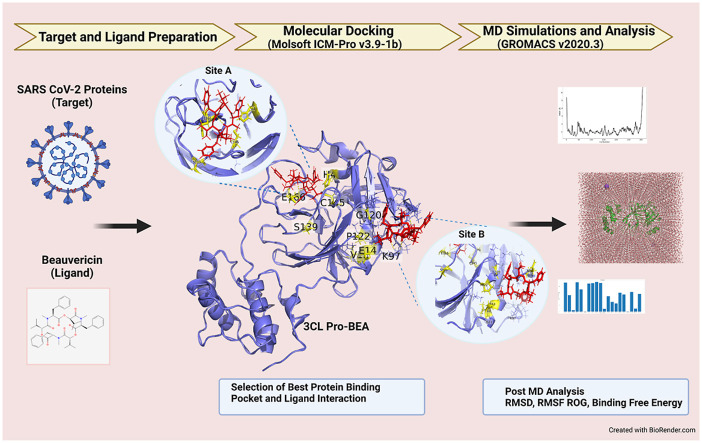
Abstract
Background
Scientists are still battling severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for the coronavirus 2019 (COVID-19) pandemic so human lives can be saved worldwide. Secondary fungal metabolites are of intense interest due to their broad range of pharmaceutical properties. Beauvericin (BEA) is a secondary metabolite produced by the fungus Beauveria bassiana. Although promising anti-viral activity has previously been reported for BEA, studies investigating its therapeutic potential are limited.
Methods
The objective of this study was to assess the potential usage of BEA as an anti-viral molecule via protein–protein docking approaches using MolSoft.
Results
In-silico results revealed relatively favorable binding energies for BEA to different viral proteins implicated in the vital life stages of this virus. Of particular interest is the capability of BEA to dock to both the main coronavirus protease (Pockets A and B) and spike proteins. These results were validated by molecular dynamic simulation (Gromacs). Several parameters, such as root-mean-square deviation/fluctuation, the radius of gyration, H-bonding, and free binding energy were analyzed. Computational analyses revealed that interaction of BEA with the main protease pockets in addition to the spike glycoprotein remained stable.
Conclusion
Altogether, our results suggest that BEA might be considered as a potential competitive and allosteric agonist inhibitor with therapeutic options for treating COVID-19 pending in vitro and in vivo validation.
Keywords: Beauvericin, Docking, Dynamic simulation spike protein, Main protease, SARS-CoV-2
Graphical abstract
1. Introduction
In December 2019, a rapidly spreading contagious pneumonia outbreak was first reported in Wuhan, Hubei Province, China. This respiratory disease quickly evolved into a pandemic that is presently affecting millions worldwide and was referred to as coronavirus 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. Multiple studies have confirmed that SARS-CoV-2 is capable of attaching to host cells via the receptor for the enzyme angiotensin-converting enzyme 2 (ACE2), which shows enhanced expression in type II alveolar cells of the lungs [2]. In addition, a “cytokine storm” elicited by the virus has channeled the interest of scientists toward reducing the inflammatory response in conjunction with developing anti-viral drugs for prevention and post-infection treatments. A key part of drug discovery is the understanding of the proteins used by this virus and potential corresponding drug candidates. Importantly, the target protein must be critical for SARS-CoV-2 infection and contain appropriate molecular regions (pockets) in which a drug can bind with both high specificity and stability. Computer-aided approaches were established and have been used for a long time in pharmaceutical research to enhance development, manufacturing, and further drug processing. These computational tools are used to systematically investigate potential lead molecules and assess the protein-ligand chemical interaction [3].
Mining the SARS-CoV-2 proteome revealed several drug targets that may play important roles in viral pathogenesis. Seven of these proteins play essential roles in viral entry and replication in the host: (1) the main protease, (2) the RNA-dependent RNA polymerase, (3) the methyltransferase, (4) the helicase, (5) the endoribonuclease, (6) the exoribonuclease, and (7) the spike protein. The main protease of the virus, the 3CL protease (3CLpro), is an indispensable enzyme for its own replication. It cleaves the translated polyprotein at 11 conserved sites, including its own N and C domains to generate among others the helicase, the single-stranded RNA-binding protein, the RNA-dependent RNA polymerase, the exoribonuclease, and the endoribonuclease of the virus [4,5]. The spike glycoprotein (S protein), which provides the crown shape to the virus by studding its surface, modulates viral entry by binding to the host ACE2 [6]. The helicase facilitates the folding and replication of SARS-CoV-2, and therefore plays a major role during the viral life cycle [7]. As for SARS-CoV-2 endoribonuclease, it was shown to play a major role in preventing the detection of the virus double-stranded RNA by the host sensor [8] while its methyltransferase enzyme displays an essential role in RNA cap formation, an indispensable progression step for viral RNA stability [9]. Finally, the RNA-dependent-RNA-polymerase is a key component for all RNA viruses; it was recognized to be the most potential pan-target for all such types of viruses including coronaviruses because of its structural and functional conservation [10,11]. SARS-CoV-2 exoribonuclease is an important regulator of nucleic acid integrity; it contributes to the maintenance of the genome by cleaving the mismatched nucleotides to provide a high-proofreading activity [12].
Currently, no specific anti-viral treatment that has proven to be specifically and highly efficient against the virus despite the approval by the food and drug administration (FDA) of three drugs as of November 2021, namely Molnupiravir, Ritonavir, and Casirivimab/Imdevimab exists [13]. As for the ongoing research, the clinical trial registry (ClinicalTrials.gov) includes only ongoing clinical trials for existing repurposed FDA-approved drugs. In contrast, researchers in basic research laboratory settings are actively testing novel small molecules for their potential therapeutical value. It is therefore imperative in the wake of the challenges caused by the virus to search for such molecules, some of which are actively synthesized in other species or kingdoms. Examples of such potential drugs are secondary metabolites produced by fungi which have been used in agriculture as pesticides and are being considered in the medical field as potential therapeutics for different diseases [14]. The entomopathogenic fungus Beauveria bassiana is known to produce several cyclodepsipeptide toxins like beauvericin (BEA) and bassianolide with insecticidal, antibiotic, antiviral, and antifungal activities [[14], [15], [16]]. BEA contains three D-hydroxyisovaleryl and three N-methylphenylalanil residues in an alternating sequence [14]. Recently, accumulated data from in vitro experiments performed mainly on cell lines have shown that BEA has a variety of bioactivities and is being considered as a potential candidate for medicinal research [17]. The antibacterial activity of BEA has been ascertained against both Gram-negative and -positive bacteria [18,19]. In addition, it was demonstrated that BEA is efficient in reducing the proliferative index of cancerous cell lines in vitro [15,20]. In parallel, it was demonstrated that BEA presents significant inhibitory activities against the purified retroviral enzyme integrase involved in the integration of the human immunodeficiency virus type 1 (HIV-1) in the host genome [21]. Remarkably, the BEA concentration used in the study was low relative to the cytotoxic concentrations against all tested human cell lines [22]. This inhibitory potential of BEA was comparable to those of baicalein and robinetin, which have previously been demonstrated to be effective antiviral compounds [23]. Finally, we have recently revealed the high efficacy of the molecule against all developmental stages of the mite Sarcoptes scabiei, which could warrant its clinical usage in the treatment of sarcoptic infections [24].
In this study, we used molecular docking, molecular dynamic simulations, and binding free energy estimations to predict the biomolecular interactions between BEA and the different SARS-CoV-2 proteins, and we demonstrated strong and stable BEA interaction with the main protease of the virus.
2. Materials and methods
2.1. Preparation of the ligand
The crystal structure of BEA was retrieved from The Toxin and Toxin Target Database (www.t3db.ca/toxins), while those of the remaining drugs were retrieved from drug bank (www.drugbank.com). The ligands were loaded in.sdf format and transformed automatically into a three-dimensional (3D) structure during the docking process.
2.2. Preparation of the viral proteins
The crystal structures of seven viral proteins encoded by SARS-CoV-2 were downloaded from RCSB PDB (https://www.rcsb.org) and used as targets. The crystal structures of all viral proteins were imported into Molsoft.icm-pro v3.9–1b [25]. The Protein Data Bank (PDB) structures of the proteins were converted into ICM objects by deleting all water molecules contained in the X-ray structures and optimizing the hydrogens (to find the best hydrogen bonding network) in addition to the amino acids histidine, proline, asparagine, glycine, cysteine (to find best orientation and protonation state). Missing side chains were treated before the receptors were set for the docking processes. To identify the binding sites and generate receptor maps, the icmPocketFinder function option was used. This method uses the protein structure to identify cavities/clefts and the “druggability” was estimated by calculating the drug-like density (DLID) score as described previously [26].
