Summary
Heart disease is the leading cause of death with no method to repair damaged myocardium due to the limited proliferative capacity of adult cardiomyocytes. Curiously, mouse neonates and zebrafish can regenerate their hearts via cardiomyocyte de-differentiation and proliferation. However, a molecular mechanism of why these cardiomyocytes can re-enter cell cycle is poorly understood. Here, we identify a unique metabolic state that primes adult zebrafish and neonatal mouse ventricular cardiomyocytes to proliferate. Zebrafish and neonatal mouse hearts display elevated glutamine levels, predisposing them to amino-acid-driven activation of TOR, and that TOR activation is required for zebrafish cardiomyocyte regeneration in vivo. Through a multi-omics approach with cellular validation we identify metabolic and mitochondrial changes during the first week of regeneration. These data suggest that regeneration of zebrafish myocardium is driven by metabolic remodeling and reveals a unique metabolic regulator, TOR-primed state, in which zebrafish and mammalian cardiomyocytes are regeneration competent.
Subject areas: Biological sciences, Tissue Engineering, Cell biology
Graphical abstract
Highlights
-
•
High glutamine levels prime the heart for regeneration
-
•
Amino-acid-driven mTOR signaling is required for cardiomyocyte proliferation
-
•
Heart injury induces mitochondrial regeneration
-
•
Wnt/β-catenin signaling is required for early heart regeneration
Biological sciences; Tissue Engineering; Cell biology
Introduction
For a short period of time in mammalian neonates, the mammalian heart can regenerate via cardiomyocyte proliferation (Bergmann et al., 2015; Senyo et al., 2012; Porrello et al., 2011). This regenerative capacity is largely absent in adults. In other organisms, including zebrafish (Danio rerio), damaged hearts can regenerate throughout their lifespans (Poss et al., 2002; Zhang et al., 2013; Uygur and Lee, 2016). Many studies have been performed to understand the mechanisms of cardiomyocyte de-differentiation and proliferation during heart regeneration (Jopling et al., 2010; Kikuchi et al., 2010); however, the underlying reason why adult zebrafish and young mammalian cardiomyocytes are primed to enter cell cycle have not been identified.
Because zebrafish are able to regenerate damaged ventricular myocardium, they serve as a good model organism to study the molecular mechanisms regulating cardiac repair (Foglia and Poss, 2016). It is currently well understood that microRNAs (miRNAs), let-7a/c and miRNA-99/100, must be repressed in order for a cardiomyocyte to de-differentiate and re-enter the cell cycle (Aguirre et al., 2014). However, the events that result in the downregulation of these miRNAs are not understood, nor is it clear how other conditions predispose zebrafish cardiomyocytes to enter regeneration even in the adult state.
Zebrafish are a regenerative model organism used to study the molecular mechanisms regulating cardiac repair, because their cardiomyocytes retain the ability to re-enter the cell cycle after injury in the adult state (Foglia and Poss, 2016; Jopling et al., 2010; Kikuchi et al., 2010). Specifically, adult zebrafish cardiomyocytes de-differentiate to re-enter the cell cycle and then repopulate the lost myocardium (Wang et al., 2011). Furthermore, the signaling events that control this regenerative process are complex, and include the cross-talk between nerves and cardiomyocytes (Mahmoud et al., 2015), notch signaling (Zhang et al., 2013), and cell-cycle regulators such as Neuregulin1 (Gemberling et al., 2015) and Msp1 (Poss et al., 2002). Interestingly, it has been recently shown that metabolic remodeling governed by Nrg1/ErbB2 signaling is important for cardiomyocyte proliferation (Honkoop et al., 2019) due to reduction of mitochondrial genes while simultaneously having an increase in glycolysis and glucose uptake (Fukuda et al., 2020). However, it is still unclear why adult zebrafish cardiomyocytes retain the potential to re-enter the cell cycle and what other signaling pathways may contribute to this process.
A plethora of signaling pathways have been implicated in nonmammalian regeneration, including activin, Bmp, fibroblast growth factor (Fgf), Sonic hedgehog, insulin-like growth factor (IGF), notch, and retinoic acid (RA) (Hirose et al., 2014; Gemberling et al., 2013). Here, we have focused on investigating the contributions of Wnt/β-catenin signaling and mechanistic target of rapamycin (mTOR) signaling to cardiomyocyte proliferation. The Wnt/β-catenin pathway is a conserved controller of cell fate and proliferation during embryonic development, whereas mTOR is a central regulator of growth and metabolism during the G1-phase of the cell cycle (Laplante and Sabatini, 2009). Interestingly, both pathways regulate zebrafish fin regeneration (Hirose et al., 2014; Stoick-Cooper et al., 2007). Currently, the roles of Wnt/β-catenin and TOR signaling have not been studied in the context of zebrafish heart regeneration. Furthermore, it is not clear whether there is an interaction between Wnt/β-catenin and mTOR signaling pathways during cardiac regeneration and if these signaling pathways can be used to stimulate human cardiomyocyte proliferation.
Here, we found through multi-omics analysis that Wnt/β-catenin and TOR signaling are upregulated during cardiac regeneration. The inhibition of either of these pathways leads to decreased cardiomyocyte proliferation. These findings implicate the Wnt-TOR signaling axis as a potent mitogen-activating pathway during zebrafish cardiac regeneration. We show the primed state of a pro-regenerative cardiomyocyte is dictated by its amino acid profile and metabolic state. Zebrafish and neonatal mouse cardiomyocytes display elevated glutamine levels, predisposing them to amino-acid-driven activation of TOR. Injury initiates Wnt/β-catenin signaling that instigates primed TOR activation and metabolic remodeling necessary for zebrafish cardiomyocyte regeneration. These studies reveal a unique TOR primed state in zebrafish and mammalian regeneration-competent cardiomyocytes.
Results
Ablated adult zebrafish hearts show early Wnt/β-catenin activity
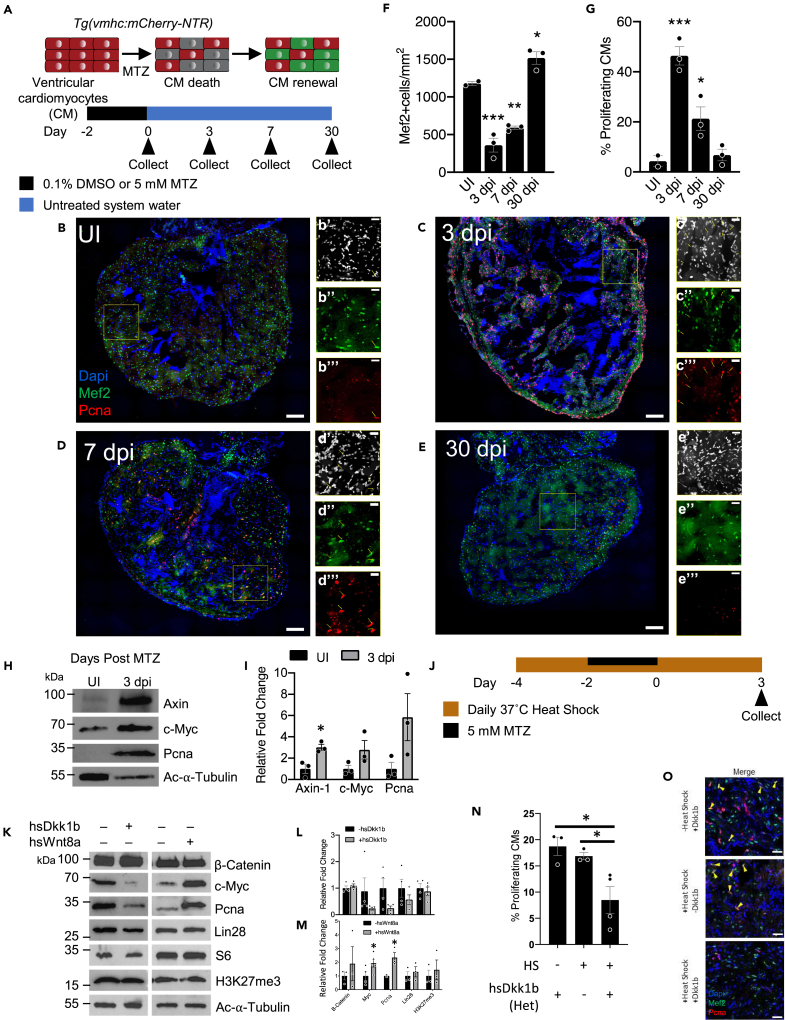
Using a transgenic chemically induced ventricular cardiomyocyte (CM) ablation model (homozygous vmhc:mCherry-NTR) (Zhang et al., 2013), we generated a regeneration paradigm in the adult zebrafish heart. This zebrafish line was engineered to express the enzyme nitroreductase (NTR) in ventricular myocytes (Figures S1A and S1B), which generates a cytotoxic reduced form of the antibiotic metronidazole (MTZ), when given as a supplement to the fish water (Figures 1A and S1C) (Zhang et al., 2013). To determine the number of days postinjury (dpi), we count the number of days once the fish are removed from the final treatment of MTZ. However, when comparing with zebrafish heart injury models in the literature, the first day of heart injury is counted as the start. Consequently, our 3 and 7 dpi in Figure 1A can also be counted as 5 and 9 days after first MTZ presentation. In the adult uninjured state (UI), we had minimal to no proliferation present in the zebrafish heart (Figure 1B). However, at 3 dpi and 7 dpi, we identified a 3-fold reduction in the cardiomyocyte number concomitantly with a 10-fold increase of proliferating cardiomyocytes compared with the uninjured control via the markers Mef2c and Pcna (Figures 1B–1D, 1F, and 1G). By 30 dpi, the regeneration process had revived the full heart, reaching the same number of cardiomyocytes as observed in the uninjured heart (Figures 1E–1G).
Figure 1.
Wnt/β-Catenin regulates the early stages of zebrafish heart regeneration
(A) Schematic of adult zebrafish heart ablation experiments.
(B–E) Immunohistochemistry of uninjured (UI), 3, 7, and 30 dpi adult zebrafish hearts. Insets show representative areas of the heart, magnified, and with channels split. Scale bar for B–E: 100 μm, area of inset: 200 μm2, scale bar in insets: 25 μm. Blue-DAPI, Red-Pcna, and Green-Mef2c. Yellow arrow heads denote proliferating cardiomyocytes that are Pcna and Mef2c positive.
(F and G) Quantification of cardiomyocyte abundance and cardiomyocyte proliferation. After injury, there is a significant increase in proliferating cardiomyocytes as seen by the double staining of Mef2c and Pcna. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA performed. n = 2–3 biological replicates.
(H) Protein analysis of UI and 3 dpi hearts for Wnt targets Axin, c-Myc, and Pcna.
(I) Quantification of protein analysis in (H). ∗p < 0.05, two-tailed t test performed. N = 3 biological replicates with each N being 3–5 pooled hearts.
(J) Schematic of adult zebrafish heart ablation and heat shock protocol for Wnt/β-catenin modulation fish.
(K) Protein analysis of Wnt/β-catenin modulation during adult zebrafish heart regeneration.
(L and M) Quantification of protein abundance in (K). ∗p < 0.05, two-tailed t test performed. N = 2–5 biological replicates with each N being 3–5 pooled hearts.
(N) Quantification of cardiomyocyte proliferation in the context of Wnt/β-catenin repression via hsDkk1b.
(O) Immunohistochemistry images of cardiomyocyte proliferation at 3 days postinjury during Wnt/β-catenin inhibition via hsDkk1b overexpression; all animals were presented with MTZ. Scale bar: 25 μm. Blue-DAPI, Red-Pcna, and Green-Mef2c. Yellow arrow heads denote proliferating cardiomyocytes that are Pcna and Mef2c positive. ∗p < 0.05, N = 3–6. One-way ANOVA was performed. Bar graphs show individual data points with error bars representing standard error of the mean. Source data are provided as a Data S2.
Because we identified a robust regenerative response at 3 dpi, we wanted to dissect the molecular mechanism that initiated this early response. Wnt/β-catenin signaling is present in the regenerating zebrafish heart as previously shown in an amputation model 5 days postinjury where cardiomyocytes along the resection plane showed active Wnt/β-catenin signaling (Zhao et al., 2019). However, by the end of the first week of regeneration, it was shown that further activation of Wnt/β-catenin signaling on day 5 and 7 postamputation (dpa), via a pan GSK3β inhibitor, (2′Z,3′E)-6-Bromoindirubin-3′-oxime (BIO), was detrimental to cardiomyocyte proliferation and heart regeneration (Zhao et al., 2019). To better understand Wnt/β-catenin signaling during early zebrafish heart regeneration, specifically during the first few days postinjury, we used the chemical cell ablation line to test whether Wnt/β-catenin signaling, a pathway known to regulate cell cycle (Niehrs and Acebron, 2012), is active shortly after injury.
