Significance
Several studies on the impact of yoga and meditation on mental and physical health have demonstrated beneficial effects. However, the potential molecular mechanisms and critical genes involved in this beneficial outcome have yet to be comprehensively elucidated. This study identified and characterized the transcriptional program associated with advanced meditation practice, and we bioinformatically integrated various networks to identify meditation-specific core network. This core network links several immune signaling pathways, and we showed that this core transcriptional profile is dysfunctional in multiple sclerosis and severe COVID-19 infection. Very importantly, we demonstrated that the meditative practice enhanced immune function without activating inflammatory signals. Together, these results make meditation an effective behavioral intervention for treating various conditions associated with a weakened immune system.
Keywords: meditation, immune, Isha yoga, Inner Engineering, COVID-19
Abstract
The positive impact of meditation on human well-being is well documented, yet its molecular mechanisms are incompletely understood. We applied a comprehensive systems biology approach starting with whole-blood gene expression profiling combined with multilevel bioinformatic analyses to characterize the coexpression, transcriptional, and protein–protein interaction networks to identify a meditation-specific core network after an advanced 8-d Inner Engineering retreat program. We found the response to oxidative stress, detoxification, and cell cycle regulation pathways were down-regulated after meditation. Strikingly, 220 genes directly associated with immune response, including 68 genes related to interferon signaling, were up-regulated, with no significant expression changes in the inflammatory genes. This robust meditation-specific immune response network is significantly dysregulated in multiple sclerosis and severe COVID-19 patients. The work provides a foundation for understanding the effect of meditation and suggests that meditation as a behavioral intervention can voluntarily and nonpharmacologically improve the immune response for treating various conditions associated with excessive or persistent inflammation with a dampened immune system profile.
Yoga and meditation are holistic disciplines that integrate both mental and physical methods for human well-being (1). These practices are growing in popularity worldwide, and according to a recent national health survey, 14% of the adult United States population used yoga or meditation within the previous year (2). Several studies have demonstrated multiple health benefits from such methods (3, 4), including reduced stress (5–8), anxiety (5, 7, 9, 10), fatigue (5, 11), depression (5, 9, 12), chronic pain (13–15), and disease severity for inflammatory bowel disease (16, 17) and cardiovascular disease (6, 18, 19). However, the mechanisms responsible for these improvements are poorly understood. These parameters are typically measured with self-reported surveys before and after meditation interventions, and such an approach may be prone to bias and subjectivity. Several studies on meditative practices have, however, shown changes in gene expression levels, demonstrating that these methods may benefit physiology at its most fundamental level (20, 21). For example, utilizing microarray technology to study the transcriptomic effects of six individuals of 38 y of twice-daily transcendental meditation practice found 200 genes differentially expressed (DE) (22). Similarly, studying the methylome of peripheral blood mononuclear cells in 17 experienced meditators after a day of intensive meditation revealed 61 differentially methylated regions (23). Studies focusing on the impact of meditation for treating irritable bowel syndrome, inflammatory bowel disease, and hypertension have observed that several genes related to fundamental pathways to be DE (24, 25). Together, several previous studies provide strong evidence for the beneficial effects of meditation by modulating the basic cellular pathways. Nevertheless, most of the previous studies are 1) cross-sectional studies (evaluating only one time point) (26–30), 2) done on highly experienced meditators (26, 27), 3) small sample sized (26–28, 30, 31), 4) tested on handpicked nonspecific biomarkers (29, 30), and 5) confounded with different lifestyle and diet (26, 29, 30).
To understand the meditative effect and to overcome these limitations, 1) we applied unbiased gene expression analyses on four time points before and after the intensive 8-d Samyama meditation (an advanced Inner Engineering program attended by ∼20,000 participants to date), and 2) we analyzed the transcriptomic changes from 388 samples obtained from 106 individuals after a residential meditation retreat including a vegan diet at the Isha Institute of Inner Sciences (McMinnville, TN). Participants spent 8 d in complete silence with more than 10 h of meditation per day. We reasoned that improvement in multiple physical and mental health conditions by this meditative practice would likely reflect significant differences in intrinsic transcriptional networks, rather than changes in expression of a few individual genes. We applied a multistaged approach to characterize the coexpression, transcriptional, and protein–protein interaction (PPI) networks associated with meditation, and we validated several network predictions using multiple approaches.
Results
Blood-based Genomic Analysis Reveals Large-Scale Genetic Changes Linked to Intense Meditation.
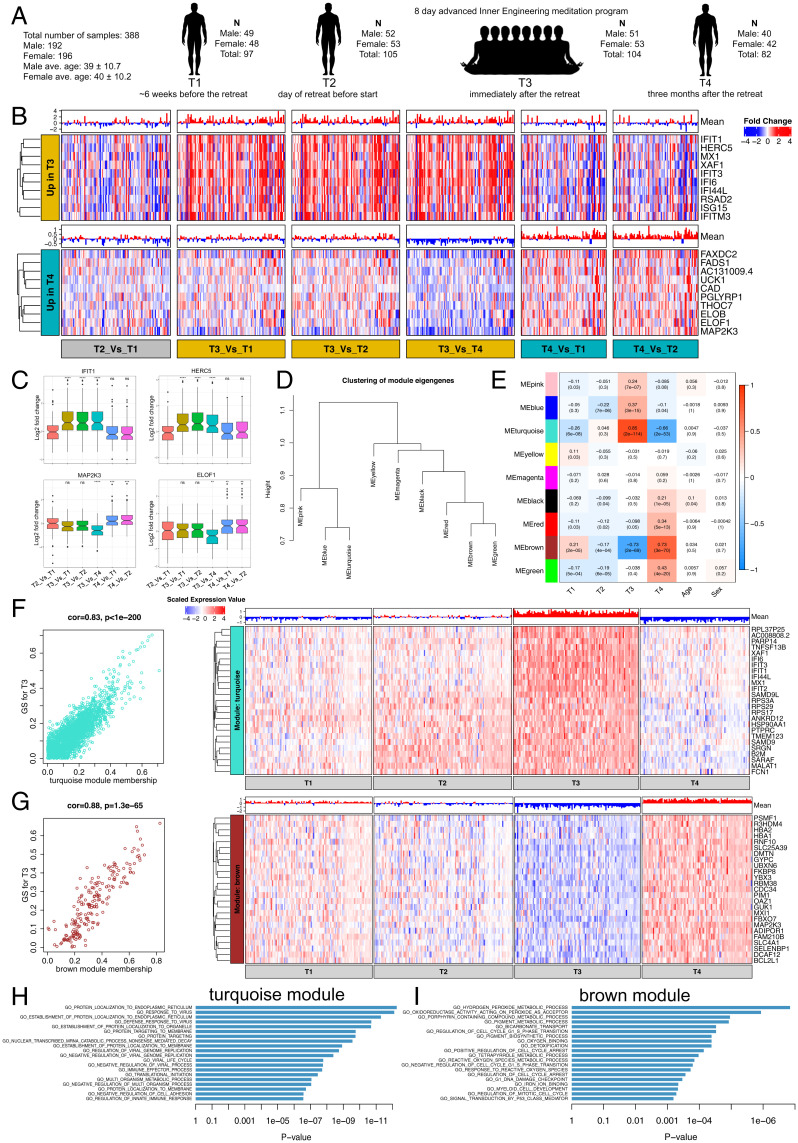
The Samyama retreat was conducted in April 2018 at the Isha Institute of Inner Sciences (McMinnville, TN) with the requirement to remain entirely silent for 8 d, with more than 10 h of meditation per day, a strict vegan diet, and a regular sleep–wake cycle. The study exclusion criteria included inability to read or comprehend the consent form; subjects with severe anemia; active use of marijuana, opioids, or related drugs; use of antibiotics or probiotic/prebiotic supplements within 60 d of enrollment; and participants living outside of the country. Blood specimens (196 from female and 192 from male) were collected at four different times from participants with an average of 40 y of age for both sexes (Fig. 1A). In total, we characterized the transcriptome profiles of whole-blood cells from 388 specimens obtained from 106 individuals before and after the meditation retreat at four time points (T1 to T4) by RNA sequencing (RNA-Seq). The T1 samples were collected 5 to 8 wk before the retreat, T2 samples were collected on the day of retreat before starting the meditation method, T3 samples were collected immediately after the retreat, and T4 samples were collected 3 mo after the retreat (T1 = 97, T2 = 105, T3 = 104, T4 = 82; N = 388) (Fig. 1A). First, we performed cellular deconvolution to identify cell types and examine their heterogeneity in composition between the time points (32). Interestingly, out of 22 different blood cell types, we observed higher neutrophil and lower CD4 and CD8 T cell relative proportions at T3 than other time points (SI Appendix, Fig. S1).
