Abstract
SARS-CoV-2 is the RNA virus responsible for COVID-19, the prognosis of which has been found to be slightly worse in men. The present study aimed to analyze the expression of different mRNAs and their regulatory molecules (miRNAs and lncRNAs) to consider the potential existence of sex-specific expression patterns and COVID-19 susceptibility using bioinformatics analysis. The binding sites of all human mature miRNA sequences on the SARS-CoV-2 genome nucleotide sequence were predicted by the miRanda tool. Sequencing data was excavated using the Galaxy web server from GSE157103, and the output of feature counts was analyzed using DEseq2 packages to obtain differentially expressed genes (DEGs). Gene set enrichment analysis (GSEA) and DEG annotation analyses were performed using the ToppGene and Metascape tools. Using the RNA Interactome Database, we predicted interactions between differentially expressed lncRNAs and differentially expressed mRNAs. Finally, their networks were constructed with top miRNAs. We identified 11 miRNAs with three to five binding sites on the SARS-COVID-2 genome reference. MiR-29c-3p, miR-21-3p, and miR-6838-5p occupied four binding sites, and miR-29a-3p had five binding sites on the SARS-CoV-2 genome. Moreover, miR-29a-3p, and miR-29c-3p were the top miRNAs targeting DEGs. The expression levels of miRNAs (125, 181b, 130a, 29a, b, c, 212, 181a, 133a) changed in males with COVID-19, in whom they regulated ACE2 expression and affected the immune response by affecting phagosomes, complement activation, and cell-matrix adhesion. Our results indicated that XIST lncRNA was up-regulated, and TTTY14, TTTY10, and ZFY-AS1 lncRN as were down-regulated in both ICU and non-ICU men with COVID-19. Dysregulation of noncoding-RNAs has critical effects on the pathophysiology of men with COVID-19, which is why they may be used as biomarkers and therapeutic agents. Overall, our results indicated that the miR-29 family target regulation patterns and might become promising biomarkers for severity and survival outcome in men with COVID-19.
Keywords: COVID-19, microRNA, lncRNA, miR-29 family
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; DEGs, differentially expressed genes; GSEA, gene set enrichment analysis; GO, gene ontology; DEmRNAs, differentially expressed mRNAs; GEO, Gene Expression Omnibus; ACE, angiotensin-converting enzyme; lncRNA, long non-coding RNA; miRNA, micro RNA
Graphical abstract
1. Introduction
COVID-19 is caused by a new beta-coronavirus known as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Bchetnia et al., 2020). It has led to the death of 5,289,826 people since the onset of its pandemic on December 8, 2019 (https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?).
Coronaviruses are enveloped positive-sense RNA viruses with genome sizes ranging from about 2 kb to 2.2 kb (Mani et al., 2020). One of the most prominent characteristics of this virus family is its genome size. Coronaviruses have the largest genome of all RNA viruses (Jafarinejad-Farsangi et al., 2020). Their genome carries a cap structure at the 5′ end and a poly (A) tail at the 3′ end, allowing genomic RNA to be used as mRNA and be translated into viral proteins by the host translation system after cell infection (Subissi et al., 2014).
Therefore, the use of host-encoded factors such as miRNAs and lncRNAs could be a useful strategy for antiviral therapy. miRNAs are single-stranded non-coding RNAs of 21–23 nucleotides that complement the mRNA in the part of the non-coding region of the 3′ end of a protein-encoding sequence and block translation (Klinge, 2018). The importance of miRNAs as an antiviral agent is that the first miRNA molecule that entered into the clinical trial was based on the application of miRNA-mediated viral silencing (Chakraborty et al., 2017).
In another study it was reported that in samples infected by influenza H5N1, miR-485 targeted the PB1 subunit of the viral RNA-dependent RNA polymerase gene, which is necessary for viral genome replication (Ingle et al., 2015).
On the other hand, the complex process of sexual differentiation can be influenced by biological factors such as non-coding-RNAs. According to previous studies, men are more susceptible to SARS-CoV-2 infection and develop more severe symptoms with worse outcomes. Genetic components may play a role in the incidence of coronavirus. A growing body of evidence indicates the role of biological agents, especially non-coding RNAs, in the differences between the sexes, but more research is needed to better explain the complexity of the interactions when contracting viral infections (Ortolan et al., 2020).
Previous studies have reported that males are more susceptible to severe COVID-19 and have a worse prognosis (Li et al., 2020a, Li et al., 2020b).
During the COVID-19 pandemic, it is crucial to study the ncRNAs involved in the interaction between SARS-CoV2 and the host. Here we predicted specific male miRNAs that target the genome of SARS-CoV-2 and miRNAs that target specific differentially expressed genes (DEGs) in males.
On the other hand, lncRNAs are non-coding transcripts of 200–100,000 nucleotides, most of which demonstrate cell-type-specific expression. In addition to gene imprinting and X chromosome inactivation, lncRNAs affect the regulation of many complex cellular and molecular processes during development. They also play a role in cancer progression (Heery et al., 2017). Many lncRNAs are involved and are expressed in spermatogenesis, but very few of them have been studied (Sahlu et al., 2020).
lncRNAs do not have an open reading frame or protein-coding sequences, but they regulate protein activities and their organization and act as precursors for small RNAs and modulate transcription patterns (Fernandes et al., 2019).
lncRNAs act as regulatory elements in various biological procedures using different mechanisms such as including splicing and transcription. Most of them have low expression levels and activity at specific times and in specific tissues; one such time is spermatogenesis (Zhang et al., 2019).
