Abstract
Cyanobacteria from the genus Arthrospira/Limnospira are considered haloalkalotolerant organisms with optimal growth temperatures around 35 °C. They are most abundant in soda lakes in tropical and subtropical regions. Here, we report the comprehensive genome-based characterisation and physiological investigation of the new strain O9.13F that was isolated in a temperate climate zone from the winter freezing Solenoye Lake in Western Siberia. Based on genomic analyses, the Siberian strain belongs to the Arthrospira/Limnospira genus. The described strain O9.13F showed the highest relative growth index upon cultivation at 20 °C, lower than the temperature 35 °C reported as optimal for the Arthrospira/Limnospira strains. We assessed the composition of fatty acids, proteins and photosynthetic pigments in the biomass of strain O9.13F grown at different temperatures, showing its potential suitability for cultivation in a temperate climate zone. We observed a decrease of gamma-linolenic acid favouring palmitic acid in the case of strain O9.13F compared to tropical strains. Comparative genomics showed no unique genes had been found for the Siberian strain related to its tolerance to low temperatures. In addition, this strain does not possess a different set of genes associated with the salinity stress response from those typically found in tropical strains. We confirmed the absence of plasmids and functional prophage sequences. The genome consists of a 4.94 Mbp with a GC% of 44.47% and 5355 encoded proteins. The Arthrospira/Limnospira strain O9.13F presented in this work is the first representative of a new clade III based on the 16S rRNA gene, for which a genomic sequence is available in public databases (PKGD00000000).
Keywords: Arthrospira, haloalkalotolerant cyanobacteria, metagenomics, phylogenomics, fatty acid
1. Introduction
Arthrospira Stizenberger ex Gomont 1892 [1]/Limnospira sensu Nowicka-Krawczyk [2] that was formerly called Spirulina [3] is a non-nitrogen fixing, filamentous cyanobacterium representing edible, haloalkalotolerant organisms. The genus Arthrospira has been observed to occur worldwide in a varied range of haloalkaline habitats, from freshwater alkaline conditions to hypersaline environments [1,4,5]. In tropical and subtropical regions, it forms exceptionally high blooms turning soda lakes into one of the world’s most productive ecosystems [6]. The occurrence of Arthrospira has been reported all around the globe (apart from polar regions) from the natural water bodies characterised by a high average temperature during summer and not entirely freezing during winter. Such environments are found throughout the world in Africa’s Rift Valley, in Central Asia, in the North American Desert, on the Andean Plateau, in the rain-shadowed regions of California and Nevada, on the Central Mexican Plateau, on a high-altitude wetland in the Chilean Altiplano, in Manitoba (Canada), in Wadi al Natrun (Egypt), on the Decan Plateau (India), as well as in salty puddles (in Baranda, Europe), in a tropical crater lake located on Mayotte Island (Comoros Archipelago, Western Indian Ocean), in Western Siberia (Russia) and in Eastern Australia [5,6,7,8,9,10,11]. In contrast, the commercial biomass of Arthrospira is cultivated only in areas with high yearly average temperatures due to economic reasons. The comparative metagenomic analyses of water, sediments and mats from Canadian, Siberian and American soda lakes revealed a common microbiome characteristic for this haloalkaline environment [8,10,12,13]. Interestingly, the genus Arthrospira/Limnospira is dominant in tropical soda lakes in Africa. In contrast, it is absent in the Southern Siberian soda lakes (Kulunda Steppe, Altai) [14] and has not been observed in this area since 1931 when N.N. Voronihin described Spirulina/Athrospira fusiformis. However, Arthrospira was detected in Solonoye Lake in Western Siberia [7].
The nutritional value of the Arthrospira biomass is commonly known and well documented. Besides its nutritional properties, Arthrospira evinces therapeutic and industrial potential. Biological activity of several Arthrospira compounds, namely, C-phycocyanin, γ-linolenic acid (GLA), calcium spirulan and immulina have all been documented in the fields of cancer-fighting, immunomodulation, cardiovascular diseases prevention and others [15]. In the era of alternative energy resources, the Arthrospira biomass is also a promising biofuel, producing mostly hydrogen [16,17,18,19]. Arthrospira sp. PCC 8005 is also a part of the fourth compartment of the Micro-Ecological Life Support System Alternative (MELiSSA), aiming to elaborate artificial ecosystems for space expeditions [20,21]. The biomass of Arthrospira is commercially available under the name ‘Spirulina’ due to a taxonomic revision proposed by Geitler in 1932 [3] that was later shown to be wrong [22]. Since then, it has been shown that many strains listed as Spirulina belong to Arthrospira [23]. However, this commercial name is not likely to be replaced by the correct one in the foreseeable future due to its practical and technological meaning [24]. The taxonomy of Arthrospira was primarily based on morphology, although ecology and the geographic origin of the organisms was also partly considered to distinguish many species [23,25]. However, the distinction of the Arthrospira species based on morphology is questionable and can be misleading, since, e.g., the helix geometry and gas vacuole presence can be influenced by physical and chemical conditions [26,27,28].
So far, for the genetic characterisation and taxonomy of Arthrospira only three genetic markers: the 16S rRNA gene, 16S-23S rRNA Internally Transcribed Spacer (ITS) and phycocyanin operon fragment cpcBA have been used. The phylogenetic studies of Arthrospira based on the 16S rRNA gene divided the genus into three clads (Comte et al. 2013). Meanwhile, only two clusters (I and II) and five subclusters (I.A, I.B, II.A, II.B, II.A/II.B) have been discriminated based on the sequence polymorphism of the ITS [29,30]. The intrageneric relatedness has also been assessed by a sequence analysis of a region of the phycocyanin operon, consisting of the intergenic spacer (IGS) and parts of the flanking cpcB and cpcA genes [5,31,32,33]. According to Papapanagiotou and Gkelis [34], the combined analysis of molecular (16S rRNA, ITS and cpcBA-IGS sequences) and phenotypic (13 morphological and morphometric) characters proposed and described by [35] data were consistent and divided the Arthrospira strains into three clusters/taxa. Unfortunately, it is difficult to assess whether the genetic relationship within the genus Arthrospira is the same for all three markers, as the phylogenies based on each of them were carried out on different sets of strains. It should be emphasised that the 16S rRNA analysis [2] was not supported by other markers that are typically used in the phylogenetic analysis of cyanobacteria, such as the ITS or phycocyanin operon fragment cpcBA. Moreover, since there are no isolates of Arthrospira jenneri strains observed in an environmental sample by [2] or deposited in culture collections, further analyses of the biochemical properties and genetic traits that would allow the comparison of both the Athrospira/Limnospira genera are not possible. Likewise, no phylogenomic analyses can be performed. Due to the lack of verified isolated strains and/or original genetic material of A. jenneri, which can be used for comparative analysis, it is debatable whether this reclassification is accurate. The modern taxonomy of prokaryotes that can be cultivated in vitro should not be based solely on the study of only one or a few genes nor exclusively based on their morphological features, which, in the case of Arthrospira, are highly variable depending on numerous factors, which have been accurately documented in the literature [36]. Therefore, in this work, in which we use genomic data for the classification of a new Siberian isolate, we retain the taxonomic name Arthrospira.
Up to now (7 October 2021), 17 Arthrospira genomes have been sequenced: Arthrospira sp. PCC 8005 [37], A. platensis NIES-39 [38], A. platensis C1 [39], A. maxima CS-328 [40], A. platensis Paraca P0 [41], A. platensis YZ [42], Arthrospira sp. TJSD091 [43], Arthrospira sp. PLM2.Bin9 [13,44], A. platensis NIES-46 [19], A. platensis FACHB-835 [45], A. fusiformis SAG 85.79 [31], Arthrospira sp. BM01 [46], A. fusiformis KN (JACGXW000000000), A. platensis FACHB-971 [45], A. platensis FACHB-439 [45], Arthrospira sp. PCC 9108 (CP066886), Arthrospira sp. SH-MAG29 [9] and Arthrospira sp. TJSD092. This publicly available genomic data can be used for comparative genomics and phylogenomic analyses allowing a reliable identification of the newly isolated Siberian strain O9.13F.
This study aimed to perform a comprehensive genetic and physiological investigation of the new Arthrospira/Limnospira sp. strain O9.13F isolated from an alkaline, winter freezing Siberian lake. The study aimed (1) to classify and determine the systematic position of the new strain, (2) to characterise physiologically the strain capable of surviving the winter period in the Siberian lake, (3) to perform a comparative genomic analysis of Siberian and tropical Arthrospira/Limnospira strains and (4) to verify whether the O9.13F strain is suitable for cultivation in the temperate climate zone.
2. Materials and Methods
2.1. Strain Isolation and Growth Conditions
On 11 September 2013, the thick cyanobacteria bloom from Solenoye Lake (Supplementary Figure S1) was collected into sterile, capped containers and transported to the laboratory. Then the biomass was resuspended in a liquid Zarrouk medium [47] and treated with trimethoprim (60 µg/mL), cycloheximide (200 µg/mL) and amphotericin B (5 µg/mL) to remove fungi. Next, a series of repassages on the solid Zarrouk medium was performed to separate cyanobacteria present in the bloom from each other and co-occurring bacteria. The axenity of the Arthrospira cultures was examined according to the procedure recommended by Rippka [48].
Moreover, to be certain that the new strains were unicyanobacterial, eight single filaments with different morphologies were isolated from the fresh biomass from an alkaline pond. From each of the filaments, pure cyanobacterial cultures (named O9.13A-H) were obtained. All new isolates were identified by sequencing of the 16S rRNA gene and screened by PCR with primers specific for the ITS subclusters [29,30] and the ITS_CL_III primer designed in this work. As the newly isolated strains were morphologically and genetically identical, only the O9.13 F strain was selected for further analyses and remained a part of the Arthrospira strains collection at the Intercollegiate Faculty of Biotechnology, University of Gdansk, Poland.
To determine the optimal growing conditions, the Arthrospira culture was grown with and without shaking at 150 rpm in the Zarrouk liquid medium pH~10–11 under a 16 h light/8 h dark photoperiod under illumination of 30–40 µmol photons m−2 s−1 provided by fluorescent lamps at four temperatures: 15 °C, 20 °C, 28 °C and 35 °C and in three salinities, NaCl concentration of 25 g/L, 50 g/L and 100 g/L, in triplicates, starting with the culture of similar optical density (3 McF in the case of strain O9.13F). Two other Arthrospira reference strains, PCC 8005 and PCC 7345, were grown in the same conditions, except temperature 15 °C at which these strains die off. The growth index was expressed as a ratio of the initial culture optical density to the final measured value of a particular sample after 8 days of incubation (averaged from all replicates). To assess the properties of the biomass, the fatty acids (FA), biosynthetic pigments and proteins were extracted from the above-mentioned cultures.
2.2. Phenotypic Characteristics of Arthrospira sp. O9.13F
2.2.1. Acclimation of Arthrospira Strains to Long-Term Stress of Low Temperature
Arthrospira, O9.13F and four strains PCC 8005, PCC 7345, SAG 49.88 and CCALA 023 were cultivated without shaking under cold room conditions in the Zarrouk medium under a constant lighting of 10 µmol photons m−2 s−1 with a daily amplitude of 5–8 °C for 10 months. After 3 and 10 months of cultivation in the cold room, all samples were inoculated onto the solid Zarrouk medium and grown at 20 °C under a light intensity of 20 µmol photons m−2 s−1 for two months. The photographic documentation was made using a Nikon 80i light microscope.
2.2.2. The Survivability of Arthrospira Strains in Different Stress Conditions
The Arthrospira strains O9.13F, PCC 8005 and PCC 7345 were subjected to stress conditions caused by the addition of different substances, cultivation in water acquired from industrial ponds or changes in medium salinity. The stress factors were selected based on several criteria: (i) They can appear in the natural pond next to the farmlands. (ii) The used stressor would be useful for the removal of an excess of associated microorganisms from the biomass before various procedures (e.g., genome sequencing). (iii) They would be beneficial in growing in numerous industrial and natural ponds. The tested stressors and conditions included: working solution of fungicide 0.35% (Curzate Cu 49.5 containing cymoxanil and copper oxychloride), working solution of herbicide 0.75 mg/L (glyphosate), working solution of insecticide 0.04% (Clothianidin), hydrogen peroxide 50, 100 and 200 mM, silver nanoparticles (AgNPs) 3 μg/mL, industrial water rich in calcium, potassium and sodium carbonate with pH = 13 (Poland); undiluted and industrial water mixed 1:1 with a Zarrouk medium, water from Gdańsk Bay (estimated 7–8‰ of salinity) and the addition of NaCl to total salinity 150 and 200 g/L. The biomass was cultivated at 28 °C under stress for one week. Afterwards, the biomass was washed with distilled water and transferred to a solid Zarrouk medium. Due to the filamentous character of the Arthrospira biomass, as well as the tendency of the biomass to create mats, a standard means of measuring bacterial growth (optical density) is not applicable for assessing the growth kinetics of this organism. Therefore, a method based on the condition, duration of the lag phase and visible increase in the number of viable filaments was developed. Here, the recovery rate was defined as to whether the trichomes survived the stress conditions and, if so, after what time the active growth was resumed upon reintroduction to physiological conditions. This process was monitored using the following criteria: 0—degradation of the filaments, 1—whole, nonfragmented filaments present, no growth observed, 2—some growth observed after two weeks of a lag phase, 3—weak growth after one week of a lag phase, 4—stable growth after a short lag phase and 5—abundant growth without a lag phase.
