Abstract
Antibody-based assays have been a cornerstone of infectious disease diagnostics for over 100 years [1]. These assays rely on the exquisite sensitivity and specificity of humoral response to almost all infections. While next-generation sequencing (NGS) has tremendous potential to improve diagnostics and uncover host-microbial relationships by directly identifying nucleic acids from infectious microbes, challenges and opportunities for new approaches remain. Here, we review a group of cutting-edge techniques that couple antibody responses with flow cytometry of antibody tagged microbes and NGS. These studies are bringing into focus the dynamic relationship between our immune systems and endogenous microbial communities, which are an important source of pathogens. For simplicity, we use the umbrella term mFLOW-Seq (microbial flow cytometry coupled to NGS) to describe these approaches.
Keywords: antibody, mFLOW-Seq, microbial flow cytometry, microbiome
The gut, skin, vagina, and lungs are colonized by trillions of microbes that provide important biological functions for the host [2]. Of these sites, the intestinal microbiome contains the largest number (up to 1012 microbes per gram of intestinal contents) and diversity of commensal microbes. Microbes from the phyla Firmicutes and Bacteroidetes are the most abundant bacteria, generally comprising ~90% of adult intestinal microbiomes, followed by Actinobacteria, Verrucomicrobia, and Proteobacteria [3]. Commensal microbes stimulate the development of a healthy immune system and provide an important source of vitamins, increased ability to break down food, and protection from other pathogenic organisms [4]. Effective barriers at these mucosal sites exclude microbes with pathogenic potential from translocating to systemic sites, while allowing beneficial microbial communities to thrive [5]. In the gut, the barrier consists of an overlying layer of protective mucus enriched with mucosal antibodies and anti-microbial peptides, a single intestinal epithelial layer, and a variety of innate and adaptive immune cells including IgA and IgM antibody-secreting B cells [6]. Some of the commensal bacteria and fungi in a healthy microbiome have the potential to cause invasive disease, notably microbes such as E. coli, Klebsiella, Enterococcus, and Candida. These potential pathogens, or pathobionts, are typically found at very low abundance (<1%) [7, 8], but during periods of illness and immune suppression, they often increase dramatically in relative abundance to become the dominant organism in the intestinal microbiome [9]. When barrier dysfunction coincides with overgrowth of potential pathogens, patients are at high risk for microbial translocation from the gut to the bloodstream leading to systemic infection [10].
IgA and IgM, the primary mucosal antibodies found in the gastrointestinal tract, help establish and maintain homeostasis with commensals at barrier sites [11]. Microbes and microbial antigens are sampled by intestinal lymphoid tissues, which induce the development of antibody-secreting B cells. These B cells produce dimeric IgA and pentameric IgM that are transported across the epithelial barrier into the gut lumen by the polymeric Ig receptor (pIgR). These mucosal immunoglobulins bind to a substantial fraction of commensal fecal microbes [12, 13]. Of note, IgA and IgM binding does not typically eradicate their targets. Rather, IgA coating has pleiotropic effects on bacteria including preventing overgrowth, averting invasion of potential pathogens, inducing transcriptional changes, inhibiting motility, localizing some microbes to specific niches, and neutralizing microbial toxins [14–16]. Less is known about the specific effects of IgM binding to commensal microbes in the intestinal environment. The interplay of the microbiota and humoral immunity is critical for maintaining a functional intestinal barrier to prevent the translocation of microbes to systemic sites [17]. Studying how host antibodies recognize and react to bacteria provides an important perspective on host-microbiota relationships.
MICROBIAL FLOW CYTOMETRY AND MFLOW-SEQ
Flow cytometry is a high-throughput technique used to analyze cells or small particles in research and clinical settings. Flow cytometry is most commonly used to study immune system cells that have been stained with fluorescently labeled antibodies specific for cell surface proteins or other markers of interest. As cells pass through the cytometer, they can be sorted based on the pattern of markers and used for downstream applications including NGS (next-generation sequencing). Flow cytometry is also an important tool to study microbes from environmental and clinical samples [18]. This technique, often referred to as microbial flow cytometry (mFLOW), can be applied to pure cultures of microbes or more complex communities to determine intrinsic physical characteristics (eg, size, shape, and granularity) or stained with fluorescent dyes to assess metabolic states [19].