2.3. Molecular docking
After conversion and selection, the binding site residues of the viral proteins were docked with the ligands (Table 1 ). At the receptor pocket, hydrogen bonding potential, van der Waals potential with carbon-, sulfur- and hydrogen-like probes, hydrophobic potential, and electrostatic potential were taken into consideration. The conformational examination of the program depends on the Biased Probability Monte Carlo (BPMC) system, which arbitrarily chooses a pose in the inside coordinate space and at that point makes a stage to another arbitrary position free of the past one, yet as indicated by a predefined constant probability distribution [27]. In this study, the thoroughness, which represents the length of the simulation, was set as 10. The ligand conformations were ranked using the ICM score [25]. The lowest ICM score refers to strong binding of protein-ligand complex, namely, the more negative score, the high interaction.
Table 1.
ICM scores for beauvericin (BEA) and different known inhibitors against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins.
| Protein ID | Pocket ID | Drug | ICM Score (kcal/mol) |
|---|---|---|---|
| 6LU7 3CL protease | A | Beauvericin | −7.24 |
| Chloroquine | −11.24 | ||
| Remdesivir | −14.37 | ||
| Luteolin | −13.57 | ||
| N3 | −20.46 | ||
| Boceprevir | −10.4 | ||
| GC376 | −20.16 | ||
| Ritonavir | −14.52 | ||
| Lopinavir | −14.35 | ||
| B | Beauvericin | −12.53 | |
| Chloroquine | −8.04 | ||
| Remdesivir | −7.9 | ||
| Luteolin | −11.15 | ||
| N3 | −2.26 | ||
| Boceprevir | −3.5 | ||
| GC376 | −4.7 | ||
| Ritonavir | 2.53 | ||
| Lopinavir | 2.09 | ||
| 6VXX Spike Glycoprotein | A | Beauvericin | −9.88 |
| Remdesivir | −2.1 | ||
| Luteolin | −20.94 | ||
| Ivermectin | 20.7 | ||
| B | Beauvericin | 2.2 | |
| Remdesivir | 10.58 | ||
| Luteolin | −23.42 | ||
| Ivermectin | 26.03 | ||
| Receptor binding domain spike-ACE2 | Beauvericin | −1.8 | |
| Remdesivir | −3.42 | ||
| Luteolin | −11.42 | ||
| Ivermectin | 7.8 | ||
| 6ZSL Helicase | A | Beauvericin | −4.82 |
| MLS001181552 | −12.24 | ||
| NPC270578 | −7.7 | ||
| B | Beauvericin | −1.25 | |
| MLS001181552 | −0.5 | ||
| NPC270578 | −10.5 | ||
| 6VWW Endoribonuclease | A | Beauvericin | −0.07 |
| Arzanol | −3.5 | ||
| RO-7 | −1.18 | ||
| NPC169474 | −10.35 | ||
| B | Beauvericin | 9.35 | |
| Arzanol | −8.211 | ||
| RO-7 | −1.5 | ||
| NPC169474 | −6.64 | ||
| 7BTF RNA-dependent-RNA-polymerase | A | Beauvericin | −3.05 |
| Remdesivir | −5.515 | ||
| NPC161224 | −14.92 | ||
| B | Beauvericin | 0.5 | |
| Remdesivir | −1.3 | ||
| NPC161224 | −15.82 | ||
| 6W4H Methyltransferase | A | Beauvericin | 13.64 |
| S-adenosylmethionine | −7.28 | ||
| Sinefungin | −6.37 | ||
| NPC226294 | −12 | ||
| B | Beauvericin | −3.94 | |
| S-adenosylmethionine | −2.1 | ||
| Sinefungin | −0.7 | ||
| NPC226294 | −12.04 | ||
| 5C8T Exoribonuclease | A | Beauvericin | 2.01 |
| MES | −15.27 | ||
| NPC137813 | −6.745 | ||
| B | Beauvericin | 1.39 | |
| MES | −14.63 | ||
| NPC137813 | −11.04 |
2.4. Molecular dynamics
Molecular docking was combined with thermodynamics-based methods to provide more reliable and accurate results. ICMpro generated the best docking poses of BEA against 3CLpro (pockets A and B) and the S protein (pocket A). The three complexes (BEA-3CL pro [pocket A], BEA-3CLpro [pocket B], and BEA- S protein) were considered as conformations for further molecular dynamics (MD) analysis. MD simulations were performed on the selected targets using GROningen MAchine for Chemical Simulations (GROMACS 2020.3). The forcefield we used to generate topologies of proteins was CHARMM36 in GROMACS format (.gro) obtained from the Mackerell lab website (http://mackerell.umaryland.edu/charmm_ff.shtml#gromacs). The topologies of BEA were obtained by the CHARMM General Force Field CGENFF and converted into.gro format using the script available at the Mackerel lab website (cgenff_charmm2gmx_py3_nx1.py). The complexes were contained in a dodecahedral unit cell shape and solvated with a single point charge (SPC) water model. All systems were neutralized by adding the appropriate number of ions using the “gmx genion” script. To release conflicting contacts, energy minimization of the complexes was done by using the steepest descending algorithm until the energy reaches less than 10 kJ/mol [28]. Afterward, the solvated energy minimized complexes were equilibrated using fixed number of atoms, volume, and temperature and fixed number of atoms, volume, and pressure (NVT and NPT ensembles, respectively). The former step was conducted for 2 ns with a constant number of particles, volume, and temperature (310 K), while the latter, was done for 8 ns with a constant number of particles, pressure, and temperature. The long-range electrostatic interactions were done by the particle mesh Ewald (PME) method with a 12 Å cut-off and 12 Å Fourier spacing [29]. Electrostatic and van der Waals (vdW) non-bonded interactions were calculated in the cut-off range of 1 nm. The bond constraint for all heavy atoms was done by using the LINCS algorithm [30]. Finally, the equilibrated systems were subjected to MD simulations for 100 ns using the leapfrog method [31]. The Root Mean Square Fluctuation (RMSF), Root Mean Square Deviation (RMSD), radius of gyration (Rg), and peptide-peptide hydrogen bond (H-bond) were calculated from the generated trajectories of the MD simulations using various scripts of GROMACS.
2.5. Free binding energy calculation
In this study, we relied on the molecular mechanics PB surface area (MM/PBSA) [32] or the calculation of binding free energy using g_mmbsa tool of gromacs [33]. The entire stable MD trajectories were used to perform MM/PBSA calculations. The binding free energy of the complex in solvent was calculated according to the formula:
in which Gcomplex is the total free energy of the complex (protein-ligand), Gprotein is the total energy of separated protein in the solvent, and Gligand is the total energy of separated ligand in the solvent. These energies were estimated by. in which x is either protein or ligand or protein-ligand complex and Emm and Gsolvation are the average molecular mechanics potential energy in vacuum and free energy of solvation, respectively.
Emm was calculated by the following formula:
Ebonded is bonded interaction including of bond, angle, dihedral, and improper interactions.
Enon-bonded is non bonded interaction including van der Waals (Evdw) and electrostatic (Eelec) interaction. ΔEbonded is considered 0.
The solvation free energy (Gsolvation) was estimated as the sum of electrostatic solvation free energy (Gpolar) and apolar solvation free energy (Gnon-polar):
Polar solvation energy (Gpolar) can be estimated using the Poisson-Boltzmann (PB) equation [32]. A linear model was used to solve the PB equation [34] to calculate the polar part of the solvation energy (Gpolar). Furthermore, the solvent accessible surface area (SASA) method was employed to calculate the nonpolar part of the solvation energy (Gnon-polar). In this model, it is assumed that Gnon-polar is linearly dependent on the SASA and is written as shown below:
in which γ is a coefficient related to surface tension of the solvent, and b is fitting parameter. The values of the constants are shown below:
γ = 0.02267 kJ/Mol/Å2 or 0.0054 Kcal/Mol/Å2
b = 3.849 kJ/Mol or 0.916 Kcal/Mol
2.6. Pharmacokinetics properties
The pharmacokinetic properties of BEA were predicted using the ADMET-SAR2 [35] and pkCSM [36] servers which are freely available online webservers for predicting the ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties of small molecules and drugs.
3. Results
3.1. Molecular docking
Molecular docking of BEA was performed with all the seven target proteins through Molsoft ICM-Pro. The ICM PocketFinder function was leveraged to generate the most likely binding pockets in the proteins, and subsequently, BEA was docked against those pockets (Supplemental Table 1). The ICM scores of each docked complex is provided in Table 1.
