Abstract
Transposable elements comprise almost half of the mammalian genome. A growing body of evidence suggests that transposable element dysregulation accompanies brain aging and neurodegenerative disorders, and that transposable element activation is neurotoxic. Recent studies have identified links between pathogenic forms of tau, a protein that accumulates in Alzheimer’s disease and related “tauopathies,” and transposable element-induced neurotoxicity. Starting with transcriptomic analyses, we find that age- and tau-induced transposable element activation occurs in the mouse brain. Among transposable elements that are activated at the RNA level in the context of brain aging and tauopathy, we find that the endogenous retrovirus (ERV) class of retrotransposons is particularly enriched. We show that protein encoded by Intracisternal A-particle, a highly active mouse ERV, is elevated in brains of tau transgenic mice. Using two complementary approaches, we find that brains of tau transgenic mice contain increased DNA copy number of transposable elements, raising the possibility that these elements actively retrotranspose in the context of tauopathy. Taken together, our study lays the groundwork for future mechanistic studies focused on transposable element regulation in the aging mouse brain and in mouse models of tauopathy and provides support for ongoing therapeutic efforts targeting transposable element activation in patients with Alzheimer’s disease.
Keywords: Alzheimer’s disease, tauopathy, aging, tau, amyloid beta, retrotransposon, endogenous retrovirus, neurodegeneration, mice
INTRODUCTION
Approximately 40% of the human genome is composed of retrotransposons1. Long and Short Interspersed Nuclear Elements (LINEs, and SINEs, respectively), and Long-Terminal Repeat (LTR)-containing elements make up the major classes of retrotransposons2. Intact LINE elements, including some LINE-1 (L1) subfamily members, are 6–8 kb and harbor two open reading frames encoding proteins with RNA binding, nucleic acid chaperone, endonuclease and reverse transcriptase activities3,4. LINE-encoded proteins function to reverse transcribe LINE and SINE RNA and subsequently insert the newly generated LINE or SINE DNA copy into genomic DNA. ERVs are structurally similar to exogenous retroviruses and are the predominant class of LTR-containing retrotransposons in mammals. Like exogenous retroviruses, the polyprotein encoded by intact ERVs encodes matrix and capsid proteins, as well as a protease, reverse transcriptase, integrase, and, on occasion, envelope proteins5,6. Reverse transcription of the ERV RNA is thought to occur inside the capsid, after which the integrase facilitates reinsertion of the new ERV DNA copy into genomic DNA7. A simplified overview of the lifecycle of an intact, autonomous LINE or LTR retrotransposon is depicted in Figure 1a.
Figure 1 |. Age-dependent increase in transposable element RNA levels in the mouse forebrain.
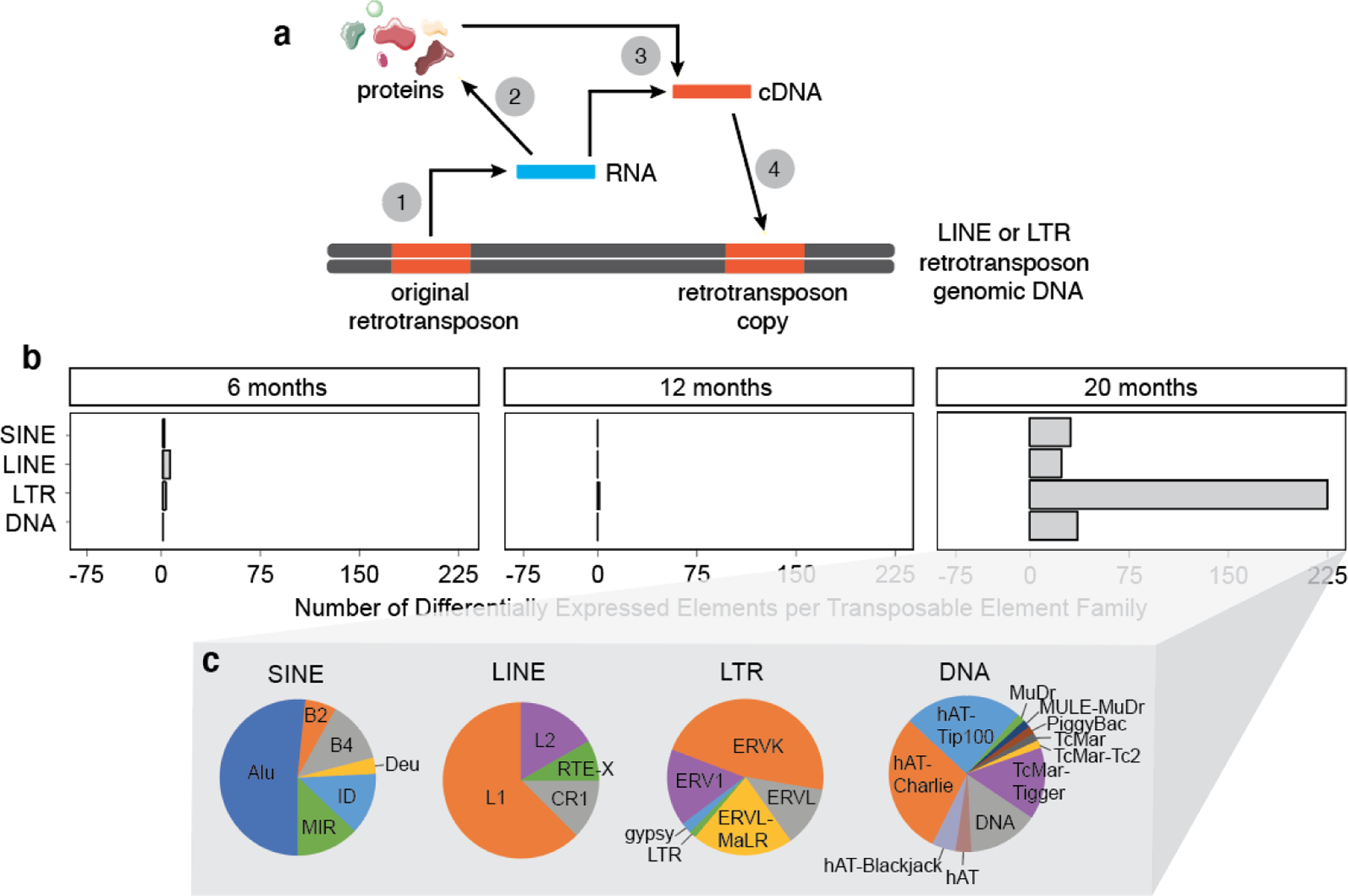
a. Lifecycle of an intact, autonomous retrotransposon: Retrotransposon DNA is transcribed into RNA (1), which encodes proteins that are needed for reverse transcription and subsequent integration into genomic DNA (2). Using retrotransposonencoded proteins, RNA is reverse transcribed into cDNA that can either exist in an episomal state (3) or can be inserted into genomic DNA (4). b. The number of differentially expressed transposable elements in mouse forebrain from six to twenty months old based on RNA-seq. Differentially expressed transposable elements were grouped into families and represented on the graph. c. Pie charts representing the proportion of subfamily members within each retrotransposon (SINE, LINE, LTR) and DNA transposon (DNA) family. 6 months: n=1 male, 7 females; 12 months: n=4 males, 4 females; 20 months: n=6 males and 5 females. An adjusted P value of <0.05 was considered significant.
It is now understood that transposition can provide an evolutionary advantage by driving genome expansion and evolutionary diversity; however, unchecked transposition can cause DNA breaks and genomic instability, genetic polymorphism, and insertional mutations that can be detrimental to the organism8. While the human and mouse genomes share a similar density of transposable elements, mice have retained a high level of transposon activity over the course of evolution, while most transposable elements in humans have become transpositionally inert1, with the exception of ~35–40 subfamilies of L1, Alu, and SVA9. In addition to mutations induced by transposition, proteins, single- and double-stranded RNAs (dsRNAs), and episomal DNA produced from retrotransposons can also impact cellular function10 (Fig. 1a). When considering potential toxicity of retrotransposon activation in a given system, retrotransposon-derived products must thus be considered in addition to consequences of retrotransposition to genomic DNA.