We first examined if Wnt/β-catenin targets were altered after chemically induced ventricular cardiomyocyte ablation. Wnt/β-catenin target protein Axin1 was significantly more abundant at 3 dpi, whereas c-Myc was on average more abundant at 3dpi (Figures 1H and 1I), suggesting that similar to surgical ventricular apex resection (Zhao et al., 2019), Wnt/β-catenin signaling was increased during early regeneration after chemically induced injury. At 3 dpi, we observed an increase in Pcna expression in cardiomyocytes, an indication of increased cell division (Figures 1C and 1G–1I). This regeneration process requires Wnt/β-catenin activity, because genetically inhibiting Wnt/β-catenin signaling through overexpressing Dickkopf 1 (Dkk1b) (heterozygous hsDkk1bGFP [Stoick-Cooper et al., 2007], heterozygous vmhc:mCherry-NTR) reduced on average c-Myc and Pcna at 3 dpi, compared with controls (Figure 1J–1L). In contrast, activating Wnt/β-catenin signaling by hsWnt8a (heterozygous hsWnt8aGFP [Weidinger et al., 2005], heterozygous vmhc:mCherry-NTR) overexpression significantly increased Pcna and c-Myc protein levels at 3 dpi compared with controls (Figures 1J, 1K, and 1M). To identify the cell type affected by these Wnt/β-catenin pathway alterations, we performed immunohistochemical analysis and found that Wnt8a overexpression led to a modest increase in cardiomyocyte proliferation, whereas Wnt/β-catenin inhibition via Dkk1b led to a significant reduction in proliferating cardiomyocyte (Figures 1N, 1O, and S2). These results suggest that proliferation required during early stages of endogenous heart regeneration is dependent on Wnt/β-catenin signaling.
Regenerating adult zebrafish hearts show metabolic remodeling
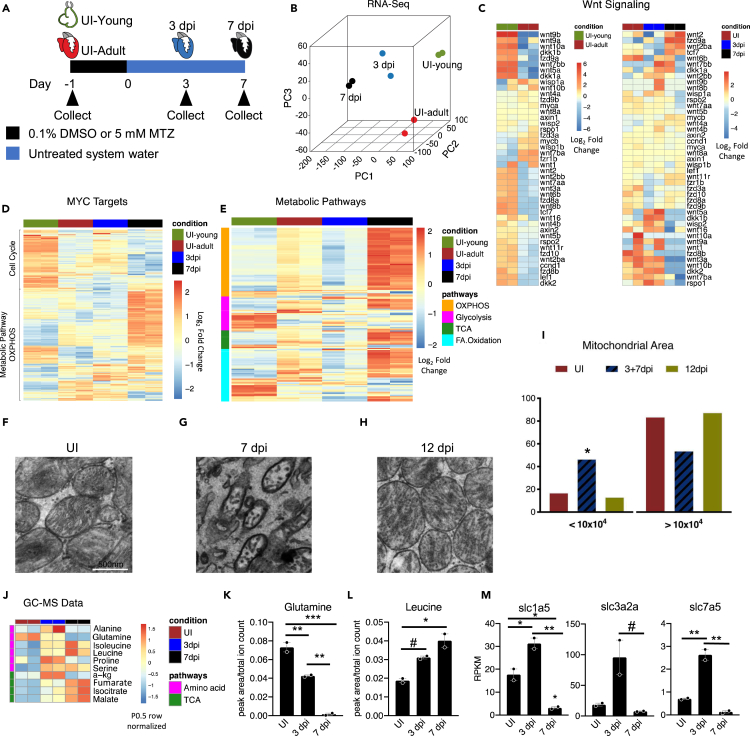
We performed RNA sequencing on young zebrafish hearts (3-day-old), adult uninjured, 3 dpi, and 7 dpi hearts (Figure 2A) in an effort to shed light on mechanisms of cardiac regeneration. We first compared RNA-sequencing datasets from our MTZ ventricular cardiomyocyte ablation model with those following cryoinjury from Bednarek et al. (Figures S3A–S3C) (Bednarek et al., 2015). Principle component analysis (PCA) demonstrated our adult uninjured heart and 3 dpi heart samples clustered with those previously published, indicating a high degree of similarity between the injury models (Figure S3C). Furthermore, we validated transcript abundance using a panel of genes by RT-qPCR (Figure S3A). We next conducted PCA to explore the clustering pattern among our samples. We found that the adult regenerating hearts (3 and 7 dpi) clustered with young hearts along PC3 (Figure 2B), supporting the previous finding that injured cardiomyocytes share a very similar transcriptome to fetal/young cardiomyocytes in zebrafish (Honkoop et al., 2019).
Figure 2.
Metabolic pathways are dynamically regulated during the early stages of zebrafish heart regeneration
(A) Schematic of zebrafish heart samples prepared for RNA sequencing.
(B) Principle component analysis of young uninjured 3-day-old hearts (UI-young), UI adult, 3 dpi, and 7 dpi adult zebrafish hearts.
(C) Heatmap of Wnt ligands and Wnt/β-catenin targets in zebrafish hearts.
(D) Heatmap of Myc targets in zebrafish hearts.
(E) Heatmap of four metabolic pathways in zebrafish hearts.
(F–H) Transmission electron microscopy of zebrafish hearts. Scale bar is 500 nm.
(I) Mitochondria area quantification in UI, 3+7, and 12 dpi zebrafish hearts. A Chi-squared test was used to assess the ratio between observed/expected. 3dpi and 7dpi showed enrichment in the area bin <10x104 nm2, whereas UI and 12 dpi has depletion in area bin <10x104 nm2. p < 2.2x10−16.
(J) Heatmap of significantly different amino acids and TCA cycle intermediates in zebrafish hearts during first week of regeneration.
(K) Abundance of glutamine in zebrafish hearts. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA performed. N = 2 with each N being n = 3–5 pooled hearts.
(L) Abundance of leucine in zebrafish hearts. ∗p < 0.05, #p = 0.058. N = 2 with each N being 3–5 pooled hearts.
(M) RNA-sequencing quantification of glutamine and essential amino acid transporters engaged during mTORC1 activation. ∗p < 0.05, ∗∗p < 0.01, #p = 0.063, one-way ANOVA performed. N = 2 with each N being 3–5 pooled hearts. Bar graphs show individual data points with error bars representing standard error of the mean. Source data are provided as a Data S2.
Because we had sequenced purified young 3-day-old hearts, we identified the genes that were up- or downregulated during regeneration that were also up- or downregulated in the 3-day-old hearts as compared with adult uninjured hearts (Figure S3D). We found that at 3 dpi, 54% of the genes that were upregulated and 82% of the genes that were downregulated reverted to a fetal/young-like expression level. At 7 dpi, the degree of fetal/young overlap decreased, 21% in common with upregulated genes and 36% in common with downregulated genes; however, the total number of differentially expressed genes had also increased at 7 dpi. When assessing the pathways these genes acted upon, we found that 3 dpi upregulated genes that were also highly expressed in the young 3-day-old hearts had gene ontology (GO) terms associated with nucleotide processes and skeletal muscle cell differentiation. The downregulated GO terms were mainly associated with various metabolic processes including generation of precursor metabolites, carboxylic acid metabolic, and ATP biosynthetic processes (Tables S1 and S2). At 7 dpi, upregulated genes that were also highly expressed in the young 3-day-old hearts had GO terms associated with RNA splicing and processing. The downregulated terms were regulation of immune system and defense response (Tables S3 and S4). These data reveal how aspects of the adult heart regeneration transcript profile are similar to a young 3-day-old heart transcriptomic profile to bring about adult heart regeneration.
Whole transcriptome analysis revealed that young hearts and regenerating hearts displayed higher transcript levels of many Wnt ligands and targets compared with adult hearts (Figure 2C). Because the Wnt/β-catenin target, c-Myc, was more abundant during the early regeneration period (Figures 1H and 1I), we assessed transcript abundance of Myc targets (Kim et al., 2008). Many Myc targets were upregulated in young hearts while being repressed in adult hearts (Figure 2D). Among these Myc-targets we identified cell-cycle pathway genes that were upregulated in young and 3 dpi hearts. Importantly, metabolic Myc-target genes, including many oxidative phosphorylation (OXPHOS)-related pathways, were dramatically upregulated in 7 dpi hearts (Figure 2D). These data suggest a role for Myc in the induction of cell proliferation and mitochondrial biogenesis or maturation (Figures 1H and 1I).
In addition to Myc-targets, we also performed genome wide global GO enrichment analyses comparing all samples and identified ten significantly changed pathways that were associated with cardiac regeneration, maturation, and metabolic regulation (Figure S3E). Many cardiac structural pathways were downregulated at 3 dpi, whereas at 7 dpi many metabolic pathways had been dramatically upregulated. In particular, OXPHOS, the citric acid (TCA) cycle, and fatty acid oxidation (FAO) were upregulated at 7 dpi to levels higher than the uninjured adult heart (Figures 2E and S3E). These data suggest that dramatic metabolic remodeling occurs during the first week of heart regeneration. Using metabolic pathway enrichment analysis, at 3 dpi we identified OXPHOS as the most downregulated metabolic pathway accompanied by FAO, glycolysis, and TCA (Figure S3F). The levels of 3 dpi metabolic gene expression were similar to the levels observed in young hearts (Figure 2E). By 7 dpi, however, OXPHOS was the highest upregulated metabolic pathway accompanied by TCA, glycolysis, and FAO (Figures 2E and S3F).
To further explore signaling and metabolic pathways during heart regeneration, we performed label-free quantitative (LFQ) proteomics to assess the proteome of adult uninjured, 3 dpi, and 7 dpi hearts (Figure S4A), where we identified and quantified 2,787 proteins across all conditions. Principle component analysis of the proteomics data revealed that regenerating hearts clustered separately from the uninjured heart on PC1 (Figure S4B). There were a number of significantly changed proteins at 3 and 7 dpi compared with uninjured adult hearts (Figure S4C). At 3 dpi, we found many GO terms associated with metabolic processes were downregulated, whereas at 7 dpi many GO terms associated with DNA packaging and assembly where upregulated (Figures S4D and S4E).
One significantly changed protein was succinate dehydrogenase complex assembly factor 3 (Sdhaf3), related to complex II of the electron transport chain (ETC). This OXPHOS protein was less abundant in early regeneration than in the uninjured heart (Figure S4F). However, on a transcript level, sdhaf3 was significantly upregulated at 7 dpi. This suggested that OXHPOS upregulation may result in the replacement of some mitochondrial proteins at this early stage of regeneration (Figure S4G). Finally, we overlapped the genes from our RNA sequencing with our proteomics dataset at 3 dpi and 7 dpi (Figure S4H). We then determined the pathways common changing genes and proteins were involved in. We found a significant enrichment of GO terms at 3 dpi associated with metabolic processes, such as generation of precursor metabolites and energy and ATP metabolic process, and metabolite synthesis processes, such as purine nucleotide metabolic process. At 7 dpi, we found protein synthesis and muscle processes as being enriched (Tables S5 and S6). These shared gene and protein expression data correlate well to help reinforce the notion that there is a strong metabolic rearrangement and substrate changes required for cell proliferation at 3 dpi. In total, system-wide RNA sequencing and LFQ proteomics analysis suggest that dynamic metabolic regulation is a key component of endogenous cardiac regeneration in zebrafish.
Our data suggest that the regeneration of zebrafish myocardium is driven by metabolic remodeling. We hypothesized that by 7 dpi, newly generated cardiomyocytes would be undergoing metabolic maturation in order to drive the primitive cardiomyocyte toward a fully matured state. This hypothesis is supported by our finding that the upregulated Wnt/β-catenin target, Myc, is associated with metabolic reorganization and promotes expression of complexes of the ETC (Brownet al., 2017; Li et al., 2005).
Mitochondrial recuperation occurs during the first week of heart regeneration
To assess mitochondrial state, we used transmission electron microscopy (TEM) to examine mitochondrial morphology during early stages of heart regeneration. Adult uninjured zebrafish hearts have large mitochondria packed against long myofibrils (Figure 2F). However, at 3 dpi and 7 dpi, we observed a significantly increased number of small mitochondria (Figure 2G). By 12 dpi the mitochondria appeared larger in size, similar to uninjured cardiomyocyte (Figures 2H and 2I). The gene expression data and the TEM analysis support the hypothesis that metabolism, and specifically mitochondrial dynamics as shown previously in cardiomyocyte maturation (Gong et al., 2015), play a key role in regeneration of the zebrafish heart.
Glutamine-enriched hearts are primed for TOR signaling and regeneration
Supporting the critical metabolism function during the first week of regeneration, we observed a dynamic change in transcripts associated with the mitochondrial TCA cycle and TCA cycle enzymes (Figure S5A). To directly identify amino acids and TCA cycle intermediates involved in heart regeneration, we performed metabolite analysis using gas chromatography-mass spectrometry (GC-MS) for adult uninjured, 3 dpi, and 7 dpi heart samples. We found that many amino acids and TCA cycle intermediates were enriched at 3 dpi or 7 dpi or both (Figure 2J). However, one amino acid, glutamine, was highly abundant at the uninjured adult state and was depleted over the first week of heart regeneration (Figure 2K). Glutamine has many roles in the cell including serving as a precursor to glutamate production and as a critical amino acid in driving mammalian target of rapamycin complex 1 (mTORC1) activation (Saxton and Sabatini, 2017). Because the TCA cycle intermediate α-ketoglutarate was enriched at 3 dpi, we considered a potential role of glutamine as a component for anaplerotic flux. However, the enzymes controlling the metabolism of glutamine to glutamate were either unchanged, gls, or significantly downregulated at 3 dpi, gls2b. Although this does not rule out anaplerotic flux, we decided to consider other possibilities, in particular TOR activation.