2.2.3. Fatty Acids Methyl Ester Analysis (FAME)
The total lipids were extracted in triplicates according to the Bligh and Dyer protocol [49] by homogenising the biomass in chloroform/methanol under ultrasonication. Following evaporation, the lipids were derivatised to FAMEs with sulphuric acid (2%) in methanol. The resulting FAMEs were extracted to hexane and analysed using a Shimadzu GC-2010 equipped with a flame ionization detector and a 60 m × 0.25 mm BPX70 column (SGE Analytical Science). Standard mixtures of fatty acids methyl esters (Supelco® 37 Component FAME Mix, 1890, cis/trans) were used to identify the major fatty acids based on their retention time. Individual FAs were expressed as percentages of the total FAs present in the chromatogram.
2.2.4. Proteins Content Analysis
The total amounts of proteins in the extracts obtained by ultrasonication of the biomass were measured following the protocol of Bradford [50]. The 1 mL samples (triplicates) of the culture of Arthrospira sp. O9.13F cultivated in various conditions were carefully washed with distilled water three times and centrifuged for 20 min at 13,200× g rpm. The cell lysis was performed with 1 mL of 0.1 M NaOH, and the cells were incubated for 30 min at 95 °C with shaking. Afterwards, the tubes were left to cool down, acidified with a 5 M solution of HCl and centrifuged for 10 min under 13,200 rpm. To each well in the 96-well plate, 250 μL of the Bradford reagent (Sigma Aldrich #B6916) and 10 μL of the samples (in triplicates) were added. The spectrophotometric measurements were done on an Envision 2105 Multimode Plate Reader (Perkin Elmer) after 30 min of incubation at room temperature without shaking in the wavelength 595 nm. The protein concentration was determined by comparison to a standard curve prepared based on a BSA solution in the range between 0.1–1.4 mg/mL.
2.2.5. The Measurements of Photosynthetic Dyes Content
The biomass obtained from 1 mL of the tested culture in triplicates was centrifuged for 20 min at 13,200× g rpm, the supernatant was removed, and 1 mL of 90% methanol was added. The biomass was further lysed by ultrasonication for 20 min and centrifuged again for 15 min at 13,200× g rpm. The absorbance was measured by the spectrophotometer at 470 nm, 665 nm and 720nm. The concentration of chlorophyll a and total carotenoids was established according to equations by [51,52]:
| Chlα [μg/mL] = 12.9447 × (A665 − A720) |
| Carotenoids [µg/mL] = 1000 × (A470 − A720) − 2.86 Chlα [µg/mL]/221 |
2.2.6. Statistical Analysis
The analysis of differences between the groups was performed using the Shapiro–Wilks test followed by Kruskal–Wallis test using the criterium of Fisher’s least significant difference (LSD) in R [53].
2.3. Microscopic Observations
2.3.1. Light Microscopy
The microscopic observations of Arthrospira sp. O9.13F filaments were performed, and the photographs were taken with a Nikon 80i microscope with a Nomarski contrast at ×400 and ×1000 magnification with the NIS Elements version D software from Nikon.
2.3.2. Transmission Electron Microscopy (TEM)
Preparation for the ultrathin sectioning and preparation for TEM were carried out as described previously [23,54,55]. Samples were fixed in 2.5% formaldehyde and 2.5% glutaraldehyde in a 0.05 M cacodylate buffer (pH 7.0) for 4 h at room temperature. Then, the material was rinsed in the same buffer and postfixed in 1% osmium tetroxide in the cacodylate buffer at 4 °C overnight. Specimens were treated with 1% uranyl acetate in distilled water for 1 h, dehydrated in an acetone series and embedded in Spurr’s resin. Serial ultrathin (60–100 nm) sections were cut with a diamond knife on a Sorvall MT2B microtome. The material on the grids was poststained with a saturated solution of uranyl acetate in 50% ethanol and 0.04% lead citrate. Observations were made using a Philips CM 100 transmission electron microscope operating at 80 kV. For the scanning electron microscope, samples from fresh cultures were fixed in 2.5% formaldehyde and 2.5% glutaraldehyde in 0.05 M Na-cacodylate buffer (pH = 7.0) for 4 h at room temperature, dehydrated in an ethanol series (30, 50, 75, 96, 100%) and critical point dried using liquid CO2 and a K850 critical point dryer (EMITECH, Ashford, England). The dried specimens were mounted on stubs with SPI carbon conductive double-sided adhesive discs, gold coated (350 Å) (SPI-Module Sputter Coater, Structure Probe, Inc., Chester, PA, USA) and examined in a Phillips XL-30 SEM operating at an accelerating voltage of 15 kV.
2.4. Molecular Characteristics
2.4.1. DNA Extraction
For DNA extraction, 10 mL of the dense Arthrospira culture cultivated at 20 °C was centrifuged, and the collected biomass was carefully washed 3 times with sterile distilled water prior to nucleic acid extraction. Afterwards, the cell lysis and nucleic acids extraction were carried out according to the protocol proposed by the Joint Genome Institute for bacterial DNA isolation using CTAB [56] followed by RNA digestion using Turbo RNase (Ambion). The DNA quantity and quality were assessed first using a NanoDrop Spectrophotometer and later with agarose gel electrophoresis.
2.4.2. PCR Amplification Sequencing and Phylogenetic Analysis
The sequences of the cpcBA, ITS and 16S rRNA gene of the new Siberian isolates were determined after cloning as previously described [57]. To confirm this assignment, one clone for each ribosomal operon variant was sequenced with primers 359F, 16S979F and 23S30R [58,59,60]. For amplification and sequencing of phycocyanin operon, fragment primers described by [32] were used. Sequencing was carried out using an ABI PRISM DNA Sequencer (Perkin-Elmer) according to the manufacturer’s manual. Both strands were sequenced using the forward and reverse PCR primers. Moreover, the clones were assigned to the different ITS clusters as described by [30] with an additional reverse primer ITS_CL_III (TTGACTATCAGAGGAAATCCG) designed by us specific for cluster III based on the sequence from Arthrospira PCC9901 (MF509176) and which was used with the primer 16S3′F [29].
Further, the obtained sequences were subjected to a BLAST sequence similarity search analysis to identify the nearest taxa. The obtained sequences of the 16S rRNA gene, cpcBA operon, ITS and sequences of the closest representative taxa that belong to cyanobacteria were aligned using the MAFFT algorithm in Geneious v9.1.8. [61]. The phylogenetic analysis was performed with the MEGA v. X software, (www.megasoftware.net (accessed on 23 October 2020)), and trees were constructed using the Neighbour Joining algorithm with a Tamura 3-parameter and Maximum Likelihood algorithm, and the Hasegawa–Kishino–Yano models selected on the model test module were implemented in MEGA. A bootstrap analysis with 1000 replications was performed to assess the robustness of the clusters.
The nucleotide sequence data reported in this paper are available under the following accession numbers: 16S rDNA, MF509165, ITS; MF509166–MF509169, cpcBA and MF509119. As the 16S rRNA gene sequences obtained for all eight clones were identical, only one was deposited in the GenBank database. A similar situation occurred with the cpcBA operon sequence. In the case of the ITS, the sequences of two different genetic variants obtained for the two clones O9.13F and O913H were submitted.
2.4.3. Whole-Genome Sequencing and Annotation
Genome sequencing was performed on an Illumina HiSeq 2000 platform in the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences, Warsaw, Poland. The genome assembly was performed using computational resources provided by the grid computer “durandal” operated by InBioS-PhytoSYSTEMS at the University of Liège (Belgium).
The raw reads were first filtered using Kraken v1.0 software [62] followed by quality and adapter trimming performed using Trimmomatic v0.32 [63]. The Kmer length and distribution were analysed using kmergenie v1.6949 [64]. Processed reads were de novo assembled using Spades v3.10.1 [65], Velvet v1.2.10 [66], MIRA v1.1.1 [67], Geneious v9.1.8 [61] and Ray v2.3.1 [68]. The resulting sets of contigs were integrated, and the final assembly was corrected by CISA v1.3 (Lin and Liao, 2013) software and finally scaffolded with SSPACE v3.0 [69]. The assembly was evaluated using Quast v4.5 software [70].
Annotation was performed by the NCBI Prokaryotic Genome Annotation Pipeline [71,72]. Functional annotations were carried out by the KEGG Automatic Annotation Server [73] and AutoFACT [74], for which the annotation was based on the COG, Smart and Pfam databases. Signal peptides and transmembrane helices were predicted with a Phobius server [75].
R-M systems were identified using the REBASE® database [76], Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) sequences were predicted using the CRISPRFinder tool [77]. CRISPR-associated genes used in comparative genomics were obtained from the MaGe platform [78]. The identification of prophage regions was performed using the PHASTER tool [79]. Plasmid sequences were detected using the PlasmidFinder server [80].
The screening for the secondary metabolite biosynthesis gene clusters in the Arthrospira sp. strain O9.13F genome sequence was assessed with AntiSMASH [81,82].
2.4.4. Phylogenomic Analyses
The phylogenomic analysis is based on the sequences of 363 of the most conserved proteins comparison using the PhyloPhlAn computational pipeline (v3) [83]. The calculation of the average nucleotide identity (ANI) was performed using the PyAni module (v0.2.10) [84] with MUMmer (v3.23) [85] as an alignment method. In silico DNA–DNA hybridisation (isDDH) values were calculated using the Genome-to-Genome Distance Calculator (v2.1) [86] with BLAST+ [87] as a local alignment tool.
3. Results
3.1. Phenotypic Characteristics of Arthrospira sp. O9.13F
3.1.1. Morphology of Arthrospira sp. O9.13F
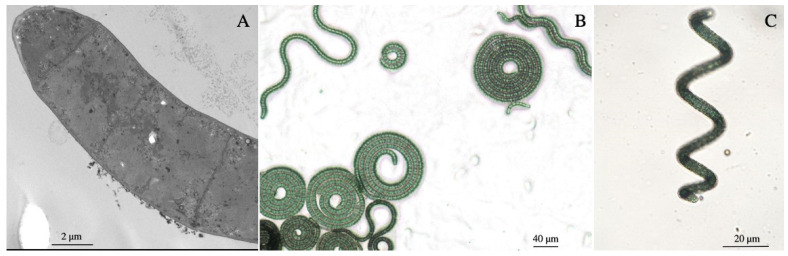
Upon introduction into the laboratory conditions, trichomes of Arthrospira sp. O9.13F were in the shape of tight left-handed open helices (Figure 1). With time and without the influence of natural UV light, the trichomes became looser. Macroscopically, the biomass of strain O9.13F either created a layer just under the surface of the medium or remained on the bottom of the bottle with a tendency to create mats. The trichomes were motile and covered with a layer consisting of exopolysaccharides. The morphology and ultrastructures of the Arthrospira sp. O9.13F were typical for representatives of this genus. The TEM (Figure 1A and Figure 2) and light microscopy show the cell dimensions and morphology of the filaments grown on a solid (Figure 1B) and liquid medium (Figure 1C).
Figure 1.
Images of Arthrospira sp. O9.13F. Photographs were taken by TEM (A) and light microscopy of Arthrospira culture grown on solid (B) and liquid Zarrouk medium (C).
