Significance
Structure-based drug design depends on the ability to predict both the three-dimensional structures of candidate molecules bound to their targets and the associated binding affinities. We demonstrate that one can substantially improve the accuracy of these predictions using easily obtained data about completely different molecules that bind to the same target without requiring any target-bound structures of these molecules. The approach we developed to integrate physical and data-driven modeling may find a variety of applications in the rapidly growing field of artificial intelligence for drug discovery.
Keywords: structural biology, drug design, artificial intelligence, antipsychotics, virtual screening
Abstract
Over the past five decades, tremendous effort has been devoted to computational methods for predicting properties of ligands—i.e., molecules that bind macromolecular targets. Such methods, which are critical to rational drug design, fall into two categories: physics-based methods, which directly model ligand interactions with the target given the target’s three-dimensional (3D) structure, and ligand-based methods, which predict ligand properties given experimental measurements for similar ligands. Here, we present a rigorous statistical framework to combine these two sources of information. We develop a method to predict a ligand’s pose—the 3D structure of the ligand bound to its target—that leverages a widely available source of information: a list of other ligands that are known to bind the same target but for which no 3D structure is available. This combination of physics-based and ligand-based modeling improves pose prediction accuracy across all major families of drug targets. Using the same framework, we develop a method for virtual screening of drug candidates, which outperforms standard physics-based and ligand-based virtual screening methods. Our results suggest broad opportunities to improve prediction of various ligand properties by combining diverse sources of information through customized machine-learning approaches.
Binding of small-molecule ligands to proteins is one of the most fundamental processes in biology, and the great majority of drugs exert their effects by binding to a target protein. Predicting properties of protein–ligand interactions—including three-dimensional (3D) structures, binding affinities, binding kinetics, selectivity, and functional effects—is critical both for the rational design of effective medicines and for addressing important questions in molecular biology. A great deal of work has thus focused on the development of computational methods to predict these properties (1, 2).
Such computational methods generally fall into two categories. “Physics-based” approaches use a 3D structure of the target protein and exploit an understanding of the physics of protein–ligand interactions (3). “Ligand-based” approaches use experimental measurements of a given property (e.g., affinity at a particular target) for many ligands and employ pattern-matching to predict the corresponding property for other ligands (4, 5).
Can one combine these two paradigms and the orthogonal sources of information they leverage in a systematic, principled manner? This has proven challenging, particularly when making predictions for ligands substantially different from those for which experimental data are available. It is especially difficult when one wishes to predict properties different from those measured experimentally (e.g., to predict ligand properties that are difficult to determine experimentally by exploiting experimental data that is easy to collect).
Here, we present a rigorous statistical framework to combine the distinct sources of information exploited by physics-based and ligand-based approaches. Using this framework, we develop ComBind, a method to improve prediction of a ligand’s binding pose at a target protein by exploiting readily available nonstructural data. We use the same framework to develop ComBindVS, a virtual-screening method that leverages both structural and nonstructural data to predict ligand binding affinities.
Determining a ligand’s binding pose—the 3D coordinates of the ligand’s atoms when bound to the target—is critical for structure-based optimization of the ligand’s pharmacological properties as well as for understanding how the ligand influences its target. Medicinal chemists have long used the binding pose of a lead compound—when available—as an intuitive guide in choosing which analogs to synthesize and assay (6–9). Binding poses also serve as starting points for computational methods that predict ligand properties such as affinity and selectivity (10–15). Indeed, knowledge of a ligand’s binding pose is so advantageous that researchers in industry and academia often spend months or years to solve an experimental structure of a particular ligand in complex with a target protein.
Because experimental structure determination is time consuming, expensive, and sometimes intractable, tremendous effort has been invested in the development of in silico “docking” methods for predicting ligand binding poses (16–25). These methods are physics based: given a structure of the target protein, they sample many candidate poses of a ligand and rank these poses using scoring functions that approximate the energetic favorability of each pose, typically by capturing interatomic interactions such as hydrogen bonds and van der Waals forces (Fig. 1A). Despite the development of dozens of docking software packages over the past five decades, binding pose predictions are typically correct less than half the time for ligands substantially different from those in the experimental structures used for docking (SI Appendix, Table S1).
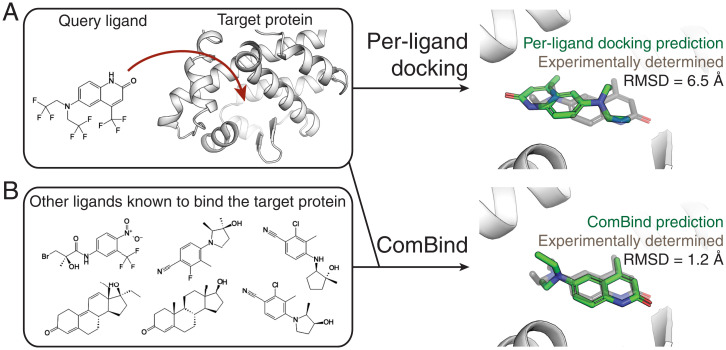
Fig. 1.
ComBind leverages nonstructural data to improve ligand binding pose predictions. (A) Standard docking methods take as input the chemical structure of the query ligand and the 3D structure of the target protein and predict a binding pose using a per-ligand scoring function. (B) ComBind additionally considers other ligands known to bind the target protein (whose binding poses are not known), resulting in more-accurate predictions. For clarity, hydrogen and fluorine atoms are omitted from the 3D renderings.
ComBind improves binding pose prediction by exploiting a widely available type of nonstructural data: the identities of other ligands known to bind the same target (Fig. 1B). Collecting such data is typically far easier than structure determination. Indeed, such data are routinely collected in drug development campaigns and are already available in public databases such as ChEMBL for most recognized drug targets (26).
How can a list of other ligands that bind to the target protein—but whose binding poses are unknown—be used to improve pose prediction? Medicinal chemists have long recognized that distinct ligands tend to bind a given protein in similar poses. Even ligands sharing no common substructure often form similar interactions with the target protein (Fig. 2A). This intuition has a sound basis in physics. For example, the energetic favorability of a protein–ligand hydrogen bond depends on the mobility of the protein atoms involved and their ability to form hydrogen bonds with water in the absence of the ligand (9). These factors contribute similarly to binding of different ligands but are difficult to predict from a static protein structure alone.
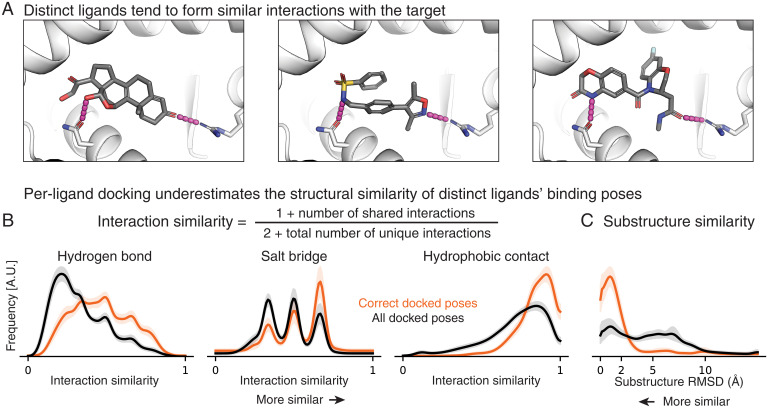
Fig. 2.
