Abstract
The large subunit of replication protein A (Rpa1) consists of three single-stranded DNA binding domains and an N-terminal domain (Rpa1N) of unknown function. To determine the essential role of this domain we searched for mutations that require wild-type Rpa1N for viability in yeast. A mutation in RFC4, encoding a small subunit of replication factor C (RFC), was found to display allele-specific interactions with mutations in the gene encoding Rpa1 (RFA1). Mutations that map to Rpa1N and confer sensitivity to the DNA synthesis inhibitor hydroxyurea, such as rfa1-t11, are lethal in combination with rfc4-2. The rfc4-2 mutant itself is sensitive to hydroxyurea, and like rfc2 and rfc5 strains, it exhibits defects in the DNA replication block and intra-S checkpoints. RFC4 and the DNA damage checkpoint gene RAD24 were found to be epistatic with respect to DNA damage sensitivity. We show that the rfc4-2 mutant is defective in the G1/S DNA damage checkpoint response and that both the rfc4-2 and rfa1-t11 strains are defective in the G2/M DNA damage checkpoint. Thus, in addition to its essential role as part of the clamp loader in DNA replication, Rfc4 plays a role as a sensor in multiple DNA checkpoint pathways. Our results suggest that a physical interaction between Rfc4 and Rpa1N is required for both roles.
Replication protein A (RPA) is a highly conserved eukaryotic single-stranded DNA (ssDNA) binding protein that was originally identified as a human factor required for simian virus 40 (SV40) DNA replication in vitro (19, 84, 86). As the eukaryotic equivalent of the Escherichia coli ssDNA binding protein (Ecssb), RPA plays multiple roles in DNA metabolism, including DNA replication, repair, and recombination (85). In addition to participating in both the initiation and elongation of DNA replication, RPA is required for nucleotide excision repair in vitro and physically interacts with XPA, XPG, and XPF (30, 39, 62, 69). RPA is also required for genetic recombination and physically interacts with Rad52 and other factors (1, 22, 47, 75).
RPA is a heterotrimeric complex. In the budding yeast Saccharomyces cerevisiae it is composed of the Rpa1 (69 kDa), Rpa2 (36 kDa), and Rpa3 (13 kDa) subunits, which are encoded by RFA1, RFA2, and RFA3, respectively. Each subunit is essential for viability in yeast, and all three subunits are required for SV40 DNA replication in vitro (12, 18, 25, 31, 32). Rpa1 exhibits strong ssDNA binding activity on its own, and a subcomplex of Rpa2 and Rpa3 binds ssDNA weakly (7, 85). We have shown that yeast Rpa1 consists of four functional domains: an 18-kDa N-terminal domain that lacks ssDNA binding activity (Rpa1N) and three tandem ssDNA binding domains (SBDs; see Fig. 1A) (11, 56). SBD-A and SBD-B are structurally homologous as determined by X-ray crystallographic analysis of human RPA (hRPA), while the C-terminal domain (SBD-C) contains a C4-type zinc-finger motif (6, 8, 11). These three ssDNA binding domains share amino acid sequence similarity with each other and with the central region of Rpa2 (11). All four domains of Rpa1, including Rpa1N, are essential for viability in yeast (11, 56).
FIG. 1.
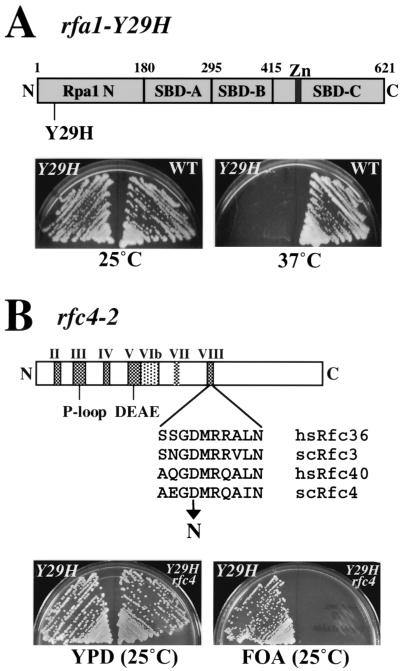
Interacting alleles rfa1-Y29H and rfc4-2. (A) A schematic diagram of Rpa1 is shown at the top. To demonstrate the ts growth phenotype caused by rfa1-Y29H, the strains HSY657 (rfa1-Y29H) and HSY679 (RFA1) were streaked onto YPD plates and incubated at 25 or 37°C, as indicated, for 3 days. (B) A schematic diagram of Rfc4 is shown together with the location of the mutation encoded by rfc4-2 and the sequences of the surrounding region obtained from several Rfc4 homologs. To demonstrate the synthetic-lethal phenotype caused by rfa1-Y29H and rfc4-2, the strains HSY657 (rfa1-Y29H) and HSY737 (rfa1-Y29H rfc4-2) were streaked onto YPD or 5-FOA plates, as indicated, and were incubated at 25°C for 3 days. SBD, ssDNA binding domain; Zn, zinc-binding domain; P-loop/DEAE, conserved motifs found in a large family of nucleoside triphosphate-binding domains.
The essential function of Rpa1N is unknown but may involve its ability to interact with other proteins. Human Rpa1N has been shown to bind DNA polymerase α, p53, T antigen, and VP16 (10, 41). Surprisingly, deletion of this domain in hRPA does not affect its ssDNA binding activity or SV40 DNA replication in vitro (25, 35). In contrast, mutations in yeast Rpa1N result in defects in DNA replication, recombination, and repair, and deletion of more than 10 amino acids from the N terminus is lethal (43, 56, 75). To determine the essential cellular role of Rpa1N, we first isolated the conditional mutation rfa1-Y29H, which maps to this domain. We then performed a synthetic-lethal screen with rfa1-Y29H under permissive conditions to identify mutations that require wild-type Rpa1N function for viability. To confirm that these synthetic-lethal mutations interacted specifically with Rpa1N, we took advantage of a large collection of Rpa1 mutants (75). The results indicate that one of the genes isolated in this screen, RFC4, displays allele-specific interactions with mutations mapping to Rpa1N.
Replication factor C (RFC), the eukaryotic clamp loader, loads the DNA polymerase δ processivity factor proliferating cell nuclear antigen (PCNA, or sliding clamp) at primer-template junctions (37, 70, 72, 88). In the presence of RPA, the loading of PCNA by RFC promotes a switch from synthesis by DNA polymerase α to processive synthesis by DNA polymerase δ (71, 74, 78, 89). RFC is a heteropentameric complex consisting of one large subunit (Rfc1 [Cdc44]) and four small subunits (Rfc2, -3, -4, and -5), all of which are essential for viability in yeast (15, 40). Mutations in RFC2 and RFC5 cause sensitivity to the DNA synthesis inhibitor hydroxyurea (HU). Consistent with this phenotype and their role in DNA replication, these mutant strains display defects in the DNA replication checkpoint pathway (50, 65, 66). The four small subunits of RFC are also associated with Rad24, one of the major components of the DNA damage checkpoint pathway (26, 27, 60). It has been proposed that the Rad24–Rfc2-5 complex is responsible for loading a PCNA-like complex of Rad17-Mec3-Ddc1 at sites of DNA repair (36, 76). Due to their association with Rad24, the small subunits of RFC might also play a role in DNA damage checkpoints. Indeed, defects in DNA damage checkpoints are exacerbated in an rfc5 rad24 double mutant (48).
In this report we describe the isolation of the rfc4-2 mutation based on its synthetic lethality with a mutation in Rpa1N. We find that the rfc4-2 mutant is sensitive to HU, is lethal in combination with Rpa1N mutations that are themselves HU sensitive, and is partially defective in the replication block checkpoint response and the intra-S DNA damage checkpoint. We also find that RFC4 functions in the RAD24 epistasis group and that RFC4 plays a role in multiple DNA damage checkpoint pathways. Taken together, the results suggest that both DNA replication and DNA checkpoint signaling require a direct physical interaction between Rfc4 and Rpa1N.
MATERIALS AND METHODS
Strains, media, and plasmid construction.
