Abstract
The serine/threonine protein kinase Akt regulates a wide range of cellular functions via phosphorylation of various substrates distributed throughout the cell, including at the plasma membrane and endomembrane compartments. Disruption of compartmentalized Akt signaling underlies the pathology of many diseases such as cancer and diabetes. However, the specific spatial organization of Akt activity and the underlying regulatory mechanisms, particularly the mechanism controlling its activity at the lysosome, are not clearly understood. We developed a highly sensitive excitation-ratiometric Akt activity reporter (ExRai-AktAR2), enabling the capture of minute changes in Akt activity dynamics at subcellular compartments. In conjunction with super-resolution expansion microscopy, we found that growth factor stimulation leads to increased colocalization of Akt with lysosomes and accumulation of lysosomal Akt activity. We further showed that 3-phosphoinositides (3-PIs) accumulate on the lysosomal surface, in a manner dependent on dynamin-mediated endocytosis. Importantly, lysosomal 3-PIs are needed for growth-factor-induced activities of Akt and mechanistic target of rapamycin complex 1 (mTORC1) on the lysosomal surface, as targeted depletion of 3-PIs has detrimental effects. Thus, 3-PIs, a class of critical lipid second messengers that are typically found in the plasma membrane, unexpectedly accumulate on the lysosomal membrane in response to growth factor stimulation, to direct the multifaceted kinase Akt to organize lysosome-specific signaling.
Short abstract
This work presents a highly sensitive fluorescent biosensor for Akt activity and reveals regulation of the kinase by lipid second messengers at lysosomes.
Introduction
The phosphoinositide 3-kinase (PI3K), Akt, and mechanistic target of rapamycin (mTOR) signaling pathway regulates a wide range of cellular processes, including cell survival, growth, and metabolism.1 Upon stimulation of receptor tyrosine kinases, activation of class I PI3K leads to production of phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2) and phosphatidylinositol (3,4,5)-triphosphate (PI(3,4,5)P3 or PIP3), which triggers the recruitment of Akt to the plasma membrane via its pleckstrin homology (PH) domain and subsequent activation of Akt.1−3 Active Akt phosphorylates various substrates distributed throughout the cell, including in the plasma membrane, endomembranes, cytosol, and nucleus, to regulate specific functions.1,2,4,5 Spatiotemporal Akt signaling plays a critical role in disease settings, including cancer and type 2 diabetes.1,2 For example, it has been suggested that phosphoinositides and the plasma membrane pool of Akt control the formation of invadopodia, plasma membrane-associated structures that play a crucial role in cancer cell migration and invasion.6 In addition, a recent study showed that PI(3,4)P2 levels in endocytic vesicles, but not at the plasma membrane, play an important role in promoting PI3K/Akt and ERK signaling in triple-negative breast cancer,7 although the precise role of PI(3,4)P2 and the relevant lipid phosphatase, inositol polyphosphate 4-phosphatase B (INPP4B), in cancer remains to be resolved.8 A thorough understanding of the spatiotemporal Akt signaling network is therefore important for our fundamental understanding of cell physiology as well as therapeutic development. However, the mechanism by which compartment-specific Akt activity is regulated to control key downstream signaling events is complex and remains poorly understood. For example, at the lysosome, which is the main degradative organelle and an emerging platform for cellular nutrient signaling,9 Akt phosphorylates TSC2 in the tuberous sclerosis complex (TSC) to relieve its inhibition of Rheb GTPase, leading to direct activation of mTOR complex 1 (mTORC1), a master regulator of cell growth and metabolism.10 Although TSC2 is localized to the lysosomal surface and phosphorylated Akt was found in the lysosomal fraction,11 it is not clear how Akt is regulated at this signaling platform and whether lysosome-specific Akt activity is required for mTORC1 activation.
Precise measurement of Akt activity is essential for understanding the regulation and functions of this central kinase as well as the intricate signaling networks in which it is embedded. Although quantitative radioactivity assay and immunoblotting remain frequently used methods for detecting protein kinase activity, fluorescent peptide sensors based on phosphorylation-sensitive chelation-enhanced fluorescence12,13 or unquenching of fluorescence,14 allow for direct, rapid, and continuous quantification of protein kinase activity in a wide range of sample types including recombinant or immunoprecipitated enzymes, cell or tissue lysates. Intracellular delivery of specifically engineered peptide-based fluorescent probes further permits dynamic measurement of Akt signaling dynamics in single cells.15 On the other hand, genetically encodable fluorescent protein-based biosensors have emerged to be powerful and indispensable tools that enable interrogation of spatiotemporal control of kinase activity in living cells.16−21 These biosensors serve as surrogate substrates of specific protein kinases, and, since they are genetically encodable, allow real-time measurement of kinase activities in living cells with high spatial and temporal resolution.17,20,22 For Akt activity, several genetically encodable sensors were developed by converting Akt-mediated phosphorylation to a change in fluorescent readout, such as changes in fluorescence resonance energy transfer (FRET) or redistribution of fluorescence from nucleus to cytosol.23−28 However, translocation-based biosensors cannot be readily adapted for probing subcellular Akt activities,25−27 and FRET-based Akt biosensors have limited sensitivity,23,24,28 which prevents reliable detection of subtle changes in Akt activities. On the basis of a newly developed excitation-ratiometric kinase activity reporter (KAR) design,29 here we report an excitation-ratiometric Akt activity reporter (ExRai-AktAR2) that enables sensitive measurement of live-cell Akt activity dynamics at subcellular locations. Using a suite of genetically encodable biosensors including ExRai-AktAR2, super-resolution expansion microscopy, as well as biochemical perturbation, we found that 3-phosphoinositides (3-PIs) accumulate on the lysosomal surface through dynamin-mediated endocytosis, and this pool of 3-PIs is critical for regulating Akt and mTORC1 on the lysosomal surface.
Results and Discussion
Development and Characterization of a Highly Sensitive Excitation-Ratiometric Akt Biosensor
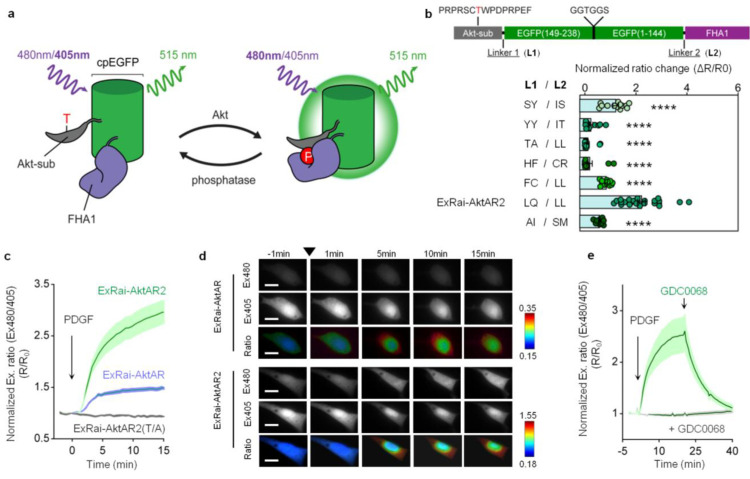
In our excitation-ratiometric KAR design,29 a circularly permuted enhanced GFP (cpEGFP), the reporting unit of the biosensor, is sandwiched between a kinase-specific substrate peptide and a phosphoamino acid binding domain, which together serve as the sensing unit of the biosensor. As shown with ExRai KARs,29 insertion of the kinase substrate and FHA1 domains is expected to “rescue” the wild-type GFP chromophore behavior, and phosphorylation-mediated conformational changes typically result in increases in the fluorescence of the deprotonated chromophore (∼480 nm) and decreases in the fluorescence of the protonated chromophore (∼405 nm), possibly due to structural changes of cpEGFP and alterations of the network surrounding the chromophore.30 The ratio of the fluorescence intensities at these two excitation wavelengths (R: Ex480/405) is used as a readout for kinase activity (Figure 1a). The first-generation excitation-ratiometric Akt activity reporter (ExRai-AktAR) developed based on this general design still has a limited dynamic range,29 although it is already enhanced from the FRET-based Akt activity reporters.28 Given that linkers at the junctions of the sensing unit and reporting unit critically influence biosensor performance,31,32 we mutated the two residues immediately preceding and following cpEGFP to identify a linker combination which would result in a large change in the excitation ratio upon Akt stimulation. We tested several linker combinations and compared the responses of biosensor variants in serum-starved NIH3T3 cells stimulated with platelet derived growth factor (PDGF) (Figure 1b). Among those biosensors, variants with linker pairs LQ-LL and SY-IS showed large responses, and the variant with the linker pair LQ-LL showed the largest response (Figure 1b). We previously found this LQ/LL mutation can result in kinase activity reporters with high sensitivity.33 This candidate with the LQ-LL linker was designated ExRai-AktAR2 and subjected to further characterization (Figure 1b).
Figure 1.
