Abstract
In an effort to find a rapid, efficient, and reliable method of screening large numbers of bacterial isolates for specific antimicrobial resistance genes, we compared conventional PCR results to the results generated using the TaqMan 5′ nuclease PCR kit in conjunction with an ABI Prism 7700 Sequence Detector for detecting the mecA gene in various species of staphylococci. DNA was extracted using two techniques. The first used a high-salt extraction method suitable for conventional PCR but resulted in a 7.2% rate of PCR inhibition with the TaqMan technique. PCR inhibition could be overcome by diluting samples 1:5 prior to testing. The second method used the Qiagen QIAamp Tissue Kit; no instances of PCR inhibition were encountered with this method. A total of 197 (96%) of the 206 samples with no inhibition showed agreement between the two methods. Eight of the nine disagreements were likely the result of low-level DNA cross contamination caused by frequent specimen handling. Target DNA in all eight of these samples was first detected in the initial tests only after >30 PCR cycles, and all were negative upon repeat testing even after 40 PCR cycles using freshly extracted DNA. Among those positive samples in agreement, target DNA was invariably detected before 30 PCR cycles. The TaqMan assay eliminated the need to load, run, stain, and read agarose gels and provided the advantage of instant detection of PCR product by laser-activated fluorescence. Thus, final results were obtained 2 h after PCR was initiated, as opposed to a requirement of 2 days to examine 96 samples by agarose gel electrophoresis.
Staphylococci are among the most frequent isolates from nosocomial infections, particularly from nosocomial bloodstream infections. They often are resistant to methicillin and other beta-lactam antimicrobial agents (3, 5, 18, 26). Methicillin-resistant Staphylococcus aureus strains are found worldwide, predominantly in hospitalized patients, and can spread rapidly from patient to patient and hospital to hospital (1–3, 7, 20). Nosocomial methicillin-resistant S. aureus strains are frequently resistant to most other classes of antimicrobial agents with the exception of glycopeptides (15); however, a few recent S. aureus strains have been reported to show intermediate levels of resistance to the glycopeptide vancomycin (11, 22).
The most frequent mechanism of methicillin resistance is mediated by the mecA gene, which produces a novel penicillin binding protein, PBP 2A, with reduced affinity for beta-lactam agents. An unusual characteristic, referred to as heterogeneous resistance, of mecA-containing staphylococci is that there may be a wide range of levels of resistance to methicillin within a cell population derived from a single isolate. While the majority of cells may show low levels of resistance, subpopulations, often only 1 in 100,000 cells, show very high resistance levels that can lead to treatment failure regardless of phenotypic expression (23). For this reason it is often desirable to use a DNA-based assay to detect the mecA gene rather than rely on phenotypic susceptibility tests. PCR is one such DNA-based test that has gained widespread use (10, 13, 24, 25). While they are very sensitive and specific, conventional PCR techniques that use agarose gel electrophoresis to detect amplification products are not well suited for rapid screening of large numbers of samples because of the limited number of samples that can be analyzed per gel and the time required for loading, running, and staining gels. To overcome these limitations, we investigated the use of the TaqMan 5′ nuclease PCR kit (12) in conjunction with an ABI 7700 Sequence Detector for rapid detection of the mecA gene in a variety of staphylococcal isolates. This system uses both PCR primers and a dye-labeled probe to detect the target sequence and provides an instantaneous reading of the PCR result. TaqMan PCR has been used successfully to detect specific DNA sequences from fungi (4), bacteria (8, 9, 19), and viruses (14, 17, 21) and to quantitate bacterial load in clinical specimens (6). The goal of this study was to compare the results of the TaqMan system to those of conventional PCR for accuracy and efficiency in detecting the mecA gene.
MATERIALS AND METHODS
Bacterial isolates.
A total of 222 staphylococcal isolates from patients in four hospitals in three states were examined. These included 76 nosocomial isolates of S. aureus and 146 coagulase-negative staphylococcal isolates from blood cultures (Table 1). Control organisms included S. aureus ATCC 43300 (mecA-positive control) and S. aureus ATCC 25923 (mecA-negative control).
TABLE 1.
Staphylococcus isolates used in this study
| Species | No. (%) mecA positivea | No. mecA negative | Total no. |
|---|---|---|---|
| S. aureus | 58 (76) | 18 | 76 |
| S. epidermidis | 52 (74) | 18 | 70 |
| S. hominis | 31 (67) | 15 | 46 |
| S. haemolyticus | 10 (71) | 4 | 14 |
| S. capitus | 0 (0) | 4 | 4 |
| S. warneri | 1 (20) | 4 | 5 |
| S. cohnii | 1 (50) | 1 | 2 |
| S. simulans | 0 (0) | 1 | 1 |
| S. auricularis | 0 (0) | 2 | 2 |
| S. lugdunensis | 0 (0) | 2 | 2 |
| Total | 153 (69) | 69 | 222 |
Conventional PCR determination.