The “transposon theory of aging” posits that the cellular control systems that keep transposable elements in check decline with age, and that loss of transposable element control contributes to age-related decline in tissue function11–14. Retrotransposon activation has been documented in the context of physiological aging of the Drosophila fat body and brain13,14. In the liver and muscle of aging mice, several retrotransposons are elevated at both the RNA and DNA levels, suggesting that age-associated transposition also occurs in somatic cells of mammals12. We and others have recently reported that retrotransposons become activated in Drosophila models of tauopathy and in post-mortem brains of patients with Alzheimer’s disease or progressive supranuclear palsy, a “primary” tauopathy15,16. Alzheimer’s disease and related tauopathies are aging-associated human neurodegenerative disorders that involve the deposition of pathological forms of tau protein17. In human Alzheimer’s disease, activation of HERV1, 2, and 3 and L1 retrotransposons are significantly associated with tau tangle burden, and HERV1, 2, and 3 activation is significantly associated with reduced cognitive performance16. Studies in fly models of tauopathy suggest that retrotransposon activation is a consequence of tau-induced breakdown of heterochromatin- and piwi-interacting RNA (piRNA)-mediated retrotransposon silencing, is a causal driver of neurodegeneration, and can be suppressed by antiretroviral therapy15.
In the current study, we report that aging- and tau-induced activation of retrotransposons occurs in the mouse brain. Our age-dependent analyses of retrotransposon activation at the RNA, protein, and DNA levels provides a foundation for future mechanistic studies, and support ongoing clinical trials and continued drug development focused on blocking retrotransposon activation in Alzheimer’s disease and related tauopathies.
MATERIALS AND METHODS
Mouse Models
rTg4510.
The founder rTg4510 and non-transgenic control mice (mixed 129S6, FVB genetic background) were obtained from the Jackson Laboratory (IMSR_JAX:024854). Mice were group-housed in a pathogen-free mouse facility with ad libitum access to food and water on a twelve-hour light/dark cycle. After anesthesia with 2% isoflurane, cardiac perfusion was performed using 2X PhosSTOP phosphatase inhibitors (Roche, Indianapolis, IN, USA) and 1X complete protease inhibitors (Roche) in PBS (Thermo Fisher Scientific, Waltham, MA, USA). All experimental procedures were performed according to the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
PS19.
The founder PS19 mice were obtained from the Jackson Laboratory (IMSR_JAX:008169). Heterozygotic offspring were bred and maintained at Baylor College of Medicine. Wild-type littermates were used as controls. Mice were housed 4–5 per cage in a pathogen-free mouse facility with ad libitum access to food and water on a twelve-hour light/dark cycle. Male mice were analyzed in this study. All procedures were performed in accordance with NIH guidelines and approval of the Baylor College of Medicine Institutional Care and Use Committee.
RNA Sequencing (RNA-seq) and Analysis
RNA was isolated from hippocampi of rTg4510 and control, and spinal cord of PS19 and control mice using the Qiagen RNeasy Mini kit and Qiagen TissueRuptor (Qiagen, Germantown, MD, USA) based on the manufacturer’s recommendations. RNA from n=17–18 rTg4510 female mice and controls per timepoint was sequenced using an Illumina HiSeq 4000 with 76 bp paired-end stranded reads at ~30 million reads per sample. RNA from 5–7 PS19 male mice was sequenced using an Illumina HiSeq 6000 with 150 bp paired-end reads at ~20 million reads per sample. For transposable element analysis in the aging mouse brain, RNA-seq data from control mice (B6C3HF1) were downloaded from AMP-AD dataset syn3157182 (n=1 male, 7 females at 6 months, n=4 males and 4 females at 12 months, and n=6 males and 5 females at 20 months). RNA-seq data from spinal cord of n=3–6 JNPL3 and Swiss Webster nontransgenic control female mice per timepoint were downloaded from AMP-AD dataset syn3157182. B6C3HF1, JNPL3 and control mice were sequenced on an Illumina HiSeq 2000 with 101 bp paired end reads at ~50 million reads per sample. RNA libraries were prepared with Illumina TruSeq library kits. Each tau model has its own set of age-matched control samples. RNA sequencing data from APP/PS1 cerebral cortex was downloaded from NCBI (GEO:GSE132177) for six and nine month timepoints for transgenic (IMSR_JAX:34832) and wild type mice (n=3 male). This dataset was generated by 150 bp, paired end, stranded sequencing using the Illumina HiSeq 4000.
FastQ files for each experiment were processed with Trimmomatic18 v.0.36 (SLIDINGWINDOW:5:20 MINLEN:50) to remove low quality reads and trim adapter sequences. FastQC19 was then used to obtain pre and post quality control processing statistics for the FastQ files. Trimmed paired reads were aligned with STAR20 v.2.7.1a to the mm10 mouse genome using parameters suggested by TEtranscripts21 (--outFilterMultimapNmax 100, --winAnchorMultimapNmax 100). Transposon counts for each sample were then obtained using TEtranscripts v.2.0.3 (default parameters) and its supplied mouse mm10 genome transposon annotation file.
Differential transposon expression analysis was completed with DESeq222. Pairwise comparisons were performed with the DESeq2 Wald test on rTg4510, JNPL3, PS19 and APP/PS1 mice and their age- and model-matched controls. To quantify transposable element RNA changes as a consequence of aging, differential transposon expression was performed as a time series analysis (test = LRT, full model = Sex + Age, reduced model = ~Sex) at six, twelve and twenty months. Differentially expressed transposons from the LRT test were clustered into similar expression patterns across the different age groups using DEGreports23. The three clusters correspond to the age at which expression of the transposable element was at its highest. Genes with an adjusted P value of <0.05 were considered significantly differentially expressed24. Sequencing data is included in Supplemental Table 1.
Barplots and heatmaps were generated using the R packages ggplot225 and pHeatmap26, respectively. Transposable elements were grouped into families and subfamilies using the TEtranscripts21 supplied annotation information.
Development of the IAP-gag Antibody
The nucleocapsid sequence corresponding to the capsid protein that is processed from the IAP-gag precursor (UniProt: P11365) was sent to GenScript for antibody development:
LTGQGAYSADQTNYHWGAYAQISSTAIRRWKGLSRAGETTGQLTKVVQGPQESFSDFVARMTEAAERIFGESEQAAPLIEQLIYEQATKECRAAIAPRKNKGLQDWLRVCRELGGPLTNAGL
The peptide was expressed in E. coli and used to immunize two rabbits for antibody production. The GeneScript proprietary OptimumAntigenTM design tool was used to measure the antigen against several protein databases to enhance the desired antibody and epitope specificity. GenScript utilized codon optimization and BacPowerTM bacterial protein expression technology and proprietary adjuvant to increase the efficiency of protein expression and animal immunization. An ELISA titer of over 1:128,000 and target protein binding were validated by immunoprecipitation and western blotting using the positive control with protein immunogen by GenScript. The final product was 0.5 ml of pre-immune serum at 1.5–6 mg/rabbit and 1 mg of the requested peptide.
Western Blotting
10–20 mgs of frozen mouse brain cortex were homogenized in 60 µl Laemmli sample buffer (Invitrogen). Total protein content was quantified with a BCA protein assay kit (Pierce) and normalized across samples. Lysates containing approximately 40 µg of protein were boiled for 10 min prior to separation by 4–20% SDS-PAGE (BIO-RAD). After transferring at 90V to a nitrocellulose membrane for 90 min at room temperature, Ponceau S staining was performed to ensure that protein loading was equal among samples prior to membrane blocking with 2% milk in PBS with 0.05% Tween (PBSTw) at room temperature for 30 min. The membrane was then incubated with the anti-IAP-gag antibody at 1:500 (GeneScript) overnight at 4°C. Membranes were washed three times with PBSTw and incubated with an HRP-conjugated secondary antibody for 2 h at room temperature. Blots were developed with an enhanced chemiluminescent substrate (Thermo Scientific). After imaging, membranes were stripped and incubated with an anti-Actin antibody at 1:1,000 dilution (#JLA20, DSHB) overnight at 4°C. The following day, membranes were washed and incubated with an HRP-conjugated secondary antibody, and imaged with an enhanced chemiluminescent substrate. Band densitometry was performed using ImageJ.
