Abstract
Selective factor Xa inhibitors effectively block coagulation cascade with a broader therapeutic window than multitargeted anticoagulants. They have evolved as a crucial part of prevention and treatment of thromboembolic diseases and in therapeutic protocols involved in many clinical trials in coronavirus disease 2019 (COVID-19) patients. Biologically-guided isolation of specific FXa inhibitors from licorice (Glycyrrhiza glabra) root extract furnished ten flavonoids. By detailed analysis of their 1H, 13C NMR and MS data, the structures of these flavonoids were established as 7,4′-dihydroxyflavone (1), formononetin (2), 3-R-glabridin (3), isoliquiritigenin (4), liquiritin (5), naringenin 5-O-glucoside (6), 3,3′,4,4′-tetrahydroxy-2-methoxychalcone (7), liquiritinapioside (8) and the two isomers isoliquiritigenin-4′-O-β-d-apiosylglucoside (9) and isoliquiritigenin-4-O-β-d-apiosylglucoside (10). All the isolated compounds were assessed for their FXa inhibitory activity using in vitro chromogenic assay for the first time. Liquirtin (5) showed the most potent inhibitory effects with an IC50 of 5.15 μM. The QikProp module was implemented to perform ADMET predictions for the screened compounds.
Biologically guided isolation of new factor Xa inhibitors from Glycyrrhiza glabra roots.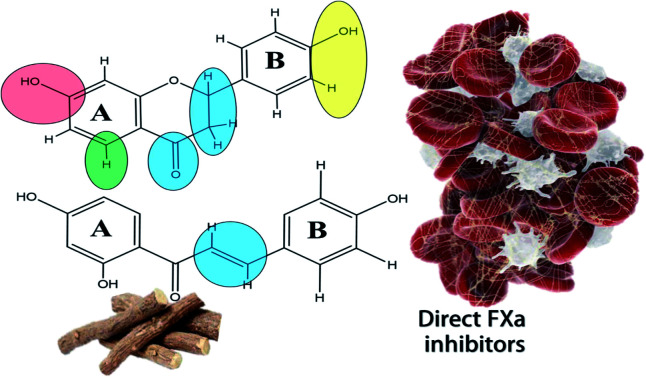
1. Introduction
Factor Xa (FXa) is a trypsin-like serine protease that acts on the basic components of the coagulation cascade through the conversion of prothrombin to thrombin catalyzing the reaction of fibrin formation and thereafter clot formation. With its position at the start of the common pathway of the extrinsic and intrinsic coagulation systems, selective factor Xa inhibitors would effectively block coagulation.1 Interestingly, it was found that inhibitors of factor Xa have a broad therapeutic window between adverse haemorrhage and thrombosis.2 The discovery of FXa inhibitors is in constant demand, this is reflected by the number of industrial and academic patented compounds targeting this specific coagulation factor.3 Most of the discovered compounds are based upon existing drugs with some structural modifications to increase efficacy and safety profile.4
With the current pandemic COVID-19 spread, research efforts have been directed to understand the disease nature, pathophysiology, and potential targets for developing therapeutic agents, vaccines and preventive measures for this ongoing threat.5,6 SARS-CoV-2 disease is manifested by many diverse symptoms involving multiple systems such as the respiratory, gastrointestinal, cardiovascular and immune systems.7 Coagulation abnormalities and thromboembolic events associated with the disease are very distinct.8 It is characterized by an increase in D-dimer level, prolonged prothrombin time PT and moderate decrease in platelet count (thrombocytopenia). These abnormalities mimic other coagulopathies as disseminated intravascular coagulation (DIC) and localised pulmonary thrombotic microangiopathy which led eventually to increased mortality rate.8,9 High level of inflammatory markers were reported to have effect in thrombosis observed in COVID-19. Tumour necrosis factor-α (TNF-α) and interleukins IL, (IL-1 and IL-6) are the main inflammatory mediators evoking coagulation and thrombin generation.8,9
Most of the guidelines and consensus documents published on behalf of professional societies focused on thrombosis and haemostasis advocate the use of anticoagulants in all patients hospitalized with COVID-19, as well as 2–6 weeks post hospital discharge in the absence of contraindications.10 Low molecular weight heparin (LMWH) or unfractionated heparin (UFH) infusions are the commonly used anticoagulants to prevent thrombotic events in COVID-19 patients.8,11,12 Direct FXa inhibitors have evolved as important part of therapeutic protocols involved in many clinical trials in (COVID-19) patients.13
Computational approaches and virtual screening techniques are progressing towards finding candidate compounds for treatment of COVID-19 disease.14,15 Recent studies showed the efficacy of natural bioflavonoid and steroidal hormones as inhibitors of SARS-CoV-2 replication using high-throughput virtual screening.16 Similarly, marine natural products of phlorotannins and flavonoid classes, pseudo peptides17 and the three approved drugs; miconazole, bedaquiline and glibenclamide were identified as promising inhibitors of the viral protease employing combined ligand-based and structure-based studies.18 The work of Zhang et al.19 that led to resolving the crystal structure of SARS-COV-2 main protease contributed to enormous increase in molecular docking studies to find therapeutic agents for the virus.20–22 Some of these studies made in silico prediction of pharmacokinetic profile of promising candidates.23 Moreover, in vitro testing of some of the obtained candidates proved their efficacy against cytokine storm in COVID-19.24
Massive research is progressing towards finding treatments for COVID-19 using drugs derived from natural sources.25–27 Recent studies suggested the beneficial use of plant extracts for COVID-19 prophylaxis and treatment. Among these plants, radix Glycyrrhizae (licorice root) pointed out as an interesting candidate.
