Abstract
Objective:
Tryptophan can be catabolized to various metabolites through host kynurenine and microbial indole pathways. We aimed to examine relationships of host and microbial tryptophan metabolites with incident type 2 diabetes (T2D), host genetics, diet, and gut microbiota.
Method:
We analyzed associations between circulating levels of 11 tryptophan metabolites and incident T2D in 9,180 participants of diverse racial/ethnic backgrounds from five cohorts. We examined host genome-wide variants, dietary intake, and gut microbiome associated with these metabolites.
Results:
Tryptophan, four kynurenine-pathway metabolites (kynurenine, kynurenate, xanthurenate, and quinolinate) and indolelactate were positively associated with T2D risk, while indolepropionate was inversely associated with T2D risk. We identified multiple host genetic variants, dietary factors, gut bacteria and their potential interplay associated with these T2D-relaetd metabolites. Intakes of fiber-rich foods, but not protein/tryptophan-rich foods, were the dietary factors most strongly associated with tryptophan metabolites. The fiber-indolepropionate association was partially explained by indolepropionate-associated gut bacteria, mostly fiber-utilizing Firmicutes. We identified a novel association between a host functional LCT variant (determining lactase persistence) and serum indolepropionate, which might be related to a host gene-diet interaction on gut Bifidobacterium, a probiotic bacterium significantly associated with indolepropionate independent of other fiber-related bacteria. Higher milk intake was associated with higher levels of gut Bifidobacterium and serum indolepropionate only among genetically lactase non-persistent individuals.
Conclusion:
Higher milk intake among lactase non-persistent individuals, and higher fiber intake were associated with a favorable profile of circulating tryptophan metabolites for T2D, potentially through the host-microbial cross-talk shifting tryptophan metabolism toward gut microbial indolepropionate production.
Introduction
Tryptophan is an essential amino acid that plays a critical role in human health and disease.[1] In addition to its role in serotonin and melatonin biosynthesis, tryptophan is the sole source for the kynurenine pathway (Figure S1),[1] in which tryptophan is first catabolized into kynurenine, mainly regulated by indoleamine 2,3-dioxygenase (IDO) and trypophan-2,3-dioxygenase (TDO), and then kynurenine is processed into several downstream metabolites, including kynurenate, xanthurenate, and quinolinate. The kynurenine pathway is involved in immune activation and inflammation regulation,[1] and has been associated with obesity and insulin resistance.[2, 3] In addition, tryptophan can be catabolized by gut microbiota, producing a variety of indole derivatives (e.g., indoleacetate, indolelactate, and indolepropionate) which have been shown to have beneficial effects on host metabolism.[4]
Emerging evidence from animal studies suggests a host-microbiota interaction on tryptophan metabolism which may affect host metabolic health.[5] In mice with genetic deficiency of IDO, tryptophan metabolism may shift from the host kynurenine pathway toward gut microbial indole derivative production, leading to an improvement in insulin sensitivity.[5] In human studies, metabolomics using a broad-spectrum of metabolites found that plasma levels of tryptophan[6] and two kynurenine-pathway metabolites (kynurenate and xanthurenate)[7] were associated with increased risk of type 2 diabetes (T2D), while a microbial metabolite of tryptophan, indolepropionate, was associated with decreased risk of T2D,[8] but relationships of other tryptophan metabolites with T2D remains unclear. Genome-wide association studies (GWAS) of the human blood metabolome identified genetic loci associated with some tryptophan metabolites and many of them might be involved in host tryptophan-kynurenine metabolism or metabolite transportation.[9, 10, 11] Dietary tryptophan is the only source of tryptophan and its catabolites for humans,[1] while several human studies found strong positive associations of fiber-rich food (e.g. fruits and vegetables) and fiber intake with circulating indolepropionate levels.[8, 12, 13] The human gut microbiome might be involved in this relationship but underlying mechanisms remain unclear, since no evidence has shown that indolepropionate can be derived from microbial catabolism of phytochemical compounds or fiber fermentation. A recent study in women reported an association between gut microbiome composition and serum indolepropionate which appeared to be independent of host dietary fiber intake.[14] To the best of our knowledge, no studies have examined host and microbial tryptophan metabolism and T2D integrating data on host genome-wide variants, dietary intake, gut microbiome and circulating levels of both host and microbial tryptophan metabolites. There is a need to integrate different layers of data to identify more relevant associations, and more importantly, potential links among these association signals, which may help better understand host-microbial cross-talk in tryptophan metabolism and its implication in human metabolic health.
In this study, we hereby examined prospective associations between circulating levels of 11 major host and microbial tryptophan metabolites and incident T2D in five epidemiological cohorts of multiple racial/ethnic groups, hypothesizing that kynurenine-pathway metabolites are associated with higher risk of T2D, while microbial indole derivatives are associated with lower risk of T2D. Furthermore, by integrating multi-omics data, we identified host genetic, dietary and gut microbial factors associated with these metabolites.
Methods
Study population
The main study population was the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), with subsequent replication analyses conducted in four additional cohorts of multiple racial/ethnic groups: the Atherosclerosis Risk in Communities Study (ARIC), the Framingham Heart Study (FHS), the Women’s Health Initiative (WHI), and a case-cohort study nested in the Prevención con Dieta Mediterránea Study (PREDIMED) (Table S1). The HCHS/SOL is a population-based cohort that recruited 16,415 Hispanic/Latino adults aged 18–74 years living in 4 US metropolitan areas.[15] A comprehensive battery of interviews and a clinical assessment with fasting blood draw were conducted at in-person clinic visits during 2008–2011 (baseline) and 2014–2017 (Visit 2). Usual dietary intake was estimated using the National Cancer Institute methodology based on two 24-h dietary recalls administered at baseline.[16] The ARIC study enrolled mostly white and black participants aged 45–64 years from four communities in the US in 1987–1989.[17] The FHS was initiated in 1971 and we included FHS participants aged 40 to 65 years who attended the 5th examination (1991–1995).[18] The WHI study was launched in 1993 enrolling US women aged 50–79 years.[19] We also used data from a case-cohort study nested in the PREDIMED study which is a multicenter trial initiated in 2008.[20, 21]
An expanded description of study populations, data collection, and statistical analyses is provided in Online Supplements. Study protocols were approved by the Institutional Review Boards at all participating institutions. All participants gave written informed consent.
Patient and Public Involvement
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research
Ascertainment of incident T2D
In all studies, participants free of diabetes at baseline who met at least one of the following criteria during follow-up visits or telephone interviews were defined as incident T2D cases: fasting time >8 hours and fasting glucose ≥7.0 mmol/L (126 mg/dL), fasting ≤8 hours and non-fasting glucose ≥11.1 mmol/L (200 mg/dL), post-OGTT glucose ≥11.1 mmol/L (200 mg/dL), HbA1C ≥6.5%, treatment with anti-diabetic medications, or self-reported physician-diagnosed diabetes.
