Abstract
Selenocysteine (Sec) tRNA (tRNA[Ser]Sec) serves as both the site of Sec biosynthesis and the adapter molecule for donation of this amino acid to protein. The consequences on selenoprotein biosynthesis of overexpressing either the wild type or a mutant tRNA[Ser]Sec lacking the modified base, isopentenyladenosine, in its anticodon loop were examined by introducing multiple copies of the corresponding tRNA[Ser]Sec genes into the mouse genome. Overexpression of wild-type tRNA[Ser]Sec did not affect selenoprotein synthesis. In contrast, the levels of numerous selenoproteins decreased in mice expressing isopentenyladenosine-deficient (i6A−) tRNA[Ser]Sec in a protein- and tissue-specific manner. Cytosolic glutathione peroxidase and mitochondrial thioredoxin reductase 3 were the most and least affected selenoproteins, while selenoprotein expression was most and least affected in the liver and testes, respectively. The defect in selenoprotein expression occurred at translation, since selenoprotein mRNA levels were largely unaffected. Analysis of the tRNA[Ser]Sec population showed that expression of i6A− tRNA[Ser]Sec altered the distribution of the two major isoforms, whereby the maturation of tRNA[Ser]Sec by methylation of the nucleoside in the wobble position was repressed. The data suggest that the levels of i6A− tRNA[Ser]Sec and wild-type tRNA[Ser]Sec are regulated independently and that the amount of wild-type tRNA[Ser]Sec is determined, at least in part, by a feedback mechanism governed by the level of the tRNA[Ser]Sec population. This study marks the first example of transgenic mice engineered to contain functional tRNA transgenes and suggests that i6A− tRNA[Ser]Sec transgenic mice will be useful in assessing the biological roles of selenoproteins.
Selenocysteine (Sec) is encoded by UGA in selenoprotein mRNAs, making Sec the 21st naturally occurring amino acid in protein (reviewed in references 6, 19, and 35). The usage of UGA as a Sec codon represents the only addition to the genetic code since the code was deciphered in the mid-1960s. Decoding of UGA as Sec, rather than termination, requires specific secondary structures in selenoprotein mRNAs, termed Sec insertion sequences or SECIS elements, several trans-acting factors, and a unique tRNA with an anticodon complementary to UGA. The tRNA is first aminoacylated with serine, which serves as the backbone for the biosynthesis of Sec. Sec tRNA is therefore designated tRNA[Ser]Sec. It is not recognized by the standard elongation factor, eEFIA, but by a specialized factor, designated eEFsec, which exhibits specificity for both the unique tRNA structure and the amino acid (16, 46). Recruitment of the Sec-tRNA-EFsec complex to the ribosome occurs via its interaction with the SECIS binding protein 2, a protein exhibiting specificity for the SECIS elements in selenoprotein mRNAs (12).
Selenoproteins typically contain only one Sec residue per polypeptide and are expressed at relatively low levels compared to most other cellular proteins. There are fewer than 10 known prokaryotic and 20 known eukaryotic selenoproteins, but they provide a selective advantage to some organisms and are essential to others (reviewed in reference 23). The central component for the synthesis of the entire class of selenoproteins is tRNA[Ser]Sec. Thus, manipulation of the gene for this tRNA provides a potential target for better understanding the biological roles of this class of proteins. tRNA[Ser]Sec gene knockout mice, however, are embryonic lethal (7). Although this observation demonstrates an essentiality of selenoprotein expression in mammals, this lethality also precludes utilization of these mice as a tool for studying selenoproteins. The expression of genetically altered tRNA[Ser]Sec genes in transgenic mice offers an alternative approach to specifically perturb selenoprotein biosynthesis and study the biological roles of selenoproteins.
In higher vertebrates, selenoprotein expression is dictated by two major isoacceptors that differ from each other by a single 2′-O-methyl group on the ribosyl moiety of the modified residue, methylcarboxymethyl-5′-uridine (mcm5U), at position 34 (reviewed in references 19 and 23). Transfer RNA[Ser]Sec methylation at position 34 results in the formation of methylcarboxymethyl-5′-uridine-2′-O-methylribose (mcm5Um). The methylation step is responsive to selenium availability (9, 15, 22) and results in a conformational change in tRNA[Ser]Sec (15). The unmethylated form, mcm5U, is therefore the precursor of the methylated form, mcm5Um (10, 30). In mammalian cells and tissues, selenium deficiency is associated with a shift in the distribution of the two isoacceptors towards the mcm5U isoform, while selenium supplementation typically results in a shift towards the mcm5Um isoform (9, 15, 22). tRNA[Ser]Sec also contains additional modified residues, including isopentenyladenosine (i6A) at position 37, pseudouridine (ψ) at position 55, and 1-methyladenosine at position 58 (14). The methylation of mcm5U to form mcm5Um does not occur if i6A is not present in the tRNA (30).
Many tRNAs that translate codons with U in their 5′ position contain i6A at position 37 (4, 5, 13). The absence of this modification dramatically reduces the efficiency of the altered tRNA in decoding nonsense codons in bacteria and yeast, and its presence apparently restricts wobble and prevents misreading (4, 5, 13). Chinese hamster ovary (CHO) cells transiently transfected with an i6A− tRNA[Ser]Sec gene showed marginal inhibition of endogenous selenoprotein synthesis but about 80% inhibition of selenoprotein type 1 deiodinase synthesized by cotransfection of an expression construct encoding the gene for this selenoprotein (47). In this same study, CHO cells treated with lovastatin, an inhibitor of the rate-limiting step in the biosynthesis of i6A, resulted in the inhibition of both general selenoprotein synthesis and type 1 deiodinase. Since all other i6A-containing tRNAs would also be expected to lack this modified base, the use of lovastatin did not distinguish between the possible effects of i6A− tRNA[Ser]Sec and other i6A-lacking tRNAs on selenoprotein synthesis.
In the present study, transgenic mice containing from 2 to 20 wild-type tRNA[Ser]Sec transgenes or from 2 to 40 i6A− tRNA[Ser]Sec transgenes were generated, and the effects of overexpression of wild-type tRNA[Ser]Sec and expression of the mutant tRNA[Ser]Sec on tRNA maturation and selenoprotein synthesis in several tissues was examined. The results of these studies are described herein.
MATERIALS AND METHODS
Materials.
[75Se]selenious acid (specific activity, 1,000 Ci/mmol) was obtained from the Research Reactor Facility, University of Missouri (Columbia, Mo.), [α-32P]dCTP and [γ-32P]ATP (specific activities, ∼6,000 Ci/mmol each) were obtained from New England Nuclear, [3H]serine (specific activity, 36 Ci/mmol) and Hybond-N+ nylon membranes were obtained from Amersham, polynucleotide kinase and reverse transcriptase were obtained from Boehringer Mannheim, the RNeasy kit was obtained from Qiagen, human β-actin cDNA probe was obtained from Clontech, and NACS PREPAC ion exchange columns, restriction endonucleases, and agarose were obtained from Gibco-BRL. All other reagents were commercial products of the highest grade available. Inbred FVB/N mice were obtained from Charles River (Frederick, Md.), and B6SJL hybrid mice were from Jackson Laboratories, Bar Harbor, Maine. The care of animals was in accordance with the National Institutes of Health institutional guidelines under the expert direction of G. Lidl (National Cancer Institute, NIH, Bethesda, Md.).
Transgenic mice and excision of tissues and organs.
A 2.17-kb StuI-PvuII fragment containing 1.93 kb of mouse DNA encoding the wild-type tRNA[Ser]Sec gene (41) and 0.24 kb of pBluescript II vector DNA was used for developing a colony of B6SJL hybrid transgenic mice carrying the wild-type transgene at the National Institute of Child Health and Human Development Transgenic Mouse Development Facility, University of Alabama. In vitro mutagenesis was used to alter a T to a C at position 9 of the tRNA[Ser]Sec gene or to alter an A to a G at the nucleotide immediately 3′ to the anticodon at position 37 using the same 2.17-kb StuI-PvuII fragment, and these fragments were used to develop additional colonies of transgenic animals encoding either the “wild type” (i.e., the T-to-C transition position at position 9) or the i6A-deficient (position 37) transgene in FVB/N mice. Transgenic mice were derived by pronuclear microinjection of fertilized eggs as previously described (8). Tissues and organs were taken from sacrificed mice, immediately placed into liquid nitrogen, and stored at −80°C until ready for use.
Southern blot analysis and gene copy number.
Genomic DNA was isolated from mouse tails (38) as modified by Promega, digested with XhoI, electrophoresed on 1% agarose gels, and transferred to a nylon membrane, and the membrane was cross-linked in an UV-Stratalinker (from Stratagene) by standard techniques. The membrane was hybridized with a 32P-labeled 240-bp fragment encoding the Bluescript II vector DNA that was integrated into the mouse genome as part of the transgene (see Fig. 1), and after obtaining an autoradiogram, the filter was stripped and then hybridized with a 32P-labeled 193-bp fragment of human DNA encoding the tRNA[Ser]Sec gene (43). Probes were labeled with [α-32P]dCTP using a random primer labeling kit (Stratagene) and used in hybridization assays, the resulting membranes were washed, and autoradiograms were prepared as described previously (40). This procedure was used to establish transgene number in mice encoding a low copy number (2 to 4 transgenes).