Radix Glycyrrhizae consists of the dried roots and rhizomes of Glycyrrhiza glabra L. and its varieties family Fabaceae.28 Licorice has a long history of use as a food flavoring in the world and is one of the common beaverges known in Egypt. The consumption of erqsoos (of which licorice is a main constituent) is a common tradition during Ramadan and is regarded as a famous traditional non-alcoholic beverage in Egypt.29,30 It is a well-known frequently used herb for a long history with safety and efficacy and has many biological activities such as anti-obesity and anti-inflammatory. Licorice extract was considered for mitigation of (COVID-19) concluded from in silico studies26,31,32 and from known activity of its major compound; glycyrrhizin through its anti-inflammatory mechanism.33
Up to 60 107 of confirmed cases were treated by Traditional Chinese Medicine in China. Ang et al. analysed the composition of all the herbal formulae used in Chinese guide-line for COVID-19 in each disease stage and found that liquorice was the herb with the highest frequency of use regardless of disease stage.34 Chinese health authorities had issued programs recommending herbs for preventing COVID-19 infection with liquorice on top of them.35
Our current research is focused on identifying natural anticoagulants that work specifically by inhibiting coagulation factor X (FXa). In this context, previous study conducted by our group described the FXa inhibitory activity of some natural sources using in silico approaches36 and highlighted the three plant extracts; Glycyrrhiza glabra, Trifolium alexandrinum and Olea europaea as candidate FXa inhibitors proved by in vitro assay. In the present work, biologically-guided isolation of secondary metabolites of licorice roots was adopted to identify the compounds responsible for FXa inhibitory action and explain some of the anticoagulant properties of the plant. In addition, ADMET profiles of the isolated compounds were computed in silico to predict their pharmacokinetic properties. It is worthy to mention that this is the first report of the biologically guided isolation of specific FXa inhibitors from licorice root.
2. Experimental
2.1. General experimental procedures
1D and 2D-NMR spectra were recorded on Bruker model AMX 500 NMR and 400 NMR spectrometers operating on a standard pulse system (Bruker, Germany) using the appropriate deuterated solvent. HRESIMS (High Resolution Electro Spray Ionization Mass Spectroscopy) spectra were recorded on a Micromass Q-Tof 2 mass spectrometer (Bruker, Rheinstetten, Germany) using negative ion mode.
HPLC (high performance liquid chromatography) separation was carried out with an Agilent 1100 HPLC system (Agilent Technologies, Waldbroon, Germany) equipped with a degasser (G1379A), quaternary pump (G13311A), auto sampler (G1313A), column oven (G1316A), and UV-diode detector (G1315B) controlled by Chemstation software. HPLC analysis was carried out on RP-C18 column (250 × 10.0 mm; particle size 10 μm; Luna) with column oven temperature set at 25 °C and using the gradient system of eluent water (A) and acetonitrile (B) for the separation of the target compounds with the addition of acetic acid as a modifier to achieve a final concentration of 0.1% in each solvent. The gradient condition was as follows: 0–2 min (10% B), 2–35 min (100% B). The flow rate of the solvent was 1.0 ml min−1, and the injection volume was 10 μl. All the analysis was carried out at wavelengths of 220 and 254 nm with a run time 35 minutes using HPLC-grade solvents.
Column chromatography was performed using silica gel 60, 0.063–0.20 mm (Merck, Darmstadt, Germany) and sephadex LH-20 (0.25–0.1 mm), (Merck, Darmstadt, Germany). Precoated TLC plates, 0.25 mm thick, (silica gel 60 F254), (Merck, Darmstadt, Germany) were used for TLC analysis.
2.2. Plant material
Roots of G. glabra were collected from Alexandria, Egypt from November to December 2018 and were kindly identified by Professor Dr Selim Zidan Heneidy, professor of Applied Ecology, Faculty of Science, Alexandria University. Voucher specimen (GG106) has been deposited in the Pharmacognosy Department, Faculty of pharmacy, Alexandria University.
2.3. Extraction and isolation procedure
The dried powdered G. glabra roots 4.7 (kg) were extracted with 70% ethanol, evaporated under reduced pressure giving residue (100 g). This was followed by re-dissolving in 90% ethanol and successive fractionation with light petroleum, methylene chloride, ethyl acetate, and n-butanol. The four fractions were evaporated under reduced pressure giving residues having weights of 7, 50, 10 and 6 gm for these fractions, respectively. This was followed by in vitro assay of the four fractions for their FXa inhibitory effect.
Since the EtOAc fraction (10 g) was the most active, it was chromatographed on a silica gel column (360 g, 4.5 × 100 cm) packed with methylene chloride. Gradient elution was performed using increased concentration of methanol to obtain 8 subfractions (A–H). Subfraction B (1.44 g) eluted with 5% CH3OH yielded a residue which was purified by repeated crystallization from methylene chloride and ethyl acetate giving compound 1 (20 mg).
Subfraction A (0.79 g) was further chromatographed on a silica gel column packed with n-hexane and eluted with increased concentration of EtOAc (5% till 30%) to obtain twenty fractions (120 ml) from (A1–A20). Fraction A5 (20 mg) was further chromatographed on a silica gel column packed with CH2Cl2 : CH3OH : HCOOH (9 : 1 : 0.1) and eluted isocratically to obtain compound 2 (2 mg). Fraction A8 eluted at 20% EtOAc in hexane afforded compound 3 (55 mg). Fraction A13 eluted at 25% EtOAc in hexane was further purified using sephadex column yielding compound 4 (20 mg).
