Genetic screening revealed a novel function for MCAD in glioblastoma, where in addition to its energetic role, it prevents toxic lipid accumulation, and targeting MCAD causes irreversible damage and cell death in glioblastoma cells, while sparing normal cells.
Abstract
Glioblastoma (GBM) is highly resistant to chemotherapies, immune-based therapies, and targeted inhibitors. To identify novel drug targets, we screened orthotopically implanted, patient-derived glioblastoma sphere-forming cells using an RNAi library to probe essential tumor cell metabolic programs. This identified high dependence on mitochondrial fatty acid metabolism. We focused on medium-chain acyl-CoA dehydrogenase (MCAD), which oxidizes medium-chain fatty acids (MCFA), due to its consistently high score and high expression among models and upregulation in GBM compared with normal brain. Beyond the expected energetics impairment, MCAD depletion in primary GBM models induced an irreversible cascade of detrimental metabolic effects characterized by accumulation of unmetabolized MCFAs, which induced lipid peroxidation and oxidative stress, irreversible mitochondrial damage, and apoptosis. Our data uncover a novel protective role for MCAD to clear lipid molecules that may cause lethal cell damage, suggesting that therapeutic targeting of MCFA catabolism may exploit a key metabolic feature of GBM.
Significance:
MCAD exerts a protective role to prevent accumulation of toxic metabolic by-products in glioma cells, actively catabolizing lipid species that would otherwise affect mitochondrial integrity and induce cell death. This work represents a first demonstration of a nonenergetic role for dependence on fatty acid metabolism in cancer.
This article is highlighted in the In This Issue feature, p. 2659
Introduction
Glioblastoma (GBM) has a median survival of 15 to 19 months (1–3). Despite extensive efforts, studies of both signal transduction inhibitors and immune-targeted agents have failed to improve the prognosis for patients with these tumors (4–8). As with most cancers, altered metabolism is a defining characteristic of glioblastoma. Dependency on aerobic glycolysis (Warburg effect), as well as on glutaminolysis, is well documented (9–11), and recent landmark studies have identified mutations in GBM that directly affect mitochondrial metabolism (12, 13). However, the molecular drivers underlying this metabolic reprogramming are only partly understood, and therapeutically actionable metabolic dependencies have not been fully determined.
In the present study, we applied an in vivo genetic screen platform (14) to orthotopically implanted, patient-derived three-dimensional glioma tumor spheres and identified mitochondrial fatty acid (FA) metabolism as an essential pathway required for tumor growth. From among the enzymes in the β-oxidation pathway that emerged as hits, we selected medium-chain acyl-CoA dehydrogenase (MCAD), a mitochondrial enzyme that catalyzes the first step of medium-chain FA (MCFA) β-oxidation (FAO), for deep functional characterization. The uptake of MCFAs lacks any negative feedback regulation; therefore, in the absence of MCAD, MCFAs accumulate to toxic levels. Still, inherited MCAD deficiency, which is screened for perinatally, is a condition compatible with normal quality of life, and afflicted individuals require only dietary adjustment to thrive (15, 16). These facts are suggestive of a therapeutic window to inhibit MCAD in lipid-addicted tumor types.
Both in vitro and in vivo, our studies showed that targeting MCAD in primary GBM models triggered massive accumulation of unmetabolized MCFAs in the mitochondria, which resulted in peroxidation of FA by-products. This overload of partially metabolized substrates generated oxidative stress that irreparably damaged the mitochondrial structure and caused tumor cell death. Thus, in GBM, the activity of MCAD does not solely support cellular energetics as part of the β-oxidation cycle, and our work demonstrates a previously unappreciated protective role for MCAD FA catabolism to clear toxic accumulations of lipid molecules that may otherwise cause lethal damage to the cell.
Results
ACADM Is an Essential Gene in Glioblastoma
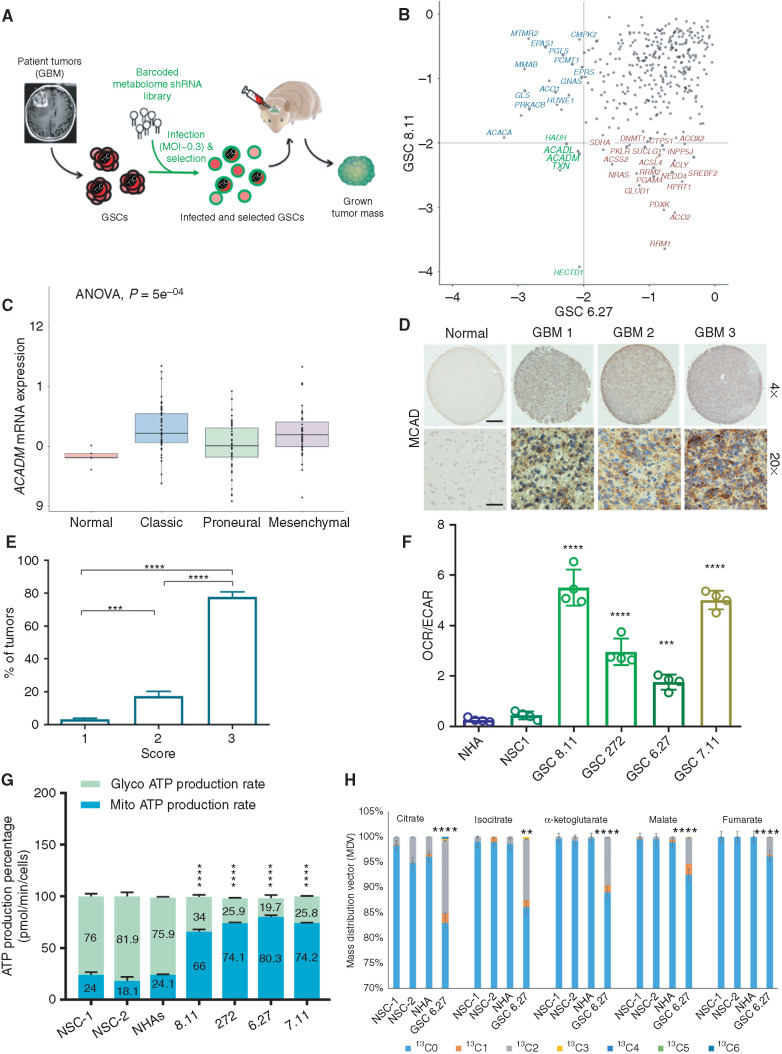
To achieve a functional output of metabolic gene functions that might be essential for GBM cell survival, we generated a prioritized list of 330 metabolism genes and created a barcoded, deep-coverage [10 short hairpin RNAs (shRNA)/gene] shRNA library that encompasses a wide range of metabolic pathways and activities (Supplementary Fig. S1A). As a disease model, we selected low-passage, three-dimensional–cultured, patient-derived glioma sphere-forming cells (GSC) established at the MD Anderson Brain Tumor Center (17–19). Among more than 70 genomically characterized GSCs available to us, we selected two carrying distinct gene expression signatures classified as belonging to the proneural (GSC 8.11) or classical (GSC 6.27; Supplementary Fig. S1B) subtypes for the genetic screen. After shRNA library transduction, cells were implanted intracranially, and tumors were grown until mice showed tumor-related neurologic symptoms (Supplementary Fig. S1C). At endpoint, tumors were excised and processed as previously described (ref. 14; Fig. 1A). Analysis of the most significantly depleted shRNAs in both GBM models uncovered the potential role of several mitochondrial enzymes involved in FA metabolism in GBM growth (Fig. 1B; Supplementary Fig. S1D–S1G). Given compelling evidence of its role in FA metabolism in brain tissues, we focused our attention on MCAD, which oxidizes medium-chain (4- to 12-carbon) FAs. MCFAs such as octanoate are normally present in plasma at significant levels (20) and readily cross the blood–brain barrier (21). It has been shown that up to 20% of brain oxidative metabolism can be attributed to octanoate use in an intact rodent physiologic system (22).
Figure 1.
ACADM emerges as a clinically relevant dependency of GBM. A, Schematics of experimental design for intracranial metabolome shRNA screens in patient-derived GSCs. The lentiviral library was transduced at a low MOI (less than one integrant/cell). B, Gene–rank analysis highlighting the behavior of genes involved in FA metabolism (ACADM, ACADL, PRKACB, and ACSL6) in in vivo screens executed in two independent GSC models: GSC 8.11 and GSC 6.27 (RSA, logP). C,ACADM mRNA levels in glioma subtypes vs. normal brain (TCGA data set). Log2 RSEM values were calculated to compare expression levels of ACADM and other genes involved in the FA synthesis pathway. Using the GBM data set (n = 167), analysis was performed comparing expression among normal tissue (n = 5), classic (n = 68), proneural (n = 46), and mesenchymal (n = 48) subtypes. ANOVA was performed across all groups to determine statistical significance, with pairwise Wilcoxon tests to identify which subtypes showed a difference. D, Immunohistochemistry for MCAD on tissue microarray (TMA) derived from normal brain and GBM tissue. Scale bars, 100 μm for ×4 and 25 μm for ×20. E, GBM percentage distribution based on MCAD expression levels in three independent TMAs. The scores 1 to 3 were independently determined using the following scoring system to approximate the percentage of cells positive for staining with the MCAD antibody: 1 = 0% to 10%, 2 = 11% to 50%, and 3 = 51% to 100%. Representative tissue scoring is presented in D. Data represent the analysis of three independent TMAs. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ***, P = 0.0002; ****, P < 0.0001. F, Bioenergetic profiling of NHA, NSC, and GSC lines using Seahorse technology. Basal oxygen consumption rate (OCR; pMoles/minute) and extracellular acidification rate (ECAR; mpH/minute) were used for calculations. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ***, P = 0.0003; ****, P = 0.0001. G, Quantification of energy production in the indicated cell lines by Seahorse XF Real-Time ATP Rate Assay. MitoATP Production Rate and glycoATP Production Rate were calculated from OCR and ECAR measurements under basal conditions. Values are expressed as mean ± SD; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ****, P < 0.0001. H, Isotopolog patterns for incorporation of 13C-labeled oleate into TCA cycle intermediates, as measured by LC/MS in NHAs, NSCs, and GSCs in basal conditions. Cells were cultured with 13C-oleate for 6 hours prior to sample collection. N = 4 biological replicates, error = ±SD. **, P = 0.0014; ****, P < 0.0001.
To explore a possible role for ACADM (the gene encoding MCAD) in GBM, we queried The Cancer Genome Atlas (TCGA) mRNA data set and found that ACADM is highly expressed across GBM subtypes compared with normal brain (Fig. 1C). Through IHC analysis, we also determined that MCAD protein levels are elevated compared with normal brain tissue in a GBM tumor microarray series (Fig. 1D and E). For in vitro studies, we selected normal neural stem cells (NSC) and normal human astrocytes (NHA) to represent normal brain cells, the latter based on previous reports of high ACADM mRNA levels relative to other brain-derived cells (ref. 23; Supplementary Fig. S1H) and also given recent findings that astrocyte-like NSCs might be the GBM cell of origin (24). We confirmed lower MCAD levels in NSCs and NHAs compared with GSCs, with NSCs showing barely detectable MCAD levels (Supplementary Fig. S2A). A similar trend was observed in the relative expression levels of other enzymes involved in β-oxidation (Supplementary Fig. S2B). Consistent with expression levels and previous reports (25), analysis of mitochondrial bioenergetics demonstrated elevated oxidative metabolism in GSCs compared with NSCs and NHAs (Fig. 1F). Further, we found that GSCs produce ATP predominantly through mitochondrial metabolism (65% to 83%), whereas NSCs and NHAs generate ATP primarily via glycolysis (∼75%; Fig. 1G), thus emphasizing the importance of oxidative metabolism in GBM. To functionally validate our findings, we performed metabolic flux analysis using 13C-oleate as the substrate. Strikingly, these data demonstrated a significant enrichment of 13C in tricarboxylic acid (TCA) cycle metabolites in GSCs compared with NSCs and NHAs (Fig. 1H; Supplementary Fig. S2C), thus supporting that FAO strongly contributes to an enhanced oxidative metabolism in GBM.