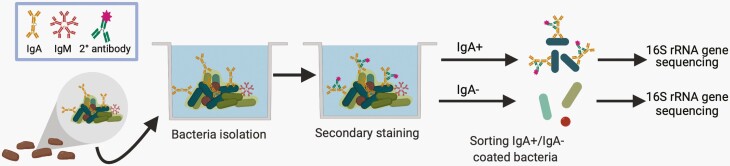
More recently, the coating of microbes with host antibodies has been used to identify the degree and type of humoral immune response to a particular microbe or group of microbes [18]. Two key observations underlie mFLOW-Seq technologies. The first is that the mucosal IgA targets and coats a subset of commensal intestinal microbes in healthy mice and humans. Early mFLOW studies in the 1990s demonstrated that mucosal IgA coats a substantial fraction (~10%-50%) of the fecal microbiota [13, 20–22]. The key technical innovation was adapting mFLOW to sort and identify specific IgA-coated microbes from the complex fecal microbial community using NGS (typically 16S rRNA gene amplicons) (Figure 1) [23]. These studies revealed that a broad range of intestinal microbes are coated with IgA at steady state, including microbes from the 5 most common bacterial phyla present in mammals: Firmicutes, Bacteroidetes, Actinobacteria, Verrucomicrobia, and Proteobacteria. Notably, IgA-targeted microbes include inflammatory and immunostimulatory microbes such as Enterobacteriaceae (eg, E. coli and Helicobacter sp.) and microbes that localize closely to the mucosal surface (eg, segmented filamentous bacteria [SFB] and Akkermansia muciniphila) [12, 15, 16, 24, 25]. Transfer of sorted and culturable consortia of IgA-coated or non-coated microbes from human patients with inflammatory bowel disease to germ-free mice demonstrated that IgA-coated microbes have an enhanced potential to induce intestinal inflammation [23]. Following these initial studies of IgA-coated microbes, similar approaches were developed by multiple research groups to identify IgA-, IgM-, and IgG-targeted intestinal microbes. In the literature, there is a growing list of these techniques (eg, IgA-Seq [12, 23], IgM-Seq [25], BugFACS [26], Fungi-Flow [27], MultiKAP [28], and mFLOW [29]). For simplicity, we propose using the umbrella term mFLOW-Seq to describe these techniques that leverage the sensitivity and specificity of host antibodies to identify which microbes are recognized by the adaptive immune system.
Figure 1.
Schematic of mFLOW-Seq for identifying fecal microbiota coated by mucosal IgA. Bacteria isolated from the feces are stained with a fluorescently labeled anti-IgA secondary antibody. Then fluorescence-activated cell sorting (FACS) is used to separate the IgA+ and IgA− bacteria. The IgA+ and IgA− populations are then analyzed by 16S rRNA gene sequencing to determine which microbes are enriched in the IgA+ fraction as an indicator of those microbes selectively bound by secretory IgA.
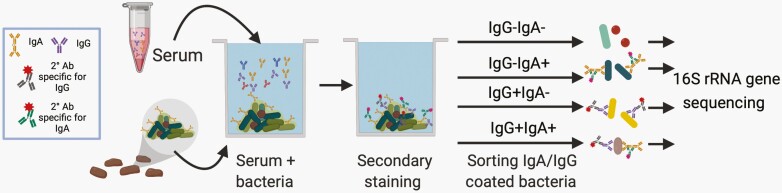
The second observation underlying mFLOW-Seq is that systemic antibodies including serum IgG, IgM, and IgA are induced by commensal microbiota and target a range of commensal microbes in healthy mice and humans [29–32]. A recent advance in mFLOW-Seq leverages these circulating anti-commensal antibodies to identify intestinal microbes that induce systemic immune responses [29]. For this technique, intestinal microbes (usually fecal microbiota) are incubated with systemic antibodies (usually obtained from serum) [29, 31, 33]. Those systemic antibodies with specificities to microbial antigens coat a subset of intestinal microbes. Fluorescently labeled secondary antibodies against IgG, IgA, or IgM then tag those microbes bound by IgG, IgA or IgM antibodies. Those tagged microbes are then isolated by flow cytometric cell sorting, and the antibody-targeted microbes are defined by NGS (Figure 2). Studies that investigate mucosal and systemic antibody binding of commensals are contributing new perspectives on commensal and host interactions at mucosal and systemic sites [28, 29, 31, 32].