All yeast strains used in this study are listed in Table 1. Plasmids used in this study are listed in Table 2. Standard genetic techniques and reagents were used in the construction, transformation, and growth of yeast (57). The starting strains for the synthetic-lethal screen, HSY657 and HSY672, were constructed by integrating pHS102, containing the rfa1-Y29H allele, at the TRP1 locus of HSY631 and HSY626, respectively. Following loss of pJM195 (RFA1/URA3/ADE3) by growth on media containing 5-fluoroorotic acid (5-FOA), these strains displayed temperature-sensitive (ts) growth. To reduce the probability of obtaining rfa2 or rfa3 mutations in a synthetic-lethal screen with rfa1-Y29H, second copies of RFA2 and RFA3 were provided by integrating pHS203 at the HIS3 locus of these strains.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypes | Reference or source |
|---|---|---|
| W303-1a | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | 68 |
| CHY125 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 | C. Hardy |
| CHY128 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 lys2 | C. Hardy |
| SBY102 | MATaade2-1 ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 rfa1-1::TRP1 [pJM114 (RFA1/URA3)] | 56 |
| HSY630 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 lys2 rfa1::loxP [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY636 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 rfa1::loxP HIS3::RFA2,RFA3 [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY657 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 rfa1::loxP HIS3::RFA2,RFA3 TRP1::rfa1-Y29H [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY672 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 lys2 rfa1::loxP HIS3::RFA2,RFA2,RFA3 TRP1::rfa1-Y29H [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY737 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 rfa1::loxP rfc4-2 HIS3::RFA2,RFA3 TRP1::rfa1-Y29H [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY740 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 rfa1::loxP rfc4-2 HIS3::RFA2,RFA3 [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY787 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 can1-100 leu2-3,112 lys2 rfa1::loxP HIS3::RFA2,RFA3 TRP1::rfa1-Y29H RFC4:LEU2 [pJM195 (RFA1/URA3/ADE3)] | This study |
| HSY1025 | MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rfc4-2:LEU2 | This study |
| HSY1027 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rfc4-2:LEU2 | This study |
| DLY408 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::HISG GAL+psi+ssd1-d2 cdc13-1 cdc15-2 | 24 |
| DLY409 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::HISG GAL+psi+ssd1-d2 cdc13-1 cdc15-2 rad9::HIS3 | 24 |
| HSY1202 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::HISG GAL+psi+ssd1-d2 cdc13-1 cdc15-2 rfc4::KAN:loxP [pHS5116 (rfc4-2/LEU2)] | This study |
| HSY1204 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::HISG GAL+psi+ssd1-d2 cdc13-1 cdc15-2 rfa1::KAN:loxP [pKU-t11 (rfa1-t11/LEU2)] | This study |
| NJY1184 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad24::KAN:loxP | This study |
| HSY1201 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad9::KAN:loxP | This study |
| Y00684 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad53-K227A:KAN | V. Geli |
TABLE 2.
Plasmids used in this study
| Plasmid | Insert/marker | Vector (copy no.) | Reference or source |
|---|---|---|---|
| pRS plasmids | 63 | ||
| pJM195 | RFA1/URA3/ADE3 | pRS416 (CEN) | This study |
| pDS1 | RFA1/LEU2 | pRS415 (CEN) | 56 |
| pHS102 | rfa1-Y29H/TRP1 | pRS404 | This study |
| pHS203 | RFA2/RFA3/HIS3 | pRS403 | This study |
| pHS118 | RFA1/LEU2 | pRS415 (CEN) | This study |
| pHS119 | RFA1/LYS2 | pRS317 (CEN) | This study |
| pHS121 | rfa1-Y29H/LEU2 | pRS415 (CEN) | This study |
| pJM116 | RFA1/LEU2 | YEp213 (2μm) | This study |
| pJM222 | RFA2/LEU2 | YEp213 (2μm) | This study |
| pKU1-t22 | rfa1-t22/LEU2 | pRS415 (CEN) | 75 |
| pKU1-t11 | rfa1-t11/LEU2 | pRS415 (CEN) | 75 |
| pKU1-t69 | rfa1-t69/LEU2 | pRS415 (CEN) | 75 |
| pKU1-t48 | rfa1-t48/LEU2 | pRS415 (CEN) | 75 |
| pKU1-t6 | rfa1-t6/LEU2 | pRS415 (CEN) | 75 |
| pKU1-t124 | rfa1-t124/LEU2 | pRS415 (CEN) | 75 |
| pHS5101 | RFC4/LEU2 | pRS415 (CEN) | This study |
| pHS5105 | RFC4 | pET11 | This study |
| pHS5116 | rfc4-2/LEU2 | pRS415 (CEN) | This study |
| pHS5119 | RAD24/TRP | pRS424 (2μm) | This study |
| YEpRFC1 | RFC1/URA3 | YEp-195 (2μm) | 50 |
| YEpRFC2 | RFC2/URA3 | YEp-195 (2μm) | 50 |
| YEpRFC3 | RFC3/URA3 | YEp-195 (2μm) | 50 |
| YEpRFC4 | RFC4/URA3 | YEp-195 (2μm) | 50 |
| YEpRFC5 | RFC5/URA3 | YEp-195 (2μm) | 50 |
| pHS4412 | RAD53/TRP | pRS424 (2μm) | This study |
Isolation of the rfa1-ts allele.
The region of the RFA1 gene encoding the promoter and amino acids 1 to 200 was amplified under mutagenic PCR conditions using plasmid pDS1 (RFA1/LEU2/CEN) as a template (56). This DNA was subjected to 35 cycles of amplification using Taq DNA polymerase and specific primers in the presence of 1 mM MnCl2. These randomly mutagenized PCR products were combined with an RFA1 plasmid vector lacking the N-terminal coding region of RFA1 (pDS1 digested with NdeI and BamHI) and were cotransformed into strain SBY102 carrying plasmid pJM114 (RFA1/URA3/CEN). The RFA1 gene was repaired by homologous recombination in vivo. Leucine prototrophs were selected and replica plated onto solid media containing 5-FOA at 25°C to shuffle out the wild-type RFA1 plasmid, pJM114. FOA-resistant colonies were replica plated to yeast extract-peptone-dextrose (YPD) plates and incubated at 25 or 37°C for 3 to 4 days. Approximately 20,000 colonies were screened for mutants that failed to grow at 37°C. The plasmids from four ts strains were rescued, and the transformation was repeated. One plasmid that was found to reproducibly confer the ts growth phenotype was sequenced and found to encode a single amino acid change at residue 29 from tyrosine to histidine. An ApaI-SpeI fragment containing the rfa1-Y29H mutation was subcloned into an otherwise wild-type RFA1 gene to confirm that the ts phenotype was due to the Y29H mutation.
Synthetic-lethal screen.
A red/white colony-sectoring assay was used to identify mutations that were synthetically lethal with rfa1-Y29H (4). To perform this screen, two rfa1-Y29H strains (HSY657 and HSY672) carrying pJM195 and showing the colony-sectoring phenotype were mutagenized with ethylmethanesulfonate to approximately 30% viability. After 7 days of growth at 25°C, approximately 100,000 colonies were screened for a nonsectoring red colony phenotype. About 200 nonsectoring mutants were obtained and rechecked for the nonsectoring phenotype by streaking on YPD and were checked for lethality on 5-FOA, which selects against uracil+ cells (i.e., cells that retain pJM195). The following four experiments were performed on the 20 strains that passed these tests. First, to determine whether these mutants required the wild-type RFA1 gene or the pJM195 plasmid (which also contains the URA3 and ADE3 marker genes), they were transformed with pHS118 (RFA1/LEU2/CEN) or pHS119 (RFA1/LYS2/CEN) and were tested for the sectoring phenotype. Mutants that showed a nonsectoring phenotype were likely due to integration of ADE3 into a chromosome and were excluded by this test. Second, the presence of rfa1-Y29H at the TRP1 locus was confirmed by both PCR and complementation testing. Genomic DNA was prepared and amplified by three sets of oligonucleotides to confirm that the rfa1 mutant gene was present at the TRP1 locus. Mutant strains were also crossed to HSY635 or HSY636. Mutants that had lost rfa1-Y29H produced diploids that required pJM195 and remained sensitive to 5-FOA due to the lack of a chromosomal RFA1 gene. Third, the synthetic-lethal mutations in these strains were shown to be recessive by backcross to HSY657 or HSY672. Lastly, each backcrossed diploid was sporulated and microdissected. The ratio of viable to inviable spores was always near unity, indicating that the synthetic lethality was caused by a single mutation. Three mutants passed these tests and were found to represent three different complementation groups. These groups were named slr51, slr157, and slr44, for synthetically lethal with rpa1.
Cloning of RFC4.
To clone the wild-type copy of slr51, the mutant strain was transformed with a yeast genomic plasmid library (LEU2/CEN). The library plasmids that complemented FOA-sensitive growth of slr51 were recovered and sequenced. The overlapping regions of these chromosomal fragments identified the complementing gene as RFC4. Genetic linkage analysis was used to confirm that a mutation in RFC4 caused the synthetic-lethal phenotype with rfa1-Y29H. Strain HSY787, which contains a LEU2 marker integrated adjacent to the RFC4 locus, was crossed to the slr51 strain, and the diploid was sporulated and microdissected. Tetrad analysis revealed that synthetic lethality and leucine auxotrophy always segregated together. This mutant allele was named rfc4-2.