Development and characterization of ExRai-AktAR2. (a) Modulation of cpEGFP fluorescence by a molecular switch dependent on Akt-mediated phosphorylation. (b) Development of ExRai-AktAR2. Domain structure (upper) and normalized ratio changes (ΔR/R0) of the linker variants in serum-starved NIH3T3 cells stimulated with 50 ng/mL of PDGF. The best-performing variant was designated ExRai-AktAR2. From top to bottom: n = 13, 13, 13, 13, 12, 24, and 14 cells from 3 independent experiments each. ****, P < 0.0001 vs LQ/LL (ExRai-AktAR2); ordinary one-way ANOVA followed by Dunnett’s multiple comparison test. (c) Average time courses of normalized excitation ratio (R: Ex480/405) in serum-starved NIH3T3 cells expressing ExRai-AktAR2 (green, n = 24), ExRai-AktAR (blue, n = 14), or ExRai-AktAR2 T/A (gray, n = 13) stimulated with 50 ng/mL of PDGF. (d) Representative fluorescence images of individual channels and pseudocolor images showing responses of ExRai-AktAR (top) and ExRai-AKAR2 (bottom) before and after PDGF stimulation in serum-starved NIH3T3 cells. Arrowhead indicates the addition of PDGF. For individual channel images, ExRai-AktAR and ExRai-AktAR2 are shown in the intensity range of 500–3500 and 500–12000, respectively. Scale bars = 5 μm. (e) Average time courses of normalized excitation ratio (R: Ex480/405) in serum-starved NIH3T3 cells expressing ExRai-AktAR2 stimulated with 50 ng/mL PDGF followed by addition of GDC0068 (1 μM) (green, n = 7) or pretreated with 1 μM GDC0068 for 20 min prior to PDGF stimulation (gray, n = 10). Solid lines in (c, e) indicate mean responses; shaded areas, SEM. Bars denote mean ± SEM (b).
As shown in Figures 1c,d and S1a,b, the response of ExRai-AktAR2 (ΔR/R0: 209 ± 22%, n = 24) is significantly enhanced compared to that of ExRai-AktAR (ΔR/R0: 49 ± 3%, n = 14) (Figure 1c). As a negative control, serum-starved NIH3T3 cells expressing ExRai-AktAR2 with a mutated phosphorylation site (T/A) showed no response to PDGF (Figure 1c). Furthermore, addition of GDC0068, a potent and selective Akt inhibitor, reversed the PDGF-induced responses to almost basal levels, whereas pretreatment with GDC0068 abolished the responses to PDGF (Figure 1e), suggesting that the biosensor is specific to Akt activity. We used Western blotting analysis to verify the phosphorylation of ExRai-AktAR2 and showed that the response of ExRai-AktAR2 was correlated with its phosphorylation upon PDGF stimulation (Figures S1c and S12). In addition, ExRai-AktAR2 is not responsive to stimulation conditions that activate protein kinase A (PKA)33 or protein kinase C (PKC),34 suggesting that ExRai-AktAR2 selectively detects Akt activity (Figure S1d,e). Furthermore, ExRai-AktAR2 was not sensitive to pH changes from 5 to 8 (Figure S2), consistent with previous studies of analogous biosensors.29 Taken together, ExRai-AktAR2 specifically reports Akt activity with a large dynamic range.
Sensitive Detection of Compartmentalized Akt Activity Using ExRai-AktAR2
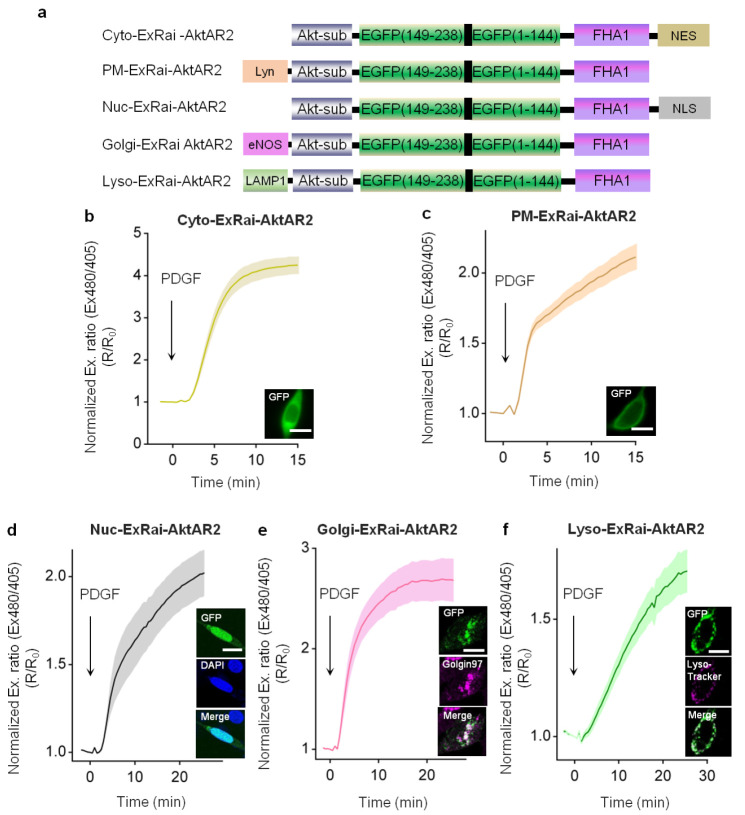
Growing evidence has suggested that Akt activity is distinctly regulated at subcellular locations, ranging from microdomains within the plasma membrane to endomembrane and nuclear compartments.23,24,35,36 ExRai-AktAR2, with high sensitivity and specificity for Akt activity, should enable sensitive detection of Akt activity at different subcellular locations. To monitor Akt activity at different subcellular locations, we fused ExRai-AktAR2 to well-established sequences for targeting to different compartments: nuclear export signal (NES) for cytosol (Cyto), Lyn kinase targeting motif (Lyn) for plasma membrane (PM), nuclear localization signal (NLS) for nucleus (Nuc), endothelial nitric oxide synthase (eNOS)-derived sequence for Golgi apparatus (Golgi),37 and lysosome-associated membrane protein 1 (LAMP1)-derived sequence for lysosome (Lyso)28,38 (Figure 2a). As expected, in the cytosol compartment and the plasma membrane, PDGF induced a 325 ± 20% (ΔR/R0, n = 20) increase and a 127 ± 11% (ΔR/R0, n = 23) increase for Cyto-ExRai-AktAR2 (Figure 2b) and PM-ExRai-AktAR2 (Figures 2c and S3a), respectively. Moreover, Nuc-ExRai-AktAR2 (Figure 2d), colocalized with nuclear marker (DAPI), exhibited a 102 ± 13% (ΔR/R0, n = 13) increase in the excitation ratio in response to PDGF, in line with previous reports on nuclear Akt activity.5,24,39 In addition, we observed PDGF-stimulated Akt activity at a previously underappreciated location, Golgi membranes. As seen in Figure 2e, Golgi-ExRai-AktAR2 colocalized with appropriate Golgi marker (Golgin-97) and responded to PDGF with a 169 ± 21% (ΔR/R0, n = 12) increase in the excitation ratio. In support of this notion, a recent study suggested that Akt and phosphoinositide-dependent kinase-1 (PDK1) are present in Golgi membranes, and Akt is regulated by S-palmitoylation,40 a protein lipidation that is reversible and often occurs in Golgi membranes.41 The specific regulation and functional role of this pool of Akt activity await further investigation.
Figure 2.
ExRai-AktAR2 enables sensitive detection of Akt activity at different subcellular compartments. (a) Domain structures of ExRai-AktAR2 targeted to the cytoplasm, plasma membrane, nucleus, Golgi, and lysosomes. (b) Average time courses of normalized excitation ratios (Ex480/405) in serum-starved NIH3T3 cells expressing Cyto-ExRai-AktAR2 (n = 20) stimulated with 50 ng/mL of PDGF. Bottom right: Image showing the localization of Cyto-ExRai-AktAR2. (c) Average time courses of the normalized excitation-ratio response (Ex480/405) in serum-starved NIH3T3 cells expressing PM-ExRai-AktAR2 (n = 23) stimulated with 50 ng/mL of PDGF. Bottom right: Image showing the localization of PM-ExRai-AktAR2. (d) Average time courses of the normalized excitation ratio response (Ex480/405) in serum-starved NIH3T3 cells expressing Nuc-ExRai-AktAR2 (n = 13) stimulated with 50 ng/mL of PDGF. Right: Images showing the localization of Nuc-ExRai-AktAR2 (green) and DAPI staining (blue), together with the merged image. (e) Average time courses of normalized excitation ratio (Ex480/405) in serum-starved NIH3T3 cells expressing Golgi-ExRai-AktAR2 (n = 12) stimulated with 50 ng/mL of PDGF. Right: Images showing the localization of Golgi-ExRai-AktAR2 (green) and Golgin-97 staining (Golgi marker) (magenta), together with the merged image. (f) Average time courses of normalized excitation ratio (Ex480/405) in serum-starved NIH3T3 cells expressing Lyso-ExRai-AktAR2 (n = 22) stimulated with 50 ng/mL of PDGF. Right: Images showing the localization of Lyso-ExRai-AktAR2 (green) and Lyso-Tracker Red staining (lysosomal marker) (magenta), together with the merged image. Solid lines in b–f indicate mean responses; shaded areas, SEM. Scale bars = 10 μm (b–f).
We next examined the Lyso-ExRai-AktAR2 response, given the growing recognition of the lysosome as a prominent subcellular location for Akt/mTOR signaling.10,11 NIH3T3 cells expressing Lyso-ExRai-AktAR2 showed fluorescence in small puncta, which were localized with Lyso-Tracker Red, a red lysosomal marker. Lyso-ExRai-AktAR2 responded to PDGF stimulation with a 70 ± 9.0% (ΔR/R0, n = 22) increase in the excitation ratio (Figure 2f), suggesting that lysosomal Akt activity is induced by PDGF. Taken together, these results suggest that ExRai-AktAR2 enables robust detection of Akt activity at various subcellular locations (Figure S1f), clearly outperforming the commonly used FRET-based AktAR2 (Figure S3b–h) and representing the most sensitive fluorescent biosensor capable of subcellular detection of Akt activity in living cells.