MCAD Depletion Impairs Glioblastoma Growth In Vitro and In Vivo
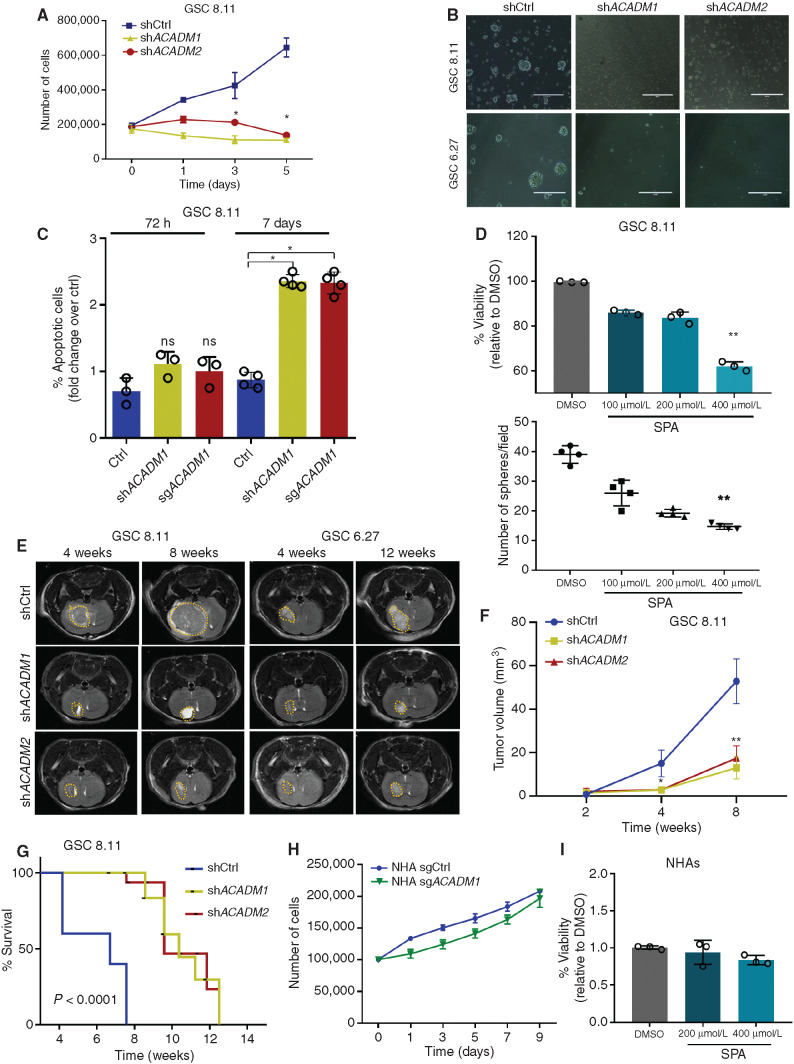
To further investigate the relevance of MCAD in this context, we validated two independent ACADM-targeting RNA interference (shRNA) constructs for their ability to downregulate MCAD protein expression (Supplementary Fig. S3A). We found that MCAD depletion dramatically impaired anchorage-independent growth in multiple GSCs representing different GBM subtypes (Fig. 2A and B; Supplementary Fig. S3C), whereas MCAD overexpression rescued viability (Supplementary Fig. S3B). Similar effects on GSC growth were obtained by ACADM deletion through CRISPR/Cas9 editing using three independent single-guide RNA (sgRNA) constructs (Supplementary Fig. S3D–S3F). To better understand the effects of ACADM downregulation on GSCs, we evaluated apoptosis in a time course experiment, which demonstrated that the growth arrest induced by ACADM ablation precedes a significant increase in apoptotic cell death (annexin V positivity) detected approximately 7 days after the end of selection in all GSCs tested (Fig. 2C; Supplementary Fig. S4A and S4B). Indeed, no significant effect on cell viability was observed at earlier time points (72 hours; Fig. 2C; Supplementary Fig. S4A). Similar effects on cell growth and sphere-forming efficiency were observed upon treating GSCs with spiropentaneacetic acid (SPA), a compound known to specifically inhibit MCAD activity (ref. 26; Fig. 2D; Supplementary Fig. S4C and S4D).
Figure 2.
MCAD is essential for in vitro and in vivo tumor growth. A, Five-day growth curve of GSC 8.11 cells upon shRNA ACADM silencing. Day 0 was defined as 48 hours postpuromycin selection. Values represent the mean ± SD of three independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P ≤ 0.02. B, Representative optical microscopy images of GSC tumor spheres 4 days after puromycin selection. Scale bar, 100 μm. C, Quantification of apoptosis in GSC 8.11 infected with anti-ACADM or scrambled shRNA by Annexin V–FITC/PI. Staining was evaluated by flow cytometry at 72 hours and on day 7 after puromycin selection. Values represent the mean ± SD of three or four independent experiments; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.04. D, Cell viability assessed by Trypan blue exclusion of GSC 8.11 treated with indicated concentration of SPA (top). Data represent mean ± SEM of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P = 0.0065. Dot plot showing GSCs 8.11 sphere formation efficiency upon SPA treatment at indicated concentrations (bottom). DMSO was used as control. Values represent the mean ± SD of four independent experiments; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P = 0.0011. E, MRI images of tumor progression after implantation of GSCs 8.11 or 6.27 at 4 and 8 weeks for GSCs 8.11 and at 4 and 12 weeks for GSC 6.27. F, Quantification of tumor progression after implantation of GSC 8.11 as measured by magnetic resonance volumetry (n = 8 mice per group). P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.04; **, P = 0.0082. G, Kaplan–Meier survival analysis after implantation of GSCs 8.11. For shCtrl, shACADM1, and shACADM2, n = 8 mice per group. P values were generated using log–rank test. H, Growth curve of NHAs infected with sgRNA targeting either ACADM or GFP. Values represent the mean ± SD of three independent experiments. I, Cell viability assessed by Trypan blue exclusion of NHAs treated with indicated concentrations of SPA or DMSO for 6 days. Mean values ± SD of three biologically independent replicates.
To determine the effect of MCAD loss in vivo, GSC 8.11 and GSC 6.27 cells harboring ACADM-targeting or nontargeting shRNA constructs were implanted into the mouse forebrain, and tumor growth was monitored by luciferase and MRI. MCAD depletion dramatically attenuated tumor growth and significantly extended survival time (Fig. 2E–G; Supplementary Fig. S5A–S5C). Similarly, MCAD depletion in tumors established through intracranial injection of GSC 8.11 cells carrying doxycycline-inducible shRNA constructs resulted in significant retardation of tumor growth (Supplementary Fig. S5D). We next generated ACADM-deleted NHAs and NSCs using our validated sgRNA guides. Interestingly, neither the cytotoxic nor the antiproliferative effects observed in GSCs were observed in NHAs and NSCs (Fig. 2H; Supplementary Fig. S5E and S5F). Likewise, pharmacologic MCAD inhibition with SPA did not result in significant toxicity in NHAs (Fig. 2I). Given the very low levels of MCAD in NSCs (Supplementary Fig. S2A), we considered NHAs more suitable to represent the normal brain counterpart for further validation experiments. Altogether, these data suggest that MCAD dependency may be a metabolic vulnerability unique to malignant cells.
ACADM Silencing Impairs Mitochondrial Function in GSCs
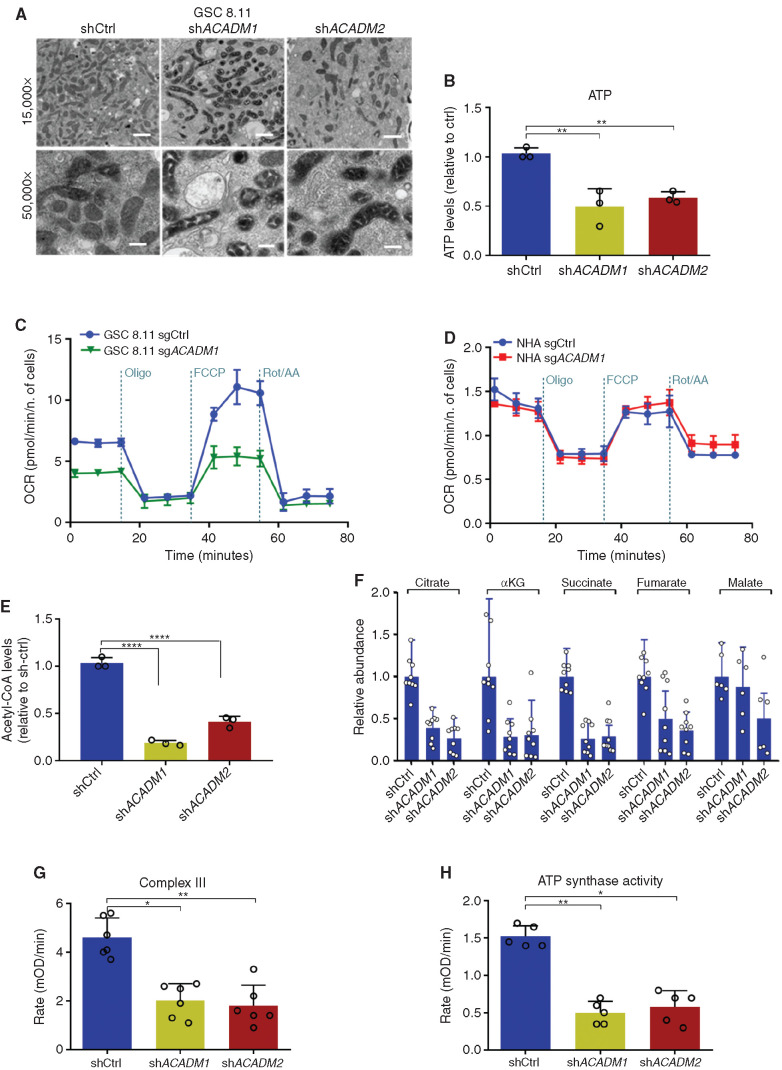
Given that GSC energetics seem to be heavily reliant on oxidative metabolism (Fig. 1F and G), we characterized the early effects of MCAD downregulation on mitochondria between 48 and 72 hours post-MCAD depletion (shRNA/sgRNA ablation). First, we acquired transmission electron microscopy (TEM) images revealing that MCAD depletion resulted in mitochondria with darker matrices and swollen cristae surrounded by vacuolar structures (Fig. 3A; Supplementary Fig. S6A). Next, a bioluminescence assay uncovered an overall decrease in ATP content in MCAD-depleted GSCs versus controls (Fig. 3B). Analysis of oxygen consumption rate (OCR) in GSCs and NHAs revealed significant decreases in basal respiration and reserve respiratory capacity in ACADM-deleted GSCs (Fig. 3C; Supplementary Fig. S6B), whereas ACADM deletion did not affect OCR in NHAs (Fig. 3D). The depletion of the acetyl-CoA pool and TCA intermediates upon ACADM downregulation (Fig. 3E and F; Supplementary Fig. S6C), together with an increased contribution of carbon skeletons derived from FAs to central carbon metabolism in GBM (Fig. 1H), confirms the critical dependence of GBM cells on oxidative metabolism, largely fueled by FAO, and the role of MCAD to support mitochondrial function in this context.
Figure 3.
ACADM silencing causes mitochondrial failure in GSCs. A, Transmission electron microscopy images of mitochondria in GSC 8.11 upon ACADM silencing (magnification ×15,000 and 50,000; scale bars, 1 μm, 300 nm). B, Measurements of mitochondrial ATP levels using a bioluminescence assay. Values are expressed as mean ± SD of three independent experiments; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P < 0.006. c and D, OCR measured in GSC 8.11 (C) and in NHAs (D) following ACADM silencing by metabolic flux assay before and after the addition of oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and rotenone/antimycin to perturb mitochondrial respiration. Values represent the mean ± SD of one representative experiment with n = 4 technical replicates. Experiments were repeated three times with similar results (see also Supplementary Fig. S6B). E, Intracellular acetyl-CoA levels in GSC 8.11 after ACADM silencing as assessed by fluorometric assay. Values are expressed as mean ± SD of three independent experiments; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ****, P = 0.0001. F, Relative abundance of TCA cycle metabolites in GSC 8.11, as detected by LC/MS analysis. Values are expressed as mean ± SD of three independent experiments. G, Mitochondrial electron transport chain complex III activity in GSC 8.11 as measured by colorimetric assay. Values are expressed as mean ± SD of six independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.03; **, P = 0.0079. H, Mitochondrial complex V (ATP synthase) activity in GSC 8.11 as measured by colorimetric assay. Values are expressed as mean ± SD of five independent experiments; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.0313; **, P = 0.0071.
To distinguish whether the observed toxicity of MCAD inactivation is triggered by mitochondrial dysfunction or by the energy deprivation resulting from the inability of GSCs to oxidize FAs, we grew ACADM-silenced GSCs in medium supplemented with acetate, which is a source of carbon molecules that can bypass MCAD activity by directly fueling the TCA cycle (acetyl-CoA), or with long-chain FAs (LCFA; e.g., linoleic acid) that are not metabolized by MCAD. Neither acetate nor LCFA supplementation rescued proliferation in MCAD-depleted GSCs (Supplementary Fig. S6D and S6E), which strongly suggests that the depletion of energy substrates following MCAD ablation may be the consequence of compromised mitochondrial function. In support of this hypothesis, we observed significant impairment of mitochondrial complex III and complex V (ATP synthase) activity in MCAD-depleted versus MCAD-proficient cells (Fig. 3G and H).