Allele specificity of rfc4-2.
To examine allele-specific interactions between RFA1 and RFC4, strain HSY740 (rfc4-2 rfa1Δ leu2 pJM195) was derived from a cross between HSY737 and HSY635. A series of rfa1 mutant alleles in plasmid pRS415 (LEU2/CEN) were previously isolated by Umezu and colleagues (63, 75). These plasmids were transformed into HSY740 and HSY636 to create 19 rfc4-2 rfa1 double mutants and 19 rfa1 single mutants, each carrying pJM195. Cells were scraped from plates and resuspended at an optical density at 600 nm of 3. Tenfold serial dilutions of cells were then prepared in a microtiter plate, and 5 μl of each dilution was transferred onto YPD plates or synthetic complete media containing 5-FOA to measure synthetic lethality.
UV, MMS, and HU sensitivity.
To measure sensitivity to UV light, cells were grown to early log phase. About 500 cells were spread on YPD plates and were irradiated with the indicated levels of UV light using a UV cross-linker (Stratagene). The number of viable cells was determined by counting colonies after 3 days of growth, and the percent viability compared to the unirradiated sample was calculated. To determine methylmethanesulfonate (MMS) sensitivity, cells were grown in liquid YPD and MMS was added to a final concentration of 0.1%. At the indicated times, aliquots were removed and neutralized with an equal volume of 10% sodium thiosulfate. The cells were then washed with water and spread on YPD plates. The number of viable cells was determined by counting colonies after 3 days of growth. To determine HU sensitivity, cells were grown in liquid YPD and HU was added to a final concentration of 200 mM. At the indicated times, aliquots were removed, washed with water, and spread onto YPD plates. Viable cells were determined as described above. HU sensitivity was also tested on solid media by replica plating. HU was added to a final concentration of 100 mM to YPD agar or to the appropriate selective media. Fresh cells were scraped from plates and were resuspended at an optical density at 600 nm of 3. Tenfold serial dilutions of cells were then prepared in a microtiter plate, and 5 μl of each dilution was transferred onto plates with and without HU.
Western blot assay for Rad53 phosphorylation.
Yeast cells were grown to early log phase at 30°C. Exponentially growing cells, cells synchronized in G1 for 2 h with α-factor (5 μg/ml), or cells arrested in G2 for 3 h with nocodazole (20 μg/ml) were released into HU (200 mM)- or MMS (0.1%)-containing media for 1 h. Alternatively, cells were irradiated with 60 J of UV light/m2 and then were incubated for 30 min. Cells treated with MMS were neutralized with 10% sodium thiosulfate. To make whole cell extracts, 5 ml of each culture was harvested, washed with water–20% trichloroacetic acid, and then resuspended in 25 μl of 20% trichloroacetic acid. An equal volume of glass beads was added, and cells were lysed by vortexing with glass beads at 4°C. Whole cell extracts were microcentrifuged at 3,000 × g for 15 min at 4°C. Protein precipitates were resuspended in 2× sodium dedocyl sulfate (SDS) loading buffer, heated for 5 min, and then centrifuged at 3,000 × g for 5 min (51). Proteins were separated by SDS–10% polyacrylamide gel electrophoresis (PAGE) and transferred to a nylon membrane. Rad53 proteins were detected with antiserum to Rad53 (90) and horseradish peroxidase-conjugated anti-rabbit secondary antibody and were visualized by a chemiluminescent developer (Amersham).
FACS analysis.
Yeast cells were grown to early log phase and were synchronized in G1 with α-factor (5 μg/ml) for 2 h. Cells were washed with water and released into YPD or YPD with 0.038% MMS. Aliquots of cells were removed every 30 min and washed with an equal volume of 10% sodium thiosulfate to neutralize the MMS. Cells were then fixed in 0.5 ml of water and 1.0 ml of 100% ethanol solution by rotating overnight. Fixed cells were washed with 1 ml of 50 mM sodium citrate, were resuspended in 0.5 ml of 50 mM sodium citrate containing 0.1 mg of RNase A/ml, and then were incubated for 2 h at 37°C. This cell suspension was added to 0.5 ml of 50 mM sodium citrate containing 50 μg of propidium iodide/ml and was incubated for at least 1 h at room temperature. DNA content was analyzed on a Coulter-Epics fluorescence-activated cell sorter (FACS) (46).
Rfc4-RPA binding assay.
Enzyme-linked immunosorbent assay (ELISA) wells (Immulon) were coated with either 0.5 μg of RPA complex, purified as described previously (13), or bovine serum albumin (BSA) in incubation buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 2 mM dithiothreitol, 5 mM MgCl2, 150 mM NaCl, 20% glycerol, 2 mM CaCl2) for 1 h at 30°C. Wells were washed three times with 1× PBST (29) and then blocked with 5% dried milk in 1× PBST for 10 min at room temperature. 35S-labeled Rfc4 protein was expressed in an in vitro transcription/translation system (Promega) and then was applied to a Superdex 75 column. 35S-Rfc4 protein was eluted in buffer A (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% NP-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) containing 0.075 M NaCl and was collected. Purified 35S-labeled Rfc4 was added into wells and incubated for 1 h at 30°C. Unbound Rfc4 protein was removed by washing three times with 1× PBST, and the bound 35S-Rfc4 was measured by scintillation counting.
RESULTS
Isolation of the conditionally lethal mutant rfa1-Y29H.
As a first step to determine the essential cellular role of the N-terminal domain of Rpa1 (Rpa1N), we isolated a conditional-lethal RFA1 mutation mapping to this domain. A strain lacking RFA1 was constructed and maintained by a copy of RFA1 on a URA3-based plasmid. This strain, SBY102 (rfa1Δ pJM114/RFA1/URA3/CEN), is unable to grow on media containing 5-FOA since selection against the URA3 plasmid is lethal in this background. Mutagenized RFA1 fragments were introduced into this strain on LEU2-based plasmids and were swapped for the RFA1 plasmid by replica plating on media containing 5-FOA at 25°C. About 20,000 colonies were screened for loss of viability at 37°C, and one recessive rfa1-ts allele was isolated. DNA sequencing of this allele revealed a mutation that changed tyrosine at residue 29 to histidine. Hereafter, we refer to this allele as rfa1-Y29H. As shown in Fig. 1A, rfa1-Y29H and wild-type cells grew well at 25°C while the rfa1-Y29H mutant showed a severe growth defect at 37°C. Additional characterization of the rfa1-Y29H mutation revealed that its function was partially compromised at the permissive temperature because the mutant strain was weakly sensitive to UV or MMS treatment at 25°C relative to the wild type (data not shown).
Isolation of rfc4-2 by its synthetic lethality with rfa1-Y29H.
To identify mutants that require wild-type Rpa1N for viability, we performed a synthetic-lethal screen with rfa1-Y29H at 25°C. HSY657 and HSY672 are ade2 ade3 strains containing a stably integrated rfa1-Y29H allele and plasmid pJM195 (RFA1/URA3/ADE3/CEN) (Table 1). These strains allow the use of a red/white colony-sectoring assay to measure plasmid stability since the starting strain is red (ade2) and plasmid loss events generate white (ade2 ade3) sectors. About 100,000 ethylmethanesulfonate-mutagenized colonies were screened for strains that require the pJM195 plasmid for viability and produce unsectored red colonies essentially as described previously (4). Three recessive slr (synthetic lethal with rpa1) complementation groups were isolated. Because the slr mutants require the wild-type RFA1 plasmid, pJM195, for viability, they acquire a 5-FOA-sensitive phenotype (Fig. 1B). The mutation in slr51 was identified by transforming the strain with a yeast genomic plasmid library and selecting strains that no longer require pJM195 by growth on 5-FOA. The library plasmids were rescued from the transformants, were sequenced, and were found to contain the RFC4 gene. Linkage analysis confirmed that a mutation in RFC4, hereafter referred to as rfc4-2, caused the synthetic-lethal phenotype with rfa1-Y29H. The rfc4-2 allele was found to have a single nucleotide change from G to A at residue 601, resulting in an amino acid change of aspartate to asparagine at residue 201. This mutation maps to the RFC box VIII/sensor2 motif, one of the conserved motifs found in all RFC subunits (Fig. 1B) (15, 21, 40).
