Abstract
Most in vivo 31P MR studies are realized on 3T MR systems that provide sufficient signal intensity for prominent phosphorus metabolites. The identification of these metabolites in the in vivo spectra is performed by comparing their chemical shifts with the chemical shifts measured in vitro on high-field NMR spectrometers. To approach in vivo conditions at 3T, a set of phantoms with defined metabolite solutions were measured in a 3T whole-body MR system at 7.0 and 7.5 pH, at 37 °C. A free induction decay (FID) sequence with and without 1H decoupling was used. Chemical shifts were obtained of phosphoenolpyruvate (PEP), phosphatidylcholine (PtdC), phosphocholine (PC), phosphoethanolamine (PE), glycerophosphocholine (GPC), glycerophosphoetanolamine (GPE), uridine diphosphoglucose (UDPG), glucose-6-phosphate (G6P), glucose-1-phosphate (G1P), 2,3-diphosphoglycerate (2,3-DPG), nicotinamide adenine dinucleotide (NADH and NAD+), phosphocreatine (PCr), adenosine triphosphate (ATP), adenosine diphosphate (ADP), and inorganic phosphate (Pi). The measured chemical shifts were used to construct a basis set of 31P MR spectra for the evaluation of 31P in vivo spectra of muscle and the liver using LCModel software (linear combination model). Prior knowledge was successfully employed in the analysis of previously acquired in vivo data.
Keywords: in vivo MR spectroscopy, 31P MRS, LCModel, liver, muscle
1. Introduction
Magnetic resonance spectroscopy (MRS) equipment and protocols have become a standard part of clinical MR systems; however, the interest is mostly focused on proton (1H) in vivo MR spectroscopy, which is used in many research and clinical applications.
Another spectroscopy technique is phosphorous (31P) MRS, which has an even longer history of in vivo applications [1]. Despite lower sensitivity, there is no overwhelming prominent signal in 31P MR spectra that would require a special suppression technique (as does the signal of water in 1H MR spectroscopy). In addition, 31P MR signals of three important compounds of energy metabolism—phosphocreatine (PCr), adenosine triphosphate (ATP), and inorganic phosphate (Pi)—show relatively high concentrations and are well-resolved in in vivo 31P MR spectra. Moreover, 31P MR spectra can be measured repeatedly with rapid succession during a physical challenge and reveal rapid changes in energy metabolites (and, e.g., pH) over time. This is dynamic 31P MRS [2]. This technique thus significantly contributes to the understanding of energy metabolism of muscles and other tissues [3].
Most of the 31P MR studies have been performed at 1.5 and 3 T systems, which provide sufficient signal for dynamic studies and are available for routine clinical examinations and research. In addition to well-resolved signals, such as PCr, Pi, and ATP in muscles, there are a number of metabolites that resonate in the narrow range between 2–8 ppm and that overlap each other and arise primarily from inorganic phosphate (Pi), phosphomonoesters (PME), and phosphodiesters (PDE) (see Figure 1). The identification of these metabolites in in vivo spectra is performed by comparing their respective chemical shifts with the chemical shifts of individual solutions of chemical compounds measured in vitro.
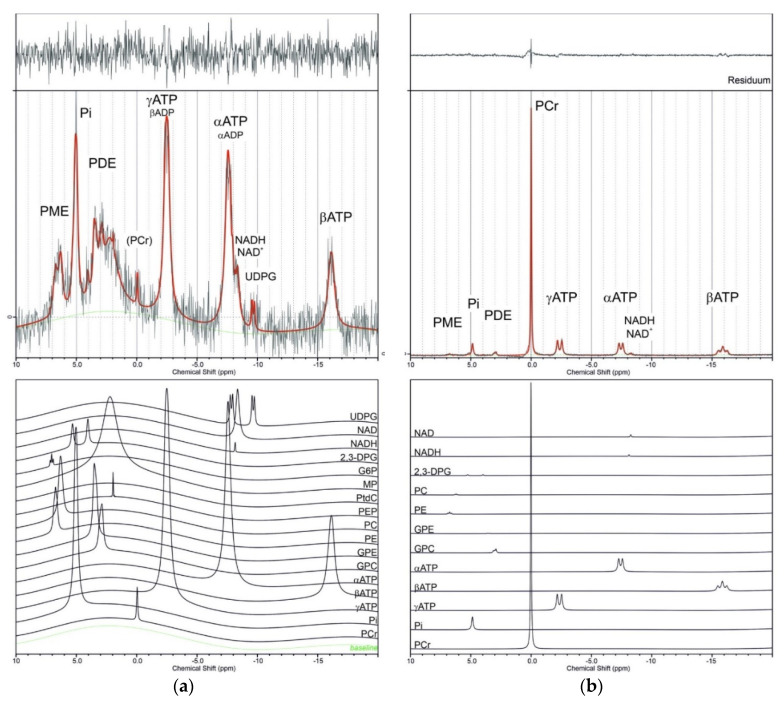
Figure 1.
Examples of in vivo 31P spectra of the liver (a) and calf muscles (b). Black lines represent measured spectra, red lines show spectra calculated by LCModel with the application of the BASISp data set, and the green line represents the calculated baseline. In the upper panels of the picture, individual contributions of metabolites to the calculated spectra are shown: UDPG—uridine-diphosphoglucose; NADH/NAD+—nicotinamide adenine dinucleotide; 2,3-DPG—2,3 diphosphoglycerate; G6P—glucose-6-phosphate; G1P—glucose-1-phosphate; MP—membrane phospholipids; PEP—phosphoenolpyruvate; PC—phosphocholine; PE—phosphoethanolamine; GPE—glycerophosphoethanolamine; GPC—glycerophosphocholine; PtdC—phosphatidylcholine (overlapped with PEP); ATP—adenosine triphosphate; ADP—adenosine diphosphate; Pi—inorganic phosphate; PCr—phosphocreatine (in the case of the liver spectrum, this signal is the contamination; it can serve as the standard for chemical shift measurement); PME—phosphomonoesters region; PDE—phosphodiesters region. (In vivo spectra of the liver and calf muscles of a young, healthy volunteer were taken from the database of the MR group IKEM. The liver spectrum was measured at 3 T TRIO MR system, 1 D ISIS, TR = 2 s, acq = 196, TE = 0.2 ms, bandwidth 3000 Hz; muscle spectrum was obtained at VIDA 3 T MR system, FID sequence, TR = 15 s, bandwidth 2000 Hz; unpublished results.
Recently, a few ideas have been suggested that have sparked new research and different interpretations. First, in the liver, the potential signals of phosphoenolpyruvate (PEP), a key component of glycolysis and gluconeogenesis, and phosphatidylcholine (PtdC) both resonate at a similar position around 2 ppm [4,5,6]. Even though PtdC is a major bile component, it could also contribute to the signal from the hepatic parenchyma, and its contribution cannot be neglected [4].
Second, applying proton decoupling at 3 T, or using ultra-high-field systems (≥7 T), the resonances of PME and PDE can easily be split into individual components of phosphocholine (PC), phosphoethanolamine (PE), glycerophosphocholine (GPC), and glycerophosphoethanolamine (GPE). Total PDEs and/or PMEs are linked to glucose homeostasis [7], overall skeletal muscle health [8], and liver cirrhosis [9]. Lately, specific levels of skeletal muscle GPC have been suggested as a biomarker for tissue-specific thyroid action [10].
Furthermore, the role of nicotinamide adenine dinucleotide (NADH) and uridine diphosphoglucose (UDPG) in energy metabolism is undisputed, and both are accessible to 31P MRS. Although their resonances are often unresolved (due to bad shim) or partially overlapped by the dominant αATP signal, the application of 1H-31P broadband decoupling can significantly improve their resolution even at 3 or 4 T magnetic fields (for details see Discussion, Section 3.4 and Section 3.5).
Another interesting task is the assignment of the second Pi skeletal muscle signal in several pathophysiological conditions [8,11,12,13]. Not only was there a significant increase observed in athletes [13] and a decrease in an obese sedentary population [11] but changes in patients with severe diabetes-related ischemia were also observed [8]. It was suggested that this signal arises from interstitia, but mitochondrial phosphate cannot be excluded. Even at 3 T, magnetic field signals in the range of 2 to 8 ppm are not well-resolved, and identification of metabolites in this range is based only on the comparison of signal maxima with in vitro data.