Metabolomic profiling
In HCHS/SOL, serum metabolomic profiling was performed using the discoveryHD4 platform at Metabolon Inc. (Durham, NC) in 3,972 participants randomly selected from the whole cohort at baseline.[22] Eleven tryptophan metabolites, including tryptophan, serotonin, five kynurenine-pathway metabolites (kynurenine, kynurenate, xanthurenate, quinolinate, and picolinate), and four indole derivatives (indoleacetate, indolelactate, indolepropionate and indoxyl sulfate) (Figure S1), were captured by an untargeted liquid chromatography-mass spectrometry (LC-MS) approach. In ARIC, seven tryptophan metabolites were available in the baseline serum metabolomics data measured by a similar LC-MS approach at Metabolon Inc.[17] In other studies, baseline plasma tryptophan metabolites (eight in FHS; five in PREDIMED; seven in WHI) were measured using LC-MS approaches at the Broad Institute (Cambridge, MA).[18, 19, 21] Metabolomic approaches at both Metabolon Inc. and the Broad Institute are semiquantitative. We performed inverse normal transformation on relative levels of metabolites and conducted analyses separately within each study.
Genome-wide genotyping and imputation
Genotyping was performed using a customized Illumina array (15041502 B3; llmina Omni 2.5M array plus ~150K custom SNPs) in HCHS/SOL,[23] the Affymetrix 6.0 chip in ARIC,[24] and the Affymetrix 500K and a 50K Human Gene Focused Panel in FHS.[9] Genome-wide imputation was carried out based on the 1000 Genomes Project phase 3 reference panel in HCHS/SOL and ARIC, and the HapMap CEU population reference panel in FHS.
Metagenomic sequencing and taxonomic profiling
Metagenomics sequencing was performed on DNA extracted from fecal samples collected by FTA card from 3,035 HCHS/SOL participants enrolled in a gut microbiome ancillary study during the HCHS/SOL Visit 2,[25] by a novel shallow-coverage method of shotgun sequencing using Illumina platforms.[26] To account for variability in sequencing depth, centered log-ratio transformation was applied to taxonomic abundances using R/microbiome. Ninety-two bacterial genera with average relative abundance≥0.01% were included in the current analyses.
Statistical analysis
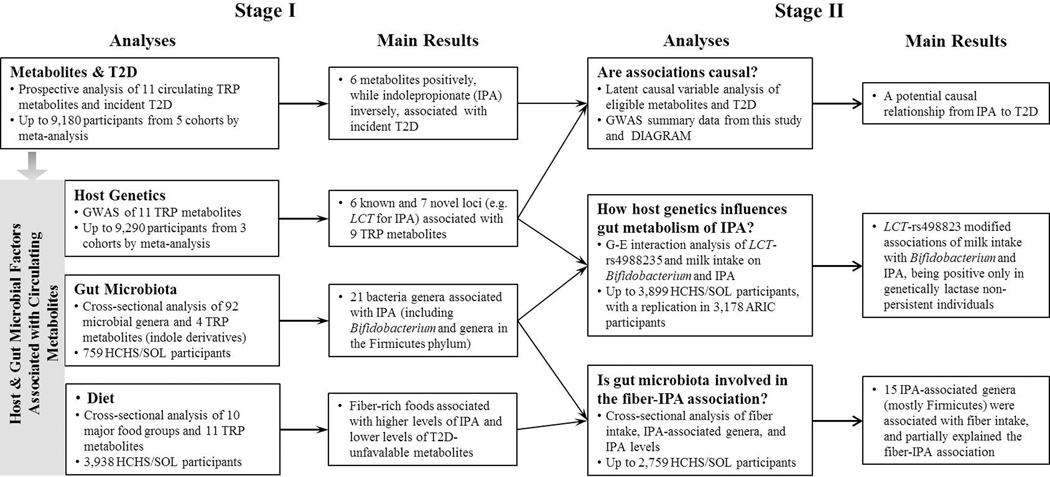
Figure 1 shows a workflow of our analysis. In Stage I, we examined associations of circulating tryptophan metabolites with incident T2D, host genetics, dietary intake and gut microbiota using data from multiple studies (Table S1). Cox regression was used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) of incident T2D per standard deviation (SD) increment in metabolites in each cohort separately, adjusting for demographic, social, behavioral, and health-related factors, and other study-specific covariates (Table S2). Results from each of the cohorts were combined using a fixed-effect meta-analysis. GWAS of standardized metabolite levels were conducted separately in 3,933 HCHS/SOL participants, 1,509 ARIC white participants, and 1,772 ARIC black participants, controlling for age, sex, population stratification and other study-specific covariates. GWAS summary statistics for metabolites in 1,438 whites from FHS, were obtained from a previous publication.[9] Meta-analyses of GWAS summary statistics were conducted using METAL.[27] Associations of serum metabolites with 10 food groups which capture commonly consumed foods, three macronutrients, and two nutrients of interest (fiber and tryptophan) were analyzed using multivariable linear regression in 3,938 HCHS/SOL participants. A medication analysis using multiple mediator models[28] was performed to examine the potential mediating effect of serum tryptophan metabolites on the association between the overall diet quality, measured by the Alternate Healthy Eating Index 2010 (AHEI-2010),[29] and incident T2D in 2,821 HCHS/SOL participants. Associations of 92 gut bacterial genera with four indole derivatives were assessed by multivariable linear regression in 759 HCHS/SOL participants.
Figure 1. Overview of the workflow integrating host genetics, diet, gut microbiota and circulating metabolites in relation to type 2 diabetes.
Eleven tryptophan (TRP) metabolites included TRP, serotonin, five kynurenine-pathway metabolites (kynurenine, kynurenate, xanthurenate, quinolinate, and picolinate), and four indole derivatives (indoleacetate, indolelactate, indolepropionate [IPA] and indoxyl sulfate). T2D, type 2 diabetes; GWAS, genome-wide association study; HCHS/SOL, Hispanic Community Health Study/Study of Latinos; ARIC, Atherosclerosis Risk in Communities Study; DIAGRAM, Diabetes Genetics Replication and Meta-analysis Consortium; LCT-rs498823, a function variant related to lactase persistence.