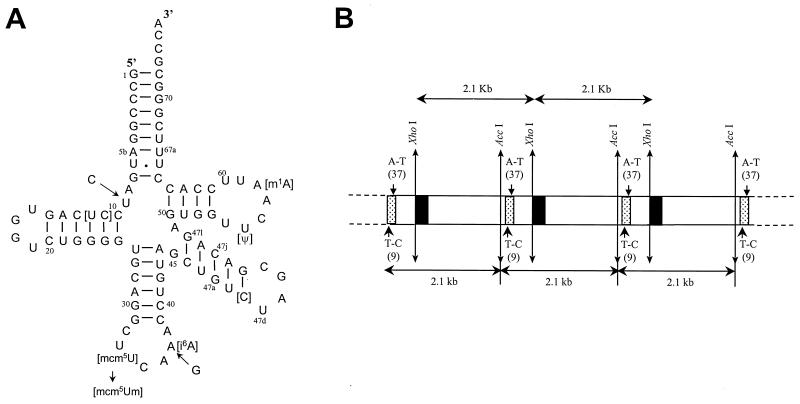
FIG. 1.
Secondary structure of tRNA[Ser]Sec and map of the construct used in making transgenic mice. (A) The cloverleaf model of tRNA[Ser]Sec is shown along with the sites of base changes at positions 9 and 37 used in this study and the sites of modified nucleosides (see the text). The numbering system for positions within tRNA[Ser]Sec is described in the text. (B) The map shows tandem 2.17-kb transgenic fragments containing 1.93 kb of mouse DNA (large open rectangles) encoding the tRNA[Ser]Sec gene (small dotted rectangles) and 0.24 kb of vector DNA (small solid rectangles). The AccI (located 48 bp upstream of the coding sequence of the tRNA[Ser]Sec gene) and XhoI (located in the multiple cloning site of the BlueScript II cloning vector) restriction sites are shown. The 3′ end of the tRNA[Ser]Sec gene is located 425 bp upstream of the 5′ end of the vector sequence. The small arrow inside the gene near the 5′ end shows the position of the T-to-C mutation at position 9 that distinguishes the two wild-type tRNA[Ser]Sec transgenes (see the text), and the other arrow inside the gene shows the position of the A-to-G mutation at position 37 that constitutes the i6A− mutant transgene.
Gene copy number in transgenic mice encoding the higher numbers of transgenes (8 to 40) was calculated using the technique employed by the National Institute of Child Health and Human Development Transgenic Mouse Development facility at the University of Alabama. Fifteen mictograms of mouse tail DNA was digested with XhoI, electrophoresed, transblotted, and hybridized with probe. The relative intensity of the resulting signal was compared to those obtained from aliquots of the 2.17-kb fragment encoding the tRNA[Ser]Sec gene and vector DNA (see Fig. 1) run on the same gel as the genomic DNA, whereby one gene copy of the fragment encoding the tRNA[Ser]Sec gene represented 5.423 pg.
Isolation and aminoacylation of tRNA, RPC-5 chromatography, and Northern blot analysis.
Total tRNA was isolated from tissues, prepared for aminoacylation, and aminoacylated with [3H]serine under limiting tRNA conditions (21), and the resulting labeled seryl-tRNA was chromatographed twice on an RPC-5 column (29), first in buffer without Mg2+ and then in buffer with Mg2+ (9, 15, 22, 40). Seryl-tRNASer is more hydrophobic than seryl-tRNA[Ser]Sec in the absence of Mg2+ and therefore elutes later on the RPC-5 column, and it is less hydrophobic in the presence of Mg2+ and therefore elutes earlier. Thus, the tRNASer and tRNA[Ser]Sec populations can be chromatographically resolved from each other and quantitated following labeling with [3H]serine as described previously (9, 15, 22, 40).
Northern blot analysis of GPX1, GPX4, D1, TR1, SPS2, and SelP mRNAs was carried out by isolating total RNA from liver and kidney using an RNeasy minikit (according to the vendor's instructions). The RNA was electrophoresed on a 1% formaldehyde–agarose gel and transblotted to a nylon membrane. Filters were probed with a 32P-labeled bovine GPX1 cDNA EcoRI-HindIII fragment (28), and several IMAGE Consortium (LLNL) cDNA clones were generated as a MluI-SalI fragment encoding the SPS2 gene (IMAGE Consortium Clone ID 791719 [accession number {AN} AA414662]) and as NotI-EcoRI fragments encoding the D1, TR1, SelP, and GPX4 genes (IMAGE Consortium Clone ID 677180 [AN AA212899], 676579 [AN AA209061], 777018 [AN AA276440], and 1364475 [AN AI006169]), respectively. Membranes were stripped and reprobed with 32P-labeled human β-actin cDNA probe.
Primer extension.
The identity of the nucleotide at position 9 in the tRNA[Ser]Sec transgene was used to distinguish the contributions of the product from the transgenes and that of the wild-type genes to the total tRNA[Ser]Sec population in transgenic mice by primer extension. An oligonucleotide, 5′-GCCTGCACCCCAGACCACTGA-3′, that was complementary to bases 12 through 32 within the tRNA[Ser]Sec gene was 5′-end labeled with [γ-32P]ATP and polynucleotide kinase, the unlabeled nucleotide was removed with a NACS PREPAC column, and the resulting labeled oligonucleotide was used as a primer in primer extension studies as described previously (40). The extension buffer included ddATP, resulting in termination at the first U in the tRNA template. Relative intensities of bands were determined using a Bio-Rad GS-710 calibrated imaging spectrophotometer.
Labeling of selenoproteins.
Transgenic and wild-type mice were injected intraperitoneally with 50 μCi of 75Se/g of body weight and sacrificed at 48 h after injection, and tissues and organs were excised and immediately placed into liquid nitrogen and stored at −80°C until ready for use. Tissues were homogenized in a solution containing 40 mM Tris-HCl (pH 7.4), 1 mM EDTA, and 0.5 mM 4-(-2-aminoethyl)benzenesulfonyl fluoride, sonicated for 2 min, and centrifuged at 4°C for 20 min. Supernatants were electrophoresed on sodium dodecyl sulfate-polyacrylamide gels, separated proteins were transferred to nylon membranes, and transblots were exposed to a PhosphorImager as described previously (20, 40). Gels were stained with Coomassie blue.
Selenoprotein assays.
GPX1 (40) and GPX3 (32) activities were assayed as previously described and measured as the nanomoles of NADPH oxidized/minute/milligram of protein using H2O2 as substrate. 5′-Deiodinase activity was measured using 125I-reverse T3 or 125I-T4 for type 1 (D1) or type 2 (D2) deiodinase, respectively, as previously described (3, 18). Thioredoxin reductase activity was determined in the presence of Escherichia coli thioredoxin using the insulin reduction method (1) in tissue extracts prepared as described below.
SelP, TR1, TR3, GPX4, and SelT were all measured by Western analysis. In addition, TR1 and TR3 were measured by 75Se labeling. SelP was measured using antibody #695 as described previously (24). For thioredoxin reductase assays, 0.8 g of 75Se-labeled mouse liver from each type of transgenic line were sonicated in 5 volumes of 25 mM Tris-HCl (pH 7.5)–1 mM EDTA–1 mM phenylmethylsulfonyl fluoride–5-μg/ml aprotinin–5-μg/ml leupeptin–5-μg/ml pepstatin A. After centrifugation, the supernatants were separately applied onto 0.5 ml ADP-Sepharose columns. The columns were washed with 0.5 ml of 25 mM Tris-HCl buffer (pH 7.5)–1 mM EDTA–0.15 M NaCl, and the proteins were eluted with 1 ml of 25 mM Tris-HCl (pH 7.5)–1 mM EDTA–1.0 M NaCl. The eluted fractions were tested for thioredoxin reductase activity and also analyzed by immunoblot assays with rabbit polyclonal antibodies specific for TR1 and TR3 (45) and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analyses followed by PhosphorImager assays to determine the 75Se content (20). All thioredoxin reductase analyses were performed in parallel for the entire set of samples.
SelT and GPx4 were analyzed with rabbit polyclonal antibodies raised against the C-terminal peptide of SelT (31) or an internal peptide of GPX4 (antibodies were kindly provided by Donna Driscoll), respectively. Crude extracts used for these assays were prepared as for the thioredoxin reductase analyses. X-ray films were quantified with a densitometer.
Blood and selenium analyses.
Blood samples were taken from mice by venal eye puncture. The serum was obtained by centrifugation and used for determining cholesterol, triglycerides, liver function (aspartate aminotransferase, alanine aminotransferase, alkaline phosphatase, total bilirubin, and total protein) and kidney function (creatinine, urea nitrogen, and uric acid) in the National Institutes of Health Clinical Center using standard techniques. The amount of selenium in plasma or soft tissues was determined by automated electrothermal atomic absorption spectrometry using a Varian Spectra AA600 (Varian Instruments, Inc., Walnut Creek, Calif.) equipped with Zeeman-effect background corrections. Soft tissues were homogenized in 10% HNO3, allowed to digest for 48 h at room temperature, and centrifuged (1,000 × g), and selenium was analyzed in the supernatant. Samples of plasma or tissue digests were mixed with 3 volumes of a matrix modifier solution (1.25% Ni[NO3]2.6H2O, 0.09% PdCl2, 0.1% Triton X-100). Absorption was measured at 196.3 nm with a 2.0-nm slit signal; peak area is calibrated automatically using aqueous solutions of Na2SeO3 as standards. The limit of detection of this method is ca. 5 pg of Se, which yields a practical detection limit of approximately 20 ng of selenium/ml of sample. Quality control was effected using multiple aliquots of exhaustively analyzed human plasma as external control samples with a coefficient of variation of >7% (for duplicate analyses) used as the criterion for acceptance of all sample results. That criterion was derived experimentally using the variance components analysis described previously (37).