In this report, 31P chemical shifts and coupling constants are presented for fifteen metabolites that can be observed in living tissue using 31P MR spectroscopy. These MR chemical shifts were determined from the measurement of individual phantom solutions in a whole-body 3 T MR system, which mimicked standard in vivo conditions (temperature 37 °C and pH~7), thus enabling a comparison of these data to the results of high-field NMR spectroscopy and other literature data. Furthermore, we used these spectra acquired in phantoms to create a 3 T spectral basis, and we included some additional literature data that can be used for deconvolution of in vivo 31P MR liver and skeletal muscle spectra by LCModel software [14] (linear combination of individual components in the frequency domain). Creating this prior knowledge allowed an approach to the analysis of previously acquired in vivo spectra.
2. Results
All metabolites were measured in a solution of pH 7.0 and 7.5, which covered a range of cytosolic, interstitial, vascular, and mitochondrial pH values. Table 1 summarizes the concentrations, multiplicity, chemical shifts, and interaction constants (J-coupling constant) of all 15 compounds in phantoms as prepared and measured in this study. PCr was used as an internal standard with a chemical shift of 0.0 ppm. See spectra in Supplementary Material.
Table 1.
Concentration of 31P MR phantoms, their chemical shifts (ppm) and coupling constants.
| Metabolite | Product Number | Concentration in Phantoms (mM) | Multiplicity | Chemical Shift at pH = 7.0 | Chemical Shift at pH = 7.5 | Chemical Shift In Vivo |
J-Coupling (Hz) |
Basis Set Deelchand | BASISp Set Present Study | |
|---|---|---|---|---|---|---|---|---|---|---|
| Phosphocreatine | P7936-5G | 1 PCr | 5 | s | 0 | 0 | 0 | - | 0 | 0 |
| Inorganic phosphate | Pi | 40 | s | 4.78 | 5.27 | ~5 | - | 4.84 | 4.78 | |
| Adenosine triphosphate | A2383-1G | αATP | 10 | d | −7.98 | −7.94 | ~7–8 | 19.5 | −7.56 | −7.53 |
| βATP | 10 | t | −18.80 | −18.58 | ~16 | 20.0 | −16.18 | −16.18 | ||
| γATP | 10 | d | −4.18 | −3.37 | ~3–4 | 19.0 | −2.53 | −2.7 | ||
| Adenosine diphosphate | 29349990900 | αADP | 10 | d | −7.28 | −7.32 | - | 19.0 | - | - |
| βADP | 10 | d | −3.50 | −3.20 | - | 19.5 | - | - | ||
| Phosphoethanolamine | P0503-1G | PE (PME) | 10 | t | 6.74 | 6.85 | 6.78 | 7.0 | 6.77 | 6.77 |
| Phospholcholine | P0378G | PC (PME) | 10 | t | 6.19 | 6.35 | 5.9 | 6.0 | 6.23 | 6.23 |
| Glucose 1-phosphate | G1259-1G | G1P | 10 | d | 4.99 | overlay with Pi (5.20) | overlay with PME | 7.5 | - | 4.99 |
| Glucose 6-phosphate | G7879-1G | G6P | 10 | t | 7.03 | 7.30 | 7.1–7.2 | 6.1 | - | 7.03 |
| Phosphoenolpyruvate | P7127-500MG | PEP (PME) | 10 | s | 2.00 | 2.27 | 2.06 | - | 2 | |
| Phosphatidylcholine | PtdC | 3 10 | t | 2.14 | 2.13 | 2.06 | - | 2.14 | ||
| Glycerol-3-phosphoryl choline | G5291-100MG | GPC (PDE) | 10 | t | 2.97 | 2.96 | 2.76 | 5.5 | 2.94 | 2.97 |
| 2.3-diphosphoglycerate | D9134-100MG | 2,3-DPG(PDE) | 2 2 | 2- d | 4.09 | 4.6 | 5.5 | 6.0 | 5.23 | 4.05 |
| 3- t | 5.338 | 5.84 | 6.3 | 9.5 | 5.71 | 5.32 | ||||
| Nicotinamide adenine dinucleotide (reduced form) | N8129-1G | NADH | 10 | s | −8.16 | −8.16 | ~8.1 | −8.13 | −8.13 | |
| Nicotinamide adenine dinucleotide (oxygenated form) | 10127981001 | NAD+ | 10 | s | −8.32 | −8.32 | ~8.3 | 6.0 | −8.31 | −8.31 |
| Uridine diphosphoglucose | U4625-500MG | UDPG | 2 2 | d | −8.11 | −8.11 | −8.1 | 10.0 | - | −7.98 |
| UDPG | 2 2 | d | −9.78 | −9.78 | −9.8 | 10.0 | - | −9.78 | ||
| 4 Membrane phospholipids | MP | 2.3 | 2.3 | |||||||
| 4 Glycerophosphoethanolamine | GPE | 3.49 | 3.49 |
1 PCr was used as an internal standard; 2 K2HPO4/NaH2PO4 to c = 10 mM; 3 40% ethanol with the addition of Triton 100 as a surfactant; 4 taken from [15]. s—singlet; d—doublet; t—triplet.
Solutions of metabolites were prepared in K2HPO4 and NaH2PO4 buffers, which guaranteed sufficient stability of the compounds for 31P MRS experiments within a few days.
Measured 31P chemical shifts of all compounds were used for the construction of a basis set for the evaluation of skeletal muscles and liver spectra, which is necessary for LCModel application. Figure 1 shows representative spectra of calf muscles and liver calculated with the newly constructed BASISp. In addition to the basis set proposed by Deelchand [15], signals of UDPG, G1P, G6P, PEP, and PtdC were appended. Some chemical shifts were optimized during testing to better suit in vivo conditions. Additionally, signals of macromolecule membrane phospholipids (MP) and glycerophosphoethanolamine were included based on the literature data [15,16]. Finally, BASISp used for the analyses contained 19 signals of 17 model spectra summarized in Table 1. It was generated internally during analysis based on the specified input parameters. ADP was omitted intentionally as the α- and β-ADP resonances are overlapped by stronger α and γ-ATP resonances. The signals of the three ATP phosphorus signals covering a significant part of the spectral range (from −2 to −17 ppm) were described by three separate data sets to accommodate different linewidths (Lorentzian signals). The chemical shifts of α, β, and γ phosphorus atoms of ATP were also modified to better suit in vivo conditions.
In addition to the representative spectra of the liver and calf muscles (Figure 1), a pilot comparison of LCModel and jMRUI calculation of signal intensities of five randomly chosen liver spectra and five muscle spectra from our database was used. Coefficients of variation were used for the comparison, and results are summarized in Table 2.
Table 2.
Pilot comparison of the LCModel and jMRUI method calculation of means of relative signal intensities (%) and relative Cramér Rao Lower Bounds (CRLB, %) of five liver and muscle spectra and means of their coefficients of variance CV (%) for the comparison. The BASISp set was applied.
| Liver | γ-ATP | α-ATP | β-ATP | GPC | PCr | Pi | |
| jMRUI | Relative signal intensity | 23.3 | 26.9 | 13.4 | 21.6 | 2.5 | 12.3 |
| Relative CRLB | 2.0 | 1.9 | 4.0 | 3.9 | 19.6 | 3.9 | |
| LCModel | Relative signal intensity | 26.0 | 34.1 | 11.3 | 15.6 | 3.3 | 9.7 |
| Relative CRLB | 3.3 | 2.7 | 4.7 | 6.0 | 11.0 | 5.0 | |
| mean CV | 11 | 23 | 16 | 37 | 33 | 23 | |
| Muscle | γ-ATP | α-ATP | β-ATP | GPC | PCr | Pi | |
| jMRUI | Relative signal intensity | 12.4 | 10.3 | 10.0 | 2.6 | 60.2 | 4.4 |
| Relative CRLB | 1.2 | 1.5 | 2.0 | 8.8 | <1 | 1.9 | |
| LCModel | Relative signal intensity | 14.6 | 12.2 | 10.5 | 3.4 | 53.6 | 5.7 |
| Relative CRLB | 1.7 | 2.0 | 2.3 | 8.7 | <1 | 2.0 | |
| mean CV | 16 | 17 | 5 | 24 | 12 | 25 |
The characterization of CV was used: CV < 10 is very good agreement, 10–20 is good agreement, 20–30 is acceptable agreement, and CV > 30 is not acceptable.