Based on findings from Stage-I analyses, we performed multiple explanatory analyses in Stage II (Figure 1 and Table S3). We used linkage disequilibrium (LD) score regression[30] to estimate genetic heritability of metabolites and their genetic correlations with T2D. We applied the latent causal variable (LCV) model, which has been recommended to distinguish genetic correlation from causation, to test potential causal relationships between metabolites and T2D, as conventional Mendelian Randomization approaches might be confounded by genetic correlations reflecting shared etiology.[31] GWAS summary statistics for metabolites in this study (up to 9,290 participants) and those for T2D obtained from the Diabetes Genetics Replication and Meta-analysis (DIAGRAM) consortium (55,005 T2D cases and 400,308 controls),[32] were used in the genetic correlation analysis and the LCV models. In HCHS/SOL, we used multivariable linear regression to examine associations between fiber intake and indolepropionate-associated bacterial genera (n=2,759), and compared associations between fiber intake and indolepropionate with and without adjustment for indolepropionate-associated bacterial genera (n=752). A mediation analysis using structural equation modelling[33] was conducted to examine whether indolepropionate-associated bacterial genera may partially explain the association between fiber intake and indolepropionate. In HCHS/SOL, we applied multivariable linear regression to examine associations of LCT-rs4988235 with milk intake (n=12,531), gut Bifidobacterium abundance (n=2,368), and serum indolepropionate (n=3,933). Multivariable linear regression was used to examine associations of milk intake with gut Bifidobacterium abundance and serum indolepropionate levels stratified by the LCT-rs4988235 genotype (lactase persistence AA/AG vs. lactase non-persistence GG), and the interaction between LCT-rs4988235 and milk intake was examined by introducing an interaction term. To validate the interaction between milk intake and LCT-rs4988235 on indolepropionate, a replication analysis was performed in ARIC (1,504 whites and 1,674 blacks).
Analyses were performed using R software unless otherwise stated. In GWAS, P<4.5×10−9 (5.0×10−8/11 metabolites) was considered as genome-wide significant, and a false discovery rate (FDR)<0.05 was considered as statistically significant for other primary analyses.
Results
Tryptophan metabolites and incident T2D
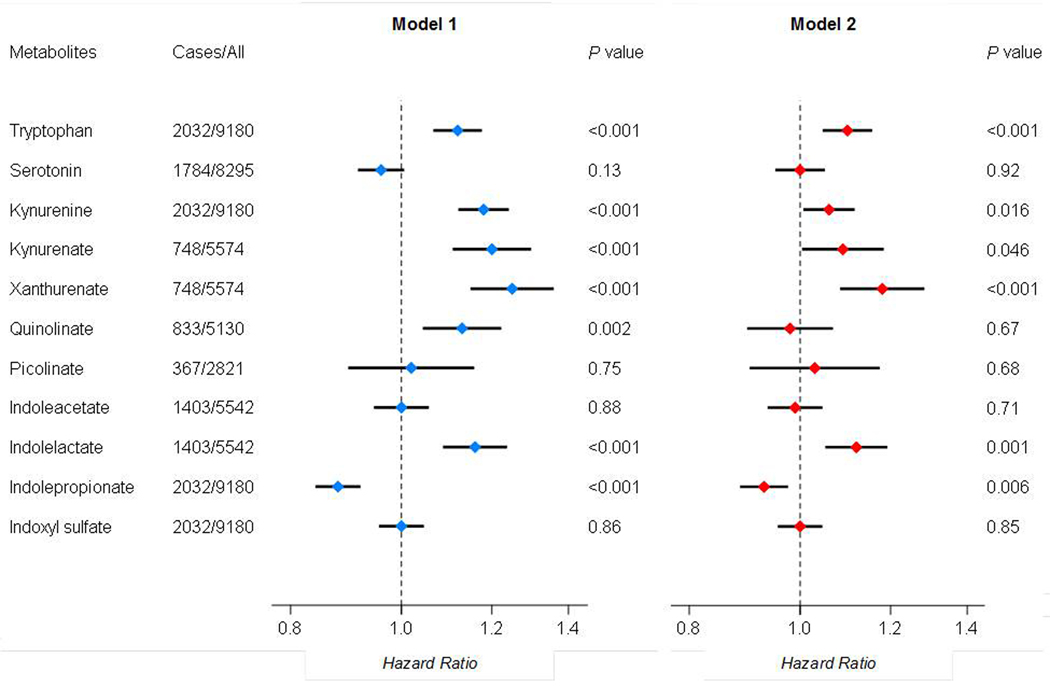
Baseline characteristics of study participants are shown in Table S4. Among 2,821 US Hispanics/Latinos without diabetes at baseline from HCHS/SOL, 367 incident T2D cases were identified during a median of 5.7 years of follow-up. Among 6,359 participants, free of diabetes at baseline, with diverse racial/ethnic backgrounds from ARIC, FHS, WHI and PREDIMED, 1,665 incident T2D cases were identified during follow-up. Of 11 metabolites, tryptophan, four kynurenine metabolites (kynurenine, kynurenate, xanthurenate and quinolinate) and indolelactate were positively associated with incident T2D, while indolepropionate was inversely associated with incident T2D after multivariable adjustment in combined analysis of all studies (all FDR<0.05; Figure 2, Model 1). Results were generally consistent across HCHS/SOL and the other four studies (Table S5). The observed associations were attenuated but remained significant after further adjusting for obesity measures including BMI and WHR, except for quinolinate (Figure 2, Model 2). Further adjustment for blood lipids, blood pressure, or physical activity and dietary quality did not materially change these associations (Table S5).
Figure 2. Associations between circulating tryptophan metabolite levels and incident type 2 diabetes.
Data are Hazard ratios and 95% confidence intervals of incident type 2 diabetes per standard deviation increment in metabolite levels, adjusted for age, sex, smoking, alcohol consumption, education, family income, family history of diabetes, self-reported hypertension and/or antihypertensive medication use, self-reported dyslipidemia and/or lipid-lowering medication us, and other study-specific covariates (Model1); and further adjusted for body mass index and waist-to-hip ratio (Model 2). Results across 5 studies were combined by fixed-effect meta-analysis.
Among 2,821 HCHS/SOL participants without diabetes at baseline, metabolites that were positively associated with T2D (i.e., tryptophan, kynurenine, kynurenate, xanthurenate, quinolinate, and indolelactate) showed weak-to-moderate correlations with each other (Spearman’s r=0.11 to 0.63) (Figure S1), and positive correlations with multiple cardiometabolic traits, especially fasting insulin, HOMA-IR and BMI (Figure S1). Indolepropionate, the only metabolite inversely associated with T2D, was not correlated with other metabolites (Spearman’s r=−0.05 to 0.06), and showed significant, albeit weak, inverse correlations with BMI and a few other cardiometabolic traits.