RESULTS
Transgenic animals.
Transgenic mice were independently generated using two tRNA[Ser]Sec gene constructs that differ from each other by a single pyrimidine transition (U→C) at position 9. C at position 9, which corresponds to wild-type chicken and Xenopus tRNA[Ser]Sec (33), permitted us to assess the levels of tRNAs derived from transgenes relative to the endogenous tRNA[Ser]Sec population. Transgenic mice were also generated by the introduction of a third tRNA[Ser]Sec gene containing a purine transition (A→G) immediately 3′ to the anticodon at position 37 (see reference 26 and references therein for numbering of tRNA[Ser]Sec nucleotide positions). The change at this position prevents the formation of the highly modified base, i6A (30), that normally occurs at this site. The cloverleaf model of tRNA[Ser]Sec, as well as the two described altered sites, are shown in Fig. 1A.
Transgenic mice were generated by introducing 2.17 kb of DNA containing 1.93 kb of mouse DNA and 0.24 kb of vector DNA (Fig. 1B). The vector sequence is located 425 bp downstream of the 3′ end of the tRNA[Ser]Sec gene and was used to monitor integration of the 2.17-kb fragment into the host genome and for determining the gene copy number. Since transgenes often integrate into genomes in a tandem, head-to-tail manner, Fig. 1B shows the expected result of integration of tandem 2.17-kb fragments into genomic DNA.
Founder mice were obtained with each of the two “wild-type” tRNA[Ser]Sec gene constructs and used to establish mouse lines containing 2 (heterozygous genotype) and 4 (homozygous genotype) tRNA[Ser]Sec transgene copies of the unaltered wild-type construct and 10 (heterozygous) to 20 (homozygous) copies of the construct carrying the U→C change at position 9. They were designated +/+/TGWT2 and +/+/TGWT2/TGWT2 and +/+TG“WT”10 and +/+/TG“WT”10/TG“WT”10, respectively (see Table 1). Three founders were obtained with the alteration at position 37 (i6A−), and the resulting mouse lines generated from these founders contained 2 to 4, 8 to 16, and 20 to 40 transgenes, respectively, and were designated +/+/TGi6A−2 and +/+/TGi6A−2/TGi6A−2, +/+/TGi6A−8 and +/+/TGi6A−8/TGi6A−8, and +/+/TGi6A−20 and +/+/TGi6A−20/TGi6A−20 (Table 1).
TABLE 1.
Transgenic mice
| Mouse straina | Transgeneb | No. of transgene copiesc (genotype) | Genotype designation |
|---|---|---|---|
| B6SJL | Wild type | 2 (heterozygous) | +/+/TGWT2 |
| 4 (homozygous) | +/+/TGWT2/TGWT2 | ||
| FVB/N | “Wild type” (T→C) at position 9d | 10 (heterozygous) | +/+/TG“WT”10 |
| 20 (homozygous) | +/+/TG“WT”10/TG“WT”10 | ||
| FVB/N | i6A− mutant (A→G) at position 37d | 2 (heterozygous) | +/+/TGi6A−2 |
| 4 (homozygous) | +/+/TGi6A−2/TGi6A−2 | ||
| 8 (heterozygous) | +/+/TGi6A−8 | ||
| 16 (homozygous) | +/+/TGi6A−8/TGi6A−8 | ||
| 20 (heterozygous) | +/+/TGi6A−20 | ||
| 40 (homozygous) | +/+/TGi6A−20/TGi6A−20 |
A single founder was generated with the B6SJL strain using the wild-type tRNA[Ser]Sec gene construct and with the FVB/N strain using the wild-type construct harboring an A→C mutation at position 9. Three founders were generated with the FVB/N strain using the i6A− mutant tRNA[Ser]Sec construct.
Constructs encoding the tRNA[Ser]Sec gene (see Materials and Methods) used for making transgenic mice.
The number of transgene copies was established as described in Materials and Methods.
The T→C mutation was generated at position 9 and the A→G mutation at position 37 as described in Materials and Methods.
Analysis of the tRNA[Ser]Sec population.
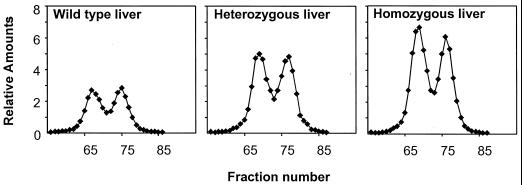
To assess changes in the tRNA[Ser]Sec population in animals bearing the above-described transgenes, tRNA was prepared from the liver, kidney, brain, and testes of transgenic wild-type and sibling mice. Isolated tRNA was aminoacylated with [3H]serine, resulting in the labeling of tRNASer and the mcm5U and mcm5Um tRNA[Ser]Sec isoforms. The amounts of the Sec isoacceptors relative to the seryl-tRNA population were determined by RPC-5 chromatography, where the unmethylated isoform, mcm5U, elutes first and the methylated form, mcm5Um, elutes second from the column (see Materials and Methods) (9, 15, 22, 40). A typical chromatographic separation of the Sec isoforms from livers of +/+, +/+/TGWT2, and +/+/TGWT2/TGWT2 sibling mice is shown in Fig. 2. The tRNA[Ser]Sec population increased with increasing wild-type gene copy numbers in the livers of transgenic animals, and the relative distributions of mcm5U and mcm5Um were altered, albeit slightly. The relative amounts of the seryl-tRNA[Ser]Sec population and the distributions of the mcm5U and mcm5Um isoacceptors from each selected organ were determined in this manner. The data from liver, kidney, brain, and testes of transgenic mice harboring wild-type tRNA[Ser]Sec transgenes are summarized in Table 2, and those from transgenic mice harboring the i6A− transgenes are summarized in Table 3.
FIG. 2.
Relative amounts of mcm5U and mcm5Um isoacceptors in livers of wild-type and heterozygous and homozygous transgenic mice bearing wild-type transgenes. Total tRNA was isolated from livers of littermates bearing +/+, +/+/TGWT2, and +/+/TGWT2/TGWT2 genotypes and aminoacylated with [3H]serine, and the resulting 3H-labeled tRNA was fractionated as described in Materials and Methods. The amounts of seryl-tRNA[Ser]Sec (mcm5U is the first eluting peak and mcm5Um is the second) found in livers of heterozygous and homozygous transgenic mice were standardized to that found in wild-type livers with the total [3H]seryl-tRNASer serving as an internal control. Sources of seryl-tRNA[Ser]Sec are shown in each graph.
TABLE 2.
Levels and distributions of wild-type tRNA[Ser]Sec isoforms in organs of transgenic micea
| Tissue | Genotype of offspring | Result for tRNA[Ser]Sec
|
||||
|---|---|---|---|---|---|---|
| % (Total)b | Relative amtc | Distributiond
|
||||
| mcm5U (%) | mcm5Um (%) | mcm5U/mcm5Ume | ||||
| Liver | +/+ | 2.4 | 1.00 | 49 | 51 | 0.97 |
| +/+/TGWT2 | 3.9 | 1.79 | 52 | 48 | 1.08 | |
| +/+/TGWT2/TGWT2 | 5.4 | 2.20 | 55 | 45 | 1.23 | |
| Liver | +/+ | 2.5 | 1.00 | 46 | 54 | 0.83 |
| +/+/TG“WT”10 | 6.4 | 2.53 | 72 | 28 | 2.60 | |
| +/+/TG“WT”10/TG“WT”10 | 9.4 | 3.69 | 81 | 19 | 4.29 | |
| Kidney | +/+ | 3.5 | 1.00 | 32 | 68 | 0.48 |
| +/+/TGWT2 | 6.8 | 1.95 | 39 | 61 | 0.63 | |
| +/+/TGWT2/TGWT2 | 7.4 | 2.13 | 39 | 61 | 0.64 | |
| Kidney | +/+ | 3.8 | 1.00 | 38 | 62 | 0.61 |
| +/+/TG“WT”10 | 12.8 | 3.67 | 63 | 37 | 1.67 | |
| +/+/TG“WT”10/TG“WT”10 | 19.8 | 5.68 | 71 | 29 | 2.45 | |
| Brain | +/+ | 3.2 | 1.00 | 26 | 74 | 0.35 |
| +/+/TGWT2 | 5.9 | 1.79 | 38 | 62 | 0.62 | |
| +/+/TGWT2/TGWT2 | 7.0 | 2.10 | 40 | 60 | 0.66 | |
| Brain | +/+ | 3.5 | 1.00 | 27 | 73 | 0.37 |
| +/+/TG“WT”10 | 13.5 | 3.82 | 59 | 41 | 1.43 | |
| +/+/TG“WT”10/TG“WT”10 | 21.6 | 6.09 | 66 | 34 | 1.91 | |
| Testes | +/+ | 8.8 | 1.00 | 67 | 33 | 2.07 |
| +/+/TGWT2 | 7.9 | 0.90 | 68 | 32 | 2.17 | |
| +/+/TGWT2/TGWT2 | 11.5 | 1.31 | 69 | 31 | 2.24 | |
| Testes | +/+ | 8.4 | 1.00 | 68 | 32 | 2.14 |
| +/+/TG“WT”10/TG“WT”10 | 19.8 | 2.31 | 76 | 24 | 3.08 | |
tRNA was isolated from liver, kidney, brain and testes tissues (column 1) of offspring (column 2) from matings of heterozygous parents (+/+/TGWT2 × +/+/TGWT2 or +/+/TG“WT”10 × +/+/TG“WT”10) and tRNA fractionated, tRNA[Ser]Sec was isolated, and the amounts of total tRNA[Ser]Sec (columns 3 and 4) and the distributions of mcm5U and mcm5Um (columns 5 to 7) were determined as described in Materials and Methods.
