Abstract
Polypurine tracts are important elements of eukaryotic promoters. They are believed to somehow destabilize chromatin, but the mechanism of their action is not known. We show that incorporating an A16 element at an end of the nucleosomal DNA and further inward destabilizes histone-DNA interactions by 0.1 ± 0.03 and 0.35 ± 0.04 kcal mol−1, respectively, and is accompanied by 1.5- ± 0.1-fold and 1.7- ± 0.1-fold increases in position-averaged equilibrium accessibility of nucleosomal DNA target sites. These effects are comparable in magnitude to effects of A16 elements that correlate with transcription in vivo, suggesting that our system may capture most of their physiological role. These results point to two distinct but interrelated models for the mechanism of action of polypurine tract promoter elements in vivo. Given a nucleosome positioned over a promoter region, the presence of a polypurine tract in that nucleosome's DNA decreases the stability of the DNA wrapping, increasing the equilibrium accessibility of other DNA target sites buried inside that nucleosome. Alternatively (if nucleosomes are freely mobile), the presence of a polypurine tract provides a free energy bias for the nucleosome to move to alternative locations, thereby changing the equilibrium accessibilities of other nearby DNA target sites.
Eukaryotic site-specific DNA binding proteins often occlude much of the entire circumference of their DNA target sites yet are able to bind to target sites that are wrapped in nucleosomes and hence inaccessible. How this is accomplished is not known. Earlier ideas that entry to such sites might be dependent on the prior action of histone acetylases or ATP-dependent chromatin remodeling machines are supplanted by recent discoveries that these factors themselves are recruited to specific chromatin regions by previously bound site-specific regulatory proteins (6, 26).
It is likely that multiple distinct mechanisms may each contribute to allowing and regulating the initial entry of regulatory proteins into their target sites in chromatin. One pathway that is not understood mechanistically but appears to be involved utilizes polypurine tracts that are incorporated into genomic DNA, near DNA target sites for upstream activator proteins. Polypurine tracts 15 to 30 bp long are overrepresented in the genomes of all eukaryotes examined (3) and are particularly enriched in promoter regions. In yeast, approximately one out of every four promoters includes an uninterrupted poly(dA) tract, and numerous additional promoters contain imperfect ones (3, 14, 33). The mechanisms by which these elements contribute to gene activation are not known, but studies of Saccharomyces cerevisiae (5, 13, 22, 35) and Candida glabrata (40) suggest that they may act to alter the stability or dynamics of nucleosomes, somehow enhancing the ability of gene activator proteins to bind to nearby DNA target sites.
Studies of DNA sequences present in isolated natural nucleosomes revealed a preference for stretches of poly(dA-dT) to occur at the ends of the nucleosomal DNA versus the middle (31), consistent with the view that poly(dA-dT) may have an intrinsic preference for adopting distinctive straight helical structures (7, 23, 24). While poly(dA-dT) elements do not necessarily exclude nucleosomes in vivo (17), longer poly(dA-dT) elements can exhibit this distinctive unbent DNA conformation in vivo and can disrupt the ordering of positioned nucleosomes in minichromosomes (33).
Several studies have investigated the effects on histone-DNA interaction affinities when poly(dA-dT) elements are incorporated into nucleosomal DNA. One study (12) reports that a 40-bp poly(dA-dT) stretch containing two 1-bp interruptions, roughly centered in a nucleosomal DNA, destabilized the nucleosome by ∼0.8 kcal mol−1 relative to a similar sequence containing alternating A-T dinucleotides in place of the poly(dA-dT). Another study (10) reports that individual 10-bp-long dA-dT stretches destabilized nucleosomes by ∼0.2 to 0.3 kcal mol−1, independent of their position in the nucleosome. However, a more recent report (21) suggested the opposite, namely, that 25-bp-long stretches of poly(dA-dT) actually stabilize the nucleosome by up to ∼1 kcal mol−1, with the amount depending on position of the (dA-dT) tract. Thus, the contributions of poly(dA-dT) elements to the affinity of histone-DNA interactions remain uncertain, and in any case it is not known how these effects on affinity would relate to changes in the behaviour of nucleosomes.
In this study, we used a purified in vitro system to examine the effects of incorporating poly(dA-dT) elements into two different locations inside nucleosomal DNA. We quantified the effects on the free energy of histone-DNA interactions and tested for and quantified effects on the equilibrium accessibility of DNA target sites inside the nucleosomes. We find that incorporating an A16 element at one end of the nucleosomal DNA and further inward destabilizes histone-DNA interactions by 0.1 ± 0.03 and 0.35 ± 0.04 kcal mol−1, respectively. This is accompanied by 1.5- ± 0.1-fold and 1.7- ± 0.1-fold increases, respectively, in position-averaged equilibrium constants for the dynamic accessibility of nucleosomal DNA target sites. These effects of A16 elements on accessibility of DNA target sites in vitro are comparable in magnitude to the effects seen in vivo, suggesting that this system may be capturing most of the physiological role of the polypurine tracts. These results point to two distinct but interrelated models for the mechanism of action of polypurine tract promoter elements in vivo.
MATERIALS AND METHODS
Preparation of DNA and histones.
Construct 601.2 (174 bp) was prepared by PCR as previously described (2). The slightly shorter (152-bp) construct 601.3 was chosen based on exonuclease III mapping data for nucleosome positioning on 601.2; it incorporates the 145-bp mapped nucleosome region of 601.2 with an additional 4 bp of 601.2 sequence on the left side and 3 bp on the right side (for the sequence as indicated in Fig. 1). Construct 601.3 and its derivatives were prepared by PCR using 601.2 DNA as the template. The primer pairs were as follows (nucleotide changes from 601.2 sequence are capitalized; changes represent the introduction of A16 elements or 16-bp segments of randomly chosen bacterial plasmid DNA sequence). For construct 601.3, the primers were 601.3 152 LE (left end; gcgggcgccctgcagaagcttg) and 601.3 152 RE (right end; gatgtatatatctgacacgtgc). For derivatives of 601.3, we used primer 601.3 152 RE for the right-hand end primer, together with the following left-hand end primers: for construct 601.3(A16End), AAAAAAAAAAAAAAAAgcttggtcccggggcc; for construct 601.3(A16Mid), gcgggcgccctgcagaagcttggtcccggggccgctAAAAAAAAAAAAAAAAgctctggatccgcttgatc; for construct 601.3(Random End), CAAGTGGACGGAGCATagcttggtcccggggccg; and for construct 601.3(Random Mid), gcgggcgccctgcagaagcttggtcccggggccgctCAAGTGGACGGAGCATgctctggatccgcttgatcg.
