Summary
I-motifs are noncanonical four-stranded DNA structures formed by C-rich sequences at acidic environment with critical biofunctions. The particular pH sensitivity has inspired the development of i-motifs as pH sensors and DNA motors in nanotechnology. However, the folding and regulation mechanisms of i-motifs remain elusive. Here, using single-molecule FRET, we first show that i-motifs are more dynamic than G4s. Impressively, i-motifs display a high diversity of six folding species with slow interconversion. Further results indicate that i-motifs can be linearized by Replication protein A. More importantly, we identified a number of helicases with high specificity to i-motifs at low pH. All these helicases directly act on and efficiently resolve i-motifs into intermediates independent of ATP, although they poorly unwind G4 or duplex at low pH. Owing to the extreme sensitivity to helicases and no need for ATP, i-motif may be applied as a probe for helicase sensing both in vitro and in vivo.
Subject areas: Biological sciences, Biochemistry, Molecular biology, Structural biology
Graphical abstract
Highlights
-
•
I-motifs display a high diversity of six folding species with slow interconversion
-
•
Replication protein A efficiently linearizes i-motifs at acidic pH
-
•
Helicases specifically bind to and unfold i-motifs at acidic pH independent of ATP
-
•
I-motif can be used as a convenient probe for helicase sensing in vitro
Biological sciences; Biochemistry; Molecular biology; Structural biology
Introduction
DNA can adopt various noncanonical structures, for instance, G-quadruplex (G4) and i-motif, in addition to the classical double-helix form (Wells, 2007). The four-stranded G4s are held together by Hoogsteen bonds and function in the initiation of DNA replication, regulation of DNA transcription, and telomere maintenance (Bochman et al., 2012). Although G4s have been extensively studied, the i-motifs in their complementary strands are still mysterious (Abou Assi et al., 2018). Different from G4s, i-motifs are formed via the stack of intercalating hemi-protonated C+: C base pairs at the acidic condition (Gehring et al., 1993). The in-cell NMR spectra showed that i-motifs are stable and persistent in the nuclei of living human cells (Dzatko et al., 2018). Besides, by using an antibody iMab, the formation of i-motifs was demonstrated in a cell-cycle and pH-dependent manner (Zeraati et al., 2018). The i-motif sequences are most enriched in gene promoters and telomeric regions, and owing to their essential roles in oncogene regulation, i-motifs have been considered as the targets for anticancer therapy (Day et al., 2014). Relying on their pH responsiveness, easy accessibility to intracellular spaces, and relatively low toxicity, i-motif sequences have also been extensively used in DNA nanotechnology (Alba et al., 2016; Dong et al., 2014). For instance, the i-motif-based nanomachines could report the spatial and temporal pH changes inside the cell (Modi et al., 2009); the i-motif nanocontainers could control the drug release in cells (Dembska, 2016). Therefore, investigating the folding mechanism of i-motifs and their regulations by specific proteins has a high significance for both the understanding and application of i-motifs.
At present, the in-depth understanding of the i-motif folding mechanism is still lacking; however, previous studies on G4s may provide valuable clues. G4s can adopt a variety of folding topologies with thermal stability mainly depending on monovalent ions (Bochman et al., 2012). During the dynamic G4 folding/unfolding process, the intermediate states like G-triplexes and G-hairpins were proposed (Hou et al., 2017). Some G4s also go through the dynamic transitions between the distinct topologies (Dai et al., 2008). However, different from G4s, the most important determinant for i-motif formation is pH (Choi et al., 2011). As the pH sensitivity makes i-motifs a powerful nanodevice (Dong et al., 2014), exploring the dynamic structural changes in i-motif DNA at different pH becomes very important. In recent years, single-molecule-level studies have captured the dynamic folding/unfolding of i-motifs in equilibrium with other unknown folding species (Kim et al., 2014; Dhakal et al., 2010; Megalathan et al., 2019; Choi et al., 2011; Paul et al., 2020; Cui et al., 2014). Unfortunately, these studies were performed under different experimental conditions and using different DNA sequences; therefore, controversies still exist about the number and the nature of diverse i-motif folding conformations, suggesting that the folding dynamics of i-motifs are far from being understood.
A large number of proteins are required to remove the folded G4 barriers and maintain the progression of DNA replication or transcription, among which single-stranded DNA binding proteins (SSBs) and specialized DNA helicases such as BLM, DHX36, and Pif1 have been extensively studied (Mendoza et al., 2016; Salas et al., 2006; Wang et al., 2021). In sharp contrast, little is known about the proteins specifically interacting with i-motifs (Abou Assi et al., 2018). HnRNP LL is one of the best-studied proteins that can bind and destabilize the i-motif in BCL2 promoter, and thereby activate BCL2 transcription (Kang et al., 2014). Together with poly-C-binding proteins hnRNP K, BmILF, αCP1-4, and αCPKL, the above-mentioned are almost all the i-motif interacting proteins that have been established so far (Yoga et al., 2012; Marsich et al., 1996, 1998; Lacroix et al., 2000; Niu et al., 2018). More importantly, how the folding conformation of i-motif DNA is regulated by these proteins remains elusive. It is worth noting that a recent study linked the formation of G4s and i-motifs in the helicase-polymerase uncoupling sites with the replicative stress and suggested the requirement of DNA repair (Amparo et al., 2020). We know that helicases participate in almost every aspect of DNA metabolism; particularly, some helicases are responsible for resolving the complicated DNA structures like G4s and Holliday junctions (Sharma, 2011). In addition, the function loss of dog-1 helicase in C. elegans was reported to produce the selective deletion mutagenesis at persistent G4s and i-motifs sites (Kruisselbrink et al., 2008). All these facts then motivated us to examine whether and how helicases may play roles in regulating i-motif DNA.
In this report, we focused on the conformational dynamics of i-motif DNA using single-molecule fluorescence resonance energy transfer (smFRET), which allows us to accurately monitor the folding/unfolding of i-motifs at the molecular level. We first looked at the intrinsic folding dynamics of i-motif DNA, and on this basis, we then examined their regulations by the specific proteins including Replication protein A (RPA) and helicases. The i-motif from BCL2 oncogene (referred to as Bcl2-IM) was used predominantly. Human telomere and insulin-linked polymorphic region (ILPR) i-motifs (referred to as hTel-IM and ILPR-IM) were also tested as they originate from different genomic regions and are linked to different biological/disease functionalities. In brief, we discovered that i-motifs are more dynamic than G4s at different pH, which is consistent with previous studies (Dai et al., 2010; Kendrick et al., 2014). Furthermore, we found that i-motifs display a high conformational diversity of up to six folding states with slow interconversion at the single-molecule level. More importantly, we identified several helicases that specifically bind to and unfold i-motifs at acidic environment in a unique ATP-independent mode. A competition assay was also performed to define the helicase behavior in the presence of both i-motif and G4 DNA. To our knowledge, this is the first report to address the interaction between i-motif and protein at the single-molecule level; therefore, it may provide new insights into our understanding about the regulation mechanisms of i-motifs and also broaden our knowledge about the functioning of DNA helicases. Finally, we showed that i-motif can be used as a sensitive probe to detect helicase in vitro and discussed some potential applications of the protein regulation of i-motif in nanotechnology.
Results and discussion
I-motifs are more dynamic than G4s from the acidic to basic environments
In DNA replication or transcription, the transient exposure of G-rich DNA sequences can fold into G4 structures. Meanwhile, the complementary C-rich strands may fold into the i-motif structures. In sharp contrast to massive knowledge about G4 DNA, how i-motif DNA dynamically fold/unfold at different pH and how they are regulated by proteins were still poorly understood. Therefore, at the beginning of this study, we carried out a comparative analysis on the folding dynamics of i-motif and G4, respectively. Phosphate buffer with 100 mM KCl was used.
