Significance
Anatomical barrier sites are subject to frequent viral infection. Antiviral immunity at these surfaces is largely regulated by the production of interferons (IFNs) that function as critical antiviral cytokines by restricting viral infection, replication, and release. Here, we show that oral epithelial cells respond to viral agonists by preferentially inducing Type III IFNs (IFN-λ). Using human gingival tissues, mouse models, and in vitro approaches, we show that IFN production and antiviral immunity are severely compromised in the presence of the oral bacterial pathogen, Porphyromonas gingivalis (Pg). Our observations show striking parallels in the molecular pathways employed by Pg to those utilized by viruses for IFN suppression.
Keywords: Interferon lambda, oral epithelial cells, Porphyromonas gingivalis, periodontitis, viral infection
Abstract
Here, we show that Porphyromonas gingivalis (Pg), an endogenous oral pathogen, dampens all aspects of interferon (IFN) signaling in a manner that is strikingly similar to IFN suppression employed by multiple viral pathogens. Pg suppressed IFN production by down-regulating several IFN regulatory factors (IRFs 1, 3, 7, and 9), proteolytically degrading STAT1 and suppressing the nuclear translocation of the ISGF3 complex, resulting in profound and systemic repression of multiple interferon-stimulated genes. Pg-induced IFN paralysis was not limited to murine models but was also observed in the oral tissues of human periodontal disease patients, where overabundance of Pg correlated with suppressed IFN generation. Mechanistically, multiple virulence factors and secreted proteases produced by Pg transcriptionally suppressed IFN promoters and also cleaved IFN receptors, making cells refractory to exogenous IFN and inducing a state of broad IFN paralysis. Thus, our data show a bacterial pathogen with equivalence to viruses in the down-regulation of host IFN signaling.
The oral mucosal barrier is heavily colonized by a complex microbiome composed of several bacterial and fungal species that interface with oral epithelial cells. A significant driver of tissue destruction at the oral epithelial surface is dysbiosis caused by specific communities of oral pathogenic bacteria that actively subvert immune pathways and suppress cytokine and chemokine generation. This immune disruption leads to pathogenic persistence, chronic inflammation, and enhanced susceptibility to secondary infections (1). Recent clinical studies show an increased prevalence of several viruses in inflamed oral tissues and sites of active periodontal inflammation (2, 3). Viruses can either directly infect oral epithelial cells that line the buccal and gingival surfaces of the oral mucosal barrier or have a transient presence in the oral cavity due to replication and release from other tissues (4). These include Herpes simplex virus (HSV-1), cytomegalovirus (CMV), varicella-zoster virus, as well as oncogenic viruses such as Epstein–Barr Virus (EBV), and human papillomaviruses (4–9). Despite the high prevalence of oral viral infections, little is known about the nature of antiviral immune responses and their regulation in the oral cavity.
Interferons (IFNs) are critical antiviral cytokines that are essential in all aspects of antiviral immunity. Type III interferons or IFN-Lambdas (IFN-λs) comprise a family of antiviral mediators that are preferentially expressed by epithelial cells at anatomic barrier sites and confer antiviral immunity without inducing the pathological inflammation typically associated with Type I IFNs (IFN-α/β). Activation of antiviral pathways by IFN-λ at the gastrointestinal (10), reproductive (11), and respiratory (12) epithelium is due to the preferential expression of the IFN-λ receptor (IFNL-R) on barrier epithelial cells and the downstream activation of interferon-stimulated genes (ISGs) that are largely similar to those induced by Type I IFNs. However, the superior role of IFN-λ at these sites is mediated by the delayed kinetics of activation and blunted recruitment and/or dampened activation of inflammatory cells (neutrophils), all of which contributes toward antiviral immunity while preventing collateral damage (13–17). Thus, Type III IFNs have emerged as the “guardians” of anatomic barriers; however, their role at the oral mucosal barrier, which often is an initial site of viral infection and functions as a portal of entry to other barrier sites, is undefined.
Recent studies show that the microbial colonizers at anatomic barrier sites can either stimulate or suppress IFN responses and ISG expression and thereby influence host susceptibility to viral infection (18–24). Detection of bacterial ligands by epithelial Toll-like receptors (TLRs) can stimulate IFN-λ expression in a manner that reinforces epithelial barrier integrity (25). Certain bacterial commensal species such as segmented filamentous bacteria play a more direct role in protecting against rotavirus infection and maintaining barrier integrity by inducing epithelial turnover and epithelial ISG expression (24). While antibiotic depletion of microbiota enhanced susceptibility to flavivirus infections (26), it protected the mice from murine norovirus infections (27). This protection was conferred by antibiotic-mediated deletion of microbial colonizers that limited the efficacy of IFN-λ–mediated antiviral pathways. Thus, the nature of colonizing bacterial species can either activate or disarm epithelial IFN responses and impact host antiviral immunity.
In this manuscript, we show that detection of viral agonists by various cytosolic and membrane-restricted pattern recognition receptors (PRRs) preferentially induced IFN-λ by gingival epithelial cells (GECs), resulting in the activation of multiple ISGs. Data from human primary tissues, cell lines, and mouse models show that IFN-λ production and downstream antiviral pathways were severely compromised in the presence of Porphyromonas gingivalis (Pg), a bacterial periodontal pathogen and resident of the subgingival microbial niche. Synergetic inactivation of multiple transcription factors, cleavage of IFN receptors, and global down-regulation of canonical IFN response genes by Pg severely compromised host IFN-mediated antiviral immunity. Our observations were specific to Pg, as other oral periodontal pathogens that are commonly associated with periodontal inflammation did not affect IFN production.
Results
Oral Mucosal Epithelial Cells Preferentially Induce IFN-λ in Response to Viral Agonists.
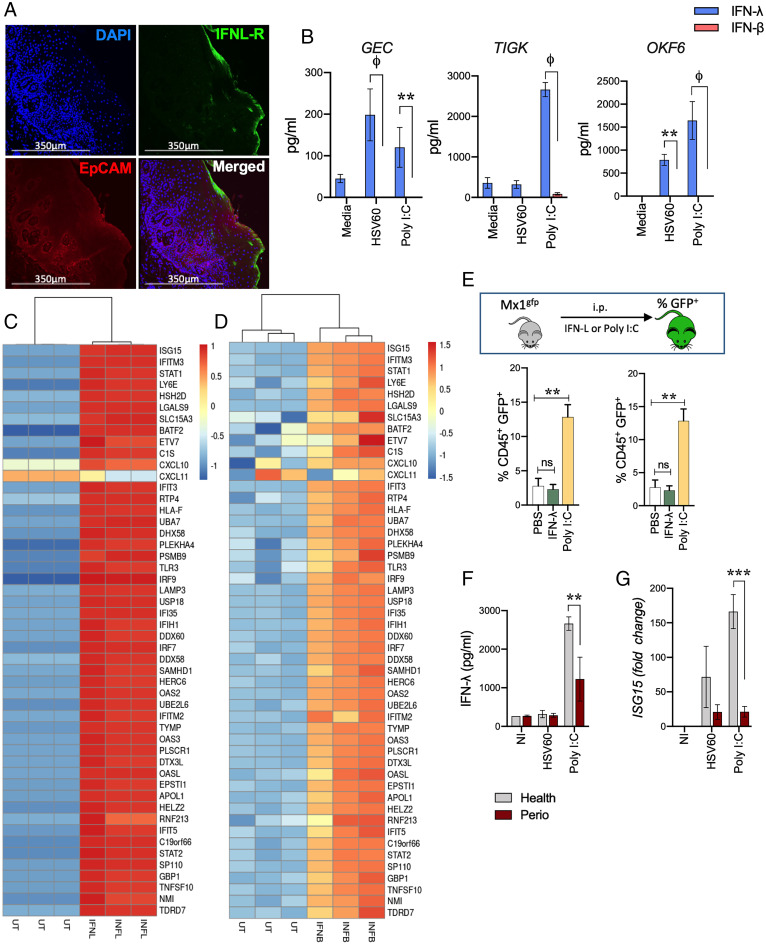
We determined levels of IFN-λ transcript as well as IFN-λ receptor (IFNL-R) expression in healthy human gingival tissues. Similar to the respiratory and intestinal epithelium (28), our data show robust expression of IFN-λ transcripts (SI Appendix, Fig. S1A) as well as IFNL-R (Fig. 1A) toward the apical side of the tissue, largely localized within the epithelial cell adhesion molecule (EpCAM)-positive region. GECs, unlike gingival fibroblasts, specifically and strongly express EpCAM (29). To determine responsiveness of viral pathogen-associated molecular patterns (PAMPs), GECs were isolated from human gingival tissues and challenged in vitro with poly I:C, a synthetic double-stranded RNA analog and TLR3 agonist, as well a 60 base pair oligonucleotide sequence derived from HSV-1 (HSV60) recognized by cytosolic DNA sensors. IFN-λ was preferentially induced in human GECs with very little IFN-β (Fig. 1B). Significantly higher levels of IFN-λ were also observed with human oral keratinocytes (OKF-6 cells) that line buccal surfaces of the mouth and also in the telomerase immortalized human gingival keratinocyte (TIGKs) cell line (30) (Fig. 1B). Thus, our observations are not limited to a single epithelial cell type. We did not detect any IFN-α or IFN-γ. In order to overcome the low yield and passage number limitations of primary GECs, we used immortalized GECs (TIGKs) for the rest of our studies. TIGKs faithfully mimic the responses observed in primary GECs cells (30), express IFNL-R (SI Appendix, Fig. S1B), and are responsive to viral agonists (Fig. 1B). RNA sequencing (RNA-seq) profiles of TIGKs stimulated with recombinant IFN-λ or IFN-β showed overlapping transcriptional signatures consistent with the induction of multiple ISGs that inhibit early viral infection, replication, and release (Fig. 1 C and D). Common and uniquely expressed transcripts from either treatment are shown in SI Appendix, Fig. S1 C–F.