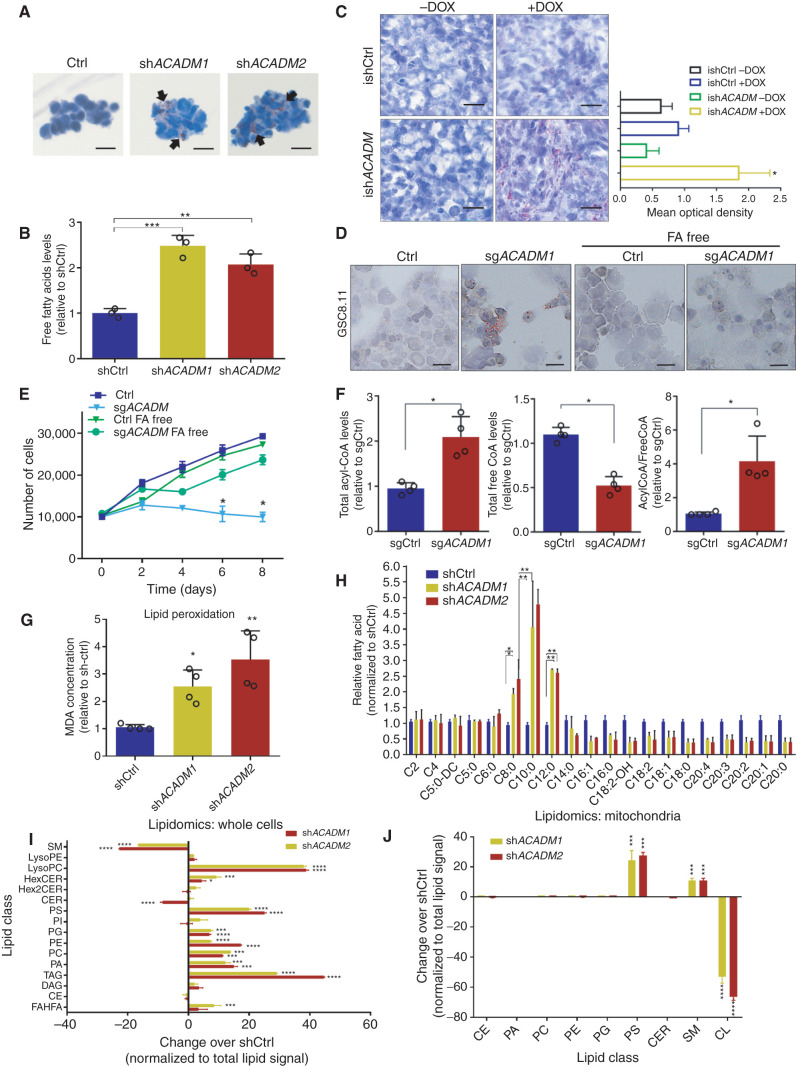
The physiologic role of MCAD is to degrade MCFAs that freely diffuse into cells (27). Thus, we hypothesized that decreased MCAD function may result in a toxic accumulation of lipids that are normally diverted to energy production, triggering mitochondrial failure. Consistent with this hypothesis, quantification of lipids in GSCs using Oil Red O lipid staining revealed massive accumulation of lipid droplets in vitro as early as 48 hours after puromycin selection in ACADM-silenced cells, as well as in GSCs pharmacologically treated with SPA (Fig. 4A; Supplementary Fig. S7A). By contrast, we did not observe lipid accumulation in NHAs (Supplementary Fig. S7A), likely because NHAs divert FAs to biosynthetic rather than oxidative pathways. Similarly, MCAD deficiency increased overall free FA content, as assessed by a colorimetric assay (Fig. 4B). Lipid accumulation was also confirmed in vivo in tumor remnants after 30 days of doxycycline-induced ACADM silencing (Fig. 4C; as in Supplementary Fig. S5D). To test whether the accumulation of FAs is causally correlated with the MCAD-deficient phenotype, we ablated MCAD in GSC 8.11 cells grown in medium supplemented with normal or FA-free B27. In FA-free conditions, MCAD ablation did not result in accumulation of lipid droplets. Moreover, cell proliferation and viability, albeit reduced, did not appear significantly affected (Fig. 4D and E; Supplementary Fig. S7B).
Figure 4.
Accumulation of MCFAs induces toxic alterations of lipid metabolism in GSCs. A, Oil Red O staining in GSC 8.11 cells infected with anti-ACADM or nontargeting shRNA in vitro. Black arrows indicate sites of lipid accumulation. Scale bar, 20 μm. B, Colorimetric determination of free FAs from GSC whole-cell extracts. Values represent the mean ± SD of three independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P = 0.0061; ***, P = 0.0003. C, Oil Red O staining in xenograft tumor tissues derived from GSC 8.11 infected with inducible shRNA (ishACADM) constructs. Doxycycline was administered approximately 20 days after cell implantation. Oil Red O staining quantification of tumor tissues shown in C was obtained using ImageJ software analysis. Scale bar, 25 μm. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.033. d and E, Oil Red O staining (D) and growth curve (E) of ACADM wild-type or null GSC 8.11 grown in normal or FA-free medium. Cells were selected with puromycin for 48 hours prior to starting the experiment. Oil Red O staining in cells after 48-hour puromycin selection. Scale bar in D, 10 μm. Data in E represent the mean ± SD of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P ≤ 0.007. F, Colorimetric analysis of acyl-CoA species in MCAD-deficient GSC 8.11 cells. Data represent the mean ± SEM of four biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.03. G, Quantification of lipid peroxidation determined by measuring the production of malondialdehyde (MDA) using the Colorimetric Microplate Assay for Lipid Peroxidation Kit. Data represent the mean ± SD of four biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.02; **, P = 0.01. H, Quantitative LC/MS-MS lipid profiling of FFA content of ACADM wild-type or null GSC 8.11 cells. Data represent mean ± SEM of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.03; **, P < 0.004. i and J, Relative amount of total lipid classes measured by mass spectrometry of whole-cell extracts (I) or mitochondria (J) from GSC 8.11 cells infected with shRNA targeting ACADM. Data are reported as fold change over control cells infected with nontargeting shRNA. Mean values ± SD of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05; ***, P = 0.001; ****, P = 0.0001. CE, cholesteryl ester; CER, ceramide; DAG, diacylglycerol; FAHFA, fatty acid ester of hydroxyl fatty acids; Hex2CER, dihexosylceramide; HexCER, hexosylceramide; LPC, lyso-phosphatidylcholine; LPE, lyso-phosphatidylethanolamine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol.
MCAD Depletion Results in Acutely Toxic MCFA Accumulation
Our data strongly suggest that lipid accumulation contributes directly to the proliferation defect observed in MCAD-deficient GSCs. To understand mechanistically how lipid accumulation may exert toxicity, we investigated the effects of MCAD depletion on coenzyme A (CoA) pools in GSCs, as conjugation of FAs to CoA as acyl-CoA is required for FAs to migrate into mitochondria and undergo oxidation. Compared with wild-type cells, MCAD-depleted cells had increased acyl-CoA and decreased free-CoA pools (Fig. 4F). Furthermore, peroxidation of accumulated lipids was increased in the absence of MCAD (Fig. 4G), which would also be expected to affect cellular structures and functions. This important finding suggests that activated FAs may enter and accumulate in the mitochondria.
To characterize the type and size of lipids accumulated in MCAD-depleted cells, we compared free FA profiles of ACADM-silenced versus control GSCs by gas chromatography/mass spectrometry (GC/MS). Here, we observed an accumulation of MCFAs and a decrease in LCFAs (Fig. 4H), consistent with a block in the metabolic pathway at the level of MCFA degradation. Next, we profiled acylcarnitines by LC/MS in GSCs grown in medium supplemented with unmodified or uniformly carbon-13 (U13C18)–labeled oleate. This experiment revealed an increase in medium-chain acylcarnitine species that would be expected to accumulate in the mitochondria (Supplementary Fig. S7C). Thus, similar to observations in patients affected by MCAD deficiency (28), the accumulation of MCFAs is observed both as free FA and acylcarnitine species, suggesting that GSCs, to some extent, continue to engage in FAO (e.g., oleate), even upon attenuated MCAD activity.
MCFAs have been shown to induce apoptosis in some cancer models (29, 30), and based on the dramatic accumulation of MCFAs in GSCs upon MCAD depletion, we speculated that MCFAs may be directly causing toxicity. Indeed, acute treatment of GSCs with varied-length FA species (between 4 and 16 carbons) revealed that C10 and C12 MCFAs negatively affected cell viability, whereas treatment with the shorter or longer FAs tested did not change viability (Supplementary Fig. S7D). Interestingly, acute MCFA exposure induced an increase in reactive oxygen species (ROS) levels that was partially rescued when cells were concomitantly grown in the presence of antioxidants, such as a cell-permeable form of glutathione–ethyl ester (GSH-EE; Supplementary Fig. S7E and S7F). Because lipid peroxidation products have been associated with ferroptosis (31), an iron-dependent form of programmed cell death, we silenced ACADM in GSCs in the presence of ferrostatin 1, an iron chelating agent. Propidium iodide staining did not reveal any improvement in viability in ACADM-silenced GSCs upon ferrostatin 1 treatment (Supplementary Fig. S7G), which suggests that the toxicity exerted by FA accumulation is iron-independent. Taken together, our data support a model wherein MCAD inhibition negatively affects cell viability through the accumulation of toxic species as a consequence of impaired FA degradation.
MCFA Accumulation Leads to Lipid Peroxidation and Structural Damage to the Mitochondria in MCAD-Depleted GSCs
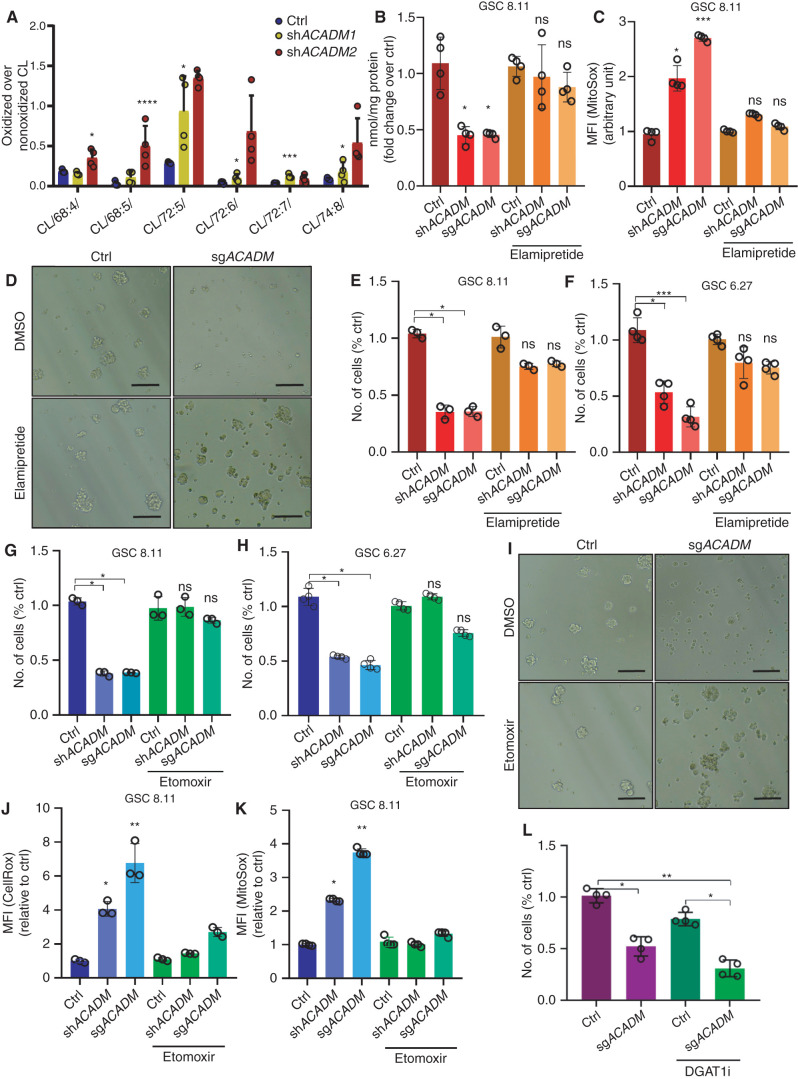
Next, to investigate the effects of MCAD depletion on cellular lipids more broadly, we conducted an LC/MS-based analysis of lipid classes on whole-cell extracts and on mitochondria isolated from GSCs. Triacylglycerols (TAG) and some phospholipid classes, such as lysophosphatidylcholine, were significantly increased in whole-cell extracts from MCAD-depleted cells compared with MCAD-proficient controls (Fig. 4I). This indicates that profound perturbations in the cellular content of complex lipids may reflect an adaptation by which MCAD-inhibited GSCs attempt to accommodate the accumulating MCFA species. Indeed, phosphatidylserine and sphingomyelin levels were approximately 20- and 10-fold higher, respectively, in MCAD-depleted versus control GSCs analyzed by LC/MS lipidomics (Fig. 4J). The most dramatic change detected in mitochondria was in cardiolipin (CL), which decreased more than 50-fold in MCAD-deficient cells compared with controls (Fig. 4J). CL is located in the inner mitochondrial membrane, where it plays a crucial role in mitochondrial bioenergetics and regulates the efficiency of the electron transport chain (ETC). Interestingly, it is well established that CL is the phospholipid most susceptible to oxidative stress due to its composition, which is enriched in unsaturated FAs (32).