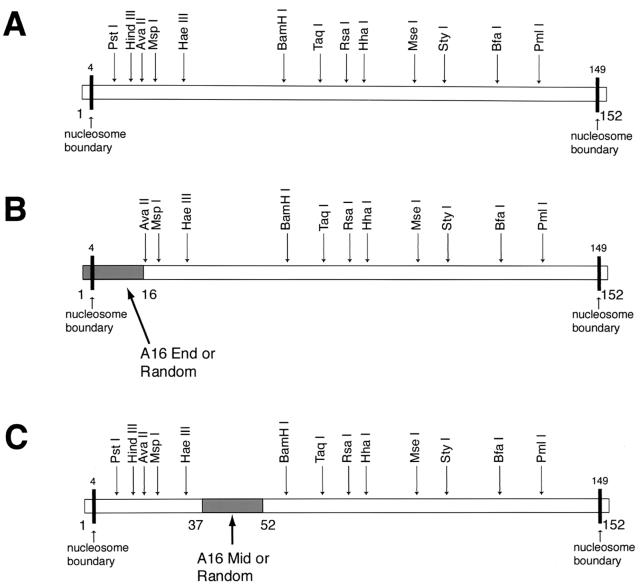
FIG. 1.
DNA constructs used. (A) Construct 601.3. The boundaries of the nucleosomal DNA as mapped on our earlier study of the slightly longer construct 601.2 are indicated by the black vertical bars; relative locations of specific restriction enzyme recognition sites are shown. The other sequences in the 601.3 series (B and C) incorporate various alterations from 601.3 that are represented as shaded boxes. (B) 601.3(A16End) and 601.3(Random End). The shaded box extends from bp 1 to 16 and represents poly(dA-dT) DNA [601.3(A16End)] or random pGEM3z DNA [601.3(Random End)]. (C) 601.3(A16Mid) and 601.3(Random Mid) DNA. The shaded box extends from bp 37 to 52 and represents poly(dA-dT) DNA [601.3(A16Mid)] or random pGEM3z DNA [601.3(Random Mid)].
All PCR products were purified by ion-exchange high-pressure liquid chromatography (HPLC) on an anion-exchange column (Mono-Q HR5/5) using a linear gradient of 0.65 M NaCl in Tris-EDTA (TE; pH 8.0, room temperature) to 0.8 M NaCl in TE over 90 min at a flow rate of 0.25 ml min−1. The purified products were concentrated on Centricon-30 filters (Millipore) and resuspended in 0.1× TE (1 mM Tris [pH 8.0], 0.1 mM EDTA). A typical yield from 10 ml of PCR synthesis was 150 μg of DNA after HPLC purification. Chicken erythrocyte histones were prepared as described elsewhere (8).
Reconstitution and purification of nucleosome core particles.
Prior to reconstitution, construct DNAs were labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs [NEB]). The 256-bp EcoRI fragment of the natural 5S gene nucleosome positioning sequence (34) used as a reference for free energy measurement (see below) was labeled by filling in the ends with the Klenow fragment of Escherichia coli DNA polymerase I, using dTTP and [α-32P]dATP. Reconstitution reaction mixtures contained 200 ng of labeled construct DNA, 19.2 μg of chicken erythrocyte core particle DNA, and 15.5 μg of chicken erythrocyte histone octamer in a 50-μl volume of 2.0 M NaCl, in 0.1× TE–0.5 mM phenylmethylsulfonyl fluoride (PMSF)–0.1 mM benzamidine (BZA). The reconstitutions were performed by a gradual stepwise salt dialysis, beginning at 2.0 M NaCl and then stepping successively to 1.5 M NaCl, 1.0 M NaCl, 0.5 M NaCl, and 5 mM NaCl, each for a minimum of 2 h and each supplemented with 0.5× TE, 0.1 mM BZA, and 0.5 mM PMSF. A final overnight dialysis step into 0.5× TE was performed before further processing. All dialyses were performed at 4°C.
Core particle samples were run on 5 to 30% (wt/vol) sucrose gradients (in 0.5× TE) at 41,000 rpm in a Beckman SW41 rotor for 24 h at 4°C. Gradients were fractionated into 0.5-ml fractions and quantified by Cerenkov counting. Fractions containing nucleosome core particles were pooled and exchanged into 0.5× TE on Centricon-30 concentrators and analyzed by native polyacrylamide gel electrophoresis.
Competitive reconstitutions and free energy measurements.
Free energies for histone binding in nucleosome reconstitution were measured using the double-dialysis competitive reconstitution procedure as described previously (18, 37). Chicken erythrocyte core particle DNA (30 μg) and tracer amounts of the gel-purified, radiolabeled DNA tracer were mixed with 2 μg of histone octamer in a total volume of 50 μl containing 10 mM Tris-HCL (pH 7.5), 1 mM EDTA, 2 M NaCl, 0.5 mM PMSF, and 1 mM BZA. The mixture was loaded into microdialysis buttons, which were then loaded into a dialysis bag containing approximately 200 ml of the same buffer. Samples were dialyzed for ≥2 h at 4°C in the starting buffer, followed by two dialyses at 4°C in 0.5× TE containing PMSF and BZA for ≥12 h. Aliquots of each competitive reconstitution were run on 5% native polyacrylamide gels containing 1/3× TBE (33 mM Tris-borate, 0.67 mM EDTA) and quantified by phosphorimager analysis. Equilibrium constants were calculated as the ratios of background subtracted counts in nucleosomal bands (or sets of bands) to counts in free DNA bands, and free energies were calculated from the relationship ΔG° = −RT ln Keq. ΔΔG°s represent differences between ΔG°s measured for a test sequence and a reference standard sequence, measured at the same time in the identical competitive environment. We used the well-characterized EcoRI fragment of the sea urchin 5S RNA gene nucleosome positioning sequence (34) as a reference tracer DNA.
Restriction enzyme assays.
Nucleosome samples were digested with the following enzymes, all at 37°C: PstI, HindIII, MspI, HaeIII, BamHI, RsaI, HhaI, MseI, StyI, BfaI, and PmlI. TaqI digestions were carried out at 65°C. All enzymes were obtained in their most concentrated commercially available form from NEB, and all digestions were carried out with the buffer supplied by NEB, supplemented with 100 μg of bovine serum albumin per ml. For digestions on naked DNA, we typically used 1 × 104- to 2 × 104-fold-lower enzyme concentration. Glycerol was added to all naked DNA digests to the same final concentration present in the corresponding core particle digestion and never exceeded 5% (vol/vol) in any reaction. The buffers used for each enzyme are as follows: 10 mM bis Tris propane-HCl–10 mM MgCl2–1 mM dithiothreitol (DTT) (pH 7.0) for RsaI and PmlI; 10 mM Tris-HCl–10 mM MgCl2–50 mM NaCl–1 mM DTT (pH 7.9) for HindIII, MspI, HaeIII, and MseI; 50 mM Tris-HCl–10 mM MgCl2–100 mM NaCl–1 mM DTT (pH 7.9) for PstI and StyI; 20 mM Tris-acetate–10 mM magnesium acetate–1 mM DTT (pH 7.9) for HhaI and BfaI; 10 mM Tris-HCl–10 mM MgCl2–100 mM NaCl (pH 8.4) for TaqI; and 10 mM Tris-HCl–10 mM MgCl2–150 mM NaCl–1 mM DTT (pH 7.9) for BamHI.