Fig. 1.
IFN-λ is strongly expressed in gingival tissues, cell lines, and activates ISG expression in oral tissues. (A) IFNL-R expression was determined in fixed human gingival tissues from healthy donors by immunofluorescent staining. (B) Oral epithelial cells were stimulated with 5 μg/mL HSV60 or 50 μg/mL poly I:C for 24 h and IFN-λ and IFN-β levels measured in cell-free supernatants by ELISA. Data are shown as mean ± SD and statistical differences determined by two-way ANOVA with Holm–Sidak multiple comparison test (**P < 0.01; ϕP < 0.001). (C) TIGKs were treated with 20 ng/mL IFN-λ1 or (D) 0.25 ng/mL IFN-β for 24 h and analyzed by RNA-seq. Hierarchical clustering heatmap (based on log [RPKM] values) of the top 50 ISGs induced by IFN-λ1 or IFN-β compared to untreated cells are shown. Color intensity denotes level of gene expression. (E) Mx1gfp mice were intraperitoneally (i.p.) injected with 50 μg poly I:C or 4 μg IFN-λ and euthanized after 36 h. GECs (EpCAM+, CD45−) and leukocytes (EpCAM−, CD45+) were analyzed by flow cytometry and % GFP-positive cells (mean ± SD) determined by flow cytometry for each population from three mice per group. Statistical differences determined by one-way ANOVA with Holm–Sidak multiple comparison test (*P < 0.05; **P < 0.01). (F) IFN-λ responses to 5 μg/mL HSV60 or 50 μg/mL poly I:C were measured by ELISA in supernatants of GECs isolated from healthy donors or periodontitis patients. (G) ISG15 expression was determined by qRT-PCR, normalized to GADPH (2-ΔΔCT), and is shown as mean ± SD. Statistical differences were determined by two-way ANOVA with Holm–Sidak multiple comparison test (ϕP < 0.001).
We determined whether viral agonists or IFN-λ activated ISG expression in the oral epithelium in vivo. Most viruses with tropism to oral epithelium are human specific and poorly or transiently infect mice. To overcome this, we used the Mx1gfp knock-in mouse model, where inducible green fluorescent protein (GFP) expression occurs specifically in response to IFN-mediated activation of the interferon-stimulated response element (ISRE) that is upstream of ISGs, such as Mx1 (31). First, we confirmed that IFNL-R is expressed in mouse gingival tissues (SI Appendix, Fig. S1G). Injection of recombinant IFN-λ strongly induced GFP expression in oral murine GECs but not in the infiltrating leukocytes in gingival tissues (Fig. 1E and SI Appendix, Fig. S1H), consistent with the preferential expression of IFNL-R in epithelial cells. In contrast, the Type I IFN receptor (IFNAR) is expressed in all nucleated cells and drives systemic responses to Type I IFN. In accordance with this, mice challenged with poly I:C, an agonist known to induce both Type I (α/β) and Type III (λ) IFNs, showed robust GFP expression in GECs as well as leukocytes (Fig. 1E). Thus, our data show that similar to respiratory and gastrointestinal barriers, IFN-λ is induced at the oral epithelial barrier and activates ISGs in barrier epithelial cells.
Periodontal Disease and Associated Dysbiosis Dampens Inducible IFN Responses and Antiviral Immunity in Oral Epithelial Tissues.
A significant predisposing factor for viral infections, or reactivation of latent viruses, in the oral cavity is chronic inflammation associated with periodontal disease (32–34). Interestingly, GECs isolated from periodontitis patients (n = 3) had significantly lower IFN-λ responses concomitant with lowered ISG expression to viral agonists compared to GECs isolated from healthy individuals (n = 4) (Fig. 1 F and G). These observations were consistent across multiple periodontitis patients and could not be attributed to differences in apoptosis, growth rates, or differences in passage numbers between donors.
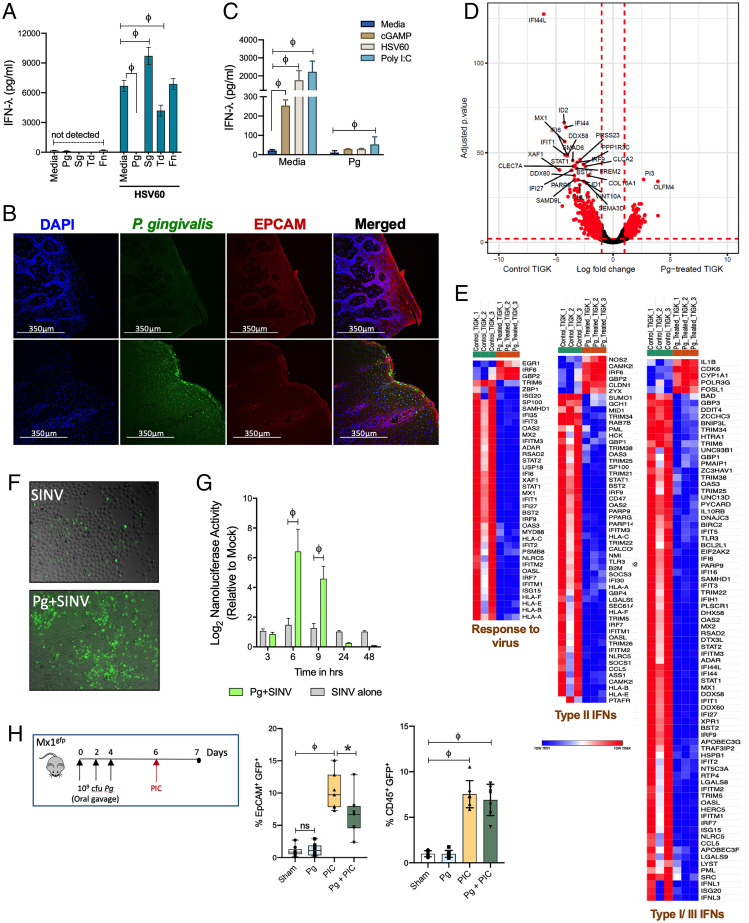
We determined whether microbial dysbiosis, which is associated with periodontal disease in humans and actively contributes to chronic inflammation, can impact IFN-λ responses. Specifically, we challenged TIGKs with HSV60 in the presence of various oral commensal and pathogenic bacterial species. Periodontal pathogens such as Fusobacterium nucleatum had no effect on HSV60-induced IFN-λ production, while Treponema denticola moderately reduced IFN-λ production but did not completely abolish it. In contrast, stimulation with the Gram-positive commensal Streptococcus gordonii (Sg) enhanced IFN-λ production (Fig. 2A). Unlike other oral colonizers, Pg completely abolished IFN-λ in response to HSV60 (Fig. 2A). Pg resides in subgingival biofilms and is highly associated with chronic periodontitis in both humans and murine models of periodontal disease (35). Interestingly, published clinical data show an increased presence of HSV-1, CMV, and EBV in patients with active periodontal lesions, which correlated with the presence of Pg in the same lesion (5, 6). As previously reported (36, 37), we found that Pg was present in gingival tissues of healthy individuals (n = 10) and periodontitis patients (n = 8). However, immunofluorescence staining showed Pg was significantly more abundant in periodontitis tissue samples (Fig. 2B and SI Appendix, Fig. S2A). We confirmed these findings using an enzyme-linked immunofluorescent assay (ELISA)–based quantitative approach to enumerate Pg in tissue homogenates and found a significant increase in total colony forming units (cfu) per milligram of tissue in patients with periodontitis (SI Appendix, Fig. S2B).
