Summary
We have produced expression profiles of all 302 neurons of the C. elegans nervous system that match the single cell resolution of its anatomy and wiring diagram. Our results suggest that individual neuron classes can be solely identified by combinatorial expression of specific gene families. For example, each neuron class expresses distinct codes of ~23 neuropeptide genes and ~36 neuropeptide receptors, delineating a complex and expansive “wireless” signaling network. To demonstrate the utility of this comprehensive gene expression catalog, we used computational approaches to (1) identify cis-regulatory elements for neuron-specific gene expression and (2) reveal adhesion proteins with potential roles in process placement and synaptic specificity. Our expression data are available at cengen.org and can be interrogated at the web application CengenApp. We expect that this neuron-specific directory of gene expression will spur investigations of underlying mechanisms that define anatomy, connectivity and function throughout the C. elegans nervous system.
In Brief
A gene expression map captures all 302 neurons in mature C. elegans deciphering the molecular basis for cell heterogeneity, connectivity and function.
Graphical Abstract
INTRODUCTION
Neurons share many common functions, yet there are a remarkable variety of different neuronal types, each with distinct features and functions. As genetic programs likely specify these differences, a comprehensive molecular model of the brain requires a gene expression map at single-cell resolution. Although profiling methods have catalogued diverse neuron types in a variety of organisms (Adorjan et al., 2019; Poulin et al., 2016; Tasic et al., 2016; Zeisel et al., 2015; Zhu et al., 2018), incomplete knowledge of the anatomy and wiring of complex nervous systems has hampered the effort to link neuron-specific functional and anatomical properties with individual molecular signatures.
To investigate the relationship between gene expression and neuroanatomy, we produced single cell RNA-Seq (scRNA-Seq) profiles for all neuron types in an entire nervous system, that of the C. elegans hermaphrodite. The complete anatomy and wiring diagram of the C. elegans nervous system were defined by serial section electron microscopy (Albertson and Thomson, 1976; Brittin et al., 2021; Cook et al., 2019; White et al., 1986; Witvliet et al., 2020). This approach identified 118 anatomically distinct classes among the 302 neurons in the mature hermaphrodite nervous system. We established the C. elegans Neuronal Gene Expression Map & Network (CeNGEN) consortium (Hammarlund et al., 2018) to generate transcriptional profiles of each neuron class, thereby bridging the gap between C. elegans neuroanatomy and the genetic blueprint that defines it. We used fluorescence activated cell sorting (FACS) to isolate neurons from L4 stage larvae for scRNA-Seq. By the L4 stage, the entire nervous system has been generated and most neurons have terminally differentiated. Our approach generated profiles of 70,296 neurons, including all 118 canonical neuron classes and thus offers a comprehensive catalog of gene expression for an entire nervous system.
We found that every neuron class is defined by distinct combinations of neuropeptide-encoding genes and neuropeptide receptors, suggesting different roles for each type of neuron in sending and receiving signals. We identified an expansive catalog of DNA and RNA sequence motifs that are correlated with cohorts of co-regulated genes. We used computational approaches to identify cell adhesion molecules associated with neuron-specific synapses and bundling. Together, our results provide a comprehensive link between neuron-specific gene expression and the structure and function of an entire nervous system. We expect that these data sets and the tools that we have developed for interrogating them will power future investigations into the genetic basis of neuronal connectivity and function.
RESULTS AND DISCUSSION
Single-cell RNA-Seq identifies all known neuron classes in the mature C. elegans nervous system.
To profile the entire C. elegans nervous system (Figure 1A), we isolated neurons at the L4 larval stage, when all neuron types have been generated (Sulston and Horvitz, 1977) and terminally differentiated to generate a functional nervous system. Initially, we used FACS to isolate neurons from a pan-neural marker strain and found that many neuron classes were either underrepresented or absent (Figure S1A–C). To overcome this limitation, we isolated cells from a series of fluorescent marker strains that labeled distinct subsets of neurons (Figure 1C, Table S1). We generated 100,955 single cell transcriptomes with a median of 928 UMIs and 328 genes/cell. Application of the Uniform Manifold Approximation and Projection (UMAP)dimensional reduction algorithm effectively segregated most of these cells into distinct groups (Figure S2A).
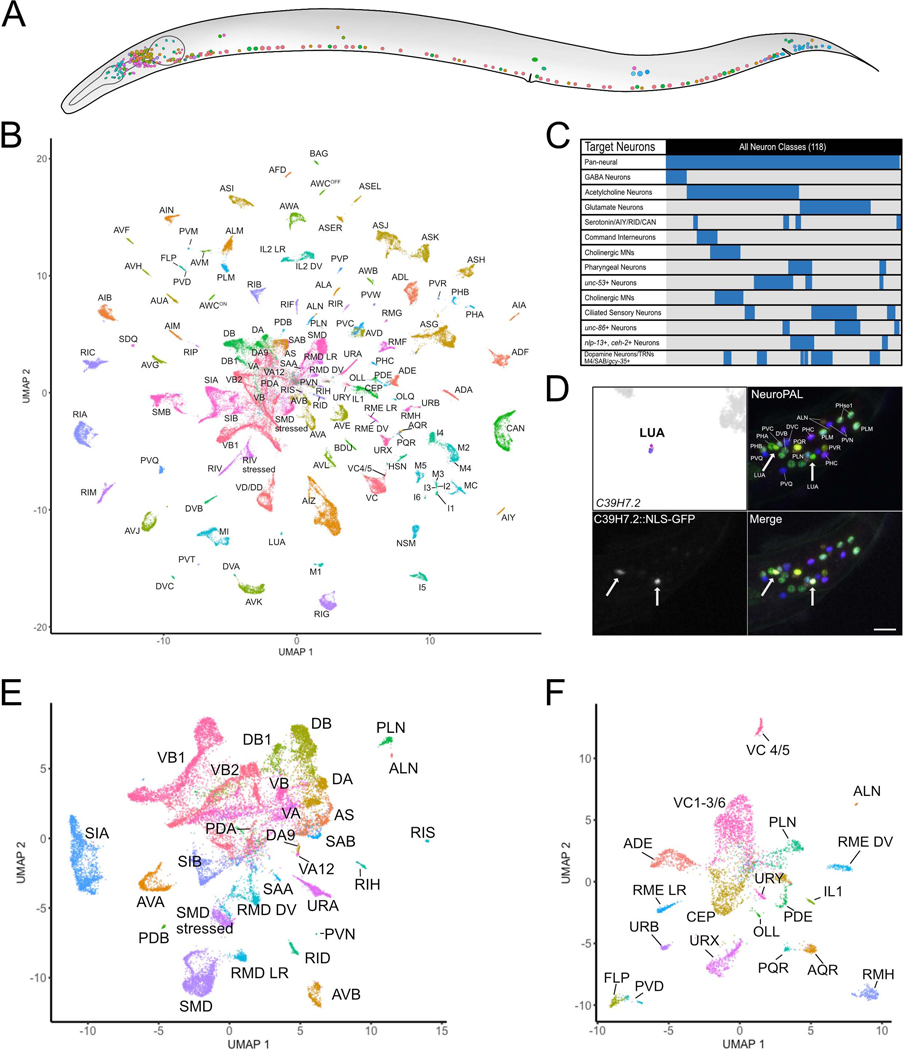
Figure 1. All known neuron types in the C. elegans nervous system are identified as individual clusters of scRNA-seq profiles.
A) All neuron types in the mature C. elegans hermaphrodite. B) UMAP projection of 70,296 neurons with all neuron types and sub-types of ten anatomically defined classes. Neuron identities were assigned based on the expression of known marker genes (Table S1, Figure S3). C) Graphical representation of neurons targeted in individual experiments. D) (top left) The LUA cluster exclusively expressed C39H7.2. Confocal image showing expression of transcriptional reporter C39H7.2::NLS-GFP in LUA neurons (LUAL and LUAR) (arrows) in tail region of NeuroPAL strain. Scale bar = 10 μm. E) Sub-UMAP of central group of cells in B. Clusters are annotated by cell types. F) Sub-UMAP of several commingled neurons in B that clearly separates closely related neuron types (e.g., FLP vs PVD) into individual clusters. See also Figure S1, S2, S3.
We separated non-neuronal cells (27,427 cells, 27.2%, Figure S2B–D) and neurons (70,296 cells, 69.6%, Figure 1A–B) into different sub-UMAPs for further annotation. Neurons had a median of 1033 UMIs and 363 genes/cell. Most neuronal UMAP clusters could be assigned to individual neuron classes based on known marker genes (Hobert et al., 2016; Reilly et al., 2020) (Figure S3A–C). For clusters that could not be so readily identified, we generated GFP transcriptional reporters for genes enriched in the target clusters for direct examination in vivo (Figures 1D, S3D–E). For example, C39H7.2 was exclusively detected in a small cluster that expressed no known distinct markers. We used the multi-colored NeuroPAL marker strain (Yemini et al., 2021) to determine that a C39H7.2::NLS-GFP transcriptional reporter was exclusively expressed in the tail interneuron LUA (Figure 1D).
Ninety of the 118 neuronal types were detected in distinct clusters in the pan-neuronal UMAP (Figure 1B). The remaining clusters contained multiple, closely related neuron classes (e.g., oxygen-sensing neurons, ventral cord motor neurons). Individual UMAP projections of these clusters facilitated the annotation of 38 additional neuron types (Figures 1E–F, S3G), including subtypes within 10 classes (see below). Only two neuron classes were inseparable, the DD and VD ventral cord GABAergic motor neurons, despite known differences in gene expression (Melkman and Sengupta, 2005; Petersen et al., 2011; Shan et al., 2005). Overall, we annotated 95.9% of the cells in the entire dataset and identified distinct clusters encompassing all of the 118 anatomically-defined neuron classes in the mature hermaphrodite nervous system (White et al., 1986).
Single-cell RNA-seq reveals transcriptionally distinct neuronal sub-types.
Reporter-based gene expression and connectivity data suggest that some of the 118 anatomically-defined neuron classes may be comprised of separate subclasses (Hobert et al., 2016; White et al., 1986). Our results confirmed this prediction by revealing 128 transcriptionally distinct neuron types, including subtypes within 10 of the 118 canonical neuron classes. Consistent with earlier findings (Cao et al., 2017; Johnston et al., 2005; Lesch et al., 2009; Packer et al., 2019; Pierce-Shimomura et al., 2001; Troemel et al., 1999; Vidal et al., 2018; Yu et al., 1997), we detected individual clusters for the bilaterally asymmetric sensory neuron pairs ASE (ASER and ASEL) and AWC (AWCON and AWCOFF) (Figures 2A, S4A). Differential gene expression analysis revealed expanded lists of subtype-specific transcripts for the ASE and AWC subclasses (Figures 2B, S4B), including asymmetric expression of receptor-type guanylyl cyclases (rGCs) (Ortiz et al., 2006) and neuropeptides (Figures 2A–B, S4A). Other than the AWC and ASE neuron pairs, we detected no other cases of molecularly separable left/right homologous cells within a neuron class.
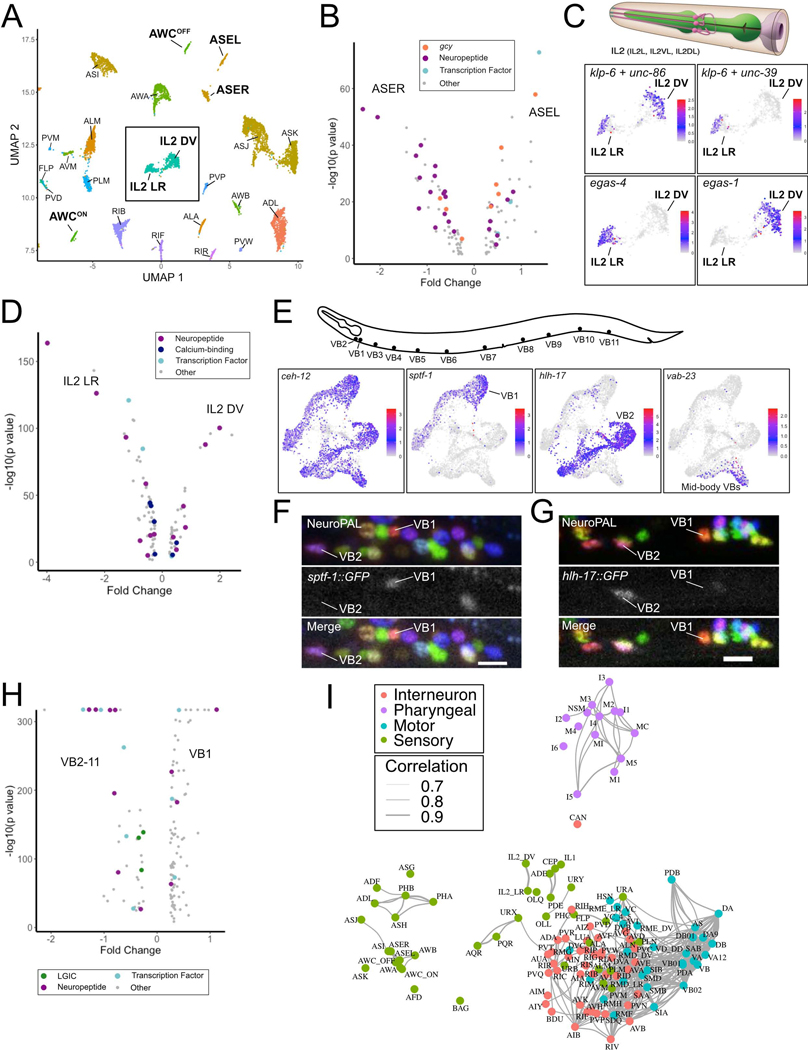
Figure 2. Identification of neuron sub-types.
A) UMAP of neurons with molecularly distinct subtypes (bold labels) from neuronal UMAP (Figure 1B). Inset denotes IL2 DV and IL2 LR clusters. B) Volcano plot of differentially expressed genes (FDR < 0.05) for ASER vs ASEL. Guanylyl cyclases (gcy), neuropeptides, and transcription factors are marked. C) (Top) 3 pairs of IL2 sensory neurons (IL2L/R, IL2VL/R, IL2DL/R) from WormAtlas. (Bottom) UMAP inset from A showing normalized expression of marker genes for all IL2 neurons (klp-6, unc-86), IL2 LR (unc-39, egas-4) and IL2 DV (egas-1). D) Volcano plot of differentially expressed genes (FDR < 0.05) between IL2 sub-types. E) (Top) VB motor neuron soma in the ventral nerve cord. (Bottom) sub-UMAPs of VB neurons highlighting VB marker (ceh-12) and genes (sptf-1, hlh-17, vab-23) expressed in specific VB sub-clusters. F) Confocal images in NeuroPAL show sptf-1::GFP expression in VB1 but not VB2 and G) selective expression of hlh-17::GFP in VB2 but not VB1. Scale bars = 10 μm. H) Volcano plot of differentially expressed genes (LGICs –ligand-gated ion channels) (FDR < 0.05) for VB1 vs all other VB neurons. I) C. elegans neuron types in a force-directed network by transcriptomic similarities. Colors denote distinct neuron modalities and widths of edges (Pearson correlation coefficients > 0.7) show strengths of transcriptome similarity between each pair of neuron types. See also Figure S4.
The remaining eight neuron classes with transcriptionally distinct subtypes are either arranged in radially symmetric groups of 4 or 6 neurons or are distributed along the anterior/posterior axis in the motor circuit. We detected distinct subclusters for two neuron classes with six-fold symmetry at the nerve ring, the inner labial IL2 neurons (Figure 2A, C) and the RMD neurons (Figures 1E, S4A). In both cases, the left/right pair of neurons (e.g., IL2L/R) segregates from the dorsal/ventral pairs (IL2DL/R and IL2VL/R). Differentially expressed genes between the IL2 clusters encode neuropeptides, ion channels, calcium binding proteins and transcription factors and point to potentially distinct functions for the subtypes (Figure 2C–D). For the GABAergic RME head motor neurons, we detected distinct dorsal/ventral (RMED/V) and left/right clusters (RMEL/R) (Figures 1F, S4A). We also identified multiple clusters for the DA, DB, VA, VB, and VC ventral nerve cord motor neuron classes. In each case, one subtype corresponded to one or two individual members of these classes. For example, VC4 and VC5, which flank the vulva, clustered independently from the other four VC neurons (Figures 1F, S4A). For A-class motor neurons (DA, VA), we detected distinct clusters corresponding to the most posterior neurons located in the pre-anal ganglion, DA9 and VA12 (Figures 1E, S4A).
Both B-class motor neuron classes (DB and VB) contained multiple independent clusters (Figures 2E, S4A). In this case, the most anterior B-class motor neurons (DB1, VB1, VB2) segregated into separate clusters. The homeodomain transcription factor CEH-12 is selectively expressed in VBs (Von Stetina et al., 2007) and marks the VB clusters (Figure 2E). We identified VB1 based on expression of a GFP reporter gene for the subcluster-specific marker sptf-1 (Figure 2E–F). The VB2 subcluster was similarly identified by the selective expression of hlh-17::GFP in VB2 among VBs in vivo (Figure 2E–G). Interestingly, all of the molecularly distinct subclasses we detected also have known differences in synaptic connectivity (Hobert et al., 2016; White et al., 1986).
We did not detect subtypes for additional classes with 3, 4, or 6-fold symmetry. This may be due to the low number of cells (< 100 for OLQ, SAA, URY, IL1, see Table S1) assigned to some of these classes. Alternatively, molecular differences among subsets of these neuron types (Hobert et al., 2016) may be limited to a small number of genes that would be insufficient to drive separation in our analyses.
Using 7,390 highly variable genes (see Methods), we generated a network describing the relative molecular relationship of the 128 identified neuron classes and subclasses (Figure 2I). This approach separated sensory and motor neurons as well as a distinct cluster of pharyngeal neurons. Interestingly, pre-motor interneurons cluster with motor neurons. Amphid/phasmid sensory neurons clearly separated from non-amphid/phasmid sensory neuron types. Within amphid/phasmid neurons, some neurons cluster according to sensory modalities. Notably, the chemorepulsive neurons ADL, ASH and PHA/PHB form their own subcluster. The CO2 sensitive BAG neuron and the CAN neuron show the least similarity to other neuron types. Thus, a systematic comparison of neuron-specific profiles confirms that neurons with shared anatomical and functional characteristics are defined by similar patterns of gene expression.
Defining gene expression across neuron types.