At various time points during the reactions, 10-μl aliquots were removed, quenched with 40 mM EDTA, and subsequently digested overnight at 37°C with 100 μg of proteinase K ml−1. Samples were analyzed on denaturing polyacrylamide gels and quantified with a phosphorimager. Background values were obtained from regions between bands on each gel and subtracted from the integrals measured for each band of interest. The substrate DNA (S) and the two products (P1 and P2) are simultaneously resolved, allowing the fraction of uncut DNA to be calculated as follows: (counts in S)/(counts in S + P1 + P2), all after background subtraction. This definition is insensitive to variations in gel loading. See references 27 and 29 for further discussion.
The data analysis was complicated by two issues, described also in references 1, 2, and 27. First, the initial time point in the nucleosomal restriction digestions exhibits an anomalously large extent of digestion. Native gels of parallel mock digestion reactions exhibit quantitatively similar fractions of apparent naked DNA. We concluded that a small fraction of the nucleosomes dissociates upon exposure to digestion conditions (elevated [Mg2+] and temperature), thus allowing a burst of digestion on the newly generated naked DNA. Due to the high concentrations of restriction enzyme utilized in the nucleosome digestions (500 to 10,000 U ml−1), the naked DNA is digested nearly instantaneously. To address and eliminate this issue, we omitted the initial time point, defined the uncut scaled fraction of the second time point as 1.0, and rescaled the uncut scaled fraction of the remaining time points accordingly. In a recent study of the effects of histone hyperacetylation on nucleosomal site exposure (1), we detected a sample-dependent (i.e., hyperacetylation-dependent) dissociation of a fraction of nucleosomes when they were brought to digestion conditions. Nevertheless, our analysis method successfully eliminates this contribution to the decay kinetics, allowing additional effects on the properties of intact nucleosomes to be revealed. And in the present case we do not detect any systematic (i.e., sample-dependent) effects on overall nucleosome stability due to the A16 elements. Taken together, these results imply that dissociation of nucleosomes prior to the restriction enzyme digestion reactions does not contribute to the rate constants (or the corresponding equilibrium accessibilities) obtained.
In addition, slow dissociation of nucleosomes during the digestions does not contribute significantly to the observed kinetics, since (i) analysis of mock digestion kinetics directly shows this not to occur at detectable levels and (ii) the digestion kinetics are strictly first order in the enzyme concentration used (28, 29; J. D. Anderson and J. Widom, submitted for publication).
A second factor complicating the quantitative analysis is that a fraction of the nucleosomes is never digested. We traced this behavior to a fraction of nucleosomes being insoluble in the digestion buffer, as measured by an ultracentrifuge-based sedimentation assay (unpublished results). This behavior has been demonstrated before in nucleosome solubility experiments (32). In our recent study of hyperacetylated nucleosomes (1), we observed a systematic sample dependence to the nucleosome solubility. In contrast, in the present study we observed no systematic dependence to solubility correlating with the presence or absence of A16 elements. In any case, variable solubility is addressed by allowing the baseline to float in the nonlinear least-squares fit of the data (1, 2, 27): we fit the scaled fraction uncut data (see above) from each digestion to a single exponential decay using the following equation: uncut scaled fraction = a0 + (1 − a0) × exp(−a1 × time), where a0 is the best-fit baseline and a1 is the best-fit rate constant. The ratio of a1 values for control nucleosomes versus experimental nucleosomes (scaled for their enzyme concentrations, which are usually identical between pairs of nucleosomal samples) yields the fold enhancement of site exposure due to the presence of a poly(dA-dT) element.
To assess the appropriateness of this analysis in an earlier study (27), several of the data sets were fit to a double exponential. Examining the resulting pairs of rate constants separately or averaging them to obtain amplitude-weighted mean values leads to results that are quantitatively similar and qualitatively equivalent to those obtained in the single exponential analysis. We therefore focused the analysis on the single exponential fits, as these have fewer adjustable parameters.
RESULTS
DNA templates.
In this study we used a family of DNA constructs that derive from a strong non-natural nucleosome positioning sequence (clone 601) isolated in an earlier SELEX (binding site selection) experiment (18). Construct 601.2 (2) introduced a number of nucleotide substitutions into 601 so as to create sites for different restriction enzymes at locations along the entire nucleosomal length. The free energy of interaction of 601.2 with histone octamer in nucleosome reconstitution, the location of the single strongly preferred nucleosome position on this template, and the position-dependent equilibrium accessibility of sites along the nucleosome length have previously been reported (2). The restriction sites are used in studies described below to quantify the dynamic equilibrium accessibility of DNA target sites contained within the nucleosomes.
Construct 601.3 (Fig. 1A) is identical to 601.2 except that it lacks a few base pairs from the short stretches of DNA extending beyond each nucleosome end of 601.2. 601.3 was modified by replacement of 16 contiguous residues with A's either at the nucleosome end, creating 601.3(A16End) (Fig. 1B), or further in toward the middle, creating 601.3(A16Mid) (Fig. 1C). We also produced variants of these two A16-containing constructs in which the A16 segments were replaced by a randomly chosen 16-bp stretch of bacterial plasmid DNA sequence, creating constructs 601.3(Random End) (Fig. 1B) and 601.3(Random Mid) (Fig. 1C). These latter constructs allow us to assess whether any effects arising from substitution of original 601.2 sequence with A16 are attributable to the presence of the A16 element or simply to loss of the corresponding patch of original 601.2 sequence.
Purification and characterization of reconstituted nucleosomes.
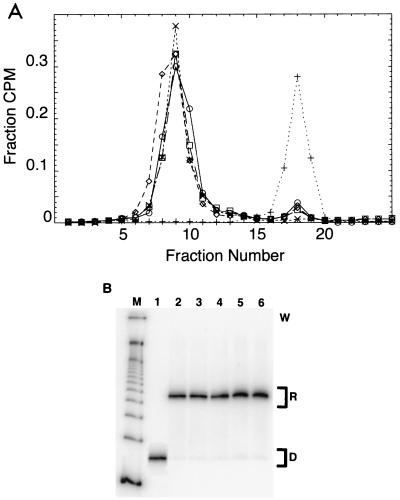
Nucleosomes were reconstituted from purified DNA and histones by salt gradient dialysis and purified by sucrose gradient ultracentrifugation. Examples of typical gradient profiles are shown in Fig. 2A. Subsequent reanalysis of the purified nucleosomes on sucrose gradient (Fig. 2A) or by native gel electrophoresis (Fig. 2B) shows them to be largely free of contaminating naked DNA and to migrate predominantly as single bands, consistent with a single strongly preferred nucleosome position. Note that multiple nucleosome positions, when these exist, can be detected and resolved by native gel electrophoresis even with DNA as short as 146 bp (9, 20). Direct mapping data and other results that imply a single strongly preferred nucleosome position on construct 601.2 are discussed in reference 2. We conclude that each of the reconstituted nucleosome samples used in the present study is strongly biased for occupancy of a single nucleosome position. Additional direct data confirming this interpretation are discussed below.