3. Discussion
The availability of ultra-high field imagers (≥3 T) also enables the study of less prominent 31P metabolites in muscles [8,11,17,18], liver [4,9,19], and brain tissue [16,20,21]. These less frequently assessed metabolites may be divided into phosphomonoester (PME) and phosphodiester (PDE) groups according to their chemical shifts ranging from approx. 5–7 and 1–5 ppm, respectively. In the PME area, signals of chemical components, such as phosphoethanolamine (PE), phosphocholine (PC), glucose-1-phosphate (G1P), and glucose-6-phosphate (G6P), can be found. In the PDE area, signals of glycerophosphoethanolamine (GPE), glycerophosphocholine (GPC), phosphatidylcholine (PtdC), and phosphoenolpyruvate (PEP) can be recognized. Another area of interest is the chemical shifts from −7 to −8.5 ppm where, e.g., signals of αATP, adenosine diphosphate (αADP), nicotinamide adenine dinucleotide (NADH/NAD+), and uridine-diphosphoglucose (UDPG) can be found. The contribution of membrane phospholipids should also be taken into consideration [16].
The exact in vivo chemical shift position of 31P metabolites may be influenced by a number of parameters. One of the most important is pH, which differs in different compartments of cells and tissues or may change under pathologic conditions. Physiological pH of body tissues usually ranges from 7.0 to 7.5. Intracellular cytosolic pH 7.0 is the same as the pH of the endoplasmic reticulum [22,23,24] (hepatocytes); the interstitial pH of the brain cortex is 7.2 [25]; and the mitochondrial matrix ranges between pH 7.5 and 8.2 in various cells and their metabolic state [26,27]. Arterial and venous pH is about 7.3–7.45, and the pH inside red cells is slightly lower, about 7.2–7.3 [28,29]. However, tissue pH may reach lower values in ischemia or after anaerobic exercise in muscle. In the brain cortex, one hour of ischemia decreases interstitial and intracellular pH from 7.24 and 7.01 to 6.43 and 6.86, respectively [25]. The pH of a resting myocyte is about 7.0, which can increase slightly during exercise (about 0.1); however, during intensive (anaerobic) exertion, the pH drops from 7.0 to 6.5 or less [17].
Most of the molecules relevant to 31P MRS evaluation are located in the cell cytosol; therefore, at a pH of 7 under physiological conditions. NAD+/NADH, ADP, and ATP are also present in the mitochondrial matrix (pH 7.5–8). G6P is transported from the cytosol to the endoplasmatic reticulum (ER) to be dephosphorylated to glucose only in the hepatocytes. However, a typical pH of the hepatocyte cytosol and the ER is the same and equals pH 7 [22,23,24]. A prominent 31P signal of blood is 2,3-diphosphoglycerate (2,3-DPG), located in red cells, therefore with a pH of 7.2. However, in red cells, the 2,3-DPG signal is not only affected by pH but also modified by its binding to hemoglobin.
In the sections that follow, further individual metabolites are discussed in detail.
3.1. Energy Metabolites—Phosphocreatine (PCr), Inorganic Phosphate (Pi), Adenosinetriphosphate (ATP), and Adenosinediphosphate (ADP)
PCr, Pi, and ATP metabolites are closely related to energy turnover and come into prominence especially in muscle metabolism studies. PCr is a dominant signal in muscles, whereas, in the liver and kidney, it is not detected. In skeletal muscles, PCr concentration is about 33 mM and Pi is about 4.5 mM [30]. The PCr signal, with a chemical shift of 0.0 ppm, is widely used as an internal standard for in vivo 31P MR experiments; in our case, it was also used as an internal reference for all our phantoms. To verify the stability of PCr frequency with pH alterations in phantoms, the signal of two phantoms with a pH of 7.0 and 7.5 were acquired in one spectrum. In the resultant spectrum, only one PCr peak was then present, which implies an identical chemical PCr shift in both conditions. However, the chemical shift position of PCr is not totally insensitive to pH change if pH changes an order of several units, as McDowell and Stewart have shown [31]. For more details, see the literature, e.g., [17,32,33,34].
Pi, which participates in energy reactions in living tissue, has been present in all phantoms as a buffering agent. The chemical shift of Pi strongly depends on pH and is controlled by the equilibrium
Frequently used equation for the calculation of pH in vivo is based on the Henderson–Hasselbalch equation in the form:
| pH = 6.75 + log ((δ − 3.27)/(5.63-δ)) |
where δ is the chemical shift frequency difference between pH-dependent Pi and pH-independent reference peak (in ppm). Usually, this reference peak is PCr, but it can also be, e.g., αATP [34] or some other metabolites [32]. In our phantoms, the δ was used to control the pH measured by a pH meter. In vivo, Pi is usually visible as a single signal, but under some pathological conditions or with improved spectral resolution due to the higher field-strength, an additional Pi signal can be visible [8,11,12].
The 31P MR spectrum of ATP is characterized by three phosphorus signals split into doublets (α, γ) and a triplet (β) due to JP-H. In vivo chemical shifts of ATP signals are significantly influenced by pH and the presence of ions, as are Zn, Cu, Mg2+, and Ca2+, which form ATP complexes with different chemical shifts. Depending on pH, ion concentration chemical shifts can be changed. The literature data show in vivo chemical shifts in the ranges: α~7–8, β ~16, and ~3–4 ppm [35,36]. It should be mentioned that only the β-ATP signal arises from pure ATP, whereas αATP includes small contributions from NAD+/NADH and αADP, and γATP usually contains a contribution from β-ADP.
In our phantoms, the chemical shifts of α and γ phosphorus are in this in vivo range; however, β-phosphorus is shifted up to 18.8 ppm. Nevertheless, this is in agreement with other findings reported in the literature [37,38,39,40].
Whereas ATP cellular concentration in muscles is assumed to be constant in young healthy individuals and is about 5.5 mmol/kg wet weight, which equals 8.3 mM cell water, ADP concentration is very low. Indeed, in resting muscle, it is only about 13 µmol [30], and thus impossible to detect by 31P MRS directly. For more information on ATP detection, see the literature [30,41,42,43].
3.2. Phosphomonoesters (PME)
Phosphomonoesters mainly represent intermediates on the phospholipid biosynthesis pathway. The main components of the PME signal are phosphocholine (PC) and phosphoethanolamine (PE), and small signal contributions are derived from sugar phosphates. PME resonate in a rather broad frequency range between 2 to 8 ppm, and thus they partly overlap with the PDE frequency region.
3.2.1. Phosphocholine (PC) and Phosphoethanolamine (PE)
Both phosphocholine (PC) and phosphoethanolamine (PE) molecules have one phosphorous atom that forms a multiple triplet structure between 6 and 7 ppm. 1H decoupling simplifies the signal shape from a triplet to a singlet. The change of chemical shift is only 0.1–0.2 ppm between pH 7.0 and 7.5. The PE chemical shift of 6.7–6.8 ppm in the phantom was in good agreement with the in vivo value (6.78 ppm), and the chemical shift of PC (6.2–6.4 ppm) was slightly higher compared to the in vivo chemical shift of 5.9 ppm [36].
In 31P MRS, signals of PC and PE often overlap; therefore, they are usually evaluated as a combined PME (phosphomonoester) signal. An increase of PC was shown using high-resolution 1H MRS, and similarly, an increased PME signal was observed in vivo in various cancers; for review, see [44]. In muscles, PME concentration is very low. However, in the liver, PME concentration is about 1–3 mM [9,19,45], and the PME signal is one of dominant signals in the spectra.
3.2.2. Phosphoenolpyruvate (PEP) and Phosphatidylcholine (PtdC)
The phosphoenolpyruvate (PEP) signal is a singlet and, in our phantom, showed a chemical shift between 2.0 and 2.27 ppm at a pH of 7.0 and 7.5, respectively. The strong dependence of the PEP signal on pH has already been described by Bierwagen [5], who measured chemical shifts of 1.53 and 2.13 ppm at a pH of 7 and 8, respectively. Similarly, Chmelik [4] reports a change in the chemical shift of PEP from −1 to 2.5 ppm in the pH range from 0 to 14 (~1.6 ppm at pH 7).
Very close to the chemical shift of PEP is the chemical shift of phosphatidylcholine (PtdC). It is one of the main compounds in the bile and can decrease in patients with cholangiopathies [46]. PtdC itself is very poorly soluble in water, and therefore, its resonance frequencies were obtained in ethanol–water mixtures (with a Triton−100 surfactant). We measured the chemical shift values between 2.14 and 2.13 ppm at pH 7.21 and 7.5, respectively. This is in accordance with the values obtained by Bierwagen [5] (chemical shift of PtdC of 2.15 and 2.16 ppm at a pH of 7 and 8) and Chmelik [4] (the phantom chemical shift of 2 ppm).