Figure 2.
Schematic of mFLOW-Seq for identifying fecal microbiota targeted by systemic antibodies obtained from serum samples. Bacteria are isolated from stool or culture, then incubated with serum antibodies as a primary stain, and then stained with fluorescently labeled anti-IgA, -IgG (or -IgM)-specific secondary antibodies. The bacteria are separated into populations based on IgG and/or IgA binding using fluorescence-activated cell sorting. The sorted populations are then analyzed by 16S rRNA gene sequencing to determine which microbes are enriched in the IgA, IgG, and unbound fractions.
MFLOW-SEQ APPLICATIONS
Bacterial Sepsis and Invasive Fungal Disease
Studies using mFLOW-Seq provide evidence that systemic IgA and IgG antibodies capable of binding commensal microbes or microbial antigens prevent sepsis from enteric microbes. Sequencing serum IgA-bound fecal bacteria demonstrated that microbes from the phylum Proteobacteria, which includes many medically important enteric pathogens, were highly targeted by systemic IgA [32]. Furthermore, serum IgA capable of binding to Proteobacteria increases survival in a murine model of bacterial peritonitis [32]. Cohousing mice with different microbiomes (which effectively transfers microbiota between the mice via coprophagia) demonstrated that the introduction of specific commensal microbes increases serum IgA concentration by stimulating IgA producing plasma cells in the bone marrow [32]. In line with these findings, a recent study demonstrated that serum IgG from healthy mice decreases mortality in another model of bacterial peritonitis [30]. Analysis of serum IgG binding to commensal microbes demonstrated that IgG directed against murein lipoprotein (MLP), a major component of the outer membrane of Gram-negative bacteria, conferred protection against endogenous (E. coli) and exogenous (Salmonella) bacterial infections [30].
In addition to the rich community of bacteria in the gut, the intestinal microbiome contains a clinically important community of fungal organisms called the mycobiome. Invasive fungal infections are a major cause of morbidity in premature infants and other immunocompromised patients. As such, it is important to study the gut mycobiota in healthy and disease states to define the mechanisms of protective immunity. Moreno-Sabater et al developed a technique (which they call Fungi-FLOW) in which systemic IgGs are incubated with a panel of cultured fungi to measure IgG antibody binding to 17 common fungal genera [27]. They reported strong responses to fungal pathobionts such as Penicillium, Malassezia, and to the budding form of Candida [27]. Recently, Doron et al developed another adaptation to mFLOW-Seq (which they call multi-kingdom antibody profiling or multiKAP) to study host responses to endogenous gut mycobiota [28]. Since fungi typically represent a very small fraction of the fecal microbiome, the authors developed a clever approach to enrich for fungi from the fecal microbiome based on size fractionation and staining of fungal cell components with calcofluor white. They analyzed the IgA-bound fraction to define the mucosal immune response to commensal fungi and then added serum IgG to delineate which commensal fungi induce a systemic antibody response. These studies reveal a strong systemic IgG response to commensal Candida albicans in mice and humans. Colonization of mice with C. albicans induces Candida-specific IgG antibodies which potently protect these mice from invasive infection by C. albicans and partially protect against infection by other species, including the highly drug-resistant emerging pathogen Candida auris [28]. These studies suggest that specific fungi induce serum IgG and IgA antibodies capable of binding to and preventing systemic fungal infection.
