Iacomini M. Van Griensven L. J. J. P. O. Anti-inflammatory properties of the medicinal mushroom Cordyceps militaris might be related to its linear (1→3)-β-d-glucan. PLoS One. 2014;9(10):e110266. doi: 10.1371/journal.pone.0110266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. P. Yang F. Q. Tsim K. W. K. Quality control of Cordyceps sinensis, a valued traditional Chinese medicine. J. Pharm. Biomed. Anal. 2006;41(5):1571–1584. doi: 10.1016/j.jpba.2006.01.046. [DOI] [PubMed] [Google Scholar]
- Nakamura K. Yoshikawa N. Yamaguchi Y. Kagota S. Shinozuka K. Kunitomo M. Antitumor effect of cordycepin (3′-deoxyadenosine) on mouse melanoma and lung carcinoma cells involves adenosine A3 receptor stimulation. Anticancer Res. 2006;26(1A):43–47. [PubMed] [Google Scholar]
- Tian X. Li Y. Shen Y. Li Q. Wang Q. Feng L. Apoptosis and inhibition of proliferation of cancer cells induced by cordycepin. Oncol. Lett. 2015;10(2):595–599. doi: 10.3892/ol.2015.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. Yin P. Lu Y. Zhou Z. Jiang C. Liu Y. Yu X. Cordycepin prevents oxidative stress-induced inhibition of osteogenesis. Oncotarget. 2015;6(34):35496–35508. doi: 10.18632/oncotarget.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y. Debnath T. Kim S.-K. Lim B. O. Anti-cancer effect and apoptosis induction of cordycepin through DR3 pathway in the human colonic cancer cell HT-29. Food Chem. Toxicol. 2013;60:439–447. doi: 10.1016/j.fct.2013.07.068. [DOI] [PubMed] [Google Scholar]
- Yoshikawa N. Yamada S. Takeuchi C. Kagota S. Shinozuka K. Kunitomo M. Nakamura K. Cordycepin (3′-deoxyadenosine) inhibits the growth of B16-BL6 mouse melanoma cells through the stimulation of adenosine A 3 receptor followed by glycogen synthase kinase-3β activation and cyclin D 1 suppression. Naunyn-Schmiedeberg's Arch. Pharmacol. 2008;377:591–595. doi: 10.1007/s00210-007-0218-y. [DOI] [PubMed] [Google Scholar]
- Cao H.-L. Liu Z.-J. Chang Z. Cordycepin induces apoptosis in human bladder cancer cells via activation of A3 adenosine receptors. Tumor Biol. 2017;39(7):1–6. doi: 10.1177/1010428317706915. [DOI] [PubMed] [Google Scholar]
- Lee D. Lee W.-Y. Jung K. Kwon Y. S. Kim D. Hwang G. S. Kim C.-E. Lee S. Kang K. S. The Inhibitory Effect of Cordycepin on the Proliferation of MCF-7 Breast Cancer Cells, and Its Mechanism: An Investigation Using Network Pharmacology-Based Analysis. Biomolecules. 2019;9(9):414. doi: 10.3390/biom9090414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. Meng X. Qiu Z. Su Y. Yu P. Qu P. Anti-tumor and anti-metastatic roles of cordycepin, one bioactive compound of Cordyceps militaris. Saudi J. Biol. Sci. 2018;25(5):991–995. doi: 10.1016/j.sjbs.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. Ling J. Zhang G. Liu F. Tao S. Han Z. Chen S. Chen Z. Le H. Cordycepin induces cell cycle arrest and apoptosis by inducing DNA damage and up-regulation of p53 in Leukemia cells. Cell Cycle. 2015;14(5):761–771. doi: 10.1080/15384101.2014.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K. Shinozuka K. Yoshikawa N. Anticancer and antimetastatic effects of cordycepin, an active component of Cordyceps sinensis. J. Pharmacol. Sci. 2015;127(1):53–56. doi: 10.1016/j.jphs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Yoon S. Y. Park S. J. Park Y. J. The anticancer properties of cordycepin and their underlying mechanisms. Int. J. Mol. Sci. 2018;19(10):3027. doi: 10.3390/ijms19103027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-J. Lin L.-C. Tsai T.-H. Pharmacokinetics of adenosine and cordycepin, a bioactive constituent of Cordyceps sinensis in rat. J. Agric. Food Chem. 2010;58(8):4638–4643. doi: 10.1021/jf100269g. [DOI] [PubMed] [Google Scholar]