PDGF Stimulation Increases Akt Colocalization with the Lysosome
Several lines of evidence suggest that the lysosome is an important site of Akt signaling.10,11 At the lysosome, activation of mTORC1 is facilitated by Akt-mediated phosphorylation of TSC2, which relieves the suppression of Rheb GTPase and directly activates mTORC1.10 Detection of lysosomal Akt activity prompted us to examine whether Akt is localized to the lysosome. The lysosome structure varies from 0.1 to 1.2 μm in size,42 and 100–500 nm in diameter, which poses a challenge for the assessment of colocalization of Akt with lysosome structures using conventional light microscopy.43 We therefore utilized two-color expansion microscopy, which was reported to provide nanoscale resolution for fluorescence imaging on conventional diffraction-limited microscopes.44−47 For sample preparation, NIH3T3 cells overexpressing GFP-fused LAMP1 (LAMP1-GFP) were fixed and then stained for total Akt following a standard expansion protocol.45,48,49 We first verified that the homogeneous expansion process yields the expected expansion ratio by showing a macroscopic expansion ratio of 2.6 (Figure S4a) and a microscopic expansion ratio of 2.7 (pre- and postexpansion) (Figure S4b–c). The close approximation of macroscopic and microscopic expansion ratios suggests the expansion is homogeneous. Moreover, the intensity distribution along the line scan of expanded NIH3T3 cells (Figure S4d–e) showed excellent contrast. With a resolution of 45 nm (resolution/expansion ratio, 120 nm/2.7), this method provided more detailed information than conventional methods, while keeping the structures well preserved.
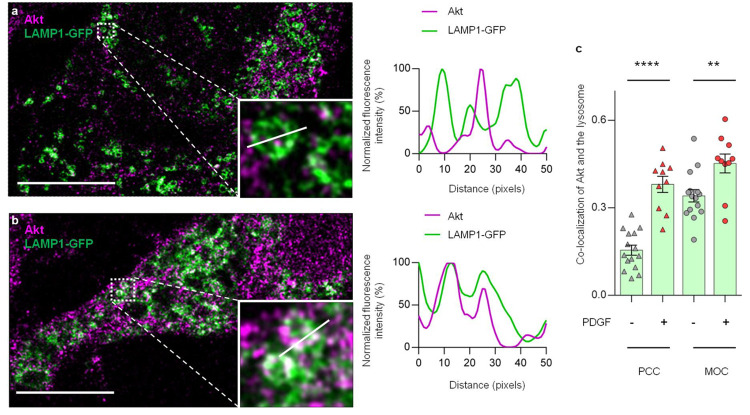
Using expansion microscopy and a validated Akt antibody (Figures S5 and S12), we analyzed the colocalization of lysosome structures and Akt signals. As shown in Figure 3a, serum-starved NIH3T3 cells expressing LAMP1-GFP exhibited punctate vesicular patterns identified as lysosomes, with little colocalization of Akt, as indicated in line scan analyses. Upon PDGF treatment, we observed an increase in colocalization of Akt and the lysosomes (Figure 3b). We then quantified the degree of colocalization between Akt and the lysosomes.50 The values of the Pearson correlation coefficient (PCC) were significantly higher for PDGF-treated samples (red triangles, 0.38 ± 0.03, n = 10), compared with control cells (gray triangles, 0.16 ± 0.02, n = 15, P < 0.0001) (Figure 3c). A similar trend was seen with the Mander’s overlap coefficient (MOC) values (red dots, 0.45 ± 0.03, n = 10; gray dots, 0.34 ± 0.02, n = 15, P < 0.01) (Figure 3c). These data suggest that PDGF stimulation induces the localization of Akt to the lysosomes. Notably, PDGF-induced changes were not detected in nonexpanded samples (P = 0.49 for PCC and P = 0.51 for MOC) (Figure S6), highlighting the capability of expansion microscopy to capture important spatial information that is lost in conventional light microscopy. We further validated this finding by proximity ligation assay (PLA). PLA signals were detected when we examined the proximity between Akt and the lysosomal marker LAMP1. These signals were significantly higher than background signals with LAMP1 antibody plus IgG control, confirming that Akt is localized at the lysosomes in nonstarved NIH3T3 cells (Figure S7).
Figure 3.
PDGF-induced Akt colocalization with lysosomes revealed by expansion microscopy. (a) Confocal images of expanded, serum-starved NIH3T3 cells expressing LAMP1-GFP (green) and stained with total Akt (magenta). Right: Line scan showing out-of-phase spatial pattern between Akt (magenta) and the lysosomes (green). Data are representative of three independent experiments. Scale bar = 10 μm. (b) Confocal image of expanded, PDGF-treated (50 ng/mL of PDGF for 30 min) NIH3T3 cells expressing LAMP1-GFP (green) and stained with total Akt (magenta). Right: Line scan showing in-phase spatial pattern between Akt (magenta) and the lysosomes (green). Data are representative of three independent experiments. Scale bar = 10 μm. (c) Quantification of Akt colocalization with lysosomes without (gray) and with (red) PDGF stimulation by Pearson correlation coefficient (PCC, triangles) and the Mander’s overlap coefficient (MOC, dots). Error bar represents the mean ± SEM ****, P < 0.0001, – PDGF vs + PDGF (PCC); **, P = 0.0065, – PDGF vs + PDGF (MOC); unpaired two-tailed Student’s t test. Bars denote mean ± SEM (c). Scale bars = 10 μm (a,b).
3-Phosphoinositides Accumulate at the Lysosomes through Endocytosis
Having demonstrated the localization and activity of Akt at the lysosomes, we next asked whether lipid second messengers are involved in Akt regulation at the lysosome. In the classical model, upon growth factor stimulation, activation of PI3K leads to production of PI(3,4)P2 and PI(3,4,5)P3 at the plasma membrane, which recruit Akt for its subsequent activation.1,2,51 We then ask whether 3-PIs are present at the lysosome to recruit Akt following growth factor stimulation. For detecting subcellular phosphoinositide dynamics, we previously constructed a genetically encodable indicator for phosphoinositides based on Akt (InPAkt)36 by sandwiching the PH domain of Akt and a “pseudoligand” between cyan and yellow fluorescent proteins. PIP3 and PI(3,4)P2, produced upon growth factor stimulation, bind to the PH domain and compete off the pseudoligand, resulting in a change in FRET between the fluorescent proteins and an increase in yellow-to-cyan emission ratio.36 We fused a LAMP1-derived lysosomal targeting motif to the N-terminus of the biosensor to generate Lyso-InPAkt. A plasma membrane-targeted InPAkt (PM-InPAkt) was used as a control (Figure 4a). As shown in Figure 4b, PM-InPAkt was responsive to PDGF stimulation, as expected, with a 7.4 ± 0.8% increase in the yellow/cyan (Y/C) emission ratio (blue, n = 22) as expected. Intriguingly, Lyso-InPAkt also showed a consistent response of 4.4 ± 0.4% to PDGF stimulation (green, n = 29). This suggests that PI(3,4)P2 and PIP3 accumulate at the lysosomal membrane in response to growth factor stimulation. Furthermore, we compared the kinetics of PI(3,4)P2 and PIP3 accumulation within these compartments by using both the lag time and T1/2, the time needed to reach 5% and 50% of the maximum amplitude, respectively (Figures 4c and S8a).52−54 We found that the kinetics of PI(3,4)P2 and PIP3 accumulation were slower at the lysosome than at the plasma membrane (P < 0.01) (Figures 4c and S8a). Additionally, accumulation of PI(3,4)P2 and PIP3 was more sustained at the lysosome compared versus the plasma membrane, quantified by the sustained activity metric at 15 min post-treatment (SAM15)55 (0.74 ± 0.03, n = 29 vs 0.63 ± 0.04, n = 22, P = 0.04) (Figures 4b and S8b). Thus, in addition to the plasma membrane, PDGF stimulates the accumulation of PI(3,4)P2 and PIP3 at the lysosome, with subcellularly distinct kinetic profiles.
Figure 4.
