Abstract
RNA helicase A (RHA) is a member of an ATPase/DNA and RNA helicase family and is a homologue of Drosophila maleless protein (MLE), which regulates X-linked gene expression. RHA is also a component of holo-RNA polymerase II (Pol II) complexes and recruits Pol II to the CREB binding protein (CBP). The ATPase and/or helicase activity of RHA is required for CREB-dependent transcription. To further understand the role of RHA on gene expression, we have identified a 50-amino-acid transactivation domain that interacts with Pol II and termed it the minimal transactivation domain (MTAD). The protein sequence of this region contains six hydrophobic residues and is unique to RHA homologues and well conserved. A mutant with this region deleted from full-length RHA decreased transcriptional activity in CREB-dependent transcription. In addition, mutational analyses revealed that several tryptophan residues in MTAD are important for the interaction with Pol II and transactivation. These mutants had ATP binding and ATPase activities comparable to those of wild-type RHA. A mutant lacking ATP binding activity was still able to interact with Pol II. In CREB-dependent transcription, the transcriptional activity of each of these mutants was less than that of wild-type RHA. The activity of the double mutant lacking both functions was significantly lower than that of each mutant alone, and the double mutant had a dominant negative effect. These results suggest that RHA could independently regulate CREB-dependent transcription either through recruitment of Pol II or by ATP-dependent mechanisms.
RNA helicase A (RHA) is a member of the DExH family of ATPases/helicases and catalyzes the displacement of both double-stranded RNA and DNA from 3′ to 5′ (32, 61, 63). Functional domains of RHA include two double-stranded RNA binding domains at the amino terminus known as dsRBD1 and dsRBD2. The catalytic core domain is located within the central region and contains a DExH motif. This core domain contains seven well-conserved motifs; one of them has an ATP binding site with the consensus GCGKT and FILDD, known as the A site the B site, respectively. The carboxyl terminus contains an RGG-rich region that is capable of binding single-strand nucleic acids (62).
RHA was originally isolated as a human homologue of Drosophila maleless protein (MLE), with which it has 50% sequence identity and 90% sequence similarity (33). In Drosophila, MLE colocalizes with acetylated histone H4 (8, 48). MLE is involved in sex-specific gene dosage compensation and elevates the level of transcription derived from a single X chromosome in male flies to a level equivalent to that derived from two X chromosome in the female (25, 29). MLE mutants are embryonic lethal to males, indicating that MLE is an essential factor in Drosophila development.
In mammals, RHA-knockout mice are embryonic lethal for homozygous RHA mutants (35). Analysis of these mice revealed that RHA is associated with differentiation of the embryonic ectoderm during gastrulation. It is possible that RHA has an important role in early embryonic development.
We previously reported that in mammalian cells, RHA functions as a bridging factor connecting the CREB binding protein (CBP) and holo-RNA polymerase II (Pol II) complexes (43). CBP is a general coactivator and plays key roles in nuclear signaling. RHA interacts with the CH3 domain of CBP via the RHA N terminus and recruits Pol II through a stretch of 410 amino acids (aa) (positions 255 to 664). RHA also recruits Pol II to the breast cancer-specific tumor suppressor protein BRCA1. BRCA1 mutants having a reduced ability to bind to RHA are observed in breast cancer. It was suggested that the weaker interaction between RHA and BRCA1 decreases the transcriptional activity of BRCA1, leading to the development of breast cancer (4). Recently, RHA was reported to be involved in human immunodeficiency virus gene expression (19) and transcriptional regulation of the p16INK4a promoter (41). These reports indicate that RHA may be an essential factor for a wide variety of transcriptional pathways.
In addition to its function as a bridging factor, the ATPase and/or helicase activity of RHA appears to be important for transactivation. With respect to CREB-dependent transcription, a lysine-to-arginine change in the ATP binding site of RHA leads to a loss of ATP binding ability and ATPase activity and results in decreased transcriptional activity (43). In Drosophila, the mutant MLE lacking ATPase activity cannot rescue dosage compensation (34). As ATPase and/or helicase activity is essential for transactivation, it is likely that both the sequence and function of these sites are conserved between RHA and MLE. It is therefore possible that RHA regulates transactivation via common mechanisms conserved from flies to humans. Investigation of these mechanisms may shed light on general transactivation mechanisms conserved in eukaryotic cells. It is not known how RHA can activate transcription via both an ATP-dependent mechanism and recruitment of Pol II. The region of RHA containing the previously reported Pol II binding domain involves helicase motifs; however, there is no motif reported specifically as a transactivation domain (54). Accordingly, in this study we have defined a 50-aa minimal transactivation domain (MTAD) and generated mutants that were not capable of interacting with Pol II. Furthermore, we also suggest that RHA could have dual roles in CREB-dependent transcription, based on a comparison with the mutant lacking ATP binding ability.
MATERIALS AND METHODS
Plasmids.