Fig. 2.
P. gingivalis infection causes IFN paralysis, characterized by the loss of basal and inducible IFN responses and ISG expression. (A) IFN-λ responses were measured by ELISA in TIGKs challenged either with P. gingivalis (Pg), T. denticola (Td), or F. nucleatum (Fn) at MOI 100 or S. gordonii (Sg) MOI 10 for 5 h, washed once with PBS, and then stimulated with 5 μg/mL HSV60 for additional 18 h. (B) P. gingivalis colonization was determined in gingival tissues by immunofluorescence staining. (C) TIGKs were either left untreated or infected with P. gingivalis (Pg) as described above with subsequent stimulation with 50 μg/mL poly I:C (TLR3 agonist), 5 μg/mL ORN06 (TLR7 agonist), 5 μg/mL HSV60, or 25 μg/mL 2’3′-cGAMP (STING agonist) for 18 h. IFN-λ levels in cell-free supernatants are shown as mean ± SD. Statistical differences were determined by two-way ANOVA (**P < 0.01; ϕP < 0.001). (D) Volcano plot of differentially expressed transcripts between GECs infected with P. gingivalis (MOI 100) and uninfected control cells. X-axis shows log-fold change between the two conditions, with positive values showing up-regulation and negative values showing down-regulated genes. Y-axis denotes P values for corresponding genes. Significantly different genes are shown highlighted in red (P < 0.001 as determined using the DESeq package in R). (E) Gene enrichment analysis of top 300 down-regulated genes was performed as described in Materials and Methods, and hierarchical clustering heatmaps (based on RPKM values) for response to virus pathway, Type II IFNs, and Type I/III IFNs pathways are shown. Color intensity denotes level of gene expression. (F) Viral growth/dissemination was measured in mock and Pg-treated GECs infected with SINV nsP3-GFP strain at an MOI of 10 PFU per cell and cultured under normal conditions for a period of 24 h prior to GFP and brightfield microscopic imaging. Data shown are representative of three independent biological replicates. (G) Viral gene expression was assessed in mock and P. gingivalis–treated GECs infected with SINV nsP3-Nanoluc at an MOI of 10 PFU per cell and cultured in the presence of ammonium chloride to limit infection to the initial single-round entry event. At the indicated time points, the cells were harvested and the level of Nanoluc activity was assayed. Statistical differences were determined by two-way ANOVA (**P < 0.01; ϕP < 0.001). (H) Mx1gfp mice were colonized by P. gingivalis before intraperitoneal (i.p.) challenge with poly I: C. GECs (EpCAM+, CD45−) and leukocytes (EpCAM−, CD45+) were analyzed by flow cytometry, and %GFP-positive cells (mean ± SD) determined by flow cytometry. Each data point represents one mouse (n = 7 to 5 per group). Statistical differences determined by one-way ANOVA (*P < 0.05; **P < 0.01).
We found that Pg infection inhibits IFN-λ induced by multiple viral PAMPs and PRRs such as poly I:C (TLR3 agonist), the TLR-7 agonist ORN06 (single-stranded RNA analog), and also 2’3′-cGAMP (stimulator of interferon genes [STING] agonist) (Fig. 2C). The cyclic GMP–AMP synthase (cGAS) and STING pathway is central to recognition of cytosolic DNA from DNA viruses such as HSV-1 (38, 39). Pg infection also blocked IFN-β responses in oral keratinocytes in response to multiple viral agonists (SI Appendix, Fig. S2 C and D), indicating that our observations were not limited to TIGKs or a singular viral agonist. We also tested other Pg strains such as W83, another common laboratory strain of Pg, as well as the clinical isolate MP-504, and found these strains also strongly inhibited IFN responses (SI Appendix, Fig. S2E). In contrast to Pg and other periodontal pathogens, stimulation with Sg slightly augmented IFN production (Fig. 2A). As Sg often cocolonizes with Pg in mixed oral biofilms, we determined whether Sg can prevent or reprogram IFN suppression induced by Pg. Costimulation of TIGKs with the two bacteria suppressed IFN-λ production, indicating that Pg is able to inhibit the stimulatory effect of Sg (SI Appendix, Fig. S2F). Further insight into IFN genes/pathways modulated by Pg was gained through RNA-seq. Pg infection led to a massive down-regulation of several genes implicated in IFN responses related to all aspects of viral infection. These include families of antiviral restriction factors essential in blocking select steps in viral replication such as ribonuclease L (RNAseL), IFIT (interferon-induced proteins with tetratricopeptide repeats) family members, ISG15 (IFN-stimulated protein of 15 kDa), protein kinase R (PKR), GTPase Mx1 (myxovirus resistance 1), and the tripartite motif (TRIM) family members (Fig. 2 D and E and SI Appendix, Table S1). IFIT proteins degrade viral RNAs during infection (40), while tetherin/BST2 protein prevents budding of virions from the plasma membrane and blocks the release of coronaviruses, herpesviruses, paramyxoviruses, and flaviviruses from infected cells. Also down-regulated was SAMHDI (SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase), which is critical in fighting retroviral infections. Significant down-regulation of transcription factors such as interferon regulatory factors (IRFs) and signal transducer and activator of transcription (STAT1, 2) was consistent with loss of IFN and ISG expression. Several genes essential in immune cell recruitment (CCL5, CXCL10) and antigen presentation (HLA) were also down-regulated. Overall, we found that the extent of IFN paralysis induced by Pg infection was broad and affected genes involved in immune responses against a wide range of RNA and DNA viruses. For example, growth/dissemination of a model positive-sense RNA virus, Sindbis virus (SINV), which is known to be highly controlled by the IFN response, was significantly elevated in Pg-infected cells (Fig. 2 F and G).
Consistent with our RNA-seq data sets, we found that Pg colonization of Mx1gfp mice resulted in a loss of IFN-inducible GFP expression in oral epithelial tissues (Fig. 2H). These data were striking for several reasons. First, we did not antibiotic treat these mice prior to colonization with Pg in order to avoid any selective advantage or disruption of the natural microbiome. Second, Pg not only successfully colonized the oral cavities of these mice (as confirmed by 16S rRNA nested PCRs using Pg-specific primers (41) but was able to specifically down-regulate IFN-induced ISG expression in vivo. These data confirm our in vitro observations and establish that the presence of Pg in the oral microbiome reduces the effectiveness of IFN responses and might mechanistically explain the clinical observations in periodontitis patients, where increased titers of EBV, HSV1, and CMV were found in deep periodontal pockets where Pg normally resides (4–6, 9).
IRF-1 Regulates Cell-Intrinsic Antiviral State by Maintaining Basal Expression of ISGs.