3.1.1. 3CL protease
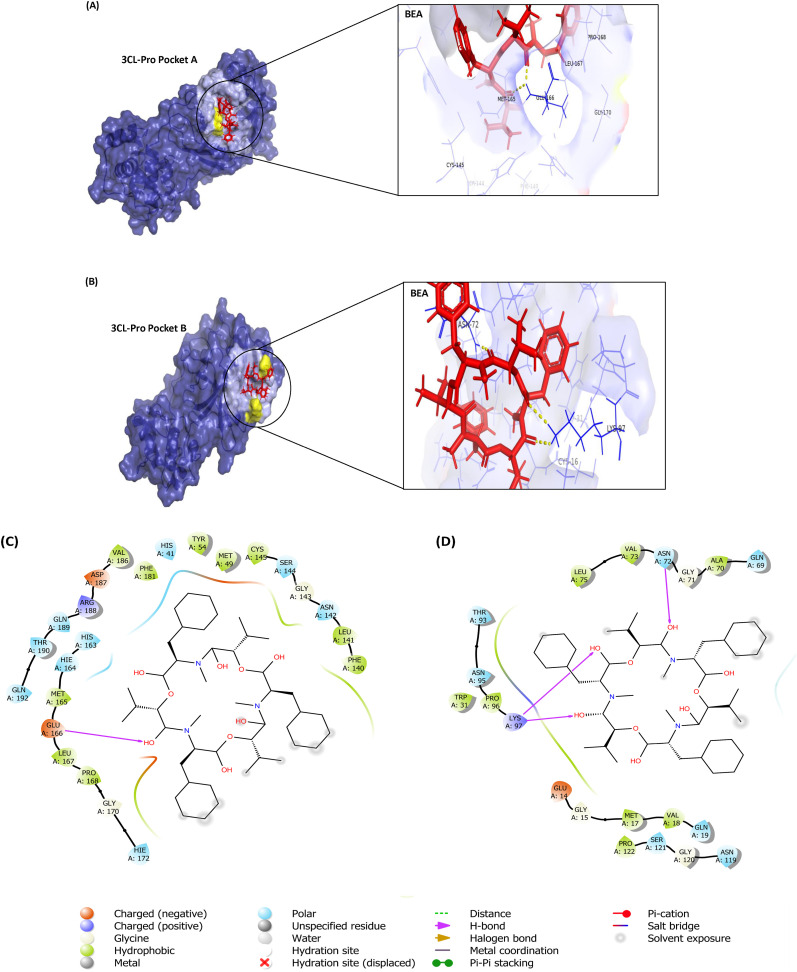
Numerous drugs with potential inhibition activities against 3CLpro (PDB: 6LU7) were used as positive controls: (1) chloroquine, an antimalarial drug found to be efficacious in treating COVID-19 infections [37,38]; (2) remdesivir, found to be effective in preventing replication of this virus [39], (3) luteolin, the main flavonoid in honeysuckle [40] found to dock nicely with the main protease of the virus [41], (4) N3 (5-Methylisoxazole-3-carboxylic acid) the inhibitor in complex with the protein crystal structure [42]), (5) boceprevir, a clinically approved anti-hepatitis C virus drug; (6) GC376 the inhibitor of feline infectious peritonitis virus for their demonstrated effectiveness against SARS-CoV-2 by targeting 3CLpro [43] and the principal components of Kaletra, (7) ritonavir; and (8) lopinavir, an inhibitor of the SARS-CoV 3CLpro [44]. The most druggable pocket (pocket A: Supplementary Table S1) found in the crystal structure consisted of the inhibitor N3 binding site. The docking score for BEA against pocket A of the 3CLpro was the highest (ICM score = −7.24 kcal/mol) when compared to the positive control ligands (Table 1).
Hydrophobic interactions were formed between BEA and M49, Y54, F140, K141, G143, C145, M165, L167, P168, G170, V186, and F181. Polar interactions were found in H41, N142, S144, H163, H164, H172, Q189, T190, and Q192. In addition, one H-bond (distance 1.78 Å) was established between BEA and E166 (Fig. 1 ). In parallel, BEA recorded the best docking conformation against Pocket B relative to the drugs that are commonly used in the clinic with the lowest ICM score of −12.53 kcal/mol and forming a strong H-bond (distance 1.91 Å) with N72 and K97, hydrophobic interactions (G15, M17, V18, W31, A70, G71, V73, P96, G120, and P122) and polar interactions (Q19, Q69, N72, T93, N95, N119, and S121) as shown in Fig. 1.
Fig. 1.
Molecular docking of beauvericin (BEA) with 3CL Pro: A) Pocket A; Surface representation of Protein in Blue and BEA is shown in red. Yellow dotted line shows H-bond with Glu166. B) Pocket B; Surface representation of protein in Blue and BEA in red. Yellow dotted lines shows H-bond with Asn72 and Lys97. C) 2D diagram of Pocket A; D) Two-dimensional (2D) diagram of Pocket B; H-bond is indicated in purple arrow while residues forming hydrophobic interactions are shown in green. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1.2. Spike glycoprotein
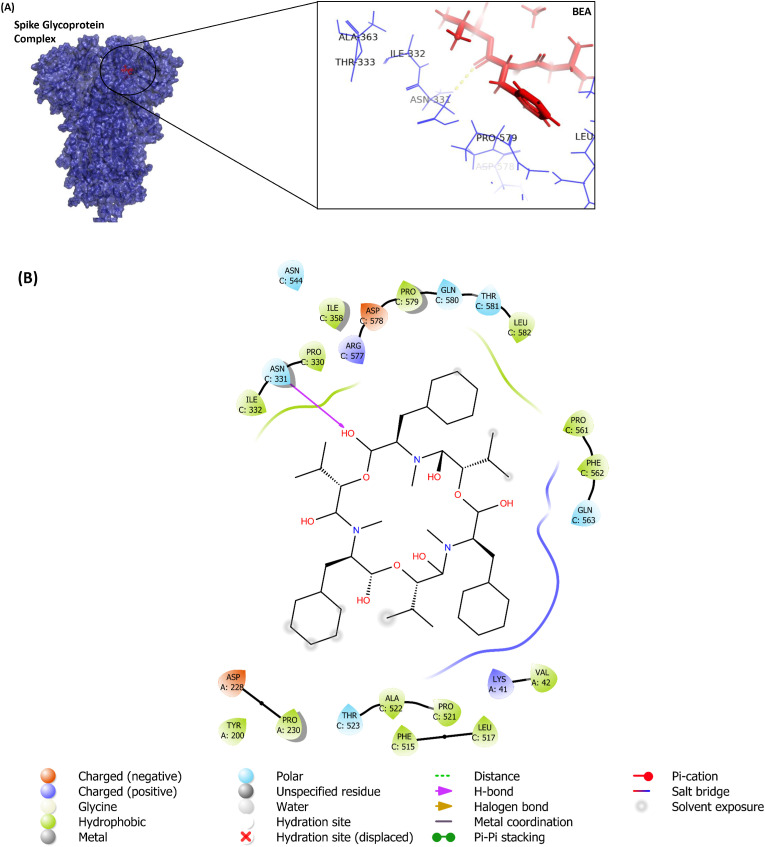
In addition to BEA, remdesivir, luteolin, and ivermectin were docked against the S protein of the SARS-CoV-2 (PDB: 6VXX). The docking results (−9.88 kcal/mol) for pocket A, the most “druggable binding site” (Supplementary Table S1), revealed that BEA mainly anchors itself to 6VXX through hydrophobic (V42, Y200, P230, P330, I332, I358, F515, L517, P521, A522, P561, F562, P579, and L582) and polar interactions (T523, N544, Q563, Q580, T581). One H-bond (distance 2.60 Å) was established with N331 (Fig. 2 ). Among all ligands, only luteolin demonstrated a lower binding affinity when compared to BEA. The BEA docked poorly (Table 1) to the second pocket (Pocket B Supplementary Table S1) as identified by the software. We further explored the pocket responsible for the binding of the S protein to the cell receptor ACE2 [45]. The ICM scores for BEA, remdesivir, luteolin, and ivermectin when docked against the receptor-binding domain were −1.8, −3.42, −11.42, and 7.8, respectively.
Fig. 2.
