Abstract
Objective
Frailty is a prevalent risk factor for adverse outcomes among patients with chronic lung disease. However, identifying frail patients who may benefit from interventions is challenging using standard data sources. We therefore sought to identify phrases in clinical notes in the electronic health record (EHR) that describe actionable frailty syndromes.
Materials and Methods
We used an active learning strategy to select notes from the EHR and annotated each sentence for 4 actionable aspects of frailty: respiratory impairment, musculoskeletal problems, fall risk, and nutritional deficiencies. We compared the performance of regression, tree-based, and neural network models to predict the labels for each sentence. We evaluated performance with the scaled Brier score (SBS), where 1 is perfect and 0 is uninformative, and the positive predictive value (PPV).
Results
We manually annotated 155 952 sentences from 326 patients. Elastic net regression had the best performance across all 4 frailty aspects (SBS 0.52, 95% confidence interval [CI] 0.49–0.54) followed by random forests (SBS 0.49, 95% CI 0.47–0.51), and multi-task neural networks (SBS 0.39, 95% CI 0.37–0.42). For the elastic net model, the PPV for identifying the presence of respiratory impairment was 54.8% (95% CI 53.3%–56.6%) at a sensitivity of 80%.
Discussion
Classification models using EHR notes can effectively identify actionable aspects of frailty among patients living with chronic lung disease. Regression performed better than random forest and neural network models.
Conclusions
NLP-based models offer promising support to population health management programs that seek to identify and refer community-dwelling patients with frailty for evidence-based interventions.
Keywords: natural language processing, machine learning, frailty, population health management, decision support systems, clinical
INTRODUCTION
Frailty is a complex condition commonly defined as a syndrome of deficits in multiple systems that results in decreased stress tolerance.1,2 Frailty is prevalent among patients with chronic lung disease and is associated with lower quality of life and higher rates of hospitalization and death.3–7 For example, among the 250 million patients worldwide with chronic obstructive pulmonary disease (COPD), the most common chronic lung disease, it is estimated that 14%–24% are frail.8,9 Therefore, reducing frailty is an important strategy for improving population health on a large scale.
Although the effects of frailty can be mitigated through evidence-based interventions,10–20 such interventions are underutilized because of logistical barriers to referral and follow up.21–27 Automated clinical decision support (CDS) tools to identify community-dwelling patients with actionable frailty syndromes are therefore needed. A key barrier to developing such automated approaches is the abstract nature of frailty syndromes. Global frailty estimates have been proposed which use structured data elements, such as claims codes and laboratory values.28–32 However, these methods are limited in their ability to guide population health management strategies because global frailty estimates do not indicate the need for specific interventions.
By contrast, the text of clinical notes contains rich information not found in traditional structured data sources.33,34 Thus, analyzing text data may assist in identifying nuanced aspects of frailty that are amenable to evidence-based interventions.35 Such unstructured text data have been used to improve detection of aspects of frailty relevant to preoperative decision making before cardiac interventions,36 and to identify geriatric syndromes37 and characterize their correlation with frailty descriptions.38 While these studies underscore the value of clinical text data to identify frailty, none were designed to support referral decisions by clinicians.
OBJECTIVE
We sought to develop and to perform a temporal external validation of a classification model incorporating unstructured data from the electronic health record (EHR) that could support a population health management program aimed at early identification of actionable aspects of frailty among community-dwelling patients living with chronic lung disease. We hypothesized that, using expertly annotated training labels, a classification model using natural language processing (NLP) could identify text suggestive of a potential benefit from pulmonary rehabilitation, physical therapy, fall reduction programs, or nutrition evaluation.
MATERIALS AND METHODS
Frailty aspects
In consultation with clinical experts in geriatric medicine, physical therapy, respiratory therapy, and pulmonary medicine, we identified the following 4 aspects of frailty with evidence-based treatments that are prevalent among patients with chronic lung disease: respiratory problems that cause functional impairment, musculoskeletal problems that cause functional impairment, risk factors for falling, and features of nutritional deficiencies. While there exist myriad definitions of frailty,1,2,39–46 these 4 aspects are common among people living with chronic lung disease and they are actionable, thus they are suited for identification in a population health management program (see Supplementary Material for further clinical details).10–25,27,47–54
Proposed use case
We present our use case for this prediction model to contextualize the methodologic choices described below (see Supplementary Material for further details). In the future, we plan to deploy a frailty classification model to support a population health management program for community-dwelling patients with chronic lung disease. The model would use EHR data from a fixed look-back period to populate a dashboard with the predicted probability of actionable aspects of frailty for each patient. A population health officer, such as a nurse, would consult the dashboard at regular intervals to review high probability patients. After manual review of a patient’s chart, the population health officer would contact the patient’s care team, the patient, and/or the patient’s caregivers to determine eligibility and appropriateness for the referrals recommended by the model. In future work, we plan to update and continuously train the model using the population health officer’s actions as a gold standard label for these frailty aspects. All software used in the project is open source, and our code repository (https://github.com/weissman-lab/frailtyclassifier) can be used to reproduce our work and implement the classifier in an EHR-based clinical setting.