Distinct ligands that bind to a given target protein often adopt similar binding poses and do so more frequently than predicted by a state-of-the-art per-ligand docking method. (A) Chemically distinct ligands share key interactions with the mineralocorticoid receptor (Protein Data Bank IDs: 2AA2, 5L7E, and 5MWP). (B) Across a set of 3,115 ligand pairs, interaction similarities are generally higher in pairs of correct poses than in pairs of poses ranked highly by a per-ligand scoring function. Shading depicts the per-target SEM. A.U.: arbitrary units. (C) Across a set of 690 ligand pairs with a shared substructure, the substructure tends to be placed more similarly in correct poses than in other poses ranked highly by a per-ligand scoring function (SI Appendix, Supplementary Text).
We use a large set of experimentally determined structures to quantify the medicinal chemist’s intuition—in particular, to determine the probability that binding poses for different ligands will share various features. We use the results to define the ComBind scoring function, which predicts the favorability of a set of binding poses comprising one pose for each ligand known to bind the target protein. By contrast, scoring functions typically utilized by docking software assign a score to the pose of a single ligand at a time; we thus refer to them as per-ligand scoring functions. The ComBind scoring function takes into account the similarities and differences between the poses of different ligands as well as the energetic favorability of each ligand’s pose, as evaluated by a per-ligand scoring function. By using this scoring function to predict the binding poses of a set of ligands simultaneously, we can predict the pose of each ligand more accurately, even when the ligands share no common scaffold and none of their binding poses are known in advance.
ComBindVS uses the same sources of information—a structure of the target protein and a set of ligands known to bind the target—for virtual screening. Here, we use the ComBind scoring function not only to predict binding poses of known binders but also to predict binding affinities of unrelated molecules.
We benchmark ComBind pose prediction by comparing its results to 248 experimentally determined ligand binding poses across 30 proteins representing all major families of drug targets. ComBind improves the pose prediction accuracy of state-of-the-art docking software for all major drug target families.
We benchmark ComBindVS for virtual screening using the Directory of Useful Decoys, Enhanced (DUD-E) benchmark set (27), which includes diverse protein targets. ComBindVS outperforms state-of-the-art structure-based docking and ligand-based virtual-screening methods, as well as approaches that combine the results of docking and ligand-based methods. ComBindVS yields performance improvements even when the candidate molecules are very different from the known binders, making it suitable for discovery of novel chemotypes.
We also illustrate the use of ComBind to predict the previously unknown binding poses of several antipsychotics at the D2 dopamine receptor (D2R), an important drug target for which experimental structure determination has proven difficult. We validate ComBind’s predictions—which differ from those of state-of-the-art docking software—using mutagenesis experiments. These results reveal a structural motif that influences the subtype selectivity of D2R-targeted drugs and may thus prove useful in optimization of these ligands. ComBindVS also enables improved prediction of the effects of ligand modifications on binding affinity.
Our approach provides a principled manner to integrate physics-based structural modeling with inference based on experimental data for other ligands, including ligands that share no common scaffold or substructure. Similar methods may prove useful in combining physics-based modeling with ligand-based approaches to improve prediction of various ligand properties by exploiting diverse sources of data.
Methods and Results
The methods that we introduce here can employ any per-ligand scoring function and pose-sampling strategy, including those implemented in any standard docking package. We use the commercial docking package Glide (20) for illustrative purposes, because it is widely used in the pharmaceutical industry and because it ranks among the most-accurate docking packages in comparative studies (28, 29).
Quantifying the Similarity of Binding Poses for Distinct Ligands.
We begin by quantifying the medicinal chemist’s intuition that different ligands tend to adopt structurally similar poses when binding the same target protein. We wish not only to measure the similarity of correct poses of different ligands but also to compare the similarity of correct poses to that of other poses ranked highly by per-ligand docking software. We consider two notions of similarity: similarity of protein–ligand interactions and similarity in position of common ligand substructures.
We compiled a set of 385 protein–ligand complex structures for 28 target proteins representing all major classes of small-molecule drug targets (SI Appendix, Table S2) (30). We docked each of the ligands using Glide (19, 20) and selected the 100 most highly ranked poses for each ligand. To reflect practical application of docking, we docked each ligand into an experimental structure solved in the presence of a ligand distinct from any of those being docked (“cross-docking”; SI Appendix, Supplementary Text).
For all pairs of ligands for each target protein, we compute the similarity between each pose of one ligand and each pose of the other ligand. We use this data to calculate a probability distribution over similarity values; we refer to this distribution as the reference distribution.
We also compute similarities between each pair of correct poses (again, one pose per ligand), in which a pose is considered correct if it is within 2.0 Å RMSD to the experimentally determined pose. We use these data to calculate a second probability distribution over similarity values, the native distribution. When calculating the native distribution, we use correct poses from the lists generate by Glide instead of using the experimentally determined poses directly, such that the similarity statistics we calculate will be most applicable to candidate poses considered during computational pose prediction.
We evaluate pose similarity separately for different types of protein–ligand interactions: hydrogen bonds, salt bridges, and hydrophobic contacts (Fig. 2B and SI Appendix, Fig. S1A). Given a pair of poses, we evaluate the similarity for each interaction type by cataloging the set of protein residues with which each ligand forms an interaction of the given type and then comparing the sizes of the intersection and union of these sets. Their ratio (the Tanimoto coefficient) (31) increases when shared interactions are formed and decreases when either ligand forms an unshared interaction. To make this metric well-defined when neither ligand forms any interactions of a particular type, we add pseudo counts. For all interaction types, the native distribution exhibits higher similarity than the reference distribution—that is, pairs of correct poses form more similar interactions than other pairs of poses ranked highly by the per-ligand scoring function (Fig. 2B).
We define substructure similarity as the RMSD of atom positions of the largest chemical substructure shared by a pair of ligands (SI Appendix, Fig. S1B). We evaluated substructure similarity for pairs of ligands that shared a substructure at least half the size of the smaller ligand. For this similarity metric, too, the native distribution exhibits higher similarity than the reference distribution, indicating that the common substructure tends to be more similarly positioned in pairs of correct poses than in other pairs of poses ranked highly by the per-ligand scoring function (Fig. 2C).
These results suggest that the similarity of correct poses is not adequately captured by a state-of-the-art per-ligand scoring function. In further support of this point, we also calculated probability distributions of similarity between the poses that the per-ligand scoring function ranks first for each ligand (SI Appendix, Fig. S2). We found that these distributions also exhibited lower similarity than the corresponding native distributions.
Derivation of a Statistical Potential for Sets of Binding Poses.