In the in vivo 31P MR spectra of the liver, the signal at 2.06 ppm was attributed to the combined signal of phosphoenolpyruvate and phosphatidylcholine, with an average concentration of 1.67 mM. Detailed studies [4,5] have shown that the main contribution comes from PtdC. In the liver parenchyma, this signal also appears and increases with decreasing distance of the VOI from the gallbladder. Therefore, the VOI for measuring the liver parenchyma should be carefully positioned to avoid unwanted signal overlap. In muscles in vivo, the PEP signal is at the noise level, so it cannot be evaluated and compared with the in vitro signal.
3.2.3. Glucose-1-Phosphate (G1P) and Glucose-6-Phosphate (G6P)
Signals of both these compounds can be found in the range of 4 to 7 ppm. The G6P chemical shift position in vitro (7.03 ppm at pH = 7) corresponds to the in vivo position of G6P = 7.1–7.2 ppm [36,47].
Without proton decoupling G1P forms a doublet and G6P forms a triplet signal. The proton decoupling technique simplifies both of them to singlet signals. In the G6P signal, two other minor signals were found, probably from other enantiomer/isomers of glucose. A pH change from 7 to 7.5 results in a chemical shift difference of ±0.2 ppm for both metabolites.
In vivo skeletal muscle concentrations of G6P were shown to be around 0.1 (mg/kg muscle) [48,49] with a blunted, insulin-stimulated increase in insulin-resistant and type 2 diabetic patients, as well as in conditions in which plasma free fatty acids are elevated [40,50]. The muscle concentration of G1P is too low to be detected by 31P MRS in vivo; moreover, its signals are overlapped by other phosphomonoesters (PME) [47].
3.3. Phosphodiesters (PDE)
Phosphodiesters resonate in a frequency range from 2 to 4 ppm and include several compounds that are difficult to separate due to insufficient resolution if a low magnetic field is used. PDE may reflect (and serve as a marker of) membrane damage. Importantly, PME and PDE signals of 31P MRS may be also used in the diagnosis of liver diseases. In cirrhosis, the PDE signal has been shown to decrease compared to healthy controls or patients with steatosis [9,50].
The overall PDE concentration in the liver reported from 31P MR spectra in vivo is in the range between 5 and 13 mM [9,19,51,52,53,54,55,56]. It depends on the number of metabolites included, spectra resolution (1.5–7 T), and corrections to relaxation times.
PDE concentration in resting muscles ranges between 2–10 mM [11,52] and is related to energy metabolism [7] or neuromuscular conditions [57]. It increases with age [58] and in overweight subjects with a sedentary lifestyle [11] and also depends on physical status [59].
3.3.1. Glycerophosphocholine (GPC)
Glycerophosphocholine (GPC) is, in addition to glycerophosphoethanolamine (GPE), the main component of the PDE signal observed in vivo. In the liver and muscles, GPC concentration is about 2–4 mM [4,11,19,56]. Recently, skeletal muscle and hepatic GPC were suggested as biomarkers of the tissue-specific action of thyroid hormones [10].
The chemical shift position of GPC was almost independent of pH. Proton decoupling simplified the triplet signal to a singlet. An in vitro chemical shift of 3 ppm was slightly different from the in vivo position of 2.76 ppm [4].
3.3.2. 2,3-Diphosphoglycerate (2,3-DPG)
In the structure of 2,3-DPG, there are two phosphorus atoms providing signal—a doublet and a triplet at positions 4.1, 5.3 ppm (pH = 7) and 4.6, and 5.8 ppm (pH = 7.5) in a phantom. Proton decoupling simplifies this multiple structure to two singlets. In blood in vivo, both signals of 2,3-DPG are singlets at positions of 5.5 and 6.3 ppm. The difference between 2,3-DPG signals in vitro in a phantom and in blood may be explained by the 2,3-DPG association with hemoglobin in erythrocytes and its paramagnetic effect that strongly influences the chemical shift of 2,3-DPG [29].
The concentration of 2,3-DPG is strongly dependent on pH and very sensitive to the energy demand of the erythrocytes. The normal concentration of 2,3-DPG is about 4.5–5.1 mM packed red cells or 10.5–16.2 µmol/g Hb. Neonatal values are about 20% lower than those in an adult [60].
The presence of 2,3-DPG in erythrocytes is the basis for a typical 31P MRS signal of blood. Its presence in the 31P MR spectra of the heart indicates contamination of the myocardial signal by the signal of blood.
3.4. Nicotinamide Adenine Dinucleotide Metabolites (NAD+, NADH)
The pair of NAD+/NADH is an electron acceptor/donor in a redox reaction involved in cellular metabolism. The 31P signals of both compounds without 1H decoupling are singlets with positions at 8.32 (NAD+) and 8.16 (NADH) ppm, both insensitive to pH change. 1H decoupling reshapes the NAD+ signal to a doublet centered at 8.32 ppm, while the NADH signal only narrows. Measured in vitro shift positions are in good agreement with other in vitro and in vivo NAD+/NADH measurements, where shifts of ~8.1 ppm for NADH and ~8.3 ppm for the center of NAD+ have been reported with 4 to 11.7 T [61,62]. Similarly, in decoupled spectra, the NADH signal had the shape of a singlet and NAD+ had the shape of a doublet/quartet.
The concentration of NAD+/NADH and their ratio can be determined in vivo by MR techniques at magnetic fields of 3–7 T. However, the resonances of NAD+ and NADH are difficult to analyze separately without 1H decoupling. The analysis allows for the determination of the NAD+/NADH redox state of the tissue, e.g., in the brain, equal to 5.7 [21]. Total intracellular NAD+/NADH concentration is approximately 0.4 mM in the human brain [20,63] or 0.5 mM in the muscle of young, healthy volunteers [64].
3.5. Uridine Diphosphoglucose (UDPG)
The UDPG molecule contains two nonequivalent phosphorus nuclei. The JP-P, as well as the JH-P interaction, strongly modulates their signal. The application of proton decoupling simplifies the signal to two doublets with center positions at −9.8 ppm (phosphorus attached to the glucose part) and −8.2 ppm (phosphorus attached to the ribose part) [20,65]. In vivo, the UDPG signal at −8.1 ppm partially overlaps with NADH (resonating at −8.16 ppm), but signal at the −9.8 ppm position is well-resolved. The concentration of UDPG is approximately 0.2–0.3 mM in the brain [20] and 2.00 ± 0.22 mM wet tissue in the liver [19]. It seems that hepatic UDPG content does not change with age or weight [55], NAFLD, or cirrhosis [6,19].
Uridine diphosphate glucose (UDP-glucose) is an “activated” form of glucose because UDP provides additional energy to the glucose molecule. As such, UDPG is a precursor of polysaccharides (glycogen), glycoproteins, lipopolysaccharides, and glycosphingolipids. UDP-glucose can also be converted into UDP-galactose and UDP-glucuronic acid used in the synthesis of polysaccharides containing galactose and glucuronic acid.
3.6. 31P Basis Set for LCModel Calculations
The commonly used quantification of in vivo MR spectra is performed in the time domain using the jMRUI software package [66], or, in the frequency domain using LCModel software [14]. Both approaches require prior knowledge of chemical shifts, interaction constants, widths, and relative signal intensities within one molecule. These parameters are obtained by an analysis of the spectra of metabolites measured in vitro or from simulated spectra. The LCModel package is mostly used to analyze 1H in vivo MR spectra in the range of chemical shifts of 5–10 ppm. In this report, we used it to evaluate 31P in vivo spectra. Its application for the analysis of 31P spectra with signals resonating in the spectral range of about 30 ppm is more difficult and is not often used. Recently, however, a successful modification of the input parameters of the calculation of 31P and 13C spectra was proposed [16,67,68,69], which enabled the correct definition of the baseline of the spectrum. We adapted these input parameters, and we used our basis set for the analysis of 31P spectra of muscles and the liver. The chemical shifts of all 15 metabolites in phantoms measured at 3 T were in agreement with published high-resolution in vitro NMR spectra and simulated datasets. In addition, small changes of chemical shifts of ATP were necessary for in vivo applications, as described above. Two other signals were added to the BASISp—the signal of membrane phospholipids and GPE based on the parameters from the literature [15]. The basis set used for the analyses contained 19 signals of 18 model spectra (Table 1)
The full BASISp set for LCModel was used in a representative calculation of the 31P MR calf muscle and liver spectra, as shown in Figure 1.