The fragments RHA1, -2, -3, and -4 were obtained from pGEX-5X-1RHA (1-250), (230-650), (630-1020), and (1000-1279), respectively (43). The fragments of other RHA deletion mutants, termed RHA2-1, RHA2-2, RHA2-3, RHA2-4, RHA2-1N, RHA2-1NL, RHA2-1CL, RHA2-1C, RHA2-1M, RHA2-1NC, MTAD, sMTAD, and cMTAD (Fig. 1A), were generated by PCR-based methods. The MTAD region of Caenorhabditis elegans (eMTAD) was amplified from a C. elegans cDNA library by PCR. An alanine scanning mutagenesis method was used to generate MTAD mutants with substitutions in each residue conserved among RHA homologues. These mutants are termed MTADw332a, MTADp334a, MTADp335a, MTADn338a, MTADw339a, MTADn340a MTADw342a, MTADn346a, MTADi347a, MTADd348a, MTADe349a, MTADl352a, MTADe358a, MTADi360a, and MTADs361a. Each of these fragments was inserted, either alone or fused to the GAL4 DNA binding domain (GAL4-DBD), into pGBT9 (Clontech) or pcDNA3 (Invitrogen) for transactivation assays in yeast or mammalian cells, respectively. Amino acids 330 to 376 were deleted from RHA2 to generate RHA2Δmtad. RHA2 mutations RHA2w339a, RHA2i347a, and RHA2matp, which contains a lysine to-arginine change at position 417 in the ATP binding site, were generated by PCR. RHA2 mutations, wild-type RHA2, and MTAD fragments were inserted into pGEX-5X-1 (Amersham Pharmacia Biotech) for glutathione: S-transferase (GST) pull down and ATP binding assays. The full-length fragment of wild-type RHA and mutant with a substitution in the ATP binding site, termed RHAwt and RHAmatp, respectively, were obtained from each RHA expression vector (43). The mutated full-length RHA fragments RHAΔmtad, RHAw339a, and RHAi347a were created by PCR. The double mutant of full-length RHA (RHAw-matp) contains the mutations generated in both RHAw339a and RHAmatp. For transient reporter and ATPase assays, full-length RHA fragments were introduced into pcDNA3-HA, which was constructed by inserting hemagglutinin antigen (HA) sequence into pcDNA3 (39). All plasmids generated by PCR were confirmed by sequence analysis. The pRc/RSVmCBP, protein kinase A (PKA), GAL4-CREB (43), and pGAL4 (39) expression vectors, CRE-Luc (59) and pG5b-Luc (60) reporter plasmids, and control plasmid RSV-β-gal (43) have been described previously.
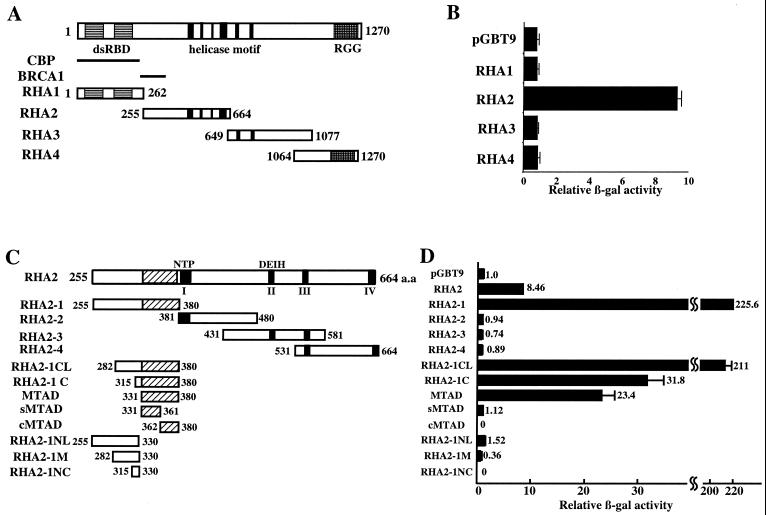
FIG. 1.
Identification of the MTAD of RHA. (A and C) Schematic representations of RHA and RHA deletion mutants used in transactivation assays. Solid boxes indicate the functional domains, dsRBD, helicase motif, and RGG-rich region. Solid bars represent the CBP binding domain (CBP) and BRCA1 binding domain (BRCA1). The hatched boxes illustrate MTAD. (B and D) Transactivation assays in yeast. Yeast strain Y190 cells were transformed with RHA deletion mutants inserted into pGBT9 or empty vector alone. The β-Gal activity derived from cells transformed with empty vector was designated 1. Each value of relative β-Gal activity represents the mean ± standard error (n = 3).
Transactivation assay.
Saccharomyces cerevisiae strain Y190 was transformed with RHA deletion mutants by a lithium acetate method, and transformants were selected by incubation on agar plates lacking tryptophan for 3 days. For the liquid β-galactosidase (β-Gal) assay, 1 ml of growth medium was inoculated with selected colonies. The assay was performed in triplicate to quantify transcriptional activity of the RHA deletion mutants as previously described (60).
Using LIPOFECTIN reagent (Gibco BRL), human embryonic kidney HEK-293T cells were transiently transfected with 50 ng of each RHA deletion mutant fused to GAL4-DBD, or the empty vector alone, and 50 ng of pG5b-Luc. Following 24 h of incubation, cells were lysed with cell lysis buffer (Toyo Ink) and luciferase activities were measured. Each measured activity was normalized for β-Gal activity using cotransfected RSV-β-gal as a control. Each experiment was performed in triplicate (14, 45).
Antibodies.
Rabbit polyclonal antibodies against the largest subunit of RNA polymerase II (N-20) were obtained from Santa Cruz Biotechnology. Mouse and rat monoclonal antibodies against the HA tag (12CA5 and 3F10) were purchased from Boehringer Mannheim.
GST pull-down assay.
The GST fusion protein of each RHA mutant was expressed in Escherichia coli strain TopXF′ (Invitrogen) and purified using glutathione-Sepharose beads (Amersham Pharmacia Biotech). Fifty micrograms of nuclear extract obtained from HEK-293T cells was incubated with 2 μg of each GST fusion protein bound to resin in 1 ml of buffer A (20 mM HEPES [pH 7.9], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.05% Tween 20, 5% glycerol, 1 mM Na3VO4, 5 mM NaF, 1 μg each of aprotinin, leupeptin, and pepstatin A per ml) for 8 h at 4°C. After washing with buffer A, bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting.
ATP binding assay.
One hundred nanograms of GST fusion protein bound with beads was incubated with [γ-32P]ATP (5,000 Ci/mmol) in 50 μl of ATP binding buffer (45 mM HEPES [pH 7.6], 0.9 mM EDTA, 0.9 mM EGTA, 4.5 mM magnesium acetate 0.14 mM KCl, 9% glycerol, 0.018% Nonidet P-40, 0.12 mg of bovine serum albumin per ml) for 10 min at room temperature. ATP was used at a concentration of 2.7 μM. After washing with ATP binding buffer, the bound ATP was measured by scintillation counting (Coulter) (27).