Percentage of tRNA[Ser]Sec population within the total seryl-tRNA population.
Amount of the tRNA[Ser]Sec population relative to that in the wild type, which was assigned a value of 1.00.
Percentages of mcm5U and mcm5Um resolved by RPC-5 chromatography from the seryl-tRNA population (see Materials and Methods).
Amount of mcm5U divided by the amount of mcm5Um.
TABLE 3.
Levels and distributions of wild-type and i6A− mutant tRNA[Ser]Sec isoforms in organs of transgenic micea
| Tissue | Genotype of offspring | Results for tRNA[Ser]Sec
|
||||
|---|---|---|---|---|---|---|
| % (Total)
|
Distribution
|
|||||
| WT | i6A− | mcm5U (%) | mcm5Um (%) | mcm5U/mcm5Um | ||
| Liver | +/+b | 2.5 | 0 | 46 | 54 | 0.85 |
| +/+/TGi6A−2 | 1.8 | 3.6 | 56 | 44 | 1.27 | |
| +/+/TGi6A−2/TGi6A−2 | 2.2 | 4.7 | 61 | 39 | 1.56 | |
| +/+/TGi6A−8 | 2.5 | 12 | 60 | 40 | 1.50 | |
| +/+/TGi6A−8/TGi6A−8 | 2.4 | 15 | 65 | 35 | 1.86 | |
| +/+/TGi6A−20 | 2.3 | 20 | 67 | 33 | 2.03 | |
| +/+/TGi6A−20/TGi6A−20 | 2.6 | 31 | 72 | 28 | 2.57 | |
| Kidney | +/+b | 3.8 | 0 | 34 | 66 | 0.52 |
| +/+/TGi6A−2 | 4.0 | 8.8 | 42 | 58 | 0.72 | |
| +/+/TGi6A−2/TGi6A−2 | 4.1 | 10.1 | 43 | 57 | 0.75 | |
| +/+/TGi6A−8 | 3.9 | 15.1 | 54 | 46 | 1.17 | |
| +/+/TGi6A−8/TGi6A−8 | 3.5 | 13.4 | 64 | 36 | 1.78 | |
| +/+/TGi6A−20 | 3.3 | 18.6 | 63 | 37 | 1.70 | |
| +/+/TGi6A−20/TGi6A−20 | 3.9 | 30.1 | 66 | 34 | 1.94 | |
| Brain | +/+b | 5.0 | 0 | 35 | 65 | 0.54 |
| +/+/TGi6A−2 | 4.7 | 7.0 | 47 | 53 | 0.89 | |
| +/+/TGi6A−2/TGi6A−2 | 4.9 | 8.5 | 49 | 51 | 0.96 | |
| +/+/TGi6A−8 | 5.1 | 14.9 | 48 | 52 | 0.92 | |
| +/+/TGi6A−8/TGi6A−8 | 4.8 | 26.0 | 54 | 46 | 1.17 | |
| +/+/TGi6A−20 | 4.7 | 34.5 | 58 | 42 | 1.38 | |
| +/+/TGi6A−20/TGi6A−20 | 4.8 | 41.2 | 60 | 40 | 1.50 | |
| Testes | +/+b | 8.4 | 0 | 68 | 32 | 2.13 |
| +/+/TGi6A−2 | 7.8 | 4.9 | 72 | 28 | 2.57 | |
| +/+/TGi6A−2/TGi6A−2 | 7.6 | 6.0 | 73 | 27 | 2.70 | |
| +/+/TGi6A−8 | 7.3 | 10.9 | 74 | 26 | 2.85 | |
| +/+/TGi6A−8/TGi6A−8 | 6.5 | 12.1 | 79 | 21 | 3.76 | |
| +/+/TGi6A−20 | 6.0 | 12.9 | 79 | 21 | 3.76 | |
| +/+/TGi6A−20/TGi6A−20 | 5.7 | 22.2 | 80 | 20 | 4.00 | |
See the footnotes to Table 2 and Materials and Methods for details. WT, wild type.
As the levels of the tRNA[Ser]Sec population and the distributions of mcm5U and mcm5Um were similar for siblings of wild-type control mice from both mouse lines (see Table 2), only tissues from wild-type mice obtained from heterozygous matings between parents containing the lowest mutant tRNA gene copy number were used to establish control values.
Overexpression of the wild-type tRNA[Ser]Sec population.
The increase in the tRNA[Ser]Sec population was clearly not directly proportional to gene copy number in transgenic animals carrying wild-type transgenes, nor was it the same in all tissues. For example, the increase was about 3.5-fold in the liver but more than 6-fold in brains of mice carrying 20 extra tRNA[Ser]Sec copies (Table 2). In addition, the relative distributions of the two tRNA[Ser]Sec isoforms in tissues of mice carrying wild-type transgenes were altered as gene copy numbers increased. As shown in Table 2, the amount of mcm5U relative to that of mcm5Um increased slightly in the TGWT2 animals, but this effect was more dramatic in the TG“WT”10 animals. These observations support the hypothesis that the methylase responsible for converting mcm5U to mcm5Um is likely to be limiting (see also references 23 and 40).
The introduced nucleotide change at position 9 in the tRNA[Ser]Sec transgene permitted us to distinguish the amount of gene product contributed to the tRNA[Ser]Sec population by the transgenes relative to that from the host genes by primer extension. Total tRNA from livers, kidneys, and testes of transgenic mice and their wild-type siblings was used as a template to extend the sequence of an oligonucleotide complementary to positions 12 through 32. Since ddATP replaced dATP in the extension buffer, primers were extended until a U was encountered in the template tRNA. The primer was therefore extended only three nucleotides when wild-type tRNA[Ser]Sec was used as a template and six nucleotides when transgene-derived tRNA[Ser]Sec was used as a template. As expected, the oligonucleotide extended only three nucleotides in tRNA samples isolated from livers, kidneys (Fig. 3A, lanes 2 and 5, respectively), and testes (data not shown) of the wild-type siblings. In contrast, total tRNA from these same tissues obtained from heterozygous and homozygous transgenic animals contained the expected extension products for tRNA transcribed from the transgenes (Fig. 3A, lanes 3, 4, 6, and 7, respectively).
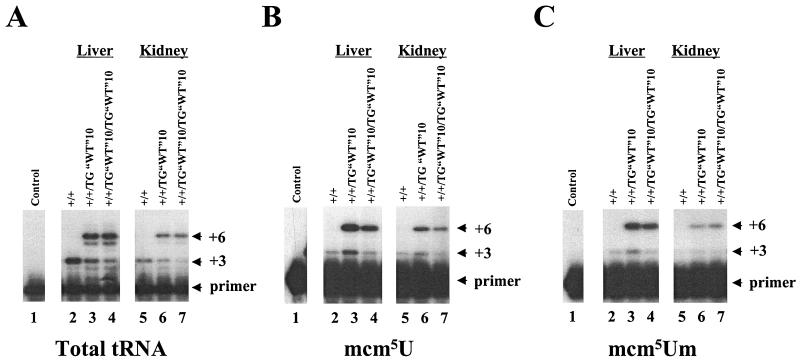
FIG. 3.
Characterization of tRNA[Ser]Sec obtained from tissues of wild-type mice and heterozygous and homozygous transgenic mice bearing transgenes with a pyrimidine transition at position 9 by primer extension. Total tRNA (A), fractionated mcm5U (B), or fractionated mcm5Um (C) was used as a template for primer extension. Column fractions were selected to minimize the overlap of the peaks representing each isoacceptor. In each panel, the order of samples is as follows: lane 1, no template; lane 2, +/+ liver; lane 3, +/+/TG“WT”10 liver; lane 4, +/+/TG“WT”10/TG“WT”10 liver; lane 5, +/+ kidney; lane 6, +/+/TG“WT”10 kidney; and lane 7, +/+/TG“WT”10/TG“WT”10 kidney. Preparation, separation, and recovery of tRNA and tRNA fractions and primer extensions were done as described in Materials and Methods. The positions of the primer and the +3 (host tRNA[Ser]Sec) and +6 (transgene tRNA[Ser]Sec) extension products are indicated.
Transfer RNA was also recovered from column fractions representing either the mcm5U or the mcm5Um isoacceptor and assayed by primer extension as described above for total tRNA. Extension products indicative of transgene origin were also present when tRNA from the earlier-eluting isoacceptor (mcm5U) (Fig. 3B) and the later-eluting isoacceptor (mcm5Um) (Fig. 3C) were used as substrates, indicating that both isoforms of the transgene-derived tRNA were capable of full maturation.