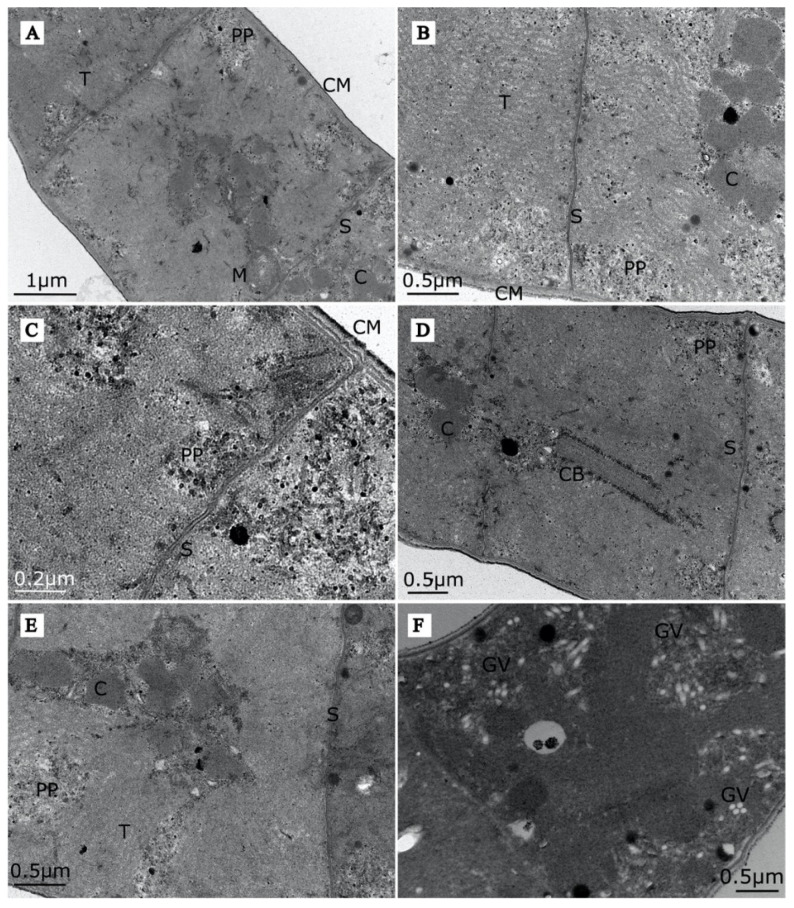
Figure 2.
TEM photography of cellular ultrastructures of Arthrospira sp. O9.13F. Panels (A–F) represent different cross-sections of cells illustrating ultrastructures recognised. Abbreviations as follows: cell membrane (CM), septum (S), gas vesicles (GV), thylakoids with bead-like ribosomes (T), mesosome (M), carboxysomes (C), cylindrical bodies (CB), polyphosphate granule (PP).
We identified all the ultrastructures recognised in other strains of Arthrospira investigated by Van Eykelenburg [54] and Tomaselli [23]—thylakoids, cylindrical bodies, mesosomes, gas vesicles, carboxysomes, cyanophycin granules, polyhydroxyalkanoate inclusions, cell membranes and septa (Figure 2A–E). Abundant gas vesicles with a diameter of 45–60 nm and a length up to 600 nm scattered throughout the cells were visible in the EM pictures of strain O9.13F (Figure 2F). The density is comparable to other strains as visible in the TEM microscopic images by [23,54].
3.1.2. Growth Conditions
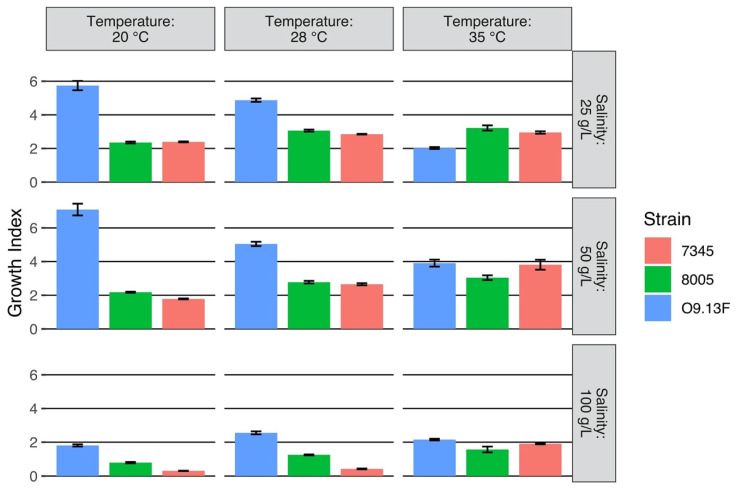
Cell growth occurred at pH 7.0–10.0, and it tolerated from 1 up to 150 (g/L) of NaCl. The growth temperature of the strain O9.13F ranged between 15 and 35 °C, with optimum growth at 20 °C (Supplementary Figure S2). The optimal growth index for O9.13F of 20 °C was lower than the optimal temperature of the tropical strains Arthrospira sp. PCC8005 (India) [30] and A. platensis PCC7345 (saline marsh Del Mar Slough, CA, USA) for which the largest increase of biomass production was observed at 28 °C (Figure 3). Two reference strains, PCC8005 and PCC7345, were not grown at a temperature of 15 °C because in this condition, both strains stopped their metabolism, and filaments started to fall apart; therefore, the measurements of proteins and fatty acids were below the sensitivity limit of the method. Interestingly, for the strain O9.13F grown at 15 °C, its growth index was higher in the medium with higher salinity. It was around 4 McF in the medium supplemented with 150–200 g/L of NaCl, while at lower concentrations of 25–50 g/L, it reached a maximum of 2.5. The highest growth was observed in the medium with a salinity of 150 g/L (Supplementary Figure S2).
Figure 3.
Effect of salinity (Zarrouk medium supplemented with NaCl to total salinity up to 25 g/L, 50 g/L, 100 g/L) on the growth of Arthrospira sp. strains, O9.13F, PCC7345 and PCC8005. Strains were grown for 8 days at different temperatures (20 °C, 28 °C and 35 °C). The optical density measurements were performed in triplicates for each strain in each of the conditions. The standard deviations are shown. The growth index is the quotient of the optical density [McF] of the culture after 8 days incubation and the optical density of the culture at the start point of the experiment.
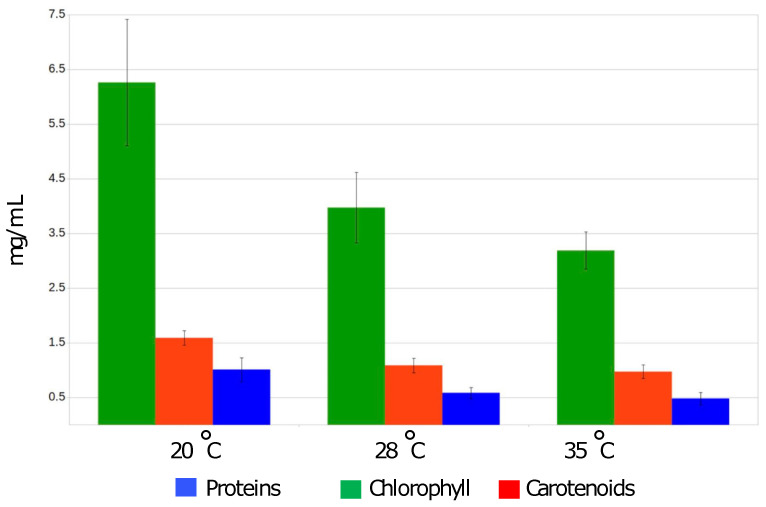
Accordingly, the highest amount of proteins (1.01 mg ± 0.22 /mL) and photosynthetic dyes: chlorophyll (6.26 ± 1.16 mg/mL) and total carotenoids (1.59 ± 0.13 mg/mL) extracted from the biomass of O9.13F, were obtained from the culture cultivated at 20 °C. Under the conditions of the highest salinity, 100 g/L, all three strains had a significantly lower biomass increase than in other salinity values, regardless of the temperature at which the cultures were grown. However, the biomass grown at 35 °C in the 100 g/L salinity medium was the lowest (Figure 4).
Figure 4.
Quantity of chlorophyll, total carotenoids and proteins [mg/mL] extracted from biomass of Arthrospira sp. O9.13F cultivated at three different temperatures, 20 °C, 28 °C and 35 °C in Zarrouk medium with a total salinity 25 g/L. Standard deviations based on three replicate analyses are shown.
3.1.3. Acclimation to Prolonged Cold Stress
As the strain was isolated from a Siberian lake that is covered by ice in the winter, we tested the tolerance in laboratory conditions at a low temperature and compared it with other strains that originated from warmer regions: PCC8005 (India), PCC7345 (USA), A. maxima SAG 49.88 (Italy) and A. fusiformis CCALA23 (Kenya). The experiment was performed in a cold room, in low light 10 µmol photons m−2 s−1 conditions, for 10 months with subculturing after three months.
The general observation was that strains lost the intensity of pigmentation, and some filaments started to decay after this period. One of the strains, A. maxima SAG 49.88, changed the morphology and began to coil filaments. After the inoculation of tested strains filaments on a solid Zarrouk medium and transfer to the higher temperature 20 °C with a change of light intensity 20 µmol photons m−2 s−1, the strains recovered to the normal pigmentation, and growth was visible after a week of incubation; with exception of the strain A. fusiformis CCALA 23 that started to grow as a visible mat of filaments on the plate after six weeks (Figure S3).
3.1.4. The Sensitivity of Arthrospira sp. O9.13F to Various Stressors
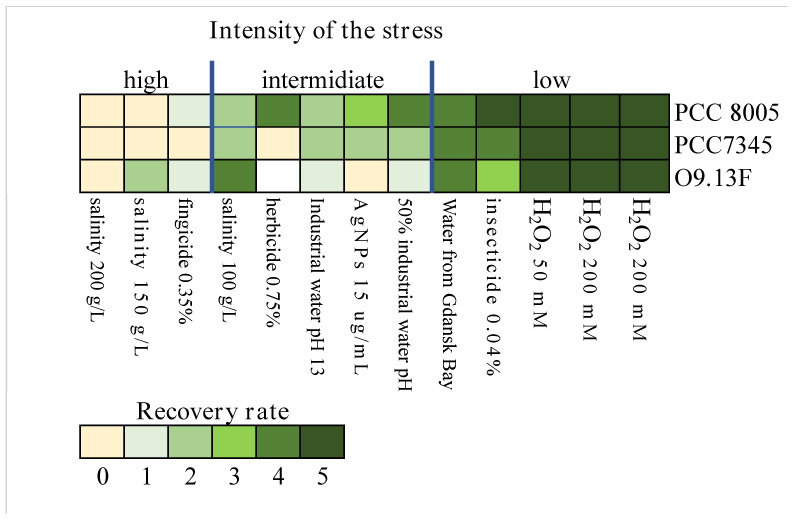
Based on the survival rate of the Arthrospira strains, the tested stress factors were divided into three levels of severity: low, intermediate and high. The best growth (low-stress severity) with no noticeable lag phase after the passage to the fresh medium was observed after the addition of hydrogen peroxide (in all three concentrations). The biomass of all tested strains was also showing minimal inhibition of the growth upon exposition to the working concentration of insecticide, water from Gdańsk Bay (lower salinity) and water from an industrial pond (pH > 13) diluted with a Zarrouk medium 1:1. The most differential and strain-dependent effect (intermediate stress severity) was observed in the case of exposition to undiluted industrial water (reach in calcium, potassium and sodium carbonate and pH > 13), silver nanoparticles (15 µg/mL) and herbicide and salinity of 100 g/L. Among the most harmful factors (high-stress severity) were high salinity, 150 and 200 g/L of total salt and a working concentration of 0.04% of fungicide. From the compilation of the results of the sensitivity of the strains to the applied stresses, we concluded that the most sensitive strain was Arthrospira sp. O9.13F (Figure 5).
Figure 5.
The heatmap presents the recovery rates of eight tested Arthrospira strains, O9.13F, PCC 7345 and PCC 8005 cultivated in stress conditions. Above the heatmap, three levels of the stress severity (low, intermediate, high) are grouped according to the overall similarity of the different strains’ responses. The colour scale given in the heatmap key corresponds to the recovery rate. The survivability of the biomass was assessed based on the condition, duration of the lag phase and visible increase in the number of viable filaments and recorded by the following system: 0—degradation of the filaments, 1—whole, nonfragmented filaments present, no growth observed, 2—growth observed after two weeks of a lag phase, 3—very weak growth recorded after one week of a lag phase, 4—stable growth after a short lag phase, 5—very abundant growth without a lag phase.
3.1.5. Fatty Acid Composition
The major fatty acids detected in the Arthrospira strains were palmitic acid (16:0), gamma-linolenic acid (18:3n6), linoleic acid (18:2n6c) and palmitoleic acid (16:1). However, the fatty acid content of the Arthrospira sp. strain O9.13F was not the same as in the strains PCC8005 and PCC7345. The most significant difference in fatty acid content in the Arthrospira biomasses was observed in the percentage of palmitic and gamma-linolenic acid (Table 1). The observed decrease of gamma-linolenic acid in favour of palmitic acid in strain O9.13F could be due to the stressful growth conditions and the fatty acid content. All tree strains were cultivated at 28 °C, which is not optimal for the growth of the Siberian strain O9.13F (Table 1). Indeed, the same observation was recorded when the fatty acid content was established for the Arthrospira sp. strain O9.13F grown at 20 °C, 28 °C and 35 °C (Supplementary Figure S4).