However, during spermatogenesis, a specific immunological process is activated in the testis to protect germ cells in autogenic and allogenic immunologic responses. Immune mechanisms in the testes are investigated based on the study of the receptor-mediated innate immune response pattern in different testicular cells (Rokade et al., 2017).
Angiotensin-converting enzyme (ACE) is effective in immunity. It is expressed in many tissues, but its expression level is high only in some tissues, such as tissues in the testes, kidneys, and lungs. ACE2 plays a key role in various kinds of disorders (Bernstein et al., 2018).
It was demonstrated that SARS-CoV-2 is present in the testis by detecting the high level of ACE2 expression in this organ. The lncRNAs and miRNAs in this organ affected ACE2 expression (Verma et al., 2020). Therefore, the present research aimed to identify lncRNAs and miRNAs in COVID-19-associated problems in different sexes using bioinformatics analysis.
To the best of our knowledge, this is the first study of non-coding RNA expression profiling based on gender that has aimed to find different gender-associated miRNA and lncRNA patterns. Identifying target genes and pathways regulated by miRNAs and lncRNA in different genders could provide important information about the biology of patients with COVID-19.
2. Materials and methods
2.1. Data collection
First, gene expression datasets of COVID-19 were searched using the keywords: ‘SARS-CoV-2 infection,’ ‘male,’ ‘female,’ and ‘Homo sapiens’ [porgn: txid9606] against the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). After a systematic review, dataset GSE157103, which had been generated using the Illumina NovaSeq 6000 (GPL24676), was used in this study. After that, using the miRBase database (version 22.1), we collected all human mature miRNA sequences; then, the human miRNA binding sites on the SARS-CoV-2 genome nucleotide sequence were predicted by the miRanda tool (version 3.3a). Finally, for the miRanda tool, the thermodynamic folding energy and alignment score thresholds of −20 kcal/mol and 150 were set; the specific alignment in the seed area was set to strict.
2.2. Data processing
Sequencing data (fastq files) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) were uploaded to and analyzed on the Galaxy web platform (usegalaxy.eu). Quality control was performed with FastQC Galaxy Version 0.72. Reads were mapped to the human reference genome (Gencode, release 32, hg38) with Hisat2 Version 2.1.0 with default parameters using the Gencode annotation file (Gencode, release 32, https://www.gencodegenes.org/human/releases.html). Reads mapped to the human reference genome were counted using featureCounts Galaxy Version 2.0.1 with default parameters using the annotation file mentioned above. R packages analyzed the output of featureCounts.
2.3. Identification of differentially expressed genes (DEG)
To identify differentially expressed mRNAs (DEmRNAs), the expression dataset was normalized and analyzed by the DeSeq2 package in Bioconductor. It was then utilized to mine statistically significant DEGs based on the difference in their expression values between samples of the COVID_female_ICU vs. COVID_male_ICU, and COVID_female_NonICU vs. COVID_male_NonICU (Brown et al., 1991). DEGs with |log2FC| ≥ 1 and adjust P-value ≤0.05 were considered to be significantly differentially expressed. Moreover, we also identified differentially expressed lncRNAs (DElncRNAs).
2.4. Gene set enrichment analysis (GSEA)
A useful approach for a biological understanding of gene expression data is gene set enrichment analysis (GSEA). We used the ToppGene database (https://toppgene.cchmc.org/) in this analysis to determine miRNAs that targeted DEGs based on the target scan database. The FDR cutoff of 0.05 was considered significant. Besides, pathways and gene ontology (GO) was defined by Metascape (https://metascape.org/gp/index.html#/main/step1), an online database that provides a robust gene list annotation for experimental biologists.
2.5. Interaction assessment between DElncRNAs, miRNA and DEmRNAs
To clearly understand the functions of DElncRNA and miRNA with DEmRNAs, we developed the network of DElncRNA-miRNA-DEmRNA interactions. RNAInter (RNA Interactome database) is a database that promotes the development of the Interactome and increases knowledge on the biological functions and molecular mechanisms of RNAs. We set categories on lncRNA, species on H. sapiens, interaction type on RNA-RNA interaction, detection method on computational prediction, and the interval of confidence score between 0.1 and 1. Using RNAInter, we identified the interactions between DElncRNAs and some DEmiRNAs that targeted DEmRNAs and had three or more binding sites on the nucleotide sequence of the SARS-CoV-2 genome. Also, the interactions between DEmRNAs, which were targeted by the mentioned DEmiRNAs and DElncRNAs, were implemented.
Finally, the lncRNA-miRNA-mRNA networks were constructed using the Cytoscape (2.7.2) software.
3. Results
3.1. Data collection and identification DEGs
In this study, GSE157103 (GPL24676) was used; the RNA-seq dataset contained 126 samples (each sample had two repetitions). Differential expression analysis of genes was firstly carried out to identify DEmRNAs and DElncRNAs using DESeq2. In COVID_female_ICU (n = 34) vs COVID_male_ICU (n = 66), and COVID_female_NonICU (n = 42) vs COVID_male_NonICU (n = 58), we identified 147 and 248 differentially expressed genes, respectively, among which 35 and 74 were up-regulated and 122 and 174 were down-regulated, respectively (Supplementary 1). We also determined DElncRNAs in this stage (Table 1).
Table 1.