Immunostaining
40 µm mouse brain sections were stored at 20°C in cryoprotectant prior to use. Individual sections were transferred to PBS in single wells of a 24 well plate. After washing three times with PBS, sections were incubated overnight with the IAP-gag primary antibody27 (1:100, a gift from Dr. Brian Cullen) and AT8 (1:200, #MN1020, Fisher) in PBS + 0.3% Triton + 2% BSA at 4°C overnight. After three washes in PBS+Triton, sections were incubated with Alexa Fluor 488- or 555-conjugated IgG (1:200, Invitrogen) at room temperature for 2 h. After washing, the sections were mounted onto microscope slides and then coverslipped with DAPI-containing Fluoromount G. Immunofluorescence was visualized on a Zeiss confocal microscope and analyzed with ImageJ. Brain sections from twelve month old rTg4510 and APP/PS1 models used for IAP-gag immunostaining were generated from mice used in a previous study28.
Digital PCR (dPCR)
dPCR probes were designed to detect active subfamily members of L1 (LINE subfamily member), ETn (ERVK subfamily member), IAP (ERVK subfamily member), and B2 (SINE subfamily member) retrotransposons by aligning subfamilies via the MAFFT alignment tool using the default parameters29 and subsequently visualized in Geneious (Geneious Prime 2021.2.2 (https://www.geneious.com)) to identify regions that are unique to active transposons within the subfamily. Target sequences were further refined by selecting regions distal from the 5’ region of transposons to account for potential 5’ truncation within novel insertions. One unique region per subfamily was selected and submitted to the IDT PrimerQuest tool. Primers were then blasted via BlastN to a custom Repbase Mouse TE database to determine specificity of primers for active elements (Supplemental Table 2).
DNA was extracted from frozen anterior cortex following the DNeasy blood & tissue kit protocol (Qiagen, Cat. No. 69504), and DNA concentration was measured using Nanodrop 8000 spectrophotometer (Thermo Scientific). To remain within the limits of detection of the dPCR chip, the quantity of total DNA loaded onto the dPCR chip was dependent upon the abundance of each element within the mouse genome. 600 ng of DNA was loaded per sample to detect L1, 30 ng was loaded per sample to detect IAP and ETn, and 5 ng was loaded to detect B2. Absolute quantification (copies/µL) was performed using the Applied Biosystems QuantStudio 3D Digital PCR system, and each target was normalized to the control, the average of which was set to one. Multiple t-tests were performed with GraphPad Prism v8.4.230.
NanoString
In collaboration with NanoString Technologies, Inc., we created a custom nCounter codeset consisting of 188 probes targeting 89 mouse transposable elements and 10 internal controls. Two probes were designed for each transposable element region and one probe was designed for each internal control (see Supplemental Table 2 for codeset sequences and data). Single copy or low copy genes were used as internal controls. To identify transposable element family members that are recognized by each probe, probe sequences were BLASTed31 to a database consisting of mouse transposable elements from Repbase32 and transposable elements present in the TEtranscripts21 mouse transposable element GTF file. Family members with >85% identity to the probe sequence and >75 bp length were considered hits. 3–5 µg of DNA was isolated from frozen cortex of 2- and 12-month-old mice according to the manufacturer’s protocol (DNeasy Blood & Tissue Kit, Qiagen, Cat No. 69504) and diluted with 10 mM Tris. Prior to hybridization, diluted DNA was processed in a Covaris Focused-ultrasonicator to produce 200 bp fragments. DNA fragmentation was confirmed using an Agilent 2100 Bioanalyzer. 300 ng of fragmented DNA was isolated via ethanol precipitation and used for NanoString nCounter Copy Number Assays. nCounter SPRINT profiler was used to quantify copy number of each target. Raw retrotransposon probe counts were normalized to ten probes recognizing invariant regions of the mouse genome (Supplemental Table 2) using nSolver 4 in order to correct for technical variability. Transposable elements with significantly different DNA copy numbers between rTg4510 and control were detected by multiple t-tests assuming equal standard deviation between samples, followed by the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli with a q value = 0.05 using GraphPad Prism v8.4.230 for MacOS (GraphPad Software, San Diego, CA USA). Volcano plots for DNA copy change were made using ggplot233.
RESULTS
Age-dependent increase of transposable element transcripts in the mouse brain
To determine if transposable elements are differentially expressed as a consequence of aging in the adult mouse brain, we analyzed transposable element transcript levels in forebrain lysates from B6C3HF1 mice aged to six, twelve and twenty months of age based on publicly available RNA-seq data available through the Accelerating Medicines Partnership – Alzheimer’s Disease (AMP-AD). Among the transposable elements that are differentially expressed in the mouse brain by twenty months, we observe that 92.7% increase with age (Fig. 1b, Supplemental Table 1). Among LINE, LTR, and DNA families of transposable elements that are increased in brains of twenty-month-old mice, we find that the LTR family is particularly affected and that ERVs are the predominant family of LTR retrotransposon that increases with age (Fig. 1c). Several retrotransposons that are elevated at the RNA level in the aging mouse brain, including but not limited to L1, IAP and IAP-E, are autonomously active elements (Supplemental Table 1).
Age-dependent increase of transposable element transcripts in brains of tau transgenic mice
Based on previous studies reporting elevated transcript levels of select retrotransposons in human tauopathies as well as Drosophila models of tauopathy15,16, we sought to determine if retrotransposon transcripts are elevated in mouse models of tauopathy. We analyzed RNA-seq datasets across several disease stages from three different mouse models of tauopathy: rTg451034, JNPL335, and PS1936.
rTg4510 mice have a Camk2a-driven 13-fold overexpression of a familial tauopathy-associated mutant form of human tau, tauP301L, on a mixed 129S6, FVB genetic background. Tau tangles can be detected by four months of age, while neuronal and synaptic loss occur by six and nine months, respectively37,38 (Fig. 2a). As females exhibit a higher degree of tau pathology and behavioral deficits than males in this model39, we analyzed transposable element transcript levels in cortical lysates from female control and rTg4510 mice at three, six, and nine months of age. We detect a general increase in transposable element transcripts by nine months of age in rTg4510 tau transgenic mice (Fig. 2b, Supplemental Table 1). Similar to our findings in the context of physiological mouse brain aging, LTR retrotransposons have the largest increase in rTg4510 mice, with ERVs as the predominant LTR that is elevated (Fig. 2c). Among elements that are transcriptionally upregulated at nine months of age, B1/B2, IAP-E and ERV-β4 subfamily members are considered to be retrotranspositionally active (Supplemental Table 1).
Figure 2 |. Age-dependent analysis of transposable element RNA levels in three different mouse models of tauopathy.