To move towards clinical application of mFLOW-Seq, additional studies are needed to evaluate whether antibody binding to endogenous bacteria or fungi predicts risk for infection in patients. For those patients requiring antibody replacement therapy, there may be benefits to testing whether specific antibody preparations bind to a patient’s endogenous microbiota to prevent invasive infections. One limitation of mFLOW-Seq approach is that antibody responses typically take 7-10 days which limits utility during the early acute phase of infection and sensitivity of the humoral response to detect acute infection is diminished in some immunocompromised patients. Further mFLOW-Seq requires time to prepare, sort, and sequence antibody-coated microbes and expertise to perform and analyze these data. Further study is needed to determine if microbe-specific serum IgA, IgM, and IgG working in concert deliver the widest range of protection in immunoglobulin replacement therapies. Clinical trials of intravenous immunoglobulin (IVIG) for sepsis treatment have produced mixed results, but have not accounted for specificity towards the species or strain causing bacteremia [34]. Intriguingly, some studies found that antibody preparations that include IgM and IgG antibody isotypes may be better suited to treat patients with sepsis [35].
Maternal Antibodies and Risk for Enteric Infection and Inflammation
Maternal factors, including in utero transfer of IgG, post-natal transfer of IgM, IgA, and IgG antibodies in breast milk, and vertical transmission of maternal microbiota, shape the early life microbiome and immune system thereby impacting susceptibility of neonates to enteric infection and sepsis [36]. A recent study demonstrated that cross-reactive or “natural” maternal IgG against a commensal Proteobacteria (Pantoea species) protects neonates against enteric infection in a murine model of enterotoxigenic E. coli infection [37]. Additionally, mFLOW-Seq was recently used to investigate whether IgA binding to commensal microbes predicts risk for neonatal enterocolitis (NEC) in premature infants [38]. Gopalakrishna et al demonstrated that enteral breast milk feeding increases IgA coating of fecal microbes, and that the fraction of the fecal microbiomes coated with IgA is inversely correlated with the risk of developing NEC. 16S rRNA gene sequencing of the IgA-bound population of microbes revealed that reduced intestinal bacterial diversity, preceded the development of NEC in a cohort of neonates and was associated with an increased proportion of IgA-uncoated microbes from the family Enterobacteriaceae. To corroborate these findings, they demonstrated that maternal breast milk IgA prevents NEC in a murine model by coating specific intestinal microbes from the family Enterobacteriaceae which includes microbes associated with NEC [38, 39]. Using mFLOW-Seq to evaluate breast milk IgA for its ability to bind intestinal microbes from premature infants, particularly to microbes from the family Enterobacteriaceae, may provide a biomarker to determine which neonates are at elevated risk for developing NEC.
In addition, prenatal probiotics or other microbial therapies may enhance IgA targeting of the intestinal microbiota [40]. This raises the possibility of matching donor milk antibody repertoires to the intestinal microbiome of high-risk infants as an innovative, noninvasive strategy to prevent NEC and early life enteric infections. Therapies that focus on enhancing the maternal antibody repertoire may ameliorate current issues with early antibiotic therapies. For example, in the case of early-onset sepsis in neonates, antibiotics prevent sepsis caused by Group B Streptococcus and E. coli, but may lead to dysbiosis and increase the risk of late-onset sepsis [40, 41]. mFLOW-Seq studies may reveal important defects in humoral targeting of microbes that contribute to the burden of neonatal enteric infections and NEC. Such findings could inform novel strategies to enhance maternal antibody targeting of bacteria, which could reduce the risk of neonatal enteric infection and sepsis without causing dysbiosis.
Primary Humoral Immunodeficiency
mFLOW-Seq has emerged as a powerful approach to explore the mechanisms by which humoral immunity supports an effective gut barrier and maintains homeostasis with commensal microbiota. For example, selective IgA deficiency (SIgAD), the most common inborn error of immunity, is an enigmatic disorder in which many subjects are asymptomatic while others have sinopulmonary infection, allergy, and/or autoimmune disease [42]. While there is no large-scale quantitative perturbation of gut microbial composition, some pathobionts expand in the absence of mucosal IgA [16]. Fadlallah et al used mFLOW-Seq to demonstrate that IgA binding to commensal microbes in adults can have differential effects on their relative abundance in the fecal microbiome, with some microbes such as E. coli apparently restrained by IgA while other microbes flourish when coated by IgA. The impact of SIgAD on the microbiomes and immune system development of pediatric patients remains an important and open question.