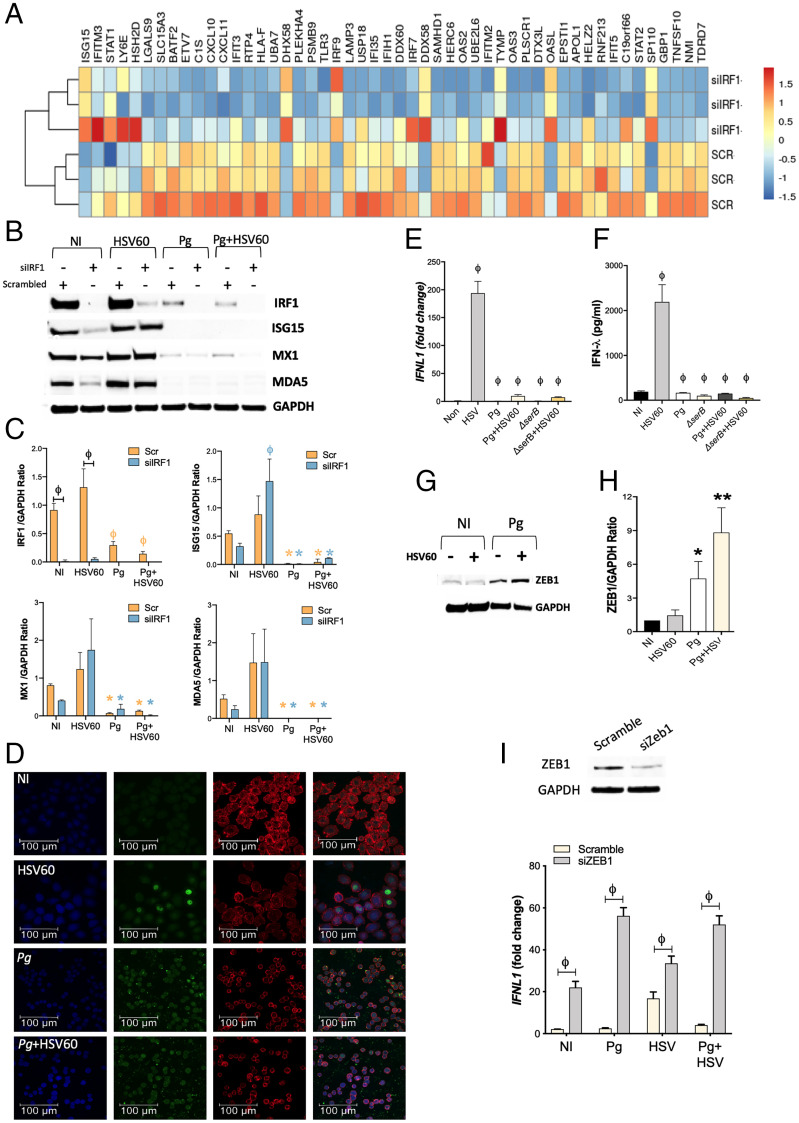
To determine how Pg blocks IFN responses in a cell-intrinsic manner, we looked at the activation of several transcription factors that either maintain the cell-intrinsic antiviral state and/or actively induce IFN expression in response to viral PAMPs. IRFs are a family of transcription factors that play critical roles in several aspects of host antiviral immunity. TLR and RIG-I-like-receptor (RLR) activation drives nuclear translocation of IRFs 1, 3, 5, and 7 and Type I IFN (42, 43) and Type III IFN production (25, 44). Recently, it was shown that unlike IRFs 3 and 7, IRF-1 expression was critical in the maintenance of constitutive or basal levels of multiple ISGs in epithelial cells independently (11, 45, 46). Thus, IRF-1 provides early protection against viral infection by maintaining a cell-intrinsic “antiviral state” (46). We previously showed that Pg transcriptionally down-regulates IRF-1 levels (47), thus we investigated whether loss of IRF-1 expression (using silencing RNA (siRNA)) was sufficient to down-regulate ISG transcripts in TIGKs. Silencing IRF-1 indeed down-regulated several IFN response genes and antiviral restriction factors (Fig. 3A and SI Appendix, Fig. S3 A and B). Select genes from the RNA-seq datasets were confirmed by qRT-PCR (SI Appendix, Fig. S3C) and immunoblotting (Fig. 3 B and C). Hence, Pg-mediated down-regulation of IRF-1 can be predicted to compromise the antiviral state in GECs by a significant reduction in basal ISG signatures.
Fig. 3.
P. gingivalis incapacitates transcription factors that positively regulate IFN-λ expression, and up-regulates ZEB1, a transcriptional repressor of IFN-λ. (A) TIGKs were transfected with siIRF1 or scrambled control siRNA and differentially expressed transcripts determined by RNA-seq. Hierarchical clustering heat map (based on log RPKM values) of the top 50 ISGs differentially expressed on IRF-1 silencing. Color intensity denotes level of expression. (B) TIGKs were transfected with siIRF1 (si) or scrambled control siRNA (scr) and infected with P. gingivalis (Pg) or stimulated with 5 μg/mL HSV60 and immunoblotted for IRF-1, ISG15, MX1, MDA5, and GAPDH. (C) Band intensities of immunoblots were determined, and ratios of IRF1, ISG15, MX1, and MDA5 normalized to GAPDH from three different blots are shown (mean ± SD). Statistical differences were determined by two-way ANOVA (ϕP < 0.001; *P < 0.05). Orange and blue symbols depict comparisons between NI (not infected) scr control and siIRF1, respectively. (D) TIGKs were infected with P. gingivalis and/or stimulated with 5 μg/mL HSV60, labeled with anti–IRF-1 antibodies (green), and analyzed by confocal microscopy. Actin was labeled with phalloidin (red) and nuclei stained with DAPI (blue). Merged images are shown on the Right. Magnification (63×) of 20 z-stacks of 0.3 μm. TIGKs were infected with P. gingivalis WT (Pg) or SerB-deficient isogenic mutant (ΔserB) for 5 h, followed by 5 μg/mL HSV60 for additional 18 h. (E) Transcript levels of IFNL1 determined by qRT-PCR, normalized to GADPH (2-ΔΔCT) and are shown as mean ± SD; (F) secreted IFN-λ in cell-free supernatants determined by ELISA and shown as mean ± SD. (G) ZEB1 and GAPDH were detected by immunoblotting. (H) Band intensities were determined, and ratios of ZEB1 to GAPDH from three different blots are shown (mean ± SD). (I) GECs were transfected with siZEB1 or scrambled control siRNA. After 48 h, media was replaced, and cells were challenged with P. gingivalis (MOI 100) for 5 h and then stimulated with 5 μg/mL HSV60 for 16 h. Transcript levels of IFNL1 were determined by qRT-PCR normalized to GADPH (2-ΔΔCT) and are shown as mean ± SD. Statistical differences were determined by two-way ANOVA (ϕP < 0.001).
During an infection, viral agonists induce IFN production, which via paracrine and autocrine signaling, reinforces antiviral defenses by driving ISG expression to levels several-fold higher than those observed in the basal state. Loss of IRF-1 did not negatively impact inducible IFN-λ expression (SI Appendix, Fig. S3D) in response to HSV60. Consistent with this, we found that stimulation with HSV60 led to activation of antiviral genes as measured by differential transcript expression in RNA-seq datasets (SI Appendix, Fig. S3E) and immunoblots (Fig. 3 B and C). ISG levels induced were comparable to those induced in TIGKs treated with scrambled control siRNA, indicating the presence of additional mechanisms of IFN-λ regulation in response to viral agonists (Fig. 3 B and C and SI Appendix, Fig. S3D). Thus, we show that while IRF-1 was essential for the maintenance of basal ISGs levels in GECs, inducible IFN production and consequent ISG expression was not affected by IRF-1 deficiency. Pg infection not only down-regulated IRF-1 levels, but it also blocked its nuclear translocation (Fig. 3D). Stimulation with HSV60 did not augment IRF-1 levels or ISG expression in HSV60-treated cells. Furthermore, in Pg-infected cells, we did not see any restoration of ISG expression even after overexpressing IRF-1 (SI Appendix, Fig. S4). Collectively, these results indicate that Pg has additional targets for antagonism of IFN-λ.
P. gingivalis Transcriptionally Represses IFNL1 by Up-Regulating ZEB1.
Multiple transcription factors, including other IRF family members (IRF-3 and IRF-7), Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB), and Activator Protein 1 (AP-1) can all cooperatively induce Type I and Type III IFN transcription independent of IRF-1 (42, 43, 48). All of the aforementioned transcription factors potentially could contribute to IFN-λ production in response to HSV60. To further investigate the mechanistic basis of Pg-mediated IFN-λ repression, we looked at other transcription factors that positively regulate IFN-λ expression. Consistent with our RNA-seq data in Fig. 2 D and E, Pg infection down-regulated IRFs 3, 7, and 9 that bind to PRD1 and ISRE elements on IFNL1 promoter to induce Type III IFN production (25, 44) (SI Appendix, Fig. S5A). Thus, we turned our attention on NF-κB, which binds to κB sites on the IFNL1 promoter (49) and also plays a role in reinforcing IFN pathways by augmenting the expression of various IRFs (50). We have previously shown that SerB, a serine phosphatase produced by Pg, dephosphorylates the serine 536 residue on the RelA subunit of NF-κB, preventing its nuclear translocation and blocking subsequent expression of NF-κB–regulated genes such as IL8/CXCL8 (51). We assessed the extent to which restoration of NF-κB activation would rescue IFN-λ responses to viral agonists. GECs were infected with either the wild-type(WT) or ΔserB strains of Pg and challenged with HSV60 DNA. IRF-1 expression was reduced on infection with either strain (SI Appendix, Fig. S5B). However, IFN-λ production remained repressed even when cells were stimulated with ΔserB (Fig. 3 E and F). These data establish that non–NF-κB-dependent mechanisms predominate in the suppression of IFN-λ production by Pg.
One such candidate is up-regulation of the zinc finger E-Box binding homeobox1 ZEB1, a transcription factor involved in epithelial–mesenchymal transition (52). ZEB1 transcriptionally represses IFNL1 by binding to E-box–like sites on the IFNL1 promoter (49) and also by epigenetically silencing IRF-1 (53). Unlike Pg, HSV60 stimulation did not significantly change ZEB1 expression (Fig. 3 G and H). Moreover, targeted knockdown of ZEB1 by siRNA significantly enhanced IFNL1 transcription in response to HSV60 and prevented Pg antagonism of IFNL1 expression (Fig. 3I). Thus, these data demonstrate that Pg represses IFN-λ both by disarming IRFs and by up-regulating the transcriptional repressor ZEB1.