Population and data collection
Patients were eligible for inclusion in the study if they carried a diagnosis of chronic lung disease and received their care in the University of Pennsylvania Health System (see Supplementary Material for detailed criteria). The date of a patient’s first inpatient or outpatient encounter during the study period was recorded as their qualifying date. Using each patient’s qualifying date as a reference, we gathered their data from the preceding 6 months and excluded all subsequent data, simulating the proposed use case of the model. All patients in the training set were sampled from qualifying dates in 2018, and all patients in the test set were randomly sampled from qualifying dates in 2019. Patients selected for inclusion in the training set were excluded from the test set.
Structured data and unstructured text from clinical notes were extracted from the EHR. Unstructured data included signed clinical notes from outpatient visits with physicians in internal medicine, family medicine, geriatrics, pulmonology, rheumatology, cardiology, or neurology. Structured data fields included demographics, routine laboratory values, vital signs, and utilization metrics, as well as indicators for missing values for each field (Supplementary Table S1 for a complete list and missingness for each element). To the structured data, we applied imputation of missing values, dimensionality reduction, and standardization (see Supplementary Material).
Text preprocessing
All eligible notes in the 6 months preceding the qualifying date were concatenated into a single document for each patient. Based on a manual review of the first 27 patients, the text of several patient survey instruments, frequently included in encounter notes, were automatically removed using regular expressions because they contained statements that did not apply directly to the patient. For example, one questionnaire asked clinicians to place a mark next to relevant statements such as “I never cough” or “I cough all the time.” We also removed most medication lists using the same approach. ScispaCy,55 was used to split notes into sentences and tokens were then converted to lowercase. Clinically relevant multi-word expressions (see Supplementary Material) were identified and joined with an underscore.56
Annotation and active learning
We created a written annotation guide to standardize the label-generation work of human annotators (see Supplementary Material for annotation guide). The guide was developed iteratively with input from clinical experts in frailty syndromes and chronic lung disease and from methodologic experts in clinical informatics and qualitative data analysis. After reviewing notes from an initial set of 13 patients, we developed a draft of the guide. As new or difficult text samples were encountered during the annotation process, new rules were added to the annotation guide to standardize our approach. The guide contained rules for annotation and criteria for positive, negative, and neutral labels for each frailty aspect. Two of 3 trained annotators (FCL, JCP, and TEK) independently labeled the full text of each note, and then 1 of 2 physicians (GEW and JAM) reviewed and adjudicated both sets of annotations. We used WebAnno version 3.6.1, a web-based annotation tool, to capture and process these annotations.57
Each sentence was labeled positive, negative, or neutral for each of the following 4 actionable frailty aspects: respiratory impairment, musculoskeletal problems, fall risk, and nutritional deficiencies. A positive sentence was defined as indicating the presence of the frailty aspect. A negative sentence was defined as indicating the absence or inverse of the frailty aspect. All other sentences, which provided no clear indication about the presence or absence of the frailty aspect, were labeled as neutral.
Notes in the training set were selected for annotation according to an active learning strategy.58–60 In active learning, we started with 139 717 unannotated clinical notes from 46 095 qualifying patients. From this pool, notes were selected for annotation in a series of rounds in which the goal is to sequentially choose only the most informative notes. In the first round, we annotated notes from a purposively sampled group of patients with words that suggested high or low probability of frailty (see Supplementary Material for list of words) and trained a multi-task neural network on these labels. We used the multi-task neural network to make predictions on all of the remaining unlabeled text and calculated entropy for each sentence’s predictions. Then, we took the mean entropy across aspects for the 50% of sentences with highest entropy in the note and selected the notes with the highest values to annotate in the subsequent round. Taking only the top 50% effectively discarded information from less-uncertain sections of notes, allowing us to choose between notes on their relatively more-uncertain sections. We made adjustments to the active learning pipeline after the second round to improve statistical efficiency and inference during the model selection process that were continued throughout all subsequent rounds (see Supplementary Material).
