Abstract
Multiple myeloma (MM) is a hematologic malignancy arising from the clonal proliferation of malignant plasma cells. tRNA-derived RNA fragments (tRFs) constitute a class of small non-coding RNAs, deriving from specific enzymatic cleavage of tRNAs. To the best of our knowledge, this is one of few studies to uncover the potential clinical significance of tRFs in MM. Total RNA was extracted from CD138+ plasma cells of MM and smoldering MM patients, and in vitro polyadenylated. First-strand cDNA synthesis was performed, priming from an oligo-dT-adaptor sequence. Next, real-time quantitative PCR (qPCR) assays were developed for the quantification of six tRFs. Biostatistical analysis was performed to assess the results and in silico analysis was conducted to predict the function of one of the tRFs. Our results showed that elevated levels of five out of six tRFs are indicators of favorable prognosis in MM, predicting prolonged overall survival (OS), while two of them constitute potential molecular biomarkers of favorable prognosis in terms of disease progression. Moreover, three tRFs could be used as surrogate prognostic biomarkers along with the R-ISS staging system to predict OS. In conclusion, tRFs show molecular biomarker utility in MM, while their mechanisms of function merit further investigation.
Keywords: tRNA-derived RNA fragment (tRF), tRNA derivative (tDR), small non-coding RNAs (sncRNAs), hematologic malignancy, plasma cell dyscrasia, bone disease, molecular biomarker, prognosis, post-transcriptional regulator, gene ontology (GO)
1. Introduction
Multiple myeloma (MM) is a hematologic malignancy, belonging to a group of disorders characterized as plasma cell dyscrasias, as it emerges from the clonal proliferation of malignant plasma cells. These malignant plasma cells reside in the bone marrow (BM) and, in the majority of cases, synthesize and secrete a monoclonal immunoglobulin (M-protein) [1]. Monoclonal gammopathy of undetermined significance (MGUS) or smoldering MM (sMM), asymptomatic states characterized by clonal proliferation of malignant plasma cells without symptoms, usually precede MM [2,3]. Symptomatic MM is characterized by the presence of end-organ damage, such as hypercalcemia, renal impairment, anemia, and bone disease, as defined by the acronym “CRAB” [4]. Among these, MM bone disease (MMBD) is a hallmark of MM, as it degenerates dramatically the quality of life of MM patients [5].
The tumor microenvironment plays a catalytic role in MM progression. MM cells interact with cell populations highly abundant in the BM microenvironment, such as mesenchymal stem cells, osteoblast and osteoclast precursors, and immune-system cells leading to cytokine production or cell to cell interactions that promote MM cell proliferation [6]. IL-6 is such a cytokine, which is produced by several cell types in the bone marrow milieu, as well as by MM cells. Moreover, it is characterized as a proliferative factor for MM cells, as it triggers several signaling pathways and also inhibits apoptosis [7,8]. Therefore, the deciphering of the molecular background of MM and MMBD is a necessity for the in-depth understanding of MM and the establishment of new therapeutic options. Regarding MM prognosis, it was until recently based on the International Staging System (ISS), which stratifies patients according to β2-microglobulin and serum albumin levels [9]. In 2015, this staging system was revised (R-ISS), so as to include lactate dehydrogenase (LDH) levels, as well as cytogenetic factors, namely t(4;14) and t(14;16) translocations, and 17p deletion [del(17p)] [10]. However, as MM remains an incurable disease, there is an imperative need for the establishment of novel prognostic biomarkers [11]. The elucidation of the MM molecular background can provide such solutions, as attempts have been made towards the establishment of molecular biomarkers in MM, a typical paradigm of which is the detection of microRNAs (miRNAs) [11,12,13,14].
tRNA derivatives constitute a class of small non-coding RNAs, emerging from explicit enzymatic cleavage of the pre- or mature tRNAs. These fragments have only recently emerged. Therefore, their function has not yet been fully elucidated [15]. Two main classes of tRNA derivatives are currently known: the tRNA halves, or tiRNAs, with a length of 35–50 nt, and tRNA-derived RNA fragments (tRFs), with a length of 15–35 nucleotides (nt). tRFs are further classified into four categories; more specifically, 5′tRFs (also known as tRF-5) incorporate the 5′ of the mature tRNA of origin, while 3′tRFs (also known as tRF-3) incorporate the 3′ of the mature tRNA of origin, and thus are characterized by the presence of the terminal -CCA sequence. tRFs may also derive from the internal part of the mature tRNA, and thus are named “internal tRFs” (i-tRFs). These tRFs might also include the anticodon. Last, tRFs may also derive from the 3′ of the pre-tRNA and thus are named 3′U-tRFs or tRF-1 [16]. Some tRFs have been reported to interact with Argonaute proteins and, similar to miRNAs, participate in the post-transcriptional regulation [17], as well as in the regulation of RNA processing and translation [18]. Moreover, they have been witnessed to exert an important role in neurodegenerative diseases, viral infections, metabolic disorders, inflammation, cancer, and hematologic malignancies [19,20,21]. In cancer and hematologic malignancies, their expression is deregulated; this fact, including their easy detection and high abundance in biological fluids, advocates their usage as molecular biomarkers [22,23]. Members of our research group have attempted to establish such fragments as molecular biomarkers in hematologic malignancies [24,25,26,27].
The present study aimed to further uncover the molecular biomarker utility of tRFs in MM, as only a few studies have elucidated their role in this disease so far [28,29,30]. Thus, we attempted to quantify six tRFs, which were previously identified by members of our research group, in CD138+ plasma cells of sMM and MM patients. We used an innovative in-house-developed quantitative PCR (qPCR) assay and examined their potential value as diagnostic and prognostic biomarkers, by performing extensive biostatistical analysis. Last, we conducted an in silico analysis, to investigate the putative function of one of the tRFs studied.
2. Materials and Methods
2.1. Sample Collection
Bone marrow aspirate (BMA) samples were collected from 94 patients. Seventy-six of them were MM patients and 18 of them were diagnosed with sMM. Adult patients with a recent diagnosis of MM or sMM were included in the study, while patients who had already received any kind of treatment or suffered from any other concomitant malignancy were excluded. The median age of MM patients was 68 years (range: 35–88 years), while the median age of sMM patients was 66 years (range: 54–81 years). The presence of high-risk cytogenetic abnormalities, namely del(17p), t(4;14), t(14;16), and (+1q) was assessed using fluorescence in situ hybridization (FISH). The MM patients were classified according to ISS and revised ISS (R-ISS) staging systems, as defined by the International Myeloma Working Group (IMWG) in 2005 and 2015, respectively [9,10]. Whole-body low-dose computed tomography (WBLDCT) was used to assess osteolysis. The characteristics of the MM patients are shown in Table 1.
Table 1.
Characteristics of the multiple myeloma (MM) patients.
| Variable | Number of Patients (%) |
|---|---|
| Gender (76/76 patients) | |
| Male | 44 (57.9%) |
| Female | 32 (42.1%) |
| M-protein isotype (75/76 patients) | |
| IgG | 44 (58.7%) |
| IgA | 17 (22.7%) |
| IgD | 2 (2.7%) |
| Kappa light chain | 7 (9.2%) |
| Lambda light chain | 3 (4.0%) |
| Not typed | 2 (2.7%) |
| del(17p) (71/76 patients) | |
| Absence | 59 (83.1%) |
| Presence | 12 (16.9%) |
| t(4;14) (70/76 patients) | |
| Absence | 62 (88.6%) |
| Presence | 8 (11.4%) |
| t(14;16) (68/76 patients) | |
| Absence | 67 (98.5%) |
| Presence | 1 (1.5%) |
| (+1q) (54/76 patients) | |
| Absence | 30 (55.6%) |
| Presence | 24 (44.4%) |
| ISS 1 stage (74/76 patients) | |
| I | 15 (20.3%) |
| II | 25 (33.8%) |
| III | 34 (45.9%) |
| R-ISS 2 stage (69/76 patients) | |
| I | 11 (15.9%) |
| II | 40 (58.0%) |
| III | 18 (26.1%) |
| Bone disease (72/76 patients) | |
| No | 22 (30.6%) |
| Yes | 50 (69.4%) |
| WBLDCT 3 osteolysis (56/76 patients) | |
| No | 18 (32.1%) |
| Yes | 38 (67.9%) |
1 International Staging System; 2 Revised International Staging System; 3 Whole-body low-dose computed tomography.
Only one patient did not receive any treatment, while the treatment of the rest 75 patients varied. More specifically, 63 out of the 75 MM patients (84.0%) were treated with bortezomib plus an immunomodulatory drug (either lenalidomide (60 cases) or thalidomide (3 cases)), 6 (8.0%) MM patients received bortezomib along with cyclophosphamide and dexamethasone, and 6 (8.0%) MM patients were treated with lenalidomide and dexamethasone. Moreover, 21 (27.6%) out of 76 MM patients were subjected to autologous stem cell transplantation following high-dose melphalan (HDM-ASCT), whereas the rest 55 (72.4%) MM patients were not candidates for bone marrow transplant, either because of being older than 65 years (48 cases) or because of severe comorbidities and/or impaired performance status (7 cases).