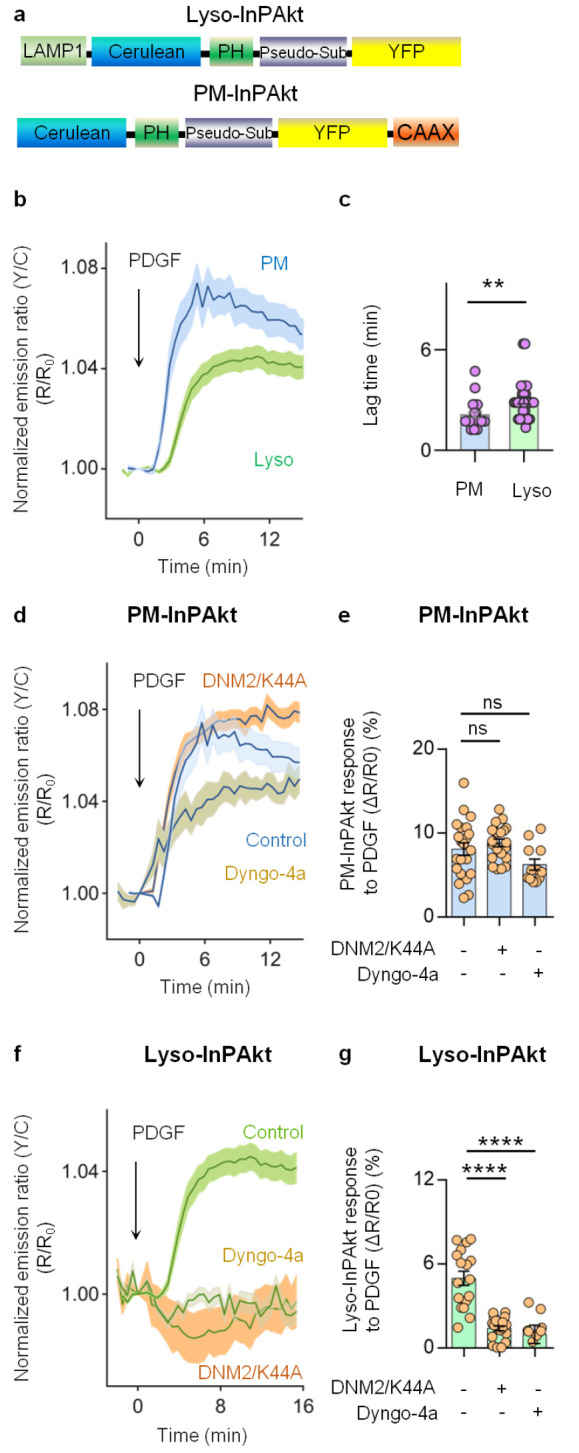
PDGF-induced 3-phosphoinositide accumulation is dependent on dynamin-mediated endocytosis. (a) Domain structure of lysosome-targeted InPAkt (Lyso-InPAkt) and plasma membrane-targeted InPAkt (PM-InPAkt). (b) Average time courses of normalized emission ratio (yellow/cyan) in serum-starved NIH3T3 cells expressing PM-InPAkt (blue, n = 22) or Lyso-InPAkt (green, n = 29) stimulated with 50 ng/mL of PDGF. (c) Comparison of lag time of PM-InPAkt (n = 22) and Lyso-InPAkt (n = 29) from three independent experiments. **, P = 0.0030; unpaired two-tailed Student’s t test with Welch’s correction. (d) Average time courses of the normalized emission ratio (yellow/cyan) in serum-starved NIH3T3 cells expressing PM-InPAkt without (blue, n = 22) or with pretreatment of 50 μM Dyngo-4a (beige, n = 12), or with overexpression of DNM2/K44A (orange, n = 21). (e) Responses of PM-InPAkt in PDGF-treated serum-starved NIH3T3 cells without (n = 22) or with pretreatment of Dyngo-4a (n = 12), or with overexpression of DNM2/K44A (n = 21). ns, not significant, P = 0.1582, control vs Dyngo-4a; ns, not significant, P = 0.6440, control vs DNM2/K44A; ordinary one-way ANOVA followed by Tukey’s multiple comparison test. (f) Average time courses of normalized emission ratio (yellow/cyan) in serum-starved NIH-3T3 cells expressing Lyso-InPAkt without (green, n = 17) or with pretreatment of 50 μM Dyngo-4a (beige, n = 9), or with overexpression of DNM2/K44A (orange, n = 20). (g) Responses of Lyso-InPAkt in PDGF-treated serum-starved NIH3T3 cells without (n = 17) or with pretreatment of Dyngo-4a (n = 9), or with overexpression of DNM2/K44A (n = 20). ****, P < 0.0001, control vs Dyngo-4a; ****, P < 0.0001, control vs DNM2/K44A; ordinary one-way ANOVA followed by Tukey’s multiple comparison test. Solid lines in (b, d, f) indicate mean responses; shaded areas, SEM. Bars denote mean ± SEM (c, e, g).
As PI(3,4)P2 was shown to internalize from the cell membrane into early endosomes,51 we examined whether lysosomal accumulation of PI(3,4)P2 and PIP3 is facilitated by endocytosis. Since a wide variety of endocytic processes requires dynamin, and dynamin-2 is ubiquitously expressed compared to dynamin-1 and −3,56−58 we perturbed endocytic pathways by expressing a dominant negative form of dynamin-2 (DNM2/K44A).59 We found that the response amplitude of PM-InPAkt (ΔR/R0: 8.1 ± 0.7%, n = 22) was not affected by overexpression of DNM2/K44A (ΔR/R0: 8.8 ± 0.4%, n = 21, P = 0.64) (Figure 4d–e). Similarly, pretreating cells with a potent dynamin inhibitor, Dyngo-4a,60,61 had little effect on the response amplitude of PM-InPAkt (ΔR/R0: 6.2 ± 0.6%, n = 12, P = 0.16), suggesting that perturbing dynamin-dependent endocytosis does not affect the maximal levels of plasma membrane PI(3,4)P2 and PIP3. However, the kinetics of PI(3,4)P2 and PIP3 accumulation at the plasma membrane were altered by dynamin inhibition. Increased SAM15 values were observed in cells overexpressing DNM2/K44A (0.88 ± 0.02, n = 21, P < 0.0001) or cells treated with Dyngo-4a (0.86 ± 0.04, n = 13, P < 0.001) compared to control cells (0.68 ± 0.04, n = 22) (Figure S8c), suggesting more sustained accumulation of 3-PIs at the plasma membrane when dynamin-dependent endocytosis was inhibited, presumably due to reduced internalization of 3-PIs. In stark contrast, the Lyso-InPAkt response (ΔR/R0: 5 ± 0.5%, n = 17) was completely abolished by inhibition of dynamin via either DNM2/K44A expression (ΔR/R0: 1.4 ± 0.2%, n = 20, P < 0.0001) or Dyngo-4a treatment (ΔR/R0: 1.0 ± 0.7%, n = 9, P < 0.0001) (Figure 4f–g), suggesting that lysosomal accumulation of PI(3,4)P2 and PIP3 is dependent on dynamin-dependent endocytosis. Moreover, given that production of PI(3,4)P2 and PIP3 at the plasma membrane is primarily mediated by class I PI3K,36,62 we would expect that lysosomal 3-phosphoinositides also depend on the same isoform. Pretreating cells with PIK-75, a highly selective inhibitor of class I PI3K,63,64 abolished the response of Lyso-InPAkt (Figure S8d), indicating that the lysosomal pool of 3-PIs indeed also depends on class I PI3K.
3-Phosphoinositides are potent lipid second messengers whose cellular levels are tightly controlled in time and space. Although PIP3 and PI(3,4)P2 are known to be predominantly produced at the plasma membrane and act as important regulators for Akt activity, there was little evidence for the presence of these two lipid products of PI3Ks at the lysosomal membrane,65 where PI(3)P and PI(3,5)P2 are considered to be abundant.66,67 Our results demonstrated that 3-PIs accumulate at the lysosomes in response to growth factor stimulation, and such accumulation is blocked by inhibition of dynamin or class I PI3K. Our findings support a model where 3-PIs are produced at the plasma membrane by class I PI3K and subsequently internalized (or transported) to the lysosome through dynamin-mediated endocytosis, consistent with previous studies showing that PI(3,4)P2 is internalized to accumulate on early endosomes in an endocytosis-dependent manner,51 although the role of other isoforms of PI3K was not directly probed here.65
Lysosomal Akt/mTORC1 Signaling Is Dependent on Endocytosis
We next asked whether blocking dynamin-dependent endocytosis inhibits Akt activity at the lysosomes. As shown in Figure 5a,b, Dyngo-4a pretreatment largely attenuated the PDGF-induced Lyso-ExRai-AktAR2 responses (gray, ΔR/R0: 3.4 ± 1.8%, n = 20) compared to controls without Dyngo-4a treatment (green, ΔR/R0: 69 ± 9.5%, n = 16, P < 0.0001). We further analyzed the effect of Dyngo-4a on phosphorylation of T1462 in TSC2. TSC2 localizes at the lysosomal surface and is a well-established Akt substrate.10,68 Consistent with the imaging data, Dyngo-4a pretreatment decreased the PDGF-induced phosphorylation of TSC2 without affecting phosphorylation of glycogen synthase kinase-3β (GSK3β), an Akt substrate localized throughout the cell (Figures S9a,b and S12).69,70 Similarly, responses of Lyso-ExRai-AktAR2 to PDGF (green, ΔR/R0: 56 ± 3.7%, n = 18) were reduced in cells overexpressing DNM2/K44A (gray, ΔR/R0: 29 ± 5.7%, n = 14, P < 0.001) (Figure S9c). These results suggest that perturbing dynamin-dependent endocytosis suppresses Akt activity at the lysosome. In contrast, PDGF-induced Akt activity at the plasma membrane (green, ΔR/R0: 95 ± 17%, n = 5) was not affected by overexpressing DNM2/K44A (gray, ΔR/R0: 105 ± 13%, n = 9, P = 0.6481) (Figure S9d). Together, these results suggest that lysosomal Akt activity is dependent on dynamin-mediated endocytosis.
Figure 5.
