Abstract
We previously described the identification of quail MafA, a novel transcription factor of the Maf bZIP (basic region leucine zipper) family, expressed in the differentiating neuroretina (NR). In the present study, we provide the first evidence that MafA is phosphorylated and that its biological properties strongly rely upon phosphorylation of serines 14 and 65, two residues located in the transcriptional activating domain within a consensus for phosphorylation by mitogen-activated protein kinases and which are conserved among Maf proteins. These residues are phosphorylated by ERK2 but not by p38, JNK, and ERK5 in vitro. However, the contribution of the MEK/ERK pathway to MafA phosphorylation in vivo appears to be moderate, implicating another kinase. The integrity of serine 14 and serine 65 residues is required for transcriptional activity, since their mutation into alanine severely impairs MafA capacity to activate transcription. Furthermore, we show that the MafA S14A/S65A mutant displays reduced capacity to induce expression of QR1, an NR-specific target of Maf proteins. Likewise, the integrity of serines 14 and 65 is essential for the MafA ability to stimulate expression of crystallin genes in NR cells and to induce NR-to-lens transdifferentiation. Thus, the MafA capacity to induce differentiation programs is dependent on its phosphorylation.
Transcription factors of the Maf family contain a conserved basic region leucine zipper (bZIP) domain, which is responsible for their dimerization and DNA binding property and is closely related to the bZIP domain of the AP1 superfamily members. Accordingly, Maf protein dimers bind to a DNA sequence motif similar to the tetradecanoyl phorbol acetate (TPA)-responsive and cyclic AMP-responsive recognition sequences for AP1 proteins: the Maf recognition element (MARE) TGCTGAC(G)TCAGCA (29). Three members of the Maf family contain the bZIP domain only and are collectively referred to as small Mafs: MafG (28), MafF, and MafK (11). In contrast, the four large Mafs (c-Maf, MafB, NRL, and MafA [also known as L-Maf]) carry an N-terminal transactivating domain which is extensively conserved and is rich in proline, serine, and threonine residues. The family prototype, c-maf (45), was discovered as an oncogene transduced in the avian AS42 retrovirus. Avian mafB, like the small members, was isolated by homology screening (27). NRL was identified in both human and murine retina (61). Finally, mafA (or L-maf) was recently isolated by two different approaches. We previously reported the identification of quail mafA by a neuroretina (NR)-specific cDNA screening based on homologies between large Maf proteins (2). The same gene, designated L-maf, was simultaneously cloned by using a functional screening for chicken proteins able to bind the promoter of the αA-crystallin gene (46). We showed that MafA expression is developmentally regulated in the NR with its highest levels coinciding with NR differentiation. MafA synthesis also correlates with the expression of an NR quiescence and differentiation marker, QR1, encoded by one of the first genes identified as a target for Maf transcription factors (52). Accordingly, we showed that MafA is able to transactivate the QR1 promoter in transient transfection assays. MafA, which is expressed in the lens placode as early as the onset of lens formation, was suggested, meanwhile, to be a master gene for lens differentiation. When expressed ectopically in chick embryos, it is able to enhance expression of the lens-specific α-, β-, and δ-crystallin genes, whose promoters contain MARE sequences, and to promote transdifferentiation of NR cells into lens fibers (46). Based on genetic analysis, murine c-Maf was also recently demonstrated to be a key regulator of lens development, probably at later stages (32, 34, 56). More generally, Maf proteins are emerging as important regulators of cell differentiation in various tissues, in particular in the eye. Besides c-Maf and MafA, NRL was suggested to be of crucial importance in the photoreceptor visual function by activating transcription of the rhodopsin gene (35, 54). In addition, MafB was shown to be implicated both in rhombencephalon development, by regulating expression of hoxa-3 and hoxb-3 genes (39, 40), and in monocytic differentiation (33, 58). Maf family members also seem to play a role in various hematopoietic lineages. One consistent example is that of the small Maf proteins which, likely as heterodimers with the p45 NF-E2 bZIP protein, are implicated in the upregulation of the β-globin locus during erythroid differentiation (24, 47).
In spite of a clearly established role of Maf family transcription factors in regulating differentiation processes, regulation of their activity remains poorly documented. In particular, nothing is known concerning their posttranslational modifications. It was reported that the NF-E2 complex is targeted by ERK1/2 mitogen-activated protein (MAP) kinase (MAPK) and protein kinase A signaling pathways during erythroid differentiation (12, 44, 62), but the molecular bases of this effect are still unclear. The activity of many transcription factors is regulated in a rapid and reversible manner by phosphorylation. Key players responsible for this process are protein kinases which act in signaling cascades initiated by extracellular stimuli. Among the pathways frequently used to transduce these signals are the highly conserved MAPK cascades. These cascades are found in all eukaryotic cells and regulate many crucial processes such as cell division, differentiation, apoptosis, and response to stress (reviewed in reference 57). Once activated, the MAPKs can translocate into the nucleus where they phosphorylate cognate nuclear targets. Many transcription factors, notably some playing a role in the eye development, have been shown to be targeted by MAPK pathways. Among these, Microphthalmia (19) and the Drosophila Yan and pointed P2 proteins of the Ets family (5) are phosphorylated by ERKs, c-Jun (21) and ATF2 (16) are phosphorylated by JNKs, and CHOP (63), Pax6 (41), and MEF2C (17) are phosphorylated by p38. Various functional consequences of phosphorylation on transcription factors have been observed, such as alteration of protein stability (43), cellular localization (8), DNA binding (64), interaction with partners (37), and transactivation capacity (3). Thus, phosphorylation cascades, elicited at the cellular membrane, have the potential to turn extracellular signals into specific and finely tuned nuclear responses.
We report here, for the first time, that the activity of a Maf transcription factor is strongly dependent on phosphorylation. We show that the MafA protein is constitutively detected as a phosphoprotein in both NR and HeLa cells and that serine 14 and serine 65 residues, which are conserved among all large Maf proteins, constitute the major phosphorylation sites. Mutation of these residues into alanine severely affected both phosphorylation of the protein and its transactivating capacity, suggesting that they are the critical sites for the functional regulation of MafA. Moreover, we show that phosphorylation of MafA is crucial for its biological activity, since mutagenesis of these residues leads to a dramatic decrease in the capacity of MafA to both activate QR1 expression and induce lens transdifferentiation of NR cells. Thus, the transcriptional capacity and, consequently, the biological function of MafA appear to be directly determined by phosphorylation on serines 14 and 65. Since these residues fit the consensus motif for phosphorylation by the kinases of the MAPK subgroup, we also investigated whether members of this family could be responsible for this regulation. We show that ERK2 but not other MAPK phosphorylated MafA serine 14 and serine 65 residues in vitro, but that activation or inhibition of the MEK/ERK cascade only slightly affected the status of MafA phosphorylation in vivo. Taken together, these results suggest that another kinase(s) is responsible for the in vivo activation of MafA protein.
MATERIALS AND METHODS
Plasmids and constructs.
A bacterial expression vector for wild-type MafA was constructed by subcloning the MafA full-length open reading frame, obtained by PCR inserting flanking EcoRI sites and Kozak consensus, into the pBluescript vector (Stratagene). This plasmid was used as a template for site-directed mutagenesis (Transformer site-directed mutagenesis kit, Clontech) to substitute serine 14 and serine 65 with alanine. The following oligonucleotides were used: GCTGCCCACCGCCCCCCTCGCC for S14 and CCGTGCCTTCCGCGCCCAGTTTC for S65 (modified bases are underlined).
pcDNA3-MafA plasmids encoding either wild-type (WT) or mutated MafA were obtained by subcloning the EcoRI wild-type or mutated inserts of the pBluescript plasmid into the EcoRI site of a pcDNA3-modified vector, in frame with the HA1 epitope coding sequence.
RCAS-MafA (kindly provided by Marc Castellazzi) and RCAS-S14A/S65A plasmids were obtained by cloning the EcoRI MafA open reading frame into the Cla12 shuttle vector and subsequent ClaI insertion into the RCAS vector (23).
For the synthesis of glutathione S-transferase (GST)-MafA recombinant proteins, the MafA EcoRI fragment, either WT or mutated, was subcloned into the EcoRI site of the pGex-4T1 vector (Amersham Pharmacia Biotech). For the synthesis of GST-Nter proteins, an EagI deletion, eliminating the bZIP domain, was performed in the previous construct. For the synthesis of GST-Cter proteins, an EagI fragment corresponding to the MafA bZIP domain was subcloned into the NotI site of pGex-4T2 (Amersham Pharmacia Biotech).
Reporter constructs, either empty (TK10) or carrying a multimerized A box (4×Abox) upstream of the thymidine kinase promoter, were previously described (52).
pcDNA3-derived vectors encoding the S222A and S218D/S222D mutants of MEK1 were previously described (50).