Subfraction F (0.73 g) was chromatographed on sephadex column eluted with methanol giving eight fractions (F1–F8). Fraction F5 (135 mg) was subjected to silica gel column using gradient elution with the system components CH2Cl2 : CH3OH : HCOOH (9 : 1 : 0.1, 8.5 : 1.5 : 0.1 and 8 : 2 : 0.1). Fraction F5.11 of the latter (eluted at polarity 8.5 : 1.5 : 0.1) was purified using sephadex column yielding compound 5 (4 mg). Fraction F6 (99 mg) was purified using silica gel column eluted isocratically using system EtOAc : CH2Cl2 : CH3OH : H2O (15 : 8 : 4 : 0.5), followed by semi preparative HPLC to obtain compound 6 (2 mg). Fraction F7 (118 mg) was purified using silica gel column eluted isocratically using system EtOAc : CH2Cl2 : CH3OH : H2O (15 : 8 : 4 : 0.5) to give compound 7 (5 mg).
Subfraction (H) (1.86 g) was chromatographed on sephadex column yielding ten fractions (H1–H10). Fraction H5 was purified using sephadex column followed by silica gel column eluted isocratically using EtOAc : CH2Cl2 : CH3OH : H2O (15 : 8 : 4 : 0.5) and yielded compound 8 (30 mg). Fraction H9 was purified using two successive sephadex columns eluted with methanol. Then, collecting very small volumes and monitoring separation using 1H-NMR. Finally, two compounds; 9 (120 mg) and 10 (40 mg) were obtained. The isolation scheme is summarized in Fig. S1.†
2.4. In vitro factor Xa inhibitory activity
Bioactivity-guided isolation was adopted in this study using in vitro factor Xa inhibitory activity. All the tested fractions, subfractions and isolates were dissolved in 50% DMSO prepared in TBS (50 mM Tris base, 150 mM NaCl, pH = 7.4) for the in vitro assay. Two concentrations of each of the main four successive fractions of liquorice (1.5 mg ml−1 and 3 mg ml−1) were used. The EtOAc subfractions (A–H) were tested using one concentration of 1 mg ml−1. The isolated compounds were tested using five different concentrations (1, 10, 40, 80 & 100 μM). The assay procedure according to Bijak et al.37 is given in details in our previous publication.36 All the experiments were carried out in a duplicate manner and results are expressed in terms of % inhibition of FXa amidolytic activity at each concentration used. The IC50 values of the tested compounds were calculated using Graphpad Prism® (Version 6.01) software.
2.5. In silico evaluation of ADMET profile of the isolated compounds
ADMET properties of the isolated compounds were evaluated using Qikprop module of Schrödinger38 to ensure the drug-likeness character of the compounds as potential orally active new oral anticoagulants (NOACs) acting on FXa and, to compare their pharmacokinetics profiles against the known synthetic drugs currently available in the market. The 3D structures of the isolated and synthetic compounds were retrieved from Pubchem database, saved as .sdf file and prepared by LigPrep module of the Schrödinger software (LLC, New York, NY)39 using OPLS3 force field algorithm. Then, optimization through energy minimization and determination of ionization states at the specified PH (7 ± 2.0) were conducted. Compounds were saved as .mae files then imported to the Qikprop wizard. The default parameters in Qikprop module were used for ADMET prediction in the normal mode.
3. Results and discussion
The four successive fractions of licorice were tested for their FXa inhibitory effects. The results (Table S1†) showed that the EtOAc and CH2Cl2 fractions were the most active with % inhibition of 97.93 ± 0.019 and 71.94 ± 0.034, respectively when tested at concentration of 3 mg ml−1. The polar subfractions of EtOAc and particularly subfraction F, (with % inhibition of 84.94 ± 0.82%) had stronger activity than the less polar subfractions (Table S2†).
3.1. Compounds identification
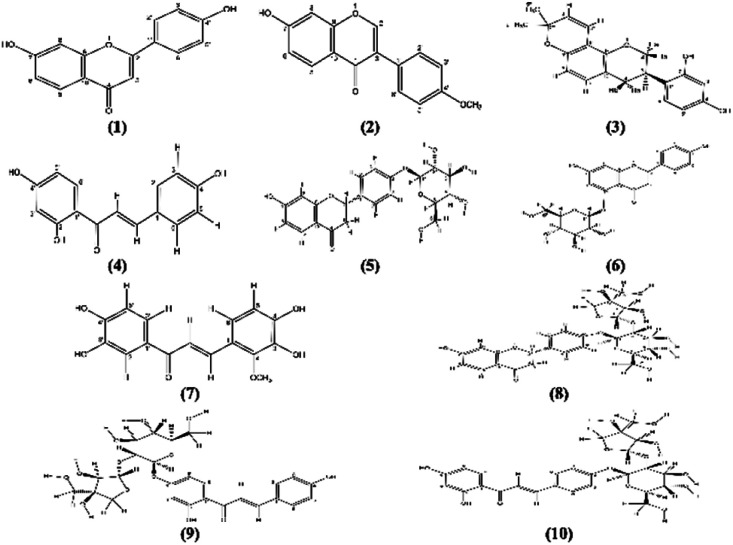
Ten known flavonoids (Fig. 1) were isolated and identified by comparing their 1H, 13C NMR spectra to those previously reported in literature (Tables S3, S4 and Fig. S2–S31; ESI†).
Fig. 1. Compounds 1–10 isolated from G. glabra roots.