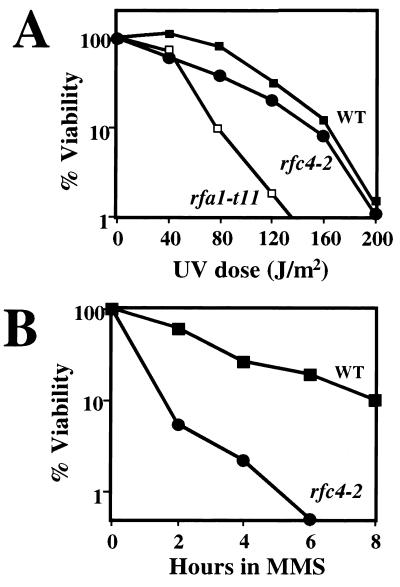
Compared to wild-type cells, the rfc4-2 single mutant showed no obvious growth defects and grew well at temperatures ranging from 25 to 37°C. Thus, DNA replication is not significantly impaired by the rfc4-2 mutation in the presence of wild-type RPA. To examine whether the rfc4-2 mutant has defects in DNA repair, we measured its sensitivity to the DNA damaging agents UV and MMS. Relative to the wild type, rfc4-2 was weakly sensitive to UV but was more resistant than the known UV-sensitive strain rfa1-t11 (75) (Fig. 2A). The rfc4-2 mutant was very sensitive to MMS treatment (Fig. 2B). We conclude that Rfc4-2 function in response to DNA damage is compromised.
FIG. 2.
The rfc4-2 mutant is sensitive to DNA damage. (A) UV sensitivity. Strains HSY636 (RFC4), HSY740 (rfc4-2), and HSY845 (rfa1-t11) were grown to early log phase in liquid culture. A volume containing about 500 cells was spread onto YPD plates and was irradiated with the indicated doses of UV light. The percent viability was determined by counting colonies following 3 days of growth. (B) MMS sensitivity. Cultures of HSY636 (RFC4) and HSY740 (rfc4-2) were grown to early log phase in liquid YPD, and MMS was added to a final concentration of 0.1%. At the indicated times an aliquot of the culture was withdrawn, the MMS was neutralized, and about 300 cells were spread onto YPD plates. The percent viability was determined as described for panel A.
Characterization of the Rfc4-Rpa1 interaction.
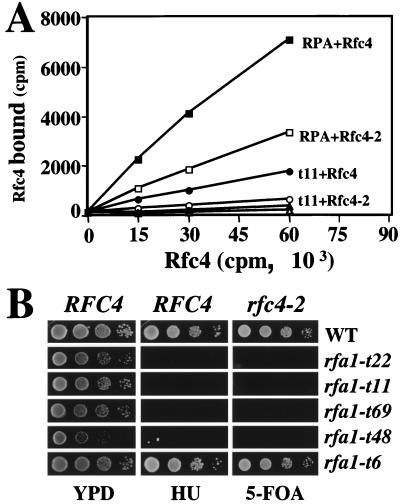
It has recently been shown that the p140, p40, and p38 subunits of human RFC bind the large subunit of human Rpa1 (89). Since yeast Rfc4 is the homolog of p40, we tested whether there was a physical interaction between Rfc4 and yeast RPA. For this experiment we used an ELISA-type assay in which wild-type or mutant RPA complex or BSA control protein was immobilized on the wells of a microtiter plate. These wells were then incubated with increasing amounts of purified 35S-labeled Rfc4 protein that was synthesized in an in vitro translation reaction. After the wells were washed, the level of bound Rfc4 was determined by scintillation counting. As shown in Fig. 3A, wells coated with wild-type RPA bound increasing amounts of wild-type 35S-Rfc4 compared to the BSA control. This result indicates that the yeast RPA complex directly interacts with yeast Rfc4 protein. When RPA was incubated with mutant 35S-Rfc4-2 protein, much less protein was retained. At maximal input only half as much Rfc4-2 protein bound RPA (Fig. 3A). As described below, we found that rfc4-2 is synthetically lethal with several previously characterized rfa1 alleles. When this assay was performed using RPA containing the rfa1-t11 mutation (RPA-t11), we found that it bound less 35S-Rfc4 than did wild-type RPA. Further, when incubated with mutant 35S-Rfc4-2 protein, the mutant RPA-t11 bound even less Rfc4 protein (Fig. 3A). This combination of mutant proteins revealed an interaction only slightly stronger than that between Rfc4 and the control BSA protein. These results suggest that the synthetic lethality between these mutant alleles is due to a compromised interaction between RPA1 and Rfc4.
FIG. 3.
An interaction between Rfc4 and RPA. (A) The physical interaction between Rfc4 and the RPA complex was tested by coating ELISA wells with 0.5 μg of either purified wild-type RPA (squares), mutant RPA-t11 (circles) or BSA (triangles) and treating the wells with increasing amounts of purified 35S-labeled wild-type Rfc4 (closed symbols) or mutant Rfc4-2 (open symbols). The counts per minute (cpm) representing bound 35S-Rfc4 was determined by scintillation counting and is plotted relative to the cpm of input 35S-Rfc4. (B) Allele-specific interaction between RFC4 and RFA1. Strains HSY636 (RFC4 rfa1Δ; left and middle columns) and HSY740 (rfc4-2 rfa1Δ; right column) carrying pJM195 (RFA1/URA3) were transformed with wild-type RFA1 or the indicated rfa1 alleles on a LEU2-based vector. Transformants were streaked onto plates lacking leucine and then were taken from the plates and serially diluted 1:10. Approximately 5 μl of each dilution was transferred to a YPD plate, a YPD plate containing HU, or a plate containing 5-FOA to measure synthetic lethality.
To test the specificity of the interaction genetically, we examined the viability of rfc4-2 in combination with a variety of rfa1 alleles. Umezu and colleagues have isolated a large group of rfa1 alleles based on their sensitivity to MMS (75). This group of alleles covers a wide range of MMS sensitivity and includes mutations covering all four functional domains of Rpa1 (75). We transformed strain HSY740 (rfc4-2 rfa1Δ pJM195/RFA1/URA3) with 19 different rfa1 alleles and tested for a synthetic-lethal phenotype by measuring growth on 5-FOA. For a quantitative comparison we spotted serial dilutions of the transformants onto plates containing 5-FOA and compared their growth to the rfa1 single mutants as controls. Of the 19 alleles tested, four grew well on YPD but showed little or no growth on 5-FOA (Fig. 3B): rfa1-t22 (F15L, M49T), -t11 (K45E), -t69 (K45E, D121G) and -t48 (L221P). By comparison, RFA1 and rfa1-t6 allowed good growth on 5-FOA (Fig. 3B). Interestingly, all four synthetic-lethal rfa1 mutations map in or near the N-terminal domain of Rpa1. In contrast, alleles with mutations that map in the ssDNA binding domains are viable in combination with rfc4-2 (data not shown). Thus, rfc4-2 displays allele-specific interactions with RFA1.
We tested whether this allele specificity correlated with other rfa1 phenotypes. Synthetic lethality did correlate with the severity of the allele as judged by MMS sensitivity. However, several rfa1 mutants that were partially MMS sensitive, temperature sensitive, or extremely defective in the repair of HO-induced DNA breakage showed no defect in the rfc4-2 background (75). When the 19 rfa1 single mutants were tested for sensitivity to HU, we found that the four synthetic-lethal rfa1 alleles also showed the strongest HU sensitivity (Fig. 3B). By comparison, rfa1-t6 was neither synthetically lethal with rfc4-2 nor sensitive to HU. We conclude that the synthetic-lethal phenotype of the rfa1 alleles correlates strongly with HU sensitivity. This suggests that these interacting genes might share defects in DNA replication checkpoint pathways (17, 49, 60, 66, 83).
RFC4 is required for the replication block checkpoint response.
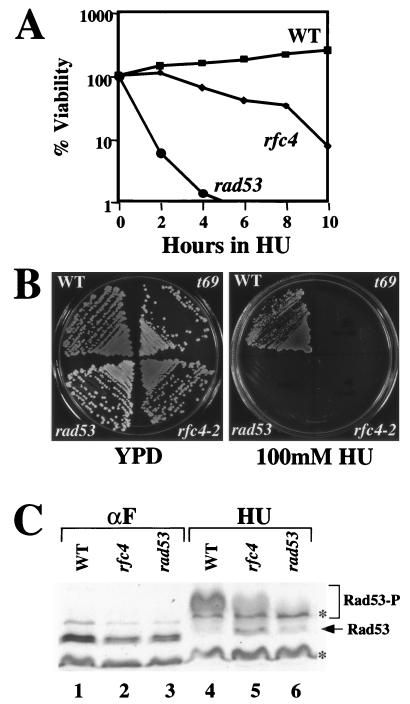
To determine whether rfc4-2 cells respond appropriately to a DNA replication block, we examined the viability of rfc4-2 mutant cells in the presence of HU. When the assay was carried out in liquid media containing 200 mM HU, rfc4-2 cells showed a mild sensitivity relative to wild-type cells (Fig. 4A). Control cells containing rad53-K227A, a kinase-defective allele of the checkpoint kinase Rad53 (2, 20), were strongly sensitive to HU. Both of these mutants displayed a severe growth defect in the presence of continuous HU exposure, although very tiny rfc4-2 colonies were were able to form on plates containing HU (Fig. 4B).