ATPase assay.
A series of HA-tagged full-length RHA polypeptides were synthesized in the Promega TNT coupled reticulocyte lysate system with [35S]-methionine and immunopurified with anti-HA antibody. Equal amounts of each HA-RHA polypeptide bound with beads were incubated at room temperature with [γ-32P] ATP (0.5 nCi/reaction, 10 μM ATP [final concentration]) in 50 μl of ATPase buffer (36 mM HEPES [pH 7.6], 0.05 mM EDTA, 0.05 mM EGTA, 3.6 mM magnesium acetate, 5% glycerol, 50 mM KCl, 0.01% Nonidet P-40). At indicated times, 2 μl of the reaction was stopped by adding 0.5 μl of 2% sodium dodecyl sulfate. One microliter of the stopped reaction was spotted onto a polyethyleneimine-cellulose thin-layer chromatography plate, previously soaked in ethanol and air dried, and subsequently developed in 0.4 M K2HPO4–0.7 M boric acid. A bioimaging analyzer (BAS2000; Fuji) was used to count radiolabeled ATP and Pi (27, 58).
Transient transfection assay.
Transfection assays were performed in HEK-293 cells by using calcium phosphate. Cells were lysed with cell lysis buffer 24 h after transfection, and reporter activities were measured. Reporter activity was induced by cotransfection with the PKA expression vector. Recorded activity was normalized to the protein level of the cell extract, quantified using the Bradford assay (Bio-Rad) and β-Gal activity from RSV-β-gal. All experiments were performed in triplicate. HEK-293T cells were transfected with 100 ng of CRE-Luc or pG5b-Luc reporter plasmid, 50 ng of wild-type or catalytically inactive PKA expression vector (PKAwt or PKAmut, respectively), 200 ng of pRc/RSVmCBP or empty vector (pRc/RSV; Invitrogen), and 0, 50, or 100 ng of full-length RHA expression vector. For assay with the pG5b-Luc reporter plasmid, cells were cotransfected with 100 ng of GAL4-CREB expression vector. Cells were cotransfected 50 ng of RSV-β-gal as a control. The total amount of expression vector was kept constant by adding appropriate amounts of the empty vector pcDNA3-HA.
RESULTS
The region between aa 331 and 380 has transcriptional activity.
We reported previously that RHA activates CREB-dependent transcription by interacting with Pol II through the region extending from aa 255 to 664 amino acids (43). To further understand the mechanism of transactivation mediated by RHA, we carried out a transactivation assay to identify the transactivation domain. First, to confirm whether the region interacting with Pol II has transcriptional activity, we fused four previously described fragments of RHA, termed RHA1 to -4 (Fig. 1A). GAL4-DBD and quantified their transcriptional activities in yeast. As expected, RHA2, which is known to interact with Pol II (43), activated transcription nine fold more than the vector alone. However, RHA1, -3, and -4 did not activate transcription (Fig. 1B).
To identify the minimal region required for transactivation, RHA2 was divided into four fragments, termed RHA2-1, RHA2-2, RHA2-3, and RHA2-4 (Fig. 1C). RHA2-1 (aa 255 to 380), RHA2-2 (aa 381 to 480), RHA2-3 (aa 431 to 581), and RHA2-4 (aa 531–664) contain the BRCA1 binding domain, helicase motif I and an ATP binding site, helicase motifs II (DEIH) and III, and helicase motifs III and IV, respectively (Fig. 1C). Among these four mutants, only RHA2-1 activated transcription 200-fold more than the empty vector. Subsequent N-terminal deletion mutants of RHA2-1 (RHA2-1CL [aa 282 to 380] RHA2-1C [aa 315 to 380], and MTAD [aa 331 to 380]) were then generated. RHA2-1CL activated transcription comparably to RHA2-1. RHA2-1C and MTAD were also capable of activating transcription 30-fold more than the empty vector, which is less than one-seventh the activation induced by RHA2-1. Independently, the N-terminal and C-terminal halves of MTAD (sMTAD [aa 331 to 361] and cMTAD [aa 362 to 380]) did not activate transcription. All of the mutants lacking the MTADs from RHA2-1, RHA2-1CL, and RHA2-1C (RHA2-1NL [aa 255 to 330], RHA2-1M [aa 282 to 330] and RHA2-1NC [aa 315 to 330], respectively did not activate transcription (Fig. 1C and D). These results suggest that the minimal region for transactivation is this 50-aa region.
To examine whether this region functions in higher eukaryotic cells as well as in yeast cells, a reporter assay was performed in HEK-293T cells. A series of deletion mutants fused to GAL4-DBD in a cytomegalovirus promoter-driven expression vector was cotransfected into HEK-293T cells with reporter plasmids containing five GAL4 binding sites upstream of a TATA element. As in yeast, the MTAD was capable of activating transcription in mammalian cells (data not shown). The expression levels of deletion mutants in each experiment were found to be comparable by Western blotting (data not shown). These results show that the minimal region for transactivation of RHA in both yeast and mammalian cells is located between aa 331 and 380.
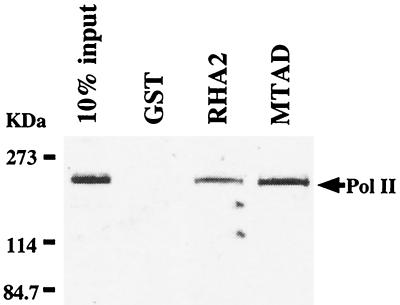
MTAD interacts with Pol II.
We previously reported that the RHA2 region interacts with Pol II (43). To test whether MTAD is in fact the Pol II binding region, a binding assay with GST fusion proteins was performed. MTAD as well as RHA2 interacted with Pol II, while GST alone did not (Fig. 2). We conclude that MTAD is the region interacting with Pol II in RHA2.
