Abstract
AF9 (MLLT3) and ENL (MLLT1) are members of the YEATS family (named after the five proteins first shown to contain this domain: Yaf9, ENL, AF9, Taf14, Sas5) defined by the presence of a YEATS domain. The YEATS domain is an epigenetic reader that binds to acetylated and crotonylated lysines, unlike the bromodomain which can only bind to acetylated lysines. All members of this family have been shown to be components of various complexes with roles in chromatin remodeling, histone modification, histone variant deposition, and transcriptional regulation. MLLT3 is a critical regulator of hematopoiesis with a role in maintaining the hematopoietic stem or progenitor cell (HSPC) population. Approximately 10% of acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL) patients harbor a translocation involving MLL (mixed lineage leukemia). In the context of MLL fusion patients with AML and ALL, MLL-AF9 and MLL-ENL fusions are observed in 34 and 31% of the patients, respectively. The intrinsically disordered C-terminal domain of MLLT3 (AHD, ANC1 homology domain) undergoes coupled binding and folding upon interaction with partner proteins AF4, DOT1L, BCOR, and CBX8. Backbone dynamics studies of the complexes suggest a role for dynamics in function. Inhibitors of the interaction of the intrinsically disordered AHD with partner proteins have been described, highlighting the feasibility of targeting intrinsically disordered regions. MLLT1 undergoes phase separation to enhance recruitment of the super elongation complex (SEC) and drive transcription. Mutations in MLLT1 observed in Wilms tumor patients enhance phase separation and transcription to drive an aberrant gene expression program.
Keywords: MLLT3, MLLT1, AF9, ENL, IDP
Graphical Abstract
Introduction
AF9 (MLLT3, Myeloid/Lymphoid Or Mixed-Lineage Leukemia; Translocated To, 3) and ENL (MLLT1, Myeloid/Lymphoid Or Mixed-Lineage Leukemia; Translocated To, 1) are members of the YEATS family. The YEATS family of proteins (named after the five proteins first shown to contain this domain: Yaf9, ENL, AF9, Taf14, Sas5) is defined by the presence of a conserved domain termed the YEATS domain [1]. In humans, there are four proteins which harbor a YEATS domain: YEATS4 (GAS41), MLLT3 (AF9), MLLT1 (ENL), and YEATS2. The YEATS domain has been shown to be an epigenetic reader that binds to acetylated as well as crotonylated lysines, unlike the bromodomain which can only bind to acetylated lysines. Structure determination of the AF9 (MLLT3) YEATS domain bound to acetylated and crotonylated lysine peptides [2, 3] showed that the YEATS domain has an “end-open” binding site unlike the “side-open” site seen in bromodomains which makes it possible for the YEATS domain to bind to the larger, more sterically demanding crotonyl modification. Structures of the YEATS domains of Taf14 and YEATS2 have also been determined [4–6]. Interestingly, whereas the YEATS domain of MLLT3 binds to crotonylated H3K9, H3K18, and H3K27, the YEATS domain of YEATS2 only binds crotonylated H3K27 and, unlike MLLT3, can accommodate binding of a 2-hydroxyisobutyryl (Khib) modification. All members of this family have been shown to be components of various complexes with roles in chromatin remodeling, histone modification, histone variant deposition, and transcriptional regulation. Indeed, both MLLT3 and MLLT1 have been shown to be integral components of the super elongation complex (SEC) [7], the AF4-ENL-P-TEFb complex (AEP) [8], and the DOT1L complex (DotCom) [9], critical regulators of transcription via phosphorylation of Pol II to facilitate productive elongation (SEC, AEP) and H3K79 methylation to maintain gene expression (DotCom).
MLLT3 is a critical regulator of hematopoiesis. Enver and co-workers first showed that MLLT3 is critical for the development of the erythroid/megakaryocyte lineage [10]. More recently, Mikkola and co-workers have clearly delineated a critical role for MLLT3 in maintaining the hematopoietic stem or progenitor cell (HSPC) population [11]. MLLT3 was localized to active promoters, enhanced levels of H3K79 methylation, and maintained a gene expression program essential for HSPCs. Daley and co-workers applied a network biology approach to identify key regulators of critical hubs for HSPCs [12], resulting in the identification of MLLT3 as a key regulator. Interestingly, loss of the homolog MLLT1 did not impact hematopoietic stem cell function, but has been shown to be essential for MLL fusion leukemia [13, 14].
The mixed lineage leukemia (MLL) protein is a histone methyltransferase that writes the histone H3 lysine 4 trimethyl (H3K4me3) mark at the promoters of target genes such as HOXA9 and MEIS1. Chromosomal translocations involving MLL lead to acute myeloid and lymphoid leukemias (AML and ALL, respectively) characterized by poor prognoses [15]. Transcriptional activation by the MLL fusion proteins is mediated by recruitment of the AEP (AF4 family/ENL family/P-TEFb) and DotCom (DOT1L–AF10 family–ENL family) complexes [8, 16]. Transcriptional activation by AF4 recruitment and transcriptional maintenance by DOT1L recruitment have been shown to be essential for MLL fusions to drive leukemia [16–18]. While more than 90 partners have been observed in MLL fusions, members of the AEP complex account for nearly 70% of MLL rearrangements [19]. These fusions constitutively activate MLL targets [8] by bypassing regulated recruitment via ENL (MLLT1) and AF9 (MLLT3) YEATS domain binding to histone H3 [2, 3]. Approximately 10% of acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL) patients harbor a translocation involving MLL [20]. MLL-AF9 and MLL-ENL fusions are observed in 34 and 31% of the patients, respectively [19]. Additionally, mutations in the YEATS domain of MLLT1 have been shown to be functionally relevant in Wilms tumor, the most common pediatric kidney tumor [21].
