Significance
Cellular and molecular mediators driving multiple sclerosis (MS) pathology have been discovered to a great extent. However, the early molecular events leading to aberrant immune responses remain largely unknown. In this study, we combined bisulfite sequencing with transcriptome profiling to characterize the epigenetic landscape of four peripheral immune cell populations isolated from newly diagnosed, untreated MS patients and healthy individuals. We demonstrate widespread hypomethylation in CD19+ B cells at clinical disease onset. Notably, this epigenetic signature is functionally linked with the overactivation of B cells. Altogether, our results pinpoint the role of aberrant DNA methylation in connecting defects in the periphery with central nervous system autoimmunity and corroborate the key role of B cells in the initial stages of MS.
Keywords: multiple sclerosis, B cell, hypomethylation
Abstract
Epigenetic changes have been consistently detected in different cell types in multiple sclerosis (MS). However, their contribution to MS pathogenesis remains poorly understood partly because of sample heterogeneity and limited coverage of array-based methods. To fill this gap, we conducted a comprehensive analysis of genome-wide DNA methylation patterns in four peripheral immune cell populations isolated from 29 MS patients at clinical disease onset and 24 healthy controls. We show that B cells from new-onset untreated MS cases display more significant methylation changes than other disease-implicated immune cell types, consisting of a global DNA hypomethylation signature. Importantly, 4,933 MS-associated differentially methylated regions in B cells were identified, and this epigenetic signature underlies specific genetic programs involved in B cell differentiation and activation. Integration of the methylome to changes in gene expression and susceptibility-associated regions further indicates that hypomethylated regions are significantly associated with the up-regulation of cell activation transcriptional programs. Altogether, these findings implicate aberrant B cell function in MS etiology.
Multiple sclerosis (MS) is a chronic inflammatory disease of the nervous system, often described as the most common cause of nontraumatic neurological disability in young adults. Although the trigger(s) remains unknown, the most accepted model of pathogenesis involves multilayered interactions between complex genetic and environmental influences (1–7). Genome-wide association studies conducted over the past decade have identified 233 genome-wide significant loci associated with MS susceptibility (1, 8, 9), collectively highlighting the importance of adaptive and innate immune responses in driving risk. Despite this notable progress, the current MS genetic map only partially accounts for the estimated disease heritability. Similar to other complex genetic diseases, hidden epigenetic effects are a reasonable explanation for the missing heritability in MS.
Epigenetic modifications can regulate gene expression in a heritable fashion without altering the DNA sequence. Moreover, environmental signals may influence epigenetic modifications, further contributing to the phenotype and potentially linking environmental risk factors to genetics. In addition to developmental abnormalities, incorrect epigenetic signatures have been associated with chronic diseases, including autoimmunity (5, 10–14). DNA methylation is the most studied epigenetic modification and refers to the addition of a methyl group to a cytosine base. In mammals, DNA methylation typically occurs in the context of CpG dinucleotides. Using the Illumina methylation assay, several studies have identified MS- and MS-subtype–associated methylation changes in bulk peripheral blood mononuclear cells (PBMCs) and sorted CD4+, CD8+, CD14+, and CD19+ cells (15–21). However, this assay has limited coverage of the CpG sites in the human genome, a shortcoming that can be addressed through genome-wide bisulfite sequencing (BS-seq) single-nucleotide mapping. Here, BS-seq was applied to generate base-resolution DNA methylome maps in four peripheral immune cell types in recently diagnosed, untreated MS patients and healthy controls. We report that the disease-associated methylation alterations are more frequent in B cells compared to T cells and monocytes, and show that these methylation alterations are associated with the activation of B cell differentiation programs.
Results
MS-Associated DNA Methylation Patterns in Four Immune Cell Types.
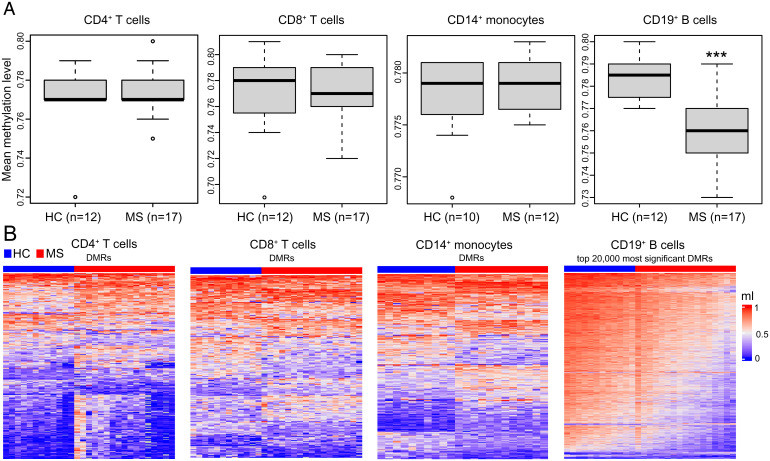
Four immune cell populations, namely CD4+ and CD8+ T cells, CD14+ monocytes, and CD19+ B cells were separated by fluorescence-activated cell sorting (FACS) from PBMC samples isolated from 17 MS cases and 12 sex/age/ancestry-matched healthy controls (SI Appendix, Fig. S1). To avoid confounding effects due to ancestral background, sex, smoking, and treatment, we limited the study to white female nonsmoker patients recruited into the study within 24 h from diagnosis. We then employed BS-seq to generate genome-wide DNA methylomes at base resolution, obtaining for each sample an average sequencing depth of ∼17× and more than 92% coverage of the reference human genome (Dataset S1). Total DNA methylation levels were computed in each cell type, comparing the average cross-sectional values between cases and controls. CD19+ B cells from MS patients displayed significantly lower levels of DNA methylation, whereas no significant differences were detected for the other three cell types analyzed (Fig. 1A). We complemented this analysis by identifying 68,209 significant differentially methylated regions (DMRs) across the genome in B cells but only 366, 230, and 331 in CD4+, CD8+ T cells, and monocytes, respectively (Fig. 1B). Consistent with the results from the global analysis, ∼96% DMRs in B cells show hypomethylation in MS patients compared to controls.
Fig. 1.
DNA methylation changes in peripheral CD4+ and CD8+ T cells, CD14+ monocytes, and CD19+ B cells in MS patients and healthy controls (HC) in the discovery cohort. (A) Boxplots represent whole genome mean methylation levels (ml) in untreated MS patients and HC for each cell type. The ml were determined by BS-seq. Differences between MS patients and HC in each cell type were assessed by two-tailed Student’s t test. ***P < 0.001. (B) Heat maps showing ml of MS patients and HC in the DMRs for each cell type. The heat map of CD19+ B cells was generated using the 20,000 most significant DMRs. The color key from blue to red represents the ml from low to high.