- Li G. Nakagome I. Hirono S. Itoh T. Fujiwara R. Inhibition of adenosine deaminase (ADA)-mediated metabolism of cordycepin by natural substances. Pharmacol. Res. Perspect. 2015;3(2):e00121. doi: 10.1002/prp2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. B. Radhi M. Cipolla E. Gandhi R. D. Sarmad S. Zgair A. Kim T. H. Feng W. Qin C. Adrower C. A novel nucleoside rescue metabolic pathway may be responsible for therapeutic effect of orally administered cordycepin. Sci. Rep. 2019;9(1):1–12. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri A. Loomis K. Smith B. Lee J.-H. Yavlovich A. Heldman E. Blumenthal R. Lipid-Based Nanoparticles as Pharmaceutical Drug Carriers: From Concepts to Clinic. Crit. Rev. Ther. Drug Carrier Syst. 2009;26(6):523–580. doi: 10.1615/CritRevTherDrugCarrierSyst.v26.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali V. Ali M. Ali J. Nanocarrier for the enhanced bioavailability of a cardiovascular agent: in vitro, pharmacodynamic, pharmacokinetic and stability assessment. Int. J. Pharm. 2011;403(1–2):46–56. doi: 10.1016/j.ijpharm.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Lehtinen J. Raki M. Bergström K. A. Uutela P. Lehtinen K. Hiltunen A. Pikkarainen J. Liang H. Pitkänen S. Määttä A.-M. Pre-targeting and direct immunotargeting of liposomal drug carriers to ovarian carcinoma. PLoS One. 2012;7(7):e41410. doi: 10.1371/journal.pone.0041410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker A. Magarkar A. Viitala T. Rational design of liposomal drug delivery systems, a review: combined experimental and computational studies of lipid membranes, liposomes and their PEGylation. Biochim. Biophys. Acta, Biomembr. 2016;1858(10):2334–2352. doi: 10.1016/j.bbamem.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Carter K. A. Shao S. Hoopes M. I. Luo D. Ahsan B. Grigoryants V. M. Song W. Huang H. Zhang G. Pandey R. K. Porphyrin–phospholipid liposomes permeabilized by near-infrared light. Nat. Commun. 2014;5(1):1–11. doi: 10.1038/ncomms4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B. Frenkel E. Nanoparticles for drug delivery in cancer treatment. Urol. Oncol.: Semin. Orig. Invest. 2008;26(1):57–64. doi: 10.1016/j.urolonc.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Zwicke G. L. Ali Mansoori G. Jeffery C. J. J. Utilizing the folate receptor for active targeting of cancer nanotherapeutics. Nano Rev. 2012;3(1):1–11. doi: 10.3402/nano.v3i0.18496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ud Din F. Aman W. Ullah I. Qureshi O. S. Mustapha O. Shafique S. Zeb A. Effective use of nanocarriers as drug delivery systems for the treatment of selected tumors. Int. J. Nanomed. 2017;12:7291–7309. doi: 10.2147/IJN.S146315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhzad O. C. Langer R. Impact of nanotechnology on drug delivery. ACS Nano. 2009;3(1):16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- Fang X. Cao J. Shen A. Technology. Advances in anti-breast cancer drugs and the application of nano-drug delivery systems in breast cancer therapy. J. Drug Delivery Sci. Technol. 2020;57:101662. doi: 10.1016/j.jddst.2020.101662. [DOI] [Google Scholar]
- Mai W. X. Meng H. Mesoporous silica nanoparticles: a multifunctional nano therapeutic system. Integr. Biol. 2012;5(1):19–28. doi: 10.1039/c2ib20137b. [DOI] [PubMed] [Google Scholar]
- Zhang L. Gu F. Chan J. Wang A. Langer R. Farokhzad O. Nanoparticles in medicine: therapeutic applications and developments. Clin. Pharmacol. Ther. 2008;83(5):761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- Gabizon A. Catane R. Uziely B. Kaufman B. Safra T. Cohen R. Martin F. Huang A. Barenholz Y. Prolonged circulation time and enhanced accumulation in malignant exudates of doxorubicin encapsulated in polyethylene-glycol coated liposomes. Cancer Res. 1994;54(4):987–992. [PubMed] [Google Scholar]