This study was approved by the Ethics Committee of the “Alexandra” General Hospital of Athens and was conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from each participant of this study.
2.2. CD138+ Plasma Cell Selection
In order to select plasma cells from BMA samples of MM and SMM patients, we performed CD138-positive selection among mononuclear cells of BMA as a method of choice. CD138, officially known as Syndecan 1, is a transmembrane (type I) heparan sulfate proteoglycan encoded by the human SDC1 gene; its expression is considered to be a hallmark of MM cells and plasma cells in the bone marrow [31,32,33]. Therefore, 10 mL of BMA were collected in tubes containing ethylenediaminetetraacetic acid (EDTA) and BMA samples were processed, immediately after collection, for CD138 enrichment. More specifically, the Ficoll-Paque technique was used to isolate mononuclear cells from BMA. Following that, a positive selection of CD138+ plasma cells was performed using anti-CD138–coated magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany).
2.3. Total RNA Extraction, In Vitro Polyadenylation, and cDNA Synthesis
Total RNA was isolated from CD138+ plasma cells, using TRI Reagent® (Molecular Research Center Inc., Cincinnati, OH, USA). The concentration of each total RNA was determined using Qubit™ 2 Fluorometer (Invitrogen™, Thermo Fisher Scientific Inc., Carlsbad, CA, USA), and its integrity was assessed by agarose gel electrophoresis. Next, 200 ng of each total extract were subjected to in vitro polyadenylation, using E. coli poly(A) polymerase (New England Biolabs Ltd., Ontario, ON, Canada) and 80 μM ATP. Each sample was incubated at 37 °C for 60 min, followed by an inactivation step at 65 °C for 10 min. Reverse transcription was then performed, using MMLV reverse transcriptase (Invitrogen™, Thermo Fisher Scientific Inc.) and an oligo-dT–adapter primer, following the manufacturer’s instructions. The oligo-dT–adapter primer sequence was 5′-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3′. Both polyadenylation and reverse transcription were conducted in a GeneAmp® PCR System 9700 (Applied Biosystems™, Thermo Fisher Scientific Inc., Foster City, CA, USA).
2.4. tRF Selection
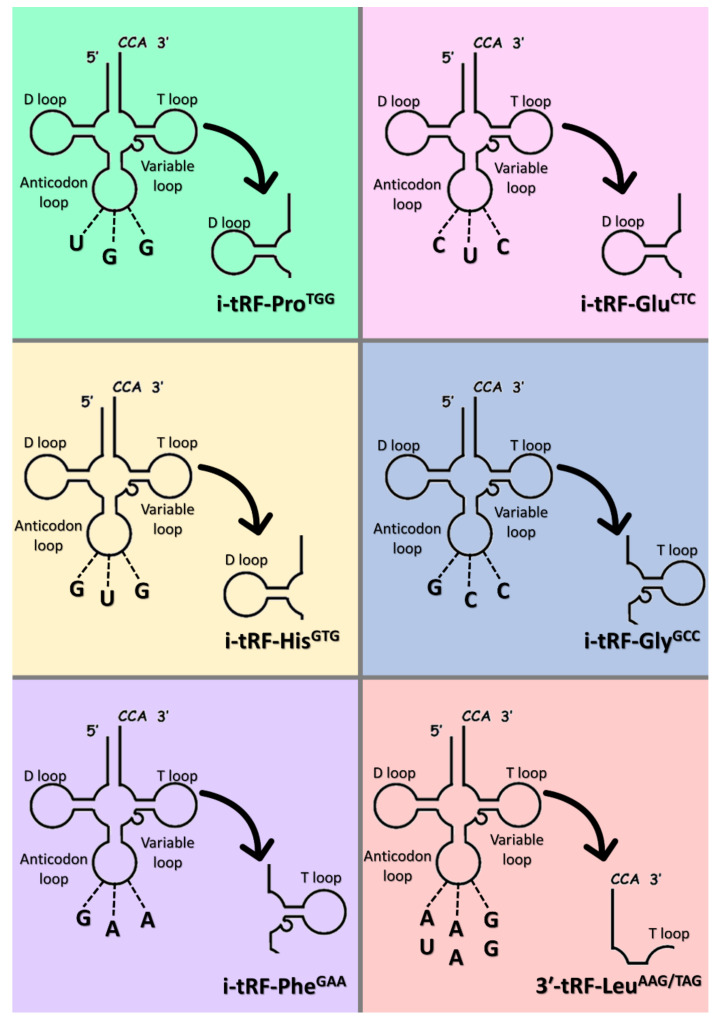
Six (6) tRFs, specifically, five (5) i-tRFs, namely i-tRF-ProTGG, i-tRF-GluCTC, i-tRF-HisGTG, i-tRF-GlyGCC, i-tRF-PheGAA, and one (1) 3′tRF, namely 3′-tRF-LeuAAG/TAG, were selected for this study. The tRFs investigated in this study are shown in Figure 1. All tRFs have previously been verified experimentally by members of our research group, after bioinformatical analysis based on publicly available RNA sequencing data. MINTbase v.2.0, one of the few databases comprising tRFs detected in human tissues, was used to align the tRFs with the respective tRNAs of origin [34]. The names of the tRFs were determined based on the tRF category, the amino acid carried by the tRNA of origin, and the respective anticodon. The sequences of these tRFs were submitted to GenBank® of NCBI. The fragment sequences, the anticodons, the localization of the tRNAs of origin, their MINTbase unique IDs, as well as their GenBank® accession numbers are shown in Table 2.
Figure 1.
Graphical illustration of the mapping of the tRNA-derived RNA fragments (tRFs) investigated in this study with their respective tRNAs of origin.
Table 2.
The tRNA-derived RNA fragments (tRFs) investigated in this study.
| tRF | Fragment Sequence | Anticodon | Localization | Accession Number | MINTbase Unique ID |
|---|---|---|---|---|---|
| i-tRF-ProTGG | 5′-GUUGGUCUAGGGGUAUGAUUCUCGG-3′ | UGG | Nucleus | MK671729 | tRF-25-78WPRLXN48 |
| i-tRF-GluCTC | 5′-GUCUAGUGGUUAGGAUUCGGCG-3′ | CUC | Nucleus | MK671728 | tRF-22-SX73V2Y8K |
| i-tRF-HisGTG | 5′-UGAUCGUAUAGUGGUUAGUACUCUGCG-3′ | GUG | Nucleus | MW650833 | tRF-27-XMSL73VL4YK |
| i-tRF-GlyGCC | 5′-GAGGCCCGGGUUCGAUUC-3′ | GCC | Nucleus | MK642309 | tRF-18-5J3KYU05 |
| i-tRF-PheGAA | 5′-UUUAGACGGGCUCACAUCACC-3′ | GAA | Mitochondrion | MK671731 | tRF-21-ZPEK45H5D |
| 3’-tRF-LeuAAG/TAG | 5′-AUCCCACCGCUGCCACCA-3′ | AAG, UAG |
Nucleus | MK671733 | tRF-18-HR0VX6D2 |
2.5. Real-Time qPCR
Specific forward primers were designed for each tRF, SNORD43 (also known as RNU43) and SNORD48 (also known as RNU48), which were used as reference genes, due to their similar size with tRFs and stable expression. Each forward primer was used along with a common universal reverse primer, complementary to the oligo-dT–adapter used during cDNA synthesis. All primers used in the qPCR assay are shown in Table 3. The 10-μL reaction mixture was composed of 0.5 μL of 10-fold diluted cDNA, 5 μL KAPA™ SYBR® FAST qPCR master mix (2X) (Kapa Biosystems Inc., Woburn, MA, USA), 1 μL of each primer to a final concentration of 200 nM each, and 2.5 μL RNase-free H2O. The qPCR assay was performed in an ABI 7500 Fast Real-Time PCR System (Applied Biosystems™), following a standard thermal protocol for cycling and melting, as previously described [26]. The levels of each tRF were determined using the comparative threshold cycle (2−ΔΔCt) method [35,36]. The normalized levels of each tRF were measured in relative quantification units (RQUs), as the ratio of tRF molecules to the geometric mean of SNORD48 and SNORD43 molecules, divided by the same ratio that had been previously determined for a cDNA pool, consisting of 5 cDNAs deriving from patient samples, which was used as a calibrator. The qPCR assay was optimized, in order to observe a unique melting curve for each amplicon, and thus be able to quantify the selected tRFs in all samples. The melting curves for each tRF, as well as those of the reference molecules, are shown in Figure S1.
Table 3.