B Cells Show DNA Hypomethylation at MS Clinical Onset.
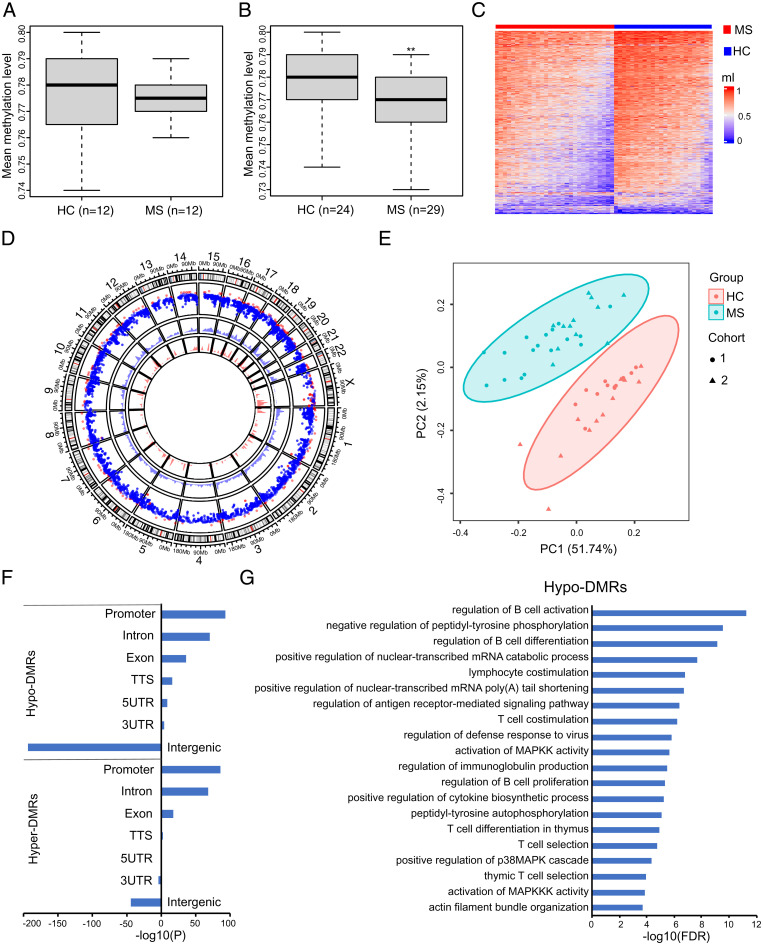
Because B cells showed the most dramatic perturbation in DNA methylation upon disease clinical onset, we focused on this immune cell population for further characterization. To confirm our initial results, we performed BS-seq profiling on B cell fractions isolated from 24 additional samples, generating a total of 53 B cell–specific genome-wide DNA methylation maps (29 MS patients and 24 healthy controls) (Dataset S2). Information for study participants is provided in Dataset S3. Methylation differences between cases and controls in the second cohort trended in the same direction as the discovery cohort (i.e., hypomethylation) but did not reach statistical significance (Fig. 2A). Study participants in both datasets were nonsmoker women, and no significant differences were measured in age and Expanded Disability Status Scale. Brain lesion load was higher in the subjects comprising the discovery cohort (23.53 versus 9.09%; SI Appendix, Fig. S2 A and B). We then computed the methylation levels between cases and controls in the pooled dataset, and, consistent with the results from the discovery cohort, the average B cell methylation levels in this expanded dataset were significantly lower in MS cases than controls (Fig. 2B). A total of 4,933 significant DMRs were detected (false discovery rate [FDR] < 0.05, greater than 10% methylation difference): 4,621 DMRs were hypomethylated and 312 DMRs were hypermethylated compared to controls (Fig. 2C). The hypo-DMRs and hyper-DMRs were uniformly distributed throughout the autosomes, whereas chromosome X was preferentially hypermethylated (Fig. 2D). Principal component analysis (PCA) of the DMRs clearly separate MS from control samples but not the two cohorts, indicating that DNA hypomethylation in B cell is a robust hallmark of early disease (Fig. 2E).
Fig. 2.
Loss of DNA methylation in CD19+ B cells of MS patients. (A) Boxplot represents whole genome mean methylation levels (ml) in CD19+ B cells in MS patients and HC from the second cohort. (B) Boxplot represents whole genome mean ml in CD19+ B cells in the combined cohorts. Significance was determined by two-tailed Student’s t test. **P < 0.01. (C) Heat map displaying significant CD19+ B cells DMRs. The color key from blue to red represents the ml from low to high. (D) Circle plot representation for DMRs location and genomic densities. The outer layer rainfall plot shows the distribution of DMRs in the genome by chromosome. The x axis corresponds to the genomic coordinate, and the y axis corresponds to the distance (log10 transformed) to its nearest DMRs. In the plot, each dot represents a DMR. Red corresponds to hyper-DMRs (gain of methylation), and blue corresponds to hypo-DMRs (loss of methylation). The two inner layer density plots display the genomic densities of the hyper-DMRs (red) and hypo-DMRs (blue), defined as the fraction of a genomic window that is covered by DMRs, respectively. (E) PCA plot of the DMRs. Each dot (cohort 1) or triangle (cohort 2) represents a sample. Ellipses show clustering of the samples. (F) Enrichment of hypo-DMRs and hyper-DMRs in different genomic regions. The P values were calculated using the Hypergeometric Optimization of Motif EnRichment (HOMER) annotatePeaks.pl function. TTS, transcription termination site. (G) Functional annotation of the hypo-DMRs by the Genomic Regions Enrichment of Annotations Tool. FDR, false discovery rate.
The found DMRs are preferentially located in promoter and gene body regions but not in intergenic regions, which is consistent with a putative regulatory activity on gene expression (Fig. 2F). The Genomic Regions Enrichment of Annotations Tool was applied to capture the biological functions associated with the DMRs. Gene ontology (GO) analysis on the genes overlapping with or located near hypo-DMRs (Dataset S4) indicated an enrichment in B cell activation and differentiation, cytokine production, and antigen presentation–related pathways, with “regulation of B cell activation” being the most significant term (Fig. 2G). On the other hand, we failed to detect any GO enrichment for hyper-DMR–associated genes. Likewise, no significant GO terms were found associated with the DMRs mapped in CD4+ and CD8+ T cells. For the monocyte population, no significant GO terms were found when running hypo-DMRs and hyper-DMRs separately, whereas the “platelet activation” pathway exceeded the statistical significance threshold when computing all DMRs together.
Specific Differentiation Programs Associated with DNA Hypomethylation in B Cells.