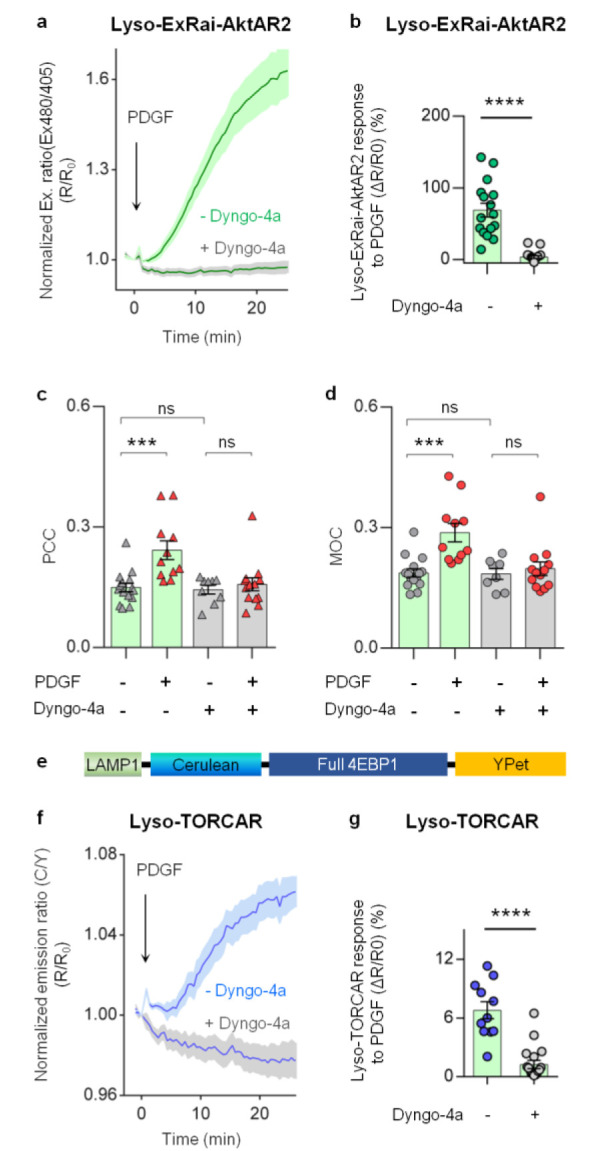
Lysosomal Akt/mTORC1 signaling activities are dependent on endocytosis. (a) Average time courses of normalized excitation ratio (R: Ex480/405) in serum-starved NIH3T3 cells expressing lysosome-targeted ExRai-AktAR2 (Lyso-ExRai-AktAR2) stimulated with 50 ng/mL of PDGF without (green, n = 16) or with 50 μM Dyngo-4a pretreatment for 10 min (gray, n = 20). (b) Responses of Lyso-ExRai-AktAR2 in PDGF-treated serum-starved NIH3T3 cells without (n = 16) or with pretreatment of Dyngo-4a (n = 20). ****, P < 0.0001; unpaired two-tailed Student’s t test with Welch’s correction. (c) Pearson correlation coefficient (PCC) analysis of Akt and lysosome colocalization in serum-starved NIH3T3 cells without (gray triangle) or with PDGF stimulation (red triangle) in the absence or presence of 50 μM Dyngo-4a. In the absence of Dyngo-4a: ***, P = 0.0006, – PDGF vs + PDGF; In the presence of Dyngo-4a: ns, not significant, P = 0.94, – PDGF vs + PDGF; Without PDGF stimulation: ns, not significant, P = 0.99, – Dyngo-4a vs + Dyngo-4a; Ordinary one-way ANOVA followed by Tukey’s multiple comparison test. (d) Mander’s overlap coefficient (MOC) analysis of colocalization of Akt and the lysosome in serum-starved NIH3T3 cells without (gray dots) or with PDGF stimulation (red dots) in the absence of presence of 50 μM Dyngo-4a. In the absence of Dyngo-4a: ***, P = 0.0003, – PDGF vs + PDGF; In the presence of Dyngo-4a: ns, not significant, P = 0.96, – PDGF vs + PDGF; Without PDGF stimulation: ns, not significant, P = 1.0, – Dyngo-4a vs + Dyngo-4a; Ordinary one-way ANOVA followed by Tukey’s multiple comparison test. (e) Domain structure of lysosome-targeted TORCAR (Lyso-TORCAR). (f) Average time courses of normalized emission ratio (cyan/yellow) in serum-starved NIH3T3 cells expressing Lyso-TORCAR stimulated with 50 ng/mL of PDGF without (blue, n = 11) or with 50 μM Dyngo-4a pretreatment for 10 min (gray, n = 18). (g) Responses of Lyso-TORCAR in PDGF-treated serum-starved NIH3T3 cells without (n = 11) or with pretreatment of Dyngo-4a (n = 18). ****, P < 0.0001; unpaired two-tailed Student’s t test. Solid lines in (a, f) indicate mean responses; shaded areas, SEM. Bars denote mean ± SEM (b–d, g).
We next asked whether inhibition of dynamin affects the colocalization of Akt with the lysosome structures. Using expansion microscopy, we found that in the presence of Dyngo-4a the increased colocalization of Akt with lysosomes induced by PDGF was effectively blunted (before PDGF, PCC = 0.14 ± 0.01, n = 9; after PDGF, PCC = 0.16 ± 0.02, n = 13, P = 0.94), whereas PDGF induced a robust increase in Akt colocalization with the lysosomes in the absence of Dyngo-4a (before PDGF, PCC = 0.15 ± 0.01, n = 15; after PDGF, PCC = 0.24 ± 0.02, n = 11, P < 0.001) (Figure 5c). Dyngo-4a did not affect the basal colocalization of Akt and the lysosomes, indicated by unchanged PCC values for cells without (0.15 ± 0.01, n = 15) or with pretreatment of Dyngo-4a (0.14 ± 0.01, n = 9, P = 0.99) (Figure 5c). Colocalization analysis using MOC agrees well with the results using PCC (Figure 5d). Taken together, our data suggest that blocking dynamin-dependent endocytosis prohibits PDGF-induced localization of Akt to the lysosomes.
mTORC1 is known to be activated at the lysosomes through Akt-mediated phosphorylation of TSC2.10 We hypothesized that localized Akt activity is required for the lysosomal activation of mTORC1. Given the demonstrated dependence of lysosomal Akt activity on dynamin-mediated endocytosis, we next examined whether perturbation of dynamin-mediated endocytosis impacts mTORC1 activity at the lysosomes. To probe mTORC1 activity, we previously constructed a genetically encodable FRET-based mTORC1 activity reporter (TORCAR) by sandwiching the mTORC1-specific substrate, eukaryotic initiation factor 4E-binding protein 1 (4EBP1), between a pair of fluorescent proteins that can undergo FRET. Upon specific phosphorylation by mTORC1, TORCAR exhibits a change in FRET, resulting in an increase in the cyan-overyellow (C/Y) emission ratio as a readout for increased mTORC1 activity.22,28 Lysosome-targeted TORCAR (Lyso-TORCAR; Figure 5e) showed a response to PDGF (blue, ΔR/R0: 6.8 ± 0.9%, n = 11), which was largely diminished in cells treated with Dyngo-4a (gray, ΔR/R0: 1.3 ± 0.4%, n = 18, P < 0.0001) (Figure 5f–g). Consistent with the Dyngo-4a data, PDGF also induced a smaller Lyso-TORCAR response in cells overexpressing DNM2/K44A (gray, ΔR/R0: 2.3 ± 0.5%, n = 17) compared to control cells (blue, ΔR/R0: 5.3 ± 0.5%, n = 19, P < 0.001) (Figure S9e), suggesting that blocking dynamin-mediated endocytosis suppresses PDGF-induced lysosomal mTORC1 activity.
Targeted Lipid Phosphatases Perturb Lysosomal Akt/mTORC1 Signaling Activity
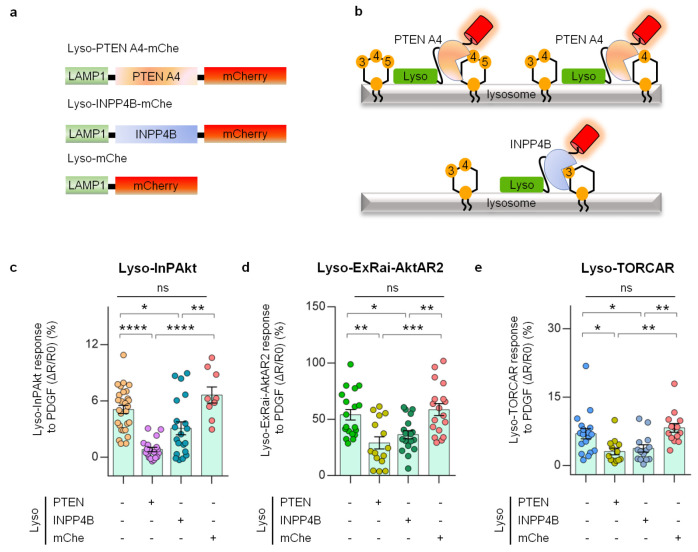
Our data so far suggest that 3-phosphoinositides generated at the plasma membrane, such as PI(3,4)P2 and PIP3, subsequently accumulate within the lysosomal membrane through dynamin-mediated lipid internalization. This leads to an increase in Akt activity at the lysosomes, which is important for mTORC1 activation at this location. To directly test our model and examine the role of lysosomal 3-PIs in regulating lysosomal Akt and its downstream target mTORC1, we set out to directly perturb 3-PIs at the lysosomes (Figure 6a). PTEN (phosphatase and tensin homologue) is a lipid phosphatase that dephosphorylates PIP3 and PI(3,4)P2 (Figure 6b).71 Mutating four residues to alanine in the C-terminal tail region (PTEN A4) prevents inhibitory phosphorylation and generates a constitutively active form of PTEN.71−73 A PTEN A4 variant localized to the lysosomes (Lyso-PTEN A4-mChe, Figure 6a) displayed proper punctate structures that colocalized with Lyso-InPAkt (Figure S10a). As shown in Figure 6c, lysosome-targeted PTEN A4 efficiently lowered lysosomal 3-PI levels, as PDGF-induced Lyso-InPAkt responses (5.1 ± 0.4%, n = 28) were robustly reduced to 0.8 ± 0.2% (n = 19, P < 0.0001), whereas no effect was observed with expression of lysosome-targeted mCherry alone (6.6 ± 0.9%, n = 9, P = 0.32). We next examined lysosomal Akt activity by measuring the responses of Lyso-ExRai-AktAR2 in the presence of lysosomal PTEN A4 (Figure S10b and Figure 6d). Lyso-ExRai-AktAR2 responses (ΔR/R0: 54 ± 4.7%, n = 19) were significantly decreased by lysosomal PTEN A4 (ΔR/R0: 29 ± 5.4%, n = 15, P < 0.01), which were unchanged by lysosomal mCherry (ΔR/R0: 59 ± 5.3%, n = 18, P = 0.89) (Figure 6d). Similar results were observed with lysosome-targeted FRET-based AktAR2 (Figure S10c–e). These results suggest that lysosomal depletion of PIP3 and PI(3,4)P2 dampens lysosomal Akt activity. Furthermore, examination of lysosomal mTORC1 activities revealed an inhibition of Lyso-TORCAR responses (ΔR/R0: 7.0 ± 1.1%, n = 18) by lysosomal PTEN A4 (ΔR/R0: 3.1 ± 0.7%, n = 13, P < 0.05), with little effect by lysosomal mCherry (ΔR/R0: 8.3 ± 0.9%, n = 14, P = 0.65) (Figure 6e). Our data suggest that the presence of 3-PIs at the lysosome is essential for lysosomal Akt and mTORC1 activities.