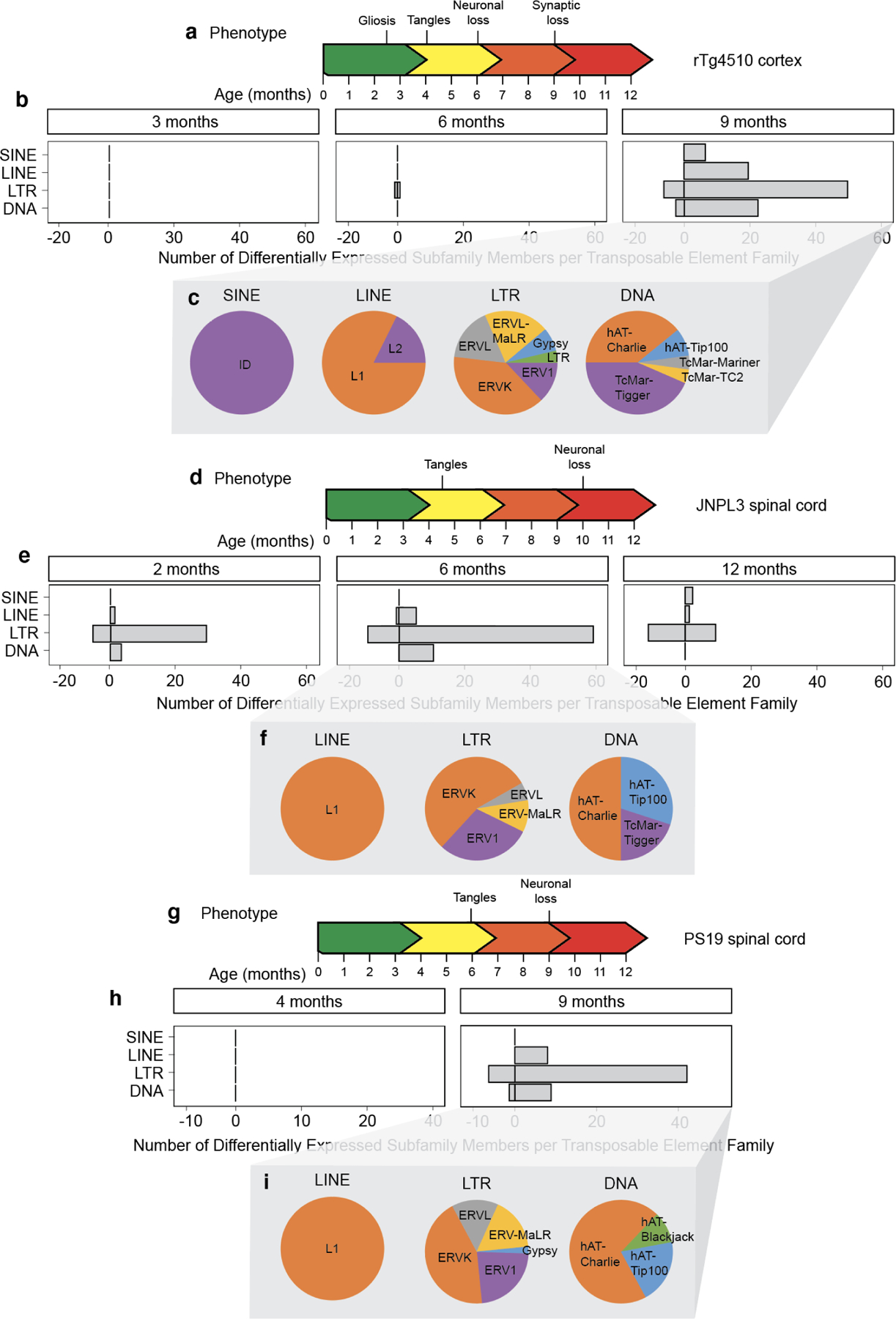
a. The rTg4510 mouse model of tauopathy. b. Differentially expressed transposable elements in the female rTg4510 mouse cortex from three to nine months based on RNA-seq. Differentially expressed transposable elements were grouped into families and represented on the graph. n=5–6 biological replicates per age. c. The proportion of different types of differentially expressed transposable element subfamilies within each transposable element family. d. The JNPL3 mouse model of tauopathy. e. Differentially expressed transposable elements in the female JNPL3 spinal cord from two to twelve months based on RNA-seq. n=3–6 biological replicates per age. f. The proportion of differentially expressed transposable element subfamilies within each transposable element family. g. The PS19 mouse model of tauopathy. h. Differentially expressed transposable elements in the male PS19 spinal cord at four and nine months based on RNA-seq. Differentially expressed transposable elements were grouped into families and represented on the graph. n=5–7 biological replicates per age. I. The proportion of differentially expressed transposable element subfamilies within each transposable element family. An adjusted P value of <0.05 was considered significant.
The JNPL3 model features Prp-driven expression of human tauP301L on a C57Bl/6, DBA/2, SW mixed genetic background. JNPL3 mice produce human tau protein at a level similar to endogenous mouse tau. As neuronal loss in the spinal cord is a predominant feature of this model35, we utilized publicly available RNA-seq data from spinal cord of homozygous JNPL3 mice at two, six, and twelve months of age. RNA-seq data was only available for females, as males exhibit a delayed and variable phenotype in this model. Tau tangles can be detected by six to seven months in the brainstem and spinal cord in female homozygotes, while loss of motor neurons occurs at approximately ten months35 (Fig. 2d). We detect an overall elevation of LTR elements as early as two months in this model, which precedes tau tangle formation. The increase in retrotransposon transcripts is elevated further at six months, after which we detect a moderate decline by twelve months (Fig. 2e, Supplemental Table 1). Again, we find ERVs to be the predominant class of LTR retrotransposons that are elevated in this model of tauopathy (Fig. 2f). Multiple L1, B1/B2, ETn, IAP, IAP-E and MULV active subfamily members are upregulated at all time points in the JNPL3 model, with the exception of one L1 subfamily member that is downregulated at six months (Supplemental Table 1).
The PS19 model features a five-fold Prp-driven overexpression of the familial tauopathy-associated tauP301S mutation on a C57Bl/6, C3H genetic background. The original publication reports presence of neurofibrillary tangles in the brain and spinal cord by six months and neuronal loss by nine months36. As reported by others40,41, our colony has delayed pathology compared to the original line, with higher levels of pathology in the spinal cord compared to the hippocampus. We analyzed RNA-seq data from spinal cord of male PS19 mice at four and nine months of age, as males generally have more consistent presentation of tau pathology in this model (Fig. 2g). While we do not detect any transposable elements that are differentially expressed at four months of age, we observe an increase in retrotransposon transcripts in the spinal cord of PS19 mice by nine months (Fig. 2h, Supplemental Table 1). Similar to our findings in human tauopathy15, physiological mouse brain aging, and rTg4510 and JNPL3 mice, we find that LTR retrotransposons are the predominant class of transposable elements that are elevated at the transcript level in PS19 spinal cord, and that ERVs are the predominant type of LTR retrotransposon that is elevated. Among active retrotransposons, subfamily members of L1, IAP-E, MULV, and MERV are upregulated in the PS19 model (Supplemental Table 1). Collectively, these data demonstrate a general age-dependent increase in retrotransposon transcript levels in tau transgenic mice despite different promoters, tissues, sexes, genetic backgrounds, and transgenic tau dosage, demonstrating that tau-induced ERV activation at the RNA level is robustly conserved among various mouse models.
To determine if retrotransposons are also activated at the transcript level in an amyloid-based mouse model of Alzheimer’s disease, we analyzed publicly-available RNA-seq data from cerebral cortex of mice carrying a chimeric mouse/human amyloid precursor protein gene harboring the Swedish mutation and human presenilin 1 lacking exon 9 (APP/PS1) on a mixed C57BL/6J/C3H/HeJ genetic background42. We analyzed transposable element expression levels at six and nine months, ages at which amyloid plaques and synaptic loss are already present43. In stark contrast to the upregulation of retrotransposon transcripts in multiple mouse models of tauopathy, we only detect three retrotransposons that are differentially expressed in this model, two of which are decreased (Supplemental Fig. 1, Supplemental Table 1).
Increase in protein encoded by an active ERV in brains of tau transgenic mice
As RNA-seq revealed an age-dependent increase in ERV retrotransposons in the context of normal aging and three different mouse models of tauopathy, we next asked if ERV-encoded proteins can be detected in the brain of tau transgenic mice. We focused on the “group antigens” (gag) polyprotein encoded by a highly active mouse ERV, IAP, which has approximately 2,800 complete or near-complete copies in the reference C57BL/6 genome44. As we do not detect a significant increase in RNA levels of ERVs prior to nine months of age in rTg4510 mice based on RNA-seq, we analyzed IAP-encoded protein at six months (prior to elevation in ERV RNA) and twelve months (after elevation in ERV RNA) using an antibody27 that detects IAP-gag. We find that IAP-gag is significantly elevated in frontal cortex of rTg4510 mice at both six and twelve months of age (Fig. 3a, b). Interestingly, the cells harboring elevated gag protein levels are not the cells harboring a disease-associated form of human tau that is phosphorylated at serine 202 and threonine 205 (AT8), suggesting that tau-induced ERV activation is non-cell autonomous. Indeed, the IAP-gag antibody detects an epitope that is present within IAP elements as well as IAPE elements, which harbor an envelope gene that encodes a membrane glycoprotein that facilitates cell-to-cell spread of IAP capsids45,46.
Figure 3 |. IAP-encoded gag protein in rTg4510 and APP/PS1 mouse models.