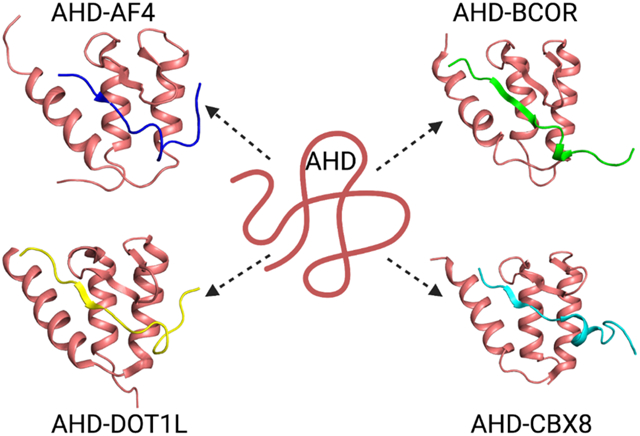
ANC1 (nuclear anchorage protein) homology domain (AHD) of MLLT3 is an intrinsically disordered protein that binds to multiple partners
PONDR analysis of both MLLT3 and MLLT1 shows predicted regions of order at the N-and C-termini of the proteins with a long intervening stretch that is predicted to be disordered. The N-terminal ordered region coincides with the YEATS domain of both proteins. The C-terminal region displays homology between the two proteins (80% identical), suggesting a functional role. Indeed, this C-terminal region, referred to as the ANC1 (nuclear anchorage protein) homology domain (AHD), mediates interactions with other proteins. The MLLT3 AHD recruits AF4 and DOT1L [22, 23], which support transcriptional activation, as well as the BCL6 corepressor (BCOR) and chromobox homolog 8 (CBX8) [24, 25], which are implicated in transcriptional repression. BCOR and CBX8 are members of variant polycomb repressive complex 1 (PRC1) complexes distinguished by their polycomb group RING finger (PCGF) components. As this C-terminal domain is the portion of MLLT3 and MLLT1 that is fused to MLL in the MLL-AF9 and MLL-ENL fusion protein drivers of leukemia, the protein-protein interactions mediated by these domains are relevant to MLL-AF9 and MLL-ENL leukemia. Based on this, we pursued characterization of the interactions of this domain as well as structure determination of the relevant complexes.
We expressed and purified the AHD from MLLT3 via fusion to an MBP tag, as the domain has limited solubility and was prone to proteolytic degradation. The MBP tag was cleaved off for biophysical studies. 15N-1H HSQC spectra of the isolated domain showed very few peaks and the peaks were collapsed to the middle of the spectrum, suggesting the domain was not folded (Figure 1A) [26]. To confirm this, CD spectra were recorded for the isolated domain at lower concentration (Figure 1B), which showed very little secondary structure present with the exception of some limited β content. This data confirmed that the MLLT3 AHD is an intrinsically disordered protein (IDP). Upon addition of a peptide corresponding to the AF4 interaction motif, the 15N-1H HSQC spectrum now shows the requisite number of peaks and very good dispersion (Figure 1C). Furthermore, CD spectroscopy of the AHD – AF4 peptide complex show the presence of both β-strand and α-helix secondary structure (Figure 1B), in stark contrast to the isolated domain. Thus, upon binding to partners the AHD undergoes a coupled binding and folding event which has been observed for a number of other IDPs [27–31]. As described earlier, the AHD has been shown to mediate interactions with additional partners (DOT1L, CBX8, BCOR). In all 4 cases, the regions in the partner proteins mediating interaction with MLLT3 are predicted to be in intrinsically disordered regions of the proteins. To explore these additional interactions, we used a bicistronic vector to co-express the AHD and selected regions of the partner proteins. In all 3 cases, we again observe well dispersed 15N-1H HSQC spectra of the resulting complexes indicative of well folded complexes [26]. In addition, the positions of the AHD residues in the spectrum is quite similar among all four complexes, indicative of significant structural homology which was confirmed by structure determinations of all 4 complexes (see below). As the AHDs of MLLT3 and MLLT1 are highly homologous (80% identical), any functional differences between the two are likely to be relevant to the differing functional roles of the two proteins as well as the phenotypic differences observed between MLL-AF9 and MLL-ENL driven leukemias. To that end, we have measured the binding of the AHDs from both proteins to binding regions from all four partner proteins using fluorescence anisotropy measurements of the binding of MBP-AHD to fluorescein-labeled peptides from each of the partners (Figure 2A) [32]. Interestingly, whereas the affinity for AF4 and DOT1L was similar, MLLT1 displays a substantially higher affinity for the corepressors CBX8 and BCOR than MLLT3, likely providing a rationale for the differing phenotypic behaviors of MLL-AF9 and MLL-ENL leukemia.
Figure 1. The MLLT3 AHD Is Intrinsically Disordered and Folds upon Binding to AF4.
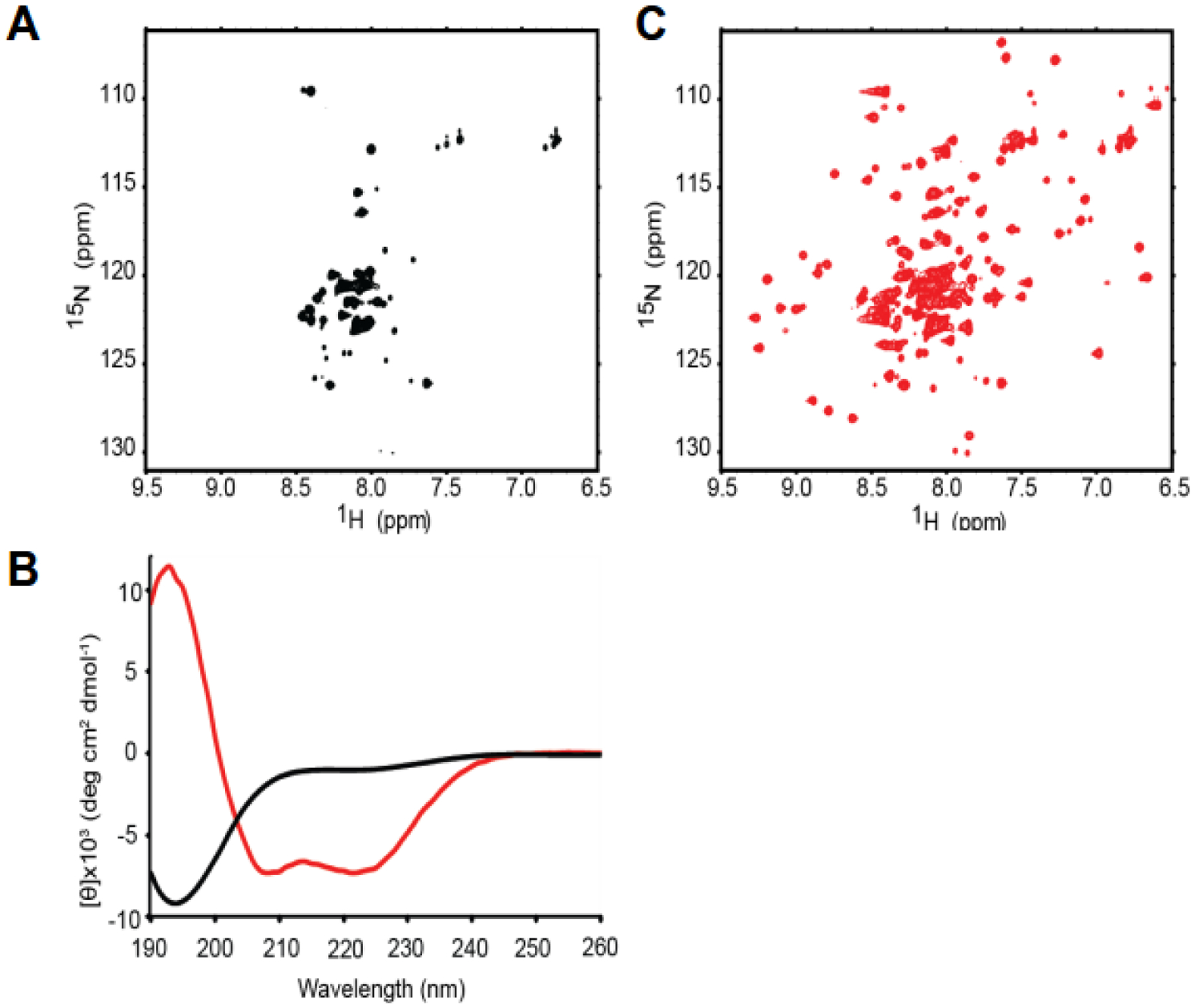
(A) 15N-1H HSQC spectrum of AF9 490–568 alone.