Figure 6.
Targeting lipid phosphatases to the lysosomes inhibits 3-phosphoinositides/Akt/mTORC1 signaling. (a) Domain structures of lysosome-targeted PTEN A4 (Lyso-PTEN A4-mChe), INPP4B (Lyso-INPP4B-mChe), and mCherry (Lyso-mChe). (b) A model depicting the conversion of 3-phosphoinositides catalyzed by lysosome-targeted PTEN A4 and INPP4B. While PTEN A4 dephosphorylates PIP3 and PI(3,4)P2, INPP4B dephosphorylates PI(3,4)P2 to PI3P. (c) Responses of Lyso-InPAkt in PDGF-treated serum-starved NIH3T3 cells without (5.1 ± 0.4%, n = 28) or with expression of Lyso-PTEN A4-mChe (PTEN, 0.8 ± 0.2%, n = 19), Lyso-INPP4B-mChe (INPP4B, 3.1 ± 0.6%, n = 20), and lysosome-targeted mCherry (mChe, 6.6 ± 0.8%, n = 9). ****, P < 0.0001, control vs Lyso-PTEN A4-mChe; ****, P < 0.0001, Lyso-mChe vs Lyso-PTEN A4-mChe; *, P = 0.0246, control vs Lyso-INPP4B-mChe; **, P = 0.0019, Lyso-mChe vs Lyso-INPP4B-mChe; ns, not significant, P = 0.3241, control vs Lyso-mChe; ordinary one-way ANOVA followed by Tukey’s multiple comparison test. (d) Responses of Lyso-ExRai-AktAR2 in PDGF-treated serum-starved NIH3T3 cells without (54 ± 4.7%, n = 19) or with expression of Lyso-PTEN A4-mChe (PTEN, 29 ± 5.4%, n = 15), Lyso-INPP4B-mChe (INPP4B, 36 ± 3.6%, n = 18), and Lyso-mChe (mChe, 58.7 ± 5.3%, n = 18). **, P = 0.003, control vs Lyso-PTEN A4-mChe; ***, P = 0.0004, Lyso-mChe vs Lyso-PTEN A4-mChe; *, P = 0.0430, control vs Lyso-INPP4B-mChe; **, P = 0.007, Lyso-mChe vs Lyso-INPP4B-mChe; ns, not significant, P = 0.89, control vs Lyso-mChe; ordinary one-way ANOVA followed by Tukey’s multiple comparison test. (e) Responses of Lyso-TORCAR in PDGF-treated serum-starved NIH3T3 cells without (7 ± 1.1%, n = 18) or with expression of Lyso-PTEN A4-mChe (PTEN, 3.1 ± 0.7%, n = 13), Lyso-INPP4B-mChe (INPP4B, 3.8 ± 0.8%, n = 14), and Lyso-mChe (mChe, 8.3 ± 0.9%, n = 14). *, P = 0.016, control vs Lyso-PTEN A4-mChe; **, P = 0.0017, Lyso-mChe vs Lyso-PTEN A4-mChe; *, P = 0.0486, control vs Lyso-INPP4B-mChe; **, P = 0.006, Lyso-mChe vs Lyso-INPP4B-mChe; ns, not significant, P = 0.65, control vs Lyso-mChe; ordinary one-way ANOVA followed by Dunnett’s multiple comparison test. Bars denote mean ± SEM (c–e).
We then ask whether the regulation of Akt/mTORC1 at the lysosome specifically involves the lipid second messenger PI(3,4)P2. INPP4B is a lipid phosphatase that specifically dephosphorylates PI(3,4)P2,74 and its localization at late endosomes/lysosomes has been recently demonstrated.75 When targeted to the lysosomes, INPP4B (Figure 6a) showed punctate structures that colocalized with Lyso-InPAkt (Figure S10a). Similar to Lyso-PTEN A4, expression of lysosome-targeted INPP4B led to a reduction in the responses of Lyso-InPAkt (3.1 ± 0.6%, n = 20, P < 0.05), which was significantly lower than the response of cells expressing Lyso-mCherry control, suggesting that targeting INPP4B to the lysosomes reduced the levels of lysosomal 3-PIs, presumably PI(3,4)P2 (Figure 6c). Lyso-INPP4B also inhibited PDGF-induced responses of Lyso-ExRai-AktAR2 (ΔR/R0: 36 ± 3.6%, n = 18, P < 0.05) (Figure S10b and Figure 6d) and the FRET-based Akt activity reporter targeted to the lysosomes (Figure S10c-e), supporting a role of lysosomal PI(3,4)P2 in promoting local Akt activity. Similarly, Lyso-TORCAR responses (7 ± 1.1%, n = 18) were suppressed by Lyso-INPP4B (3.8 ± 0.8%, n = 14, P < 0.05) but not by lysosomal mCherry (Figure S10f and Figure 6e).
Taken together, these results suggest that 3-PIs in the lysosomal membrane, specifically PI(3,4)P2, are required for lysosomal Akt activity, which in turn activates mTORC1 on this signaling platform (Figure S11). 3-PIs critically regulate membrane recruitment and activation of Akt.76 As these lipid second messengers are dynamically modulated within specific membranes through redistribution of lipids and regulation of lipid enzymes,66 their spatial compartmentalization is expected to critically contribute to the spatiotemporally distinct activation patterns of Akt.51,76,77 At the molecular level, how do lysosomal 3-PIs upregulate Akt activity at the lysosome? Akt may be directly recruited to lysosomal membrane sites of 3-PI accumulation via engagement of the Akt PH domain. 3-PI accumulation may also drive the lysosomal membrane recruitment of proteins containing PH domains such as PDK1 and the Sin1 component within the mTORC2 complex for localized Akt activation.2 In addition, Akt association with 3-PIs at the lysosomal membrane may prevent Akt dephosphorylation and deactivation.77 Alternatively, activated and phosphorylated Akt could depart from the plasma membrane78 and relocalize to the lysosomal surface via binding to 3-PIs.
Conclusions
In summary, we present ExRai-AktAR2, which exhibited high specificity and sensitivity with an improved dynamic range compared with both ExRai-AktAR (Figure 1) and the widely used FRET-based Akt activity reporter.28 Because of its subcellular targetability, ExRai-AktAR2 also outperforms translocation-based biosensors25−27 and represents the most sensitive fluorescent biosensor capable of measuring subcellular Akt activity in living cells. This new molecular tool enabled us to detect Akt activity in distinct membrane compartments, including the Golgi membrane. Complementary to imaging biochemical activities, expansion microscopy remarkably improved the spatial resolution of subcellular imaging of Akt with standard microscopes.45,46 This strategy of combining kinase biosensors for tracking subcellular activities with super-resolution imaging for mapping the spatial distribution of the kinase itself should be a generalizable and powerful approach to probe the biochemical activity architecture of the cell.16,17
Our results revealed that 3-PIs, a class of critical lipid second messengers that are predominantly present in the plasma membrane, accumulate on the lysosomal surface in response to growth factor stimulation in an endocytosis-dependent manner. The lysosomal accumulation of 3-PIs specifically promotes Akt/mTOR signaling activities at the lysosome. Given that dysregulation of this pathway underlies the pathology of many diseases such as cancer and diabetes, spatial compartmentation adds another intricate layer to the complex PI3K/Akt signaling pathway and may also provide new opportunities for selective targeting.79
Acknowledgments
This work was supported by NIH Grants R35 CA197622 and R01 GM111665 (J.Z.), and NS047101 to UCSD Microscopy Core. M.C. was supported in part by the UCSD Graduate Training Program in Cellular and Molecular Pharmacology through an institutional training grant from the National Institute of General Medical Sciences, T32 GM007752. We thank Sohum Mehta for constructive suggestions, editing, and helping create Figure 1a. We thank Ayse Z Sahan for critical reading of the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00919.