A key consideration for scRNA-Seq data is accurately determining whether a detected signal (UMI) for a given gene is actual expression in a cell type (rather than noise). We addressed this question quantitatively by thresholding aggregated data for each cell type using a ground-truth dataset of high-confidence gene expression results across the entire nervous system (mostly fosmid-based reporters and/or reporter-tagged endogenous genes; see Methods, Figure S5). We selected 4 threshold levels (designated as 1–4) offering different compromises between the risk of false positives and false negatives. We used threshold 2 for subsequent analyses. With this threshold, we estimate a true positive detection rate of 0.81 and a false discovery rate of 0.14 (see Methods). The number of genes detected per neuron type (median 5842, range = 1371 [ALN] to 7542 [ASJ]) was positively correlated with the number of cells sequenced per neuron type (median 352, range = 12 [M4] to 3189 [AIZ]; Figure S5I, Spearman rank correlation = 0.783, p < 2.2e-16) and with the true positive rate (Figure S5J, Spearman rank correlation = 0.6776, p < 2.2e-16). Neurons with fewer cells and fewer detected genes were concentrated in the anterior and pre-anal ganglia (Figure S5H), possibly reflecting bias in the dissociation procedure. Nine neuron classes with the fewest detected genes and lowest true positive rates compared to ground truth are labeled in Figure S5J. These cell types are likely to have the highest rates of false negatives, as we estimate the true mean number of genes expressed per neuron type to be ~6550 (see Methods).
We examined the distribution of genes encoding ribosomal proteins to test whether our thresholding approach would preserve a predicted ubiquitous pattern of gene expression. Our results show that 65 of the 78 ribosomal genes (83%) are detected in ≥ 98% of neuron types, with 53 (68%) expressed in all but one cell type (ALN, Figure 3A). Overall, these results indicate that our thresholding approach accurately identifies expressed genes for most cell types in the C. elegans nervous system.
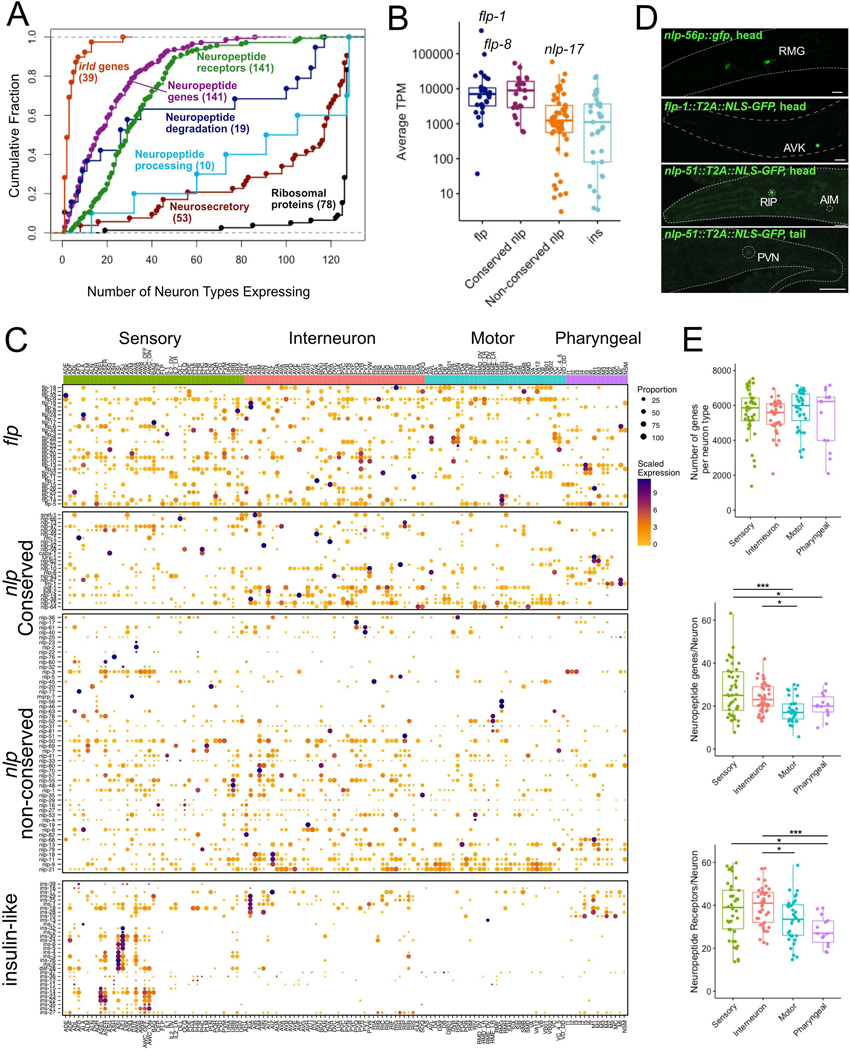
Figure 3. Expression of neuropeptide signaling genes.
A) Cumulative distribution plot of neuron types expressing different classes of neuropeptide signaling genes. Each dot is a gene, genes expressed in the same number of neuron types overlap. Numbers in parentheses denote the sum of genes in each category. B) Average expression (TPM) for neuropeptide subfamilies across neuron types. flp-1, flp-8, nlp-17 are highly expressed. Boxplot spans 25th percentile, median and 75th percentile. C) Heatmap (rows) for flp (FMRFamide-related peptide), nlp (neuropeptide-like protein) and ins (insulin-like peptide) subfamilies across 128 neuron types (columns) grouped by functional/anatomical modalities (Sensory, Interneuron, Motor, Pharyngeal). Conserved nlp genes are shown separately. Rows are clustered within each family. Circle diameter denotes the proportion of neurons in each cluster that expresses a given gene. D) GFP reporters confirm selective expression of nlp-56 (promoter fusion) in RMG, flp-1 (CRISPR reporter) in AVK, and nlp-51 (CRISPR reporter) in RIP, with weaker expression in PVN and AIM. Scale bars = 10 μm. E) Number of all genes (top), neuropeptides (middle) and neuropeptide receptors (bottom) per neuron, grouped by neuron modality. Boxes are interquartile ranges. ANOVA, with Tukey post-hoc comparisons for neuropeptide receptors, Kruskal-Wallis test for other comparisons. *p < 0.05, ***p < 0.001. See also Figure S6, Data S1.
Neuron-specific codes of neuropeptide signaling genes.
We used the thresholded dataset (threshold 2) to probe expression of selected gene families known to be involved in various aspects of neuron function and development (Data S1) and provide highlights of this analysis here in the main text. Neuropeptide-encoding genes (31 FMRFamide-like peptides [flp], 33 insulin-related peptides [ins] and 77 neuropeptide-like proteins [nlp]genes, total of 141 genes) were detected in every neuron class (a minimum of 6, maximum of 62 per neuron) (Figure 3). Consistently, neuropeptide processing genes were broadly expressed throughout the nervous system (Figure 3A). Strikingly, each neuron class expressed a distinct combination of neuropeptides, averaging 23 genes. Sensory neurons and interneurons expressed more neuropeptide genes than motor neurons (Figure 3E). Further, neuropeptide encoding genes are among the most highly expressed transcripts in our data set, similar to reports from Hydra, Drosophila and mouse neurons (Siebert et al., 2019; Allen et al., 2020; Smith et al., 2019). Moreover, the subset of 25 nlp genes with homologs in other species (Husson et al., 2009; Koziol et al., 2016; Mirabeau and Joly, 2013), along with the flp family genes, were detected at higher levels than ins and non-conserved nlp genes (Figure 3B).
Whereas several neuropeptide-encoding genes (flp-9, flp-5, nlp-21) were widely expressed, we also detected neuropeptides with expression restricted to just one or two neuron types, including exclusive expression of flp-1 in AVK, flp-23 in HSN, nlp-56 in RMG, nlp-2 and nlp-23 in AWA and ins-13 in RMED/V (Figure 3C). We validated the restricted expression of nlp-56 in the RMG cluster and flp-1 in AVK with CRISPR/Cas9-engineered reporter alleles (Figure 3D) (see also Figure S6).
Of the more than 140 neuropeptide receptors, most show highly restricted expression, with a few notable exceptions (Figure 3A). The predicted neuropeptide receptors pdfr-1, npr-23 and F59D12.1 were expressed in over 100 neuron types. daf-2, the only insulin/IGF receptor-like tyrosine kinase in C. elegans, was detected in 103 of 128 neuron types. Most other neuropeptide receptor genes were expressed in a restricted subset of neurons; half were expressed in 29 or fewer cell types (Figure 3A). Each individual neuron type expressed a distinct set of neuropeptide receptors, averaging 36 genes. Sensory neurons and interneurons expressed more neuropeptide receptor genes than pharyngeal neurons (Figure 3E). With on-going efforts to match neuropeptide GPCRs to their cognate ligands (https://worm.peptide-gpcr.org/project/), these expression data for all neuropeptide genes and receptors provide a basis for establishing a nervous-system wide map of modulatory neuropeptide signaling.
Signaling complexity across the nervous system is also determined by diverse ionotropic neurotransmitter receptor expression. Each neuron expresses on average 20 ionotropic neurotransmitter receptors, and each individual neuron type expresses a distinct combination of these genes (Data S1). The expression pattern of ionotropic neurotransmitter receptors also suggests extensive non-synaptic volume transmission (Gendrel et al., 2016), further illustrating the complexity of information flow in the C. elegans nervous system. The tunability of individual C. elegans neurons is illustrated by the wide-spread and complex expression of potassium channels (Data S1). For example, each individual neuron expresses 1 to 18 distinct two-pore TWK-type ion channels.
Differential expression of gene regulatory factors.
We interrogated gene families involved in gene regulation, including all predicted transcription factors (TFs) [wTF 3.0, (Fuxman Bass et al., 2016)] and RNA-binding proteins (Tamburino et al., 2013) (Figure 4A–C, Data S1). 705 of 941 (75%) of predicted transcription factors and 497 of 587 (86%) of predicted RNA-binding proteins were detected in at least one neuron type. Overall, transcription factors were more restricted in their expression than RNA-binding proteins (Figure 4C).
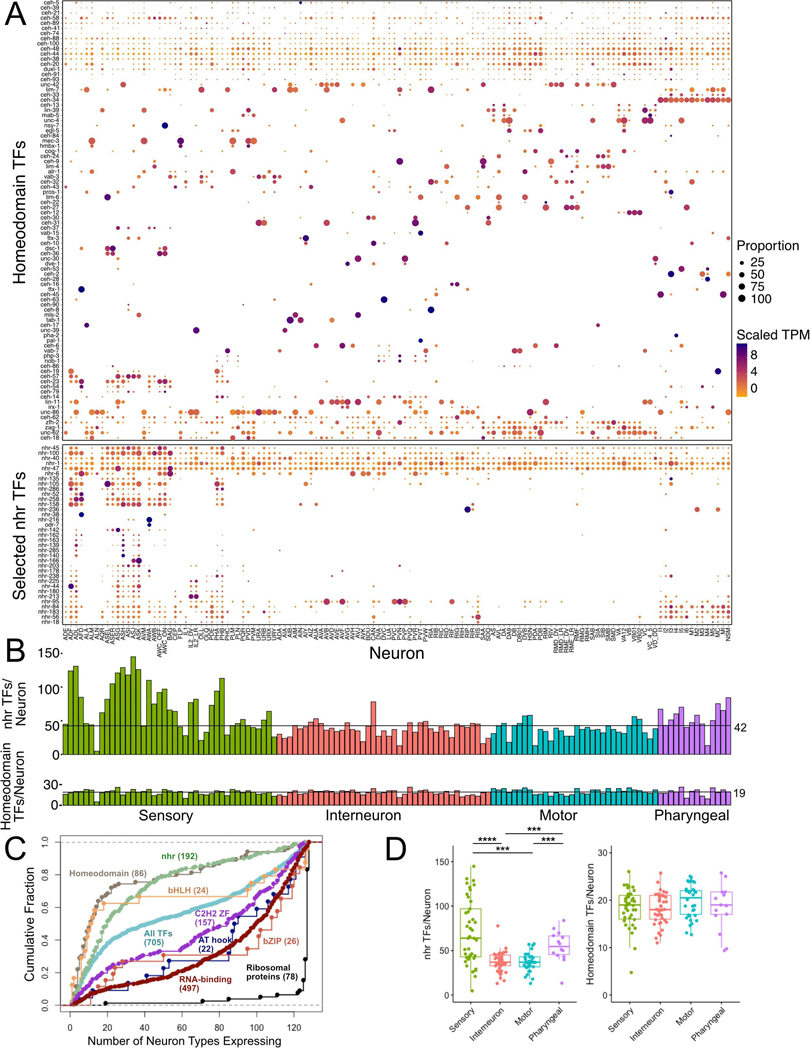
Figure 4. Expression of transcription factor families.
A) Heatmap of homeodomain and representative subset of nuclear hormone receptor (nhr) transcription factors (TFs) across 128 neuron types (columns) grouped by neuron modality. TFs are clustered for each subfamily. Circle diameter represents the proportion of neurons in each cluster that expresses a given gene. B) Bar graphs of number of nhr and Homeodomain TFs in each neuron type, grouped by neuron modality. C) Cumulative distribution of number of neuron types expressing Homeodomain, bHLH, nhr, C2H2 ZF (Zinc Finger), AT hook, bZIP transcription factor (TF) families, RNA binding proteins and ribosomal proteins (see also Figure 3A). D) Quantitative comparison of TFs per neuron for nhr (left) and Homeodomain TFs (right) shows enrichment in sensory neurons for nhrs, but no differences for Homeodomains. Boxplots are median and interquartile range (25th – 75th percentile), Kruskal-Wallis. ***p < 0.001, ****p < 0.0001. See also Data S1.
We analyzed expression of all TF classes that contain more than 15 members (homeodomain, nuclear hormone receptor [nhr], helix-loop-helix [bHLH], C2H2 zinc finger, bZIP, AT hook and T-box genes) and found distinct themes for individual gene families. At one extreme are T-box genes, only two of which are expressed in postembryonic neurons (Data S1). In contrast, AT hook and bZIP genes are expressed broadly throughout the nervous system. Individual bHLH and C2H2 TF genes show a combination of broad and selective expression in the nervous system (Figure 4C). Each neuron expressed multiple different nhr TFs, but sensory and pharyngeal neurons expressed many more nhr TFs than either motor neurons or interneurons (Figure 4A–D). Each amphid and phasmid sensory neuron expressed more than 90 nhr TFs. Notably, ASJ expressed 144 nhr TFs, 75% of the 191 nhr TFs detected in the entire neuronal dataset (Figure 4A, B). Abundant expression of a broad array of nhr genes in sensory neurons is suggestive of specific roles in mediating transcriptional responses to sensory stimuli.
Homeobox gene expression profiles are distinct from that of other TF families. In agreement with a recent report (Reilly et al., 2020), the majority of homeodomain TFs are sparsely expressed in the nervous system. Most individual homeodomain TFs are selectively expressed in subsets of neuron classes (Figure 4A, B). In addition, each neuron class expressed a unique combination of homeodomain transcription factors.
Single neuron-expressed genes
Between 160 (threshold 1, covering 44/128 neuron types) to 1348 (threshold 4, covering 112/128 neuron types) genes are exclusively detected in a single neuron type (Table S3). The single-neuron specificities of many of these genes are validated by published, fosmid-based reporter gene analysis. For example, fosmid-based reporters for the ceh-63 (DVA), ceh-28 (M4) and ceh-8 (RIA) homeobox genes match the neuron specificity of our scRNA-Seq results (Reilly et al., 2020). The cis-regulatory control regions of these genes are candidate drivers for genetic access to individual cells in the nervous system (Lorenzo et al., 2020). Neurons not covered by single neuron-specific drivers can be genetically accessed by the intersection of drivers that are more broadly expressed.
Bulk RNA-sequencing confirms scRNA-Seq results and detects additional classes of non-coding RNAs.
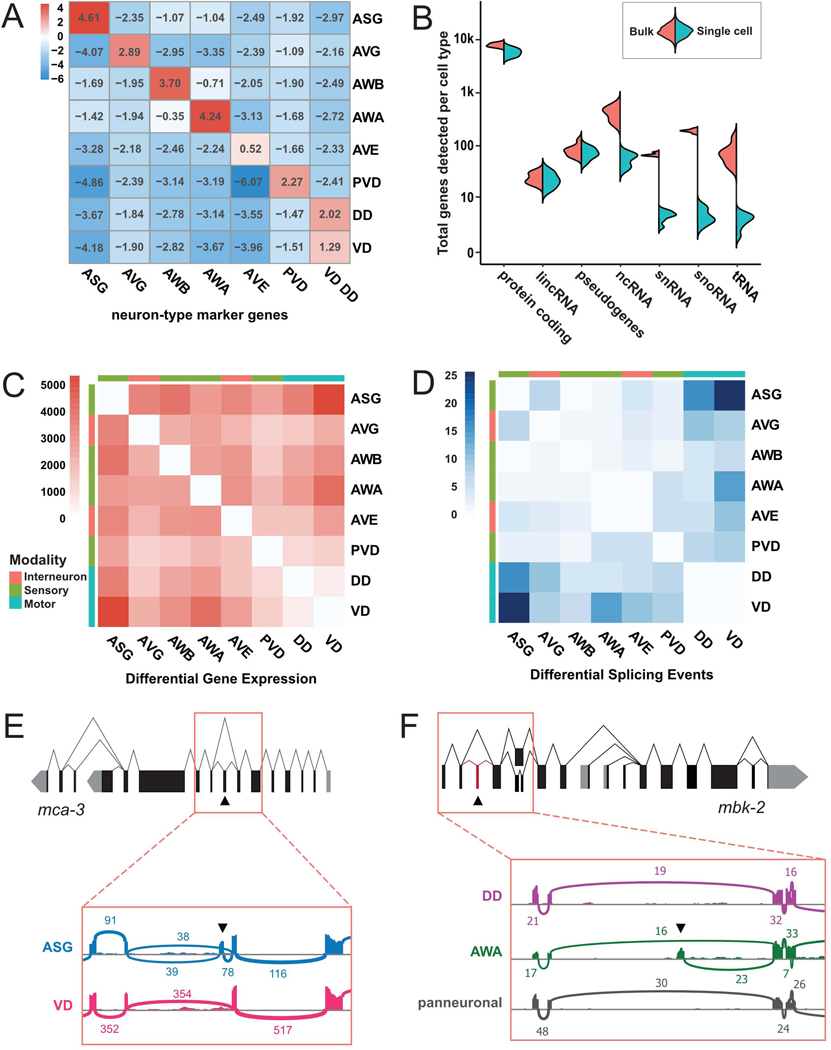
To validate our scRNA-Seq dataset with an orthogonal approach, we used FACS to generate bulk RNA-Seq profiles for eight neuron types: ASG, AVE, AVG, AWA, AWB, PVD, VD, and DD (Spencer et al., 2014) (Methods). Genes enriched in the single-cell clusters of these neurons (i.e., “marker genes”) were also most enriched in the corresponding bulk profiles (Figure 5A). For example, ASG marker genes from scRNA-Seq (left column) are enriched ~24-fold (24.61) in the ASG bulk RNA-Seq profile (top left cell) compared to a pan-neuronal bulk reference. By contrast, markers for other cells are depleted in ASG bulk data (remainder of top row). Thus, independently-derived single cell and bulk RNA-Seq data sets yielded consistent gene expression profiles. Consistent with their commingling in the scRNA-Seq data, VD and DD GABAergic motor neurons had the fewest differentially expressed genes among all neuron pairs (Figure 5C). These results suggest that DD and VD GABAergic neurons are more closely related than are other pairs of different neuron types and that methods for distinguishing neuron types in single cell data are relatively insensitive to small differences in gene expression.