Primers used in real-time quantitative PCR (qPCR) for the relative quantification of the tRFs in all samples.
| Amplified Molecule | Primer Sequence (5′→3′) | Direction | Length (nt 1) | Tm (°C) |
|---|---|---|---|---|
| i-tRF-ProTGG | GTTGGTCTAGGGGTATGATTCTCGGA | Forward | 26 | 62 |
| i-tRF-GluCTC | GTCTAGTGGTTAGGATTCGGCGA | 23 | 61 | |
| i-tRF-HisGTG | TGATCGTATAGTGGTTAGTACTCTGCG | 27 | 59 | |
| i-tRF-GlyGCC | GAGGCCCGGGTTCGATTC | 18 | 62 | |
| i-tRF-PheGAA | TTTAGACGGGCTCACATCACC | 21 | 59 | |
| 3’-tRF-LeuAAG/TAG | ATCCCACCGCTGCCACCA | 18 | 66 | |
| SNORD43 | ACTTATTGACGGGCGGACA | 19 | 59 | |
| SNORD48 | TGATGATGACCCCAGGTAACTCT | 23 | 59 | |
| Universal reverse | GCGAGCACAGAATTAATACGAC | Reverse | 22 | 56 |
1 Nucleotides.
2.6. Biostatistical Analysis
Data analysis was performed using SPSS® (v.26) software (IBM Corporation, Armonk, NY, USA). Due to the non-canonical distribution of the levels of each tRF in the patient cohorts, non-parametric tests were conducted. The X-tile algorithm was used to generate optimal cut-off points [37], which were used to classify the patients in distinct subgroups, based on the levels of each tRF. The Mann–Whitney U test was used to assess the significance of differences in tRF levels between distinct groups. Putative associations between each tRF expression and other categorical variables were assessed with the chi-square test or Fischer’s exact test. Regarding survival, Kaplan–Meier curves, concerning overall (OS) and progression-free (PFS) survival of MM patients, were built, and differences between the curves were assessed using the Mantel–Cox (log-rank) test [38]. Last, bootstrap (1000 samples) univariate and multivariate Cox regression analyses were carried out, estimating bias-corrected and accelerated (BCa) 95% confidence intervals (CI) of each hazard ratio (HR). Statistical results were considered significant only when the p value score was less than or equal to 0.05.
2.7. In Silico Analysis for tRF Target Prediction and Gene Ontology (GO) Enrichment Analysis
Two databases, namely tRFTar [39] and tRFtarget [40] were used to predict the target genes of each tRF. tRFTar uses cross-linking immunoprecipitation-sequencing (CLIP-seq) data to provide putative targets, while tRFtarget uses prediction tools. In tRFtarget, the default parameters were used (free energy ≤−10 Kcal, and maximum complementary length >8). Binding sites in the whole transcript were chosen in both databases. However, none of the databases provided information about the targets of the i-tRFs included in this study, but only for 3′-tRF-LeuAAG/TAG. The observed common target genes of 3′-tRF-LeuAAG/TAG were used for GO enrichment analysis; this analysis categorized the target genes based on the cellular component they act, the biological process in which they are implicated, and the molecular function they exert and provides an enrichment score, which shows if a set of genes associated with a specific cellular component/biological process/molecular function is over-presented among the others.
3. Results
3.1. Differences in tRF Levels of CD138+ Plasma Cells between sMM and MM Patients, as Well as among MM Patients’ Subgroups
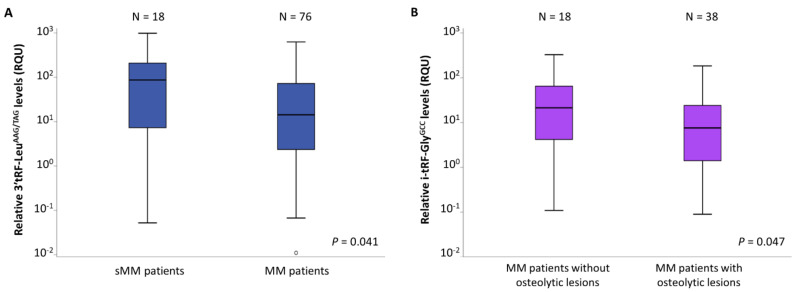
A mean ± standard error (S.E.) of 181.1 ± 62.5 RQU was observed for 3′-tRF-LeuAAG/TAG levels in sMM patients’ CD138+ plasma cells, while a mean ± S.E. of 51.5 ± 10.5 RQU was observed for the levels of the same tRF in MM patients’ CD138+ plasma cells. As revealed by the Mann–Whitney U test, this difference is statistically significant (p = 0.041); these results are shown in Figure 2A. The levels of the other five tRFs did not differ significantly between sMM and MM patients.
Figure 2.
Boxplots, showing the differences of relative 3′-tRF-LeuAAG/TAG levels between multiple myeloma (MM) and smoldering MM (sMM) patients (A), and of relative i-tRF-GlyGCC levels between MM patients without and with osteolytic lesions (B).
In order to examine potential relationships between tRFs and MM features, we checked whether the tRF levels differed between distinct subgroups of MM patients, including those with different M-protein isotype as well as patients with or without osteolytic lesions. No statistically significant differences were observed in the levels of each tRF among MM patients with different M-protein isotype (IgG and IgA). On the other hand, the Mann–Whitney U test revealed significantly higher levels of i-tRF-GlyGCC in the CD138+ plasma cells of patients without osteolytic lesions (mean ± S.E. = 47.8 ± 18.3), compared to those with osteolytic lesions (mean ± S.E. = 17.7 ± 5.1), as shown in Figure 2B (p = 0.047).
3.2. tRFs as Promising Molecular Indicators of Favorable Prognosis in MM