(B) Far-UV Circular dichroism spectrum of AF9 490–568 alone (black) and AF9 490–568 + AF4 761–774 (red).
(C) 15N-1H HSQC spectrum of AF9 490–568 + unlabeled AF4 761–774.
Data from reference 26.
Figure 2. Structures of MLLT3 AHD - AF4, DOT1L, BCOR, CBX8 complexes and binding affinity.
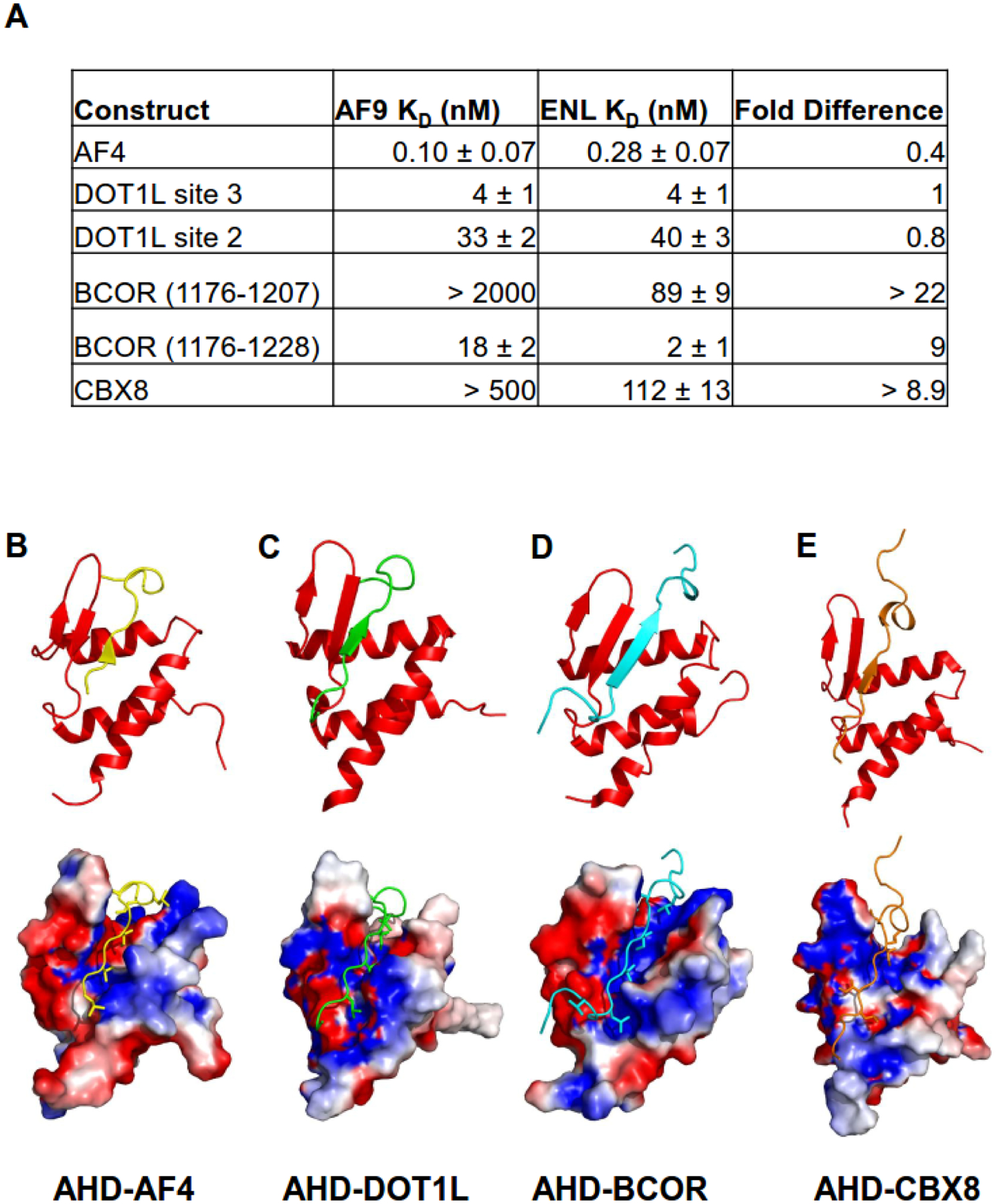
(A) Results of fluorescence polarization based measurements of the binding of the MLLT3 and MLLT1 AHDs to AF4, DOT1L, BCOR, and CBX8 peptides.
(B–E) Ribbon representations (top) and surface representations (bottom) colored according to electrostatic potential with bound peptides indicated for the (B) AF9-AF4 (PDB code 2LM0), (C) AF9-DOT1L (PDB code 2MV7), (D) AF9-BCOR (PDB code 6B7G), and (E) AF9-CBX8 (PDB code 2N4Q) complexes.
Structures of MLLT3 AHD - partner complexes (AF4, DOT1L, CBX8, BCOR)
Using NMR spectroscopy, we solved the structure of the MLLT3 AHD-AF4 complex [26]. The MLLT3 AHD-AF4 complex forms a mixed α-β structure where the interacting residues from AF4 make critical contributions to the formation of the hydrophobic core of the complex (Figure 2B). This fold was novel at the time, as a Dali search did not yield any structural homologs. A β-hairpin formed by MLLT3 residues 535–546 is the likely source of the β secondary structure observed in the CD spectrum of the isolated AHD. This β-hairpin combines with AF4 residues 761–766 to form a three-stranded antiparallel β sheet. The remainder of the AHD folds around the AF4 peptide in the form of 3 α-helices. Following the AF4 β-strand is a conserved LXXL motif (L767–L770) which forms a turn and packs behind the end of the hairpin to form a key hydrophobic cluster. The last four structured residues (771–775) of AF4 make contacts with the MLLT3 AHD, although these are not as intimate as in the remainder of the peptide. The interaction region between AF4 and the MLLT3 AHD is extensive and hydrophobic. A number of aliphatic residues from AF4 are observed to be deeply buried in the hydrophobic core (Figure 2B). In particular, V763 and I765 pack into the interface of the hairpin and the α1 and α3 helices and appear to stabilize the tertiary fold of the complex. An electrostatic interaction between AF4 K764 and MLLT3 D544 likely contributes to the specificity of AF4 binding. The extensive hydrophobic interface between AF4 and MLLT3 AHD provides an explanation for how the significant entropic cost of folding of the complex can be overcome upon binding. Clearly, the hydrophobic core of MLLT3 AHD is not sufficiently extensive to maintain an independently folded structure, providing a rationale for its IDP behavior.