We used the similarity distributions described in the previous section to derive a statistical potential that—instead of acting on features of a single pose, as in previous docking software—acts on a set of hypothesized poses, one for each ligand known to bind the target protein:
where n is the total number of ligands, is a constant, is the frequency of the observed pose pair similarity in the native distribution, and is the frequency of the observed pose pair similarity in the reference distribution.
The first component evaluates the energetic favorability of each ligand’s pose individually using a per-ligand scoring function (e.g., a scoring function used in Glide or another docking software package). The summation is over ligands known to bind the target protein, with “pose” referring to the hypothesized pose for each ligand. The constant factor depends on the per-ligand scoring function employed and can be determined as described in SI Appendix, Supplementary Text. For Glide, we set it to 1.
The second component rewards sets of poses with a degree of similarity that is more often observed in correct poses than in other poses ranked highly by the per-ligand scoring function. Here, the outer summation is over pairs of distinct ligands known to bind the target protein, and the inner summation is over the similarity measures shown in Fig. 2 B and C: hydrogen bond similarity, salt bridge similarity, hydrophobic contact similarity, and substructure similarity. “Pose pair similarity” refers to the calculated similarity value of the given type for the hypothesized poses of the given ligand pair. The “native distribution” and “reference distribution” for each similarity type are determined as described above. The resulting negative log likelihood ratios have the mathematical properties of an energy, namely that an additive decrease in energy corresponds to a multiplicative increase in likelihood ratio, allowing for straightforward integration with standard per-ligand docking scores, which are typically in units of energy (SI Appendix, Fig. S3). For pairs of ligands that do not share a substructure at least half the size of the smaller ligand, the substructure similarity term is not included in the summation.
The second component acts as a correction to the first. If the per-ligand scoring function were perfect, in the sense that it could perfectly distinguish correct poses from incorrect ones, the terms in the second component would consistently assume values of zero. Because per-ligand scoring functions remain imperfect—and in particular tend to underpredict the likelihood that a set of ligands will adopt similar poses—the second component typically assumes nonzero values.
ComBind: Structure Prediction Informed by Nonstructural Data.
The ComBind pose prediction method identifies a set of binding poses—one for each of a set of ligands known to bind the target protein—that minimizes the ComBind potential. More specifically, given a target protein and a query ligand whose binding pose we wish to predict, we proceed as follows:
-
1.
Compile a set of other ligands known to bind the target protein (e.g., from a public database such as ChEMBL or from ligands tested as part of a drug discovery project). We refer to these as helper ligands.
-
2.
Dock the query ligand and each helper ligand individually to the target protein (with a per-ligand docking software package), generating many candidate poses and associated docking scores for each ligand.
-
3.
Determine the set of poses—one per ligand—that minimizes the ComBind potential. We use an expectation–maximization algorithm for this purpose (SI Appendix, Supplementary Text).
As an illustrative example, we apply ComBind to predict binding poses for ligands at the β1-adrenergic receptor (β1AR), the primary target of the beta blocker drugs that are widely used to treat heart attack, heart failure, and hypertension. We selected 11 diverse ligands known to bind β1AR, including both beta blockers and beta agonists. We docked 11 distinct ligands to a crystallographic β1AR structure using Glide, producing up to 100 candidate poses for each ligand. We then solved for a set of poses—one per ligand—that minimizes the ComBind potential (Fig. 3A). The crystallographic β1AR structure used for docking was determined in complex with a ligand distinct from any of the 11 docked ligands. Crystallographic ligand poses were not used in any way by Glide or ComBind.
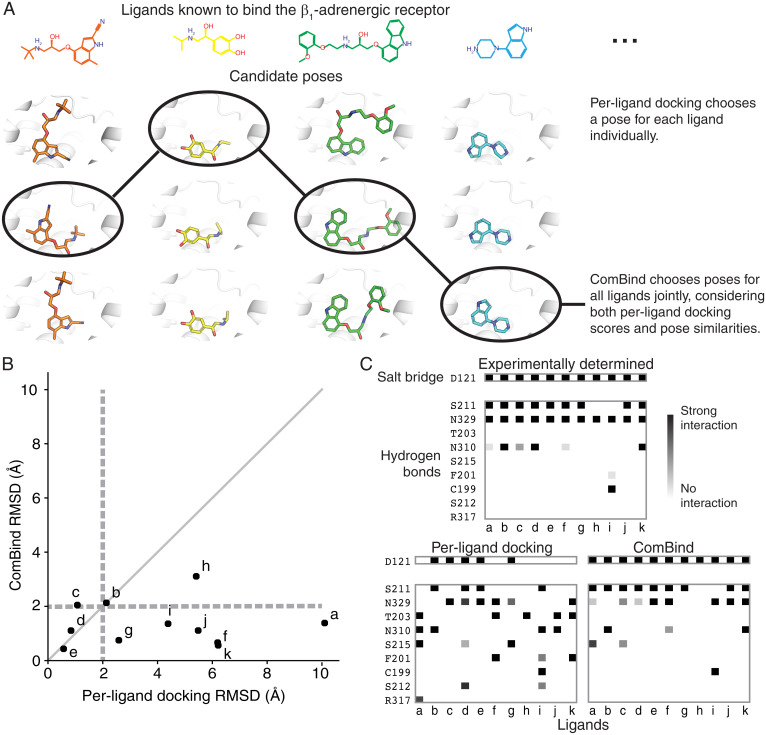
Fig. 3.
ComBind discovers and rewards key interactions shared by distinct ligands. (A) Whereas per-ligand docking considers each ligand individually, ComBind jointly selects poses for all ligands, optimizing for poses that are individually favorable according to a per-ligand scoring function and together form a coherent set of protein–ligand interactions. (B) We tested the ability of ComBind to predict the poses of 11 ligands that bind the β1-adrenergic receptor. Each dot corresponds to a single ligand, with the dot’s position indicating the error in the predicted pose (RMSD from the experimentally determined pose) for ComBind and for state-of-the art per-ligand docking software (Glide). A pose is considered correct if its RMSD is ≤2.0 Å (dashed lines). ComBind predicts a substantially more accurate pose than Glide for seven of the 11 ligands. (C) The set of residues with which each ligand forms salt bridges or hydrogen bonds when positioned in its experimentally determined pose (Top), the pose predicted by per-ligand docking (Left), and the pose predicted by ComBind (Right).
Glide’s top-ranked pose was correct for four of 11 ligands, whereas the pose selected by ComBind was correct for 10 of 11 ligands (Fig. 3B). In ComBind’s selected poses—as in experimentally determined poses—most of the ligands form a salt bridge with D121 and hydrogen bonds with S211 and N329 (Fig. 3C). In comparison, the poses ranked most highly by Glide’s per-ligand scoring function exhibited more varied hydrogen bonds and salt bridges (Fig. 3C).
We emphasize that ComBind does not require that all ligands adopt similar poses or form similar interactions. ComBind correctly predicts, for example, that two of these β1AR ligands do not form a hydrogen bond with S211.
ComBind Outperforms a State-of-the-Art Method on a Diverse Benchmark Set.