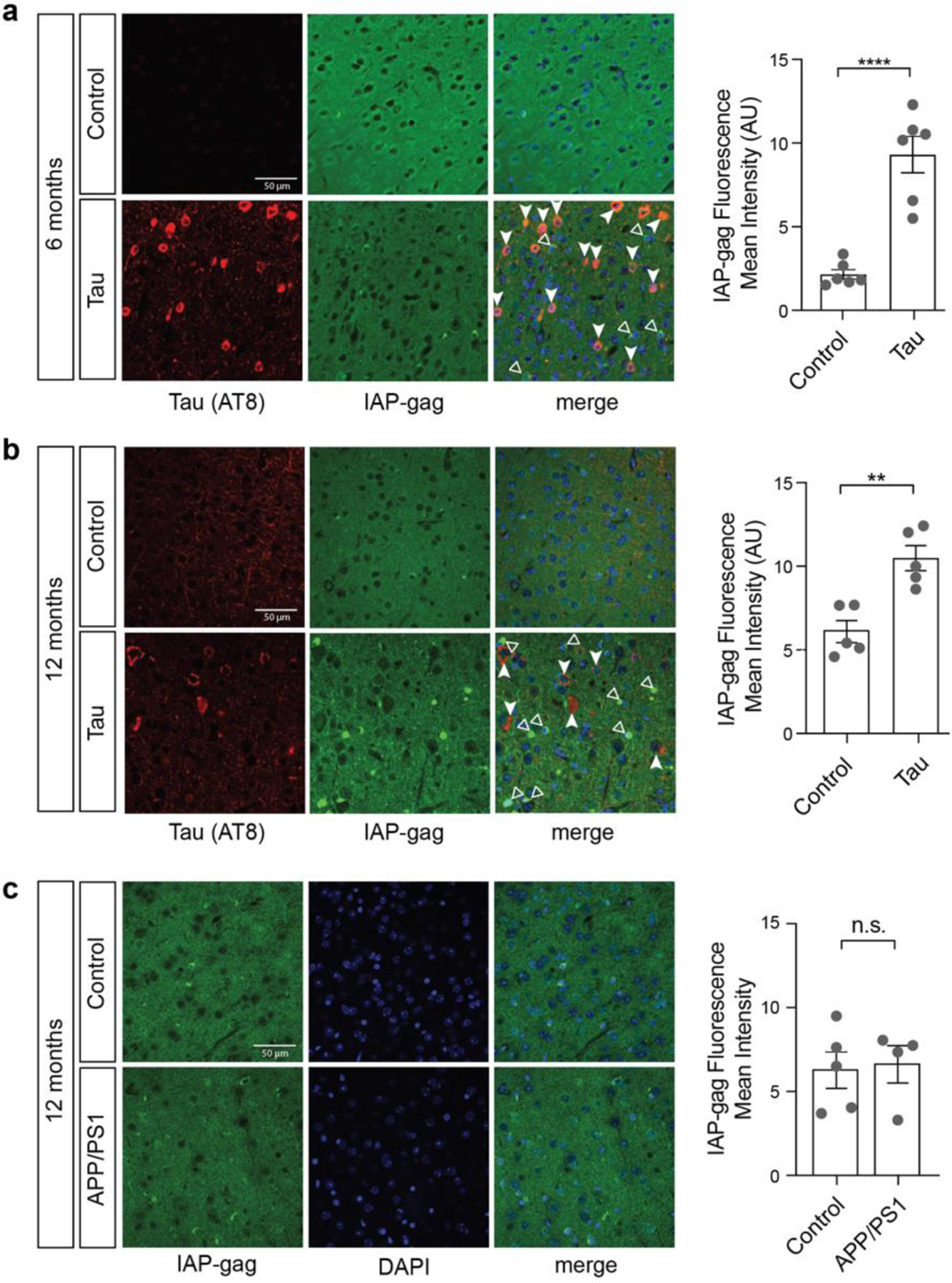
Frontal cortices of rTg4510 mice at six (a) and twelve (b) months were stained with antibodies recognizing IAP-encoded gag and tau AT8. c. Frontal cortices of APP/PS1 mice were stained with antibodies recognizing IAP-encoded gag. n=4–6 biological replicates per genotype, per age. Filled-in arrows indicate tau AT8-positive cells, while empty arrows indicate IAP-gag-positive cells. **p<0.01, ****p<0.0001, unpaired t-test. Error bars=SEM.
We next visualized IAP-gag protein in brains of APP/PS1 mice. Consistent with the lack of retrotransposon activation at the RNA level in this model, we do not detect differences in IAP-encoded gag at twelve months of age in cortex of APP/PS1 mice (Fig. 3c). Along with our RNA-seq analysis in APP/PS1 mice and previous studies in Drosophila models of tauopathy, these data strongly suggest that the retrotransposon activation present in brains of human Alzheimer’s disease patients15,16 is a consequence of pathogenic forms of tau rather than amyloid beta.
The IAP-encoded gag polyprotein is processed to a mature form in brains of tau transgenic mice
Intact copies of IAP encode a gag-pro-pol polyprotein precursor47. The IAP-encoded gag-pro-pol polyprotein undergoes a series of proteolytic cleavages by the pro-encoded protease. Gag is further processed to generate capsid, matrix, and nucleocapsid proteins48 (Fig. 4a).
Figure 4 |. IAP polyprotein processing is increased in brains of rTg5410 mice at twelve months of age.
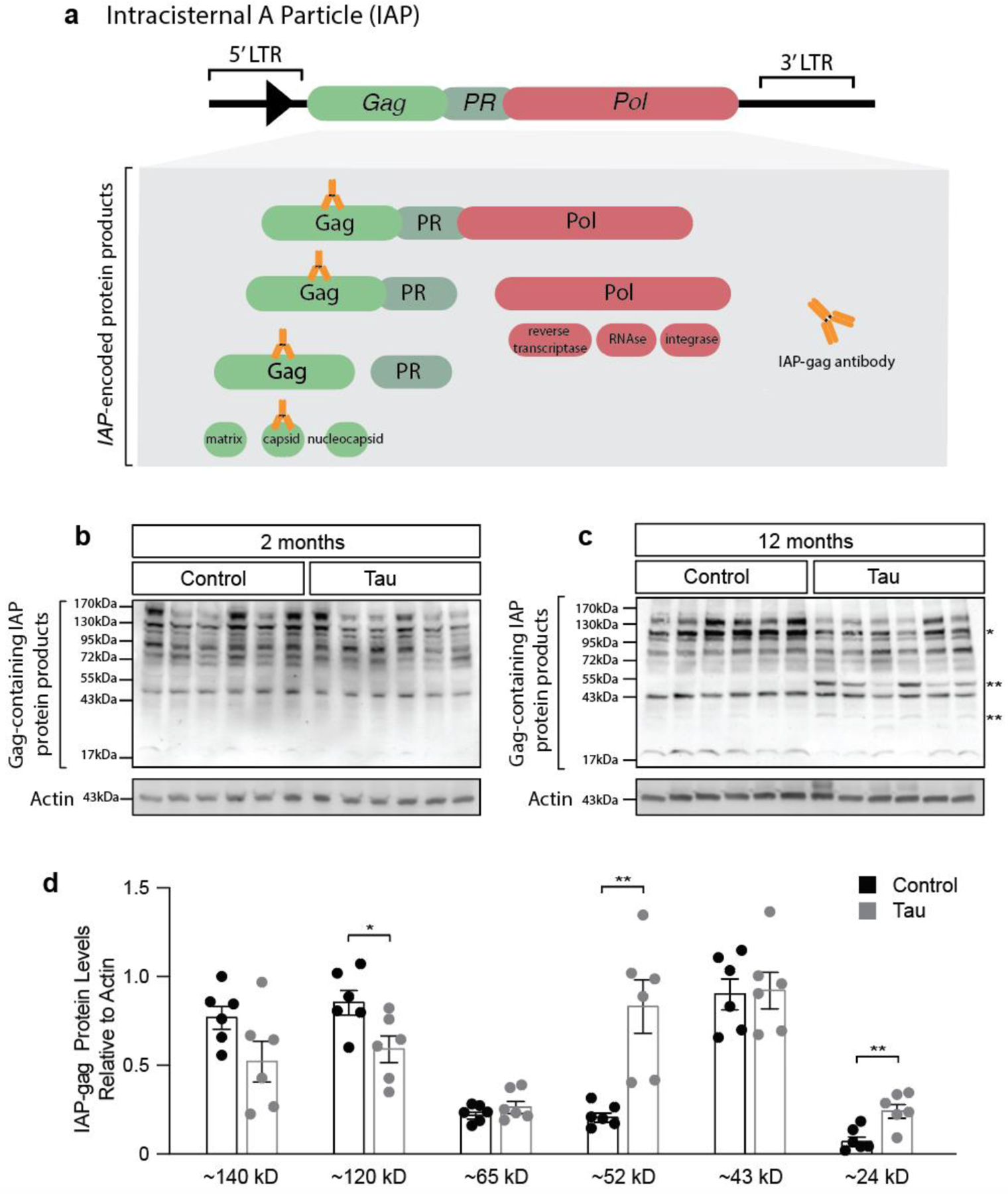
a. Intact copies of the IAP gene encode a gag-pro-pol precursor. The gag-pro-pol precursor is processed into multiple protein products. The antibody used for western blotting recognizes the capsid protein that is produced from gag. IAP polyprotein processing based on immunoblotting with an antibody recognizing the gag-encoded capsid protein in lysates from rTg4510 mouse brain compared to control at two (b) and twelve (c) months of age. d. At twelve months of age, rTg4510 mice have significantly decreased levels of a band corresponding to a ~120 kD gag-pro-pol precursor, and significantly increased levels of ~52 kD and ~24 kD gag-containing proteins. n=6 biological replicates, two-sided t-tests shown on the same graph for visual ease. *p<0.05, **p<0.01. Error bars=SEM.