In addition, the comparison of the LCModel and jMRUI methods was performed on a group of five randomly selected 31P liver spectra and five spectra from the calf muscle (at rest) from our internal database, see Table 2. The modified BASISp set with only six prominent signals in both measured tissues was used. The calculated relative intensities are similar for most signals, as shown by the coefficients of variation (CV) of the metabolite relative intensity signals in Table 2. Concerning muscle spectra, both methods give similar results with the highest but still acceptable CV for GPC and Pi. As expected in the liver due to lower signal to noise, the highest CV values (CV > 30, unacceptable difference) were obtained for GPC signal, which represents different PDE metabolites. In addition, PCr signal was also used in this case for comparison, although it represents only contamination from the surrounding tissue, see Table 2. CVs in this pilot test show that both LCModel and jMRUI methods often reach similar results; the difference originates from different mathematical principles of signal intensity calculation, as described in the original literature [14,66].
Regarding the calculation of the concentration of metabolites, a number of papers have been published concerning the application of LCModel frequency domain approach to 1H MRS (mostly brain), and most of the 31P MRS reports used the AMARES based jMRUI time–domain fitting approach. Besides the historical reason for this difference, there was no available complete basis set for muscle or liver 31P MRS that could be used for LCModel. In line with our experience from 1H MRS data processing, the LCModel is more useful for routine calculation of larger data sets than jMRUI. The AMARES-based jMRUI approach, on the other hand, can be used more easily to characterize and analyze a single spectrum. Based on our experience and the result of this study, we think that it is useful to combine both methods for evaluating spectra.
4. Materials and Methods
4.1. Phantom Preparation
All chemicals from Table 1 were purchased from the Sigma-Aldrich Company. Structural formulas are depicted in Supplementary Materials. MgCl2, K2HPO4, and NaH2PO4 were obtained from Lachema, Brno, Czech Republic.
Phantoms of 100 mL volume consisted of water solutions: MgCl2 (concentration CMgCl2 = 0.35 mM), K2HPO4, and NaH2PO4 1:1 (concentration Csum = 40 mM) and phosphocreatine (concentration CPCr = 5 mM) (as a standard). To this solution, metabolites were added to reach a final concentration of 10 mM; see details in Table 1. UDPG and 2,3-DPG phantoms were prepared with a lower concentration of c ~2 mM. In these phantoms, the concentration of K2HPO4/NaH2PO4 was reduced to c = 10 mM.
MgCl2 was added to phantoms to stabilize solutions because Mg2+ ions often form complexes with 31P metabolites that may affect the shape and chemical shift of the 31P metabolites [37,70]. The K2HPO4 and NaH2PO4 were used to buffer pH and prevent cleavage of Pi from 31P metabolites.
Two phantoms with a pH of 7.03 and 7.50 were prepared for each metabolite. pH was adjusted by titration of NAOH/HCl in solutions at 37 °C and slightly fluctuated about ±0.05 during adjustment. The pH of phantoms was also verified during MR measurement from differences in the chemical shift between the PCr and Pi signal.
In addition, a blood phantom for in vitro measurement of human venous blood was prepared for the comparison of 2,3-DPG solutions. The blood phantom consisted of 50 mL human blood with heparin.
Two phantoms were prepared without other specific metabolites containing PCr (CPCr = 5 mM), MgCl2, and K2HPO4/NaH2PO4 with a pH = 7.03 and 7.5 to test the pH dependency of the chemical shift position of PCr on pH.
4.2. 31P MR Spectroscopy
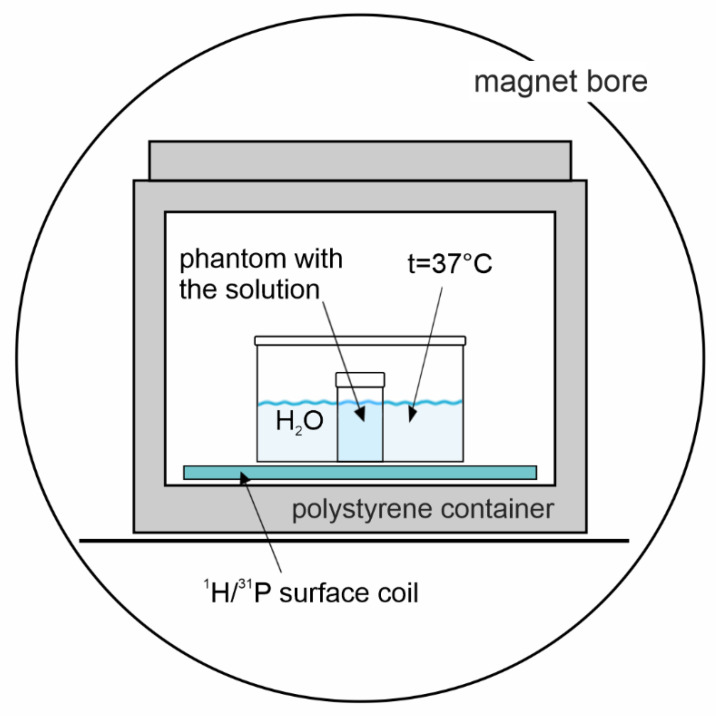
Measurements were performed using a 3T MR system, VIDA (Siemens Healthineers, Erlangen, Germany) equipped with a dual-channel 1H/31P surface coil (Rapid Biomedical, Rimpar, Germany) with a diameter of 11 cm and an interface device; see Figure 2.
Figure 2.
Arrangement of the phantom 31P MRS experiment. A plastic bucket filled with water keeps the temperature and loading of the coil.
An MRI localizer sequence was used to verify the position of the phantom and adjustment volume, which was defined as a cube (35 × 35 × 35 mm) in the center of the phantom. Automatic and manual adjustment of magnetic field homogeneity were performed at the water signal; an excellent half-width of the water signal (3–4 Hz) was achieved for all phantoms in defined adjustment volume (a critical step in achieving excellent magnetic field homogeneity was to align the water level in the phantom and temperature reservoir to the same position before measurement).
Two free induction decay (FID)-based non-localized 31P MR spectroscopic sequences were applied in each phantom with the same parameters, as follows: repetition time (TR) of 15 s; acquisition delay (TE*) = 0.4 ms, number of acquisitions (acq) = 64; flip angle (FA) = 90° (BIR-4 adiabatic pulse); vector size = 2048; and bandwidth = 1000 Hz (20 ppm) to cover chemical shift of studied metabolites. Transmitter frequency was set to that of the PCr signal except for the ATP measurement; it was shifted to the signal of αATP in this case to excite all ATP signals sufficiently. Non-slice selective proton decoupling (the WALTZ4 decoupling scheme) was applied in the second sequence to cancel the JP-H interaction (standard Siemens setting—decoupling pulse duration 1 ms, decoupling duration 1000 ms, decoupling flip angle 90 or 180°, offset frequency for 1H—0 ppm, duty cycle 50%).
4.3. Spectra Evaluation
Spectra were loaded into jMRUI software (version 6.0). Manual correction of the phase (zero- and first-order) was performed to obtain the absorption shape of phosphorous signals, and then the position of the PCr and second metabolite were read according to their maxima. The BASISp set for 31P LCModel version 6.3–0 G was constructed from phantom chemical shifts presented in Table 1.
To analyze in vivo data, the spectra were phased manually in the jMRUI software to obtain reliable prior knowledge of the zero- and first-order phase parameters used further in the LCModel analysis (starting points and standard deviations, DEGZER = −96, SDDEGZ = 10, DEGPPM = 7, SDDEGP = 4). In addition, the following LCModel input parameters were used: DKNTMN = 2*99; XSTEP = 5; RFWHM = 3; FWHMBA = 0.049; NREFPK(2) = 1; PPMREF(1,2) = 0; and DESDSH = 0.01. The default values that define the spline baseline function were also adjusted to take into account the larger spectral range of 31P compared to 1H: ALPBMN = 7.8 × 10−10; ALPBMX = 3.9 × 10−7; ALPBPN = 9.8 × 10−10; and ALPBST = 1.2 × 10−9, as mentioned previously [71]. A representative evaluation of in vivo 31P MR spectra of the liver and calf muscles using LCModel software and the BASISp basis set is shown in Figure 1 and Table 2.