Figure 5. Comparison of bulk and single-cell RNA-Seq.
A) Heatmap for enrichment of scRNA-Seq neuron-type marker genes (Methods) (columns) in bulk RNA-Seq data for each neuron type (ASG, AVG, AWB, AWA, AVE, PVD, DD, VD) vs expression in all neurons. P-values < 0.001 for all comparisons except for AVE markers (all comparisons p-value > 0.05). B) Split violin plot quantifying detection of different RNA classes in bulk and scRNA-seq data sets for neuron types in A. C-D) Heatmaps showing the number of differentially expressed genes (C) and differential splicing events (D) in pairwise comparisons of bulk RNA-seq datasets. E) Gene model and alternative splicing for mca-3. Inset, Sashimi plot shows alternative splicing of specific exon (arrowhead) in ASG vs VD. F) Gene model and alternative splicing of mbk-2. Inset, Sashimi plot shows detection of previously undescribed, alternatively spliced exon (arrowhead) in AWA but not in DD or pan neuronal bulk RNA-Seq. For Sashimi plots in E and F, vertical bars represent exonic reads and arcs indicate the number of junction-spanning reads. See also Table S4.
Protein coding genes, lincRNAs and pseudogenes show similar coverage in both bulk and scRNA-Seq data sets. However, as expected, non poly-adenylated ncRNAs, snRNAs, and snoRNAs are rarely detected in our scRNA-seq data (possibly due to spurious priming) but are abundant in bulk RNA-Seq samples derived from rRNA-depleted total RNA (Figure 5B). The smallest species of ncRNAs, miRNAs and piRNAs, are excluded from our bulk profiles due to a size exclusion step in library preparation, and their characterization awaits further studies.
Widespread differential splicing between neuron types
Differential splicing plays a critical role in the development and function of the nervous system (Raj and Blencowe, 2015; Vuong et al., 2016) and has been reported for individual neuron types in C. elegans (Moresco and Koelle, 2004; Norris et al., 2014; Thompson et al., 2019; Tomioka et al., 2016). Because the 3’ bias of the 10x Genomics scRNA-Seq method limits its use for detecting alternatively spliced transcripts (Arzalluz-Luqueángeles and Conesa, 2018; Dehghannasiri et al., 2020; Patrick et al., 2019), we leveraged the bulk RNA-Seq profiles to identify differentially spliced transcripts among C. elegans neurons.
We discovered 111 high confidence occurrences of differential use of splicing sites between 8 neuron classes (Figure 5D–F, Table S4). Most neuron pairs displayed some differential use of splicing sites (Figure 5D), with wide variations between pairs. For example, we detected 16 differential splicing events between ASG and VD, and only 2 differences between ASG and AWA.
In addition, we detected 63 previously unannotated exons (Table S4, see Methods). For example, the mbk-2 transcript in AWA includes an additional 77 nt sequence corresponding to an alternative 5’ exon that is not expressed in the other seven neuron types in our data set (Figure 5F). This mbk-2 exon is predicted by GenemarkHMM (Pavy et al., 1999) but its expression was not detected by whole-worm RNA-Seq (Tourasse et al., 2017). Thus, our data underscore the capacity of bulk RNA-Seq of single neuron types to detect differential splicing events that could not be reliably detected either by whole animal bulk RNA-Seq or by 10x Genomics scRNA-Seq.
Analysis of cis-regulatory elements reveals a rich array of 5’ and 3’ motifs
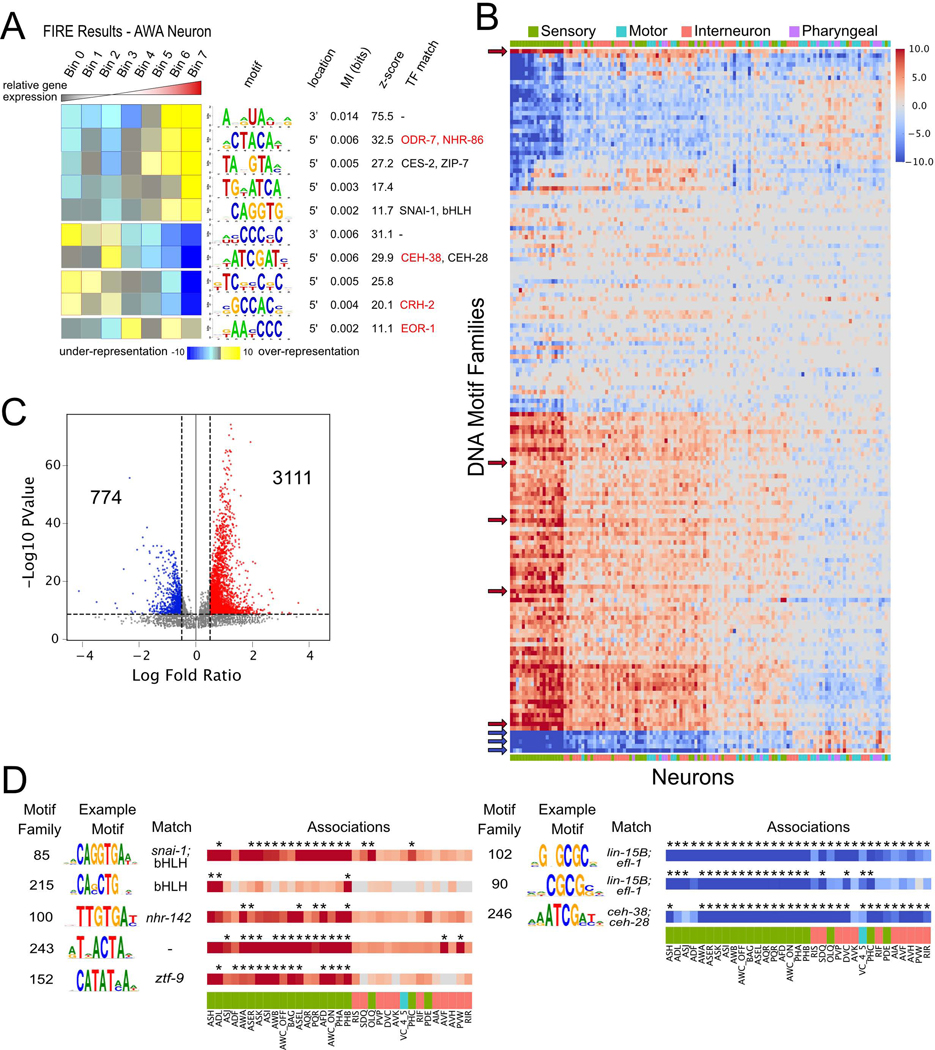
To identify candidate cis-regulatory elements that underlie the distinct patterns of gene expression among neuron types, we used the FIRE motif discovery algorithm. FIRE detects DNA motifs within promoter sequences and linear RNA motifs in 3’ untranslated regions (UTRs) among cohorts of similarly regulated genes (Elemento et al., 2007). FIRE detects motifs that are significantly informative of relative gene expression in each neuron type (Figure 6A). Motifs of positive regulators, for example, should be significantly over-represented (yellow squares, red borders) in genes with high relative expression in the neuron (right columns). A subset of 5’ DNA motifs matched known transcription factor DNA binding preferences (Khan et al., 2018; Weirauch et al., 2014). For example, a motif corresponding to the DNA binding sequence (CTACA) of several nhr transcription factors, including ODR-7, is over-represented in genes that are highly enriched in the AWA neuron (Figure 6A). Notably, ODR-7 is exclusively expressed in AWA where it regulates neuron identity (Colosimo et al., 2003; Sengupta et al., 1994, 1996).
Figure 6. Cis-regulatory elements in neuronal transcriptomes.
A) FIRE results for AWA neuron, featuring the motif logo, location (5’ or 3’), mutual information, z-scores from randomization-based statistical test and matching transcription factors. Genes were grouped into seven bins based on relative expression from lowest (left) to highest (right). Heatmap denotes over-representation (yellow) or under-representation (blue) of each motif (rows) in genes within each bin. Significant over-representation is indicated by red outlines, whereas significant under-representation is indicated by blue outlines. Transcription factors in red are expressed in AWA. B) Heatmap for enrichment of clustered motifs (rows) in each neuron class (columns). Red denotes enrichment in genes with highest relative expression, whereas blue indicates enrichment in genes with lowest relative expression (see Methods). Color intensity represents log10(p-value) from hypergeometric test. Motif families and neurons are ordered by similarity. Color bar across x-axis indicates neuron modality. Arrows denote motif families featured in panel D. C) Volcano plot showing log fold ratio and -log10 p-value for all motif family-neuron associations. Significant associations with p-value < 1e-5 and log fold ratio > 0.5 (3111) or < −0.5 (774) are noted. D) Eight selected motif families with significant associations with neurons from panel C: Motif families: E-box motifs (85 and 215), motifs for nhrs (100), homeodomains (246), and a previously undescribed motif (243). Asterisks denote significant associations. See also Figure S7.
We clustered all discovered motifs (see Methods), resulting in 159 distinct DNA and 65 RNA motif families. 101 of 159 DNA motif families showed similarity to DNA binding sequences from available databases. For example, FIRE discovered a DNA motif family (TAATCC) which corresponds to the core DNA binding sequence of K50 class homeodomain transcription factors (Driever and Nüsslein-Volhard, 1989; Treisman et al., 1989) in genes with high relative expression in ASEL, ASER, AWCON, AWCOFF, BAG, and AWA neurons (Figure S7A). The TAATCC sequence matches in vitro-derived binding motifs for C. elegans K50 class homeodomain genes that are expressed in these neurons (ceh-36 in ASE and AWC, ceh-37 in BAG and AWA; Figure S7A) and are required for their development (Chang et al., 2003; Koga and Ohshima, 2004; Lanjuin et al., 2003; Serrano-Saiz et al., 2013). These results indicate that our approach has the potential to reveal functionally relevant regulatory elements.
To limit false positives, the FIRE algorithm uses stringent criteria for motif discovery and therefore generates conservative results. Although each motif family was discovered in an average of 5 neurons, we reasoned that the identified motif families might also regulate gene expression in additional neuron types. We therefore generated motif-neuron associations for each motif family (see Methods, Figures 6B–C, S7C). We detected an average of 9 significant neuron associations for each motif family (log fold change > 0.5 and p-value < 1e-5). This additional analysis significantly expanded the list of associations for neurons with previously established co-regulated genes. For example, motif family 184 matches the X-box sequences bound by DAF-19, which regulates cilia formation in all 28 ciliated neuron types (Efimenko et al., 2005; Swoboda et al., 2000). This X-box motif was initially discovered by FIRE in 10 ciliated neurons, but was significantly associated with another 12 ciliated sensory neurons by our additional analysis (Figure S7E).
Our approach also points to previously undetected roles for TFs in neuron-specific gene regulation. For example, motif family 85 corresponds to the E-box motif CAGGTG and is strongly associated with most amphid and phasmid neurons (Figure 6D). This particular E-box sequence is enriched in hlh-4 target genes in the nociceptive sensory neuron ADL (Masoudi et al., 2018), but can also bind at least 10 distinct bHLH dimers (Grove et al., 2009). Interestingly, motif family 215 contained a different E-box sequence which was positively associated only with the chemorepulsive sensory neurons ADL, ASH, and PHB (Figure 6D). Based on the expression patterns of bHLH TFs in the adult nervous system, motif 215 may be a target of a HLH-2 homodimer (Masoudi et al., 2018).
Intriguingly, a substantial number of the motifs with strong positive associations with sensory neurons match TFs with uncharacterized roles in the nervous system or do not match any known TFs (Figure 6D). For example, motif family 100 showed a strong association with several sensory neurons and is similar to the binding site of the nuclear hormone receptor protein, NHR-142. nhr-142 is almost exclusively expressed in a subset of amphid sensory neurons (Figure 4A), and the binding domain of nhr-142 is closely related to several other nhr TFs (Lambert et al., 2019) which are expressed primarily in sensory neurons (nhr-45, nhr-213, nhr-18, nhr-84, nhr-178), suggesting roles for these nhr TFs in sensory neuron function. Additionally, several motifs showed strong negative associations with enriched genes across many neurons (Figure 6D, right), indicating possible cis-regulatory elements of transcriptional repressors.
RNA motif analysis revealed that most RNA motif families showed positive associations with many neurons (indicating over-representation of RNA motifs in the enriched genes for each neuron type). Similar to DNA motifs, the strongest effects for RNA motifs were seen in sensory neurons (Figure S7F). In contrast to all other RNA motif families, motif family 23 showed negative associations with most neuron types. This motif family corresponds to a poly-C sequence (Figure S7G). A subclass of KH-domain RNA binding proteins interacts with poly-C regions in RNA and microRNAs (Choi et al., 2009). The C. elegans poly-C binding protein HRPK-1 positively regulates the function of several microRNA families, including those that act in the nervous system (Li et al., 2019). The over-representation of the poly-C motif family in depleted genes in most neurons indicates a potential role for this motif in microRNA-mediated repression. Overall, our analysis of neuron-specific gene expression identified over 200 cis-regulatory elements that could be sites for trans-acting factors such as transcription factors, RNA-binding proteins and microRNAs.
Cell adhesion molecules (CAMs) are differentially expressed among neurons that are synaptically connected and that define anatomically distinct fascicles in the nerve ring.
We compared our transcriptomic data to the C. elegans connectome to identify candidate genetic determinants of neurite bundling and synaptic connectivity. For this analysis, we utilized the nerve ring (Figure 7A), the largest expanse of neuropil in the C. elegans nervous system, because electron microscope reconstructions from multiple animals have detailed both membrane contacts and synapses in this region (Brittin et al., 2021; Cook et al., 2019; Witvliet et al., 2020). We limited our analyses to putative cell adhesion molecules (CAMs), which have documented roles in axon pathfinding, fasciculation and synapse formation (Bruce et al., 2017; Colón-Ramos et al., 2007; Kim and Emmons, 2017; Shen and Bargmann, 2003; Siegenthaler et al., 2015; Sperry, 1963). 141 CAMs ((Cox et al., 2004; Hobert, 2013), see Table S3) were detected in neurons in our scRNA-Seq dataset.
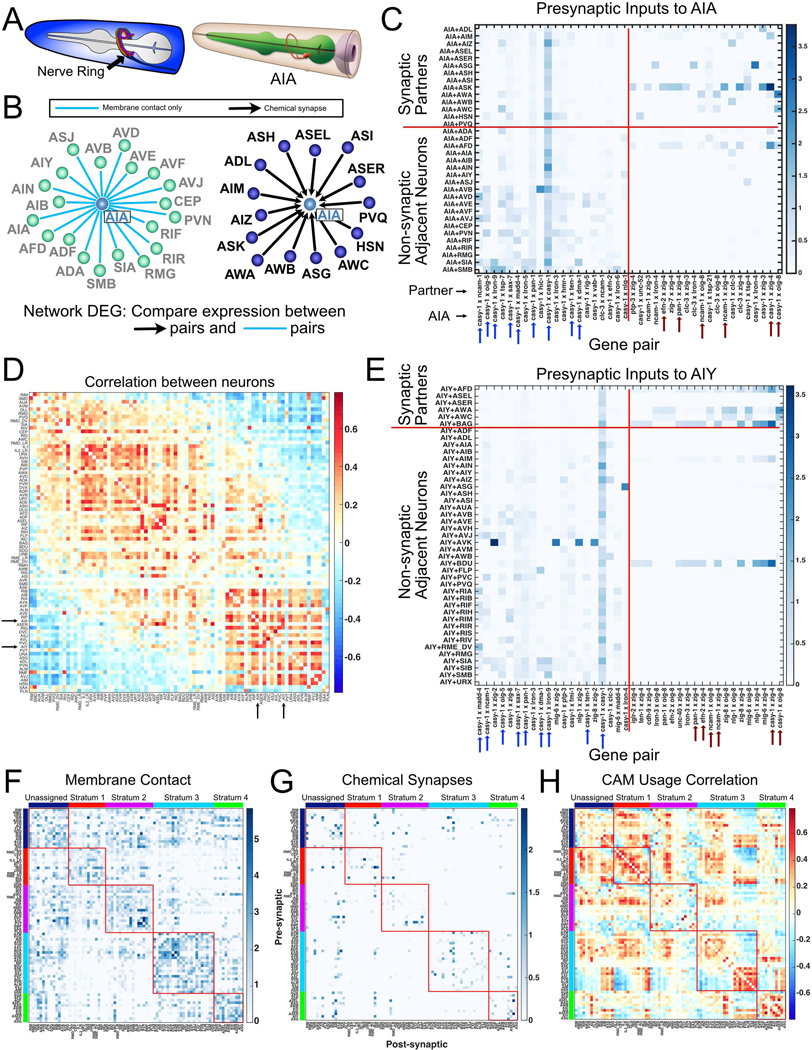
Figure 7. Differential expression of cell adhesion molecules among neurons and their presynaptic partners.
A) (Left) The C. elegans nerve ring. (Right) AIA ring interneuron. From WormAtlas. B) Neurons with presynaptic input to AIA (right) and neurons with membrane contact but no synapses with AIA (left). C) Heatmap of 20 cell adhesion molecule (CAM) gene pairs with highest log fold change in AIA + presynaptic inputs vs AIA + non-synaptic adjacent neurons (right of vertical red line). 20 CAM gene pairs with highest log fold change in AIA + non-synaptic adjacent neurons vs AIA + presynaptic partners (left of vertical red line). Arrows denote gene pairs common for AIA and AIY (panel E). D) Correlation matrix for CAM usage (see text) across all neurons in the nerve ring (84 neuron types). Arrows indicate AIA and AIY (correlation = 0.568). E) Heatmap as in C, for AIY. Arrows denote gene pairs common for AIA and AIY. F) Membrane adjacency matrix was grouped by nerve ring strata (each outlined with red box) (Moyle et al., 2021). Within each stratum, neurons were ordered according to CAM usage correlations (see panel H). G) Strata ordering as in F was imposed upon the chemical connectome revealing that most synapses are detected between neurons within the same stratum. H) The CAM usage correlation matrix (as in D) was grouped by strata, then sorted by similarity within each stratum. CAM usage is broadly shared for neurons in strata 1 and 4. Stratum 3 shows two distinct populations. See also Methods S1.
Recent computational analysis revealed a modular structure for the nerve ring, with four distinct neurite bundles or “strata” as well as a fifth group of unassigned neurons that contacts neurons in multiple strata (Moyle et al., 2021) (Figure S8A). See also (Brittin et al., 2021). Nerve ring formation begins in the embryo, but this structure is also modified throughout larval development as additional axons extend into the nerve ring and form synapses (Moyle et al., 2021; Witvliet et al., 2020). Together, these results point to the importance of both periodic as well as sustained expression of genetic determinants that initiate, modify or maintain the overall structure of the nerve ring and its connectome.