Subsequent work by our lab led to the determination of the structures of the MLLT3 AHD bound to peptides from DOT1L, CBX8, and BCOR [18, 32]. As shown in Figure 2C–E, the structures are very similar with the partners binding in the same site on the structure in all 4 cases. This is consistent with previous biochemical studies which had suggested the partners bind mutually exclusively [8, 25]. The mechanism of exchange between different partners is not known at this time, but it is interesting to note that in cases where intrinsically disordered proteins (IDPs) compete for binding to a common site, coupled folding and binding may allow one IDP to displace another without its prior dissociation, facilitating rapid exchange between high affinity partners [33]. Comparison of the binding constants for the 4 partner proteins to the MLLT3 AHD (Figure 2A) shows that binding to AF4, DOT1L, and BCOR is high affinity whereas that to CBX8 is significantly weaker. There is not a great deal of sequence identity among the binding motifs for the different partners with the exception of the conservation of hydrophobic residues at specific positions, all of whom make specific contacts with the MLLT3 AHD. Interestingly, CBX8 deviates from this at the second conserved position with an Ala at this position (A335) whereas all 3 other partners have a Val at this position. Indeed, mutation of this Ala to Val resulted in a KD of 12 nM [32], consistent with the other high affinity interactions.
Dynamics of MLLT3 AHD - partner complexes could have functional implications
The MLLT3 AHD binds to 4 different partner proteins which begs the question how the exchange process between partners occurs. Clearly, the selection of binding partners will be dependent to a large degree on the local effective concentrations of the partner proteins at specific sites in the genome. However, with the high affinities observed for binding to AF4, DOT1L, and BCOR in particular, it would be expected that the half-lives of such complexes could be quite long and potentially too long to make exchange among partners possible on a functionally useful timescale. As mentioned above, in cases where intrinsically disordered proteins (IDPs) compete for binding to a common site, coupled folding and binding may allow one IDP to displace another without its prior dissociation, facilitating rapid exchange between high affinity partners [33]. However, to do so likely requires dynamic behavior of the protein that can lower the activation barrier for this to occur. A protein that is dynamically “quiet” is unlikely to be capable of such behavior. Indeed, such increased dynamic behavior, particularly at the interface, has been demonstrated in a molecular dynamics (MD) study of another IDP complex [34].To probe the backbone dynamics of the MLLT3 AHD complexes with AF4, CBX8, and BCOR, we recorded data to delineate 15N{1H} heteronuclear NOE, 15N transverse relaxation rates (R2), and 15N longitudinal relaxation rates (R1). In order to analyze backbone dynamics, we calculated the product of R1 and R2, which cancels effects of anisotropic motions on nuclear relaxation and allows direct evaluation of both slow and fast timescale motions [35]. Conformational exchange on the μs-ms timescale results in elevation of the R1*R2 product, whereas fast exchange (ps-ns), results in decreased values of R1*R2. R1*R2 data for all four complexes is shown in Figure 3.
Figure 3. Backbone dynamics of the MLLT3 AHD - AF4, DOT1L, BCOR, CBX8 complexes measured by NMR spectroscopy.
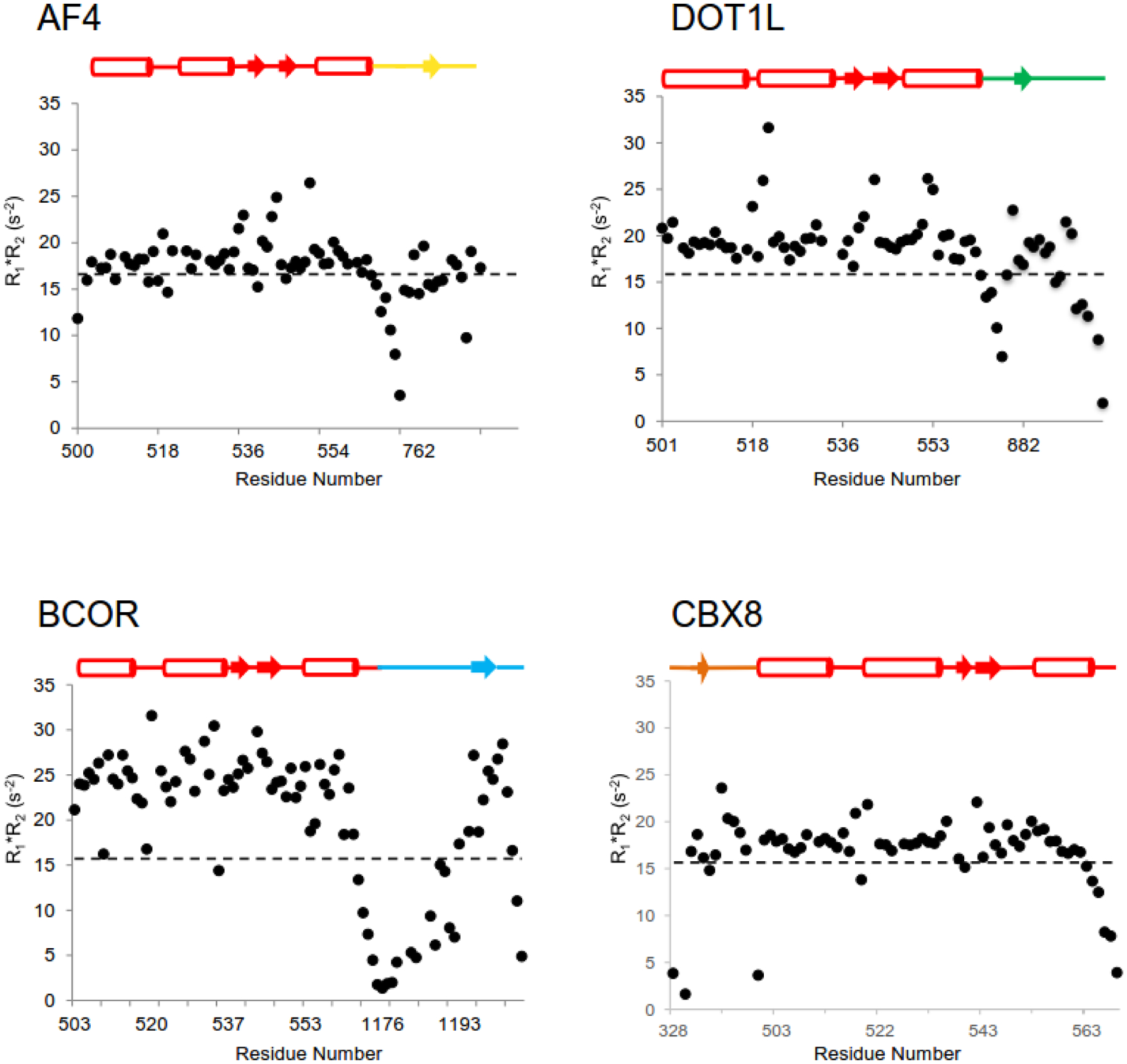
15N R1*R2 values are plotted for each of the four complexes. The schematic above each plot indicates the secondary structure of the AHD as well as the location of data for the AHD and the binding partner in each plot. The dashed line represents the expected value at 14.1 T field strength in the absence of chemical exchange using S2=0.87.