The expression vector for HA1-ERK2 (20) was kindly provided by Michael J. Weber. Expression vectors for HA1-JNK1 (9) and HA1-p38α (4) were kindly provided by Roger J. Davis. GST-Jun/1–79 (21) and GST-ATF2/1–109 (16) recombinant proteins were a kind gift from Joël Raingeaud. Expression vectors for HA1-ERK5 and for the synthesis of GST-MEF2C were kind gifts from Silvio Gutkind (7). The expression vector for MEK5DD was kindly provided by Melanie Cobb (10).
Antibodies.
To identify mafA gene products we prepared a rabbit polyclonal antiserum raised against a MafA-specific peptide encompassing residues 131 to 153. The corresponding sequence was obtained by PCR and subcloned into the pLC24 vector (55), in frame with the bacteriophage MS2 polymerase. The bacterially expressed fusion protein was purified and used for immunization. Antiserums directed against chicken α-, β-, and δ-crystallins were kindly provided by Joram Piatigorsky (42, 48). QR1-directed antiserum was previously described (6).
Cell culture and transfection assays.
HeLa cells were maintained and transfected in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). For TPA stimulation experiments, cultures were starved for 6 to 8 h in DMEM containing 0.1% FBS and stimulated by the addition of 160 μM TPA (Sigma Aldrich) for 5 min, in the presence or not of 30 μM PD98059 (Alexis Biochemicals; added 2 h prior to the addition of TPA).
Quail and chicken NR cells were dissected from 7- and 8-day-old embryos, respectively, dissociated, and put in culture as previously described (49). NR cell cultures were maintained and transfected in basal medium of Eagle supplemented with 10% FBS. HeLa and NR cell cultures were transfected according to the standard calcium phosphate coprecipitation technique, as previously described (52). Twenty-four hours prior to transfection, NR and HeLa cells were seeded at a density of 8 × 106 to 10 × 106 and 1.5 × 106 cells per 100-mm dish, respectively. Transfection was carried out with a total of 20 μg of DNA. When pcDNA3-MafA constructs were cotransfected with expression vectors for MEK1, a 1/2 MafA/MEK1 ratio was used. For HeLa cells, the precipitate was left 24 h and then the medium was changed and the cells were lysed 24 h later. For NR cells, the precipitate was left 5 h, the cultures were washed 4 to 5 times with phosphate-buffered saline (PBS), and new medium was then added.
Cell extracts were prepared in lysis buffer (140 mM NaCl, 20 mM Tris [pH 8.0], 2 mM EDTA, 1/10 [vol/vol] glycerol, 1/100 [vol/vol] NP-40, supplemented with protease and phosphatase inhibitors) and centrifuged at 21,000 × g (20 min, 4°C). The concentration of total soluble proteins in the supernatant was quantified using the Bradford reagent (Bio-Rad).
CAT assays.
MafA transcriptional activity was examined in quail embryo NR cells transfected with tsNY68, a temperature-sensitive variant of Rous sarcoma virus. We previously showed that transcription of the QR1 gene is shut down in infected quail embryo NR cells maintained at 37°C and restored when the cultures are shifted to 41°C, the temperature at which the v-Src oncoprotein is inactivated (14, 15). Actively proliferating cells were seeded at 9 × 105 cells per 100-mm-diameter dish and transfected 48 h later by the calcium phosphate technique, as described previously (52). Cultures were shifted to 41°C 2 h after transfection. Each transfection mixture contained 1 μg of the respective reporter plasmids, 3 μg of MafA expression vectors, and 2 μg of β-actin–lacZ (1) internal control. Cells were harvested 48 h later, and chloramphenicol acetyltransferase (CAT) activity was measured (13) after normalization according to β-galactosidase activity. Activity of distinct chloramphenicol substrate and products, following thin-layer chromatography, was determined by using the PhosphorImager instrument (Molecular Dynamics).
Metabolic labeling.
HeLa cultures transfected with pcDNA3-MafA plasmids were left 4 h in phosphate-free DMEM (ICN) containing 2% dialyzed newborn calf serum and then labeled for 5 h by adding 1 m Ci of [32P]- or [33P]orthophosphate (Amersham Pharmacia Biotech) per ml. The dishes were placed on ice and washed twice in ice-cold PBS, and the cells were harvested by scrapping in radioimmunoprecipitation assay (RIPA) buffer (PBS containing 20% sodium dodecyl sulfate [SDS], 0.5 M EDTA, 1% aprotinine, 250 mM deoxycholic acid, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 1 mM orthovanadate, 25 mM β-glycerophosphate) and centrifuged at 21,000 × g (20 min, 4°C). The concentration of total soluble proteins in the supernatant was quantified using the Bradford reagent (Bio-Rad).
Immunoprecipitation and phosphatase treatment.
Cellular extract (250 μg of total proteins) was incubated (2 h, 4°C) with 2 μl of anti-MafA or preimmune serum and proteine A-Sepharose (Sigma Aldrich), centrifuged, washed in RIPA buffer, and resuspended in 2× SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer.
For λ-phosphatase treatment, washed immunoprecipitates were split in two and washed twice in 10 mM Tris-HCl (pH 7.4)–150 mM NaCl–1 mM EDTA buffer and the beads were resuspended in 50 μl of λ-phosphatase buffer (New England Biolabs). The reaction was carried out (30 min, 30°C) in one of the tubes by addition of 80 U of λ-phosphatase (New England Biolabs) and stopped by addition of 2× SDS-PAGE loading buffer.
Western blot analysis.
Immunoprecipitates resuspended in loading buffer or samples (100 to 300 μg) of total protein extracts were run on SDS-10 to 12% polyacrylamide gels and transferred to Immobilon-p membranes (Millipore). The membranes were incubated with the following primary antibodies (overnight, 4°C) diluted in blocking buffer TBST (Tris-buffered saline containing 0.2% Tween 20, supplemented with 5% nonfat dry milk): anti-HA1 12CA5 monoclonal antibody (Roche; 100 ng/ml), anti-MafA (dilution, 1/1,000), anti-GST (Amersham Pharmacia Biotech; 500 ng/ml), anti-β-actin (Sigma Aldrich; 1/5,000), anti-ERK1 (Santa Cruz Biotechnology; 1/2,000), anti-phospho-ERK (New England Biolabs; 1/1,000), anti-α-, β-, or δ-crystallin (1/1,000 to 1/2,000), and anti-QR1 (1/8,000). After four washes in TBST buffer, membranes were incubated with the following secondary antibodies conjugated to horseradish peroxidase and diluted in blocking buffer: swine anti-mouse antibody for anti-HA1 and anti-β-actin (Amersham Pharmacia Biotech; 1/10,000); donkey anti-rabbit antibody (Amersham Pharmacia Biotech; 1/20,000) for anti-MafA, anti-ERK1, anti-phospho-ERK, and anti-QR1; rabbit anti-goat antibody (Sigma Aldrich; 1/8,000) for anti-GST; and monoclonal anti-goat or -sheep antibody (Sigma Aldrich; 1/10,000) for anti-α-, β-, and δ-crystallin antibodies. Membranes were further processed using the chemiluminescence Super Signal Ultra reagent (Pierce).
Phosphoamino acid analysis.
Phosphoamino acid analysis of MafA protein excised from Western blot membrane and fixed in methanol was performed by acid hydrolysis (6 M HCl, 1 h, 110°C). The hydrolyzed protein was dried out and resuspended in 10/100/1,890 pyridine-acetic acid-H2O (pH 3.5) and subjected to thin-layer electrophoresis.
In vitro protein kinase assays.
GST fusion recombinant proteins were purified from Escherichia coli DH5α extracts using glutathione-Sepharose beads according to the manufacturer's recommendations (Amersham Pharmacia Biotech). Proteins were eluted by addition of 50 mM glutathione and stored at −80°C. HeLa cells were transiently transfected with expression vectors encoding either HA1-tagged ERK2, JNK1, or p38α MAPKs or cotransfected with expression vectors for MEK5DD and HA1-tagged ERK5. Twenty-four hours after transfection, ERK2-expressing cells were starved for 2 h and were treated for 10 min with TPA (160 μM) prior to lysis. JNK1- and p38α-expressing cells were starved for 2 h and left untreated or treated for 30 s with UV-C (80 J/m2) and then incubated another 40 min at 37°C prior to lysis. HA1-tagged MAPKs were immunoprecipitated from cell extracts by incubation for 2 h at 4°C with 12CA5 anti-HA1 antibody (Roche) bound to protein A-Sepharose beads (Sigma Aldrich). The immune complexes were washed in kinase buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM dithiothreitol, 1 mM orthovanadate), and equal amounts were added to 0.5 μg of recombinant proteins, 1 mM ATP and 10 μCi of [γ-32P]ATP. The reaction was carried out 30 min at 30°C, stopped by addition of 2× SDS-PAGE loading buffer, and subjected to Western blot analysis and autoradiography. Kinase assays (see Fig. 4B) were carried out with 5 U of recombinant activated ERK2 (New England Biolabs).