We benchmarked ComBind on a set of 248 protein–ligand complexes representing all major families of drug targets. We took several steps to mimic a real-world use case. First, when predicting the binding pose of a query ligand with ComBind, we used helper ligands selected from the public ChEMBL database (26). We did not use any experimental information on poses of helper ligands; indeed, for nearly all helper ligands selected, poses have not been determined experimentally. Second, we never used a target protein structure determined in the presence of a ligand that shares a chemical scaffold with the query ligand in order to avoid self-docking and other “easy” cases in which one could predict the pose of the query ligand by overlaying it on the crystallographic ligand pose (SI Appendix, Supplementary Text). Finally, when predicting ligand binding poses at a given target protein, we omitted all structures involving that protein when constructing the distributions used to define the ComBind potential.
We evaluated two ways of choosing, from ChEMBL, helper ligands for use in ComBind: 1) a diverse set of ligands with the highest binding affinity (“high affinity”) and 2) the ligands sharing the largest substructure with the query ligand (“congeneric”). Both of these selection criteria lead to substantial performance improvements over Glide (Fig. 4 and SI Appendix, Fig. S4), indicating that ComBind could be applied effectively using either a diverse set of ligands identified from a high-throughput screen or a congeneric series of ligands generated during lead optimization.
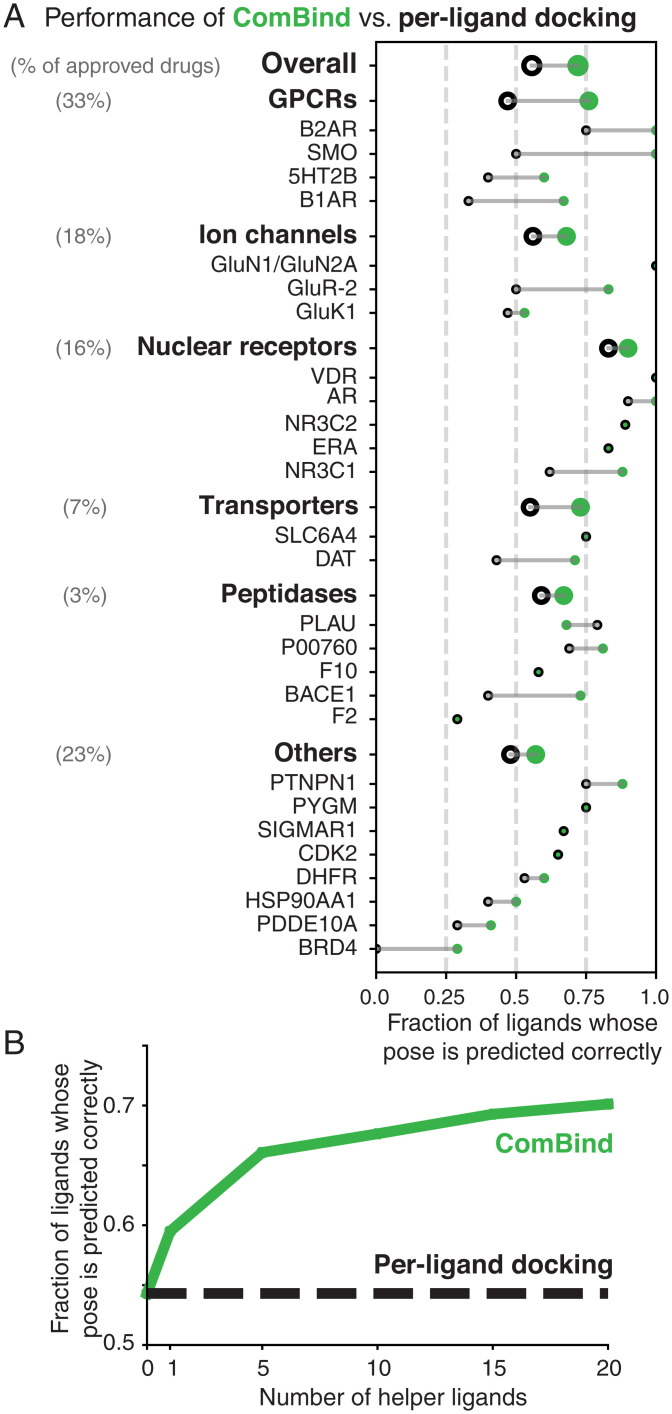
Fig. 4.
ComBind outperforms per-ligand docking on a diverse benchmark set. Performance of ComBind, as compared to a per-ligand scoring function, using helper ligands selected automatically from ChEMBL. All results are for “cross-docking” (the query ligand is docked into a structure determined in the presence of a distinct ligand). (A) Performance per target protein, target protein family (GPCRs, ion channels, etc.), and overall. Green disks and black circles indicate performance for ComBind and a state-of-the art per-ligand docking software package (Glide), respectively. (B) Performance as a function of the number of helper ligands. When using no helper ligands, ComBind is equivalent to per-ligand docking.
ComBind’s performance improves with the use of more helper ligands (up to 20, the maximum number that we tested) (Fig. 4B and SI Appendix, Fig. S4B). Interestingly, ComBind substantially outperforms Glide even when using only a single helper ligand.
In the ComBind benchmark results described below (Fig. 4A), we used 20 helper ligands for each query ligand, selected from ChEMBL by the high-affinity criterion. When computing overall results, we averaged across target families, weighted the performance for each family by the fraction of US Food and Drug Administration–approved drugs targeting that family (30).
On average, ComBind selects a correct pose for 57% of all ligands and 70% of ligands for which at least one correct pose was included among the list of candidates considered—a 30% improvement over Glide in both cases (SI Appendix, Table S1). ComBind improves pose prediction performance for all target families considered. Even at the individual target level, we find that use of ComBind hardly ever degrades performance: ComBind only reduced performance for one of the 30 targets considered, and this performance reduction was minor. ComBind increased pose prediction accuracy both for targets with shallow, poorly formed binding pockets and for targets with deep, well-formed binding pockets (SI Appendix, Fig. S12).
ComBindVS: Deep Integration of Physics-Based and Ligand-Based Modeling for Virtual Screening and Binding Affinity Prediction
We used the same statistical framework to develop ComBindVS. Given a structure of the target protein, a set of ligands known to bind the target (helper ligands), and a library of candidate molecules to screen, ComBindVS proceeds as follows (SI Appendix, Supplementary Text):
-
1.
ComBind is used to predict poses for all of the helper ligands.
-
2.
For each candidate molecule, a pose is selected that minimizes the ComBind score with respect to the helper ligands.
-
3.
The ComBind score of each candidate molecule in its predicted pose is used as a prediction of its affinity relative to other molecules. For virtual screening, the candidate molecules are ranked by this score.
Notably, ComBindVS integrates physics-based and ligand-based modeling not only to predict poses of candidate molecules but also to predict relative affinities of these molecules given their predicted poses. Virtual-screening campaigns using per-ligand docking methods often search for candidate molecules forming particular interactions believed to be important for binding based on experimentally determined ligand poses (32). ComBindVS estimates the importance of interactions automatically from the helper ligands without requiring any information on their binding poses.