Table 1.
Total FA composition (%) of Arthrospira strains O9.13F, PCC 7345 and PCC 8005 cultivated at 28 °C.
| Fatty Acid | O9.13F | PCC 8005 | PCC 7345 |
|---|---|---|---|
| 16:0 | 36.71 ± 3.93 | 30.81 ± 1.2 | 31.64 ± 3.87 |
| 16:1 | 8.80 ± 1.1 | 10.29 ± 0.65 | 9.97 ± 1.13 |
| 18:0 | 1.12 ± 0.33 | 0.87 ± 0.16 | 1.62 ± 0.68 |
| 18:1n9c | 2.40 ± 0.97 | 2.59 ± 0.32 | 1.54 ± 0.66 |
| 18:1n11c | 1.67 ± 0.3 | 1.29 ± 0.04 | 1.32 ± 0.35 |
| 18:2n6c | 18.86 ± 0.34 | 18.78 ± 0.9 | 19.66 ± 2.43 |
| 18:3n6 | 30.45 ± 4.45 | 35.37 ± 1.98 | 34.26 ± 6.16 |
n = 3; 16:0—Palmitic acid; 16:1—Palmitoleic acid; 18:0—Stearic acid; 18:1n9c—Oleic acid; 18:1n11c—Vaccenic acid; 18:2n6c—Linoleic acid; 18:3n6—γ-Linolenic acid. The data are means ± standard deviations based on three replicate analyses.
3.2. Taxonomy of Arthrospira sp. Strain O9.13F
The new Siberian isolate of Arthrospira was identified based on the 16S rRNA gene, ITS and the phycocyanin operon fragment cpcBA sequencing and three different genomic analyses: ANI, isDDH and core proteome-based phylogeny.
The cloning and sequencing of the amplified 16S and cpcBA revealed that all eight isolates O9.13A-H (each originated from a single filament) have identical (100%) sequences. As a result of the ITS cloning and sequencing, we obtained two different genetic variants representing the ITS clusters I and III according to the numbering proposed by [30,88], respectively. These observations are in agreement with the results of the detection of the ITS variants by PCR with subcluster-specific primers described by Baurain et al. [30] and the reverse primer ITS_CLIII designed by us.
As the newly isolated strains were morphologically and genetically identical, only strain O9.13 F was used for further analyses. The phylogenetic analyses based on the 16S rRNA gene, ITS and phycocyanin operon fragment cpcBA were performed on the same dataset.
The obtained 16S rRNA gene sequence of the isolate O9.13F was 99.91% similar to A. erdosensis ‘Inner Mongolia’ (JN831261), Arthrospira sp. PCC 9901 (MG777134) and 99.72% similar to the other Arthrospira sp. strains TJSD092 (CP028914) and PCC 8005 (FO818640).
The phylogenetic analysis based on the small ribosomal subunit sequence showed that the Siberian strain groups with the strain PCC 9901 belonging to the new III clade described by Comte et al. [88] (Supplementary Figure S5). However, it is worth noting that the sequence 16S rRNA gene of the BM01 strain is only 92.5% similar to the sequences of other Arthospira strains, and it was excluded from the analysis.
Likewise, in 16S rRNA-based phylogeny, the analysis of the ITS region showed only two phylogroups within the Arthrospira genus called ITS clusters and five different genotypes named subclusters according to Baurain et al. [30]. ITS cluster I gathers strains belonging to clades I and III described based on the 16S rRNA gene, while ITS cluster II consists of all strains from the 16S clade II. Strains belonging to 16S clades I and III formed clearly separated subclusters within ITS cluster I. The results of the cloning and sequencing of ITS from strain O9.13F showed that two different genetic variants (genotypes) had been detected within the chromosome. The ITS sequence of clone I is grouped together with strains from 16S clade I, while the ITS clone III is grouped together with strains from 16S clade III (Supplementary Figure S6).
Based on the sequence of the phycocyanin operon fragment cpcBA, the newly described strain O9.13F is grouped with strains that belong to clade I (based on the 16S rRNA gene). However, unlike the results of ITS-based phylogeny, strains carrying the 16S gene sequences belonging to clade III appear to be more closely related to strains with the ITS cluster II.B. Moreover, strains belonging to the ITS subcluster II.A described by Baurain et al. [30] are more related with strains cluster I (Supplementary Figure S7).
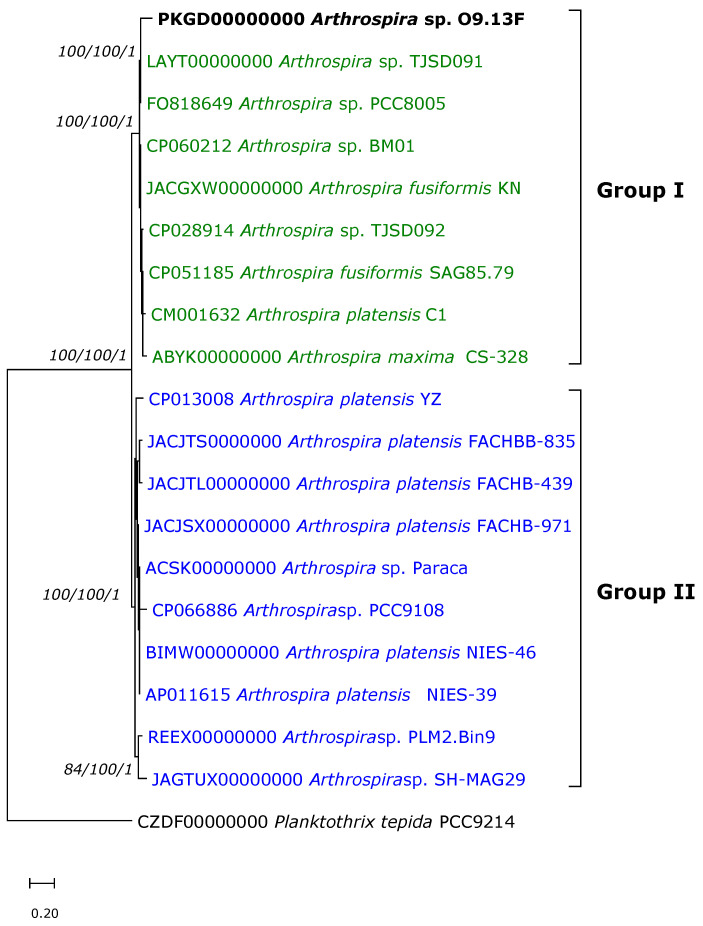
The phylogenomic analysis is based on 363 of the most conserved proteins retrieved from 17 Arthrospira strains for which the genomic sequences are available in databases. The use of the most evolutionarily conserved proteins whose genes occur in a single copy in the cell and an analysis based on genomic sequences reduce the impact of intragene or operons recombination on the results of phylogenomic analyses. The topology of the ML tree based on core protein showed that the Arthrospira genus consists of only two groups, and the O9.13F strain clusters together with the Arthrospira strains PCC8005, TJSD091, TJSD092 and CS-328, all of which are representatives of 16S clade I and ITS cluster I (Figure 6). The presence of two phylogenetic groups revealed by our phylogenomic analyses is consistent with the analyses described above and with the previously described analyses of ITS sequences by Baurain et al. [30]. Phylogenesis based on the analysis of core proteins enabled the precise classification of all strains for which the genomes are known. Also, for strains for which the identification relying on single locus, 16S rRNA gene, was doubtful, e.g., Arthrospira sp. BM01. The same goes for strains A. platensis Paraca and Arthrospira sp. PLM2.Bin9 that were ungrouped when the fragment of the cpcBA phycocyanin operon was analysed [31].
Figure 6.
Maximum Likelihood tree indicating the current relatedness of Arthrospira sp. O9.13F.
The phylogenetic tree establishing Arthrospira sp. O9.13F’s phylogenetic position is built based on the sequences of 363 of the most conserved proteins extracted from Arthrospira proteomes that are available in the Genbank database. The Maximum Likelihood tree was constructed using the PhyloPhlAn computational pipeline (Segata et al. 2013). The distance tree was inferred using Geneious software v 9.1.8. The gene sequences of Planktothrix tepida PCC9214 (CZDF00000000) were used as an outgroup. The bootstrap consensus was inferred from 1000 replicates. Bootstrapping values <50% were cut off. Numbers above branches indicate the bootstrap value for ML, the NJ method and the posterior probabilities for the Bayesian inference method, respectively. The sequence of strain O9.13F is indicated in bold font. The colours correspond to the 16S clades: green -clade 1, blue—clade II (according to [30]) and red—clade III (according to [88]).
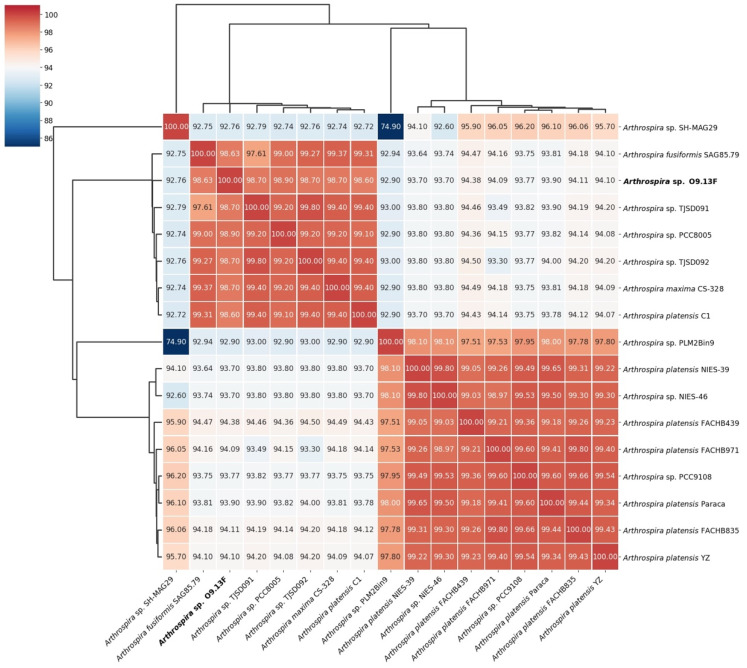
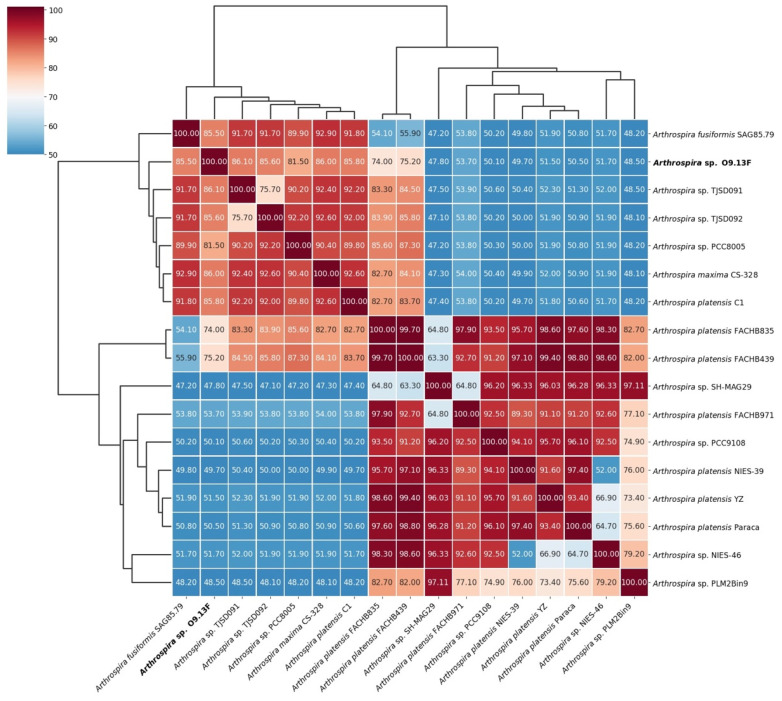
To compare the relatedness of the Arthrospira sp. O9.13F to other available genomes from the Arthrospira genus, we analysed the Average Nucleotide Identity (Figure 7) and in silico DNA–DNA hybridisation (Figure 8). Both methods confirmed the assignment of the strain to the Arthrospira genus, presenting similarity to the available genomes between 92.9% and 99.4% based on ANI and between 48.1% and 86.1% based on isDDH (Supplementary Table S1). Moreover, the presented results for all genomes were consistent with Arthrospira division into two main clusters based on core protein phylogeny and ITS sequence [30].