Differentially expressed lncRNAs (DElncRNAs).
| COVID_female_ICU vs COVID_male_ICU |
COVID_female_NonICU vs COVID_male_NonICU |
||
|---|---|---|---|
| Up regulated lnCRNAs |
Up regulated lnCRNAs |
||
| Symbol | Full name | Symbol | Full name |
| XIST | X-inactive specific transcript | XIST | X-inactive specific transcript |
| TSIX | TSIX transcript, XIST antisense RNA | IFNG-AS1 | IFNG antisense RNA 1 |
| Down regulated lnCRNAs | Down regulated lnCRNAs | ||
| TTTY14 | Testis-specific transcript, Y-linked 14 | TTTY14 | Testis-specific transcript, Y-linked 14 |
| TTTY10 | Testis-specific transcript, Y-linked 10 | TTTY10 | Testis-specific transcript, Y-linked 10 |
| ZFY-AS1 | ZFY antisense RNA 1 | ZFY-AS1 | ZFY antisense RNA 1 |
| LINC00278 | Long intergenic non-protein coding RNA 278 | LINC00629 | Long intergenic non-protein coding RNA 629 |
| LINC01505 | Long intergenic non-protein coding RNA 1505 | AOAH-IT1 | AOAH intronic transcript 1 |
| LINC00298 | Long intergenic non-protein coding RNA 298 | LINC00278 | Long intergenic non-protein coding RNA 278 |
| EXTL3-AS1 | EXTL3 antisense RNA 1 | NFE4 | Nuclear factor, erythroid 4 |
| TPRG1-AS1 | TPRG1 antisense RNA 1 | ||
| CLRN1-AS1 | CLRN1 antisense RNA 1 | ||
| LINC00604 | Long intergenic non-protein coding RNA 604 | ||
| MAFA-AS1 | MAFA antisense RNA 1 | ||
| OVCH1-AS1 | OVCH1 antisense RNA 1 | ||
For identification of the binding sites of human miRNAs on the coronavirus 2 reference genome, 2565 mature miRNAs were collected, 444 miRNAs were detected among all the 2654 human mature miRNAs at various locations with direct binding sites throughout the SARS-CoV-2 reference genome (Supplementary 2). We searched for the perfect matching interactions at the seed region and sorted out 160 miRNAs. Fifteen miRNAs with more than three binding sites were detected among them (Table 2).
Table 2.
List miRNAs that have more than three seed binding site on SARS-COVID-2 genome reference.
| Three binding site |
| hsa-miR-770-5p, hsa-miR-103a-3p, hsa-miR-4772-3p, hsa-miR-624-5p, hsa-miR-3130-3p, hsa-miR-497-5p, hsa-miR-6715b-5p, hsa-miR-761, hsa-miR-320b, hsa-miR-107, hsa-miR-7845-5p |
| Four binding site |
| hsa-miR-21-3p, hsa-miR-6838-5p, hsa-miR-29c-3p |
| Five binding site |
| hsa-miR-29a-3p |
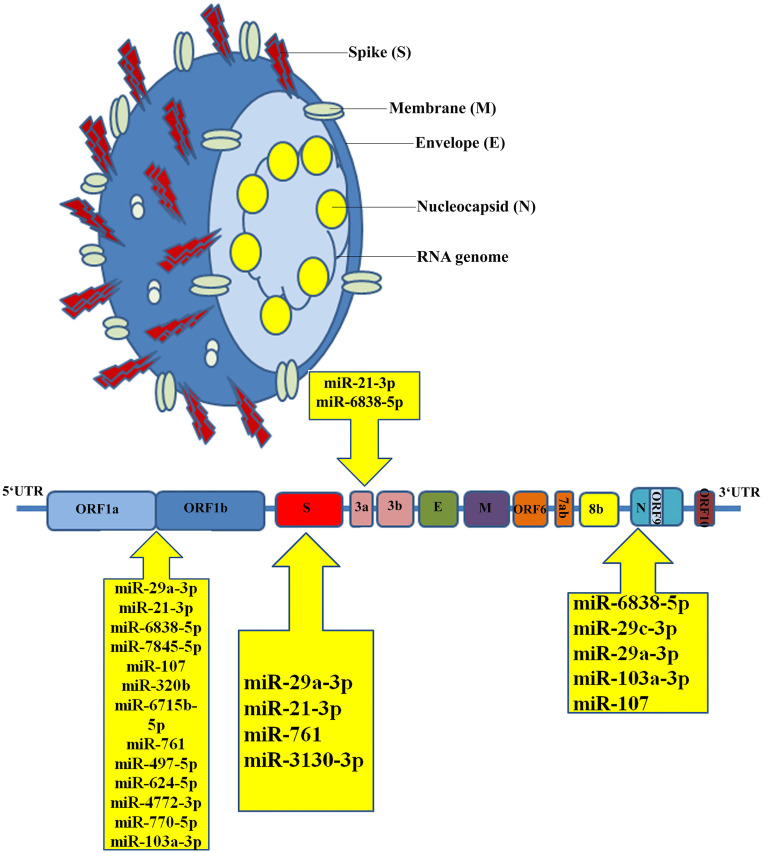
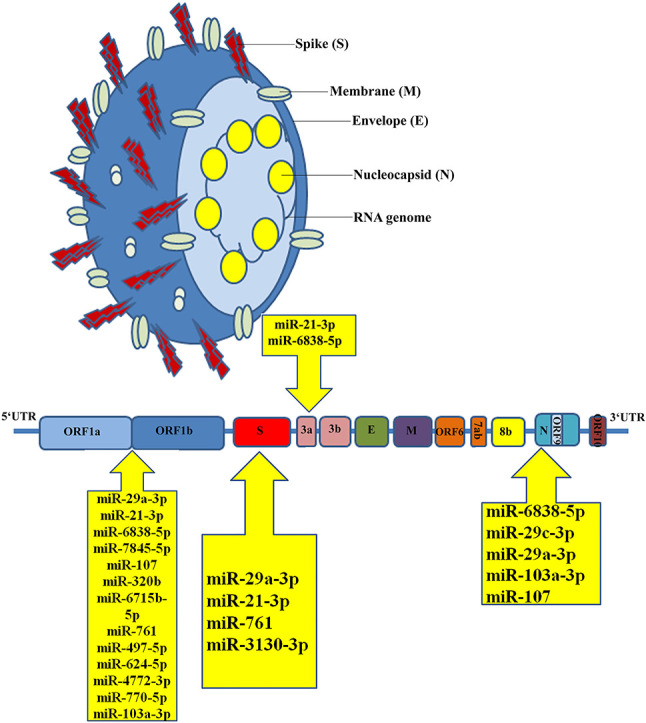
Our findings suggest that the miR-29 family (miR-29a, miR-29b, and miR-29c) had the maximum binding sites (11 sites). We also discovered miRNAs with more than three binding sites on the SARS-CoV-2 genome sequence (Fig. 1) and reported ORF1ab, spike, nucleocapsid, membrane, ORF3a, and ORF7a coding regions with strong binding capacity with human host miRNAs. The most binding sites belonged to ORF1ab, nucleocapsid, and spike sequences. Among the miRNAs, the various binding sites on ORF1ab, nucleocapsid, and spike sequences were presented by miR-29. ORF1ab, spike, and ORF3a include binding sites of miR-21. For viral entry, the spike region that codes for the spike protein is required and is a potential target for antiviral therapy.