DNA extraction.
DNA extraction was performed by two methods. The first was the salting-out procedure modified from that described by Miller et al. (16), in which bacterial cells approximating one-quarter of a 10-μl loop were scraped from an overnight blood agar plate, suspended in 455 μl of TEN buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 100 mM NaCl) containing 3 μg of lysostaphin per ml, and incubated at 37°C for 15 min. Forty-five microliters of a solution containing 0.8% Sarkosyl and 100 μg of proteinase K per ml was added, and the tube was incubated at 37°C for 1.5 h. One hundred microliters of 6 M NaCl was added and mixed by vortexing for 15 s before centrifugation at 750 × g for 15 min. The supernatant was transferred to a fresh tube, two volumes of absolute ethanol were added, and the mixture was centrifuged at 16,000 × g for 15 min. The DNA precipitate was rinsed with 70% ethanol, centrifuged at 16,000 × g, decanted, dried, resuspended in 400 to 600 μl of 10 mM Tris–1 mM EDTA (pH 8.0), treated with RNase at 37°C for 30 min, and stored at −20°C until use.
Isolates were later recultivated, and the DNA was extracted by a second method that utilized the Qiagen DNeasy 96 Tissue Kit with modifications for DNA extraction from staphylococci. Briefly, using a 10-μl loop, one-fourth of a loop of cells from an overnight blood agar plate was placed in a 1.5-ml tube containing 180 μl of buffer (29 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1.2% Triton), 200 μg of lysozyme per ml, and 20 μg of lysostaphin per ml. The suspensions were incubated for 1.5 h at 37°C, 25 μl of proteinase K and 200 μl of kit buffer AL were added and the suspension was mixed by vortexing. The tubes were placed in a centrifuge block and incubated at 70°C for 30 min, and then 210 μl of ethanol was added to each tube. The tubes were centrifuged in the block for 10 s at 1,450 × g, and 615-μl portions of the lysates were transferred to a DNeasy 96 plate, sealed with tape, and centrifuged at 5,800 × g for 10 min. The tape was removed, and 500 μl of kit buffer AW2 was added to each well. The plate was resealed and centrifuged at 5,800 × g for 5 min. The tape was removed, and the DNeasy 96 plate was placed on top of a collection microtube rack and incubated at 70°C for 15 min in a dry oven. DNA was eluted by adding 200 μl of kit buffer AE preheated to 70°C, resealing, and centrifuging at 5,800 × g for 2 min. The DNA solution was treated with RNase at 37°C for 30 min and stored at −20°C until use.
Conventional PCR.
PCR for detection of the mecA gene was performed using primers M1 (TGG CTA TCG TGT CAC AAT CG) and M2 (CTG GAA CTT GTT GAG CAG AG), described by Vannuffel et al. (25). One microliter of template DNA was added to 24 μl of reaction mixture consisting of Perkin-Elmer GeneAmp PCR buffer II, 0.2 mM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 25 pmol of each mecA primer, 25 pmol of each rRNA universal primer (16SRRI [CAG CAG CCG CGG TAA TAC] and 16SRRII [CCG TCA ATT CCT TTG AGT TT]) to check for presence of template, and 2 U of Taq polymerase. Amplification was performed in a Perkin-Elmer 9600 thermocycler. The cycle program was 94°C for 3 min, 25 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 3 min. PCR product was detected by addition of 5 μl of sample mixed with 1 μl of loading buffer and dye to a 1.5% agarose gel in 0.5× Tris-borate-EDTA and electrophoresis at 200V in a submarine gel unit with 18°C circulating coolant. Electrophoresis was continued until the bromophenol blue dye line migrated 10 cm. Gels were stained with ethidium bromide and photographed.
TaqMan PCR.