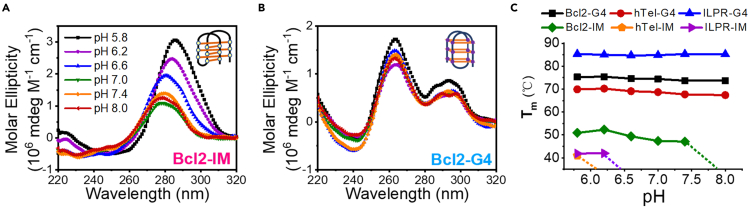
We first examined the formation of i-motifs and the corresponding G4s from pH 5.8 to 8.0 by CD spectrum. Figure 1A shows that, at acidic condition, Bcl2-IM folds into the typical i-motif topology with a peak at ∼285 nm and a valley at ∼255 nm, which is identical to the previous studies (Wright et al., 2017; Ruggiero et al., 2019; Nguyen et al., 2017) but different from either the mutant Bcl2-IM sequence (referred to as Bcl2-Mut) or the random ssDNA (Figures S1A and S1B). With the increases in pH, the peak positions shift to the left accompanied by the decreases in magnitudes, suggesting the gradual disassembly of i-motif structures. The spectrum and changes in hTel-IM and ILPR-IM are consistent with Bcl2-IM (Figures S2A and S2B). However, only mild changes can be observed in G4 DNA (Figures 1B, S2C, and S2D), reflecting the stable folding of G4 structures. The thermal stability of these i-motifs and G4s were further determined by the FRET-melting assay (De Cian et al., 2007). Figure 1C indicates that G4s are far more stable than i-motifs in the same buffer. More importantly, G4s display very little Tm change over a wide pH range; however, the stability of i-motifs decreases with the increases in pH. In particular, the Tm of hTel-IM and ILPR-IM can only be determined at very acidic conditions.
Figure 1.
I-motif and G4 DNA respond differently to the changes in pH
(A and B) CD spectrum of i-motif DNA and G4 DNA formed at different pH with the sequences from BCL2 promoter.
(C) Tm values determined by the FRET-melting assay. The dashed line indicates that the Tm cannot be calculated above this pH value, since at higher pH levels, there may not be well-folded i-motif structures and consequently, the transition between the folded and the unfolded state is lost.
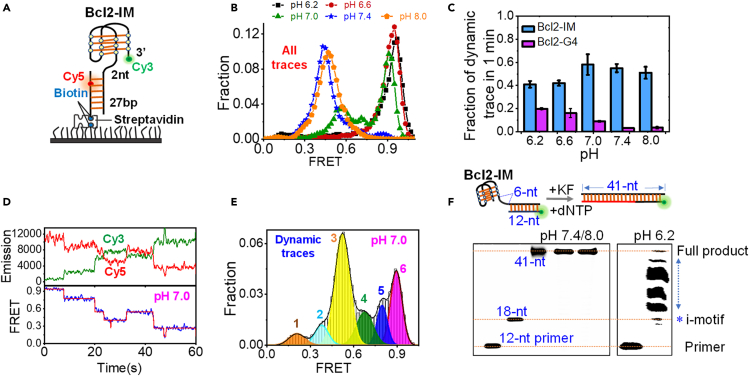
Then we further addressed the dynamic folding/unfolding of Bcl2-IM at the single-molecule level. The schematic design of the substrate was shown in Figure 2A. Cy3 was attached to the 3′-end of i-motif and Cy5 was located at the sixth nucleotide inside the duplex. The fluorophores were so spaced that the FRET signal can sensitively report the conformational change of i-motif. At pH 6.2, the FRET distribution from all Bcl2-IM traces displays a single peak at ∼ E0.95, reflecting the uniform folding of i-motif structures (Figure 2B). With the increases in pH, the FRET distributions significantly shift to the left and finally center at ∼E0.45, suggesting the disassembly of i-motif structures at higher pH. The representative traces at different pH were shown in Figure S3. As a control, Bcl2-Mut has little change with the increases in pH (Figures S1C and S1D). The Bcl2-G4 also shows the insensitivity to pH (Figure S4). Then we further analyzed the fractions of FRET traces showing the dynamic changes within 1 min in the above structures (Figure 2C). Most G4s are static from pH 6.2 to 8.0; however, 40%–60% i-motifs are dynamic, particularly at neutral or alkaline conditions. Altogether, the above results demonstrate that i-motif structures are more dynamic than the complementary G4s.
Figure 2.
I-motifs are more dynamic than the complementary G4s
(A) The schematic design of smFRET experiment for Bcl2-IM.
(B) FRET distributions of Bcl2-IM at different pH. All traces were included whether it was static or dynamic.
(C) The fractions of FRET traces showing the dynamic changes during the 1-min recording time. Data are represented as mean ± SEM.
(D) The representative dynamic traces of Bcl2-IM at pH 7.0. The emission intensity was in arbitrary unit.
(E) FRET distribution of the dynamic Bcl2-IM traces at pH 7.0.
(F) Analysis of the KF replication reactions with Bcl2-IM template at different pH. DNA template was 1 μM, and 200 nM KF was used. The bottom line shows the position of the 12-nt primer. The other two lines are the 18- and 41-nt FAM labeled sequences that mimic the polymerization of 6-nt poly-T linker only and the full product, respectively (Table S1); therefore, the asterisk should indicate the position of the stalled product at i-motif. At pH 7.4 and pH 8.0, Bcl2-IM can be completely polymerized by 200 nM KF. However, at pH 6.2, four intermediate bands can be observed.
I-motifs display the high conformational diversity of up to six folding states
To further probe the probable folding status of i-motif DNA, we then selected the dynamic Bcl2-IM traces at pH 7.0 as shown in Figure 2D, in which multiple FRET states can be distinguished with a slow transition at the ∼10 s level. Then, we combined all those dynamic traces and plot the FRET distribution in Figure 2E. As a result, six FRET peaks that may reflect the six folding states of i-motif DNA can be identified. Therefore, besides the completely folded i-motif state at the highest FRET and the ssDNA state at the lowest FRET, four intermediate states may exist. Figure S5 further indicates that possessing multiple folding states is a common phenomenon in different i-motif sequences.
We also employed Klenow fragment (KF) polymerase stop assay to dissect the Bcl2-IM folding status. In the template (Figure 2F), Bcl2-IM was linked to the region complementary to the FAM-labeled primer (Takahashi et al., 2017), enabling us to discriminate the stalled product from the unreacted primer and the full-length product. The concentration of DNA template was 1 μM and 200 nM KF was used. The top band in the gel should be the full product and the asterisk indicates the stop site at i-motif. At the i-motif destabilizing condition pH 7.4 and pH 8.0, Bcl2-IM was completely polymerized by KF; however, at pH 6.2, four middle bands show up, suggesting the blocking of KF by the partial i-motif structures. The analysis on hTel-IM and ILPR-IM were also performed (Figures S6A and S6B). At pH 6.2, 200 nM KF almost completely overcomes and polymerizes those two structures possibly owing to the lower stability than Bcl2-IM. When the KF concentration was decreased to 50 or 100 nM, four intermediate bands can be observed in ILPR-IM, in agreement with the result of Bcl2-IM. Fewer middle bands appear in hTel-IM, consistent with its lowest stability among the three i-motifs.
To probe the possible forms of intermediate states, we designed a series of partial i-motif sequences with two or three C-tracts (Figure S7A). The sequences from human telomere were selected owing to the uniform loop lengths. In general, higher-order structures were formed and five FRET states can be identified totally (Figures S7B and S7C). Except that the i-motif state at E0.95 is missing, other states at ∼ E0.8, ∼E0.7, ∼E0.5, ∼E0.4, ∼E0.25 can all be found in both the original i-motif and the modified sequences (Figures S7B and S5D), in which the ∼E0.25 should reflect the ssDNA form. According to the consistency in FRET values, we speculate that the i-motif folding intermediates may be formed by those partial i-motif sequences via C+: C base pairs. We also noticed that, besides the ssDNA state at ∼ E0.25, there are two peaks in each FRET histogram, indicating that different types of C-hairpins are formed in the same sequence. Although we could not determine exactly what C-hairpin each FRET state may represent, based on G-hairpins (Stadlbauer et al., 2015, 2019), some speculated C-hairpins were shown in Figure S7D. One may query whether these different proposed conformations may stall elongation in the KF stop assay in the same way as the natural sequences. For this purpose, we selected the hTel-m2 and hTel-m4 for test (Figures S6C and S6D). Indeed, some middle bands show up below the full-length product and above the stop site after the polymerization of 6-nt linker in both substrates. These evidences suggest that higher-order structures may form in the partial i-motif sequences, consistent with the smFRET observations in Figure S7.
Taken together, the above-mentioned results indicate that i-motif sequences may fold into a variety of different conformations in a pH-dependent manner. Our recently published results further showed that i-motif sequences fold into different status at different ion concentrations (Gao and Hou, 2021). All those evidences suggest that i-motif is a kind of nucleic acid structure very sensitive to buffer conditions. Therefore, it is likely that the i-motif sequences inside the cell may adopt various conformations such as complete i-motif, different forms of C-hairpins, and ssDNA based on the ambient environment.