Fig. 1.
Schematic representation of human miRNA-binding sites on the SARS-CoV-2 genome.
3.1.1. Gene set enrichment analysis (GSEA) and interaction analysis
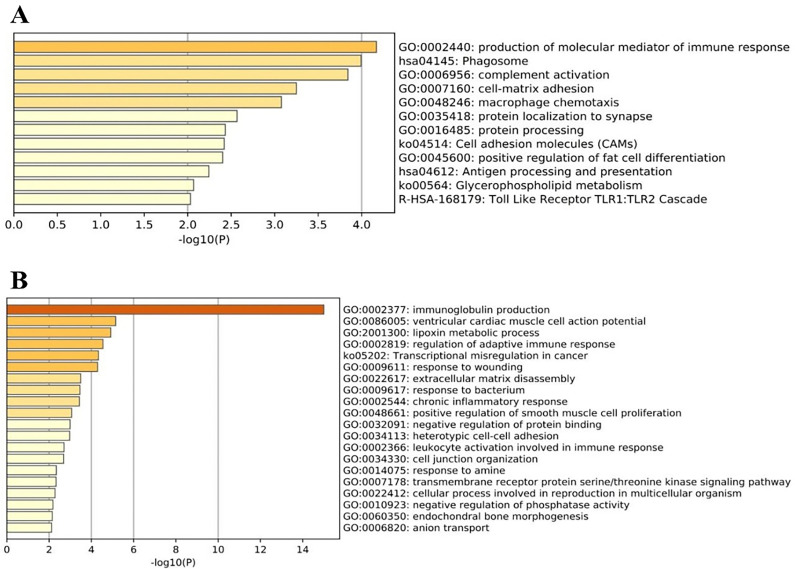
Enrichment analysis of differentially expressed genes has been shown in Fig. 2 . Pathways and functional gene ontology analysis of DEGs was conducted. Production of molecular mediator of immune response, Phagosome, complement activation, cell-matrix adhesion and macrophage chemotaxis were most prevalent in DEGs related to COVID_female_ICU vs COVID_male_ICU. In addition, immunoglobulin production, ventricular cardiac muscle cell action potential, lipoxin metabolic process, regulation of adaptive immune response and Transcriptional misregulation in cancer were more significant in DEGs related to COVID_female_NonICU vs COVID_male_NonICU (Fig. 2).
Fig. 2.
GSEA of all DEGs (up and down regulated) in response to SARS-COV-2 infection for COVID_female_ICU vs COVID_male_ICU (A), and COVID_female_NonICU vs COVID_male_NonICU (B).
Moreover, we recognized miRNAs that target DEGs by ToppGene, among all miRNAs, hsa-miR-29a-3p and hsa-miR-29c-3p in addition to having five and four binding site respectively in SARS-CoV-2 genome, regulated some DEGs (Table 3 ).
Table 3.
Differentially expressed mRNAs that were targeted by hsa-miR-29a-3p & hsa-miR-29c-3p.