We therefore investigated whether the dynamic changes of glutamine during regeneration correlated with regulation of TOR in zebrafish hearts. Amino acid activation of TOR has been shown to be a result of glutamine import and then export (Nicklin et al., 2009). Specifically, in mammalian cells, glutamine can be imported via SLC1A5, loading a cell with cytosolic glutamine. Then, this facilitates the activity of a complex between SLC3A2 and SLC7A5, referred to as the large neutral amino acid transporter (LAT1). LAT1 then imports leucine while reciprocally exporting glutamine (Beaumatin et al., 2019). Finally, cytosolic leucine is able to specifically inhibit the protein Sestrin2, an inhibitor of the mTORC1 pathway. Therefore, leucine inhibition of Sestrin2 activates the mTORC1 pathway (Nicklin et al., 2009; Wolfson et al., 2016). We found leucine abundance was increased during the first week of zebrafish heart regeneration (Figure 2L). Furthermore, the transporters governing these amino acids' import and export were dynamically and tightly regulated during the first week of regeneration. The glutamine transporter was upregulated already in uninjured adult heart, possibly contributing to an explanation of the high levels of glutamine at this stage. The glutamine/leucine exchange transporter complex (LAT1) on the other hand was dramatically upregulated at 3 dpi, suggesting that the increased abundance of leucine at 3 and 7 dpi was a result of the export of glutamine (Figure 2M). These data suggest the baseline abundance of glutamine may prime the cell for the rapid exportation of glutamine for the importation of leucine after injury has occurred. High leucine levels may play a role in activating the TOR pathway in zebrafish regenerating adult hearts (Saxton et al., 2016).
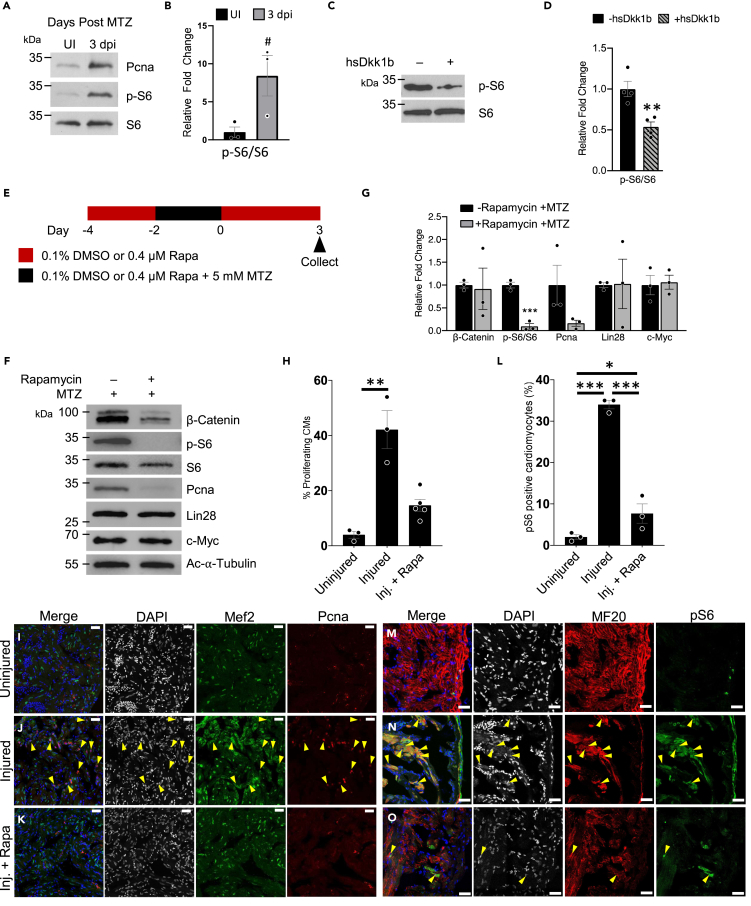
We next examined the regulation of TOR and asked whether it was required during the early stages of adult zebrafish heart regeneration by analyzing TOR targets and inhibiting the pathway in vivo by rapamycin treatment. We found that the phosphorylation sites ser235/236 (pS6) of S6, a target and readout of TOR activity, were increased over the course of early heart regeneration following the trend of Pcna abundance (Figures 3A, 3B, 3H–3K, and S6). Another target of mTORC1 is the serine/threonine kinase Unc-51 like kinase 1 (ULK1). ULK1 phosphorylation at Ser757 disrupts the interaction between ULK1 and 5′AMP-activated protein kinase (AMPK) resulting in cell growth (Kim et al., 2011; Egan et al., 2011). We found pULK1 was more abundant on average at 3 dpi compared with uninjured control (Figures S6A and S6B). This suggests that TOR is activated by 3 dpi and corroborates that TOR might be activated by the dynamic flux of glutamine and leucine due to the upregulation of amino acid transporters at 3 dpi (Figures 2K–2M). Interestingly, in mammals, mTORC1 can be activated by both amino acids and by the crosstalk between signaling pathways, including Wnt/β-catenin signaling (Shimobayashi and Hall, 2014; Inoki et al., 2006). Active Wnt/β-catenin signaling can activate mTOR by inhibiting GSK3 and does not involve beta-catenin-dependent transcription. GSK3 normally inhibits the mTOR pathway by phosphorylating TSC2 in a manner dependent on AMPK-priming phosphorylation (Inoki et al., 2006). We tested the effects of TOR activation in zebrafish via Wnt/β-catenin signaling by inhibiting Wnt/β-catenin signaling via overexpression of Dkk1b. Repression of Wnt/β-catenin signaling by Dkk1b led to reduced pS6 at 3 dpi (Figures 3C and 3D), suggesting TOR activation is downstream of Wnt/β-Catenin signaling in this context.
Figure 3.
TOR regulates the early stages of zebrafish heart regeneration
(A) Protein analysis of TOR activity monitored via the phosphorylation of S6 and proliferation via Pcna in regenerating adult zebrafish hearts.
(B) Quantification of pS6/S6 in (A) #p = 0.054, two-tailed t test performed. N = 3 biological replicates with each N being 3–5 pooled hearts.
(C) Protein analysis of TOR activity monitored via the phosphorylation of S6 in regenerating adult zebrafish hearts that have Wnt/β-catenin inhibition through the expression Dkk1b.
(D) Quantification of westerns in (C) ∗∗p < 0.01, two-tailed t test performed. N = 4 biological replicates with each N being 3–5 pooled hearts.
(E) Schematic of adult zebrafish heart ablation and TOR inhibition via rapamycin.
(F) Protein analysis of TOR inhibited regenerating adult zebrafish hearts.
(G) Quantification of protein abundance in (F). ∗∗∗p < 0.001, two-tailed t test performed. N = 3 biological replicates with each N being 3–5 pooled hearts.
(H) Quantification of proliferating cardiomyocytes (Mef2c and Pcna positive cells). One-way ANOVA with pairwise multiple comparison using a Holm-Sidak method were performed. ∗∗p < 0.01. N = 3–5.
(I–K) Immunohistochemistry assessing the number of proliferating cardiomyocytes in the adult zebrafish heart 3 dpi after being treated with no rapamycin and no MTZ, uninjured (I), injured (+MTZ) with no rapamycin (J), or injured (+MTZ) with rapamycin (K). Images have scale bars with a 25 μm length. Blue-DAPI, Red-Pcna, and Green-Mef2c. Yellow arrow heads denote proliferating cardiomyocytes, Pcna and Mef2c positive.
(L) Quantification of TOR positive cardiomyocytes (MF20 and pS6 positive cells). One-way ANOVA with pairwise multiple comparison using a Holm-Sidak method was performed. N = 3, ∗p < 0.05, ∗∗∗p < 0.001. Immunohistochemistry assessing the number of TOR positive cardiomyocytes in the adult zebrafish heart 3 dpi after being treated with no rapamycin and no MTZ, uninjured (M), injured (+MTZ) with no rapamycin (N), or injured (+MTZ) with rapamycin (O). Scale bars: 25 μm. Bar graphs show individual data points with error bars representing standard error. Source data are provided as a Data S2.
To specifically test if TOR is required for regeneration, we used rapamycin, a selective mTORC1 inhibitor in mammals (Li et al., 2014), to assess whether TOR activation was necessary for in vivo adult zebrafish cardiac cell proliferation after injury (Figure 3E). We found that rapamycin significantly inhibited pS6 and greatly reduced Pcna abundance in injured adult zebrafish hearts at 3 dpi (Figures 3F and 3G). Furthermore, rapamycin did not inhibit Myc, β-catenin, or Lin28 (Aguirre et al., 2014) abundance, suggesting that inhibition of TOR does not affect Wnt/β-catenin activity. To identify the cell type TOR is activated in during zebrafish heart regeneration, we analyzed Pcna and pS6 expression in cardiomyocytes after injury using immunocytochemistry. We found rapamycin-treated zebrafish had fewer proliferating cardiomyocytes as compared with non-rapamycin-treated zebrafish (no significant difference between uninjured and rapamycin-treated injured heart, Figures 3H–3K and S6C–S6E). Finally, we assessed whether the TOR pathway was engaged in cardiomyocytes in the regenerating zebrafish heart by examining a mTORC1 target, phosphorylation of S6. There was a significant increase in pS6 protein in cardiomyocytes (as determined via MF20 expression or mCherry-NTR expression) upon injury (Figures 3L–3O, S6F–S6I, and S6K–S6M). Rapamycin treatment significantly reduced pS6 staining in the injured ventricle to levels similar to the uninjured heart. Interestingly, we also observed cells along the border of the heart that were negative for our cardiac markers, MF20 or mCherry expression driven by the ventricular myosin heavy chain promoter. These non-myocytes were positive for pS6. Because these cells were along the outermost boarder of the heart, we postulated that these may be epicardial cells that also have TOR activated during the regeneration process (Figures S6J and S6N).
Because ventricular cardiomyocytes strongly activated the TOR pathway during heart regeneration, it is interesting to postulate that ventricular cardiomyocytes may be primed via glutamine to activate the TOR pathway upon injury. Finally, these results suggest that TOR is necessary for ventricular zebrafish heart regeneration, and that, potentially, TOR acts in both epicardial cells and ventricular CMs during in vivo zebrafish heart regeneration.
scRNA sequencing reveals a multicellular response during the heart regeneration process
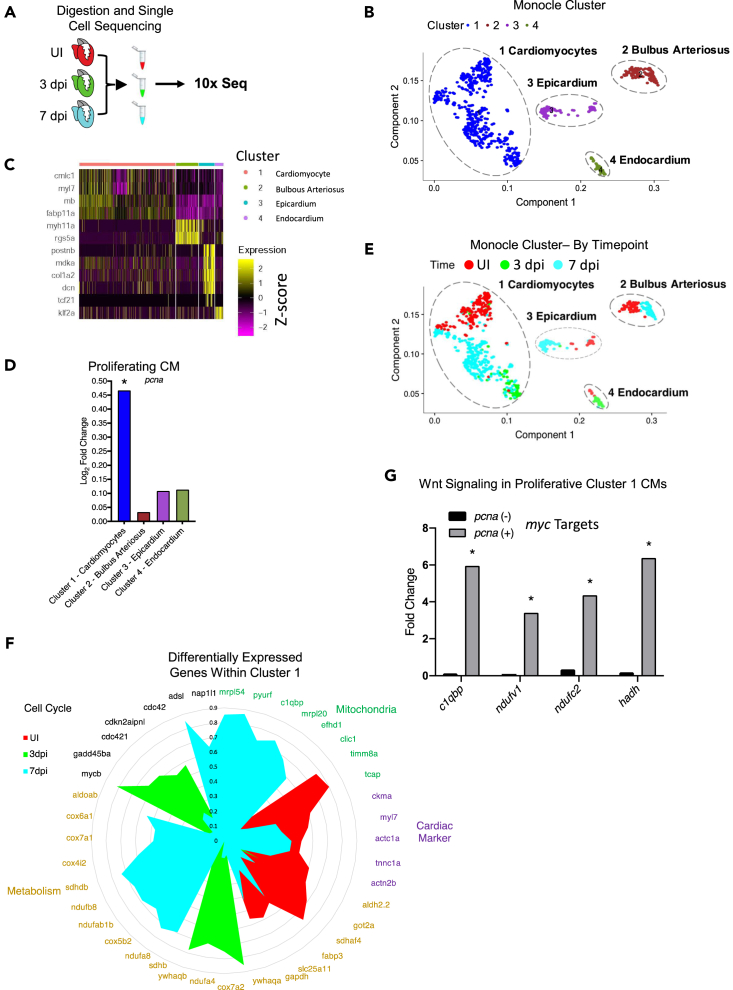
In order to gain better insight into the cells involved in regeneration after injury of the zebrafish heart, we performed single-cell RNA sequencing (scRNA-seq) on adult uninjured, 3 dpi, and 7 dpi zebrafish hearts (Figure 4A). Using tSNE analysis, we plotted the clusters generated by the monocle software package at all time points (Figure 4B) and separated by time point (Figure S7A). Four clusters were identified (Figure 4B) and further characterized in a heatmap with known gene markers of these cell types (Figure 4C). Interestingly, we found some of these clusters represented transient cell states and activated subpopulations that arose during zebrafish heart regeneration (Figure 4C).