FIG. 4.
The rfc4-2 mutant is defective in the DNA replication block checkpoint response. (A) HU sensitivity of rfc4-2 cells. Strains W303-1a (wild type), HSY1025 (rfc4-2), and Y00684 (rad53-K227A) were grown to early log phase in liquid YPD, and HU was added to a final concentration of 200 mM. At the indicated times an aliquot was removed and a volume containing approximately 300 cells was spread onto a YPD plate. The percent viability was determined by counting colonies after 3 days. (B) Sensitivity of rfc4-2 cells to continuous HU exposure. The three strains used in panel A, along with the rfa1-t69 mutant, were streaked onto YPD plates, with or without 100 mM HU, and were incubated at 30°C for 3 days. (C) Phosphorylation of Rad53 in response to HU treatment. Strains W303-1a (WT), HSY1027 (rfc4), and Y00684 (rad53) were synchronized by treatment with α-factor (lanes 1, 2, and 3) and were released into liquid YPD medium containing 200 mM HU for 1 h (lanes 4, 5, and 6). The cells were harvested, and whole cell extracts were prepared. Extracts were resolved by SDS–10% PAGE and were Western blotted using an antiserum against Rad53. Asterisks (∗) denote nonspecific bands. Rad53-P denotes phosphorylated forms of Rad53.
To test whether the HU sensitivity of rfc4-2 cells reflects a defect in the DNA replication block checkpoint response, we assayed Rad53 phosphorylation in the presence of HU. RAD53 encodes a protein kinase and is an essential transducer in checkpoint pathways together with MEC1 (2, 5, 33, 58, 64, 67, 87, 90). Rad53 is phosphorylated in response to both DNA damage and inhibition of DNA replication, and the phosphorylation of Rad53 is an essential step in the signaling of checkpoints (16, 55, 58). Wild-type and mutant cells were synchronized in G1 by treatment with α-factor, were released into YPD media containing 200 mM HU, and then were analyzed for Rad53 phosphorylation. In the absence of HU, Rad53 was not phosphorylated in G1-arrested wild-type or mutant cells (Fig. 4C, lanes 1, 2, and 3). Following HU treatment, Rad53 was completely phosphorylated in wild-type cells; all unphosphorylated forms of Rad53 disappeared (Fig. 4C, lane 4). In contrast, the phosphorylation of Rad53 was reduced in the rfc4-2 mutant following HU treatment. Both unphosphorylated and phosphorylated forms of Rad53 were present in rfc4-2 mutant cells after replication block (Fig. 4C, lane 5). In rad53-K227A control cells, both forms of Rad53 disappeared after HU treatment as observed previously (77) (Fig. 4C, lane 6). Thus, rfc4-2 cells are partially defective in the activation of the replication block checkpoint pathway.
RFC4 is required for the intra-S checkpoint.
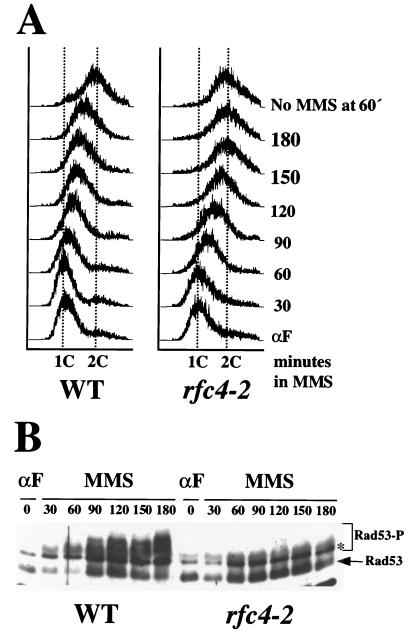
DNA damage by continuous exposure to MMS significantly extends the length of the S phase in wild-type cells (9, 52, 53, 61, 80). This response is distinguished from the replication block pathway in that it requires the function of RAD9 and other genes (53). To determine whether RFC4 is required for the intra-S checkpoint, cells were synchronized in G1 with α-factor and were released into YPD media containing 0.038% MMS, and aliquots of cells were removed every 30 min and treated for microfluorometric analysis. The rate of S-phase progression in wild-type and rfc4-2 mutant cells was then monitored by FACS. In the absence of MMS, wild-type and rfc4-2 mutant cells finished DNA synthesis 60 min after release from the α-factor block (Fig. 5A, top two histograms). In the presence of MMS, S-phase length was extended in both wild-type and rfc4-2 cells. However, rfc4-2 cells finished S phase faster than the wild type; rfc4-2 cells completed replication 150 min after α-factor release, whereas wild-type cells were still in S phase even at 180 min (Fig. 5A, left).
FIG. 5.
The rfc4-2 mutant is defective in the intra-S checkpoint. (A) W303-1a (WT) and HSY1027 (rfc4-2) cells were synchronized in G1 phase with α-factor and were released into YPD containing 0.038% MMS. Aliquots were removed every 30 min, and the MMS was neutralized. Cells were then fixed, stained, and analyzed by FACS. As a control, cells of each type were released into YPD in the absence of MMS and were similarly analyzed. The DNA content of untreated control cells 60 min after α-factor release is presented in the top histograms as representative of cells having completed S phase. (B) Wild-type and rfc4-2 cells were treated as above, and aliquots were removed every 30 min. To analyze Rad53 phosphorylation, whole cell extracts were prepared, were resolved by SDS–10% PAGE, and were Western blotted using an antiserum against Rad53. The asterisk (*) denotes nonspecific bands. Rad53-P denotes phosphorylated forms of Rad53.
The phosphorylation of Rad53 was analyzed in wild-type and rfc4-2 cells to see whether defects in regulating the rate of S-phase progression correlate with defects in Rad53 phosphorylation. Wild-type cells showed significant levels of phosphorylated Rad53 while inducing S-phase delay in response to MMS exposure (Fig. 5B, left panel). On the other hand, Rad53 was only partially phosphorylated in rfc4-2 cells in response to MMS treatment (Fig. 5B, right panel). Compared to the wild type, the ratio of phosphorylated to unphosphorylated forms of Rad53 was greatly reduced in rfc4-2 cells at 180 min. This rfc4-2 phenotype contrasts slightly with that of rfc5-1, where cells entered G2 phase 90 min after release, and Rad53 was phosphorylated and then dephosphorylated in response to MMS (65). Taken together, we conclude that rfc4-2 cells are partially defective in the intra-S checkpoint and Rad53 phosphorylation.
RFC4 is required for the DNA damage checkpoint pathway.
As mentioned previously, Rad53 phosphorylation is also required for the activation of the DNA damage checkpoint pathway. To examine whether RFC4 is involved in DNA damage checkpoint control, we analyzed Rad53 phosphorylation in response to MMS and UV irradiation. MMS was added to exponentially growing cells to a final concentration of 0.1%, and the phosphorylation of Rad53 was detected by Western blotting. In wild-type cells, unphosphorylated forms of Rad53 completely disappeared and phosphorylated forms of Rad53 with slower mobility accumulated after MMS treatment (Fig. 6A, lane 4). However, Rad53 phosphorylation was greatly reduced in rfc4-2 mutant cells (Fig. 6A, lane 5). As above, Rad53 disappeared in rad53-K227A cells following MMS treatment (Fig. 6A, lane 6). We conclude that RFC4 is required for the DNA damage checkpoint pathway.
FIG. 6.
The rfc4-2 mutant is defective in the DNA damage checkpoint. (A) Exponentially growing cells of the indicated genotype were incubated with or without 0.1% MMS for 1 h. Both MMS-treated (lanes 4, 5, and 6) and -untreated (lanes 1, 2, and 3) cells were analyzed for Rad53 phosphorylation by Western blotting. (B) Exponentially growing wild-type and rfc4-2 cells were treated with or without UV light (60 J/m2), were incubated for 30 min, and then were analyzed for Rad53 phosphorylation by Western blotting (lanes 1 and 2). In addition, wild-type and rfc4-2 cells were synchronized in G1 or G2 with α-factor or nocodazole, respectively, and were treated and analyzed as above (lanes 3 to 6). Asterisks (∗) denote nonspecific bands. (C) The rfc4-2 mutant was tested for G2/M arrest in response to lesions caused by cdc13. Four cdc13 cdc15 strains, DLY408 (WT), DLY409 (rad9), HSY1202 (rfc4-2), and HSY1204 (rfa1-t11), were synchronized in G1 with α-factor at 23°C. Cells were then released from the G1 block, shifted to 36°C, and incubated for 3.5 h. Cells were fixed and stained, and their morphology was analyzed by fluorescence microscopy. The percentage of cells arrested at G2/M (large budded cell with a single nucleus at the neck) or at the cdc15 arrest point (large budded cell with two nuclei) is shown in the table in the lower right panel. Cells with nuclear DNA stretched between the mother and daughter compartments are not represented in the table and account for the percentages summing to less than 100.