Because of the need for optimal matching of primer and probe annealing temperatures, a set of primers different from the conventional PCR primers was prepared for TaqMan PCR. 5′ nuclease PCR primers and the DNA probe for the mecA gene were selected using Primer Express software. The primers and probe were prepared by the Centers for Disease Control and Prevention Molecular Biology Core Facility as follows: Forward primer, 5′ TGCTAAAGTTCAAAAGAGTATTTATAACAACA 3′; reverse primer, 5′ TGTGCTTACAAGTGCTAATAATTCACC 3′; and probe, 5′ATTATGGCTCAGGTACTGCTATCCACCCTCAAA 3′. The PCR mixture was prepared using TaqMan Universal PCR Master Mix and TaqMan Exogenous IPC reagents according to the manufacturer's instructions, except that the final PCR mixture volume was 25 μl instead of 50 μl. Five microliters of template DNA was added to each test well. Amplification was performed on an ABI Prism 7700 Sequence Detector programmed to hold at 50°C for 2 min, hold at 95°C for 10 min, and complete 40 cycles of 95°C for 15 s and 60°C for 60 s. The data were analyzed in both the real-time and plate read modes.
RESULTS AND DISCUSSION
Of the 222 staphylococci isolates extracted by the high-salt method and examined by conventional PCR, all produced the expected 409-bp fragment when universal primers were used, indicating the presence of template DNA. However, 16 (7.2%) DNA samples extracted with the salt method and processed with the TaqMan assay showed PCR inhibition as detected by the TaqMan internal positive control reagents (Table 2). This inhibition could be overcome by diluting the sample 1:5 in H2O or by preparing a fresh DNA extraction with the Qiagen kit. No Qiagen kit-extracted DNA showed PCR inhibition in either the PCR or TaqMan assay (Table 2). The binding and washing steps in the Qiagen procedure result in a cleaner product than the salting-out method, which uses only relatively low-speed centrifugation to separate protein and DNA. This likely accounts for the difference in PCR inhibition between the two procedures.
TABLE 2.
Inhibition and false-positive results observed with initial PCR testing and repeat testing results after 1:5 dilution or fresh DNA extractiona
| Conventional PCR result | No. of isolates with the following TaqMan resultb:
|
|||
|---|---|---|---|---|
| Positive | Negative | No amplification | Total | |
| Positive | 140 (153) | 1 (0) | 12 (0) | 153 (153) |
| Negative | 8 (0) | 57 (69) | 4 (0) | 69 (69) |
| No amplification | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Total | 148 (153) | 58 (69) | 16 (0) | 222 (222) |
1:5 dilutions were prepared for the 16 samples that originally showed TaqMan inhibition, and fresh DNA extractions were prepared for the 8 samples that were conventional PCR negative and TaqMan positive.
Data in parentheses are repeat testing results.
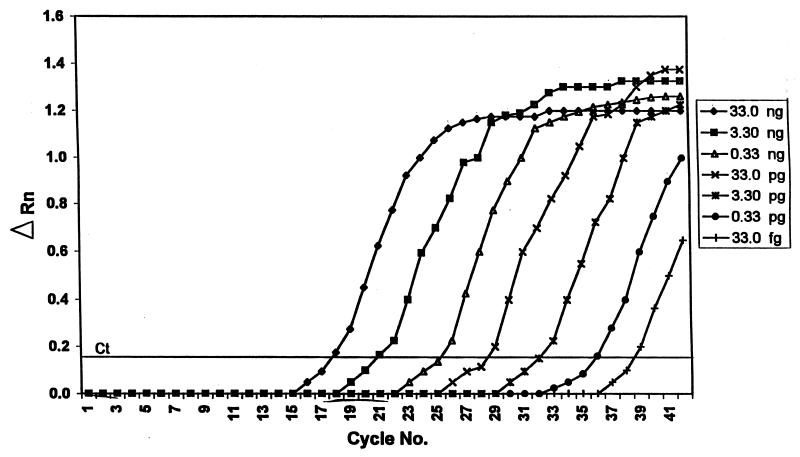
Initial testing included DNA samples from tubes that had been entered several times, thus providing the opportunity for low-level cross contamination of minute quantities of DNA that would not be detected by conventional PCR because of the low cycle amplification number (25 cycles) and relative insensitivity of ethidium bromide for detecting DNA in agarose. Using the TaqMan assay in plate read mode, the ABI 7700 takes only one reading of test and control wells after all PCR cycles are completed and reports each test well as positive or negative after comparing the fluorescence intensities measured in the test wells with those of the control wells. The TaqMan assay is potentially much more sensitive than conventional PCR using an agarose gel detection method, because the TaqMan assay employs 40 amplification cycles, measures changes in fluorescence intensity, and can detect template concentrations in the femtogram range (Fig. 1). This enhanced sensitivity may explain why the eight samples that were negative using the conventional PCR assay tested positive by the TaqMan PCR assay. All eight samples were negative by TaqMan when fresh DNA extracts were prepared. The single sample that was positive by conventional PCR but negative by the TaqMan assay during initial testing was positive in the TaqMan assay upon repeat testing (Table 2).