Fig. 1.
Coexpression network analysis of whole-blood transcriptome profile changes after meditation. (A) Longitudinal blood collection at four time points (T1 to T4) from advanced Inner Engineering meditation program participants. (B) Heatmaps are depicting the expression of the top ten genes (rows) across samples (columns) for six different time point comparisons (red corresponds to gene up-regulation and blue to down-regulation). Mean gene expression levels are shown as a bar plot. (Top and Bottom) The top 10 genes up-regulated at T3 (after meditation) and T4 (3 mo after meditation) are shown. (C) Boxplot represents the variability in the expression levels of the top two genes up-regulated at T3 (Top) and T4 (Bottom). (D) Hierarchical clustering of WGCNA module eigengenes. (E) Module-trait associations. Each row corresponds to a module eigengene and column to a trait. Each cell contains the corresponding correlation and P value. Red and blue color denote positive and negative correlation with gene expression, respectively. (F and G) A scatter plot of gene significance (GS) for T3 versus the module membership (MM) in the turquoise (Top) and brown (Bottom) module. Intramodular analysis of the genes found in these two modules contains genes that have a high correlation with meditation, with P < 1.3e−65 and correlation >0.8. Heatmaps are depicting the expression of top 25 hub genes (rows) across samples (columns) for four time points (red corresponds to gene up-regulation and blue to down-regulation) in the turquoise (Top) and brown (Bottom) module. Mean gene expression levels are shown as a bar plot. Hub genes have a high GS and MM value with T3. (H and I) GSEA of gene ontology terms calculated by Fisher's exact test for the turquoise and brown module.
To study transcriptome changes linked to blood cells, we eliminated any confounding effects associated with this cell type heterogeneity. We used the cell type proportion estimates to adjust the data for all analyses (see Materials and Methods). Next, we performed differential expression analyses on batch and cell type composition corrected data in all four time points (T1, T2, T3, T4) (see Materials and Methods) by pairwise comparison of all six permutation combinations and identified 1,649 genes DE in total (Dataset S1). We observed distinct early (T3) versus late (T4) genetic alterations in blood cells “after” meditation. Out of 1,649 DE genes, we observed almost 44% (719) of DE genes exclusively in the time point immediately after meditation (T3), followed by 30% (496) differentially expressed at the 3-mo follow-up (T4) compared with premeditation time points (T1 or T2). We observed 116 genes DE only in T3 (Dataset S1), and the top 10 DE genes (IFIT1, HERC5, MX1, XAF1, IFIT3, IFI6, IFI44L, RSAD2, ISG15, IFITM3) in T3 alone were directly related to immune response including antiviral defense function (Fig. 1 B and C). This suggests a previously unknown influence of meditation intervention on the genetic make-up of blood cells with the early wave–associated immune-related changes (Dataset S1 and Fig. 1B). We also observed long-term effects due to meditation; 58 genes were DE only in T4 (3 mo after meditation), which consisted of several up-regulated genes involved in catalytic activity (CAD, FADS1, FAXDC2, MAP2K3, PGLYRP1, and UCK1), localized to the mitochondrial membrane (CISD2, NDUFAF1, and MRPS18A), and involved in translation elongation factor activity (ELOF1 and ELOB) (Fig. 1 B and C). Together, these findings suggest that meditation has an immediate impact on immune cells and genes, which are transient in nature and followed by a dynamically altered effect observed 3 mo after the meditation retreat.
Gene Coexpression Network Analysis Identifies Modules Associated with Meditation.
We next performed weighted gene coexpression network analysis (WGCNA) (33, 34), a powerful method for understanding the modular network structure of the transcriptome, to organize the meditation-related transcriptional changes in an unbiased manner. WGCNA permits the identification of modules of highly coexpressed genes whose grouping reflects shared biological functions and key functional pathways and key hub genes within the modules and has been widely applied to understanding disease-related transcriptomes (35, 36). Using WGCNA (34, 36), we identified nine robust coexpression modules (Fig. 1 D and E and Dataset S2) for the datasets generated at four time points before and after meditation. Based on the significant module-trait (time points) relationships, we identified two modules (turquoise and brown) strongly associated with T3 (Fig. 1 D and E). To further validate turquoise and brown modules, we calculated gene significance (correlation of a gene expression profile with a sample trait) with T3 and observed significant correlation (Fig. 1 F and G). Based on the eigengene-based intramodular connectivity (module membership), we observed that genes present in the turquoise module were significantly up-regulated and genes present in the brown module were significantly down-regulated following meditation (Fig. 1 H and I). These two modules with a high correlation between gene significance and module membership could represent pathways associated with meditation. Gene ontology enrichment analysis revealed that several categories related to immune function, protein targeting, and localization were up-regulated at T3. The up-regulated RNA coexpression network (turquoise module) included many previously identified genes known to regulate the immune system and related pathways, including 220 genes DE after meditation, which are directly related to the immune response (Dataset S3). This list included 68 genes related to interferon (IFN) signaling. Interestingly, the top 10 hub genes in the up-regulated module included well-known genes directly associated with type I IFN signaling pathway, namely IFI6, IFIT1, IFIT2, IFIT3, IFI44L, MX1, and XAF1 (Fig. 1F). The down-regulated coexpression module was enriched for hydrogen peroxide, detoxification, and cell cycle process, including glutathione peroxidase 1 (GPX1) and peroxiredoxin 2 and 5 (PRDX2 and PRDX5) (Dataset S3). GPX1 catalyzes the reduction of hydrogen peroxide by glutathione and thereby protects cells against oxidative damage. PRDX2 and PRDX5 play a role in cell protection against oxidative stress by detoxifying intracellular peroxides. These results suggest that meditation leads to differential expression of several genes related to oxidative stress, in agreement with a previous study (37). The down-regulated module also included genes regulating the cell cycle, namely BCL2-like 1 (BCL2L1), F-box protein 7 (FBXO7), Pim-1 proto-oncogene serine/threonine kinase (PIM1), and RNA binding motif protein 38 (RBM38). BCL2L1 is known to contribute to programmed cell death (38); PIM1 is known to control cell growth, differentiation, and apoptosis (39); RBM38 is shown to regulate cell cycle arrest (40); and FBXO7 is shown to activate cell cycle regulators (41). Down-regulation of these genes suggests that meditation may exert effects on cell cycle regulation through transcriptional regulation.
Coexpressed Genes after Meditation Are Not Influenced by Diet and Circadian Changes.
Diet has been shown to influence gene expression levels directly (42, 43). Since this meditation retreat required about 2 mo of vegan diet before the retreat, we examined whether this diet regime influenced the gene expression changes. First, we compared the expression levels of the top 25 vegetarian diet–specific genes (43) in our samples before and after meditation for the time point comparisons, T3 (after meditation) versus T2 (before meditation) showed no significant changes across samples (SI Appendix, Fig. S2). Next, based on the survey data from these participants, a vegan diet was reported in 90.3% (176/195) of participants at T2 (44). By looking at the number of genes that are overlapping between two comparisons, T2 versus T1 and T2 versus T3, we found only 12 genes in common (Dataset S1). Hence, the majority of gene expression changes we observed at T3 are not due to the vegan diet since T1 or T2 versus T3 changes were not observed in the T1 versus T2 comparison. To examine whether the gene expression changes after meditation were influenced by the time of sample collection and sleep pattern, we examined the expression levels of all the genes involved in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway circadian rhythm (hsa04710) before and after meditation. We observed no significant changes across samples (SI Appendix, Fig. S3). Together, these results suggest most gene expression changes at T3 (after meditation) are due to meditation intervention with minimal effect due to the diet or the time of sample collection.
Transcriptional Regulatory Network Analyses Identify Critical Drivers Associated with Meditation.