I-motif structures can be easily disrupted by RPA into the linear state
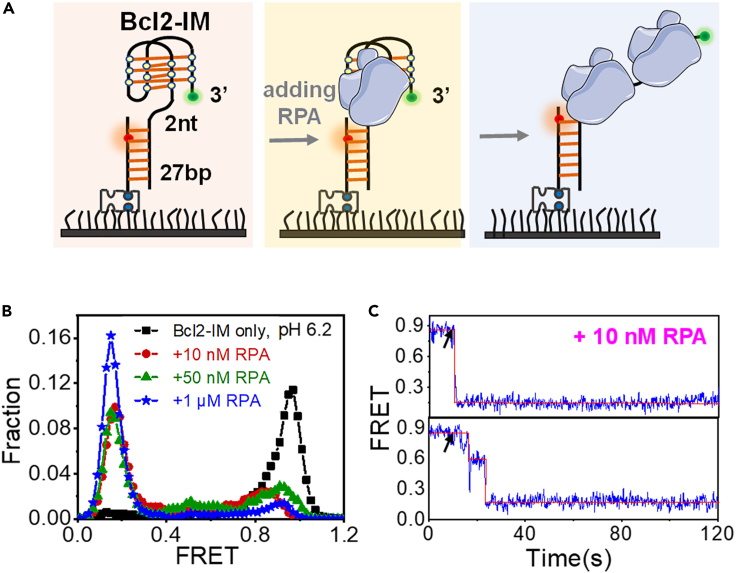
After characterizing the intrinsic folding dynamics of i-motif DNA, we next aimed to look for the proteins that can finely regulate the folding status of i-motif. As the most abundant and ubiquitous single-strand DNA binding protein inside the eukaryotic cell, RPA has been reported to unfold the human telomeric G4 DNA (Salas et al., 2006). Therefore, we then examined the interaction between RPA and Bcl2-IM using the substrate in Figure 3A. We prepared the DNA substrate at pH 6.2 under which condition the Bcl2-IM can be well folded in all the following experiments. To check the DNA binding activity of RPA at the acidic condition, we first performed analysis on the interaction between Bcl2-Mut and RPA at pH 6.2 as a control. As shown in Figure S8, after adding RPA, the FRET distribution thoroughly shifted to the left at ∼ E0.15, reflecting that even 10 nM RPA can bind to and stretch Bcl2-Mut at low pH. Next, different concentrations of RPA were added to Bcl2-IM. At 10 nM RPA, the FRET distribution of Bcl2-IM shifted significantly to the left from ∼E0.95 to ∼ E0.15, indicating the unfolding and linearization of i-motif DNA (Figure 3B). As the RPA concentration increased to 1 μM, most of i-motifs were disrupted. Figure 3C further demonstrated the selective FRET traces of Bcl2-IM with the addition of 10 nM RPA. Both one-step (69.4%) and two-step (30.6%) decreases can be observed, and the final FRET level was maintained at ∼E0.15 reflecting the stable unfolding of i-motifs. Since the sequence length of Bcl2-IM is ∼20 nt, one or two RPA molecules may sequentially associate with the unfolded ssDNA sequence, displaying the stepwise decreases in FRET traces (Wang et al., 2019).
Figure 3.
RPA effectively disrupts the well-folded Bcl2-IM
(A) The experimental design to characterize the interaction between i-motif and RPA. The concentration of DNA substrate in the microfluidic chamber was 50 pM. After DNA immobilization, 10 nM, 50 nM, or 1 μM RPA was added. Therefore, the minimum ratio between RPA and DNA was about 1:200. However, as the DNA was anchored at the coverslip surface with less degree of freedom, the interaction efficiency between RPA and DNA on the surface may be lower than in the free solution.
(B) FRET distributions of Bcl2-IM at pH 6.2 before and 4 min after the addition of RPA.
(C) The representative FRET traces with the addition of 10 nM RPA. The black arrows indicate the addition of protein.
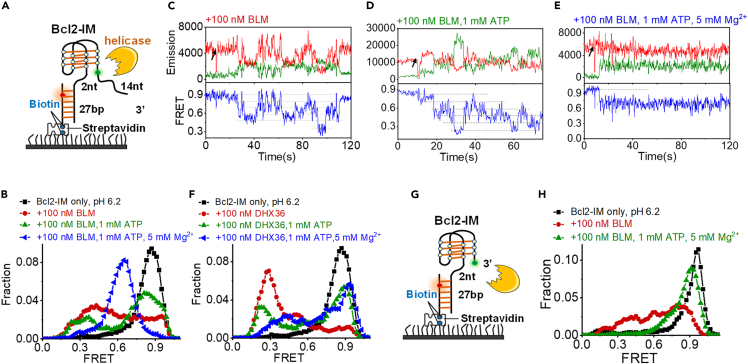
Helicases show much higher specificity to i-motif than to other DNA structures at the acidic environment
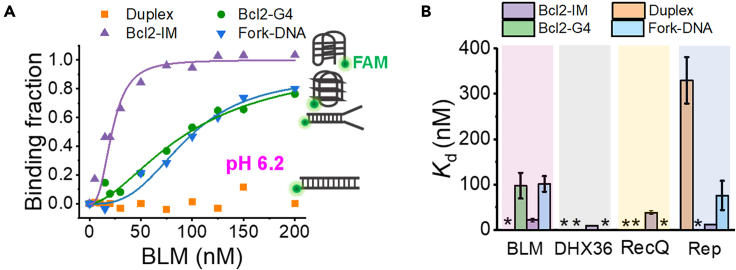
After addressing the interaction between RPA and i-motifs, we further examined whether helicases can regulate the folding status of i-motif DNA. To this end, the binding affinity between i-motif structures and the 3′-5′ helicases was measured first using the fluorescence anisotropy method (Dou and Xi, 2010). Four different types of DNA constructs including i-motif, G4, fork DNA, and duplex DNA were tested. To our surprise, BLM helicase binds to the i-motif structure with a much higher affinity than to other DNA structures at low pH (Figure 4A). Specifically, the Kd value of BLM to i-motif was 22.0 nM, much lower than the 97.7 nM to G4 and the 101.4 nM to fork DNA (Figure 4B). The binding of BLM to duplex DNA even cannot be detected. In addition to BLM, several other helicases, including DHX36, RecQ, and Rep, display the high specificity to i-motif structure at the acidic pH (Figures 4B and S9); however, they poorly recognize other types of DNA structures. The above evidences suggest that these DNA helicases may potentially interact with i-motif structures and regulate the folding status of i-motif DNA.
Figure 4.
Helicases bind to i-motif with high specificity at the acidic environment
(A) The changes of DNA binding fractions with the increases in BLM concentrations. The binding curve was fitted by the Hill equation: y = [protein]n/(Kdn+[protein]n), where y is the binding fraction, n is the Hill coefficient, and Kd is the apparent dissociation constant.
(B) The Kd resulted from the fitting. Data are represented as mean ± SEM. ∗ denotes the binding curves were not available or could not be fitted owing to the poor affinity. The corresponding values were placed in Table S2.
Helicases efficiently unfold i-motif structures into intermediates in an ATP-independent mode at low pH
We further examined whether the above helicases that specifically recognize i-motifs can also resolve this complex structure. For this purpose, we designed the substrate shown in Figure 5A and recorded the folding/unfolding of Bcl2-IM in the presence of helicases. There was an additional 3′-ssDNA tail placed adjacent to Bcl2-IM for the helicases to associate with based on the traditional translocation model of DNA helicases (Lohman and Bjornson, 1996).
Figure 5.
Helicases mediate the unfolding of Bcl2-IM at low pH
(A) The experimental design to characterize the unfolding of Bcl2-IM by the 3′-5′ DNA helicases.
(B) FRET distributions of Bcl2-IM before and 4 min after the addition of BLM at the apo, ATP-binding, and ATP hydrolysis states.
(C–E) The representative fluorescence emission and FRET traces of Bcl2-IM. (F) FRET distributions of Bcl2-IM before and 4 min after the addition of DHX36 at different ATP states.
(G) The substrate design of Bcl2-IM without the ssDNA loading tail.
(H) FRET distributions of Bcl2-IM before and 4 min after the addition of BLM at the apo and ATP hydrolysis states.