We took a biochemical approach to determine if the IAP-encoded polyprotein is processed to a mature form in brains of rTg4510 mice. As we detected elevated levels of IAP-encoded gag in rTg4510 mice by six months of age, we extended our earlier timepoint to two months of age. We generated an antibody that recognizes the polypeptide sequence corresponding to the capsid protein cleaved from the gag precursor. While this epitope is present in multiple cleavage products of the gag-pro-pol precursor, and will thus detect multiple stages of processing, proteins generated by cleavage of the precursor can be differentiated by size (Fig. 4a). While IAP-encoded polyprotein processing at two months does not differ between control and tau transgenic mice (Fig. 4b), twelve-month-old rTg4510 mice have a significant decrease in a ~120 kD band, with a concomitant increase in bands at ~52 kD and ~24 kD, consistent with protease-mediated cleavage of an IAP-encoded polyprotein and generation of a mature gag capsid protein (Fig. 4c, d). These data suggest that the polyprotein encoded by IAP is processed to a greater extent in the rTg4510 model, consistent with activation.
Transposable element DNA copy number is elevated in tau transgenic mice
As retrotransposition occurs through a copy-and-paste mechanism, each mobilization event increases the total number of DNA copies of a given element by one. Having identified active retrotransposon subfamily members that are elevated at the RNA and protein levels in brains of tau transgenic mice, we next asked if retrotransposon DNA copy number is elevated in brains of rTg4510 mice. We note that this approach will identify elements that are actively reverse transcribed into DNA, but cannot discriminate between retrotransposon DNA that has integrated into the genome versus retrotransposon DNA that exists in an episomal state. We first created dPCR probes to recognize individual active members of retrotransposon subfamilies: L1 (LINE subfamily member), IAP (ERVK subfamily member), ETn (ERVK subfamily member), and B2 (SINE subfamily member). These probes were specifically designed to detect active elements and their specificity was confirmed by BLAST analysis (see Methods and Supplemental Table 2). While L1, IAP, ETn, and B2 DNA copy number are unchanged in brains of tau transgenic mice at two months of age (Fig. 5a), all of these elements with the exception of B2 are significantly increased in brains of tau transgenic mice compared to control by twelve months (Fig. 5b).
Figure 5 |. dPCR and NanoString-based transposable element DNA copy number analysis in the cortex of rTg4510 mice.
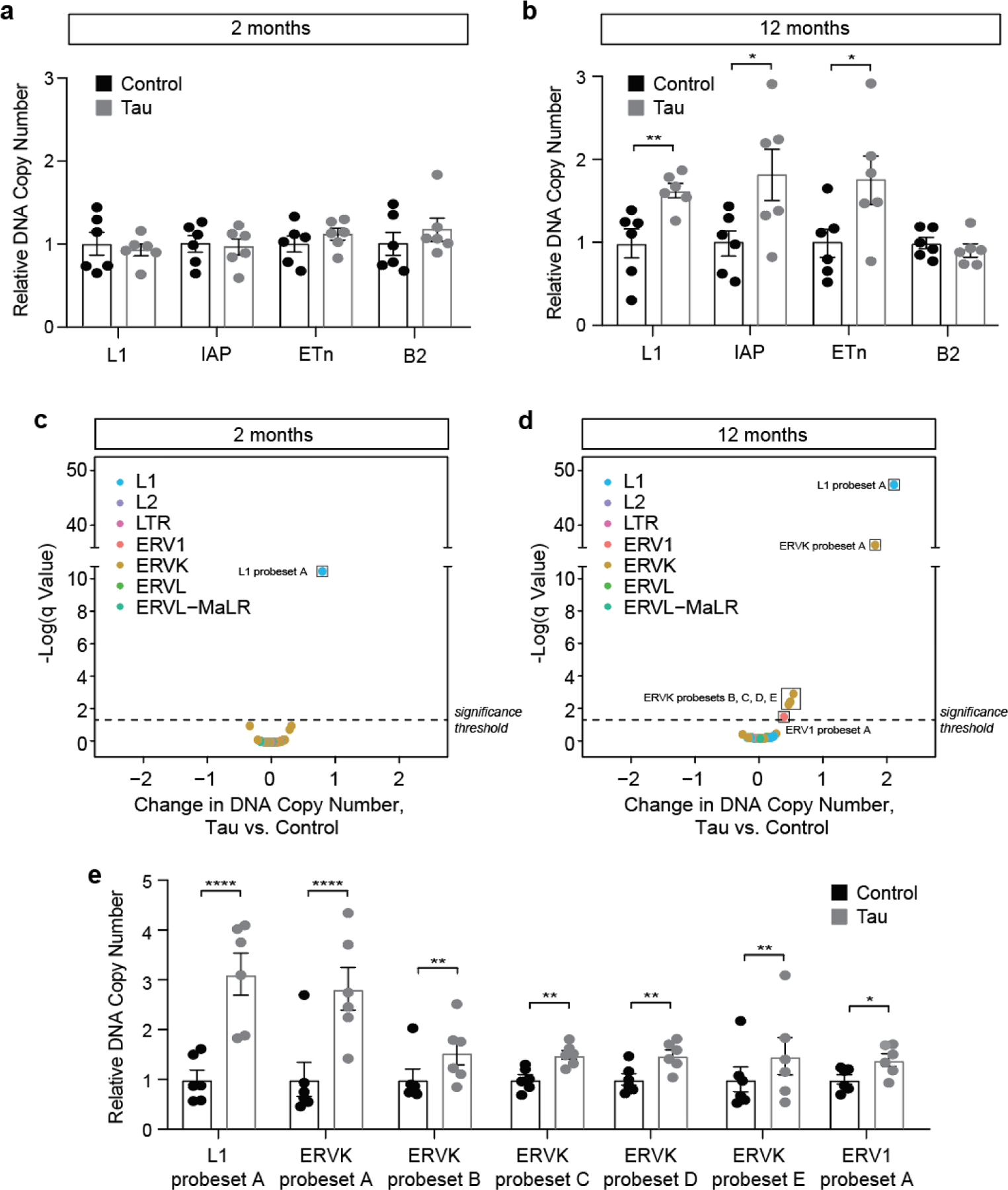
dPCR-based quantification of retrotransposon DNA copy number at two (a) and twelve (b) months of age in cortex of rTg4510 vs. control mouse brain. DNA copy number is normalized to the average of controls for each element. NanoString-based quantification of retrotransposon DNA copy number at two (c) and twelve (d) months of age in cortex of rTg4510 vs. control mouse brain. DNA copy number is normalized to the average of controls for each element. Elements that are significantly different between rTg4510 and control are boxed. False-discovery rate, q=1%. e. Individual data points for elements that are significantly increased in rTg4510 mouse brain at twelve months of age compared to control. n=6 females per genotype, per age. Error bars=SEM. Multiple t-tests, *q<0.05, **q<0.01, ****q<0.000001.