Figure 4.
Single-cell sequencing reveals a Wnt responding regenerating cardiomyocyte population during the first week of zebrafish heart regeneration
(A) Schematic of the scRNA-sequencing set-up.
(B) Principle component analysis of the sequenced cells for UI, 3 dpi, and 7 dpi zebrafish hearts.
(C) A heatmap of the genes that highlight the identity of the cell type in each cluster.
(D) Transcript levels of the proliferation marker pcna in each of the clusters identifies cluster 1 as a main regenerating cluster.
(E) Principle component analysis of the sequenced cells for UI, 3 dpi, and 7 dpi zebrafish hearts separated by cluster and labeled by time.
(F) Circle plot of significantly changed genes in dynamically changed pathways during the first week of zebrafish heart regeneration.
(G) Transcript levels of Myc target genes (electron transport chain genes) that are significantly upregulated in the subpopulation of cluster 1 cardiomyocytes that are proliferating (pcna positive). ∗p < 0.05. Source data are provided as a Data S2.
Cluster 1 was present at all time points and was identified as cardiomyocytes due to the markers cmlc1 and myl7 (Figure 4C). Cluster 1 had hallmark pathway term associated with myogenesis and GO biological processes associated with morphogenesis, striated and cardiac muscle cell proliferation, and DNA replication (Tables S7 and S8). Although cluster 1 cells were positive for cardiomyocyte markers cmlc1 and myl7, a subset of the cells was positive for the ventricular myocyte markers hspb11 and vmhcl (Figure S7B) (Singh et al., 2016). Moreover, cluster 1 cardiomyocytes at 7 dpi showed a striking upregulation of lamtor5, a component of the Ragulator complex and a positive regulator of TOR (Cai et al., 2017), supporting our previous finding of the activation of TOR signaling during the first week of heart regeneration (Figure 3, S6, and S7C). Consequently, cluster 1 encompassed cardiomyocytes from all time points, suggesting that cluster 1 also held the regenerating cardiomyocyte population.
Cluster 2 was present at uninjured and 7 dpi time points and was identified as the bulbus arteriosus due to the markers myh11a and rgs5a (Figure S7D) (Singh et al., 2016). Cluster 2 had upregulated hallmark pathway term associated with epithelial mesenchymal transition (EMT) (Table S9) (Singh et al., 2016). At 7 dpi, many genes associated with cell migration and differentiation were upregulated, and interestingly, the gene tgfb3, a secreted TGF ligand, was significantly upregulated (Figure S7E). It has been previously shown that TGFβ signaling is important for zebrafish cardiomyocyte proliferation and extracellular matrix remodeling (Choi et al., 2013; Chablais and Jazwinska, 2012). These data elucidate a potential role of the bulbus arteriosus as part of the TGFβ signaling that occurs during heart regeneration.
Cluster 3 was present at mainly the uninjured and 7 dpi time points and was identified as epicardial cells due to the marker tcf21 (Figure S7F). At 7 dpi, cluster 3 had an activated epicardium signature with the expression of postnb and mdka (Figure S7G), and fibroblast-like markers, col1a2 and dcn, arose at 7 dpi (Figure S7H). The top hallmark pathway in cluster 3 was epithelial mesenchymal transition (Table S10), and the top GO term was extracellular matrix organization (Table S11). The identified cell type in the uninjured adult heart, nonactivated epicardial cell, and the identified cell type at 7 dpi, an activated epicardial cell and transient fibroblasts, matched well with prior findings that showed epicardial cells (tcf21 positive) undergo cell division, migration into the injured myocardium, and then differentiation into a transient fibroblast population (col1a2 positive), which promoted cardiomyocyte proliferation during zebrafish heart regeneration (Figure S7H) (Sanchez-Iranzo et al., 2018; Cao et al., 2016; Marin-Juez et al., 2019). Furthermore, we identified the upregulation of mdka, an early regenerating signal that is expressed in epicardial cells (Figure S7G) (Lien et al., 2006). These data identify cluster 3 cells as quiescent epicardium (uninjured heart) and activated epicardium and fibroblasts (7 dpi) (Gonzalez-Rosa et al., 2012).
Cluster 4 was present at the uninjured and 3 dpi time points and was identified as endocardium at uninjured and then activated endocardium at 3 dpi. The top hallmark pathway in cluster 4 was hypoxia, and the top GO term was blood vessel development, suggesting the endocardium was responding to the disrupted myocardium and vasculature (Tables S12 and S13). Cluster 4 had klf2a, a marker of differentiating endocardium (Figure S7I) (Palencia-Desai et al., 2015). These data identify cluster 4 cells as quiescent (uninjured) and activated (3 dpi) endocardium.
scRNA sequencing reveals Myc targets in proliferating cardiomyocytes
We compared the major signaling pathways known during zebrafish heart regeneration against our scRNA-seq data (Gonzalez-Rosa et al., 2017). We found that the majority of signaling pathways were identified in our dataset, and because we have cell-specific clusters, we were able to identify which cell types were upregulating these specific genes (Table S14). Finally, we did not identify atrial cardiomyocytes due to the isolation procedure (Figure S7J). Consequently, our data identify the majority of cell types found in the zebrafish heart and their molecular signature during regeneration.
One of the main events that occurs during zebrafish heart regeneration is the proliferation of cardiomyocytes. We identified cluster 1 as having the strongest upregulation of pcna, indicating cellular proliferation (Figure 4D). Cluster 1 was present at all time points and was identified as having multiple states, as seen by the different transcriptomic signatures (the cells shift along PC1 and PC2) during the course of zebrafish heart regeneration (Figure 4E). Within cluster 1, we wanted to distinguish the different molecular signatures of the cardiomyocytes at the different days of regeneration. We found that at the uninjured adult state there was a strong expression of mature cardiac genes (Figure 4F). However, in cardiomyocytes at 3 dpi and 7 dpi, with single-cell resolution, we were able to re-affirm our previous findings showing that indeed, cardiomyocytes first go through an upregulation of cell-cycle genes (3 dpi) and then later have a strong upregulation of metabolic and mitochondrial genes (7 dpi) (Figure 4F).
Finally, although we showed Wnt/β-catenin signaling was necessary for heart regeneration (Figures 1J–1O), we did not have the resolution to identify which cell types were secreting Wnt ligands and which cells were receiving those ligands. Importantly, the scRNA-seq data identified the activated endocardium, cluster 4, as the main cell type secreting a Wnt ligand, wnt11r, at 3 dpi (Figure S7K). We previously showed that a target of Wnt/β-catenin signaling, Myc, had a large number of predicted gene targets, specifically ones with metabolic relevance, change during zebrafish heart regeneration (Figure 2D). We have now identified the proliferating cardiomyocytes, pcna positive, in cluster 1 as having Myc targets related to ETC/metabolism being upregulated (Figure 4G) (Morrish and Hockenbery, 2014). These data suggest that proliferating cardiomyocytes are recipients of Wnt/β-catenin signaling via the activation of Myc resulting in the upregulation of genes associated with ETC and metabolism during the first week of zebrafish heart regeneration.
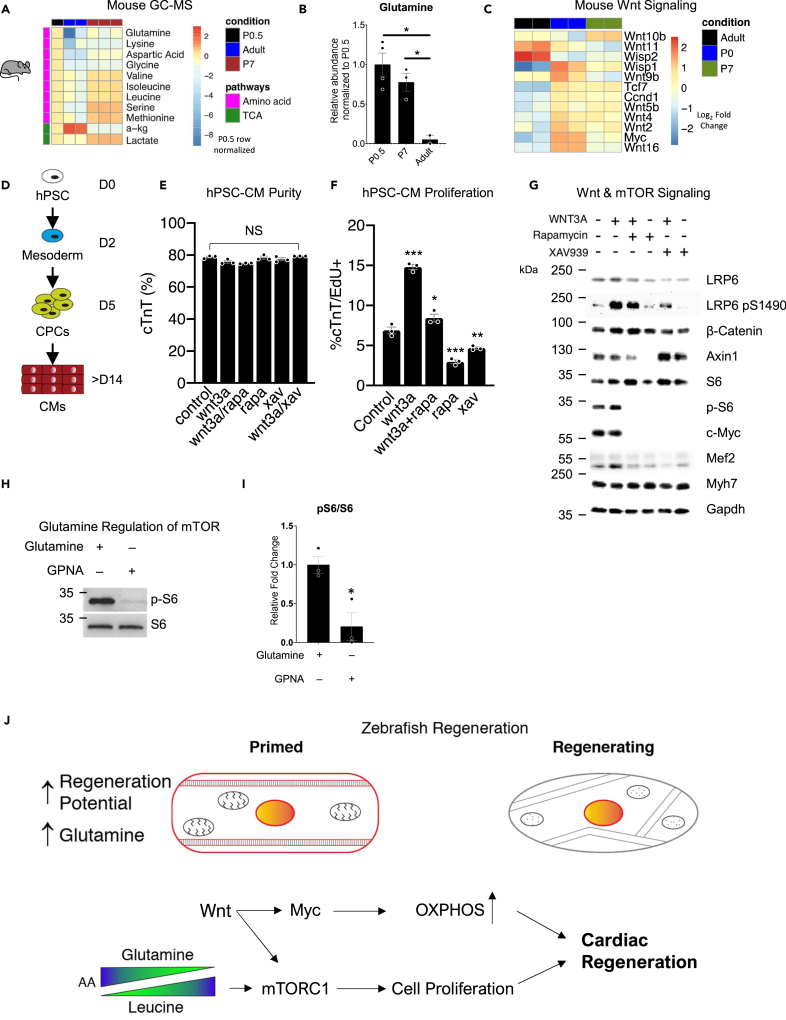
Glutamine enrichment is found in the proregenerative neonatal mouse heart
Because injured adult zebrafish hearts use Wnt/β-catenin and amino acids to stimulate TOR signaling during regeneration, we next examined these components in young mouse hearts, which, similar to zebrafish, are able to regenerate when damaged. Strikingly, the zebrafish heart can regenerate into adulthood, whereas the mouse heart can regenerate only during the first week of life. Indeed, through metabolic analysis we showed that, similar to the primed proregenerative adult zebrafish heart, proregenerative young P0.5 mouse hearts possessed higher amounts of glutamine, suggesting that they are also primed for regeneration. This correlation between glutamine and regeneration is also evident by examining adult mouse hearts that have lost their ability to regenerate and that display lower amounts of glutamine (Figures 5A and 5B). We postulate that high levels of glutamine keep the neonatal heart in a primed state for regeneration so that activation of mTORC1 via the export of glutamine for the import of leucine happens readily leading to heart regeneration.
Figure 5.
Wnt/β-catenin and mTORC1 are active in regenerating mouse and human cardiomyocytes
(A) Heatmap of significantly different amino acids and TCA cycle intermediates in mouse hearts.
(B) Abundance of glutamine in mouse hearts. ∗p < 0.05 versus control, one-way ANOVA performed. N = 2–4 biological replicates.
(C) Heatmap of Wnt ligands and targets in mouse hearts.
(D) Schematic of stem-cell-derived cardiomyocyte (hPSC-CM) generation.
(E) Purity of cardiomyocytes after Wnt activation, inhibition, and/or mTORC1 inhibition as determined by FACs. One-way ANOVA performed. N = 3 biological replicates.
(F) Percentage of proliferating hESC-CMs after Wnt activation, inhibition, and/or mTORC1 inhibition as determined by FACs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA performed. N = 3 biological replicates.
(G) Protein analysis of hPSC-CMs after Wnt activation, inhibition, and/or mTORC1 inhibition.
(H) Protein analysis of hPSC-CMs mTORC1 activation after glutamine transporter inhibition via GPNA. N = 3 biological replicates.
(I) Quantification of p-S6/S6 blots in (H). ∗p < 0.05, two-tailed t test performed.
(J) Schematic of amino acid primed proregenerative cardiomyocyte and the signaling cascade that Wnt/β-catenin drives to turn on mTORC1, Lin28, and Myc to bring about zebrafish heart regeneration. Bar graphs show individual data points with error bars representing standard error of the mean. Source data are provided as a Data S2.