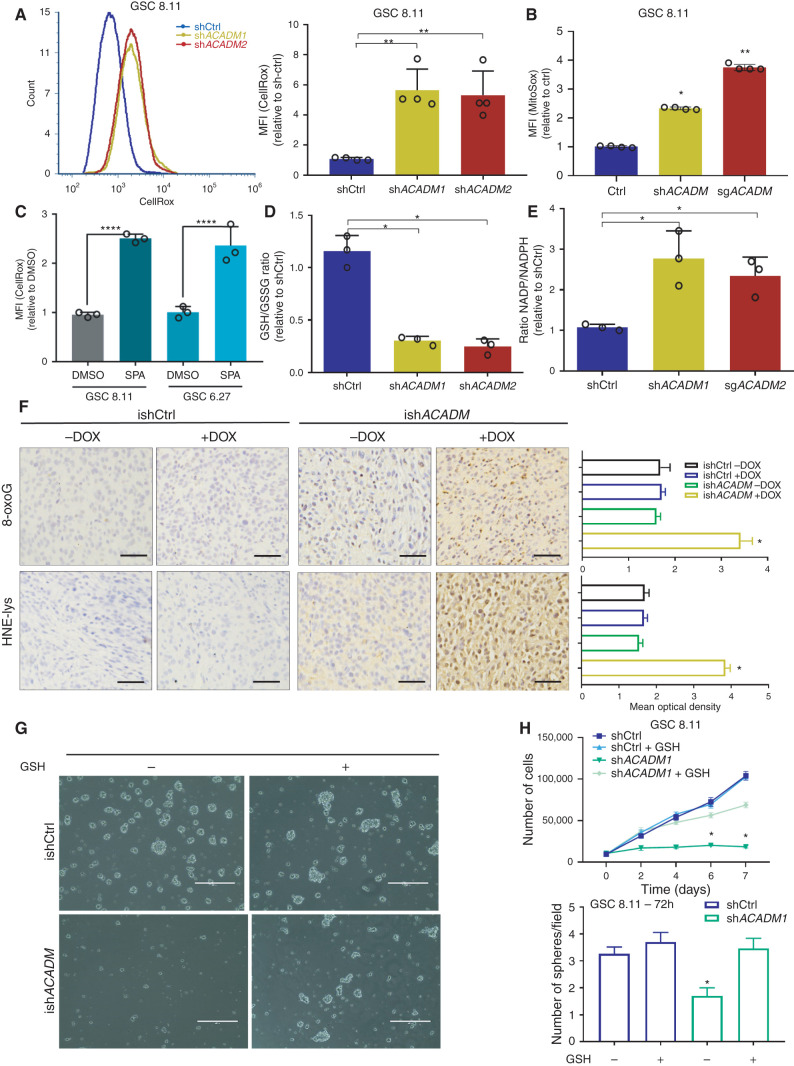
To evaluate whether these changes in mitochondrial lipids may relate to mitochondrial dysfunction, we measured ROS levels in MCAD-deficient GSCs. Similar to MCFA treatment (Supplementary Fig. S7D–S7F), MCAD depletion using ACADM-targeting shRNA or SPA treatment altered the redox state in GSCs, as evidenced by increased ROS levels as well as decreased ROS scavengers and NADPH levels in all GSCs tested, but did not induce a similar increase in ROS in NHAs (Fig. 5A–E; Supplementary Fig. S8A–S8D). This correlates with our previous finding of high levels of lipid peroxidation in this context (Fig. 4G). Notably, as evidenced by staining with Mitosox, the ROS identified upon MCAD ablation were specifically generated in mitochondria (Fig. 5B). We also found high levels of 8-oxoguanine and 4-HNE-lysine, two indicators of oxidative damage and lipid peroxidation, which supports that MCAD depletion similarly affects redox balance in tumors in vivo (Fig. 5F; Supplementary Fig. S8E). To investigate the impact of redox state on viability, we grew ACADM-silenced GSCs in suspension culture in the presence of a cell-permeable form of glutathione, GSH-EE. GSH-EE exposure transiently rescued the growth inhibition observed upon MCAD depletion; however, it did not enable MCAD-depleted spheres to expand to the same extent as control spheres at later time points (Fig. 5G and H; Supplementary Fig. S8F and S8G). Thus, despite the central role of ROS in driving the observed phenotype, our data convincingly demonstrate that oxidative damage is only one aspect of how deficient MCAD function affects GSCs. In strong support of this, culturing MCAD-depleted GSCs in FA-free medium preserved the proliferative phenotype (Fig. 4E) and eliminated the increase in ROS induced by ACADM silencing (Supplementary Fig. S8H). To determine whether the oxidative stress induced by FA overload in the mitochondria might be directly damaging the organelle structure, we first looked for oxidative damage of CL, as this phospholipid was determined to be significantly depleted upon MCAD inhibition (Fig. 4J). As mentioned, the localization and the particular composition of CL make it a prime target for oxidative damage (33–35). We focused on the most abundant (C18:2)–containing CL species and searched for products of linoleic acid with molecular weight increased by 48 Da; that is, +48 amu (atomic mass unit) adducts that would result from the addition of three oxygen atoms during the mitochondria-driven peroxidation of linoleic acid. Indeed, the formation of +48 amu compounds has been largely reported as the result of the mitochondria-mediated peroxidation of linoleic acid (36, 37). Interestingly, the proportion of oxidized CL markedly increased in GSCs following MCAD knockdown compared with control cells (Fig. 6A). We identified six abundant CL species that underwent oxidation, resulting in signals with m/z of 723.54 (m/z 699.6 + 8 + 16 = m/z 723.6, CL 68:4), 722.60 (m/z 698.6, CL 68:5), 750.60 (m/z 726.6, CL 72:5), 749.58 (m/z 725.6, CL 72:6), 748.60 (m/z 724.6, CL 72:7), and 761.59 (m/z 737.59, CL 74:8). These data confirm previous findings (38) and further support the hypothesis that peroxidation of FA by-products trapped in mitochondria following MCAD ablation leads to severe damage of important structures, such as CL, which ultimately results in mitochondrial functional impairment.
Figure 5.
MCAD knockdown triggers ROS-related damage in GSCs in vitro and in vivo. A, ROS production as measured by CellROX Green in flow cytometry (left) and quantification of fluorescence intensity (right) in GSC 8.11 harboring anti-ACADM or nontargeting shRNA. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P < 0.002. B, Mitochondrial ROS quantification by MitoSox staining intensity in GSC 8.11 cells harboring anti-ACADM shRNA or sgRNA 72 hours after gene silencing. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.046; **, P = 0.0033. C, ROS production quantification by CellRox Green staining intensity in GSC 8.11 and 6.27 upon 72-hour exposure to SPA (400 μmol/L) or DMSO. Values represent the mean ± SD of three independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ****, P = 0.0001. d and E, GSH/GSSG and NADP/NADPH ratios in GSC 8.11 measured by colorimetric assay. D, *, P ≤ 0.03. E, *, P ≤ 0.03. Values represent the mean ± SD of three independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. F, Immunostaining for 8-oxoguanine (showing oxidized DNA, top) or hydroxynonenal (HNE) adducts (showing lipid peroxidation and oxidative protein damage, bottom) and relative quantification of GSC 8.11–derived tumors from experiment shown in Supplementary Fig. S5D; scale bar, 50 μm. Quantification was conducted with ImageJ software, six images per condition, where each condition was represented by three biological replicates. Values are expressed as mean ± SD; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P ≤ 0.02. G, Representative images of the rescue effect of GSH-EE on GSC 8.11 tumor spheres with doxycycline-induced MCAD knockdown (scale bar, 400 μm). H, Top, growth curve of GSC 8.11 cells infected with ACADM or nontargeting shRNA grown in the presence or absence of GSH-EE. Values are expressed as mean ± SD; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P ≤ 0.03. Bottom, quantification of the number of spheres formed by GSC 8.11 harboring ACADM or nontargeting shRNAs grown for 72 hours in the presence or absence of GSH-EE, as indicated. Values represent the number of spheres per field expressed as mean ± SD. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.04.
Figure 6.
Overloading of mitochondria with FAs triggers structural and functional alterations upon MCAD ablation and leads to GSCs' death. A, Relative amount of (C18:2)–containing CL species with molecular weight increased by 48 Da (+48 amu adducts) measured by mass spectrometry in mitochondria from GSC 8.11 cells infected with shRNA targeting ACADM. Data are reported as fold change over native (nonoxidized) CL. Mean values ± SD of three biologically independent replicates. P values were generated using one-way ANOVA. *, P < 0.05; ***, P = 0.001; ****, P = 0.0001. B, Cardiolipin content per milligram of protein as determined by a fluorometric assay 72 hours following ACADM silencing in GSC 8.11 mitochondria. Cells were grown in normal medium or supplemented with elamipretide as indicated. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05. C, Quantification of mitochondrial ROS (mean of fluorescence intensity) as measured by MitoSox staining in GSC 8.11 cells harboring anti-ACADM shRNA or sgRNA 72 hours after gene silencing in the presence or in the absence of elamipretide. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05; ***, P < 0.001. D, Representative optical microscopy images of GSC 8.11 tumor spheres 6 days after ACADM silencing in the presence or absence of elamipretide (20 μmol/L). Scale bar, 40 μm. e and F, GSC 8.11 and 6.27 cells were grown with elamipretide (20 μmol/L) for 1 week before genetic ACADM silencing. The number of viable cells (as percentage of the ctrl) was assessed at 96 hours after gene silencing by Trypan blue exclusion. Values represent the mean ± SD of three or four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05; ***, P < 0.001. G–K, GSC 8.11 and 6.27 cells were pretreated with etomoxir (2.5 μmol/L) for 48 hours before genetic ACADM silencing. g and H, The number of viable cells (as percentage of the ctrl) was assessed upon genetic ACADM silencing by Trypan blue exclusion. Values represent the mean ± SD of three or four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05. I, Representative optical microscopy images of GSC 8.11 tumor spheres 6 days after genetic ACADM silencing in the presence or absence of etomoxir (2.5 μmol/L). Scale bar, 40 μm. j and K, ROS quantification (mean of fluorescence intensity) as measured by CellRox Green (J) and MitoSox (K) staining in GSC 8.11 cells harboring anti-ACADM shRNA or sgRNA 72 hours after genetic silencing. Values represent the mean ± SD of three or four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05; **, P < 0.01. l, The number of viable cells (as percentage of the ctrl) was assessed 96 hours after genetic ACADM silencing by Trypan blue exclusion. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05; **, P < 0.01.
To investigate whether CL oxidation and its consequent decreased representation in mitochondria may be responsible for the observed mitochondrial failure and cell death, we grew GSCs in medium supplemented with 20 mmol/L elamipretide, a small tetrapeptide designed to specifically target mitochondria by binding CL, thus protecting its structure from oxidative stress (39). Strikingly, sparing CL from peroxidation restored the overall CL content (Fig. 6B) and significantly decreased the amount of mitochondrial ROS (Fig. 6C), likely due to both the preservation of mitochondrial structures and the antioxidant properties of elamipretide. As expected, the effect of MCAD inhibition on GSC growth in the presence of elamipretide was significantly attenuated (Fig. 6D–F), which was likely driven by the protective effect of elamipretide on CL, although some contribution of the drug's ROS-quenching activity could not be excluded. Together with our demonstration that growing MCAD-depleted GSCs in medium deprived of lipids preserves cell growth and restores ROS to control levels (Fig. 4D and E; Supplementary Figs. S7B and S8H), this observation further supports that the critical physiologic consequence of targeting MCAD is the overload of MCFAs trapped in mitochondria as partially metabolized substrates, where they generate oxidative stress and trigger cell death.