We first determined CAMs that were differentially expressed between strata (Figure S8B–C). Six CAMs were significantly enriched in the neurons in one stratum compared to the neurons in all other strata (Figure S8C). Notably, the transcript for MADD-4/punctin, a secreted protein that has been shown to direct process outgrowth as well synaptic placement (Zhou and Bessereau, 2019), is significantly enriched in stratum 1. tsp-7, a homolog of the human protein CD63, a member of the tetraspanin superfamily, is highly expressed in stratum 2. Tetraspanins interact with integrins and have been implicated in membrane trafficking and synaptogenesis (Murru et al., 2018; Pols and Klumperman, 2009). lron-5 and lron-9 (extracellular leucine rich repeat proteins) are selectively expressed in a subset of neurons in stratum 2 which could be indicative of roles in organizing these specific fascicles (Figure S8B). Thus, our approach has identified candidate genes that can now be experimentally tested for roles in organizing and maintaining structurally and functionally distinct domains of the nerve ring.
In addition to mediating axon fasciculation, we reasoned that specific CAMs might contribute to synaptic maintenance in the mature nervous system. We surmised that CAMs mediating synaptic stability are more highly expressed in synaptically connected neurons than in adjacent neurons with membrane contacts but no synapses. We generated high-confidence membrane adjacency and chemical synaptic connectomes by retaining only contacts and synapses that are preserved across animals in EM reconstructions of the nerve ring (see Methods, Table S5) (Brittin et al., 2021; Cook et al., 2019; White et al., 1986; Witvliet et al., 2020). These datasets include 84 of the 128 neuron classes. The importance of genetic determinants of connectivity in this circuit is underscored by the observation that membrane contacts between neurons in the nerve ring are much more numerous than synapses; on average, in the nerve ring, each neuron synapses with only 15% of the neurons it contacts (means of 6.42 presynaptic inputs, 6.42 postsynaptic outputs, 42 contacted cells) (Brittin et al., 2021; White et al., 1986).
For each neuron, we compared the expression of all possible combinations of pairs of CAMs in the neuron and its synaptic partners relative to the neuron and its non-synaptic adjacent neurons (Figure 7B–C). Two independent comparisons were generated, one for presynaptic partners (Figure 7C) and a second result for postsynaptic neurons (Methods S1). Our analysis revealed multiple CAM gene pairs with enrichment in synaptically connected neurons compared to adjacent but not synaptically connected neurons. A representative example for presynaptic inputs to the interneuron AIA shows that CAM pairs enriched in synaptically connected neurons were not uniform for the different presynaptic partners of AIA (Figure 7C). For example, AIA and its presynaptic partner, ASK, show strong enrichment for casy-1 (Calsyntenin) and zig-4 (secreted 2-Ig domain protein) whereas the AIA-ASG pair is enriched for casy-1 (Calsyntenin) and lron-4 (extracellular leucine rich repeat protein). This finding is consistent with the prediction that distinct combinatorial codes of CAMs could be required for patterning connectivity between individual pairs of neurons (Kim and Emmons, 2017). Additionally, we identified distinct CAM pairs that are enriched in adjacent, not synaptically connected neurons (Figure 7C). This observation indicates that some CAM interactions may functionally inhibit either the formation or maintenance of synapses between neurons. Anti-synaptic effects have been documented for the axon guidance molecules netrin, sema-5B and their cell surface receptors (O’Connor et al., 2009; Poon et al., 2008; Tran et al., 2009).
To examine patterns across the nerve ring, we restricted our analysis to gene pairs with a log fold change > 0.2 in either synaptically connected or in adjacent but not connected neurons for at least one neuron type. We refer to this pattern of CAM pairs enriched in synaptic or solely adjacent neurons as “CAM usage.” Of 19,881 possible CAM pairs, 439 pairs passed our log fold change threshold for presynaptic connections, whereas 443 pairs showed > 0.2 log fold change for postsynaptic connections (Methods S1). To identify neurons with similar patterns of presynaptic CAM usage, we generated correlation matrices from pairwise comparisons of all neurons and sorted neurons by similarity using multidimensional scaling (Figure 7D). For example, CAM usage for presynaptic inputs to AIA and AIY is strongly correlated (correlation 0.568) due to the co-occurrence for each neuron of multiple shared combinations of CAMs (Figure 7E, blue and red arrows). This analysis also separated neurons into two main groups based on CAM usage which could be indicative of underlying shared roles for CAMs among these distinct sets of neurons.
We sought to understand the relationship between stratum membership and synaptic CAM usage for nerve ring neurons. Both membrane contact and chemical synapses are denser among neurons within strata than across strata (Figure 7F–G), a finding also observed for an independent assessment of nerve ring axon bundles (Brittin et al., 2021). We sorted neurons by CAM usage within each stratum (Figure 7H) to assess intra-stratum correlations. This approach revealed high correlations among neurons within strata. Additionally, neurons in some strata split into distinct groups based on CAM usage (Stratum 3, Figure 7H, see Methods S1). This observation suggests that CAM usage at synaptic connections is likely distinct from CAMs that may be involved in strata formation and/or maintenance. Although CAM usage correlations were often elevated among neurons within strata, high correlations were also detected among neurons in different strata that are not synaptically connected and with minimal contacts, thus suggesting roles for CAMs in nerve ring architecture and connectivity likely depend on additional factors. We suggest that that the overall results of our analysis point to specific CAMs that can now be investigated for roles in the formation and maintenance of synapses as well as fasciculation between specific neurons in the C. elegans nerve ring.
Data interface
We developed a web application, CengenApp (http://cengen.shinyapps.io/CengenApp) to facilitate analysis of these scRNA-Seq data. Users can generate gene expression profiles by neuron class or by gene at different thresholds, and perform differential gene expression analysis between either individual neurons or between groups of neuron types. In addition, an interactive graphical interface is available for generating heat map representations (e.g., Figure 3C) of gene expression across the nervous system. Raw data are available at Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) (single cell data at GSE136049, bulk data at GSE169137). The data and additional supporting files can be downloaded from the CeNGEN website, www.cengen.org and code is available at Github, www.github.com/cengenproject.
CONCLUSIONS
We have produced a gene expression map for the entire C. elegans nervous system, complementing earlier partial profiles of the C. elegans nervous system at embryonic and early larval stages (Cao et al., 2017; Packer et al., 2019). This catalog of gene expression provides an essential foundation for a comprehensive exploration of transcriptional and gene regulatory patterns that lead to neuronal diversity, connectivity and function. C. elegans is the first organism in which a complete anatomical map of its nervous system is matched with a nervous system-wide molecular map, therefore providing new opportunities to investigate neuronal development and function.
We developed a thresholding approach for single-cell data to generate high confidence profiles for each neuron type. Multiple findings indicate that neuropeptide signaling is widely utilized and likely crucial for a variety of functions. First, neuropeptide-encoding genes are among the most abundantly detected genes in the dataset. Second, at the most stringent threshold examined, each neuron expresses at least four different neuropeptide-encoding genes. Third, each neuron expresses a distinct combination of both neuropeptide genes and putative neuropeptide receptors. Recent reports show abundant and widespread neuropeptide expression in Hydra (Siebert et al., 2019), Drosophila (Allen et al., 2020) and mouse cortical neurons (Smith et al., 2019), indicating that these salient features of neuropeptide signaling are conserved among diverse species.
Our analysis of transcription factor expression reveals that different transcription factor families appear to have segregated into distinct functions during cellular differentiation. Some families are underrepresented in the mature nervous system (T-box genes), others show broad expression patterns in the nervous system (Zn finger), whereas others are sparsely expressed and appear to exquisitely track with neuronal identity (homeodomains) (Reilly et al., 2020). The nuclear hormone receptors (nhrs) may have acquired a unique function, as inferred by their striking enrichment in sensory neurons. The identification of enriched cis-regulatory motifs in neuronal gene batteries provides an opportunity for future experiments to dissect the mechanisms of gene regulation in the nervous system.
Finally, we devised computational strategies that exploit our gene expression profile of the C. elegans nervous system to reveal the genetic underpinnings of neuron-specific process placement and connectivity. Previous computational efforts to forge a link between neuron-specific gene expression and the C. elegans wiring diagram have been hampered by incomplete and largely qualitative expression data (Barabási and Barabási, 2020; Baruch et al., 2008; Kaufman et al., 2006; Kovacs et al., 2020; Varadan et al., 2006). Here, we leveraged our nervous-system wide catalog of gene expression to deduce combinatorial codes for cell adhesion molecules (CAMs) that likely contribute to the maintenance and formation of this complex neuropil. Importantly, this analysis can now be extended to specific groups of neurons and to any gene family to generate specific hypotheses of process placement and connectivity for direct experimental validation.
We expect that these data will be useful for future studies of individual genes, neurons, and circuits, as well as global analyses of an entire nervous system and the development of scRNA-Seq analysis methods. Coupled with the fully described cell lineages (Sulston and Horvitz, 1977; Sulston et al., 1983), neuronal anatomy (Albertson and Thomson, 1976; Brittin et al., 2021; Cook et al., 2019; White et al., 1986; Witvliet et al., 2020), and powerful functional analyses, such as pan-neuronal calcium imaging and neuronal identification (Kato et al., 2015; Nguyen et al., 2016; Venkatachalam et al., 2016; Yemini et al., 2021), our dataset provides the foundation for discovering the genetic programs underlying neuronal development, connectivity and function.
Limitations of the Study
Although we provide gene expression profiles of every neuron class in the C. elegans hermaphrodite, these neuron-specific transcriptomes are incomplete for several reasons:
Some neuron classes are under-represented, likely due to biases in the dissociation procedure, thus resulting in incomplete detection of expressed transcripts in the corresponding scRNA-Seq data set (Figure S5I–L).
Our scRNA-Seq library construction method largely excluded non-coding RNAs that are not poly-adenylated (Figure 5B).
Alternative splicing is rarely detected in our scRNA-Seq data set due to short reads and the 3’ bias of the library construction method (Figure 5D–F).
Additional approaches, such as isolation of individual neuron types for bulk RNA-Seq (Figure 5A), single-nuclei RNA-Seq, long-read sequencing and alternative RNA-Seq library preparation methods could be used in future studies to produce a more comprehensive description of the C. elegans neuronal transcriptome.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Requests for resources and reagents should be directed to the Lead Contact, David Miller (david.miller@vanderbilt.edu)
Materials Availability
The strains generated in this study are available at the Caenorhabditis Genetics Center or by request from the lead contact.
Data and Code Availability
The raw data are available at GEO (single cell data: Accession Number GSE136049, bulk sequence data: Accession Number GSE169137). The full and neuron only datasets are available at www.cengen.org. Analysis code is available at github https://github.com/cengenproject.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Preparation of larvae and dissociation
Worms were grown on 8P nutrient agar 150 mm plates seeded with E. coli strain NA22. To obtain synchronized cultures of L4 worms, embryos obtained by hypochlorite treatment of adult hermaphrodites were allowed to hatch in M9 buffer overnight (16–23 hours at 20° C) and then grown on NA22-seeded plates for 45–48 hours at 23° C. The developmental age of each culture was determined by scoring vulval morphology (>75 worms) (Mok et al., 2015). Single cell suspensions were obtained as described (Kaletsky et al., 2016; Spencer et al., 2014; Zhang et al., 2011) with some modifications. Worms were collected and separated from bacteria by washing twice with ice-cold M9 and centrifuging at 150 rcf for 2.5 minutes. Worms were transferred to a 1.6 mL centrifuge tube and pelleted at 16,000 rcf for 1 minute. 250 μL pellets of packed worms were treated with 500 μL of SDS-DTT solution (20 mM HEPES, 0.25% SDS, 200 mM DTT, 3% sucrose, pH 8.0) for 2–4 minutes. In initial experiments, we noted that SDS-DTT treatment for 2 minutes was sufficient to dissociate neurons from the head and tail, but longer times were required for effective dissociation of neurons in the mid-body and ventral nerve cord. The duration of SDS-DTT was therefore selected based on the cells targeted in each experiment. For example, NC3582, OH11746, and juIs14 L4 larvae were treated for 4 minutes to ensure dissociation and release of ventral cord motor neurons. NC3579, NC3580 and NC3636 L4 larvae were treated with SDS-DTT for 3 minutes. All other strains were incubated in SDS-DTT for 2 minutes. Following SDS-DTT treatment, worms were washed five times by diluting with 1 mL egg buffer and pelleting at 16,000 rcf for 30 seconds. Worms were then incubated in pronase (15 mg/mL, Sigma-Aldrich P8811, diluted in egg buffer) for 23 minutes. During the pronase incubation, the solution was triturated by pipetting through a P1000 pipette tip for four sets of 80 repetitions. The status of dissociation was monitored under a fluorescence dissecting microscope at 5-minute intervals. The pronase digestion was stopped by adding 750 μL L-15 media supplemented with 10% fetal bovine serum (L-15–10), and cells were pelleted by centrifuging at 530 rcf for 5 minutes at 4 C. The pellet was resuspended in L-15–10, and single-cells were separated from whole worms and debris by centrifuging at 100 rcf for 2 minutes at 4 C. The supernatant was then passed through a 35-micron filter into the collection tube. The pellet was resuspended a second time in L-15–10, spun at 100 rcf for 2 minutes at 4 C, and the resulting supernatant was added to the collection tube.
METHOD DETAILS
FACS isolation of neuron types for RNA-Seq
Fluorescence Activated Cell Sorting (FACS) was performed on a BD FACSAria™ III equipped with a 70-micron diameter nozzle. DAPI was added to the sample (final concentration of 1 μg/mL) to label dead and dying cells. To prepare samples for scRNA-sequencing, our general strategy used fluorescent reporter strains to isolate subgroups of cells. For example, we used an eat-4::mCherry reporter (OH9625) to target glutamatergic neurons and an ift-20::NLS-TagRFP reporter (OH11157) to label ciliated sensory neurons. We used an intersectional labeling strategy with a nuclear-localized pan-neural marker (otIs355 [rab-3(prom1)::2xNLS-TagRFP] IV) to exclude cell fragments labeled with cytosolic GFP markers (NC3582). In other cases, we used an intersectional strategy to exclude non-neuronal cells. For example, stIs10447 [ceh-34p::HIS-24::mCherry] is expressed in pharyngeal muscles, pharyngeal neurons and coelomocytes. To target pharyngeal neurons, we generated strain NC3583 by crossing stIs10447 [ceh-34p::HIS-24::mCherry] with the pan-neural GFP marker evIs111 to isolate cells that were positive for both mCherry and GFP. Non-fluorescent N2 (wild-type reference strain) (Brenner, 1974) standards and single-color controls (in the case of intersectional labeling approaches) were used to set gates to exclude auto-fluorescent cells and to compensate for bleed-through between fluorescent channels. For two experiments, single-cell suspensions from separate strains were combined (OH16003 plus PS3504 and nIs175, NC3635 plus NC3532) prior to FACS. In some cases, we expanded FACS gates to encompass a wide range of fluorescent intensities to ensure capture of targeted cell types. This less stringent approach may contribute to the presence of non-neuronal cells in our dataset (see Results). Cells were sorted under the “4-way Purity” mask.
For 10X Genomics single-cell experiments, sorted cells were collected into L-15–33 (L-15 medium containing 33% fetal bovine serum), concentrated by centrifugation at 500 rcf for 12 minutes at 4° C, and counted on a hemocytometer. Single-cell suspensions used for 10x Genomics single-cell sequencing ranged from 300–900 cells/μL.
For bulk RNA-sequencing of individual cell types, sorted cells were collected directly into TRIzol LS. At ~15-minute intervals during the sort, the sort was paused, and the collection tube with TRIzol was inverted 3–4 times to ensure mixing. Cells in TRIzol LS were stored at −80° C for RNA extractions (see below).
Single-cell RNA sequencing
Each sample (targeting 5,000 or 10,000 cells per sample) was processed for single cell 3’ RNA sequencing utilizing the 10X Chromium system. Libraries were prepared using P/N 1000075, 1000073, and 120262 following the manufacturer’s protocol. The libraries were sequenced using the Illumina NovaSeq 6000 with 150 bp paired end reads. Real-Time Analysis software (RTA, version 2.4.11; Illumina) was used for base calling and analysis was completed using 10X Genomics Cell Ranger software (v3.1.0). Most samples were processed with 10x Genomics v2 Chemistry, except for samples from juIs14, NC3583, NC3636, CX5974, OH16003, PS3504, nIs175, NC3635 and NC3532, which were processed with v3 Chemistry. Detailed experimental information is found in Table S1.
Single-cell RNA-Seq Mapping
Reads were mapped to the C. elegans reference transcriptome from WormBase, version WS273. Due to the possibility that 3’ untranslated region (UTR) annotations in the reference transcriptome may be too short (Packer et al., 2019), we dynamically extended the 3’ UTR of each gene to its optimal length, thereby enabling the additional mapping of reads to the 3’ extremity of the gene body. We generated eight versions of gene annotations based on WormBase WS273 annotation, with 3’ UTRs in each version elongated by 50, 100, 150, 200, 250, 300, 400 and 500 base pairs (bps), respectively. Elongation of genes which overlapped with other genes during the extension process was terminated before encountering an adjacent exon. Subsequently, eight custom genome indexes, which respectively combined the C. elegans WS273 reference genome with the eight extended gene annotation versions, were generated using CellRanger (version 3.1.0).
All sequenced reads from each of the 17 single-cell samples were mapped to the eight reference genomes using the CellRanger pipeline. We next selected the best UTR extension length of each annotated gene independently for the 17 samples, as a number of genes were heavily enriched in specific samples. First, we calculated the total number of mapped reads for each of the expressed genes in each sample, resulting in eight mapped-read values representing the eight gene annotation versions. To discard the UTR extension intervals which harbor sparse additional reads, as well as to allow for the intervals which harbor fewer reads but are surrounded by read-enriched intervals, we took advantage of the trimming algorithm in Burrows-Wheeler Alignment (Li and Durbin, 2009) to find the best extension. Specifically, a cutoff of 20 reads was applied to each extension interval (50, 50, 50, 50, 50, 100, and 100 bps). Cumulative sums from 3’ to 5’ end were then calculated after subtracting the cutoff in each interval, and the smallest sum of less than 0 was located as the trimming point for a given sample. Considering all 17 samples, the trimming point agreed by most samples (or at least two samples if one gene is expressed in limited samples) was chosen as the ultimate one. Consequently, we extended the UTRs for 1,012 C. elegans genes, encompassing 40, 216, 175, 113 and 468 genes with UTRs extended by 150, 200, 250, 300 and 400 bps at the 3’ end, respectively. Lastly, with the gene annotation file containing the optimal extension length for each gene, we remapped and quantified the gene expression in all 17 samples using CellRanger.