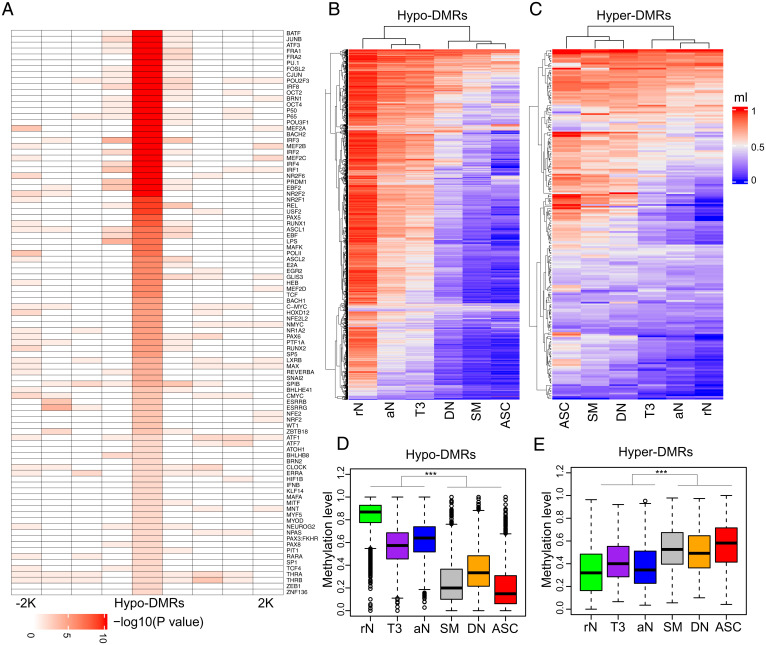
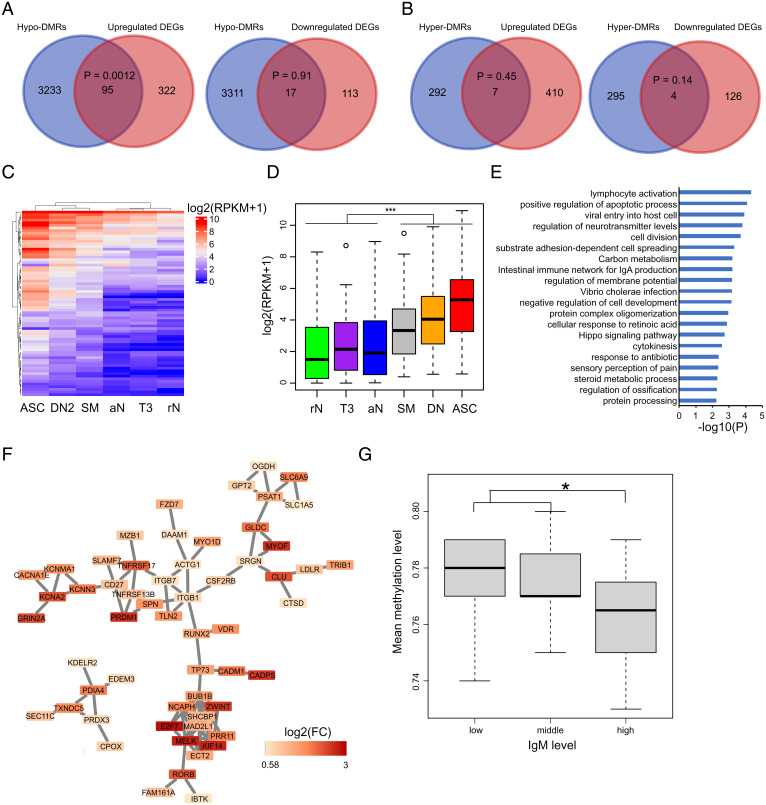
The transcription factor DNA-binding motifs enriched in the DMRs were examined to impute the transcription factors affected by the methylation changes in B cells from MS patients. Hypo-DMRs significantly overlapped with consensus motifs of transcription factors important for B cell activation and differentiation such as AP-1 (e.g., BATF), OCT2, MEF2B, PU.1, BACH2, PRDM1, and RUNX1 (Fig. 3A). To further validate these associations, we analyzed the transcription factor chromatin immunoprecipitation sequencing (ChIP-seq) dataset of a B cell line (GM12878) made available by the Encyclopedia of DNA Elements (ENCODE) Consortium. The binding peaks of these transcription factors significantly overlapped with the hypo-DMRs (Dataset S5). Notably, the regions demethylated in germinal center B cells or bone marrow plasma cells relative to naïve follicular B cells have been reported to be enriched for AP-1, OCT2, MEF2B, PU.1, BACH2, and PRDM1 binding motifs (22). On the other hand, the MS hyper-DMRs were enriched for the basic helix–loop–helix factors such as the ETS family of transcription factors and TCF12 as well as homeodomain transcription factors (Dataset S6). The ETS and TCF12 factors were reported to be enriched in the regions hypermethylated along the B cell differentiation process (22). Interestingly, we identified one hypo-DMR located in the intron region of BATF and one hypo-DMR mapped to the promoter region of PRDM1, suggesting that DNA methylation changes not only affect transcription factor binding to DNA but may also regulate their own expression.
Fig. 3.
MS-associated methylation changes are linked to B cell differentiation and activation. (A) Transcriptional factor binding motif enrichment between hypo-DMRs and neighboring regions. The hypo-DMRs and every 500 base pair (bp) window in ±2 kilobase (kb) flanking hypo-DMRs were used to calculate the enrichment P values. The motif enrichment was determined by binomial distribution using HOMER. (B and C) Heat maps displaying the methylation patterns of hypo-DMRs (B) and hyper-DMRs (C) in each B cell subset. The methylation levels (ml) were calculated by RRBS data in the Gene Expression Omnibus repository (GSE118255). In the heatmaps, each column represents the mean ml of all subjects in the cell type. (D and E) Box plots of ml of hypo-DMRs (D) and hyper-DMRs (E) for each cell type. Significance was determined by two-tailed Student’s t test. ***P < 0.001. rN, resting naïve; aN, activated naïve; T3, transitional T3; DN, double-negative; SM, switched memory; ASC, antibody-secreting cell.
DNA hypomethylation is a major feature of B cell differentiation (23, 24). Given the results described in the previous paragraph, we sought to further understand how MS-associated DNA methylation changes are linked to B cell differentiation. A recent study combined reduced-representation BS-seq (RRBS) with assays for transposase-accessible chromatin using sequencing and RNA sequencing (RNA-seq) to describe the epigenetic states of B cell subsets, including resting naïve, transitional, activated naïve, isotype-switched memory and double-negative B cells, and antibody-secreting cells (ASCs) in subjects with systemic lupus erythematosus and healthy controls (25). We build on the granularity achieved in that study to further characterize the MS-associated DNA methylation patterns at the subset level by quantifying the mean methylation level of each MS-associated DMR covered by the RRBS dataset (2,050 hypo-DMRs and 208 hyper-DMRs) in healthy controls across each B cell subpopulation as reported in Scharer et al. (25). We found the regions that lose methylation in MS also become hypomethylated along the normal differentiation process from naïve B cells to ASCs (Fig. 3 B and D). An analogous pattern was found for the hypermethylated regions (Fig. 3 C and E). Altogether, the data provide evidence for the epigenetic activation of B cell differentiation in MS.