Since IgM antibodies can be secreted into the gut and coat a range of commensal microbes, mucosal IgM is thought to compensate for some of the functions of IgA in SIgAD [16, 25]. For example, patients with combined variable immune deficiency (CVID), who lack IgA and IgM, have reduced fecal microbial diversity [16]. In healthy humans, mucosal IgM has been found to coat a significant proportion of intestinal microbes in some studies [43, 44] but not in others [16, 25]. Magri et al report that mucus-embedded microbes are coated by both IgM and IgA, suggesting some degree of cooperative binding and a role in localizing microbes to this niche [43]. While studies using mFLOW-Seq are defining the specific bacteria targeted by IgA and IgM, more research is needed to determine whether IgM is redundant to IgA in the gut or serves unique functions. These studies demonstrate that mFLOW-Seq can be used directly with accessible human samples to understand changes in host and microbial interactions in patients with primary immunodeficiency.
Fecal Microbiota Transplantation
Fecal microbiota transplantation (FMT) effectively treats recurrent C. difficile infections for many, but not all patients. For those patients who are refractory to FMT, there may be an underlying dysregulated immune response to FMT microbes [45]. Current measures for selecting fecal donors mostly focus on pathogen screening, common laboratory studies, and health questionnaires [46]. Using mFLOW-Seq, Huus et al demonstrated that analyzing IgA targeting of the microbiota of the donor can potentially predict how the recipient’s IgA will target the new microbiota. After successful FMT, IgA coating of commensal microbes shifted from predominantly IgA targeting E. coli (pre-FMT) to diverse IgA coating of commensal species (post-FMT) [47]. This study is in line with others that demonstrated an intrinsic ability of intestinal microbiota to induce distinct host IgA responses [48]. Furthermore, with this new marker for engraftment of a healthy FMT microbiome, Huus et al determined that both capsule and colonoscopy delivery of the FMT led to this positive change in IgA targeting. Key next steps include studies to evaluate whether the donor’s or recipient’s pre-FMT IgA targeting of commensal microbes predicts the efficacy of FMT. mFLOW-Seq provides a new tool to evaluate FMT donors and recipients, which may help optimize donor selection.
Biomarker of Intestinal Dysfunction
Sequencing the antibody-bound fecal microbial population reveals patterns of targeting that reflect intestinal disease. For example, intestinal inflammation is associated with increased IgA coating of Proteobacteria such as E. coli [49]. Furthermore, Rengarajan et al demonstrated that specific patterns of IgG and IgA binding to fecal microbiota were associated with inflammatory bowel disease (IBD), and that antibody coating of fecal microbes such as Gemella, Peptostreptococcus, and Streptococcus species, that typically colonize the oropharynx, may be a biomarker for disease severity [50]. These finding suggest that anatomic mislocalization of commensal microbes reflects intestinal inflammation and dysfunction.
Continuing to define what factors influence antibody recognition of commensals will be an important step towards effective mFLOW-Seq-based diagnostics. For example, Kau et al demonstrated that malnutrition in children correlated with IgA targeting of specific bacterial taxa including microbes from the family Enterobacteriaceae [26]. Addition of the IgA-targeted strains to germ-free mice induced diet-dependent enteropathy, suggesting that IgA targeting may reflect an important component of the pathogenesis of malnutrition [26]. Furthermore, Huus et al showed that malnutrition in mice can lead to a loss of IgA targeting of Lactobacillus species [51]. This loss of IgA and microbe interactions was mediated through adaptation of bacterial glycans in response to nutrient limitation [51]. Together, these studies demonstrate how changes in nutritional state can impact IgA targeting of intestinal microbiota.
As we continue to expand the clinical studies defining immunoglobulin-bound microbes, we may discover that antibody binding to specific microbes is both a biomarker and driver for healthy or disease states. For example, loss of IgA or IgM targeting of enteric pathobionts (eg, Enterobacteriaceae or Enterococcus) may predict the risk of sepsis from these specific organisms in immunocompromised patients. Taken together, there is a tantalizing opportunity to use mFLOW-Seq to better predict which patients are at risk of bacteremia and which bacteria are likely to be unchecked and cause sepsis. These studies may inform prophylactic or empiric antibiotic choices and the use of immunoglobulin replacement therapy.