Downstream Processing
We distinguished cells from empty droplets, corrected background RNA expression and generated quality control metrics for each sample independently, then merged the files together into one dataset. The default barcode filtering algorithm in CellRanger can fail to capture cells in some conditions, especially with cells with variable sizes and RNA content (Lun et al., 2019). Neurons in particular tend to have lower UMI counts than other cell types and can be missed by the default algorithm (Packer et al., 2019). We therefore used the EmptyDrops method (with a threshold of 50 UMIs for determining empty droplets) from the R package DropletUtils (Lun et al., 2019) to determine which droplets contained cells. This approach detected significantly more cells than the CellRanger method, and we were able to confidently annotate these additional cells as neurons.
The SoupX R package (Young and Behjati, 2020) was used to correct for background RNA. We used a more conservative threshold for determining background RNA for SoupX than for EmptyDrops to exclude low-quality cells in the background correction. We therefore set a threshold of droplets with fewer than 25 UMIs to estimate the background RNA. Genes with patterns of strong expression in restricted sets of cells (from the literature or from preliminary clustering analysis for each single-cell experiment) were selected for each dataset (Table S1). SoupX uses these genes, preliminary clustering, and the calculated background RNA profile (from droplets with fewer than 25 UMIs) to estimate the percent of contamination in each sample. The estimated background contamination ranged from 4.15–13.56%, with a mean of 8.01%. For the ceh-28_dat-1 experiment, no combination of genes tested resulted in satisfactory performance, so the contamination was set manually to 10.00%. SoupX uses the calculated contamination level to correct the expression of genes that are abundant in the background RNA profile, and returns a corrected gene by cell count matrix. The background corrected count matrices produced by SoupX were rounded to integer counts and used for subsequent downstream processing.
Following background correction, quality control metrics were calculated for each dataset with the R package scater (McCarthy et al., 2017), using the percentage of UMIs from the mitochondrial genes nduo-1, nduo-2, nduo-3, nduo-4, nduo-5, nduo-6, ctc-1, ctc-2, ctc-3, ndfl-4, atp-6, and ctb-1. Droplets with greater than twenty percent of UMIs coming from mitochondrial genes were removed. Datasets from individual experiments were merged using Seurat (v3) (Stuart et al., 2019). Genes detected in fewer than five cells were removed. Log-normalized expression matrices were then used for downstream analysis using monocle (2.99.3), monocle3 (0.2.1) (Cao et al., 2019; Qiu et al., 2017a, 2017b; Trapnell et al., 2014) and Seurat (v3) packages.
Dimensionality reduction and batch correction
We imported the merged dataset into monocle3, and reduced the dimensionality of the dataset with PCA (135 principal components, based on examination of an elbow plot showing the variance explained by each principal component), followed by the Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2019; McInnes et al., 2018) algorithm in monocle3 (reduce_dimension function, parameters were default other than: umap.min_dist = 0.3, umap.n_neighbors = 75). We then clustered cells using the leiden algorithm in monocle3 (res = 3e-4). Batch correction between experiments was performed using the align cds function (Cao et al., 2019; Haghverdi et al., 2018). We processed the neuron-only dataset with the following parameters (125 PCs, umap.min_dist = 0.3, umap.n_neighbors = 75, alignment_k (for align_cds) = 5, clustering resolution 3e-3).
Cell Identification
We assigned tissue and cell identity to the majority of cells in our dataset based on a manually compiled list of reported gene expression profiles with an average of > 20 molecular markers per neuron type (Hobert et al., 2016), and a recently described protein expression atlas of >100 homeodomain proteins (Reilly et al., 2020) (Table S1). Most of the neuronal UMAP clusters could be readily assigned to an individual neuron type on the basis of these known markers. We manually excluded clusters we identified as doublets due to co-expression of cell-type specific markers. We manually merged multiple clusters that corresponded to the same neuron type. We noted that coelomocytes were most abundant in experiments using strains expressing mCherry (otIs292 and otIs447). This effect likely results from neurons shedding mCherry+ exophers, which are then taken up by coelomocytes (Melentijevic et al., 2017), causing them to be isolated along with mCherry-labeled neurons.
Some clusters in the initial global dataset appeared to contain multiple closely related neuron types (i.e., cholinergic motor neurons, dopaminergic neurons, oxygen sensing neurons AQR, PQR, URX and pharyngeal neurons). Additional analysis of these separate clusters (i.e., reapplication of PCA, UMAP, and clustering to just these clusters) separated these cell types into individual clusters (Figure 1E–F). Finally, we identified separate clusters for the neuron classes RIV and SMD. In both of these instances, however, one of the putative clusters showed strong expression of stress-related transcripts rather than sub-type specific markers and therefore likely correspond to a subset of RIV and SMD neurons damaged by the isolation protocol. These two aberrant clusters were excluded from further analyses.
In the complete dataset, cells had a median of 928 UMIs/cell and 328 genes/cell. In the neuron only dataset, neurons had a median of 1033 UMIs/cell and 363 genes/cell. We note that these metrics are lower than generally observed for Drosophila or mouse 10X experiments (10X Genomics, 2017; Davie et al., 2018). We believe that this is likely due to the lower RNA content in C. elegans neurons (~2 um in diameter) compared to Drosophila (2–6 um) or mouse (10–30 um) neurons.
Neuron network analysis
The neuron network containing all neuron types was constructed on the basis of the transcriptome similarity between each pair of neuron types. We obtained the transcriptional profile of each neuron type by averaging gene expression across all cells within the given type, resulting in the gene expression trajectory for each neuron type. We next calculated transcriptome similarity (after log transformation) as the Pearson correlation coefficient between pairwise neuron types, using 7,390 highly variable genes identified by Seurat based on their variance and mean expression. The neuron network in a graphopt layout was constructed by the package “igraph” (Csárdi and Nepusz 2006) in R using the force-directed graphopt algorithm based on the above similarity matrix.
Gene expression analyses
Averaged gene expression profiles for each neuron class were generated as described (Cao et al., 2017). Quantitative expression data for a subset of genes are distorted by overexpression from fosmid reporters or co-selectable markers (lin-15A, lin-15B, pha-1, rol-6, unc-119, dpy-20, cho-1), the promoter regions used for marking cell types (unc-53, unc-47, gcy-35, C30A5.16, saeg-2, F38B6.2, C30F8.3, cex-1) or from a gene-specific 3’ UTR included in fluorescent reporter constructs (eat-4, unc-54). These genes are annotated in the CengenApp web application.
For visualization of gene expression data in the web application and for differential gene expression tests, data were imported into Seurat (v3) and raw counts were normalized using the variance stabilizing transformation (VST) implemented in the function sctransform with default parameters and regressing out the percent of mitochondrial reads (Hafemeister and Satija, 2019; Stuart et al., 2019). Differential gene expression tests used the Seurat v3 default Wilcoxon rank sum test with default parameters (a gene must be detected in > 10% of the cells in the higher-expressing cluster and have an adjusted p-value < 0.05).
Stress-induced genes
The dissociation procedure used to isolate single cells can induce cellular stress responsive pathways (Van Den Brink et al., 2017; Kaletsky et al., 2016). To identify likely stress-induced genes, we examined the distribution in our data of a list of 199 stress-induced genes, including heat shock protein (hsp) family genes and additional genes from the literature (Van Den Brink et al., 2017; Brunquell et al., 2016; Kaletsky et al., 2016) (Table S1). 20 of these genes showed abundant and broad expression across the entire nervous system. We generated a stress index for each single cell by calculating the percent of UMIs mapping to these 20 genes. We then tested the correlation of each gene’s expression pattern with the stress index to identify additional putative stress-responsive genes. We identified a total of 49 genes featuring correlations > 0.1 with the stress index and which were detected in at least 75 neuron types as likely stress responsive genes (Table S1).
Thresholding
The wealth of known gene expression data in C. elegans from fluorescent reporter strains provides a unprecedented opportunity to set empirical thresholds for our scRNA-Seq data based on ground truth. We first compiled a ground truth dataset of 160 genes with expression patterns across the nervous system previously determined with high confidence fosmid fluorescent reporters, CRISPR strains or other methods (Bhattacharya et al., 2019; Harris et al., 2020; Reilly et al., 2020; Stefanakis et al., 2015; Yemini et al., 2021) (Table S2). For each gene, we then aggregated expression across the single cells corresponding to each neuron type and calculated several metrics, including the total UMI count, the number of single cells of each neuron type in which each gene was detected with at least one UMI, the proportion of single cells of each neuron type in which gene was detected with at least one UMI and a normalized transcripts per million (TPM) expression value (Packer et al., 2019). We generated receiver operating characteristic (ROC) and precision recall (PR) curves for each metric by thresholding the data across a range of values, and calculated true positive, false positive, and false discovery rates by comparing the single-cell data to the ground truth. We used the area under the curve to decide which metric to use for thresholding. The proportion of cells in which a gene was detected performed the best (had the highest AUC) and was thus used to establish gene-level thresholds.
We first set initial thresholds to retain ubiquitously-expressed genes and to remove non-neuronal genes. Genes detected in ≥ 1% of the cells in every neuron cluster were considered expressed in all neuron types (193 genes), whereas transcripts detected in ≤ 2% of the cells in every neuron cluster were considered non-neuronal (4806 genes; no genes were detected in ≥ 1 % and ≤ 2 % of the cells in every neuron). As most genes displayed different levels of expression, we found that a single threshold failed to reliably capture expression for all genes. Thus, we applied percentile thresholding for each gene individually. For example, the AFD cluster showed the highest proportion of cells (76.3%, Figure S5A) expressing the homeodomain transcription factor ttx-1. For unc-25/GAD, the VD_DD cluster had the highest proportion of cells (94.4%, Figure S5G), whereas for the homeodomain transcription factor ceh-13, the DA neuron cluster had the highest proportion (13.4%, not shown). Thresholds were calculated as a fraction of the highest proportion of cells for each individual gene. For example, a threshold of 0.04 results in different absolute cut-offs for each gene. For ttx-1, with a highest proportion of 76.3%, we scored ttx-1 as “not expressed” in clusters in which it was detected in < 3.05% of cells (0.04*76.3 = 3.05%). For unc-25, with a highest proportion of expressing cells of 94.4%, we scored unc-25 as “not expressed” in clusters in which it was detected in < 3.77% of cells (0.04*94.4 = 3.77%). Similarly, and we scored ceh-13 as “not expressed” in clusters in which it was detected in < 0.536% of cells (0.04*13.4 = 0.536%).
For each threshold percentile, we generated 5,000 stratified bootstraps of the ground truth genes using the R package boot (Canty and Ripley, 2019; Davison and Hinkley, 1997) and computed the True Positive Rate (TPR), False Positive Rate (FPR) and False Discovery Rate (FDR) for the entire dataset as well as for each neuron type. We estimated 95% confidence intervals with the adjusted percentile (BCa) method, and plotted the ROC and PR curves (Figure S5C, D). Finally, we selected 4 thresholds of increased stringency (1–4, see Table S2 for statistics for each neuron type). Threshold 2 was used for analyses profiling gene expression across all neuron types and across gene families.
Estimating coverage for individual neurons
We used threshold 2 to model the relationship between the number of cells in each neuron type cluster and the number of genes detected with the expression:
| (Eq. 1) |
Where GN is the number of genes detected, Gmax is the maximal number of genes detected with an infinite number of cells, NC is the number of cells of a given type, and b is the number of cells at which GN = half of Gmax. Using 1000 bootstrapped samples, we estimate 6550 ± 7 genes for Gmax and 34.22 ± 0.3 for b (Figure S5I). In other words, this finding suggests that single cell sequencing would detect an average of ~6,500 transcripts per neuron type if an infinite number of cells were sampled and that sampling of ~30 cells/neuron type is sufficient to capture 50% of these genes.
To address the possibility that transcript complexity could vary across neuron types, we used a down-sampling strategy to model the relationship between genes detected vs the number of cells sampled for each neuron class. We performed 60–100 iterations of down-sampling for each neuron type to generate plots of numbers of cells vs numbers of genes for each cell type at threshold 2 (Figure S5K). Fitting equation 1 to each plot predicts a maximal number of genes detected at an infinite number of cells for each neuron type (Figure S5L, Table S2). Estimates for some neuron types are less confident due to undersampling of cells. However, we also see a wide range of predicted values among well-represented cell types, suggesting that these estimates could be indicative of biological variation in the genetic complexity of individual neuron types across the nervous system (Table S2).
Determining distinct combinations of gene sets
Expression matrices of selected gene families from threshold 2 were binarized. Genes were clustered following default parameters in the R package hclust. We determined if neurons expressed a distinct combinatorial code for given gene families by determining whether any two columns (neurons) of the binarized expression matrix were identical. For analyzing expression of gene regulatory families, we treated C2H2 zinc finger proteins as transcription factors and removed them from the list of RNA-binding proteins. We also removed ribosomal proteins from the RNA-binding protein list.
Connectivity Analysis
To determine neurons postsynaptic to either ACh or glutamate-releasing neurons, we used the C. elegans hermaphrodite chemical connectome data from (Cook et al., 2019). For this analysis, we scored synapses as connections detected in more than 3 electron micrograph sections.
Reporter strains
GFP reporters for the neuropeptide genes flp-33, nlp-17, nlp-42, nlp-52 and nlp-56 were created by PCR Fusion (Hobert, 2002) whereby the 5’ intergenic region of the gene of interest and the coding sequence of GFP with 3’ UTR of unc-54 were fused in subsequent PCR reactions. We used the entire intergenic region of the genes of interest: 1519 bp for flp-33 (forward primer: aggaagttgataaacttgcttgttttaatg, reverse primer: ggtagggggaccctggaag), 372 bp for nlp-17 (forward primer: tcatctaaaatatattttcaaaacgattttctgtgc, reverse primer: attttctgtgaaaaagcctgactttttc), 3250 bp for nlp-42 (forward primer: ttgtctgaaaatatgggttttgcatgg, reverse primer: tttacctgaaaatttgcaatttttcagatttttac), 3731 bp for nlp-52 (forward primer: ttgcttgcattttctgaaataagatgg, reverse primer: ttttgggaagaggtacctggaac), and 2954 bp for nlp-56 (forward primer: ggttcactggaataaatatatgcactgtatc, reverse primer: ctggaagagttgaatcatatggtttagaag). Reporters were injected directly into NeuroPAL pha-1 strain (OH15430 pha-1(e2123); otIs669[NeuroPAL 15]) (Yemini et al., 2021) as a complex array with OP50 DNA (linearized with ScaI) and pBX [pha-1 (+)] (Granato et al., 1994) as a co-injection marker. For flp-33 and nlp-52, the reporter, pBX [pha-1 (+)] and OP50 DNA were injected at concentrations of 7.75 ng/μl, 6.2 ng/μl, 99.96 ng/μl, respectively. For nlp-42, the reporter, pBX [pha-1 (+)] and OP50 DNA were injected at 11.80 ng/μl, 8.7 ng/μl and 88.86 ng/μl. For nlp-17, the reporter, pBX [pha-1 (+)] and OP50 DNA were injected at 10 ng/μl, 6.2 ng/μl and 99.96 ng/μl. For nlp-56, the reporter, pBX [pha-1 (+)] and OP50 DNA were injected at concentrations of 9.5 ng/μl, 5.2 ng/μl and 94.9 ng/μl. After injection, animals were kept at 25°C for selection of the array positive worms and maintained for at least three generations before imaging (see below). CRISPR reporter strains for flp-1 and nlp-51 were generated by engineering a T2A::3xNLS::GFP cassette into the respective gene loci just before the stop codons. The nspc-1 promoter fusion reporter was constructed using the entire 713 bp intergenic region upstream of npsc-1 fused driving GFP.
Sequences of C39H7.2 and nhr-236 were acquired from C. elegans BioProject PRJNA13758 browser (via WormBase). We combined 1447 bp upstream of the C39H7.2 sequence (forward primer: Gtatggtctgcaggagtatc, reverse primer: Gcccatggaagtgtcgaatt) with 2044 bp of UberPN::3xNLS-intronGFP (forward primer: CCCAAAGgtatgtttcgaat, reverse primer: AACTGTTTCCTACTAGTCGG) via overlap PCR. For nhr-236, we combined 802 bp immediately upstream of the ATG sequence of the first exon of nhr-236 (forward primer: Tcttgaagggcacgccgatt, reverse primer: Gctctgtgtcggtattccgg) with 2044 bp of UberPN::3xNLS-intronGFP (primers as above) via overlap PCR. The resulting overlap PCR products were injected with 50 ng/μl of pha-1 rescue construct pBX [pha-1 (+)] and 1Kb+ladder (Promega Corporation, G5711) into GE24 [pha-1(e2123) III]. The injected lines were grown at 25 C for selection of the pha-1+ worms and were maintained for at least five generations before imaging with a Spinning Disk Confocal microscope (Nikon). The images were analyzed using Volocity Imaging Software and also crossed into the NeuroPAL strain otIs669 to identify the neurons expressing the reporters.
Imaging
Confocal images were obtained on either a Nikon A1R confocal laser scanning microscope or a Zeiss LSM 880 microscope using 20x or 40x oil immersion objectives. Brightness and contrast adjustments were performed with FIJI.
RNA Extraction
Cell suspensions in TRIzol LS (stored at −80° C) were thawed at room temperature. Chloroform extraction was performed using Phase Lock Gel-Heavy tubes (Quantabio) according to the manufacturer’s protocol. The aqueous layer from the chloroform extraction was combined with an equal volume of 100% ethanol and transferred to a Zymo-Spin IC column (Zymo Research). Columns were centrifuged for 30 sec at 16,000 rcf, washed with 400 μL of Zymo RNA Prep Buffer and centrifuged for 16,000 rcf for 30 sec. Columns were washed twice with Zymo RNA Wash Buffer (700 μL, centrifuged for 30 sec, followed by 400 μL, centrifuged for 2 minutes). RNA was eluted by adding 15 μL of DNase/RNase-Free water to the column filter and centrifuging for 30 sec. A 2 μL aliquot was submitted for analysis using the Agilent 2100 Bioanalyzer Picochip to estimate yield and RNA integrity and the remainder stored at −80° C.
Bulk sequencing and mapping
Each bulk RNA sample was processed for sequencing using the SoLo Ovation Ultra-Low Input RNAseq kit from Tecan Genomics according to manufacturer instruction, modified to optimize rRNA depletion for C. elegans (Barrett et al., 2021). Libraries were sequenced on the Illumina Hiseq 2500 with 75 bp paired end reads. Reads were mapped to the C. elegans reference transcriptome from WormBase (version WS274) using STAR version 2.7.0. Duplicate reads were removed using SAMtools (version 1.9), and a counts matrix was generated using the featureCounts tool of SubRead (version 1.6.4).