We observed that loops of the MLLT3 AHD that are located near the AF4 peptide were undergoing conformational exchange [26]. One of these is the loop formed by residues 539–542 at the tip of the hairpin which is located next to the turn formed by AF4 residues 767–770. A second is the preceding loop formed by residues 531–535 which makes contacts with C-terminal residues of AF4. Interestingly, the β-strand formed by MLLT3 residues 761–765 and parts of the helix adjacent to it exhibit dynamic behavior on a fast (nanosecond to picosecond) time scale, which has been shown to correlate with side chain conformational entropy [36]. This dynamic behavior of the AHD for residues in proximity to the AF4 peptide is likely to play a functional role in the ability of the AHD to exchange partners. Indeed, MD studies have suggested that IDPs facilitate rapid dissociation [37], a property that is likely to be relevant for AHD partner exchange. Notably, we observe that the methyl resonances in the core of the protein, particularly those adjacent to the aromatic rings of MLLT3 F543 and F545, show surprisingly poor chemical shift dispersion, indicative of internal dynamics within the hydrophobic core. Thermal equilibrium unfolding measurements monitored by far UV CD spectroscopy showed the MLLT3 AHD-AF4 complex unfolds with a broad transition, quite different from the highly cooperative transitions observed for rigid domains with stably packed hydrophobic cores. These data suggest that aliphatic residues at the AF4-MLLT3 AHD interface retain a significant amount of conformational entropy, which may partially compensate for the loss of conformational entropy during coupled folding and binding and may also be required for dynamic exchange between binding partners. Figure 3 also shows R1*R2 data for the DOT1L, CBX8, and BCOR complexes. To an even greater extent than seen for the AF4 complex, these complexes show evidence of significant conformational exchange, particularly for the BCOR complex which may be a product of the much weaker binding affinity of BCOR (1176–1207) used for the structural and dynamics studies. One intriguing question from this data is whether the different dynamic behavior of the complexes makes specific orders of partner exchange more rapid than others, an effect that could have significant functional implications. For example, does the increased conformational exchange of the corepressor complexes (CBX8, BCOR) versus the activator complex (AF4) suggest exchange of CBX8 or BCOR for AF4 (perhaps DOT1L) is preferred? Such a scenario would be useful in settings where rapid transcriptional upregulation is required to respond to environmental alterations or differentiation cues. Studies of the kinetics of binding of partners to various complexes has the potential to address these interesting questions. Studies of such effects in cells will require a facile way to initiate the process of exchange in a synchronized manner.
The BCOR complex presents clear evidence of functioning as a fuzzy complex [38]. Our studies of this complex [32] showed that a somewhat longer BCOR peptide binds with high affinity to the MLLT3 AHD (18 nM) whereas a shorter peptide in line with the length used for the other partners binds with much weaker affinity (>2000 nM) (Figure 2A). However, NMR spectra of the MLLT3 AHD complexed to the longer peptide are missing a large fraction of the peaks expected in the spectrum, likely due to exchange broadening. However, CD spectroscopy comparison of the short and long BCOR peptide complexes indicates the additional portion of BCOR in the longer complex has α-helical secondary structure. Thus, on average this region is helical and interacting with the AHD, however the interaction is fuzzy despite the substantial binding energy being contributed by this region. Such behavior has been linked to frustration of a more ordered complex that makes the fuzzy complex more energetically favorable [39]. In addition, such behavior is likely to contribute to the specificity of such interactions. We compared 15N-1H HSQC spectra of the short and long BCOR complexes to gain insight into where on the AHD this fuzzy region may be interacting. The residue with the largest chemical shift change between the two complexes was E531, indicating this residue is at the interface with the fuzzy BCOR region. Indeed, we introduced a charge reversal mutation at this site (E531R) and showed this reduced the affinity of BCOR binding more than 167-fold (KD > 3,000 nM). Interestingly, this mutation had little to no effect on the binding of the other partners, so it appears to be unique for the BCOR interaction. The functional significance of this fuzzy complex behavior remains to be elucidated.
MLLT3 AHD-partner interactions are essential for MLL-AF9 leukemia
The determination of the structures of the MLLT3 AHD bound to AF4, DOT1L, CBX8, and BCOR provided the foundation to develop point mutations that could selectively, or at least preferentially, disrupt specific interactions and thereby provide insights into their role in driving MLL-AF9 leukemia. This effort is confounded by the binding of all the partners to the same site on the MLLT3 AHD. The use of these mutants of the AHD made it possible to show that recruitment of AF4 and DOT1L played a critical role in MLL-AF9 driven leukemia and that there was a specific subset of MLL-AF9 target genes where loss of DOT1L binding altered the H3K79 methylation profile [17, 18]. It is worth noting that the mutants used for these studies also impact BCOR binding, so their effects are not solely on AF4 and DOT1L binding. A subsequent study by Nikolovska-Coleska and co-workers also showed a critical role for DOT1L recruitment in MLL-AF9 fusion leukemia as well as demonstrating that loss of MLLT3 binding to DOT1L did not impact normal hematopoiesis, establishing this protein-protein interaction as a valid target for inhibitor development [40]. For the binding of CBX8, we used a mutation in CBX8 to probe the role of its recruitment and showed it does not impact MLL-AF9 driven transformation. For the BCOR interaction, as described above, we identified a point mutation on the AHD which interacts with the fuzzy complex portion of BCOR and used this to show that direct recruitment of BCOR was essential for MLL-AF9 driven leukemia largely via alteration of MYC levels [32]. All of this data highlighted the critical role these protein-protein interactions play in driving MLL-AF9 leukemia and validate them as potential targets for drug development to treat MLL-AF9 leukemia.