MM patients were subgrouped based on the levels of each tRF, based on the cut-off points determined for prognostic purposes using the X-tile software, as aforementioned. The frequencies of MM patient subgroups are shown in Table S1. Interestingly, positive i-tRF-GlyGCC status was found to be associated with the presence of chromosomal aberration (+1q) (Table S2). Associations between each tRF status and del(17p), t(4;14) or t(14;16) could not be checked, due to the limited number of MM patients bearing at least one of these chromosomal aberrations.
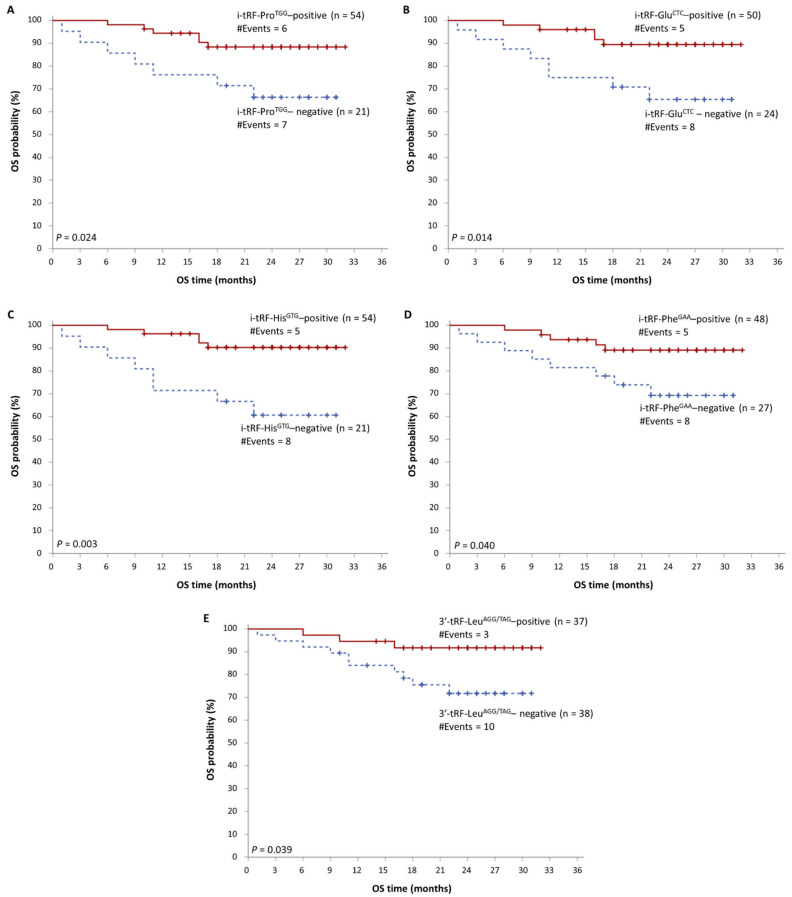
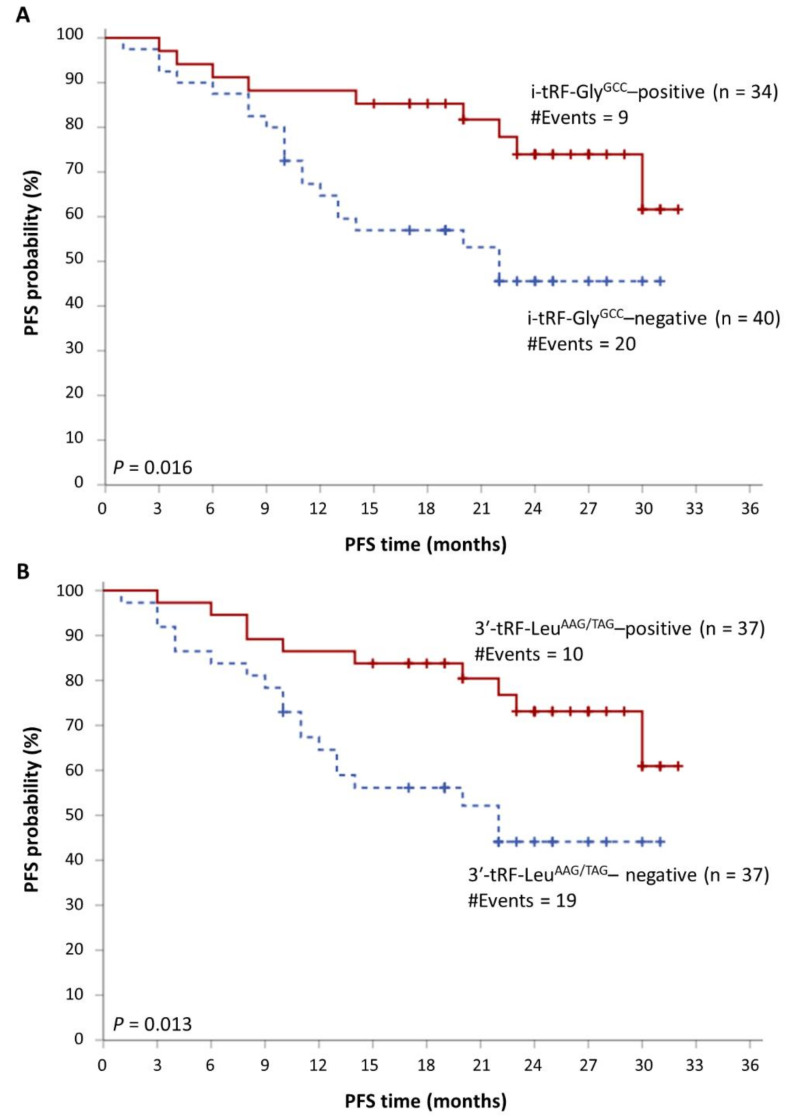
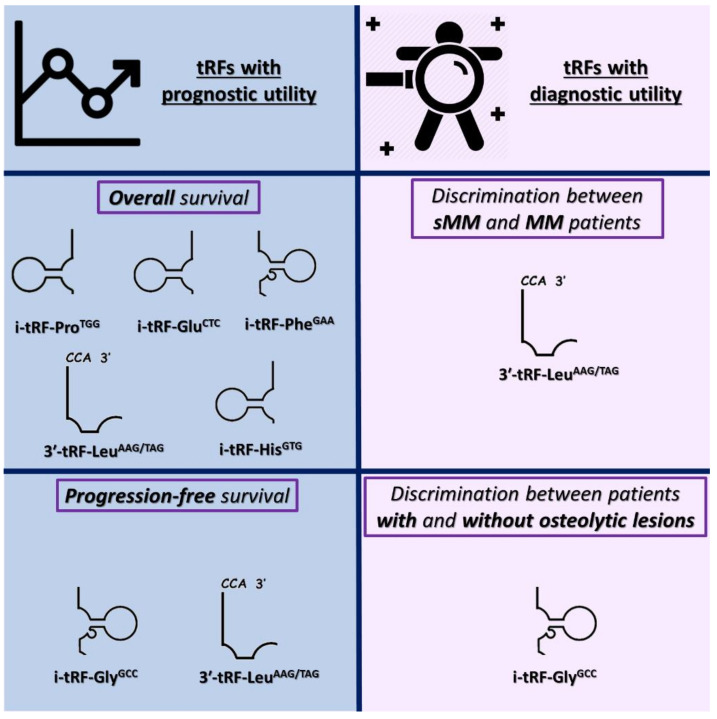
The median follow-up time of MM patients was 24 months. During the accrual period, 29 MM patients showed disease progression, while 13 MM-related deaths were also recorded. As revealed by Kaplan–Meier survival analysis, MM patients with high i-tRF-ProTGG, i-tRF-GluCTC, i-tRF-HisGTG, i-tRF-PheGAA, or 3′-tRF-LeuAAG/TAG levels show significantly prolonged OS, compared to those with lower levels of each of these molecules (p = 0.024, 0.014, 0.003, 0.040, and 0.039, respectively). These data are presented in Figure 3. Additionally, i-tRF-GlyGCC and 3′-tRF-LeuAAG/TAG can predict disease progression in MM patients. The PFS intervals were significantly longer in MM patients with high i-tRF-GlyGCC or 3′-tRF-LeuAAG/TAG levels (p = 0.016 and 0.013, respectively). These results are presented in Figure 4, while the putative biomarker utility of all tRFs is summarized in Figure 5.
Figure 3.
Kaplan–Meier overall survival (OS) curves, showing the differences in the survival intervals of MM patients with high levels of i-tRF-ProTGG (A), i-tRF-GluCTC (B), i-tRF-HisGTG (C), i-tRF-PheGAA (D), and 3′-tRF-LeuAAG/TAG (E), compared to patients with lower levels of these molecules.
Figure 4.
Kaplan–Meier progression-free survival (PFS) curves, showing the differences in the survival intervals of MM patients with high levels of i-tRF-GlyGCC (A), and 3′-tRF-LeuAAG/TAG (B), compared to patients with lower levels of these molecules.
Figure 5.
Graphical summary of the putative molecular biomarker utility of each tRF.
Moreover, patients who underwent HDM-ASCT and had high i-tRF-ProTGG, i-tRF-GluCTC, or i-tRF-HisGTG levels show significantly prolonged OS, compared to those who received HDM-ASCT and had lower levels of each of these molecules (p = 0.008, 0.045, and 0.019, respectively). Furthermore, patients who did not receive HDM-ASCT and had high i-tRF-HisGTG levels showed significantly longer OS intervals compared to patients with low i-tRF-HisGTG levels (p = 0.033) These results are presented in Figure S2. Regarding PFS, patients who received HDM-ASCT and had high i-tRF-GlyGCC or 3′-tRF-LeuAAG/TAG levels, show significantly prolonged PFS intervals in comparison with patients who received HDM-ASCT and had lower i-tRF-GlyGCC or 3′-tRF-LeuAAG/TAG levels (p = 0.039 and 0.022, respectively). These results are presented in Figure S3.
Intriguingly, bootstrap multivariate Cox regression analysis revealed that the prognostic significance of i-tRF-ProTGG, i-tRF-GluCTC, and i-tRF-HisGTG regarding MM patients’ OS is independent of that of R-ISS staging. Moreover, the prognostic significance of i-tRF-GlyGCC and 3′-tRF-LeuAAG/TAG is independent of ISS staging. These results are presented in Table 4.
Table 4.
Multivariate Cox regression analysis, regarding MM patients overall and progression-free survival.
| Covariate | HR 1 | 95% CI 2 | p Value 3 | BCa 4 Bootstrap 5 95% CI 2 | Bootstrap 5 p Value 3 | |
|---|---|---|---|---|---|---|
| Overall survival (OS) | i-tRF-ProTGG status | |||||
| Positive | 1.00 | |||||
| Negative | 4.06 | 1.28–12.82 | 0.0 17 | 0.98–46.70 | 0.011 | |
| R-ISS 6 (ordinal) | 3.39 | 1.28–8.96 | 0.0 14 | 0.93–38.24 | 0.024 | |
| i-tRF-GluCTC status | ||||||
| Positive | 1.00 | |||||
| Negative | 5.87 | 1.75–19.63 | 0.0 04 | 1.02–5.58 × 105 | 0.001 | |
| R-ISS 6 (ordinal) | 3.98 | 1.50–10.57 | 0.006 | 0.58 –7.39 × 105 | 0.010 | |
| i-tRF-HisGTG status | ||||||
| Positive | 1.00 | |||||
| Negative | 6.49 | 1.94–21.74 | 0.0 02 | 0.97–7.32 × 105 | 0.001 | |
| R-ISS 6 (ordinal) | 3.77 | 1.44–9.91 | 0.0 07 | 0.87–4.42 × 105 | 0.008 | |
| Progression-free survival (PFS) | i-tRF-GlyGCC status | |||||
| Positive | 1.00 | |||||
| Negative | 3.06 | 1.33–7.00 | 0.008 | 1.12–12.63 | 0.0 07 | |
| ISS 7 (ordinal) | 2.22 | 1.25–3.95 | 0.0 07 | 1.20–7.71 | 0.008 | |
| 3′-tRF-LeuAAG/TAG status | ||||||
| Positive | 1.00 | |||||
| Negative | 2.94 | 1.32–6.55 | 0.008 | 1.28–8.76 | 0.005 | |
| ISS 7 (ordinal) | 2.16 | 1.22–3.80 | 0.008 | 1.21–6.37 | 0.008 |
1 Hazard ratio; 2 Confidence interval; 3 Italics indicate a significant p value; 4 Bias-corrected and accelerated; 5 Based on 1000 bootstrap samples; 6 Revised International Staging System; 7 International Staging System.