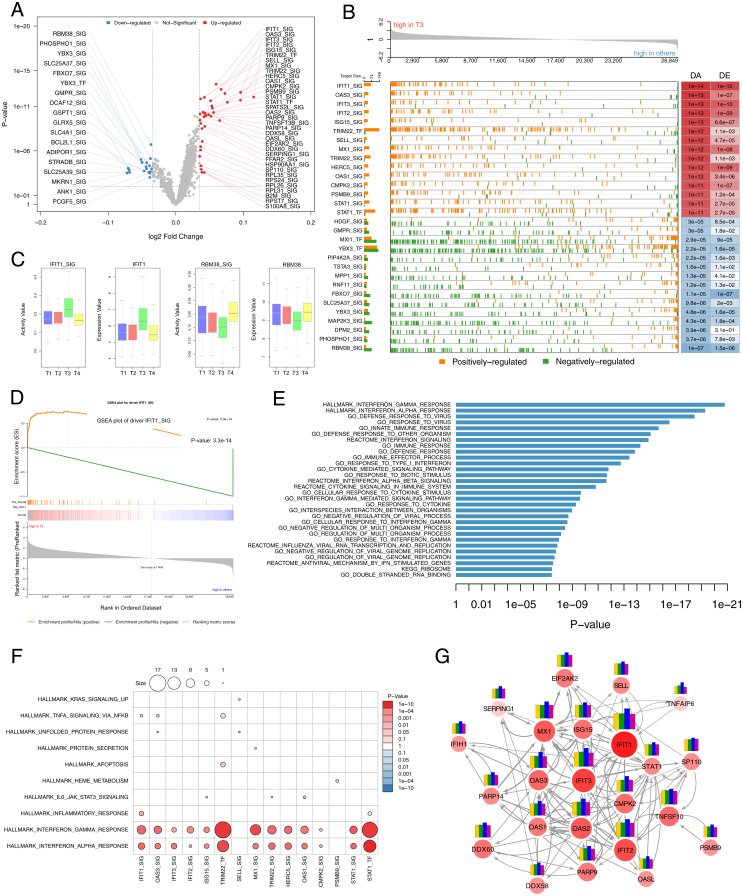
To identify critical regulators related to meditation, we performed NetBID (Network-based Bayesian Inference of Drivers) analysis, a data-driven network-based Bayesian inference that identifies key drivers (transcriptional factors [TFs] or signaling factors) in a given transcriptome (45). First, we reverse engineered a meditation-specific regulatory network using the SJARACNe (scalable solution of ARACNe algorithm–Algorithm for the Reconstruction of Accurate Cellular Networks) algorithm (46). We then applied the activity inference algorithm in NetBID to identify drivers whose network activity is significantly different before versus after meditation. We identified 90 drivers with significant differential activity (DA) compared with T3 versus other time points (Fig. 2A and Dataset S4). To interpret the significance of these drivers we evaluated the expression pattern of their targets by plotting them on a ranked gene list (Fig. 2B). We observed that positively regulated target genes of the up-regulated DA drivers tended to have higher fold-change expression values in T3 compared with other time points. We observed the opposite pattern for the targets regulated by the down-regulated DA drivers, validating the significance of these drivers and their targets (Fig. 2 B–D). Next, we performed gene set enrichment analysis (GSEA) against the collection of annotated gene sets from MSigDB (Molecular Signatures Database) (47) to elucidate the functional relevance of these drivers. Strikingly, we observed several functional categories associated with immune function significantly enriched for up-regulated drivers (Fig. 2E and Dataset S5). Among the top enriched functional categories included IFN-alpha and gamma response and other direct immune-related categories (Fig. 2E). Next, we examined the enriched functional categories associated with the target genes of these drivers utilizing the hallmark gene sets from MSigDB. Remarkably, we observed that the targets of these drivers were significantly enriched in IFN-alpha and gamma response gene sets (Fig. 2F). We also observed 24 of these up-regulated DA drivers were also present within the hallmark IFN-alpha and gamma response gene sets from MSigDB (Fig. 2G). These 24 drivers included IFN-induced protein with tetratricopeptide repeats (IFIT1, IFIT2, and IFIT3), IFN-stimulated gene 15 (ISG15), and IFN-induced MX dynamin-like GTPase 1 (MX1) with known antiviral activity. MX1 gene protects mice against influenza strains pandemic 1918 and the highly lethal human H5N1 (48). Other drivers included IFN-induced production of the 2′-5′-oligoadenylate synthetase family (OAS1, OAS2, and OAS3) and DExD/H-box helicase 58 and 60 (DDX58 and DDX60). RNA helicases DDX58 are known to activate kinases, which phosphorylate the IFN regulatory factor IRF7 to induce IFN-alpha and IFN-beta IFNs (49, 50). Similarly, DDX60 is known to positively regulate IFIH1-dependent type I IFN signaling (51). Together, these data suggest that immune function pathways (particularly IFN related) are enhanced immediately after meditation.
Fig. 2.
Transcriptional regulatory network analyses identify critical signaling pathways and drivers associated with meditation. (A) Volcano plot displaying top drivers [TFs or signaling factors (SIG)] with significant DA in T3 when compared with other time points. The red dots represent the up-regulated drivers; the green dots represent the down-regulated drivers after meditation. (B) GSEA plot shows the significance of top DA drivers with their expression and regulation of their target genes. Positively regulated target genes (orange) of the up-regulated DA drivers (red) tend to have higher fold-change expression values in T3 than other time points. The opposite pattern is observed for the targets regulated by the down-regulated DA drivers (blue). The number of target genes for each driver (Left) and DA and DE values (Right) are shown. (C) Boxplot representation of the variability in the activity (Left) and expression (Right) values of top driver up-regulated (IFIT1) and down-regulated (RBM38) after meditation. (D) GSEA enrichment plot of top driver IFIT1 shows significant enrichment for positively regulated target genes with higher fold-change expression values in T3 than other time points. (E) Biological function enrichment of top DA drivers. Bar charts show significantly enriched gene sets in the gene MSigDB database for the top drivers. (F) Biological function enrichment of the target genes of top 15 DA drivers against curated hallmark gene sets from MSigDB. (G) Subnetwork of the reverse-engineered meditation-specific regulatory network showing up-regulated DA drivers directly associated with IFN-alpha and gamma response. Nodes correspond to genes and edges to mutual information (MI). Edge size corresponds to MI value. Larger nodes correspond to differential expression (Z-score). Node color represents up-regulation (red) or down-regulation (blue). Bar plot depicting the gene expression levels at different time points (Left to Right; T1 to T4), showing consistent elevation at T3 for all drivers.
Transcription Factor Binding Site Enrichment in Meditation-Specific Coexpression Modules.
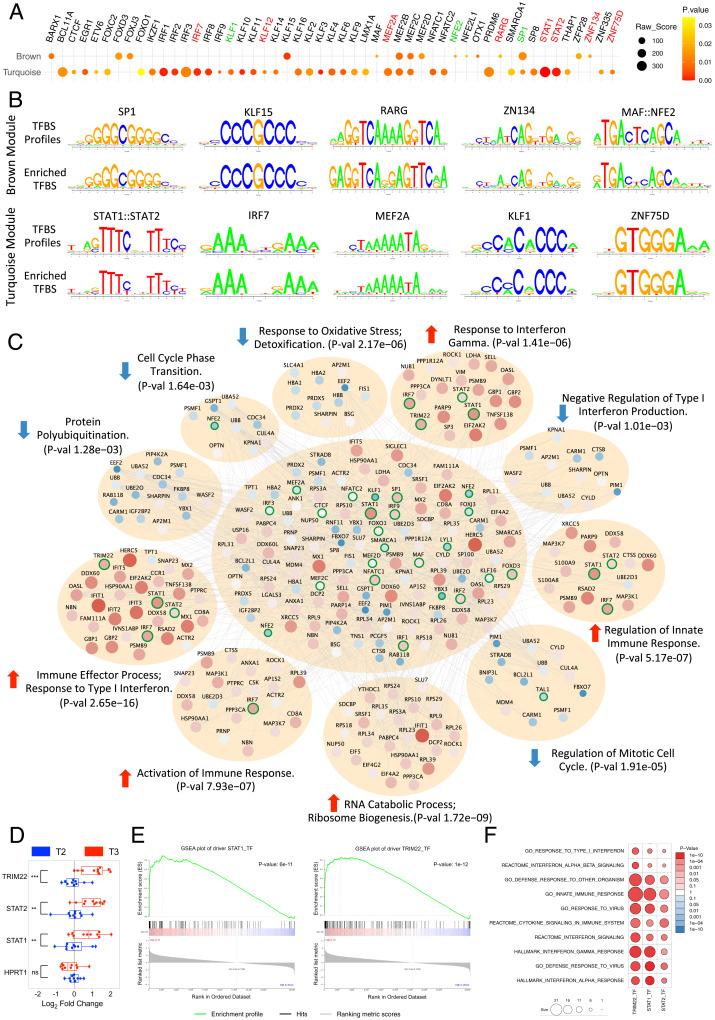
From network analyses, we observed 220 genes directly associated with immune response in the turquoise module (the major up-regulated module after meditation), which contained 68 genes related to IFN signaling including the 24 IFN driver genes (Fig. 2G). To uncover the potential regulatory network contributing to the consistent coexpression of multiple immune-related genes after meditation, we performed sequence-based TF-binding site (TFBS) enrichment analysis for both turquoise and brown coexpression modules (see Materials and Methods). To avoid confounders and identify the most statistically robust sites, we used three different background datasets (1,000-bp sequences upstream of all human genes, human CpG islands, and the human chromosome 20 sequence). We identified 51 TFs (see Materials and Methods) significantly enriched in these two coexpression modules (Fig. 3 A and B and Dataset S6). Remarkably, 6 IRF genes were over-represented in the turquoise module, the major up-regulated module after meditation (Fig. 3A). Next, we examined each TF’s association with IFN signaling from published literature by testing association with the keywords “interferon signaling” or “interferon pathway” in the PubMed database for every TF (see Materials and Methods). This analysis identified 16 TFs (31%) that were strongly associated with IFN signaling, which included signal transducer and activator of transcription 1 and 2 (STAT1, STAT2) and several IFN genes: IRF1, 2, 3, 7, 8, and 9 (Fig. 3A and Dataset S7). Notably, three of these over-represented TFs (STAT1, STAT2, and IRF7) are present in the turquoise module, the major up-regulated module after meditation (Fig. 3 A and B and Dataset S6), suggesting their critical role in activating the enriched pathways in the turquoise module. Interaction of IRFs with STAT1-STAT2 heterodimers or with STAT2 homodimers in response to IFNs redirects these complexes to a distinct group of target genes harboring the IFN-stimulated response element to regulate their expression levels involved in IFN signaling (52). Together, these results suggest critical involvement of these TFs in IFN signaling after meditation.