Hypomethylated Regions in B Cells Overlap with MS Risk Genes.
Genetic and epigenetic mechanisms may jointly affect MS susceptibility (18). Therefore, we tested the overlap between DMRs and MS susceptibility genes. DMRs were mapped to the nearest protein-coding genes, and these significantly overlapped with the current catalog of candidate MS risk genes (1) (P = 6.2E-11, SI Appendix, Fig. S3A). When analyzing the hypo- and hyper-DMRs separately, only the hypo-DMR–coupled genes overlapped significantly with MS risk genes (P = 3.4E-12, SI Appendix, Fig. S3A). Next, hypo-DMRs located within ±100-kb transcriptional start site (TSS) flanking regions were analyzed. Based on the distance from the nearest TSS, hypo-DMRs were assigned to the relative coding genes and classified as TSS-proximal (3 kb upstream and downstream of the TSS), TSS-distal 10 kb (10 kb upstream and downstream of the TSS), and TSS-distal 100 kb (100 kb upstream and downstream of the TSS). We found the hypo-DMR–coupled coding genes in all three categories significantly overlap with MS risk genes, with TSS-proximal hypo-DMRs having the lowest P value (P = 2E−10, SI Appendix, Fig. S2B and Dataset S7). Overall, the distance to TSS negatively correlates with the overlap significance (R2 = 0.63, P = 0.006) (SI Appendix, Fig. S3C), consistent with the colocalization of genetic and epigenetic regulation in MS. The GO enrichment analysis of TSS-proximal hypo-DMR–coupled risk genes that are both genetically and epigenetically regulated highlights “regulation of leukocyte activation” as the most significant associated pathway (SI Appendix, Fig. S3D). Interestingly, the “regulation of B cell differentiation” category was specifically enriched in TSS-proximal hypo-DMR–coupled risk genes compared to TSS-distal hypo-DMR–coupled genes (SI Appendix, Fig. S3E).
Transcriptomic Analysis of B Cells at Disease Onset.
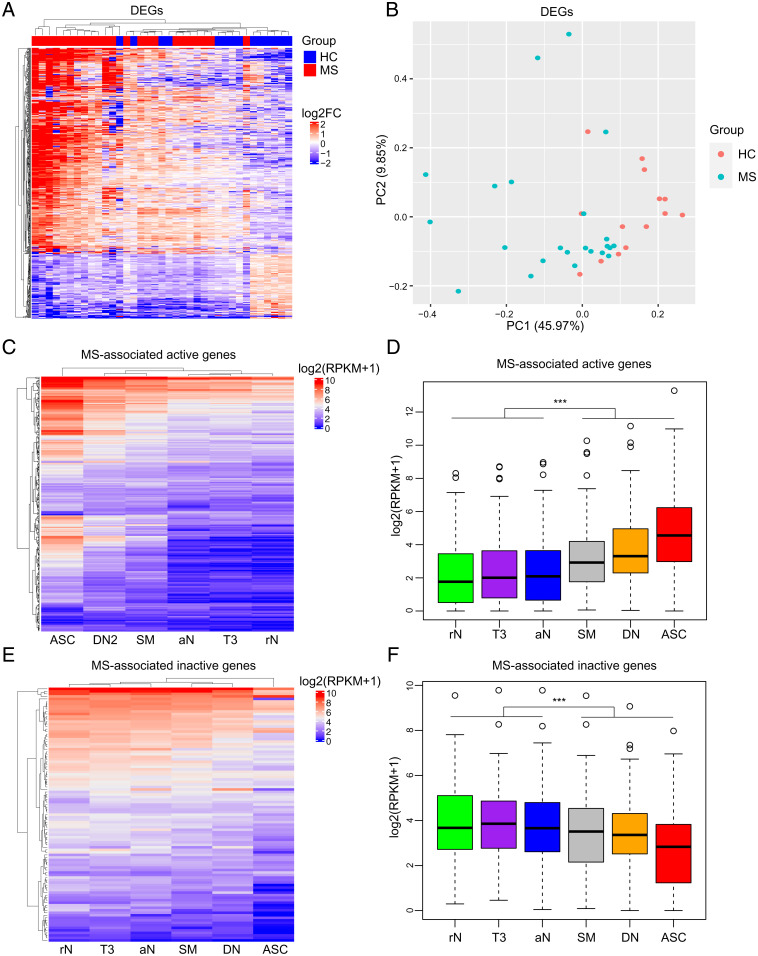
To better understand the functional consequences of B cell DNA hypomethylation, sorted CD19+ B cell samples from the combined epigenetic cohort were subjected to messenger RNA-seq (mRNA-seq). Comparative analysis identified 547 differentially expressed genes (DEGs) (417 up-regulated and 130 down-regulated) between MS cases and controls (Fig. 4A and Dataset S8). PCA of the DEGs was able to separate MS from controls (Fig. 4B), albeit with lower resolution compared to DMRs (Fig. 2E). GO enrichment on the up-regulated DEGs highlighted “complement activation, classical pathway” as the most significant category along with other pathways implicated in B cell differentiation and activation (26–28) (SI Appendix, Fig. S4A and Dataset S9). The down-regulated genes are most significantly enriched in “nucleosome assembly,” “regulation of apoptotic signaling pathway,” and “regulation of autophagy” pathways (SI Appendix, Fig. S4B and Dataset S10). The gene expression profiles were compared to recent transcriptomic signatures obtained from different B cell subsets (25). We found that the genes up-regulated upon MS are also activated as B cells differentiate toward ASCs (Fig. 4 C and D), mimicking the epigenetic process at the transcriptional level. Likewise, the genes repressed upon disease are down-regulated along the B cell maturation pathway (Fig. 4 E and F). Intriguingly, 17 DEGs are annotated as epigenetic factors according to the EpiFactors database (29) (Dataset S11). Among them, APBB1, GFI1B, KDM5B, RNF168, AURKB, SETD7, TTK, PRDM1, and CIT function on histone modification process; HJURP and ASF1B act as histone chaperones; and CDC6, TOP2A, KEAP1, and VDR can play on chromatin remodeling process (29). The known transcription factor FOXO1 can regulate the chromatin states (30). Moreover, the DNA cytosine deaminase APOBEC3B was among the up-regulated genes. The dysregulation of these epigenetic factors may contribute to the hypomethylation.