The compounds are 7,4′-dihydroxyflavone (1),40,41 7-hydroxy-4′-methoxyisoflavone (2) commonly known as formononetin,42–45 3-R-glabridin (3),46–50 isoliquiritigenin (4),51–53 liquiritin (5),50,54–56 naringenin 5-O-glucoside (6),57–59 3,3′,4,4′-tetrahydroxy-2-methoxychalcone (7),60,61 liquiritigenin-4′-O-β-d-apiosyl(1-2)-β-D-glucoside known as liquiritinapioside (8),50,52,54,55,62 and the two isomers isoliquiritigenin-4′-O-β-d-apiosylglucoside (9) and isoliquiritigenin-4-O-β-d-apiosylglucoside (10) commonly known as licraside & isoliquirtin apioside, respectively.63,64 All the isolated flavonoids are of common existence in G. glabra roots and belong to different classes namely; flavones (1) & flavanone (5, 6 and 8), isoflavones and isoflavans (2 and 3) and chalcones (4, 7, 9 and 10). Moreover, flavonoids (5, 8 and 10) are commonly distributed in the plant roots in addition to the species-specific biomarker 3-R-glabridin (3).49
HRESIMS spectra provided further identification of the compounds where in negative ion mode; compound 1 showed pseudomolecular ion peak [M − H]− at m/z 253.051 and its dimer [2M − H]− at m/z 507.107. HRESIMS (−) of 2 had pseudomolecular ion peak [M − H]− at m/z 267.070 and for 3, the pseudomolecular dimer [2M − H]− and the pseudomolecular ion peak [M − H]− were observed at m/z 647.265 and 323.129. The same spectrum for 4 showed pseudomolecular ion peak [M − H]− at m/z 255.066 and for 5, [M − H]− was observed at m/z 417.124. Compounds 6 and 7 showed pseudomolecular ion peak [M − H]− in HRESIMS (−) at m/z 433.119 and 301.0688, respectively. The HRESIMS (−) spectra of 8, 9 and 10 were very similar showing pseudomolecular ion peaks [M − H]− at m/z 549.1549, 549.1554, 549.1562, respectively.
3.2. FXa inhibitory activity of the isolated compounds
All the isolated compounds were assessed for their FXa inhibitory activity (Table 1). Liquirtin (5) being the most active with IC50 of 5.15 μM. Besides, the three compounds, 3,3′,4,4′-tetrahydroxy-2-methoxychalcone (7), naringenin-5-O-glucoside (6) and 7-hydroxy-4′-methoxyisoflavone (2) had also potent effects as concluded from their IC50 values, (10.43, 12.5 and 17.71 μM, respectively). Moderate activity was observed for glabridin (3), 7,4′-dihydroxyflavone (1) and isoliquiritigenin (4). On the other hand, compounds 8, 9 and 10 had very weak effects with IC50 values in mM range. The effects of the isolated compounds on FXa amidolytic activity are given in ESI (Fig. S32†).
IC50 values of the isolated compounds from G. glabra EtOAc subfraction in FXa inhibitory assay.
| Compound | IC50a (μM) |
|---|---|
| Liquiritin | 5.15 ± 0.09 |
| 3,3′,4,4′-Tetrahydroxy-2-methoxychalcone | 10.43 ± 0.52 |
| Naringenin 5-O-glucoside | 12.5 ± 0.52 |
| 7-Hydroxy-4′-methoxyisoflavone | 17.71 ± 0.28 |
| 3-R-Glabridin | 35.30 ± 1.03 |
| 7,4′-Dihydroxyflavone | 67.09 ± 1.75 |
| Isoliquiritigenin | 74.43 ± 1.29 |
| Isoliquiritigenin 4-O-β-d-apiosylglucoside (isoliquirtin apioside) | 0.46 × 103 ± 2.53 |
| Liquiritinapioside | 1.72 × 103 ± 0.79 |
| Isoliquiritigenin 4′-O-β-d-apiosylglucoside (licraside) | 48.54 × 103 ± 0.15 |
| Rivaroxaban (positive control)b | 1.066b ± 0.01 |
Results are expressed as mean ± SD of duplicate measurements.
IC50 in nM.
These findings further supported the results provided in many studies that polyphenols and polyphenol-rich extracts possess anticoagulant properties besides their antioxidant effects.65 Moreover, targeting specific coagulation factor by licorice compounds was also observed for chalcones and glycyrrhizin representing new class of direct thrombin inhibitors.66
In addition to glycyrrhetinic that showed direct FXa inhibitory action.67 Interestingly, some of the tested compounds were identified as naturally occurring thrombin inhibitors e.g., isoliquiritigenin and isoliquiritin apioside having IC50 of 17.95 and 37.85 μM, respectively. Meanwhile liquiritin, glabridin and liquiritin apioside showed negligible thrombin inhibitory effects in the same study.66 All together, these finding suggest that flavonoids having direct FXa inhibitory action may not be effective in targeting thrombin as potent FXa inhibitors in our study; liquirtin was found to be ineffective as direct thrombin inhibitors.
It was found that coagulation factor X has multimodal pathologic effects in coronavirus infection. These effects begin with the viral invasion of the host cells as FX is one of the cleavage proteins for the virus allowing its cellular infection. FXa is involved in local and systemic inflammation and coagulation. It is expressed in many cells types as, alveolar, and bronchiolar epithelium, cardiac myocytes, and macrophages where it may play a role in both cardiac dysfunction and acute and chronic pulmonary inflammation. Patients with preexisting conditions, including pulmonary disease, cardiovascular disease and diabetes are at high risk of developing severe COVID-19 symptoms due to the higher baseline expression of FX in addition to other coagulation and inflammatory mediators.13,68
Based on these observations, inhibitors of FXa may have a potential role in the prophylaxis and treatment of COVID-19. This was explained by their ability to reduce cell infectivity, as well as reducing systemic inflammation and coagulation. This was reflected by the number of clinical studies that include direct FXa inhibitors as part of treatment protocol in COVID-19 patients.13,69 Consequently, licorice flavonoids inhibiting FXa as evidenced in our study are speculated to explain the plant beneficial effects in COVID-19 disease.