FIG. 2.
Sucrose gradient purification and reanalysis by sucrose gradient and native gel electrophoresis. Nucleosomes are reconstituted by gradual salt dialysis and separated from naked DNA on 5 to 30% (wt/vol) sucrose gradients (A). Purified nucleosomes are further analyzed on a second sucrose gradient (A) and by native gel electrophoresis (B) +, naked 601.3 DNA; □, preparative run of reconstituted 601.3 nucleosomes; ○, preparative run of reconstituted 601.3(A16End) nucleosomes; ◊, preparative run of reconstituted 601.3(A16Mid) nucleosomes; ×, reanalysis of gradient purified 601.3 nucleosomes. (B) Native gel analysis. W indicates the location of the loading wells; R indicates the mobility of the reconstituted nucleosomes; D indicates the mobility of naked DNA. Lane M, 100-bp DNA marker; lane 1, naked 601.3 DNA; lane 2, purified 601.3 nucleosomes; lane 3, purified 601.3(A16End) nucleosomes; lane 4, purified 601.3(A16Mid); lane 5, purified 601.3(Random End) nucleosomes; lane 6, purified 601.3(Random Mid) nucleosomes. Phosphorimager analysis of the gel reveals contamination by free DNA and other nonnucleosomal aggregates to be ≤0.5%. This small level of naked DNA does not contribute to the observed kinetics because it is digested to completion within the first time point, which we omit from the kinetic analysis.
Free energy measurements.
We used a standard competition method (18, 19, 37) to measure the differences in free energy of histone-DNA interactions in nucleosome reconstitution that result from substitution of 16-bp-long stretches of 601.3 sequence with 16 A's or with random sequence 16-mers (Fig. 3). In this assay, tracer quantities of radiolabeled test DNA competes with a large excess of unlabeled arbitrary-sequence competitor DNA for limiting amounts of histone octamer. The ratio of nucleosomal to free tracer DNA defines an equilibrium constant (affinity) and a corresponding free energy for the tracer that is valid for that competitive environment. To facilitate comparisons between studies, we report differences in free energies (ΔΔG°s), measuring the free energy of the tracer relative to that of a reference sequence measured at the same time in the identical competitive environment; we used a fragment of the sea urchin 5S rRNA gene nucleosome positioning sequence (34) as a reference tracer DNA. Note that the use of this (or any) reference molecule does not influence the ΔΔG°s obtained for comparison between differing DNA samples measured in the same competitive environment.
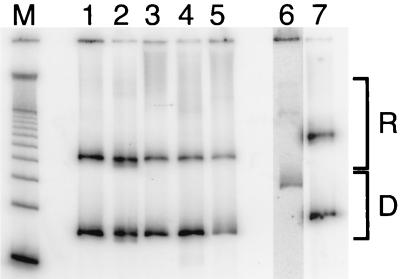
FIG. 3.
Native gel analysis of competitive reconstitution assays. Radiolabeled tracer competes with a large excess of unlabeled natural nucleosome core particle DNA for limiting quantities of histone octamer in dialysis from concentrated NaCl. Lane M, 100-bp DNA marker; lane 1, 601.3; lane 2, 601.3(A16End); lane 3, 601.3(A16Mid); lane 4, 601.3(Random End); lane 5, 601.3(Random Mid); lane 6, the 256-bp EcoRI fragment of the well-characterized natural nucleosome-positioning sequence from sea urchin 5S rRNA gene (34); lane 7, 601.2; D, mobility of naked DNA; R, mobilities of reconstituted nucleosomes. The diverse mobilities represent reflect a range of positionings on the different molecules. The 5S derivative yields several distinct nucleosomal positions, whereas 601.2 and the 601.3 series show one predominant position. The raw data show that 601.2 and the 601.3 series compete much more effectively for the limiting histone octamer than does the 5S sequence (greater ratio of counts in band R versus counts in band D). The contrast in lane 6 was increased to a larger degree than the rest of the gel due to the presence of fewer counts in that lane (see Materials and Methods). Note that this lane is included only for comparison with other studies; the use of this (or any) reference molecule does not influence the ΔΔG°s obtained for comparison between differing DNA samples measured in the same competitive environment.
The results of one such experiment are illustrated in Fig. 3 for the 601.3 sequence (lane 1) and the 5S reference molecule (lane 6). The results from many such experiments are summarized quantitatively in Table 1. The free energy of sequence 601.3(A16End) (lane 2) is 0.11 kcal mol−1 greater (i.e., lower affinity) than that of the parent 601.3 sequence, and that of 601.3(A16Mid) variant (lane 3) is 0.35 kcal mol−1 greater.
TABLE 1.
Quantitative free energy measurements
| DNA sequence | Free energy (kcal mol−1)
|
|
|---|---|---|
| ΔΔG°a (n) | ΔΔG601.3b | |
| 601.2 | −0.97 ± 0.13 (6) | −0.02 ± 0.07 |
| 601.3 | −0.95 ± 0.12 (6) | 0 |
| 601.3(A16End) | −0.84 ± 0.08 (6) | +0.11 ± 0.06 |
| 601.3(A16Mid) | −0.60 ± 0.10 (6) | +0.35 ± 0.06 |
| 601.3(Random End) | −0.93 ± 0.07 (7) | +0.02 ± 0.06 |
| 601.3(Random Mid) | −0.84 ± 0.15 (6) | +0.11 ± 0.08 |
Measured relative to the 5S reference sequence (34) and calculated as ΔG°sample − ΔG°5S. ΔG° (= −RT ln Keq) values were obtained using the indicated radiolabeled tracer with chicken erythrocyte core particle DNA as competitor. Values are expressed as means ± standard deviations; n indicates the number of independent measurements from a series of experiments such as that shown in Fig. 3. Whereas ΔG°s are specific to particular competitive environments, ΔΔG°s reflect intrinsic properties of the molecules independent of the particular competitive environment.
Free energy difference relative to sequence 601.3, shown for convenience, calculated from the ΔΔG° values. Values are expressed as differences ± standard error. The free energy differences reflect the effects of each set of sequence modifications present in the A16 or random DNA series.
To test whether the observed small effects on free energy were due to inherent A16 characteristics and not simply the loss of native 601.3 sequence, a randomly chosen sequence from plasmid pGEM3z was used to replace the poly(dA-dT) sequences [601.3(Random End), lane 4; 601.3(Random Mid), lane 5]. The plasmid sequence restored most of the lost affinity, implying that most of the small destabilization is an active consequence of the presence of the A16 elements.
Restriction enzyme digestion kinetics assay for position-dependent equilibrium accessibility of nucleosomal DNA target sites.