Comparing scRNA-Seq and bulk RNA data
Differential gene expression comparing sorted cell samples with sorted pan-neuronal samples was performed using TMM-normalized counts in edgeR (version 3.28.1). Two to five replicates per cell type were used in each sample (ASG: 4, AVE: 3, AVG: 3, AWA: 4, AWB: 5, DD: 3, PVD: 2, VD: 4, pan-neuronal: 5). Marker genes from the single cell dataset were selected using a Wilcoxon test in Seurat v3, calling enriched genes by comparing individual neuronal clusters to all other neuronal clusters. Marker genes were defined as genes with a log fold change >2, and adjusted p-value < 0.001. To examine marker gene enrichment in each bulk cell type, pairwise Wilcoxon tests were performed in R comparing the corresponding bulk cell type’s enrichment against the enrichment in all other bulk cell types.
To compare the overlap of gene detection between bulk and single cell datasets, bulk TMM counts were normalized to gene length, and the true positive rate (TPR) for detecting ground truth markers (see Thresholding) was calculated for a range of length normalized TMM values. At each expression threshold, if > 65% of samples showed expression equal to or higher than the threshold, the gene was called expressed. TPR, FPR, and FDR rates were calculated with 5,000 stratified bootstraps of the ground truth genes, which were generated using the R package boot (Canty and Ripley, 2019; Davison and Hinkley, 1997). We used a threshold of 5.7 length normalized TMM, to match the TPR (0.81) of the single cell Threshold 2. To calculate the relationship between single cell cluster size and the overlap between bulk and single cell gene expression, only protein coding genes were considered. Classifications from WormBase were used to define each gene’s RNA class.
Alternative Splicing
Alternative splicing events were detected using the software SplAdder (Kahles et al., 2016). The common splicing graph was built based on all 32 individual samples and each pair of neurons was tested for differential use of AS events (with confidence level of 3 and parameters –ignore-mismatches, --validate-sg and sg_min_edge_count=3). The resulting tables were loaded in R to adjust the p-value for multiple testing, and events with FDR > 0.1 were discarded. Sashimi plots for the genes mca-3 and mbk-2 were generated using the Integrated Genomics Viewer (Robinson et al., 2011).
For the previously unannotated exons, the splicing graph generated by SplAdder was recovered. It consisted of 197,576 exons; of these, 3,860 were not annotated in WormBase WS274. To avoid counting exons resulting from intron retention events or imprecise annotation of neighboring exons, we filtered out exons sharing their start and end positions with annotated exons, to keep 2,142 exons displaying an unannotated start or end. As many of these had extensive overlap with annotated exons, we further filtered the set to keep 63 exons, 42 of them displaying no overlap with annotated exons, and 21 exons having less than 90% of their sequence overlapping with annotated exons.
Generating connectivity matrices
We compiled membrane contact and chemical synapse matrices from published electron microscope reconstructions, N2U (Cook et al., 2019; White et al., 1986) and Adults 7 and 8 (Witvliet et al., 2020). Membrane contact data are available for N2U and Adult 8. Chemical synapse data was obtained from three adult animals (N2U, Adult 7 and Adult 8). These sources contain data for each individual neuron (e.g., for each of the six IL2 neurons). Data were summed across the individual neurons corresponding to each neuron type in the single-cell data (e.g., IL2DL, IL2DR, IL2VL, IL2VR were summed for the IL2_DV class, IL2L and IL2R were summed for IL2_LR). Only contacts and synapses present across all animals were retained to generate high confidence sets of invariant contacts and synapses.
Regulatory patterns of neuron transcriptomes
In order to identify distinct regulatory patterns for the transcriptome of each neuron, log-transformed expression values were converted to z-scores from the distribution of expression across all neurons for each gene. A high (low) z-score for a particular gene in a specific neuron type indicates an up-regulated (down-regulated) gene relative to the expression in other neurons. For motif discovery in promoters and 3’UTRs, gene z-scores were mapped to their isoform transcripts. Unique isoforms were maintained by applying a simple duplicate removal procedure, which guarantees that no pair of promoters and no pair of 3’UTRs will have a Blast local alignment with E-value < 10−10 (Elemento et al., 2007). For promoter sequences we considered sequences 1KB upstream of the transcriptional start site of each isoform, while for 3’UTRs we considered 1KB within the from the start of each annotated 3’UTR sequence (or 1KB downstream of the stop codon for transcripts without annotated 3’UTRs). To identify expression patterns of co-regulated transcripts, z-score values across all neuron types were clustered using hierarchical clustering with three different cut-offs (python/scipy fcluster implementation, cosine metric, criterion=‘distance’, cophenetic threshold= 1.2, 1.25, 1.37). We chose these thresholds to provide clustering of the data ranging from coarse to fine (16, 48, and 76 transcript clusters). For individual neurons, transcripts were categorized into bins with high to low z-scores based on the distribution of all z-scores across transcripts and neuron types. Z-score bin intervals were defined considering the following percentiles of the overall distribution of z-scores: 2.5%, 5%, 10%, 20%, 80%, 90%, 95%, 97.5%. For each neuron type, the top bin included transcripts with z-scores above the 97.5th percentile, the second to top included z-scores between the 95th and 97.5th percentile, etc. The bottom bin included transcripts with z-scores below the 2.5th percentile, the second to bottom included z-scores between the 2.5th and 5th percentile, etc. To avoid poorly populated bins, any given category containing less than 350 transcripts was merged with the next closest bin towards the center of the distribution.
Cis-regulatory element discovery
To systematically explore the regulatory effect of short DNA and RNA cis-regulatory elements, we utilized FIRE, a computational framework for de novo discovery of linear motifs in DNA and RNA whose presence or absence in a transcript’s promoter and 3’UTR regions is informative of regulatory patterns. We ran FIRE in discrete mode including transcript identifiers (Wormbase transcript IDs) along with either their z-score bin categories (for individual neurons) or transcript cluster IDs (for patterns of co-regulated genes). Over representation (yellow) and under representation (blue) patterns are shown for each discovered motif within each category (bin or cluster) of transcripts as well as mutual information (MI) values and z-scores associated with a randomization-based statistical test. All discovered motifs pass a three-fold jackknifing test more than 6 out of 10 times. Each time one-third of the transcripts was randomly removed and the statistical significance of the MI value of the motif was reassessed. For each of the 10 tests, the remaining two-thirds of the transcripts was shuffled 10,000 times and the motif was deemed significant if its MI was greater than all 10,000 MI scores from the randomized sets (Elemento et al., 2007). For every motif identified through FIRE, we defined the regulon for that motif as the collection of transcripts that harbored instances of the motif in their promoters (DNA motifs) or 3’UTRs (RNA motifs).
Motif families
Motifs with similar nucleotide compositions and regulons were discovered across individual neurons and gene expression patterns. We sought to identify the extent of redundancy between individual motifs and group them into motif families based on their similarity. We included additional motifs in this analysis for known transcription factors (CIS-BP, JASPAR), RNA binding proteins (CISBP-RNA) and miRNA 6-mer seeds (5’ extremity of known miRNA sequences of C. elegans). To quantify the similarity between nucleotide compositions between motifs we applied TOMTOM (MEME version 5.0.5). For each motif, we used its IUPAC motif sequence to convert it into a MEME formatted motif (iupac2meme function) as input to TOMTOM and compared it against all other discovered and known motifs. We specified a minimum overlap of 5, and an E-value threshold of 10 to identify significant matches. To quantify the extent of overlap between two motif modules, we defined a similarity measure between a module A and B as S(A, B) = (GA ∩ GB)/min(GA, GB), where GK is the set of transcripts in module K. We calculated TOMTOM and module similarity scores for all motif pairs. Module similarity scores were deemed significant if p<10−4 (hypergeometric test). To ensure that motifs are considered redundant only when they are similar both in nucleotide and module composition, we set the module similarity scores to 0 if either the TOMTOM or the module similarity scores were not significant. We clustered the motifs into motif families based on the masked similarity measures of all motif pairs using hierarchical clustering (python/scipy fcluster implementation, cosine metric, criterion=‘distance’, cophenetic threshold= 0.9). We set out to identify potential known regulators that represent a given motif family. To this end, we applied TOMTOM to match the motif family members with the binding preferences of known regulators. For each motif family, we counted all the significant TOMTOM scores for every family member compared to a known regulator. We considered a known regulator as a potential match for the motif family, if it had a significant TOMTOM score for more than 2/3 of the family members.
Associations of motif families and neurons
We set out to assess the regulatory potential of each motif family on each neuron type. Motifs with positive regulatory potential should have consistent patterns across the z-score bins, i.e., predominantly over-represented in genes with high z-scores or under-represented in genes with low z-scores. On the other hand, motifs with negative regulatory potential should be over-represented in genes with low z-scores or under-represented in genes with high z-scores. For each neuron type and each motif, we considered the frequency of transcripts carrying the motif in the top two z-score bins combined (ft), as well as the bottom two z-score bins (fb). To consider a positive association of the motif with the neuron type we required that the motif is: over-represented in the top two bins (p<0.005) and not over-represented in the bottom two bins (p>0.05), or, under-represented in the bottom (p<0.005) two bins and not under-represented in the top two bins (p>0.05). To consider a negative association of the motif with the neuron type we required that the motif is: over-represented in the bottom two bins (p<0.005) and not over-represented in the top two bins(p>0.05),or, under-represented in the top two bins (p<0.005) and not under-represented in the bottom two bins (p>0.05). We calculated a Log2-fold ratio (log2[R] =log2) and an associated p-value (hypergeometric test) between the two categories. We reported significant associations (|log2[R]| > 0.5 and p<10−5). For each motif family, we report the Log2-fold ratio and signed p-value (−sgn(log2(R))* log10(p)) for the motif member with the lowest p-value.
Cell adhesion molecule by stratum analysis
Given a set of gene expression profiles for the neurons classes in the nerve ring and their memberships in different strata, we can execute standard differential gene expression (DGE) analysis (Soneson and Delorenzi, 2013) to determine which genes are enriched in members of particular strata. Standard DGE analysis involves performing univariate t-tests between the gene expression levels of members of a particular stratum versus the members of all the remaining strata. The visual representation of this test can be seen in Methods S1. In detail, the DGE model involves fitting a regression model where the response variables are the gene expression levels for every neuron and the design matrix is a vector of 1s and −1s corresponding to the neurons in the two groups that are being compared. The gene expression is logarithm transformed to Gaussianize count-based data (Love et al., 2014). The output of this test is a vector of t-statistics and log-fold changes for every single gene in which this tuple of information can be visualized via volcano plots (Figure S8C). We deem that genes that pass the Bonferroni threshold for multiple comparisons (q<0.05) are significantly enriched or depleted in particular strata.
Network differential gene expression analysis
Whereas standard DGE analysis is useful for delineating univariate differences between groups of neurons, here we introduce a generalization of DGE, termed “network” DGE (nDGE), to establish the genetic determinants of synaptic formation and maintenance. Unlike DGE where gene expression levels of disjoint groups of neurons are compared, in nDGE, the multiplicative co-expression of genes, between sets of pairs of neurons (representing edges in a network) is compared. The visual representation of the nDGE statistical model can be seen in Methods S1. In nDGE, the response variables are the pairwise co-expression of all genes in all pairs of neurons. On the other hand, the design matrix captures two sets of pairs of neurons, one for each group. Similar to standard DGE, the output of this test is a set of t-statistics and log-fold changes for gene associations. However, unlike standard DGE, the t-statistics and log-fold changes in nDGE capture the effect of co-expression of pairs of genes, one corresponding to the gene observed in the pre-synaptic neuron partner and the other corresponding to the gene observed in the post-synaptic one. To deem a pair of genes significant under nDGE analysis, we also utilize the Bonferroni correction for p-values. However, the number of comparisons in nDGE is the square of the number of genes interrogated.
Since nDGE is a generalization of standard DGE, it enables the testing of a variety of hypotheses in addition to what is testable in standard DGE. The types of hypotheses that are tested are encoded in the design matrix of nDGE of which several examples are displayed in Methods S1. Methods S1 shows how standard DGE can be executed through nDGE, by placing 1s and −1s in the diagonal of the design matrix corresponding to the neuron groups. Three other types of hypotheses that can be tested are whether particular gene pairs have global effects of synaptic formation across all the neurons, whether there are differential gene co-expression differences in the synapses of two different neurons, or which gene co-expression patterns are implicated in the synapses of an individual neuron. In these scenarios, the design matrix has 1s where there is a synapse and a −1 where there is membrane contact, but no synapse, restricted to the sets of neurons of interest (all, pair, or one, respectively).
The main caveat in nDGE is the lack of independence of samples that are compared between groups. Since “samples” in nDGE are the co-expression of genes in pairs of neurons, the information from a particular neuron will inevitably be represented multiple times and possibly in different groups e.g., the gene expression from neuron AIA is represented in multiple synaptic gene co-expression values for all synaptic partners of AIA as well as the non-synaptic adjacent partners of AIA (Figure 7B). This lack of independence in the test samples can falsely inflate/deflate the sample variance, which can introduce excess false positives and false negatives. To accurately estimate the null distribution of the nDGE test statistics, we generate randomized “pseudoconnectomes” that respect the topology of the original connectome. Specifically, the pseudoconnectomes preserve the same number of synaptic partners for each neuron and the shuffled synaptic partners are confined to be neurons that have membrane contact (Milo et al., 2003). The latter constraint prevents infeasible pseudoconnectomes where synapses exist between neurons that do not share a membrane contact. Examples of pseudoconnectomes that are generated using the chemical connectome and membrane contact adjacency matrices are displayed in Methods S1. We execute nDGE analysis with the design matrices corresponding to 1000 pseudoconnectomes and compute a t-statistic using the mean and variance of the resulting null distribution.
While the nDGE technique introduced here is a generalization of standard DGE, interrogating the contribution of pairs of genes in the formation and maintenance of synapses between pairs of neurons, nDGE can only account for a single co-expressed gene in either of the two synaptic terminals (pre/post). For this reason, the nDGE model will tend to underestimate the effects of trimer (or higher-order) proteins in the formation and maintenance of synapses. Therefore, it is imperative to keep in mind that lack of significant hits for a particular neuron might not mean that there are no genes implicated in the formation of synapses for that neuron, but rather that higher-order gene interactions might be at play. Conceptually, it is straightforward to extend the model to higher-order gene interactions, but the prohibitive number of combinatorial gene co-expression enumeration is a computational bottleneck.
Another feature of nDGE is that it is a mass-univariate method, which does not take into account the possibility of interaction of different co-expressed genes in forming or inhibiting synapses. Therefore, the significance results output by nDGE tends to be very conservative with strict control of type 1 errors. This is in contrast with multivariate methods for explaining the genetic bases of connectivity (Kovacs et al., 2020). Due to the relatively high dimensionality of the gene expression data compared to the number of synapses in the chemical connectome, multivariate models tend to overfit and introduce type 1 errors.
QUANTIFICATION AND STATISTICAL ANALYSIS
Details of quantification and statistical testing, sample size, center and dispersion are found in the figure legends and STAR Methods Method Details section for individual analyses.
ADDITIONAL RESOURCES
Data files and information about the CeNGEN Consortium can be found at www.cengen.org. Single cell RNA-Seq data can be explored, analyzed and downloaded at the CengenAPP, found at cengen.shinyapps.io/CengenApp.
Supplementary Material
(A) Confocal images of the pan-neural marker, rab-3::TagRFP (otIs355), ventral cord cholinergic motor neurons marked with acr-2::GFP (juIs14) and neurons in the head region dual-labeled with the GABA neuron-specific reporter, unc-47::GFP (oxIs12) and the pan neural marker strain, rab-3::TagRFP. Scale bars = 10 μm. L4 animals were treated with SDS-DTT and dissociated with pronase to produce single-cell suspensions. Targeted subgroups of neurons were isolated by Fluorescence Activated Cell Sorting (FACS, red boxes) and collected for scRNA-Seq using the 10x Genomics 3’ platform. B) Results from 17 separate profiling experiments were submitted to a series of processing steps to produce a final merged data set (see Methods). Right panel shows UMAP projection of all 100,955 cells, colored by cluster. C) Graphical depiction of relative abundance of each neuron class in cells isolated from the pan-neural marker strain (rab-3::TagRFP). The fraction of observed cells for each neuron type was divided by the expected ratio (i.e., # neuron type/302) and annotated for each cell type according to heat map index. Note under-representation (dark blue/black) of neurons in the anterior pharynx, anterior ganglion and pre-anal ganglion (dashed lines). * For pairs ASE and AWC, the individual neurons are treated separately.
A) UMAP projection of 100,955 single cell profiles, colored by cluster. B) UMAP insets from A showing known markers that identify clusters of non-neuronal cells (unc-122 + galt-1, coelomocytes; myo-3 + pat-10, body wall muscles; sqt-3 and col-12, epidermis). Solid and dashed outlines correspond to similarly boxed sub-regions in A. C) UMAP projection of all cells as in panel A showing the neuropeptide processing gene sbt-1 is expressed in all neuronal clusters (blue-magenta) and largely absent from non-neuronal clusters (gray). D) Sub-UMAP of all non-neuronal cells, labeled by cell type. E) nspc-1, a member of the nematode-specific peptide c (nspc) gene family, is restricted to a single non-neuronal cluster (arrow). F) The transcriptional reporter nspc-1::GFP is exclusively expressed in the excretory gland cell (yellow). DIC (Differential Interference Contrast). Scale bar = 10 μm.
A-C) Higher magnification regions of neuronal UMAP from Figure 1B showing selective co-expression of genes used to assign cell identity to individual clusters. A) Co-expression of the glutamate receptor genes glr-3 and glr-6 demarcate the RIA cluster. B) Co-expression of the homeodomain transcription factors ceh-10 and lim-4 label the RID cluster. C) Co-expression of the transient receptor potential channel (trp) gene ocr-2 and tryptophan hydroxylase tph-1 label the sensory neuron ADF. D) Neuronal UMAP as in Figure 1B showing normalized expression of the nuclear hormone receptor nhr-236. nhr-236 is primarily detected in three clusters, corresponding to RIP, M2, and MC. E) Z-projection of confocal stack showing nhr-236::NLS-GFP expression in RIP and MC in a NeuroPAL strain. M2 (not shown) also consistently expressed GFP. Scale bar = 5 μm. F) Diagram denoting neurons (blue) in pharynx (gray). G) Sub-UMAP of all neuronal clusters expressing the pharyngeal neuron marker ceh-34 revealed independent clusters for each known pharyngeal neuron type. H-J) Higher magnification regions of pharyngeal UMAP in G showing expression of marker genes used to identify pharyngeal neuron classes. H) The M4-specific homeodomain transcription factor, ceh-28 is exclusively detected in a small but distinct group of 12 cells. I) ceh-2, a marker for I3, M3 and NSM pharyngeal neurons, is restricted to 3 clusters. J) Restricted expression of gur-3, a marker for I2 and I4 pharyngeal neurons.