Protecting Mitochondria from FA Accumulation Prevents Oxidative Damage and Restores Viability in MCAD-Depleted GSCs
To delineate the role of the accumulation of mitochondrial FA by-products in the cytotoxic effects triggered by MCAD ablation, we grew GSCs 8.11 and 6.27 in medium containing 2.5 μmol/L etomoxir, which, at low concentrations, is known to block the transport of LCFAs into mitochondria by inhibiting CPT1. Cells were exposed to etomoxir beginning 48 hours before ACADM silencing through the end of the experiment. Etomoxir robustly rescued GSCs from the effects of MCAD ablation, as evidenced by no increase in mitochondrial ROS and restored cell viability (Fig. 6G–K). Conversely, targeting diacylglycerol O-acyltransferase 1 (DGAT1), a key enzyme in lipid droplet formation that catalyzes the terminal and only committed step in triacylglycerol synthesis by using diacylglycerol and fatty acyl-CoA as substrates, with a commercially available inhibitor (A-922500) decreased GSC 8.11 growth by an additional 20% upon ACADM silencing (Fig. 6L; Supplementary Fig. S8I), thus accelerating the detrimental effects described above. It is well known that one of the main roles of lipid droplets is to buffer cellular levels of potentially toxic lipids to prevent lipotoxicity and oxidative stress (40), and the importance of DGAT1 in the context of FA metabolism in GBM has been previously reported (41). As expected, A-922500 (DGAT1i) treatment effectively inhibited lipid droplet formation in MCAD-depleted GSCs (Supplementary Fig. S8J), suggesting that inhibition of the machinery that enables GSCs to counteract lipotoxicity induced by MCAD ablation further worsens the observed phenotype.
Taken together, our functional characterization of MCAD in GBM models uncovers a novel, protective role wherein MCAD function is required for continuous degradation of molecular species that would otherwise accumulate and cause mitochondrial dysfunction, oxidative stress, and, eventually, cell death.
Discussion
In this study, we interrogated metabolic dependencies in in vivo GBM tumors using our previously described PILOT platform (14). This screen and our subsequent studies confirmed MCAD, a mitochondrial enzyme that catalyzes the first step of MCFA FAO as a node of metabolic vulnerability for GBM tumor growth and maintenance. Specifically, MCAD depletion in primary GBM models triggered massive accumulation of unmetabolized MCFAs, lipid peroxidation, and oxidative stress, which resulted in irreversible mitochondrial damage and apoptosis. Altogether, this work uncovered an irreversible cascade of detrimental metabolic effects induced by MCAD depletion, and it represents the first demonstration of a nonenergetic role for dependence on FA metabolism in cancer.
MCAD deficiency is a human-inherited autosomal recessive disorder caused by inactivation of the ACADM gene, and the presence of ACADM mutations is screened perinatally. Because the uptake of MCFAs lacks any negative feedback regulation, affected individuals must follow appropriate dietary recommendations and, in doing so, can lead normal lives (15, 16). The role of ACADM in the pathogenesis of MCAD deficiency has been confirmed by the generation of an Acadm-deficient mouse model that shows similar clinical manifestations and histopathologic characteristics (42). Given these facts and our demonstration that depleting MCAD leads to toxic accumulation of MCFAs and cell death specifically in tumor cells, we expect that an actionable therapeutic window may exist to target MCAD in patients with GBM.
Previous reports have indicated that the upregulation of FAO confers a survival advantage to GBM cells, at least in part by fueling oxidative metabolism to sustain cellular energetic requirements (43). The significance of FAO to fuel cancer cell survival beyond brain cancer is rapidly gaining recognition (44). Several studies have reported that blocking LCFA oxidation, primarily through CPT1 inhibition, exerts robust antitumor effects in preclinical models, but CPT1-targeted therapies have not yet proved to be clinically viable due to significant toxicity (45–49).
This work demonstrates that the sustained influx of FAs in GBM cells can become toxic when lipids are not properly metabolized. We show that accumulated MCFAs are particularly damaging to GSCs compared with FAs of different chain lengths. Thus, our data support that targeting MCAD may have advantages over inhibitors of CPT1 because partial oxidation of LCFA to MCFA may contribute to the overall accumulation of toxic lipid species in contexts where MCFA metabolism is inefficient. Owing to the toxicity associated with C10 and C12 lipid accumulation and the lack of a feedback mechanism to prevent continued MCFA accumulation, we posit that, in a lipid-rich environment such as the brain, overexpression of MCAD or enzymes capable of diverting potentially harmful lipid pools to energy production may confer a significant proliferation advantage to cancer cells. This may explain the high reliance of GSCs on sustained FA metabolism, and it suggests that targeting MCAD in FA-embedded tumor types may be especially effective.
We describe a novel role for MCAD to protect GSCs from accumulating toxic levels of lipids in the mitochondria, which can induce apoptosis. These data offer a new therapeutic concept for cancer cell metabolism, which has proved difficult to exploit with targeted drugs. Although tumor cells have a documented, remarkable adaptability to shunt substrates to different metabolism pathways to survive metabolic perturbations, impairing MCFA catabolism in GSCs irreparably damages mitochondria and is acutely toxic. These findings, in combination with our data in normal human brain models and the known physiology of congenital MCAD deficiency, prompt further studies to translate this cancer cell–specific dependency into a clinical opportunity for patients with GBM.
Methods
Human Samples, Primary Cells, and Cell Lines
GSCs were isolated from specimens from patients with GBM who had undergone surgery at the University of Texas MD Anderson Cancer Center (MDACC; Houston, TX). The models used in this research are abbreviated GSC 8.11, GSC 6.27, GSC 7.11, and GSC 272 but elsewhere are published using the nomenclature MDA-GSC 8–11, MDA-GSC 6–27, MDA-GSC 7–11, and MDA-GSC 272, respectively. The diagnosis of GBM was established by histologic examination by the World Health Organization classification. Samples derived from patients were obtained with the written informed consent of patients to an ethically approved institutional review board protocol that is compliant with the principles of the Belmont Report [LAB04–0001, chaired by F. F. Lang (MDACC)] and are registered in the MDACC Cell Line Registry. Tumor specimens were dissociated and resuspended in DMEM-F12 (Gibco) supplemented with B27 (×1; Invitrogen), basic fibroblast growth factor (bFGF; 20 ng/mL; PeproTech), and EGF (20 ng/mL; PeproTech). Cells were cultured as neurospheres and passaged every 5 to 7 days on the basis of sphere size.
GSC authentication and Mycoplasma testing were performed by the MDACC Cell Line Authentication Core every 2 to 3 months. Authentication was performed by short tandem repeat (STR) DNA profiling, also known as DNA fingerprinting, which is based on screening regions of microsatellite instability with defined tri- or tetrad-nucleotide repeats located throughout the chromosomes. PCR reactions using primers on nonrepetitive sequences flanking those regions will generate PCR products of different sizes based on the number of repeats in the region; the size of these PCR products is determined by capillary electrophoresis. Between 8 and 16 STR loci, such as D5S818, D13S317, D7S820, D16S539, vWA, TH01, TPOX, and CSF1PO, are combined to uniquely identify a sample. The Core assay screens 16 loci using the Promega Powerplex 16 HS kit. The test also includes matching the STR profiles against an internal database comprising public profiles and profiles that are unique to cell lines developed by MDACC investigators. The current database MDACC has more than 4,000 profiles. The Lonza Mycoalert Mycoplasma Detection kit was used for Mycoplasma testing. The length of time or number of passages between collection or thawing and use is described in the described experiments. All GSCs were used within 20 to 30 passages.
NHAs were obtained from Sciencell Research Laboratories (NC0273812)and grown in Astrocyte Medium (#1801) according to instructions. Neural stem cells 1 (NSC-1; also referred as neural progenitor cells) were purchased from Lonza (PT-2599). Neural stem cells 2 (NSC-2; also referred as iPSC-derived NSCs) were obtained from iXCells Biotechnologies (NC1831429). T98G cells were obtained through ATCC and grown under standard tissue-culture conditions. NHAs, NSC-1, NSC-2, and T98G were authenticated by the manufacturer using specific markers. NHAs, NSC-1, NSC-2, and T98G cells tested negative for Mycoplasma per the manufacturer's certificate of analysis provided for every cell lot, and these cells were used within eight passages from thawing.
GSC Treatments, Proliferation, and Sphere Formation Assays
Sodium butyrate (156–54–7), sodium hexanoate (C4026), sodium octanoate (C5038), sodium decanoate (C4151), and sodium dodecanoate (L9755) were obtained from Sigma-Aldrich. BSA-Palmitate (102720–100) was purchased from Agilent Technologies. SPA was synthesized by the Institute of Applied Cancer Science Chemistry Core Facility at MD Anderson. For each treatment, GSCs were disaggregated, and 50,000 cells were plated in 6-well plates. Cells were disaggregated at different time points (as indicated in the figures); cell number and viability were determined by Trypan blue exclusion assay. For sphere formation assays, approximately 2 × 104 cells from disaggregated GSC spheres were resuspended in a 0.8% methylcellulose semisolid DMEM-F12 and seeded in 6-well plates. At different time points, the spheres were counted in at least four fields per well.
Library Design and Construction
A custom library was composed by 338 genes specifically belonging to key metabolic pathways (Kyoto Encyclopedia of Genes and Genomes; see Supplementary Fig. S2A). This library was constructed using chip-based oligonucleotide synthesis and cloned into the pRSI16 lentiviral vector (Cellecta) as a pool. The shRNA library is constituted by 338 genes with a coverage of 10 shRNAs/gene. The shRNA includes two G/U mismatches in the passenger strand, a 7-nt loop and a 21-nt targeting sequence. Targeting sequences were designed using a proprietary algorithm (Cellecta). The oligo corresponding to each shRNA was synthesized with a unique molecular barcode (18 nucleotides) for measuring representation by next-generation sequencing.
In Vivo shRNA Screens
In vivo shRNA screens were performed using adapted procedures previously described in (14). In brief, concentrated lentiviral particles (transducing units) from libraries or single plasmids were either purchased by Cellecta or produced by transfecting 293T cells according to the protocol in the Cellecta User Manual. Precisely calculated lentiviral particles, together with 2 μg/mL polybrene (Millipore), were added to single-cell dissociated GSCs to achieve a multiplicity of infection (MOI) = 0.3. Forty-eight hours after infection, the medium was replaced, and puromycin (2 μg/mL) was added for 96 hours. For in vitro/in vivo validation studies, GSCs were infected at MOI = 3, with single shRNA knocking down specific target genes. Transduction efficiency was determined sample by sample as the percentage of GFP-positive cells 2 days after infection as measured by FACS analysis. For the in vivo experiments, each intracranial injection was performed with 1 × 106 cells to ensure a coverage of ∼300 cells/barcode. Upon brain collection, tumors displaying green fluorescence signal were precisely dissected, weighted, and quickly snap-frozen. Genomic DNA extraction, barcode amplification, and next-generation sequencing were performed according to the Cellecta user manual for shRNA libraries processing (details about PCR primers, PCR conditions, and reads counting in Carugo and colleagues, ref. 14).
Bioinformatics Analysis: Hit Identification
Multiple replicates (N = 3) were sequenced for in vitro samples for GSC 627 (N Ref = 2) and GSC811 (N Ref = 1) using Illumina HiSeq2500 and processed using CASAVA (v.1.8.2) to derive FASTQ files. The sequencing read consists of two 18-bp barcodes separated by a 4-bp spacer (CGAA) between the two barcodes. Reads were filtered for the 4-bp spacer and aligned with the 23- to 40-bp barcode using Bowtie (2.0.2) and counted using SAMtools.
Read counts were normalized for the amount of sequencing reads retrieved for each sample by using library size normalization (to 1 million reads). For each sample, log2 fold change (FC) was calculated compared with the reference pellet before injection. A summary measure per condition was derived using the median of quantile-transformed log2 FC across replicates. Thereafter, a modified version of the redundant shRNA activity (RSA) algorithm was used to derive the gene-level summary measure per condition. Specifically, we controlled for the factor that at least two hairpins were used when calculating the minimum P value (in RSA). In addition, hairpins that ranked above luciferase controls were not used in choosing the minimum P value. Quantile rank of luciferase control barcodes was evaluated across all experiments. On average, luciferase barcodes ranked >0.6 (on the quantile transformed log2 FC scale). Therefore, hairpins with quantile-transformed log2 FC >0.6 were not used for gene-level RSA scores (log value; ref. 50).