3.3. In Silico Functional Analysis of 3′-tRF-LeuAAG/TAG
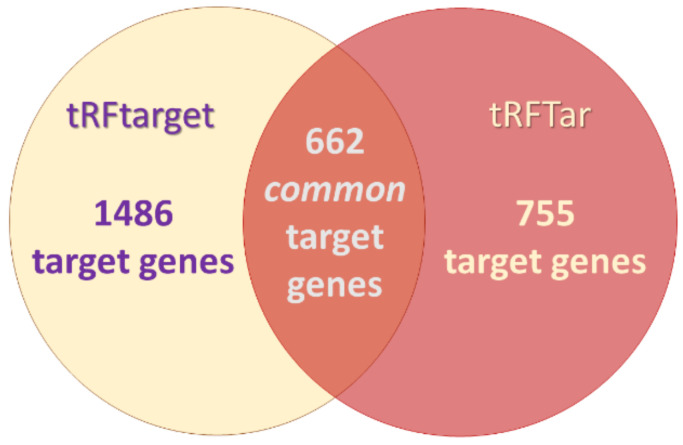
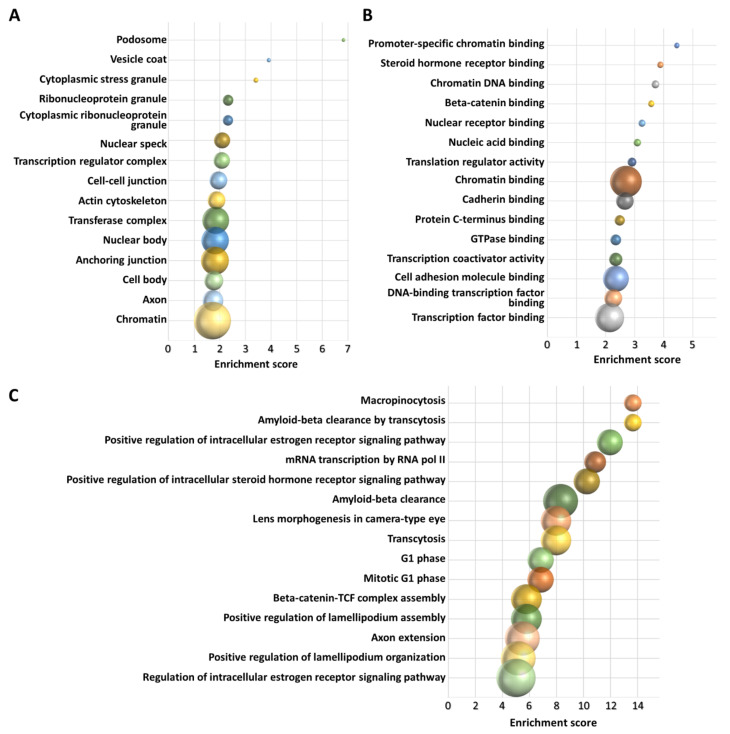
tRFTar and tRFtarget databases revealed 755 and 1486 target genes for 3′-tRF-LeuAAG/TAG, respectively. We preferred to perform the downstream analysis using the common target genes (662) provided by the two databases, as shown in Figure 6. The subsequent GO analysis of this gene list revealed 48 significantly enriched cellular components, 268 significantly enriched biological processes, and 63 significantly enriched molecular functions. Each enrichment score was considered as significant only when the p value was less or equal to 0.05. Based on these results, 3′-tRF-LeuAAG/TAG was found to be implicated among others in the regulation of gene expression, WNT signaling pathway, and steroid hormone signaling. In Figure 7, the cellular components, molecular functions, and biological processes showing the highest enrichment scores are graphically presented.
Figure 6.
Venn chart showing the number of putative 3′-tRF-LeuAAG/TAG targets obtained from each database and their intersection, which was used for the Gene Ontology (GO) analysis.
Figure 7.
The results of functional GO analysis for 3′-tRF-LeuAAG/TAG. The cellular components (A), molecular functions (B), and biological processes (C) showing the highest enrichment scores are shown in the charts. The size of each bubble indicates the number of genes implicated. Each chart is drawn in scale.
4. Discussion
By virtue of small RNA sequencing and other novel approaches, tRNA derivatives represent a study field that although it is still in its infancy, has arisen scientific interest. Their involvement in various molecular and cellular processes, including transcriptional regulation, cell growth, and differentiation, has highlighted their potential utility as biomarkers and therapeutic targets against cancer, as these processes are directly linked to cancer development [41]. Their deregulated levels in cancer have been variously reported, and numerous studies have attempted to associate this deregulation with the malignant state [42]. However, to the best of our knowledge, only a handful of research studies have attempted to investigate the role of tRFs in MM [28,29,30].
Firstly, our study proved the abundance of tRFs in the CD138+ plasma cells of MM and sMM patients. Moreover, we showed that 3′-tRF-LeuAAG/TAG was significantly overexpressed in sMM cases compared to MM patients. Nowadays, many imaging modalities that make bone disease detection extremely exact and differential diagnosis between sMM and MM simple and accurate are available. However, in some ambiguous cases, in which setting a diagnosis is really important [43], there is an unmet need for the discovery of molecular biomarkers able to distinguish between sMM and MM patients. Given that, 3′-tRF-LeuAAG/TAG most likely represents a novel molecular biomarker that could prove useful for the differential diagnosis of MM from sMM. Thus, further research in larger cohorts is required in order to establish such a role of this molecule. In the same context, i-tRF-GlyGCC, which is overexpressed in patients without osteolytic lesions, may serve as a biomarker able to distinguish between patients with and without bone disease.
Our study revealed a putative prognostic value in MM for all six molecules studied. More specifically, higher levels of i-tRF-ProTGG, i-tRF-GluCTC, i-tRF-HisGTG, and i-tRF-PheGAA were associated with superior OS, higher levels of i-tRF-GlyGCC were associated with prolonged PFS, while higher levels of 3′-tRF-LeuAAG/TAG correlated both with prolonged OS and PFS in the total population. Currently, the R-ISS staging system is universally used as a tool able to predict survival in MM patients. R-ISS staging consists of three subgroups with different survival rates; R-ISS I (5-year OS rate: 82%), R-ISS II (5-year OS rate: 62%), and R-ISS III (5-year OS rate: 40%) [10]. Our study revealed that three of the investigated tRFs, namely i-tRF-ProTGG, i-tRF-GluCTC, and i-tRF-HisGTG have a prognostic value independent of R-ISS staging. This means that those molecules may be able to serve in the future as additional prognostic biomarkers in MM that could even be incorporated or combined with R-ISS. It would be interesting to evaluate their prognostic significance and their ability to subclassify R-ISS stage II patients, who represent the larger and most heterogeneous R-ISS class (62% of the total population was classified as R-ISS II in the relevant publication) [10]. Moreover, all tRFs retained or showed a tendency for retaining their prognostic significance in patients who were subjected to HDM-ASCT. In the subgroup of patients not subjected to HDM-ASCT, i-tRF-HisGTG retained its prognostic significance regarding OS. Three out of the six tRFs investigated in this study, namely i-tRF-GlyGCC, i-tRF-PheGAA, and 3′-tRF-LeuAAG/TAG have already been proposed as biomarkers in CLL by previous works of members of our research group [24,25,27]. tRFs deriving from tRNAGluCTC have been detected in exosomes of cancer patients [44], while i-tRFs deriving from tRNAProTGG and tRNAHisGTG were shown to have prognostic value in squamous cell carcinoma of the head and neck [45]. These data highlight their implication in cancer progression and their utility as molecular biomarkers.
i-tRFs represent the least investigated tRF class. This may happen due to the fact that the cleavage position for i-tRF emergence varies, generating a large number of i-tRFs, some of which have subtle differences and subsequently, are difficult to be distinguished. A problem deriving from the poor investigation of i-tRFs is that an in silico functional analysis could not be carried out for the five i-tRFs included in this study, due to the lack of CLIP-seq or prediction data [30]. On the contrary, 3′tRFs represent the most well-investigated tRF class. This tRF category shares functional similarities to miRNAs, as they interact with Argonaute (AGO) proteins, and scientific evidence shows that they probably have a seed sequence, located at the same sequence positions as the respective seed sequence of the miRNAs [46].