FIG. 1.
Assay sensitivity using a 10× dilution scheme. ΔRN = (Rn+) − (Rn−), where Rn+ = (emission intensity of reporter dye)/(emission intensity of passive reference dye) in PCR with template and Rn− = (emission intensity of reporter dye)/(emission intensity of passive reference dye) in PCR without template or early cycles of a real-time reaction. Ct = threshold cycle, i.e., cycle at which a statistically significant increase in ΔRn is first detected.
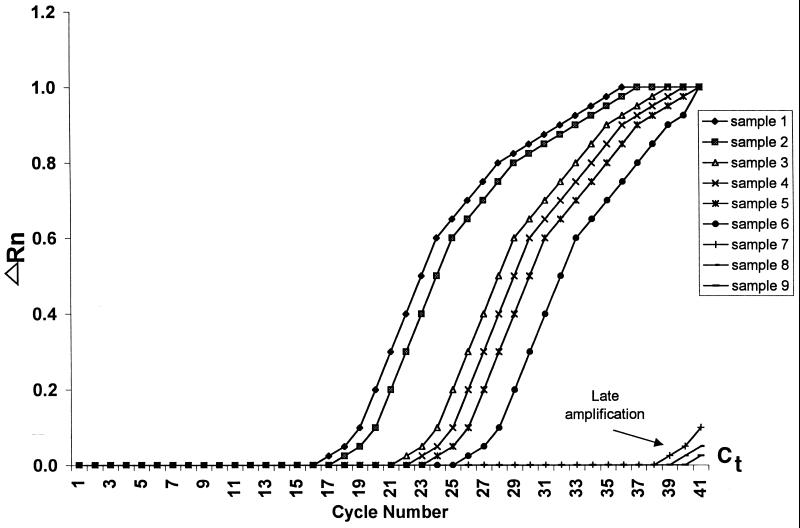
In the real-time mode, the ABI 7700 instrument reads each well every few seconds and computes a mean baseline reading for early PCR cycles. Real-time results are reported as cycle threshold (Cj) values, the cycle at which fluorescence readings exceed the mean baseline readings by 10 standard deviations. Of the 140 samples that were positive for mecA by conventional PCR, 139 had Ct values of <30 cycles. The one exception had a Ct value well below 30 upon retesting. In comparing results, it was observed that the eight samples that were positive in plate read mode but with a real-time Ct value of >30 (Fig. 2) were invariably negative upon retesting using template DNA that was freshly extracted.
FIG. 2.
PCR amplification in real-time mode. ΔRN = (Rn+) − (Rn−), where Rn+ = (emission intensity of reporter dye)/(emission intensity of passive reference dye) in PCR with template and Rn− =(emission intensity of reporter dye)/(emission intensity of passive reference dye) in PCR without template or early cycles of a real-time reaction. Ct = threshold cycle, i.e., cycle at which a statistically significant increase in ΔRn is first detected.
Using the Qiagen DNeasy kit, two 96-well blocks can be processed in 1 day for a total of 192 DNA extractions, compared to about 40 isolates per day using the salt method. After the DNA was extracted, conventional PCR and gel electrophoresis using one gel box required a full day to obtain results for 40 samples. Because the gel electrophoresis step is eliminated with the ABI Prism 7700 sequencing detector, the TaqMan PCR and analysis can be performed on DNA samples in a 96-well plate in 2 h.
The cost for materials and reagents for DNA extraction with the Qiagen DNeasy kit was approximately $2.00 per sample, and the cost of reagents for the TaqMan procedure was $2.90 per sample, for a total cost per isolate of $4.90 (excluding labor). The list price for the ABI 7700 sequence detector is approximately $95,000.00.
The combination of DNA extraction with the Qiagen DNeasy kit and use of the TaqMan assay kit is well suited to large-scale screening for the mecA gene in staphylococci. In our laboratory, it increased output at least threefold. The TaqMan procedure appears to be much more sensitive than our conventional PCR method for target detection. When using relatively high copy numbers of target DNA as provided by the extraction techniques, low-level cross contamination can be a problem upon repeated entry into a sample. Samples should be aliquoted to avoid this, and samples that are positive in plate read mode with a real-time Ct value of >30 should be reevaluated.
ACKNOWLEDGMENTS
We thank Susie Hubert for providing strains and Chris Steward for helpful discussions.