A) High magnification regions of neuronal UMAPs (Figure 1B for ASE, AWC, RMD; Figure 1F for VC and RME; Figure 1E for DA, VA, DB) showing expression of known markers for neuron sub-types for ASE (gcy-3, ASER and gcy-6, ASEL), AWC (str-2, AWCON and srsx-3, AWCOFF), RMD (unc-42 + unc-46, RMD DV and unc-42 + cog-1, RMD LR), RME (unc-25 + lim-6, RME LR and unc-25 + slt-1, RME DV), VC (vab-7 + unc-4, VC1–3,6 and cat-1 + unc-4, VC4–5), DA9 (unc-53 + egl-5), VA12 (bnc-1 + egl-5) and DB (vab-7, DB1–7 and vab-7 + sptf-1, DB1). B) Volcano plots of genes that are differentially expressed between neuron subtypes. Inset depicts color coding for selected gene families.
A) Jitter plot of normalized ttx-1 expression (Y-axis) in all neuronal clusters (X-axis) shows strongest expression in AFD, M2, RIB and RIP. B) Confocal image of ttx-1crispr::GFP shows expression in AFD, M2, RIB and RIP neurons and in glia and epidermal cells.C) Receiver-Operator Characteristic (ROC) curve of True Positive Rate (TPR) vs False Positive Rate (FPR) for a range of thresholds (1–4) (red dots) compared to ground truth expression data (see Methods). Increased stringency diminishes both the TPR and FPR. Grey shading represents 95% confidence intervals. D) Thresholds 1–4 (red dots) plotted on Precision-Recall (PR) Curve of Recall (TPR) vs Precision [1 – False Discovery Rate (FDR)]. Grey shading represents 95% confidence intervals. E) The proportion of cells in each neuron-specific cluster expressing ttx-1. Inset shows expanded view of boxed region. Thresholds (1–4) are set to different proportions of ttx-1-expressing cells in each cluster (see Methods). Note that neurons RIB and M2, that show expression of native ttx-1crispr::GFP, are excluded by the thresholds 3 and 4. Numbers (1–4) correspond to thresholds in C and D. F) Jitter plot shows strong expression of unc-25/GAD in seven known GABAergic neuron types (AVL, DVB, RIB, RIS, RME, DD, VD) but scattered unc-25/GAD expression is also detected in other cell types. G) Bar graph plotting the proportion of cells in each neuron type that express unc-25/GAD. Thresholds of increasing stringency (1–4) are set to different proportions of cells in a given cluster that express unc-25/GAD. Note that threshold 2 distinguishes known unc-25/GAD-positive neurons from other neuron types with lower detected levels of unc-25/GAD (see Methods). H) Cell soma for each neuron type in the head, mid-body and tail regions are colored according to the number of genes detected using threshold 2. Neuron types in the anterior ganglion (dashed line) are among those with the fewest cells and also lower numbers of detected genes. I) Number of genes detected with threshold 2 for each neuron type plotted against the number of cells in each neuron-type cluster. Spearman’s rank correlation = 0.783, p < 2.2e-16). J) Number of genes detected with threshold 2 plotted against the True Positive Rate (TPR) for each neuron type. Spearman’s rank correlation = 0.678, p < 2.2e-16. Neurons with fewest cells, low TPR and number of genes/type are denoted (red). Related to Figure 4, 5, S9-S12. K) Number of genes detected plotted against the number of cells from down-sampling of ASJ cluster. L) Number of genes detected plotted against the number of cells from down-sampling for ASJ (red), NSM (green) and RIA (blue), with corresponding model fits.
A) Diagrams depicting upstream regions (black lines) in transcriptional GFP reporter genes. B) Heatmap showing single cell RNA-Seq expression of four selected neuropeptides (nlp-42, nlp-17, nlp-52, flp-33) and (C) corresponding confocal micrographs. Neuron types expressing each neuropeptide reporter were determined by co-localization with NeuroPAL markers (not shown). nlp-42p::gfp is robustly expressed in AIN and RMF with faint expression in NSM and PHB. nlp-17 expression at threshold 2 was 480x stronger in PVQ than for six additional neuron types and the nlp-17p::gfp reporter was selectively detected in PVQ. GFP reporters for nlp-52 and flp-33 (C) are largely congruent with single-cell RNA-Seq results (B). All scale bars = 10 μm.
A) Left: Motif family 109 includes highly similar motifs discovered in independent FIRE analyses from individual neurons. Center: Motifs from the CIS-BP database for six different K50 homeodomain transcription factors (CEH-36, CEH-37, TTX-1, CEH-53, UNC-30, CEH-45) match motif family 109. Right) Expression (TPM or transcripts per million) of two transcription factors (ceh-36, ceh-37) in our scRNA-Seq data corresponds to the neurons in which the cognate motif was discovered. B) The TAATCC motif (red boxes) in the 5’ regions of genes enriched in ASEL, ASER, AWCON or AWCOFF, including genes expressed in all four neurons (top rows) as well as genes expressed in subsets of these neurons. Expression of eat-4, tax-2, gcy-6, gcy-7 and srt-2 is ceh-36-dependent in these neurons. C) Schematic showing the z-score bins tested for relative enrichment for motif-neuron associations (see Methods). D) Motif-neuron associations (from FIRE Main Figure B for motif family 109 in a subset of neurons) showing correspondence to expression of K50 homeodomain proteins (black arrows). Colors indicate -log10(p-value) for positive associations and log10(p-value) for negative associations. Asterisks denote significant associations (pval < 1e-5, log fold change > 0.5). E) Top: Two similar motifs discovered in FIRE runs of individual neurons IL2_DV and ADL are members of motif family 184 that matches the DAF-19 motif from CIS-BP. Other members of the DAF-19 motif family were discovered in FIRE analysis of 8 additional individual neurons (black boxes). Bottom: Motif family 184 showed significant positive associations (asterisks) with 22 ciliated sensory neuron types (green boxes) and significant negative associations with some motor neurons (blue boxes with asterisks). F) Heatmap of log-transformed p-values showing motif-neuron associations for RNA motif families (columns) across all neuron types (rows). Note the strong positive signal in sensory neurons, and the one motif family with largely negative associations with neurons (blue arrow). G) Representative RNA motif (from ADL FIRE run) of family 23 showing a poly-C RNA sequence in the 3’ UTR.
A) Cartoon representation of nerve ring strata. Adapted with permission from Nature, Structural and developmental principles of neuropil assembly in C. elegans, Moyle et al., 2021. B) Heatmap of CAM expression (columns) in neurons grouped by strata (rows). Black arrows indicate four significantly enriched CAMs with the highest log fold changes between strata. Colors indicate log-transformed expression values. Red lines separate strata. C) Volcano plots showing log-fold change (x-axis) by -log10 p-value (y-axis) for each CAM in the neurons in each stratum compared to all other neurons. The six labeled genes were significantly enriched in the strata shown.
CellRanger and SoupX details for individual experiments, genes used to assign cell identity to clusters, counts of cell types by experiment, and stress-related transcripts.
Ground truth matrix used for thresholding, thresholding metrics (true positive rate, false positive rate, false discovery rate, including 95% confidence limits for each neuron), and parameters from model fitting to estimate the number of missing genes.
A list of the genes included in each gene family examined, and subsequent tabs with genes detected in a single neuron at each of the four thresholds.
Differential splicing results and previously unannotated exons.
Membrane contact matrix with nerve ring contacts common between two adult animals. Chemical synapse matrix for nerve ring synapses common between two adult animals.
Highlights.
Gene expression profiles of all 118 neuron classes in the C. elegans hermaphrodite
Each neuron type expresses a distinct code of neuropeptide genes and receptors
Expression profiles enable discovery of cell-type specific cis-regulatory sequences
Cell adhesion molecules correlate with neuron-specific connectivity
Acknowledgements
We thank the CeNGEN Advisory Board for guidance, M. Zhen for ZM9592, and H. Sun for imaging neuropeptide reporters. Funded by NIH R01NS100547 to MH, OH, DMM, NS, R01 NS110391 to OH, and by Vanderbilt TIPs to DMM. OH is an Investigator with the Howard Hughes Medical Institute. FACS [Flow Cytometry Shared Resource, supported by Ingram Cancer Center (P30 CA68485), DDRC (DK058404)], scRNA-Seq [VANTAGE, supported by CTSA (5UL1 RR024975-03), Ingram Cancer Center (P30 CA68485), Vision Center (P30 EY08126), NIH/NCRR (G20 RR030956)] and confocal imaging [Cell Imaging Shared Resource (NIH CA68485, DL20593, DK58404, DK59637, EY08126)] were performed at Vanderbilt. Strains were provided by the CGC, (NIH P40 OD010440A). G.S. was supported by “la Caixa” Foundation (LCF/BQ/PI19/11690010, ID 100010434) and by Ministerio de Ciencia e Innovación, Spain (PID2019-104700GA-I00).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 10X Genomics (2017). Application Note - Transcriptional Profiling of 1.3 Million Brain Cells with the Chromium Single Cell 3’ Solution.
- Adorjan I, Tyler T, Bhaduri A, Demharter S, Finszter CK, Bako M, Sebok OM, Nowakowski TJ, Khodosevich K, Møllgård K, et al. (2019). Neuroserpin expression during human brain development and in adult brain revealed by immunohistochemistry and single cell RNA sequencing. J. Anat. 235, 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, and Thomson JN (1976). The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 275, 299–325. [DOI] [PubMed] [Google Scholar]
- Allen AM, Neville MC, Birtles S, Croset V, Treiber CD, Waddell S, Goodwin SF, and Mann RS (2020). A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. Elife 9, e54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzalluz-Luqueángeles, and Conesa A. (2018). Single-cell RNAseq for the study of isoform-show is that possible? Genome Biol. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási DL, and Barabási AL (2020). A Genetic Model of the Connectome. Neuron 105, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A, McWhirter R, Taylor SR, Weinreb A, Miller DM, and Hammarlund M. (2021). An optimized ribodepletion approach for C. elegans RNA-sequencing libraries. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioRxiv. Baruch L, Itzkovitz S, Golan-Mashiach M, Shapiro E, and Segal E. (2008). Using expression profiles of Caenorhabditis elegans neurons to identify genes that mediate synaptic connectivity. PLoS Comput. Biol. 4, e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Aghayeva U, Berghoff EG, and Hobert O. (2019). Plasticity of the Electrical Connectome of C. elegans. Cell 176, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C, and Van Oudenaarden A. (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936. [DOI] [PubMed] [Google Scholar]
- Brittin CA, Cook SJ, Hall DH, Emmons SW, and Cohen N. (2021). A multi-scale brain map derived from whole-brain volumetric reconstructions. Nature 591, 105–110. [DOI] [PubMed] [Google Scholar]
- Bruce FM, Brown S, Smith JN, Fuerst PG, and Erskine L. (2017). DSCAM promotes axon fasciculation and growth in the developing optic pathway. Proc. Natl. Acad. Sci. U. S. A. 114, 1702–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunquell J, Morris S, Lu Y, Cheng F, and Westerheide SD (2016). The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics 17, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty A, and Ripley B. (2019). boot: Bootstrap R (S-Plus) Functions. R package version; 1.3–24. [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, and Hobert O. (2003). A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of c. elegans. Genes Dev. 17, 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Song KY, Law PY, Wei LN, and Loh HH (2009). Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun. 380, 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Ramos DA, Margeta MA, and Shen K. (2007). Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science 318, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo ME, Tran S, and Sengupta P. (2003). The Divergent Orphan Nuclear Receptor ODR-7 Regulates Olfactory Neuron Gene Expression via Multiple Mechanisms in Caenorhabditis elegans. Genetics 165, 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LTH, Bayer EA, Duerr JS, et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EA, Tuskey C, and Hardin J. (2004). Cell adhesion receptors in C. elegans. J. Cell Sci. 117, 1867–1870. [DOI] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, Bravo González-Blas C, et al. (2018). A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 982–998.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AC, and Hinkley DV (1997). Bootstrap Methods and their Application (Cambridge: Cambridge University Press; ). [Google Scholar]
- Dehghannasiri R, Olivieri JE, and Salzman J. (2020). Specific splice junction detection in single cells with SICILIAN. BioRxiv 10.1101/2020.04.14.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. (2002). A new family of putative insulin receptor-like proteins in C. elegans. Curr. Biol. 12, R155–7. [DOI] [PubMed] [Google Scholar]
- Driever W, and Nüsslein-Volhard C. (1989). The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337, 138–143. [DOI] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mark HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, and Swoboda P. (2005). Analysis of xbx genes in C. elegans. Development 132, 1923–1934. [DOI] [PubMed] [Google Scholar]
- Elemento O, Slonim N, and Tavazoie S. (2007). A Universal Framework for Regulatory Element Discovery across All Genomes and Data Types. Mol. Cell 28, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Pons C, Kozlowski L, Reece-Hoyes JS, Shrestha S, Holdorf AD, Mori A, Myers CL, and Walhout AJ (2016). A gene-centered C. elegans protein– DNA interaction network provides a framework for functional predictions. Mol. Syst. Biol. 12, 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Atlas EG, and Hobert O. (2016). A cellular and regulatory map of the GABAergic nervous system of C. elegans. Elife 5, e17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, Schnabel H, and Schnabel R. (1994). pha-1, a selectable marker for gene transfer in C.elegans. Nucleic Acids Res. 22, 1762–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, and Walhout AJM (2009). A Multiparameter Network Reveals Extensive Divergence between C. elegans bHLH Transcription Factors. Cell 138, 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S, Singer E, Waring D, and Jin Y. (2000). The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development 127, 4239–4252. [DOI] [PubMed] [Google Scholar]
- Hammarlund M, Hobert O, Miller DM, and Sestan N. (2018). The CeNGEN Project: The Complete Gene Expression Map of an Entire Nervous System. Neuron 99, 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, et al. (2020). WormBase: A modern Model Organism Information Resource. Nucleic Acids Res. 48, D762–D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Galvin BD, and Horvitz HR (2010). Six and eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 107, 15479–15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2002). PCR fusion-based approach to create reporter Gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728–730. [DOI] [PubMed] [Google Scholar]
- Hobert O. (2013). The neuronal genome of Caenorhabditis elegans. WormBook 1–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Glenwinkel L, and White J. (2016). Revisiting Neuronal Cell Type Classification in Caenorhabditis elegans. Curr. Biol. 26, R1197–R1203. [DOI] [PubMed] [Google Scholar]
- Husson SJ, Lindemans M, Janssen T, and Schoofs L. (2009). Comparison of Caenorhabditis elegans NLP peptides with arthropod neuropeptides. Trends Parasitol. 25, 171–181. [DOI] [PubMed] [Google Scholar]
- Inoue T, Sherwood DR, Aspöckb G, Butler JA, Gupta BP, Kirouac M, Wang M, Lee PY, Kramer JM, Hope I, et al. (2002). Gene expression markers for Caenorhabditis elegans vulval cells. Mech. Dev. 119. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Chang S, Etchberger JF, Ortiz CO, and Hobert O. (2005). MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. U. S. A. 102, 12449–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahles A, Ong CS, Zhong Y, and Rätsch G. (2016). SplAdder: Identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics 32, 1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, and Murphy CT (2016). The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Kaplan HS, Schrödel T, Skora S, Lindsay TH, Yemini E, Lockery S, and Zimmer M. (2015). Global Brain Dynamics Embed the Motor Command Sequence of Caenorhabditis elegans. Cell 163, 656–669. [DOI] [PubMed] [Google Scholar]
- Kaufman A, Dror G, Meilijson I, and Ruppin E. (2006). Gene expression of Caenorhabditis elegans neurons carries information on their synaptic connectivity. PLoS Comput. Biol. 2, e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk SY, Kratsios P, Hart M, Mourao R, and Hobert O. (2017). Diversification of C. elegans Motor Neuron Identity via Selective Effector Gene Repression. Neuron 93, 80–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, Van Der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. (2018). JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, and Emmons SW (2017). Multiple conserved cell adhesion protein interactions mediate neural wiring of a sensory circuit in C. elegans. Elife 6, e29257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, and Ohshima Y. (2004). The C. elegans ceh-36 Gene Encodes a Putative Homemodomain Transcription Factor Involved in Chemosensory Functions of ASE and AWC Neurons. J. Mol. Biol. 336, 579–587. [DOI] [PubMed] [Google Scholar]
- Kovacs IA, Barabási DL, and Barabási AL (2020). Uncovering the genetic blueprint of the C. elegans nervous system. BioRxiv 10.1101/2020.05.04.076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol U, Koziol M, Preza M, Costábile A, Brehm K, and Castillo E. (2016). De novo discovery of neuropeptides in the genomes of parasitic flatworms using a novel comparative approach. Int. J. Parasitol. 46, 709–721. [DOI] [PubMed] [Google Scholar]
- Lambert SA, Yang AWH, Sasse A, Cowley G, Albu M, Caddick MX, Morris QD, Weirauch MT, and Hughes TR (2019). Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 51, 981–989. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, and Sengupta P. (2003). Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell 5, 621–633. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Gehrke AR, Bulyk ML, and Bargmann CI (2009). Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev. 23, 345–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Veksler-Lublinsky I, and Zinovyeva A. (2019). HRPK-1, a conserved KH-domain protein, modulates microRNA activity during Caenorhabditis elegans development. PLoS Genet. 15, e1008067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sánchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, et al. (2009). Analysis of Cell Fate from Single-Cell Gene Expression Profiles in C. elegans. Cell 139, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo R, Onizuka M, Defrance M, and Laurent P. (2020). Combining single-cell RNA-sequencing with a molecular atlas unveils new markers for Caenorhabditis elegans neuron classes. Nucleic Acids Res. 48, 7119–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, and Marioni JC (2019). EmptyDrops: Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoudi N, Tavazoie S, Glenwinkel L, Ryu L, Kim K, and Hobert O. (2018). Unconventional function of an Achaete-Scute homolog as a terminal selector of nociceptive neuron identity. PLoS Biol. 16, e2004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, and Großberger L. (2018). UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3. [Google Scholar]
- Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, Taub D, Parker JA, Neri C, Gabel CV, et al. (2017). C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkman T, and Sengupta P. (2005). Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development 132, 1935–1949. [DOI] [PubMed] [Google Scholar]
- Milo R, Kashtan N, Itzkovitz S, Newman MEJ, and Alon U. (2003). On the uniform generation of random graphs with prescribed degree sequences. ArXiv https://arxiv.org/abs/cond-mat/0312028. [Google Scholar]
- Mirabeau O, and Joly JS (2013). Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. U. S. A. 110, E2028–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DZL, Sternberg PW, and Inoue T. (2015). Morphologically defined sub-stages of C. Elegans vulval development in the fourth larval stage. BMC Dev. Biol. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco JJ, and Koelle MR (2004). Activation of EGL-47, a Gαo-coupled receptor, inhibits function of hermaphrodite-specific motor neurons to regulate Caenorhabditis elegans egg-laying behavior. J. Neurosci. 24, 8522–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle MW, Barnes KM, Kuchroo M, Gonopolskiy A, Duncan LH, Sengupta T, Shao L, Guo M, Santella A, Christensen R, et al. (2021). Structural and developmental principles of neuropil assembly in C. elegans. Nature 591, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murru L, Moretto E, Martano G, and Passafaro M. (2018). Tetraspanins shape the synapse. Mol. Cell. Neurosci. 91, 76–81. [DOI] [PubMed] [Google Scholar]