Fig. 3.
TFBS enrichment and convergent pathway analyses. (A) Dot plot showing 51 enriched TFs for the brown (Top) and turquoise (Bottom) module. The dot's size corresponds to the raw enrichment score obtained from the Clover algorithm, and the color of the dot corresponds to enrichment significance. Highlighted names of the over-represented TFs denote their presence in the brown (green) or turquoise (red) module. (B) Sequence logo plots of reference TFBS profiles (JASPAR/HOCOMOCO) and identified position weight matrix for selected TFs significantly over-represented and present in the brown (Top) or turquoise (Bottom) module. (C) PPI network represented by the genes from both turquoise and brown modules along with over-represented TFs and all drivers with significant DA. Significantly enriched functional pathways (corrected P value < 0.05 from Fisher's exact test) in the PPI network are shown. Nodes correspond to genes and edges to PPI. Larger nodes correspond to differential expression (Z-score). Node color represents up-regulation (red) or down-regulation (blue). Nodes with highlighted border (green) correspond to enriched or driver TFs. (D) Quantitative PCR validation of three critical up-regulated TFs after meditation. Boxplot representation of the expression levels of three critical TFs from 10 random individuals for each time point. Expression levels of HPRT1 are shown as endogenous control. Paired t test was performed to calculate the statistical significance (**P ≤ 0.01, ***P ≤ 0.001). (E) GSEA enrichment plot of STAT1 and TRIM22 showing significant enrichment for positively regulated target genes with higher fold-change expression values in T3 compared with other time points. (F) Biological function enrichment of the target genes of STAT1, STAT2, and TRIM22 against curated gene sets from MSigDB showing enrichment for several gene sets related to immune and IFN pathways.
Coregulated Genes after Meditation Represent Convergent Pathways.
To extend this work to the level of specific proteins and identify potential conserved protein-functional pathways represented by the meditation coexpression modules, we determined the PPI network represented by the genes in both turquoise and brown modules (see Materials and Methods). We reasoned that this would not only provide independent validation of the relationships inferred by RNA coexpression and regulatory networks but that the PPIs would allow us to dissect significant functional pathways impacted by meditation. We screened experimentally validated PPIs among all possible combinations of gene pairs present in the coexpressed modules, over-represented TFs (Fig. 3A), and all drivers with significant DA (Fig. 2A), obtaining a PPI network consisting of 143 unique nodes and 495 edges (Fig. 3C). We observed enrichment of critical functional pathways related to the response to oxidative stress, detoxification, regulation of cell cycle, protein polyubiquitination, and negative regulation of type I IFN production to be down-regulated after meditation (Fig. 3C). Likewise, we also observed enrichment of several critical functional pathways that contribute to the immune function to be up-regulated after meditation, which included immune effector process, response to type I IFN, RNA catabolic process, activation of immune response, regulation of innate immune response, and response to IFN-gamma (Fig. 3C). These results validate the network-based approaches, as immune-related pathways were consistently enriched in coexpression, regulatory, and PPI network analyses. By integrating these three different network analyses, we found three critical TFs—STAT1, STAT2, and TRIM22 (tripartite motif–containing 22)—that are DE after meditation (Fig. 3D) and regulate several genes that are enriched for gene sets related to IFN signaling (Fig. 3 E and F). All three TFs are known to be IFN-induced and play a role in restricting infection from diverse viruses by activating immune function (53, 54). Together, these results suggest that meditation as a behavioral intervention could have important implications for targeting diseases with impaired IFN response.
Comparison of Transcriptional Profiles Obtained from COVID-19 and Multiple Sclerosis Patient Samples.
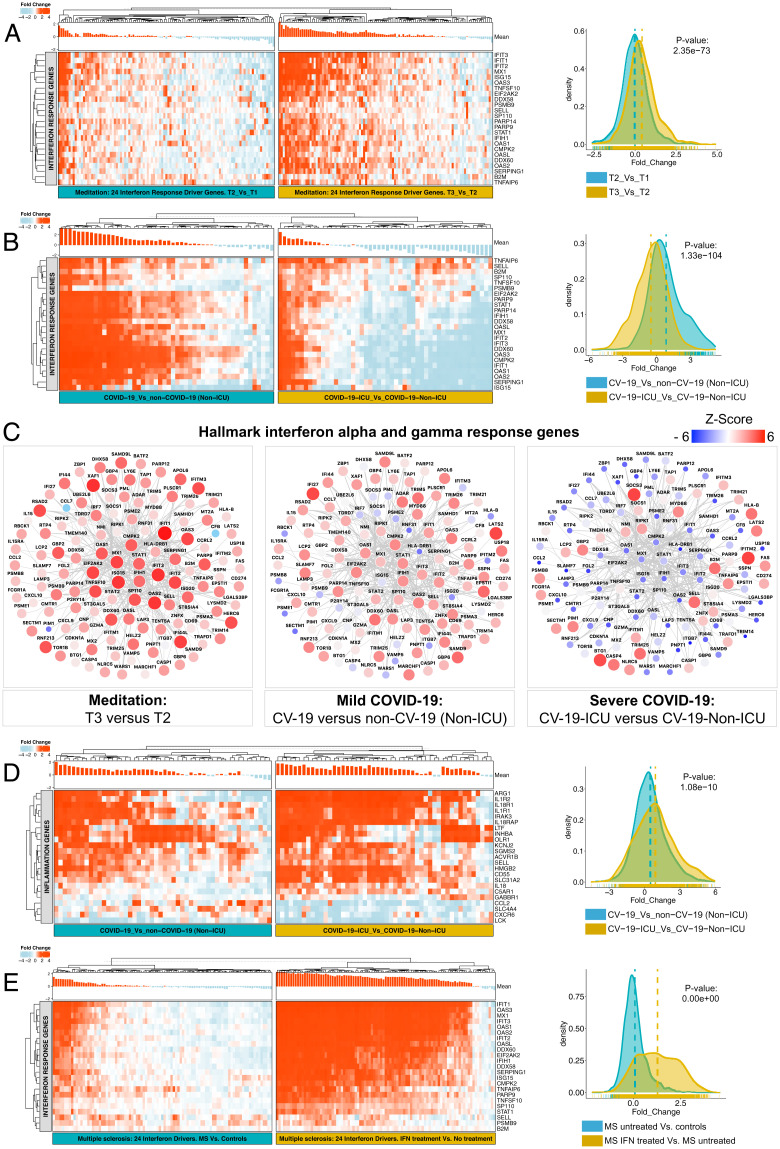
Impaired IFN response and exacerbated inflammatory response are generally observed in viral infections and are the hallmark of severe SARS-CoV-2 infection (55). Hence, we compared the transcriptional network obtained from a publicly available dataset of leukocytes from COVID-19 patients with our meditation dataset to directly examine their expression pattern. First, we examined the expression levels of well-established viral infection marker genes (PTX3, CXCL10, CD46) in both datasets. As expected, we observed up-regulation of these marker genes in the COVID-19 dataset, but not in the meditation dataset (SI Appendix, Fig. S4). Next, we examined the expression levels of 24 meditation-associated DA drivers (Fig. 2G), which are hallmark IFN-alpha and gamma response genes. The comparison of T3 (after meditation) versus T2 (before meditation) revealed significant up-regulation of these drivers when compared with T2 versus T1 (baseline) (Fig. 4A). The comparison of COVID-19 patients [non-intensive care unit (ICU)] versus non-COVID-19 patients (non-ICU) also revealed up-regulation of several of these drivers (Fig. 4B). Strikingly, the comparison of severe COVID-19 patients (ICU) versus mild COVID-19 patients (non-ICU) revealed down-regulation of these drivers (Fig. 4B). This suggests impairment of these 24 critical drivers regulating IFN signaling in severe COVID-19 patients, which are elevated after meditation. Next, we examined the expression of the targets of these 24 drivers, which are also part of the hallmark IFN-alpha and gamma response gene sets from MSigDB. For this analysis, we merged the reverse-engineered regulatory network generated for both COVID-19 and meditation datasets to examine the changes. We observed that 97% of these genes were up-regulated after meditation, 76% of these genes were up-regulated in mild COVID-19 patients, and strikingly, only 31% of IFN response genes were up-regulated in severe COVID-19 patients (for whom the majority of the genes were down-regulated) (Fig. 4C). We also found eight up-regulated DA drivers (ARG1, CD55, HMGB2, IL18R1, IL1R1, IL1R2, IRAK3, LTF) associated with cytokine and inflammation signaling in COVID-19 regulatory network. By examining the expression of the targets of these drivers present in the hallmark inflammatory response gene set from MSigDB, we observed significant up-regulation of these inflammation-related genes in severe COVID-19 patients compared with mild COVID-19 patients (Fig. 4D). The comparison of T1 (baseline), T2 (before meditation), and T3 (after meditation) revealed no significant changes in these inflammation-related genes (SI Appendix, Fig. S5A). Previous studies have shown that exercise also improves immune competency (56). By comparing the whole-blood transcriptome datasets of exercise training with the meditation dataset, we did not observe up-regulation of the 24 IFN driver genes after exercise training suggesting a unique gene signature associated with meditation (SI Appendix, Fig. S5 B and C). These results suggest that a unique gene signature associated with meditation enhances immune function without activating inflammatory signals impaired in severe COVID-19 patients.
Fig. 4.