P. gingivalis–Mediated Cleavage of IFN Receptors Enforces IFN Paralysis.
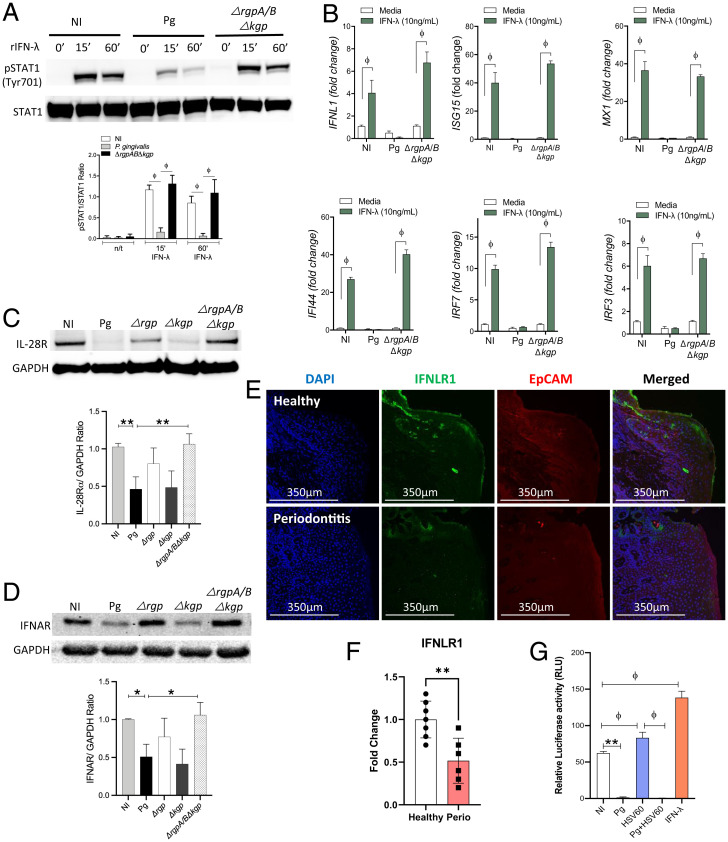
In vivo, Pg-induced IFN-λ paralysis could be circumvented by IFN-λ secreted from noninfected epithelial cells or from additional cell types. Hence, we determined whether exogenously added IFN-λ would restore ISG expression. The heterodimeric IFN-λ receptor (IFNL-R) is composed of the IL-28 receptor chain and the IL-10 receptor β-chain and on binding to IFN-λ family members, induces phosphorylation of signal transducer and activator of transcription (STAT) proteins STAT1 and STAT2. Phosphorylated STATs along with IRF-9 form the heterotrimeric transcription factor complex ISGF3 (ISG factor 3) that binds to the ISRE within the promoters of interferon-inducible genes (14, 54). GECs incubated with recombinant IFN-λ rapidly phosphorylated STAT1; however, Pg infection blocked STAT1 phosphorylation in the presence of exogenously added IFN-λ, suggestive of an additional component to the Pg antagonistic process (Fig. 4A).
Fig. 4.
P. gingivalis infection reduces responsiveness to exogenous IFN-λ. TIGKs were infected with P. gingivalis WT (Pg) or gingipain-null triple mutant (ΔrgpABΔkgp) for 1 h and then stimulated with 20 ng/mL IFN-λ for indicated timepoints. (A) Phospho-STAT1 (pSTAT1) and total STAT1 expression was determined by Western blotting. Band intensities were determined, and ratios of pSTAT1 to total STAT1 from three different blots are shown (mean ± SD). Statistical differences were determined by one-way ANOVA (*P < 0.05; **P < 0.01). (B) Changes in transcript levels (mean ± SD) of ISG15, MX1, IFI44, IRF7, IRF3, and IFNL1 were determined by qRT-PCR and normalized to GAPDH (2-ΔΔCT). Data are shown as mean ± SD, and statistical differences were determined by two-way ANOVA (ϕP < 0.001). TIGKs were infected with P. gingivalis WT (Pg) or gingipain mutant strains Δkgp, ΔrgpA/B, and ΔrgpABΔkgp for 1 h, and (C) IFNL-R (IL-28R), (D) IFNAR, and GAPDH expression was determined in protein lysates by Western blotting. GAPDH was used as a loading control. Densitometry ratios for IL-28R and IFNAR from three different blots are shown as mean ± SD. Statistical differences were determined by one-way ANOVA (*P < 0.05; **P < 0.01). (E) IFNL-R (IL-28R) was detected in human gingival tissues from periodontitis patients and healthy controls. (F) Mean fluorescence intensity for IFNL-R within the EpCAM-positive region was calculated using Imaris software for seven control (healthy) donors and six periodontitis patients (perio). Statistical differences were determined by Student’s t test (**P < 0.01). (G) Dual luciferase reporter analysis of ISRE-Luc activities in GECs under various stimulation conditions. Statistical differences were determined by one-way ANOVA (ϕP < 0.05; **P < 0.01).
Pg produces several cysteine proteases, the arginine-specific gingipains A and B (RgpA and RgpB) as well as the lysine-specific gingipain (Kgp), that proteolytically degrade cellular proteins and attenuate signal transduction pathways (55–58). We examined the role of gingipains in preventing IFN-λ–induced activation of STAT1 by infecting GECs with a triple mutant strain (ΔrgpABΔkgp) of Pg lacking expression of all gingipains. Loss of gingipain activity prevented attenuation of STAT1 phosphorylation by Pg upon IFN-λ stimulation (Fig. 4A) and also restored expression of ISGs (Fig. 4B). Even in GECs overexpressing STAT1 protein, phosphorylation levels remained low in cells infected with Pg (SI Appendix, Fig. S6). We hypothesized that the loss of phosphorylation following Pg challenge was mediated by an upstream target sensitive to gingipain-mediated proteolytic cleavage. Thus, we tested the ability of Pg to cleave interferon receptors and thereby limit STAT1 phosphorylation. GECs were incubated with the parental strain of Pg or mutants lacking either Kgp, RgpA/B, or the triple mutant. Our data show that cleavage of IFNL-R (IL-28R) was mediated predominantly by the arginine-specific gingipains RgpA/B (Fig. 4C). Although Type I and III interferons bind to distinct receptors, they activate overlapping downstream signaling pathways leading to ISG expression (14). Thus, we also looked for cleavage of the Type I interferon receptor (IFNAR) by Western blotting. Pg arginine gingipains also cleaved IFNAR (Fig. 4D). Interestingly, IFNL-R was also cleaved in periodontitis patients (Fig. 4 E and F), with cleavage evident toward the apical epithelial side that interfaces with colonizing bacteria such as Pg (Fig. 2B). At later timepoints, Pg also cleaved STAT1 protein (SI Appendix, Fig. S7), compromising the formation of ISGF3, the heterotrimeric transcription factor complex that binds to the ISRE sites found on promoters of several ISGs. Additionally, viral agonists (HSV60) as well as exogenously added IFN-λ failed to stimulate the ISRE promoter element in Pg-infected GECs (Fig. 4G), further enforcing IFN paralysis.
Discussion
Here, we report a previously unknown role for IFN-λ in modulating ISG expression and antiviral immunity at the oral mucosal epithelium of humans and mice. Modulation of viral infection by the indigenous microbiota is well-established (9, 19–24), and one recently documented mechanism involves microbial priming of antiviral interferon pathways (21, 24, 27). We find here a mechanism by which an indigenous bacterial colonizer of the oral cavity can suppress IFN-λ responses and increase susceptibility to viral infection. Interestingly, the concerted action of multiple Pg virulence factors blocked IFN pathways (summarized in SI Appendix, Fig. S8) using molecular strategies that are similar to those used by certain viruses. For example, Pg infection suppressed the activation of multiple IFN-driving transcription factors such as IRFs 1, 3, 7, and 9, NF-κB, and STAT-1, subsequently blocking ISGF3-driven ISG expression. Additionally, an increase in ZEB1, a negative regulator of IFNL1, synergistically blocked transcription of IFN-λ. The targeting of IFN-activating transcription factors by Pg is similar to the strategies by various viral pathogens to antagonize IFN transcription and IFN receptor–mediated activation of ISGs. For example, viral proteins can directly bind to IRF-3 and -7 in a manner that blocks IFN transcription either due to dephosphorylation of IRFs themselves or by preventing their interaction with other binding partners required for IFN transcription (59). NS5 from the yellow fever virus, upon binding to STAT2, prevents the formation of the ISGF3 complex and ISG induction (60). In addition to misdirection of signaling, viral proteases can also cleave and degrade host proteins by targeting them for proteasomal degradation (61). In our model, IFN paralysis was further reinforced by Pg via proteolytic degradation of INFL-R and IFNAR making GECs insensitive to exogenous IFNs produced by other cells such as plasmacytoid dendritic cells in tissues. The NS5 protein of West Nile and tick-borne encephalitis virus inhibits the expression of these receptors on the plasma membrane (62). Thus, there are a number of parallels between Pg-mediated antagonism of IFN responses and mechanisms used by viral pathogens.