Fig. 4.
Up-regulation of the B cell differentiation transcriptional program in MS. (A) Unsupervised hierarchical clustering of differentially expressed genes (DEGs, fold change ±1.5) between MS patients and healthy controls. The distances between samples were calculated using the Euclidean distance method. Genes were clustered based on log2FC (each sample to the mean expression level of controls). FC, fold change. (B) PCA plot of the DEGs. Each point represents a sample. The colors indicate the sample groups. (C) Heat map showing the expression patterns of MS-associated active genes (up-regulated in MS patients) in B cell subsets. The gene expression values for each cell type were obtained from the RNA-seq data in the Gene Expression Omnibus repository (GSE118254). In the heatmap, each column represents the mean expression value of all subjects in the cell type. (D) Box plot of gene expression levels of MS-associated active genes for each cell type. (E) Heat map showing the expression patterns of MS-associated inactive genes (down-regulated in MS patients) in each cell type. In the heatmap, each column represents the mean expression value of all subjects in the cell type. (F) Box plot of gene expression levels of MS-associated inactive genes for each cell type. Significance was determined by two-tailed Student’s t test. ***P < 0.001. rN, resting naïve; aN, activated naïve; T3, transitional T3; DN, double-negative; SM, switched memory; ASC, antibody-secreting cell.
Hypo-DMRs in Patients with MS Are Associated with Gene Activation.
DNA methylation profiles were integrated with gene expression data to reconstruct the MS-associated functional epigenetic landscape of B cells. A total of 112 overlaps between hypo-DMR–associated genes and DEGs were identified (95 up-regulated DEGs and 17 down-regulated DEGs) (Fig. 5A and SI Appendix, Fig. S5A), while 11 overlaps were found between hyper-DMR–associated genes and DEGs (7 up-regulated DEGs and 4 down-regulated DEGs) (Fig. 5B). Only the overlap between the up-regulated genes and hypo-DMRs reached statistical significance, in agreement with the notion that DNA methylation generally inhibits gene expression (Fig. 5A and SI Appendix, Fig. S5A). Notably, these 95 genes are up-regulated as B cells differentiate to ASCs (Fig. 5 C and D). Among them, SLAMF7, KCNN3, CSF2RB, LDLR, PLXNC1, PMVK, ARHGAP31, and ZBTB38 are nominated MS susceptibility genes (1). GO enrichment analysis identified “lymphocyte activation” as the top pathway associated with these 95 genes (Fig. 5E and Dataset S12), whereas network analysis highlighted that the proteins encoded by these genes physically interact with each other (P = 1.8 × 10−13) (Fig. 5F).
Fig. 5.
Loss of DNA methylation in B cells from MS patients is associated with gene activation. (A) Venn diagrams showing the overlap between hypo-DMR-coupled genes and up- (Left) or down-regulated (Right) genes. The statistical significance of the overlap was assessed by Fisher’s exact test using the GeneOverlap package. (B) Venn diagrams showing the overlap between hyper-DMR–coupled genes and up- (Left) or down-regulated (Right) genes. (C) Heat map showing the expression patterns of 95 overlapping genes in A (Left) in B cell subsets. Gene expression values for each cell type were downloaded by RNA-seq data in the Gene Expression Omnibus repository (GSE118254). In the heatmap, each column represents the mean expression value of all subjects in the cell type. (D) Box plot of gene expression levels of the 95 genes in each B cell subset. Significance was determined by two-tailed Student’s t test. ***P < 0.001. rN, resting naïve; aN, activated naïve; T3, transitional T3; DN, double-negative; SM, switched memory; ASC, antibody-secreting cell. (E) Histograms showing the most significant GO terms and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways for the 95 genes. (F) Protein–protein interaction network using the 95 genes as input. (G) Whole genome mean methylation levels of subjects with different IgM levels. Subjects were clustered into three groups according to their IgM levels. Significance was determined by two-tailed Student’s t test. *P < 0.05.
To further explore the mechanistic correlation between DNA methylation and gene expression, the analysis of DMRs mapping within gene promoter regions was carried out. In total, 848 hypo-DMRs and 117 hyper-DMRs located within 3-kb up- and downstream regions of the TSS were identified. The hypo-DMRs and hyper-DMRs can be assigned to 761 and 116 nearest protein-coding genes, respectively. Of the 761 hypo-DMR–associated genes, 31 are up-regulated and 2 are down-regulated in MS (SI Appendix, Fig. S5B). Of the 117 hyper-DMR–associated genes, one is up-regulated and another one is down-regulated in MS (SI Appendix, Fig. S5C). Fisher’s exact test revealed a significant association of gene expression up-regulation with hypo-DMRs within promoter regions (SI Appendix, Fig. S5B). Network analysis highlighted that the proteins encoded by the genes interact with each other (P = 0.0017), and the main network is significantly enriched in genes from the “lymphocyte activation” pathway (SI Appendix, Fig. S5D). The inverse correlation between methylation levels at promoters and gene expression was experimentally validated by means of luciferase assays. The promoter regions from five randomly selected DMR-overlapping DEGs, namely DCAF12, PRDM1, SEM4A, TNFRSF17, and ZWINT, were cloned into a CpG-free luciferase reporter vector and methylated in vitro. The different methylated constructs were then transfected into HeLa cells, and their luciferase activities were compared to cells expressing the nonmethylated counterparts. Consistent with the sequencing data, promoter methylation resulted per se in a statistically significant decrease in luciferase transcription (SI Appendix, Fig. S5E).
Overactivation of B Cells in MS.
To access the consequences of the epigenetic changes described above, we assessed IgM and IgG blood levels and selected cytokines in MS patients and controls. There was no significant difference in IgM (fold change = 1.25, P = 0.55; SI Appendix, Fig. S6A) or IgG (fold change = 1.02, P = 1; SI Appendix, Fig. S6A) levels between cases and controls. However, when subjects were stratified according to IgM levels, a significant association emerged between IgM levels and hypomethylation (Fig. 5G). Furthermore, patients with more than 20 brain lesions had significantly higher levels of IgM compared to patients with fewer than 10 brain lesions (SI Appendix, Fig. S6B). No differences were detected for IgG levels (SI Appendix, Fig. S6B). No differences in the levels of TRAIL, BAFF, IL6, and IL10 between patients and controls were detected (Dataset S13).