Comparison of IFN and inflammatory response in meditation, COVID-19– and MS-specific transcriptional profiles. (A) Heatmaps are depicting the expression of the 24 IFN response driver genes (rows) across meditation samples (columns) for two different time point comparisons, T2 (before meditation) versus T1 (baseline) and T3 (after meditation) versus T2. (B) Heatmaps comparing the expression of the 24 IFN driver genes in COVID-19 samples for two different comparisons: non-ICU COVID-19 patients versus non-ICU non-COVID-19 patients (Left, mild patients) and ICU COVID-19 patients versus non-ICU COVID-19 patients (Right, severe patients). (C) Subnetwork of the reverse-engineered meditation-specific and COVID-19–specific regulatory network showing targets of the 24 IFN drivers directly associated with IFN-alpha and gamma response, displaying dramatic down-regulation in severe COVID-19 patients. Nodes correspond to genes and edges to MI. Larger nodes correspond to differential expression (Z-score). Node color represents up-regulation (red) or down-regulation (blue). (D) Heatmaps comparing the expression of the 23 inflammatory response genes in COVID-19 samples for two different comparisons as described in B, displaying significant up-regulation in severe COVID-19 patients. (E) Heatmaps comparing the expression of the 24 meditation-specific IFN driver genes in multiple sclerosis samples for two different comparisons: MS patients versus controls (Left) and MS patients after IFN treatment versus MS untreated patients (Right). For all heatmaps, the red color corresponds to gene up-regulation and blue to down-regulation. Mean gene expression levels are shown as a bar plot on top of each heatmap. All the heatmaps are supplemented with density plots showing the distribution of log2 fold change, and the significance of the variability in the expression levels between the two groups are calculated by a two-sample t test.
Next, we compared the transcriptional changes in a publicly available multiple sclerosis (MS) dataset. MS is characterized by autoimmune inflammation and subsequent neuronal degeneration. IFN treatment was the first disease-modifying therapy available to treat MS, preventing and reducing relapse rates and reducing new brain lesions, progression, and cognitive loss (57). Hence, we examined the expression levels of 24 meditation-associated IFN response driver genes in the whole-blood gene expression dataset obtained from MS patients undergoing β-IFN therapy. Firstly, we observed that the up-regulated meditation-associated IFN response driver genes were significantly down-regulated in MS versus control comparison (SI Appendix, Fig. S5D). Strikingly, the comparison of MS IFN-treated versus MS untreated patients revealed dramatic up-regulation of these drivers compared with MS untreated versus controls (Fig. 4E). These results revealed that up-regulation of these 24 critical drivers regulating IFN signaling in MS patients after IFN disease-modifying treatment is also elevated after meditation. Together, these results support the hypothesis that meditation contributes to improving multiple health conditions by regulating various critical pathways directly related to the disease pathogenesis.
Discussion
Yoga and meditation techniques have improved health outcomes and quality of life for patients with many disease conditions. No comprehensive study had previously been conducted to evaluate how meditation impacts biological processes that are directly involved in disease pathogenesis. We evaluated an advanced-level 8-d meditation practice and applied a comprehensive systems genomics approach, including gene expression profiling combined with multilevel bioinformatic analyses and validation of network predictions. We hypothesized that meditative practices would likely impact intrinsic transcriptional networks directly related to the disease pathogenesis rather than solely change the expression of a few individual genes. Pathways related to immune system activity are particularly relevant in this context, given that immune function has been implicated in several major health conditions, including asthma, cardiovascular disease, certain types of cancer, depression, metabolic disorders, MS, neurodegenerative disorders, obesity, osteoporosis, posttraumatic stress disorder, rheumatoid arthritis, and sepsis (58, 59). In this study, using an integrative approach we found several immune-related and other fundamental cellular pathways were altered after the meditation retreat. This work provides a key proof of principle for the power of the systems genomic approaches in tackling complex problems by demonstrating that meditation practices enhance gene networks associated with distinct pathways.
In this study, we uncovered an immunomodulatory function of meditation via the activation of IFN pathways and observed higher neutrophil relative proportions after meditation, which accompanied the elevated IFN pathway activity. Our gene coexpression network analysis identified up-regulated and down-regulated modules with a significant correlation with meditation. The up-regulated RNA coexpression network included many previously identified genes known to regulate the immune system and related pathways, which included IFN signaling. The IFN signaling pathway plays a critical role in immune function by triggering a complex regulatory system of innate and adaptive immune responses to defend against pathogens and diseases. Likely, countering a dysregulated immune system profile with meditation could theoretically function to improve health outcomes by enhancing immune defenses that protect against viral and bacterial infection and various diseases with diminished immune function. The down-regulated coexpression module showed enrichment for hydrogen peroxide, detoxification, and cell cycle process. These results suggest meditation differentially expresses several genes related to oxidative stress in agreement with a previous study (37). The down-regulated module also included genes regulating the cell cycle, suggesting that meditation may exert effects on cell cycle regulation through transcriptional regulation, in agreement with previous studies (20).
We also observed long-term effects due to meditation; 58 genes were DE only in T4 (3 mo after meditation), consisting of several up-regulated genes involved in catalytic activity, localized to the mitochondrial membrane, and involved in translation elongation factor. These results indicate meditation may exert long-term changes in the transcriptional profiles related to the most fundamental cellular pathways. Interestingly, most of the immune-related up-regulated genes at T3 were reversed at T4, suggesting acute but not chronic immune activation due to meditation. Chronic immune activation is known to cause immune hyporesponsiveness, anergy (60), and malignancy (61). We identified 90 drivers with significant DA, which regulates this robust acute immune activation after meditation. These drivers comprised 24 up-regulated DA drivers associated with IFN-alpha and gamma response signaling, including IFIT1-3, ISG15, MX1, OAS1-3, and DDX58 and 60. MX1 gene has been shown to protect mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses (48). DDX58 and DDX60 are known to regulate IRF7 and IFIH1 to induce IFN-alpha and beta signaling (49–51). Remarkably, IRF7 and IFIH1 are also up-regulated after meditation, and TFBS enrichment analyses also revealed an over-representation of 16 TFs strongly associated with IFN signaling, which included IRF7, suggesting an active role of IRF7 and IFIH1 in IFN signaling after meditation. Other TFs included STAT1, STAT2, and several IFN regulatory factor genes: IRF1, 2, 3, 7, 8, and 9. Interestingly, STAT1 and STAT2 are present in the turquoise module, the major up-regulated module after meditation, suggesting critical involvement of these TFs in IFN signaling after meditation.