Molecular docking of BEA with S protein: A) Surface representation of protein in blue and BEA is shown in red. Yellow dotted line shows H-bond with Asn331. B) 2D diagram of interacting residues; H-bond is indicated by purple arrow while residues forming hydrophobic interactions are shown in green. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1.3. Helicase
MLS001181552, the inhibitor of helicase, and astilbin (NPC270578), a plant flavonol, were used as references for their demonstrated capabilities of docking strongly with the helicase of SARS-CoV-2 (PDB: 6ZSL) [46]. The SARS-CoV-2 helicase protein (6ZSL) demonstrated a docking score of −4.82 kcal/mol with BEA. The anchoring included hydrophobic residues P406, P408, R409, L412, T413, K414, L417, T552, A553, and H554 and hydrogen bond, Y515, to maintain an optimal binding. The BEA docked effectively against Pocket A (Supplementary Table S1); however, it docked poorly against the remaining pocket of the SARS-CoV-2 helicase (Table 1).
3.1.4. Endoribonuclease
RO-7 and arzanol were used as positive controls in our docking experiment because of their capabilities to neutralize endoribonucleases (PDB: 6VWW) [47]. Two different receptor sites were selected for ligand docking (Pocket A and B Supplementary Table S1). The BEA docked poorly against all selected receptors. The best conformation (−0.07 kcal/mol, Table 1) was recorded against pocket A.
3.1.5. RNA-dependent-RNA-polymerase (RdRp)
The SARS-CoV-2 RdRp (PDB: 7BTF) was used for docking against BEA, remdesivir, and dATP [48]. In the case of pocket A, BEA showed significant affinity (−3.05 kcal/mol) for the RdRp of SARS-CoV-2. However, this affinity was lower than the positive control drugs which are being proposed as candidates against SARS-CoV-2 (Table 1). BEA formed hydrophobic interactions with L172, R249, T252, F321, P322, S255, F396, P461, and hydrogen bonds with R349. In the case of pocket B, BEA showed low affinity for RdRp when compared with the known molecules used as controls.
3.1.6. Methyltransferase
For the docking to methyltransferase (PDB: 6W4H), S-adenosyl-l-methionine, the natural ligand of the protein, already bound to the 6W4H crystal structure, sinefungin, a natural nucleoside related to S-adenosyl-l-methionine, with demonstrated antiviral activity [49], and 2-(3,4-dihydroxyphenyl)-5,6-dihydroxy-7-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6- ethyloxan-2-]yloxychromen-4-one (NPC226294) were used as controls [46]. The binding receptor site (Pocket A) with the highest DLID was shown to be the cleft bonding of the natural ligand S-adenosyl-l-methionine. BEA docked poorly against the active site of this protein, and the binding energy was the highest among all docked ligands. In parallel, BEA docked well with pocket B (with the second highest DLID score) as shown in Supplementary Table S1. However, the binding energy was still the lowest when compared to the controls (Table 1). The methyltransferase (6W4H) demonstrated a docking score of −3.94 kcal/mol. BEA was mainly bound to the protein across seven hydrophobic interactions and two hydrogen bonds involving G6837, Q4289, P4290, T4292, T4354, T4355, A4357, and N4358 in addition to K6836.
3.1.7. Exoribonuclease
The two best positive controls used for the docking against SARS-CoV-2 exoribonuclease (5C8T) were 2-(N-Morpholino)-ethanesulfonic acid, as a pan-inhibitor of exonucleases [50], and 1-O-(2-Methoxy-4-Acetylphenyl)-6-O-(E-Cinnamoyl)-Beta-d-Glucopyranoside (NPC137813), a promising drug against the SARS-CoV-2 exoribonuclease [46]. BEA docked poorly with all identified pockets, but the recorded binding energy was the highest when compared to both controls (Table 1).
3.2. Molecular dynamics
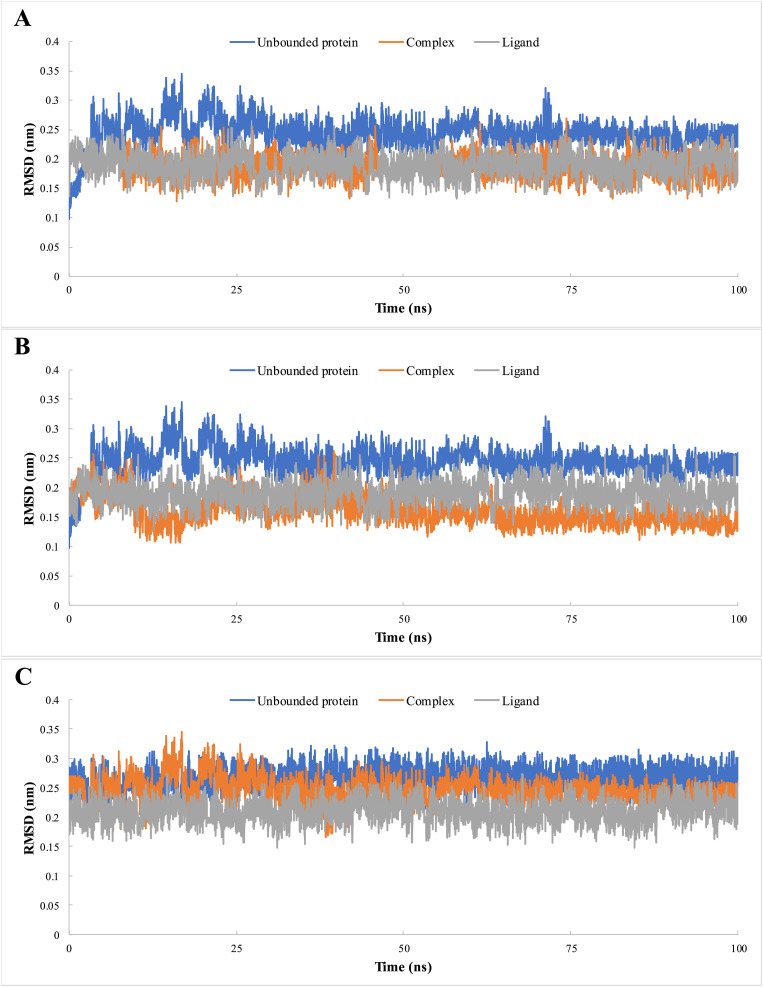
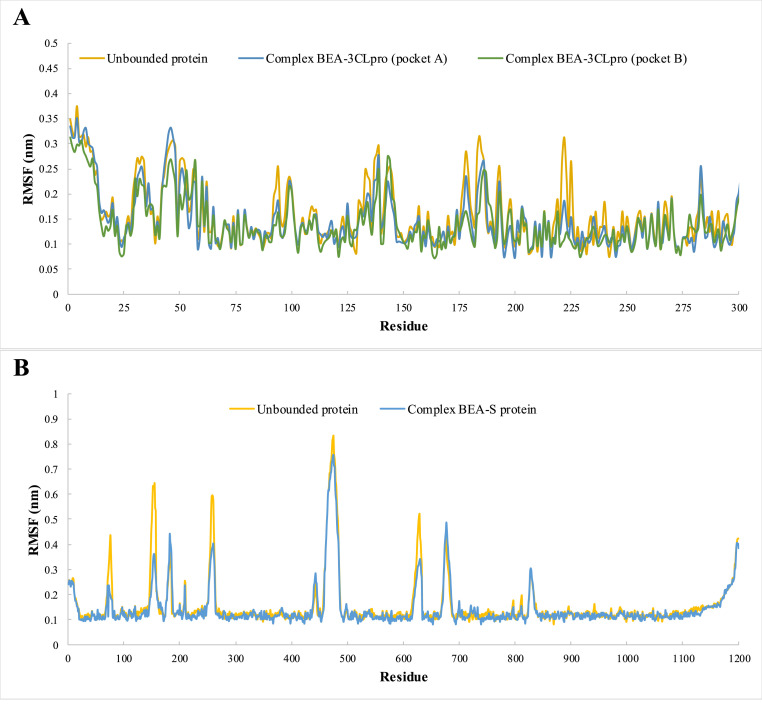
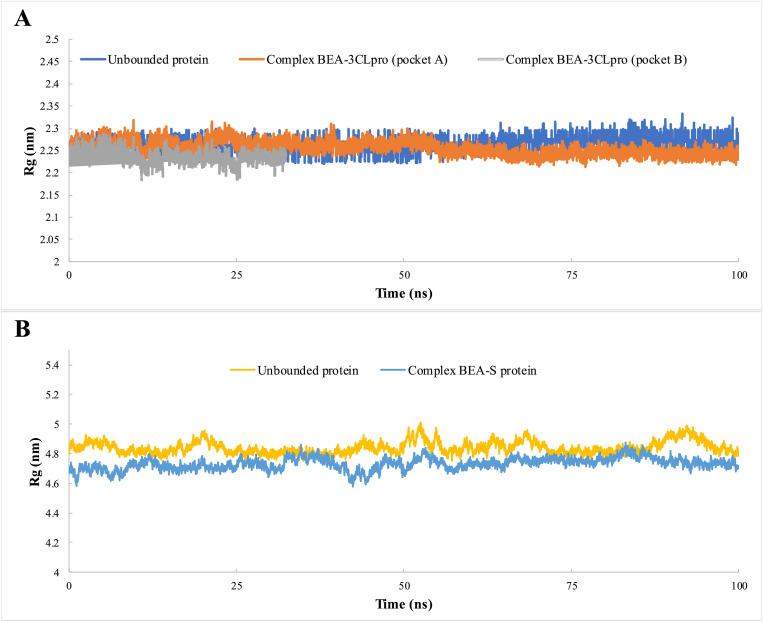