Our findings are of broad relevance not only for oral health but also for host responses to a wide range of viral pathogens that infect various mucosal surfaces. Translocation of Pg to other mucosal surfaces such as the gut and surfaces of the digestive tract is associated with dysbiosis and other complications including gastric and esophageal cancers (63, 64); translocation to the respiratory epithelial tissues exacerbated aspiration-induced pneumonia (65–67) in patients and mouse models. In periodontitis patients, increased titers of EBV, HSV1, and CMV were observed in deep periodontal pockets (4–6, 9), the predominant niche of Pg. While the contribution of these viruses to periodontitis is unclear, Pg-mediated IFN paralysis might promote reactivation or replication of these and other viruses within the subgingival epithelium. Extension of this work in vivo to test specific viral pathogens will require technological advances, as genetically tractable mouse models that phenocopy the oral epithelial tropism of these viruses as seen in human cells have not been described. However, data from our human tissues and the Mx1gfp reporter mice strongly correlates with our observations in multiple oral epithelial cell lines.
IFN-λ has been investigated for the treatment of viral diseases given its focused role on epithelial cells. In patients with hepatitis C infection, IFN-λ treatment was efficacious in lowering viral titers, similar to IFN-α but with lower side effects and toxicity in comparison to IFN-α (68). Pegylated IFN-λ is also being investigated as a broad-spectrum antiviral to provide immediate or early protection to healthcare workers against the SARS-CoV2 outbreak (69). While it is premature to state that Pg infection might be a risk factor in exacerbating clinical symptoms of COVID-19, our data do demonstrate that it can certainly enhance susceptibility by compromising the “antiviral state.”
Materials and Methods
Bacteria.
P. gingivalis 33277, W83, and MP4-504 were cultured in trypticase soy broth (TSB) supplemented with hemin (5 μg/mL) and menadione (1 μg/mL) and 1 mg/mL yeast extract. Isogenic mutants were cultured in supplemented TSB with the appropriate antibiotics: ΔrgpA, Δkgp, and ΔserB had 10 μg/mL erythromycin; ΔrgpAB had 10 μg/mL erythromycin and 1 μg/mL tetracycline; ΔrgpABΔkgp had 10 μg/mL erythromycin, 1 μg/mL tetracycline, and 20 μg/mL chloramphenicol (70). F. nucleatum 25866 was cultured in brain heart infusion broth with yeast extract (1 mg/mL), hemin (5 μg/mL), and menadione (1 μg/mL). S. gordonii DL1 was cultured in TSB with yeast extract. T. denticola 35405 was cultured in new oral spirochete media. All strains were grown anaerobically (85% N2, 10% H2, and 5% CO2) at 37 °C.
Processing of Human Gingival Tissues.
Deidentified gingival tissue specimens were obtained from healthy individuals that needed gingival tissue excision for aesthetic or functional purposes and periodontal disease patients undergoing periodontal surgeries. All studies were approved by the University of Louisville’s Institutional Review Board (IRB No. 15.0163). Tissues were subdivided and fixed in formalin as well as cryopreserved in OCT for histology and immunofluorescence staining. A total of 10 to 50 mg tissue was also homogenized in phosphate-buffered saline (PBS) containing protease inhibitors using a Bead Beater and the lysing matrix D (MP Biomedicals). Tissue homogenates were used for ELISA.
Cell Culture.
Primary GECs were isolated from human gingival specimens as previously described (30). Briefly, tissues were treated with 5 mg/mL Dispase II overnight at 4 °C, followed by mechanical separation of the epithelial layer from the underlying connective tissue. Tissues were further dissociated by enzymatic digestion with Trypsin-EDTA at 37 °C for 10 min and then minced to obtain single-cell suspension. Cells were cultured in keratinocyte basal serum-free media with Gentamicin (30 mg/mL) and Amphotericin B (15 ng/mL) and used for assays between passages 2 and 6. GECs (TIGKs) (30) and human oral keratinocyte (OKF6) cell lines were cultured in keratinocyte basal serum-free media (Invitrogen) supplemented with 0.4 mM calcium chloride, 25 μg/mL bovine pituitary extract, and 0.2 ng/mL epidermal growth factor at 37 °C and 5% CO2.
Immunofluorescence and Confocal Laser Scanning Microscopy.
GECs were grown on glass coverslips in 24-well plates. Cells were fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 10 min at room temperature (RT). After blocking with 5% bovine serum albumin (20 min), cells were incubated with either anti-human IRF-1 (Cell Signaling Technologies) primary antibody 1:200 dilution (0.165 μg/mL) overnight at 4 °C and Alexa fluor 488–conjugated anti-rabbit secondary (Invitrogen) antibody at 1:1,000 dilution (2 μg/mL) at RT for 30 min or anti-human ZEB1 primary antibody (Novus Biologicals) 1:100 dilution (100 μg/mL) overnight at 4 °C and Alexa fluor 488–conjugated anti-mouse secondary antibodies at 1:2,000 dilution (1 μg/mL) at RT for 30 min. Cells were labeled with Texas Red-phalloidin for 40 min at RT. OCT embedded gingival tissues were sectioned at 5 μm in a cryotome and fixed with ice-cold 100% methanol. Slides were permeabilized with 0.4% Triton X-100 and blocked with 5% goat serum and stained with anti-human IFNL-R antibodies. The presence of Pg in gingival tissues was measured using IgG isolated from rabbits immunized with Pg 33277 (52). After 2 h of staining at RT, slides were washed three times with PBS, and then incubated with Alexa Fluor 488–conjugated anti-rabbit secondary antibody (1:1,000) for 1 h. Following this, slides were counterstained with DAPI (4’6-diamidino-2-phenylindole). Formalin-fixed paraffin-embedded gingival tissues were sectioned at 5 μm in a microtome, dewaxed, and rehydrated. Unmasking of antigens was performed using citrate-based antigen unmasking solution (Vector Laboratories). Tissue sections on slides were then blocked with blocking buffer (2.5% goat serum, 1% bovine serum albumin (BSA), 0.05% Tween 20, and 0.05% Triton X-100). Slides were stained with APC-conjugated mouse anti-human EpCAM (Biolegend) at 1:50 dilution, anti-human IFNL-R 1:50 dilution, or rabbit anti-Pg antibody (52) at 1:500 dilution overnight at 4 °C. Slides were washed three times with PBS and then incubated further with Alexa Fluor 488–conjugated anti-rabbit secondary antibody at 1:1,000 dilution for 1 h. Slides were counterstained with DAPI (4’6-diamidino-2-phenylindole) and mounted using ProLong antifade mounting media. Images were visualized using LAS X Life Science software (Leica Microsystems) and analyzed using Imaris software (OXFORD instruments). Mean fluorescence intensities determined by enumerating positive pixel intensity as described by Shihan et al. (71).
RNA Scope.
Transcript expression was determined in formalin-fixed paraffin-embedded gingiva tissues (sectioned at 5 μm) using transcript-specific probes and RNAscope Fluorescent Multiplex Assay version 2 kit (Advanced Cell Diagnostics Inc.) as per the manufacturer’s protocol.
Oral Infection of Mice.
Mx1gfp mice (31) were purchased from Jackson Laboratory and maintained in specific pathogen-free conditions. Male and female mice, 8 to 12 wk old, were orally infected with 109 cfu Pg 33277 suspended in 2% carboxymethylcellulose (CMC) on alternate days for a total of three infection cycles. The bacterial suspension was directly applied to the gingival margin of each mouse under brief isoflurane anesthesia. Sham-infected mice received 2% CMC alone. At 48 h after last inoculation, mice received either 50 μg poly I:C or 40 μg IFN-λ in 100 μL PBS intraperitoneally and euthanized after 36 h. The maxilla and mandible were surgically dissected from each mouse and tissues digested using the tumor dissociation Kit (Miltenyi Biotech) as per the manufacturer’s instructions. Soft tissue was harvested and minced through a 70-μm strainer to obtain a cell suspension. Cells were then washed and stained for flow cytometry using antibodies against CD45-BV605 (1:400) and EpCAM-APC (1:400). Cells were processed using FACSCelesta, and data were analyzed using FlowJo Software. Alternatively, total RNA/protein was isolated from dissected gingival tissues from maxillary molars using the Nucleospin dual extraction kit (Takara Biosciences) as per manufacturer’s instructions.
Western Blots.