Host genetics and tryptophan metabolites
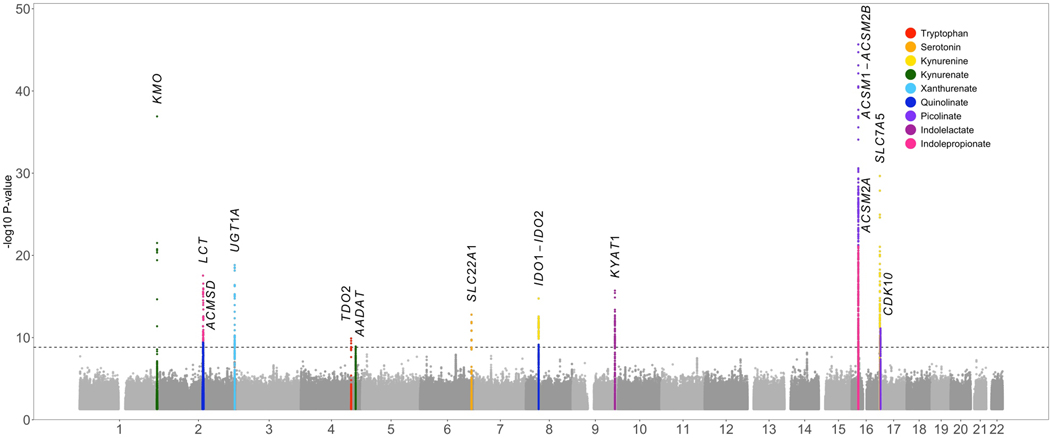
Our genome-wide meta-analyses (n=up to 9,290) identified 21 independent signals at 13 loci associated with nine of 11 tryptophan metabolites (P<4.5×10−9) (Figure 3 and Table S6). Genetic variants at seven loci have not been previously associated with the corresponding metabolites, including those in or near SLC22A, IDO1-IDO2, AADAT, ACMSD, ACSM2B-ACSM1, CDK10, and LCT. We confirmed known genetic associations at six loci.[9, 10, 11] When the threshold of significance was relaxed to traditional genome-wide significance (P<5.0×10−8), we found 16 additional loci associated with tryptophan metabolites (Table S6). Many newly identified and confirmed signals reside in genomic regions harboring genes involved in host kynurenine pathway metabolism (e.g. TDO2, IDO1-IDO2, KMO, AADAT and ACMSD) or transportation of tryptophan metabolites (e.g., SLC7A5, SLC22A1, and SLC16A10).
Figure 3. Manhattan plot for GWAS of circulating tryptophan metabolite levels.
Meta-analyses of GWAS in up to 9,290 individuals from HCHS/SOL, ARIC, and FHS identified 13 loci for 9 tryptophan metabolites (color indicated in inset). The significant P-value threshold is 4.5×10–9 (indicated by a dash line).
Based on GWAS summary statistics from our meta-analysis, genome-wide SNP-based heritability (h2) was estimated at 13.0% (SE=4.9%) for serotonin, 10.7% (5.8%) for indolepropionate, 7.4% (4.8%) for kynurenine, and 0–7.0% for other metabolites (Table S7). As expected, these genome-wide SNP-based heritability estimates were much lower than those estimated using the classical twin model and were generally higher than those estimated based on a few genome-wide significant variants in previous studies (Table S7).[9, 10, 11] We then examined potential causal relationships between three metabolites (serotonin, indolepropionate and kynurenine), which had heritability estimates meeting the criteria for LCV models,[31] and T2D using GWAS summary statistics for metabolites in our study (n=up to 9,290) and those for T2D obtained in the DIAGRAM (55,005 T2D cases and 400,308 controls).[32] Indolepropionate showed a potential causal relationship with T2D (genetic causality proportion=76%, P=1.6×10−24) (Table S7).
Dietary intake and tryptophan metabolites
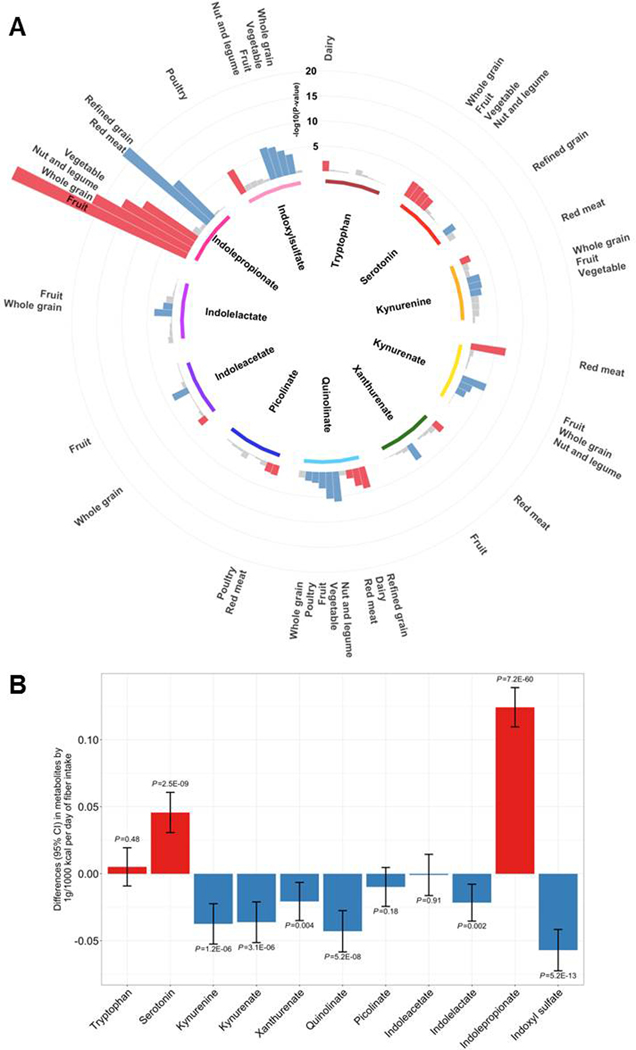
In 3,938 HCHS/SOL participants, we observed significant associations of higher intakes of vegetables, fruits, whole grains, nuts and legumes, and lower intakes of refined grains and red meat, with higher serum indolepropionate levels (Figure 4A). Intakes of some fiber-rich foods which were positively associated with indolepropionate showed inverse associations with other indole derivatives and most kynurenine-pathway metabolites. Mutual adjustment for other food groups did not materially change the results (Table S8). Consistently, higher fiber intake was associated with higher indolepropionate (P=7.3×10−60), and with lower levels of other indole derivatives and most kynurenine-pathway metabolites (Figure 4B). These associations were independent of intakes of macronutrients and tryptophan (Table S9). Intakes of some protein-rich foods (e.g., red meat, poultry, and dairy) and tryptophan were positively associated with serum levels of tryptophan, most kynurenine-pathway metabolites, and indoxylsulfate (Figure 4A, Table S8 and Table S9). Our mediation analysis in 2,821 HCHS/SOL participants without diabetes at baseline indicated a significant meditating effect of these tryptophan metabolites on the association between the overall diet quality (i.e., AHEI-2010) and incident T2D (proportion mediated=61.5%; P=0.01).
Figure 4. Dietary intake and serum tryptophan metabolite levels.
(A) Polar plot for associations of 10 major food groups with serum tryptophan metabolites in the HCHS/SOL. Red: positive associations (FDR<0.05); Blue, inverse associations (FDR<0.05). (B) Differences (95% CI) in serum tryptophan metabolite levels (inverse normal transformed) associated with 1g/1000Kcal per day of dietary fiber intake in the HCHS/SOL.