The column profile analysis indicated that the increase in tRNA[Ser]Sec obtained by increasing the transgene copy number from 10 to 20 did not result in a comparable doubling of tRNA[Ser]Sec levels (Table 2); and this observation was verified by the primer extension data presented in Fig. 3A. To determine the relative contributions of transgenes and host genes to the observed increase in the tRNA[Ser]Sec population, we took advantage of our experimental design, which permitted the independent quantitation of endogenous and transgene-derived tRNA[Ser] by primer extension. Quantitation of the lower bands (endogenous) by densitometry indicated that the levels of the host tRNA[Ser]Sec declined by 30 to 75% with an increasing tRNA[Ser]Sec gene copy number for all tissues examined, and clear dose responses were observed for the kidney and liver (Fig. 3A).
Expression of the i6A− transgenes.
The tRNA[Ser]Sec population was examined in selected organs of transgenic mice containing tRNA[Ser]Sec genes engineered to be incapable of forming the i6A modification. The i6A− tRNA[Ser]Sec elutes earlier from the RPC-5 column than the endogenous i6A-containing tRNA[Ser]Sec population (10, 30, 42, 47) while retaining its ability to be aminoacylated (42, 47). These observations facilitated the analysis of each of the tRNA[Ser]Sec species for the liver, kidney, brain, and testes. The total amount of the host tRNA[Ser]Sec population remained virtually unchanged in livers, kidneys and brains of these mice, even though the i6A− form increased to levels as high as about 30 to 40% of the total tRNASer population in mice carrying the highest transgene copy number (Table 3). In testes, the level of endogenous tRNA declined slightly as the level of i6A− tRNA increased. Examination of the distributions of mcm5U and mcm5Um in animals expressing i6A− tRNA[Ser]Sec indicated that there was an increase in mcm5U with a proportional decline in mcm5Um in the livers, kidneys, and brains of these animals. The data also suggest that the levels of wild-type tRNA[Ser]Sec and i6A− tRNA[Ser]Sec are regulated independently of each other and that the level of the tRNA[Ser]Sec population is determined in these tissues, at least in part, by a feedback mechanism governed by the isoforms containing i6A at position 37 (see Discussion).
Protein synthesis.
Selenoprotein biosynthesis was assessed in the transgenic mice described above by injection with 75Se. Proteins from livers, kidneys, testes, brains, muscles, and hearts of these and control animals were isolated following labeling with 75Se and examined by gel electrophoresis. Coomassie blue-stained gels of total proteins from these tissues showed only minor differences in protein patterns in transgenic mice containing 10 to 20 wild-type transgenes compared to results for their wild-type, nontransgenic siblings (data not shown). PhosphorImaging, used specifically to detect 75Se-labeled selenoproteins, also failed to detect significant differences between transgenic and corresponding control tissues. Therefore, the higher levels of tRNA[Ser]Sec resulting from the expression of 2 to 4 and 10 to 20 wild-type transgene copies had little or no effect on either general protein synthesis or selenoprotein levels (data not shown).
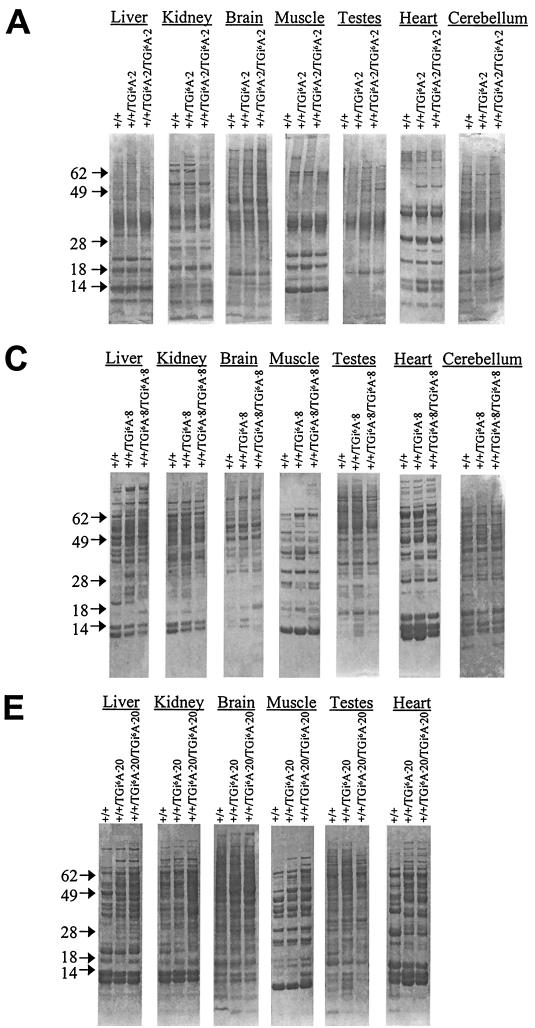
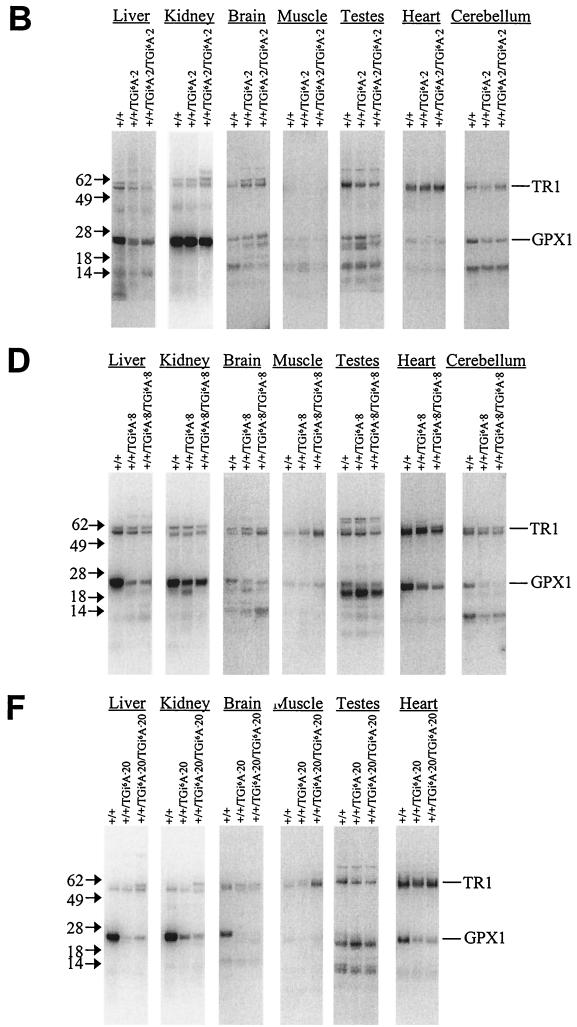
In contrast to the data presented above, the presence of i6A− tRNA[Ser]Sec caused considerable changes in selenoprotein synthesis. Seven tissues, including the cerebellum, were excised from 75Se-labeled mice, and the resulting protein extracts were electrophoresed. Coomassie blue staining of total protein within gels showed some variations in tissue extracts from either wild-type or heterozygous and homozygous i6A− tRNA[Ser]Sec transgenic mice (Fig. 4A, C, and E). No consistent differences were observed, however, when duplicate samples of sibling mice were analyzed (data not shown). In contrast, 75Se-labeled proteins were significantly altered in the tissues of these mice, with selenoprotein patterns being different in each of the seven tissues examined (Fig. 4B, D, and F). For example, there was an apparent decrease in GPX1 in the livers of transgenic mice carrying two or more i6A− tRNA[Ser]Sec transgenes (Fig. 4B, D and F) and in kidneys of mice carrying eight or more i6A− tRNA[Ser]Sec transgenes (Fig. 4D and F). GPX1 levels appeared to be less affected in kidney than liver in mice with genotype +/+/TGi6A−8 or +/+/TGi6A−8/TGi6A−8 (Fig. 4D).
FIG. 4.
Protein and selenoprotein analysis in tissues of wild-type and sibling heterogeneous and homogenous i6A-deficient transgenic mice. Littermates were labeled with 75Se, and proteins were extracted from the different tissues, electrophoresed, and transblotted onto a membrane; the membrane was stained with Coomassie blue. Total protein of +/+, +/+/TGi6A−2, and +/+/TGi6A−2/TGi6A−2 (A), +/+, +/+/TGi6A−8, and +/+/TGi6A−8/TGi6A−8 (C), and +/+, +/+/TGi6A−20, and +/+/TGi6A−20/TGi6A−20 (E) mice and 75Se-labeled proteins of +/+, +/+/TGi6A−2, and +/+/TGi6A−2/TGi6A−2 (B), +/+, +/+/TGi6A−8, and +/+/TGi6A−8/TGi6A−8 (D), and +/+, +/+/TGi6A−20, and +/+/TGi6A−20/TGi6A−20 (F), mice were detected with a PhosphorImager as described in Materials and Methods. Protein marker sizes are shown on the left of each panel as indicated by the arrows.