Cells were lysed in radio immunoprecipitation (RIPA) buffer containing protease inhibitor mixture, phophoStop phosphatase inhibitors (Roche), and 50 μM gingipain inhibitor Nα-Tosyl-L-Lysine chloromethyl ketone hydrochloride (Sigma Aldrich). Protein concentration was estimated by a bicinchoninic acid assay. Samples were separated by SDS-PAGE and electroblotted onto nitrocellulose (0.2 μm) membranes. Membranes were blocked in PBS-Tween (PBST) containing 5% nonfat dried milk and incubated with primary antibodies. Anti-human IL-28R (IFNL-R), IFNAR, phospho-STAT1 (Y701), STAT1, MX1, ISG15, and GAPDH antibodies were purchased from Cell Signaling Technologies. ZEB1 and IRF-1 antibodies were purchased from ThermoFisher. MDA5 and IFNAR antibodies were purchased from ENZO Life Sciences and Santa Cruz Biotechnology, respectively. For mouse gingival tissue lysates, IFNL-R expression was determined using anti-mouse IL-28R antibody from Lifespan Biosciences.
ELISA.
Interferon-λ (IL-29 and IL-28), IFN-β, and IL-8 ELISA kits were from R&D. IFN-α ELISA kit was from PBL Assay Science, and it detects 14 out of 15 IFN-α subtypes. Cytokine levels were measured in cell-free supernatants as per the manufacturer’s instructions. Pg abundance in human gingival tissue homogenates was determined by ELISA. Briefly, gingival tissues were weighed and then homogenized in sterile PBS containing protease inhibitors. Immunosorbent plates (Nunc) were coated with gingival tissue lysates (0.2 mg/mL). A standard curve of Pg homogenates obtained from serial dilutions of 109 cfu Pg 33277 was used to compare calculate Pg cfu per milligram of tissue. Plates were subsequently blocked with 1% BSA in PBS and then incubated with anti-Pg (4 μg/mL) antibody (52) overnight followed by biotinylated anti-rabbit secondary antibody (200 ng/mL), 2 h incubations. Streptavidin-HRP–conjugated reagent was used to detect the secondary antibody and developed with colorimetric substrate (R&D). OD was measured at 450 nm.
qPCR.
Total RNA was extracted using the RNAeasy kit (Qiagen) and converted to cDNA using the high-capacity cDNA reverse transcription kit (ThermoFisher). Transcript expression was determined by TaqMan assays using TaqMan mastermix. All prevalidated primer sets and probes were purchased from ThermoFisher.
Dual Luciferase Assay.
GECs were transfected with pGL4.45-luc2P/ISRE Vector (Promega) containing five copies of the ISRE element that drives that drives the transcription of the luc2P firefly luciferase reporter gene using Lipojet (SignaGen). Cells were also transfected with the pRL vector that provides constitutive expression of Renilla luciferase as an internal control. At 24 h after transfection, cells were stimulated with 10 µg/mL HSV60 with or without Pg infection. Cells were also stimulated with 100 ng/mL recombinant IFN-λ. Cells were lysed 24 h poststimulation, after which dual luciferase reporter assays were performed using Stop & Glo dual luciferase reporter kit from Promega as per the manufacturer’s instructions. Luciferase activity was measured using a 10-s integration time in a Luminometer (Molecular Devices). Firefly luciferase activity was normalized over Renilla luciferase activity from the same lysates.
Transfection Studies: siRNA and Overexpression Vectors.
GECs were transfected with siRNA (75 μM) against IRF-1 or ZEB1 or scrambled control RNA (Life Technologies) using Lipojet. At 24 h posttransfection, cells were stimulated with Pg (multiplicity of infection [MOI] 100) for 5 h, washed once with PBS, and then stimulated with HSV60 for additional 18 h. Immunoblots confirmed siRNA-mediated knockdown. For overexpression, pcDNA-GFP-STAT1 (Addgene No. 11987) was a gift from Alan Steven Johnson, and pcDNA3.2 (72) vector control was a gift from Jan Rehwinkel (Addgene No. 120833). pCMV-IRF-1 (47) and pCMV empty vector control were from Panomics.
RNA-seq.
RNA was extracted using the RNAqueous-Micro Total RNA Isolation kit (ThermoFisher Scientific). The TruSeq Stranded Total RNA with RiboZero Gold kit (Illumina) was used to generate a sequencing library from 1 µg total RNA. Paired-end sequencing was performed on an Illumina NextSeq 500 at the University of Louisville core, using the NextSeq 500 High-Output Kit (150 cycles) (Illumina). Base calls were made using the BaseSpace FastQ version 1.0.0 application (Illumina, Inc.). For the analysis of differentially expressed genes, demultiplexed paired-end fastq files were aligned to reference GRCh38 by top-level assembly with STAR (version 2.6.1). Gene counts were produced by RSEM (version 1.3.1). We used DESeq2 R/Bioconductor package to obtain differential expression between Pg-treated and control samples (n = 3 per sample). DESeq2 guidelines were used to identify differentially expressed genes, and all P values were adjusted for testing multiple genes (Benjamini–Hochberg procedure, alpha = 0.1). For gene set enrichment analysis, we used fgsea Multilevel function from fgsea R/Bioconductor package (https://doi.org/10.1101/060012). RNA-seq datasets have been deposited in the NCBI GEO database under Accession Nos. GSE184456 and GSE184463.
Viral Assays.
To determine if Pg infection influenced viral replication, GECs were seeded in 24-well plates at 80% confluency. Cells were infected with Pg WT or the gingipain-deficient mutant at MOI of 10 for 5 h. GECs were washed once with PBS and fresh media added. Cells were then infected with Toto1101-derived SINV strains containing either nsP3-GFP (nsP3-green fluorescent protein) or nsP3-Nanoluc at an MOI of 10 plaque-forming units (PFU) per cell (as determined in BHK-21 cells) (73). After a 1 h adsorption period, the inoculum was removed, and the cells were washed twice with 1× PBS to remove unbound viral particles and incubated at 37 °C in a humidified incubator in the presence of 5% CO2. For assaying viral susceptibility and permissivity (which together amount to infectability), the cells were examined at 24 h postinfection via a GFP-capable epifluorescence microscope. The assessment of viral gene expression was identical to that described; however, after the removal of any unbound viral particles, the infectious process was limited to a single round via the addition of media supplemented with 40 µM ammonium chloride. At the indicated times postinfection, the cells were lysed by the addition of 1× PBS supplemented with 0.5% Triton X-100. The lysate was frozen until the completion of the experimental time course. The samples were thawed and clarified by centrifugation at 16,000 × g for 5 min, and equal cell volumes of the nanoluciferase samples were processed using a Nano-Glo nanoluciferase assay system (Promega) according to the manufacturer’s instructions. Luminescence was recorded using a Synergy H1 microplate reader (BioTek).
Statistics.
Statistical analyses utilized GraphPad Prism 6.0 (GraphPad). A P value <0.05 was considered statistically significant. A detailed description of the statistical tests used is stated in each figure legend.
Supplementary Material
Acknowledgments
These studies were funded by National Institute of Dental and Craniofacial Research (NIDCR) Grant Nos. DE028031 and DE028296 to J.B.; National Institute of General Medical Sciences grant GM125504 to J.B., K.J.S., and R.J.L.; and NIDCR grants DE011111, DE012505, DE023193, and DE017921 to R.J.L.; Part of this work was performed with the assistance of the University of Louisville Genomics Facility, which is supported by NIH P20GM103436 (KY IDeA Networks of Biomedical Research Excellence), the J. G. Brown Cancer Center, and user fees. K.Z. was supported by Government of Russian Federation (Grant No. 08-08).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105170118/-/DCSupplemental.
Data Availability
RNA-seq datasets have been deposited in the publicly available NCBI GEO database (GSE184456 and GSE184463).