Material and methods and Figures S1–S12. (Figure S1) ExRai-AktAR2 enables selective detection of Akt kinase activity; (Figure S2) ExRai-AktAR2 is insensitive to pH change; (Figure S3) Single-cell traces of PM-ExRai-AktAR2 demonstrate cell heterogeneity, and ExRai-AktAR2 outperforms FRET-based AktAR2 in detecting Akt activities at various subcellular compartments; (Figure S4) Determination of macroscopic and microscopic expansion ratios; (Figure S5) Validation of the Akt antibody; (Figure S6) Nonexpanded NIH3T3 cells fail to show the spatial correlation information between Akt and the lysosomes; (Figure S7) Proximity ligation assay (PLA) confirms that Akt is localized to the lysosome; (Figure S8) Accumulation of 3-phosphoinositides at the plasma membrane and the lysosome upon PDGF stimulation; (Figure S9) Endocytosis blockade suppresses the activities of Akt and mTORC1 at the lysosome; (Figure S10) Lysosome-targeted lipid phosphatases perturb lysosomal Akt/mTORC1 signaling activities; (Figure S11) A model showing that lysosomal Akt/mTOR signaling activities are regulated by lysosomal 3-PIs; (Figure S12) Uncropped Western blots (PDF)
Author Contributions
# M.C. and T.S. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Sugiyama M. G.; Fairn G. D.; Antonescu C. N. Akt-ing Up Just About Everywhere: Compartment-Specific Akt Activation and Function in Receptor Tyrosine Kinase Signaling. Front. Cell Dev. Biol. 2019, 7, 7. 10.3389/fcell.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B. D.; Toker A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169 (3), 381. 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole P. A.; Chu N.; Salguero A. L.; Bae H. AKTivation mechanisms. Curr. Opin. Struct. Biol. 2019, 59, 47. 10.1016/j.sbi.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudushkin I. Getting the Akt Together: Guiding Intracellular Akt Activity by PI3K. Biomolecules 2019, 9 (2), 67. 10.3390/biom9020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli A. M.; Tabellini G.; Bressanin D.; Ognibene A.; Goto K.; Cocco L.; Evangelisti C. The emerging multiple roles of nuclear Akt. Biochim. Biophys. Acta, Mol. Cell Res. 2012, 1823 (12), 2168. 10.1016/j.bbamcr.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H.; Yoshida S.; Muroi E.; Yoshida N.; Kawamura M.; Kouchi Z.; Nakamura Y.; Sakai R.; Fukami K. Phosphoinositide 3-kinase signaling pathway mediated by p110alpha regulates invadopodia formation. J. Cell Biol. 2011, 193 (7), 1275. 10.1083/jcb.201009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Paddock M. N.; Wang H.; Murphy C. J.; Geck R. C.; Navarro A. J.; Wulf G. M.; Elemento O.; Haucke V.; Cantley L. C.; et al. The INPP4B Tumor Suppressor Modulates EGFR Trafficking and Promotes Triple-Negative Breast Cancer. Cancer Discovery 2020, 10 (8), 1226. 10.1158/2159-8290.CD-19-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D. E.; Shokat K. M. INPP4B and PTEN Loss Leads to PI-3,4-P2 Accumulation and Inhibition of PI3K in TNBC. Mol. Cancer Res. 2017, 15 (6), 765. 10.1158/1541-7786.MCR-16-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R. E.; Zoncu R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21 (2), 133. 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- Menon S.; Dibble C. C.; Talbott G.; Hoxhaj G.; Valvezan A. J.; Takahashi H.; Cantley L. C.; Manning B. D. Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome. Cell 2014, 156 (4), 771. 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E.; Koga H.; Diaz A.; Mocholi E.; Patel B.; Cuervo A. M. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol. Cell 2015, 59 (2), 270. 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Olivares D.; Kaoud T. S.; Zeng L.; Pridgen J. R.; Zhuang D. L.; Ekpo Y. E.; Nye J. R.; Telles M.; Anslyn E. V.; Dalby K. N. Quantification of ERK Kinase Activity in Biological Samples Using Differential Sensing. ACS Chem. Biol. 2020, 15 (1), 83. 10.1021/acschembio.9b00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stains C. I.; Tedford N. C.; Walkup T. C.; Lukovic E.; Goguen B. N.; Griffith L. G.; Lauffenburger D. A.; Imperiali B. Interrogating signaling nodes involved in cellular transformations using kinase activity probes. Chem. Biol. 2012, 19 (2), 210. 10.1016/j.chembiol.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oien N. P.; Nguyen L. T.; Jernigan F. E.; Priestman M. A.; Lawrence D. S. Long-wavelength fluorescent reporters for monitoring protein kinase activity. Angew. Chem., Int. Ed. 2014, 53 (15), 3975. 10.1002/anie.201309691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S.; Li Z.; Cheng H.; Wang S.; Perkins N. G.; Sarkar P.; Wei W.; Xue M. A Chemical Approach for Profiling Intracellular AKT Signaling Dynamics from Single Cells. J. Am. Chem. Soc. 2018, 140 (42), 13586. 10.1021/jacs.8b08931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Mehta S.; Zhang J. Genetically Encodable Fluorescent and Bioluminescent Biosensors Light Up Signaling Networks. Trends Biochem. Sci. 2020, 45 (10), 889. 10.1016/j.tibs.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S.; Zhang J. Biochemical Activity Architectures Visualized-Using Genetically Encoded Fluorescent Biosensors to Map the Spatial Boundaries of Signaling Compartments. Acc. Chem. Res. 2021, 54 (10), 2409. 10.1021/acs.accounts.1c00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt D. L.; Mehta S.; Zhang J. Illuminating the kinome: Visualizing real-time kinase activity in biological systems using genetically encoded fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2020, 54, 63. 10.1016/j.cbpa.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryu G.; Miura H.; Uda Y.; Komatsubara A. T.; Matsuda M.; Aoki K. Live-cell Imaging with Genetically Encoded Protein Kinase Activity Reporters. Cell Struct. Funct. 2018, 43 (1), 61. 10.1247/csf.18003. [DOI] [PubMed] [Google Scholar]
- Greenwald E. C.; Mehta S.; Zhang J. Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 2018, 118 (24), 11707. 10.1021/acs.chemrev.8b00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolbat A.; Schultz C. Recent developments of genetically encoded optical sensors for cell biology. Biology of the cell 2017, 109 (1), 1. 10.1111/boc.201600040. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Li S.; Zhang J. Tracking the Activity of mTORC1 in Living Cells Using Genetically Encoded FRET-based Biosensor TORCAR. Current protocols in chemical biology 2016, 8 (4), 225. 10.1002/cpch.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.; Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol. Biol. Cell 2008, 19 (10), 4366. 10.1091/mbc.e08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel M. T.; Ni Q.; Tsien R. Y.; Zhang J.; Newton A. C. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 2005, 280 (7), 5581. 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S. M.; Rotwein P. Akt signaling dynamics in individual cells. J. Cell Sci. 2015, 128 (14), 2509. 10.1242/jcs.168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S. M.; Rotwein P. Mapping growth-factor-modulated Akt signaling dynamics. J. Cell Sci. 2016, 129 (10), 2052. 10.1242/jcs.183764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryu G.; Matsuda M.; Aoki K. Multiplexed Fluorescence Imaging of ERK and Akt Activities and Cell-cycle Progression. Cell Struct. Funct. 2016, 41 (2), 81. 10.1247/csf.16007. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Clister T. L.; Lowry P. R.; Seldin M. M.; Wong G. W.; Zhang J. Dynamic Visualization of mTORC1 Activity in Living Cells. Cell Rep. 2015, 10 (10), 1767. 10.1016/j.celrep.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S.; Zhang Y.; Roth R. H.; Zhang J. F.; Mo A.; Tenner B.; Huganir R. L.; Zhang J. Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat. Cell Biol. 2018, 20 (10), 1215. 10.1038/s41556-018-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509. 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Hires S. A.; Zhu Y.; Tsien R. Y. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (11), 4411. 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L.; Hires S. A.; Mao T.; Huber D.; Chiappe M. E.; Chalasani S. H.; Petreanu L.; Akerboom J.; McKinney S. A.; Schreiter E. R.; et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 2009, 6 (12), 875. 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. F.; Liu B.; Hong I.; Mo A.; Roth R. H.; Tenner B.; Lin W.; Zhang J. Z.; Molina R. S.; Drobizhev M.; et al. An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nat. Chem. Biol. 2021, 17 (1), 39. 10.1038/s41589-020-00660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross B. L; Tenner B.; Markwardt M. L; Zviman A.; Shi G.; Kerr J. P; Snell N. E; McFarland J. J; Mauban J. R; Ward C. W; Rizzo M. A; Zhang J. Single-color, ratiometric biosensors for detecting signaling activities in live cells. eLife 2018, 7, 7. 10.7554/eLife.35458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H.; Matsuda M.; Aoki K. Development of a FRET biosensor with high specificity for Akt. Cell Struct. Funct. 2014, 39 (1), 9. 10.1247/csf.13018. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan B.; Ni Q.; Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (42), 15081. 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos L. L.; Kunkel M. T.; Newton A. C. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J. Biol. Chem. 2006, 281 (41), 30947. 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- Miyamoto T.; Rho E.; Sample V.; Akano H.; Magari M.; Ueno T.; Gorshkov K.; Chen M.; Tokumitsu H.; Zhang J.; et al. Compartmentalized AMPK signaling illuminated by genetically encoded molecular sensors and actuators. Cell Rep. 2015, 11 (4), 657. 10.1016/j.celrep.2015.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Zhong Y.; Molinar-Inglis O.; Kunkel M. T.; Chen M.; Sun T.; Zhang J.; Shyy J. Y.-J.; Trejo J.; Newton A. C.; Zhang J.; et al. Location specific Inhibition of Akt reveals regulation of mTORC1 activity in the nucleus. Nat. Commun. 2020, 11, 6088. 10.1038/s41467-020-19937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein M.; Piegari E.; Martinez Calejman C.; Vila A.; Amante A.; Manese M. V.; Zeida A.; Abrami L.; Veggetti M.; Guertin D. A.; et al. Akt Is S-Palmitoylated: A New Layer of Regulation for Akt. Front. Cell Dev. Biol. 2021, 9, 626404. 10.3389/fcell.2021.626404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A. M.; Toomre D.; Bogan J. S. Acylation - A New Means to Control Traffic Through the Golgi. Front. Cell Dev. Biol. 2019, 7, 109. 10.3389/fcell.2019.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnel W.Color Atlas of Cytology, Histology, and Microscopic Anatomy, 4th ed.; Thieme, 2003. [Google Scholar]