In addition to assessing selenoprotein levels by quantifying 75Se-labeled proteins, several selenoproteins were analyzed directly by enzymatic assay or Western analyses. These assays confirmed that the levels of several selenoproteins were dramatically reduced in all tissues examined, while others were selectively reduced in one tissue but not in another. At least one selenoprotein, TR3, appeared to be more highly expressed, while GPX1 activity was decreased in every tissue examined (Table 4). A dose-dependent effect was observed for GPX1 activities, since tissues from mice expressing less of the i6A− tRNA[Ser]Sec exhibited more GPX1 activity than those expressing more of the i6A− tRNA[Ser]Sec (Table 4). A similar dose-dependent effect was observed for the inhibition of GPX3, which is synthesized in the kidney and secreted into plasma. The effects of the expression of i6A− tRNA[Ser]Sec on GPX4, deiodinase 1 (D1 synthesized in the liver), deiodinase 2 (D2, synthesized in the pituitary gland), and SelP (synthesized primarily in the liver and secreted to plasma), TR1, and SelT (31) are also presented in Table 4. The tissue specificity of these effects is particularly apparent in the testes, where as much as an 80% reduction in GPX1 was observed while the levels of GPX4, TR3, and SelT were either unaffected or stimulated.
TABLE 4.
Levels of selenoproteins in i6A− mutant tRNA[Ser]Sec transgenic mice and their siblingsa
| Selenoprotein | Tissue | Level of selenoproteins (% of wild type)b
|
|||||
|---|---|---|---|---|---|---|---|
| TGi6A−2 | TGi6A−2/TGi6A−2 | TGi6A−8 | TGi6A−8/TGi6A−8 | TGi6A−20 | TGi6A−20/TGi6A−20 | ||
| GPX1c | Liver | 46 | 22 | 10 | 10 | 5 | 3 |
| Kidney | 30 | 36 | 28 | 24 | 8 | 4 | |
| Brain | 39 | 56 | 47 | 43 | ND | ND | |
| Testes | 99 | 100 | 45 | 40 | 41 | 19 | |
| GPX3c | Plasma (kidney) | 57 | 61 | 24 | 16 | 22 | 20 |
| GPX4d | Liver | NDf | ND | 8 | <5 | 28 | <5 |
| Testes | ND | ND | 100 | 100 | 99 | 86 | |
| D1c | Liver | 39 | 15 | 25 | 21 | 15 | 17 |
| D2c | Pituitary | ND | ND | 36 | 41 | 60 | 13 |
| SelPd | Plasma (liver) | ND | ND | ND | ND | 66 | 45 |
| TR1d | Liver | 80 | 60 | 100 | 50 | 60 | 30 |
| TR3d | Liver | 102 | 126 | 87 | 111 | 176 | 162 |
| TR1/TR3c | Liver | 65 | 42 | 63 | 67 | 33 | 63 |
| TR1/TR3e | Liver | ND | ND | 66 | 89 | 52 | 64 |
| SelTd | Liver | ND | ND | 57 | 40 | 56 | 43 |
| Kidney | ND | ND | 38 | 21 | 42 | 31 | |
| Brain | ND | ND | 63 | 56 | 53 | 42 | |
| Testes | ND | ND | 111 | 107 | 105 | 113 | |
Selenoproteins were analyzed by enzyme activity or Western analysis (see footnotes b and c) as described in Materials and Methods.
Enzyme activities, immunoblot levels, or 75Se PhosphorImager signals observed in wild-type siblings were considered to represent 100% of selenoprotein expression, and the values observed in transgenic mice harboring mutant transgenes are reported as the percentage of that found in the corresponding wild-type tissues.
Activity determined by direct assay.
Western analysis.
PhosphorImager analysis.
ND, not determined.
Northern blot analysis.
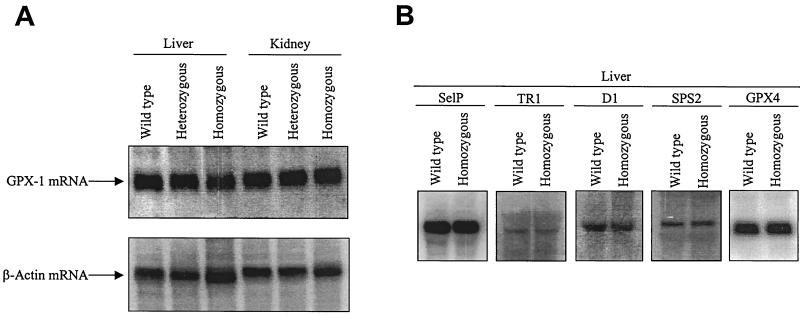
The data presented above cataloging the reduced amounts of selenoproteins in organs of transgenic mice could be explained by effects on either transcription or translation. Therefore, we examined the mRNA levels of several selenoproteins by Northern analysis. GPX1 activity was dramatically reduced in i6A− tRNA[Ser]Sec-expressing animals, as judged by both 75Se labeling and direct enzyme assay. GPX1 mRNA levels were measured by Northern blot analysis in liver and kidney tissue of transgenic mice, where it was apparent that the amount of GPX1 mRNA was virtually unchanged in these two tissues, with the possible exception in livers of mice carrying the highest number of mutant tRNA[Ser]Sec transgenes (Fig. 5A). We also examined several other selenoprotein mRNAs, which included SelP, TR1, D1, SPS2, and GPX4 from livers of mice expressing i6A− tRNA transgenes (Fig. 5B). In general, any differences observed in selenoprotein mRNA levels in organs from i6A− mice were insufficient to account for the reduction observed in the corresponding selenoproteins. The data therefore indicate that the defect in selenoprotein biosynthesis caused by expression of the i6A-deficient tRNA[Ser]Sec occurs at the translation step. Warner et al. (47) have also shown that CHO cells transfected with an i6A− tRNA[Ser]Sec gene exhibited reduced type 1 deiodinase levels without affecting steady-state levels of the corresponding mRNA.
FIG. 5.
Northern analysis of several selenoprotein mRNAs. (A) mRNA levels of GPX1 in liver and kidney of heterozygous and homozygous transgenic mice carrying the highest number of mutant transgenes and their wild-type siblings are shown. (B) mRNA levels of Se1P, TR1, D1, SPS2, and GPX4 in liver tissue of homozygous transgenic mice carrying the highest number of mutant transgenes and their wild-type siblings are shown. mRNA was extracted from livers and kidneys of mice harboring 20 or 40 i6A− mutant tRNA[Ser]Sec transgenes and their wild-type siblings, electrophoresed, and transblotted onto membranes. The membranes were hybridized with 32P-labeled probes complementary to each mRNA shown in both panels, their levels were quantitated by phosphoimagery, and the filters were stripped and rehybridized with β-actin as described in Materials and Methods.
Selenium levels and blood chemistries.
Selenium levels in livers, kidneys, brains, testes, and serum of selected transgenic mice and their wild-type siblings were analyzed (Table 5). Dramatic differences in selenium levels between the i6A− mice and their sibling controls were observed in plasma (30 to 60%), liver, kidney, and heart, while the difference was less apparent in the testes and brain.
TABLE 5.
Selenium levels in tissues of i6A− mutant transgenic and wild-type micea
| Tissue | Genotype | Selenium levela (ppb) | % Decreaseb |
|---|---|---|---|
| Plasma | +/+ | 349.2 | |
| +/+/TGi6 A−2 | 236.0 | 32.4 | |
| +/+/TGi6A−2/TGi6A−2 | 211.3 | 39.5 | |
| +/+ | 360.7 | ||
| +/+/TGi6A−8 | 168.2 | 53.4 | |
| +/+/TGi6A−8/TGi6A−8 | 141.6 | 60.7 | |
| +/+ | 310.7 | ||
| +/+/TGi6A−20 | 163.4 | 47.4 | |
| +/+/TGi6A−20/TGi6A−20 | 159.2 | 48.8 | |
| Liver | +/+ | 777.5 | |
| +/+/TGi6A−20/TGi6A−20 | 254.4 | 67.3 | |
| Kidney | +/+ | 649.5 | |
| +/+/TGi6A−20/TGi6A−20 | 390.2 | 39.9 | |
| Heart | +/+ | 188.9 | |
| +/+/TGi6A−20/TGi6A−20 | 109.9 | 42.8 | |
| Brain | +/+ | 86.6 | |
| +/+/TGi6A−20/TGi6A−20 | 69.3 | 19.6 | |
| Testes | +/+ | 512.0 | |
| +/+/TGi6A−20/TGi6A−20 | 344.2 | 32.8 |
Selenium levels were determined in i6A− tRNA[Ser]Sec mice and their wild-type siblings as described in Materials and Methods. Weights of wild-type and homozygous animals used for selenium organ analysis were 22.4 and 22.8 g, respectively.
Percent decrease in selenium level in i6A− tRNA[Ser]Sec tissues from wild-type level.
No overt signs of ill health were observed in animals overexpressing either wild-type or i6A− transgenes as evidenced by their phenotypic appearance or behavior. Their blood chemistries (see Materials and Methods) fell within the normal ranges of their wild-type siblings regarding cholesterol, serum triglycerides, and liver and kidney function tests (data not shown).
DISCUSSION
There is a growing appreciation of the diverse biological roles for selenoproteins and the central role for tRNA[Ser]Sec in their synthesis. To study the regulation of selenoprotein biosynthesis, we generated transgenic mice that expressed elevated levels of either wild-type or i6A-deficient tRNA[Ser]Sec and examined the consequences of these manipulations on selenoprotein and tRNA[Ser]Sec levels. The generation of the animals described in this study provides the first example of the production of transgenic mice in which multiple copies of a tRNA gene are stably introduced and expressed.