Unexpectedly, after adding only BLM without ATP, FRET distribution of Bcl2-IM significantly shifted to the left with a broad band (Figure 5B), reflecting the complete or partial unfolding of Bcl2-IM. Figure 5C shows the quick transitions of Bcl2-IM between the multiple intermediate states in the presence of BLM. The dashed lines further marked the six discrete states of i-motif DNA. Both these two evidences suggested that BLM can unfold i-motif structures in an ATP-independent mode. In addition to BLM, the addition of DHX36, RecQ, or Rep also induces a significant left shift in the FRET distributions of Bcl2-IM (Figures 5F and S10), reflecting a similar ATP-independent unfolding mode. The traces in the left panels in Figures S11A–S11C further reveal that in the presence of DHX36, RecQ, and Rep, Bcl2-IM makes conversions between several discrete states, which is similar to the transition of Bcl2-IM in BLM.
As ATP is not necessary for unfolding i-motifs by the above-shown helicases, we then aimed to address whether these helicases directly act on the i-motif structures without ATP-driven translocation on the loading tail (Figure 5G). Figures 5H and S12A–S12D revealed that the 3′-ssDNA tail is dispensable for the above-mentioned helicases to resolve the i-motif structures, as the shift of FRET distributions to the lower band can be observed in the presence of BLM, DHX36, RecQ, and Rep. The representative traces in Figures S12F–S12I also show the similar fluctuations as that in Bcl2-IM with the ssDNA tail. Therefore, helicases may directly act on i-motifs, inducing the disruption of i-motifs by the conformational changes in proteins during the binding process. A previous study on hnRNP LL suggested that i-motif is the most kinetically favorable conformation for protein binding (Kang et al., 2014) and the unfolding of i-motif upon protein binding provides the more thermodynamically favorable single-stranded form. Therefore, we speculated that the ATP-independent mode may be a common mechanism shared by the i-motif binding proteins possibly related to the peculiar folding topology of i-motif.
ATP differentially regulates the activity of different helicases on i-motif DNA
Helicases are molecular motors that use the energy of nucleoside triphosphate hydrolysis to unwind nucleic acid structures (Patel and Donmez, 2006). Generally, their DNA unwinding activity depends on the hydrolysis of ATP in the presence of Mg2+ (Patel and Donmez, 2006). For different helicases, the most suitable concentration of Mg2+ may not be the same. To maintain the consistency of the experimental conditions, 5 mM Mg2+ was used for all the helicases. As no obvious change can be observed in the Tm of Bcl2-IM with or without the 5 mM Mg2+ (Figure S13), we speculate that the addition of Mg2+ may not cause the changes the i-motif structure and the phenomena we observed should be caused by the different ATP state of the helicases. Based on the variation of FRET distributions, the binding of ATP to helicases mildly weakens their activity to resolve the i-motif structures (Figures 5 and S10). However, with the hydrolysis of ATP mediated by Mg2+, we observed different phenomena. In the presence of BLM, the FRET distribution and FRET traces were stabilized at ∼E0.7 (Figures 5B and 5E), which may reflect one of the partial folding states of i-motif DNA (Figure 2E). Previous studies reported that the ATP-dependent G4s unfolding by BLM displays low processivity compared with other G4 destabilizers such as Pif1 (Budhathoki et al., 2015). BLM either remained bound in the vicinity of the G4/ssDNA junction or dissociated from the substrate before it can unfold G4 structures in the ATP hydrolysis state (Budhathoki et al., 2015). Therefore, the possible reason here may also be the low processivity of BLM to resolve the higher-order structures in the ATP-driven translocation mode.
In the presence of DHX36, Bcl2-IM shows frequent fluctuations (Figure S11A, right panel), reflecting the very quick unfolding/refolding in contrast with the relatively stable unfolding mediated by only DHX36. It is worth noting that, in the study by the Myong group, DHX36 was shown to unfold G4 structures without ATP (Tippana et al., 2019). However, G4s refold frequently once DHX36 was in the ATP hydrolysis state. This is consistent with our results in Figure S11A. The possible reason is that DHX36 has an annealing activity with the hydrolysis of ATP (Tippana et al., 2019). Alternatively, DHX36 may easily dissociate from the substrate as the ATP hydrolysis changes the distance between the RecA1 and RecA2 domains (Chen et al., 2018). In the above two situations, ATP hydrolysis negatively regulates the helicases-mediated unfolding of i-motifs. However, ATP hydrolysis positively drives RecQ and Rep to unfold the i-motifs and then efficiently unwind the downstream duplex DNA (Figures S10, S11B, and S11C, right panels).
Figures 5H and S12B further show that, when no loading strand is present, BLM and DHX36 in their ATP hydrolysis state almost lost their function to resolve i-motifs. However, in Figures 5B and 5F, the complete or partial unfolding of i-motif structures can still be observed when BLM and DHX36 are in their ATP hydrolysis state. This discrepancy suggests that, except for the ATP-independent mode, BLM and DHX36 may also have their ATP-dependent mode on i-motifs relying on the presence of ATP and loading tail. However, it is worth noting that, not every helicase can resolve i-motif, such as WRN (Figures S12E and S12J).
In addition to the 3′-5′ helicases, we also characterized the interaction between i-motifs and the 5′-3′ Pif1 family helicases (Paeschke et al., 2013). Similar ATP-independent i-motif unfolding was observed, and ATP hydrolysis induced the efficient unwinding of duplex DNA downstream of i-motif (Figure S14). However, the Bacteroides sp. 3_1_23 Pif1 cannot resolve the i-motif structures.
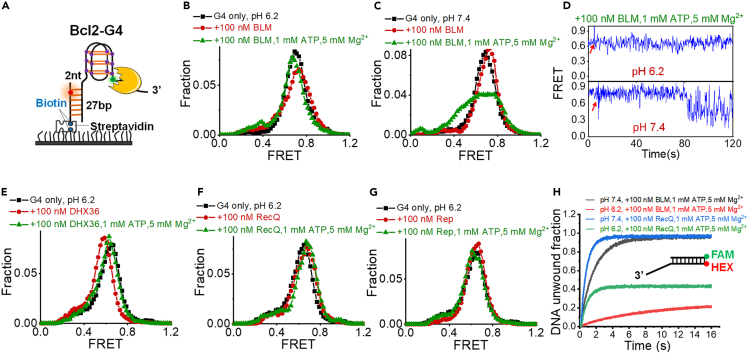
In contrast to i-motif, G4 and duplex DNA are poorly unwound by helicases at the acidic environment
These results demonstrate that specific helicases can potently resolve i-motifs at low pH in the ATP-independent mode. However, this kind of unwinding mode cannot be applied to G4 structures at the acidic condition (Figure 6A), as the FRET distributions display very little change in the presence of BLM (Figure 6B). In the normal cellular condition, the pH is near 7.4 (Brinkman and Sharma, 2021); therefore, we then performed analysis at this condition. The FRET distributions in Figure 6C and traces in Figure 6D confirmed that BLM at the ATP hydrolysis state can indeed induce the repetitive unfolding of G4s at neutral pH, consistent with our previous study (Wu et al., 2015); however, G4 structures have no obvious change in acidic buffer. In addition, DHX36, RecQ, and Rep also demonstrate very poor unfolding activity toward G4s at the acidic condition (Figures 6E–6G). Then we further selected BLM and RecQ and compared their duplex DNA unwinding activities at different pH by the stopped-flow assay (Zhang et al., 2006). Both helicases show much lower unwinding activity at pH 6.2 than pH 7.4 (Figure 6H). We speculated that the low unwinding efficiency may be because the ATP hydrolysis activity of helicases is decreased at low pH. In contrast, these helicases display a unique ATP-independent mode that can efficiently disrupt the i-motif at the acidic environment.
Figure 6.
Helicases poorly unfold G4 or unwind duplex DNA at the acidic condition
(A) The experimental design to characterize the unfolding of G4 structures by the 3′-5′ DNA helicases.
(B and C) FRET distributions of G4 structures before and 4 min after the addition of BLM at pH 6.2 and pH 7.4.
(D) The representative FRET traces with the addition of helicases at the ATP hydrolysis state at pH 6.2 and pH 7.4.
(E–G) FRET distributions of G4 structures before and 4 min after the addition of DHX36, RecQ, and Rep at pH 6.2.
(H) The impacts of pH on the duplex DNA unwinding activity of helicases measured by stopped-flow assay. The substrate with a 15-nt ssDNA loading tail and a 16-bp duplex region was used.