Figure 7.
Average Nucleotide Identity (ANI) of 17 Arthrospira genomes available in the GenBank database as calculated using PyAni v0.2.10 [84], with MUMer v3.23 [85] as the alignment method. Depicted linkage is calculated as UPGMA, distance is estimated using the Euclidean method.
Figure 8.
In silico DNA–DNA hybridization of 17 Arthrospira genomes available in the GenBank calculated with GGDC 2.1 [86] with BLAST+ as a local alignment tool.
3.3. General Features of the Arthrospira sp. O9.13F Genome
The genome of Arthrospira sp. O9.13F is deposited in the GenBank database under accession number PKGD00000000, bioproject PRJNA384118. The draft genome of Arthrospira sp. O9.13F consisted of 928 scaffolds of a total length of 4 945 448 bp and a G + C content of 44.47%. A total of 5355 genes were annotated of which 4688 (87.5%) were protein-coding genes, 625 (11.67%) were pseudogenes and 42 (0.78%) genes were encoding RNA and three CRISPR clusters (Table 2). A total of 3278 (69.92%) of predicted genes were assigned to COGs. Most genes were assigned to protein families responsible for replication, recombination and repair as well as signal transduction. The distribution of genes into COGs is presented in Table 3. The additional annotation of the genome with the KEGG Automatic Annotation Server (https://www.genome.jp/kegg/ (accessed on 3 January 2019)) ascribed 1287 genes into 189 pathways, the highest numbers of genes were mapped to the following processes: ABC transporters (55), porphyrin and chlorophyll metabolism (41), photosynthesis (40), purine metabolism (39), two-component system (33), pyrimidine metabolism (32) and oxidative phosphorylation (31).
Table 2.
Genome statistics of Arthrospira sp. O9.13F.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,945,448 | 100 |
| DNA coding (bp) | 3,759,108 | 76.01 |
| DNA G + C (bp) | 2,198,162 | 44.5 |
| DNA scaffolds | 928 | - |
| Total genes | 5355 | 100 |
| Protein coding genes | 4688 | 87.5 |
| RNA genes | 42 | 0.78 |
| Pseudogenes | 625 | 11.67 |
| Genes in internal clusters | - | - |
| Genes with function prediction | 3363 | 62.80 |
| Genes assigned to COGs | 3278 | 69.92 |
| Genes with Pfam domains | 3237 | 69.05 |
| Genes with signal peptides | 400 | 8.53 |
| Genes with transmembrane helices | 862 | 18.39 |
| CRISPR repeats | 3 | 0.06 |
Table 3.
The number of genes associated with general COG functional categories.
| Code | Value | % of Total * | Description |
|---|---|---|---|
| J | 146 | 4.45 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 145 | 4.42 | Transcription |
| L | 311 | 9.49 | Replication, recombination and repair |
| B | 2 | 0.06 | Chromatin structure and dynamics |
| D | 25 | 0.76 | Cell cycle control, cell division, chromosome partitioning |
| V | 84 | 2.56 | Defence mechanisms |
| T | 308 | 9.4 | Signal transduction mechanisms |
| M | 209 | 6.38 | Cell wall/membrane biogenesis |
| N | 39 | 1.19 | Cell motility |
| U | 44 | 1.34 | Intracellular trafficking and secretion |
| O | 156 | 4.76 | Posttranslational modification, protein turnover, chaperones |
| C | 174 | 5.31 | Energy production and conversion |
| G | 140 | 4.27 | Carbohydrate transport and metabolism |
| E | 195 | 5.95 | Amino acid transport and metabolism |
| F | 58 | 1.77 | Nucleotide transport and metabolism |
| H | 115 | 3.51 | Coenzyme transport and metabolism |
| I | 73 | 2.23 | Lipid transport and metabolism |
| P | 138 | 4.21 | Inorganic ion transport and metabolism |
| Q | 95 | 2.90 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 499 | 15.22 | General function prediction only |
| S | 322 | 9.82 | Function unknown |
| - | 1811 | 33.82 | Not in COGs |
* The % of total is based on the total number of protein coding genes in the genome.
3.4. Comparative Genome Analyses
A comparative analysis of the Arthrospira sp. O9.13F genome with other Arthrospira genomes revealed the absence of genetic determinants related to the synthesis of cyanobacterial toxins, such as microcystins, nodularin or cylindrospermopsin [89]. The secondary metabolites analysis of the Arthrospira sp. O9.13F genome revealed the presence of five different secondary metabolite gene clusters that were present in all of the studied Arthrospira genomes: four cyanobactin/bacteriocin gene clusters and one phytoene synthase. The presence of phytoene is obvious because there are different carotenoids in the Arthrospira cells, and a complete pathway for carotenoids synthesis was present within the genome of this cyanobacterium. The secondary metabolites analysis did not indicate the presence of siderophore biosynthetic gene clusters, which agreed with the negative results of the CAS plate assay [90].
The sequences of genes involved in gliding motility present in A. platensis C1 and A. platensis NIES-39 were compared with the Arthrospira sp. O9.13F genome. Genes encoding the S-layer gliding motility protein (oscillin) [91], as well as genes encoding type IV pilus-related proteins involved in twitching motility (pilA, pilC-like, pilD and pilT2 genes) are present in the Arthrospira sp. O9.13F genome.
The buoyancy of Arthrospira sp. O9.13F is regulated by gas vesicles (Figure 2F) encoded by certain identified genes: gvpA, gvpC, gvpK, gvpFL, gvpV, gvpW, gvpN and gvpJ. GvpA proteins form the vesicle core, while GvpC proteins provide structural support and influence the shape of the gas vesicles [92]. The proteins GvpK, GvpFL and GvpN possibly stabilise the structure of the gas vesicle. The GvpJ protein is considered to determine the shape of the gas vesicle. The functions of the GvpV and GvpW proteins remain unknown [92,93].
No plasmid sequences were observed during the DNA extraction and analysis of the genomic data for the Arthrospira sp. O9.13F isolate.
One incomplete prophage region was identified in the Arthrospira genome of 10.6 kb, with 10 total proteins, including seven phage hits and three hypothetical proteins. All 10 proteins are common in other Arthrospira genomes. The phage with the highest protein similarity rate to the identified region was Citrobacter phage Margaery (NC 028755.1).
We checked the existence of loci coding RM and CRISPR/Cas systems in the genome of the analysed cyanobacterium. The genome sequence of the Arthrospira sp. O9.13F strain contains three sets of the type I R-M systems (hsdM, hsdR and hsdS genes), and isoschizomers of Hin1I, Eco88I, Mph1103I, BshNI and Pfl23II complete the type II R-M systems. Genes encoding two isoschizomers of the type II solitary methyltransferases, namely, BspPI and Bsp143I, were also identified.
A total of 26 different CRISPR/Cas system direct repeats (DR) sequences were found in the Arthrospira sp. O9.13F genome, with six confirmed and 20 questionable. It also contains CRISPR/Cas essential cas1 and cas2 genes as well as the cas6 gene and CRISPR-associated protein csx3 gene. Repeat Associated Mysterious Protein (RAMP) genes, involved in CRISPR/Cas defence processes, were also identified, namely, cmr2, cmr4, cmr6, csm1, csm2, csm3, csm4 and csm5. The organisation of the CRISPR/Cas operons is shown in Supplementary Figure S8.
3.4.1. Genes Responsible for the Stress Adaptation
Cyanobacteria of the Arthrospira genus are known to be resistant to temperatures up to 40 °C, alkaline pH and high salt concentration. Numerous genes responsible for their adaptation capacity have been detected in the genomes of these cyanobacteria. So far, the genes encoding Na+/H+ antiporters, CO2 uptake (NDH−1) as well as genes responsible for the biosynthesis of three compatible solutes (sucrose, trehalose, glucosylglycerol or glucosylglycerate) have been described [15,94,95]. To verify whether the Siberian strain O9.13F differs from tropical ones, the presence of 68 genes related to the stress response was checked in its genome (Supplementary Table S3). We confirmed the presence of 60 genes. The sequences of these genes are almost identical or differ only in single polymorphisms between O9.13F and other Arthrospira strains. It is worth emphasising that the sequences of the Siberian strain that persists during the winter period in a freezing lake are the same as for the tropical strains. What is more, it should be noted that the absence of eight genes in the genome of the O.M13F strain does not necessarily mean that some pathways are incomplete because the sequence of its genome is not closed, and its size of approximately 4.95 Mbp is smaller than in the case of most Arthrospira genomes (from 5.75 to 6.78 Mbp) that are available in the GenBank database (Supplementary Table S2).
3.4.2. Arthrospira sp. O9.13F Unique Genes
A comparative genomic analysis of Arthrospira sp. O9.13F and other Arthrospira genomes revealed 24 genes that are unique for this strain (Table 4). Among them, we found 14 genes encoding hypothetical proteins, RNA-binding S4 domain-containing protein as well as two genes encoding four helix bundle proteins. Moreover, unique CDs encode proteins involved in the immunity system, N-6 DNA methylase, which is most similar with Methylase AflIII, the DUF433 domain-containing protein that might be a part of the putative toxin/antitoxin system ([96]) as well as the WYL domain-containing protein, which is a regulator of the Type VI-D CRISPR-Cas system [97] and thus might regulate the response to environmental stresses [98] (Table 4).
Table 4.
Arthrospira sp. O9.13F-specific CDs.
| ORF | Description | The Most Similar Sequence in Genbank | Identity | |
|---|---|---|---|---|
| 1 | B9S53_26655 | hypothetical protein | - | - |
| 2 | B9S53_26345 | hypothetical protein | AI-2E family transporter [Natronoflexus pectinivorans] WP_165921875.1 | 61% |
| 3 | B9S53_26040 | IS91 family transposase | transposase [Lentisphaerae bacterium] MBM4144559.1 | 73% |
| 4 | B9S53_26035 | hypothetical protein | WYL domain-containing protein [Lentisphaerae bacterium] MBM4165248.1 | 83% |
| 5 | B9S53_25925 | hypothetical protein | - | - |
| 6 | B9S53_25920 | RNA-binding protein | RNA-binding S4 domain-containing protein [Flavobacteriales bacterium] TVQ76998.1 | 78% |
| 7 | B9S53_25790 | hypothetical protein | - | - |
| 8 | B9S53_25595 | four helix bundle protein | four helix bundle protein [Chloroflexi bacterium] HFC09004.1 | 67% |
| 9 | B9S53_25590 | hypothetical protein | four helix bundle protein [Chloroflexi bacterium] HFC09004.1 | 53% |
| 10 | B9S53_25580 | four helix bundle protein | - | |
| 11 | B9S53_24895 | hypothetical protein | N-6 DNA methylase [Sphaerospermopsis sp. FACHB-1094] WP_190648063.1 | 59% |
| 12 | B9S53_23130 | hypothetical protein | ROK family protein [Pseudanabaena sp. FACHB-1277] WP_190353199.1 | 43% |
| 13 | B9S53_22765 | hypothetical protein | - | - |
| 14 | B9S53_21915 | hypothetical protein | - | - |
| 15 | B9S53_21010 | IS4 family transposase | transposase [Phormidium sp. FACHB-1136] WP_199324687.1 | 75% |
| 16 | B9S53_20985 | hypothetical protein | - | - |
| 17 | B9S53_20435 | hypothetical protein | hypothetical protein [Cylindrospermopsis raciborskii] WP_071249329.1 | 67% |
| 18 | B9S53_19635 | hypothetical protein | - | - |
| 19 | B9S53_12695 | hypothetical protein | - | - |
| 20 | B9S53_12545 | hypothetical protein | - | - |
| 21 | B9S53_11425 | hypothetical protein | aldolase [Chloroflexi bacterium] MBT7081734.1 | 50% |
| 22 | B9S53_11330 | IS4 family transposase | transposase, partial [Leptolyngbya sp. PCC 6406] WP_155834780.1 | 75% |
| 23 | B9S53_06535 | DUF2281 domain-containing protein | DUF2281 domain-containing protein [Geitlerinema sp. P-1104] NMG59147.1 | 94% |
| 24 | B9S53_01945 | hypothetical protein | hypothetical protein [Nodularia sp. LEGE 06071] WP_193996843.1 | 32% |
4. Discussion
The temperature of the water in Solenoye Lake varies between 0 and 30 °C during vegetation season. The water salinity level varies from mesohaline during spring to polyhaline in the remaining seasons, and seasonal pH variation is in the range of 8.94–9.94 [7]. The lake is located on salt clay, and it freezes only on the surface during the winter. The ice cover remains for three months, from December to the beginning of March. The surface water temperature in these months ranges between −3 °C and −15 °C, with an average of approximately −10 °C. In winter, brine, which has a large heat accumulation capacity, may keep a higher temperature on the lower water layers. In addition, the salinity of the water lowers the temperature of the ice cover formation. The ice formation on the lake’s surface increases salinity in the lower water layers and allows the medium to remain liquid [99]. The cyanobacteria blooms in Solenoye Lake are observed annually. The composition of the lake’s phytoplankton has been investigated since 2007 [7]. During the entire ice-free period, cyanobacteria abundantly develop in the lake as does the species of the Arthrospira genus. Arthrospira forms the bloom in the warmer season, although its vegetation is noted in the lake even in the winter under the ice. The presence of Arthrospira in Solenoye Lake was noted even a few decades ago when there was a resort where the healing properties of the microflora in the lake were used to treat a variety of human diseases [7].