| COVID_female_ICU vs COVID_male_ICU |
COVID_female_NonICU vs COVID_male_NonICU |
||
|---|---|---|---|
| hsa-miR-29a-3p & hsa-miR-29c-3p |
hsa-miR-29a-3p & hsa-miR-29c-3p |
||
| Symbol | Full name | Symbol | Full name |
| HRK | Harakiri, BCL2 interacting protein | ASIC1 | Acid sensing ion channel subunit 1 |
| PER1 | Period circadian regulator 1 | FERMT2 | Fermitin family member 2 |
| FIGN | Fidgetin, microtubule severing factor | SDK1 | Sidekick cell adhesion molecule 1 |
| ENHO | Energy homeostasis associated | HBEGF | Heparin binding EGF like growth factor |
| MAPK10 | Mitogen-activated protein kinase 10 | COL27A1 | Collagen type XXVII alpha 1 chain |
| ID1 | Inhibitor of DNA binding 1, HLH protein | ||
| C1orf226 | Chromosome 1 open reading frame 226 | ||
| LPL | Lipoprotein lipase | ||
| DDX3Y | DEAD-box helicase 3 Y-linked | ||
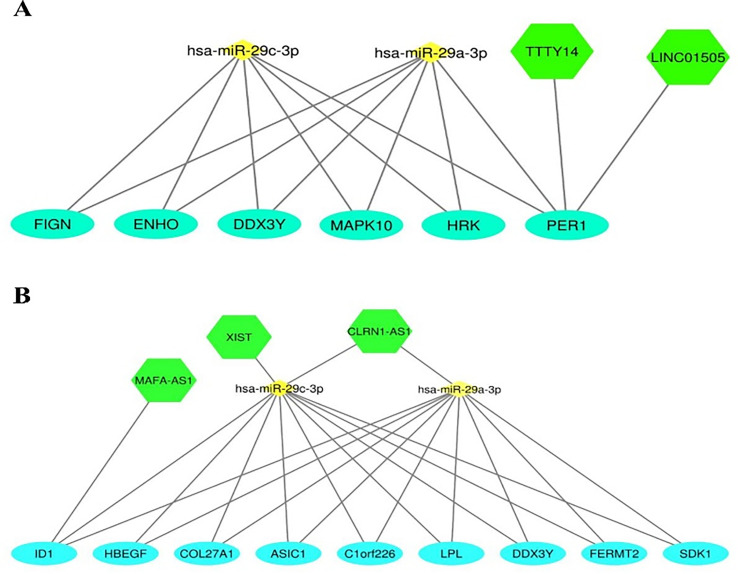
At last the DElncRNAs that had interaction with hsa-miR-29a-3p, hsa-miR-29c-3p and their targets was identified (supplementary 3). lncRNAs-miRNAs-mRNAs networks have been shown in Fig. 3.
Fig. 3.
Networks were constructed between lncRNA-miRNA-mRNA for COVID_female_ICU vs COVID_male_ICU (A), and COVID_female_NonICU vs COVID_male_NonICU (B). Green hexagonal represents DElnRNAs, small yellow diamonds represent top miRNAs and light blue circles represent DEmRNAs respectively. We found TTY14 and LINC01505 as DElncRNAs which targeted PER1 (A). other DElncRNAs such as CLRN1-AS1 had interaction with both top miRNAs, but XIST targeted one miRNA (has-miR-29c-3p), in addition MAFA-AS1 targeted ID1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
COVID-19 is an infectious disease, which results in slightly more severe outcomes in men. This study aimed to analyze the correlation between characteristics of ICU and non-ICU COVID-19 patients and non-coding RNAs based on bioinformatics analysis to predict the probable symptoms of males with COVID-19. Besides, due to the high incidence of COVID-19 and the low effectiveness of conventional therapies, the development of new therapies seems necessary (Verma et al., 2020). Because of the yet unknown mechanism of virus action, different treatments may be necessary. However, miRNAs affect gene regulation, and there is much hope in using them for therapeutic purposes. Identification of miRNA target proteins or metabolic pathways is among the main purposes of using them as targeted inhibitors (Ritchie, 2017). Bioinformatics predictions play a key role in various studies using computational predictions of miRNAs. Since the onset of the COVID-19 pandemic, several studies have been conducted on human miRNA and virus genome interaction based on miRTarget computing tools and artificial intelligence (Ivashchenko et al., 2020, Demirci and Adan, 2020).
One main method in regulating gene expression is achieved through miRNAs that prevent the expression of one or more protein-coding genes. It is estimated that these molecules control the expression of one-third of all coding genes. Several studies have identified the role of these molecules in various diseases and pathological processes, such as cell death, differentiation, migration, cycle, autophagy, and apoptosis (MacFarlane, 2010).
On the other hand, the process of sexual differentiation leads to the difference between males and females. Finding the target genes and pathways regulated by miRNAs in men and women can provide new knowledge on the biology and pathogenesis of coronavirus based on gender (Ortolan et al., 2019). This study focused on interactions between the SAR-COV-2 virus genome and host miRNAs that bind to specific genes in males but not in females. Using miRTtarBase, we illustrated the miRNAs bound to specific sequences at the 3′ UTR and evaluated the interaction of miRNAs with all genes of the SARS-CoV-2 genome.
It has been proposed that miRNAs can act as prognostic and therapeutic agents. Previous studies used various platforms to detect the interaction of miRNAs with COVID-19. Arison et al. reported that seven miRNAs have binding sites on SARS-COV-2. In another study, the miRTarget prediction database showed that miR-5197-3p binds effectively to the SARS-COV-2 genome. A total of thirteen host miRNAs can influence the MERS-CoV genome (Arisan et al., 2020).
Our results showed miR-29's binding sites within the ORF1ab, spike, and nucleocapsid coding regions of SARS-CoV-2. Spike proteins are necessary for host-receptor binding before the virus enters the cell. The members of the miR-29 family have common seed sequences that are expected to target overlapping sets of genes. MiR-29 s also bind to a specific set of genes in men, exhibiting various binding sites along the coding regions of the male genome sequence and targeting a large group of functionally related genes. MiR-29 attaches to 16 genes of the extracellular matrix. It has been found that miR-29 s have anti-fibrotic effects in the kidney, heart, and some other organs. Additionally, the miR-29 family has been demonstrated to be proapoptotic and implicated in cell differentiation. Therefore, miR-29 s have functional effects on some diseases (Kriegel et al., 2012). It was found that the miR-29 s have binding sites on the host A20/TNFAIP3 transcript in response to influenza A. They reported the subsequent modulation of antiviral and pro-inflammatory responses to influenza (Zhang et al., 2014). Donyavi et al., 2021 reported that miR-29a-3p, and -146a-3p as novel biomarkers in COVID-19 severity. They discussed about the difference in miRNA expression pattern between COVID-19 patients and healthy controls, as well as between acute and post-acute COVID-19 (Donyavi et al., 2021). Nevertheless, our results showed that miR-29a-3p and miR-29c-3p were the top miRNAs targeting DEGs. The effect of the host miR-29 family on the regulation of viral activities is related to their binding to the host transcripts or the virus genome.