- Wu P.-K. Tao Z. Ouyang Z. Cao J.-Y. Geng D. Liu J. Wang C.-M. The anti-tumor effects of cordycepin-loaded liposomes on the growth of hepatoma 22 tumors in mice and human hepatoma BEL-7402 cells in culture. Drug Dev. Ind. Pharm. 2016;42(9):1424–1433. doi: 10.3109/03639045.2016.1141930. [DOI] [PubMed] [Google Scholar]
- Bi Y. Zhou Y. Wang M. Li L. Lee R. J. Xie J. Teng L. Targeted delivery of cordycepin to liver cancer cells using transferrin-conjugated liposomes. Anticancer Res. 2017;37(9):5207–5214. doi: 10.21873/anticanres.11944. [DOI] [PubMed] [Google Scholar]
- Perillo B. Di Donato M. Pezone A. Di Zazzo E. Giovannelli P. Galasso G. Castoria G. Migliaccio A. ROS in cancer therapy: the bright side of the moon. Exp. Mol. Med. 2020:192–203. doi: 10.1038/s12276-020-0384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano J. M. de Paula E. Pickholz M. Molecular dynamics simulations to study drug delivery systems. Molecular Dynamics. 2018:74–90. [Google Scholar]
- Kavyani S. Dadvar M. Modarress H. Amjad-Iranagh S. Molecular perspective mechanism for drug loading on carbon nanotube–dendrimer: a coarse-grained molecular dynamics study. J. Phys. Chem. B. 2018;122(33):7956–7969. doi: 10.1021/acs.jpcb.8b04434. [DOI] [PubMed] [Google Scholar]
- Nisoh N. Jarerattanachat V. Karttunen M. Wong-Ekkabut J. Formation of aggregates, icosahedral structures and percolation clusters of fullerenes in lipids bilayers: the key role of lipid saturation. Biochim. Biophys. Acta, Biomembr. 2020;1862(9):183328. doi: 10.1016/j.bbamem.2020.183328. [DOI] [PubMed] [Google Scholar]
- Hashemzadeh H. Javadi H. Darvishi M. Study of Structural stability and formation mechanisms in DSPC and DPSM liposomes: a coarse-grained molecular dynamics simulation. Sci. Rep. 2020;10(1):1837. doi: 10.1038/s41598-020-58730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonnoy P. Karttunen M. Wong-ekkabut J. Alpha-tocopherol inhibits pore formation in oxidized bilayers. Phys. Chem. Chem. Phys. 2017;19(8):5699–5704. doi: 10.1039/C6CP08051K. [DOI] [PubMed] [Google Scholar]
- Khuntawee W. Karttunen M. Wong-ekkabut J. A molecular dynamics study of conformations of beta-cyclodextrin and its eight derivatives in four different solvents. Phys. Chem. Chem. Phys. 2017;19(35):24219–24229. doi: 10.1039/C7CP04009A. [DOI] [PubMed] [Google Scholar]
- Yong T. Chen S. Xie Y. Chen D. Su J. Shuai O. Jiao C. Zuo D. Cordycepin, a characteristic bioactive constituent in Cordyceps militaris, ameliorates hyperuricemia through URAT1 in hyperuricemic mice. Front. Microbiol. 2018;9:58. doi: 10.3389/fmicb.2018.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tania M. Shawon J. Saif K. Kiefer R. Khorram M. S. Halim M. A. Khan M. Cordycepin downregulates Cdk-2 to interfere with cell cycle and increases apoptosis by generating ROS in cervical cancer cells: in vitro and in silico study. Curr. Cancer Drug Targets. 2019;19(2):152–159. doi: 10.2174/1568009618666180905095356. [DOI] [PubMed] [Google Scholar]
- Kongsema M. Wongkhieo S. Khongkow M. Lam E. W.-F. Boonnoy P. Vongsangnak W. Wong-Ekkabut J. Molecular mechanism of Forkhead box M1 inhibition by thiostrepton in breast cancer cells. Oncol. Rep. 2019;42(3):953–962. doi: 10.3892/or.2019.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-Q. Wu D. Jiang K.-M. Zhang D. Zheng X. Wan C.-P. Zhu H.-Y. Xie X.-G. Jin Y. Lin J. Preparation, spectroscopy and molecular modelling studies of the inclusion complex of cordycepin with cyclodextrins. Carbohydr. Res. 2015;406:55–64. doi: 10.1016/j.carres.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Uusitalo J. J. Ingólfsson H. I. Akhshi P. Tieleman D. P. Marrink S. J. Computation. Martini coarse-grained force field: extension to DNA. J. Chem. Theory Comput. 2015;11(8):3932–3945. doi: 10.1021/acs.jctc.5b00286. [DOI] [PubMed] [Google Scholar]
- Jahn A. Vreeland W. N. Gaitan M. Locascio L. E. Controlled vesicle self-assembly in microfluidic channels with hydrodynamic focusing. J. Am. Chem. Soc. 2004;126(9):2674–2675. doi: 10.1021/ja0318030. [DOI] [PubMed] [Google Scholar]