Competition assay on the helicase behavior in the presence of both i-motif and G4 DNA
Although a lot of studies have focused on the i-motif and G4 structures separately in single-stranded DNA fragments, it is still arguable what happens to the G4 counterpart when i-motif is present and vice versa. Based on previous investigations (Tun and Jean-Louis, 2002), at acidic pH, the 1:1 mixture of the G-rich and C-rich sequences produced the folded G4 and i-motif, respectively; however, at pH 7.0 and 100 mM NaCl, duplex was formed predominantly. In our recent study, we also showed that, at acidic conditions, free G4 sequences had little impact on i-motif folding; however, at neutral pH, the formation of duplex DNA was favored (Gao and Hou, 2021). The above-mentioned results were all obtained by mixing the folded G4 and i-motif sequences together. When complementary G4 and i-motif appear in the opposite strands in a duplex DNA, Sun and Hurley reported that both structures were present at the same time with slight displacement from each other (Sun and Hurley, 2015). Meanwhile, Mao group showed that they were mutually exclusive by steric hindrance; however, both structures showed up when the sequences were offset in the two strands (Cui et al., 2016). All these evidences suggest that the influences of i-motif structure on G4 may depend on the pH, ion strength, and the position of these two sequences relative to each other. However, it is still unknown what happens to G4 or i-motif when the counterpart is unfolded by proteins. Therefore, we further performed the FRET assay to define the interaction between G4s and i-motif DNA in the presence of helicases.
In Figures S15A–S15D, the Bcl2-IM substrate with FRET labeling was anchored onto the surface. In the first two experiments, Bcl2-G4 or BLM was added separately to Bcl2-IM as a control. In the third experiment, Bcl2-G4 and BLM were mixed and then added simultaneously. At low pH, G4 has little effects on i-motif, likely owing to the well folding of both structures. Although BLM can disrupt the i-motif at pH 6.2, once BLM was mixed with G4 and added together to Bcl2-IM, the disruptive effects on Bcl2-IM decreases. This might be caused by the prior association of BLM with G4 to some extent. At pH 7.4, Bcl2-IM prefers to form duplexes with Bcl2-G4, displaying a low FRET peak at ∼E0.2. Interesting, after the addition of BLM to Bcl2-IM, there are higher FRET populations in the FRET distribution, quite different from the interaction between BLM and Bcl2-IM at pH 6.2. The selective traces in Figure S15D show that Bcl2-IM makes dynamic transitions between multiple states including that with higher FRET levels, highlighting the different behaviors of BLM on i-motif structure at different pH. When BLM was mixed with Bcl2-G4 and added together to Bcl2-IM at pH 7.4, duplex DNA was still formed predominantly, i.e., at neutral pH, with or without BLM, Bcl2-IM and Bcl2-G4 are easy to anneal with each other and exist as the duplex, consistent with our previous study (Gao and Hou, 2021). In Figures S15E and S15F, the Bcl2-G4 substrate was anchored onto the surface. At pH 6.2, BLM displays little disruptive effects on Bcl2-G4 regardless of the absence or presence of Bcl2-IM. At pH 7.4, Bcl2-IM and Bcl2-G4 prefer to anneal and form duplex DNA; therefore, we did not perform the additional competition assay on Bcl2-G4.
Taken together, the competition assays suggest that, at low pH, Bcl2-IM and Bcl2-G4 can exist stably with the existence of each other. BLM binds to and repetitively unfolds Bcl2-IM without G4s; however, the presence of G4s may inhibit the interaction between BLM and Bcl2-IM to a certain degree. At neutral pH, these two structures prefer to form duplex DNA. Therefore, there might be other proteins like nucleolin and LARK that can promote the folding of G4s to regulate the status of those secondary DNA structures together with helicases (Gonzalez et al., 2009); otherwise, they may anneal with each other easily at neutral pH. In the future, we wish to report more information about the dynamic interaction between G4s and i-motif DNA in the presence of proteins and ligands.
The possible biological effects of the interaction between helicase and i-motif DNA at the cellular level
In a recent study, Takahashi et al. suggested that i-motif formation can block DNA replication and, in turn, may result in genome instability (Takahashi et al., 2017). In addition, Amparo et al. discovered that the formation of non-B structures including G4 and i-motif could induce the helicase-polymerase uncoupling, causing the replicative stress (Amparo et al., 2020). In cells, helicases assist DNA polymerase to replicate the structured DNAs, and a number of helicases including Pif1, BLM, DHX36, and WRN have been shown to resolve G4s both in vivo and in vitro. Although helicases that can unfold i-motif structures have not been identified yet, our in vitro results possibly suggest that the interaction between helicases and i-motifs may have potential significance for rescuing the stalled replication forks and maintaining the genomic stability in the cell.
Furthermore, a number of studies show that i-motifs function as molecular switches in the regulation of DNA transcription (Brown and Kendrick, 2021). Depending on the specific promoter region, the unique i-motif sequence, and the associated transcription factors, i-motifs have the potential to act as an activator or repressor of transcription. In the activation mode, an i-motif serves as a scaffold for transcription factor binding and leads to the transcription activation. For instance, the transcription factor hnRNP LL binds to the Bcl2 i-motif and initiates unfolding of the structure upon binding, consequently leading to the increased transcription (Kang et al., 2014). In contrast, in the repressor mode, the i-motif functions as the initial binding site of the transcription factor; however, the persistence of i-motif structures prevents the transcription activation (Sutherland et al., 2016). In this condition, helicase is likely to help transcription factors to resolve the i-motif structures, so that transcription factors can start the subsequent process. In the future, further study will be needed to identify the interacting helicases of i-motifs in cell, as well as to address their biological functions.
The potential applications of helicases-regulated i-motif DNA in nanotechnology
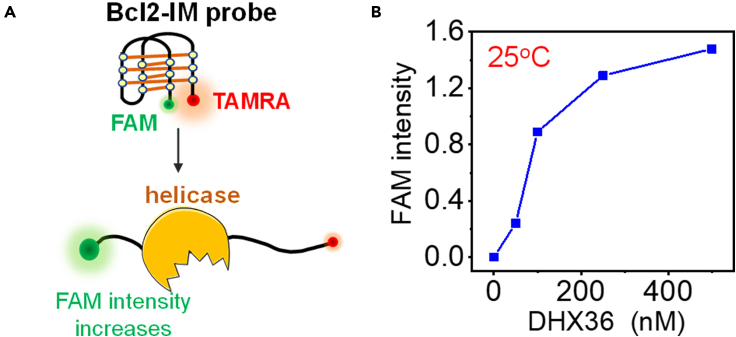
According to the aforementioned findings, an i-motif probe for helicase sensing was designed by labeling FRET pairs FAM and TAMRA at the ends (Figure 7A). The i-motif concentration was 250 nM, and DHX36 was selected for testing. Upon the addition of only 50–100 nM helicase (Figure 7B), the FAM intensity increases significantly, reflecting the extreme sensitivity of i-motif to this helicase. In addition, the FAM signal continues to rise with the further increases in helicase concentration, indicating that the probe has a relatively wide detection range. Based on previous reports, we can roughly estimate the concentration of some helicases in E. coli and S. cerevisiae. For example, in E. coli, Rep has 50 molecules (Scott and Kornberg, 1978) and the concentration would be ∼80 nM (50/6.02×1023/10−15 M/L). DNA helicase II is 8–13 μM (UvrD) and DNA helicase I is 0.8–1.2 μM, respectively (Ishihama et al., 2008; Klinkert et al., 1980). In S. cerevisiae, Pif1 and Sgs1, which is most closely related to human BLM, are ∼2.3 and ∼1.8 μM (Ho et al., 2018). Although we have not found out the number of helicases in mammalian cell from the literature, we speculate that the concentration might be similar to the homologous protein in S. cerevisiae. From these above-mentioned evidences we see that the concentration of helicase in the cell is usually in the range of 0.1 to several micromolar; therefore, the cell-friendly i-motif probe we presented here may be applied as a biosensor to detect the helicase concentrations both in vitro and in vivo. This i-motif probe may also be used to screen the helicase inhibitors in solution with high throughput.
Figure 7.
i-Motif can be used as a convenient and sensitive probe to detect helicase concentration
(A) The experimental design. The i-motif probe (250 nM) and helicase were mixed in phosphate buffer (pH 6.2) with 100 mM KCl at 25°C for 5 min, and then FAM intensity (in arbitrary unit) was recorded.
(B) FAM intensity increases significantly with the addition of DHX36 helicase, reflecting the efficient unfolding of Bcl2-IM by helicase.