Bioinformatics Analysis: TCGA Data
Level 3 TCGA GBM and lower-grade glioma data for gene expression and mutations were downloaded from GDAC Firehose (https://gdac.broadinstitute.org). Data analysis using the TCGA cohort was performed using R.
Log2 RSEM values were calculated to compare expression levels of ACADM and other genes involved in the FA synthesis pathway. Using the GBM data set (n = 167), analysis was performed comparing expression between normal tissue (n = 5), classic (n = 68), proneural (n = 46), and mesenchymal (n = 48) subtypes (The Cancer Genome Atlas Network, 2015). ANOVA was performed across all groups to determine statistical significance, with pairwise Wilcoxon tests performed to identify which subtypes showed a difference. This was conducted using the “ggpubr” R package [R Core Team, 2017; Alboukadel Kassambara (2020). ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.4.0. https://CRAN.R-project.org/package=ggpubr]. Tukey HSD was also calculated in R to interrogate pairwise differences driving significance with ANOVA.
Animal Studies
Male athymic nude mice (nu/nu) were purchased from The Jackson Laboratories. All procedures performed in this study were approved by MDACC. All animal manipulations were performed in the veterinary facilities in accordance with institutional, state, and federal laws and ethics guidelines under an approved protocol. Intraperitoneal injections of ketamine (100 mg/kg)/xylazine (10 mg/kg) were used to anesthetize animals in all experiments. For intracranial xenografts of GSC 8.11 and 6.27, 106 cells expressing a PLX304–mCherry–LUC vector (5-μL cell suspension) were implanted using a guide screw and a multiport microinfusion syringe pump (Harvard Apparatus; refs. 51, 52). For subcutaneous experiments, GSCs were injected subcutaneously (100-μL cell suspension) into the left flank. For all bioluminescence imaging, d-luciferin (150 mg/kg) was administered by subcutaneous injection to mice 10 minutes before imaging. In all experiments, mice were monitored daily for signs of illness and were euthanized when they reached endpoints. For in vivo studies of tumor maintenance, inducible shRNAs TRCN0000028530 and TRCN0000028509 cloned in a Tet–pLKO–puro vector were obtained from Sigma-Aldrich. Doxycycline (2 mg/mL) was administered in drinking water to mice. The starting point for doxycycline administration corresponded to the first day tumors were detected (approximately 20 days postinjection) and was maintained until study completion.
For MRI, a 7T Bruker Biospec (BrukerBioSpin), equipped with 35-mm inner-diameter volume coil and 12-cm inner-diameter gradients, was used. A fast acquisition with relaxation enhancement sequence with TR/TE of 2,000/39 ms, matrix size of 256 × 192, resolution of 156 μmol/L, 0.75-mm slice thickness, 0.25-mm slice gap, 40 × 30 cm FOV, 101-kHz bandwidth, and 4 NEX was used to acquire multislice T2-weighted images in coronal and axial geometries. To reduce the respiratory motion, the axial scan sequences were respiratory gated. The brain lesions' volumes were quantified manually using ImageJ software. All animal imaging, preparation, and maintenance were carried out in accordance with MD Anderson's Institutional Animal Care and Use Committee policies and procedures.
ETC Activity
The Mitochondria Isolation Kit (ab110170; Abcam) was used to isolate mitochondria from MCAD silenced and control GSCs, which were then used to test the activity of complex III and V. Activity of mitochondrial complex III was analyzed using the Mitochondrial Complex III Activity Assay Kit (K520–100; Biovision). Cytochrome C was added to the reaction, and its reduction through the activity of complex III was measured at 550 nm. ATP synthase activity was measured by using the ATP Synthase Specific Activity Microplate Assay Kit (ab109716; Abcam). The antibody for ATP synthase was precoated in the wells of the microplate, and samples containing 50 μg mitochondrial extracts were added to wells. In this assay, the conversion of ATP to ADP by ATP synthase was coupled to the oxidative reaction of NADH to NAD+. The formation of NAD+ resulted in a decrease in absorbance at 340 nm. Subsequently, in these same wells, the quantity of ATP synthase was determined by adding an ATP synthase-specific antibody conjugated with alkaline phosphatase. This phosphatase changed the substrate pNPP from colorless to yellow (OD 405 nm) in a time-dependent manner proportional to the amount of protein captured in the wells. Absorbance was read by a Benchmark microplate reader (Bio-Rad). All tests were done in triplicate.
ATP Quantification
ATP was quantified by using the luminescence ATP detection assay from Abcam (ab113849). The assay quantifies the amount of light emitted by luciferin when oxidized by luciferase in the presence of ATP and oxygen. Cells were plated on the same day into a 96-well plate in DMEM-F12 (100 μL) without glucose and supplemented with B27, EGF, and bFGF (20 ng/mL), as well as galactose (10 mmol/L), to obtain the ATP amount generated by mitochondrial activity. ATP standards were loaded on the same plate as references. The assay was performed according to the manufacturer's instructions.
Oxygen Consumption Rate and Extracellular Acidification Rate Measurements
The functional status of mitochondria in MCAD-deficient GSCs 8.11 and NHA was determined by analyzing multiple parameters of oxidative metabolism using the XF96 Extracellular Flux Analyzer (Agilent), which measures extracellular flux changes of oxygen and protons. Cells were plated in XF96-well microplates (30,000 cells per well) in a final volume of 80 μL DMEM-F12 medium (17.5 mmol/L glucose, 2.5 mmol/L glutamine) supplemented with B27, EGF (20 ng/mL), and bFGF (20 ng/mL). For the mitochondrial stress test, cells were incubated at 37°C in the absence of CO2 and in 180 μL DMEM-F12 (17.5 mmol/L glucose, 2.5 mmol/L glutamine) supplemented with B27, EGF (20 ng/mL), and bFGF (20 ng/mL XF-Mito–MEM) per well for 1 hour before the assay. The ports of the sensor cartridge were sequentially loaded with 20 μL per well of the appropriate compound: the ATP coupler oligomycin (O4876; Sigma-Aldrich), the uncoupling agent carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (C2920; Sigma), and the complex I inhibitor rotenone (R8875; Sigma-Aldrich).
Immunoblotting
Protein lysates were resolved on 4% to 15% gradient polyacrylamide SDS gels and transferred onto nitrocellulose membranes according to standard procedures. Membranes were incubated with indicated primary antibodies, washed, and probed with horseradish peroxidase–conjugated secondary antibodies. The detection of bands was carried out upon chemiluminescence reaction followed by film exposure. The following primary antibodies were used: anti-ACADM antibody (ab92461), anti–oxoguanine 8 antibody (ab206461), anti–4 hydroxynonenal antibody (HNEJ-2; ab48506), antivinculin antibody (VIN-54; ab130007), anti-GAPDH antibody (EPR6256; ab128915), and anti–β-actin antibody (ab8224).
TEM
TEM was performed at the MDACC High Resolution Electron Microscopy Facility. Samples were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 mol/L cacodylate buffer, pH 7.3, for 1 hour. After fixation, the samples were washed and treated with 0.1% Millipore-filtered cacodylate buffered tannic acid, postfixed with 1% buffered osmium tetroxide for 30 minutes, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, then infiltrated and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for 2 days. Ultrathin sections were cut using a Leica Ultracut microtome, stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV. Digital images were obtained using an AMT Imaging System (Advanced Microscopy Techniques Corp.).
Oxidative Stress Detection
Reduced and oxidized forms of glutathione (GSH and GSSG, respectively) were measured in GSC 8.11 and GSC 6.27 using a GSH/GSSG Ratio Detection Assay Kit (ab138881; Abcam) according to the manufacturer's protocol. Data from three separate experiments were averaged for the results. NADP+ and NADPH levels were measured from GSC 8.11 and 6.27 using a NADP/NADPH Assay Kit (ab65349; Abcam) according to the manufacturer's instructions. NADP and NADPH levels in total lysate were calculated by comparison with the standard curve.
Flow Cytometry
Cellular apoptosis was detected using a FITC Annexin V Apoptosis Detection Kit I (BD Biosciences), according to the manufacturer's instructions. Following virus infection with shACADM or shScr and 48 hours of puromycin selection, cells were harvested and resuspended in cold PBS. Subsequent to centrifugation at 1,000 rpm for 5 minutes at 4°C, the cells were resuspended with 500 μL binding buffer and mixed with 5 μL annexin V-FITC. The cells were subsequently incubated with 5 μL propidium iodide (PI) in the dark at room temperature for 5 to 15 minutes. Excitation was at 488 nm, and the emission filters used were 515 to 545 BP (green, FITC) and 620 LP (red, PI). All assays were performed in triplicate.
To detect ROS, 2 × 105 cells were stained with CellRox Green (100 nmol/L; Molecular Probes) for 20 minutes, washed twice, and resuspended in PBS. Excitation was at 488 nm, and the emission filters used were 515 to 545 BP (green, FITC). ROS were induced by 4-hydroxynonenal treatment (10 μmol/L) for positive controls. For mitochondrial ROS, cells were stained with MitoSOX Red (2.5 μmol/L; Molecular Probes) for 10 minutes at 37°C, washed twice, and resuspended in Hank's Balanced Salt Solution/Ca/Mg. Excitation was at 510 nm, and the emission filters used were 580 BP (red). Gating strategies to exclude doublets and dead cells (DAPI) were always employed. After staining, samples were acquired using a BD FACSCantoII flow cytometer. Data were analyzed by BD FACSDiva or FlowJo (Tree Star).
Immunohistochemistry and Immunocytochemistry
For immunohistochemistry staining, tumor samples were fixed in 4% formaldehyde for 2 to 4 hours on ice, moved in 70% ethanol for 12 hours, and then embedded in paraffin (ASP300S; Leica). After cutting (RM2235; Leica), baking, and deparaffinization, slides were treated with Citra-Plus Solution (BioGenex) according to specifications. Endogenous peroxidases were inactivated by 3% hydrogen peroxide. Nonspecific signals were blocked using 3% BSA, 10% goat serum, and 0.1% Triton. Tumor samples were stained with primary antibodies. ImmPress and ImmPress-AP (Vector Laboratories) were used as secondary antibodies; Nova RED, Vector BLUE, and DAB were used for detection (Vector Lab). Images were captured with a Nikon DS-Fi1 digital camera using a wide-field Nikon EclipseCi microscope. For Oil Red O lipid staining of cells, GSCs were grown on coverslips and GSCs were attached to tissue slides by cytospin. For Oil Red O lipid staining of tumors, tumor tissue was 4% paraformaldehyde fixed, cryoprotected with 30% sucrose, and OCT embedded and sectioned (5 μm thick). Oil Red O lipid staining was performed according to the manufacturer instructions (ab150678; Abcam).