Based on the obtained results, the miR-29 s bind to the coding regions of SARS-CoV-2. These related proteins of SARS-CoV-2 have been evaluated as targets for SARS-CoV-2 antiviral drug development.
Moreover, 15 differentially expressed miRNAs (DEmiRNAs) were found with three, four, and five seed binding sites on the SARS-COVID-2 genome reference. Tang et al., 2019 reported that three of our predicted microRNAs (miR-4772-3p, miR-624-5p, and miR-21) could be used as signatures or prognostic indicators for cancer. It has been predicted that combinations of different miRNAs might be more sensitive and useful than single miRNAs. Thus these miRNAs should be considered for application by further studies in the future. In another study, it was shown that miR-770 affects cell proliferation and induces apoptosis using the Wnt/β-catenin signaling pathway (Zhang et al., 2018). Besides, miR-103a is also related to colorectal carcinoma through the Wnt signaling pathway (Fasihi et al., 2018). MiR-4772-3p was found as a predictor of cancer recurrence (Liu et al., 2016). Previous findings showed that the blood level of miR-320b could promote ischemic heart disease, which is also one of the major problems of men with COVID-19 (Wakabayashi et al., 2020). It was demonstrated that miR-6838-5p promotes cell growth and migration through the Wnt/β-Catenin signaling pathway (Zhou et al., 2020). Our findings showed that miR-6838-5p has four seed binding sites on the SARS-COVID-2 genome reference. Also, it has been illustrated that the expression of miR-21-3p had a high chance of binding to the SARS-COVID-2 genome and being up-regulated in mouse lungs with SARS-CoV infection. In this study, it was demonstrated that miR-21-3p had the potential to bind to the SARS-COVID-2 genome reference. Furthermore, it was shown that different genomic positions (various mRNAs) could act as miR-21-3p targets. MiR-21-3p and miR-29a-3p have four and three seed binding sites on the SARS-COVID-2 genome reference, respectively.
MiR-761 inhibited metastasis and suppressed the nasal mucosa remodeling via LCN2 and the LCN2/Twist1 signaling pathway inhibition (Cheng et al., 2020). Our findings showed that miR-761 had three seed binding sites on the SARS-COVID-2 genome, and it may cause the shortness of breath and severe problems that occur after COVID-19 infection in the lung bronchi.
MiR-6715b was reported as a biomarker to predict cancer stages (Xu et al., 2020). Our results showed that miR-6715b participates in the progression of COVID-19 in males.
It was reported the conserved sequence for the evolutionary miR-15/107 family, including miRNA-6838-5p (Wang et al., 2019). In the present study, we observed that miR-6838-5p was significantly correlated in men with COVID-19. This miRNA can be a signature and has the potential to be used as a factor in the treatment of COVID-19 patients.
Besides, Angiotensin-converting enzyme 2 (ACE2), which helps SARS-CoV-2 inter into the host cells, is expressed in the testis. Additionally, miRNAs in this organ affect ACE2 expression (Olaniyan et al., 2020).
The results of the present study indicated that the antiviral therapies and diagnostics can be developed differentially on the basis of gender, but it is obvious that, many variables, such as hormone-specific reactions and the activation of X-linked genes may influence the innate and adaptive immune response to viral infection and contribute to the discrepancy in COVID-19 outcomes.
Our results indicated changes in the expression of miRNAs (125, 181b, 130a, 29a, b, c, 212, 181a, 133a) in males with COVID-19. Previous studies have shown that they play a fundamental role in the regulation of ACE2 expression (Olaniyan et al., 2020). Understanding the role of these elements in angiogenesis can help us design therapeutic agents in antiviral therapy for SARS-CoV-2 infection based on gender. Additionally, MiR-29a is up-regulated by oxidative stress (Lin et al., 2020), suggesting that miR-29a mediates the regulation of ACE2 and may play a fundamental role in COVID-19 infection.
In another study, Li et al., 2020 reported that the expression pattern of microRNAs were different in the peripheral blood from human patients with COVID-19 and during viral infection, it may control immunological responses and viral replication.
Besides, the testis has a crucial role in the immunological response, and there is a high level of ACE2 expression in the testis (Olaniyan et al., 2020). Therefore, coronaviruses may localize in the testis, and thus, men with COVID-19 show worse symptoms than women.
The immune system affects testis development and acts during spermatogenesis. The immune system is often observed, with macrophages and T-cells, which secrete some pro-inflammatory cytokines (Olaniyan et al., 2020). In fact, non-coding RNAs are crucial elements regulating these activities. Thus non-coding RNAs may play an important role in the appearance of severe symptoms in males with COVID-19.
Accordingly, in several men with SARS-CoV-2 infection, viral RNA was found not only in the lungs but also in the testes and kidneys (Fan et al., 2020). Given ACE2 expression in the testis, it is predictable that the SARS-CoV-2 can directly affect males.
The current study demonstrated that miRNAs and lncRNAs have a regulating effect on immune responses. Therefore, ncRNA-regulated angiogenesis can be involved in the pathogenesis of COVID-19 in men. Our findings provided evidence for the implication of lncRNAs and miRNAs in key processes related to cellular pathways associated with immune response and severe symptoms in men with COVID-19.
lncRNA XIST has the potential of interacting with some proteins and miRNAs such as miR-29a (Brown et al., 1991). On the other hand, miR-29a has five seed binding sites on the SARS-COVID-2 genome reference. Thus some serious problems in men with COVID-19 may be related to the interaction between lncRNA XIST and miR-29a.