Having found that three of four candidate retrotransposons tested were elevated at the DNA level in brains of tau transgenic mice, we extended our analysis to a larger set of retrotransposons that includes both autonomous and non-autonomous elements. We worked with NanoString to create a custom codeset that detects DNA copy number of L1, L2, ERV1, ERVK, ERVL, ERVL-MaLR and solo LTR subfamily members. As members within a transposable element subfamily share a high degree of sequence similarity, many of the NanoString probes recognize multiple subfamily members (both active and inactive) based on BLAST analysis (Supplemental Table 2). DNA copy number was normalized to ten different invariant regions of the mouse genome (Supplemental Table 2).
At two months of age, rTg4510 have a significant increase in L1 probeset A compared to controls based on NanoString (Fig. 5c, Supplemental Table 2). At twelve months of age, we detect a further three-fold elevation of elements recognized by L1 probeset A, as well as significant elevation of ERVK probesets A-E and ERV1 probeset A in brains of rTg4510 mice compared to controls (Fig. 5d, e, Supplemental Table 2). Retrotransposon DNA copy number does not significantly differ between two and twelve months in control mice (Supplemental Table 2). Taken together, these data indicate that a subset of retrotransposon RNAs are actively reverse transcribed into DNA in brains of tau transgenic mice, and that tau-induced increase in retrotransposon DNA copy number is age-dependent.
DISCUSSION
Multiple previous reports suggest that retrotransposons are activated in the brain as a consequence of physiological aging in Drosophila13,14, and that pathogenic forms of tau also activate retrotransposons in Drosophila models of tauopathy and in postmortem human brain tissue from patients with tauopathies, including Alzheimer’s disease15,16. In the current study, we investigate the extent of retrotransposon activation from young to old age in the mouse brain and over the course of disease in three different mouse models of tauopathy.
We find that the directionality of differentially expressed retrotransposons is predominantly increased in the context of both aging and tauopathy in the mouse brain. This is in slight contrast to previous studies that identify retrotransposons that are both increased and decreased with aging and tauopathy in the Drosophila brain, as well as in postmortem human brains of patients with late-stage Alzheimer’s disease or progressive supranuclear palsy15,16. Similar to human Alzheimer’s disease and tau transgenic Drosophila, we find an overrepresentation of ERVs among the classes of transposable elements that are elevated at the transcript level in the context of aging and tauopathy in the mouse brain. Increased transcript levels of ERVs are also present in human disorders including but not limited to amyotrophic lateral sclerosis24,49, multiple sclerosis50, and various types of cancers51. Given that transposable element fragments are present within introns of many “normal” protein-coding genes, a caveat of transcriptomic analyses is that transposable elements embedded with introns of pre-mRNAs could be mistakenly attributed to independent transposable element transcripts. As we detect a rather high degree of similarity between transposable element families elevated at the transcript level in RNA-seq data from polyA-enriched and non-polyA-enriched datasets, we do not think that the bulk of the differentially expressed elements derive from introns of “normal” genes. Nevertheless, this limitation could be resolved in future studies by long-read sequencing of RNA extracted from various tauopathy conditions.
In addition to identifying retrotransposons that are elevated at the transcript level in the context of tauopathy, we find that the gag capsid protein encoded by IAP, a highly active mouse Class II ERV, is elevated in the brain of rTg4510 tau transgenic mice. While it is possible that differential IAP processing precedes differential expression of IAP, we speculate that our ability to detect elevated levels of IAP-encoded protein prior to our ability to detect elevated IAP transcript levels in brains of rTg4510 mice is a consequence of sequencing depth and cellular heterogeneity of the samples.
Having established that ERVs are elevated at the RNA and protein levels in tau transgenic mice, we next asked if brains of rTg4510 tau transgenic mice have increased retrotransposon DNA content using two complementary approaches. Both approaches identify LINE and ERV subfamily members that are elevated at the DNA level in brains of tau transgenic mice at 12 months of age. While use of both dPCR and NanoString to analyze retrotransposon DNA content contributes to the robustness of our study, each assay has its own inherent limitations. Given the high copy number of transposable elements in the mouse brain that would otherwise saturate dPCR and NanoString assays, the input DNA used was lower than what would be used to analyze a single copy gene. Low DNA input can negatively affect the precision of dPCR and can contribute to variation for NanoString. In addition, NanoString probesets recognize multiple members (active and inactive) of transposon subfamilies due to the high degree of sequence similarity between members of a subfamily. Differences in the absolute copy number of retrotransposons in dPCR versus NanoString-based CNV analyses likely result from the ability of dPCR probes to recognize specific active targets versus the redundant detection of subfamily members by NanoString. We also note that neither assay discriminates between genomic vs. episomal DNA. We thus do not currently know whether the extra retrotransposon DNA copies are integrated into the genome and/or exist in an episomal state. Determining the proportion of extra retrotransposon copies that are genomic versus episomal is an important next step, as genomic insertions generate novel mutations, while episomal DNA could drive a viral response as described in the context of aging, senescence and activation of LINE-1 elements in somatic tissues52.
Cells have multiple mechanisms to safeguard the genome against transposable element activation. Transposable element transcription is limited by enrichment of heterochromatin around retrotransposon DNA, while retrotransposon transcripts are degraded by small RNAs termed piwi-interacting RNAs and endogenous small interfering RNAs53. We have previously reported that pathogenic forms of tau disrupt heterochromatin- and piRNA-mediated silencing of retrotransposons in the Drosophila brain15. While our current study does not address mechanistic links between pathogenic forms of tau and retrotransposon activation in the mouse brain, depletion of heterochromatin protein 1 in motor neurons of the spinal cord in the JNPL3 mouse model of tauopathy54 is consistent with our overall hypothesis that pathogenic tau-induced heterochromatin decondensation drives retrotransposon activation. In addition, studies in tau knockout mice suggest that maintaining the integrity of pericentromeric heterochromatin is a physiological function of tau55.
Much emphasis in human transposon biology is placed on young, active elements that retain full mobilization potential in the human genome, as they would produce novel insertions if they were to retrotranspose. L1 activation is also reported to induce DNA double-strand breaks56 and somatic deletions57. In addition to consequences of complete and/or failed retrotransposition, the RNAs, dsRNAs, protein products, and episomal DNA generated from retrotransposons can also affect cellular function. For example, bidirectional transcription of retrotransposons and subsequent dsRNA formation can induce an interferon response through the RNA-sensing innate immune network58–60, and ERV-encoded proteins can drive autoimmunity60 and motor neuron disease61. We are currently investigating links between transposable activation and induction of the innate immune response in laboratory models of tauopathy and are analyzing de novo transposable element insertions in human tauopathy using long-read sequencing.
CONCLUSIONS
It has been 17 years since the last drug was approved for the symptomatic treatment of Alzheimer’s disease. Treatment pipelines for primary tauopathies are in their infancy, with only a few having disease-modifying potential. Pathogenic forms of tau and downstream cellular mediators of neuronal death represent potential therapeutic targets that have been largely unexploited. Due to the similarities between retroviruses and retrotransposons, antiretroviral drugs that interfere with HIV-1 replication have recently gained momentum as therapeutic approaches in neurodegenerative disorders, with promising results from a Phase 2 clinical trial for amyotrophic lateral sclerosis (NCT02868580)62 and recent initiation of Phase 1 (NCT04500847) and Phase 2 trials for Alzheimer’s disease (NCT04552795). Taken together, our data confirms that aging- and tau-induced transposable element activation is conserved in the mouse central nervous system, and further strengthens the case for transposable element activity as a therapeutic target for human tauopathies.
Supplementary Material
HIGHLIGHTS.
Retrotransposon transcripts increase in the aging brain and in three mouse models of tauopathy
Retrotransposon-encoded protein is elevated in brains of tau transgenic mice
DNA copy number of mobile retrotransposons is elevated in brains of tau transgenic mice
Acknowledgements
The IAP-gag antibody used for immunofluorescence was a kind gift of Dr. Bryan Cullen.
Funding
This study was supported by the Rainwater Foundation (BF, SD, BH), the National Institute of Neurological Disorders and Stroke RF1 NS112391 (BF), the National Institute on Aging RF1 AG057587 (WC), T32 AG00022 (SLD), the National Institute of General Medical Sciences R25 GM095480 (PR) and T32 GM113896 (GZ). Acknowledgement is made to the donors of Alzheimer’s Disease Research, a program of BrightFocus Foundation, for support of this research (WS, SLD). WJR and VBP are supported by the Neurodegeneration Consortium, the Belfer Family foundation and the Oskar Fisher Project.