The total sample size for the training set was determined by visualizing a plateau in the learning curve of the multi-task neural network model’s cross-validated performance on the training data. We used Monte Carlo simulation to estimate a test set sample size of 80 000 sentences to detect a 0.1 difference in the multiclass SBS with 80% power at an alpha of 0.05 (see Supplementary Material).
Model training and selection
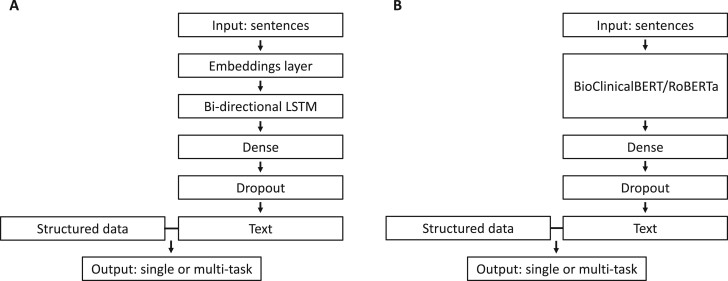
4We compared the performance of 4 model types: multinomial elastic net regression, random forests, single-task neural networks, and multi-task neural networks. Models classified sentences as positive, negative, or neutral for each of the 4 frailty aspects. Elastic net regression, random forests, and single-task neural networks were fit separately for each frailty aspect. Multi-task neural networks were fit to predict all 4 frailty aspects in a single model. Structured data and text features were concatenated as inputs for elastic net regression and random forests. The architecture for single-task and multi-task neural networks was the same except for the output layer (Figure 1). All model types were trained after each round of annotations, but only the best multi-task neural network was used to select the next batch of notes for annotation in the active learning pipeline.
Figure 1.
We compared 2 strategies for modeling text input in neural network models: (A) word2vec embeddings input into a bidirectional long short-term memory (bi-LSTM) layer and (B) a transformer-based language model (BioClinicalBERT or RoBERTa). Each model had either 1 or 2 dense layers with regularization and 256 or 64 units followed by dropout. In the penultimate layer, structured data was concatenated with the output of the final dense layer. In the output layer, single-task neural networks predicted a single frailty aspect and multi-task neural networks predicted all 4 aspects.
Hyperparameters for elastic net regression, random forests, and neural networks were selected using a complete grid search strategy (see Supplementary Material for list of hyperparameters for each model). Three-times repeated 10-fold cross-validation was used to quantify a model’s performance with each set of hyperparameters. The hyperparameters with the best mean cross-validated performance were selected for the final model.
Text featurization
We compared the performance of 3 different types of word embeddings in our models—word2vec, BioClinicalBERT, and RoBERTa. We used 300-dimensional word2vec embeddings that were previously trained on a publicly available medical corpus.56 In the random forests and elastic net regression models, we calculated the minimum, maximum, and mean of each dimension of the word2vec embeddings across the tokens in each sentence and concatenated these values as inputs in the model. In the neural network models, word2vec embeddings were loaded into a nontrainable embeddings layer which was followed by a bidirectional long short-term memory (bi-LSTM) layer (Figure 1A). When using BioClinicalBERT and RoBERTa, we generated 768-dimensional representations of each sentence in our dataset, and trained models using these as the input (Figure 1B). With BioClinicalBERT, a BERT model pretrained on clinical and biomedical text, we specifically used the CLS token.61–63 With RoBERTa, we used an average of token-level embeddings because the RoBERTa CLS token, without further fine-tuning, does not represent a summary of the input sentence.64 For random forest and elastic net regression models, we also used 300-dimensional and 1000-dimensional truncated SVD of n-grams weighted by term frequency-inverse document frequency (TF-IDF).65
Model validation and performance metrics
For each model type, a final model was fit using all sentences in the training set and then used to make predictions on sentences in the test set. We used the multi-class scaled Brier Score (SBS) as the primary performance metric for all models. We chose the multi-class SBS because it is: (1) a composite measure of both discrimination and calibration, (2) intuitive to interpret in that a positive value indicates that a model is better than guessing the event rate, (3) similar in interpretation to the proportion of explained variance (R2), and (4) a strictly proper scoring rule in that it is only maximized by predictions that reflect the true probability distribution.66–69 We calculated a multi-class SBS for each frailty aspect, and then calculated the macro averaged multi-class SBS across all 4 frailty aspects. We also calculated a SBS for each class (positive, negative, and neutral) of each frailty aspect as a dichotomous outcome, and we calculated the macro averaged SBS for each class across all 4 frailty aspects.