FIG. 2.
Interaction of RHA with Pol II in vitro. Nuclear extracts from HEK-293T cells were incubated with GST or deletion mutants of RHA fused to GST. Bound Pol II was visualized by Western blot analysis using the antibody against the largest subunit of Pol II. The input lane represents 10% of the total volume of nuclear extract used in each reaction.
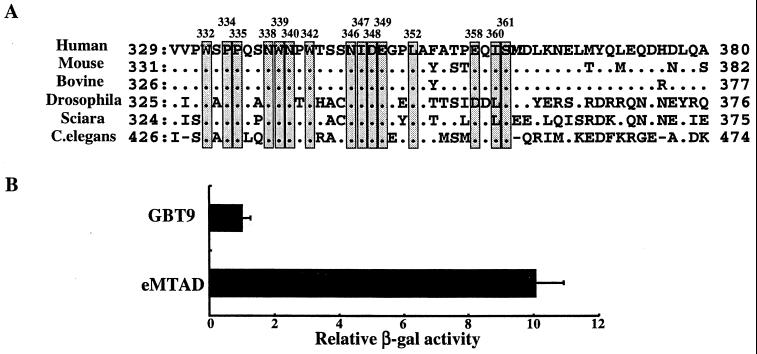
MTAD is well conserved among the RHA homologues.
Homologues of RHA have been reported in six species from C. elegans to humans. Comparison of the amino acid sequences of human MTAD and these homologues showed them to be well conserved, particularly within the N-terminal half (Fig. 3A). This prompted us to examine whether the function of MTAD was also conserved among RHA homologues. The transcriptional activity of the C. elegans MTAD (eMTAD), which is the least conserved homologue of RHA, was examined in both yeast and mammalian cells (data not shown). Transcriptional activity of eMTAD fused to GAL4-DBD was 10-fold greater than that of GAL4 alone (Fig. 3B), suggesting a conserved role for MTAD among RHA homologues.
FIG. 3.
Features of MTAD and the RHA homologues. (A) Alignment of MTAD sequences of RHA homologues from C. elegans to humans. The conserved residues among RHA homologues are indicated with shaded boxes. Mutants with each conserved residue changed to alanine were generated. (B) Transactivation assay with the region in C. elegans corresponding to MTAD. Yeast strain Y190 cells were transformed with eMTAD inserted into pGBT9 or empty vector alone. The β-Gal activity derived from cells transformed with empty vector was designated 1. Each value of relative β-Gal activity represents the mean ± standard error (n = 3).
We attempted to identify other proteins that contain this domain. However, using the BLAST sequence alignment program, MTAD was not found in any other proteins, suggesting that MTAD is unique to RHA and distinct from transactivation domains reported previously.
Hydrophobic residues are important for transactivation.
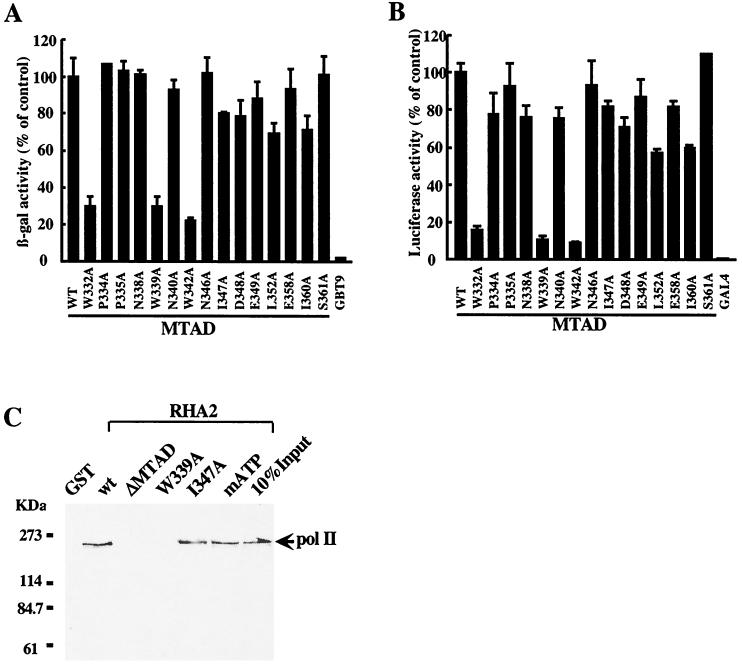
The transactivation domains of some transcriptional factors analyzed previously tend to be classified according to amino acid composition. Domains can be classified as acidic, glutamine rich, or proline rich (54). The MTAD of RHA cannot be classified in this way, as it has few acidic residues and is not glutamine or proline rich. To identify the amino acid residues that are important for transactivation in MTAD, we generated 15 mutants in which residues conserved between RHA homologues were replaced by alanine by site-directed mutagenesis. These mutants are referred to as W332A, P334A, P335A, N338A, W339A, N340A, W342A, N346A, I347A, D348A, E349A, L352A, E358A, I360A, and S361A (Fig. 3A). These MTAD mutations were then fused to GAL4-DBD to examine their effects on transactivation. They are referred to as MTADw332a, MTADp334a, MTADp335a, MTADn338a, MTADw339a, MTADn340a MTADw342a, MTADn346a, MTADi347a, MTADd348a, MTADe349a, MTADl352a, MTADe358a, MTADi360a, and MTADs361a. As shown in Fig. 1, wild-type MTAD activated transcription 50-fold more than the empty vector in yeast cells and 140-fold more in mammalian cells. Three mutants with substituted tryptophan residues, MTADw332a, MTADw339a, and MTADw342a, were approximately 20% as transcriptionally active as wild-type MTAD in both yeast and mammalian cells, whereas the other mutants had levels of activity comparable to that of wild-type MTAD (Fig. 4A and B). These results show that the hydrophobic residues in MTAD play an important role in transactivation.