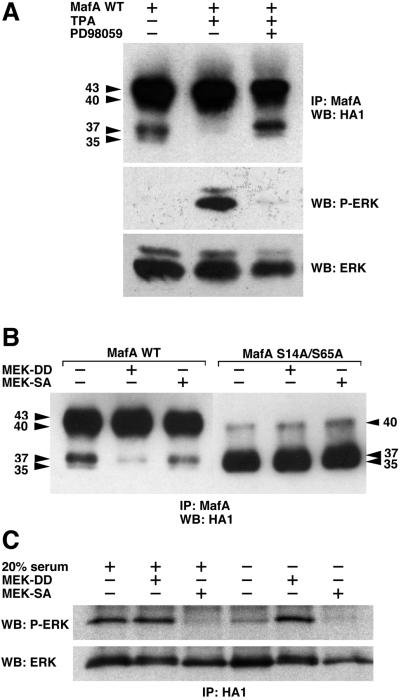
FIG. 4.
MafA serines 14 and 65 can be phosphorylated in vivo in response to activation of the MEK/ERK pathway. (A) HeLa cells were transiently transfected with pcDNA3-derived vectors encoding WT MafA and either serum-starved (−), treated with TPA (TPA), or treated with TPA in the presence of PD98059 (PD98059). Cellular extracts were immunoprecipitated with MafA-directed antiserum and analyzed by Western blotting with anti-HA1 antibody. The status of ERK activation was controlled as follows: protein extracts from identical cultures were analyzed by Western blotting with anti-phospho-ERK antibody, stripping of the membranes, and reprobing with anti-ERK antibody. (B) HeLa cells were transiently transfected with pcDNA3-derived vectors encoding either WT MafA (WT) or S14A/S65A MafA (S14A/S65A) and with expression vectors for MEK1S218D/S222D (MEK-DD), MEK1S222A (MEK-SA), or empty vector (−). Cell lysates were treated as described for panel A. (C) Control of the constitutively active and dominant negative property of the respective S218D/S222D and S222A MEK1 mutants. Cultures of HeLa cells were transfected as described for panel B, with the addition of an HA-ERK2 expression plasmid in the transfection mix. The status of ERK activation in the presence of either MEK-DD or MEK-SA was analyzed under two experimental conditions. In the first three lanes (left side), serum-starved cultures were stimulated for 10 min by addition of medium containing 20% FBS. In the last three lanes (right side), cells were maintained in culture without any additional stimulation, as in panel B. Cellular extracts were immunoprecipitated with HA1-directed antibody and analyzed by Western blotting with anti-phospho-ERK antibody, stripping of the membranes, and reprobing with anti-ERK antibody.
Gel shift analysis.
MafA proteins encoded by the pcDNA3 vector were obtained by in vitro transcription and translation in reticulocyte lysates (TNT-coupled reticulocyte lysate system; Promega) and used in electrophoretic mobility shift assays. Double-stranded oligonucleotide corresponding to the 48-bp A box region was labeled with polynucleotide kinase and [γ-32P]ATP and incubated with MafA proteins 20 min at 20°C in binding buffer. Complexes were resolved on 5% polyacrylamide gels and subjected to autoradiography.
RT-PCR.
Total RNA was prepared using the RNA Now extraction kit (In Vitrogen). Reverse transcription (RT) was carried out with 1 μg of total RNA by using the Promega reverse transcription system. PCR was performed with Taq DNA polymerase (Promega) in the presence of 200 μM deoxynucleoside triphosphate and 1 μM primers. Primer pairs, number of cycles, and annealing temperatures were as follows: MafA (5′TTCCACCCCTCTCAGCAC and 5′CTCCCGAACCGACATACT; 26 cycles; 58°C); αA-crystallin (5′GCCTTTGTTCTCCTCCACTATCAG and 5′GTGGAACTCACGAGAGATGTAGC; 26 cycles; 56°C); βB1-crystallin (5′AGCAGCTGCCCAGACCCGAG and 5′GCTGACGATGACACTGCCCAC; 26 cycles; 60°C); δ1-crystallin (5′CTGAGCTGGAGAAGATCCTGAG and 5′TCCACCAGGGTCTTGATGAGC; 23 cycles; 56°C); and S17 (5′TACACCCGTCTGGGCAACGAC and 5′CCGCTGGATGCGCTTCATCAG; 23 cycles; 58°C).
Immunofluorescence.
Chicken NR cells or HeLa cells were transfected, respectively, with RCAS- or pcDNA3-derived plasmids encoding WT or mutant MafA proteins and were seeded on coverslips precoated with poly-l-lysine. Forty-eight hours (HeLa cells) or 6 days (NR cells) after transfection, cells were fixed in 4% paraformaldehyde for 20 min and permeabilized in 0.25% Triton X-100 for 15 min. Coverslips were then blocked with 5% PBS-FBS for 1 h and incubated with primary antibodies (1/1,000) for 1 h at room temperature. After washing with PBS, cells were incubated with secondary antibodies for 1 h at room temperature: rhodamine-conjugated donkey anti-rabbit antibody for MafA (Jackson ImmunoResearch Laboratories, Inc.; 1/150), fluorescein isothiocyanate-conjugated donkey anti-goat antibody for crystallins (Sigma Aldrich; 1/150) and fluorescein isothiocyanate-conjugated goat anti-mouse antibody for HA1 epitope (Sigma Aldrich; 1/200). Preparations were then washed in PBS and mounted in Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories).
RESULTS
MafA is a phosphoprotein.
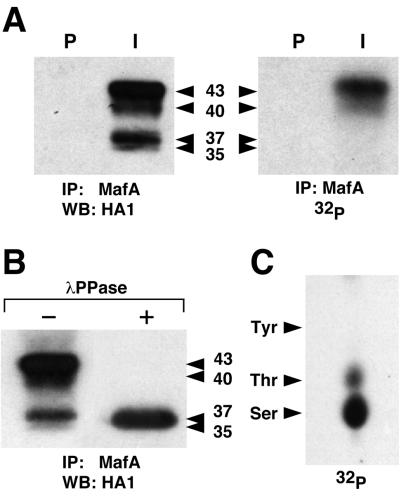
To investigate the phosphorylation status of MafA, we transfected HeLa cells with a pcDNA3-derived vector encoding an HA1 epitope-tagged MafA and labeled the cells with [32P]orthophosphate. Cellular lysates, prepared 48 h after transfection, were immunoprecipitated with an antiserum raised against a MafA-specific region located between the two histidine repeats (see Materials and Methods) and analyzed by Western blotting with anti-HA1 antibody (Fig. 1A). MafA products could be resolved into four major bands: two upper ones of apparent molecular mass of 40 and 43 kDa and two lower ones of 35 and 37 kDa. Autoradiography of the same blot showed that only the two upper bands contained [32P]phosphate, whereas we could not detect a radioactive signal in the lower ones. To confirm that the distinct MafA migrating bands visualized by Western blot were due to phosphorylation, we treated MafA immunoprecipitates with λ phage phosphatase. In this case only one band, comigrating with the 35-kDa lower band, could be detected (Fig. 1B). Thus, at least the 37-, 40-, and 43-kDa migrating forms of MafA appear to be generated by phosphorylation. Phosphoamino acid analysis showed that MafA immunoprecipitated from HeLa cells is phosphorylated predominantly on serine and, to a lesser extent, on threonine but not on tyrosine residues (Fig. 1C). These results indicated that overexpressed MafA protein was phosphorylated in HeLa cells. Phosphorylation markedly affected its electrophoretic mobility and presumably involved serine/threonine kinases.
FIG. 1.
MafA is phosphorylated in HeLa cells. (A) HeLa cells transfected with pcDNA3-MafA were metabolically labeled with [32P]orthophosphate, and cellular extracts were immunoprecipitated with either preimmune (P) or MafA-specific (I) serum. Immunoprecipitates were analyzed by Western blotting followed either by incubation with anti-HA1 antibody (left panel) or by direct autoradiography (right panel). (B)MafA immunoprecipitates from HeLa cells were treated with λ phosphatase (+) or left untreated (−) and analyzed by Western blotting with anti-HA1 antibody. Apparent molecular masses (kDa) of MafA bands are indicated. (C) Phosphoamino acid analysis of 32P-labeled MafA immunoprecipitated from HeLa cells. The migration of phosphoamino acid standards is indicated.
Serine 14 and serine 65 are the major MafA phosphorylation sites.