Interesting results were obtained from the functional analysis concerning 3′-tRF-LeuAAG/TAG. This tRF was shown to be implicated in processes related to oncogenesis. More specifically, this tRF was found to interact with molecules that exert their function in the podosome and actin cytoskeleton. Podosomes are actin-rich structures anchored to the extracellular matrix [47], which have been shown to drive osteoclast-dependent bone resorption [48], a typical characteristic of MMBD [5]. Another cellular component showing a high enrichment score is the vesicle coat; MM cell-derived extracellular vesicles (EVs) play a significant role in MM proliferation, by providing the BM healthy cell populations with a cargo that facilitates malignant cell proliferation [49,50].
Regarding the possible implication of 3′-tRF-LeuAAG/TAG in specific biological processes, macropinocytosis is intriguingly interesting, as this process represents an amino acid supply mechanism for cancer cells [51]. In MM, macropinocytosis is also a mechanism for EV internalization. In MM cells bearing a mutation in RAS, a common MM patients’ mutation, this process was also shown to promote MM cell growth and survival, through glutamine provision [52].
Other significantly enriched biological processes related to MM are positive regulation of the estrogen receptor pathway and in general steroid hormone receptor pathway regulation. Glucocorticosteroids, one of the first treatments for MM, remain the cornerstone of MM treatment, even in the era of new agents, where glucocorticosteroids, especially dexamethasone, are a part of the majority of anti-myeloma treatment regimens [45,53]. The cytotoxic effect of dexamethasone in MM cells is well known and the mechanism behind that seems to be the upregulation of pro-apoptotic genes and downregulation of anti-apoptotic genes [54,55]. Decreased glucocorticoid receptor expression has been reported in MM patients, leading to resistance to glucocorticoid therapy [56]. Even more interestingly, 3′-tRF-LeuAAG/TAG was found to interact with genes that bind to steroid hormone receptors, which presents the most significantly enriched molecular function. Concerning the estrogen receptor, its activation inhibits IL-6–mediated MM cell growth, by inducing PIAS3 and leading to blockage of STAT3-induced signaling [57]. PIAS3 also blocks RANKL (also known as TNFSF11) expression, leading to inhibition of the RANK (also known as TNFRSF11A) receptor signaling and subsequently to MMBD reduction, as this pathway is directly linked to the genesis of osteolytic lesions [58].
Furthermore, the GO analysis showed that 3′-tRF-LeuAAG/TAG contributes to beta-catenin-TCF complex assembly. Beta-catenin (also known as CTNNB1) is the main mediator of WNT signaling; its interaction with the transcription factor TCF enhances the transcription of its target genes and consequently enhances bone formation. Moreover, MM cells secrete WNT inhibitors, leading to aberrant WNT-mediated signaling, an aspect associated with MM pathogenesis [59]. The high enrichment score of this biological process comes in line with the fact that the molecular function “Beta-catenin binding” is one of the molecular functions with the highest enrichment scores.
Most of the highly enriched molecular functions, combined with the also highly enriched biological process “mRNA transcription by RNA polymerase II”, could be explained by the regulatory role imputed to 3′tRFs, regarding gene expression. These molecular functions include promoter-specific chromatin binding, steroid hormone receptor binding, chromatin DNA binding, translation regulator activity, nucleic acid binding, translation regulator activity, chromatin binding, protein C-terminus binding, transcription coactivator activity, DNA-binding transcription factor binding, and transcription factor binding. All these data indicate that investigation of the mechanistic role of 3′-tRF-LeuAAG/TAG in MM could reveal new aspects, concerning the MM molecular background; however, due to the lack of experimental validation, we cannot be sure about the implication of this fragment in one, some or all of the processes described above. Thus, experimental verification is a necessity to provide a clearer perspective regarding the implication of 3′-tRF-LeuAAG/TAG in specific molecular processes and how these processes are connected.
Our study has some limitations. Firstly, the cohort size is not very large; nevertheless, it is important to mention that our study population is not subjected to age or performance status limitations and is quite indicative of the real-world MM population. Due to the small sample size, our study can only present some evidence about the clinical utility of the aforementioned tRFs as molecular biomarkers in MM. For the same reason, no associations between tRF levels and features of patients with rare MM types, such as IgD and non-secretory myeloma type, could be studied. Moreover, the results regarding the association of positive i-tRF-GlyGCC status with the presence of (+1q) need to be validated in a larger cohort of MM patients. Secondly, the median follow-up time of the patients included in the current study is only 2 years; however, a longer follow-up time is needed to uncover the emerging potential of particular tRFs as important molecular biomarkers in MM. Additionally, a cohort consisting of normal controls is not available, as BMA is a painful procedure with rare but still possible complications, performed only when the presence of particular hematologic diseases is suspected. However, unprocessed bone marrow samples could be purchased and used for comparison purposes. The inclusion of such samples could strengthen our conclusions about the diagnostic and prognostic utility of these tRFs in MM. Moreover, a weakness of our qPCR assays developed for tRF quantification is that fragments with slight sequence differences generated from the same tRNAs of origin are considered as a single one and are hence cumulatively quantified. Lastly, another limitation is that the selected CD138+ plasma cells included not only malignant plasma cells but also normal ones [60]. Furthermore, a subpopulation of malignant plasma cells, which are more primitive and which have a higher proliferative potential than CD138+ plasma cells, are CD138- [61], and hence not included in the positively selected plasma cells used in our study. In fact, current consensus recommendations include CD38, CD138, and CD45 as the best combination of backbone markers for the identification and selection of plasma cells; the addition of CD19 enables efficient distinction between normal/reactive and clonal plasma cell compartments based on their most frequent aberrant phenotypes [62,63,64]. However, multi-marker immunomagnetic enrichment of malignant plasma cells from BMA samples would be very laborious and most probably result in an insufficient cell number [65]. Cell selection based on this antibody panel for immunophenotyping could properly be performed by flow cytometry [66]; however, the amount of total RNA extracted by sorted malignant cells would be a limiting factor by itself for our experiments.
Summarizing the key findings of this work, we proved the abundance of tRFs in CD138+ plasma cells of MM and sMM patients. Moreover, we showed that some of these molecules represent putative molecular biomarkers that could distinguish between MM and sMM or between patients with and without osteolytic lesions. Regarding MM patients’ prognosis, all the investigated tRFs correlated with MM patients’ prognosis as analytically described above. All these results merit validation in a larger cohort. Additionally, the in silico functional analysis carried out for 3′-tRF-LeuAAG/TAG revealed putative new aspects in the regulation of the MM molecular background.
Our future goals include the deciphering of the molecular mechanisms underlying the function of these tRFs in MM. This could reveal novel therapeutic strategies concerning MM treatment; such an attempt has been also suggested for miRNAs, but there are still important limitations, including impaired miRNA stability, side effects, and incapability of targeted miRNA delivery. These problems will be probably faced if similar attempts are made with tRFs. Additionally, the detection of these molecules in MM-derived EVs could contribute to our deeper understanding of their involvement in MM.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biomedicines9121811/s1, Table S1. Subgrouping of the 76 MM patients based on the levels of each tRF in CD138+ plasma cells. Table S2. Associations between the status of each tRF and the chromosomal aberration (+1q). Figure S1: Melting curves of i-tRF-ProTGG (A), i-tRF-GluCTC (B), i-tRF-HisGTG (C), i-tRF-GlyGCC (D), i-tRF-PheGAA (E), 3′-tRF-LeuAAG/TAG (F), SNORD43 (G), and SNORD48 (H). Figure S2: Kaplan–Meier OS curves, showing the differences in the survival intervals of MM patients who had received autologous stem cell transplantation following high dose melphalan (HDM-ASCT) and showed high levels of i-tRF-ProTGG (A), i-tRF-GluCTC (B), and i-tRF-HisGTG (C), and in the survival intervals of MM patients who had not received HDM-ASCT and showed high levels of i-tRF-HisGTG (D), compared to patients with lower levels of these molecules. Figure S3: Kaplan–Meier PFS curves, showing the differences in the survival intervals of MM patients who had received HDM-ASCT and showed high levels of i-tRF-GlyGCC (A), and 3′-tRF-LeuAAG/TAG(B), compared to patients with lower levels of these molecules.