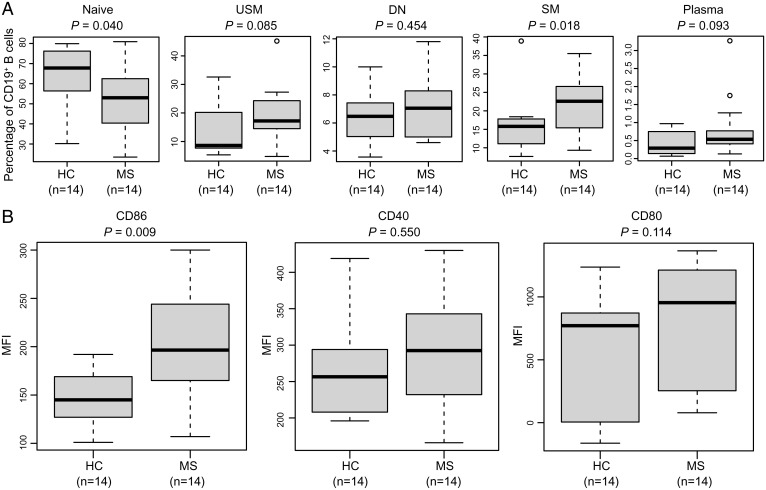
Next, different B cell subpopulations were immunophenotyped by flow cytometry. Five circulating human B cell subsets were analyzed: naïve B cells (CD19+CD27−IgD+), unswitched memory B cells (CD19+CD27+IgD+), switched memory B cells (CD19+CD27+IgD−), double-negative B cells (CD19+CD27−IgD−), and plasma cells (CD19+IgD−CD27hiCD38hi) (SI Appendix, Fig. S7 A and B). Naïve B cells were significantly decreased in MS (P = 0.040), whereas the switched memory B cells were significantly increased (P = 0.018) in MS patients compared to controls (Fig. 6A). We observed a trend toward increased percentages of unswitched memory B cells (P = 0.085) and plasma cells (P = 0.093) in MS patients compared to controls (Fig. 6A). Furthermore, naïve B cells were significantly decreased in subjects with lower methylation levels, whereas the unswitched memory, switched memory, and double-negative B cells were significantly increased in these subjects (SI Appendix, Fig. S8A), supporting the association between B cell differentiation and hypomethylation. Notably, expression of CD86 in CD19+ B cells was significantly up-regulated in MS patients (Fig. 6B). Consistent with this observation, the mRNA-seq analysis showed the up-regulation of CD86 mRNA levels in MS patients (P = 0.049; fold increase: 1.3). In addition, expression of CD86 in CD19+ B cells was significantly up-regulated in the subjects with lower methylation levels (SI Appendix, Fig. S8B). Altogether, the data provide evidence for the B cell overactivation phenotype in MS at the time of clinical onset.
Fig. 6.
B cell activation phenotype in MS patients. (A) Box plots showing the percentage of each B cell subset between MS and HC subjects. Statistical tests were performed using the Mann–Whitney U test. USM, unswitched memory; DN, double-negative; SM, switched memory. (B) Box plots showing the mean fluorescence intensity of CD86, CD40, and CD80 expression on CD19+ B cells between MS and HC subjects. Statistical tests were performed using the Mann–Whitney U test.
Discussion
The combined quantifiable impact of genetic and environmental factors strongly suggests that epigenetic mechanisms including DNA methylation, histone modifications, and microRNAs play important roles in MS heritability and pathogenesis. DNA methylation is the most studied epigenetic mechanism in MS, and distinct methylation signatures have been reported affecting multiple cell types, including B cells (15–21). However, the results are not entirely consistent and are difficult to interpret because of assay limitations, absence of cellular specificity, disease heterogeneity, and loose study inclusion criteria that ignored disease-modifying treatments, smoking history, and other confounders of epigenetic signatures. In this study, we performed BS-seq in four immune cell types associated with MS pathogenesis isolated from newly diagnosed, untreated MS patients, and sex/age/ancestry-matched healthy individuals. Using consistent experimental conditions and a robust analysis pipeline, we found global hypomethylation in MS compared to controls in CD19+ B cells with the number of DMRs reaching ∼185-fold relative to other cell types. The global hypermethylation of the X chromosome was the remarkable exception. In contrast, no significant global differences in the DNA methylation status were detected in CD4+ and CD8+ T cells, and CD14+ monocytes isolated in the same conditions. Consistent with the present results, a recent study employing the Illumina array-based methodology has similarly identified MS-associated hypomethylation changes in CD19+ B cells compared to the other three immune cell types (19).
For decades, the role of T cells was considered central in MS pathogenesis, often overlooking the functions of other immune cell types in mediating the disease (31–33). However, growing evidence supports the mechanistic involvement of multiple immune cell populations in initiating and sustaining the autoimmune injury in the central nervous system (CNS) (34, 35). Among them, it is now clear that B cells are central actors in the pathogenesis of MS, operating through antigen presentation, secretion of proinflammatory cytokines and soluble neurotoxic factors, and possibly autoantibody production (36, 37). The importance of B cells in MS has been firmly established by the high degree of effectiveness of anti-CD20 monoclonal antibodies against all forms of the disease (38–40). Despite this consistent body of experimental data, however, the specific molecular pathways leading to B cell dysfunction in MS are largely unknown. Here, we show that the MS-associated methylation signatures in B cells connect with specific genetic programs underlying B cell differentiation and activation. Previous studies support the notion of B cell aberrant activation in the CNS of MS patients (41–44). Our results add to this collective body of evidence highlighting peripheral B cell overactivation in early MS, putting forward this cell population as a main trigger of MS pathology.
DNA demethylation, a process through which methylated cytosines are reversed to unmodified cytosines, can be achieved by two means. The first is the passive DNA demethylation, in which the methylated cytosines are diluted during DNA replication because of reduced activity or absence of DNA maintenance methyltransferase DNMT1. Another is active DNA demethylation, in which the ten-eleven translocation (TET) enzymes can oxidize 5-methylcytosine with conversion to unmodified cytosine by DNA replication–dependent dilution or thymine DNA glycosylase–dependent base excision repair. Recent studies suggest that the deaminases activation-induced cytidine deaminase (AID)/APOBEC family is also involved in the active DNA demethylation via deamination (45–47). Here, we found that APOBEC3B was among the up-regulated genes in MS (fold increase: 2, P = 0.0004, FDR = 0.015), whereas TET1 was up-regulated in MS at nominal significance (fold increase: 2, P = 0.01, FDR = 0.1). Collectively, these findings support active DNA demethylation mediated by TET and APOBEC enzymes as the preferential mechanism for B cell hypomethylation in early MS. The significant overlap between hypomethylated regions and MS risk genes suggests that the DNA hypomethylation may be regulated by MS-associated genetic factors.