ComBindVS Outperforms Physics-Based and Ligand-Based Modeling at Virtual Screening.
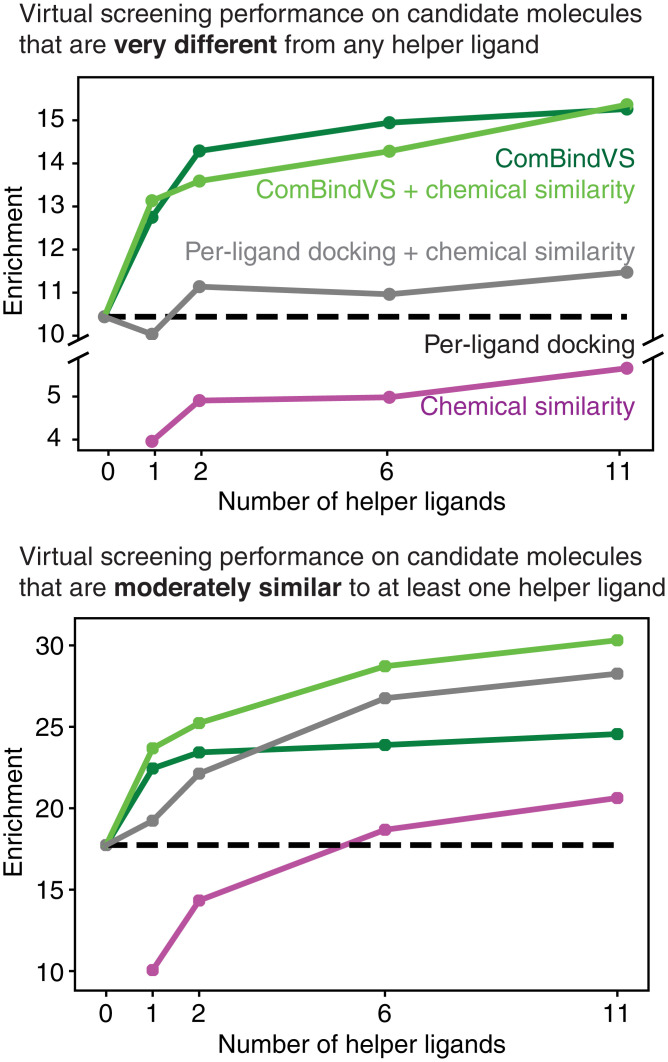
We compared the performance of ComBindVS to per-ligand docking, a state-of-the-art ligand-based method (“chemical similarity”), and several strategies for integrating the results of the two, on the DUD-E dataset (27) (SI Appendix, Supplementary Text). For each of 102 diverse target proteins, this dataset includes a structure of the target, a list of molecules known to bind the target (“true binders”), and a list of decoy molecules meant to be hard to distinguish from the true binders. We evaluated the ability of each method to pick out the true binders from the decoys as quantified by the enrichment factor 1%: the number of true binders that the method ranks in the top 1% divided by the expected number given a random ranking.
We first considered cases in which the candidate molecules are restricted to be very different from any of the helper ligands (maximum Tanimoto coefficient < 0.2) in order to reflect the use of virtual screening to discover novel chemotypes. Similar filters are commonly applied in real virtual-screening campaigns to avoid “rediscovering” variants of known binders (32). In this regime, ComBindVS outperforms per-ligand docking by more than 20% with a single helper ligand and by nearly 50% with 11 helper ligands. The ligand-based chemical similarity method consistently underperforms both per-ligand docking and ComBindVS and provides little benefit when combined with either of them (Fig. 5A and SI Appendix, Fig. S9E).
Fig. 5.
On virtual screening, ComBindVS outperforms per-ligand docking, a ligand-based method (“chemical similarity”), and a combination of the two. Performance on DUD-E benchmark set when restricting to candidate molecules that have less than 0.2 Tanimoto similarity to all helper ligands (Top) or a maximum of between 0.2 and 0.3 Tanimoto similarity to any helper ligand (Bottom). “Chemical similarity” is a state-of-the-art ligand-based method (SI Appendix, Supplementary Text).
When considering ligands that are moderately similar to one or more helper ligands (0.20 < maximum Tanimoto coefficient < 0.30), chemical similarity information becomes more valuable, and the combination of ComBindVS and chemical similarity gave the best results (Fig. 5B and SI Appendix, Fig. S9E). ComBindVS on its own substantially outperforms chemical similarity, but the two are complementary, because ComBindVS evaluates ligands using their intermolecular interactions, whereas the chemical similarity method considers the presence of specific chemical groups.
Predicting Binding Poses and Affinities of Antipsychotics at the D2R.
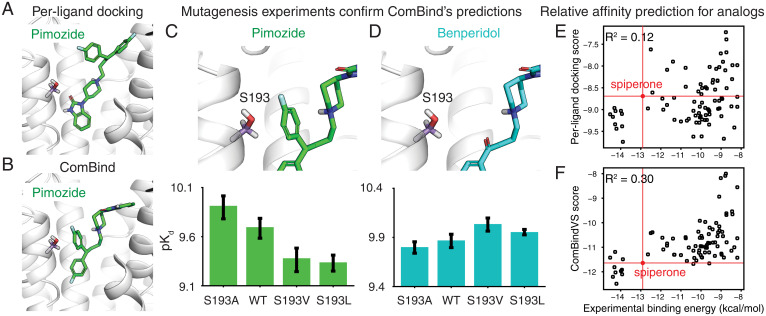
To further illustrate the practical application of ComBind, we predicted the binding poses of three antipsychotic drugs—pimozide, benperidol, and spiperone—at their target, D2R. Knowledge of these binding poses could aid ongoing efforts to develop antipsychotics with improved pharmacological properties, including ligands that bind selectively to D2R over other dopamine receptors (33, 34). Solving experimental structures of D2R has proven difficult, despites decades of effort (35, 36). At the time that we made these predictions, the only available D2R structure was for D2R bound to risperidone (35), a ligand substantially different from those whose poses we wished to predict.
We predicted binding poses for pimozide, benperidol, and spiperone as well as the tool compound mespiperone using both Glide and ComBind (SI Appendix, Supplementary Text). For spiperone and mespiperone, ComBind and Glide predict similar poses. For pimozide and benperidol, however, ComBind’s predictions are different from Glide’s: a fluorobenzene ring of each compound is positioned near the top of the binding pocket by Glide and near the bottom by ComBind (Fig. 6 A and B and SI Appendix, Fig. S6 A and B).
Fig. 6.
Prediction and validation of the binding poses of antipsychotics at D2R. (A and B) Glide (A) and ComBind (B) predict very different binding poses for pimozide (and for benperidol; SI Appendix, Fig. S6). (C) Mutagenesis experiments validate ComBind’s predictions. In ComBind’s predicted pose for pimozide, its “extra” ring is uncomfortably close to S193, such that decreasing the size of residue 193 (S193A) increases pimozide’s binding affinity and increasing the size of residue 193 (S193V and S193L) decreases pimozide’s binding affinity. WT represents the wild-type (unmutated) receptor. (D) As a control, we verified that benperidol—which lacks this “extra” ring but is otherwise identical to pimozide—does not exhibit the same trend. Error bars show SEM. Refer to SI Appendix, Fig. S6 for additional data. (E) Comparison of experimental binding energies and per-ligand docking scores for analogs of spiperone. The red circle denotes the experimental binding energy and per-ligand docking score for spiperone. (F) Same but for ComBindVS scores instead of per-ligand docking scores.