Our findings showed a complex mapping of disease-related factors and ncRNAs significantly affecting the pathways (Table 2). We also identified miRNAs involved in immune response; Phagosome, complement activation, cell-matrix adhesion, and macrophage chemotaxis were most prevalent in DEGs related to COVID_female_ICU vs. COVID_male_ICU. Moreover, immunoglobulin production, ventricular cardiac muscle cell action potential, lipoxin metabolic process, adaptive immune response regulation, and transcriptional misregulation were more significant in DEGs based on gender. However, upon viral infection, it is possible to change the expression of certain miRNAs to regulate the corresponding pathway. Therefore, many pathological pathways may be developed by protein-processing problems or aberrant post-translational modifications in response to the SARS-CoV-2 infection. Regulation of gene expression can affect post-translational modification; thus, permanent and temporary arrests can happen. Virus-encoded miRNAs can control host immune response to viral infection and enhance virus replication, while cellular miRNAs can affect some specific pathways and gene expression in males, especially in leucocyte activation involved in immune response (Fig. 2).
It was revealed that particular genetic variants of chromosome Y affect susceptibility to type A influenza virus infection in the mouse model and accelerate immune response in the lungs (Krementsov et al., 2017).
Our results demonstrated various lncRNA expressions in men with COVID-19. The results indicated that XIST lncRNA was up-regulated, and TTTY14, TTTY10, and ZFY-AS1 lncRNAs were down-regulated in both ICU and non-ICU males with COVID-19. lncRNA XIST is a necessary regulator for X inactivation in mammals. XIST plays a role in cancer progression, clearly, because of the gene dysregulation caused by epigenetic instability (Weakley et al., 2011).
Clinical research has shown that men with COVID-19 have a worse prognosis than women and differ in the frequency of their histopathological changes. Our investigations revealed the effects of non-coding RNA profiling in men with COVID-19. We found that non-coding RNAs could reliably predict the prognosis of men with COVID-19. Nevertheless, further experiments are necessary to investigate the biological functions of these factors in males and females.
In conclusion, COVID-19 infection is a complex disorder, and more experiments, such as ncRNA studies, are necessary to find the molecular mechanisms of SARS-CoV-2 infection based on gender. Using ncRNAs in the management and control of diseases is one of the new strategies in gene therapy because they can play an important role by influencing cellular processes and controlling gene expression regulation. ncRNAs can develop promising strategies both as biomarkers and therapeutic agents in COVID-19 based on gender.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.105195.
Appendix A. Supplementary data
Supplementary 1
Supplementary 2
Supplementary 3
Supplementary 4
References
- Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S., Uysal-Onganer P. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. doi: 10.3390/v12060614. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bchetnia M., Girard C., Duchaine C., Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review of the current global status. J. Infect. Public Health. 2020;13(11):1601–1610. doi: 10.1016/j.jiph.2020.07.011. Aug 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K.E., Khan Z., Giani J.F., Cao D.Y., Bernstein E.A., Shen X.Z. Angiotensin-converting enzyme in innate and adaptive immunity. Nat. Rev. Nephrol. 2018;14(5):325. doi: 10.1038/nrneph.2018.15. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. Jan. [DOI] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Sharma G., Doss C.G., Lee S.S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;15(8):132–143. doi: 10.1016/j.omtn.2017.06.005. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Chen J., Zhao Y., Yang J., Xue K., Wang Z. MicroRNA-761 suppresses remodeling of nasal mucosa and epithelial–mesenchymal transition in mice with chronic rhinosinusitis through LCN2. Stem Cell Res Ther. 2020;11 doi: 10.1186/s13287-020-01598-7. Dec. 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci M.D., Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. doi: 10.7717/peerj.9369. Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donyavi T., Bokharaei-Salim F., Baghi H.B., Khanaliha K., Janat-Makan M.A., Karimi B., Nahand J.S., Mirzaei H., Khatami A., Garshasbi S., Khoshmirsafa M. Acute and post-acute phase of COVID-19: analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021;97:107641. doi: 10.1016/j.intimp.2021.107641. Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Li K., Ding Y., Lu W.L., Wang J. ACE2 expression in kidney and testis may cause kidney and testis damage after 2019-nCoV infection. MedRxiv. 2020 doi: 10.3389/fmed.2020.563893. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasihi A., Soltani B.M., Atashi A., Nasiri S. Introduction of hsa?miR?103a and hsa?miR?1827 and hsa?miR?137 as new regulators of Wnt signaling pathway and their relation to colorectal carcinoma. J. Cell. Biochem. 2018;119(7):5104–5117. doi: 10.1002/jcb.26357. Jul. [DOI] [PubMed] [Google Scholar]
- Fernandes J.C., Acuña S.M., Aoki J.I., Floeter-Winter L.M., Muxel S.M. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Non-Coding RNA. 2019;5(1):17. doi: 10.3390/ncrna5010017. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery R., Finn S.P., Cuffe S., Gray S.G. Long non-coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells. Cancers. 2017;9(4):38. doi: 10.3390/cancers9040038. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle H., Kumar S., Raut A.A., Mishra A., Kulkarni D.D., Kameyama T., Takaoka A., Akira S., Kumar H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015;8(406):ra126. doi: 10.1126/scisignal.aab3183. Dec 8. [DOI] [PubMed] [Google Scholar]
- "Ivashchenko A, Rakhmetullina A, Aisina D. How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV, and MERS-CoV (2020).".