Nucleosomes in vitro are in rapid dynamic equilibrium with alternative conformational states in which the nucleosomal DNA is partially unwrapped off the histone surface. Thus, DNA target sites that in the time average are buried inside the nucleosome and inaccessible are nevertheless transiently freely accessible to a binding protein R or nuclease E (27–30). Such site exposure processes are occurring constantly yet transiently in a rapid preequilibrium. Site exposure is nondissociative: one side of the DNA remains bound, while the other side is exposed. In vitro, site exposure occurs via partial uncoiling of the nucleosomal DNA, rather than by translocation of the histone octamer (Anderson and Widom, submitted). The equilibrium constants for site exposure Keqconf (the equilibrium fraction of the time that nucleosomal DNA target sites are freely accessible, as though they were naked DNA) decrease from the end of the nucleosomal DNA inward toward the middle (2). The apparent equilibrium affinities of proteins binding to nucleosomal target sites, and—equivalently—the observed rate constants for digestion of nucleosomal DNA by restriction endonucleases or nonspecific exonucleases, are reduced from their values on naked DNA by a factor equal to the position-dependent value of Keqconf. For further discussion of the site exposure model, see references 27 to 29.
We used the restriction enzyme digestion kinetics assay (1, 2, 27, 29) to measure relative values Keqconf at target sites throughout the nucleosome. The goal of this study is to compare Keqconf values at sites throughout the nucleosome for A16-containing sequences versus the same sequences lacking the A16 element. Consequently, rather than measuring absolute values of Keqconf (which requires parallel analyses of naked DNA), we measured relative values, obtained from the ratio of digestion rates on the pairs of nucleosomal samples in parallel digestions in identical conditions.
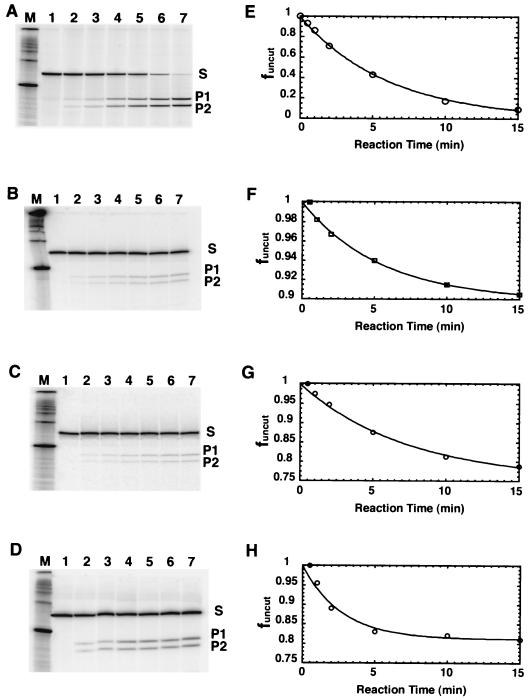
Parallel digestions of native nucleosomes (containing DNA construct 601.3) and test nucleosomes (containing a 601.3 derivative) were carried out in identical solution conditions initiated by the addition of enzyme. Aliquots were removed as a function of time and quenched. Samples were analyzed via denaturing polyacrylamide gel electrophoresis. Representative results of one such experiment, probing the HhaI recognition sequence spanning bp 76 to 79 from the edge of the nucleosome, are illustrated in Fig. 4A to D for naked 601.3 DNA, 601.3 nucleosomes, 601.3(A16End), and 601.3(A16Mid), respectively. Figure 4E to H show the corresponding quantitative analyses for Fig. 4A to D, respectively.
FIG. 4.
Representative kinetic analysis, probing site exposure at the HhaI site, 76 to 79 bp pairs from the 5′ end of the predominant core particle position. (A to D) Denaturing polyacrylamide gel analysis of the time course of digestion. Lanes 1 through 7 in all digestion gels are samples removed at 0, 0.5, 1, 2, 5, 10, and 15 min from reaction initiation. In each case, the substrate (S; 152 nucleotides [nt] for all 601.3 constructs) is converted over time to two products (82 nt [P1] and 72 nt [P2] for all 601.3 constructs). The sizes of S, P1, and P2 expected from the DNA sequence are confirmed against the 100-bp DNA markers in lane M. (A) Naked 601.3 DNA, digested with HhaI at 0.1 U ml−1; (B) 601.3 nucleosomes, digested with HhaI at 2,000 U ml−1; (C) 601.3(A16End) nucleosomes, digested with HhaI at 2,000 U ml−1; (D) 601.3(A16Mid) nucleosomes, digested with HhaI at 2,000 U ml−1. (E to H) Quantitative analyses of the time course of digestion from the data in panels A to D, respectively. The fraction of DNA remaining uncut is corrected for a small initial extent of nucleosome dissociation (which did not correlate with the presence or absence of A16 elements, in contrast to another case in which nucleosome stability was dependent on the acetylation state of the histones [1]) and is plotted versus time. The superimposed lines represent the results of fits to a single exponential decay. See Materials and Methods for further discussion of the kinetic analysis. Note that a 20,000-fold-lower enzyme concentration was used for the digestion on naked DNA.
The raw data in the gel phosphorimages themselves show that (i) all nucleosomal samples have equilibrium accessibilities much lower than those for naked DNA (note that the digestions on naked DNA utilize 20,000-fold-lower enzyme concentration); and (ii) the equilibrium accessibility (relative rate of cleavage) on 601.3(A16Mid) is detectably greater than that on 601.3 or 601.3(A16End).
The increased accessibilities due to the A16 elements are properties of actual A16-containing nucleosomes, not artifacts arising from A16 element-dependent decreased overall stability of the nucleosomes. The analysis method quantifies and accounts for any nucleosome dissociation caused by initial adjustment of the sample to the elevated [Mg2+] and temperature needed for the restriction enzyme digestions (see Materials and Methods), revealing and eliminating contributions of sample-dependent decreased overall stability when this does in fact occur (1). In fact, such dissociation occurred only at low levels and was not systematically dependent on the sample (in contrast to our findings in a recent study of the effects of histone hyperacetylation [1]). Moreover, subsequent additional slow nucleosome dissociation does not contribute significantly to the observed kinetics, since direct analysis of mock digestions shows no significant slow dissociation beyond the small extent of dissociation which occurs immediately on elevation of [Mg2+] and temperature and also since the reactions occur as first-order ones in the enzyme concentration used (see Materials and Methods).
The results from many such experiments are summarized quantitatively in Table 2. The presence of an A16 element at the end of the nucleosomal DNA, from bp 1 to 16 on the construct, changes Keqconf 0.7- to 2.2-fold, depending on position, with an average increase of 1.5- ± 0.1-fold (mean ± standard error). Placing the A16 element further in toward the middle of the nucleosome, from bp 37 to 52 (approximately 24 bp from the nucleosomal dyad), increases Keqconf 1.0- to 2.3-fold, with an average increase of 1.7- ± 0.1-fold. The significance of these changes is discussed below.
TABLE 2.