Inhibitor development targeting the MLLT3 AHD
The idea of targeting an IDP such as the MLLT3 AHD for inhibitor development is likely to be viewed with significant skepticism. Certainly, the concept of a small molecule binding with specificity to an unstructured protein is a strong departure from traditional concepts of pharmacology. However, there is good evidence that this skepticism may be misplaced. Certainly, the opportunity to drug the large part of the proteome that is intrinsically disordered makes such efforts highly relevant. Indeed, 79% of cancer associated proteins and 57% of cardiovascular disease associated proteins contain significant regions of intrinsic disorder [41, 42]. A recent computational analysis of ligandable cavities of IDPs used the structures of IDP complexes to assess their druggability [43]. Strikingly, their findings indicate that IDPs are predicted to have more binding cavities than structured proteins of a similar length. The MLLT3 AHD was one of fourteen IDPs the authors identified as having druggable cavities, with the MLLT3 AHD displaying five such druggable cavities, supporting the concept that it may well be possible to develop inhibitors targeting this IDP.
Nikolovska-Coleska and co-workers took a peptidomimetic approach to the development of inhibitors of AHD-partner binding [44]. They identified a 7 mer peptide derived from the most potent of the DOT1L binding motifs that has a KI of 160 nM in a fluorescence polarization based peptide displacement assay with the MLLT3 AHD. Using the structure of the DOT1L – AHD complex [18] and systematic variation of three portions of the peptide, they were able to derive a peptidomimetic inhibitor with a KI of 20 nM in the FP assay, similar to the 19 nM KD observed for a 10 mer peptide derived from the highest affinity DOT1L binding motif. Using cell lysates, they also showed disruption of the protein-protein interaction with endogenous full-length proteins. No cellular activity for this compound was reported. The large size (> 800 Da) and highly peptide-like nature of the compound may make it challenging to achieve cellular and in vivo efficacy. However, the clear structure-activity relationships observed and the ability to develop a potent inhibitor of this protein-protein interaction certainly supports the notion that it is druggable. Furthermore, as we have shown that interaction with AF4, DOT1L, and BCOR is critical for the activity of MLL-AF9 fusion proteins, such an approach to disrupt all of these interactions is well-validated.
MLLT1 undergoes phase separation to facilitate transcriptional activation – basis for role of MLLT1 mutations in Wilms tumor
A recent study by Lin and co-workers elucidated a role for MLLT1 mediated phase separation in regulation of transcriptional elongation via concentration of the super elongation complex (SEC) to enhance transcriptional elongation [45]. Promoter proximal pausing of Pol II is used by cells to effect rapid but synchronous expression of genes in response to specific stimuli [46, 47]. Release from this state into productive elongation is driven by P-TEFb (complex of CDK9 and CCNT1) mediated phosphorylation of Pol II [48, 49]. The authors showed that MLLT1 colocalizes with phase separated SEC component AFF4. They further showed that the intrinsically disordered regions (IDRs) of both AFF4 as well as MLLT1 can mediate phase separation. Importantly, they showed that the MLLT1 IDR could concentrate CDK9 in live cells, providing a phase separation based mechanism for the Pol II phosphorylation necessary for productive transcriptional elongation. Indeed, knockdown of MLLT1 reduced the rapid formation of CDK9 puncta at the FOS gene and reduced the transcriptional induction rate of the FOS gene. Only full length MLLT1 could drive FOS expression. A deletion mutant lacking the IDR was unable to do so. Interestingly, they also found that the MLL-ENL fusion protein resulted in an increase in the number of nuclear puncta harboring SEC components, possibly providing a phase separation based rationale for the increased transcription seen at specific MLL-ENL target genes. A recent study found that the YEATS family member Taf14 also undergoes phase separation [50], suggesting this may be a property shared by all the YEATS family members.
As mentioned above, mutations in the YEATS domain of MLLT1 are found in Wilms tumor patients, the most common pediatric kidney cancer. Allis and co-workers examined the effect of these mutations on transcription [21]. They found increased occupancy of these mutant forms of MLLT1 at a set of critical developmental genes including members of the HOX cluster. The location of the mutations in the YEATS domain is outside the site where acylated histone peptides bind and they showed these mutations did not affect binding of acylated histone peptides. They hypothesized that the mutations may have a role in self-association of MLLT1 to generate the higher occupancy they observed at select genes. Indeed, expression of fluorescently tagged MLLT1 showed the mutant proteins form discrete puncta in the nucleus whereas the wildtype protein did not display this behavior. Elegant imaging studies subsequently showed that the puncta formed by the mutant forms of MLLT1 were spherical, undergo fusion on contact, and are highly dynamic, i.e. they display all the hallmarks of a phase separation driven condensate [51–53]. Deletion of the intrinsically disordered region between the YEATS domain and the AHD reduced the ability of mutant MLLT1 proteins to undergo self-association, identifying a critical role for this region in the self-association process. In contrast, deletion of the AHD in the mutant MLLT1 proteins had a limited effect on self-association but had profound effects on gene expression, consistent with the recruitment of AF4 and DOT1L by this domain. This study clearly highlights the disease relevance of altered phase separation (Figure 4), a phenomenon that likely has implications in many disease settings.
Figure 4.