This study is the first comprehensive, physiological and genomic description of the Arthrospira strain O9.13F isolated from Solenoye Lake in September 2013. It is the first well-characterised isolate of Arthrospira found in Siberia. The described strain O9.13F is cultivable in laboratory conditions and showed the highest relative growth index and highest accumulation of proteins and photosynthetic pigments in the biomass upon cultivation at 20 °C. The observed optimal growth temperature is significantly lower than the temperature of 35 °C reported in the literature as optimal for most Arthrospira strains that typically originate from tropical areas. The earlier study of Kumar et al. 2011 showed that tropical strains of Arthrospira could not effectively grow and accumulate biopigments at 20 °C. We obtained similar observations during our research. Two reference strains, PCC 8005 and PCC 7345, grew less efficiently at 20 °C than the Siberian strain (Figure 4). While at the temperature of 15 °C, both strains stopped their metabolism, and filaments started to fall apart; therefore, the measurements of proteins and fatty acids were below the sensitivity limit of the method.
Nevertheless, the O9.13F strain was able to grow at 15 °C. Interestingly, we observed the highest growth index in the Zarrouk medium at this temperature with the most elevated tested salinity of 200 g/L. Furthermore, we showed that Arthrospira O9.13F could persist in temperatures around 9 °C for several months. They were unable, however to survive freezing at −20 °C in laboratory conditions. The waters of Lake Solenoje do not freeze to the bottom. Considering the properties of the O9.13F strain and the fact that in the ice-covered lake, the water in the deeper layers is more saline and warmer than those closer to the surface [99], we can hypothesise that the Siberian Arthrospira strain can survive the winter periods on the bottom of the lake and cause annual blooms observed in the summer.
The fatty acid content of the biomass of the Siberian strain does not deviate much from the biomass of two other analysed strains originating from tropical climates when each was cultivated in their optimal temperatures: 20 °C for O9.13F and 28 °C for PCC 8005 and PCC 7345. The same trend of a decrease of gamma-linolenic acid in favour of palmitic acid caused by a change of the temperature was observed for the Siberian strain as for the tropical ones. A drop in temperature from 30 to 20 °C led to a decrease in palmitic acid accompanied by an increase in linoleic, and the gamma-linolenic acid content was observed earlier by [100], suggesting an increase in membrane fluidity in response to the temperature change. Colla et al. [101] observed that elevation of the growth temperature from 30 °C to 35 °C increased the palmitoleic and linoleic acid concentration but decreased gamma-linolenic acid. Thus, the observed reduced amount of gamma-linolenic acid in the biomass of the Siberian strain after the elevation of the growth temperature from 20 °C to 28 °C confirm its acclimation to cooler than tropical climate.
Additionally, we concluded that it is possible to recover the Arthrospira strains after long incubation periods at a low temperature (9 °C) and in low light conditions (10 µmol photons m−2 s−1). This observation is valuable in case of storage problems with this species in culture collections as the Arthrospira strains are known to not survive the cryopreservation and lyophilisation procedures [102].
Based on this study’s detailed genetic characteristics, the newly isolated Siberian strain O9.13F belongs to the Arthrospira/Limnospira genus. On the genomic level, the Siberian strain O9.13F is highly similar to 17 other Arthrospira/Limnospira genomes that are available in the GenBank. Undoubtedly it is not a new species, and based on genomic analyses it belongs to the group gathering strains A. maxima CS-328, A. platensis C1, Arthrospira sp. PCC 8005, TJSD091, TJSD092, BM1 and Arthrospira fusiformis KN and SAG 85.79. The second group consists of the strains A. platensis Paraca, NIES-39, NIES-46, YZ, FACHBB-835, FACHBB-439, FACHBB-971 and Arthrospira sp. PCC9108, PLM2.Bin9 and SH-MAG29. Presently published Arthrospira genomes belong to the I.A, I.B and II.A ITS subclusters distinguished by Baurain et al. The strain Arthrospira sp. O9.13F presented in this work is not only the first strain isolated from a climate other than a tropical or subtropical one (Siberia, Russia) but is also a representative of a new clade III based on the 16S rRNA gene (according to Comte et al. 2013), for which the genomic sequence is available in public databases (PKGD00000000).
What is more, here we report that the Siberian strain O9.13F has two different ITS variants in contrast to most Arthrospira strains that possess only one variant of the ITS within the ribosomal operon, except for strain PCC 7345 that was shown by [30].
The discrepancy between the phylogenies based on ITS and cpcBA has already been observed in the works of [5,32], in which the possibility of recombination has been postulated. Our results of the cpcBA sequence analysis do not agree with the data presented by [31,34]. These authors only used the noncoding IGS fragment between the cpcBA genes. It could be caused by the fact that we used, apart from the IGS region, also fragments of the flanking cpcB and cpcA genes, similar to Manen and Falquet 2002 and Dadheech et al., 2010 earlier [5,32].
The results of the phylogenetic analyses based on the 16S rRNA gene, ITS and phycocyanin operon fragment cpcBA are not consistent, indicating that these three genetic markers, which are typically used in the phylogenetic analysis of cyanobacteria, are not appropriate for the classification of the new isolates of Arthrospira. They do not allow for establishing a taxonomy of this genus as their results are not congruent. Furthermore, the position of the PCC9901 strain, which represents the 16S clade III, is undetermined.
Nevertheless, the results obtained in this study reveal the presence of only two groups within the genus Arthrospira. Finally, the strain O9.13F constantly merges with the same group of strains from the first clad 16S and the first ITS cluster. And these observations correlate with the results of genomic analyses, ANI, isDDH and core protein-based phylogeny.
The genomic comparisons showed that the O9.13F strain, like other members of this genus, possesses extensive R-M and CRISPR/Cas systems that enhance their protection system against exogenic invasive genetic elements, such as plasmids and viruses as well as intragenomic rearrangements. In addition, the gas vesicle genes operon of strain O9.13F is similar to other Arthrospira strains. The virtual analysis agreed with the TEM analysis, and both confirmed that the Siberian strain produces gas vesicles to regulate buoyancy depending on the light intensity.
Comparative genomics revealed no apparent characteristic genes that may be directly responsible for the adaptation of the Siberian strain to low temperatures of its habitat during the winter period. However, among unique CDs, we found those which encode proteins that might regulate the response to environmental stresses, such as methylase, the putative toxin/antitoxin system or a regulator of the CRISPR-Cas system [96,97,98]. What is more, strain O9.13F did not possess a different set of genes associated with the stress response than tropical strains.
The observed discrepancy between different physiological characteristics (observed acclimatisation) with the simultaneous lack of significant genetic differences (no signs of adaptation in the form of unique genes or different genes conditioning the survival of the low-temperature period) remains an intriguing observation that requires further research. Furthermore, in further studies aimed at explaining the mechanisms that allow the Siberian strain to survive the winter period, the influence of other microorganisms that coexist in the waters of Lake Solonye should be considered.
Considering all the data we collected; we can conclude that the genus Arthrospira has a much broader acclimation capacity than previously reported. The observed physiological properties of the Siberian strain indicate their usefulness in terms of production in temperate climate zones with the use of local water resources, such as water from the Baltic Sea.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/cells10123411/s1, Figure S1: The cyanobacterial bloom, Lake Solenoye, Figure S2: The growth index quantity of chlorophyll, total carotenoids extracted from biomass of Arthrospira sp. O9.13F cultivated at different temperatures and salt concentrations, Figure S3: Acclimation of Arthrospira strains to long-term cold stress, Figure S4: The percentage of the total fatty acids in the biomass of Arthrospira sp. O9.13F cultivated at 20, 28 and 37 °C, Figure S5: Phylogenetic tree of Arthrospira strains based on 16S rRNA partial sequences, Figure S6: Phylogenetic tree of Arthrospira strains based on ITS rRNA partial sequences, Figure S7: Phylogenetic tree of Arthrospira strains based on cpcBA sequences, Figure S8: Organisation of CRISPR/Cas operons in Arthrospira sp. O9.13F genome, Table S1. ANI and isDDH values between Arthospira sp. O9.13F and other 16 Arthrospira strains for which genomes are available in the GenBank, Table S2: Summary of statistics of Arthrospira genomes available on NCBI. Table S3: Presence of genes responsible for the stress adaptation in Arthrospira genomes. References [103,104] are refered to in Supplementary Materials.