Recently, some new methods that could cause pH changes were developed in i-motif nanotechnology. For instance, Wang et al. reported “NOR” and “NAND” logic operations by combining the conformational change of i-motif DNA with the enzymatic reactions, which can produce an acid gradient (Wang et al., 2015). The authors used acetylcholinesterase plus glucose oxidase as the inputs for NOR and invertase plus glucose oxidase as the inputs for NAND. We discovered the convenient biological method to regulate i-motif DNA by proteins. With different helicases and ATP states, i-motif DNA displays different types of structural changes. Inspired by Wang et al., we then speculated that the helicases plus ATP may be used together with i-motif DNA as the molecular logic gates in the future. One another important application of i-motif DNA is the molecular motor used in intelligent surfaces, nanopores, and controlled release devices. The i-motif motor has an open state and a close state switched by pH, and the operating speed of this motor is only limited by the intramolecular chain movement. As shown in Figures 2D and S3, the i-motif intrinsic structural changes are slow (at ∼10 s level). However, proteins can speed up the movement in i-motif DNA and generate a faster i-motif motor (right panel in Figure S11A, movement in less than 1 s level). Therefore, RPA or helicases may be applied in the future to control the i-motif molecular motors particularly for the in vivo purpose.
Conclusions
Our results first indicated that i-motifs are more dynamic than the complementary G4s from acidic to basic pH. In fact, i-motifs make interconversions between up to six folding states and different forms of C-hairpins are likely involved. Further experiments showed that the i-motif structures can be easily linearized by RPA. We discovered several helicases that not only associated with i-motif in high specificity at the acidic pH but also efficiently unfolded i-motifs in an ATP-independent mode. The adjacent ssDNA loading tail was dispensable for the helicases, suggesting that they directly act on i-motif structures. Besides, ATP differentially regulates the activity of different helicases on i-motif DNA. In contrast to i-motif, G4 or duplex DNA are poorly unwound by helicases at the acidic environment. These results may broaden our understanding about the biological activity of i-motifs and benefit the study to regulate i-motifs as nanodevices inside cells.
Limitations of the study
Although here we provided the in vitro evidences for the specific binding to and unfolding of i-motif DNA by helicases at acidic pH, further study will be needed to identify the interacting helicases of i-motifs in vivo, as well as to address their biological functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli strain BL21 (DE3) | NEB | Cat#C2527H |
| E. coli strain rosetta (DE3) | Boule and Zakian (2007) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| D-glucose | Sigma-Aldrich | Cat#G8270-100g |
| Glucose oxidase | Sigma-Aldrich | Cat#G2133-50KU |
| Catalase | Sigma-Aldrich | Cat#C9322-5g |
| Trolox | Sigma-Aldrich | Cat#238813-1g |
| Streptavidin | Invitrogen | Cat#43-4301 |
| mPEG-SC | Laysan bio | Cat#mPEG-SC-5000 |
| Biotin-PEG-SC | Laysan bio | Cat#Biotin-mPEG-SC-5000 |
| Oligonucleotides | ||
| CCCGCCCCCTTCCTCCCGCGCCC; see Table S1 for sequences used in CD, FRET-melting, DNA polymerase stop assay, stopped-flow assay, binding assay and smFRET. | This paper | N/A |
| CCCTAACCCTAACCCTAACCC; see Table S1 | This paper | N/A |
| CCCCACACCCCTGTCCCCACACCCC; see Table S1 | This paper | N/A |
| GGGCGCGGGAGGAAGGGGGCGGG; see Table S1 | This paper | N/A |
| GGGTTAGGGTTAGGGTTAGGG; see Table S1 | This paper | N/A |
| GGGGTGTGGGGACAGGGGTGTGGGG; see Table S1 | This paper | N/A |
| GTGTGGTGTTGGGCCCGCGC; see Table S1 | This paper | N/A |
| CCCTAACCCTTTTTTGTACATCAAATC; see Table S1 | This paper | N/A |
| CCCTAACCCTAATTTTTTT TTGTACATCAAATC; see Table S1 |
This paper | N/A |
| CCCTAATTTTAACCCTT TTTTGTACATCAAATC; see Table S1 |
This paper | N/A |
| CCCTAACCCTAACCCT TTTTTGTACATCAAATC; see Table S1 |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: p11d-tRPA | Henricksen et al. (1994) | N/A |
| Plasmid: PET-15b-SUMO | Wu et al. (2015) | N/A |
| Plasmid: PET-28a | Boule and Zakian (2007) | N/A |
| Software and algorithms | ||
| smCamera | Lee at al., 2019 | http://ha.med.jhmi.edu/resources/#1464200861600-0fad9996-bfd4 |
| Matlab2019b | MathWorks | https://www.mathworks.com/ |
| Origin2019b | OriginLab | https://www.originlab.com/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Xi-Miao Hou (houximiao@nwsuaf.edu.cn).
Materials availability
Materials and the information used for the experiments are available upon reasonable request.
Experimental model and subject details
E. coli strains for in vitro studies
The E. coli strain Rosetta (DE3) was used to express Pif1 protein. The E. coli strain BL21 (DE3) was used to express RPA and all other helicase proteins. These e. coli cells were incubated at 37°C in LB medium for 4 h, and then the protein expression was induced by IPTG at 18°C for 16 h.
Method details
DNA constructs
All oligonucleotides required to make the DNA substrates were purchased from Sangon Biotech (Shanghai, China). Sequences and labeling positions of all the oligonucleotides were listed in Table S1. For DNA constructs used in smFRET measurements, DNA was annealed with a 1:2 mixture of the stem and i-motif strands by incubating the mixture at 95°C for 10 min, then slowly cooling down to room temperature in about 7 h. The strand without biotin was used in excess to reduce the possibility of anchoring non-annealed strands on the coverslip surface. The concentration of the stem strand was 5 nM.
Buffers
Unless otherwise specified, 50 mM phosphate buffer at different pH with 100 mM KCl was used. For single-molecule FRET measurements, 0.8% D-glucose, 1 mg/mL glucose oxidase (266,600 units/g, Sigma), 0.4 mg/mL catalase (2000-5000 units/mg, Sigma) and 4 mM Trolox were added to prevent the photobleaching and photoblinking.
Protein expression and purification
The expression and purification of human BLM-core, WRN, Bos DHX36, E. coli RecQ, E. coli Rep were based on the previous studies (Wu et al., 2015, 2017; Chen et al., 2018; Teng et al., 2020). BLM-core was expressed in E. coli strain BL21 (DE3). Starter cultures were grown at 37°C and induced with 1 mM IPTG at 18°C for 16 h. Then purified by fast protein liquid chromatography (FPLC) with sequential chromatography on Ni-NTA (GE Healthcare, Chicago, IL, USA) and Superdex200 10/300 GL column (GE Healthcare). WRN was expressed in E. coli ER2566 cell (NEB) and induced with 0.3 mM IPTG at 18°C overnight. After centrifugation, WRN was purified by FPLC with sequential chromatography on Ni-NTA and Heparin (GE Healthcare). Bos DHX36 was expressed in E. coli ER2566 cell (NEB). Starter cultures were grown at 37°C and induced with 1 mM IPTG at 20°C and grown overnight. Then loaded on a Ni-NTA column and further purified on a Superdex 200 PG column (GE Healthcare). As for RecQ and Rep, in brief, RecQ was only need to purified through Ni-NTA column. And Rep was purified by FPLC with sequential chromatography on Ni-NTA and cationic exchange column Hi-Trap SP (GE Healthcare). Human RPA was expressed as described previously (Wang et al., 2019). E. coli strain BL21 (DE3) was transformed with a plasmid p11d-tRPA for recombinant human RPA that permits the co-expression of RPA1, RPA2, and RPA3 at 18°C for 16 h (Henricksen et al., 1994). Then, RPA was purified with Affi-Gel blue, hydroxyapatite (BioRad), and Q-sepharose chromatography columns (GE healthcare). Pif1 family helicases were expressed and purified essentially according to Boule and Zakian (Boule and Zakian, 2007). The protein was expressed in the E. coli strain Rosetta (DE3) at 18°C for 16 h and then purified by FPLC with sequential chromatography on Ni-NTA and Hi-Trap SP column.
Circular dichroism
Circular dichroism (CD) experiments were performed with a Bio-Logic MOS450/AF-CD optical system (BioLogic Science Instruments, France), using a quartz cell with a 1-mm path length. A 3 μM solution of DNA was prepared by incubating in phosphate buffer at 95°C for 10 min, then slowly cooling down to room temperature. CD spectra were recorded in the 220-320 nm regions in 0.75 nm increments at 25°C.
FRET-melting assay
FRET-melting experiments were conducted with FAM-TAMRA dual labeled oligomers listed in Table S1 using a Rotor-Gene Q real-time PCR machine (Qiagen) according to the previous study (De Cian et al., 2007). Oligonucleotides were tested at 0.25-0.5 μM strand concentration in 50 mM phosphate buffer at different pH. The emission of FAM fluorophore was normalized between 0 and 1, and the melting temperature Tm was determined as the temperature at which the normalized emission equals 0.5.