Restriction enzyme kinetic studies on nucleosomes containing poly(dA-dT) DNA
| Restriction enzyme | Region probed (approx bp from nucleosome end) | Keqconf (A16End)/ Keqconf (601.3) | Keqconf (A16Mid)/ Keqconf (601.3) |
|---|---|---|---|
| PstI | 6–11 | 2.15 ± 0.28 | |
| HindIII | 12–17 | 1.93 ± 0.77 | |
| AvaII | 18–22 | 1.76 ± 1.73 | 1.78 ± 0.34 |
| MspI | 22–25 | 1.41 ± 0.04 | 1.23 ± 0.26 |
| HaeIII | 26–29 | 2.24 ± 0.31 | 2.28 ± 1.08 |
| BamHI | 54–59 | 1.26 ± 0.28 | 1.77 ± 0.26 |
| TaqI | 66–69 | 0.95 ± 0.50 | 1.09 ± 0.09 |
| RsaI | 72–75 | 0.70 ± 0.43 | 0.99 ± 0.06 |
| HhaI | 76–79 | 1.28 ± 0.52 | 2.34 ± 0.27 |
| MseI | 93–96 | ND | 1.26 ± 0.30 |
| StyI | 100–105 | 1.51 ± 0.90 | 1.87 ± 1.09 |
| BfaI | 116–119 | 1.90 ± 0.97 | 1.87 ± 1.44 |
| PmlI | 128–133 | 1.58 ± ND | 1.99 ± ND |
| Avg | 1.46 ± 0.14 | 1.74 ± 0.12 |
Fold enhancements of DNA site accessibility (averages ± 1 standard deviation) for sites throughout each construct from multiple independent experiments. Fold enhancements for each independent experiment are obtained from ratios of best-fit rate constants for digestions of test nucleosomes (i.e., with any 601.3 derivative) versus control nucleosomes (i.e., with 601.3 DNA), scaled for the enzyme concentrations used (27–29). ND, not determined.
Additional studies (not shown) were carried out to compare Keqconf at various sites for the new construct 601.3 versus 601.2, which we have extensively characterized (2). The results for the two constructs are identical within experimental error, consistent with the results of a detailed analysis of the dependence of Keqconf on DNA length (Anderson and Widom, submitted). These observations allow us to relate the present relative measurements of Keqconf for A16-containing derivatives of 601.3 versus 601.3 itself to the absolute measurements of the position-dependent values of Keqconf for 601.2 measured in our earlier work. The resulting position-dependent values for Keqconf for constructs 601.2, 601.3(A16Mid), and 601.3(A16End) are summarized in Fig. 5.
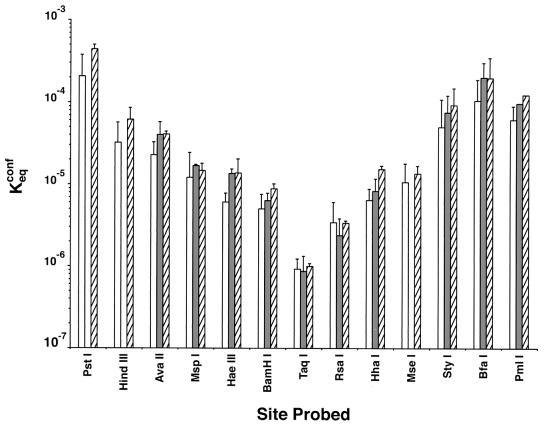
FIG. 5.
Effects of poly(dA-dT) elements on position-dependent equilibrium accessibilities of nucleosomal DNA target sites. Results for poly(dA-dT)-containing constructs are determined relative to those for reference construct 601.2, which were measured on an absolute scale (2). See Fig. 1 for nucleosomal locations of the different restriction sites. Open bars, 601.2 reference; shaded bars, 601.3(A16End); hatched bars, 601.3(A16Mid). Note the log scale for Keqconf.
DISCUSSION
Keqconf values are strongly dependent on position in the nucleosome.
The approximately 1.5- to 1.7-fold average increases in equilibrium accessibility (Keqconf) detected here (Table 2) are superimposed on a strong dependence of Keqconf on position within the nucleosome: Keqconf decreases progressively by 2 to 3 orders of magnitude with distance from either nucleosome end in toward the middle (dyad axis) of the nucleosomal DNA (Fig. 5 and references 2 and 28). Thus, the small effects on Keqconf arising from the incorporation of an A16 element are a positive demonstration of small changes, not a negative finding that might have resulted were the assay unable to detect large changes in protection when these in fact exist.
The strong dependence of Keqconf on position inside the nucleosome, together with the substantial protection afforded to even the most accessible regions at the ends of the nucleosomal DNA, provides additional evidence that mispositioning of nucleosomes did not contribute significantly to the results obtained here. Given the known protection factors, any nonnucleosomal DNA protruding beyond an end of a mispositioned nucleosome would be digested to completion within the first time point and therefore would not contribute to the present analysis, since we fit the disappearance of full-length reactant subsequent to the first time point (see Materials and Methods). Actually, however, we find that the fraction of total template DNA digested within the first time point is quite small and moreover is closely similar to the small fraction of naked DNA present in mock-digested samples as assessed by native gel electrophoresis and sucrose gradient ultracentrifugation (data not shown). Thus, we conclude that any mispositioned nucleosomes are present in at most small amounts, not detectable in the assays used here (and therefore also not contributing to the results obtained), consistent with the presence of single bands in native gel electrophoresis.
Poly(dA-dT) elements decrease the affinity of histone-DNA interactions in nucleosomes and increase the equilibrium accessibility of nucleosomal DNA target sites.
Placing an A16 element at bp 1 to 16 or 37 to 52 decreases the favorable free energy of histone-DNA interactions by approximately 0.1 or 0.35 kcal mol−1, respectively. While small, these differences are significant, as the corresponding standard errors are ∼0.06 kcal mol−1. These measured differences in free energies are consistent with some, but not all, earlier reports of the effects of poly(dA-dT) elements (see the introduction), although differences in the details of the experiments prohibit an exact comparison. We attribute the one report of opposite energetic effects from poly(dA-dT) elements (see the introduction) to a likely failure of that study to reach equilibrium. The differences in free energies that we report are attributable to the A16 elements themselves and not simply to loss of 16-bp stretches of the original high-affinity positioning sequence 601.3, since replacement of the A16 elements by randomly chosen bacterial plasmid sequence restores most or all of the original free energy for nucleosome formation of 601.3.
The decreased favorable free energy of histone-DNA interactions is accompanied by corresponding increases in Keqconf averaged over the full set of measurable locations in the nucleosomes, of 1.5- and 1.7-fold, respectively, for the same two A16-containing constructs. These increases are significant, as the corresponding standard errors are only 0.14 and 0.12, respectively.
The measured changes in Keqconf are not uniform across the nucleosome and even include a small number of apparent decreases. The results are consistent between the two constructs in showing an apparent absence of any enhancement in site accessibility around the nucleosome dyad (nearest the RsaI site). The results for both constructs are also in agreement in showing, within experimental error, that enhanced accessibility extends to both sides of the nucleosomal DNA—that is, to the side that does not contain the poly(dA-dT) element as well as to the side that does. This would imply that there is free energy coupling across the nucleosome. We consider that we cannot state with confidence that these position-dependent fluctuations are real, as each of them is based on relatively fewer measurements with a correspondingly greater standard error. For this reason, we focus the present analysis on the average across the full set of measurements for each construct, which is much more robust. However, in other studies (of the effects of changed DNA length [Anderson and Widom, submitted]) we detect no such systematic changes in a set of measurements made across the nucleosome, suggesting that the elevated accessibilities detected here for both sides versus the middle may in fact be real.