CONCLUSIONS AND OUTLOOK
Mucosal and systemic antibodies are not only a means by in which we defend ourselves from pathogens, but also how we communicate and maintain our relationships with commensals. By combining metagenomic sequencing and mFLOW, human and murine studies have uncovered new perspectives on antibody-mediated, host-microbe interactions, yet important questions remain. We need to better understand how mucosal and systemic antibody networks work together to prevent the translocation of endogenous microbes and the factors that regulate the development of these anti-commensal antibody systems in pediatric patients. Combining the strengths of NGS with specific antibody responses against endogenous microbes has advanced our understanding of host-pathogen and host-commensal interactions and may well become a clinically relevant tool for the infectious disease field. Looking forward, mFLOW-Seq is poised to develop personalized profiles of IgM, IgA, and IgG antibody binding to endogenous pathobionts. Clinical studies are needed to evaluate whether mFLOW-Seq data can help predict infection in pediatric patients and then be applied to develop personalized antibody replacement therapies.
Notes
Acknowledgments. Thank you to the members of the Silverman laboratory for helpful discussions and J.B. Lubin, John Deschaine, and Isaiah Rozich for comments on this manuscript.
Financial support. This work was supported by the National Institutes of Health [grant number 1R21AI153956-01] and Juvenile Diabetes Research Foundation (JDRF) [grant number 5-CDA-2020-946-S-B] to M.A.S.
Supplement sponsorship. This supplement was sponsored by Illumina and IDbyDNA.
Potential conflicts of interest. Both authors: No potential conflicts of interest.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fison ET. Widal’s sero-diagnosis of typhoid fever. Br Med J 1897; 2:266–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007; 449:804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arumugam M, Raes J, Pelletier E, et al. ; MetaHIT Consortium . Enterotypes of the human gut microbiome. Nature 2011; 473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 2017; 356:j831. [DOI] [PubMed] [Google Scholar]
- 5. Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol 2021; 39:449–79. [DOI] [PubMed] [Google Scholar]
- 6. Rogier EW, Frantz AL, Bruno ME, Kaetzel CS. Secretory IgA is concentrated in the outer layer of colonic mucus along with Gut Bacteria. Pathogens 2014; 3:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005; 308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jochum L, Stecher B. Label or concept – what is a pathobiont? Trends Microbiol 2020; 28:789–92. [DOI] [PubMed] [Google Scholar]
- 9. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ubeda C, Taur Y, Jenq RR, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen K, Magri G, Grasset EK, Cerutti A. Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat Rev Immunol 2020; 20:427–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bunker JJ, Flynn TM, Koval JC, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015; 43:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut 1996; 38:348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weis AM, Round JL. Microbiota-antibody interactions that regulate gut homeostasis. Cell Host Microbe 2021; 29:334–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bunker JJ, Bendelac A. IgA responses to microbiota. Immunity 2018; 49:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fadlallah J, El Kafsi H, Sterlin D, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med 2018; 10:1–15. [DOI] [PubMed] [Google Scholar]
- 17. Fine RL, Manfredo Vieira S, Gilmore MS, Kriegel MA. Mechanisms and consequences of gut commensal translocation in chronic diseases. Gut Microbes 2020; 11:217–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moor K, Fadlallah J, Toska A, et al. Analysis of bacterial-surface-specific antibodies in body fluids using bacterial flow cytometry. Nat Protoc 2016; 11:1531–53. [DOI] [PubMed] [Google Scholar]
- 19. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013; 152:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van der Waaij LA, Mesander G, Limburg PC, van der Waaij D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry 1994; 16:270–9. [DOI] [PubMed] [Google Scholar]
- 21. Kroese FG, de Waard R, Bos NA. B-1 cells and their reactivity with the murine intestinal microflora. Semin Immunol 1996; 8:11–8. [DOI] [PubMed] [Google Scholar]