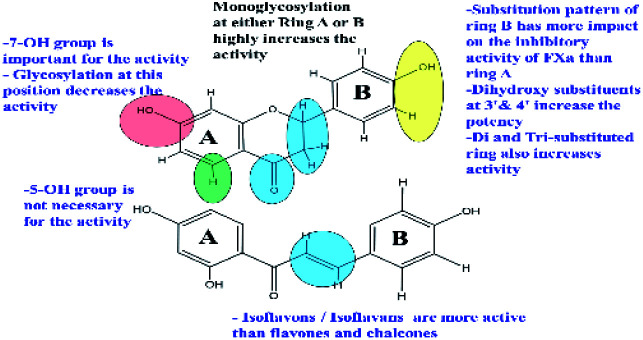
3.3. Structure activity relationship (SAR) of the compounds
Structure observations of the tested flavonoids (Fig. 1) showed some differences that could be correlated to the obtained FXa inhibitory activity. Free aglycones, isoflavones/isoflavanes manifested by 7-hydroxy-4′-methoxyisoflavone and glabridin have more inhibitory activity than both flavones and chalcones.
The mono-glycosylated flavanone as liquiritin and naringenin 5-O-glucoside had the most potent FXa inhibitory action. Meanwhile, glycosylated flavonoids; (in particular, the disaccharides) have very weak activity compared to the free aglycones. This was also observed in a previous study for the inhibitory effects of flavonoids on thrombin.70 In addition, glycosylated chalcones with disaccharide moiety are more effective than the corresponding flavanone glycosides e.g.: isoliquirtin apioside vs. liquiritinapioside (Table 1).
Examining the influence of substitution pattern of rings, A & B, and glycosylation of flavonoids on the FXa inhibitory activity showed that for ring A, the presence of free OH group at 7-position; (equivalent to 4′-position of chalcone) is important for activity.
This was concluded from the very weak activity observed for licraside (IC50 48.54 mM) compared to isoliquirtin apioside which was explained because of substitution with disaccharide moiety at this position. The 5-OH group doesn't strongly influence the activity since compounds having OH group at this position showed similar activity to those lacking it (Table 1).
In case of ring B, the substitution pattern has more impact on the flavonoid potency and inhibitory activity of FXa than ring A. This is well demonstrated from the fact that among potent compounds; tetrahydroxy methoxy chalcone had three substituents at ring B (positions 2, 3 & 4 of chalcone) in comparison to isoliquirtigenin. In addition, ring B of glabridin had two hydroxyl groups at positions 2′ and 4′ which showed also high activity. These observations came in agreement with previous studies that explained the FXa inhibitory activity of flavonoids having dihydroxy substitution pattern at ring B (positions 3′ & 4′).37
Glycosylation of flavonoids showed strong relationship to FXa inhibitory action where monoglycosylated flavanones (in either ring A or B) have the highest FXa inhibitory effects compared to the bulkier flavonoid glycosides with disaccharide moiety and flavonoids having disubstituted pattern in ring B. These SAR are summarized in (Fig. 2).
Fig. 2. SAR of flavonoids responsible for FXa inhibitory activity.
3.4. ADMET profiles
QikProp module generates physically relevant descriptors and uses them to perform ADMET predictions for the screened compounds.71 Analysis of these descriptors (Table S5†) shows that all compounds are free from violations of Lipinski's rule72 except for three glycosides with disaccharide moiety (8, 9 and 10). The predicted pharmacokinetic properties were found to be within the standardized range defined for human use. The results of this in silico ADMET prediction indicated that seven of the investigated compounds follow the criteria for orally active drugs. These compounds possess small molecular weight less than 1000 Da that is considered a major advantage over larger proteins and peptides isolated from animals and leeches and acting as FXa inhibitors.73–75 In addition, the predicted pharmacokinetic properties were found to be within the standardized range defined for human use. These findings suggest the appropriateness for oral administration. This is of great importance as the currently available synthetic FXa inhibitors are administered orally.
Toxicity prediction showed that none of the tested compounds was toxic in nature as they didn't possess any blockage effect of HERG K+ (human ether-a-go-go related gene-potassium ion) channel. Hence, the tested compounds are free from cardiac toxicity.76
Moreover, most of the tested compounds showed better predicted oral absorption and less metabolic interactions than the synthetic drug “Edoxaban”. The two compounds: 7-hydroxy-4′-methoxyisoflavone and glabridin had the highest oral absorption (100%). Thus, representing very good candidates for developing orally active direct FXa inhibitors new oral anticoagulants (NOAC) since both compounds showed strong activity (IC50 of 17.1 and 35.30 μM, respectively).