It is evident from our analyses of the tRNA[Ser]Sec population in several tissues from these transgenic mice that they can tolerate relatively high levels of tRNA[Ser]Sec, since some tissues exhibited increases of more than 6-fold in wild-type tRNA[Ser]Sec or more than 12-fold in i6A− tRNA[Ser]Sec without apparent ill effects on their health. The use of primer extension permitted the independent examination of endogenous tRNA[Ser]Sec and that derived from the wild-type transgenes, and the data indicated that both contribute to the resulting tRNA population. Using this approach, we observed a reduction in endogenous tRNA[Ser]Sec with increasing amounts of tRNA[Ser]Sec derived from wild-type transgenes (Fig. 3). Levels of i6A-deficient tRNA[Ser]Sec could be directly evaluated by RPC-5 chromatography, and quantities were correlated with gene copy number. Levels of i6A-deficient tRNA[Ser]Sec as high as 40% of the total tRNASer population did not affect the level of tRNA[Ser]Sec expressed from the corresponding endogenous genes. The distributions of mcm5U and mcm5Um, however, were dramatically affected with increasing amounts of i6A− tRNA[Ser]Sec. Their distributions mimicked those seen in liver and kidney tissue of selenium-deficient rats and mice and in mammalian cells grown in selenium-deficient medium, where mcm5Um was significantly reduced (9, 15, 22). Collectively, these observations support the existence of a feedback mechanism of control that limits the amounts of tRNA[Ser]Sec found in particular tissues. The effector(s) involved in this feedback control likely requires the i6A modification at position 37. The data also show that the presence of the i6A− tRNA[Ser]Sec results in an inhibition of the maturation process.
For mice overexpressing wild-type tRNA[Ser]Sec, the increase was largely restricted to the mcm5U isoform in the tissues examined. There was no apparent effect on selenoprotein synthesis. These data are consistent with the methylation of tRNA[Ser]Sec on the ribosyl moiety at position 34 being a limiting step in tRNA maturation (40). As noted previously in both cell culture (10, 22) and animal models (9, 15), the distribution of the two major isoforms of mammalian tRNA[Ser]Sec responds to increased selenium availability with a characteristic increase in mcm5Um and translational induction of GPX1. This, and the restrictions observed in the conversion to the mcm5Um seen in vivo, suggest that the balance between tRNA[Ser]Sec isoacceptors serves a biologically significant purpose that is yet to be defined.
In contrast to the data obtained with the overexpression of wild-type tRNA[Ser]Sec, selenoprotein synthesis was dramatically affected in mice expressing i6A− tRNA[Ser]Sec. This was demonstrated by 75Se labeling of selenoproteins and by direct enzyme assay and/or Western blot analyses. The expression of several selenoproteins was reduced substantially in all tissues examined (e.g., GPX1), with a dose-dependent effect being evident. All selenoproteins examined in this study appeared to be reduced in the livers of i6A− mice, with the exception of TR3, which was either unaffected or stimulated. The expression of other selenoproteins, such as GPX4 and SelT, was also inhibited in the liver but unaffected in the testes by increasing amounts of i6A− tRNA[Ser]Sec. The lack of an effect in the testes may be due to the fact that the tRNA[Ser]Sec population in the testes (>7% of the total serine tRNA population) is substantially higher than that in the liver (∼2.5% of the total serine tRNA population). Therefore, the i6A-deficient tRNA[Ser]Sec population represented a much lower proportion of the tRNA[Ser]Sec population in the testes than in the liver (Table 3). Translation of GPX1 appeared to be particularly sensitive to the presence of i6A− tRNA[Ser]Sec, consistent with the possibility that its synthesis may be more dependent on the mcm5Um isoform than is that of other selenoproteins (9).
A hierarchy exists with regard to the effects of selenium deficiency on the maintenance of individual selenoproteins as well as selenium retention by different organs (2, 24, 34, 36, 39). For example, GPX1 activity was reduced to 1% in liver tissue and about 4 to 9% in kidney, heart and lung tissues during selenium deficiency in rats. GPX4 activity, on the other hand, was reduced only about 25 to 50% in these tissues but was unaffected in the testes. A similar hierarchy of selenium maintenance may be occurring in response to the expression of i6A− tRNA[Ser]Sec. As seen in Table 4, the levels of each of the selenoproteins analyzed in i6A− mice were reduced to different degrees, and the extent of the reduction differed depending on the organ examined.
The greater sensitivity of GPX1 activity to selenium deficiency was attributed in large part to increased mRNA turnover (11, 34, 44). In contrast, the reduction in GPX1 observed with increasing amounts of i6A-deficient tRNA is not likely due to mRNA turnover, since GPX1 mRNA stability was not significantly altered in kidneys of i6A− mice. The stem-loop structure in the 3′ untranslated region of mammalian selenoprotein mRNAs, designated the SECIS element (35), has also been shown to play a role in establishing a selenoprotein hierarchy (36). Thus, it would appear that there are several levels of regulation involved in determining the priority of selenoprotein synthesis under various biological conditions.
Under conditions of selenium deprivation in the diets of rats and mice, the levels of this element are reduced in the liver and kidneys, while the brain and testes retain most of their selenium (2, 25). Transfer RNA[Ser]Sec maturation and selenoprotein synthesis are responsive to selenium status, and thus these two parameters are more affected by change in selenium status in the liver and kidneys than in the testes and brain (15, 25). Selenoprotein biosynthesis was most affected in the liver, kidneys, and brain in the presence of i6A− tRNA[Ser]Sec and least affected in the testes, while selenium losses were highest in the liver and kidneys and lowest in the testes and brain. The losses in selenium in the brain were the lowest of the tissues examined, and this reduction may reflect only an inhibition in selenoprotein biosynthesis. Observations showing a differential loss in selenium retention and differential rates in selenoprotein biosynthesis suggest that the i6A− tRNA[Ser]Sec transgenic mice can be used as a model system to better understand the hierarchy in selenium retention by different tissues.
There is an abundance of literature supporting selenium as a protective agent against a variety of mutagens, carcinogens, and viruses in laboratory animals, namely rats and mice (reviewed in reference 19). Animals given slightly elevated levels of selenium in their diets have the greatest protection upon exposure to these environmental stresses. Studies with humans also support a role of selenium as a beneficial micronutrient in the diet (17). It is unclear whether these beneficial effects of selenium on health are due to selenium-containing, low-molecular-weight compounds (see references 17 and 27 and references therein) or to selenium-containing proteins (reviewed in reference 19). Since transgenic i6A− tRNA[Ser]Sec mice have reduced selenoprotein levels, it will be of considerable interest to determine if slightly elevated levels of dietary selenium will afford these transgenic mice the same protection from environmental stress as their wild-type siblings have.
Since the i6A− tRNA[Ser]Sec mice selectively express selenoproteins, then by controlling the mutant tRNA[Ser]Sec gene dosage, these animals may be used to provide a useful model for resolving the roles of individual selenoproteins as well as their overall influence on health. It is of interest that adult animals made selenium deficient through diet appear to be as healthy as their control counterparts maintained on selenium-sufficient diets unless they are challenged or stressed environmentally. The i6A− tRNA[Ser]Sec transgenic mice also appear to be as healthy as their wild-type littermates. By the selective use of individual carcinogens, mutagens, and viruses and careful monitoring of the rates of malignant change at target sites, these transgenic mice may be used as a powerful tool for determining the role of individual selenoproteins in protecting against specific environmental stresses.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants GM616603 (V.N.G.), CAA81153 (A.M.D.), DK47320 (M.J.B.) and ES02497 (R.F.B. and K.E.H.) and a grant 99A026 from the American Institute for Cancer Research (A.M.D.) and from the Molecular Medicine Research Group Program, Ministry of Science and Technology of Korea (B.J.L.).
The authors express their sincere appreciation to Glen Merlino for his advice and helpful discussions throughout the course of this study.
M.E.M. and B.A.C. contributed equally to this project.