FIG. 4.
Critical effects of the tryptophan residues in MTAD. Transactivation assays were performed with yeast (A) and mammalian (B) cells transformed with conserved-residue MTAD mutants. Yeast strain Y190 cells were transformed with MTAD mutants or empty vector alone. HEK-293T cells were transfected with 50 ng of MTAD mutants fused to GAL4-DBD or empty vector and 50 ng of pG5b-Luc reporter plasmid. The β-Gal or luciferase activity of each mutant was compared to that of the wild-type MTAD, which was designated 100%. Each value of relative β-Gal or luciferase activity represents the mean ± standard error (n = 3). (C) Binding activities of the RHA2 mutants to Pol II in GST pull-down assay. Nuclear extracts from HEK-293T cells were incubated with the RHA2 mutants fused to GST or GST alone. Western blot analysis was performed with the antibody against Pol II. The input lane represents 10% of the total volume of nuclear extract used in each reaction.
We then examined whether tryptophan residues in MTAD contributed to Pol II binding. A GST pull-down assay was performed with a series of GST-RHA2 mutants. In addition, a mutant lacking ATP binding ability was also used in this assay to assess the importance of the ATP binding site in Pol II binding activity. This mutant, RHA2matp, has an arginine substitution for lysine at position 417. Both RHA2w339a and RHA2Δmtad, which have the region between aa 330 and 376 deleted, did not interact with Pol II (Fig. 4C). Mutants with other substituted tryptophan residues, RHA2w332a and RHA2w342a, gave the same result as RHA2w339a (data not shown), while one mutant (RHA2i347a) which activated transcription comparably to wild-type MTAD in the transactivation assay, and another (RHA2matp) that had the same activity as the wild type both interacted with Pol II. These results show that tryptophan residues in MTAD are important for associating with Pol II.
RHA activates CREB-dependent transcription through MTAD.
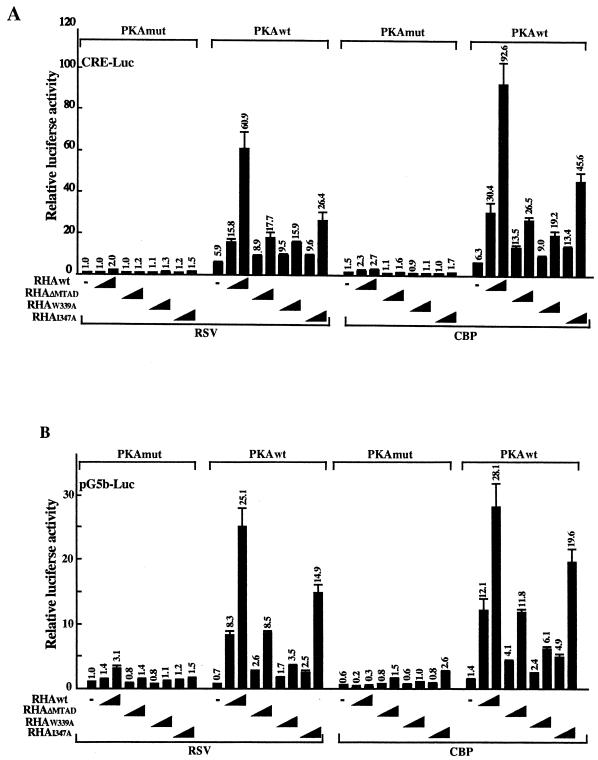
To confirm the contribution of MTAD to RHA-mediated transcription, the effects of MTAD on CREB-dependent transcription were examined using reporter assays with full-length RHA mutants (RHAΔmtad, RHAw339a, and RHAi347a). These mutants were cotransfected with the CRE-Luc reporter and CBP expression plasmid into HEK-293 cells. In PKA-stimulated cells, RHAwt activated CRE-Luc 60-fold, and the activity was further enhanced approximately 1.5-fold with CBP as described previously (43). In contrast, RHAΔmtad enhanced activity 18-fold, which is 30% of the activity shown by RHAwt. Mutants RHAw339a and RHAi347a reduced activity to 30 and 50% of levels induced by RHAwt (Fig. 5A). The activity induced by RHAw339a was the same as that induced by RHAΔmtad. It is therefore clear that MTAD, especially the tryptophan residues, contributes to CRE-dependent transcription. These data suggest that the capacity of MTAD to interact with Pol II is critical to its ability to fully activate transcription.
FIG. 5.
Effects of MTAD and the tryptophan residue on CREB-dependent transcription, determined by transient reporter assays with the CRE-Luc (A) or pG5b-Luc (B) reporter. HEK-293 cells were transfected with 100 ng of CRE-Luc or pG5b-Luc reporter plasmid, 50 ng of PKAwt or PKAmut expression vector, 200 ng of pRc/RSVmCBP or empty vector alone, and 0, 50, or 100 ng of each mutant of full-length RHA expression vector. For assay with the pG5b-Luc reporter, cells were cotransfected with 100 ng of GAL4-CREB expression vector. The total amount of expression vector was kept constant with the addition of empty vector (pcDNA3-HA). Luciferase activity was normalized to the protein level of cell extract quantified by the Bradford assay. The luciferase activity of cells transfected with empty vector alone and PKAmut was designated 1. Each value of relative luciferase activity represents the mean ± standard error (n = 3).
Some transcriptional factors, for example, ATF and CREM, are known to bind to the CRE sequence and regulate CRE-dependent transcription (18, 21, 37). To rule out possible interference from CRE binding proteins other than CREB, a reporter assay was performed with pG5b-Luc. Instead of CRE-Luc, CREB fused to GAL4-DBD and pG5b-Luc were cotransfected with RHA into HEK-293 cells. The ability of RHAΔmtad and RHAw339a to activate transcription was reduced (Fig. 5B), consistent with results previously reported for CRE-Luc. Western blot analysis showed that the expression level of these mutants was comparable to the wild-type level (data not shown). These results suggest that MTAD has an important role in CREB-dependent transcription.