4. Conclusions
In the present study, licorice flavonoids were proved for the first time to have direct inhibitory activity on coagulation factor Xa when tested in vitro. The four compounds, liquiritin, 3,3′,4,4′-tetrahydroxy-2-methoxychalcone, naringenin 5-O-glucoside and 7-hydroxy-4′-methoxyisoflavone pointed out as promising candidate natural anticoagulant agents for the design of FXa inhibitors. In silico prediction of ADMET properties of these compounds suggested safety and good oral bioavailability and can be considered as good candidates for developing orally active direct FXa inhibitors (NOAC). Moreover, the compounds 7-hydroxy-4′-methoxyisoflavone, liquiritin and 3,3′,4,4′-tetrahydroxy-2-methoxychalcone showed predicted oral absorption characters superior to the synthetic drug, edoxaban. These results present promising candidates that need further in vitro, in vivo and clinical studies to introduce these natural FXa inhibitors as marketed drugs.
With the current pandemic COVID-19 spread, direct FXa inhibitors have evolved as an important part of therapeutic protocols involved in many clinical trials in coronavirus diseased (COVID-19) patients. Introducing these four licorice flavonoids with the potent FXa inhibitory activity together with their good pharmacokinetic profile provided a justification for the use of licorice root extract in prophylaxis and treatment of some complications of the (COVID-19) disease.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The Egyptian Missions sector in “Ministry of Higher Education and Scientific Research” and “National Center for Natural products Research (NCNPR), School of Pharmacy, University of Mississippi, USA” are really appreciated. Since, part of the work accomplished in this thesis was performed at NCNPR research center as a part of a short-term scholarship funded by the Ministry of Higher Education and Scientific Research. The authors are grateful to Dr Mohamed Ali Ibrahim, Research Scientist in (NCNPR), School of Pharmacy, University of Mississippi, USA, for performing HPLC purification of some isolated compounds. The assistance of Dr Mona H. Haron, Research Scientist in (NCNPR), School of Pharmacy, University of Mississippi, USA in providing lab facilities to perform the in vitro testing is highly appreciated.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00359c
References
- Perzborn E. Strassburger J. Wilmen A. Pohlmann J. Roehrig S. Schlemmer K. H. Straub A. J. Thromb. Haemostasis. 2005;3:514–521. doi: 10.1111/j.1538-7836.2005.01166.x. [DOI] [PubMed] [Google Scholar]
- Wong P. C. Crain E. J. Watson C. A. Xin B. J. Thromb. Haemostasis. 2009;7:1313–1320. doi: 10.1111/j.1538-7836.2009.03503.x. [DOI] [PubMed] [Google Scholar]
- Khadse A. N. Sharma M. K. Murumkar P. R. Rajput S. J. Yadav M. R. Mini-Rev. Med. Chem. 2018;18:1332–1353. doi: 10.2174/1389557518666180424120726. [DOI] [PubMed] [Google Scholar]
- Pinto D. J. Smallheer J. M. Cheney D. L. Knabb R. M. Wexler R. R. J. Med. Chem. 2010;53:6243–6274. doi: 10.1021/jm100146h. [DOI] [PubMed] [Google Scholar]
- Liu C. Zhou Q. Li Y. Garner L. V. Watkins S. P. Carter L. J. Smoot J. Gregg A. C. Daniels A. D. Jervey S. Albaiu D. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K. Sharun K. Tiwari R. Dadar M. Malik Y. S. Singh K. P. Chaicumpa W. Hum. Vaccines Immunother. 2020;16:1232–1238. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E. Ntanasis-Stathopoulos I. Elalamy I. Kastritis E. Sergentanis T. N. Politou M. Psaltopoulou T. Gerotziafas G. Dimopoulos M. A. Am. J. Hematol. 2020;95:834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Samkari H. Karp Leaf R. S. Dzik W. H. Carlson J. C. T. Fogerty A. E. Waheed A. Goodarzi K. Bendapudi P. K. Bornikova L. Gupta S. Leaf D. E. Kuter D. J. Rosovsky R. P. Blood. 2020;136:489–500. doi: 10.1182/blood.2020006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M. Thachil J. Iba T. Levy J. H. Lancet Haematol. 2020;7:e438–e440. doi: 10.1016/S2352-3026(20)30145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasecka A. Borovac J. A. Guerreiro R. A. Giustozzi M. Parker W. Caldeira D. Chiva-Blanch G. Cardiovasc. Drugs Ther. 2020:1–15. doi: 10.1007/s10557-020-07084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obi A. T. Barnes G. D. Wakefield T. W. Brown S. Eliason J. L. Arndt E. Henke P. K. J. Vasc. Surg. Venous Lymphat. Disord. 2020;8:526–534. doi: 10.1016/j.jvsv.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo M. Bertinato E. M. Birocchi S. Brizio C. Malavolta D. Manzoni M. Muscarella G. Orlandi M. Thromb. Haemostasis. 2020;120:1230–1232. doi: 10.1055/s-0040-1712097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Horani R. A. Am. J. Cardiovasc. Drugs. 