We used molecular dynamics to analyze the stability of 3CLpro (pockets A and B) and S protein (pocket A) with BEA. The RMSD values of the complexes (ligand bounded proteins) and free (unbounded) proteins were 0.18 and 0.16 nm for pockets A and B, respectively, in the case of 3CLpro and 0.24 and 0.26 nm for pockets A and B, respectively, in the spike protein (Fig. 3 ). To ensure the binding stability of BEA to the different viral proteins, ligand positional RMSD was also examined. After forming a complex with 3CLpro (pocket A), 3CLpro (pocket B), and S protein, BEA remained stable throughout the simulation (Fig. 3). RMSF analysis was calculated and plotted against the residue number to calculate the residual mobility for the complexes and free proteins. Elevated fluctuations in BEA-3CLpro (pocket A) were observed in amino acids around the positions 1–14, 34, 46, 94, 133, 139, 143, 178, 186, 193, 222, 225, 283, and 296–306. However, the fluctuation is reduced in the residues of the substrate-binding domain. In the case of BEA-3CLpro (pocket B), 1–14, 34, 46, 94, 133, 139, 143, 178, 186, 193, 222, 225, 283, and 296–306 were also among the most flexible amino acids, while the content of helical and β-sheet were among the most stable. In contrast to the BEA-3CL complex (pocket A), residues interacting with substrates showed high fluctuation. The RMSF plot indicates that minimal amino acid fluctuations were present in most parts of the BEA-S protein complex. Nevertheless, ambiguous fluctuations were recorded around residues 1–18, 76, 154, 183, 259, 442, 473, 629, 478, 829, and 1138–1200. A relative constant value of approximately an Rg of 2.26 nm was recorded for unbound 3CL over time indicating a minimal compression (Fig. 4 ). In comparison with free proteins, Rg values dropped around 2.25 and 2.22 nm after the binding with BEA in pocket A and pocket B, respectively. Similarly, the S protein in complex with BEA (Rg = 4.73 nm) presented more compactness relative to the unbound protein (Rg = 4.84 nm) as shown in Fig. 5 .
Fig. 3.
Root mean square deviation (RMSD) of the 3CLpro (pocket A) (A) 3CLpro (pocket B) and S protein (C) of SARS-CoV-2 in complex with BEA, during 100 ns of the molecular dynamic (MD) simulation period.
Fig. 4.
Root mean square fluctuation (RMSF) of 3CLpro (A) and S protein (B) as unbounded proteins and forming complexes with BEA, during the simulation period.
Fig. 5.
Radius of gyration of 3CLpro (A) and S protein (B) as unbounded proteins and forming complexes with BEA, during 100 ns of the MD simulation period.
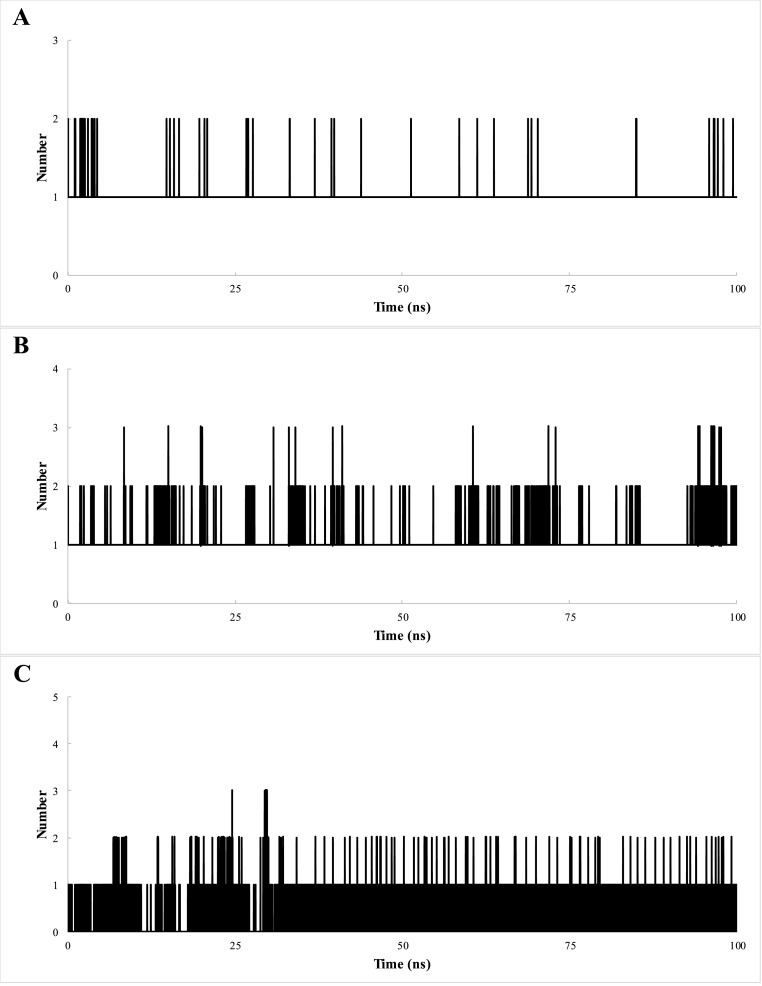
For 3CLpro, a mean of 1–2 and 1–3 hydrogen bonds were found after the binding of BEA in pockets A and B, respectively, during the simulation period. In the case of the S protein, a mean of 0–4 hydrogen bond was found with BEA through the entire 100 ns simulation (Fig. 6 ).
Fig. 6.
Total number of hydrogen bonds interactions between BEA and proteins 3CLpro pocket A (A) 3CLpro pocket B (B) and S protein (C).
MM/PBSA based binding energy calculations were performed for the three selected protein-ligand complexes. The results indicated that BEA-3CLpro (pocket B) possessed the highest negative energy of −355.71 kJ/mol followed by BEA-3CLpro (pocket A) and BEA-S protein with energies of −313.48 and −289.66 kJ/mol, respectively. Van der Waals forces were the primary contributors to the binding energy in all BEA-protein interactions (Table 2 ).
Table 2.
Average binding free energies calculated from the molecular dynamic (MD) simulation in triplicate.
| Complex | Van der Waals (kJ/mol) | Electrostatic (kJ/mol) | Polar energy (kJ/mol) | Non-polar energy (kJ/mol) | Binding energy (kJ/mol) |
|---|---|---|---|---|---|
| BEA-3CLpro (pocket A) | −332.48 ± 12.3 | −12.51 ± 1.1 | 44.22 ± 3.1 | −12.71 ± 0.8 | −313.48 ± 10.3 |
| BEA-3CLpro (pocket B) | −378.52 ± 15.7 | −11.26 ± 1 | 41.87 ± 2.8 | −11.8 ± 0.8 | −355.71 ± 11.1 |
| BEA-S protein | −311.66 ± 10.4 | −9.31 ± 0.8 | 38.85 ± 2.4 | −10.53 ± 0.7 | −289.66 ± 9.9 |
3.3. Pharmacokinetics properties
To better apprehend the potential clinical usage of beauvericin, we used the ADMET-SAR and pkCSM online servers to characterize its molecular (Table 3 ) and pharmacokinetics properties (Table 4 ). With a molecular weight of 783.9 KDa and LogP value of 4.9 (Table 3), BEA has low GI absorption and low bioavailability; however, it can cross the blood-brain barrier (Table 4). Moreover, the compound is a substrate for P-glycoproteins and CYP3A4 and can have interactions with their inhibitors. Finally, carcinogenicity was not observed, and the compound did not exhibit AMES Toxicity but level III (Table 4).
Table 3.
Molecular characteristics of BEA.
| 2D representation | SMILES | Molecular Weight | LogP | Surface area |
|---|---|---|---|---|
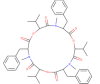 |
CC(C)C1C(=O)N(C(C(=O)OC(C(=O)N(C(C(=O)OC(C(=O)N(C(C(=O)O1)CC2=CC=CC=C2)C)C(C)C)CC3=CC=CC=C3)C)C(C)C)CC4=CC=CC=C4)C | 783.963 | 4.9125 | 335.982 |
Table 4.