ABBREVIATIONS
- ERV
endogenous retrovirus
- LINE
Long Interspersed Nuclear Element
- SINE
Short Interspersed Nuclear Elements
- LTR
Long-Terminal Repeat
- DNA
Deoxyribonucleic acid
- piRNA
Piwi-interacting RNA
- RNA-seq
Ribonucleic acid sequencing
- IAP
Intracisternal A-particle
- APP
Amyloid Precursor Protein
- PS1
Presenilin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
Consent for publication
Yes
ETHICS DECLARATIONS
Ethics approval and consent to participate
All experimental procedures were performed according to the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center or the Baylor College of Medicine Institutional Care and Use Committee.
Availability of data and material
Alignment files (mm10 genome transposon annotation) for RNA-seq based transposon counts for each sample will be deposited in the Gene Expression Omnibus upon acceptance.
REFERENCES
- 1.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Platt RN, Vandewege MW & Ray DA Mammalian transposable elements and their impacts on genome evolution. Chromosome Research (2018) doi: 10.1007/s10577-017-9570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin SL et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 Protein. J. Mol. Biol (2005) doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Feng Q, Moran JV, Kazazian HH & Boeke JD Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell (1996) doi: 10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 5.Hughes JF & Coffin JM A novel endogenous retrovirus-related element in the human genome resembles a DNA transposon: Evidence for an evolutionary link? Genomics (2002) doi: 10.1016/S0888-7543(02)96856-4. [DOI] [PubMed] [Google Scholar]
- 6.Nelson PN et al. Demystified … Human endogenous retroviruses. Journal of Clinical Pathology - Molecular Pathology (2003) doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns KH Our Conflict with Transposable Elements and Its Implications for Human Disease. Annual Review of Pathology: Mechanisms of Disease (2020) doi: 10.1146/annurev-pathmechdis-012419-032633. [DOI] [PubMed] [Google Scholar]
- 8.Hancks DC & Kazazian HH Roles for retrotransposon insertions in human disease. Mobile DNA (2016) doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills RE, Bennett EA, Iskow RC & Devine SE Which transposable elements are active in the human genome? Trends in Genetics (2007) doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Beck CR, Garcia-Perez JL, Badge RM & Moran JV LINE-1 elements in structural variation and disease. Annual Review of Genomics and Human Genetics (2011) doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray V Are transposons a cause of ageing? Mutat. Res. DNAging (1990) doi: 10.1016/0921-8734(90)90011-F. [DOI] [PubMed] [Google Scholar]
- 12.De Cecco M et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany. NY) (2013) doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci (2013) doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood JG et al. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci 113, 11277–11282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun W, Samimi H, Gamez M, Zare H & Frost B Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci 21, 1038–1048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo C et al. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep 5, 2874–2880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arendt T, Stieler JT & Holzer M Tau and tauopathies. Brain Res. Bull 126, 238–292 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews S, Krueger F, Seconds-Pichon A, Biggins F & Wingett S FastQC. A quality control tool for high throughput sequence data. Babraham Bioinformatics. Babraham Institute (2015). [Google Scholar]
- 20.Dobin A et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics (2013) doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y, Tam OH, Paniagua E & Hammell M TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics (2015) doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantano L et al. DEGreport: Report of DEG analysis (2021).
- 24.Tam OH et al. Postmortem Cortex Samples Identify Distinct Molecular Subtypes of ALS: Retrotransposon Activation, Oxidative Stress, and Activated Glia. SSRN Electron. J (2019) doi: 10.2139/ssrn.3364349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ginestet C ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A (Statistics Soc (2011) doi: 10.1111/j.1467-985x.2010.00676_9.x. [DOI] [Google Scholar]
- 26.Kolde R Package `pheatmap’. Bioconductor (2012). [Google Scholar]
- 27.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK & Cullen BR APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res (2006) doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeVos SL et al. Tau reduction in the presence of amyloid-β prevents tau pathology and neuronal death in vivo. Brain (2018) doi: 10.1093/brain/awy117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K & Standley DM MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Krieger AM & Yekutieli D Adaptive linear step-up procedures that control the false discovery rate. Biometrika (2006) doi: 10.1093/biomet/93.3.491. [DOI] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol (1990) doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Bao W, Kojima KK & Kohany O Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA (2015) doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham H ggplot2. ggplot2 (2009). doi: 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
- 34.Santacruz K et al. Medicine: Tau suppression in a neurodegenerative mouse model improves memory function. Science (80-. ) (2005) doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis J et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L)tau protein. Nat. Genet (2000) doi: 10.1038/78078. [DOI] [PubMed] [Google Scholar]
- 36.Yoshiyama Y et al. Synapse Loss and Microglial Activation Precede Tangles in a P301S Tauopathy Mouse Model. Neuron (2007) doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Spires TL et al. Region-specific dissociation of neuronal loss and neurofibrillary pathology in a mouse model of tauopathy. Am. J. Pathol (2006) doi: 10.2353/ajpath.2006.050840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopeikina KJ et al. Synaptic alterations in the rTg4510 mouse model of tauopathy. J. Comp. Neurol (2013) doi: 10.1002/cne.23234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue M, Hanna A, Wilson J, Roder H & Janus C Sex difference in pathology and memory decline in rTg4510 mouse model of tauopathy. Neurobiol. Aging (2011) doi: 10.1016/j.neurobiolaging.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Iba M et al. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of alzheimer’s-like tauopathy. J. Neurosci (2013) doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B et al. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and alzheimer-like pathology in an interventional study with aged tau transgenic mice. J. Neurosci (2012) doi: 10.1523/JNEUROSCI.4922-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minkeviciene R et al. Age-related decrease in stimulated glutamate release and vesicular glutamate transporters in APP/PS1 transgenic and wild-type mice. J. Neurochem 105, 584–594 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Jankowsky JL et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: Evidence for augmentation of a 42-specific γ secretase. Human Molecular Genetics (2004) doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 44.Gagnier L, Belancio VP & Mager DL Mouse germ line mutations due to retrotransposon insertions. Mobile DNA (2019) doi: 10.1186/s13100-019-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuss FU & Schaller HC cDNA sequence and genomic characterization of intracisternal A-particle-related retroviral elements containing an envelope gene. J. Virol 65, 5702–5709 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribet D et al. An infectious progenitor for the murine IAP retrotransposon: Emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res (2008) doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuff EL & Lueders KK The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res 51, 183–276 (1988). [DOI] [PubMed] [Google Scholar]
- 48.Welker R, Janetzko A & Krausslich HG Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J. Virol (1997) doi: 10.1128/jvi.71.7.5209-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Douville R, Liu J, Rothstein J & Nath A Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol 69, 141–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt K et al. Comprehensive Analysis of Human Endogenous Retrovirus Group HERV-W Locus Transcription in Multiple Sclerosis Brain Lesions by High-Throughput Amplicon Sequencing. J. Virol (2013) doi: 10.1128/jvi.02388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kassiotis G Endogenous Retroviruses and the Development of Cancer. J. Immunol (2014) doi: 10.4049/jimmunol.1302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Cecco M et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodier JL Restricting retrotransposons: A review. Mobile DNA (2016) doi: 10.1186/s13100-016-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost B, Hemberg M, Lewis J & Feany MB Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci 17, 357–366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansuroglu Z et al. Loss of Tau protein affects the structure, transcription and repair of neuronal pericentromeric heterochromatin. Sci. Rep (2016) doi: 10.1038/srep33047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasior SL, Wakeman TP, Xu B & Deininger PL The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol 357, 1383–1393 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erwin JA et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci 19, 1583–1591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuellar TL et al. ---Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia--. J. Cell Biol 11, 3535–3549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roulois D et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volkman HE & Stetson DB The enemy within: endogenous retroelements and autoimmune disease. Nat. Immunol 15, 415–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med 7, 307ra153–307ra153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gold J et al. Safety and tolerability of Triumeq in amyotrophic lateral sclerosis: the Lighthouse trial. Amyotroph. Lateral Scler. Front. Degener (2019) doi: 10.1080/21678421.2019.1632899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