Biological significance of 1.5- and 1.7-fold increases in the equilibrium accessibility of nucleosomal DNA target sites; relation to studies in vivo.
Do 1.5- or 1.7-fold changes in equilibrium accessibility of nucleosomal DNA target sites have any biological significance? Studies of the roles of poly(dA-dT) elements in the expression of the HIS3 gene in S. cerevisiae (13) and the AMT1 gene in C. glabrata (40) provide some appropriate comparisons. The natural HIS3 promoter contains an imperfect 17-bp-long poly(dA-dT) element adjacent to a binding site for the protein Gcn4, the upstream activator protein for that gene. Replacing the endogenous element with a perfect 42-bp-long one leads to 1.6- and 1.7-fold increases in the accessibility of two adjacent sites to the restriction enzyme HinfI expressed in vivo, which in turn correlates with a 3- to 11-fold increase in steady-state transcript levels. One of the two HinfI sites is actually contained within the Gcn4 site and serves to monitor accessibility precisely at that site. In another experiment, the imperfect 17-bp element was either deleted altogether or replaced with a perfect 17-mer. The perfect 17-mer gave a threefold increase in steady-state transcript levels compared to the strain in which the element was deleted. A more recent study from this laboratory (22) further investigated the contributions of poly(dA-dT) tracts and other promoter elements to HinfI accessibility in vivo. This new study suggests that increased site accessibility of the promoter DNA in vivo cannot be attributed to any particular promoter element [such as a poly(dA-dT) tract] but rather may be due to some more global features of the promoter DNA sequence, such as its base composition. However, the data themselves reveal that each of the two poly(dA-dT) elements that flank the HinfI site contributes ∼2-fold increases to the HinfI site accessibility. Thus, the results of that study show that individual poly(dA-dT) elements do indeed contribute importantly to site accessibility, consistent with their earlier results.
A different study investigates the role of a perfect 16-bp-long poly(dA-dT) element on Cu-dependent activation of the AMT1 gene in C. glabrata (40). These investigators conclude that the A16 element plays a critical role in the ability of Amt1 protein to bind to its target site and stimulate transcription of its own gene. Deletion of the A16 element or its replacement by a random sequence 16-mer substantially abrogated activation of the AMT1 gene, by ∼10-fold at early times and ∼2-fold at later times. The diminished ability of the random sequence-containing promoter construct to allow gene activation was accompanied by ∼2.3- and ∼1.5-fold reductions in the rate of digestion of nearby restriction sites in permeabilized spheroplasts.
Taken together, these studies show that the effect of poly(dA-dT) elements in vivo is to cause very modest changes in DNA accessibility as measured by restriction enzymes; these changes correlate with increased accessibility to gene-activating proteins, which in turn cause modest increases in steady-state transcript levels that are of critical biological importance to the living cells. Importantly, the 1.5- and 1.7-fold increases in Keqconf caused by the incorporation of A16 elements found in this study approximate the ∼1.5-, 1.6-, 1.7-, ∼2-, and 2.3-fold effects found for the experiments in vivo mentioned above, suggesting that our in vitro system may be capturing most of the physiological role of the poly(dA-dT) elements.
These poly(dA-dT) elements act in combination with other sequence motifs or elements that remain to be defined (22). These other elements too make small but significant contributions to the overall accessibility of the promoter region, allowing for an overall larger increase in accessibility.
Finally, as another measure of the significance of quantitatively similar effects on transcription, we compare the modest effects attributable to poly(dA-dT) elements in vitro and in vivo with the consequences of the phenomenon of dosage compensation in Drosophila. In that system, many different small effects on transcription combine to yield an overall twofold increase in X-chromosome transcription in males (36).
Two interrelated models for the mechanisms of action of poly(dA-dT) elements in vivo.
The present results support two interrelated but distinct models for the mechanism of action of poly(dA-dT) elements. To understand the distinction between the two models, we must recall that this study was carried out with nucleosomes that are constrained to exist in a fixed position along the DNA. This constraint is provided in the forms of a driving force for a particular positioning (18, 19), the use of a DNA sequence that is barely longer than the core particle DNA length (strongly disfavoring alternative positionings), and a kinetic barrier that effectively prevents movement away from this position in the time scale of these studies (Anderson and Widom, submitted).
One model for the mechanism of action of poly(dA-dT) elements in vivo is directly implied by the present results. Given nucleosomes that are constrained to a fixed location along the DNA, we show here that incorporation of poly(dA-dT) elements destabilize the wrapping of DNA on the histone core, thereby increasing the accessibility of DNA target sites to transactivating factors that must bind to sites contained within the same nucleosome. The present results show this to be true in vitro; such a mechanism would apply also in vivo if nucleosomes are similarly immobile in vivo or, even if nucleosomes are mobile in vivo, if sufficiently large forces exist to strongly bias the time-averaged positioning of nucleosomes. Examples of forces that are likely to be relevant to nucleosome positioning in vivo, and their corresponding magnitudes, are discussed elsewhere (39).
A second model recognizes that nucleosomes may be rapidly mobile in vivo. Many ATP-dependent factors have been discovered that are capable of catalyzing nucleosome mobility in vitro and are linked to gene regulation in vivo (11, 15, 16, 25, 26, 38). If nucleosomes are indeed freely mobile, it follows that poly(dA-dT) elements, by disfavoring packaging into nucleosomes (by the modest but significant free energy penalties summarized above), would bias nucleosome positioning so as to favor (in the time average) positions in which the poly(dA-dT) elements lie outside the nucleosome. Such nucleosome positions may in turn favor binding of transactivating factors, by placing the binding sites off of the nucleosomes altogether (where equilibrium accessibility is greatest) or by placing the binding sites only short distances inside the nucleosome, where equilibrium accessibilities are less than in linker DNA yet still orders of magnitude greater than when the sites are located near the nucleosome middle (2, 28).
These two models can in principle both be operative in vivo. Actually, the two models are closely related: they are simply two different manifestations of the same basic behavior of nucleosomes. This is because the position-dependent free energy of histone-DNA interactions is an important determinant of both nucleosome positioning (19) and site exposure (2). Indeed, the two effects can be quantitatively linked, with Keqconf values for poly(dA-dT) element-containing nucleosomes increasing by the factor exp(−ΔΔG°/RT), where ΔΔG° is the magnitude of the free energy penalty for incorporating the poly(dA-dT) element (J. Widom, unpublished results). In this case, the present measured ΔΔG° of ∼0.35 kcal mol−1 for construct 601.3(A16Mid) would yield a ∼1.8-fold increase in equilibrium accessibility. This is close to the ∼1.7-fold average increase that we observe experimentally, highlighting the relatedness of these two seemingly different models.
Other nucleosome-destabilizing DNA sequence elements.