Gut microbiota and indole derivatives
As indole pathway is carried out mostly by gut microbiota,[4] we examined associations between 92 gut microbial genera and serum levels of four indole derivatives in 759 HCHS/SOL participants. We focused on indolepropionate and indolelactate as these two indole derivatives were significantly associated with incident T2D in our study, and identified 21 genera significantly associated with indolepropionate (FDR<0.05) but none associated with indolelactate (Table S10). In addition, five bacterial genera were significantly associated with indoleacetate and 11 were associated with indoxyl sulfate.
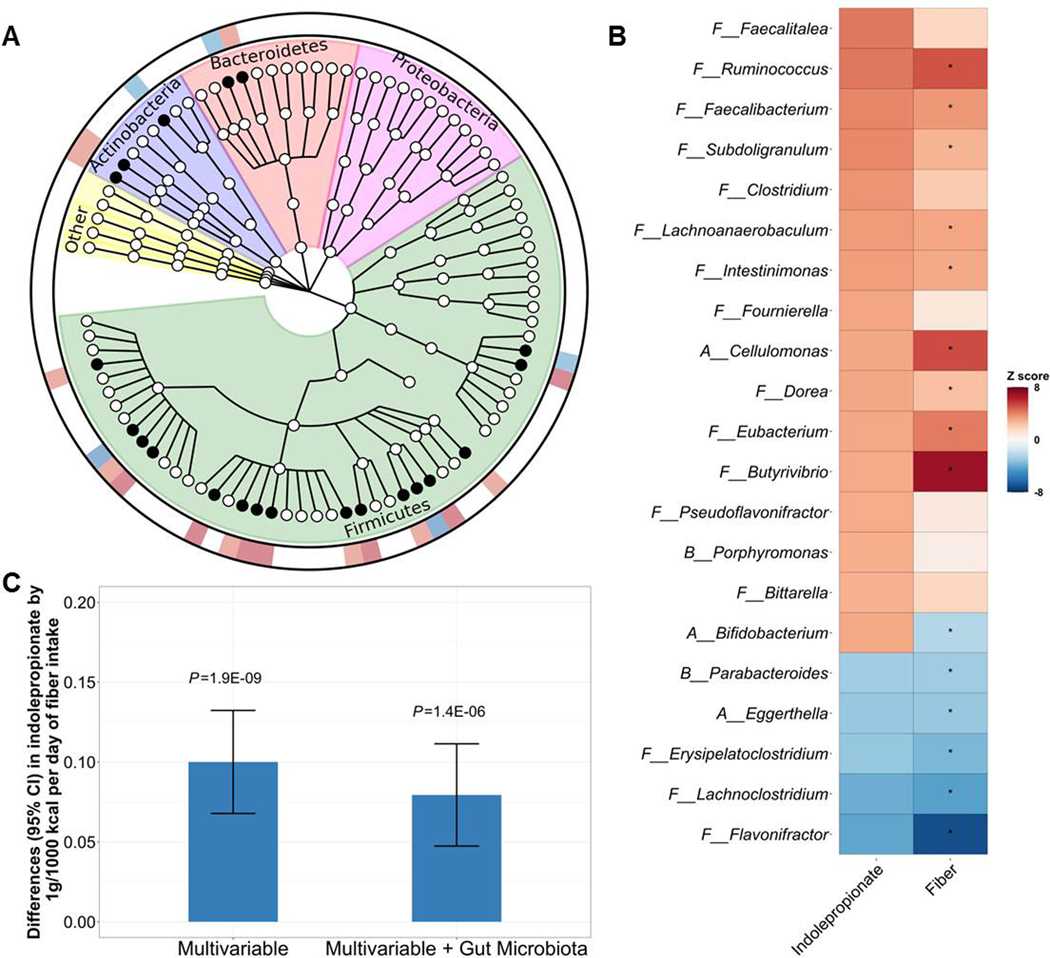
The 21 indolepropionate-associated genera span 3 phyla (Firmicutes, n=16; Actinobacteria, n=3; and Bacteroidetes, n=2) (Figure 5A). When we included all 21 genera in the linear regression model on indolepropionate simultaneously, associations for these genera (especially those in Firmicutes) were greatly attenuated or abolished, while the association between Bifidobacterium and indolepropionate did not change (Figure S1).
Figure 5. Dietary fiber intake, gut microbiota and serum indolepropionate.
(A) Phylogenetic tree of taxonomic features in association with host serum indolepropionate levels in the HCHS/SOL. A total of 21 gut microbial genera significantly associated with serum indolepropionate (FDR<0.05) are indicated by solid circles. Data showing in the outer ring are effect sizes (positive, red; inverse, blue) of gut microbiota genera on serum indolepropionate. (B) Associations of 21 indolepropionate-assocaited gut microbial genera with dietary fiber intake in the HCHS/SOL. To show comparable estimates for the associations of gut microbial genera with indolepropionate and fiber intake, data are presented as Z-scores (regression coefficients/standard errors). *FDR<0.05 for the associations between dietary fiber intake and gut microbial genera. (C) Associations between dietary fiber intake and serum indolepropionate levels with and without adjustment for gut microbiota (20 indolepropionate-associated gut microbial genera) in the HCHS/SOL. Bifidobacterium, which showed opposite associations with indolepropionate and fiber intake, was not included.
Fiber intake, gut microbiota, and indolepropionate
In 2,759 HCHS/SOL participants with diet and gut microbiome data, all indolepropionate-associated bacterial genera were associated with fiber intake (15 genera showing FDR<0.05) with the same directions as those associations between bacterial genera and indolepropionate, except for Bifidobacterium (Figure 5B). In 752 HCHS/SOL participants with diet, metabolomics, and gut microbiome data, the association between fiber intake and indolepropionate was attenuated after further adjustment for the 20 indolepropionate-associated bacterial genera excluding Bifidobacterium (Figure 5C). The attenuation was similar when including Bifidobacterium in the model. We also found a potential mediating effect of these 20 indolepropionate-associated bacterial genera on the association between fiber intake and indolepropionate (proportion mediated=22.3%; P=0.003). These results suggested that these 20 indolepropionate-associated bacterial genera may partially explain the association between fiber intake and indolepropionate, while Bifidobacterium may be involved in other pathways related to indolepropionate.