The multi-class SBS is based on the multi-class Brier score (BSmc) which in turn is adapted from the original Brier score (BS).70 The original BS is equivalent to the mean-squared error of the predicted probabilities against a binary outcome and is given by:
for pairs of observed outcomes () and predicted probabilities (). This definition is extended such that:
for the case of multiple classes (). The BS ranges from 0 to 1 but is insensitive to the event rate. However, the SBS is defined as:
where is the BS calculated where every prediction is the event rate (). This is:
in the multiclass scenario. Thus, the multi-class SBS ranges from to 1. In summary, the SBS is equal to 1 for perfect predictions, equal to 0 when the model performs equivalently to guessing the event rate, and negative when it performs worse than this guess.
We secondarily assessed the positive predictive value (PPV) over a range of thresholds, the area under the precision recall curve (PR AUC), the receiver operating characteristic area under the curve (ROC AUC), the F1 score, and visual inspection of calibration plots to obtain a full view of each model’s performance. For each statistic calculated on the test set, we used the basic bootstrap method to calculate 2 sided, nonparametric confidence intervals from 1000 bootstrapped replicates.71
This study was deemed exempt by the institutional review board of the University of Pennsylvania. We adhered to the TRIPOD checklist for reporting of prediction models (see Supplementary Material).
Error analysis
To understand the error patterns in our final models and guide refinement of future models for clinical deployment, we selected false positive and false negative cases from the test set for manual review. For this analysis, we chose a classification threshold for each frailty aspect such that the sensitivity was 80%.
Algorithmic equity
Clinical prediction models risk reinforcing existing biased practices when deployed.72 Therefore, we sought to evaluate how well our models performed on historically marginalized groups by sex (male and female) and race (white and nonwhite). Sex, as recorded in the EHR, does not distinguish between karyotypic, anatomic, or other definitions, and is a limited proxy for self-reported gender. We selected the best-performing model in the test set and calculated the SBS and estimated the PPV over a range of thresholds within each subgroup, consistent with previous algorithmic equity evaluations.73,74
RESULTS
We manually annotated 155 861 sentences in the encounter notes of 326 patients to create the training and test sets. Among the patients, 182 (56%) were female and the median age was 69.4 years (IQR 62.9 to 76.9) (Table 1). We performed 5 rounds of active learning until cross-validated training performance plateaued (Supplementary Figure S1). The prevalence of sentences that were positive or negative for any frailty aspect was 9.5% (14 771 sentences). The training and test sets contained a similar proportion of labels for each frailty aspect (Table 2).
Table 1.
Selected demographics, laboratory values, medications, and utilization metrics for patients in the training and test sets
| Training (N = 178) | Test (N = 148) | |
|---|---|---|
| Age, median (IQR) | 68.7 (62.8–76.6) | 71.9 (63.5–77.1) |
| Female, n (%) | 96 (54%) | 86 (58%) |
| Race | ||
| White, n (%) | 120 (67%) | 96 (65%) |
| Black, n (%) | 47 (26%) | 41 (28%) |
| Other/multi-racial, n (%) | 6 (3%) | 7 (5%) |
| Missing, n (%) | 5 (3%) | 4 (3%) |
| BMI, median (IQR) | 27.1 (23.5–31.4) | 28.18 (24.1–33.3) |
| Max supplemental O2 (LPM), median (IQR) | 4.0 (3.0–11.0) | 4.0 (2.0–6.0) |
| Albumin, median (IQR) | 3.9 (3.46–4.17) | 4.1 (3.82–4.2) |
| CO2, median (IQR) | 27.0 (24.0–28.6) | 26.2 (24.9–28.8) |
| Creatinine, median (IQR) | 1.01 (0.84–1.31) | 0.97 (0.80–1.19) |
| Hemoglobin, median (IQR) | 12.2 (10.3–13.8) | 13.2 (11.8–14.4) |
| Number of encounters, median (IQR) | 7.0 (4.0–16.0) | 7.0 (3.8–15.0) |
| Number of ED visits, median (IQR, range) | 0.0 (0.0–0.0, 0.0–8.0) | 0.0 (0.0–0.0, 0.0–14.0) |
| Number of admissions, median (IQR, range) | 0.0 (0.0–0.0, 0.0–22.0) | 0.0 (0.0–0.0, 0.0–3.0) |
| Days hospitalized, median (IQR, range) | 0.0 (0.0–0.7, 0.0–111.5) | 0.0 (0.0–0.01, 0.0–20.2) |
| Unique medications, median (IQR) | 10.0 (4.0–34.0) | 9.0 (5.0–24.0) |
| Number of comorbidities, median (IQR) | 8.0 (4.0–12.0) | 10.0 (5.8–16.3) |
| Elixhauser score, median (IQR) | 2.0 (1.0–4.0) | 2.0 (1.0–4.0) |
Note: See Supplementary Table S2 for the remaining structured data elements not included in this table.