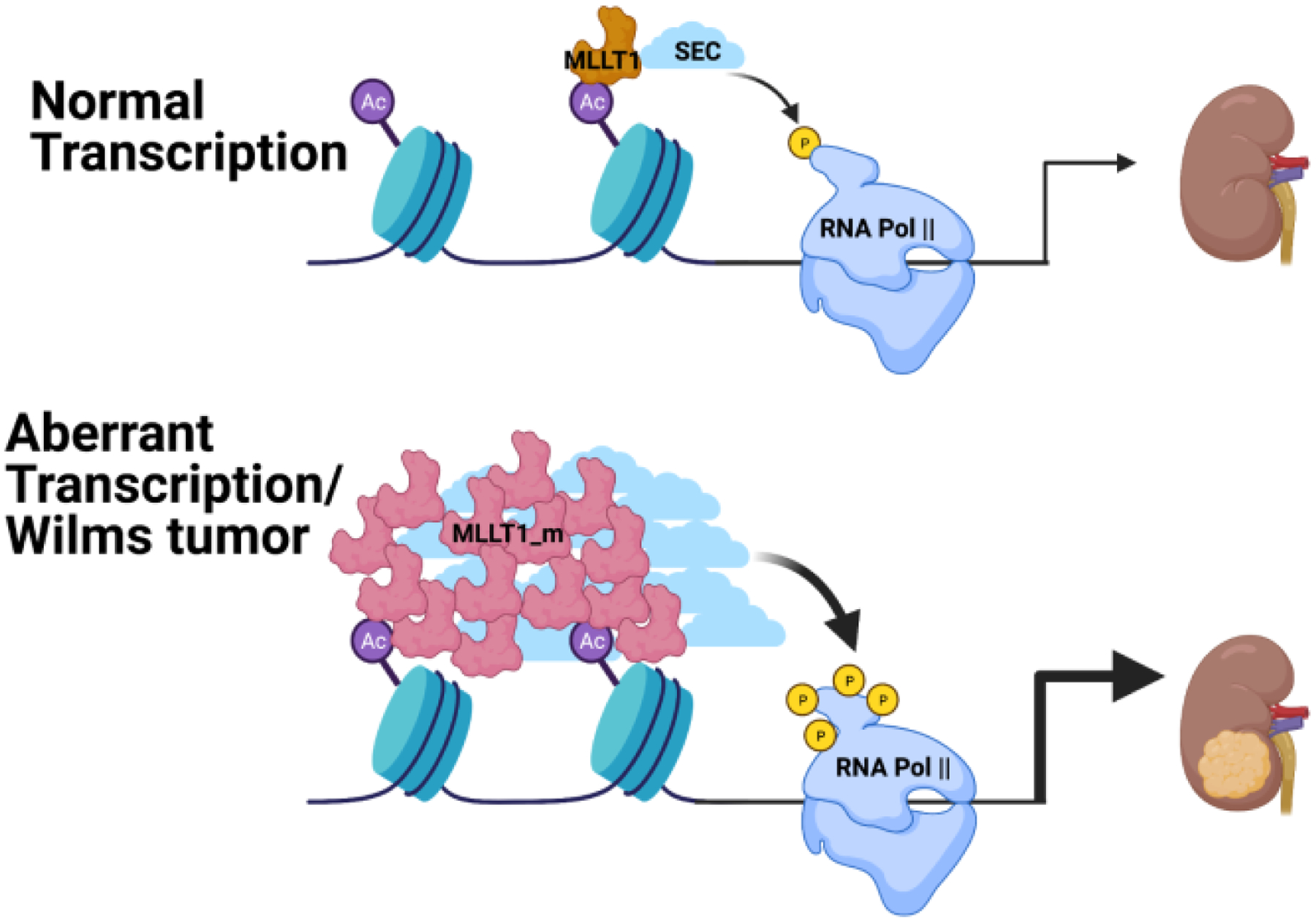
Schematic representation of the functional effects of MLLT1 mutations that enhance self-association on transcription in Wilms tumor.
Conclusions and Perspectives
The functional role of MLLT3 and MLLT1 in regulation of gene transcription remains an active area of research with many open questions. Furthermore, the roles of the AHD of MLLT3 and MLLT1 in MLL-AF9 and MLL-ENL fusion proteins, which harbor this domain, is also an active area of research with many mechanistic questions remaining to be elucidated. The roles of the intrinsically disordered AHD in mediating critical protein-protein interactions and of the intrinsically disordered mid-region of MLLT1 (and presumably MLLT3 as well) in mediating phase separation clearly highlight the functional importance of these intrinsically disordered regions in function. This represents a far cry from the not so distant view that such regions only serve as flexible linkers with no inherent function themselves.
Several YEATS family members have now been shown to engage in phase separation. Whether this behavior is found in other members, MLLT3 for example, and how that behavior modulates function remains an open question. Recent studies that highlight the critical role of clusters of aromatic residues in phase separation of IDRs [54] is likely to provide the knowledge to manipulate this phase behavior with a minimal set of mutations and thereby provide the high quality biological reagents necessary to explore the functional results more effectively. Indeed, as pointed out by Tijan and co-workers, the characterization of such liquid-liquid phase separation versus other possible mechanisms for compartmentalization is still an evolving field and requires careful studies of proteins expressed at endogenous levels in cells using state-of-the-art imaging approaches to achieve a robust assessment [55]. Interestingly, we noted in our studies of the interaction of DOT1L with the MLLT3 AHD that there are 3 binding sites for the AHD in DOT1L [18]. Such polyvalent interaction could assist in the nucleation necessary to drive phase separation. The AHD of MLLT3 and MLLT1 interacts with multiple partner proteins, most of whom bind with high affinity. We have observed that different partner interactions seem to have effects at specific subsets of MLL-AF9 target genes that largely don’t overlap with each other. The manner in which this multi-modal switch is bound to specific partners at specific sites and what the functional effects of this are remains an open question. Furthermore, with the high affinity of the interactions, there may be a question of how the domain can exchange partners on a functionally relevant time scale, whether partners can enhance the dissociation of one another, and whether there may be kinetically preferable pathways for such exchange events that help to define the order of events, if there is one. Finally, our results have clearly demonstrated that the protein-protein interactions between the AHD of MLL-AF9 and AF4, DOT1L, and BCOR is a valid therapeutic target. Certainly, the targeting of IDPs challenges our conventional concepts about drug-protein interaction, but emerging data strongly suggests such targeting may well be possible. The possibility of opening up the very large fraction of the proteome which is intrinsically disordered to drug development certainly justifies significant effort in this area. Drug development in this realm certainly presents numerous challenges, not the least of which is how to effectively validate binding of compounds to such regions. The recent emergence of 13C- and 15N-direct detection based NMR methods [56, 57] provides a potential path forward. The observation of meaningful chemical shift perturbations by a compound in such spectra would provide the robust validation data needed to pursue an optimization effort on an initial lead compound. The degree to which such optimization efforts can derive a potent inhibitor remains to be demonstrated.
Research Highlights:
MLLT3 (AF9) and MLLT1 (ENL) mediate multiple mutually exclusive protein-protein interactions via coupled binding and folding of their intrinsically disordered C-terminal domains (AHD).
Fusion proteins (MLL-AF9 and MLL-ENL) with MLL are drivers of leukemia via the protein-protein interactions of the AF9 and ENL AHD portions of the fusions.
Inhibitors of the intrinsically disordered AF9 AHD protein-protein interactions have been developed.
Mutations in MLLT1 (ENL) drive Wilms tumor via enhanced phase separation and transcription at target genes.