The W339A mutant has ATP binding and ATPase activities.
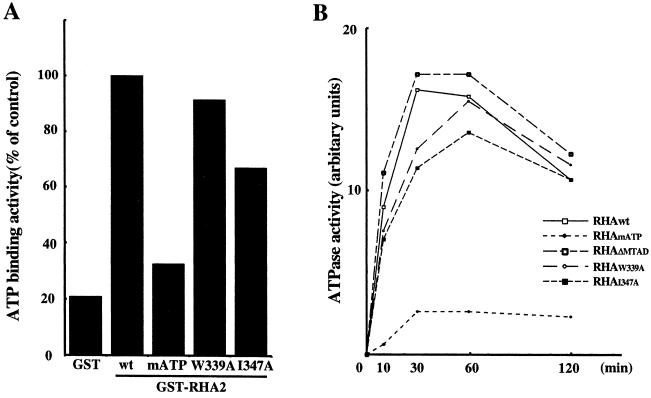
As reported previously, ATPase and/or helicase activities of RHA are required for CREB-dependent transactivation (43). It is possible that mutation of hydrophobic residues in MTAD could affect the ATPase and helicase activities of RHA. Therefore, ATP binding and ATPase activities of mutants were examined. An ATP binding assay showed that RHA2matp had 30% of the ATP binding activity of RHAwt, while RHA2w339a and RHA2i347a had ATP binding activity comparable to that of RHAwt (Fig. 6A).
FIG. 6.
Influence of hydrophobic residue mutations in MTAD on ATP-dependent activities of RHA. (A) ATP binding assay. RHA2 mutants fused to GST were incubated with [γ-32P]ATP, and bound ATP was measured. The ATP binding activity of mutants was compared to that of wild-type RHA2 (wt), which was designated 100%. (B) ATPase assay. Full-length RHA mutants were synthesized using an in vitro translation system and subsequently immunopurified with anti-HA antibody. The immunocomplexes were incubated with [γ-32P]ATP. ATP hydrolysis was monitored over time by separation of ATP and Pi by thin-layer chromatography. hnRNP A1 was used as a negative control, and its ATPase activity was subtracted from each value.
ATPase activity was measured with HA-tagged RHA mutants synthesized using an in vitro translation system and immunopurified with anti-HA antibody. The negative control was hnRNP AI, which does not belong to the ATPase family. While RHAmatp could not hydrolyze ATP, RHAw339a and RHAΔmtad hydrolyzed ATP at a level comparable to that of RHAwt (Fig. 6B). These results show that the mutations in MTAD had no effect on ATP binding and ATPase activities of RHA.
RHA has dual effects on CREB-dependent transcription.
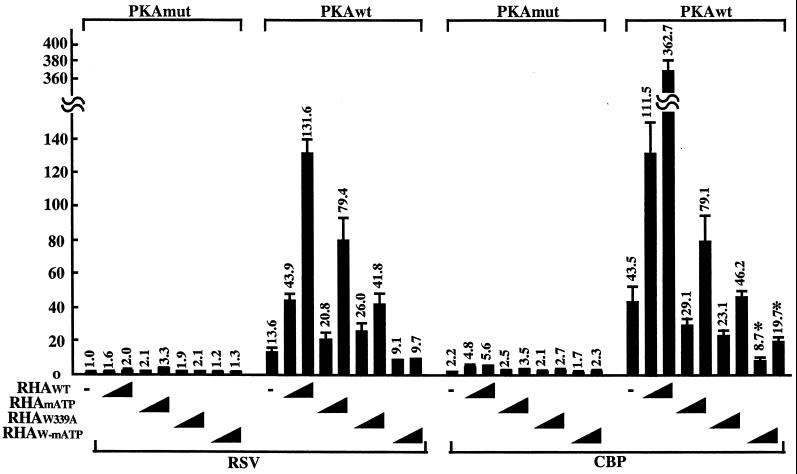
To test whether the role of the tryptophan residue at position 339 in MTAD can be distinguished from ATPase or helicase activity in CREB-dependent transcription, a double mutant (RHAw-matp) with a mutated ATP binding site and an alanine substitution for tryptophan at position 339 was generated. The activity of RHAw-matp was measured in a reporter assay. As described above, RHAwt enhanced CREB-dependent transcription in PKA-stimulated cells. Mutants RHAmatp and RHAw339a reduced CREB-dependent transcription to 50 and 30%, respectively, of the levels obtained with RHAwt. The double mutant induced transcription to only 5% of the wild-type level, which is lower than for each individual mutant. Furthermore, the transcriptional activity of the double mutant was significantly lower than that of the empty vector. The double mutant appeared to have a dominant negative effect on transcription (Fig. 7). Western blot analysis confirmed that the expression level of each of these mutants is comparable to the wild-type level (data not shown). These results suggest that RHA could independently activate CREB-dependent transcription either through interaction with Pol II or by ATP-dependent mechanisms.
FIG. 7.
Dual roles of RHA on CREB-dependent transcription. Transient reporter assays were performed using the CRE-Luc reporter with the RHA double mutant containing the mutations RHAw339a and RHAmatp. HEK-293 cells were transfected with 100 ng of CRE-Luc reporter plasmid, 50 ng of PKAwt or PKAmut, 200 ng of pRc/RSVmCBP or empty vector alone, and 0, 50, or 100 ng of each mutant of the full-length RHA expression vector. The total amount of expression vector was kept constant with the addition of empty vector (pcDNA3-HA). Luciferase activity (shown on the y axis) was normalized to the protein level of cell extract and β-Gal activity from cotransfected RSV-β-gal control plasmid. The luciferase activity from the empty vector alone (PKAmut) was designated 1. Each value of relative luciferase activity represents the mean ± standard error (n = 3). (∗, P < 0.035, RHAw-matp versus empty vector).