IQR: interquartile range.
Table 2.
Distribution of sentences for each frailty aspect in the training and test sets
| Training | Test | |
|---|---|---|
| Sentences, n | 73 010 | 82 851 |
| Sentences per patient, median (IQR) | 197.5 (86–571) | 396 (195–703) |
| Encounter notes per patient, median (IQR) | 2.0 (1.0–3.0) | 2.5 (1.8–4.0) |
| Respiratory impairment | ||
| Positive, n (%) | 2426 (3.3%) | 2324 (2.8%) |
| Negative, n (%) | 1183 (1.6%) | 1189 (1.4%) |
| Musculoskeletal problem | ||
| Positive, n (%) | 842 (1.2%) | 1095 (1.3%) |
| Negative, n (%) | 582 (0.8%) | 574 (0.7%) |
| Fall risk | ||
| Positive, n (%) | 724 (1.0%) | 586 (0.7%) |
| Negative, n (%) | 1171 (1.6%) | 1044 (1.3%) |
| Nutritional deficiency | ||
| Positive, n (%) | 140 (0.2%) | 93 (0.1%) |
| Negative, n (%) | 399 (0.6%) | 399 (0.5%) |
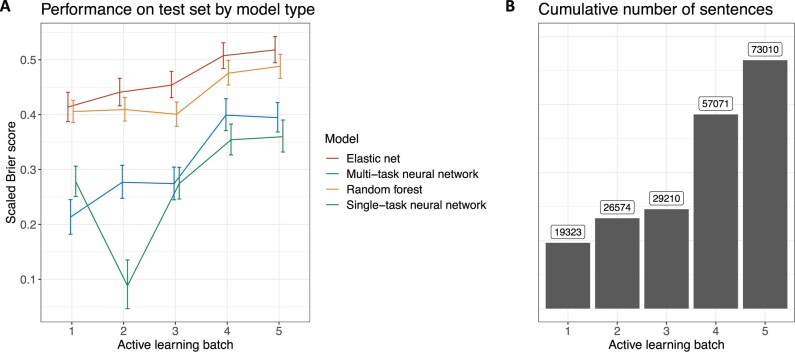
Elastic net regression using the word2vec embeddings had the best performance on the test set in each of the 5 rounds of active learning followed closely by random forests (Figure 2A). The largest gain in performance happened from active learning batch 3 to active learning batch 4, corresponding to the largest addition of training sentences (Figure 2B). Multi-task neural networks generally outperformed single-task neural networks except in the first round of active learning where the training dataset was smallest.
Figure 2.
Model performance on the test set by active learning batch, as measured by the mean of the multi-class scaled Brier scores for each frailty aspect (A). Error bars represent 95% confidence intervals. Elastic net regression had the best performance in each of the 5 rounds of active learning. Random forests also outperformed neural networks in each round. Multi-task neural networks outperformed single-task neural networks in all but the first and third rounds of active learning. Performance varied the most in early rounds when the training sample is the smallest. The cumulative number of sentences increased the most from active learning round 3 to active learning round 4 (B), corresponding to the largest increase in model performance.
After 5 rounds of active learning, elastic net regression had the best average performance for all 4 aspects of frailty followed by random forests, multi-task neural networks, and single-task neural networks (Table 3). Elastic net regression also had the best performance in the positive, negative, and neutral class, followed by random forests, multi-task neural networks, and single-task neural networks (see Supplementary Table S3 for performance for each frailty aspect).