- Nguyen JP, Shipley FB, Linder AN, Plummer GS, Liu M, Setru SU, Shaevitz JW, and Leifer AM (2016). Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 113, E1074–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Gao S, Norris ML, Ray D, Ramani AK, Fraser AG, Morris Q, Hughes TR, Zhen M, and Calarco JA (2014). A Pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell 54, 946–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TP, Cockburn K, Wang W, Tapia L, Currie E, and Bamji SX (2009). Semaphorin 5B mediates synapse elimination in hippocampal neurons. Neural Dev. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C, Etchberger J, Posy S, Frokjaer-Jensen C, Lockery S, Honig B, and Hobert O. (2006). Searching for neuronal left/right asymmetry: Genome wide analysis of nematode receptor-type guanylyl cyclases. Genetics 173, 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J, et al. (2019). A lineage-resolved molecular atlas of C. Elegans embryogenesis at single-cell resolution. Science 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick R, Humphreys D, Oshlack A, Ho JWK, Harvey RP, and Lo KK (2019). Sierra: Discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Rombauts S, Déhais P, Mathé C, Ramana DVV, Leroy P, and Rouzé P. (1999). Evaluation of gene prediction software using a genomic data set: Application to Arabidopsis thaliana sequences. Bioinformatics 15, 887–899. [DOI] [PubMed] [Google Scholar]
- Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, et al. (2015). A cellular and regulatory map of the cholinergic nervous system of C. Elegans. Elife 4, e12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SC, Watson JD, Richmond JE, Sarov M, Walthall WW, and Miller DM (2011). A transcriptional program promotes remodeling of GABAergic synapses in Caenorhabditis elegans. J. Neurosci. 31, 15362–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Faumont S, Gaston MR, Pearson BJ, and Lockery SR (2001). The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410, 694–698. [DOI] [PubMed] [Google Scholar]
- Pols MS, and Klumperman J. (2009). Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315, 1584–1592. [DOI] [PubMed] [Google Scholar]
- Poon VY, Klassen MP, and Shen K. (2008). UNC-6/netrin and its receptor UNC-5 locally exclude presynaptic components from dendrites. Nature 455, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, and Awatramani R. (2016). Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C. (2017a). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C. (2017b). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, and Blencowe BJ (2015). Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 87, 14–27. [DOI] [PubMed] [Google Scholar]
- Reilly MB, Cros C, Varol E, Yemini E, and Hobert O. (2020). Unique homeobox codes delineate all the neuron classes of C. elegans. Nature 584, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Colbert HA, and Bargmann CI (1994). The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell 79, 971–980. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, and Bargmann CI (1996). odr-10 Encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84, 899–909. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, and Hobert O. (2013). Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan G, Kim K, Li C, and Walthall WW (2005). Convergent genetic programs regulate similarities and differences between related motor neuron classes in Caenorhabditis elegans. Dev. Biol. 280, 494–503. [DOI] [PubMed] [Google Scholar]
- Shen K, and Bargmann CI (2003). The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112, 619–630. [DOI] [PubMed] [Google Scholar]
- Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, and Juliano CE (2019). Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler D, Enneking EM, Moreno E, and Pielage J. (2015). L1CAM/Neuroglian controls the axon-axon interactions establishing layered and lobular mushroom body architecture. J. Cell Biol. 208, 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Smbül U, Graybuck LT, Collman F, Seshamani S, Gala R, Gliko O, Elabbady L, Miller JA, Bakken TE, et al. (2019). Single-cell transcriptomic evidence for dense intracortical neuropeptide networks. Elife 8, e47889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, and Delorenzi M. (2013). A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, McWhirter R, Miller T, Strasbourger P, Thompson O, Hillier LDW, Waterston RH, and Miller DM (2014). Isolation of specific neurons from C. Elegans larvae for gene expression profiling. PLoS One 9, e112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry RW (1963). Chemoaffinity in theorderly growth of nerve fiber paterns and connections. Proc. Natl. Acad. Sci. USA 50, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanakis N, Carrera I, and Hobert O. (2015). Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina SE, Fox RM, Watkins KL, Starich TA, Shaw JE, and Miller DM (2007). UNC-4 represses CEH-12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans. Genes Dev. 21, 332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Swoboda P, Adler HT, and Thomas JH (2000). The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. Elegans. Mol. Cell 5, 411–421. [DOI] [PubMed] [Google Scholar]
- Tamburino AM, Ryder SP, and Walhout AJM (2013). A compendium of Caenorhabditis elegans RNA binding proteins predicts extensive regulation at multiple levels. G3 Genes, Genomes, Genet. 3, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Bixby R, Dalton R, Vandenburg A, Calarco JA, and Norris AD (2019). Splicing in a single neuron is coordinately controlled by RNA binding proteins and transcription factors. Elife 8, e46726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Naito Y, Kuroyanagi H, and Iino Y. (2016). Splicing factors control C. elegans behavioural learning in a single neuron by producing DAF-2c receptor. Nat. Commun. 7, 11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse NJ, Millet JRM, and Dupuy D. (2017). Quantitative RNA-seq meta-analysis of alternative exon usage in C. elegans. Genome Res. 27, 2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, and Kolodkin AL (2009). Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature 462, 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J, Gönczy P, Vashishtha M, Harris E, and Desplan C. (1989). A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell 59, 553–562. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, and Bargmann CI (1999). Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398. [DOI] [PubMed] [Google Scholar]
- Tursun B, Patel T, Kratsios P, and Hobert O. (2011). Direct conversion of C. elegans germ cells into specific neuron types. Science 331, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan V, Miller DM, and Anastassiou D. (2006). Computational inference of the molecular logic for synaptic connectivity in C. elegans. Bioinformatics 22, e497–506. [DOI] [PubMed] [Google Scholar]
- Venkatachalam V, Ji N, Wang X, Clark C, Mitchell JK, Klein M, Tabone CJ, Florman J, Ji H, Greenwood J, et al. (2016). Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 113, E1082–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal B, Aghayeva U, Sun H, Wang C, Glenwinkel L, Bayer EA, and Hobert O. (2018). An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 16, e2004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, and Zheng S. (2016). The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 17, 265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Li B, Nelson CE, and Nabavi S. (2019). Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics 20, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Southgate E, Thomson JN, and Brenner S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. London. B, Biol. Sci. 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Witvliet D, Mulcahy B, Mitchell JK, Meirovitch Y, Berger DR, Wu Y, Liu Y, Koh WX, Parvathala R, Holmyard D, et al. (2020). Connectomes across development reveal principles of brain maturation in C. elegans. BioRxiv 10.1101/2020.04.30.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemini E, Lin A, Nejatbakhsh A, Varol E, Sun R, Mena GE, Samuel ADT, Paninski L, Venkatachalam V, and Hobert O. (2021). NeuroPAL: A Multicolor Atlas for Whole-Brain Neuronal Identification in C. elegans. Cell 184, 272–288.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, and Behjati S. (2020). SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. Gigascience 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, and Garbers DL (1997). Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc. Natl. Acad. Sci. U. S. A. 94, 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lönnerberg P, Manno G. La, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang S, Banerjee D, and Kuhn JR (2011). Isolation and culture of larval cells from C. elegans. PLoS One 6, e19505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, and Maricq AV (1999). Neuronal Control of Locomotion in C. elegns is Modified by a Dominant Mutation in the GLR-1 Ionotropic Glutamate Receptor. Neuron 24, 347–361. [DOI] [PubMed] [Google Scholar]
- Zhou X, and Bessereau JL (2019). Molecular Architecture of Genetically-Tractable GABA Synapses in C. elegans. Front. Mol. Neurosci. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrández-Peral L, Gulden FO, et al. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Confocal images of the pan-neural marker, rab-3::TagRFP (otIs355), ventral cord cholinergic motor neurons marked with acr-2::GFP (juIs14) and neurons in the head region dual-labeled with the GABA neuron-specific reporter, unc-47::GFP (oxIs12) and the pan neural marker strain, rab-3::TagRFP. Scale bars = 10 μm. L4 animals were treated with SDS-DTT and dissociated with pronase to produce single-cell suspensions. Targeted subgroups of neurons were isolated by Fluorescence Activated Cell Sorting (FACS, red boxes) and collected for scRNA-Seq using the 10x Genomics 3’ platform. B) Results from 17 separate profiling experiments were submitted to a series of processing steps to produce a final merged data set (see Methods). Right panel shows UMAP projection of all 100,955 cells, colored by cluster. C) Graphical depiction of relative abundance of each neuron class in cells isolated from the pan-neural marker strain (rab-3::TagRFP). The fraction of observed cells for each neuron type was divided by the expected ratio (i.e., # neuron type/302) and annotated for each cell type according to heat map index. Note under-representation (dark blue/black) of neurons in the anterior pharynx, anterior ganglion and pre-anal ganglion (dashed lines). * For pairs ASE and AWC, the individual neurons are treated separately.
A) UMAP projection of 100,955 single cell profiles, colored by cluster. B) UMAP insets from A showing known markers that identify clusters of non-neuronal cells (unc-122 + galt-1, coelomocytes; myo-3 + pat-10, body wall muscles; sqt-3 and col-12, epidermis). Solid and dashed outlines correspond to similarly boxed sub-regions in A. C) UMAP projection of all cells as in panel A showing the neuropeptide processing gene sbt-1 is expressed in all neuronal clusters (blue-magenta) and largely absent from non-neuronal clusters (gray). D) Sub-UMAP of all non-neuronal cells, labeled by cell type. E) nspc-1, a member of the nematode-specific peptide c (nspc) gene family, is restricted to a single non-neuronal cluster (arrow). F) The transcriptional reporter nspc-1::GFP is exclusively expressed in the excretory gland cell (yellow). DIC (Differential Interference Contrast). Scale bar = 10 μm.
A-C) Higher magnification regions of neuronal UMAP from Figure 1B showing selective co-expression of genes used to assign cell identity to individual clusters. A) Co-expression of the glutamate receptor genes glr-3 and glr-6 demarcate the RIA cluster. B) Co-expression of the homeodomain transcription factors ceh-10 and lim-4 label the RID cluster. C) Co-expression of the transient receptor potential channel (trp) gene ocr-2 and tryptophan hydroxylase tph-1 label the sensory neuron ADF. D) Neuronal UMAP as in Figure 1B showing normalized expression of the nuclear hormone receptor nhr-236. nhr-236 is primarily detected in three clusters, corresponding to RIP, M2, and MC. E) Z-projection of confocal stack showing nhr-236::NLS-GFP expression in RIP and MC in a NeuroPAL strain. M2 (not shown) also consistently expressed GFP. Scale bar = 5 μm. F) Diagram denoting neurons (blue) in pharynx (gray). G) Sub-UMAP of all neuronal clusters expressing the pharyngeal neuron marker ceh-34 revealed independent clusters for each known pharyngeal neuron type. H-J) Higher magnification regions of pharyngeal UMAP in G showing expression of marker genes used to identify pharyngeal neuron classes. H) The M4-specific homeodomain transcription factor, ceh-28 is exclusively detected in a small but distinct group of 12 cells. I) ceh-2, a marker for I3, M3 and NSM pharyngeal neurons, is restricted to 3 clusters. J) Restricted expression of gur-3, a marker for I2 and I4 pharyngeal neurons.
A) High magnification regions of neuronal UMAPs (Figure 1B for ASE, AWC, RMD; Figure 1F for VC and RME; Figure 1E for DA, VA, DB) showing expression of known markers for neuron sub-types for ASE (gcy-3, ASER and gcy-6, ASEL), AWC (str-2, AWCON and srsx-3, AWCOFF), RMD (unc-42 + unc-46, RMD DV and unc-42 + cog-1, RMD LR), RME (unc-25 + lim-6, RME LR and unc-25 + slt-1, RME DV), VC (vab-7 + unc-4, VC1–3,6 and cat-1 + unc-4, VC4–5), DA9 (unc-53 + egl-5), VA12 (bnc-1 + egl-5) and DB (vab-7, DB1–7 and vab-7 + sptf-1, DB1). B) Volcano plots of genes that are differentially expressed between neuron subtypes. Inset depicts color coding for selected gene families.
A) Jitter plot of normalized ttx-1 expression (Y-axis) in all neuronal clusters (X-axis) shows strongest expression in AFD, M2, RIB and RIP. B) Confocal image of ttx-1crispr::GFP shows expression in AFD, M2, RIB and RIP neurons and in glia and epidermal cells.C) Receiver-Operator Characteristic (ROC) curve of True Positive Rate (TPR) vs False Positive Rate (FPR) for a range of thresholds (1–4) (red dots) compared to ground truth expression data (see Methods). Increased stringency diminishes both the TPR and FPR. Grey shading represents 95% confidence intervals. D) Thresholds 1–4 (red dots) plotted on Precision-Recall (PR) Curve of Recall (TPR) vs Precision [1 – False Discovery Rate (FDR)]. Grey shading represents 95% confidence intervals. E) The proportion of cells in each neuron-specific cluster expressing ttx-1. Inset shows expanded view of boxed region. Thresholds (1–4) are set to different proportions of ttx-1-expressing cells in each cluster (see Methods). Note that neurons RIB and M2, that show expression of native ttx-1crispr::GFP, are excluded by the thresholds 3 and 4. Numbers (1–4) correspond to thresholds in C and D. F) Jitter plot shows strong expression of unc-25/GAD in seven known GABAergic neuron types (AVL, DVB, RIB, RIS, RME, DD, VD) but scattered unc-25/GAD expression is also detected in other cell types. G) Bar graph plotting the proportion of cells in each neuron type that express unc-25/GAD. Thresholds of increasing stringency (1–4) are set to different proportions of cells in a given cluster that express unc-25/GAD. Note that threshold 2 distinguishes known unc-25/GAD-positive neurons from other neuron types with lower detected levels of unc-25/GAD (see Methods). H) Cell soma for each neuron type in the head, mid-body and tail regions are colored according to the number of genes detected using threshold 2. Neuron types in the anterior ganglion (dashed line) are among those with the fewest cells and also lower numbers of detected genes. I) Number of genes detected with threshold 2 for each neuron type plotted against the number of cells in each neuron-type cluster. Spearman’s rank correlation = 0.783, p < 2.2e-16). J) Number of genes detected with threshold 2 plotted against the True Positive Rate (TPR) for each neuron type. Spearman’s rank correlation = 0.678, p < 2.2e-16. Neurons with fewest cells, low TPR and number of genes/type are denoted (red). Related to Figure 4, 5, S9-S12. K) Number of genes detected plotted against the number of cells from down-sampling of ASJ cluster. L) Number of genes detected plotted against the number of cells from down-sampling for ASJ (red), NSM (green) and RIA (blue), with corresponding model fits.
A) Diagrams depicting upstream regions (black lines) in transcriptional GFP reporter genes. B) Heatmap showing single cell RNA-Seq expression of four selected neuropeptides (nlp-42, nlp-17, nlp-52, flp-33) and (C) corresponding confocal micrographs. Neuron types expressing each neuropeptide reporter were determined by co-localization with NeuroPAL markers (not shown). nlp-42p::gfp is robustly expressed in AIN and RMF with faint expression in NSM and PHB. nlp-17 expression at threshold 2 was 480x stronger in PVQ than for six additional neuron types and the nlp-17p::gfp reporter was selectively detected in PVQ. GFP reporters for nlp-52 and flp-33 (C) are largely congruent with single-cell RNA-Seq results (B). All scale bars = 10 μm.
A) Left: Motif family 109 includes highly similar motifs discovered in independent FIRE analyses from individual neurons. Center: Motifs from the CIS-BP database for six different K50 homeodomain transcription factors (CEH-36, CEH-37, TTX-1, CEH-53, UNC-30, CEH-45) match motif family 109. Right) Expression (TPM or transcripts per million) of two transcription factors (ceh-36, ceh-37) in our scRNA-Seq data corresponds to the neurons in which the cognate motif was discovered. B) The TAATCC motif (red boxes) in the 5’ regions of genes enriched in ASEL, ASER, AWCON or AWCOFF, including genes expressed in all four neurons (top rows) as well as genes expressed in subsets of these neurons. Expression of eat-4, tax-2, gcy-6, gcy-7 and srt-2 is ceh-36-dependent in these neurons. C) Schematic showing the z-score bins tested for relative enrichment for motif-neuron associations (see Methods). D) Motif-neuron associations (from FIRE Main Figure B for motif family 109 in a subset of neurons) showing correspondence to expression of K50 homeodomain proteins (black arrows). Colors indicate -log10(p-value) for positive associations and log10(p-value) for negative associations. Asterisks denote significant associations (pval < 1e-5, log fold change > 0.5). E) Top: Two similar motifs discovered in FIRE runs of individual neurons IL2_DV and ADL are members of motif family 184 that matches the DAF-19 motif from CIS-BP. Other members of the DAF-19 motif family were discovered in FIRE analysis of 8 additional individual neurons (black boxes). Bottom: Motif family 184 showed significant positive associations (asterisks) with 22 ciliated sensory neuron types (green boxes) and significant negative associations with some motor neurons (blue boxes with asterisks). F) Heatmap of log-transformed p-values showing motif-neuron associations for RNA motif families (columns) across all neuron types (rows). Note the strong positive signal in sensory neurons, and the one motif family with largely negative associations with neurons (blue arrow). G) Representative RNA motif (from ADL FIRE run) of family 23 showing a poly-C RNA sequence in the 3’ UTR.
A) Cartoon representation of nerve ring strata. Adapted with permission from Nature, Structural and developmental principles of neuropil assembly in C. elegans, Moyle et al., 2021. B) Heatmap of CAM expression (columns) in neurons grouped by strata (rows). Black arrows indicate four significantly enriched CAMs with the highest log fold changes between strata. Colors indicate log-transformed expression values. Red lines separate strata. C) Volcano plots showing log-fold change (x-axis) by -log10 p-value (y-axis) for each CAM in the neurons in each stratum compared to all other neurons. The six labeled genes were significantly enriched in the strata shown.
CellRanger and SoupX details for individual experiments, genes used to assign cell identity to clusters, counts of cell types by experiment, and stress-related transcripts.
Ground truth matrix used for thresholding, thresholding metrics (true positive rate, false positive rate, false discovery rate, including 95% confidence limits for each neuron), and parameters from model fitting to estimate the number of missing genes.
A list of the genes included in each gene family examined, and subsequent tabs with genes detected in a single neuron at each of the four thresholds.
Differential splicing results and previously unannotated exons.
Membrane contact matrix with nerve ring contacts common between two adult animals. Chemical synapse matrix for nerve ring synapses common between two adult animals.
Data Availability Statement
The raw data are available at GEO (single cell data: Accession Number GSE136049, bulk sequence data: Accession Number GSE169137). The full and neuron only datasets are available at www.cengen.org. Analysis code is available at github https://github.com/cengenproject.