DISCUSSION
In this study, we defined a stretch of 50 amino acid residues between aa 331 and 380 as the MTAD. This domain has transcriptional activity in both yeast and mammalian cells (Fig. 1) and interacts with Pol II (Fig. 2). Sequence analysis revealed that this domain contains six conserved hydrophobic residues unique to the MTAD of RHA homologues (Fig. 3). Mutational analyses indicated that the tryptophan residues are essential for transactivation and Pol II binding of MTAD.
Transactivation domains can generally be classified as acidic, glutamine rich, and proline rich on the basis of amino acid composition. In some cases, it has been shown that hydrophobic residues are important for transcriptional activity (54). Alanine scanning mutagenesis has revealed a role for hydrophobic residues in other acidic transactivation domains from transcription factors such as VP16 (13, 49, 56), RelA (p65) (6, 51), p53 (31, 36, 57), glucocorticoid receptor (2, 3), and GCN4 (24). For VP16 and p53, it was reported that the aromatic residues in particular were important for transactivation, as determined by mutational analysis. Structural analysis also revealed that tryptophan at position 23 in p53 (31) and phenylalanine at position 479 in VP16 (49) were important for interaction with MDM2 and TATA binding protein-associated factor TAFII31, respectively. The importance of tryptophan residues was also reported for Spl and CREB (20, 50). It was suggested that the alternating glutamine and hydrophobic amino acid sequence motifs, which include tryptophan residues, could represent a surface for interaction with TAFs. The MTAD of RHA is neither acidic nor glutamine rich (Fig. 3A), and mutational analysis revealed that a few acidic amino acids were not important for transactivation (Fig. 4A, and B). Therefore, we expect that MTAD may be a novel transactivation domain utilizing hydrophobic residues unique to RHA. Previous studies have reported the presence of CBP and RHA within a holo-Pol II complex (12, 42, 44). In yeast, holo-Pol II complexes contain several components, including Pol II, general transcription factors, the mediator and SWI-SNF complexes, and other proteins (23, 40). Some complexes in mammals including Pol II are homologues of complexes found in yeast (5, 11, 38). In this study, MTAD fused to GAL4-DBD activated transcription in yeast and mammalian cells, even though RHA is not found in yeast. Therefore, we expect that this particular transactivation domain could recruit the general component of holo-Pol II conserved in eukaryotes.
Three tryptophan residues in MTAD are critical for transactivation and interaction with Pol II (Fig. 4). It remains to be clarified whether it is hydrophobicity or the aromatic side chain of tryptophan that is critical for interaction with Pol II. In general, hydrophobic residues are located within proteins, while polar and charged residues prefer surfaces. In contrast, tryptophan residues are found on surfaces and in the interior with nearly identical frequencies. Although tryptophan residues are comparatively rare, they are statistically most likely to be found at interfaces (7, 55). This is because tryptophan can contribute aromatic π interactions, is a hydrogen donor, and has a large hydrophobic surface (7). It is possible that MTAD has three tryptophan residues, instead of polar or charged residues, that enable it to act as an interface on the surface of RHA. However, the role of the tryptophan residues in MTAD is not understood. The tryptophan residues may be necessary for maintaining the structure of MTAD or may be directly involved in the interaction with Pol II. These questions can be addressed by probing the structure of MTAD, using spectroscopic or crystallographic techniques, or by exploring the association of MTAD with putative target proteins within holo-Pol II complexes.
Our results indicate that the minimal region required for transactivation and Pol II binding is MTAD. However in transactivation assay, RHA2-1 and RHA2-1CL are more active than MTAD. Experiments with RHA2-1NL, RHA2-1M, and RHA2-1NC revealed that there is no additional transactivation domain in RHA2-1 region and activation by RHA2-1 depends on MTAD. There are several explanations, not mutually exclusive, for the higher activity of RHA2-1 and RHA2-1CL. First, the N-terminal region of RHA2-1CL might play a role in stabilization of interaction between MTAD and Pol II. Second, additional factors with no transcriptional activity might bind to the N-terminal region of RHA2-1CL and stabilize the formation of the complex or enhance transcriptional activation by MTAD. Structural analyses of the MTAD and Pol II complex, in combination with investigation into the component of holo-Pol II that interacts with MTAD, are expected to clarify this question.
In a previous study, we showed that RHA could activate CREB-dependent transcription with its ATPase and/or helicase activity (43). Members of the ATPase/helicase family play important roles in many transcriptional processes. They are essential for initiation and transcription-coupled repair and may have important roles in elongation, termination, and transcript stability (16). For example, ATPases/helicases such as TFIIH and the chromatin remodeling complexes participate in transcription, especially in initiation and preinitiation. The enzymatic activities of XPB contained in TFIIH are required for promoter opening (15, 53). It is suggested that the chromatin remodeling complexes, such as SWI-SNF and NURF (nucleosome remodeling factor), could alter chromatin structure with the energy of ATP hydrolysis (22, 26, 52). In addition, MLE, the Drosophila homologue of RHA located within the X chromosome, may be utilized for chromatin remodeling of X-linked genes (34). In a preliminary study, we observed that RHA interacts specifically with the hypophosphorylated form of Pol II (Pol IIA) (S. Aratani and T. Nakajima, unpublished data). It is possible that RHA is involved in preinitiation and/or initiation of transcription. It is tempting to speculate that RHA may contribute to chromatin remodeling and/or transcriptional initiation. However, it is not known which transcriptional activation processes require RHA.