- 22. Bos NA, Bun JC, Popma SH, et al. Monoclonal immunoglobulin A derived from peritoneal B cells is encoded by both germ line and somatically mutated VH genes and is reactive with commensal bacteria. Infect Immun 1996; 64:616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bunker JJ, Erickson SA, Flynn TM, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017; 358:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Catanzaro JR, Strauss JD, Bielecka A, et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep 2019; 9:13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kau AL, Planer JD, Liu J, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 2015; 7:276ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreno-Sabater A, Autaa G, Sterlin D, et al. Systemic anti-commensal response to fungi analyzed by flow cytometry is related to gut mycobiome ecology. Microbiome 2020; 8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doron I, Leonardi I, Li XV, et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell 2021; 184:1017–1031.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koch MA, Reiner GL, Lugo KA, et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 2016; 165:827–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeng MY, Cisalpino D, Varadarajan S, et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016; 44:647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fadlallah J, Sterlin D, Fieschi C, et al. Synergistic convergence of microbiota-specific systemic IgG and secretory IgA. J Allergy Clin Immunol 2019; 143:1575–1585.e4. [DOI] [PubMed] [Google Scholar]
- 32. Wilmore JR, Gaudette BT, Gomez Atria D, et al. Commensal microbes induce serum IgA responses that protect against Polymicrobial Sepsis. Cell Host Microbe 2018; 23:302–311.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ansaldo E, Slayden LC, Ching KL, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019; 364:1179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almansa R, Tamayo E, Andaluz-Ojeda D, et al. The original sins of clinical trials with intravenous immunoglobulins in sepsis. Crit Care 2015; 19:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kakoullis L, Pantzaris ND, Platanaki C, et al. The use of IgM-enriched immunoglobulin in adult patients with sepsis. J Crit Care 2018; 47:30–5. [DOI] [PubMed] [Google Scholar]
- 36. Atyeo C, Alter G. The multifaceted roles of breast milk antibodies. Cell 2021; 184:1486–99. [DOI] [PubMed] [Google Scholar]
- 37. Zheng W, Zhao W, Wu M, et al. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 2020; 577:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gopalakrishna KP, Macadangdang BR, Rogers MB, et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med 2019; 25:1110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silverman MA, Konnikova L, Gerber JS. Impact of antibiotics on necrotizing enterocolitis and antibiotic-associated diarrhea. Gastroenterol Clin North Am 2017; 46:61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singer JR, Blosser EG, Zindl CL, et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat Med 2019; 25:1772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuppala VS, Meinzen-Derr J, Morrow AL, Schibler KR. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J Pediatr 2011; 159:720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yel L. Selective IgA deficiency. J Clin Immunol 2010; 30:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Magri G, Comerma L, Pybus M, et al. Human secretory IgM emerges from plasma cells clonally related to gut memory b cells and targets highly diverse commensals. Immunity 2017; 47:118–134.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Janzon A, Goodrich JK, Koren O, Group TS, Waters JL, Ley RE. Interactions between the gut microbiome and mucosal immunoglobulins A, M, and G in the developing infant gut. mSystems 2019; 4:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Littmann ER, Lee JJ, Denny JE, et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat Commun 2021; 12:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bibbo S, Settanni CR, Porcari S, et al. Fecal microbiota transplantation: screening and selection to choose the optimal donor. J Clin Med 2020; 9:1757–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huus KE, Frankowski M, Pučić-Baković M, et al. Changes in IgA-targeted microbiota following fecal transplantation for recurrent Clostridioides difficile infection. Gut Microbes 2021; 13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Planer JD, Peng Y, Kau AL, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 2016; 534:263–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Viladomiu M, Kivolowitz C, Abdulhamid A, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 2017; 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rengarajan S, Vivio EE, Parkes M, et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes 2020; 11:405–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huus KE, Bauer KC, Brown EM, et al. Commensal bacteria modulate immunoglobulin A binding in response to host nutrition. Cell Host Microbe 2020; 27:909–921.e5. [DOI] [PubMed] [Google Scholar]