ACADM shRNAs and sgRNAs
The hairpin RNA interference plasmid for human ACADM, pLKO.1 ACADM TRCN0000028530 (shAC1), TRCN0000028509 (shAC2), and the scramble control pLKO.1-Puro plasmid (shScr) were obtained from Sigma-Aldrich. The sequence of shRNAs is as follows:
shAC1, CCGGGCTGGCTGAAATGGCAATGAACTCGAGTTCATTGCCATTTCAGCCAGCTTTTT
shAC2, CCGGGTGCAGATACTTGGAGGCAATCTCGAGATTGCCTCCAAGTATCTGCACTTTTT
The inducible shRNA was obtained from Cellecta by cloning into a pRSIT16–U6Tet–sh-CMV–TetRep-2A–TagRFP-2A–Puro vector the following hairpin sequence:
ACCGGGAGTTCACTGAACAGTAGAAAGTTAATATTCATAGCTTTCTGCTGTTCGGTGAACTCTTTT
To generate ACADM sgRNAs, three pairs of oligonucleotides were designed and used as follows:
| Sg1: SgACADM1F | CACCGAAGATGTGGATAACCAACGG |
| SgACADM1R | AAACCCGTTGGTTATCCACATCTTC |
| Sg2: SgACADM2F | CACCGACACACATTCCAGAGAACTG |
| SgACADM2R | AAACCAGTTCTCTGGAATGTGTGTC |
| Sg3: SgACADM3F | CACCGATTGGCTTATGGATGTACAG |
| SgACADM3R | AAACCTGTACATCCATAAGCCAATC |
All sgRNAs were validated for their knockout efficiency. sgACADM1 was selected for further studies to knockout ACADM. To specifically amplify ACADM in genomic DNA, the following oligonucleotides were used:
| ACADM1F | TATTCAAGGCTTATTGTGTAACAGAACC |
| ACADM1R | CTGAACTGTTTATAATCTCCTTTAGGCC |
| ACADM2F | AATAATTTTCCCTTAGAGTTCACCGAAC |
| ACADM2R | GACTGAGTAGAGTTCCACAATTTTTCTT |
| ACADM3F | CAAGTTAGAACAGGAATACAGGTTCAAA |
| ACADM3R | CTTTAAAGAAAACTGTGTGTACCCACTA |
Lipidomics of Total Cell Extracts and Mitochondria
After adding equal volumes of dichloromethane/methanol/PBS (1:1:1), samples were centrifuged (2,000 rpm; 5 minutes) to collect the organic phase. The extraction was repeated twice. After drying using a gentle stream of N2 gas, samples were dissolved in 8 mmol/L ammonium fluoride dichloromethane/isopropyl alcohol/methanol (2:1:1) and sonicated (5 minutes) before an internal standard was added. Lipids were measured using the shotgun lipidomics by directly infusing a modified Blight–Dyer extract into a SCIEX Triple TOF 5600+ mass spectrometer for MS/MS⁁ALL analysis as previously described (53). Acquisition was performed once in positive and once in negative ion mode. The phospholipid species were identified based on their characteristic m/z value, fragmentation analysis, and precursor ion or neutral loss scans. Using precursor ion scanning techniques, negative precursors of m/z 241 and 196 identified parents of phosphatidylinositol, phosphatidylethanolamine, and lysophosphatidylethanolamine. Positive precursors of m/z 184 yielded parents of sphingomyelin, phosphatidylcholine, and lysophosphatidylcholine. Neutral loss scans of 141 and 185 yielded parents of phosphatidylethanolamine and phosphatidylserine, respectively. Neutral lipids, including triacylglycerides, diacylglycerides, and cholesterylesters, were identified based on their FA neutral loss in positive ion mode using the Lipid Maps database (http://www.lipidmaps.org/). The same extraction method was used for lipidomics on purified mitochondria. Cardiolipin peaks were identified as their [M-2H]2− ions in the negative-ionization mode by PI scanning of m/z 153. The intensity of each peak was normalized to the total lipid signal and to the internal standard. The normalized data relative to each lipid species were summed to give the intensity of each class, which was reported as a percentage of all lipids.
Lentivirus Production
Lentivirus was produced by polyethyleneimine transfection of 293T cells. Four million cells were transfected with 30 μg transfer vector, 19 μg packaging plasmid pCMV–Δ8.74, and 9.5 μg envelope plasmid pMD2G-VSVG. After 16 hours, medium was replaced and viral particles in the medium were collected 72 hours later by ultracentrifugation at 23,000 × g for 3 hours.
Cell Infection
Cells were infected with lentiviral particles expressing shRNAs, inducible shRNAs or sgRNAs, as indicated for each experiment. Then, 4 × 106 GSCs were infected with 4 × 106 lentiviral particles and selected for 48 hours with puromycin (3 μg/mL). In the experiments with NHAs and T98G, 1 × 106 cells were infected with 1 × 106 lentiviral particles.
T7 Endonuclease 1 Assay
Seventy-two hours posttransfection or 1 week after lentiviral infection of guide and nuclease, cell genomic DNA was extracted with the Qiagen DNeasy blood and tissue kit per the manufacturer's protocol. Genomic DNA was used for the template of PCR with NEB Onetaq mastermix per the manufacturer's protocol. The product was first denatured at 98°C for 5 minutes, then slowly annealed to 75°C at 1°C/s and eventually to 25°C at 0.1°C/s. In total, 5 U T7 endonuclease 1 was used for digestion of less than 300 ng annealed product for 30 minutes. The digested product was subjected to 2% TAE gel electrophoresis.
Colorimetric/Fluorometric Assays
Free FA levels (MAK044; Sigma-Aldrich), acetyl-CoA (ab87546; Abcam), coenzymeA/acyl-CoA levels (MAK034; Sigma-Aldrich), and MDA concentrations (MAK085; Sigma-Aldrich) were determined according to the manufacturer's instructions. A PHERAstar FS (BMG Labtech) microplate reader was used for quantifications.
FA Analysis
MCAD silenced or control GSCs were grown for 24 hours after puromycin selection in complete DMEM-F12. After 24 hours, cells were harvested and the pellets were placed at –80°C. Microtubes containing cell pellets were removed from –80°C storage and maintained on wet ice throughout the processing steps. Then, 20 μL of 0.75 μg/μL D27 myristic acid was spiked in as an internal standard (IS). Next, 1 mL methanol was added to each sample, subjected to sonication for 2 minutes, and centrifuged at 4,000 rpm for 10 minutes at 4°C. Supernatant was transferred into a new tube and dried under N2. For medium samples, 500 μL of sample was spiked with 20 μL D27 myristic acid and prepared as above. Stock solutions of hexanoic acid, octanoic acid, and decanoic acid were prepared at 28 μg/μL, 11 μg/μL, and 11 μg/μL, respectively, in methanol. The D27 myristic acid IS was spiked into standards at 0.75 μg/μL. Cell lysates, media samples, and standards were then derivatized, as follows. First, 100 μL N-methyl-N-trimethylsilyl trifluoroacetamide (MSTFA; Pierce) with 1% trimethylchlorosilane (1% TMCS; Thermo Scientific) was added to react for 30 minutes at 60°C. Then, 1 μL of sample was injected into GC/MS for detection. GC/MS analysis was performed using an Agilent 7890A GC equipped with a 15-m DB-5MS+DG capillary column and a Leap CTC PAL ALS as the sample injector. The GC was connected to an Agilent 5975C quadrupole MS operating under positive electron impact ionization at 70 eV. All tunings and data acquisition were done with an HP PC with Win 7 professional OS that included the ChemStation E.02.01, PAL Loader 1.1.1, Agilent Pal Control Software Rev A, and Pal Object Manager updated firmware. MS tuning parameters were in default settings. GC injection port was set at 250°C, and GC oven temperature was held at 60°C for 5 minutes and increased to 220°C at 10°C/minute, then held for 10 minutes under constant flow with initial pressure of 10.91 psi. MS source and quadrupole were held at 230 and 159°C, respectively, and the detector was run in scanning mode, recording ion abundance in the range of 26 to 600 m/z with solvent delay time of 2 minutes. The data set was translated into .D format. The extraction was done with Agilent MassHunter WorkStation Software GCMS Quantitative Analysis Version B.07. Identification was performed by searching NIST2011. A one-point calibration was performed in this study, and data were normalized by IS to generate the final report.
13C Tracer Metabolomics
Cells were grown in complete DMEM-F12 supplemented with13C-oleate/13C-glucose. After 6/12 hours, cells were harvested and the cell pellets were dissolved in 50 μL water/methanol (50:50), and 3 μL was injected onto a Waters Acquity UPLC BEH TSS C18 column (2.1 × 100 mm, 1.7 μm) into an Agilent 1260 UHPLC system with mobile phase A consisting of 0.5 mmol/L NH4F and 0.1% formic acid in water and mobile phase B consisting of 0.1% formic acid in acetonitrile. Gradient program: mobile phase B was held at 1% for 1.5 minutes, increased to 80% in 15 minutes and then to 99% in 17 minutes, and held for 2 minutes before going to the initial condition and held for 10 minutes. The column was at 40°C, and 3 μL of sample was injected into an Agilent 6520 Accurate–Mass Q-TOF LC/MS. The LC/MS flow rate was 0.2 mL/minute. Calibration of TOF MS was achieved through Agilent ESI–Low Concentration Tuning Mix. In negative acquisition mode, key parameters were as follows: mass range, 100 to 1,200 Da; gas temperature, 350°C; fragmentor, 150 V; skimmer, 65 V; drying gas, 10 L/minute; nebulizer flow at 20 psi and Vcap 3,500 V; reference ions at 119.0363 and 980.01637 Da; and ref nebulizer at 20 psi. In positive acquisition mode, key parameters were as follows: mass range, 100 to 1,200 Da; gas temperature, 350°C; fragmentor, 150 V; skimmer, 65 V; drying gas, 10 L/minute; nebulizer flow at 20 psi and Vcap 3,500 V; reference ions at 121.050873 and 922.009798 da; and ref nebulizer at 20 psi. Agilent Mass Hunter Workstation Software LC/MS Data Acquisition for 6200 series TOF/6500 series Q-QTOF Version B.06.01 was used for calibration and data acquisition.
Acylcarnitine Profiling
GSC 8.11 were grown for 24 hours in complete DMEM-F12 supplemented with 13C-oleate. After 24 hours, cells were harvested and the pellets were placed at −80°C. Microtubes containing cell pellets were removed from −80°C storage and maintained on wet ice throughout the processing steps. To initiate protein precipitation, 0.3 mL of a chilled mixture of methanol and chloroform (8:2; EMD) was added to each sample, and the mixture was vortexed briefly and allowed to incubate on ice for 10 minutes. Postincubation, the vortex step was repeated, and samples were centrifuged at 14,000 rpm for 10 minutes in 4°C. Postcentrifugation, 100 μL of supernatant was transferred to an autosampler vial for LC/MS analysis. From the remaining supernatant from each sample, a small aliquot was transferred to a new microtube to create a pooled sample for quality control purposes.
LC/MS analysis was performed on an Agilent system consisting of a 1290 UPLC module coupled with a 6490 QqQ mass spectrometer (Agilent Technologies). A 1-μL injection of acylcarnitine metabolites was separated on an Acquity HSS-T3 1.8-μmol/L, 2.1-mm × 50-mm column (Waters) maintained at 40°C, using 10 mmol/L ammonium acetate in water, adjusted to pH 9.9 with ammonium hydroxide, as mobile phase A and acetonitrile as mobile phase B. The flow rate was 0.25 mL/minute, and the gradient was linear 0% to 80% A over 7 minutes, then 80% to 100% over 1.5 minutes, followed by isocratic elution at 100% A for 5 minutes. The system was returned to starting conditions for 3 minutes to allow for column reequilibration before injecting another sample. The mass spectrometer was operated in ESI– mode with the following instrument settings: gas temperature, 275°C; flow, 15 L/minute; nebulizer, 35 psi; capillary, 3,500 V; sheath gas, 250°C; and sheath gas flow, 11 L/minute. The ion funnel high/low pressure RF settings were 150/60 V, respectively. Acylcarnitine transitions were monitored for the 85-Da product ion that is common to each carnitine species. Metabolites were identified by matching the retention time and mass (±10 ppm) to authentic standards. Isotope peak areas were integrated using MassHunter Quantitative Analysis vB.07.00 (Agilent Technologies). Peak areas were corrected for natural isotope abundance using an in-house written software package based on the method of Fernandez and colleagues (54), and the residual isotope signal was reported. Data were normalized to cell protein content prior to analysis of metabolite fluxes for central carbon, acylcarnitine, and FA metabolites.