Supportingly, RNA sequencing of young P0 mouse heart showed higher Wnt/β-catenin activity than an adult mouse heart (Figure 5C) (O'Meara et al., 2015). At the cusp of potential mouse heart regeneration, P7, we see a downward trend of Wnt ligand and Wnt/β-catenin target gene expression until it reaches the adult levels (Figure 5C). These data suggest that, just as in regeneration-competent adult zebrafish, a young mouse heart is primed for mTORC1 activity jointly through amino acid and Wnt/β-catenin signaling. This hypothesis is further supported by comparing RNA-sequencing data using a time course of ex vivo cardiomyocyte de-differentiation assay from O'Meara et al. (Figure S8A) (O'Meara et al., 2015). As adult murine cardiomyocytes de-differentiated, they showed greater proliferation and Wnt/β-catenin signaling activity, coupled with a depressed metabolic signature (Figures S8B and S8C), similar to 3 dpi zebrafish hearts. These data suggest that a mammalian cardiomyocyte requires Wnt/β-catenin signaling, priming of mTORC1, and remodeling of several metabolic pathways to de-differentiate and re-enter the cell cycle.
Wnt/β-catenin and mTORC1 signaling pathways regulate proliferation in human cardiomyocytes
We next sought to understand how Wnt/β-catenin and mTORC1 signaling pathways interacted to regulate cardiomyocyte proliferation. For these studies we used human embryonic stem-cell-derived cardiomyocytes (hESC-CM) as a model system (Figure 5D). We first showed that stimulation of Wnt/β-catenin signaling by recombinant Wnt3A increased hESC-CM proliferation, whereas inhibiting the pathway with XAV939 decreased hESC-CM proliferation. Moreover, mTORC1 inhibition through rapamycin treatment inhibited hESC-CM proliferation, which was rescued by increased Wnt/β-catenin signaling (Figures 5E and 5F). Immunoblotting for key protein targets of Wnt/β-catenin and mTORC1 signaling identified that Wnt3A induced phosphorylation of the co-receptor LRP6 and stimulated pS6 and c-Myc. Moreover, inhibition of mTORC1 by rapamycin inhibited pS6 and c-Myc, which was also observed by inhibiting Wnt/β-catenin signaling by XAV939 (Figure 5G). Furthermore, we showed that this activation of mTORC1 in hiPSC-CMs is in part due to amino acid signaling and that using a glutamine transport inhibitor (γ-L-Glutamyl-p-nitroanilide hydrochloride, GPNA) (Nicklin et al., 2009) we could inhibit pS6 (Figures 5H, 5I and S8D). Together, these data indicate that the Wnt/β-catenin and mTORC1 signaling axes cooperate to promote cardiomyocyte cell-cycle re-entry following terminal differentiation.
Discussion
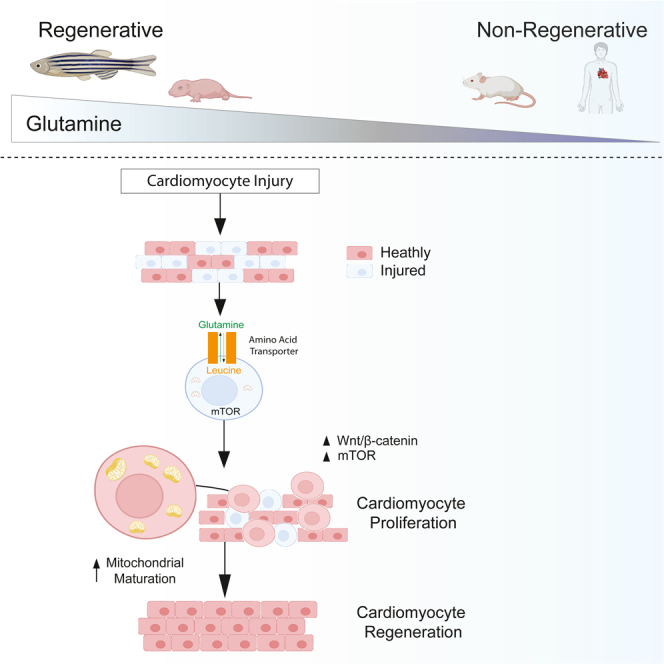
Myocardial infarction (MI) as a result of cardiovascular disease continues to be a top cause of mortality in the United States (Go et al., 2013). Due to the limited ability of the mammalian heart to regenerate, there has been considerable interest in developing strategies to repair the heart. For example, human stem-cell-derived cardiomyocyte cell therapies are being developed to remuscularize the heart (Laflamme and Murry, 2011; Chong et al., 2014), and progress has been made toward transdifferentiating scar tissue into cardiomyocytes (Chen et al., 2012; Song et al., 2012) and advancement in microRNA-based endogenous cardiomyocyte proliferation therapies (Gabisonia et al., 2019). In addition, much work has been done in understanding the pathways that turn on during cardiac regeneration in model organisms (Han et al., 2019), yet the initial events that start this process are not well understood, hence hard to recapitulate in mammalian therapies (Tzahor and Poss, 2017). We now show through metabolic and functional analysis that adult zebrafish cardiomyocyte regeneration of the heart is a result of amino acid primed TOR activation and early stage Wnt/β-catenin signaling (Figure 5J).
Previous findings have shown that in order for heart regeneration to commence in the zebrafish, miRNAs Let7a/c and 99/100 must be repressed (Aguirre et al., 2014). Wnt/β-catenin signaling has been shown in some context to stimulate Lin28 expression, which is a known repressor of the Let7 family of microRNAs (Yao et al., 2016). We now show that Wnt/β-catenin signaling is an early responder after zebrafish heart injury. One possibility is that Wnt/β-catenin signaling is activated during the early stages of zebrafish heart regeneration as a direct consequence to a loss of cell-cell junctions. After cell-cell junctions are lost in response to cardiomyocyte ablation, p-120 catenin may no longer be localized to intercalated disks (Gutstein et al., 2003) and is free to bind to ZTB33 (Kaiso), a known repressor of Wnt/β-catenin signaling and of β-catenin target genes such as Myc (Park et al., 2005; Del Valle-Perez et al., 2011). Interestingly, it has been previously shown that inhibition of Wnt/β-catenin signaling was necessary to promote cardiomyocyte proliferation at later stages of zebrafish heart regeneration, starting the Wnt/β-catenin inhibition at 5 days postamputation (Zhao et al., 2019). In this study, we found that activating Wnt/β-catenin before and during the initial 3 days of regeneration was beneficial to adult zebrafish cardiomyocyte proliferation. Although the two injury models were different, it is exciting to postulate that careful Wnt/β-catenin activation and subsequent repression is required during the first week of heart regeneration to orchestrate a full regenerative response [12]. In fact, this exact biphasic Wnt/β-catenin signaling, activation, and then repression is shown to be essential for zebrafish and mouse cardiac development (Ueno et al., 2007; Naito et al., 2006) and has thereby been adapted in well-defined hPSC-CM differentiation protocols (Lian et al., 2013).
We also reveal a strong correlation between metabolic remodeling and zebrafish heart regeneration. In particular, using single-cell sequencing, metabolomics, and microscopy we show dramatic mitochondrial and metabolomic remodeling during the early stages of regeneration. Glutamine/leucine levels and the amino acid transporters that control mTORC1 activation are dramatically changed during the early regeneration process. Furthermore, mTORC1 activity is essential for epicardial activation and cardiomyocyte regeneration. We propose that Wnt/β-catenin signaling, in conjunction with amino acid priming, activates the mTORC1 pathway to initiate epicardial activation and CM proliferation (Inoki et al., 2006), leading to the initiation of heart regeneration. It is plausible that mTORC1 activity in turn results in Wnt/β-catenin pathway inhibition, observed previously in later stages of regeneration (Zeng et al., 2018; Zhao et al., 2019).
Similar to zebrafish, we showed that regeneration-competent neonatal mouse cardiomyocytes also display elevated glutamine levels, predisposing them to amino-acid-driven activation of mTORC1. Because activating Wnt/β-catenin and mTORC1 signaling in human cardiomyocytes also leads to a proliferative state, these data reveal a common mTORC1 primed stage as the prerequisite for heart regeneration in zebrafish and mammals and paves a way to new regenerative therapies.
Limitations of the study
In this manuscript we identify key metabolic changes in zebrafish hearts 3 days postinjury. This analysis does not distinguish the cell type in the heart in which the observed metabolic changes occur. Future methods development is required to allow metabolic analysis on a single-cell level. Another technical limitation is the inhibition of glutamine transport in 3 days postinjury fish hearts. In this manuscript, we identified glutamine as a key amino acid for heart regeneration as it primes a cardiomyocyte for mTORC1 pathway activation. To prove this concept, we applied glutamine transporter inhibitor (γ-L-glutamyl-p-nitroanilide hydrochloride, GPNA) in hiPSC and observed mTORC1 inhibition by pS6 reduction. We attempted to prove glutamine necessity by GPNA injection prior to fish heart injury; however, our trials failed due to high mortality of the fish at the time of injury. We believe future generation of genetic mutations in glutamine transporters are required to solve this issue. Nevertheless, the technical caveats presented here have not deterred from the key findings in the manuscript.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| Anti-Rabbit MEF2 | Santa Cruz | SC-313 | |
| Anti-Mouse PCNA | Santa Cruz | sc-56 | |
| Anti-Rabbit p-S6(Ser235/236) (IHC) | Cell Signaling | 4857 | |
| Anti-Mouse MF20 | DSHB | P13538 | |
| Anti-Rabbit S6 | Cell Signaling | 2217S | |
| Anti-Rabbit c-MYC | Cell Signaling | 5605S | |
| Anti-Rabbit Axin1 | Cell Signaling | 2087S | |
| Anti-Rabbit H3K27Me3 | Active Motif | 39158 | |
| Anti-Rabbit P-S6 (Ser235/236) (Western) | Cell Signaling | 4858S | |
| Anti-Rabbit GAPDH | Cell Signaling | 5174S | |
| Anti-Rabbit β-Actin | Cell Signaling | 4970S | |
| Anti-Rabbit β-Catenin | Cell Signaling | 9562S | |
| Anti-Rabbit α-Tubulin | Cell Signaling | 2144S | |
| Anti-Mouse Total OXPHOS (Complex I) | MitoSciences | MS604 | |
| Anti-Rabbit LRP6 | Cell Signaling | 3395S | |
| Anti-Rabbit LRP6 pS1490 | Cell Signaling | 2568S | |
| Anti-Mouse MYH7 | Santa Cruz | sc-53090 | |
| Anti-Rabbit SDHAF3 (ACN9) | ThermoFisher | PA5-24526 | |
| Anti-Rabbit Lin28 | Abcam | ab46020 | |
| Goat anti-mouse IgG HRP | Bio-Rad | 1706516 | |
| Goat anti-rabbit IgG HRP | Bio-Rad | 1706515 | |
| Anti-Rabbit p-ULK1 | Cell Signaling | 14202 | |
| Chemicals, peptides, and recombinant proteins | |||
| Rapamycin | Sigma-Aldrich | 553210 | |
| metrodinazole (MTZ) | Sigma-Aldrich | M3761-100G | |
| Dapi | Thermo Fisher Scientific | D1306 | |
| Trizol | Thermo Fisher Scientific | 15596026 | |
| Matrigel | Corning | 356231 | |
| DMEM | Thermo Fisher Scientific | 11995065 | |
| B27 supplement | Thermo Fisher Scientific | 12587-010 | |
| GlutaMAX | Thermo Fisher Scientific | 35050-061 | |
| b-mercaptoethanol | Thermo Fisher Scientific | 21985-023 | |
| Rocki Y27632 | R & D Systems (R&D) | 1254 | |
| FGF4 | Life technologies | PHG0263 | |
| BSA | New England Biolabs | B9000S | |
| Versene | Invitrogen (Gibco/BRL Life Tech) | 15040-066 | |
| syber | Fisher Scientific | 4367659 | |
| PMSF | Fisher Scientific | PI-36978 | |
| Lysis Buffer | Our Lab | N/A | |
| EtOH | Fisher Scientific | 04-355-222 | |
| chloroform | Fisher Scientific | C298500 | |
| isopropanol | Fisher Scientific | BP2618-4 | |
| PFA | Electron Microscopy Science | 15710-S | |
| BSA | VWR Scientific | 0332-100G | |
| PBS | Genesee Scientific | 18-116 | |
| Triton | Sigma-Aldrich | T9284 | |
| Click-iT™ EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 488 dye | ThermoFisher Scientific | C10337 | |
| GPNA (L-Glutamic acid γ-(p-nitroanilide) hydrochloride) | MP Biomedicals | 02151495-CF | |
| Critical commercial assays | |||
| Click-iT™ Plus TUNEL Assay for In Situ Apoptosis Detection, Alexa Fluor™ 488 dyeGreen features | ThermoFisher Scientific | C10617 | |
| Deposited data | |||
| RNA-Seq GEO | GSE188243 | This Study | |
| RNA-Seq Cryoablation Zebrafish Heart | GSE71755 | Bednarek et al. (2015) | |
| Proteomics Zebrafish Heart Regeneration | PXD011791 | This Study | |
| Metabolomics Data | Data S2 | This Study | |
| RNA-Seq juvenile mouse hearts | GSE64403 | O'Meara et al. (2015) | |
| Single Cell RNA-Seq | GSE184914 | This Study | |
| Experimental models: Cell lines | |||
| WTC-11 | Coriell Institute | GM25256 | |
| RUES2 | Rockefeller University | hESC-09-0013 | |
| Experimental models: Organisms/strains | |||
| Mice C57BL/6 | Charles River | N/A | |
| Zebrafish AB/Wild Type | Zebrafish International Resource Center, Eugene, OR, USA | N/A | |
| Zebrafish hsWnt8:GFP Tg(hsp70l:Wnt8a-GFP)w34 | Randall Moon's Lab | N/A | |
| Zebrafish hsDKK1:GFP [Tg(hsp70l:DKK1b-GFP)w32 | Randall Moon's Lab | N/A | |
| Zebrafish vmhc:mCherry-NTR [Tg(vmhc:mCherry-Eco.NfsB) | Neil Chi’s Lab | N/A | |
| Oligonucleotides | |||
| gata4 | GGCTCCTCTGAAGGTCAGTC | CAGGCTGTTCCACACTTCAC | |
| nkx2.5 | GGGATGGTAAACCGTGTCTG | TTGCTGTTGGACTGTGAAGG | |
| β-actin | AAGCAGGAGTACGATGAGTC | TGGAGTCCTCAGATGCATTG | |
| pdk2a | AGTTTGTCGCTGCTCTGGTC | ACTGGATTTGTGGCTCCATC | |
| pdk2b | TGATAGAGGCGGTGGAGTTC | TACAGACGGGAGATGGGAAG | |
| pdk3a | ATTCGTCAGCTCCAACATCC | ACACCATCTTAGCGGTTTCG | |
| pdk4 | TATCGACCCAAACTGTGACG | TCCACAGTTGCTCTCATTGC | |
| myl7 | ACCGGGATGGAGTTATCA | CTCCTGTGGCATTAGGG | |
| cpt1b | ATGCTGCAGTATCGCCGTAA | GGAAACTTCTGAGTGTCATCTAGGA | |
| cd36 | GTCCAAACCTGTGTTGGTGC | CTAGGCTCAAAGGTGGCTCC | |
| Software and algorithms | |||
| R | The R Foundation | https://www-r-project-org.offcampus.lib.washington.edu/ | |
| Image J | NIH | https://imagej-nih-gov.offcampus.lib.washington.edu/ij/ | |
| Microsoft office | Microsoft | https://products.office.com | |
| Github | Yuliang Wang's GitHub for Zebrafish Heart Regeneration Project | https://github.com/yuliangwang/zebrafish-heart-regeneration-expression-data-analysis- | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hannele Ruohola-Baker (hannele@u.washington.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
Zebrafish
Wild-type (AB; Zebrafish International Resource Center, Eugene, OR, USA), , hsWnt8:GFP [Tg(hsp70l:Wnt8a-GFP)w34] and hsDKK1:GFP [Tg(hsp70l:DKK1b-GFP)w32] (Stoick-Cooper et al., 2007) and vmhc:mCherry-NTR [Tg(vmhc:mCherry-Eco.NfsB)] (Zhang et al., 2013) were used and maintained using standard procedures (Westerfield, 2000) in accordance with the Institutional Animal Care and Use Committee-approved protocols IACUC 2057-01 & 4364-02. Reverse osmosis system water is used and the water chemistry was adjusted to a temperature of 27.5°C, pH of 7.5, conductivity of 800 μS, hardness of 140 ppm, alkalinity of 35 ppm, dissolved oxygen content of 7.8 mg/L, and a total gas pressure of 101%. The average nitrate level is 55 ppm, the average nitrite level is 0.05 ppm, and the average ammonium level is 0.01 ppm. Larval feed starting at 5 days post fertilization (dpf) is a rotifer polyculture supplemented with the Zeigler larval diet, at 12 dpf live artemia is added to the diet, and at 60-90 dpf the fish are started on the Zeigler adult diet supplemented with live artemia once per day. The health status of the colony showed low presence of pseudoloma neurophilia and an extremely low presence of mycobacterium spp. with no major clinical or subclinical findings of significance.