5. Conclusions
The 31P chemical shifts of 15 metabolites were measured under conditions close to those of an in vivo examination at 3T and were compared and supplemented with published values for in vitro and in vivo conditions. Based on all these data, a set of 19 signals necessary for the successful quantification of in vivo liver and muscle spectra by LCModel software was generated. The tabulated values of chemical shifts can also be used for other methods of in vivo spectra analysis, e.g., jMRUI software and other analytical procedures.
Supplementary Materials
The following are available online: spectra and chemical structures of measured metabolites.
Author Contributions
Conceptualization, M.H., P.S., M.K. and M.D.; methodology, M.H. and M.B.; software, P.S. and M.B.; validation, T.D. and M.D.; formal analysis, M.D.; investigation, P.S., T.D. and M.B.; resources, M.H., P.S. and M.K.; data curation, M.D. and T.D.; writing—original draft preparation, M.H. and M.D.; writing—review and editing, M.D. and M.K.; visualization, M.D. and P.S.; supervision, M.H.; project administration, P.S.; funding acquisition, M.H., M.D. and M.K. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by Ministry of Health, Czech Republic—conceptual development of research organization (“Institute for Clinical and Experimental Medicine—IKEM, IN 00023001”) and by the Austrian Ministry for Science, Research, and Economy (Scientific & Technological Cooperation Austria-Czech Republic 2018–2019 CZ08/2018)/Czech Ministry of Education, Youth and Sports, Mobility Czech Republic-Austria, project No. 8J18AT023.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
On demand to: petr.sedivy@ikem.cz, monika.dezortova@ikem.cz.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bottomley P.A., Hart H.R., Edelstein W.A., Schenck J.F., Smith L.S., Leue W.M., Mueller O.M., Redington R.W. Nmr Imaging Spectroscopy System to Study Both Anatomy and Metabolism. Lancet. 1983;2:273–274. doi: 10.1016/S0140-6736(83)90250-7. [DOI] [PubMed] [Google Scholar]
- 2.Šedivý P., Kipfelsberger M.C., Dezortová M., Krššák M., Drobný M., Chmelík M., Rydlo J., Trattnig S., Hájek M., Valkovič L. Dynamic 31P MR spectroscopy of plantar flexion: Influence of ergometer design, magnetic field strength (3 and 7 T), and RF-coil design. Med. Phys. 2015;42:1678–1689. doi: 10.1118/1.4914448. [DOI] [PubMed] [Google Scholar]
- 3.Valkovič L., Chmelík M., Krššák M. In-vivo (31)P-MRS of skeletal muscle and liver: A way for non-invasive assessment of their metabolism. Anal. Biochem. 2017;529:193–215. doi: 10.1016/j.ab.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chmelík M., Valkovič L., Wolf P., Bogner W., Gajdošík M., Halilbasic E., Gruber S., Trauner M., Krebs M., Trattnig S., et al. Phosphatidylcholine contributes to in vivo (31)P MRS signal from the human liver. Eur. Radiol. 2015;25:2059–2066. doi: 10.1007/s00330-014-3578-y. [DOI] [PubMed] [Google Scholar]
- 5.Bierwagen A., Begovatz P., Nowotny P., Markgraf D., Nowotny B., Koliaki C., Giani G., Klüppelholz B., Lundbom J., Roden M. Characterization of the peak at 2.06 ppm in (31) P magnetic resonance spectroscopy of human liver: Phosphoenolpyruvate or phosphatidylcholine? NMR Biomed. 2015;28:898–905. doi: 10.1002/nbm.3323. [DOI] [PubMed] [Google Scholar]
- 6.Sevastianova K., Hakkarainen A., Kotronen A., Cornér A., Arkkila P., Arola J., Westerbacka J., Bergholm R., Lundbom J., Lundbom N., et al. Nonalcoholic fatty liver disease: Detection of elevated nicotinamide adenine dinucleotide phosphate with in vivo 3.0-T 31P MR spectroscopy with proton decoupling. Radiology. 2010;256:466–473. doi: 10.1148/radiol.10091351. [DOI] [PubMed] [Google Scholar]
- 7.Szendroedi J., Schmid A.I., Chmelik M., Krssak M., Nowotny P., Prikoszovich T., Kautzky-Willer A., Wolzt M., Waldhäusl W., Roden M. Skeletal Muscle Phosphodiester Content Relates to Body Mass and Glycemic Control. PLoS ONE. 2011;6:e21846. doi: 10.1371/journal.pone.0021846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedivy P., Dezortova M., Drobny M., Dubsky M., Dusilova T., Kovar J., Hajek M. Origin of the (31) P MR signal at 5.3 ppm in patients with critical limb ischemia. NMR Biomed. 2020;33:e4295. doi: 10.1002/nbm.4295. [DOI] [PubMed] [Google Scholar]
- 9.Dezortova M., Taimr P., Skoch A., Spicak J., Hajek M. Etiology and functional status of liver cirrhosis by 31P MR spectroscopy. World J. Gastroenterol. 2005;11:6926–6931. doi: 10.3748/wjg.v11.i44.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beiglbock H., Wolf P., Pfleger L., Caliskan B., Fellinger P., Zettinig G., Anderwald C.H., Kenner L., Trattnig S., Kautzky-Willer A., et al. Effects of Thyroid Function on Phosphodiester Concentrations in Skeletal Muscle and Liver: An In Vivo NMRS Study. J. Clin. Endocrinol. Metab. 2020;105:e4866–e4874. doi: 10.1210/clinem/dgaa663. [DOI] [PubMed] [Google Scholar]
- 11.Valkovič L., Chmelík M., Ukropcová B., Heckmann T., Bogner W., Frollo I., Tschan H., Krebs M., Bachl N., Ukropec J., et al. Skeletal muscle alkaline Pi pool is decreased in overweight-to-obese sedentary subjects and relates to mitochondrial capacity and phosphodiester content. Sci. Rep. 2016;6:20087. doi: 10.1038/srep20087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kan H.E., Klomp D.W.J., Wong C.S., Boer V.O., Webb A.G., Luijten P.R., Jeneson J.A. In vivo 31P MRS detection of an alkaline inorganic phosphate pool with short T1 in human resting skeletal muscle. NMR Biomed. 2010;23:995–1000. doi: 10.1002/nbm.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Oorschot J.W.M., Schmitz J.P.J., Webb A., Nicolay K., Jeneson J.A.L., Kan H.E. 31P MR Spectroscopy and Computational Modeling Identify a Direct Relation between Pi Content of an Alkaline Compartment in Resting Muscle and Phosphocreatine Resynthesis Kinetics in Active Muscle in Humans. PLoS ONE. 2013;8:e76628. doi: 10.1371/journal.pone.0076628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Provencher S.W. Estimation of Metabolite Concentrations from Localized in-Vivo Proton Nmr-Spectra. Magn. Reson. Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 15.Deelchand D.K., Nguyen T.M., Zhu X.H., Mochel F., Henry P.G. Quantification of in vivo (3)(1)P NMR brain spectra using LCModel. NMR Biomed. 2015;28:633–641. doi: 10.1002/nbm.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S., van der Veen J.W., An L., Stolinski J., Johnson C., Ferraris-Araneta M., Victorino M., Tomar J.S., Shen J. Cerebral phosphoester signals measured by 31P magnetic resonance spectroscopy at 3 and 7 Tesla. PLoS ONE. 2021;16:e0248632. doi: 10.1371/journal.pone.0248632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley K.E., Ali A.S., Flores B., Jubrias S.A., Shankland E.G. Mitochondrial NAD(P)H In vivo: Identifying Natural Indicators of Oxidative Phosphorylation in the (31)P Magnetic Resonance Spectrum. Front. Physiol. 2016;7:45. doi: 10.3389/fphys.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyerspeer M., Boesch C., Cameron D., Dezortová M., Forbes S.C., Heerschap A., Jeneson J.A.L., Kan H.E., Kent J., Layec G., et al. 31P magnetic resonance spectroscopy in skeletal muscle: Experts’ consensus recommendations. NMR Biomed. 2021;34:e4246. doi: 10.1002/nbm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purvis L.A.B., Clarke W.T., Valkovič L., Levick C., Pavlides M., Barnes E., Cobbold J.F., Robson M.D., Rodgers C.T. Phosphodiester content measured in human liver by in vivo (31) P MR spectroscopy at 7 tesla. Magn. Reson. Med. 2017;78:2095–2105. doi: 10.1002/mrm.26635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Graaf R.A., De Feyter H.M., Brown P.B., Nixon T.W., Rothman D.L., Behar K.L. Detection of cerebral NAD(+) in humans at 7T. Magn. Reson. Med. 2017;78:828–835. doi: 10.1002/mrm.26465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peeters T.H., van Uden M.J., Rijpma A., Scheenen T.W.J., Heerschap A. 3D (31) P MR spectroscopic imaging of the human brain at 3 T with a (31) P receive array: An assessment of (1) H decoupling, T(1) relaxation times, (1) H-(31) P nuclear Overhauser effects and NAD(+) NMR Biomed. 2019;34:e4169. doi: 10.1002/nbm.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollock A.S. Intracellular pH of hepatocytes in primary monolayer culture. Am. J. Physiol. 1984;246:F738–F744. doi: 10.1152/ajprenal.1984.246.5.F738. [DOI] [PubMed] [Google Scholar]