Table 3.
Performance of the best model of each type and featurization approach selected by cross-validation across a range of tuning parameters after 5 rounds of active learning
| Model | Text features | Multiclass SBS (95% CI) | Positive class SBS (95% CI) | Negative class SBS (95% CI) | Neutral class SBS (95% CI) |
|---|---|---|---|---|---|
| Elastic net | word2vec | 0.52 (0.49–0.54) | 0.33 (0.28–0.38) | 0.55 (0.52–0.58) | 0.54 (0.52–0.57) |
| BioClinicalBERT | 0.48 (0.45–0.51) | 0.23 (0.18–0.29) | 0.57 (0.54–0.6) | 0.5 (0.47–0.53) | |
| RoBERTa | 0.46 (0.44–0.49) | 0.22 (0.17–0.28) | 0.55 (0.52–0.59) | 0.49 (0.46–0.51) | |
| TF-IDF 1000-d | 0.4 (0.37–0.42) | 0.21 (0.19–0.24) | 0.47 (0.44–0.5) | 0.42 (0.39–0.44) | |
| TF-IDF 300-d | 0.27 (0.25–0.29) | 0.09 (0.08–0.11) | 0.36 (0.33–0.4) | 0.28 (0.26–0.3) | |
| Random forest | word2vec | 0.49 (0.47–0.51) | 0.32 (0.28–0.35) | 0.53 (0.5–0.56) | 0.51 (0.49–0.53) |
| BioClinicalBERT | 0.44 (0.41–0.46) | 0.23 (0.2–0.27) | 0.55 (0.52–0.58) | 0.45 (0.42–0.47) | |
| RoBERTa | 0.41 (0.39–0.43) | 0.2 (0.17–0.23) | 0.52 (0.49–0.55) | 0.42 (0.4–0.44) | |
| TF-IDF 1000-d | 0.38 (0.36–0.41) | 0.18 (0.14–0.22) | 0.5 (0.47–0.54) | 0.39 (0.36–0.41) | |
| TF-IDF 300-d | 0.36 (0.34–0.38) | 0.15 (0.12–0.18) | 0.49 (0.46–0.53) | 0.36 (0.34–0.39) | |
| Single-task neural network | word2vec | 0.32 (0.29–0.36) | 0.07 (0.02–0.13) | 0.39 (0.35–0.43) | 0.37 (0.34–0.41) |
| BioClinicalBERT | 0.36 (0.33–0.39) | 0.01 (−0.04–0.06) | 0.52 (0.49–0.56) | 0.39 (0.36–0.42) | |
| RoBERTa | 0.32 (0.29–0.35) | −0.11 (−0.17–−0.03) | 0.47 (0.44–0.51) | 0.36 (0.33–0.39) | |
| Multi-task neural network | word2vec | 0.36 (0.33–0.39) | 0.13 (0.09–0.19) | 0.48 (0.44–0.52) | 0.38 (0.35–0.41) |
| BioClinicalBERT | 0.39 (0.37–0.42) | 0.07 (0.02–0.12) | 0.54 (0.51–0.57) | 0.42 (0.39–0.45) | |
| RoBERTa | 0.29 (0.27–0.32) | −0.14 (−0.2–−0.06) | 0.49 (0.46–0.52) | 0.33 (0.3–0.36) |
Note: Each scaled Brier score is the macro average across all 4 frailty aspects. Our primary performance metric is the multiclass scaled Brier score. We also report the scaled Brier score for each class (positive, negative, or neutral) separately and evaluated as a binary outcome against the other 2 classes.
SBS: scaled Brier score.
Among the elastic net regression and random forest models, we observed the highest multi-class SBS point estimate using word2vec embeddings pretrained on clinical text, although differences between word2vec, BioClinicalBERT, and RoBERTa were small. All 3 embedding strategies outperformed TF-IDF weighted n-grams. Performance of the best elastic net regression model with word2vec embeddings was similar with and without patient-level structured data (see Supplementary Material). Among neural network models, we observed the highest multi-class SBS point estimate using BioClinicalBERT, although differences between the 3 embedding strategies were also small.