DNA methylation can influence gene expression by affecting the interactions with both chromatin proteins and specific transcription factors. Here, MS-associated hypo-DMRs are significantly enriched in B cell activation– and differentiation–related transcription factor binding sites, including motifs for AP-1 (e.g., BATF), OCT2, BACH2, and PRDM1. BATF can regulate the class-switch recombination process in B cells by directly controlling the expression of AID (48). OCT2 is known to be required for B cell receptor and Toll-like receptor 4 signaling (49), and BACH2 plays important roles in determining the fate of activated B cells such as the initiation of B cell differentiation and generation of memory B cells (50, 51). Finally, PRDM1 is known for its role in controlling the differentiation of mature B cells into ASCs (52, 53). A recent study in mice has shown that ectopic expression of PRDM1 in B cells can enhance the production of self-reactive plasma cells and eventually cause autoimmune disease (54). It is generally accepted that DNA methylation impairs the binding of transcription factors to their target sequences. Therefore, the hypomethylated status of B cells upon MS is likely to promote their differentiation by enhancing the transcriptional activity of the aforementioned factors. In this regard, our mRNA-seq analysis clearly showed the up-regulation of genes involved in B cell differentiation. Furthermore, the MS-associated and normal B cell differentiation hypomethylated regions overlap, further corroborating the notion that the MS-associated methylation signatures are linked to an early activation of genes involved in B cell differentiation. Intriguingly, one of these genes, SLAMF7, is a known MS susceptibility gene on chromosome 1q23.3 coding for a regulator of both innate and adaptive immune responses. Elotuzumab, a monoclonal antibody directed against SLAMF7, is approved for treatment of multiple myeloma, and, in light of the current data, the potential value of this therapeutic approach for MS should be explored.
Using flow cytometry, we corroborated the previously reported increase of switched memory B cells (CD19+CD27+IgD−) in the peripheral blood of untreated MS patients (55). Memory B cells are key players to investigate in MS because of their unique features, including long-term survival, antigen experience, and rapid response upon second stimulation. In addition to being antigen-experienced precursors to ASCs, memory B cells are highly efficient antigen-presenting cells. Many disease-modifying treatments for MS (including anti-CD20 monoclonal antibodies) have been proven to affect memory B cells (56), and one recent study showed that memory B cells in the peripheral blood can be used for monitoring the B cell depletion therapy in MS (57). When subjects were stratified according to methylation levels, a significant association emerged between B cell differentiation and hypomethylation. Consistent with this observation, subjects with higher serum IgM levels had significantly lower methylation levels. Finally, the transcriptome and flow cytometry analyses confirmed the up-regulation of CD86, a classical activation marker of B cells and a costimulatory molecule important for T cell activation.
In summary, our study provides evidence of global hypomethylation of MS CD19+ B cells at time of diagnosis at a genome-wide single base resolution. We report the identification of hypo-DMRs reflecting B cell activation and dysfunction, posing a mechanistic link to the remarkable clinical efficiency of the anti-CD20 antibody treatments for this disease. The analysis also suggests that hypomethylation changes in B cells could potentially serve as an ancillary biomarker for MS diagnosis. Further studies will be required to systematically address the heterogeneity of the B cell compartment and carry out DNA methylation analysis in more restricted functional subsets such as the B naïve and memory populations.
Materials and Methods
All studies were approved by the University of California San Francisco Institutional Review Board, and informed consent was obtained from all subjects. Details on the subjects in this study, as well as on the sample preparation for FACS, are provided in the SI Appendix. Details on DNA extraction, BS-seq library preparation and sequencing, RNA extraction and mRNA-seq, publicly available B cell dataset analysis, overlap with MS susceptibility genes, cell lines, plasmids, luciferase assays, enzyme-linked immunosorbent assays, lymphocyte immunophenotyping, and bioinformatics analyses are provided in the SI Appendix.
Supplementary Material
Acknowledgments
This study was supported by NIH Grants R01NS102153 (to J.R.O.) and R35NS111644 (to S.L.H.) and by the Valhalla Foundation. The collection of DNA samples at the University of California San Francisco was supported by a grant from the National Multiple Sclerosis Society (SI-2001-35701 to J.R.O.). The authors acknowledge the contributions of Rosa Guerrero for sample processing, Adam Santaniello and Adam Renschen for assistance with data management, Rita Loudermilk and Jessa Alexander for assistance with flow cytometry, and Dr. Michael Rehli for the kind gift of the CpG-free firefly luciferase reporter vectors.
Footnotes
The authors declare no competing interest.
A complete list of the University of California San Francisco MS-EPIC Team can be found in the SI Appendix.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111920118/-/DCSupplemental.
Contributor Information
Collaborators: Jessa Alexander, Riley Bove, Sergio Baranzini, Bruce A. C. Cree, Eduardo Caverzasi, Richard Cuneo, Stacy J. Caillier, Tiffany Cooper, Ari J. Green, Chu-Yueh Guo, Jeffrey M. Gelfand, Refujia Gomez, Sasha Gupta, Jill Hollenbach, Meagan Harms, Roland G. Henry, Stephen L. Hauser, Myra Mendoza, Jorge R. Oksenberg, Nico Papinutto, Sam Pleasure, Adam Santaniello, Joseph J. Sabatino, Jr., William A. Stern, Michael R. Wilson, and Scott Zamvil
Data Availability
The sequencing data reported in this study have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession GSE173790. The RNA-seq and BS-seq analysis steps are included in the SI Appendix.