Pharmacokinetics properties of BEA.
| Absorption | |
|---|---|
| Water solubility (log mol/L) | −2.984 |
| Caco2 permeability (log Papp in 10-6 cm/s) | 0.919 |
| Intestinal absorption (human) (% Absorbed) | 63.474 |
| Skin Permeability (log Kp) | −2.735 |
| P-glycoprotein substrate | Yes |
| P-glycoprotein I inhibitor | Yes |
| P-glycoprotein II inhibitor | Yes |
|
Distribution | |
| VDss (human) (log L/kg) | −0.402 |
| Fraction unbound (human) (Fu) | 0.121 |
| BBB permeability (log BB) | −0.831 |
| CNS permeability (log PS) | −2.408 |
|
Metabolism | |
| CYP2D6 substrate | No |
| CYP3A4 substrate | Yes |
| CYP1A2 inhibitor | No |
| CYP2C19 inhibitor | No |
| CYP2C9 inhibitor | No |
| CYP2D6 inhibitor | No |
| CYP3A4 inhibitor | Yes |
|
Excretion | |
| Total Clearance (log ml/min/kg) | 0.981 |
| Renal OCT2 substrate | No |
|
Toxicity | |
| AMES toxicity | No |
| Acute Oral Toxicity Class | III |
| Skin Sensitization | No |
| Hepatotoxicity | Yes |
| Carcinogenicity | No |
4. Discussion
Awaiting in vitro and pre-clinical experiments, a parallel approach using in silico docking experiments was undertaken to investigate the potential targets of BEA on SARS-CoV-2. We have opted for ICM-pro, the most accurate predictive tool of binding geometry, to find possible conformations of BEA against seven of the drug–target proteins of the SARS-CoV-2. The icmPocketFinder tool was used to identify buried and open cavities within the 3D structure of the viral proteins (Supplementary Table 1). It is important to note the accuracy and reliability of this tool in predicting “ligandable” orthosteric and allosteric interaction sites.
Of all seven SARS-CoV-2 proteins, BEA has the highest affinity towards the main protease (Table 1). The host cell entry of the virus can be attributed to the high affinity between S protein and the catalytic domain of the ACE2 receptor [2]. In this study, it was demonstrated that BEA does not bind with high affinity to the receptor-binding domain. Therefore, it can be speculated that BEA would not impede the interaction between S protein of the virus and ACE2 receptor. It has been demonstrated that ivermectin possesses antiviral activity against SARS-CoV-2-infected Vero-hSLAM cells [51]. Furthermore, data have emerged about the beneficial administration of ivermectin amid COVID-19 infection in hospitalized patients, a finding that could account for lower mortality rates. Previously, it was noted that ivermectin could dock effectively with the receptor-binding domain and interfere with the attachment of the S protein to the receptor membrane [52]. Our results do not support this observation, and the use of ivermectin is still controversial as its mechanism of action against SARS-CoV-2 remains elusive despite speculations on an indirect inhibitory action over the importin protein IMPα/β1 [53]. Using computational methods, more recent evidence highlights the inhibitory effect of BEA against the RdRp, one of the most important enzymes involved in SARS-CoV-2 in viral replication [54]. The authors conducted a sequence of “blind” and active site targeted docking, and BEA scored the second-best binding energy when compared to 99 other potential inhibitory fungal metabolites. The blind docking method suffers from several pitfalls. On one hand, it has been demonstrated that pocket-targeting docking has a better hit ratio and accuracy [55]. On the other hand, it will be necessary that the grid box search the entire surface of the protein of interest; therefore, the calculation must be exhaustive [55]. This drawback has been moderately overcome by using the icmPocketFinder, which can facilitate the docking experiment and enhance the accuracy by predicting the binding sites of target proteins. However, we showed that BEA could only bind with low and moderate affinities to the most druggable pockets of the enzyme (Table 1). Interestingly, after targeting the active site of RdRp, BEA formed bonds with D760 and D761, which play major roles in the catalytic mechanism of the enzyme [54]. Taken together, these results lend further support to the potential antiviral activity of BEA against the key proteins of the SARS-CoV-2.
Interestingly, the most druggable pocket found within the 3CLpro was identical to the natural inhibitor binding site that was identified experimentally from the crystal structure in the PDB. This result has further strengthened our confidence that icmPocketFinder identifies clefts that are most likely to contain ligands. 3CLpro is considered a promising antiviral drug target for its involvement in processing viral polyproteins. The crystal structure of the SARS-CoV-2 3CLpro shared a highly conserved substrate-binding pocket when compared to SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus (HCoV-HKU1), bat coronavirus (BtCoV-HKU4), mouse hepatitis virus (MHV-A59), porcine epidemic diarrhea virus (PEDV), feline infectious peritonitis virus (FIPV), transmissible gastroenteritis virus (TGEV), HCoV-NL63, HCoV-229E, and infectious bronchitis virus (IBV) [56]. The residues interacting with the substrates in the active site of SARS-CoV-2 include T24, T25, H41, C44, M49, Y54, F140, N142, G143, C145, H163, H164, M165, E166, L167, P168, D187, R188, Q189, and T190. In this study, molecular docking analysis revealed that BEA can block several of these important residues for substrate binding (Fig. 1). Most importantly, the inhibition of the catalytic dyad residues H41 and C145 could impact the catalytic activity in addition to the conformational fold of the enzyme. BEA binds to the enzymatic catalytic cleft of the 3CLpro located between domains I and II. Evidently, BEA can interrupt the chemical pathway of 3CLpro by competitive inhibition and thus strongly interact at the same active site as the substrate. It is noteworthy that BEA established a strong H-bonding with E166, a key residue for the dimerization of 3CLpro [57]. The interaction between the N-finger and E166 of each monomer is behind the structure and function of the dimer in addition to the formation of the S1 pocket of the substrate-binding site [58]. Moreover, we showed that BEA can dock strongly to a different site of the enzyme (pocket B). Allosteric inhibition could take place due to the presence of an allosteric site on the protein structure that is not located at the active site. Of note, the contact involves G15, G120, and P122 that establish crucial residue–residue interactions on the dimer interface and contribute well to the dimerization of 3CLpro [59,60]. In the light of these observations, two different strategies could be adopted for the development of BEA as an inhibitor of SARS-CoV-2 3CLpro: (1) direct neutralization of the active/catalytic site by blocking the substrate-binding site and (2) reduction of the catalytic activity by targeting the dimer interface of the enzyme.
It would be interesting to cross-compare the results of BEA to those of the Pfizer FDA-approved molecule ritonavir (Paxlovid) in this regard since other known ligands (chloroquine, luteolin, remdesivir, N3, boceprevir, and GC376) used as positive control in our docking experiments showed high binding affinity toward 3CLpro in accordance with previously published studies but with a lower score compared to BEA [41,43].