To test ComBind’s predictions, we designed mutagenesis experiments. First, we tested a series of mutations of Ser193 (S193), which is positioned uncomfortably close to the second fluorobenzene ring of pimozide in ComBind’s predicted pose but not in Glide’s (Fig. 6C). Indeed, mutating S193 to a larger residue (Val or Leu) decreases pimozide’s affinity, while mutating S193 to a smaller residue (Ala) increases pimozide’s affinity. Such effects are not observed for benperidol, which is identical to pimozide except that it lacks the fluorobenzene ring that contacts S193 in pimozide (Fig. 6D). Indeed, benperidol’s affinity actually increases when S193 is mutated to a larger residue. These results are consistent with ComBind’s predicted poses but not with Glide’s: Glide predicts that pimozide and benperidol position nearly identical chemical groups in essentially identical positions near S193. Additional experiments involving mutation of residues surrounding the top and bottom of the binding pocket also support ComBind’s predictions (SI Appendix, Fig. S6C).
Shortly before submission of this manuscript, a haloperidol-bound D2R crystal structure appeared (37). Haloperidol shares a common substructure with the ligands that we considered, and this substructure is positioned similarly in in the crystal structure and in ComBind’s predictions, further supporting the accuracy of these predictions.
Our predicted poses suggest that a previously unrecognized structural motif contributes to selective binding to D2R. The antipsychotics that we studied have picomolar affinity at D2R and bind more tightly to D2R than to the D3 dopamine receptor (D3R). Haloperidol, by contrast, binds with weaker (nanomolar) affinity and is not selective for D2R over D3R. Comparison of the binding poses reveals that the primary difference in the protein–ligand interactions is that the antipsychotics we studied—but not haloperidol—place a ring structure in the “extracellular vestibule,” located above the orthosteric site where dopamine binds. The extracellular vestibule has much higher sequence diversity among the different dopamine receptors than does the orthosteric site, supporting the hypothesis that ligand interactions with this region contribute to selectivity. Optimizing ligands to strengthen these interactions could lead to drugs with greater selectivity for D2R.
We also assessed the accuracy of ComBindVS in predicting the binding affinities of a set of spiperone analogs. Using only our four original ligands as helper ligands, ComBindVS predicted the experimentally measured relative binding affinities significantly more accurately than per-ligand docking (R2 = 0.30 and 0.12, respectively, P = 0.001; Fig. 6 E and F). For 94% of the 83 analogs, ComBindVS correctly predicts whether the affinity is higher or lower than that of spiperone, whereas per-ligand docking makes this prediction correctly for 48% of the analogs. ComBindVS may therefore provide a useful guide to optimizing such ligands for affinity or to avoiding a substantial loss of affinity while optimizing for other properties.
Inspection of individual ligands suggests that ComBindVS’s improved performance stems both from more-accurate binding pose predictions and from directly leveraging the interactions formed by the helper ligands for scoring (SI Appendix, Fig. S10). Interestingly, ComBindVS correctly predicts that addition of hydrophobic groups to the secondary amine of spiperone increases ligand affinity, even though this addition decreases interaction similarity to all the helper ligands (SI Appendix, Fig. S11).
Discussion
We have introduced a statistical potential that acts on a set of structures for different ligands in complex with a given protein rather than on a single structure. We have used this potential to develop ComBind and ComBindVS, methods that improve the accuracy of binding pose prediction and affinity prediction by leveraging the knowledge that certain other ligands bind the target, even when these other ligands have unknown binding poses and are very different from the molecules of interest.
Importantly, ComBind and ComBindVS do not assume that all ligands considered bind in similar poses. Instead, they consider both the favorability of each individual ligand’s pose, as evaluated by a per-ligand scoring function, and the tendency of different ligands to adopt similar poses, as determined by analysis of hundreds of experimental structures. ComBind often predicts correctly that two ligands position their common scaffold differently or that they form substantially different interactions with the binding pocket (SI Appendix, Figs. S7 and S8).
Applicability and Robustness.
ComBind and ComBindVS are broadly applicable. For most major drug targets, numerous binders have already been identified. Even for a completely novel target, several binders would typically be identified in the very early stages of a drug discovery project by high-throughput screening. Both methods achieve significant improvements in accuracy even when given very few known binders.
Binding pose and affinity prediction are important in many areas beyond drug discovery. These include the study of biological phenomena such as cellular signaling (e.g., binding of hormones and neurotransmitters), sensation (e.g., binding of odorants and flavorants), enzyme function (e.g., binding of nutrients and other metabolic substrates), and defense mechanisms (e.g., binding of toxins and antibiotics) as well as understanding the effects of genetic variation on responses to both naturally occurring ligands and drugs, which is essential to personalized medicine (38). In each of these cases, multiple ligands are typically known to bind the targets of interest, so ComBind and ComBindVS may prove useful.
ComBind’s robustness is illustrated by its accuracy in our benchmarks, which used helper ligands selected automatically according to approximate affinity values listed in the ChEMBL database. These data are noisy, not only because ligand affinities were measured by many laboratories using different assays but also because the data often includes values that were inputted incorrectly (39, 40). In addition, ligands selected automatically from ChEMBL sometimes bind to completely different binding pockets on the same target.
ComBind generally produces an accurate prediction for the query ligand even when no correct candidate poses are generated for many helper ligands. SI Appendix, Table S3 shows an example in which the majority of ligands considered had no correct candidate pose; ComBind nevertheless outperformed per-ligand docking.
The per-ligand docking software used to generate and score individual ligand poses in our current implementations of ComBind and ComBindVS treat the protein as rigid. Nevertheless, ComBind and ComBindVS generally prove effective even when considering a set of ligands that bind diverse protein conformations. For example, the β1AR ligands considered in Fig. 3 include both agonists, which bind preferentially to the protein’s active conformation, and inverse agonists, which bind preferentially to its inactive conformation (SI Appendix, Table S4).
Relationship to Previous Work.
ComBind and ComBindVS build upon several methods that combine ligand-based and physics-based information in more limited settings. Three-dimensional quantitative structure–activity relationship techniques, including field-based methods and 3D pharmacophore methods, are ligand-based approaches that consider potential 3D conformations of many ligands (41–43). These methods attempt to align ligands in three dimensions, but they do not require a structure of the target protein, and even when such a structure is available, it is typically used only in a limited way (e.g., to define excluded volume) (44). These methods require data for a large number of binders and are generally not applied to pose prediction. Several previous virtual-screening approaches perform docking and ligand-based screening independently and then combine the results (45, 46) or combine the results of docking and pharmacophore modeling (47, 48).