Author Contributions
Conceptualization, M.W., K.F.W. and O.B.; methodology M.W., K.F.W., O.B., A.E.M., M.F., M.M.W. and M.D.; validation M.W., K.F.W., O.B., A.E.M., M.F., M.M.W. and M.D.; formal analysis, M.W., K.F.W., A.E.M. and M.M.W.; investigation, M.W., K.F.W., O.B., A.E.M., M.F., M.M.W. and M.D.; resources, M.W. and K.F.W.; data curation, M.W., K.F.W. and M.M.W.; writing—original draft preparation, M.W., K.F.W., M.M.W., A.E.M. and M.D.; visualization, M.W., K.F.W., A.E.M., M.F. and M.M.W.; supervision, M.W., K.F.W. and O.B.; project administration M.W. and K.F.W.; funding acquisition, M.W., K.F.W. and O.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the University of Gdansk project 531-N107-D801-21. Sample collection was funded by the “European Union—European Social Fund” (UDA-POKL.04.01.01-00-017/10) (2012–2013), University of Gdansk, Gdansk, Poland. Genome sequencing was supported by the Ministry of Science and Higher Education Republic of Poland from the quality-promoting subsidy under the Leading National Research Centre (KNOW) program 2012–2017 Faculty of Pharmacy– [dec. MNiSW-DS-6002-4693-23/WA/12] and the National Natural Science Foundation of China (32071480).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors (K.F.W. and M.W.), upon request. The draft genome sequence of Arthrospira sp. O9.13F has been deposited in NCBI/Genbank under accession number PKGD00000000. The nucleotide sequence data reported in this paper are available under the following accession numbers: 16S rDNA; MF509165, ITS; MF509166–MF509169, cpcBA and MF509119.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Castenholz R.W., Rippka R., Herdman M., Wilmotte A. Form- Arthrospira. In: Staley J.T., Bryant M.P., Pfenning N., Holt J.G., editors. Bergey’s Manual of Systematic Bacteriology. Volume 3. William & Wilkins; Baltimore, MD, USA: 1989. pp. 1774–1775. [Google Scholar]
- 2.Nowicka-Krawczyk P., Muhlsteinova R., Hauer T. Detailed Characterization of the Arthrospira Type Species Separating Commercially Grown Taxa into the New Genus Limnospira (Cyanobacteria) Sci. Rep. 2019;9:694. doi: 10.1038/s41598-018-36831-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geitler L. Dr. L. Rabenhorst’s Kryptogamen-Flora von Deutschland, und der Schweiz. Zweite, vollständig neu bearbeitete Auflage; Vierzehnter Band. Die Algen herausgegeben von Prof. Dr. R. Kolkwitz, Berlin, Cyanophyceae von Europa unter Berücksichtigung der anderen Kontinente/von Dr. Lothar Geitler; Kryptogamen-Flora von Deutschland, und der Schweiz. Zweite, vollständig neu bearbeitete Auflage. 2nd ed. Volume 14 Akademische Verlagsgesellschaft m. b. H.; Leipzig, Germany: 1930. [Google Scholar]
- 4.Ciferri O. Spirulina, the Edible Microorganism. Microbiol. Rev. 1984;47:551–578. doi: 10.1128/mr.47.4.551-578.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dadheech P.K., Ballot A., Casper P., Kotut K., Novelo E., Lemma B., Pröschold T., Krienitz L. Phylogenetic Relationship and Divergence among Planktonic Strains of Arthrospira (Oscillatoriales, Cyanobacteria) of African, Asian and American Origin Deduced by 16S–23S ITS and Phycocyanin Operon Sequences. Phycologia. 2010;49:361–372. doi: 10.2216/09-71.1. [DOI] [Google Scholar]
- 6.Linhart C., Schagerl M. Seasonal Succession of the Travertine-Forming Desmid Oocardium Stratum. J. Phycol. 2015;51:1055–1065. doi: 10.1111/jpy.12345. [DOI] [PubMed] [Google Scholar]
- 7.Bazhenova O., Konovalova O. Phytoplankton of Lake Solenoye (Omsk) as a Promising Source of Bioresources. Contemp. Probl. Ecol. 2012;5:275–280. doi: 10.1134/S199542551203002X. [DOI] [Google Scholar]
- 8.Leboulanger C., Agogué H., Bernard C., Bouvy M., Carré C., Cellamare M., Duval C., Fouilland E., Got P., Intertaglia L., et al. Microbial Diversity and Cyanobacterial Production in Dziani Dzaha Crater Lake, a Unique Tropical Thalassohaline Environment. PLoS ONE. 2017;12:e0168879. doi: 10.1371/journal.pone.0168879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro-Severyn J., Pardo-Esté C., Mendez K., Fortt J., Marquez S., Molina F., Castro-Nallar E., Remonsellez F., Saavedra C. Living to the High Extreme: Unraveling the Composition, Structure, and Functional Insights of Bacterial Communities Thriving in the Arsenic-Rich Salar de Huasco Altiplanic Ecosystem. Microbiol. Spectr. 2021;9:e0044421. doi: 10.1128/Spectrum.00444-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorokin D.Y., Berben T., Melton E.D., Overmars L., Vavourakis C.D., Muyzer G. Microbial Diversity and Biogeochemical Cycling in Soda Lakes. Extremophiles. 2014;18:791–809. doi: 10.1007/s00792-014-0670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fužinato S., Fodora A., Subakov-Simić G. Arthrospira Fusiformis (Voronichin) Komarek et Lund (Cyanoprokaryota) A New Species for Europe. Algol. Stud. 2010;134:17–24. doi: 10.1127/1864-1318/2010/0134-0017. [DOI] [Google Scholar]
- 12.Vavourakis C., Mehrshad M., Balkema C., van Hall R., Andrei A.-S., Ghai R., Sorokin D., Muyzer G. Metagenomes and Metatranscriptomes Shed New Light on the Microbial-Mediated Sulfur Cycle in a Siberian Soda Lake. BMC Biol. 2019;17:69. doi: 10.1186/s12915-019-0688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zorz J., Sharp C., Kleiner M., Gordon P., Pon R., Dong X., Strous M. A Shared Core Microbiome in Soda Lakes Separated by Large Distances. Nat. Commun. 2019;10:4230. doi: 10.1038/s41467-019-12195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant W.D., Sorokin D.Y. Distribution and Diversity of Soda Lake Alkaliphiles. In: Horikoshi K., editor. Extremophiles Handbook. Springer; Tokyo, Japan: 2011. pp. 27–54. [Google Scholar]
- 15.Furmaniak M., Misztak A., Franczuk M., Wilmotte A., Waleron M., Waleron K. Edible Cyanobacterial Genus Arthrospira: Actual State of the Art in Cultivation Methods, Genetics, and Application in Medicine. Front. Microbiol. 2017;8:2541. doi: 10.3389/fmicb.2017.02541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananyev G., Carrieri D., Dismukes G. Optimization of Metabolic Capacity and Flux through Environmental Cues to Maximize Hydrogen Production by the Cyanobacterium “Arthrospira (Spirulina) Maxima”. Appl. Environ. Microbiol. 2008;74:6102–6113. doi: 10.1128/AEM.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ainas M., Hasnaoui S., Bouarab R., Abdi N., Drouiche N., Mameri N. Hydrogen Production with the Cyanobacterium Spirulina Platensis. Int. J. Hydrogen Energy. 2017;42:4902–4907. doi: 10.1016/j.ijhydene.2016.12.056. [DOI] [Google Scholar]
- 18.Krishnan A., Qian X., Ananyev G., Lun D., Dismukes G. Rewiring of Cyanobacterial Metabolism for Hydrogen Production: Synthetic Biology Approaches and Challenges. Adv. Exp. Med. Biol. 2018;1080:171–213. doi: 10.1007/978-981-13-0854-3_8. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki S., Yamaguchi H., Kawachi M. The Draft Genome of a Hydrogen-Producing Cyanobacterium, Arthrospira Platensis NIES-46. J. Genom. 2019;7:56–59. doi: 10.7150/jgen.38149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badri H., Monsieurs P., Coninx I., Ruddy W., Leys N. Molecular Investigation of the Radiation Resistance of Edible Cyanobacterium Arthrospira Sp. PCC 8005. MicrobiologyOpen. 2015;4:187–207. doi: 10.1002/mbo3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrickx L., De Wever H., Hermans V., Mastroleo F., Morin N., Wilmotte A., Janssen P., Mergeay M. Microbial Ecology of the Closed Artificial Ecosystem MELiSSA (Micro-Ecological Life Support System Alternative): Reinventing and Compartmentalizing the Earth’s Food and Oxygen Regeneration System for Long-Haul Space Exploration Missions. Res. Microbiol. 2006;157:77–86. doi: 10.1016/j.resmic.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Gugliemi G., Rippka R., Tandeau De Marsac N. Main Properties That Justify the Different Taxonomic Position of Spirulina spp. and Arthrospira spp. among Cyanobacteria. Bull. Inst. Oceanogr. Monaco. 1993;12:13–23. [Google Scholar]
- 23.Tomaselli L. Morphology, Ultrastructure and Taxonomy of Arthrospira (Spirulina Maxima and Arthrospira (Spirulina) Platensis. In: Vonshak A., editor. Spirulina platensis (Arthrospira): Physiology, Cell-Biology and Biotechnology. Taylor & Francis Ltd.; London, UK: 1997. pp. 1–15. [Google Scholar]
- 24.Komárek J., Lund J.W.G. What Is «Spirulina Platensis» in Fact? Algol. Stud. Hydrobiol. Suppl. Vol. 1990;58:1–13. [Google Scholar]
- 25.Komárek J., Anagnostidis K. Süßwasserflora von Mitteleuropa, Bd. 19/2: Cyanoprokaryota: Bd. 2/Part 2: Oscillatoriales. Elsveire GmbH; Munchen, Germany: 2005. [Google Scholar]
- 26.Bai N., Seshadri C.V. On Coiling and Uncoiling of Trichomes in the Genus Spirulina. Algol. Stud. 1980;26:32–47. [Google Scholar]
- 27.Bai N. Competitive Exclusion or Morphological Transformation? A Case Study with Spirulina Fusiformis. Arch. Hydrobiol. 1985;71:191–199. [Google Scholar]
- 28.Mühling M., Harris N., Belay A., Whitton B. Reversal of Helix Orientation in the Cyanobacterium Arthrospira. J. Phycol. 2003;39:360–367. doi: 10.1046/j.1529-8817.2003.01246.x. [DOI] [Google Scholar]
- 29.Scheldeman P., Baurain D., Bouhy R., Scott M., Mühling M., Whitton B., Belay A., Wilmotte A. Arthrospira (‘Spirulina’) Strains from Four Continents Are Resolved into Only Two Clusters, Based on Amplified Ribosomal DNA Restriction Analysis of the Internally Transcribed Spacer. FEMS Microbiol. Lett. 1999;172:213–222. doi: 10.1111/j.1574-6968.1999.tb13471.x. [DOI] [PubMed] [Google Scholar]
- 30.Baurain D., Renquin L., Grubisic S., Scheldeman P., Belay A., Wilmotte A. Remarkable Conservation of Internally Transcribed Spacer Sequences of Arthrospira (“Spirulina”) (Cyanophyceae, Cyanobacteria) Strains from Four Continents and of Recent and 30-Year-Old Dried Samples from Africa. J. Phycol. 2002;38:384–393. doi: 10.1046/j.1529-8817.2002.01010.x. [DOI] [Google Scholar]
- 31.Hicks M., Tran-Dao T.-K., Mulroney L., Bernick D. De-Novo Assembly of Limnospira Fusiformis Using Ultra-Long Reads. Front. Microbiol. 2021;12:657995. doi: 10.3389/fmicb.2021.657995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manen J.-F., Falquet J. The CpcB-CpcA Locus as a Tool for the Genetic Characterization of the Genus Arthrospira (Cyanobacteria): Evidence for Horizontal Transfer. Int. J. Syst. Evol. Microbiol. 2002;52:861–867. doi: 10.1099/00207713-52-3-861. [DOI] [PubMed] [Google Scholar]
- 33.Ballot A., Dadheech P., Krienitz L. Phylogenetic Relationship of Arthrospira, Phormidium and Spirulina Strains from Kenyan and Indian Waterbodies. Algol. Stud. 2004;113:37–56. doi: 10.1127/1864-1318/2004/0113-0037. [DOI] [Google Scholar]
- 34.Papapanagiotou G., Gkelis S. Taxonomic Revision of Commercially Used Arthrospira (Cyanobacteria) Strains: A Polyphasic Approach. Eur. J. Phycol. 2019;54:595–608. doi: 10.1080/09670262.2019.1624832. [DOI] [Google Scholar]
- 35.Mühling M., Somerfield P., Harris N., Belay A., Whitton B. Phenotypic Analysis of Arthrospira (Spirulina) Strains (Cyanobacteria) Phycologia. 2006;45:148–157. doi: 10.2216/05-21.1. [DOI] [Google Scholar]
- 36.Ogato T., Kifle D., Fetahi T., Sitotaw B. Evaluation of Growth and Biomass Production of Arthrospira (Spirulina) Fusiformis in Laboratory Cultures Using Waters from the Ethiopian Soda Lakes Chitu and Shala. J. Appl. Phycol. 2014;26:2273–2282. doi: 10.1007/s10811-014-0251-4. [DOI] [Google Scholar]
- 37.Janssen P., Morin N., Mergeay M., Leroy B., Ruddy W., Vallaeys T., Waleron K., Waleron M., Wilmotte A., Quillardet P., et al. Genome Sequence of the Edible Cyanobacterium Arthrospira Sp. PCC 8005. J. Bacteriol. 2010;192:2465–2466. doi: 10.1128/JB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujisawa T., Narikawa R., Okamoto S., Ehira S., Yoshimura H., Suzuki I., Masuda T., Mochimaru M., Takaichi S., Awai K., et al. Genomic Structure of an Economically Important Cyanobacterium, Arthrospira (Spirulina) Platensis NIES-39. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes. 2010;17:85–103. doi: 10.1093/dnares/dsq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheevadhanarak S., Paithoonrangsarid K., Prommeenate P., Kaewngam W., Musigkain A., Tragoonrung S., Tabata S., Kaneko T., Chaijaruwanich J., Sangsrakru D., et al. Draft Genome Sequence of Arthrospira Platensis C1 (PCC9438) Stand. Genom. Sci. 2012;6:43–53. doi: 10.4056/sigs.2525955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrieri D., Ananyev G., Lenz O., Bryant D., Dismukes G. Contribution of a Sodium Ion Gradient to Energy Conservation during Fermentation in the Cyanobacterium Arthrospira (Spirulina) Maxima CS-328. Appl. Environ. Microbiol. 2011;77:7185–7194. doi: 10.1128/AEM.00612-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefort F., Calmin G., Crovadore J., Falquet J., Hurni J.-P., Østerås M., Haldemann F., Farinelli L. Whole-Genome Shotgun Sequence of Arthrospira Platensis Strain Paraca, a Cultivated and Edible Cyanobacterium. Genome Announc. 2014;2:e00751-14. doi: 10.1128/genomeA.00751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu T., Qin S., Hu Y., Song Z., Ying J., Li P., Dong W., Zhao F., Yang H., Bao Q. Whole Genomic DNA Sequencing and Comparative Genomic Analysis of Arthrospira Platensis: High Genome Plasticity and Genetic Diversity. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes. 2016;23:325–338. doi: 10.1093/dnares/dsw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong S., Chen J., Wang S., Wu Y., Hou H., Li M., Yan C. Draft Genome Sequence of Cyanobacteria Arthrospira Sp. TJSD091 Isolated from Seaside Wetland. Mar. Genom. 2015;24:197–198. doi: 10.1016/j.margen.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Sharp C., Urschel S., Dong X., Brady A., Slater G., Strous M. Robust, High-Productivity Phototrophic Carbon Capture at High PH and Alkalinity Using Natural Microbial Communities. Biotechnol. Biofuels. 2017;10:84. doi: 10.1186/s13068-017-0769-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M.-Y., Teng W., Zhao L., Hu C.-X., Zhou Y., Han B.-P., Song L.-R., Shu W.-S. Comparative Genomics Reveals Insights into Cyanobacterial Evolution and Habitat Adaptation. ISME J. 2020;15:211–227. doi: 10.1038/s41396-020-00775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maghembe R., Angelina M., Harish A., Nyandoro S., Lyantagaye S., Hati-Kaul R. Draft Genome Sequence of Limnospira Sp. Strain BM01, Isolated from a Hypersaline Lake of the Momela Ecosystem in Tanzania. Microbiol. Resour. Announc. 2021;10:e00132-21. doi: 10.1128/MRA.00132-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarrouk C., Zarrouk C. Contribution à l´Étude d´une Cyanophycée Influence de Divers Facteurs Physiques et Chimiques sur la Croissance et la Photosynthése de Spirulina Maxima Geitler. Academie de Paris; Paris, France: 1966. [Google Scholar]
- 48.Rippka R. Isolation and Purification of Cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 49.Bligh E.L.G., Dyer W.J.A. A Rapid Method of Total Lipid Extraction And Purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 50.Bradford M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Ritchie R.J. Consistent Sets of Spectrophotometric Chlorophyll Equations for Acetone, Methanol and Ethanol Solvents. Photosynth. Res. 2006;89:27–41. doi: 10.1007/s11120-006-9065-9. [DOI] [PubMed] [Google Scholar]
- 52.Wellburn A. The Spectral Determination of Chlorophylls a and b, as Well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 1994;144:307–313. doi: 10.1016/S0176-1617(11)81192-2. [DOI] [Google Scholar]
- 53.Pinheiro J., Bates D., DebRoy S., Sarkar D., R Core Team nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-153, 2021. [(accessed on 1 October 2020)]. Available online: https://CRAN.R-project.org/package=nlme.