Young hearts were obtained from 3-day old larvae. Adult heart regeneration studies were performed on fish 3 months to 2 years of age. A mix of genders was used.
Mouse
All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of the University of Washington. Mice with C57BL/6 background were purchased from Charles River. Mice were maintained on standard chow diet and water was available ad libitum in a vivarium with a 12-hr light-dark cycle at 22°C. Adult mouse hearts were males with an age of 3-12 months. Neonatal mice hearts were generated from a mix of genders.
Cell culture
Human embryonic stem cells (RUES2, Rockefeller University; hESC-09-0013) and hiPSC line WTC #11, previously derived in the Conklin laboratory (Hofsteen et al., 2016; Palpant et al., 2015; Kreitzer et al., 2013).
Method details
Chemical ventricular cardiomyocyte ablation of the heart ventricle and young 3-day old heart isolation
All chemical ventricular cardiomyocyte (VCM) ablation were conducted on adult Zebrafish (∼5-12 months old). For chemical mediated VCM ablation, fish were treated with 5 mM Metronidiazole (MTZ; Sigma) containing 0.1% dimethyl sulfoxide (DMSO) or vehicle control (0.1% DMSO) for 48 hours protected from light and refreshed every 24 hours. For Wnt and DKK heat shock heterozygous animals, 7.5 mM MTZ was used. Following chemical treatment fish were placed back in original aquaria and hearts were collected at 0 (prior to MTZ), 3, 7, 12 and 30 days post injury (dpi). Larval hearts (72 hpf) were isolated by repeatedly expelling juvenile fish through a 19-gauge needle, 1.5 inches long, regular bevel and a 6mL syringe into a 1.5mL microcentrifuge tube (Burns and MacRae, 2006).
Zebrafish heat shock
Heterozygous Tg(hsp70l:DKK1b-GFP)w32 and Tg(hsp70l:Wnt8a-GFP)w34 x Tg(vmhc:mCherry-NTR)+/+ fish were used to inhibit and induce canonical Wnt/β-catenin signaling respectively (Stoick-Cooper et al., 2007; Ueno et al., 2007; Weidinger et al., 2005). Wnt8a and Dkk1b induction was achieved by exposing fish to water slowly increased from 28°C to 37°C. Fish remained at 37°C for 60 minutes and were slowly brought back to normal water temperature. Fish were exposed to this heat shock protocol for 2 days prior to MTZ treatment. For the MTZ treatment, fish were heat shocked for 60 minutes, transferred to MTZ overnight and transferred back to heat shock tanks for the 2 days of treatment. Fish were subsequently heat shocked for 3 days post MTZ treatment and hearts were harvested at 3 dpi for analysis.
Rapamycin treatment of zebrafish
Homozygous vmhc:mCherry-NTR fish were treated with rapamycin to determine how TOR inhibition affected heart regeneration. An immersion solution of system water and a final concentration of DMSO (vehicle control) or rapamycin at 0.4 μM was prepared. Fish were put in the water for 48 hours, replacing the water and rapamycin or DMSO every 24 hours. On day 3, the fish were exposed to DMSO or rapamycin and a final concentration of 5 mM MTZ. The water was replaced, and the same conditions were repeated for day 4. On day 5, fish were treated with DMSO or rapamycin with fresh water and drug replaced every 24 hours. At 3 dpi hearts were harvested.
Mouse heart isolation for GC-MS
Adult mice were anesthetized with sodium pentobarbital (150 mg/kg intraperitoneally) prior to heart isolation. Ventricles were removed and freeze clamped in liquid nitrogen. P0.5 and P7 mice were euthanized by decapitation. The hearts were removed and trimmed of atrial tissue, rinsed with PBS to remove blood, and snap frozen in liquid nitrogen. The hearts were stored at −80°C until analysis.
Gas chromatography-mass spectrometry (GC-MS)
Frozen zebrafish heart tissue (20–25 mg) specimens were mixed with a 1.2 mL mixture of cold methanol and chloroform (1:2 v/v; 4°C) and homogenized using tissue teAror homogenizer and sonicated for 20 seconds (Marney et al., 2013). A further 800 μL cold chloroform/distilled water solution (1:1 v/v) was added, the sample was then vortexed and the set aside for 30 minutes on ice to separate the solvent layers. Next, after centrifugation (2000 rpm), the aqueous (top) layer was separated and filtered using 1.5 mL 0.2 μm syringe filter and freeze dried.
To prepare the samples for GC/MS analysis, 30 uL of 20 mg/mL methoxyamine hydrochloride dissolved in pyridine was added to the dried aqueous metabolites, and samples were incubated at 37°C for 90 minutes. MTBSTFA (70 uL) was subsequently added and incubated at 37°C for 30 minutes. Samples were run on an Agilent 5977A Series GC/MSD system.
N = 2 for adult mouse heart, N = 3 for P7 mouse heart and N = 4 for P0.5 mouse heart.
N = 2 for adult uninjured zebrafish heart, N = 2 for 3 dpi and N = 2 for 7 dpi. Each sample for zebrafish hearts was a pool of n = 4–6 hearts. One-way ANOVA was performed to determine significantly changed metabolites.
Bulk RNA-sequencing and bioinformatics
RNA-sequencing samples generated by this study and Bednarek et al. ((Bednarek et al., 2015), accession number GSE71755) were aligned to Ensembl GRCz10 using Tophat ((Trapnell et al., 2009), version 2.0.13). Gene-level read counts were quantified using htseq-count (Anders et al., 2015) using Ensembl GRCz10 gene annotations. Genes with at least 10 normalized read counts summed across RNA-seq samples were kept for further analysis. prcomp function from R was used to for Principal Component Analysis. DESeq (Anders and Huber, 2010) was used for differential gene expression analysis. Genes with Benjamin- Hochberg adjusted False Discovery Rate <0.1 and fold change >2 were considered differentially expressed. topGO R package (Alexa et al., 2006) was used for Gene Ontology enrichment analysis.
For young, 3-day old zebrafish heart samples, we sequenced N = 2 with about n = 500 hearts pooled. For the adult heart samples, we sequenced N = 2 hearts at UI, 3 and 7 dpi with each n = 4–6 pooled hearts. A negative binomial test was performed to determine significantly changed genes. Around 20,000 genes were found. These data can be found in the Data S1.
Single cell RNA-sequencing and bioinformatics
Cells were resuspended in PBS containing 0.04% BSA at a concentration of 2600 cells/33.8 μL solution. In total, 2600 cells per sample were loaded into each well of a 10X Chromium single cell capture chip with a recovery of 272 cells for uninjured heart, 141 cells for 3 days post injury and 368 cells for 7 days post injury sample. The captured cells then underwent lysis, reverse transcription, cDNA amplification, and library preparation with indexing per the manufacturer's protocol (10X Genomics). The libraries were sequenced together on an Illumina HiSeq 4000 (Illumina Inc., San Diego, CA) using a high output 150 cycle kit with read lengths recommended by 10X Genomics. Raw data processing: Cellranger V1.2 was used to process the raw fastq files and to generate unique molecular identifiers (UMI) counts for each gene in each cell.
Monocle version 3 alpha was used for single cell RNA-seq data analysis and visualization (Trapnell et al., 2014). We kept cells with at least 200 expressed genes, and 1000 Unique Molecular Identifiers (UMIs), and less than 40% reads mapped to mitochondrial transcripts. We used preprocessCDS function to preprocess single cell RNA-seq data before clustering, with parameter num_dim = 50. reduceDimension function was used to produce low dimensional representation of the single cell RNA-seq data, and the UMAP option was used. UMAP was recent shown to have superior performance compared to tSNE (Becht et al., 2018). clusterCells function was used for clustering, with method = ’louvian’, resolution = 1e-6. differentialGeneTest function was used to identify differentially expressed genes. Genes with adjusted p-value <0.1 were defined as differentially expressed. Hallmark gene sets (Liberzon et al., 2015) and topGO (Alexa et al., 2006) were used for pathway enrichment testing.
6 hearts, 3 males and 3 females, were isolated from Adult UI, 3 dpi and 7 dpi zebrafish. Each group had 2 sets of samples of 3 hearts each for the digestion process. Each set of 3 hearts were single celled by pooling three harvested adult hearts into a microcentrifuge tube with a digestion buffer (12.5μM CaCl2 plus collagenase II and IV at 5mg/mL each) for 2 hours at 32°C shaking at 800 rpm. After digestion, flick the tube to break apart any pieces of large tissue. Next, wash the cells in 7 subsequent buffers of increasing CaCl2 concentration by gently pipetting up and down with a p1000 and spinning for 5 minutes at 4°C between each buffer (Sander et al., 2013). All cells per group were then pooled and sent for sequencing. The number of cells sequenced per group was: Adult UI - 272 cells; 3 dpi - 114 cells; 7 dpi - 368 cells. 754 cells in total. A negative binomial test was used to determine significant genes.