To determine if this defect is specific to the G1/S or G2/M checkpoint, Rad53 phosphorylation was analyzed in G1- or G2-arrested wild-type and rfc4-2 mutant cells in response to UV irradiation. Exponentially growing cells were arrested in G1 with α-factor or in G2 with nocodazole and were irradiated with UV at 60 J/m2, and the phosphorylation of Rad53 was analyzed by Western blotting. In wild-type exponential cells, Rad53 was completely phosphorylated following UV irradiation (Fig. 6B, top panel, lane 2). In contrast, the phosphorylation of Rad53 was greatly reduced in exponential-phase and G1- and G2-arrested rfc4-2 mutant cells in response to UV. The rfc4-2 cells, regardless of their phase of the cell cycle, retain a significant amount of unphosphorylated Rad53 in response to UV irradiation (Fig. 6B, bottom panel, lanes 2, 4, and 6). The defect in Rad53 phosphorylation in response to DNA damage appears to be more pronounced than that obtained during replication block with HU.
To confirm that the G2/M checkpoint defects were not limited to Rad53 phosphorylation, we assayed the ability of cells to arrest growth at G2 in response to DNA damage. Cells containing the cdc13 mutation undergo DNA damage upon shift to 36°C and arrest in G2 with a large bud and a single nucleus. In contrast, checkpoint-defective cdc13 cells at 36°C continue into the next cell cycle. We used a cdc13 cdc15 background to measure this effect since checkpoint-defective cells are unable to exit mitosis and arrest with two nuclei (the cdc15 phenotype) (24). When an otherwise wild-type cdc13 cdc15 mutant was shifted to 36°C, all of the cells arrested at the G2 block. In contrast, a large percentage (87%) of rad9 cdc13 cdc15 cells arrested with two nuclei (Fig. 6C). When rfc4-2 cdc13 cdc15 cells were shifted to 36°C, 40% of the cells had passed the G2 arrest point, as indicated by the presence of two nuclei. These data confirm a G2/M checkpoint defect in rfc4-2 cells, although the effect is not as pronounced as in rad9 cells. Mutations in Rpa1 have been reported to have G1/S and intra-S checkpoint defects but no defect in G2/M (42). When rfa1-t11 cdc13 cdc15 mutants were shifted to 36°C we again observed about 40% of the cells arresting with two nuclei (Fig. 6C). We conclude that both RFC4 and RFA1 are required for wild-type G2/M checkpoint response.
Interactions between RFC4, RAD24, and RFA1.
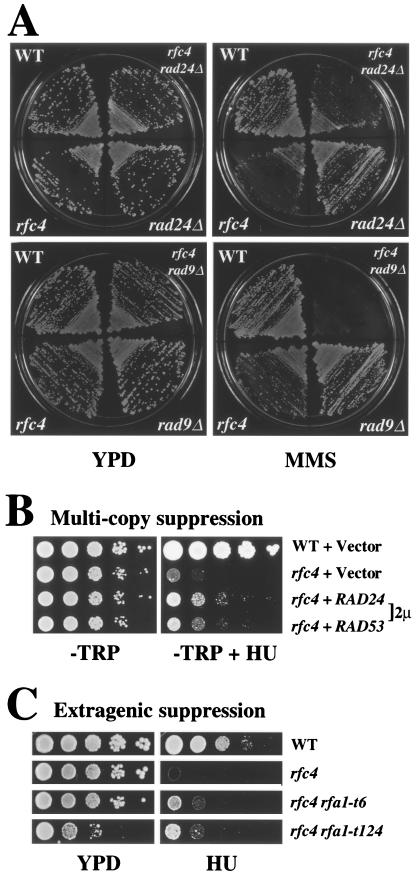
DNA damage checkpoints are controlled by the RAD9 and RAD24 epistasis groups (16, 45, 81, 82). To determine whether RFC4 belonged to either or both of these pathways, we constructed the following double mutants and tested their response to MMS treatment: rfc4-2 rad24Δ, rfc4-2 rad9Δ, and rfc4-2 rad53-K227A. The rfc4-2 rad9Δ strain showed an enhanced sensitivity to MMS whereas the MMS sensitivity of the other two double mutants was no greater than either single mutant (Fig. 7A and data not shown). These results indicate that RFC4 functions as part of the RAD24 epistasis group and not RAD9. This result is consistent with the fact that Rad24 is associated with all four small subunits of the RFC complex (26, 27).
FIG. 7.
Genetic interactions between checkpoint genes. (A) RAD24 and RFC4 are epistatic with respect to MMS sensitivity. Strains of the indicated genotype were tested for MMS sensitivity by streaking onto YPD plates with or without 0.012% MMS. (B) Suppression of rfc4-2 HU sensitivity by high-copy-number RAD24 or RAD53. Strain HSY1027 (rfc4-2) was transformed with RAD24 or RAD53 genes on a 2μm plasmid. Cells were serially diluted 1:10, and 5 μl was replica plated to solid media lacking tryptophane. (C) Extragenic suppression of rfc4-2 HU sensitivity by rfa1 mutant alleles. Strain HSY740 (rfc4-2 rfa1Δ) containing plasmid pJM195 (RFA1/URA3/ADE3) was transformed with pHS118 (RFA1), pKU1-t6 (rfa1-t6), and pKU1-t124 (rfa1-t124). Transformants were then streaked onto plates containing 5-FOA to remove pJM195. The resulting strains were serially diluted 1:10, and 5 μl was replica plated onto YPD plates with or without 100 mM HU. The resulting strain genotypes are shown at the right of the figure. Strain HSY636 was used as a wild-type control.
We next examined whether RAD24 and rfc4-2 interacted in the replication block checkpoint by testing if overexpression of RAD24 or RAD53 can suppress the HU sensitivity of the rfc4-2 mutant. We transformed the rfc4-2 mutant strain with high-copy-number RAD24 or high-copy-number RAD53 plasmids and transferred serial dilutions of each transformant onto a plate containing HU. As observed for the rfc5-1 mutant (60), the HU sensitivity of rfc4-2 was partially suppressed by overexpression of RAD24 (Fig. 7B). In contrast, high-copy-number plasmids of the other four RFC subunits showed no effect on the HU sensitivity of the rfc4-2 strain (data not shown). RAD53 overexpression also partially suppressed the HU sensitivity of rfc4-2. Although other explanations are possible, the simplest interpretation of this genetic result is that RFC4 functions upstream of RAD53 (Fig. 7B).
Given the interaction between RFC4 and RFA1, we examined whether the HU sensitivity of rfc4-2 could be suppressed by overexpressing RFA1 or RFA2. These genes on high-copy-number plasmids failed to suppress this phenotype (data not shown). On the other hand, as part of our analysis of synthetic interactions between rfc4-2 and rfa1, we observed that the HU sensitivity of rfc4-2 could be suppressed by two specific rfa1 alleles out of the 19 alleles tested. As shown in Fig. 7C, the growth of an rfc4-2 strain in the presence of HU is improved when it carries rfa1-t6 or rfa1-t124 instead of wild-type RFA1. These rfa1 alleles result from amino acid changes mapping to the ssDNA binding domains, not the N terminus. This extragenic suppression further suggests that the interaction between Rpa1 and Rfc4 is likely to be important for checkpoint signaling.
DISCUSSION
A genetic interaction between RFA1 and RFC4.
The large subunit of replication protein A (Rpa1) contains three ssDNA binding domains and an amino-terminal domain of approximately 180 amino acids (Rpa1N). Although Rpa1N is dispensable for DNA replication in vitro, it is likely to play an important role in vivo because it is essential for viability in yeast (11, 25, 35, 56, 85). Moreover, the most severe mutations identified in a random mutagenesis to create MMS-sensitive rfa1 alleles map to Rpa1N as opposed to the ssDNA binding domains (75). To investigate the essential function of this domain, we searched for mutations that are lethal in the presence of the rfa1-Y29H mutation. This screen identified an allele of RFC4 (rfc4-2) which encodes a subunit of the eukaryotic sliding clamp loader. Since human Rpa1N is known to bind a number of proteins (10, 41), we considered the possibility of a direct interaction between Rpa1N and Rfc4. The results of binding assays and allele-specific lethality observed in a collection of rfa1 rfc4-2 double mutants are consistent with this notion. The simplest interpretation of these results is that an interaction between Rpa1N and Rfc4 is essential for DNA replication in vivo.