Taken together with the results of earlier studies, our new results suggest that poly(dA-dT) elements act in combination with other sequence motifs or elements to decrease the probability of nucleosomes being located along a given region of promoter DNA or to destabilize the wrapping of DNA in a nucleosome that is positioned on the promoter DNA. The results of Mai et al. (22) suggest that these additional elements are distributed throughout the length of the promoter region. The absence of dependence on known histone acetyltransferases or other chromatin-remodeling enzymes suggests that these sequence elements may, like the poly(dA-dT) elements, act through a direct effect on the positioning or stability of nucleosomes.
At present we can only speculate as to the nature of these hypothetical sequence elements that repel or destabilize nucleosomes. However, two recent studies provide some examples of DNA sequence motifs that do have such behavior, showing that this viewpoint is plausible. A negative selection experiment carried out with nonnatural DNA led to the isolation of sequences that have anomalously low affinity for the histone octamer (4). The most prevalent of these sequence motifs include repeats of TGGA, TGA, or CA, suggesting that such sequence motifs may be unfavorable (in comparison to arbitrary sequence DNA) for incorporation in nucleosomes. Another study from our own laboratory, investigating sequences that favor incorporation into nucleosomes (18), revealed at the same time certain sequence motifs that were systematically underrepresented (i.e., actively selected against) and which thus apparently disfavor nucleosomal packaging. Interestingly, these include the dinucleotide steps AT and CA (=TG), which are all featured in the longer repeating motifs isolated in the negative selection. This implies that these dinucleotides themselves and the longer motifs that contain them do in fact disfavor incorporation into nucleosomes. New studies will be required to assess whether these particular small sequence motifs contribute to gene regulation in vivo.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH (to J.W.) and by an NIH Cell and Molecular Basis of Disease Traineeship (to J.D.A.).
We acknowledge with gratitude the use of instruments in the Keck Biophysics Facility. We thank the members of our group for valuable discussions and comments on the manuscript.
REFERENCES
- 1.Anderson J D, Lowary P T, Widom J. Effects of histone acetylation on the dynamic equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2001;307:977–985. doi: 10.1006/jmbi.2001.4528. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J D, Widom J. Sequence- and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 3.Behe M J. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao H, Widlund H R, Simonsson T, Kubista M. TGGA-repeats impair nucleosome formation. J Mol Biol. 1998;281:253–260. doi: 10.1006/jmbi.1998.1925. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Tabor S, Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987;50:1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 6.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 7.DiGabriele A D, Steitz T A. A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. J Mol Biol. 1993;231:1024–1039. doi: 10.1006/jmbi.1993.1349. [DOI] [PubMed] [Google Scholar]
- 8.Feng H-P, Scherl D S, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering- test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 9.Flaus A, Luger K, Tan S, Richmond T J. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc Natl Acad Sci USA. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Getts R C, Behe M J. Isolated oligopurine tracts do not significantly affect the binding of DNA to nucleosomes. Biochemistry. 1992;31:5380–5385. doi: 10.1021/bi00138a020. [DOI] [PubMed] [Google Scholar]
- 11.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent nucleosome sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 12.Hayes J, Bashkin J, Tullius T D, Wolffe A P. The histone core exerts a dominant constraint on the structure of DNA in a nucleosome. Biochemistry. 1991;30:8434–8440. doi: 10.1021/bi00098a022. [DOI] [PubMed] [Google Scholar]
- 13.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlin S, Blaisdell B E, Sapolsky R J, Cardon L, Burge C. Assessments of DNA inhomogeneities in yeast chromosome III. Nucleic Acids Res. 1993;21:703–711. doi: 10.1093/nar/21.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 16.Langst G, Bonte E J, Corona D F V, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 17.Losa R, Omari S, Thoma F. Poly(dA) · poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 1990;18:3495–3502. doi: 10.1093/nar/18.12.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowary P T, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 19.Lowary P T, Widom J. Nucleosome packaging and nucleosome positioning of genomic DNA. Proc Natl Acad Sci USA. 1997;94:1183–1188. doi: 10.1073/pnas.94.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luger K, Rechsteiner T J, Flaus A J, Waye M M Y, Richmond T J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 21.Mahloogi H, Behe M J. Oligoadenosine tracts favor nucleosome formation. Biochem Biophys Res Commun. 1997;235:663–668. doi: 10.1006/bbrc.1997.6858. [DOI] [PubMed] [Google Scholar]
- 22.Mai X, Chou S, Struhl K. Preferential accessibility of the yeast his3promoter is determined by a general property of the DNA sequence, not by specific elements. Mol Cell Biol. 2000;20:6668–6676. doi: 10.1128/mcb.20.18.6668-6676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson H C, Finch J T, Luisi B F, Klug A. The structure of an oligo(dA) · oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 24.Packer M J, Dauncey M P, Hunter C A. Sequence-dependent DNA structure: tetranucleotide conformational maps. J Mol Biol. 2000;295:85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 25.Pazin M J, Bhargava P, Geiduschek E P, Kadonaga J T. Nucleosome mobility and the maintenance of nucleosome positioning. Science. 1997;276:809–812. doi: 10.1126/science.276.5313.809. [DOI] [PubMed] [Google Scholar]
- 26.Peterson C L, Workman J L. Promoter pargeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 27.Polach K J, Lowary P T, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 2000;298:211–223. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- 28.Polach K J, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 29.Polach K J, Widom J. Restriction enzymes as probes of nucleosome stability. Methods Enzymol. 1999;304:278–298. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 30.Protacio R U, Polach K J, Widom J. Coupled enzymatic assays for the rate and mechanism of DNA site-exposure in a nucleosome. J Mol Biol. 1997;274:708–721. doi: 10.1006/jmbi.1997.1440. [DOI] [PubMed] [Google Scholar]
- 31.Satchwell S C, Travers A A. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 1989;8:229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz P M, Felthauser A, Fletcher T M, Hansen J C. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu M, Mori T, Sakurai T, Shindo H. Destabilization of nucleosomes by an unusual DNA conformation adopted by poly(dA):poly(dT) tracts in vivo. EMBO J. 2000;19:3358–3365. doi: 10.1093/emboj/19.13.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson R T, Stafford D W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci USA. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuckenholz C, Kageyama Y, Kuroda M I. Guilt by association: non-coding RNAs, chromosome-specific proteins and dosage compensation in Drosophila. Trends Genet. 1999;15:454–458. doi: 10.1016/s0168-9525(99)01855-7. [DOI] [PubMed] [Google Scholar]
- 37.Thåström A, Lowary P T, Widlund H R, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 38.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 39.Yao J, Lowary P T, Widom J. Twist constraints on linker DNA in the 30-nm chromatin fiber: implications for nucleosome phasing. Proc Natl Acad Sci USA. 1993;90:9364–9368. doi: 10.1073/pnas.90.20.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z, Thiele D J. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell. 1996;87:459–470. doi: 10.1016/s0092-8674(00)81366-5. [DOI] [PubMed] [Google Scholar]