2020;20:525–533. doi: 10.1007/s40256-020-00438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. J. Chem. Inf. Model. 2020;60:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed K. Yazdanpanah N. Saghazadeh A. Rezaei N. Bioorg. Chem. 2021;106:104490. doi: 10.1016/j.bioorg.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olubiyi O. O. Olagunju M. Keutmann M. Loschwitz J. Strodel B. Molecules. 2020;25:3193. doi: 10.3390/molecules25143193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile D. Patamia V. Scala A. Sciortino M. T. Piperno A. Rescifina A. Mar. Drugs. 2020;18:225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraz W. R. Gomes R. A. Al S. N. Goulart Trossini G. H. Future Med. Chem. 2020;12:1815–1828. doi: 10.4155/fmc-2020-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Lin D. Sun X. Curth U. Drosten C. Sauerhering L. Becker S. Rox K. Hilgenfeld R. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo S. T. Quynh Anh Pham N. Thi Le L. Pham D. H. Vu V. V. J. Chem. Inf. Model. 2020;60:5771–5780. doi: 10.1021/acs.jcim.0c00491. [DOI] [PubMed] [Google Scholar]
- Shahinshavali S. Hossain K. A. Kumar A. Reddy A. G. Kolli D. Nakhi A. Rao M. V. B. Pal M. Tetrahedron Lett. 2020;61:152336. doi: 10.1016/j.tetlet.2020.152336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar M. Ahmed H. A. Aljohani G. Alhaddad O. A. Int. J. Mol. Sci. 2020;21:3922. doi: 10.3390/ijms21113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexpandi R. De Mesquita J. F. Pandian S. K. Ravi A. V. Front. Microbiol. 2020;11:1796. doi: 10.3389/fmicb.2020.01796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemboli R. Kapavarapu R. Deepti K. Prasad K. R. S. Reddy A. G. Kumar A. V. D. N. Rao M. V. B. Pal M. J. Mol. Struct. 2021;1230:129868. doi: 10.1016/j.molstruc.2020.129868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antonio A. Wiedemann L. S. M. Veiga-Junior V. F. RSC Adv. 2020;10:23379–23393. doi: 10.1039/D0RA03774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. H. Wu K. L. Zhang X. Deng S. Q. Peng B. J. Integr. Med. 2020;18:152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacka K. Witek-Krowiak A. Skrzypczak D. Mikula K. Mlynarz P. J. Funct. Foods. 2020;73:104146. doi: 10.1016/j.jff.2020.104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W. H. Organization, WHO monographs on medicinal plants commonly used in the Newly Independent States (NIS), Libros Digitales-World Health Organization (WHO), 2010 [Google Scholar]
- Hassan-Wassef H. East. Mediterr. Health J. 2004;10:898–915. [PubMed] [Google Scholar]
- Omar H. R. Komarova I. El-Ghonemi M. Fathy A. Rashad R. Abdelmalak H. D. Yerramadha M. R. Ali Y. Helal E. Camporesi E. M. Ther. Adv. Endocrinol. Metab. 2012;3:125–138. doi: 10.1177/2042018812454322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawky E. Nada A. A. Ibrahim R. S. RSC Adv. 2020;10:27961–27983. doi: 10.1039/D0RA05126H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathy H. Abdelhady W. Ibrahim R. Int. J. Pharm. Sci. Rev. Res. 2020:197–202. [Google Scholar]
- Murck H. Front. Immunol. 2020;11:1239. doi: 10.3389/fimmu.2020.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L. Lee H. W. Choi J. Y. Zhang J. Soo Lee M. Integr. Med. Res. 2020;9:100407. doi: 10.1016/j.imr.2020.100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H. Tang Q. L. Shang Y. X. Liang S. B. Yang M. Robinson N. Liu J. P. Chin. J. Integr. Med. 2020;26:243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim R. S. Mahrous R. S. R. Fathy H. M. Omar A. A. Abu El-Khair R. M. Med. Chem. Res. 2020;29:707–726. doi: 10.1007/s00044-020-02516-5. [DOI] [Google Scholar]
- Bijak M. Ponczek M. B. Nowak P. Int. J. Biol. Macromol. 2014;65:129–135. doi: 10.1016/j.ijbiomac.2014.01.023. [DOI] [PubMed] [Google Scholar]
- Schrödinger L., QikProp, version 3.5, New York, NY: 2012 [Google Scholar]
- Schrödinger L., LigPrep, version 2.3, New York, NY: 2009 [Google Scholar]
- Park Y. Moon B. H. Lee E. Lee Y. Yoon Y. Ahn J. H. Lim Y. Magn. Reson. Chem. 2007;45:674–679. doi: 10.1002/mrc.2010. [DOI] [PubMed] [Google Scholar]
- Hatano T. Kagawa H. Yasuhara T. Okuda T. Chem. Pharm. Bull. 1988;36:2090–2097. doi: 10.1248/cpb.36.2090. [DOI] [PubMed] [Google Scholar]
- Zhao S. Zhang L. Gao P. Shao Z. Food Chem. 2009;114:869–873. doi: 10.1016/j.foodchem.2008.10.026. [DOI] [Google Scholar]
- Chang Y.-C. Nair M. G. Santell R. C. Helferich W. G. J. Agric. Food Chem. 1994;42:1869–1871. doi: 10.1021/jf00045a007. [DOI] [Google Scholar]
- Du X. Bai Y. Liang H. Wang Z. Zhao Y. Zhang Q. Huang L. Magn. Reson. Chem. 2006;44:708–712. doi: 10.1002/mrc.1806. [DOI] [PubMed] [Google Scholar]