By integrating PPIs with the identified networks, we characterized a core meditation-specific network highly enriched in critical pathways. From this core network analysis, we found three TFs—STAT1, STAT2, and TRIM22, which are DE after meditation—regulate several genes enriched for gene sets related to IFN signaling. It is well known that by interacting with their specific receptors, IFNs activate STAT complexes. This STAT-containing complex binds to an IFN-stimulated response element in IFN-stimulated gene promoters to driver type I IFN–stimulated transcriptional activation, which is an essential primary barrier for virus infection and for activation of innate and adaptive immune responses (53). Both STAT1- and STAT2-deficient mice were highly sensitive to diverse viruses and bacterial pathogens, including vesicular stomatitis virus, influenza virus, herpes simplex virus, dengue virus, and others, highlighting the importance of these TFs in antiviral immunity (53). Likewise, we found that the IFN-induced tripartite motif protein TRIM22 is known to play a role in the restriction of infection by diverse viruses, including HIV, encephalomyocarditis virus, influenza A virus, hepatitis B virus, and hepatitis C virus (54). Together, up-regulation of STAT1, STAT2, and TRIM22 by meditation may function as a host antiviral factor induced by IFNs to restrict infection by diverse viruses. Together, these results support the hypothesis that meditation contributes to improving multiple health conditions by regulating various critical pathways directly related to the disease pathogenesis.
We also showed that the meditative practice enhanced immune function without activating inflammatory signals. This suggests that meditation, as a behavioral intervention, may be an effective component in treating diseases characterized by increased inflammatory responsiveness with a weakened immune system. It has been shown that dysregulation of IFN response, coupled with exuberant inflammatory cytokine production, is the defining and driving feature of COVID-19 pathogenesis (55). It has been widely suggested that a preventive measure by the administration of IFNs to elicit a preexisting antiviral state may block viral infection at the very early stage. As of May 5, 2021, more than 40 clinical studies of various formulations of IFN treatment plans have been registered on clinicaltrials.gov for COVID-19. Interesting, early IFN treatment before peak viral replication protects mice from lethal severe acute respiratory syndrome coronaviruses (SARS-CoV) or the Middle East respiratory syndrome-coronavirus (MERS-CoV) challenge, whereas late IFN treatment failed to effectively inhibit virus replication and aggravated immunopathology (62, 63). Daily IFN-alpha nasal drops along with standard personal protective equipment were shown to protect at-risk health-care workers from COVID-19 over 28 d (64). These results demonstrate the benefits of early intervention and that activation of the IFN response can prevent susceptible healthy people from acquiring COVID-19. By comparing the reverse-engineered meditation-specific and COVID-19–specific regulatory network, we showed that the meditation-associated up-regulated IFN-related drivers and their targets were significantly down-regulated in severe COVID-19 patients. We also observed the opposite trend for cytokine and inflammation signaling genes, for which we observed significant up-regulation in severe COVID-19 patients compared with mild patients and no significant changes after meditation. In MS patients, IFN treatment was the first disease-modifying therapy available, reducing relapse rates and disease progression (57). By comparing the transcriptional changes in MS patient samples, we show these critical meditation-specific drivers regulating IFN signaling were also elevated in MS patients after IFN disease-modifying treatment (but not in untreated MS patients). We also observed that meditation did not elevate bona fide viral infection marker genes and displayed distinct gene signature activation compared with the gene profiles after exercise training, which is known to improve immune competency (56). Taken together, we speculate that early and short-term nonpharmacological intervention by meditation may voluntarily provoke and enhance the immune response before immunotherapy for many conditions, including MS and COVID-19 vaccination. Immune dysfunction is also commonly associated with several neurological and mental disorders. Several recent studies have shown that the peripheral immune response may influence neuronal function, behavior such as spatial learning and memory, and normal brain function (65, 66). Thus, targeting the peripheral immune system by meditation as an intervention may indicate a new era of developing behavioral therapies to maintain brain health and modify currently irreversible neurological diseases. Future studies will be needed in healthy and diseased subjects to test these scenarios to examine the associations between meditation, immune function, and disease symptomatology.
The study design of this research has some limitations. We could not randomize retreat participants, as all interventions (2 mo of prerequisite practices, vegan diet, and 8 d of Samyama meditation program) are required aspects of the retreat program. 1) The prerequisite practices offered by the Isha Institute of Inner Sciences have been demonstrated to have numerous benefits in several previous studies (), and 2) the diet patterns are known to change gene expression levels (42, 43). The reported gene expression changes after meditation in this study might be due to the outcome of the combined synergistic benefit of all these interventions. However, in our in-depth analyses, we found robust evidence to state that the significant impact was due to meditation since we observed very weak gene expression changes between the first two time points before the retreat when more than 90% of the participants were on a vegan diet and following their prerequisite practices (44). Similarly, we also observed a minimal impact on the gene expression changes due to sleep patterns and the time of sample collection by examining the circadian rhythm pathway genes. Together, these results suggest most gene expression changes after meditation are due to meditation intervention with minimal effect due to the diet or the time of sample collection. These results provide a platform for starting randomized clinical trials to validate the role of meditation in delivering the beneficial effects for several diseased conditions.
In conclusion, we identified and characterized the transcriptional program associated with advanced yoga and meditation practices, and we bioinformatically integrated various networks to identify a meditation-specific core network and validate several network predictions. This core network links several immune signaling pathways via a known set of drivers, and we showed that this core transcriptional profile is dysfunctional in MS and severe COVID-19 infection. Very importantly, we demonstrated that these yoga and meditative practices enhanced immune function without activating inflammatory signals. This present proof-of-principle study demonstrated that the immune system can be voluntarily influenced by nonpharmaceutical interventions like yoga and meditation. This suggests that meditation as a behavioral intervention could have important implications for treating various conditions associated with excessive or persistent inflammation with a dampened immune system profile.
Materials and Methods
Subject Recruitment.
The Isha Institute of Inner Sciences (McMinnville, TN) provided a list of registrants for the April 2018 Samyama Program (8-d advanced Inner Engineering meditation retreat). Invitation letters with study information, including a link to the online survey, were sent electronically to all registrants 2 to 3 mo prior to the program. Study eligibility criteria included advanced meditation program participants of at least 18 y of age. Study exclusion criteria were inability to read or comprehend the consent form; subjects with medical conditions in which a blood draw would be contraindicated (e.g., severe anemia); active use of marijuana, opioids, or related drugs; use of antibiotics or probiotic/prebiotic supplements within 60 d of enrollment; participants living outside of the country. Potential subjects were provided a study information sheet at the beginning of the online survey. They were given the following options: 1) survey-only participation (no blood sampling); 2) survey and blood sampling (requiring two blood samples prior to the Samyama program and two blood samples upon completion of the program); and 3) no participation. Analyses of the survey data from these participants have been previously published elsewhere (44). This study was reviewed and approved by the Institutional Review Board (IRB) of the Indiana University School of Medicine (IRB #1801728792). The genome-wide transcriptome analyses part of the study was also reviewed and approved by the IRB of the University of Florida (IRB #201802368). This study was registered at clinicaltrials.gov (NCT04366544) and complied with the Consolidated Standards of Reporting Trials.
Details of Samyama Program.
The Samyama retreat was conducted in April 2018 at the Isha Institute of Inner Sciences (McMinnville, TN) with a strict vegan diet and sleep–wake cycle. Sixty days before the program, Samyama participants began preparatory training and dietary restrictions including vegan diet and elimination of substances such as garlic, onion, chili, eggplant, asafoetida, coffee, tea, alcohol, cigarettes, and other stimulants; intake of minimum 50% natural foods; and focus on “living” and uncooked foods. The preparatory daily practices included Hata yoga (Surya Kriya and Yogasanas; asanas, or postures, meant to knead and strengthen the body), Kriyas (Shakti Chalana Kriya and Shambhavi Mahamudra; combinations of posture, breath, and sound that are meant to purify and enhance the flow of one’s energies while simultaneously increasing general stability), Shoonya (conscious nondoing meant to bring stillness and stimulate the release of physical, mental, and emotional blocks), Pranayama (Sukha Kriya; regulation of breath, meant to facilitate overall stability, balance, and steady attention), and Ardha Siddhasana (an asana in which one sits cross legged with the heel of the left foot placed at the foundational junction of one’s energy channels, meant to help participants sit in stillness with spine erect for longer durations). Study participants had at least 1 y of meditation experience (not Samyama) and had a daily yoga and meditation practice of 1 to 2 h per day before the retreat. During the Samyama retreat, individuals were expected to remain entirely silent for 8 d, with more than 10 h of meditation per day.