Equilibrium DNA-binding assay with helicases
The binding of helicases to DNA was analyzed by a fluorescence polarization assay using Infinite F200 PRO (Tecan group, Switzerland) at 25°C. Various amounts of protein were added to a 150-μL aliquot of binding buffer (50 mM phosphate buffer at pH 6.2 with 100 mM KCl) containing 5 nM DNA. Each sample was allowed to equilibrate in the solution for 5 min, after which the fluorescence polarization was measured.
Single-molecule fluorescence data acquisition and analysis
The smFRET assay was performed as described previously (Hou et al., 2015). Streptavidin (10 μg/mL) was added to the microfluidic chamber made of the PEG-coated coverslip and incubated for 10 min. After washing, 50 pM DNA was added to the chamber and allowed to be immobilized for 10 min. Then free DNA was removed by washing with the imaging buffer (50 mM phosphate buffer, 100 mM KCl, 0.8% D-glucose, 1 mg/mL glucose oxidase, 0.4 mg/mL catalase, and 4 mM Trolox). We used an exposure time of 100 ms for all single-molecule measurements at a constant temperature of 22°C.
DNA polymerase stop assay
The polymerase stop assay was performed as previously described (Teng et al., 2017). 50-200 nM Klenow Fragment (KF) was incubated with 1 μM FAM-labeled DNA template in a standard reaction mixture with 100 μM dNTPs at 22°C. The reactions were stopped with 2×loading buffer (8 M urea, 0.05% xylene cyanole) and then heated for 10 min at 98°C. These samples were subjected to 15% polyacrylamide/8 M urea gel electrophoresis.
Stopped-flow unwinding assay
Fluorescence stopped-flow assay was performed as previously described (Zhang et al., 2006). Briefly, unwinding kinetics were measured in a two-syringe mode, where helicases and fluorescently labeled DNA substrate were pre-incubated at 25°C in one syringe for 5 min and the unwinding reaction was initiated by rapidly mixing ATP from another syringe. The final concentration of DNA and proteins were 5 nM and 100 nM, respectively. The unwinding buffer contained 100 mM KCl in 50 mM phosphate buffer at pH 7.4 and pH 6.2.
Quantification and statistical analysis
FRET distributions
The numbers of the FRET traces used in each FRET distribution was shown in Table S3. All those traces from the same experiment were collected by smCamera software (Lee et al., 2019) and combined together. The FRET efficiency was calculated using E = IA/(ID + IA), where ID and IA represent the intensities of the donor and acceptor. Basic data analysis including transition density plot (TDP) was carried out by scripts written in MATLAB. All data fitting was conducted with Origin 2019b. An automated step-finding method (from http://bio.physics.illinois.edu/HaMMy.asp) was used to characterize the dynamic transitions of i-motif DNA between different states.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32071225, 31870788 and 32071291), the Natural Science Basic Research Program of Shaanxi (2020JQ-251), and the Chinese Universities Scientific Fund (Z109021718).
Author contributions
X.-M.H. designed and supervised the study. B.G., Y.-T.Z., and A.-M.S. conducted the experiments and analyzed the data. X.-M.H. and B.G. wrote the manuscript with input from all authors.
Declaration of interests
The authors declare no competing financial interest.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103575.
Supplemental information
Data and code availability
This study does not generate any deposited data sets.
This study does not generate any deposited code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abou Assi H., Garavis M., Gonzalez C., Damha M.J. i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 2018;46:8038–8056. doi: 10.1093/nar/gky735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba J.J., Sadurni A., Gargallo R. Nucleic acid i-motif structures in analytical chemistry. Crit. Rev. Anal. Chem. 2016;46:443–454. doi: 10.1080/10408347.2016.1143347. [DOI] [PubMed] [Google Scholar]
- Amparo C., Clark J., Bedell V., Murata-Collins J.L., Martella M., Pichiorri F., Warner E.F., Abdelhamid M.A.S., Waller Z.A.E., Smith S.S. Duplex DNA from sites of helicase-polymerase uncoupling links non-B DNA structure formation to replicative stress. Cancer Genomics Proteomics. 2020;17:101–115. doi: 10.21873/cgp.20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule J.B., Zakian V.A. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman J.E., Sharma S. StatPearls; 2021. Physiology, Metabolic Alkalosis. [PubMed] [Google Scholar]
- Brown S.L., Kendrick S. The i-motif as a molecular target: more than a complementary DNA secondary structure. Pharmaceuticals (Basel) 2021;14:96. doi: 10.3390/ph14020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhathoki J.B., Stafford E.J., Yodh J.G., Balci H. ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity. Nucleic Acids Res. 2015;43:5961–5970. doi: 10.1093/nar/gkv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.C., Tippana R., Demeshkina N.A., Murat P., Balasubramanian S., Myong S., Ferre-D'Amare A.R. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018;558:465–469. doi: 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Kim S., Tachikawa T., Fujitsuka M., Majima T. pH-induced intramolecular folding dynamics of i-motif DNA. J. Am. Chem. Soc. 2011;133:16146–16153. doi: 10.1021/ja2061984. [DOI] [PubMed] [Google Scholar]
- Cui Y., Koirala D., Kang H., Dhakal S., Yangyuoru P., Hurley L.H., Mao H. Molecular population dynamics of DNA structures in a bcl-2 promoter sequence is regulated by small molecules and the transcription factor hnRNP LL. Nucleic Acids Res. 2014;42:5755–5764. doi: 10.1093/nar/gku185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Kong D., Ghimire C., Xu C., Mao H. Mutually exclusive formation of G-quadruplex and i-motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016;55:2291–2299. doi: 10.1021/acs.biochem.6b00016. [DOI] [PubMed] [Google Scholar]
- Dai J., Hatzakis E., Hurley L.H., Yang D. I-motif structures formed in the human c-MYC promoter are highly dynamic–insights into sequence redundancy and I-motif stability. PLoS One. 2010;5:e11647. doi: 10.1371/journal.pone.0011647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.X., Carver M., Yang D.Z. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day H.A., Pavlou P., Waller Z.A. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014;22:4407–4418. doi: 10.1016/j.bmc.2014.05.047. [DOI] [PubMed] [Google Scholar]
- De Cian A., Guittat L., Kaiser M., Sacca B., Amrane S., Bourdoncle A., Alberti P., Teulade-Fichou M.P., Lacroix L., Mergny J.L. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Dembska A. The analytical and biomedical potential of cytosine-rich oligonucleotides: a review. Anal. Chim. Acta. 2016;930:1–12. doi: 10.1016/j.aca.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Dhakal S., Schonhoft J.D., Koirala D., Yu Z.B., Basu S., Mao H.B. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J. Am. Chem. Soc. 2010;132:8991–8997. doi: 10.1021/ja100944j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Yang Z., Liu D. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014;47:1853–1860. doi: 10.1021/ar500073a. [DOI] [PubMed] [Google Scholar]
- Dou S.X., Xi X.G. Fluorometric assays for characterizing DNA helicases. Methods. 2010;51:295–302. doi: 10.1016/j.ymeth.2010.04.017. [DOI] [PubMed] [Google Scholar]
- Dzatko S., Krafcikova M., Hansel-Hertsch R., Fessl T., Fiala R., Loja T., Krafcik D., Mergny J.L., Foldynova-Trantirkova S., Trantirek L. Evaluation of the stability of DNA i-motifs in the nuclei of living mammalian cells. Angew. Chem. Int. Ed. Engl. 2018;57:2165–2169. doi: 10.1002/anie.201712284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Hou X.M. Opposite effects of potassium ions on the thermal stability of i-motif DNA in different buffer systems. ACS Omega. 2021 doi: 10.1021/acsomega.0c06350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring K., Leroy J.L., Gueron M. A tetrameric DNA-structure with protonated Cytosine.Cytosine base-pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez V., Guo K., Hurley L., Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Ho B., Baryshnikova A., Brown G.W. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 2018;6:192–205.e3. doi: 10.1016/j.cels.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Hou X.M., Fu Y.B., Wu W.Q., Wang L., Teng F.Y., Xie P., Wang P.Y., Xi X.G. Involvement of G-triplex and G-hairpin in the multi-pathway folding of human telomeric G-quadruplex. Nucleic Acids Res. 2017;45:11401–11412. doi: 10.1093/nar/gkx766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.M., Wu W.Q., Duan X.L., Liu N.N., Li H.H., Fu J., Dou S.X., Li M., Xi X.G. Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 2015;466:189–199. doi: 10.1042/BJ20140997. [DOI] [PubMed] [Google Scholar]
- Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F.U., Kerner M.J., Frishman D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.J., Kendrick S., Hecht S.M., Hurley L.H. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014;136:4172–4185. doi: 10.1021/ja4109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick S., Kang H.J., Alam M.P., Madathil M.M., Agrawal P., Gokhale V., Yang D., Hecht S.M., Hurley L.H. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 2014;136:4161–4171. doi: 10.1021/ja410934b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.E., Lee I.B., Hyeon C., Hong S.C. Destabilization of i-motif by submolar concentrations of a monovalent cation. J. Phys. Chem. B. 2014;118:4753–4760. doi: 10.1021/jp500120d. [DOI] [PubMed] [Google Scholar]
- Klinkert M.Q., Klein A., Abdel-Monem M. Studies on the functions of DNA helicase I and DNA helicase II of Escherichia coli. J. Biol. Chem. 1980;255:9746–9752. [PubMed] [Google Scholar]
- Kruisselbrink E., Guryev V., Brouwer K., Pontier D.B., Cuppen E., Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Lacroix L., Lienard H., Labourier E., Djavaheri-Mergny M., Lacoste J., Leffers H., Tazi J., Helene C., Mergny J.L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000;28:1564–1575. doi: 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y., Mcnerney C., Myong S. G-quadruplex and protein binding by single-molecule FRET microscopy. Methods Mol. Biol. 2019;2035:309–322. doi: 10.1007/978-1-4939-9666-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- Marsich E., Piccini A., Xodo L.E., Manzini G. Evidence for a HeLa nuclear protein that binds specifically to the single-stranded d(CCCTAA)(n) telomeric motif. Nucleic Acids Res. 1996;24:4029–4033. doi: 10.1093/nar/24.20.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsich E., Xodo L.E., Manzini G. Widespread presence in mammals and high binding specificity of a nuclear protein that recognises the single-stranded telomeric motif (CCCTAA)(n) Eur. J. Biochem. 1998;258:93–99. doi: 10.1046/j.1432-1327.1998.2580093.x. [DOI] [PubMed] [Google Scholar]
- Megalathan A., Cox B.D., Wilkerson P.D., Kaur A., Sapkota K., Reiner J.E., Dhakal S. Single-molecule analysis of i-motif within self-assembled DNA duplexes and nanocircles. Nucleic Acids Res. 2019;47:7199–7212. doi: 10.1093/nar/gkz565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M., Mergny J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S., Swetha M.G., Goswami D., Gupta G.D., Mayor S., Krishnan Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnology. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Fraire C., Sheardy R.D. Linking pH, temperature, and K(+) concentration for DNA i-motif formation. J. Phys. Chem. B. 2017;121:7872–7877. doi: 10.1021/acs.jpcb.7b06317. [DOI] [PubMed] [Google Scholar]
- Niu K.K., Zhang X.J., Deng H.M., Wu F., Ren Y.D., Xiang H., Zheng S.C., Liu L., Huang L.H., Zeng B.J., et al. BmILF and i-motif structure are involved in transcriptional regulation of BmPOUM2 in Bombyx mori. Nucleic Acids Res. 2018;46:1710–1723. doi: 10.1093/nar/gkx1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.S., Donmez I. Mechanisms of helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- Paul S., Hossain S.S., Samanta A. Insights into the folding pathway of a c-MYC-promoter-based i-motif DNA in crowded environments at the single-molecule level. J. Phys. Chem. B. 2020;124:763–770. doi: 10.1021/acs.jpcb.9b10633. [DOI] [PubMed] [Google Scholar]
- Ruggiero E., Lago S., Sket P., Nadai M., Frasson I., Plavec J., Richter S.N. A dynamic i-motif with a duplex stem-loop in the long terminal repeat promoter of the HIV-1 proviral genome modulates viral transcription. Nucleic Acids Res. 2019;47:11057–11068. doi: 10.1093/nar/gkz937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas T.R., Petruseva I., Lavrik O., Bourdoncle A., Mergny J.L., Favre A., Saintome C. Human replication protein a unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.F., Kornberg A. Purification of the rep protein of Escherichia coli. An ATPase which separates duplex DNA strands in advance of replication. J. Biol. Chem. 1978;253:3292–3297. [PubMed] [Google Scholar]
- Sharma S. Non-B DNA secondary structures and their resolution by RecQ helicases. J. Nucleic Acids. 2011;2011:724215. doi: 10.4061/2011/724215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer P., Kührová P., Banáš P., Koča J., Bussi G., Trantírek L., Otyepka M., Šponer J. Hairpins participating in folding of human telomeric sequence quadruplexes studied by standard and T-REMD simulations. Nucleic Acids Res. 2015;43:9626–9644. doi: 10.1093/nar/gkv994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer P., Kuhrova P., Vicherek L., Banas P., Otyepka M., Trantirek L., Sponer J. Parallel G-triplexes and G-hairpins as potential transitory ensembles in the folding of parallel-stranded DNA G-Quadruplexes. Nucleic Acids Res. 2019;47:7276–7293. doi: 10.1093/nar/gkz610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Hurley L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2015;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C., Cui Y., Mao H., Hurley L.H. A mechanosensor mechanism controls the G-quadruplex/i-motif molecular switch in the MYC promoter NHE III1. J. Am. Chem. Soc. 2016;138:14138–14151. doi: 10.1021/jacs.6b09196. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Brazier J.A., Sugimoto N. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc. Natl. Acad. Sci. U S A. 2017;114:9605–9610. doi: 10.1073/pnas.1704258114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F.Y., Hou X.M., Fan S.H., Rety S., Dou S.X., Xi X.G. Escherichia coli DNA polymerase I can disrupt G-quadruplex structures during DNA replication. FEBS J. 2017;284:4051–4065. doi: 10.1111/febs.14290. [DOI] [PubMed] [Google Scholar]
- Teng F.Y., Wang T.T., Guo H.L., Xin B.G., Sun B., Dou S.X., Xi X.G., Hou X.M. The HRDC domain oppositely modulates the unwinding activity of E. coli RecQ helicase on duplex DNA and G-quadruplex. J. Biol. Chem. 2020;295:17646–17658. doi: 10.1074/jbc.RA120.015492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippana R., Chen M.C., Demeshkina N.A., Ferre-D'Amare A.R., Myong S. RNA G-quadruplex is resolved by repetitive and ATP-dependent mechanism of DHX36. Nat. Commun. 2019;10:1855. doi: 10.1038/s41467-019-09802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun P.A., Jean-Louis M. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang G., Zhang D. Enzyme-driven i-motif DNA folding for logic operations and fluorescent biosensing. Chem. Commun. (Camb) 2015;51:3812–3815. doi: 10.1039/c4cc09905b. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Yang Y.T., Wang Y.R., Gao B., Xi X.G., Hou X.M. Human replication protein A induces dynamic changes in single-stranded DNA and RNA structures. J. Biol. Chem. 2019;294:13915–13927. doi: 10.1074/jbc.RA119.009737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.R., Guo T.T., Zheng Y.T., Lai C.W., Sun B., Xi X.G., Hou X.M. Replication protein A plays multifaceted roles complementary to specialized helicases in processing G-quadruplex DNA. iScience. 2021;24:102493. doi: 10.1016/j.isci.2021.102493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R.D. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Wright E.P., Huppert J.L., Waller Z.A.E. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017;45:2951–2959. doi: 10.1093/nar/gkx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.Q., Hou X.M., Li M., Dou S.X., Xi X.G. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015;43:4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.Q., Hou X.M., Zhang B., Fosse P., Rene B., Mauffret O., Li M., Dou S.X., Xi X.G. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 2017;7:43954. doi: 10.1038/srep43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoga Y.M., Traore D.A., Sidiqi M., Szeto C., Pendini N.R., Barker A., Leedman P.J., Wilce J.A., Wilce M.C. Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides. Nucleic Acids Res. 2012;40:5101–5114. doi: 10.1093/nar/gks058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeraati M., Langley D.B., Schofield P., Moye A.L., Rouet R., Hughes W.E., Bryan T.M., Dinger M.E., Christ D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018;10:631–637. doi: 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- Zhang X.D., Dou S.X., Xie P., Hu J.S., Wang P.Y., Xi X.G. Escherichia coli RecQ is a rapid, efficient, and monomeric helicase. J. Biol. Chem. 2006;281:12655–12663. doi: 10.1074/jbc.M513089200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study does not generate any deposited data sets.
This study does not generate any deposited code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.