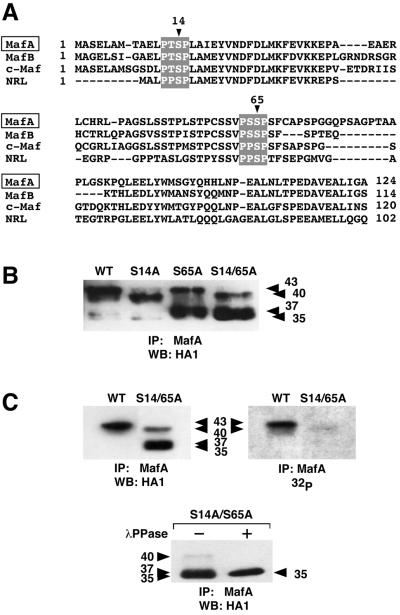
The N-terminal region of the large Maf proteins is remarkably conserved and rich in proline, serine, and threonine residues. Two putative phosphorylation sites (serine 14 and serine 65 in MafA) for proline-directed kinases, fitting the PXS/TP consensus (60), were conserved among all large Maf amino-terminal regions (Fig. 2A), raising the interesting possibility that they could be targeted by common regulatory mechanisms. To assess whether these two sites were phosphorylated in MafA, we mutated the serines individually or together into alanine residues. WT or mutated (S14A, S65A, or S14A/S65A) HA1-tagged proteins were expressed in HeLa cells, and their phosphorylation status was examined on the basis of their electrophoretic mobility. The three MafA mutants were differentially affected in their migration, since each of them specifically generated one major band or doublet (Fig. 2B). Thus, the S14A mutant essentially migrated as a major 40-kDa band and a minor 35-kDa one, whereas the S65A mutant generated a major band at 37 kDa and a minor one at 43 kDa. In contrast, the S14A/S65A double mutant essentially migrated as a 35- to 37-kDa doublet with a minor 40-kDa form. Noteworthily, the major bands generated by the S14A/S65A double mutant migrated at the lowest apparent molecular mass, a pattern comparable to that of phosphatase-treated WT MafA. We then examined the phosphorylation status of the S14A/S65A mutant by metabolic labeling in comparison to that of WT MafA protein. Immunoprecipitated S14A/S65A protein generated a faint radioactive signal, at an apparent molecular mass of 40 kDa, that was barely detectable compared to that of the 40- to 43-kDa doublet of WT MafA (Fig. 2C). Moreover, the major bands generated by S14A/S65A MafA, migrating with an apparent molecular mass of 35 to 37 kDa in immunoblots, could not be detected following metabolic labeling. Phosphatase treatment of immunoprecipitated S14A/S65A protein further generated a unique 35-kDa band, similar to that of WT MafA, indicating that part of the S14A/S65A protein was still phosphorylated, but to a much lower level since the mutant protein was barely detectable upon 32P labeling. These results show that integrity of serine 14 and serine 65 residues is necessary to achieve full MafA phosphorylation in HeLa cells. They also suggest that phosphorylation generates major conformational changes of MafA protein leading to substantial shifts in electrophoretic mobility and which could support functional modifications.
FIG. 2.
MafA serine 14 and serine 65 are major phosphorylation sites. (A) Amino acid sequence comparison of the TAD of large Maf proteins (quail MafA, chicken MafB and c-Maf, and human NRL). Conserved sequences fitting the proline-directed sequence consensus and surrounding serines 14 and 65 in MafA are highlighted. (B) Western blot analysis of WT and mutated MafA. pcDNA3-derived vectors encoding either WT, S14A, S65A, or S14A/S65A MafA were transfected into HeLa cells, and cellular extracts were analyzed by immunoprecipitation with MafA-directed serum and Western blotting with anti-HA1 antibody. (C) HeLa cells transfected with either pcDNA3-WT or pcDNA3-S14A/S65A were metabolically labeled with [32P]orthophosphate, and cellular extracts were immunoprecipitated with MafA-directed serum. Immune complexes were analyzed by Western blotting followed by either incubation with anti-HA1 antibody (left panel) or direct autoradiography (right panel). In the lower panel, S14A/S65A MafA immunoprecipitates were treated with λ phosphatase (+) or left untreated (−) and analyzed by Western blotting with anti-HA1 antibody.
MafA is phosphorylated in vitro by MAPKs ERK2 and p38α but not by JNK1 and ERK5.
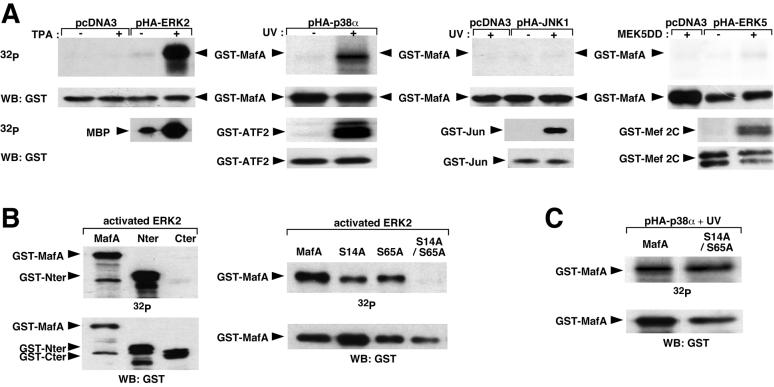
Since serine 14 and serine 65 residues were lying in a proline-directed kinase consensus, we considered the possibility that MafA could be a substrate for serine/threonine kinases of the MAPK family, a subgroup of proline-directed kinases known to phosphorylate transcription factors of the AP1/bZIP family (26). We first examined the ability of MafA to be phosphorylated in vitro by MAPKs ERK2, JNK1, p38α, and ERK5. Therefore, we transfected HeLa cells with expression vectors encoding each of the HA1-tagged kinases, and we activated the different kinases with their appropriate stimuli. Activated kinases were then immunoprecipitated with HA1-specific antibody and incubated with either GST-MafA protein or with their cognate control substrates in the presence of [γ-32P]ATP. ERK2 displayed a strong ability to phosphorylate MafA upon activation by TPA (Fig. 3A). Likewise, activated p38α was able to substantially phosphorylate MafA. In contrast, JNK1 or ERK5 had no effect on MafA phosphorylation compared to their respective effects on GST-Jun and GST-MEF2C positive controls. To further characterize the region of MafA that was phosphorylated by ERK2, we performed similar experiments with GST fusion proteins containing either the transactivating domain (TAD) of MafA (GST-Nter) or its bZIP domain (GST-Cter). The MafA N-terminal part could be efficiently phosphorylated by MAPK ERK2 whereas the C-terminal domain was not phosphorylated (Fig. 3B). Since serine 14 and serine 65 residues were located in the TAD, we performed kinase assays with activated ERK2 on GST-MafA fusion proteins carrying the S14A and/or the S65A mutation in order to map the phosphorylation sites in MafA. Each of the single mutants showed reduced phosphorylation when compared to the WT protein. Moreover, phosphorylation of the S14A/S65A mutant by ERK2 was no longer detectable (Fig. 3B). Taken together, these results demonstrated that activated ERK2 phosphorylated MafA TAD in vitro and that the S14 and S65 residues were its major and specific targets. We performed similar experiments to determine if the same residues were phosphorylated by p38α (Fig. 3C). GST-MafA recombinant proteins, either WT or carrying the S14A/S65A mutation, appeared to be similarly phosphorylated by activated p38α, indicating that S14 and S65 residues were not the major targets for MafA phosphorylation by p38. In conclusion, although both ERK2 and p38 MAPKs phosphorylate MafA in vitro, only ERK2 appears to be able to phosphorylate MafA on S14 and S65.
FIG. 3.
Differential phosphorylation of MafA by members of the MAPK family in vitro. (A) HeLa cells were transfected with expression vectors for ERK2, p38α, JNK1, or ERK5 HA1-tagged MAPKs and stimulated, respectively, with TPA, UV, UV, or by cotransfection with an expression plasmid for MEK5DD, an activated version of MEK5. Kinases were immunoprecipitated with anti-HA1 antibody and incubated with [γ-32P]ATP and GST-MafA recombinant protein, followed by Western blot and either direct autoradiography or incubation with anti-GST antibody. Positive controls (myelin basic protein [MBP] for ERK2, GST-ATF2 for p38α, GST-Jun for JNK1, and GST-MEF2C for ERK5) were treated likewise. (B) GST-MafA fusion proteins containing either truncated (left panels) or mutated (right panels) forms of MafA were subjected to kinase assay with recombinant activated ERK2. MafA, GST fused to full-length MafA; Nter, GST fused to the N-terminal half of MafA; Cter, GST fused to the C-terminal half of MafA; S14A, S65A, and S14A/S65A, GST fused to full-length MafA carrying the respective mutations. In vitro kinase assays were analyzed as described for panel A. (C) GST-MafA, either WT or carrying the S14A/S65A mutation, was subjected to in vitro kinase assay with activated p38α immunoprecipitated from HeLa cells as described for panel A.
MafA can be phosphorylated by the ERK1/2 MAPKs in vivo.