Proteomics
Proteins were suspended in 1M urea, 50mM ammonium bicarbonate, pH 7.8, and heated to 50°C for 20 minutes. Denatured proteins were reduced with 2 mM DTT, alkylated with 15 mM iodoacetamide, and digested overnight with trypsin. The resulting peptides were desalted on Waters Sep-Pak C18 cartridges. Peptides were separated using a heated 50°C 30 cm C18 columns in a 180 min gradient of 1% to 45% (vol/vol) acetonitrile with 0.1% (vol/vol) formic acid. Peptides were measured on a Thermo Scientific Lumos Orbitrap operated in data-dependent mode with the following settings: 60000 resolution, 400-1600 m/z full scan, Top Speed of 3 seconds, and a 1.8 m/z isolation window. Identification and label free quantification of proteins was done with MaxQuant 1.5 using a 1% false discovery rate (FDR) against the Danio rerio proteome dataset downloaded from Uniprot on July 1st, 2016. Peptides were searched using a 5 ppm mass error and a match between run window of 2 minutes. 2787 proteins were identified and quantified across 2 biological samples per condition (each biological sample was a pool of 8 hearts), and 3 technical replicates per each biological sample. Proteins that were significantly regulated between conditions were identified in Perseus 1.4.1.3 using either a two-way ANOVA with a permutation-based FDR of 5% for PCA plots, or a two-sided permutation-based Student’s t-test with an S0-1 for volcano plots. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD011791.
Cryopreservation
Adult zebrafish hearts were collected at 0, 3, 7, and 30 dpi, rinsed in cold fish fix buffer (recipe below). Hearts were fixed in fish fix buffer with 4% paraformaldehyde (PFA) at 4°C overnight, rinsed with fish fix buffer without PFA and incubated in 30% sucrose in PBS overnight. Hearts were then frozen in Tissue Freezing MediumTM (General Data Healthcare) and sectioned on a Leica CM1850 Cryostat at −26°C. 10 μm sections were placed on SuperfrostTM slides (Fisher Scientific) and stored at −80°C.
Immunofluorescence
Sections were allowed to thaw at room temperature and circled with a hydrophobic pen (Fisher Scientific). Slides were soaked in 10% SDS (Sigma-Aldrich) and rinsed in PBS containing 0.2% Triton-X 100 (PBS-T, Sigma-Aldrich). Slides were blocked at room temperature for 1 hour in blocking buffer: a solution of PBS-T containing 5% normal goat serum (NGS) (MP Biomedicals) and 1% Bovine Serum Albumin (BSA, VWR). Slides were incubated with primary antibodies overnight at 4°C in blocking buffer at the concentrations listed in Table S15. Slides were rinsed with PBS-T and incubated with secondary antibodies: DAPI (0.02 μg/mL, Molecular Probes), goat anti-rabbit 488, and goat anti-mouse 647 (1:500, Molecular Probes) for 2 hours at room temperature in blocking buffer. (NTR was visualized using endogenous mCherry expression). Slides were mounted with Vectashield® (Vector Labs) and imaged using a Leica TCS-SPE confocal microscope at 10x ,40x, or 63x. Quantification of Mef2c and PCNA positive nuclei were performed using ImageJ. Antibodies used can be found in Table S15.
Images were opened in Image J as a maximum projection of Z planes. Images were smoothed to reduce noise and split into separate channels. Each channel was made binary using the threshold tool to allow for quantification of particles. Co-localization was assessed by multiplying binary images to achieve a new binary image with values for overlapping pixels. Nuclei were quantified in the multiplied binary image by using the automated particle analysis tool in ImageJ. Particles less than 5 sq μm and greater than 100 sq μm were eliminated. The area of the ventricle was analyzed by drawing a region of interest around the ventricle and measuring the area in ImageJ.
For MTZ only experiments N=2 hearts for Adult UI, 3 hearts for 3 dpi, 7 dpi, and 30 dpi. 2-3 images per heart were quantified and a one-way ANOVA was used to determine significance.
For rapamycin Mef2c and Pcna experiments quantification was done using image semi automation calculations on ImageJ and verified by manual hand counts of sampled data. For rapamycin pS6 and mCherry experiments hand counting was performed. N=5 Hearts for adult 3 dpi - average of 2 images/heart were quantified. N=1 heart for adult UI – 3 images/heart were quantified.
For whole mount heart images, spinning disc imaging was used for zebrafish vmhc:mCherry-NTR expression. 6-12 months old zebrafish were euthanized using standard operation procedure. Hearts were harvested and washed 3 times in 1x PBS. Finally, hearts were submerged in 1x PBS in optical glass bottom chamber and imaged using a Live-Cell Inverted Widefield/Spinning Disk Confocal microscope using 2x magnification. Images were processed using image J software. The same 6–12-month-old zebrafish hearts that were imaged for ventricular mCherry-NTR expression were also imaged using a standard stereoscope under bright light.
Cell culture and cardiomyocyte directed differentiation
hESCs were plated on matrigel (BD) coated tissue culture plates and maintained with irradiated mouse embryonic fibroblast conditioned media containing 5 ng/mL human bFGF (Peprotech, 100-18B). High density hESC directed differentiation towards cardiomyocyte was conducted. hiPSCs were cultured on Matrigel growth factor-reduced basement membrane matrix (Corning) in mTeSR media (StemCell Technologies). A monolayer-based directed differentiation protocol was followed to generate hESC-CMs and hiPSC-CMs, as done previously (Palpant et al., 2017). Cardiomyocytes from a single line were harvested on day 25 of the protocol, pooled and randomly re-plated in a 24-well tissue culture plates at a seeding density of 200k cells/well in 1 mL of RMPI (Invitrogen) containing B27 supplement. Cardiomyocytes were allowed to recover for 4 days until they were exposed to recombinant Wnt3A (100 ng/mL, R&D Systems) or CHIR99021 (5μM, Cayman Chemicals), Rapamycin (200 nM, Selleck Chemicals), and XAV-939 (5 μM, Tocris) for 2 days prior to collection for flow cytometry or protein.
Reverse transcriptase-quantitative PCR (RT-qPCR)
RNA was isolated from young (3 days old), adult (∼6 months old), and regenerating (3 and 7 dpi) zebrafish hearts as per manufacturers' protocol (TRIzol, ThermoFisher). Complementary DNA (cDNA) was synthesized using the Superscript III enzyme kit (Invitrogen). RT-qPCR was conducted using the sensimix SYBR PCR Kit (Bioline) on a 7900HT Fast-Real-Time PCR system (Applied Biosystems). Transcript abundance were normalized to the house keeping gene β-actin and primers are listed in Table S16.
Western blotting
Adult zebrafish hearts were collected at adult UI and 3 dpi, washed in cold phosphate buffered saline (PBS), and frozen using dry ice. Human embryonic stem cell (hESC) derived cardiomyocytes (CM) were washed with ice cold PBS prior to collection and flash freezing. Hearts and hESC-CMs were lysed using ice cold cell lysis buffer (Cell Signaling, 9803) containing protease/phosphatase inhibitor cocktail (Cell Signaling, 5872) and cellular debris were separated by centrifugation. Protein concentration was quantified and normalized using a BCA Protein Assay Kit (ThermoFisher). Primary and secondary antibodies are listed in Table S17.
Western blots were quantified using the ImageJ gel analysis toolbar. The area under the curve for peak intensity was normalized to Ac-α-Tubulin or β-actin for each sample, with the exception of pS6 which was normalized to S6 for all samples. Samples were divided by the mean of the controls (-MTZ, -hsDkk1b, -hsWnt8a, -Rapamycin) to calculate fold change relative to control. A two-tailed student’s t-test was performed to determine significance between control and experimental groups for each protein.
Western blots of adult zebrafish UI and 3 dpi consisted of N = 3 with each n = 4–6 pooled hearts. hsWnt8a and hsDkk1b N = 3–4 with each n = 4 pooled hearts. Rapamycin experiments used N = 3 with each n = 3 pooled hearts. Proteomics validation of Sdhaf3 consisted of UI, 3 dpi, and 7 dpi N = 1 with each n = 4–6 pooled hearts. hPSC-CM protein consisted of N = 2 for Figure 5G and N = 3 for Figure 5H.
Cardiomyocyte purity assay
Cells were labeled for flow cytometry using cardiac troponin T (Thermo Scientific) or an IgG corresponding isotype control. Cells were analyzed using a BD FACSCANTO II (Beckton Dickinson, San Jose, CA) with FACSDiva software (BD Biosciences). Instrument settings were adjusted to avoid spectral overlap. Data analysis was performed using FlowJo (Tree Star, Ashland, Oregon). Only CM with greater than 70% purity were used.
Glutamine starvation assay
hESC-CMs were cultured in RPMI + B27 insulin supplement media until day 30. Cells were then cultured for 24 hours in RPMI + B27 no insulin media. Next cells were starved for 6 hours using RPMI that had no glutamine and no B27 supplement. Finally, cells were treated for 2 hours with each of the test conditions 1) RPMI media containing glutamine + B27 insulin supplement + vehicle (DMSO), 2) RPMI media with no glutamine and no supplement with the drug GPNA (L-Glutamic acid γ-(p-nitroanilide) hydrochloride, MP Biomedicals) at a final concentration of 5mM and 3) media containing RPMI no glutamine and no supplement + vehicle (DMSO). Following the 2 hour treatment, cells were harvested for protein.
EdU (5-ethynyl-2'-deoxyuridine) assay
Cardiomyocytes were exposed to 10 μM EdU (DMSO) for 24 hours one day prior to collection for flow cytometry. Samples were trypsinized to obtain single cells and fixed for 10 minutes with 4% paraformaldehyde. Staining procedures for EdU incorporation were performed using the Click-iT EdU Flow Cytometry Cell Proliferation Assay kit using an Alexa Fluor 647 antibody (ThermoFisher).
Transmission electron microscopy (TEM)
Hearts were fixed in 4% glutaraldehyde in sodium cacodylate buffer, post fixed in osmium tetroxide, en bloc stained in 1% uranyl acetate, dehydrated through a series of ethanol, and embedded in Epon Araldite. 70 nm sections were cut on a Leica EM UC7 ulta microtome and viewed on a JEOL 1230 TEM. Statistical analysis of mitochondrial area was performed using a Chi-squared test.
TUNEL assay
6–12-month-old zebrafish were treated with MTZ or DMSO (control) for 2 days and were immediately collected using standard euthanizing protocol. Hearts were harvested, rinsed in cold fish fix buffer and sectioned exactly as described above. To detect cardiomyocyte cell death, we used Click-iT™ Plus TUNEL Assay for In Situ Apoptosis Detection, Alexa Fluor™ 488 dye (Thermofisher C10617). Sections were treated following the manufacturer’s protocol. Briefly, sections were permeabilized using a Proteinase K solution for 45 minutes followed by a 2-hour terminal deoxynucleotidyl transferase (TdT) reaction mixture incubation. Sections were then treated with 1x Click-iT™ Plus reaction solution for 1h, washed and finally imaged using SP8 confocal at 64x magnification. Images were processed using image J software.
Quantification and statistical analysis
All of the statistical methods for each experiment can be found in the Figure legends as well as in the method details section. P-values and individual data points for the main figures can be found in the Data S2.
Acknowledgments
We thank the ISCRM community for helpful discussions during this work. This work is supported by grants from the National Institutes of Health 1P01GM081619 for RM, CM, and HR-B and R01GM097372, R01GM97372-03S1 and R01GM083867 and the NHLBI Progenitor Cell Biology Consortium (U01HL099997; UO1HL099993), R01HL135143, R01HL146436, P30AR074990 and R44HL149566 for HR-B. This work was supported in part by the University of Washington’s Proteomics Resource (UWPR95794). LA was supported by the Bioengineering Cardiovascular Training Grant T32-EB1650. PH was supported by the Experimental Pathology of Cardiovascular Disease Training Grant T32-HL007312. JWM is an ISCRM Fellow, supported by funds from Washington State, Wellstone Muscular Dystrophy Cooperative Research Center: U54AR065139 and the NSERC Alexander Grahm Bell Graduate Scholarship. SL is a WRF and ISCRM Fellow, supported by fund from Washington state. EC is supported by NSF. We would like to thank the Mike and Lynn Garvey Cell Imaging Lab at the Institute for Stem Cell and Regenerative Medicine (UW), the Vision Core for their TEM services (P30 EY001730). The Cell Analysis Facility Flow Cytometry and Imaging Core in the Department of Immunology at the University of Washington (ISCRM) and the ISCRM Aquatics core for zebrafish husbandry and maintenance.
Author contributions
Conceptualization, J.W.M., S.L., P.H., and H.R.-B.; Software, Y.W.; Investigation, J.W.M., S.L., P.H., D.I.M., E.C., A.M.R., G.S., L.A., J.M.G., I.P., and Y.W.; Writing—Original Draft, J.W.M., S.L., P.H., and H.R.-B.; Writing—Review and Editing, J.W.M., S.L., P.H., Y.W., and H.R.-B.; Supervision, J.M., A.M., M.T.C., R.T., C.E.M., R.T.M., and H.R.-B.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. We worked to ensure diversity in experimental samples through the selection of the cell lines. We worked to ensure diversity in experimental samples through the selection of the genomic datasets. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103574.
Supplemental information
Data and code availability
RNA-sequencing, single cell RNA-sequencing, and proteomics data have been deposited at GEO and are publicly available as of the date of publication. This paper also analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. GC-MS metabolite data are available in the source data file. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at GitHub and is publicly available as of the date of publication.
DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aguirre A., Montserrat N., Zacchigna S., Nivet E., Hishida T., Krause M.N., Kurian L., Ocampo A., Vazquez-Ferrer E., Rodriguez-Esteban C., et al. In vivo activation of a conserved MicroRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumatin F., O'prey J., Barthet V.J.A., Zunino B., Parvy J.P., Bachmann A.M., O'prey M., Kania E., Gonzalez P.S., Macintosh R., et al. mTORC1 activation requires DRAM-1 by facilitating lysosomal amino acid efflux. Mol. Cell. 2019;76:163–176 e8. doi: 10.1016/j.molcel.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., Mcinnes L., Healy J., Dutertre C.A., Kwok I.W.H., NG L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bednarek D., Gonzalez-Rosa J.M., Guzman-Martinez G., Gutierrez-Gutierrez O., Aguado T., Sanchez-Ferrer C., Marques I.J., Galardi-Castilla M., De Diego I., Gomez M.J., et al. Telomerase is essential for zebrafish heart regeneration. Cell Rep. 2015;12:1691–1703. doi: 10.1016/j.celrep.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S.L., Szewczykowska M., Jackowska T., Dos Remedios C., et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Brown K., Yang P., Salvador D., Kulikauskas R., Ruohola-Baker H., Robitaille A.M., Chien A.J., Moon R.T., Sherwood V. WNT/beta-catenin signaling regulates mitochondrial activity to alter the oncogenic potential of melanoma in a PTEN-dependent manner. Oncogene. 2017;36:3119–3136. doi: 10.1038/onc.2016.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.G., MacRae C.A. Purification of hearts from zebrafish embryos. BioTechniques. 2006;40 274, 276, 278 passim. [PubMed] [Google Scholar]
- Cai X., Cao C., Li J., Chen F., Zhang S., Liu B., Zhang W., Zhang X., Ye L. Inflammatory factor TNF-alpha promotes the growth of breast cancer via the positive feedback loop of TNFR1/NF-kappaB (and/or p38)/p-STAT3/HBXIP/TNFR1. Oncotarget. 2017;8:58338–58352. doi: 10.18632/oncotarget.16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Navis A., Cox B.D., Dickson A.L., Gemberling M., Karra R., Bagnat M., Poss K.D. Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–243. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F., Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development. 2012;139:1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- Chen J.X., Krane M., Deutsch M.A., Wang L., Rav-Acha M., Gregoire S., Engels M.C., Rajarajan K., Karra R., Abel E.D., et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ. Res. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.Y., Gemberling M., Wang J., Holdway J.E., Shen M.C., Karlstrom R.O., Poss K.D. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140:660–666. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Palpant N.J., Gantz J.A., et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle-Perez B., Casagolda D., Lugilde E., Valls G., Codina M., Dave N., DE Herreros A.G., Dunach M. Wnt controls the transcriptional activity of Kaiso through CK1epsilon-dependent phosphorylation of p120-catenin. J. Cell Sci. 2011;124:2298–2309. doi: 10.1242/jcs.082693. [DOI] [PubMed] [Google Scholar]
- Egan D.F., Shackelford D.B., Mihaylova M.M., Gelino S., Kohnz R.A., Mair W., Vasquez D.S., Joshi A., Gwinn D.M., Taylor R., et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglia M.J., Poss K.D. Building and re-building the heart by cardiomyocyte proliferation. Development. 2016;143:729–740. doi: 10.1242/dev.132910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., Marin-Juez R., El-Sammak H., Beisaw A., Ramadass R., Kuenne C., Guenther S., Konzer A., Bhagwat A.M., Graumann J., Stainier D.Y. Stimulation of glycolysis promotes cardiomyocyte proliferation after injury in adult zebrafish. EMBO Rep. 2020;21:e49752. doi: 10.15252/embr.201949752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabisonia K., Prosdocimo G., Aquaro G.D., Carlucci L., Zentilin L., Secco I., Ali H., Braga L., Gorgodze N., Bernini F., et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418–422. doi: 10.1038/s41586-019-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Bailey T.J., Hyde D.R., Poss K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611–620. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Karra R., Dickson A.L., Poss K.D. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4:e05871. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go A.S., Mozaffarian D., Roger V.L., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., et al. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G., Song M., Csordas G., Kelly D.P., Matkovich S.J., Dorn G.W., 2nd Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa J.M., Burns C.E., Burns C.G. Zebrafish heart regeneration: 15 years of discoveries. Regeneration (Oxf). 2017;4:105–123. doi: 10.1002/reg2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa J.M., Peralta M., Mercader N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012;370:173–186. doi: 10.1016/j.ydbio.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Gutstein D.E., Liu F.Y., Meyers M.B., Choo A., Fishman G.I. The organization of adherens junctions and desmosomes at the cardiac intercalated disc is independent of gap junctions. J. Cell Sci. 2003;116:875–885. doi: 10.1242/jcs.00258. [DOI] [PubMed] [Google Scholar]
- Han Y., Chen A., Umansky K.B., Oonk K.A., Choi W.Y., Dickson A.L., Ou J., Cigliola V., Yifa O., Cao J., et al. Vitamin D stimulates cardiomyocyte proliferation and controls organ size and regeneration in zebrafish. Dev. Cell. 2019;48:853–863.e5. doi: 10.1016/j.devcel.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Shiomi T., Hozumi S., Kikuchi Y. Mechanistic target of rapamycin complex 1 signaling regulates cell proliferation, cell survival, and differentiation in regenerating zebrafish fins. BMC Dev. Biol. 2014;14:42. doi: 10.1186/s12861-014-0042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsteen P., Robitaille A.M., Chapman D.P., Moon R.T., Murry C.E. Quantitative proteomics identify DAB2 as a cardiac developmental regulator that inhibits WNT/β-catenin signaling. Proc. Natl. Acad. Sci. U S A. 2016;113:1002–1007. doi: 10.1073/pnas.1523930113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkoop H., De bakker D.E., Aharonov A., Kruse F., Shakked A., Nguyen P.D., De Heus C., Garric L., Muraro M.J., Shoffner A., et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife. 2019;8:e50163. doi: 10.7554/eLife.50163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Marti M., Raya A., Izpisua Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J.E., Werdich A.A., Anderson R.M., Fang Y., Egnaczyk G.F., Evans T., Macrae C.A., Stainier D.Y., Poss K.D. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J.H., Iyer V.R. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer F.R., Salomonis N., Sheehan A., Huang M., Park J.S., Spindler M.J., Lizarraga P., Weiss W.A., So P.L., Conklin B.R. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am. J. Stem Cells. 2013;2:119–131. [PMC free article] [PubMed] [Google Scholar]
- Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O'donnell K.A., Kim J.W., Yustein J.T., Lee L.A., Dang C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kim S.G., Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C.L., Schebesta M., Makino S., Weber G.J., Keating M.T. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud A.I., O'meara C.C., Gemberling M., Zhao L., Bryant D.M., Zheng R., Gannon J.B., Cai L., Choi W.Y., Egnaczyk G.F., et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev. Cell. 2015;34:387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Juez R., El-Sammak H., Helker C.S.M., Kamezaki A., Mullapuli S.T., Bibli S.I., Foglia M.J., Fleming I., Poss K.D., Stainier D.Y.R. Coronary revascularization during heart regeneration is regulated by epicardial and endocardial cues and forms a scaffold for cardiomyocyte repopulation. Dev. Cell. 2019;51:503–515 e4. doi: 10.1016/j.devcel.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marney L.C., Kolwicz S.C., Tian R., synovec R.E. Sample preparation methodology for mouse heart metabolomics using comprehensive two-dimensional gas chromatography coupled with time-of-flight mass spectrometry. Talanta. 2013;108:123–130. doi: 10.1016/j.talanta.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F., Hockenbery D. MYC and mitochondrial biogenesis. Cold Spring Harb. Perspect. Med. 2014;4:a014225. doi: 10.1101/cshperspect.a014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A.T., Shiojima I., Akazawa H., Hidaka K., Morisaki T., Kikuchi A., Komuro I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl. Acad. Sci. U S A. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C., Acebron S.P. Mitotic and mitogenic Wnt signalling. EMBO J. 2012;31:2705–2713. doi: 10.1038/emboj.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara C.C., Wamstad J.A., Gladstone R.A., Fomovsky G.M., Butty V.L., Shrikumar A., Gannon J.B., Boyer L.A., Lee R.T. Transcriptional reversion of cardiac myocyte fate during mammalian cardiac regeneration. Circ. Res. 2015;116:804–815. doi: 10.1161/CIRCRESAHA.116.304269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia-Desai S., Rost M.S., Schumacher J.A., Ton Q.V., Craig M.P., Baltrunaite K., Koenig A.L., Wang J., Poss K.D., Chi N.C., et al. Myocardium and BMP signaling are required for endocardial differentiation. Development. 2015;142:2304–2315. doi: 10.1242/dev.118687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant N.J., Hofsteen P., Pabon L., Reinecke H., Murry C.E. Cardiac development in zebrafish and human embryonic stem cells is inhibited by exposure to tobacco cigarettes and E-cigarettes. PLoS One. 2015;10:e0126259. doi: 10.1371/journal.pone.0126259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant N.J., Pabon L., Friedman C.E., Roberts M., Hadland B., Zaunbrecher R.J., Bernstein I., Zheng Y., Murry C.E. Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nat. Protoc. 2017;12:15–31. doi: 10.1038/nprot.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.I., Kim S.W., Lyons J.P., Ji H., Nguyen T.T., Cho K., Barton M.C., Deroo T., Vleminckx K., Moon R.T., Mccrea P.D. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Sanchez-Iranzo H., Galardi-Castilla M., Sanz-Morejon A., Gonzalez-Rosa J.M., costa R., Ernst A., Sainz DE Aja J., Langa X., Mercader N. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc. Natl. Acad. Sci. U S A. 2018;115:4188–4193. doi: 10.1073/pnas.1716713115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander V., Sune G., Jopling C., Morera C., Izpisua Belmonte J.C. Isolation and in vitro culture of primary cardiomyocytes from adult zebrafish hearts. Nat. Protoc. 2013;8:800–809. doi: 10.1038/nprot.2013.041. [DOI] [PubMed] [Google Scholar]
- Saxton R.A., Knockenhauer K.E., Wolfson R.L., Chantranupong L., Pacold M.E., Wang T., Schwartz T.U., Sabatini D.M. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M., WU T.D., Guerquin-Kern J.L., Lechene C.P., Lee R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2012;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M., Hall M.N. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- Singh A.R., Sivadas A., Sabharwal A., Vellarikal S.K., Jayarajan R., Verma A., Kapoor S., Joshi A., Scaria V., Sivasubbu S. Chamber specific gene expression landscape of the zebrafish heart. PLoS One. 2016;11:e0147823. doi: 10.1371/journal.pone.0147823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper C.L., Weidinger G., Riehle K.J., Hubbert C., Major M.B., Fausto N., Moon R.T. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor E., Poss K.D. Cardiac regeneration strategies: staying young at heart. Science. 2017;356:1035–1039. doi: 10.1126/science.aam5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A.D., Golob J.L., Pabon L., Reinecke H., Moon R.T., Murry C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uygur A., Lee R.T. Mechanisms of cardiac regeneration. Dev. Cell. 2016;36:362–374. doi: 10.1016/j.devcel.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Panakova D., Kikuchi K., Holdway J.E., Gemberling M., Burris J.S., Singh S.P., Dickson A.L., Lin Y.F., Sabeh M.K., et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G., Thorpe C.J., Wuennenberg-Stapleton K., Ngai J., Moon R.T. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr. Biol. 2005;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Westerfield M. Univ. of Oregon Press; 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- Wolfson R.L., Chantranupong L., Saxton R.A., Shen K., Scaria S.M., Cantor J.R., Sabatini D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K., Qiu S., Tian L., Snider W.D., Flannery J.G., Schaffer D.V., Chen B. Wnt regulates proliferation and neurogenic potential of Muller Glial cells via a lin28/let-7 miRNA-dependent pathway in adult mammalian retinas. Cell Rep. 2016;17:165–178. doi: 10.1016/j.celrep.2016.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Lu B., Zamponi R., Yang Z., Wetzel K., Loureiro J., Mohammadi S., Beibel M., Bergling S., Reece-Hoyes J., et al. mTORC1 signaling suppresses Wnt/beta-catenin signaling through DVL-dependent regulation of Wnt receptor FZD level. Proc. Natl. Acad. Sci. U S A. 2018;115:E10362–E10369. doi: 10.1073/pnas.1808575115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.F., Ocorr K., Kang G., Chen J., Stainier D.Y., et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013;498:497–501. doi: 10.1038/nature12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ben-Yair R., Burns C.E., burns C.G. Endocardial notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through wnt pathway antagonism. Cell Rep. 2019;26:546–554 e5. doi: 10.1016/j.celrep.2018.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing, single cell RNA-sequencing, and proteomics data have been deposited at GEO and are publicly available as of the date of publication. This paper also analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. GC-MS metabolite data are available in the source data file. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at GitHub and is publicly available as of the date of publication.
DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.