Some factors, e.g., transcriptional regulators such as CREB, CBP, and TFIIH, regulate transcription through dual mechanisms. CREB has bipartite transactivation domains consisting of constitutive and inducible activators, termed Q2 and kinase-inducible domain (KID), respectively (9, 47). The glutamine-rich Q2 domain engages the transcriptional apparatus via a constitutive interaction with human TAFII130 (17, 42). By contrast, the KID region modulates CREB activity via a phosphorylation-dependent association with CBP (10, 46). CBP activates transcription with its histone acetyltransferase activity and functions as a molecular platform for transcriptional activators. It is reported that CREB and STAT1 require both functions of CBP, while nuclear receptors do not require the histone acetyltransferase activity of CBP (28, 30). TFIIH contains two helicases (XPB and XPD) and a kinase (cdk7). Promoters exhibit differential requirements for the kinase activity of cdk7 (1, 53). As described above, it is possible that these transcriptional factors utilize each mechanism differentially according to the situation. In this study, we show that RHA may have dual roles for transactivation. The double mutant lacking the Pol II binding ability and ATP-dependent activity reduced transcription significantly less than each individual mutant (Fig. 7). These results raise the possibility that each function of RHA may be utilized independently in transactivation. In addition, RHA could participate in transcription mediated by several factors. The dual transactivation mechanisms may allow RHA to regulate a broad range of transcription exquisitely in different situations.
ACKNOWLEDGMENTS
We thank Noriyuki Hatae for critical discussions. We also thank Yukiko Okada and Megumi Fujita for technical assistance.
This work was supported by grants from the Japanese Ministry of Education, Science, Culture, and Sports (10480196 and 10177230), Japanese Ministry of Health and Welfare, JST (PRESTO), and Human Health Science Foundation and by funds from the Memorial Yamanouchi Foundation, Kaken Pharmaceutical Co. Ltd., and Santen Pharmaceutical Co. Ltd.
REFERENCES
- 1.Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 2.Almlof T, Gustafsson J A, Wright A P. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Mol Cell Biol. 1997;17:934–945. doi: 10.1128/mcb.17.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almlof T, Wallberg A E, Gustafsson J A, Wright A P. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor tau 1-core activation domain and target factors. Biochemistry. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson S F, Schlegel B P, Nakajima T, Wolpin E S, Parvin J D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 5.Bjorklund S, Almouzni G, Davidson I, Nightingale K P, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. doi: 10.1016/s0092-8674(00)80586-3. [DOI] [PubMed] [Google Scholar]
- 6.Blair W S, Bogerd H P, Madore S J, Cullen B R. Mutational analysis of the transcription activation domain of ReIA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogan A A, Thorn K S. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 8.Bone J R, Lavender J, Richman R, Palmer M J, Turner B M, Kuroda M I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 9.Brindle P, Linke S, Montminy M. Protein-kinase-A-dependent activator in transcription factor CREB reveals new role for CREM repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 10.Brindle P, Nakajima T, Montminy M. Multiple protein kinase A-regulated events are required for transcriptional induction by cAMP. Proc Natl Acad Sci USA. 1995;92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 12.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 14.Dorris D R, Struhl K. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol Cell 2Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 16.Eisen A, Lucchesi J C. Unraveling the role of helicases in transcription. Bioessays. 1998;20:634–641. doi: 10.1002/(SICI)1521-1878(199808)20:8<634::AID-BIES6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 17.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foulkes N S, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 19.Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J Biol Chem. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 20.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hai T W, Liu F, Coukos W J, Green M R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1990;4:682. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 22.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 23.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 24.Jackson B M, Drysdale C M, Natarajan K, Hinnebusch A G. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley R L, Kuroda M I. Equality for X chromosomes. Science. 1995;270:1607–1610. doi: 10.1126/science.270.5242.1607. [DOI] [PubMed] [Google Scholar]
- 26.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 27.Klemm R D, Austin R J, Bell S P. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 28.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda M I, Kernan M J, Kreber R, Ganetzky B, Baker B S. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell. 1991;66:935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 31.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 32.Lee C G, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J Biol Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 33.Lee C G, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 34.Lee C G, Chang K A, Kuroda M I, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;16:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C G, da Costa Soares V, Newberger C, Manova K, Lacy E, Hurwitz J. RNA helicase A is essential for normal gastrulation. Proc Natl Acad Sci USA. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 37.Maekawa T, Sakura H, Kanei-Ishii C, Sudo T, Yoshimura T, Fujisawa J, Yoshida M, Ishii S. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J. 1989;8:2023–2028. doi: 10.1002/j.1460-2075.1989.tb03610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 39.Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. Regulation of lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J Biol Chem. 2000;275:35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- 40.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 41.Myohanen S, Baylin S B. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 43.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 44.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nevado J, Gaudreau L, Adam M, Ptashne M. Transcriptional activation by artificial recruitment in mammalian cells. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn P G. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 48.Rastelli L, Kuroda M I. An analysis of maleless and histone H4 acetylation in Drosophila melanogaster spermatogenesis. Mech Dev. 1998;71:107–117. doi: 10.1016/s0925-4773(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 49.Regier J L, Shen F, Triezenberg S J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojo-Niersbach E, Furukawa T, Tanese N. Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- 51.Schmitz M L, dos Santos Silva M A, Altmann H, Czisch M, Holak T A, Baeuerle P A. Structural and functional analysis of the NF-kappa B p65 C terminus. An acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 52.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 53.Tirode F, Busso D, Coin F, Egly J M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 54.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 55.Tsai C J, Lin S L, Wolfson H J, Nussinov R. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 57.Uesugi M, Verdine G L. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc Natl Acad Sci USA. 1999;96:14801–14806. doi: 10.1073/pnas.96.26.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 59.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida E, Aratani S, Itou H, Miyagishi M, Takiguchi M, Osumu T, Murakami K, Fukamizu A. Functional association between CBP and HNF4 in trans-activation. Biochem Biophys Res Commun. 1997;241:664–669. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 63.Zhang S S, Grosse F. Purification and characterization of two DNA helicases from calf thymus nuclei. J Biol Chem. 1991;266:20483–20490. [PubMed] [Google Scholar]