Host LCT, gut Bifidobacterium, and indolepropionate
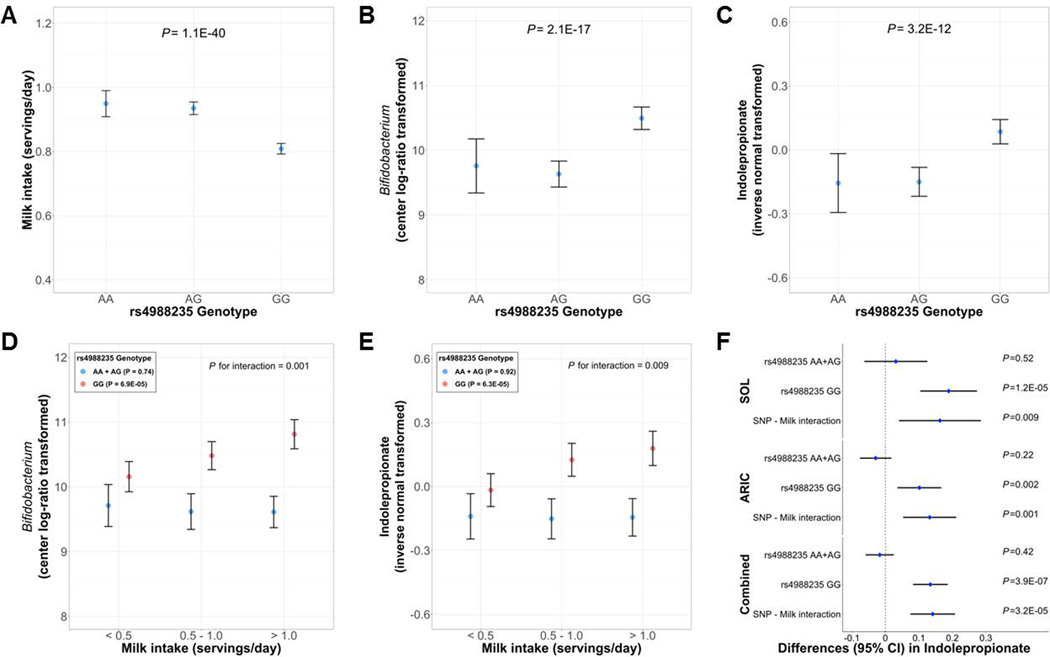
We then focused on gut Bifidobacterium in association with indolepopionate, as gut Bifidobacterium abundance has been related to a host functional LCT variant (rs4988235)[34, 35] and our GWAS also identified LCT as a novel locus for indolepropionate. LCT rs4988235 is a known variant which determines lactase persistence in adulthood (AA/AG is related to lactase persistence and GG is related to lactase non-persistence).[36] In line with previous evidence,[34, 35, 36] the rs4988235-G allele was associated with lower milk intake (P=1.1×10−40; n=12,531) (Figure 6A) and higher gut Bifidobacterium abundance (P=2.1×10−17; n=2,368) (Figure 6B) in HCHS/SOL. In our GWAS, rs4988235-G allele was associated with higher circulating indolepropionate levels (P=3.2×10−17 in meta-analysis, n=9,290; P for heterogeneity=0.51) (P=3.2×10−12 in HCHS/SOL, n=3,933; Figure 6C). When we included both LCT-rs4988235 and Bifidobacterium in the multivariable linear regression model on indolepropionate (n=752), Bifidobacterium, but not LCT-rs4988235, was significantly associated with indolepropionate.
Figure 6. Host LCT genotype, milk intake, gut Bifidobacterium and serum indolepropionate.
(A) Adjusted means and 95% confidence intervals (CIs) of milk intake (servings/day) according to LCT-rs49883235 genotypes in the HCHS/SOL. (B) Adjusted means and 95% CIs of gut Bifidobacterium abundance (center log-ratio transformed) according to LCT-rs49883235 genotypes in the HCHS/SOL. (C) Adjusted means and 95% CIs of serum indolepropnate levels (inverse normal transformed) according to LCT-rs49883235 genotypes in the HCHS/SOL. (D) Adjusted means and 95% CIs of gut Bifidobacterium abundance (center log-ratio transformed) according to milk intake stratified by the LCT-rs49883235 genotype in the HCHS/SOL. (E) Adjusted means and 95% CIs of serum indolepropnate levels (inverse normal transformed) according to milk intake stratified by the LCT-rs49883235 genotype in the HCHS/SOL. (F) Differences and 95% CIs in serum indolepropnate levels (inverse normal transformed) associated with one serving per day of milk intake according to the LCT-rs49883235 genotype in the HCHS/SOL and ARIC separately, and combined by meta-analysis.
Consistent with prior evidence,[34, 35] we found that milk intake was positively associated with gut Bifidobacterium abundance only among lactase non-persistent participants (rs4988235 GG, P=1.5×10−7) but not among those with lactase persistence (rs4988235 AG+GG; P=0.49) in HCHS/SOL (P-interaction=0.001; n=2,342) (Figure 6D). Paralleling the LCT-milk interaction on gut Bifidobacterium, we identified a novel interaction between milk intake and LCT genotype on serum indolepropionate (P-interaction=0.009; n=3,899). Milk intake was positively associated with serum indolepropionate levels only among lactase non-persistent individuals (P=6.3×10−5) but not in those with lactase persistence (P=0.92) (Figure 6E). This significant interaction was replicated in ARIC (P-interaction=0.001; n=3,178) (Figure 6F). LCT-rs4988235 did not show significant associations with intakes of dairy products low in lactose (e.g., yogurt, cheese) or significant interactions with other dairy products on gut Bifidobacterium abundance or serum indolepropionate levels (data not shown).
Discussion
In large-scale populations with diverse racial/ethnic backgrounds, our study demonstrated that circulating levels of kynurenine-pathway metabolites, a group of host tryptophan catabolites, including kynurenine, quinolinate, kynurenate and xanthurenate[7] were associated with increased risk of T2D. We also found that higher intakes of animal-based, protein-rich foods and lower intakes of plant-based, fiber-rich foods were associated with higher circulating levels of kynurenine-pathway metabolites, but the associations between kynurenine-pathway metabolites and T2D did not change after further adjustment for diet quality score. This suggests that these metabolites could be potential mediators linking unhealthy diets with increased risk of T2D rather than simple biomarkers reflecting adverse dietary effects. Moreover, these kynurenine-pathway metabolites were positively correlated with obesity measures and insulin resistance, and obesity may partially explain our observed associations between these metabolites and T2D. These findings are in line with previous evidence and support the notion that activation of the kynurenine pathway by obesity and related inflammation may affect insulin signaling and contribute to increased risk of T2D.[2, 3, 7, 21]
Indole derivatives, a group of microbial tryptophan catabolites, are generally beneficial for human health.[4] Higher circulating indolepropionate has been associated with lower risk of T2D,[8, 12] but it was argued that this association might just reflect beneficial effects of dietary fiber intake on T2D.[14] Our study documented the beneficial association between indolepropionate and T2D and further suggested potential causality. This is consistent with the potential role of indolepropionate in anti-oxidation, anti-inflammation, and amelioration of glucose metabolism.[4]
As little evidence suggests that indolepropionate can be derived from fiber fermentation, the strong positive association between fiber intake and circulating indolepropionate is intriguing[8, 12, 14], but may be explained, in part, by a potential novel pathway suggested by our integrative analysis. Tryptophan is the sole source for indolepropionate production which is suggested as completely gut microbiota-dependent in mice,[37] involving bacterial species mostly in the Clostridium genus.[38] Consistently, a majority of identified gut bacterial genera in our study, including Clostridium,[38] showed positive associations with indolepropionate. Most of these genera are members of Firmicutes, a phylum that includes many species use dietary fiber as main energy source.[39] Catabolism of aromatic amino acids including tryptophan has been demonstrated in Firmicutes but not in other phyla.[38] We also found several indolepropionate-bacterial genera in other phyla which might be related to fiber intake, although it is unknown whether they are involved in the indolepropionate production. For example, Cellulomonas, a genus in Actinobacteria, is known to degrade cellulose,[40] a type of fiber found in plant cell walls. These findings suggest that higher fiber intake may increase populations of fiber-utilizing bacteria,[39] some of which may have the capability to produce indolepropionate or its substrates from tryptophan,[4] thus shifting host tryptophan-to-kynurenine catabolism more towards gut microbial indolepropionate production. However, it should be noted that the association between fiber intake and indolepropionate was not fully explained by the identified bacteria in our study. Gut bacteria involved in this pathway might not be fully captured by our fecal metagenomics. A notable limitation of our study is that the assessments of diet and serum metabolites preceded fecal sample collection by a median of seven years. Although the human gut microbiome was found to be notably stable over a long period, [41] the 7-year time lag might attenuate the associations of the gut microbiota with diet and metabolites in this study. It is possible that we would observe stronger associations of gut microbiota with fiber intake and serum indolepropionate with concurrently collected data. Nevertheless, our findings suggest indolepropionate production, in addition to short-chain fatty acid production,[39] as a potential novel microbial metabolite pathway for beneficial effects of dietary fiber on human cardiometabolic health.