The MD simulation is considered an emerging computational approach in the study of the conformational flexibility and dynamics of ligand-protein complexes. In reference to the unbounded 3CLpro and S protein, it was shown that the systems reached equilibrium after 30 and 10 ns, respectively, thus indicating that the physical and chemical properties of the system no longer change as a function of time (Fig. 3). These results noted that the simulation time and the simulating structures are comparatively sufficient and stable. The complex that recorded a lower mean RMSD value when compared to the free protein was considered stable [61]. This criterion was fulfilled in all formed complexes. Moreover, no fluctuations were observed in the BEA positional RMSD, a finding that indicates a stable binding in the active site. Altogether, it can be assumed that BEA did not disturb the structural stability and remained firmly bound to the binding site of these proteins (Fig. 3). The loops in the crystal structure of 3CLpro were indicated by noticeable RMSF fluctuations (Fig. 4). It is important to note that in the case of 3CLpro, residues interacting with BEA binding site in both pockets A and B were among the most stable residues. In addition, the binding of BEA to pocket B of 3CLpro caused a significant change in the fluctuation values of the residues involved in substrate binding thus inhibiting the formation of the viral polyproteins. The docking of BEA induced remarkable conformational rearrangements in the catalytic dyad and substrate-binding pocket of the SARS-CoV-2 3CLpro. Concomitantly, the generation of functionally active viral replication complex could be disrupted. The drugs that can lead to destabilization of the 3CLpro dimerization and in parallel, enhance the propensity for the active/drug-insensitive conformation are gaining particular interest [62]. From this standpoint, it can be safely assumed that BEA fits in the orthosteric/active site in which it competes with the natural substrate for its attachment on the active site and allosteric site in which it disturbs the dimer interface and in some way, brings about a change in the shape of the enzyme so that the active site of the enzyme becomes unfit for forming a complex with its substrate. In the case of the spike protein, significant fluctuations that occurred in the crystal structure belonged to missing residues. In contrast to 3CLpro, the binding of BEA in the pocket A did not induce any changes in the receptor-binding domain in such a way that a substrate cannot recognize it. Therefore, it can be concluded that pocket A is not an allosteric site that allows BEA to inhibit the enzyme activity. The decrease in Rg indicates the compression of protein targets after binding with BEA. In addition, no abrupt fluctuations in Rg values were recorded, denoting 3CLpro and S protein folding stabilities after binding with BEA (Fig. 5). It has been widely documented that the secondary structure of the protein can be stabilized by H-bonding. H-bonds between a protein and its ligand provide binding affinity and specificity of interaction [63]. A moderate mean number of H-bonds was exhibited between BEA and 3CLpro-pocket A (1–2), 3CLpro-pocket B (1–3), and S protein (0–4) as shown in Fig. 6. These results are in line with those obtained by Ebrahimi et al. [54] in which the authors showed that BEA formed only one stable H-bond with C813 which is located at the motif E of RdRp. It can be hypothesized that the stability of BEA is mainly due to hydrophobic interactions revealed from the docking analysis. Beyond molecular docking, the MM/PBSA is a profitable procedure that yields a more accurate estimation of drug–target affinities and therefore, removes possible false-positive results generated by the standard molecular docking [32,33]. The results obtained in this study showed that BEA has the highest affinity to pocket B of 3CLpro. On one hand, van der Waals, electrostatic, and non-polar solvation contribute negatively to overall binding energy; on the other hand, polar solvation energy positively contributes to the total interaction energy. Expectedly, the interaction provided by van der Waals was greater when compared to that by electrostatic and non-polar energy (Table 2). Therefore, this finding further supports a dominant role for hydrophobic interactions in stabilizing the BEA–viral proteins complexes. It was demonstrated that BEA may neutralize proteins involved in key developmental stages of SARS-CoV-2, and we hypothesize that due to its potential activity against key proteins, different BEA molecules would have a synergistic effect and thus higher efficacy over existing drugs. We have shown that even within one protein, BEA could strongly bind to the orthosteric and allosteric sites. Multi-targets molecules can provide a higher therapeutic index combined with a minimal side effect when compared to drugs with single targets [64]. Polypharmacology is considered a novel and appealing paradigm in drug development, and this area is paving the way for rational design of the next generation of more effective but less toxic therapeutic agents [65]. Despite the encouraging results, one major hurdle for any drug to overcome is linked to its pharmacokinetics in a living organism. As such, absorption, distribution, metabolism, and excretion are decisive characteristics not only for the efficacy but also the safety of the drug, a critical step for clinical trials. The pharmacokinetics properties showed that BEA violates Lipinski's Rule of 5 [66] for molecular weight ≤500 g/mol; and therefore, a lower chance of exhibiting oral bioavailability could be expected. Contrary to the violations of RO5, the compound can be suggested as a Beyond Rule of 5 (bRO5) drug. Numerous drugs on the market that have been approved by the FDA violate Ro5 but are considered bRo5, indicating potential therapeutics [67]. The compounds are mostly natural compounds with high molecular weight >500 g/mol and can have a high binding affinity on complex hotspots of the target, inducing a more inhibitory effect [68]. The high molecular weight does seem to significantly increase the protein–ligand interface area and improve binding affinity, especially if the interacting surface is hydrophobic. In addition, Egbert and his colleagues reported that 97.8% of potent inhibitor drugs that could bind to HIV-protease with high affinity are large having a large molecular weight (>500 g/mol) [68]. Preliminary studies demonstrate that no health effect was observed on animals after acute and chronic exposure to BEA [69]. However, extensive in vivo toxicity data are needed to perform a human risk assessment. Therefore, it is suggested that to attain the medicinal effects of the molecule, a cautious drug administration dosage should be monitored in clinical studies. Previous studies have proven that BEA possesses anti-inflammatory properties and that the BEA could significantly inhibit the nuclear translocation of the NF-κB pathway subunits p65 and p50 in RAW264.7 cells without inducing cellular toxicity [70]. The inhibition of the nuclear factor light (NF-κB) pathway is a hallmark of the studies implicated in the cytokine storm depicted in the severe cases of SARS-CoV-2 [71]. This cellular protection against toxicity when using BEA in vitro was also corroborated in vivo in mice. The combination of BEA (0.5 mg/kg) and ketoconazole (0.5/kg) was shown to prolong the survival of Candida parapsilosis-infected mice even when compared to subjects treated with high doses of ketoconazole alone (50 mg/kg) with no side effects [15]. At this stage of understanding, we believe that BEA may also enhance the therapeutic index of drugs; therefore, it can be speculated that BEA could be used alone or in combination for treating COVID-19 infection.
5. Conclusion
Based on the docking scores combined with the molecular dynamics simulation results on both the 3CLpro and S protein and taking into consideration the safety reports on the usage of BEA so far, we hypothesize that BEA might be effective in pre-clinical and potential clinical trials. In the world of pandemic, the “toxin” could be repurposed into a “drug” when the biological and ethical contexts are there to support saving human lives.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or non-profit organizations.
Conflicts of interest
None.
Ethical approval
Not required.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgement
The authors would like to thank the insightful comments of the anonymous reviewers of this article. The authors would like to thank the American Manuscript Editors and Ms. Cynthia Elia for proofreading this study. The schematic illustration in the graphical abstract was created using Biorender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.105171.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W., Jr. Computational methods in drug discovery. Pharmacol. Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 6.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Habtemariam S., Nabavi S.F., Banach M., Berindan-Neagoe I., Sarkar K., Sil P.C., Nabavi S.M. Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy? Arch. Med. Res. 2020;51:733–735. doi: 10.1016/j.arcmed.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkataraman S., Prasad B., Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses. 2018:10. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2020;93:300–310. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 12.Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan A.T., Gukasyan J., Arabian S., Wang S., Neeki M.M. Emergent inpatient Administration of Casirivimab and imdevimab antibody cocktail for the treatment of COVID-19 pneumonia. Cureus. 2021;13 doi: 10.7759/cureus.15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Xu L. Beauvericin, a bioactive compound produced by fungi: a short review. Molecules. 2012;17:2367–2377. doi: 10.3390/molecules17032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Yan K., Zhang Y., Huang R., Bian J., Zheng C., Sun H., Chen Z., Sun N., An R., Min F., Zhao W., Zhuo Y., You J., Song Y., Yu Z., Liu Z., Yang K., Gao H., Dai H., Zhang X., Wang J., Fu C., Pei G., Liu J., Zhang S., Goodfellow M., Jiang Y., Kuai J., Zhou G., Chen X. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4606–4611. doi: 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L., Wang J., Zhao J., Li P., Shan T., Wang J., Li X., Zhou L. Beauvericin from the endophytic fungus, Fusarium redolens, isolated from Dioscorea zingiberensis and its antibacterial activity. Nat. Prod. Commun. 2010;5:811–814. [PubMed] [Google Scholar]