- Mitscher L. A. Park Y. H. Clark D. Beal J. L. J. Nat. Prod. 1980;43:259–269. doi: 10.1021/np50008a004. [DOI] [PubMed] [Google Scholar]
- Simmler C. Pauli G. F. Chen S. N. Fitoterapia. 2013;90:160–184. doi: 10.1016/j.fitote.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T. Kajiyama K. Hiraga Y. Takahashi K. Tamura Y. Mizutani K. Heterocycles. 1996;3:653–664. [Google Scholar]
- Jirawattanapong W. Saifah E. Patarapanich C. Arch. Pharmacal Res. 2009;32:647–654. doi: 10.1007/s12272-009-1501-x. [DOI] [PubMed] [Google Scholar]
- Saitoh T. Kinoshita T. Shibata S. Chem. Pharm. Bull. 1976;24:752–755. doi: 10.1248/cpb.24.752. [DOI] [Google Scholar]
- Pastorino G. Cornara L. Soares S. Rodrigues F. Oliveira M. Phytother. Res. 2018;32:2323–2339. doi: 10.1002/ptr.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S. Li Z. Song W. Wang Y. Liang W. Li K. Tang S. Wang Q. Qiao X. Zhou D. Yu S. Ye M. J. Nat. Prod. 2016;79:281–292. doi: 10.1021/acs.jnatprod.5b00877. [DOI] [PubMed] [Google Scholar]
- Fenwick G. R. Lutomski J. Nieman C. Food Chem. 1990;38:119–143. doi: 10.1016/0308-8146(90)90159-2. [DOI] [Google Scholar]
- Chin Y. W. Jung H. A. Liu Y. Su B. N. Castoro J. A. Keller W. J. Pereira M. A. Kinghorn A. D. J. Agric. Food Chem. 2007;55:4691–4697. doi: 10.1021/jf0703553. [DOI] [PubMed] [Google Scholar]
- Nakanishi T. Inada A. Kambayashi K. Yoneda K. Phytochemistry. 1985;24:339–341. doi: 10.1016/S0031-9422(00)83548-7. [DOI] [Google Scholar]
- Fu B. Li H. Wang X. Lee F. S. Cui S. J. Agric. Food Chem. 2005;53:7408–7414. doi: 10.1021/jf051258h. [DOI] [PubMed] [Google Scholar]
- Nomura T. and Fukai T., in Fortschritte der Chemie organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products, ed. W. Herz, G. W. Kirby, R. E. Moore, W. Steglich and C. Tamm, Springer Vienna, Vienna, 1998, pp. 1–140, 10.1007/978-3-7091-6480-8_1 [DOI] [PubMed] [Google Scholar]
- Hendra R. Willis A. Keller P. A. Nat. Prod. Res. 2019;33:1997–2003. doi: 10.1080/14786419.2018.1483922. [DOI] [PubMed] [Google Scholar]
- Wijaya S. Jin K. T. Nee T. K. Wiart C. J. Complementary Integr. Med. 2012;9:11. doi: 10.1515/1553-3840.1615. [DOI] [PubMed] [Google Scholar]
- Ragab E. A. Hosny M. Kadry H. A. Ammar H. A. J. Nat. Prod. 2010;3:35–46. [Google Scholar]
- Hatano T. Takagi M. Ito H. Yoshida T. Chem. Pharm. Bull. 1997;45:1485–1492. doi: 10.1248/cpb.45.1485. [DOI] [Google Scholar]
- Birari R. B. Gupta S. Mohan C. G. Bhutani K. K. Phytomedicine. 2011;18:795–801. doi: 10.1016/j.phymed.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Hatano T. Takagi M. Ito H. Yoshida T. Phytochemistry. 1998;47:287–293. doi: 10.1016/S0031-9422(97)00560-8. [DOI] [Google Scholar]
- Kitagawa I. Hori K. Uchida E. Chen W. Z. Yoshikawa M. Ren J. Chem. Pharm. Bull. 1993;41:1567–1572. doi: 10.1248/cpb.41.1567. [DOI] [PubMed] [Google Scholar]
- Nomura T. and Fukai T., in Fortschritte der Chemie organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products, Springer, 1998, pp. 1–140 [DOI] [PubMed] [Google Scholar]
- Bijak M. Saluk J. Szelenberger R. Nowak P. Chem.-Biol. Interact. 2016;257:35–45. doi: 10.1016/j.cbi.2016.07.022. [DOI] [PubMed] [Google Scholar]
- Shi C.-C. Chen T.-R. Zhang Q.-H. Wei L.-H. Huang C. Zhu Y.-D. Liu H.-B. Bai Y.-K. Wang F.-J. Guo W.-Z. RSC Adv. 2020;10:3626–3635. doi: 10.1039/C9RA09203J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. Wang Q. Shen S. Xiao T. Li Y. Thromb. Res. 2014;133:501–506. doi: 10.1016/j.thromres.2013.12.025. [DOI] [PubMed] [Google Scholar]
- Frydman G. H. Streiff M. B. Connors J. M. Piazza G. TH Open. 2020;4:e288–e299. doi: 10.1055/s-0040-1718415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kow C. S. Hasan S. S. J. Thromb. Thrombolysis. 2021;51:29–30. doi: 10.1007/s11239-020-02200-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedinak A. Maliar T. Grancai D. Nagy M. Phytother. Res. 2006;20:214–217. doi: 10.1002/ptr.1836. [DOI] [PubMed] [Google Scholar]
- Ntie-Kang F. Springerplus. 2013;2:353. doi: 10.1186/2193-1801-2-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A. Lombardo F. Dominy B. W. Feeney P. J. Adv. Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Lee W. Lee H. Kim M. A. Choi J. Kim K. M. Hwang J. S. Na M. Bae J. S. Sci. Rep. 2017;7:7934. doi: 10.1038/s41598-017-08330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuszynski G. P. Gasic T. B. Gasic G. J. J. Biol. Chem. 1987;262:9718–9723. doi: 10.1016/S0021-9258(18)47993-8. [DOI] [PubMed] [Google Scholar]
- Waxman L. Smith D. E. Arcuri K. E. Vlasuk G. P. Science. 1990;248:593–596. doi: 10.1126/science.2333510. [DOI] [PubMed] [Google Scholar]
- Aronov A. M. Drug Discovery Today. 2005;10:149–155. doi: 10.1016/S1359-6446(04)03278-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.