REFERENCES
- 1.Aires de Sousa M, Sanches I S, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayliffe G A J. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24(Suppl. 1):S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 3.Boyce J M. Methicillin-resistant Staphylococcus aureus in hospitals and longterm care facilities: microbiology, epidemiology, and preventive measures. Infect Control Hosp Epidemiol. 1992;13:725–737. doi: 10.1086/648346. [DOI] [PubMed] [Google Scholar]
- 4.Brandt M E, Padhye A A, Mayer L W, Holloway B P. Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus. J Clin Microbiol. 1998;36:2057–2062. doi: 10.1128/jcm.36.7.2057-2062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cockerill F R, III, Hughes J G, Vetter E A, Mueller R A, Weaver A L, Ilstrup D M, Rosenblatt J E, Wilson W R. Analysis of 281,797 consecutive blood cultures performed over an eight-year period: trends in microorganisms isolated and the value of anaerobic culture of blood. Clin Infect Dis. 1997;24:403–418. doi: 10.1093/clinids/24.3.403. [DOI] [PubMed] [Google Scholar]
- 6.Desjardin L E, Chen Y, Perkins M D, Teixeira L, Cave M D, Eisenbach K D. Comparison of the ABI 7700 System (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment for tuberculosis. J Clin Microbiol. 1998;36:1964–1968. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez M A, de Lancaster H, Linares J, Tomasz A. Spread of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J Clin Microbiol. 1994;32:2081–2087. doi: 10.1128/jcm.32.9.2081-2087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett K D E, Hornung L J, Andersen A A. Rapid detection of the Chlamydiaceae and other families in the order Chlamydiales: three PCR tests. J Clin Microbiol. 1999;37:575–580. doi: 10.1128/jcm.37.3.575-580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins J A, Ezzell J, Hinnebusch J, Shipley M, Henchal E A, Ibrahim M S. 5′ nuclease PCR assay to detect Yersinia pestis. J Clin Microbiol. 1998;36:2284–2288. doi: 10.1128/jcm.36.8.2284-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Kihara H, Yokota T. Analysis of borderline-resistant strains of methicillin-resistant Staphylococcus aureus using polymerase chain reaction. Microbiol Immunol. 1992;36:445–453. doi: 10.1111/j.1348-0421.1992.tb02043.x. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 12.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi N, Wu H, Kajima K, Taniguchi K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect. 1994;113:259–266. doi: 10.1017/s0950268800051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laue T, Emmerich P, Schmitz H. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan Automated Amplification System. J Clin Microbiol. 1999;37:2543–2547. doi: 10.1128/jcm.37.8.2543-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 16.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris T, Robertson B, Gallagher M. Rapid reverse transcription PCR detection of hepatitis C virus RNA in serum by using the TaqMan fluorogenic detection system. J Clin Microbiol. 1996;34:2933–2936. doi: 10.1128/jcm.34.12.2933-2936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) report, data summary from October 1986–April 1998, issued June 1998. Am J Infect Control. 1998;26:522–533. doi: 10.1016/s0196-6553(98)70026-4. [DOI] [PubMed] [Google Scholar]
- 19.Pusteria N, Hunder J B, Leutenegger C M, Braun U, Madigan J E, Lutz H. Quantitative real-time PCR for detection of members of the Ehrlichia phagocytophila genogroup in host animals and Ixodes ricinus ticks. J Clin Microbiol. 1999;37:1329–1331. doi: 10.1128/jcm.37.5.1329-1331.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roman R S, Smith J, Walker M, Byrne S, Ramotar K, Dycjk B, Kabani A, Nicolle L E. Rapid geographic spread of a methicillin-resistant Staphylococcus aureus strain. Clin Infect Dis. 1997;25:698–705. doi: 10.1086/513758. [DOI] [PubMed] [Google Scholar]
- 21.Ryncarz A, Goddard J, Wald A, Huang M-L, Roizman B, Corey L. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J Clin Microbiol. 1999;37:1941–1947. doi: 10.1128/jcm.37.6.1941-1947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, McAllister S A, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unal S, Hoskins J E, Iokowitsch C Y E, Preston D A, Skatrud P E. Detection of methicillin-resistant staphlococci by using the polymerase chain reaction. J Gen Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vannuffel P, Gigi J, Ezzedine H, Vanderman B, Delmee M, Wauters G, Gala J-L. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstein M P, Towns M L, Quartey S M, Mirrett S, Reimer L G, Parmigiani G, Reller L B. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis. 1997;24:584–602. doi: 10.1093/clind/24.4.584. [DOI] [PubMed] [Google Scholar]