Another novel finding of this study is that a lactase persistence-determining variant at LCT was associated with circulating indolepropionate, through an apparent interaction with milk intake. This might be related to an indolepropionate-associated gut bacterium identified in this study, Bifidobacterium, which has been associated with host LCT and milk intake.[34, 35] Compared to lactase persistent individuals, lactase non-persistent individuals cannot hydrolyze lactose after consuming milk and thus have more lactose in the gut as an energy source for Bifidobacterium growth,[34, 35] which may then contribute to higher indolepropionate production. Indeed, although it is unknown whether Bifidobacterium has the capability to produce indolepropionate, many strains in the Bifidobacterium genus have been found to produce indolelactate,[42, 43] a substrate for indolepropionate. Moreover, both human[44, 45, 46, 47] and animal studies[48] suggested a potential protective role of gut Bifidobacterium in T2D. Taken together, our observations extend the previously identified host gene-diet interaction on gut microbiota[34, 35] to microbiota-produced metabolites in host circulation, and suggest microbial indole derivative production as a potential mechanism through which gut Bifidobacterium is associated with T2D. Due to limitations of shallow shotgun sequencing data,[26] we did not examine Bifidobacterium species or strains, or functional features for indole derivative production which need to be clarified in future studies.
The other two indole derivatives, indoleacetate and indolelactate, have been shown to act through aryl hydrocarbon receptor activation,[4] which could reduce inflammation and insulin resistance.[5] However, we did not find beneficial associations of these two metabolites with T2D. In contrast, indolelactate was associated with increased risk of T2D in our study, and inconsistent associations between indolelactate and insulin resistance were also reported in previous studies.[49, 50] Interestingly, we found that serum indolelactate was more closely correlated with kynurenine-pathway metabolites than other indole derivatives, and host factors (e.g., genetic variants in KYAT1,[10] a gene involved in host tryptophan-kynurenine metabolism) rather than gut microbial factors were associated with circulating indolelactate levels. Further studies are warranted to clarify the relationship between circulating and fecal indole derivatives and their associations with T2D.
In summary, circulating tryptophan, several kynurenine-pathway metabolites and indolelactate showed adverse associations with incident T2D, while indolepropionate showed a beneficial association with incident T2D. We identified multiple host genetic, dietary and gut microbial factors associated with these metabolites. In particular, higher fiber intake, and milk intake (only among genetically lactase non-persistent individuals) were associated with higher circulating levels of indolepropionate possibly through the host-microbial cross-talk shifting tryptophan metabolism toward gut microbial indolepropionate production. It should be noted that our study is unable to make causal inference due to its observational nature, although our findings may have strong biological plausibility. These findings contribute to our understanding of the host-microbial cross-talk in tryptophan metabolism and its implications in human metabolic health and disease, and may help to identify high-risk individuals based on circulating metabolite profiles for targeted interventions through dietary intervention and gut microbiota modification.
Supplementary Material
Significance of this study.
What is already known on this subject?
Tryptophan can be catabolized to various metabolites through host kynurenine and microbial indole pathways.
Evidence from animal studies suggests a host-microbiota interaction on tryptophan metabolism which may affect host metabolic health.
Circulating levels of some tryptophan metabolites have been associated with risk of type 2 diabetes in human studies.
Genetic variants located on genes that are involved in the host tryptophan-kynurenine pathway and dietary factors have been associated with circulating tryptophan metabolites, but the role of gut microbiome and its interplay with host genetics and diet in tryptophan metabolism remain unclear in humans.
What are the new findings?
In large-scale populations with diverse racial/ethnic backgrounds, circulating levels of tryptophan and several kynurenine-pathway metabolites were positively associated with risk of type 2 diabetes, while a microbial indole derivative, indolepropionate, was inversely associated with risk of type 2 diabetes. The indolepropionate-T2D association was suggested to be potentially causal by the latent causal variable model.
Intakes of fiber-rich foods, but not protein/tryptophan-rich foods, were the dietary factors most strongly associated with circulating tryptophan metabolites. The fiber-indolepropionate association can be partially explained by indolepropionate-associated gut bacteria (mostly fiber-utilizing Firmicutes bacteria).
We identified a novel genetic association between a host functional LCT variant (determining lactase persistence) and serum indolepropionate, which might be a result of host gene-diet interaction on gut Bifidobacterium. Higher milk intake was associated with higher levels of gut Bifidobacterium and serum indolepropionate only among genetically lactase non-persistent individuals.
How might it impact on clinical practice in the foreseeable future?
These findings contribute to our understanding of the host-microbial cross-talk in tryptophan metabolism and its implications in human metabolic health and disease, and may help to identify high-risk individuals based on circulating metabolite profiles for targeted interventions through dietary intervention and gut microbiota modification.