REFERENCES
- 1.Arner E S, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 2.Behne D, Hilmet H, Scheid S, Gessner H, Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim Biophys Acta. 1988;996:12–21. doi: 10.1016/0304-4165(88)90123-7. [DOI] [PubMed] [Google Scholar]
- 3.Berry M J, Kieffer J D, Harney J W, Larsen P R. Selenocysteine confers the biochemical properties characteristic of the type I iodothyronine deiodinase. J Biol Chem. 1991;266:14155–14158. [PubMed] [Google Scholar]
- 4.Bjork G R. Modified nucleosides at positions 34 and 37 of tRNAs and their predicted coding capacities. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: American Society for Microbiology; 1998. pp. 577–581. [Google Scholar]
- 5.Bjork G R, Rasmuson T. Links between tRNA modification and metabolism and modified nucleosides as tumor markers. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: American Society for Microbiology; 1998. pp. 471–492. [Google Scholar]
- 6.Böck A. Biosynthesis of selenoproteins—an overview. Biofactors. 2000;11:77–78. doi: 10.1002/biof.5520110122. [DOI] [PubMed] [Google Scholar]
- 7.Bosl M R, Takadu K, Oshima M, Nishimura S, Taketo M M. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster R L, Chen H Y, Trumbauer M E, Yagle M K, Palmiter R D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chittum H S, Hill K E, Carlson B A, Lee B J, Burk R F, Hatfield D L. Replenishment of selenium deficient rats with selenium results in redistribution of the selenocysteine tRNA population in a tissue specific manner. Biochim Biophys Acta. 1997;1359:25–34. doi: 10.1016/s0167-4889(97)00092-x. [DOI] [PubMed] [Google Scholar]
- 10.Choi I S, Diamond A M, Crain P F, Kolker J D, McCloskey J A, Hatfield D L. Reconstitution of the biosynthetic pathway of selenocysteine tRNAs in Xenopus oocytes. Biochemistry. 1994;33:601–605. doi: 10.1021/bi00168a027. [DOI] [PubMed] [Google Scholar]
- 11.Christensen M J, Burgener K W. Dietary selenium stabilized glutathione peroxidase mRNA in rat liver. J Nutr. 1992;122:1620–1626. doi: 10.1093/jn/122.8.1620. [DOI] [PubMed] [Google Scholar]
- 12.Copeland P R, Fletcher J E, Carlson B A, Hatfield D L, Driscoll D M. Identification of a novel mammalian RNA binding protein required for the cotranslational incorporation of selenocysteine. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran J F. Modified nucleosides in translation. In: Groesjean H, Benne R, editors. Modification and editing of tRNA. Washington, D.C.: American Society for Microbiology; 1998. pp. 493–516. [Google Scholar]
- 14.Diamond A M, Dudock B, Hatfield D L. Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981;25:497–506. doi: 10.1016/0092-8674(81)90068-4. [DOI] [PubMed] [Google Scholar]
- 15.Diamond A M, Choi I S, Crain P F, Hashizume T, Pomerantz S C, Cruz R, Steer C, Hill K E, Burk R F, McCloskey J A, Hatfield D L. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec) J Biol Chem. 1993;268:14215–14223. [PubMed] [Google Scholar]
- 16.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganther H E. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis (London) 1999;20:1657–1666. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- 18.Gereben B, Bartha T, Tu H M, Harney J W, Rudas P, Larsen P R. Cloning and expression of the chicken type 2 iodothyronine deiodinase. J Biol Chem. 1999;274:13768–13776. doi: 10.1074/jbc.274.20.13768. [DOI] [PubMed] [Google Scholar]
- 19.Gladyshev V N, Martin-Romero F J, Xu X-M, Kumaraswamy E, Carlson B A, Hatfield D L, Lee B J. Molecular biology of selenium and its role in cancer, AIDS and other human diseases. Recent Res Dev Biochem. 1999;1:145–167. [Google Scholar]
- 20.Gladyshev V N, Stadtman T C, Hatfield D L, Jeang K T. Levels of major selenoproteins in T cells decrease during HIV infection and low molecular mass selenium compounds increase. Proc Natl Acad Sci USA. 1999;96:835–839. doi: 10.1073/pnas.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatfield D L, Mathews C R, Rice M. Aminoacyl-transfer RNA populations in mammalian cells, chromatographic profiles and patterns of codon recognition. Biochim Biophys Acta. 1979;564:414–423. doi: 10.1016/0005-2787(79)90032-7. [DOI] [PubMed] [Google Scholar]
- 22.Hatfield D L, Lee B J, Hampton L, Diamond A M. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 1991;19:939–943. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatfield D L, Gladyshev V N, Park J M, Park S I, Chittum H S, Huh J R, Carlson B A, Kim M, Moustafa M E, Lee B J. Biosynthesis of selenocysteine and its incorporation into protein as the 21st amino acid. In: Kelly J W, editor. Comprehensive natural products chemistry. Vol. 4. Oxford, England: Elsevier Science Ltd.; 1999. pp. 353–380. [Google Scholar]
- 24.Hill K E, Chittum H S, Lyons P R, Boeglin M E, Burk R F. Effect of selenium on selenoprotein P expression in cultured liver cells. Biochim Biophys Acta. 1996;1313:29–34. doi: 10.1016/0167-4889(96)00047-x. [DOI] [PubMed] [Google Scholar]
- 25.Hill K E, Lyons P R, Burk R F. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem Biophys Res Commun. 1992;185:260–263. doi: 10.1016/s0006-291x(05)80984-2. [DOI] [PubMed] [Google Scholar]
- 26.Hubert N, Sturchler C, Westhof E, Carbon P, Krol A. The 9/4 secondary structure of eukaryotic selenocysteine tRNA: more pieces of evidence. RNA. 1998;4:1029–1033. doi: 10.1017/s1355838298980888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ip C, Thompson H J, Zhu A, Ganther H E. In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer prevention. Cancer Res. 2000;60:2882–2886. [PubMed] [Google Scholar]
- 28.Jung J E, Karoor V, Sandbaken M G, Lee B J, Ohama T, Gesteland R F, Atkins J F, Mullenbach G T, Hill K E, Wahba A J, Hatfield D L. Utilization of selenocysteyl-tRNA[Ser]Sec and seryl-tRNA[Ser]Sec in protein synthesis. J Biol Chem. 1994;269:29739–29745. [PubMed] [Google Scholar]
- 29.Kelmers A D, Heatherly D E. Columns for rapid chromatographic separation of small amounts of tracer-labeled transfer ribonucleic acids. Anal Biochem. 1971;44:486–495. doi: 10.1016/0003-2697(71)90236-3. [DOI] [PubMed] [Google Scholar]
- 30.Kim L K, Matsufuji T, Matsufuji S, Carlson B A, Kim S S, Hatfield D L, Lee B J. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA. 2000;6:1306–1315. doi: 10.1017/s1355838200000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kryukov G V, Kryukov V M, Gladyshev V N. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine sequence elements. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence R A, Burk R F. Glutathione peroxidase activity in selenium-deficient rat liver. Biochem Biophys Res Commun. 1976;71:952–958. doi: 10.1016/0006-291x(76)90747-6. [DOI] [PubMed] [Google Scholar]
- 33.Lee B J, Rajagopalan M, Kim Y S, You K H, Jacobson K B, Hatfield D L. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol Cell Biol. 1990;10:1940–1949. doi: 10.1128/mcb.10.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei X G, Evenson J K, Thompson K M, Sunde R A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 35.Low S C, Berry M J. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–207. [PubMed] [Google Scholar]
- 36.Low S C, Grundner-Culemann E, Harney J W, Berry M J. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McShane L M, Clark L C, Combs G F, Jr, Turnbull B W. Reporting the accuracy of biochemical measurements for epidemiologic and nutrition studies. Am J Clin Nutr. 1991;53:1354–1360. doi: 10.1093/ajcn/53.6.1354. [DOI] [PubMed] [Google Scholar]
- 38.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell J H, Nicol F, Beckett G J, Arthur J R. Selenium and iodine deficiencies: effects on brain and brown adipose tissue selenoenzyme activity and expression. J Endocrinol. 1997;155:255–263. doi: 10.1677/joe.0.1550255. [DOI] [PubMed] [Google Scholar]
- 40.Moustafa M E, El-Saadani M A, Kandeel K M, Mansur D B, Lee B J, Hatfield D L, Diamond A M. Overproduction of selenocysteine tRNA in Chinese hamster ovary cells following transfection of the mouse tRNA[Ser]Sec gene. RNA. 1998;4:1436–1443. doi: 10.1017/s1355838298981043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohama T, Choi I S, Hatfield D L, Johnson K R. Mouse selenocysteine tRNA[Ser]Sec (Trsp) and its localization on chromosome 7. Genomics. 1994;19:595–596. doi: 10.1006/geno.1994.1116. [DOI] [PubMed] [Google Scholar]
- 42.Ohama T, Jung J-E, Park S I, Clouse K A, Lee B J, Hatfield D. Identification of new selenocysteine tRNA[Ser]Sec isoacceptors in human cell lines. Biochem Mol Biol Int. 1995;36:421–427. [PubMed] [Google Scholar]
- 43.O'Neill V A, Eden F C, Pratt K, Hatfield D L. A human opal suppressor tRNA gene and pseudogene. J Biol Chem. 1985;260:2501–2508. [PubMed] [Google Scholar]
- 44.Saedi M S, Smith C G, Frampton J, Chambers I, Harrison P R, Sunde R A. Effect of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem Biophys Res Commun. 1988;153:855–861. doi: 10.1016/s0006-291x(88)81174-4. [DOI] [PubMed] [Google Scholar]
- 45.Sun Q-A, Wu Y, Zappacosta F, Jeang K-T, Lee B J, Hatfield D L, Gladyshev V N. Redox regulation of cell signaling by selenocysteine in mammalian thioredoxin reductases. J Biol Chem. 1999;274:24522–24530. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- 46.Tujebajeva R M, Copeland P R, Xu X, Carlson B A, Harney J W, Driscoll D M, Hatfield D L, Berry M J. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warner G J, Berry M J, Moustafa M E, Carlson B A, Hatfield D L, Faust J R. Inhibition of selenoprotein synthesis by selenocysteine tRNA[Ser]Sec lacking isopentenyladenosine. 2000. J Biol Chem. 2000;36:28110–28119. doi: 10.1074/jbc.M001280200. [DOI] [PubMed] [Google Scholar]