- Jafarinejad-Farsangi S., Jazi M.M., Rostamzadeh F., Hadizadeh M. High affinity of host human microRNAs to SARS-CoV-2 genome: an in silico analysis. Non-Coding RNA Res. 2020;5(4):222–231. doi: 10.1016/j.ncrna.2020.11.005. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge C.M. Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr. Relat. Cancer. 2018;25(4):R259–R282. doi: 10.1530/ERC-17-0548. Apr 1. [DOI] [PubMed] [Google Scholar]
- Krementsov D.N., Case L.K., Dienz O., Raza A., Fang Q., Ather J.L., Poynter M.E., Boyson J.E., Bunn J.Y., Teuscher C. Genetic variation in chromosome Y regulates susceptibility to influenza A virus infection. Proc. Natl. Acad. Sci. 2017;114(13):3491–3496. doi: 10.1073/pnas.1620889114. Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel A.J., Liu Y., Fang Y., Ding X., Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genomics. 2012;44(4):237–244. doi: 10.1152/physiolgenomics.00141.2011. Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Yang J., Zhao F., Zhi L., Wang X., Liu L., Bi Z., Zhao Y. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020;109(5):531–538. doi: 10.1007/s00392-020-01626-9. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Hu X., Li L., Li J.H. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020;34(10):e23590. doi: 10.1002/jcla.23590. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.Y., Yang Y.L., Wang P.W., Wang F.S., Huang Y.H. The emerging role of microRNAs in NAFLD: highlight of microRNA-29a in modulating oxidative stress, inflammation, and beyond. Cells. 2020;9(4):1041. doi: 10.3390/cells9041041. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Eng C., Shen J., Lu Y., Takata Y., Mehdizadeh A., Chang G.J., Rodriguez-Bigas M.A., Li Y., Chang P., Mao Y. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016;7(46):76250. doi: 10.18632/oncotarget.12841. Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane L.A., Murphy P.R. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. Nov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020:197989. doi: 10.1016/j.virusres.2020.197989. Apr 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaniyan O.T., Dare A., Okotie G.E., Adetunji C.O., Ibitoye B.O., Bamidele O.J., Eweoya O.O. Testis and blood-testis barrier in Covid-19 infestation: role of angiotensin-converting enzyme 2 in male infertility. J. Basic Clin. Physiol. Pharmacol. 2020;31(6) doi: 10.1515/jbcpp-2020-0156. Oct 5. [DOI] [PubMed] [Google Scholar]
- Ortolan A., Lorenzin M., Felicetti M., Doria A., Ramonda R. Does gender influence clinical expression and disease outcomes in COVID-19? A systematic review and meta-analysis. Int. J. Infect. Dis. 2020;1(99):496–504. doi: 10.1016/j.ijid.2020.07.076. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W. microRNA target prediction. InCancer Gene Netw. 2017:193–200. doi: 10.1007/978-1-4939-6539-7_13. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Rokade S., Kishore U., Madan T. Surfactant protein D regulates murine testicular immune milieu and sperm functions. Am. J. Reprod. Immunol. 2017;77(3):e12629. doi: 10.1111/aji.12629. Mar. [DOI] [PubMed] [Google Scholar]
- Sahlu B.W., Zhao S., Wang X., Umer S., Zou H., Huang J., Zhu H. Long noncoding RNAs: new insights in modulating mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2020;11(1):1–2. doi: 10.1186/s40104-019-0424-8. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antivir. Res. 2014;1(101):122–130. doi: 10.1016/j.antiviral.2013.11.006. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Saksena S., Sadri-Ardekani H. ACE2 receptor expression in testes: implications in coronavirus disease 2019 pathogenesis. Biol. Reprod. 2020;103(3):449–451. doi: 10.1093/biolre/ioaa080. Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi I., Eguchi R., Sotoda Y., von Lewinski D., Sourij H., Daimon T., Groschner K., Rainer P.P. Blood levels of microRNAs associated with ischemic heart disease differ between Austrians and Japanese: a pilot study. Sci. Rep. 2020;10(1):1–2. doi: 10.1038/s41598-020-69332-0. Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhu W., Xu J., Guo Y., Yan J., Meng L., Jiang C., Lu S. Interpreting the MicroRNA-15/107 family: interaction identification by combining network based and experiment supported approach. BMC Med. Genet. 2019;20(1) doi: 10.1186/s12881-019-0824-9. Dec. 1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weakley S.M., Wang H., Yao Q., Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J. Surg. 2011;35(8):1751–1756. doi: 10.1007/s00268-010-0951-0. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Xie J., Liu Y., Tang F., Long Z., Wang Y., Luo J., Li J., Li G. MicroRNA expression profiling and target gene analysis in gastric cancer. Medicine. 2020;99(37) doi: 10.1097/MD.0000000000021963. Sep 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dong C., Sun X., Li Z., Zhang M., Guan Z., Duan M. Induction of the cellular miR-29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochem. Biophys. Res. Commun. 2014;450(1):755–761. doi: 10.1016/j.bbrc.2014.06.059. Jul 18. [DOI] [PubMed] [Google Scholar]
- Zhang J.F., Zhang J.S., Zhao Z.H., Yang P.B., Ji S.F., Li N., Shi Q.D., Tan J., Xu X., Xu C.B., Zhao L.Y. MicroRNA-770 affects proliferation and cell cycle transition by directly targeting CDK8 in glioma. Cancer Cell Int. 2018;18(1):1–3. doi: 10.1186/s12935-018-0694-9. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z., Shen F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019;20(22):5573. doi: 10.3390/ijms20225573. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ding X., Jin P., Li P. miR-6838-5p affects cell growth, migration, and invasion by targeting GPRIN3 via the Wnt/?-catenin signaling pathway in gastric cancer. Pathobiology. 2020;87(6):327–337. doi: 10.1159/000511691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary 1
Supplementary 2
Supplementary 3
Supplementary 4