References
- 1.International Multiple Sclerosis Genetics Consortium, Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Or A., et al. , Epstein-Barr virus in multiple sclerosis: Theory and emerging immunotherapies. Trends Mol. Med. 26, 296–310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mora J. R., Iwata M., von Andrian U. H., Vitamin effects on the immune system: Vitamins A and D take centre stage. Nat. Rev. Immunol. 8, 685–698 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsson T., Barcellos L. F., Alfredsson L., Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 13, 25–36 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Mendelson M. M., et al. , Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: A Mendelian randomization approach. PLoS Med. 14, e1002215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson S. Jr., Blizzard L., Otahal P., Van der Mei I., Taylor B., Latitude is significantly associated with the prevalence of multiple sclerosis: A meta-analysis. J. Neurol. Neurosurg. Psychiatry 82, 1132–1141 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Jacobs B. M., et al. , Gene-environment interactions in multiple sclerosis: A UK biobank study. Neurol. Neuroimmunol. Neuroinflamm. 8, e1007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranzini S. E., Oksenberg J. R., The genetics of multiple sclerosis: From 0 to 200 in 50 years. Trends Genet. 33, 960–970 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Multiple Sclerosis Genetics Consortium, Low-frequency and rare-coding variation contributes to multiple sclerosis risk. Cell 175, 1679–1687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen K., et al. , Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 26, 103–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao J., et al. , Genome-wide DNA methylation profile analysis identifies differentially methylated loci associated with ankylosis spondylitis. Arthritis Res. Ther. 19, 177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., et al. , Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCartney D. L., et al. , Epigenetic prediction of complex traits and death. Genome Biol. 19, 136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teschendorff A. E., et al. , Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 20, 440–446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves M. C., et al. , Methylation differences at the HLA-DRB1 locus in CD4+ T-cells are associated with multiple sclerosis. Mult. Scler. 20, 1033–1041 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Maltby V. E., et al. , Genome-wide DNA methylation profiling of CD8+ T cells shows a distinct epigenetic signature to CD4+ T cells in multiple sclerosis patients. Clin. Epigenetics 7, 118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maltby V. E., et al. , Genome-wide DNA methylation changes in CD19+ B cells from relapsing-remitting multiple sclerosis patients. Sci. Rep. 8, 17418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kular L., et al. , DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun. 9, 2397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewing E., et al. , Combining evidence from four immune cell types identifies DNA methylation patterns that implicate functionally distinct pathways during multiple sclerosis progression. EBioMedicine 43, 411–423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roostaei T., et al. , Impact of genetic susceptibility to multiple sclerosis on the T cell epigenome: Proximal and distal effects. bioRxiv [Preprint] (2020) 10.1101/2020.07.11.198721 (Accessed 12 July 2020). [DOI] [PMC free article] [PubMed]
- 21.Bos S. D., et al. , Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis. PLoS One 10, e0117403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barwick B. G., et al. , B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat. Commun. 9, 1900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scharer C. D., Barwick B. G., Guo M., Bally A. P. R., Boss J. M., Plasma cell differentiation is controlled by multiple cell division-coupled epigenetic programs. Nat. Commun. 9, 1698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulis M., et al. , Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat. Genet. 47, 746–756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scharer C. D., et al. , Epigenetic programming underpins B cell dysfunction in human SLE. Nat. Immunol. 20, 1071–1082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll M. C., Isenman D. E., Regulation of humoral immunity by complement. Immunity 37, 199–207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laidlaw B. J., Cyster J. G., Transcriptional regulation of memory B cell differentiation. Nat. Rev. Immunol. 21, 209–220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gass J. N., Gifford N. M., Brewer J. W., Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277, 49047–49054 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Medvedeva Y. A., et al. , FANTOM Consortium, EpiFactors: A comprehensive database of human epigenetic factors and complexes. Database (Oxford) 2015, bav067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatta M., Cirillo L. A., Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J. Biol. Chem. 282, 35583–35593 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Zamvil S., et al. , T-cell clones specific for myelin basic protein induce chronic relapsing paralysis and demyelination. Nature 317, 355–358 (1985). [DOI] [PubMed] [Google Scholar]
- 32.Fletcher J. M., Lalor S. J., Sweeney C. M., Tubridy N., Mills K. H., T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin. Exp. Immunol. 162, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zamvil S. S., et al. , Encephalitogenic T cell clones specific for myelin basic protein. An unusual bias in antigen recognition. J. Exp. Med. 162, 2107–2124 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baecher-Allan C., Kaskow B. J., Weiner H. L., Multiple sclerosis: Mechanisms and immunotherapy. Neuron 97, 742–768 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Dendrou C. A., Fugger L., Friese M. A., Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15, 545–558 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Comi G., et al. , Expert Panel of the 27th Annual Meeting of the European Charcot Foundation, Role of B cells in multiple sclerosis and related disorders. Ann. Neurol. 89, 13–23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamvil S. S., Hauser S. L., Antigen presentation by B cells in multiple sclerosis. N. Engl. J. Med. 384, 378–381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kappos L., et al. , Ocrelizumab in relapsing-remitting multiple sclerosis: A phase 2, randomised, placebo-controlled, multicentre trial. Lancet 378, 1779–1787 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Hauser S. L., et al. , OPERA I and OPERA II Clinical Investigators, Ocrelizumab versus interferon beta-1a in relapsing multiple sclerosis. N. Engl. J. Med. 376, 221–234 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Hauser S. L., et al. , ASCLEPIOS I and ASCLEPIOS II Trial Groups, Ofatumumab versus teriflunomide in multiple sclerosis. N. Engl. J. Med. 383, 546–557 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Machado-Santos J., et al. , The compartmentalized inflammatory response in the multiple sclerosis brain is composed of tissue-resident CD8+ T lymphocytes and B cells. Brain 141, 2066–2082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankoti J., et al. , In multiple sclerosis, oligoclonal bands connect to peripheral B-cell responses. Ann. Neurol. 75, 266–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serafini B., Rosicarelli B., Magliozzi R., Stigliano E., Aloisi F., Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 14, 164–174 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramesh A., et al. , University of California, San Francisco MS-EPIC Team, A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 117, 22932–22943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teperek-Tkacz M., Pasque V., Gentsch G., Ferguson-Smith A. C., Epigenetic reprogramming: Is deamination key to active DNA demethylation? Reproduction 142, 621–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y., et al. , DNA cytosine and methylcytosine deamination by APOBEC3B: Enhancing methylcytosine deamination by engineering APOBEC3B. Biochem. J. 471, 25–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schutsky E. K., Nabel C. S., Davis A. K. F., DeNizio J. E., Kohli R. M., APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic Acids Res. 45, 7655–7665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ise W., et al. , The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12, 536–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emslie D., et al. , Oct2 enhances antibody-secreting cell differentiation through regulation of IL-5 receptor alpha chain expression on activated B cells. J. Exp. Med. 205, 409–421 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ochiai K., Muto A., Tanaka H., Takahashi S., Igarashi K., Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int. Immunol. 20, 453–460 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Shinnakasu R., et al. , Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17, 861–869 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Nutt S. L., Fairfax K. A., Kallies A., BLIMP1 guides the fate of effector B and T cells. Nat. Rev. Immunol. 7, 923–927 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Turner C. A. Jr., Mack D. H., Davis M. M., Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77, 297–306 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Bönelt P., et al. , Precocious expression of Blimp1 in B cells causes autoimmune disease with increased self-reactive plasma cells. EMBO J. 38, e100010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cunill V., et al. , Relapsing-remitting multiple sclerosis is characterized by a T follicular cell pro-inflammatory shift, reverted by dimethyl fumarate treatment. Front. Immunol. 9, 1097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiSano K. D., Gilli F., Pachner A. R., Memory B cells in multiple sclerosis: Emerging players in disease pathogenesis. Front. Immunol. 12, 2002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novi G., et al. , Tailoring B cell depletion therapy in MS according to memory B cell monitoring. Neurol. Neuroimmunol. Neuroinflamm. 7, e845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this study have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession GSE173790. The RNA-seq and BS-seq analysis steps are included in the SI Appendix.