- 17.Wu Q., Patocka J., Nepovimova E., Kuca K. A review on the synthesis and bioactivity aspects of beauvericin, a Fusarium mycotoxin. Front. Pharmacol. 2018;9:1338. doi: 10.3389/fphar.2018.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meca G., Sospedra I., Soriano J.M., Ritieni A., Moretti A., Manes J. Antibacterial effect of the bioactive compound beauvericin produced by Fusarium proliferatum on solid medium of wheat. Toxicon. 2010;56:349–354. doi: 10.1016/j.toxicon.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H., Ruan C., Bai X., Zhang M., Zhu S., Jiang Y. Isolation and identification of the antimicrobial agent beauvericin from the endophytic Fusarium oxysporum 5-19 with NMR and ESI-MS/MS. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/1084670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilos D., Rodriguez-Carrasco Y., Englinger B., Timelthaler G., van Schoonhoven S., Sulyok M., Boecker S., Sussmuth R.D., Heffeter P., Lemmens-Gruber R., Dornetshuber-Fleiss R., Berger W. The natural fungal metabolite beauvericin exerts anticancer activity in vivo: a pre-clinical pilot study. Toxins. 2017;9 doi: 10.3390/toxins9090258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin C.G., An D.G., Song H.H., Lee C. Beauvericin and enniatins H, I and MK1688 are new potent inhibitors of human immunodeficiency virus type-1 integrase. J. Antibiot. (Tokyo) 2009;62:687–690. doi: 10.1038/ja.2009.102. [DOI] [PubMed] [Google Scholar]
- 22.Olleik H., Nicoletti C., Lafond M., Courvoisier-Dezord E., Xue P., Hijazi A., Baydoun E., Perrier J., Maresca M. Comparative structure-activity analysis of the antimicrobial activity, cytotoxicity, and mechanism of action of the fungal cyclohexadepsipeptides enniatins and beauvericin. Toxins. 2019:11. doi: 10.3390/toxins11090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fesen M.R., Pommier Y., Leteurtre F., Hiroguchi S., Yung J., Kohn K.W. Inhibition of HIV-1 integrase by flavones, caffeic acid phenethyl ester (CAPE) and related compounds. Biochem. Pharmacol. 1994;48:595–608. doi: 10.1016/0006-2952(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 24.Al Khoury C., Nemer N., Nemer G., Kurban M., Bernigaud C., Fischer K., Guillot J. In vitro activity of beauvericin against all developmental stages of Sarcoptes scabiei. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02118-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neves M.A., Totrov M., Abagyan R. Docking and scoring with ICM: the benchmarking results and strategies for improvement. J. Comput. Aided Mol. Des. 2012;26:675–686. doi: 10.1007/s10822-012-9547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheridan R.P., Maiorov V.N., Holloway M.K., Cornell W.D., Gao Y.D. Drug-like density: a method of quantifying the "bindability" of a protein target based on a very large set of pockets and drug-like ligands from the Protein Data Bank. J. Chem. Inf. Model. 2010;50:2029–2040. doi: 10.1021/ci100312t. [DOI] [PubMed] [Google Scholar]
- 27.Salvati M.E., Balog A., Shan W., Wei D.D., Pickering D., Attar R.M., Geng J., Rizzo C.A., Gottardis M.M., Weinmann R., Krystek S.R., Sack J., An Y., Kish K. Structure based approach to the design of bicyclic-1H-isoindole-1,3(2H)-dione based androgen receptor antagonists. Bioorg. Med. Chem. Lett. 2005;15:271–276. doi: 10.1016/j.bmcl.2004.10.085. [DOI] [PubMed] [Google Scholar]
- 28.Hirshman S.P., Whitson J.C. Steepest‐descent moment method for three‐dimensional magnetohydrodynamic equilibria. Phys. Fluid. 1983;26:3553–3568. [Google Scholar]
- 29.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 31.Van Gunsteren W.F., Berendsen H.J.C. A leap-frog algorithm for stochastic dynamics. Mol. Simulat. 1988;1:173–185. [Google Scholar]
- 32.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D.A., Cheatham T.E., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 33.Kumari R., Kumar R., Open C. Source Drug Discovery, A. Lynn, g_mmpbsa -a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 34.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- 36.Pires D.E., Blundell T.L., Ascher D.B., pkCSM Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 38.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mothay D., Ramesh K.V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. Virusdisease. 2020:1–6. doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C., Zang C., Nie Q., Yang B., Zhang B., Duan S. Simultaneous determination of seven flavonoids, two phenolic acids and two cholesterines in Tanreqing injection by UHPLC-MS/MS. J. Pharm. Biomed. Anal. 2019;163:105–112. doi: 10.1016/j.jpba.2018.08.058. [DOI] [PubMed] [Google Scholar]
- 41.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 43.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nukoolkarn V., Lee V.S., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J. Theor. Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 46.Naik B., Gupta N., Ojha R., Singh S., Prajapati V.K., Prusty D. High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. Int. J. Biol. Macromol. 2020;160:1–17. doi: 10.1016/j.ijbiomac.2020.05.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones J.C., Marathe B.M., Lerner C., Kreis L., Gasser R., Pascua P.N., Najera I., Govorkova E.A. A novel endonuclease inhibitor exhibits broad-spectrum anti-influenza virus activity in vitro. Antimicrob. Agents Chemother. 2016;60:5504–5514. doi: 10.1128/AAC.00888-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang K.W., Hsu K.C., Chu L.Y., Yang J.M., Yuan H.S., Hsiao Y.Y. Identification of inhibitors for the DEDDh family of exonucleases and a unique inhibition mechanism by crystal structure analysis of CRN-4 bound with 2-Morpholin-4-ylethanesulfonate (MES) J. Med. Chem. 2016;59:8019–8029. doi: 10.1021/acs.jmedchem.6b00794. [DOI] [PubMed] [Google Scholar]
- 51.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehrer S., Rheinstein P.H. Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2. In Vivo. 2020;34:3023–3026. doi: 10.21873/invivo.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choudhary R., Sharma A.K. Potential use of hydroxychloroquine, ivermectin and azithromycin drugs in fighting COVID-19: trends, scope and relevance. N. Microbes N. Infect. 2020;35 doi: 10.1016/j.nmni.2020.100684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebrahimi K.S., Ansari M., Hosseyni Moghaddam M.S., Ebrahimi Z., Salehi Z., Shahlaei M., Moradi S. In silico investigation on the inhibitory effect of fungal secondary metabolites on RNA dependent RNA polymerase of SARS-CoV-II: a docking and molecular dynamic simulation study. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghersi D., Sanchez R. Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. Proteins. 2009;74:417–424. doi: 10.1002/prot.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banerjee R., Perera L., Tillekeratne L.M.V. Potential SARS-CoV-2 main protease inhibitors. Drug Discov. Today. 2021;26:804–816. doi: 10.1016/j.drudis.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silvestrini L., Belhaj N., Comez L., Gerelli Y., Lauria A., Libera V., Mariani P., Marzullo P., Ortore M.G., Palumbo Piccionello A., Petrillo C., Savini L., Paciaroni A., Spinozzi F. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci. Rep. 2021;11:9283. doi: 10.1038/s41598-021-88630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S., Zhang J., Hu T., Chen K., Jiang H., Shen X. Residues on the dimer interface of SARS coronavirus 3C-like protease: dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 2008;143:525–536. doi: 10.1093/jb/mvm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Abranyi-Balogh P., Brandao-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keseru G.M., Klein H.F., Martin M.P., Noble M.E.M., O'Brien P., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu K., Watanabe E., Kokubo H. Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations. J. Comput. Aided Mol. Des. 2017;31:201–211. doi: 10.1007/s10822-016-0005-2. [DOI] [PubMed] [Google Scholar]
- 62.El-Baba T.J., Lutomski C.A., Kantsadi A.L., Malla T.R., John T., Mikhailov V., Bolla J.R., Schofield C.J., Zitzmann N., Vakonakis I., Robinson C.V. Allosteric inhibition of the SARS-CoV-2 main protease: insights from mass spectrometry based assays. Angew Chem. Int. Ed. Engl. 2020;59:23544–23548. doi: 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menendez C.A., Accordino S.R., Gerbino D.C., Appignanesi G.A. Hydrogen bond dynamic propensity studies for protein binding and drug design. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W., Bai Y., Wang Y., Xiao W. Polypharmacology in drug discovery: a review from systems pharmacology perspective. Curr. Pharmaceut. Des. 2016;22:3171–3181. doi: 10.2174/1381612822666160224142812. [DOI] [PubMed] [Google Scholar]
- 65.Anighoro A., Bajorath J., Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. J. Med. Chem. 2014;57:7874–7887. doi: 10.1021/jm5006463. [DOI] [PubMed] [Google Scholar]
- 66.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Tyagi M., Begnini F., Poongavanam V., Doak B.C., Kihlberg J. Drug syntheses beyond the rule of 5. Chemistry. 2020;26:49–88. doi: 10.1002/chem.201902716. [DOI] [PubMed] [Google Scholar]
- 68.Egbert M., Whitty A., Keseru G.M., Vajda S. Why some targets benefit from beyond rule of five drugs. J. Med. Chem. 2019;62:10005–10025. doi: 10.1021/acs.jmedchem.8b01732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.E.P.o.C.i.t.F.C. (CONTAM) Scientific Opinion on the risks to human and animal health related to the presence of beauvericin and enniatins in food and feed. EFSA J. 2014;12:3802. [Google Scholar]
- 70.Yoo S., Kim M.Y., Cho J.Y. Beauvericin, a cyclic peptide, inhibits inflammatory responses in macrophages by inhibiting the NF-kappaB pathway. KOREAN J. PHYSIOL. PHARMACOL. 2017;21:449–456. doi: 10.4196/kjpp.2017.21.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El-Hachem N., Eid E., Nemer G., Dbaibo G., Abbas O., Rubeiz N., Zeineldine S., Matar G.M., Bikorimana J.P., Shammaa R., Haibe-Kains B., Kurban M., Rafei M. Integrative transcriptome analyses empower the anti-COVID-19 drug arsenal. iScience. 2020;23 doi: 10.1016/j.isci.2020.101697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.