- 23.Strazzabosco M., Poci C., Spirlì C., Zsembery A., Granato A., Massimino M.L., Crepaldi G. Intracellular pH regulation in Hep G2 cells: Effects of epidermal growth factor, transforming growth factor-alpha, and insulinlike growth factor-II on Na+/H+ exchange activity. Hepatology. 1995;22:588–597. [PubMed] [Google Scholar]
- 24.Kim J.H., Johannes L., Goud B., Antony C., Lingwood C.A., Daneman R., Grinstein S. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl. Acad. Sci. USA. 1998;95:2997–3002. doi: 10.1073/pnas.95.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nedergaard M., Kraig R.P., Tanabe J., Pulsinelli W.A. Dynamics of interstitial and intracellular pH in evolving brain infarct. Am. J. Physiol. 1991;260:R581–R588. doi: 10.1152/ajpregu.1991.260.3.R581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhmedov D., Braun M., Mataki C., Park K.S., Pozzan T., Schoonjans K., Rorsman P., Wollheim C.B., Wiederkehr A. Mitochondrial matrix pH controls oxidative phosphorylation and metabolism-secretion coupling in INS-1E clonal beta cells. FASEB J. 2010;24:4613–4626. doi: 10.1096/fj.10-162222. [DOI] [PubMed] [Google Scholar]
- 27.Santo-Domingo J., Demaurex N. The renaissance of mitochondrial pH. J. Gen. Physiol. 2012;139:415–423. doi: 10.1085/jgp.201110767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labotka R.J., Kleps R.A. A phosphate-analogue probe of red cell pH using phosphorus-31 nuclear magnetic resonance. Biochemistry. 1983;22:6089–6095. doi: 10.1021/bi00295a008. [DOI] [PubMed] [Google Scholar]
- 29.Labotka R.J. Measurement of intracellular pH and deoxyhemoglobin concentration in deoxygenated erythrocytes by phosphorus-31 nuclear magnetic resonance. Biochemistry. 1984;23:5549–5555. doi: 10.1021/bi00318a026. [DOI] [PubMed] [Google Scholar]
- 30.Kemp G.J., Meyerspeer M., Moser E. Absolute quantification of phosphorus metabolite concentrations in human muscle in vivo by 31P MRS: A quantitative review. NMR Biomed. 2007;20:555–565. doi: 10.1002/nbm.1192. [DOI] [PubMed] [Google Scholar]
- 31.McDowell R.W., Stewart I. Peak assignments for phosphorus-31 nuclear magnetic resonance spectroscopy in pH range 5-13 and their application in environmental samples. Chem. Ecol. 2005;21:211–226. doi: 10.1080/02757540500211590. [DOI] [Google Scholar]
- 32.Moon R.B., Richards J.H. Determination of intracellular pH by 31P magnetic resonance. J. Biol. Chem. 1973;248:7276–7278. doi: 10.1016/S0021-9258(19)43389-9. [DOI] [PubMed] [Google Scholar]
- 33.Sedivy P., Drobny M., Dezortova M., Herynek V., Roztocil K., Cermakova H., Nemcova A., Dubsky M., Hajek M. 31P-MR spectroscopy in patients with mild and serious lower limb ischemia. Int. Angiol. 2018;37:293–299. doi: 10.23736/S0392-9590.18.03943-3. [DOI] [PubMed] [Google Scholar]
- 34.Rata M., Giles S.L., deSouza N.M., Leach M.O., Payne G.S. Comparison of three reference methods for the measurement of intracellular pH using 31P MRS in healthy volunteers and patients with lymphoma. NMR Biomed. 2014;27:158–162. doi: 10.1002/nbm.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung W.I., Widmaier S., Seeger U., Bunse M., Staubert A., Sieverding L., Straubinger K., van Erckelens F., Schick F., Dietze G., et al. Phosphorus J coupling constants of ATP in human myocardium and calf muscle. J. Magn. Reson. B. 1996;110:39–46. doi: 10.1006/jmrb.1996.0005. [DOI] [PubMed] [Google Scholar]
- 36.De Graaf R.A. In Vivo NMR Spectroscopy, Principles and Techniques. 2nd ed. John Wiley&Sons Ltd.; Chichester, UK: 2007. pp. 1–570. [Google Scholar]
- 37.Turner B.L., Mahieu N., Condron L.M. Phosphorus-31 nuclear magnetic resonance spectral assignments of phosphorus compounds in soil NaOH-EDTA extracts. Soil Sci. Soc. Am. J. 2003;67:497–510. doi: 10.2136/sssaj2003.4970. [DOI] [Google Scholar]
- 38.Cade-Menun B.J. Improved peak identification in 31P-NMR spectra of environmental samples with a standardized method and peak library. Geoderma. 2015;257–258:102–114. doi: 10.1016/j.geoderma.2014.12.016. [DOI] [Google Scholar]
- 39.Cohn M., Hughes T.R., Jr. Nuclear magnetic resonance spectra of adenosine di- and triphosphate. II. Effect of complexing with divalent metal ions. J. Biol. Chem. 1962;237:176–181. doi: 10.1016/S0021-9258(18)81382-5. [DOI] [PubMed] [Google Scholar]
- 40.Szabo Z. Multinuclear NMR studies of the interaction of metal ions with adenine-nucleotides. Coord. Chem. Rev. 2008;252:2362–2380. doi: 10.1016/j.ccr.2008.03.002. [DOI] [Google Scholar]
- 41.Hernández E., Kahl S., Seelig A., Begovatz P., Irmler M., Kupriyanova Y., Nowotny B., Nowotny P., Herder C., Barosa C., et al. Acute dietary fat intake initiates alterations in energy metabolism and insulin resistance. J. Clin. Investig. 2017;127:695–708. doi: 10.1172/JCI89444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hultman E., Nilsson L.H., Sahlin K. Adenine nucleotide content of human liver. Normal values and fructose-induced depletion. Scand. J. Clin. Lab. Investig. 1975;35:245–251. doi: 10.3109/00365517509095736. [DOI] [PubMed] [Google Scholar]
- 43.Traussnigg S., Kienbacher C., Gajdošík M., Valkovič L., Halilbasic E., Stift J., Rechling C., Hofer H., Steindl-Munda P., Ferenci P., et al. Ultra-high-field magnetic resonance spectroscopy in non-alcoholic fatty liver disease: Novel mechanistic and diagnostic insights of energy metabolism in non-alcoholic steatohepatitis and advanced fibrosis. Liver Int. 2017;37:1544–1553. doi: 10.1111/liv.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng M., Bhujwalla Z.M., Glunde K. Targeting Phospholipid Metabolism in Cancer. Front. Oncol. 2016;6:266. doi: 10.3389/fonc.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosner Z., Dezortova M., Tintera J., Hajek M. Application of two-dimensional CSI for absolute quantification of phosphorus metabolites in the human liver. Magma. 2001;13:40–46. doi: 10.1007/BF02668649. [DOI] [PubMed] [Google Scholar]
- 46.Pfleger L., Halilbasic E., Gajdošík M., Benčíková D., Chmelík M., Scherer T., Trattnig S., Krebs M., Trauner M., Krššák M. Concentration of Gallbladder Phosphatidylcholine in Cholangiopathies: A Phosphorus-31 Magnetic Resonance Spectroscopy Pilot Study. J. Magn. Reson. Imaging. 2021 doi: 10.1002/jmri.27817. [DOI] [PubMed] [Google Scholar]
- 47.De Haan J.H., Klomp D.W., Tack C.J., Heerschap A. Optimized detection of changes in glucose-6-phosphate levels in human skeletal muscle by 31P MR spectroscopy. Magn. Reson. Med. 2003;50:1302–1306. doi: 10.1002/mrm.10630. [DOI] [PubMed] [Google Scholar]