- Weiss S. Shattering the diffraction limit of light: a revolution in fluorescence microscopy?. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (16), 8747. 10.1073/pnas.97.16.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chozinski T. J.; Halpern A. R.; Okawa H.; Kim H. J.; Tremel G. J.; Wong R. O.; Vaughan J. C. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 2016, 13 (6), 485. 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg P. W.; Chen F. Expansion Microscopy: Scalable and Convenient Super-Resolution Microscopy. Annu. Rev. Cell Dev. Biol. 2019, 35, 683. 10.1146/annurev-cellbio-100818-125320. [DOI] [PubMed] [Google Scholar]
- Wassie A. T.; Zhao Y.; Boyden E. S. Expansion microscopy: principles and uses in biological research. Nat. Methods 2019, 16 (1), 33. 10.1038/s41592-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.; Tillberg P. W.; Boyden E. S. Optical imaging. Expansion microscopy. Science (Washington, DC, U. S.) 2015, 347 (6221), 543. 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Tong Z.; Ye Q.; Sun T.; Hong Z.; Zhang L.; Bortnick A.; Cho S.; Beuzer P.; Axelrod J.; et al. Molecular organization of mammalian meiotic chromosome axis revealed by expansion STORM microscopy. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (37), 18423. 10.1073/pnas.1902440116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S. M.; Gao R.; Wassie A. T.; Tillberg P. W.; Chen F.; Boyden E. S. Expansion Microscopy: Protocols for Imaging Proteins and RNA in Cells and Tissues. Current protocols in cell biology 2018, 80 (1), e56 10.1002/cpcb.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaron J. S.; Taylor A. B.; Chew T. L.. Image co-localization - co-occurrence versus correlation. J. Cell Sci. 2018, 131 ( (3), ). 10.1242/jcs.211847 [DOI] [PubMed] [Google Scholar]
- Liu S. L.; Wang Z. G.; Hu Y.; Xin Y.; Singaram I.; Gorai S.; Zhou X.; Shim Y.; Min J. H.; Gong L. W.; et al. Quantitative Lipid Imaging Reveals a New Signaling Function of Phosphatidylinositol-3,4-Bisphophate: Isoform- and Site-Specific Activation of Akt. Mol. Cell 2018, 71 (6), 1092. 10.1016/j.molcel.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunemann M.; Frank M.; Lohse M. J. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (26), 16077. 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H.; Chung M.; Dobrzynski M.; Fey D.; Blum Y.; Lee S. S.; Peter M.; Kholodenko B. N.; Jeon N. L.; Pertz O. Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 2015, 11 (11), 838. 10.15252/msb.20156458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R.; Wu J.; Ouyang M.; Lei L.; Wei J.; Peng Q.; Harrison R.; Wu Y.; Cheng B.; Li K.; Zhu C.; Tang L.; Wang Y.; Lu S. Biophysical basis underlying dynamic Lck activation visualized by ZapLck FRET biosensor. Science advances 2019, 5 (6), eaau2001 10.1126/sciadv.aau2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes J.; Ganesan A.; Molinar-Inglis O.; Hamidzadeh A.; Zhang J.; Ling M.; Trejo J.; Levchenko A.; Zhang J. Signaling diversity enabled by Rap1-regulated plasma membrane ERK with distinct temporal dynamics. eLife 2020, 9, 9. 10.7554/eLife.57410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M.; Chen P. H.; Srinivasan S.; Danuser G.; Schmid S. L. Regulation of Clathrin-Mediated Endocytosis. Annu. Rev. Biochem. 2018, 87, 871. 10.1146/annurev-biochem-062917-012644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E.; Ferreira A. P.; Almeida-Souza L.; Debard S.; Vallis Y.; Howard G.; Bertot L.; Sauvonnet N.; McMahon H. T. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 2015, 517 (7535), 460. 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- Casamento A.; Boucrot E. Molecular mechanism of Fast Endophilin-Mediated Endocytosis. Biochem. J. 2020, 477 (12), 2327. 10.1042/BCJ20190342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H.; Baba T.; Warnock D. E.; Schmid S. L. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 1994, 127 (4), 915. 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey A.; Daniel J. A.; Hadzic G.; Chau N.; Clayton E. L.; Mariana A.; Whiting A.; Gorgani N. N.; Lloyd J.; Quan A.; et al. Building a better dynasore: the dyngo compounds potently inhibit dynamin and endocytosis. Traffic (Oxford, U. K.) 2013, 14 (12), 1272. 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M. J.; Deane F. M.; Robinson P. J.; McCluskey A. Synthesis of Dynole 34–2, Dynole 2–24 and Dyngo 4a for investigating dynamin GTPase. Nat. Protoc. 2014, 9 (4), 851. 10.1038/nprot.2014.046. [DOI] [PubMed] [Google Scholar]
- Czech M. P. Dynamics of phosphoinositides in membrane retrieval and insertion. Annu. Rev. Physiol. 2003, 65, 791. 10.1146/annurev.physiol.65.092101.142522. [DOI] [PubMed] [Google Scholar]
- Jethwa N.; Chung G. H.; Lete M. G.; Alonso A.; Byrne R. D.; Calleja V.; Larijani B. Endomembrane PtdIns(3,4,5)P3 activates the PI3K-Akt pathway. J. Cell Sci. 2015, 128 (18), 3456. 10.1242/jcs.172775. [DOI] [PubMed] [Google Scholar]
- Hayakawa M.; Kaizawa H.; Kawaguchi K.; Ishikawa N.; Koizumi T.; Ohishi T.; Yamano M.; Okada M.; Ohta M.; Tsukamoto S.; et al. Synthesis and biological evaluation of imidazo[1,2-a]pyridine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg. Med. Chem. 2007, 15 (1), 403. 10.1016/j.bmc.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Marat A. L.; Wallroth A.; Lo W. T.; Muller R.; Norata G. D.; Falasca M.; Schultz C.; Haucke V. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science (Washington, DC, U. S.) 2017, 356 (6341), 968. 10.1126/science.aaf8310. [DOI] [PubMed] [Google Scholar]
- Dickson E. J.; Hille B. Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J. 2019, 476 (1), 1. 10.1042/BCJ20180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallroth A.; Haucke V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 2018, 293 (5), 1526. 10.1074/jbc.R117.000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B. D.; Tee A. R.; Logsdon M. N.; Blenis J.; Cantley L. C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 2002, 10 (1), 151. 10.1016/S1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Cross D. A.; Alessi D. R.; Cohen P.; Andjelkovich M.; Hemmings B. A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995, 378 (6559), 785. 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Beurel E.; Grieco S. F.; Jope R. S. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114. 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F.; Ramaswamy S.; Nakamura N.; Sellers W. R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 2000, 20 (14), 5010. 10.1128/MCB.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. H.; Gericke A. Phosphorylation keeps PTEN phosphatase closed for business. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (5), 1297. 10.1073/pnas.0812473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar M.; Inoue T.; Meyer T.; Zhang J.; Vazquez F.; Devreotes P. N. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (2), 480. 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulden B. D.; Pacheco J.; Dull A.; Zewe J. P.; Deiters A.; Hammond G. R. V. A high-avidity biosensor reveals plasma membrane PI(3,4)P2 is predominantly a class I PI3K signaling product. J. Cell Biol. 2019, 218 (3), 1066. 10.1083/jcb.201809026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers S. J.; Ooms L. M.; Oorschot V. M. J.; Schittenhelm R. B.; Nguyen E. V.; Hamila S. A.; Rynkiewicz N.; Gurung R.; Eramo M. J.; Sriratana A.; Fedele C. G.; Caramia F.; Loi S.; Kerr G.; Abud H. E.; Ramm G.; Papa A.; Ellisdon A. M.; Daly R. J.; McLean C. A.; Mitchell C. A. INPP4B promotes PI3Kalpha-dependent late endosome formation and Wnt/beta-catenin signaling in breast cancer. Nat. Commun. 2021, 12 (1), 3140. 10.1038/s41467-021-23241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. R.; Downes C. P.; Gigg R.; Grove S. J.; Holmes A. B.; Alessi D. R. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem. J. 1996, 315 (3), 709. 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner M.; Lucic I.; Leonard T. A.; Yudushkin I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol. Cell 2017, 65 (3), 416. 10.1016/j.molcel.2016.12.028. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan B.; Fosbrink M.; Rahdar M.; Zhang J. Live-cell molecular analysis of Akt activation reveals roles for activation loop phosphorylation. J. Biol. Chem. 2007, 282 (50), 36634. 10.1074/jbc.M706227200. [DOI] [PubMed] [Google Scholar]
- Davis W. J.; Lehmann P. Z.; Li W. Nuclear PI3K signaling in cell growth and tumorigenesis. Front. Cell Dev. Biol. 2015, 3, 24. 10.3389/fcell.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.