Author Contributions
Conceptualization, A.S., E.T. and C.K.K.; methodology, P.K., P.I.A. and C.K.K.; software, P.K.; validation, P.K., A.-M.P. and P.I.A.; formal analysis, P.K.; investigation, P.K., P.I.A., A.-A.L. and M.-A.K.; resources, A.-M.P., C.-I.L., D.P., N.M.-K., I.N.-S., M.G., E.K., M.-A.D. and E.T.; data curation, C.K.K.; writing—original draft preparation, P.K. and A.-M.P.; writing—review and editing, P.I.A. and C.K.K.; visualization, P.K.; supervision, C.K.K.; project administration, A.S. and E.T.; funding acquisition, M.-A.D. and E.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the General Hospital of Athens “Alexandra” (protocol code 859/24-10-2017 and date of approval: 25 October 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical issues.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Landgren O., Kyle R.A., Pfeiffer R.M., Katzmann J.A., Caporaso N.E., Hayes R.B., Dispenzieri A., Kumar S., Clark R.J., Baris D., et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: A prospective study. Blood. 2009;113:5412–5417. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajkumar S.V., Landgren O., Mateos M.V. Smoldering multiple myeloma. Blood. 2015;125:3069–3075. doi: 10.1182/blood-2014-09-568899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajkumar S.V. Multiple myeloma: 2018 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2018;93:981–1114. doi: 10.1002/ajh.25117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terpos E., Ntanasis-Stathopoulos I., Gavriatopoulou M., Dimopoulos M.A. Pathogenesis of bone disease in multiple myeloma: From bench to bedside. Blood Cancer J. 2018;8:7. doi: 10.1038/s41408-017-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombo M., Giannandrea D., Lesma E., Basile A., Chiaramonte R. Extracellular Vesicles Enhance Multiple Myeloma Metastatic Dissemination. Int. J. Mol. Sci. 2019;20:3236. doi: 10.3390/ijms20133236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthes T., Manfroi B., Huard B. Revisiting IL-6 antagonism in multiple myeloma. Crit. Rev. Oncol. 2016;105:1–4. doi: 10.1016/j.critrevonc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Guillerey C., Nakamura K., Vuckovic S., Hill G.R., Smyth M.J. Immune responses in multiple myeloma: Role of the natural immune surveillance and potential of immunotherapies. Cell Mol. Life Sci. 2016;73:1569–1589. doi: 10.1007/s00018-016-2135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greipp P.R., San Miguel J., Durie B.G., Crowley J.J., Barlogie B., Bladé J., Boccadoro M., Child J.A., Avet-Loiseau H., Kyle R.A., et al. International staging system for multiple myeloma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 10.Palumbo A., Avet-Loiseau H., Oliva S., Lokhorst H.M., Goldschmidt H., Rosinol L., Richardson P., Caltagirone S., Lahuerta J.J., Facon T., et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta N., Sharma A., Sharma A. Emerging biomarkers in Multiple Myeloma: A review. Clin. Chim. Acta. 2020;503:45–53. doi: 10.1016/j.cca.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 12.Papanota A.M., Tsiakanikas P., Kontos C.K., Malandrakis P., Liacos C.I., Ntanasis-Stathopoulos I., Kanellias N., Gavriatopoulou M., Kastritis E., Avgeris M., et al. A Molecular Signature of Circulating MicroRNA Can Predict Osteolytic Bone Disease in Multiple Myeloma. Cancers. 2021;13:3877. doi: 10.3390/cancers13153877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papadimitriou M.A., Papanota A.M., Adamopoulos P.G., Pilala K.M., Liacos C.I., Malandrakis P., Mavrianou-Koutsoukou N., Patseas D., Eleutherakis-Papaiakovou E., Gavriatopoulou M., et al. miRNA-seq and clinical evaluation in multiple myeloma: miR-181a overexpression predicts short-term disease progression and poor post-treatment outcome. Br. J. Cancer. 2021 doi: 10.1038/s41416-021-01602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Artemaki P.I., Letsos P.A., Zoupa I.C., Katsaraki K., Karousi P., Papageorgiou S.G., Pappa V., Scorilas A., Kontos C.K. The Multifaceted Role and Utility of MicroRNAs in Indolent B-Cell Non-Hodgkin Lymphomas. Biomedicines. 2021;9:333. doi: 10.3390/biomedicines9040333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna S., Raghavan S., DasGupta R., Palakodeti D. tRNA-derived fragments (tRFs): Establishing their turf in post-transcriptional gene regulation. Cell Mol. Life Sci. 2021;78:2607–2619. doi: 10.1007/s00018-020-03720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magee R., Rigoutsos I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020;48:9433–9448. doi: 10.1093/nar/gkaa657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuscu C., Kumar P., Kiran M., Su Z., Malik A., Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093–1105. doi: 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzzi N., Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020;17:1214–1222. doi: 10.1080/15476286.2020.1732694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong F., Hu Z., Jiang K., Lei B., Wu Z., Yuan G., Luo H., Dong C., Tang B., Zheng C., et al. Complement C3 activation regulates the production of tRNA-derived fragments Gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res. 2019;29:548–561. doi: 10.1038/s41422-019-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cosentino C., Toivonen S., Diaz Villamil E., Atta M., Ravanat J.L., Demine S., Schiavo A.A., Pachera N., Deglasse J.P., Jonas J.C., et al. Pancreatic beta-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018;46:10302–10318. doi: 10.1093/nar/gky839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu P., Yu J., Zhou P. Role of tRNA-derived fragments in cancer: Novel diagnostic and therapeutic targets tRFs in cancer. Am. J. Cancer Res. 2020;10:393–402. [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng T., Hua Y., Sun C., Zhang Y., Yang F., Yang M., Yang Y., Li J., Huang X., Wu H., et al. Relationship between tRNA-derived fragments and human cancers. Int. J. Cancer. 2020;147:3007–3018. doi: 10.1002/ijc.33107. [DOI] [PubMed] [Google Scholar]
- 23.Papadimitriou M.A., Avgeris M., Levis P., Papasotiriou E.C., Kotronopoulos G., Stravodimos K., Scorilas A. tRNA-Derived Fragments (tRFs) in Bladder Cancer: Increased 5’-tRF-LysCTT Results in Disease Early Progression and Patients’ Poor Treatment Outcome. Cancers. 2020;12:3661. doi: 10.3390/cancers12123661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karousi P., Adamopoulos P.G., Papageorgiou S.G., Pappa V., Scorilas A., Kontos C.K. A novel, mitochondrial, internal tRNA-derived RNA fragment possesses clinical utility as a molecular prognostic biomarker in chronic lymphocytic leukemia. Clin. Biochem. 2020;85:20–26. doi: 10.1016/j.clinbiochem.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Karousi P., Katsaraki K., Papageorgiou S.G., Pappa V., Scorilas A., Kontos C.K. Identification of a novel tRNA-derived RNA fragment exhibiting high prognostic potential in chronic lymphocytic leukemia. Hematol. Oncol. 2019;37:498–504. doi: 10.1002/hon.2616. [DOI] [PubMed] [Google Scholar]
- 26.Katsaraki K., Artemaki P.I., Papageorgiou S.G., Pappa V., Scorilas A., Kontos C.K. Identification of a novel, internal tRNA-derived RNA fragment as a new prognostic and screening biomarker in chronic lymphocytic leukemia, using an innovative quantitative real-time PCR assay. Leuk Res. 2019;87:106234. doi: 10.1016/j.leukres.2019.106234. [DOI] [PubMed] [Google Scholar]
- 27.Katsaraki K., Adamopoulos P.G., Papageorgiou S.G., Pappa V., Scorilas A., Kontos C.K. A 3’ tRNA-derived fragment produced by tRNA(LeuAAG) and tRNA(LeuTAG) is associated with poor prognosis in B-cell chronic lymphocytic leukemia, independently of classical prognostic factors. Eur. J. Haematol. 2021;106:821–830. doi: 10.1111/ejh.13613. [DOI] [PubMed] [Google Scholar]
- 28.Xu C., Fu Y. Expression Profiles of tRNA-Derived Fragments and Their Potential Roles in Multiple Myeloma. OncoTargets Ther. 2021;14:2805–2814. doi: 10.2147/OTT.S302594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojek A.E., Katanski C.D., Stefka A., Derman B.A., Jakubowiak A., Pan T. Abstract 2394: tRNA expression and tRFs in multiple myeloma: Progression from monoclonal gammopathies to relapsed/refractory disease. Cancer Res. 2021;81:2394. doi: 10.1158/1538-7445.Am2021-2394. [DOI] [Google Scholar]
- 30.Xu C., Liang T., Zhang F., Liu J., Fu Y. tRNA-derived fragments as novel potential biomarkers for relapsed/refractory multiple myeloma. BMC Bioinform. 2021;22:238. doi: 10.1186/s12859-021-04167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridley R.C., Xiao H., Hata H., Woodliff J., Epstein J., Sanderson R.D. Expression of syndecan regulates human myeloma plasma cell adhesion to type I collagen. Blood. 1993;81:767–774. doi: 10.1182/blood.V81.3.767.767. [DOI] [PubMed] [Google Scholar]