- 48.Roden M., Krssak M., Stingl H., Gruber S., Hofer A., Furnsinn C., Moser E., Waldhausl W. Rapid impairment of skeletal muscle glucose transport/phosphorylation by free fatty acids in humans. Diabetes. 1999;48:358–364. doi: 10.2337/diabetes.48.2.358. [DOI] [PubMed] [Google Scholar]
- 49.Rothman D.L., Shulman R.G., Shulman G.I. P-31 Nuclear-Magnetic-Resonance Measurements of Muscle Glucose-6-Phosphate—Evidence for Reduced Insulin-Dependent Muscle Glucose-Transport or Phosphorylation Activity in Non-Insulin-Dependent Diabetes-Mellitus. J. Clin. Investig. 1992;89:1069. doi: 10.1172/JCI115686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cline G.W., Petersen K.F., Krssak M., Shen J., Hundal R.S., Trajanoski Z., Inzucchi S., Dresner A., Rothman D.L., Shulman G.I. Impaired glucose transport as a cause of decreased insulin-stimulated muscle glycogen synthesis in type 2 diabetes. N. Engl. J. Med. 1999;341:240. doi: 10.1056/NEJM199907223410404. [DOI] [PubMed] [Google Scholar]
- 51.Noren B., Dahlqvist O., Lundberg P., Almer S., Kechagias S., Ekstedt M., Franzén L., Wirell S., Smedby O. Separation of advanced from mild fibrosis in diffuse liver disease using 31P magnetic resonance spectroscopy. Eur. J. Radiol. 2008;66:313–320. doi: 10.1016/j.ejrad.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Buchli R., Meier D., Martin E., Boesiger P. Assessment of absolute metabolite concentrations in human tissue by 31P MRS in vivo. Part II: Muscle, liver, kidney. Magn. Reson. Med. 1994;32:453–458. doi: 10.1002/mrm.1910320405. [DOI] [PubMed] [Google Scholar]
- 53.Li C.W., Negendank W.G., Murphy-Boesch J., Padavic-Shaller K., Brown T.R. Molar quantitation of hepatic metabolites in vivo in proton-decoupled, nuclear Overhauser effect enhanced 31P NMR spectra localized by three-dimensional chemical shift imaging. NMR Biomed. 1996;9:141–155. doi: 10.1002/(SICI)1099-1492(199606)9:4<141::AID-NBM403>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 54.Chmelík M., Schmid A.I., Gruber S., Szendroedi J., Krššák M., Trattnig S., Moser E., Roden M. Three-dimensional high-resolution magnetic resonance spectroscopic imaging for absolute quantification of 31P metabolites in human liver. Magn. Reson. Med. 2008;60:796–802. doi: 10.1002/mrm.21762. [DOI] [PubMed] [Google Scholar]
- 55.Laufs A., Livingstone R., Nowotny B., Nowotny P., Wickrath F., Giani G., Bunke J., Roden M., Hwang J.-H. Quantitative liver 31 P magnetic resonance spectroscopy at 3T on a clinical scanner. Magn. Reson. Med. 2014;71:1670–1675. doi: 10.1002/mrm.24835. [DOI] [PubMed] [Google Scholar]
- 56.Pfleger L., Gajdošík M., Wolf P., Smajis S., Fellinger P., Kuehne A., Krumpolec P., Trattnig S., Winhofer Y., Krebs M., et al. Absolute Quantification of Phosphor-Containing Metabolites in the Liver Using (31)P MRSI and Hepatic Lipid Volume Correction at 7T Suggests No Dependence on Body Mass Index or Age. J. Magn. Reson. Imaging. 2019;49:597–607. doi: 10.1002/jmri.26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hooijmans M.T., Doorenweerd N., Baligand C., Verschuuren J., Ronen I., Niks E.H., Webb A.G., Kan H.E. Spatially localized phosphorous metabolism of skeletal muscle in Duchenne muscular dystrophy patients: 24-month follow-up. PLoS ONE. 2017;12:e0182086. doi: 10.1371/journal.pone.0182086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krumpolec P., Klepochova R., Just I., Tusek Jelenc M., Frollo I., Ukropec J., Ukropcova B., Trattnig S., Krssak M., Valkovic L. Multinuclear MRS at 7T Uncovers Exercise Driven Differences in Skeletal Muscle Energy Metabolism Between Young and Seniors. Front. Physiol. 2020;11:644. doi: 10.3389/fphys.2020.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klepochova R., Valkovic L., Hochwartner T., Triska C., Bachl N., Tschan H., Trattnig S., Krebs M., Krssak M. Differences in Muscle Metabolism Between Triathletes and Normally Active Volunteers Investigated Using Multinuclear Magnetic Resonance Spectroscopy at 7T. Front Physiol. 2018;9:300. doi: 10.3389/fphys.2018.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roper D., Layton M. Investigation of the hereditary haemolytic anaemias: Membrane and enzyme abnormalities. In: Lewis S., Bain B., Bates I., editors. Dacie and Lewis Practical Haematology. 10th ed. Elsevier; Amsterdam, The Netherlands: 2006. [DOI] [Google Scholar]
- 61.Lu M., Zhu X.H., Zhang Y., Chen W. Intracellular Redox State Revealed by In Vivo P-31 MRS Measurement of NAD(+) and NADH Contents in Brains. Magn. Reson. Med. 2014;71:1959–1972. doi: 10.1002/mrm.24859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu M., Zhu X.H., Chen W. In vivo P-31 MRS assessment of intracellular NAD metabolites and NAD(+)/NADH redox state in human brain at 4 T. NMR Biomed. 2016;29:1010–1017. doi: 10.1002/nbm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chouinard V.A., Kim S.Y., Valeri L., Yuksel C., Ryan K.P., Chouinard G., Cohen B.M., Du F., Öngür D. Brain bioenergetics and redox state measured by (31)P magnetic resonance spectroscopy in unaffected siblings of patients with psychotic disorders. Schizoph. Res. 2017;187:11–16. doi: 10.1016/j.schres.2017.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henriksson J., Katz A., Sahlin K. Redox state changes in human skeletal muscle after isometric contraction. J. Physiol. 1986;380:441–451. doi: 10.1113/jphysiol.1986.sp016296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarma R.H., Lee C.H., Hruska F.E., Wood D.J. H-1, H-1-[P-31], P-31, and P-31-[H-1] fast Fourier transform NMR study of solution conformation of cofactors involved in glycogen synthesis: Adenosinediphosphoglucose and uridinediphosphoglucose. FEBS Lett. 1973;36:157–162. doi: 10.1016/0014-5793(73)80359-X. [DOI] [PubMed] [Google Scholar]
- 66.Stefan D., Cesare F.D., Andrasescu A., Popa E., Lazariev A., Vescovo E., Strbak O., Williams S., Starcuk Z., Cabanas M., et al. Quantitation of magnetic resonance spectroscopy signals: The jMRUI software package. Meas. Sci. Technol. 2009;20:104035. doi: 10.1088/0957-0233/20/10/104035. [DOI] [Google Scholar]
- 67.Gajdosik M., Landheer K., Swanberg K.M., Juchem C. INSPECTOR: Free software for magnetic resonance spectroscopy data inspection, processing, simulation and analysis. Sci. Rep. 2021;11:2094. doi: 10.1038/s41598-021-81193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deelchand D.K., Ugurbil K., Henry P.G. Investigating brain metabolism at high fields using localized 13C NMR spectroscopy without 1H decoupling. Magn. Res. Med. 2006;55:279–286. doi: 10.1002/mrm.20756. [DOI] [PubMed] [Google Scholar]
- 69.Skupienski R., Do K.Q., Xin L. In vivo 31P magnetic resonance spectroscopy study of mouse cerebral NAD content and redox state during neurodevelopment. Sci. Rep. 2020;10:15623. doi: 10.1038/s41598-020-72492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hájek M., Palyzová D., Korínek M., Kurková D. Concentrations of free mg2+, pH and 31P MR metabolite ratios in calf muscles of healthy controls and patients with primary juvenile hypertension. Physiol. Res. 2002;51:159–167. [PubMed] [Google Scholar]
- 71.Henry P.G., Oz G., Provencher S., Gruetter R. Toward dynamic isotopomer analysis in the rat brain in vivo: Automatic quantitation of 13C NMR spectra using LCModel. NMR Biomed. 2003;16:400–412. doi: 10.1002/nbm.840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
On demand to: petr.sedivy@ikem.cz, monika.dezortova@ikem.cz.