Acknowledgment
This work is supported by R01-DK119268 (Dr. Qi) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), R01-MD011389 (Drs. Kaplan, Burk and Knight) from the National Institute on Minority Health and Health Disparities, and R01-HL060712 (Drs. Qi and Hu) from the National Heart, Lung, and Blood Institute (NHLBI). Other funding sources for this study include UM1-HG008898 from the National Human Genome Research Institute; R01-HL140976, and R01-HL136266 from the NHLBI; and R01-DK120870, R01 DK126698 and the New York Regional Center for Diabetes Translation Research (P30DK111022) from NIDDK. Support for metabolomics data was graciously provided by the JLH Foundation (Houston, Texas). Dr. Li is supported by 9-17-CMF-011 from the American Diabetes Association (ADA); K99-DK122128 from NIDDK; and a pilot and feasibility grant from the NIDDK-funded Boston Nutrition Obesity Research Center (P30-DK046200). Dr. Yu is supported by the American Heart Association (17SDG33661228); and R01-HL141824 and R01-HL142003 from NHLBI. Drs. Meigs and Dupuis are supported by U01-DK078616 from NIDDK. Dr. Merino is supported by a postdoctoral fellowship funded by the European Commission Horizon 2020 program; Marie Skłodowska-Curie actions (H2020-MSCA-IF-2015-703787); and P30DK040561 from NIDDK. Funding and acknowledgment information for each participating studies is provided in Online Supplements.
Disclosures: All authors have completed the ICMJE uniform disclosure form and declare: no support from any organisation for the submitted work; no financial relationships with any organisations that might have an interest in the submitted work in the previous three years, no other relationships or activities that could appear to have influenced the submitted work.
Footnotes
Dissemination declaration: We stated that dissemination of the results to study participants is not applicable.
Reference
- 1.Badawy AA. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int J Tryptophan Res 2017;10:1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Favennec M, Hennart B, Caiazzo R, Leloire A, Yengo L, Verbanck M, et al. The kynurenine pathway is activated in human obesity and shifted toward kynurenine monooxygenase activation. Obesity (Silver Spring) 2015;23:2066–74. [DOI] [PubMed] [Google Scholar]
- 3.Oxenkrug G. Insulin resistance and dysregulation of tryptophan-kynurenine and kynurenine-nicotinamide adenine dinucleotide metabolic pathways. Mol Neurobiol 2013;48:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun 2018;9:3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurans L, Venteclef N, Haddad Y, Chajadine M, Alzaid F, Metghalchi S, et al. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat Med 2018;24:1113–20. [DOI] [PubMed] [Google Scholar]
- 6.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011;17:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vangipurapu J, Fernandes Silva L, Kuulasmaa T, Smith U, Laakso M. Microbiota-Related Metabolites and the Risk of Type 2 Diabetes. Diabetes Care 2020. [DOI] [PubMed] [Google Scholar]
- 8.de Mello VD, Paananen J, Lindstrom J, Lankinen MA, Shi L, Kuusisto J, et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci Rep 2017;7:46337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee Eugene P, Ho Jennifer E, Chen M-H, Shen D, Cheng S, Larson Martin G, et al. A Genome-wide Association Study of the Human Metabolome in a Community-Based Cohort. Cell Metabolism 2013;18:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet 2014;46:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet 2017;49:568–78. [DOI] [PubMed] [Google Scholar]
- 12.Tuomainen M, Lindstrom J, Lehtonen M, Auriola S, Pihlajamaki J, Peltonen M, et al. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr Diabetes 2018;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallister T, Jennings A, Mohney RP, Yarand D, Mangino M, Cassidy A, et al. Characterizing Blood Metabolomics Profiles Associated with Self-Reported Food Intakes in Female Twins. PLoS One 2016;11:e0158568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menni C, Hernandez MM, Vital M, Mohney RP, Spector TD, Valdes AM. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut Microbes 2019;10:688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorlie PD, Aviles-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Annals of epidemiology 2010;20:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siega-Riz AM, Sotres-Alvarez D, Ayala GX, Ginsberg M, Himes JH, Liu K, et al. Food-group and nutrient-density intakes by Hispanic and Latino backgrounds in the Hispanic Community Health Study/Study of Latinos. Am J Clin Nutr 2014;99:1487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebholz CM, Yu B, Zheng Z, Chang P, Tin A, Kottgen A, et al. Serum metabolomic profile of incident diabetes. Diabetologia 2018;61:1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merino J, Leong A, Liu C-T, Porneala B, Walford GA, von Grotthuss M, et al. Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetologia 2018;61:1315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paynter Nina P, Balasubramanian R, Giulianini F, Wang Dong D, Tinker Lesley F, Gopal S, et al. Metabolic Predictors of Incident Coronary Heart Disease in Women. Circulation 2018;137:841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estruch R, Ros E, Salas-Salvadó J, Covas M-I, Corella D, Arós F, et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. New England Journal of Medicine 2018;378:e34. [DOI] [PubMed] [Google Scholar]
- 21.Yu E, Papandreou C, Ruiz-Canela M, Guasch-Ferre M, Clish CB, Dennis C, et al. Association of Tryptophan Metabolites with Incident Type 2 Diabetes in the PREDIMED Trial: A Case-Cohort Study. Clin Chem 2018;64:1211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen GC, Chai JC, Yu B, Michelotti GA, Grove ML, Fretts AM, et al. Serum sphingolipids and incident diabetes in a US population with high diabetes burden: the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Am J Clin Nutr 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 2016;98:165–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009;2:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan RC, Wang Z, Usyk M, Sotres-Alvarez D, Daviglus ML, Schneiderman N, et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol 2019;20:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, et al. Evaluating the information content of shallow shotgun metagenomics. Msystems 2018;3:e00069–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preacher KJ, Hayes AF. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav Res Methods 2008;40:879–91. [DOI] [PubMed] [Google Scholar]
- 29.Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr 2012;142:1009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connor LJ, Price AL. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 2018;50:1728–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nature Genetics 2018;50:1505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosseel Y. lavaan: An R Package for Structural Equation Modeling. J Stat Softw 2011;48. [Google Scholar]
- 34.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet 2016;48:1407–12. [DOI] [PubMed] [Google Scholar]
- 35.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016;19:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet 2002;30:233–7. [DOI] [PubMed] [Google Scholar]
- 37.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 2009;106:3698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017;551:648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makki K, Deehan EC, Walter J, Backhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018;23:705–15. [DOI] [PubMed] [Google Scholar]
- 40.Chaudhary P, Kumar NN, Deobagkar DN. The glucanases of Cellulomonas. Biotechnol Adv 1997;15:315–31. [DOI] [PubMed] [Google Scholar]
- 41.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res 2013;57:523–35. [DOI] [PubMed] [Google Scholar]
- 43.Aragozzini F, Ferrari A, Pacini N, Gualandris R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol 1979;38:544–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, et al. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog 2017;111:362–9. [DOI] [PubMed] [Google Scholar]
- 45.Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr 2016;116:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol 2010;61:69–78. [DOI] [PubMed] [Google Scholar]
- 47.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017;23:850–8. [DOI] [PubMed] [Google Scholar]
- 48.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020;51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–81. [DOI] [PubMed] [Google Scholar]
- 50.Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 2019;569:663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.