Blood Sampling and Storage.
Study subjects who elected blood analysis had ∼10 mL peripheral blood taken at four time points (T1, T2, T3, T4). T1 samples were collected 5 to 8 wk before the retreat, T2 samples were collected on the day of the retreat before starting the meditation method, T3 samples were collected immediately after the retreat, and T4 samples were collected 3 mo after the retreat. At T1 and T4, blood draws were performed at home by an in-home phlebotomist or at an Isha Center group meditation by study personnel. The remaining samples were collected at the Isha Institute of Inner Sciences before/after the meditation retreat. Individuals who did not submit baseline blood samples were allowed to participate at T2 and T3 (but not T4). Peripheral blood was collected in PAXgene tubes and frozen before RNA extraction. Samples were stored frozen at −80 °C until analysis was performed.
Transcriptome Profiling by RNA-Seq.
The human blood samples collected into PAXgene Blood RNA Tubes were removed from the −80 °C freezer and incubated overnight at room temperature. A total of 389 samples from 106 individuals at four time points (T1 = 98, T2 = 105, T3 = 104, T4 = 82) were included, and samples were randomized before RNA extraction to eliminate any effects based on time point, age, sex, or batch. The manufacturer’s protocol was followed for manual purification of total RNA from human whole blood (Qiagen, catalogue no. 762164). RNA was eluted with RNase-free water, and the concentration and purity (A260/A280 ratios >1.8) were assessed using a spectrophotometer. RNA was also tested for suitable mass (RiboGreen) and integrity (Agilent TapeStation), reverse transcribed to complementary DNA (Lexogen QuantSeq 3′ FWD), and sequenced on a HiSeq 4000 instrument (Illumina) in the UCLA Neuroscience Genomics Core laboratory following the manufacturers' standard protocols. Sequencing targeted mean 3 million 65-nt single-stranded reads per sample, which were mapped to the human transcriptome and quantified as transcripts per million mapped reads using the STAR aligner. Briefly, all sequencing data were uploaded to Illumina’s BaseSpace in real time for downstream analysis of quality control. Raw Illumina (fastq.gz) sequencing files were downloaded from BaseSpace and uploaded to Bluebee’s genomics analysis platform (https://www.bluebee.com) to align reads against the human genome and to obtain raw read counts.
Supplementary Material
Acknowledgments
This work was supported by the Department of Pediatrics, University of Florida. The authors appreciate support provided by the Isha Institute of Inner Sciences, McMinnville, TN, for allowing us to conduct this research on participants of the advanced Inner Engineering meditation retreat program. The authors appreciate support provided by the volunteers of the Isha Institute of Inner Sciences for sample collection. The authors also thank thoughtful review and constructive feedback from Nagarajan Kannan, PhD, Department of Laboratory Medicine and Pathology, Mayo Clinic School of Medicine. The authors acknowledge Janelle S. Renschler, DVM, PhD (Department of Anesthesia, Indiana University, Indianapolis, IN) for assistance with medical writing funded by Indiana University in accordance with Good Publication Practice guidelines.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110455118/-/DCSupplemental.
Data Availability
RNA-Seq data have been deposited in GEO (Gene Expression Omnibus) (GSE174083). Previously published data were used for this work [GEO accession numbers for the publicly available datasets analyzed in this study are GSE157103 (COVID-19), GSE41850 (MS), and GSE111554 and GSE111553 (exercise training)]. The WGCNA package can be found online (71). The NetBID package can be found in GitHub (72). The TFBS enrichment package can be found online (73). All analyses were carried out following user manual tutorials; the code for WGCNA analyses can be found online (74). The NetBID analyses code can be found online (75). All R packages or other software used are given in Materials and Methods for each relevant analysis.
References
- 1.Dossett M. L., Fricchione G. L., Benson H., A new era for mind-body medicine. N. Engl. J. Med. 382, 1390–1391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke T. C., Barnes P. M., Black L. I., Stussman B. J., Nahin R. L., Use of yoga, meditation, and chiropractors among U.S. adults aged 18 and over. NCHS Data Brief, 1–8 (2018). [PubMed] [Google Scholar]
- 3.Birdee G. S., Ayala S. G., Wallston K. A., Cross-sectional analysis of health-related quality of life and elements of yoga practice. BMC Complement. Altern. Med. 17, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross A., Friedmann E., Bevans M., Thomas S., National survey of yoga practitioners: Mental and physical health benefits. Complement. Ther. Med. 21, 313–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalsen A., et al. , Rapid stress reduction and anxiolysis among distressed women as a consequence of a three-month intensive yoga program. Med. Sci. Monit. 11, CR555–CR561 (2005). [PubMed] [Google Scholar]
- 6.Schneider R. H., et al. , Stress reduction in the secondary prevention of cardiovascular disease: Randomized, controlled trial of transcendental meditation and health education in Blacks. Circ. Cardiovasc. Qual. Outcomes 5, 750–758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith C., Hancock H., Blake-Mortimer J., Eckert K., A randomised comparative trial of yoga and relaxation to reduce stress and anxiety. Complement. Ther. Med. 15, 77–83 (2007). [DOI] [PubMed] [Google Scholar]
- 8.van der Kolk B. A., et al. , Yoga as an adjunctive treatment for posttraumatic stress disorder: A randomized controlled trial. J. Clin. Psychiatry 75, e559–e565 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Hofmann S. G., Sawyer A. T., Witt A. A., Oh D., The effect of mindfulness-based therapy on anxiety and depression: A meta-analytic review. J. Consult. Clin. Psychol. 78, 169–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javnbakht M., Hejazi Kenari R., Ghasemi M., Effects of yoga on depression and anxiety of women. Complement. Ther. Clin. Pract. 15, 102–104 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Duong N., et al. , Mind and body practices for fatigue reduction in patients with cancer and hematopoietic stem cell transplant recipients: A systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 120, 210–216 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Woolery A., Myers H., Sternlieb B., Zeltzer L., A yoga intervention for young adults with elevated symptoms of depression. Altern. Ther. Health Med. 10, 60–63 (2004). [PubMed] [Google Scholar]
- 13.Garfinkel M. S., et al. , Yoga-based intervention for carpal tunnel syndrome: A randomized trial. JAMA 280, 1601–1603 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Herman P. M., et al. , Cost-effectiveness of mindfulness-based stress reduction versus cognitive behavioral therapy or usual care among adults with chronic low back pain. Spine 42, 1511–1520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolasinski S. L., et al. , Iyengar yoga for treating symptoms of osteoarthritis of the knees: A pilot study. J. Altern. Complement. Med. 11, 689–693 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Gerbarg P. L., et al. , The effect of breathing, movement, and meditation on psychological and physical symptoms and inflammatory biomarkers in inflammatory bowel disease: A randomized controlled rrial. Inflamm. Bowel Dis. 21, 2886–2896 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Hood M. M., Jedel S., Mindfulness-based interventions in inflammatory bowel disease. Gastroenterol. Clin. North Am. 46, 859–874 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Bharshankar J. R., Bharshankar R. N., Deshpande V. N., Kaore S. B., Gosavi G. B., Effect of yoga on cardiovascular system in subjects above 40 years. Indian J. Physiol. Pharmacol. 47, 202–206 (2003). [PubMed] [Google Scholar]
- 19.Yogendra J., et al. , Beneficial effects of yoga lifestyle on reversibility of ischaemic heart disease: Caring heart project of International Board of Yoga. J. Assoc. Physicians India 52, 283–289 (2004). [PubMed] [Google Scholar]
- 20.Venditti S., et al. , Molecules of silence: Effects of meditation on gene expression and epigenetics. Front. Psychol. 11, 1767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buric I., Farias M., Jong J., Mee C., Brazil I. A., What is the molecular signature of mind-body interventions? A systematic review of gene expression changes induced by meditation and related practices. Front. Immunol. 8, 670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenuganen S., et al. , Transcriptomics of long-term meditation practice: Evidence for prevention or reversal of stress effects harmful to health. Medicina (Kaunas) 57, 218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaix R., et al. , Differential DNA methylation in experienced meditators after an intensive day of mindfulness-based practice: Implications for immune-related pathways. Brain Behav. Immun. 