- Shashidhar G. Manohar B. Nanocharacterization of liposomes for the encapsulation of water soluble compounds from Cordyceps sinensis CS1197 by a supercritical gas anti-solvent technique. RSC Adv. 2018;8:34634–34649. doi: 10.1039/C8RA07601D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malde A. K. Zuo L. Breeze M. Stroet M. Poger D. Nair P. C. Oostenbrink C. Mark A. E. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 2011;7(12):4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- Lins R. D. Hunenberger P. H. A new GROMOS force field for hexopyranose-based carbohydrates. J. Comput. Chem. 2005;26(13):1400–1412. doi: 10.1002/jcc.20275. [DOI] [PubMed] [Google Scholar]
- López C. A. de Vries A. H. Marrink S. J. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLoS Comput. Biol. 2011;7(3):e1002020. doi: 10.1371/journal.pcbi.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard R., The dictionary of substances and their effects, BMJ Publishing Group Ltd, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straatsma T. Berendsen H. J. T. Free energy of ionic hydration: analysis of a thermodynamic integration technique to evaluate free energy differences by molecular dynamics simulations. J. Chem. Phys. 1988;89(9):5876–5886. doi: 10.1063/1.455539. [DOI] [Google Scholar]
- Straatsma T. Berendsen H. Postma J. Free energy of hydrophobic hydration: a molecular dynamics study of noble gases in water. J. Chem. Phys. 1986;85(11):6720–6727. doi: 10.1063/1.451846. [DOI] [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F. and Hermans J., Interaction models for water in relation to protein hydration, in Intermolecular Forces, ed. B. Pullman, Springer Netherlands, 1981, vol. 14, pp. 331–342 [Google Scholar]
- Bussi G. Donadio D. Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Bussi G. Zykova-Timan T. Parrinello M. Isothermal-isobaric molecular dynamics using stochastic velocity rescaling. J. Chem. Phys. 2009;130(7):074101. doi: 10.1063/1.3073889. [DOI] [PubMed] [Google Scholar]
- Bennett C. H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976;22(2):245–268. doi: 10.1016/0021-9991(76)90078-4. [DOI] [Google Scholar]
- Abraham M. J. Murtola T. Schulz R. Páll S. Smith J. C. Hess B. Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Hendrich A. B. Michalak K. Lipids as a Target for Drugs Modulating Multidrug Resistance of Cancer Cells. Curr. Drug Targets. 2003;4(1):23–30. doi: 10.2174/1389450033347172. [DOI] [PubMed] [Google Scholar]
- Merchanf T. E. Diamantis P. M. Lauwers G. Haida T. Kasimos J. N. Guillem J. Glonek T. G. Minsky B. D. Characterization of malignant colon tumors with 31p nuclear magnetic resonance phospholipid and phosphatic metabolite profiles. Cancer. 1995;76(10):1715–1723. doi: 10.1002/1097-0142(19951115)76:10<1715::AID-CNCR2820761007>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Patra M. Karttunen M. Hyvönen M. T. Falck E. Vattulainen I. Lipid bilayers driven to a wrong lane in molecular dynamics simulations by subtle changes in long-range electrostatic interactions. J. Phys. Chem. B. 2004;108(14):4485–4494. doi: 10.1021/jp031281a. [DOI] [Google Scholar]
- Darden T. York D. Pedersen L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- Essmann U. Perera L. Berkowitz M. L. Darden T. Lee H. Pedersen L. G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- Karttunen M. Rottler J. Vattulainen I. Sagui C. Electrostatics in biomolecular simulations: where are we now and where are we heading? Curr. Top. Membr. 2008;60:49–89. [Google Scholar]
- Hess B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008;4(1):116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- Parrinello M. Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- Wong-Ekkabut J. Miettinen M. S. Dias C. Karttunen M. Static charges cannot drive a continuous flow of water molecules through a carbon nanotube. Nat. Nanotechnol. 2010;5(8):555–557. doi: 10.1038/nnano.2010.152. [DOI] [PubMed] [Google Scholar]