ComBind also draws inspiration from previous methods that predict binding poses of multiple known binders simultaneously. Some of these methods consider a congeneric series of ligands and require that the shared scaffold is similarly placed (49, 50). Others use either the number of similarly placed functional groups (51) or the number of shared interactions (52) between a set of docked ligands as a scoring function, assuming that the ligands adopt maximally similar poses. ComBind goes beyond these techniques in that it not only applies to any set of ligands but also provides a principled method to combine information from per-ligand docking scores with information on pose similarity across multiple ligands. This is essential to ComBind’s success in cases in which ligands form substantially different interactions or position shared substructures very differently (53). Likewise, ComBind provides a principled method to combine multiple metrics of pose similarity. Indeed, ComBind’s performance drops substantially if one omits per-ligand docking scores, substructure similarity, or interaction similarity from its scoring function (SI Appendix, Fig. S5).
A great deal of innovative recent work has explored machine-learning methods—particularly deep learning methods—for predicting ligand properties (54). These methods, which promise to make a substantial impact in drug discovery, generally fall into two categories. Some use 3D structures of target proteins and learn per-ligand scoring functions for general protein–ligand interactions (55–57). Others are ligand-based methods that learn a direct relationship between small-molecule chemical structures and their properties at particular targets (58–60). ComBind is also a machine-learning method, but it is orthogonal to these innovations in that it integrates structure-based and ligand-based modeling. To enable this combination, we designed a machine-learning framework different from neural networks and other traditional machine-learning architectures.
Performance.
Our extensive benchmarks show that ComBind outperforms a state-of-the-art per-ligand pose prediction method across all major families of drug targets. For individual targets, ComBind often substantially improves pose prediction accuracy and hardly ever degrades it. Across a broad range of targets, ComBindVS often substantially improves virtual-screening performance, while almost always avoiding substantial performance degradation (SI Appendix, Fig. S9E). Using ComBind or ComBindVS thus has a substantial upside and little downside.
For G protein-coupled receptors (GPCRs), the largest family of drug targets, ComBind selects a correct binding pose over 60% more frequently than per-ligand docking, increasing the probability of correct prediction from 47 to 76% for ligands that do not share a chemical scaffold with the ligand present in the protein structure used for docking. This improvement is particularly noteworthy, not only because GPCRs represent the targets of one-third of all approved drugs—and a very large fraction of current drug discovery efforts—but also because experimentally determining structures of GPCRs in complex with lead compounds is often extremely difficult (61).
Performance of ComBind and ComBindVS could be improved through use of curated or in-house data. In particular, a careful human curator could 1) identify ligands that can most confidently be classified as binders (e.g., based on multiple reports or on particularly reliable data sources), 2) identify ligands demonstrated to bind in the same binding pocket (e.g., by competition binding assays), and 3) remove data that was inputted incorrectly to a database. For a major drug discovery project focused on a particular target, a substantial amount of additional in-house data will often be available on ligands found to bind the target, and that data will typically have been collected in a more uniform and consistent manner than data extracted from multiple publications.
Extensibility and Future Work.
Because ComBind and ComBindVS can use any per-ligand docking method for pose generation and scoring of individual ligands, they will be able to take advantage of improvements to these methods. For example, several recent machine-learning methods show promise in fitting more-accurate per-ligand scoring functions (56, 62, 63), and other methods allow for binding pocket flexibility when generating candidate poses (18, 64, 65).
Likewise, ComBind and ComBindVS can be used with any pairwise pose similarity metric or combination thereof. Their performance could potentially be improved by using more fine-grained interaction descriptors (66, 67) or by using similarity metrics based on field-based methods developed for virtual screening (43, 68).
The formulation of the ComBind potential is sufficiently general that it could be extended to incorporate other types of data, ranging from multiple experimental structures of the protein in complex with different ligands to effects of protein mutation on ligand binding. Likewise, future work might exploit the affinity of each known binder; we have not done so here to avoid obscuring the general applicability of our method, as the affinity estimates available in public databases are often determined by different techniques and thus difficult to compare to one another.
Our work suggests rich opportunities to improve prediction of diverse ligand properties by combining physics-based and ligand-based modeling. Ligand-based and physics-based modeling have both found widespread use for decades, but ligand-based approaches are generally limited in their ability to predict affinities of molecules very different from those for which experimental data are available, while physics-based approaches are generally limited to properties whose physical basis is known a priori and typically require use of various approximations that introduce error. A careful combination of the two approaches, perhaps exploiting the ComBind statistical potential, might prove effective for predicting properties including functional activity, selectivity, or binding kinetics. Further work will be necessary to explore these possibilities.
Supplementary Material
Acknowledgments
We thank B. Kelly, N. Latorraca, A. Venkatakrishnan, and N. Sohoni for advice and guidance in the early stages of the project, all members of the R.O.D. laboratory for insightful comments, and J. Javitch for providing materials for mutagenesis experiments. This work was supported by NIH grants R01GM127359 (to R.O.D.), R01GM083118 (to R.K.S.), and U19GM106990 (to R.K.S.) and by a Stanford Graduate Fellowship (to J.M.P.).
Footnotes
Competing interest statement: Stanford University has filed a patent application related to this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112621118/-/DCSupplemental.
Data Availability
Implementations of ComBind and ComBindVS are available in GitHub at https://github.com/drorlab/combind.