Authors' Disclosures
F. Puca reports grants from National Cancer Institute R01 and grants from American-Italian Cancer Foundation (AICF) during the conduct of the study. C. Bartolacci reports grants from CPRIT #RP140672 #RP150519 during the conduct of the study. P. Pettazzoni is currently an employee of Hoffmann La Roche. F.F. Lang reports other support from DNAtrix, Inc outside the submitted work. E.P. Sulman reports grants, personal fees, and nonfinancial support from Novocure; personal fees and nonfinancial support from Merck; personal fees and nonfinancial support from Zai Lab; and personal fees from Physician's Education Resource outside the submitted work. C.A. Lyssiotis reports personal fees from Astellas Inc outside the submitted work; in addition, C.A. Lyssiotis has a patent for Methods for Diagnosing and Treating Oncogenic Kras-Associated Cancer (US Patent No: 2015126580-A1, 05/07/2015) and a patent for Targeting the Glutamine to Pyruvate Pathway for Treatment of Oncogenic Kras-Associated Cancer (US Patent No: 20190136238, 05/09/2019, International Patent No: WO2013177426-A2, 04/23/2015). G.F. Draetta reports grants from NCI-NIH during the conduct of the study; personal fees from Tessa Pharmaceuticals, personal fees from Metabomed, personal fees from Helsinn, personal fees from Nurix, personal fees from Alligator Biosciences, personal fees from Karyopharm, personal fees from Forma Therapeutics, personal fees from Taiho Pharmaceutical Co, personal fees from Blueprint Medicines, personal fees from BiovelocITA, personal fees from Orionis, and personal fees from Frontier Medicines outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures S1 through S3
Acknowledgments
We thank Dr. Jason Huse from the MD Anderson Department of Anatomical Pathology for advice regarding GBM classification; Jay Dunn at Seahorse Bioscience for support; Kenneth Dunner Jr. and the High Resolution Electron Microscopy Facility at MDACC for TEM (NCI CA16672); the MDACC Flow Cytometry and Cellular Imaging (FCCI) Core Facility (NCI P30CA16672) for flow cytometers and FACS; Robert Nguyen for lab management; and the MD Anderson veterinary medicine staff, Shan Jiang, and Ivonne Flores for support with in vivo experiments. F. Puca, J. Liu, C.A. Lyssiotis, A. Viale, and G.F. Draetta were supported by NCI R01 CA218139 01 A1. G.F. Draetta was also supported by the Sewell Family Chairmanship in Genomic Medicine. C.A. Lyssiotis was also supported by NCI R37CA237421, R01CA248160, R01CA244931 and UMCCC Core Grant P30CA046592. P.P. Scaglioni was supported by Cancer Prevention and Research Institute of Texas grants RP140672 and RP150519. F. Puca and P. Petazzoni received support from the American-Italian Cancer Foundation. F. Yu was supported by the CPRIT Training Program RP170067. S. Srinivasan was supported by the CPRIT Graduate Scholar Fellowship. F.F. Lang was supported by NCI 2P50CA127001, The University of Texas MD Anderson Cancer Center Moon Shots Program, The Broach Foundation for Brain Cancer Research, and The Elias Family Fund to F.F. Lang. Metabolomics studies performed at the University of Michigan were supported by NIH grant DK097153, the Charles Woodson Research Fund, and the UM Pediatric Brain Tumor Initiative. This research was also supported by NCI Core Facility Grant CA16672 and the NCI Cancer Center Support Grant to MD to MD Anderson P30CA16672.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
F. Puca: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. F. Yu: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, project administration, writing–review and editing. C. Bartolacci: Data curation, investigation, methodology, writing–review and editing. P. Pettazzoni: Conceptualization, data curation, formal analysis, investigation, methodology. A. Carugo: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–review and editing. E. Huang-Hobbs: Resources, investigation, methodology, writing–review and editing. J. Liu: Investigation. C. Zanca: Investigation, methodology. F. Carbone: Investigation, methodology. E. Del Poggetto: Formal analysis, investigation. J. Gumin: Resources, formal analysis, investigation. P. Dasgupta: Resources, investigation. S. Seth: Formal analysis, investigation. S. Srinivasan: Formal analysis, writing–review and editing. F.F. Lang: Resources, supervision. E.P. Sulman: Resources, supervision. P.L. Lorenzi: Investigation, methodology. L. Tan: Investigation, methodology. M. Shan: Investigation, methodology. Z.P. Tolstyka: Investigation, methodology. M. Kachman: Investigation, methodology. L. Zhang: Investigation, methodology. S. Gao: Funding acquisition, project administration, writing–review and editing. A.K. Deem: Conceptualization, supervision, funding acquisition, project administration, writing–review and editing. G. Genovese: Conceptualization, resources, supervision, methodology, writing–review and editing. P. Scaglioni: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft. C.A. Lyssiotis: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. A. Viale: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. G.F. Draetta: Conceptualization, resources, data curation, supervision, funding acquisition, validation, project administration, writing–review and editing.
References
- 1. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJet al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 2. Liebelt BD, Shingu T, Zhou X, Ren J, Shin SA, Hu J. Glioma stem cells: signaling, microenvironment, and therapy. Stem Cells Int 2016;2016:7849890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ru P, Williams TM, Chakravarti A, Guo D. Tumor metabolism of malignant gliomas. Cancers 2013;5:1469–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hou LC, Veeravagu A, Hsu AR, Tse VC. Recurrent glioblastoma multiforme: a review of natural history and management options. Neurosurg Focus 2006;20:E5. [DOI] [PubMed] [Google Scholar]
- 5. Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz Met al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol 2015;129:679–93. [DOI] [PubMed] [Google Scholar]
- 6. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol 2017;14:434–52. [DOI] [PubMed] [Google Scholar]
- 7. Gan HK, van den Bent M, Lassman AB, Reardon DA, Scott AM. Antibody-drug conjugates in glioblastoma therapy: the right drugs to the right cells. Nat Rev Clin Oncol 2017;14:695–707. [DOI] [PubMed] [Google Scholar]
- 8. Yang J, Yan J, Liu B. Targeting EGFRvIII for glioblastoma multiforme. Cancer Lett 2017;403:224–30. [DOI] [PubMed] [Google Scholar]
- 9. DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli Set al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A 2007;104:19345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Portais JC, Voisin P, Merle M, Canioni P. Glucose and glutamine metabolism in C6 glioma cells studied by carbon 13 NMR. Biochimie 1996;78:155–64. [DOI] [PubMed] [Google Scholar]
- 11. Warburg O. On the origin of cancer cells. Science 1956;123:309–14. [DOI] [PubMed] [Google Scholar]
- 12. Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SRet al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carugo A, Genovese G, Seth S, Nezi L, Rose JL, Bossi Det al. In vivo functional platform targeting patient-derived xenografts identifies WDR5-Myc association as a critical determinant of pancreatic cancer. Cell Rep 2016;16:133–47. [DOI] [PubMed] [Google Scholar]
- 15. Schatz UA, Ensenauer R. The clinical manifestation of MCAD deficiency: challenges towards adulthood in the screened population. J Inherit Metab Dis 2010;33:513–20. [DOI] [PubMed] [Google Scholar]
- 16. Vishwanath VA. Fatty acid beta-oxidation disorders: a brief review. Ann Neurosci 2016;23:51–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomas JG, Parker Kerrigan BC, Hossain A, Gumin J, Shinojima N, Nwajei Fet al. Ionizing radiation augments glioma tropism of mesenchymal stem cells. J Neurosurg 2018;128:287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hossain A, Gumin J, Gao F, Figueroa J, Shinojima N, Takezaki Tet al. Mesenchymal stem cells isolated from human gliomas increase proliferation and maintain stemness of glioma stem cells through the IL-6/gp130/STAT3 pathway. Stem Cells 2015;33:2400–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter Det al. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro-oncol 2018;20:380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mamunes P, Devries GH, Miller CD, David RB, Maurer HM. Fatty acids in Reye's syndrome. Pediatr Res 1974;8:436. [Google Scholar]
- 21. Spector R. Fatty acid transport through the blood-brain barrier. J Neurochem 1988;50:639–43. [DOI] [PubMed] [Google Scholar]
- 22. Ebert D, Haller RG, Walton ME. Energy contribution of octanoate to intact rat brain metabolism measured by 13C nuclear magnetic resonance spectroscopy. J Neurosci 2003;23:5928–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe Set al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014;34:11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJet al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 2018;560:243–7. [DOI] [PubMed] [Google Scholar]
- 25. Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou Met al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci U S A 2011;108:16062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tserng KY, Jin SJ, Hoppel CL. Spiropentaneacetic acid as a specific inhibitor of medium-chain acyl-CoA dehydrogenase. Biochemistry 1991;30:10755–60. [DOI] [PubMed] [Google Scholar]
- 27. Marten B, Pfeuffer M, Schrezenmeir J. Medium-chain triglycerides. Int Dairy J 2006;16:1374–82. [Google Scholar]
- 28. Smith EH, Thomas C, McHugh D, Gavrilov D, Raymond K, Rinaldo Pet al. Allelic diversity in MCAD deficiency: the biochemical classification of 54 variants identified during 5 years of ACADM sequencing. Mol Genet Metab 2010;100:241–50. [DOI] [PubMed] [Google Scholar]
- 29. Lappano R, Sebastiani A, Cirillo F, Rigiracciolo DC, Galli GR, Curcio Ret al. The lauric acid-activated signaling prompts apoptosis in cancer cells. Cell Death Discov 2017;3:17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fauser JK, Matthews GM, Cummins AG, Howarth GS. Induction of apoptosis by the medium-chain length fatty acid lauric acid in colon cancer cells due to induction of oxidative stress. Chemotherapy 2013;59:214–24. [DOI] [PubMed] [Google Scholar]
- 31. Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol 2016;26:165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paradies G, Paradies V, Ruggiero FM, Petrosillo G. Oxidative stress, cardiolipin and mitochondrial dysfunction in nonalcoholic fatty liver disease. World J Gastroenterol 2014;20:14205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O'Brien PJ, Little C. Intracellular mechanisms for the decomposition of a lipid peroxide. II. Decomposition of a lipid peroxide by subcellular fractions. Can J Biochem 1969;47:493–9. [DOI] [PubMed] [Google Scholar]
- 34. Sevanian A, Hochstein P. Mechanisms and consequences of lipid peroxidation in biological systems. Annu Rev Nutr 1985;5:365–90. [DOI] [PubMed] [Google Scholar]
- 35. Sevanian A, McLeod LL. Cholesterol autoxidation in phospholipid membrane bilayers. Lipids 1987;22:627–36. [DOI] [PubMed] [Google Scholar]
- 36. Iwase H, Takatori T, Nagao M, Iwadate K, Nakajima M. Monoepoxide production from linoleic acid by cytochrome c in the presence of cardiolipin. Biochem Biophys Res Commun 1996;222:83–9. [DOI] [PubMed] [Google Scholar]
- 37. Imagawa T, Kasai S, Matsui K, Nakamura T. Methyl hydroperoxy-epoxy-octadecenoate as an autoxidation product of methyl linoleate: a new inhibitor-uncoupler of mitochondrial respiration. J Biochem 1982;92:1109–21. [DOI] [PubMed] [Google Scholar]
- 38. Buratta M, Castigli E, Sciaccaluga M, Pellegrino RM, Spinozzi F, Roberti Ret al. Loss of cardiolipin in palmitate-treated GL15 glioblastoma cells favors cytochrome c release from mitochondria leading to apoptosis. J Neurochem 2008;105:1019–31. [DOI] [PubMed] [Google Scholar]
- 39. Birk AV, Liu S, Soong Y, Mills W, Singh P, Warren JDet al. The mitochondrial-targeted compound SS-31 re-energizes ischemic mitochondria by interacting with cardiolipin. J Am Soc Nephrol 2013;24:1250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 2019;20:137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng X, Geng F, Pan M, Wu X, Zhong Y, Wang Cet al. Targeting DGAT1 ameliorates glioblastoma by increasing fat catabolism and oxidative stress. Cell Metab 2020;32:229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tolwani RJ, Hamm DA, Tian L, Sharer JD, Vockley J, Rinaldo Pet al. Medium-chain acyl-CoA dehydrogenase deficiency in gene-targeted mice. PLoS Genet 2005;1:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin H, Patel S, Affleck VS, Wilson I, Turnbull DM, Joshi ARet al. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro Oncol 2017;19:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer 2013;13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aires CC, Ijlst L, Stet F, Prip-Buus C, de Almeida IT, Duran Met al. Inhibition of hepatic carnitine palmitoyl-transferase I (CPT IA) by valproyl-CoA as a possible mechanism of valproate-induced steatosis. Biochem Pharmacol 2010;79:792–9. [DOI] [PubMed] [Google Scholar]
- 46. Sayed-Ahmed MM, Shouman SA, Rezk BM, Khalifa MH, Osman AM, El-Merzabani MM. Propionyl-L-carnitine as potential protective agent against adriamycin-induced impairment of fatty acid beta-oxidation in isolated heart mitochondria. Pharmacol Res 2000;41:143–50. [DOI] [PubMed] [Google Scholar]
- 47. He L, Kim T, Long Q, Liu J, Wang P, Zhou Yet al. Carnitine palmitoyltransferase-1b deficiency aggravates pressure overload-induced cardiac hypertrophy caused by lipotoxicity. Circulation 2012;126:1705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cabrero A, Merlos M, Laguna JC, Carrera MV. Down-regulation of acyl-CoA oxidase gene expression and increased NF-kappaB activity in etomoxir-induced cardiac hypertrophy. J Lipid Res 2003;44:388–98. [DOI] [PubMed] [Google Scholar]
- 49. Haynie KR, Vandanmagsar B, Wicks SE, Zhang J, Mynatt RL. Inhibition of carnitine palymitoyltransferase1b induces cardiac hypertrophy and mortality in mice. Diabetes Obes Metab 2014;16:757–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks Eet al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods 2009;6:569–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakamizo A, Marini F, Amano T, Khan A, Studeny M, Gumin Jet al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res 2005;65:3307–18. [DOI] [PubMed] [Google Scholar]
- 52. Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg 2000;92:326–33. [DOI] [PubMed] [Google Scholar]
- 53. Padanad MS, Konstantinidou G, Venkateswaran N, Melegari M, Rindhe S, Mitsche Met al. Fatty acid oxidation mediated by acyl-CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep 2016;16:1614–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom 1996;31:255–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1 through S3