- Wong-Ekkabut J. Karttunen M. The good, the bad and the user in soft matter simulations. Biochim. Biophys. Acta, Biomembr. 2016;1858(10):2529–2538. doi: 10.1016/j.bbamem.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Boonnoy P. Karttunen M. Wong-ekkabut J. Does α-tocopherol flip-flop help to protect membranes against oxidation? J. Phys. Chem. B. 2018;122(45):10362–10370. doi: 10.1021/acs.jpcb.8b09064. [DOI] [PubMed] [Google Scholar]
- Mendes M. Cova T. Basso J. Ramos M. L. Vitorino R. Sousa J. Pais A. Vitorino C. Hierarchical design of hyaluronic acid-peptide constructs for glioblastoma targeting: combining insights from NMR and molecular dynamics simulations. J. Mol. Liq. 2020;315:113774. doi: 10.1016/j.molliq.2020.113774. [DOI] [Google Scholar]
- Nalakarn P. Boonnoy P. Nisoh N. Karttunen M. Wong-ekkabut J. Dependence of fullerene aggregation on lipid saturation due to a balance between entropy and enthalpy. Sci. Rep. 2019;9(1):1037. doi: 10.1038/s41598-018-37659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Ekkabut J. Karttunen M. Molecular dynamics simulation of water permeation through the alpha-hemolysin channel. J. Biol. Phys. 2016;42(1):133–146. doi: 10.1007/s10867-015-9396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W. Dalke A. Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Torrie G. M. Valleau J. P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977;23(2):187–199. doi: 10.1016/0021-9991(77)90121-8. [DOI] [Google Scholar]
- Kumar S. Rosenberg J. M. Bouzida D. Swendsen R. H. Kollman P. A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13(8):1011–1021. doi: 10.1002/jcc.540130812. [DOI] [Google Scholar]
- Hub J. S. de Groot B. L. van der Spoel D. g_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J. Chem. Theory Comput. 2010;6(12):3713–3720. doi: 10.1021/ct100494z. [DOI] [Google Scholar]
- Dana P. Bunthot S. Suktham K. Surassmo S. Yata T. Namdee K. Yingmema W. Yimsoo T. Ruktanonchai U. R. Sathornsumetee S. Active targeting liposome-PLGA composite for cisplatin delivery against cervical cancer. Colloids Surf., B. 2020;196:111270. doi: 10.1016/j.colsurfb.2020.111270. [DOI] [PubMed] [Google Scholar]
- Nunes A. S. Barros A. S. Costa E. C. Moreira A. F. Correia I. J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019;116(1):206–226. doi: 10.1002/bit.26845. [DOI] [PubMed] [Google Scholar]
- Piboonprai K. Khumkhrong P. Khongkow M. Yata T. Ruangrungsi N. Chansriniyom C. Iempridee T. J Anticancer activity of arborinine from Glycosmis parva leaf extract in human cervical cancer cells. Biochem. Biophys. Res. Commun. 2018;500(4):866–872. doi: 10.1016/j.bbrc.2018.04.175. [DOI] [PubMed] [Google Scholar]
- Zhang L. Bennett W. D. Zheng T. Ouyang P.-K. Ouyang X. Qiu X. Luo A. Karttunen M. Chen P. Effect of cholesterol on cellular uptake of cancer drugs pirarubicin and ellipticine. J. Phys. Chem. B. 2016;120(12):3148–3156. doi: 10.1021/acs.jpcb.5b12337. [DOI] [PubMed] [Google Scholar]
- Thakur R. Das A. Chakraborty A. The fate of anticancer drug, ellipticine in DPPC and DMPC liposomes upon interaction with HSA: a photophysical approach. J. Photochem. Photobiol., B. 2014;130:122–131. doi: 10.1016/j.jphotobiol.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Wong-ekkabut J., Vongsangnak W., Khuntawee W., Yata T. and Namdee K.. Microencapsulation of Cordyceps Extract and Production Proces, Thailand Patent 1703002400, October 1, 2019
- Chibowski E. Szcześ A. Zeta potential and surface charge of DPPC and DOPC liposomes in the presence of PLC enzyme. Adsorption. 2016;22(4–6):755–765. doi: 10.1007/s10450-016-9767-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.