- 32.Kawano Y., Fujiwara S., Wada N., Izaki M., Yuki H., Okuno Y., Iyama K., Yamasaki H., Sakai A., Mitsuya H., et al. Multiple myeloma cells expressing low levels of CD138 have an immature phenotype and reduced sensitivity to lenalidomide. Int. J. Oncol. 2012;41:876–884. doi: 10.3892/ijo.2012.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wijdenes J., Vooijs W.C., Clement C., Post J., Morard F., Vita N., Laurent P., Sun R.X., Klein B., Dore J.M. A plasmocyte selective monoclonal antibody (B-B4) recognizes syndecan-1. Br. J. Haematol. 1996;94:318–323. doi: 10.1046/j.1365-2141.1996.d01-1811.x. [DOI] [PubMed] [Google Scholar]
- 34.Pliatsika V., Loher P., Magee R., Telonis A.G., Londin E., Shigematsu M., Kirino Y., Rigoutsos I. MINTbase v2.0: A comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018;46:D152–D159. doi: 10.1093/nar/gkx1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 37.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 38.Papageorgiou S.G., Kontos C.K., Kotsianidis I., Karousi P., Symeonidis A., Galanopoulos A., Bouchla A., Hatzimichael E., Repousis P., Zikos P., et al. Effectiveness of 5-Azacytidine in older patients with high-risk myelodysplastic syndromes and oligoblastic acute myeloid leukemia: A retrospective analysis of the Hellenic (Greek) MDS Study Group. J. Geriatr. Oncol. 2020;11:121–124. doi: 10.1016/j.jgo.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y., Peng H., Cui Q., Zhou Y. tRFTar: Prediction of tRF-target gene interactions via systemic re-analysis of Argonaute CLIP-seq datasets. Methods. 2021;187:57–67. doi: 10.1016/j.ymeth.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Li N., Shan N., Lu L., Wang Z. tRFtarget: A database for transfer RNA-derived fragment targets. Nucleic Acids Res. 2021;49:D254–D260. doi: 10.1093/nar/gkaa831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu M., Lu B., Zhang J., Ding J., Liu P., Lu Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020;13:121. doi: 10.1186/s13045-020-00955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S.Q., Sun B., Xiong Z.P., Shu Y., Zhou H.H., Zhang W., Xiong J., Li Q. The dysregulation of tRNAs and tRNA derivatives in cancer. J. Exp. Clin. Cancer Res. 2018;37:101. doi: 10.1186/s13046-018-0745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimopoulos M.A., Moreau P., Terpos E., Mateos M.V., Zweegman S., Cook G., Delforge M., Hajek R., Schjesvold F., Cavo M., et al. Multiple Myeloma: EHA-ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-up. Hemasphere. 2021;5:e528. doi: 10.1097/HS9.0000000000000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu L., Li J., Gong Y., Wu Q., Tan S., Sun D., Xu X., Zuo Y., Zhao Y., Wei Y.Q., et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer. 2019;18:74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu X., Wang L., Coates P.J., Boldrup L., Fahraeus R., Wilms T., Sgaramella N., Nylander K. Transfer-RNA-Derived Fragments Are Potential Prognostic Factors in Patients with Squamous Cell Carcinoma of the Head and Neck. Genes. 2020;11:1344. doi: 10.3390/genes11111344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy D.A., Courtneidge S.A. The ‘ins’ and ‘outs’ of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgess D., Machuca-Gayet I., Blangy A., Jurdic P. Podosome organization drives osteoclast-mediated bone resorption. Cell Adh. Migr. 2014;8:191–204. doi: 10.4161/cam.27840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y., Tu C., Zhang J., Wang J. Inhibition of multiple myelomaderived exosomes uptake suppresses the functional response in bone marrow stromal cell. Int J. Oncol. 2019;54:1061–1070. doi: 10.3892/ijo.2019.4685. [DOI] [PubMed] [Google Scholar]
- 50.De Luca L., Laurenzana I., Trino S., Lamorte D., Caivano A., Musto P. An update on extracellular vesicles in multiple myeloma: A focus on their role in cell-to-cell cross-talk and as potential liquid biopsy biomarkers. Expert Rev. Mol. Diagn. 2019;19:249–258. doi: 10.1080/14737159.2019.1583103. [DOI] [PubMed] [Google Scholar]
- 51.Commisso C. The pervasiveness of macropinocytosis in oncological malignancies. Philos. Trans. R. Soc. B Biol. Sci. 2019;374:20180153. doi: 10.1098/rstb.2018.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masoumi Moghaddam S., Spencer A. KRAS-Mutated Myeloma Cells Exploit Autophagy and Macropinocytosis to Tolerate Glutamine-Deprived Conditions. Blood. 2017;130:1786. doi: 10.1182/blood.V130.Suppl_1.1786.1786. [DOI] [Google Scholar]
- 53.Alexanian R., Dimopoulos M.A., Delasalle K., Barlogie B. Primary dexamethasone treatment of multiple myeloma. Blood. 1992;80:887–890. doi: 10.1182/blood.V80.4.887.887. [DOI] [PubMed] [Google Scholar]
- 54.Gehring U., Mohit B., Tomkins G.M. Glucocorticoid action on hybrid clones derived from cultured myeloma and lymphoma cell lines. Proc. Natl. Acad. Sci. USA. 1972;69:3124–3127. doi: 10.1073/pnas.69.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chauhan D., Auclair D., Robinson E.K., Hideshima T., Li G., Podar K., Gupta D., Richardson P., Schlossman R.L., Krett N., et al. Identification of genes regulated by dexamethasone in multiple myeloma cells using oligonucleotide arrays. Oncogene. 2002;21:1346–1358. doi: 10.1038/sj.onc.1205205. [DOI] [PubMed] [Google Scholar]
- 56.Thomas A.L., Coarfa C., Qian J., Wilkerson J.J., Rajapakshe K., Krett N.L., Gunaratne P.H., Rosen S.T. Identification of potential glucocorticoid receptor therapeutic targets in multiple myeloma. Nucl. Recept. Signal. 2015;13:e006. doi: 10.1621/nrs.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L.H., Yang X.Y., Mihalic K., Xiao W., Li D., Farrar W.L. Activation of estrogen receptor blocks interleukin-6-inducible cell growth of human multiple myeloma involving molecular cross-talk between estrogen receptor and STAT3 mediated by co-regulator PIAS3. J. Biol. Chem. 2001;276:31839–31844. doi: 10.1074/jbc.M105185200. [DOI] [PubMed] [Google Scholar]
- 58.Papanota A.M., Karousi P., Kontos C.K., Ntanasis-Stathopoulos I., Scorilas A., Terpos E. Multiple Myeloma Bone Disease: Implication of MicroRNAs in Its Molecular Background. Int. J. Mol. Sci. 2021;22:2375. doi: 10.3390/ijms22052375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Andel H., Kocemba K.A., Spaargaren M., Pals S.T. Aberrant Wnt signaling in multiple myeloma: Molecular mechanisms and targeting options. Leukemia. 2019;33:1063–1075. doi: 10.1038/s41375-019-0404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S., Kimlinger T., Morice W. Immunophenotyping in multiple myeloma and related plasma cell disorders. Best Pract. Res. Clin. Haematol. 2010;23:433–451. doi: 10.1016/j.beha.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reid S., Yang S., Brown R., Kabani K., Aklilu E., Ho P.J., Woodland N., Joshua D. Characterisation and relevance of CD138-negative plasma cells in plasma cell myeloma. Int. J. Lab. Hematol. 2010;32:e190–e196. doi: 10.1111/j.1751-553X.2010.01222.x. [DOI] [PubMed] [Google Scholar]
- 62.Harada H., Kawano M.M., Huang N., Harada Y., Iwato K., Tanabe O., Tanaka H., Sakai A., Asaoku H., Kuramoto A. Phenotypic difference of normal plasma cells from mature myeloma cells. Blood. 1993;81:2658–2663. doi: 10.1182/blood.V81.10.2658.2658. [DOI] [PubMed] [Google Scholar]
- 63.Paiva B., Almeida J., Perez-Andres M., Mateo G., Lopez A., Rasillo A., Vidriales M.B., Lopez-Berges M.C., Miguel J.F., Orfao A. Utility of flow cytometry immunophenotyping in multiple myeloma and other clonal plasma cell-related disorders. Cytom. B Clin. Cytom. 2010;78:239–252. doi: 10.1002/cyto.b.20512. [DOI] [PubMed] [Google Scholar]
- 64.Rawstron A.C., Orfao A., Beksac M., Bezdickova L., Brooimans R.A., Bumbea H., Dalva K., Fuhler G., Gratama J., Hose D., et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica. 2008;93:431–438. doi: 10.3324/haematol.11080. [DOI] [PubMed] [Google Scholar]
- 65.Beasley A.B., Acheampong E., Lin W., Gray E.S. Multi-Marker Immunomagnetic Enrichment of Circulating Melanoma Cells. Methods Mol. Biol. 2021;2265:213–222. doi: 10.1007/978-1-0716-1205-7_16. [DOI] [PubMed] [Google Scholar]
- 66.Van Dongen J.J., Lhermitte L., Böttcher S., Almeida J., van der Velden V.H., Flores-Montero J., Rawstron A., Asnafi V., Lécrevisse Q., Lucio P., et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–1975. doi: 10.1038/leu.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical issues.