The interaction between an ssDNA binding protein and its cognate clamp loader appears to be conserved from bacteria to higher eukaryotes. In E. coli, the γ complex physically interacts with Ecssb. The χ subunit has been shown to mediate the binding of the γ complex to the C terminus of Ecssb (34). In the human system, RPA and RFC are known to interact in multiple ways. RPA targets RFC to the primer-template junction by inhibiting its nonspecific ssDNA binding activity (73). RPA also plays an essential role in the DNA polymerase switch by loading the primer-recognition complex, which then blocks the ability of DNA polymerase α to extend the primer (74). More recently, it was shown that RPA is specifically needed for RFC and PCNA to remain on primed DNA and inhibit DNA polymerase activity; inhibition is lost if Ecssb is substituted for RPA (89). These investigators also showed that the p140, p40, and p38 subunits of human RFC are capable of binding directly to the large subunit of human Rpa1 (89).
The results reported here indicate that RFC and RPA interact via Rfc4 and Rpa1N. Based on these results, we propose the following model to explain rfc4-2 defects in DNA replication and the DNA damage sensing mechanism. RPA is required at the initiation of DNA replication and at each Okazaki fragment to bind ssDNA and stimulate DNA polymerase α (10, 71, 78). As shown in Fig. 8, RFC competes with DNA polymerase α for RPA, which results in the loading of RFC and PCNA and a switch to processive synthesis by DNA polymerase δ (73, 74, 89). Although Rfc4-2 and Rfa1-Y29H functions are compromised, they are individually capable of performing this step. In combination, however, their defects are exacerbated, resulting in impaired DNA synthesis and lethality. This interaction is likely conserved during the loading of Rad24–Rfc2-5 in DNA damage repair (Fig. 8, right). In this example, UV-induced DNA damage is first recognized by Rad14 (XPA in human cells) and RPA (39, 79). Following removal of the damaged DNA, additional RPA binds to the exposed ssDNA lesion and recruits the Rad24–Rfc2-5 complex to the primer-template junction. The PCNA-like complex of Rad17-Mec3-Ddc1 is subsequently loaded at the primer-template junction. This protein assembly on the ssDNA lesion, combined with MEC1-dependent Ddc1 hyperphosphorylation, then activates the signal transduction pathways leading to cell cycle arrest (51). We propose that Rfc4-2 and Rfa1-Y29H are partially impaired in their ability to load the Rad24–Rfc2-5 complex, leading to a partial defect in the DNA damage checkpoint response.
FIG. 8.
Role of the Rfc4-Rpa1 interaction in DNA replication and DNA damage checkpoint signaling. (Left) The DNA polymerase switch during lagging-strand synthesis is shown. The model proposes that a direct interaction between Rpa1N, bound to the lagging strand, and Rfc4 is required to target RFC to the primer-template junction. Bound RFC then displaces polymerase α, loads the PCNA sliding clamp, and allows processive DNA synthesis by polymerase δ. This reaction fails in the presence of Rfc4-2 and Rfa1-Y29H, resulting in synthetic lethality. (Right) The DNA damage response at a UV-induced lesion is shown. Again, a direct interaction between Rpa1N, bound at the processed lesion, and Rfc4 is required to target the Rad24–Rfc2-5 complex to the primer-template junction. This complex loads the Rad17-Mec3-Ddc1 sliding clamp and activates the signal transduction pathway. This reaction is compromised in the presence of either Rfc4-2 or Rpa1-Y29H, resulting in partially defective checkpoint signaling.
RFC4 as a sensor in checkpoint control.
Examination of the rfc4-2 phenotype revealed a mild sensitivity to HU, consistent with a defect in the DNA replication block checkpoint. The RAD53 protein kinase is an essential transducer in checkpoint pathways and is phosphorylated in response to DNA damage and inhibition of DNA replication (16, 55, 58). G1-arrested rfc4-2 cells show only partial Rad53 phosphorylation following release into HU; approximately half of the Rad53 remains unphosphorylated. In contrast, when wild-type cells are subjected to this treatment all Rad53 protein is found in the phosphorylated form (Fig. 4). Thus, rfc4-2 cells have a partial defect in the replication block checkpoint response consistent with the mild HU sensitivity of the mutant. A more pronounced defect in Rad53 phosphorylation is observed in response to DNA damage in exponentially growing, G1- or G2-arrested rfc4-2 mutant cells. The significant amount of unphosphorylated Rad53 that is detected in UV-irradiated rfc4-2 cells is notable given that the mutant is not strongly sensitive to UV irradiation (Fig. 6B). The reason for this discrepancy is unclear, although the DNA damage response may simply require a lower threshold of Rad53 phosphorylation. We also cannot rule out the possibility of additional pathways for DNA damage signaling. Together, these findings indicate that RFC4 is a common component of the checkpoint response to both unreplicated DNA and DNA damage.
Wild-type cells exhibit an intra-S checkpoint which greatly extends the length of S phase in the presence of low levels of MMS. Although rfc4-2 cells extend S phase under these conditions, they complete S phase well before wild-type cells do (Fig. 5A). Similarly, with respect to the G2/M checkpoint, essentially all wild-type cells are able to arrest growth at G2 in response to cdc13-induced damage. In contrast, about half the rfc4-2 cells fail to arrest under these conditions (Fig. 6C). Thus, rfc4-2 cells are partially defective in the intra-S and G2/M checkpoints consistent with the partial defect in DNA damage signaling. Based on these results we propose that RFC4 plays an essential role as a sensor, upstream of RAD53, in all the checkpoints tested in this study.
A number of DNA replication proteins, including several RFC subunits, have been identified as sensors in checkpoint pathways (3, 49, 50, 59, 65, 66). This raises the question of the role of the individual subunits in sensing DNA defects versus the role of the complex as a whole. If the overall integrity of the RFC complex was important, one might expect mutations in the small subunits to behave similarly. Indeed, these subunits share significant sequence similarity and are expected to be structurally redundant. Although two of the small RFC subunits, Rfc5 in S. cerevisiae (scRfc5) and Rfc3 in Schizosaccharomyces pombe (spRfc3; orthologous to scRfc3 and human p36), are required for wild-type checkpoint function, mutations in these subunits produce somewhat different phenotypes (59, 65). In a wild-type RAD24 background, scrfc5-1 appears to compromise the replication block checkpoint more than the DNA damage checkpoint (48, 65). In contrast, mutant sprfc3 produces noticeable defects in both the replication and DNA damage checkpoints, similar to the scrfc4-2 mutant reported here (59). An scrfc2 mutant, which is defective in the S/M replication block checkpoint, is sensitive to DNA damage although it has not been tested for DNA damage checkpoint function (50). While the phenotypic variation between rfc3, rfc4, and rfc5 mutants might reflect allele-specific differences, all three mutations appear to map to the same region of these homologs. The scrfc4-2 and sprfc3 mutations map to RFC box VIII (sensor 2 motif) while scrfc5-1 maps to RFC box II (P loop) (59, 66). Based on the X-ray crystallographic analysis of the δ′ subunit of the E. coli clamp loader, these two motifs are predicted to be very close to one another in tertiary structure (28). Mutations in these motifs might therefore be expected to have similar effects on their structure and activity.
The fact that each of the small RFC subunits is essential for viability clearly indicates that these subunits have distinct functions. In addition, a number of differences between the small subunits have been detected biochemically. As mentioned above, the human p37 and p36 subunits (Rfc2 and Rfc3, respectively) do not biochemically interact with the p70 subunit of human RPA, whereas the p40 and p38 subunits (Rfc4 and Rfc5, respectively) do (89). In addition, Cai and coworkers have studied the role of the conserved ATP-binding domains of each of the individual subunits of human RFC. These investigators have shown that mutation of p38 (Rfc5) has no effect on the ability of RFC to support in vitro DNA synthesis. In contrast, the identical mutation in the other four subunits inhibits the ATPase activity of RFC as well as its ability to support in vitro DNA synthesis (14). Differences between the small subunits are also revealed by genetic suppression experiments. The HU and temperature sensitivity of the scrfc2-1 mutant is suppressible by overexpressing RFC5 (50), whereas we observed no suppression of the HU sensitivity of rfc4-2 by overexpressing any of the other four subunits. The failure to suppress the rfc4-2 defect is consistent with the notion that Rfc4-2 is defective in its interaction with Rpa1 and not with the other four RFC subunits. In fact, partial suppression of the HU sensitivity of rfc4-2 is obtained with specific rfa1 alleles. Taking these findings together, we suspect that the individual RFC subunits not only have specific activities but that they interact with distinct sets of proteins. Defects in these interactions might explain the phenotypic differences due to mutations in RFC2-5.