To investigate whether MafA could also be a target for ERK1/2 kinases in vivo, we transfected HeLa cells with an expression vector encoding WT HA1-tagged MafA protein and examined changes in mobility following stimulation or inhibition of the MEK/ERK cascade. Transfected cultures were maintained in medium containing 0.1% FBS during 8 h and subsequently treated with TPA in the presence or absence of PD98059, a specific inhibitor of the MEK/ERK pathway. Cellular lysates were then immunoprecipitated with MafA-specific antiserum and subjected to HA1-specific imunoblot. Serum-deprived HeLa cells predominantly contained the MafA 40- to 43-kDa forms and, in lower amounts, the 35- and 37-kDa forms (Fig. 4A). These two lower bands were no longer observed after TPA treatment and became again detectable when cells were treated with TPA in the presence of PD98059, indicating that activated ERK1/2 could phosphorylate the lower migrating forms. To confirm these results, we transfected the MafA expression vector together with plasmids encoding dominant negative (S222A) or constitutively activated (S218D/S222D) MEK1, the upstream specific activator of ERK1/2. Again, the lower 35- to 37-kDa bands were no longer detectable when the ERK1/2 pathway was activated in the presence of S218D/S222D MEK1 (Fig. 4B). However, cotransfection with S222A MEK1 did not affect MafA electrophoretic mobility, indicating that MafA phosphorylation could not be significantly altered by inhibiting the MEK/ERK cascade (Fig. 4B). Activation or inhibition of the ERK cascade had no effect on the S14A/S65A mutant MafA (Fig. 4B). Thus, MafA could be phosphorylated in response to activation of the MEK/ERK cascade in vivo, presumably on S14 and S65 residues. However, contribution of this pathway to MafA phosphorylation in vivo appears to be moderate.
Mutation of S14 and S65 residues severely impairs MafA transcriptional activity.
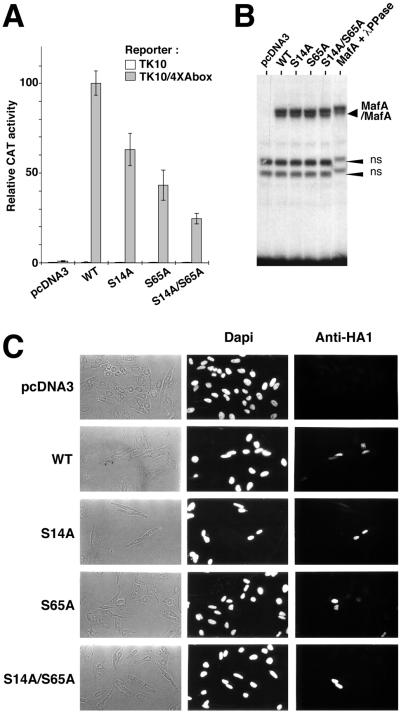
To evaluate the functional importance of MafA phosphorylation, we examined the transactivating capacity of MafA proteins mutated at S14, S65, or at both residues. We previously reported that the promoter of the NR-specific QR1 gene carries a Maf responding sequence, the A box, which contains two hemipalindromes of the Maf recognition element (52). We showed that the QR1 promoter, as well as the multimerized A box alone, is transactivated by members of the large Maf protein subfamily, such as c-Maf, MafB, and MafA (2, 52). Therefore, we compared the transcriptional capacity of WT and mutant MafA proteins by using a reporter construct containing four copies of the A box sequence cloned upstream of the tk promoter and CAT reporter gene (52). Quail NR cells infected with a temperature-sensitive mutant of Rous sarcoma virus were transfected with reporter constructs and pcDNA3-derived expression vectors for mafA alleles and transferred to 41°C, the nonpermissive temperature for cell division, at which QR1 expression is induced in response to v-Src inactivation (14, 51). Forty-eight hours later, cellular extracts were assayed for CAT activity. In contrast to the strong transactivating potential of WT MafA on the A box-containing reporter, the S14A, S65A, and S14A/S65A mutant proteins all displayed a significant decrease (by 30, 55, and 75%, respectively) in transcriptional activating capacity (Fig. 5A). Similar effects were observed when we used the QR1 promoter reporter plasmid (data not shown). Thus, the transcriptional capacity of MafA strongly relies upon the integrity of the S14 and S65 phosphorylation sites.
FIG. 5.
Serines 14 and 65 are required for MafA transcriptional activity. (A) Quail NR cells infected with tsNY68 virus were transiently transfected with the TK10/4×Abox reporter or empty TK10 plasmid and pcDNA3-derived vectors either empty or encoding WT, S14A, S65A, or S14A/S65A versions of MafA. CAT activity was measured at 41°C and expressed as a relative value with respect to the TK10/4×Abox and pcDNA3-WT transfection. Results represent mean values from four experiments. Expression levels for WT and mutant MafA proteins were controlled by Western blotting in each experiment (data not shown). (B) Serine 14 and serine 65 residues are not required for MafA DNA binding. WT or mutated MafA proteins encoded by pcDNA3-derived plasmids were obtained by in vitro transcription and translation in reticulocyte lysates and subjected to gel-shift analysis with a 32P-labeled A box probe. MafA + λPPase indicates that the WT MafA-containing lysate was incubated with λ phosphatase prior to DNA binding. MafA/MafA indicates MafA homodimers bound to the probe. ns, nonspecific bands. (C) Integrity of serine 14 and serine 65 residues is not required for MafA nuclear localization. HeLa cells were transiently transfected with pcDNA3-derived vectors encoding WT, S14A, S65A, or S14A/S65A forms of MafA and analyzed by immunofluorescence with anti-HA1 antibody.
The marked reduction in transcriptional activity of the MafA mutants was not due to their decreased ability to form homodimers and to bind DNA, since these proteins synthesized in vitro retained their ability to bind the A box fragment (Fig. 5B). Furthermore, the binding capacity of all Maf proteins was unaffected by phosphatase treatment (Fig. 5B and data not shown), suggesting that phosphorylation was not an absolute requirement for dimer formation and interaction with the target DNA sequence.
To assess whether the mutation of S14 and S65 residues affected MafA subcellular localization, we expressed HA1-tagged MafA proteins in HeLa cells and analyzed their localization by immunofluorescence with HA1-specific antibody. WT and each MafA mutant protein were detected exclusively in the nucleus (Fig. 5C), indicating that mutation of serines 14 and 65 and subsequent protein underphosphorylation did not impair nuclear localization. In conclusion, our results show that S14 and S65, the major determinants of MafA phosphorylation, are also major determinants of its transcriptional property. Since mutation of these sites severely impaired transactivating potential without affecting DNA binding and nuclear localization, it is likely that their phosphorylation controls the activity of the TAD per se.
Integrity of S14 and S65 residues is required to activate QR1 expression in NR cells.
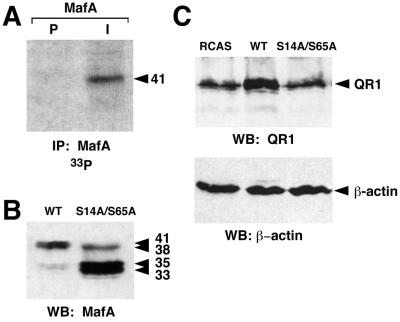
The QR1 gene product is a secreted glycoprotein, associated with the extracellular matrix, that belongs to the SPARC family. It is specifically expressed in Müller glial cells of postmitotic and differentiating NR and could play a role in the differentiation of photoreceptor cells (6, 14). To investigate the importance of MafA phosphorylation in regulating the expression of NR-specific target genes, we first examined whether MafA was also phosphorylated in these cells. Therefore, we transfected quail embryonic NR cells with an RCAS-derived retroviral vector encoding WT MafA (RCAS-MafA). After three passages, we metabolically labeled the cells with [33P]orthophosphate and immunoprecipitated cellular lysates with MafA-specific serum. The presence of a protein with an apparent molecular mass of 41 kDa specifically recognized by MafA but not by preimmune serum was detected by autoradiography, indicating that MafA was also phosphorylated in NR cells (Fig. 6A). Interestingly, Western blot analysis of NR cells transfected by RCAS-MafA or by a retroviral vector expressing S14A/S65A MafA (RCAS-S14A/S65A) showed a protein migration pattern similar to that observed in HeLa cells (Fig. 6B): a major 38- to 41-kDa doublet for WT and a major 33- to 35-kDa doublet and minor 38-kDa band for S14A/S65A. Thus, MafA phosphorylation in NR cells appears also to principally rely on the integrity of serine 14 and serine 65 residues.
FIG. 6.
MafA phosphorylation is required to activate QR1 expression in NR cells. (A) MafA is phosphorylated in NR cells. Quail embryo NR cells were transfected with an RCAS-derived vector encoding WT MafA and metabolically labeled with [33P]orthophosphate. Cellular lysates were immunoprecipitated with either preimmune (P) or MafA-directed (I) serum and subjected to Western blotting and autoradiography. (B) Chicken embryo NR cells were transfected with RCAS-derived vectors encoding WT or S14A/S65A MafA. Cell extracts were analyzed by direct Western blotting with MafA-directed serum. (C) Integrity of serines 14 and 65 is required for activation of QR1 expression by MafA. Chicken embryo NR cells were transfected with empty RCAS (RCAS), RCAS-MafA (WT), or RCAS-S14A/S65A (S14A/S65A), and cellular lysates were analyzed by direct Western blotting with anti-QR1 serum (upper panel) and with anti-β-actin antibody as a loading control (lower panel).