Acknowledgements
The work described was supported by grants from the NCI (R01 CA155328 and R01 CA233749; to N.J. Zeleznik-Le and J.H. Bushweller) and an ASH Bridge Grant (to N.J. Zeleznik-Le). The 800 MHz NMR spectrometer at the University of Virginia used in these studies was purchased with funds from NIH S10RR023035.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:65–75. [DOI] [PubMed] [Google Scholar]
- [2].Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159:558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Molecular cell. 2016;62:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang W, Zhang J, Zhang X, Xu C, Tu X. Solution structure of the Taf14 YEATS domain and its roles in cell growth of Saccharomyces cerevisiae. The Biochemical journal. 2011;436:83–90. [DOI] [PubMed] [Google Scholar]
- [5].Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nature chemical biology. 2016;12:396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, et al. YEATS2 is a selective histone crotonylation reader. Cell research. 2016;26:629–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular cell. 2010;37:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer cell. 2010;17:198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes & development. 2010;24:574–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pina C, May G, Soneji S, Hong D, Enver T. MLLT3 regulates early human erythroid and megakaryocytic cell fate. Cell stem cell. 2008;2:264–73. [DOI] [PubMed] [Google Scholar]
- [11].Calvanese V, Nguyen AT, Bolan TJ, Vavilina A, Su T, Lee LK, et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature. 2019;576:281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McKinney-Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, Curran M, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell stem cell. 2012;11:701–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 2017;543:270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature. 2017;543:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meyer C, Schneider B, Jakob S, Strehl S, Attarbaschi A, Schnittger S, et al. The MLL recombinome of acute leukemias. Leukemia. 2006;20:777–84. [DOI] [PubMed] [Google Scholar]
- [16].Okuda H, Stanojevic B, Kanai A, Kawamura T, Takahashi S, Matsui H, et al. Cooperative gene activation by AF4 and DOT1L drives MLL-rearranged leukemia. The Journal of clinical investigation. 2017;127:1918–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lokken AA, Achille NJ, Chang MJ, Lin JJ, Kuntimaddi A, Leach BI, et al. Importance of a specific amino acid pairing for murine MLL leukemias driven by MLLT1/3 or AFF1/4. Leukemia research. 2014;38:1309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kuntimaddi A, Achille NJ, Thorpe J, Lokken AA, Singh R, Hemenway CS, et al. Degree of recruitment of DOT1L to MLL-AF9 defines level of H3K79 Di- and tri-methylation on target genes and transformation potential. Cell reports. 2015;11:808–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Meyer C, Burmeister T, Groger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews Cancer. 2007;7:823–33. [DOI] [PubMed] [Google Scholar]
- [21].Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature. 2020;577:121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Erfurth F, Hemenway CS, de Erkenez AC, Domer PH. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia. 2004;18:92–102. [DOI] [PubMed] [Google Scholar]
- [23].Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. The Journal of biological chemistry. 2006;281:18059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hemenway CS, de Erkenez AC, Gould GC. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene. 2001;20:3798–805. [DOI] [PubMed] [Google Scholar]
- [25].Srinivasan RS, de Erkenez AC, Hemenway CS. The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene. 2003;22:3395–406. [DOI] [PubMed] [Google Scholar]
- [26].Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure. 2013;21:176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–5. [DOI] [PubMed] [Google Scholar]
- [28].Dogan J, Mu X, Engstrom A, Jemth P. The transition state structure for coupled binding and folding of disordered protein domains. Scientific reports. 2013;3:2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Giri R, Morrone A, Toto A, Brunori M, Gianni S. Structure of the transition state for the binding of c-Myb and KIX highlights an unexpected order for a disordered system. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rogers JM, Oleinikovas V, Shammas SL, Wong CT, De Sancho D, Baker CM, et al. Interplay between partner and ligand facilitates the folding and binding of an intrinsically disordered protein. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chemes LB, Sanchez IE, de Prat-Gay G. Kinetic recognition of the retinoblastoma tumor suppressor by a specific protein target. Journal of molecular biology. 2011;412:267–84. [DOI] [PubMed] [Google Scholar]
- [32].Schmidt CR, Achille NJ, Kuntimaddi A, Boulton AM, Leach BI, Zhang S, et al. BCOR Binding to MLL-AF9 Is Essential for Leukemia via Altered EYA1, SIX, and MYC Activity. Blood Cancer Discov. 2020;1:162–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].De Guzman RN, Martinez-Yamout MA, Dyson HJ, Wright PE. Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites. The Journal of biological chemistry. 2004;279:3042–9. [DOI] [PubMed] [Google Scholar]
- [34].Huang Y, Liu Z. Smoothing molecular interactions: the “kinetic buffer” effect of intrinsically disordered proteins. Proteins. 2010;78:3251–9. [DOI] [PubMed] [Google Scholar]
- [35].Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. Journal of the American Chemical Society. 2002;124:1852–3. [DOI] [PubMed] [Google Scholar]
- [36].Gagne SM, Tsuda S, Spyracopoulos L, Kay LE, Sykes BD. Backbone and methyl dynamics of the regulatory domain of troponin C: anisotropic rotational diffusion and contribution of conformational entropy to calcium affinity. Journal of molecular biology. 1998;278:667–86. [DOI] [PubMed] [Google Scholar]
- [37].Umezawa K, Ohnuki J, Higo J, Takano M. Intrinsic disorder accelerates dissociation rather than association. Proteins. 2016;84:1124–33. [DOI] [PubMed] [Google Scholar]
- [38].Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends in biochemical sciences. 2008;33:2–8. [DOI] [PubMed] [Google Scholar]
- [39].Gianni S, Freiberger MI, Jemth P, Ferreiro DU, Wolynes PG, Fuxreiter M. Fuzziness and Frustration in the Energy Landscape of Protein Folding, Function, and Assembly. Accounts of chemical research. 2021;54:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Grigsby SM, Friedman A, Chase J, Waas B, Ropa J, Serio J, et al. Elucidating the Importance of DOT1L Recruitment in MLL-AF9 Leukemia and Hematopoiesis. Cancers (Basel). 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. Journal of molecular biology. 2002;323:573–84. [DOI] [PubMed] [Google Scholar]
- [42].Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45:10448–60. [DOI] [PubMed] [Google Scholar]
- [43].Zhang Y, Cao H, Liu Z. Binding cavities and druggability of intrinsically disordered proteins. Protein science : a publication of the Protein Society. 2015;24:688–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Du L, Grigsby SM, Yao A, Chang Y, Johnson G, Sun H, et al. Peptidomimetics for Targeting Protein-Protein Interactions between DOT1L and MLL Oncofusion Proteins AF9 and ENL. ACS medicinal chemistry letters. 2018;9:895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guo C, Che Z, Yue J, Xie P, Hao S, Xie W, et al. ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Sci Adv. 2020;6:eaay4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes & development. 2011;25:1486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nature reviews Molecular cell biology. 2012;13:543–7. [DOI] [PubMed] [Google Scholar]
- [49].Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annual review of biochemistry. 2012;81:119–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen G, Wang D, Wu B, Yan F, Xue H, Wang Q, et al. Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation. Nature communications. 2020;11:4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology. 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. 2020;367:694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes & development. 2019;33:1619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cook EC, Usher GA, Showalter SA. The Use of (13)C Direct-Detect NMR to Characterize Flexible and Disordered Proteins. Methods in enzymology. 2018;611:81–100. [DOI] [PubMed] [Google Scholar]
- [57].Gibbs EB, Kriwacki RW. Direct detection of carbon and nitrogen nuclei for high-resolution analysis of intrinsically disordered proteins using NMR spectroscopy. Methods. 2018;138–139:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]