References
- 1.Lamont R. J., Hajishengallis G., Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonoyan L., Chevalier M., Vincent-Bugnas S., Marsault R., Doglio A., Detection of Epstein-Barr virus in periodontitis: A review of methodological approaches. Microorganisms 9, E72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puletic M., Popovic B., Jankovic S., Brajovic G., Detection rates of periodontal bacteria and herpesviruses in different forms of periodontal disease. Microbiol. Immunol. 64, 815–824 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Christersson L. A., Fransson C. L., Dunford R. G., Zambon J. J., Subgingival distribution of periodontal pathogenic microorganisms in adult periodontitis. J. Periodontol. 63, 418–425 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Imbronito A. V., Okuda O. S., Maria de Freitas N., Moreira Lotufo R. F., Nunes F. D., Detection of herpesviruses and periodontal pathogens in subgingival plaque of patients with chronic periodontitis, generalized aggressive periodontitis, or gingivitis. J. Periodontol. 79, 2313–2321 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Imbronito A. V., et al. , Detection of Epstein-Barr virus and human cytomegalovirus in blood and oral samples: Comparison of three sampling methods. J. Oral Sci. 50, 25–31 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Kamma J. J., Contreras A., Slots J., Herpes viruses and periodontopathic bacteria in early-onset periodontitis. J. Clin. Periodontol. 28, 879–885 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Passariello C., et al. , Evaluation of microbiota associated with Herpesviruses in active sites of generalized aggressive periodontitis. Ann. Stomatol. (Roma) 8, 59–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C., Feng P., Slots J., Herpesvirus-bacteria synergistic interaction in periodontitis. Periodontol. 2000 82, 42–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pott J., et al. , IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U.S.A. 108, 7944–7949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caine E. A., et al. , Interferon lambda protects the female reproductive tract against Zika virus infection. Nat. Commun. 10, 280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klinkhammer J., et al. , IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 7, e33354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broggi A., Tan Y., Granucci F., Zanoni I., IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat. Immunol. 18, 1084–1093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazear H. M., Schoggins J. W., Diamond M. S., Shared and distinct functions of Type I and Type III interferons. Immunity 50, 907–923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells A. I., Coyne C. B., Type III interferons in antiviral defenses at barrier surfaces. Trends Immunol. 39, 848–858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galani I. E., et al. , Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 46, 875–890.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Forero A., et al. , Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by Type I and Type III interferons. Immunity 51, 451–464.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li N., Ma W. T., Pang M., Fan Q. L., Hua J. L., The commensal microbiota and viral infection: A comprehensive review. Front. Immunol. 10, 1551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker F. C., Baldridge M. T., Interactions between noroviruses, the host, and the microbiota. Curr. Opin. Virol. 37, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grau K. R., et al. , The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol. 5, 84–92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingle H., et al. , Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat. Microbiol. 4, 1120–1128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan E., Baldridge M. T., Norovirus encounters in the gut: Multifaceted interactions and disease outcomes. Mucosal Immunol. 12, 1259–1267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotenko S. V., Rivera A., Parker D., Durbin J. E., Type III IFNs: Beyond antiviral protection. Semin. Immunol. 43, 101303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Z., et al. , Segmented filamentous bacteria prevent and cure rotavirus infection. Cell 179, 644–658.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odendall C., et al. , Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thackray L. B., et al. , Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 22, 3440–3453.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldridge M. T., et al. , Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347, 266–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linden J., et al. , Interferon-λ receptor expression: Novel reporter mouse reveals within- and cross-tissue heterogeneity. J. Interferon Cytokine Res. 40, 292–300 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Hyun S. Y., et al. , Amelogenic transcriptome profiling in ameloblast-like cells derived from adult gingival epithelial cells. Sci. Rep. 9, 3736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffatt-Jauregui C. E., et al. , Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J. Periodontal Res. 48, 713–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uccellini M. B., García-Sastre A., ISRE-Reporter mouse reveals high basal and induced Type I IFN responses in inflammatory monocytes. Cell Rep. 25, 2784–2796.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contreras A., et al. , Relationship between herpesviruses and adult periodontitis and periodontopathic bacteria. J. Periodontol. 70, 478–484 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Contreras A., Nowzari H., Slots J., Herpesviruses in periodontal pocket and gingival tissue specimens. Oral Microbiol. Immunol. 15, 15–18 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Naqvi A. R., Shango J., Seal A., Shukla D., Nares S., Herpesviruses and microRNAs: New pathogenesis factors in oral infection and disease? Front. Immunol. 9, 2099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajishengallis G., et al. , Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borisy G. G., Valm A. M., Spatial scale in analysis of the dental plaque microbiome. Periodontol. 2000 86, 97–112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ximénez-Fyvie L. A., Haffajee A. D., Socransky S. S., Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27, 648–657 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Hornung V., Hartmann R., Ablasser A., Hopfner K. P., OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 14, 521–528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi O., Akira S., Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Fensterl V., Sen G. C., Interferon-induced Ifit proteins: Their role in viral pathogenesis. J. Virol. 89, 2462–2468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller D. P., et al. , Genes contributing to Porphyromonas gingivalis fitness in abscess and epithelial cell colonization environments. Front. Cell. Infect. Microbiol. 7, 378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda K., Takaoka A., Taniguchi T., Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Jefferies C. A., Regulating IRFs in IFN driven disease. Front. Immunol. 10, 325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffiths S. J., et al. , A systematic analysis of host factors reveals a Med23-interferon-λ regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog. 9, e1003514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamane D., et al. , Basal expression of interferon regulatory factor 1 drives intrinsic hepatocyte resistance to multiple RNA viruses. Nat. Microbiol. 4, 1096–1104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair S., Poddar S., Shimak R. M., Diamond M. S., Interferon regulatory factor 1 protects against Chikungunya virus-induced immunopathology by restricting infection in muscle cells. J. Virol. 91, e01419-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jauregui C. E., et al. , Suppression of T-cell chemokines by Porphyromonas gingivalis. Infect. Immun. 81, 2288–2295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuyama T., et al. , Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75, 83–97 (1993). [PubMed] [Google Scholar]
- 49.Siegel R., Eskdale J., Gallagher G., Regulation of IFN-λ1 promoter activity (IFN-λ1/IL-29) in human airway epithelial cells. J. Immunol. 187, 5636–5644 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Iwanaszko M., Kimmel M., NF-κB and IRF pathways: Cross-regulation on target genes promoter level. BMC Genomics 16, 307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi H., et al. , The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-κB RelA/p65. PLoS Pathog. 9, e1003326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sztukowska M. N., et al. , Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell. Microbiol. 18, 844–858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J., Tian B., Sun H., Garofalo R. P., Brasier A. R., Epigenetic silencing of IRF1 dysregulates type III interferon responses to respiratory virus infection in epithelial to mesenchymal transition. Nat. Microbiol. 2, 17086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye L., Schnepf D., Staeheli P., Interferon-λ orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 19, 614–625 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Barth K., Genco C. A., Microbial degradation of cellular kinases impairs innate immune signaling and paracrine TNFα responses. Sci. Rep. 6, 34656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakayama M., Inoue T., Naito M., Nakayama K., Ohara N., Attenuation of the phosphatidylinositol 3-kinase/Akt signaling pathway by Porphyromonas gingivalis gingipains RgpA, RgpB, and Kgp. J. Biol. Chem. 290, 5190–5202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stafford P., et al. , Gingipain-dependent degradation of mammalian target of rapamycin pathway proteins by the periodontal pathogen Porphyromonas gingivalis during invasion. Mol. Oral Microbiol. 28, 366–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y., et al. , Noncanonical activation of β-catenin by Porphyromonas gingivalis. Infect. Immun. 83, 3195–3203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang S., et al. , Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5, 166–178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laurent-Rolle M., et al. , The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16, 314–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant A., et al. , Zika virus targets human STAT2 to inhibit Type I interferon signaling. Cell Host Microbe 19, 882–890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lubick K. J., et al. , Flavivirus antagonism of Type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe 18, 61–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakajima M., et al. , Oral administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of Enterobacteria to the liver. PLoS One 10, e0134234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kageyama S., et al. , Characteristics of the salivary microbiota in patients with various digestive tract cancers. Front. Microbiol. 10, 1780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benedyk M., et al. , Gingipains: Critical factors in the development of aspiration pneumonia caused by Porphyromonas gingivalis. J. Innate Immun. 8, 185–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terpenning M. S., et al. , Aspiration pneumonia: Dental and oral risk factors in an older veteran population. J. Am. Geriatr. Soc. 49, 557–563 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Okuda K., et al. , Involvement of periodontopathic anaerobes in aspiration pneumonia. J. Periodontol. 76(11, suppl.)2154–2160 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Muir A. J., et al. ; EMERGE study group, A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 61, 1238–1246 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Prokunina-Olsson L., et al. , COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 217, e20200653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J. Y., et al. , Maturation of the Mfa1 Fimbriae in the Oral Pathogen Porphyromonas gingivalis. Front. Cell. Infect. Microbiol. 8, 137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shihan M. H., Novo S. G., Le Marchand S. J., Wang Y., Duncan M. K., A simple method for quantitating confocal fluorescent images. Biochem. Biophys. Rep. 25, 100916 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hertzog J., et al. , Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur. J. Immunol. 48, 1120–1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokoloski K. J., et al. , Identification of interactions between Sindbis virus capsid protein and cytoplasmic vRNA as novel virulence determinants. PLoS Pathog. 13, e1006473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets have been deposited in the publicly available NCBI GEO database (GSE184456 and GSE184463).