To further correlate MafA phosphorylation with transcriptional stimulation of its QR1 target gene, we compared the levels of endogenous QR1 protein 6 days after transfection of chicken embryonic NR cells with either RCAS-MafA or RCAS-S14A/S65A. We found that the amount of detected QR1 protein was significantly increased in RCAS-MafA infected cells compared to that of cells infected with the RCAS control virus (Fig. 6C). In contrast, we did not detect a variation in the level of QR1 in NR cells expressing the S14A/S65A protein. Thus, the ability of MafA to activate QR1 transcription and subsequent protein expression in NR cells was dependent on the integrity of S14 and S65 residues, demonstrating that phosphorylation of MafA was required for this biological property.
Impairment of lens-inductive properties in the S14A/S65A MafA mutant.
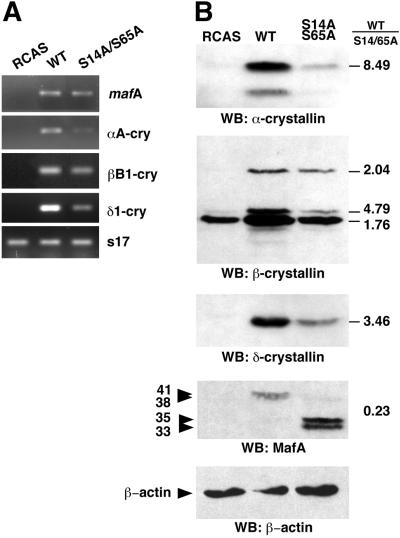
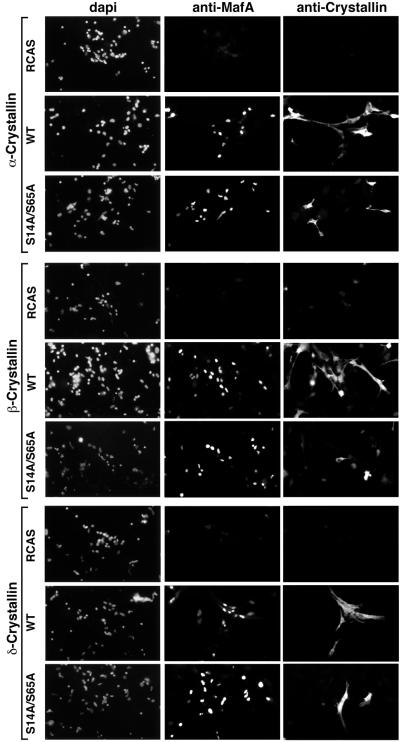
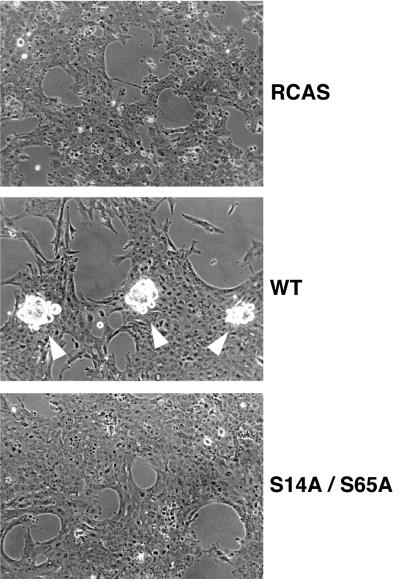
MafA was previously reported to be a major inducer of lens differentiation, since it was able to induce NR cells to acquire a number of phenotypic characteristics of lens fiber cells, a process termed neuroretina-to-lens transdifferentiation that occurred spontaneously in culture, albeit with slower kinetics (46, 59). Therefore, we examined the importance of MafA phosphorylation at S14 and S65 residues for its ability to induce lens differentiation in NR cells based on two criteria: the short-term induction of crystallin expression as monitored by RT-PCR, Western blotting, and cellular immunofluorescence, and the long-term formation of lentoid bodies. The level of specific crystallin mRNAs (αA-, δ1-, and, to a lesser extent, βB1-crystallin), detectable by RT-PCR, was significantly reduced in S14A/S65A-expressing NR cells compared to that detected in WT MafA-containing cultures (Fig. 7A). In addition, the overall levels of crystallin proteins detected by Western blot analysis were strikingly reduced in cells containing S14A/S65A MafA (Fig. 7B). This was correlated with a critical decrease in the number of cells that were positive for both S14A/S65A MafA and crystallin expression, compared to that of cultures expressing WT MafA, in immunofluorescence experiments (Fig. 8). Thus, only a minority of S14A/S65A MafA-positive cells were doubly stained, whereas doubly stained cells represented 65 to 95% of WT MafA-positive cells (Table 1). The functional implication of MafA phosphorylation in this process was further revealed when we examined the formation of lentoid bodies in long-term-transfected NR cultures. We observed the presence of abundant and large lentoid bodies in WT MafA-containing cultures, passaged two or three times, whereas only a few and small ones could be seen in S14A/S65A MafA chronically infected cells (Fig. 9). Typically, we found a 50:1 ratio of lentoid bodies between the two types of cultures. These results demonstrate that MafA phosphorylation strongly determines its capacity to initiate genetic programs leading to lens differentiation in NR cells.
FIG. 7.
Activation of crystallin expression by MafA requires the integrity of serines 14 and 65. Chicken embryo NR cells were transfected with empty RCAS (RCAS), RCAS-MafA (WT), or RCAS-S14A/S65A (S14A/S65A) plasmids. (A) Transcription of αA-, βB1-, and δ1-crystallin genes was analyzed 6 days after transfection by RT-PCR. MafA- and ribosomal s17-specific PCR were performed as controls. (B) Cellular lysates prepared 6 days after transfection were analyzed by Western blotting with α-crystallin-, β-crystallin-, δ-crystallin-, or MafA-directed sera. Relative amounts of proteins expressed in the WT versus S14A/S65A MafA-containing cultures were determined according to densitometry of autoradiograms and are indicated on the right. A β-actin Western blot was performed as a loading control.
FIG. 8.
Analysis of crystallin expression in MafA-containing cells by immunofluorescence. Chicken embryo NR cells were transfected with empty RCAS (RCAS), RCAS-MafA (WT), or RCAS-S14A/S65A (S14A/S65A) plasmids. Induction of crystallin expression was analyzed 6 days after transfection by double staining with α-, β-, or δ-crystallin-directed antibodies and anti-MafA serum. Quantification of the results is presented in Table 1.
TABLE 1.
Efficiency of crystallin induction in MafA-overexpressing NR cells
| Overexpressed MafA | % of MafA-positive cells expressing:
|
||
|---|---|---|---|
| α-Crystallin | β-Crystallin | δ-Crystallin | |
| WT | 90 | 95 | 65 |
| S14A/S65A | 20 | 35 | 5 |
FIG. 9.
NR-to-lens transdifferentiation induced by MafA requires the integrity of serine 14 and serine 65 residues. Formation of lentoid bodies was examined, after three passages, in chicken NR cultures transfected by RCAS-derived vectors encoding WT or S14A/S65A MafA. Lentoid bodies are indicated by white arrowheads.
DISCUSSION
Previous studies have suggested that Maf proteins are involved in regulation of various developmental processes and that their function is modulated by interaction with heterologous proteins through the formation of leucine zipper dimers or distinct complexes (22, 30, 38, 58). Unexpectedly, little is known about their regulation by covalent posttranslational modifications, such as phosphorylation, unlike the numerous reports on other bZIP proteins of the AP1 superfamily. In this work we show, for the first time, that MafA (also known as L-Maf), a member of the large Maf protein subgroup, undergoes functional regulation by phosphorylation. We have identified two serine residues (S14 and S65) located in the MafA TAD that are of critical importance not only for overall MafA phosphorylation but also for its transcriptional capacity and potential to control expression of differentiation programs in NR cells. Interestingly, these serine residues and their neighboring amino acids are conserved between all large Mafs. Significantly, the MafA serine 65 corresponds in NRL to serine 50, whose mutation was implicated in the occurrence of retinitis pigmentosa (D. A. Bessant, A. M. Payne, K. P. Mitton, Q. L. Wang, P. K. Swain, C. Plant, A. C. Bird, D. J. Zack, A. Swaroop, and S. S. Bhattacharya, Letter, Nat. Genet. 21:355–356), suggesting that these conserved residues are important for biological function of Maf proteins.
MafA is a phosphoprotein.