- 54.Eykelenburg C. The Ultrastructure of Spirulina Platensis in Relation to Temperature and Light Intensity. Antonie Leeuwenhoek. 1979;45:369–390. doi: 10.1007/BF00443277. [DOI] [PubMed] [Google Scholar]
- 55.Hayat M.A. Principles and Techniques of Electron Microscopy: Biological Applications. Edward Arnold; London, UK: 1981. [Google Scholar]
- 56.William S., Feil H., Copeland A. Bacterial Genomic DNA Isolation Using CTAB 2012. [(accessed on 1 October 2019)]. Available online: http://1ofdmq2n8tc36m6i46scovo2e.wpengine.netdna-cdn.com/wp-content/uploads/2014/02/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf.
- 57.Waleron M., Waleron K., Vincent W.F., Wilmotte A. Allochthonous Inputs of Riverine Picocyanobacteria to Coastal Waters in the Arctic Ocean. FEMS Microbiol. Ecol. 2007;59:356–365. doi: 10.1111/j.1574-6941.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- 58.Hrouzek P., Ventura S., Lukesova A., Mugnai M., Turicchia S., Komárek J. Diversity of Soil Nostoc Strains: Phylogenetic and Phenotypic Variability. Algol. Stud. 2005;117:251–264. doi: 10.1127/1864-1318/2005/0117-0251. [DOI] [Google Scholar]
- 59.Taton A., Grubisic S., Brambilla E., De Wit R., Wilmotte A. Cyanobacterial Diversity in Natural and Artificial Microbial Mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): A Morphological and Molecular Approach. Appl. Environ. Microbiol. 2003;69:5157–5169. doi: 10.1128/AEM.69.9.5157-5169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilmotte A., Van der Auwera G., Wachter R. Structure of the 16 S Ribosomal RNA of the Thermophilic Cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus Laminosus HTF’) Strain PCC7518, and Phylogenetic Analysis. FEBS Lett. 1993;317:96–100. doi: 10.1016/0014-5793(93)81499-P. [DOI] [PubMed] [Google Scholar]
- 61.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinform. Oxf. Engl. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood D., Salzberg S., Wood D.E., Salzberg S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignment. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolger A., Lohse M., Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinform. Oxf. Engl. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chikhi R., Medvedev P. Informed and Automated K-Mer Size Selection for Genome Assembly. Bioinform. Oxf. Engl. 2013;30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 65.Bankevich A., Nurk S., Antipov D., Gurevich A., Dvorkin M., Kulikov A., Lesin V., Nikolenko S., Pham S., Prjibelski A., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zerbino D., Birney E. Velvet: Algorithms for De Novo Short Read Assembly Using De Bruijn Graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chevreux B., Wetter T., Suhai S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information; Proceedings of the Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB’99); Braunschweig, Germany. 7–10 October 1999; p. 56. [Google Scholar]
- 68.Boisvert S., Laviolette F., Corbeil J. Ray: Simultaneous Assembly of Reads from a Mix of High-Throughput Sequencing Technologies. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boetzer M., Henkel C., Jansen H., Butler D., Pirovano W. Scaffolding Pre-Assembled Contigs Using SSPACE. Bioinform. Oxf. Engl. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 70.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinform. Oxf. Engl. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E., Zaslavsky L., Lomsadze A., Pruitt K., Borodovsky M., Ostell J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016;44:gkw569. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haft D., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M., Gonzales N., et al. RefSeq: An Update on Prokaryotic Genome Annotation and Curation. Nucleic Acids Res. 2017;46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moriya Y., Itoh M., Okuda S., Yoshizawa A., Kanehisa M., Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koski L., Gray M., Lang B., Burger G. AutoFACT: An Automatic Functional Annotation and Classification Tool. BMC Bioinform. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Käll L., Krogh A., Sonnhammer E. Advantages of Combined Transmembrane Topology and Signal Peptide Prediction—The Phobius Web Server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roberts R., Vincze T., Posfai J., Macelis D. REBASE-A Database for DNA Restriction and Modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: A Web Tool to Identify Clustered Regularly Interspaced Short Palindromic Repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vallenet D., Calteau A., Dubois M., Amours P., Bazin A., Beuvin M., Burlot L., Bussell X., Fouteau S., Gautreau G., et al. MicroScope: An Integrated Platform for the Annotation and Exploration of Microbial Gene Functions through Genomic, Pangenomic and Metabolic Comparative Analysis. Nucleic Acids Res. 2019;48:D579–D589. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arndt D., Grant J., Marcu A., Sajed T., Pon A., Liang Y., Wishart D. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016;44:gkw387. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carattoli A., Zankari E., Garcia-Fernandez A., Larsen M., Lund O., Villa L., Aarestrup F., Hasman H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weber T., Blin K., Duddela S., Krug D., Kim T.Y., Bruccoleri R., Lee S.Y., Fischbach M., Müller R., Wohlleben W., et al. AntiSMASH 3.0-A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blin K., Medema M., Kottmann R., Lee S.Y., Weber T. The AntiSMASH Database, a Comprehensive Database of Microbial Secondary Metabolite Biosynthetic Gene Clusters. Nucleic Acids Res. 2016;45:gkw960. doi: 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Segata N., Börnigen D., Morgan X., Huttenhower C. PhyloPhlAn Is a New Method for Improved Phylogenetic and Taxonomic Placement of Microbes. Nat. Commun. 2013;4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pritchard L., Glover R., Humphris S., Elphinstone J., Toth I. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods. 2015;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 85.Kurtz S., Phillippy A., Delcher A., Smoot M., Shumway M., Antonescu C., Salzberg S. Versatile and Open Software for Comparing Large Genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meier-Kolthoff J., Auch A., Klenk H.-P., Göker M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Comte K., Coursin T., Carre-Mlouka A. A New Genotype in the Genus Arthrospira (Oscillatoriales, Cyanobacteria) Revealed by a Mosaic-like Structure of the 16S-23SrRNA Intergenic Spacer Region in Strain PCC 9901. Phycologia. 2013;52:333–337. doi: 10.2216/12-063.1. [DOI] [Google Scholar]
- 89.Burns B., Saker M., Moffitt M., Neilan B. Molecular Detection of Genes Responsible for Cyanobacterial Toxin Production in the Genera Microcystis, Nodularia, and Cylindrospermopsis. Methods Mol. Biol. 2004;268:213–222. doi: 10.1385/1-59259-766-1:213. [DOI] [PubMed] [Google Scholar]
- 90.Waleron M., Misztak A., Waleron M., Furmaniak M., Mrozik A., Waleron K. Arthrospiribacter Ruber Gen. Nov., Sp. Nov., a Novel Bacterium Isolated from Arthrospira Cultures. Syst. Appl. Microbiol. 2020;43:126072. doi: 10.1016/j.syapm.2020.126072. [DOI] [PubMed] [Google Scholar]
- 91.Hoiczyk E., Baumeister W. Oscillin, an Extracellular, Ca2+-Binding Glycoprotein Essential for the Gliding Motility of Cyanobacteria. Mol. Microbiol. 1997;26:699–708. doi: 10.1046/j.1365-2958.1997.5971972.x. [DOI] [PubMed] [Google Scholar]
- 92.Miklaszewska M., Waleron M., Morin N., Calusinska M., Wilmotte A., Marsac N., Rippka R., Waleron K. Elucidation of the Gas Vesicle Gene Clusters in Cyanobacteria of the Genus Arthrospira (Oscillatoriales, Cyanophyta) and Correlation with ITS Phylogeny. Eur. J. Phycol. 2012;47:233–244. doi: 10.1080/09670262.2012.692817. [DOI] [Google Scholar]
- 93.Carre-Mlouka A., Comte K., Castets A.-M., Bouchier C., Marsac N. The Gas Vesicle Gene Cluster from Microcystis Aeruginosa and DNA Rearrangements That Lead to Loss of Cell Buoyancy. J. Bacteriol. 2004;186:2355–2365. doi: 10.1128/JB.186.8.2355-2365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hagemann M. Molecular Biology of Cyanobacterial Salt Acclimation. FEMS Microbiol. Rev. 2011;35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 95.Hagemann M. Chapter Two—Genomics of Salt Acclimation: Synthesis of Compatible Solutes among Cyanobacteria. In: Chauvat F., Cassier-Chauvat C., editors. Advances in Botanical Research. Volume 65. Academic Press; Cambridge, MA, USA: 2013. pp. 27–55. Genomics of Cyanobacteria. [Google Scholar]
- 96.Matelska D., Ginalski K. Comprehensive Classification of the PIN Domain-like Superfamily. Nucleic Acids Res. 2017;45:6995–7020. doi: 10.1093/nar/gkx494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang H., Dong C., Li L., Wasney G., Min J. Structural Insights into the Modulatory Role of the Accessory Protein WYL1 in the Type VI-D CRISPR-Cas System. Nucleic Acids Res. 2019;47:5420–5428. doi: 10.1093/nar/gkz269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koonin E., Zhang F. Coupling Immunity and Programmed Cell Suicide in Prokaryotes: Life-or-Death Choices. BioEssays. 2016;39:1–9. doi: 10.1002/bies.201600186. [DOI] [PubMed] [Google Scholar]
- 99.Stappen G., Litvinenko L., Litvinenko A., Boyko E., Marden B., Sorgeloos P. A Survey of Artemia Resources of Southwest Siberia (Russian Federation) Rev. Fish. Sci. 2009;17:117–148. doi: 10.1080/10641260802590095. [DOI] [Google Scholar]
- 100.Mühling M., Belay A., Whitton B. Variation in Fatty Acid Composition of Arthrospira (Spirulina) Strains. J. Appl. Phycol. 2005;17:137–146. doi: 10.1007/s10811-005-7213-9. [DOI] [Google Scholar]
- 101.Colla L.M., Bertolin T.E., Costa J.A.V. Fatty Acids Profile of Spirulina Platensis Grown Under Different Temperatures and Nitrogen Concentrations. Z. Nat. C. 2004;59:55–59. doi: 10.1515/znc-2004-1-212. [DOI] [PubMed] [Google Scholar]
- 102.Shiraishi H. Cryopreservation of the Edible Alkalophilic Cyanobacterium Arthrospira Platensis. Biosci. Biotechnol. Biochem. 2016;80:2051–2057. doi: 10.1080/09168451.2016.1189320. [DOI] [PubMed] [Google Scholar]
- 103.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 104.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors (K.F.W. and M.W.), upon request. The draft genome sequence of Arthrospira sp. O9.13F has been deposited in NCBI/Genbank under accession number PKGD00000000. The nucleotide sequence data reported in this paper are available under the following accession numbers: 16S rDNA; MF509165, ITS; MF509166–MF509169, cpcBA and MF509119.