84, 36–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo B., et al. , Genomic and clinical effects associated with a relaxation response mind-body intervention in patients with irritable bowel syndrome and inflammatory bowel disease. PLoS One 10, e0123861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhasin M. K., et al. , Specific transcriptome changes associated with blood pressure reduction in hypertensive patients after relaxation response training. J. Altern. Complement. Med. 24, 486–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q. Z., Li P., Garcia G. E., Johnson R. J., Feng L., Genomic profiling of neutrophil transcripts in Asian Qigong practitioners: A pilot study in gene regulation by mind-body interaction. J. Altern. Complement. Med. 11, 29–39 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Ravnik-Glavač M., Hrašovec S., Bon J., Dreo J., Glavač D., Genome-wide expression changes in a higher state of consciousness. Conscious. Cogn. 21, 1322–1344 (2012). Correction in: Conscious. Cogn. 21, 1626 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Black D. S., et al. , Yogic meditation reverses NF-κB and IRF-related transcriptome dynamics in leukocytes of family dementia caregivers in a randomized controlled trial. Psychoneuroendocrinology 38, 348–355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irwin M. R., et al. , Tai chi, cellular inflammation, and transcriptome dynamics in breast cancer survivors with insomnia: A randomized controlled trial. J. Natl. Cancer Inst. Monogr. 2014, 295–301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bower J. E., et al. , Mindfulness meditation for younger breast cancer survivors: A randomized controlled trial. Cancer 121, 1231–1240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu S., Olafsrud S. M., Meza-Zepeda L. A., Saatcioglu F., Rapid gene expression changes in peripheral blood lymphocytes upon practice of a comprehensive yoga program. PLoS One 8, e61910 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman A. M., et al. , Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geschwind D. H., Konopka G., Neuroscience in the era of functional genomics and systems biology. Nature 461, 908–915 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikshak N. N., et al. , Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandran V., et al. , A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89, 956–970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S. B., Yadav R., Yadav R. K., Tolahunase M., Dada R., Telomerase activity and cellular aging might be positively modified by a yoga-based lifestyle intervention. J. Altern. Complement. Med. 21, 370–372 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Li M., Wang D., He J., Chen L., Li H., Bcl-XL: A multifunctional anti-apoptotic protein. Pharmacol. Res. 151, 104547 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Brasó-Maristany F., et al. , PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat. Med. 22, 1303–1313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Léveillé N., et al. , Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat. Commun. 2, 513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laman H., Fbxo7 gets proactive with cyclin D/cdk6. Cell Cycle 5, 279–282 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Ulven S. M., et al. , Using metabolic profiling and gene expression analyses to explore molecular effects of replacing saturated fat with polyunsaturated fat-a randomized controlled dietary intervention study. Am. J. Clin. Nutr. 109, 1239–1250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen J. J., et al. , Associations between dietary patterns and gene expression pattern in peripheral blood mononuclear cells: A cross-sectional study. Nutr. Metab. Cardiovasc. Dis. 30, 2111–2122 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Sadhasivam S., et al. , Isha yoga practices and participation in Samyama Program are associated with reduced HbA1C and systemic inflammation, improved lipid profile, and short-term and sustained improvement in mental health: A prospective observational study of meditators. Front. Psychol. 12, 659667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du X., et al. , Hippo/Mst signalling couples metabolic state and immune function of CD8α+ dendritic cells. Nature 558, 141–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khatamian A., Paull E. O., Califano A., Yu J., SJARACNe: A scalable software tool for gene network reverse engineering from big data. Bioinformatics 35, 2165–2166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liberzon A., et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tumpey T. M., et al. , The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 81, 10818–10821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y., et al. , Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat. Commun. 8, 15138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cadena C., et al. , Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177, 1187–1200.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyashita M., Oshiumi H., Matsumoto M., Seya T., DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 31, 3802–3819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivashkiv L. B., Donlin L. T., Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Au-Yeung N., Mandhana R., Horvath C. M., Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAK-STAT 2, e23931 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lian Q., Sun B., Interferons command Trim22 to fight against viruses. Cell. Mol. Immunol. 14, 794–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Acharya D., Liu G., Gack M. U., Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 20, 397–398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell J. P., Turner J. E., Debunking the myth of exercise-induced immune suppression: Redefining the impact of exercise on immunological health across the lifespan. Front. Immunol. 9, 648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lacy M., et al. , The effects of long-term interferon-beta-1b treatment on cognitive functioning in multiple sclerosis: A 16-year longitudinal study. Mult. Scler. 19, 1765–1772 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Ndisang J. F., Rastogi S., Vannacci A., Immune and inflammatory processes in obesity, insulin resistance, diabetes, and related cardiometabolic complications. J. Immunol. Res. 2014, 579560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slavich G. M., Understanding inflammation, its regulation, and relevance for health: A top scientific and public priority. Brain Behav. Immun. 45, 13–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borkow G., et al. , Chronic immune activation associated with intestinal helminth infections results in impaired signal transduction and anergy. J. Clin. Invest. 106, 1053–1060 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Byrne K. J., Dalgleish A. G., Chronic immune activation and inflammation as the cause of malignancy. Br. J. Cancer 85, 473–483 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Channappanavar R., et al. , Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Channappanavar R., et al. , IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 129, 3625–3639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng Z., et al. , An experimental trial of recombinant human interferon alpha nasal drops to prevent COVID-19 in medical staff in an epidemic area. medRxiv [Preprint] (2020). 10.1101/2020.04.11.20061473 (Accessed 2 June 2021). [DOI] [PubMed]
- 65.Croese T., Castellani G., Schwartz M., Immune cell compartmentalization for brain surveillance and protection. Nat. Immunol. 22, 1083–1092 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Filiano A. J., et al. , Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peterson C. T., Bauer S. M., Chopra D., Mills P. J., Maturi R. K., Effects of Shambhavi Mahamudra kriya, a multicomponent breath-based yogic practice (pranayama), on perceived stress and general well-being. J. Evid. Based Complementary Altern. Med. 22, 788–797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sadhasivam S., et al. , Inner engineering practices and advanced 4-day Isha yoga retreat are associated with cannabimimetic effects with increased endocannabinoids and short-term and sustained improvement in mental health: A prospective observational study of meditators. J. Evid. Based Complementary Altern. Med. 2020, 8438272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braboszcz C., et al. , Plasticity of visual attention in Isha yoga meditation practitioners before and after a 3-month retreat. Front. Psychol. 4, 914 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cahn B. R., Goodman M. S., Peterson C. T., Maturi R., Mills P. J., Yoga, meditation and mind-body health: Increased BDNF, cortisol awakening response, and altered inflammatory marker expression after a 3-month yoga and meditation retreat. Front. Hum. Neurosci. 11, 315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.P. Langfelder, S. Horvath, WGCNA: Weighted correlation network analysis. R version 1.70-3. https://cran.r-project.org/web/packages/WGCNA/index.html. Accessed 6 December 2021. [Google Scholar]
- 72.Yu Laboratory, NetBID Package. https://github.com/jyyulab/NetBID. Accessed 6 December 2021. [Google Scholar]
- 73.V. Chandran, S. Azariah, D. H. Geschwind, Transcription factor and micro RNA enrichment pipeline. https://tfenrichment.semel.ucla.edu/. Accessed 6 December 2021. [Google Scholar]
- 74.P. Langfelder, S. Horvath, Tutorials for the WGCNA package. https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/. Accessed 6 December 2021. [Google Scholar]
- 75.Yu Laboratory, NetBID user guide. https://jyyulab.github.io/NetBID/docs/user_guide. Accessed 6 December 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been deposited in GEO (Gene Expression Omnibus) (GSE174083). Previously published data were used for this work [GEO accession numbers for the publicly available datasets analyzed in this study are GSE157103 (COVID-19), GSE41850 (MS), and GSE111554 and GSE111553 (exercise training)]. The WGCNA package can be found online (71). The NetBID package can be found in GitHub (72). The TFBS enrichment package can be found online (73). All analyses were carried out following user manual tutorials; the code for WGCNA analyses can be found online (74). The NetBID analyses code can be found online (75). All R packages or other software used are given in Materials and Methods for each relevant analysis.