In this study we present evidence that MafA is phosphorylated in two different cell types, HeLa and NR cells. Metabolic labeling, Western blotting, and phosphatase treatment experiments performed with HeLa cell extracts show that the multiple MafA bands detected in Western blots reflect various levels of MafA phosphorylation. Surprisingly, the high level of basal protein phosphorylation under standard culture conditions in HeLa cells could not be efficiently modified by extracellular changes such as serum starvation, suggesting that the kinase(s) that phosphorylates MafA is constitutively active in these cells. We identified serine 14 and serine 65 as the major determinants of MafA phosphorylation. Mutagenesis of these two sites into alanine leads to dramatic changes in the overall distribution of migrating forms, with a significant increase in underphosphorylated forms, as could be concluded from the decrease in the in vivo phosphate content of mutant proteins. This strongly suggests that phosphorylation of S14 and S65 residues determines the overall phosphorylation state of MafA in vivo, either because they are the most efficiently phosphorylated sites or because the protein undergoes sequential phosphorylation. For example the phosphorylation, or at least integrity, of S14 and S65 could be required to ensure proper docking or recognition of other kinases which are responsible for the phosphorylation of distinct sites. In addition, our data show that the S14A/S65A MafA mutant remains slightly phosphorylated and phosphoamino acid analysis indicates that WT MafA is also phosphorylated on threonine residues. Thus, MafA is likely to undergo multiple phosphorylations and could be regulated by various signaling networks.
It is interesting to note that the migration patterns of WT and S14A/S65A MafA are very similar in HeLa and NR cells, as well as in other cell types tested (data not shown), suggesting that the phosphorylation process of MafA is essentially similar in these cell types and that phosphorylation implicating S14 and S65 residues constitutes a general mechanism for regulating MafA.
The biological properties of MafA strongly depend on phosphorylation.
We evaluated the importance of MafA phosphorylation at two distinct levels: by its capacity to activate transcription of reporter genes and its ability to activate expression of an NR-specific target gene and to drive a complete gene expression program. The MafA capacity to activate transcription of a promoter containing a MARE-related sequence was strongly affected by the double mutation. Thus, the presence of underphosphorylated MafA clearly correlates with a sharp decrease in transcriptional capacity, demonstrating that this activity is strictly dependent on MafA phosphorylation. This could not be explained by a change in subcellular localization, since all MafA mutants were localized in the nucleus. Similarly, we found that these mutations did not alter the binding of MafA to DNA, although we cannot exclude that MafA phosphorylation could influence its interaction with target sequences. Likewise, our results indicated that MafA phosphorylation did not significantly affect the stability of the protein, since the amounts of WT and S14A/S65A MafA proteins did not markedly differ in Western blots (Fig. 2B and C and data not shown). Thus, MafA phosphorylation is likely to control the transactivating capacity of MafA per se. It could either modulate MafA's ability to interact with the basal transcription machinery or affect its capacity to interact with critical partners, as was demonstrated in the case of the MyoD transcription factor (37). Likewise, phosphorylation of JunB, on threonine 102 and threonine 104, by JNK was suggested to favor interaction with c-Maf dimers and to consequently elicit synergistic activation of the interleukin-4 promoter by both transcription factors (38). The presence of these phosphorylated residues could also be required for the recruitment of more general coactivators, such as histone acetylases of the p300/CBP family, as in the case of the CREB bZIP (36) and the melanocyte-specific Mi (53) transcription factors.
In a second set of experiments we established that MafA underphosphorylation could be correlated with the loss of its property to activate endogenous QR1 expression. This result is in agreement with the failure of S14A/S65A MafA to transactivate both the A box and the complete promoter of QR1 in transcription assays. We further showed that the mutation also impaired MafA capacity to induce lens phenotypic transformation in NR cells, as concluded from the activation of target crystallin genes expression and the formation of lentoid bodies. These results clearly demonstrate that MafA phosphorylation is of critical importance in the context of these biological processes. Moreover, they underline that lens differentiation may be induced by signaling cascades targeting the MafA transcription factor. Signals originating from the contiguous optic vesicle have been suggested to drive the transformation of the lens placode during embryonic development (for review, see reference 25). It is possible that MafA is involved in this process, since its expression could be detected in the lens placode at the stage of embryonic development at which the optic vesicle contacts the head ectoderm (46). Thus, phosphorylation of MafA in response to such signals could strongly enhance its function. Likewise, NRL was suggested to play a key role in the terminal differentiation of retina photoreceptors by turning on expression of specific genes, including that encoding rhodopsin (18). It is noteworthy that mutation of NRL serine 50, which is equivalent to MafA serine 65, was implicated in a genetic disease affecting photoreceptors (Bessant et al., letter). Thus, it would be interesting to determine whether a phosphorylation process similar to the one regulating MafA function also accounts for NRL and, more generally, for other Maf proteins.
ERK2 phosphorylates MafA in vitro but is not responsible for its high basal phosphorylation in vivo.
Localization of MafA S14 and S65 residues in a perfect MAPK consensus strongly suggested that they might be targeted by kinases of this family. Indeed, we show that MafA can be phosphorylated in vitro by ERK2 and p38α but not by JNK1 or ERK5. We also show that p38α phosphorylates MafA, likely on residues distinct from S14 and S65. Interestingly, the control of rat αB-crystallin gene expression by stress-related signals involving p38 has been reported (31). Thus, the p38 MAPK cascade could affect expression of crystallins through an effect on MafA. Several putative sites for proline-directed kinases, in addition to serines 14 and 65, are present in MafA. It should be noted, moreover, that MafA is phosphorylated on threonine residues in HeLa cells. Thus, further studies are required to identify the residues phosphorylated by p38 and to determine whether their phosphorylation is involved in regulating transcriptional activity.
While activated ERK2 phosphorylates MafA on S14 and S65 residues in vitro, the MEK/ERK pathway appears to contribute only partially to MafA phosphorylation in vivo. Indeed, either transient or constitutive activation of the MEK/ERK pathway leads to detectable modification of MafA electrophoretic mobility, an effect which was correlated with an increase in MafA phosphate content (data not shown). Implication of the MEK/ERK pathway in the control of MafA activity appears relevant regarding several physiological aspects. Thus, differentiation of lens fibers during eye development was reported to be controlled by fibroblast growth factor signaling (for review, see reference 25), which notably targets the ERK/MAPK pathway. However, we could not significantly decrease the level of MafA background phosphorylation by using dominant negative alleles or inhibitors of the MEK1 kinase. Furthermore, attempts to modulate MafA transcriptional activity on reporter genes by activating the MEK/ERK cascade did not yield significant results (data not shown), probably because of the high basal phosphorylation of S14 and S65 residues. Taken together, our results indicate that while stimulation of the MEK/ERK cascade can lead to phosphorylation of MafA at S14 and S65 sites, it does not appear to be responsible for the basal phosphorylation of the protein. Thus, the actual kinase(s), likely a proline-directed kinase, responsible for the extensive phosphorylation of S14 and S65 residues remains to be identified. Because of the importance of Maf proteins in regulating various differentiation processes, identification of this kinase(s) should prove essential to our understanding of the signals that regulate Maf protein activity.
ACKNOWLEDGMENTS
We thank Melanie Cobb, Marc Castellazzi, Roger Davis, Silvio Gutkind, Joram Piatigorsky, Joël Raingeaud, and Mike Weber for providing reagents used in this study. We also thank Joël Raingeaud for helpful discussion and Odile Lecoq for preparing the MafA-directed antiserum.
This work was supported by the Centre National de la Recherche Scientifique, the Institut Curie, the Association pour la Recherche sur le Cancer (grant 5276), and Retina France. S.B. and S.P. were supported by fellowships from Retina France, the Ligue Nationale Contre le Cancer (Comité de l'Essonne), the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Fondation pour la Recherche Médicale, and the Société Française du Cancer.