RFC4 and RAD24 checkpoint control.
Two epistasis groups have been identified that control DNA damage checkpoints in yeast: the RAD24 group, which includes RAD17, MEC3, and DDC1, and the RAD9 group, which contains no other members (16, 45). RAD24 encodes a 76-kDa protein with limited sequence similarity to the RFC proteins that has been shown to be associated with the four small subunits of RFC (26, 27, 60). Consistent with these findings, our results place RFC4 in the RAD24 epistasis group and extend this group to include genes involved in DNA replication. Rad24 is known to compete with Rfc1 for Rfc2-5, as overexpression of RAD24 exacerbates the growth defect of the rfc1 mutant (44). Surprisingly, we find that RAD24 overexpression partially complements the HU sensitivity of the rfc4-2 mutant, implying that RAD24 plays a role in the replication block checkpoint. Previous studies have not revealed a role for RAD24 in the S/M replication block checkpoint, although there is suggestive evidence for such a role (23). In the present case, increased levels of Rad24/Rfc2-5 mutant complexes may have improved the efficiency of the HU response directly by participating in Rad53 phosphorylation. Alternatively, increased Rad24 protein may have further compromised the Rfc1/Rfc2-5 mutant complex and indirectly improved the HU response by activating an alternate checkpoint pathway. Thus, while the present data suggest that RAD24 functions in the replication block checkpoint, we cannot rule out an indirect mechanism for the suppression of the HU sensitivity of the rfc4-2 mutant.
Like Rfc4, Rpa1 appears to be required for the DNA damage checkpoint at all phases of the cell cycle. Rpa1 was previously shown to be required for the G1/S and intra-S-phase checkpoints (42), and our results confirm that Rpa1 is also required for the G2/M checkpoint (Fig. 6C). Recently, Rpa1 was shown to play an additional role in DNA damage signaling. The rfa1-t11 mutation leads to premature adaptation and suppresses the permanent G2/M arrest of cells containing two double-strand breaks (38). The rfa1 mutants that display synthetic lethality with rfc4-2 share a number of phenotypes with rfc4-2. For example, like rfc4-2 cells, rfa1-Y29H cells display reduced levels of Rad53 phosphorylation in response to UV irradiation at the nonpermissive temperature (data not shown). Likewise, rfa1-t11 cells show reduced Rad53 phosphorylation in response to double-strand breaks (54). Thus, rfc4 and rfa1 mutants appear to be defective in the same steps of the DNA damage checkpoint response. Although the specific structure that initiates the checkpoint and adaptation response is unknown, these results suggest that Rpa1N and Rfc4 might cooperate to establish this signal.
ACKNOWLEDGMENTS
We are grateful to Richard Kolodner for generously providing RFA1 mutants, to Christina DeCoste for assistance with FACS analysis, and to Vincent Geli, Suzanne Shanower, David Stern, Akio Sugino, and Ted Weinert for strains, plasmids, proteins, and antibodies. We also thank Nancy Walworth and laboratory members for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant GM55583.
REFERENCES
- 1.Alani E, Thresher R, Griffith J D, Kolodner R D. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Araki H, Leem S H, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II (epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender A, Pringle J R. Use of a screen for synthetic lethal and multicopy suppressor mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 7.Bochkareva E, Frappier L, Edwards A M, Bochkarev A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- 8.Bochkareva E, Korolev S, Bochkarev A. The role for zinc in replication protein A. J Biol Chem. 2000;275:27332–27338. doi: 10.1074/jbc.M000620200. [DOI] [PubMed] [Google Scholar]
- 9.Boddy M N, Russell P. DNA replication checkpoint control. Front Biosci. 1999;4:D841–D848. doi: 10.2741/boddy. [DOI] [PubMed] [Google Scholar]
- 10.Braun K A, Lao Y, He Z, Ingles C J, Wold M S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 11.Brill S J, Bastin-Shanower S. Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol Cell Biol. 1998;18:7225–7234. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brill S J, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 13.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 14.Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O'Donnell M, Hurwitz J. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci USA. 1998;95:11607–11612. doi: 10.1073/pnas.95.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Torre-Ruiz M A, Green C M, Lowndes N F. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohrmann P R, Oshiro G, Tecklenburg M, Sclafani R A. RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics. 1999;151:965–977. doi: 10.1093/genetics/151.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erdile L F, Heyer W D, Kolodner R, Kelly T J. Characterization of a cDNA encoding the 70-kDa single-stranded DNA-binding subunit of human replication protein A and the role of the protein in DNA replication. J Biol Chem. 1991;266:12090–12098. [PubMed] [Google Scholar]
- 19.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fay D S, Sun Z, Stern D F. Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr Genet. 1997;31:97–105. doi: 10.1007/s002940050181. [DOI] [PubMed] [Google Scholar]
- 21.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firmenich A A, Elias-Arnanz M, Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frei C, Gasser S M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner R, Putnam C W, Weinert T. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes X V, Wold M S. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 26.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths D J, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. Crystal structure of the delta′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 29.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 30.He Z, Henricksen L A, Wold M S, Ingles C J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 31.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 32.Heyer W D, Rao M R, Erdile L F, Kelly T J, Kolodner R D. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoekstra M F. Responses to DNA damage and regulation of cell cycle checkpoints by the ATM protein kinase family. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- 34.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand—the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D K, Stigger E, Lee S H. Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J Biol Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 36.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong X P, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 38.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R D, Haber J E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Lu X, Peterson C A, Legerski R J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Burgers P M. Cloning and characterization of the essential Saccharomyces cerevisiae RFC4 gene encoding the 37-kDa subunit of replication factor C. J Biol Chem. 1994;269:21880–21884. [PubMed] [Google Scholar]
- 41.Lin Y L, Chen C, Keshav K F, Winchester E, Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 42.Longhese M P, Neecke H, Paciotti V, Lucchini G, Plevani P. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res. 1996;24:3533–3537. doi: 10.1093/nar/24.18.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longhese M P, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol Cell Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 45.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 46.Maniar H S, Wilson R, Brill S J. Roles of replication protein-A subunits 2 and 3 in DNA replication fork movement in Saccharomyces cerevisiae. Genetics. 1997;145:891–902. doi: 10.1093/genetics/145.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles C J, Edwards A M, Chazin W J. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 48.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navas T A, Zhou Z, Elledge S J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 50.Noskov V N, Araki H, Sugino A. The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4914–4923. doi: 10.1128/mcb.18.8.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 53.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pellicioli A, Lee S E, Lucca C, Foiani M, Haber J E. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 55.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore P P, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Philipova D, Mullen J R, Maniar H S, Lu J, Gu C, Brill S J. A hierarchy of SSB promoters in replication protein A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 57.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 58.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 59.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimoda C, Nojima H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 62.Shivji M K, Podust V N, Hubscher U, Wood R D. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 63.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;12:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stern D F, Zheng P, Beidler D R, Zerillo C. Spk1, a new kinase from Saccharomyces cerevisiae, phosphorylates proteins on serine, threonine, and tyrosine. Mol Cell Biol. 1991;11:987–1001. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 68.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 69.Tsurimoto T, Fairman M P, Stillman B. Simian virus 40 DNA replication in vitro: identification of multiple stages of initiation. Mol Cell Biol. 1989;9:3839–3849. doi: 10.1128/mcb.9.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsurimoto T, Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989;9:609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 74.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 75.Umezu K, Sugawara N, Chen C, Haber J E, Kolodner R D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Venclovas C, Thelen M P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 79.Wang M, Mahrenholz A, Lee S H. RPA stabilizes the XPA-damaged DNA complex through protein-protein interaction. Biochemistry. 2000;39:6433–6439. doi: 10.1021/bi000472q. [DOI] [PubMed] [Google Scholar]
- 80.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 81.Weinert T, Hartwell L. Control of G2 delay by the RAD9 gene of Saccharomyces cerevisiae. J Cell Sci Suppl. 1989;12:145–148. doi: 10.1242/jcs.1989.supplement_12.12. [DOI] [PubMed] [Google Scholar]
- 82.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 83.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 84.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wold M S. RPA: a heterotrimeric, single-stranded DNA binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–91. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 86.Wold M S, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wright J A, Keegan K S, Herendeen D R, Bentley N J, Carr A M, Hoekstra M F, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yao N, Turner J, Kelman Z, Stukenberg P T, Dean F, Shechter D, Pan Z Q, Hurwitz J, O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 89.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zheng P, Fay D S, Burton J, Xiao H, Pinkham J L, Stern D F. SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol Cell Biol. 1993;13:5829–5842. doi: 10.1128/mcb.13.9.5829. [DOI] [PMC free article] [PubMed] [Google Scholar]