References
- 1.Yu W., A. D. MacKerell, Jr, Computer-aided drug design methods. Methods Mol. Biol. 1520, 85–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sliwoski G., Kothiwale S., Meiler J., E. W. Lowe, Jr, Computational methods in drug discovery. Pharmacol. Rev. 66, 334–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J., Fu A., Zhang L., An overview of scoring functions used for protein-ligand interactions in molecular docking. Interdiscip. Sci. 11, 320–328 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Leelananda S. P., Lindert S., Computational methods in drug discovery. Beilstein J. Org. Chem. 12, 2694–2718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherkasov A., et al. , QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 57, 4977–5010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sweeney Z. K., et al. , Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase. J. Med. Chem. 51, 7449–7458 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Hellmann J., et al. , Structure-based development of a subtype-selective orexin 1 receptor antagonist. Proc. Natl. Acad. Sci. U.S.A. 117, 18059–18067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hübner H., et al. , Structure-guided development of heterodimer-selective GPCR ligands. Nat. Commun. 7, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bissantz C., Kuhn B., Stahl M., A medicinal chemist’s guide to molecular interactions. J. Med. Chem. 53, 5061–5084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyu J., et al. , Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cournia Z., et al. , Rigorous free energy simulations in virtual screening. J. Chem. Inf. Model. 60, 4153–4169 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Hollingsworth S. A., Dror R. O., Molecular dynamics simulation for all. Neuron 99, 1129–1143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCorvy J. D., et al. , Structure-inspired design of β-arrestin-biased ligands for aminergic GPCRs. Nat. Chem. Biol. 14, 126–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalut J., et al. , Rational design of novel benzisoxazole derivatives with acetylcholinesterase inhibitory and serotoninergic 5-HT4 receptors activities for the treatment of Alzheimer’s disease. Sci. Rep. 10, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira L. G., Dos Santos R. N., Oliva G., Andricopulo A. D., Molecular docking and structure-based drug design strategies. Molecules 20, 13384–13421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Platzer K. E. B., Momany F. A., Scheraga H. A., Conformational energy calculations of enzyme-substrate interactions. II. Computation of the binding energy for substrates in the active site of -chymotrypsin. Int. J. Pept. Protein Res. 4, 201–219 (1972). [DOI] [PubMed] [Google Scholar]
- 17.Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E., A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 161, 269–288 (1982). [DOI] [PubMed] [Google Scholar]
- 18.Jain A. N., Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 46, 499–511 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Friesner R. A., et al. , Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49, 6177–6196 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Friesner R. A., et al. , Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones G., Willett P., Glen R. C., Leach A. R., Taylor R., Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Rarey M., Kramer B., Lengauer T., Klebe G., A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 261, 470–489 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Allen W. J., et al. , DOCK 6: Impact of new features and current docking performance. J. Comput. Chem. 36, 1132–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatachalam C. M., Jiang X., Oldfield T., Waldman M., LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 21, 289–307 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Gaulton A., et al. , The ChEMBL database in 2017. Nucleic Acids Res. 45 (D1), D945–D954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mysinger M. M., Carchia M., Irwin J. J., Shoichet B. K., Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., et al. , Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: The prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 18, 12964–12975 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Pagadala N. S., Syed K., Tuszynski J., Software for molecular docking: A review. Biophys. Rev. 9, 91–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos R., et al. , A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajusz D., Rácz A., Héberger K., Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 7, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein R. M., et al. , Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Butiniet al., . Polypharmacology of dopamine receptor ligands. Prog. Neurobiol. 142, 68–103 (2016). [DOI] [PubMed] [Google Scholar]
- 34.A. E. Moritz, R. B. Free, D. R. Sibley, Advances and challenges in the search for D2 and D3 dopamine receptor-selective compounds. Cell. Signal. 41, 75–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., et al. , Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Yinet al., . Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane. Nature 584, 125–129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. Fanet al., . Haloperidol bound D2 dopamine receptor structure inspired the discovery of subtype selective ligands. Nat. Commun. 11, 1074 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauser A. S., et al. , Pharmacogenomics of GPCR drug targets. Cell 172, 41–54.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer C., Kalliokoski T., Gedeck P., Vulpetti A., The experimental uncertainty of heterogeneous public K(i) data. J. Med. Chem. 55, 5165–5173 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Papadatos G., Gaulton A., Hersey A., Overington J. P., Activity, assay and target data curation and quality in the ChEMBL database. J. Comput. Aided Mol. Des. 29, 885–896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma J., Khedkar V. M., Coutinho E. C., 3D-QSAR in drug design--A review. Curr. Top. Med. Chem. 10, 95–115 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Cramer R. D., Patterson D. E., Bunce J. D., Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 110, 5959–5967 (1988). [DOI] [PubMed] [Google Scholar]
- 43.Sastry G. M., Dixon S. L., Sherman W., Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J. Chem. Inf. Model. 51, 2455–2466 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Alam S., Khan F., 3D-QSAR, Docking, ADME/Tox studies on Flavone analogs reveal anticancer activity through Tankyrase inhibition. Sci. Rep. 9, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cleves A. E., Jain A. N., Structure- and ligand-based virtual screening on DUD-E+: Performance dependence on approximations to the binding pocket. J. Chem. Inf. Model. 60, 4296–4310 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Sastry G. M., Inakollu V. S., Sherman W., Boosting virtual screening enrichments with data fusion: Coalescing hits from two-dimensional fingerprints, shape, and docking. J. Chem. Inf. Model. 53, 1531–1542 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Vass M., et al. , Molecular interaction fingerprint approaches for GPCR drug discovery. Curr. Opin. Pharmacol. 30, 59–68 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Jiang L., Rizzo R. C., Pharmacophore-based similarity scoring for DOCK. J. Phys. Chem. B 119, 1083–1102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu D. Y., Meiler J., RosettaLigandEnsemble: A small-molecule ensemble-driven docking approach. ACS Omega 3, 3655–3664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieth M., Cummins D. J., DoMCoSAR: A novel approach for establishing the docking mode that is consistent with the structure-activity relationship. Application to HIV-1 protease inhibitors and VEGF receptor tyrosine kinase inhibitors. J. Med. Chem. 43, 3020–3032 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Wallach I., Lilien R., Predicting multiple ligand binding modes using self-consistent pharmacophore hypotheses. J. Chem. Inf. Model. 49, 2116–2128 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Renner S., Derksen S., Radestock S., Mörchen F., Maximum common binding modes (MCBM): Consensus docking scoring using multiple ligand information and interaction fingerprints. J. Chem. Inf. Model. 48, 319–332 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Malhotra S., Karanicolas J., When does chemical elaboration induce a ligand to change its binding mode? J. Med. Chem. 60, 128–145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleming N., How artificial intelligence is changing drug discovery. Nature 557, S55–S55 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Eismann S., et al. , Hierarchical, rotation-equivariant neural networks to select structural models of protein complexes. Proteins 89, 493–501 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Ragoza M., Hochuli J., Idrobo E., Sunseri J., Koes D. R., Protein- ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feinberg E. N., et al. , PotentialNet for molecular property prediction. ACS Cent. Sci. 4, 1520–1530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altae-Tran H., Ramsundar B., Pappu A. S., Pande V., Low data drug discovery with one-shot learning. ACS Cent. Sci. 3, 283–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramsundar B., et al. , Is multitask deep learning practical for pharma? J. Chem. Inf. Model. 57, 2068–2076 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Yang K., et al. , Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59, 3370–3388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim J., et al. , Predicting drug-target interaction using a novel graph neural network with 3D structure-embedded graph representation. J. Chem. Inf. Model. 59, 3981–3988 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Chupakhin V., Marcou G., Baskin I., Varnek A., Rognan D., Predicting ligand binding modes from neural networks trained on protein-ligand interaction fingerprints. J. Chem. Inf. Model. 53, 763–772 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Sherman W., Day T., Jacobson M. P., Friesner R. A., Farid R., Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 49, 534–553 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Ravindranath P. A., Forli S., Goodsell D. S., Olson A. J., Sanner M. F., AutoDockFR: Advances in protein-ligand docking with explicitly specified binding site flexibility. PLOS Comput. Biol. 11, e1004586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Da C., Kireev D., Structural protein-ligand interaction fingerprints (SPLIF) for structure-based virtual screening: Method and benchmark study. J. Chem. Inf. Model. 54, 2555–2561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gainza P., et al. , Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 17, 184–192 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Cleves A. E., Jain A. N., Quantitative surface field analysis: Learning causal models to predict ligand binding affinity and pose. J. Comput. Aided Mol. Des. 32, 731–757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Implementations of ComBind and ComBindVS are available in GitHub at https://github.com/drorlab/combind.