S. Benkhelifa and S. Provot contributed equally to this work.
REFERENCES
- 1.Beddington R S, Morgernstern J, Land H, Hogan A. An in situ transgenic enzyme marker for the midgestation mouse embryo and the visualization of inner cell mass clones during early organogenesis. Development. 1989;106:37–46. doi: 10.1242/dev.106.1.37. [DOI] [PubMed] [Google Scholar]
- 2.Benkhelifa S, Provot S, Lecoq O, Pouponnot C, Calothy G, Felder-Schmittbuhl M P. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene. 1998;17:247–254. doi: 10.1038/sj.onc.1201898. [DOI] [PubMed] [Google Scholar]
- 3.Binetruy B, Smeal T, Karin M. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature. 1991;351:122–127. doi: 10.1038/351122a0. [DOI] [PubMed] [Google Scholar]
- 4.Brunet A, Pouyssegur J. Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science. 1996;272:1652–1655. doi: 10.1126/science.272.5268.1652. [DOI] [PubMed] [Google Scholar]
- 5.Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- 6.Casado F J, Pouponnot C, Jeanny J C, Lecoq O, Calothy G, Pierani A. QRI, a retina-specific gene, encodes an extracellular matrix protein exclusively expressed during neural retina differentiation. Mech Dev. 1996;54:237–250. doi: 10.1016/0925-4773(95)00482-3. [DOI] [PubMed] [Google Scholar]
- 7.Chiariello M, Marinissen M J, Gutkind J S. Multiple mitogen-activated protein kinase signaling pathways connect the Cot oncoprotein to the c-jun promoter and to cellular transformation. Mol Cell Biol. 2000;20:1747–1758. doi: 10.1128/mcb.20.5.1747-1758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 9.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylated the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 10.English J M, Pearson G, Hockenberry T, Shivakumar L, White M A, Cobb M H. Contribution of the ERK5/MEK5 pathway to Ras/Raf signaling and growth control. J Biol Chem. 1999;274:31588–31592. doi: 10.1074/jbc.274.44.31588. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara K T, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- 12.Garingo A D, Suhasini M, Andrews N C, Pilz R B. cAMP-dependent protein kinase is necessary for increased NF-E2.DNA complex formation during erythroleukemia cell differentiation. J Biol Chem. 1995;270:9169–9177. doi: 10.1074/jbc.270.16.9169. [DOI] [PubMed] [Google Scholar]
- 13.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guermah M, Crisanti P, Laugier D, Dezelee P, Bidou L, Pessac B, Calothy G. Transcription of a quail gene expressed in embryonic retinal cells is shut off sharply at hatching. Proc Natl Acad Sci USA. 1991;88:4503–4507. doi: 10.1073/pnas.88.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guermah M, Gillet G, Michel D, Laugier D, Brun G, Calothy G. Down regulation by p60v-src of genes specifically expressed and developmentally regulated in postmitotic quail neuroretina cells. Mol Cell Biol. 1990;10:3584–3590. doi: 10.1128/mcb.10.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 18.He L, Campbell M L, Srivastava D, Blocker Y S, Harris J R, Swaroop A, Fox D A. Spatial and temporal expression of AP-1 responsive rod photoreceptor genes and bZIP transcription factors during development of the rat retina. Mol Vision. 1998;4:32. [PubMed] [Google Scholar]
- 19.Hemesath T J, Price E R, Takemoto C, Badalian T, Fisher D E. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 20.Her J H, Lakhani S, Zu K, Vila J, Dent P, Sturgill T W, Weber M J. Dual phosphorylation and autophosphorylation in mitogen-activated protein (MAP) kinase activation. Biochem J. 1993;296:25–31. doi: 10.1042/bj2960025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 22.Ho I C, Hodge M R, Rooney J W, Glimcher L H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 23.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 25.Jean D, Ewan K, Gruss P. Molecular regulators involved in vertebrate eye development. Mech Dev. 1998;76:3–18. doi: 10.1016/s0925-4773(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 26.Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 27.Kataoka K, Fujiwara K T, Noda M, Nishizawa M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol Cell Biol. 1994;14:7581–7591. doi: 10.1128/mcb.14.11.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataoka K, Igarashi K, Itoh K, Fujiwara K T, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kataoka K, Noda M, Nishizawa M. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos and small Maf proteins. Oncogene. 1996;12:53–62. [PubMed] [Google Scholar]
- 31.Kato K, Ito H, Kamei K, Iwamoto I. Selective stimulation of Hsp27 and alphaB-crystallin but not Hsp70 expression by p38 MAP kinase activation. Cell Stress Chaperones. 1999;4:94–101. [PMC free article] [PubMed] [Google Scholar]
- 32.Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, Yasuda K, Yamamoto M. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274:19254–19260. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- 33.Kelly L M, Englmeier U, Lafon I, Sieweke M H, Graf T. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J I, Li T, Ho I C, Grusby M J, Glimcher L H. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar R, Chen S, Scheurer D, Wang Q L, Duh E, Sung C H, Rehemtulla A, Swaroop A, Adler R, Zack D J. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J Biol Chem. 1996;271:29612–29618. doi: 10.1074/jbc.271.47.29612. [DOI] [PubMed] [Google Scholar]
- 36.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 37.Lenormand J L, Benayoun B, Guillier M, Vandromme M, Leibovitch M P, Leibovitch S A. Mos activates myogenic differentiation by promoting heterodimerization of MyoD and E12 proteins. Mol Cell Biol. 1997;17:584–593. doi: 10.1128/mcb.17.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Tournier C, Davis R J, Flavell R A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manzanares M, Cordes S, Ariza-McNaughton L, Sadl V, Maruthainar K, Barsh G, Krumlauf R. Conserved and distinct roles of kreisler in regulation of the paralogous Hoxa3 and Hoxb3 genes. Development. 1999;126:759–769. doi: 10.1242/dev.126.4.759. [DOI] [PubMed] [Google Scholar]
- 40.Manzanares M, Cordes S, Kwan C T, Sham M H, Barsh G S, Krumlauf R. Segmental regulation of Hoxb-3 by kreisler. Nature. 1997;387:191–195. doi: 10.1038/387191a0. [DOI] [PubMed] [Google Scholar]
- 41.Mikkola I, Bruun J A, Bjorkoy G, Holm T, Johansen T. Phosphorylation of the transactivation domain of Pax6 by extracellular signal-regulated kinase and p38 mitogen-activated protein kinase. J Biol Chem. 1999;274:15115–15126. doi: 10.1074/jbc.274.21.15115. [DOI] [PubMed] [Google Scholar]
- 42.Milstone L M, Piatigorsky J. Rates of protein synthesis in explanted embryonic chick lens epithelia: differential stimulation of delta-crystallin synthesis. Dev Biol. 1975;43:91–100. doi: 10.1016/0012-1606(75)90133-5. [DOI] [PubMed] [Google Scholar]
- 43.Musti A M, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 44.Nagai T, Igarashi K, Akasaka J, Furuyama K, Fujita H, Hayashi N, Yamamoto M, Sassa S. Regulation of NF-E2 activity in erythroleukemia cell differentiation. J Biol Chem. 1998;273:5358–5365. doi: 10.1074/jbc.273.9.5358. [DOI] [PubMed] [Google Scholar]
- 45.Nishizawa M, Kataoka K, Goto N, Fujiwara K T, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci USA. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- 47.Onodera K, Shavit J A, Motohashi H, Yamamoto M, Engel J D. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J. 2000;19:1335–1345. doi: 10.1093/emboj/19.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostrer H, Beebe D C, Piatigorsky J. Beta-crystallin mRNAs: differential distribution in the developing chicken lens. Developmental Biology (Orlando) 1981;86:403–408. doi: 10.1016/0012-1606(81)90198-6. [DOI] [PubMed] [Google Scholar]
- 49.Pessac B, Calothy G. Transformation of chick embryo neuroretinal cells by Rous sarcoma virus in vitro: induction of cell proliferation. Science. 1974;185:709–710. doi: 10.1126/science.185.4152.709. [DOI] [PubMed] [Google Scholar]
- 50.Peyssonnaux C, Provot S, Felder-Schmittbuhl M, Calothy G, Eychène A. Induction of postmitotic neuroretina cell proliferation by distinct Ras downstream signaling pathways. Mol Cell Biol. 2000;20:7068–7079. doi: 10.1128/mcb.20.19.7068-7079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierani A, Pouponnot C, Calothy G. Transcriptional downregulation of the retina-specific QR1 gene by pp60v-src and identification of a novel v-src-responsive unit. Mol Cell Biol. 1993;13:3401–3414. doi: 10.1128/mcb.13.6.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pouponnot C, Nishizawa M, Calothy G, Pierani A. Transcriptional stimulation of the retina-specific QR1 gene upon growth arrest involves a Maf-related protein. Mol Cell Biol. 1995;15:5563–5575. doi: 10.1128/mcb.15.10.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price E R, Ding H F, Badalian T, Bhattacharya S, Takemoto C, Yao T P, Hemesath T J, Fisher D E. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem. 1998;273:17983–17986. doi: 10.1074/jbc.273.29.17983. [DOI] [PubMed] [Google Scholar]
- 54.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack D J, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Remaut E, Stanssens P, Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981;15:81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- 56.Ring B, Cordes S, Overbeek P, Barsh G. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- 57.Schaeffer H J, Weber M J. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sieweke M H, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 59.Simonneau L, Crisanti P, Lorinet A M, Alliot F, Courtois Y, Calothy G, Pessac B. Crystallin gene expression and lentoid body formation in quail embryo neuroretina cultures transformed by the oncogenic retrovirus Mill Hill 2 or Rous sarcoma virus. Mol Cell Biol. 1986;6:3704–3710. doi: 10.1128/mcb.6.11.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swaroop A, Xu J Z, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Versaw W K, Blank V, Andrews N M, Bresnick E H. Mitogen-activated protein kinases enhance long-range activation by the beta-globin locus control region. Proc Natl Acad Sci USA. 1998;95:8756–8760. doi: 10.1073/pnas.95.15.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 64.Yang S H, Shore P, Willingham N, Lakey J H, Sharrocks A D. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 1999;18:5666–5674. doi: 10.1093/emboj/18.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]