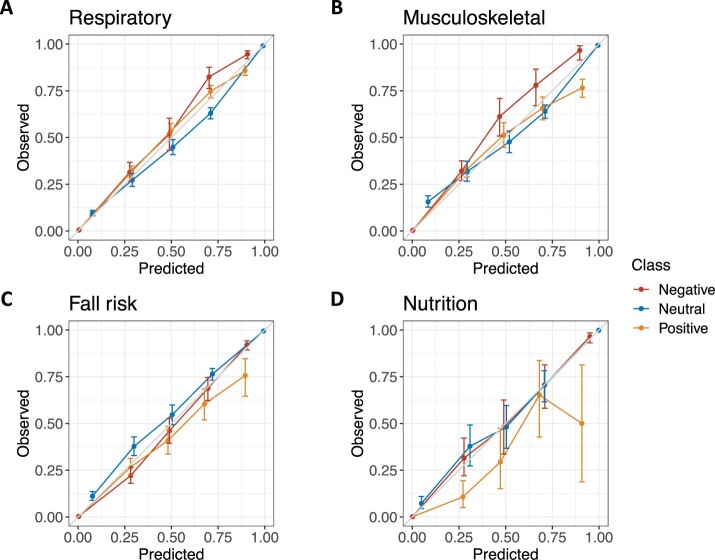
Calibration for the best elastic net model with word2vec embeddings was strong for fall risk, respiratory impairment, and musculoskeletal problem predictions (Figure 3). Calibration was poor for the positive class of nutritional deficiencies. Among random forest (Supplementary Figure S2), single-task neural network (Supplementary Figure S3), and multi-task neural network (Supplementary Figure S4) models, calibration was moderate overall but was not consistent across all aspects and classes.
Figure 3.
Calibration plots for the positive (orange), negative (red), and neutral (blue) class of respiratory impairment (A), musculoskeletal problem (B), fall risk (C), and nutritional deficiency (D) for the best performing elastic net model with word2vec embeddings. Error bars represent 95% confidence intervals.
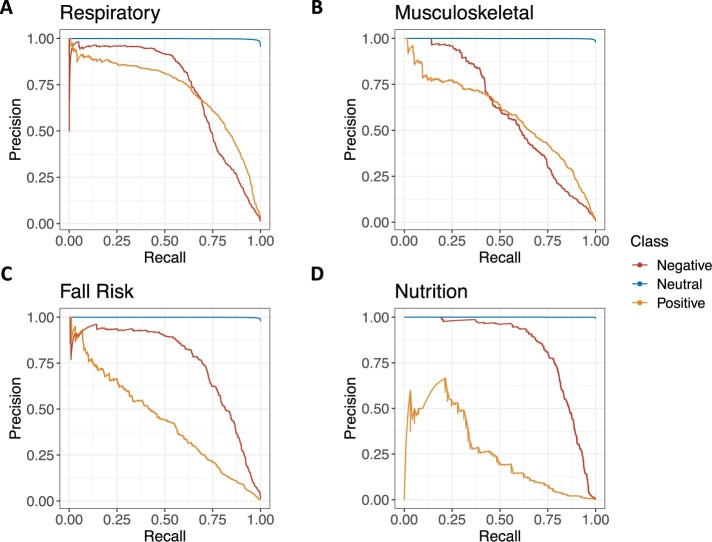
Precision-recall curves show the discrimination of the best elastic net model with word2vec embeddings (Figure 4). Precision-recall curves for the best random forest, and neural network models can be found in Supplementary Figures S5–S7. The PR AUC was 0.71 (95% CI 0.69–0.73) for respiratory impairment positive, 0.72 (95% CI 0.70–0.75) for respiratory impairment negative, 0.44 (95% CI 0.39–0.48) for fall risk positive, 0.75 (95% CI 0.73–0.78) for fall risk negative, 0.58 (95% CI 0.55–0.61) for musculoskeletal problem positive, 0.60 (95% CI 0.57–0.64) for musculoskeletal problem negative, 0.26 (95% CI 0.15–0.34) for nutritional deficiency positive, and 0.83 (95% CI 0.79–0.86) for nutritional deficiency negative (see Supplementary Tables S4 and S5 for complete PR AUC and ROC AUC results for all models). At a threshold with a sensitivity of 80%, the PPV of the elastic net model for each class was as follows: respiratory impairment positive 54.8% (95% CI 53.2%–56.6%), respiratory impairment negative 36.1% (95% CI 34.3%–38.0%), musculoskeletal problem positive 37.5% (95% CI 35.6%–39.6%), musculoskeletal problem negative 21.0% (95% CI 19.1%–22.6%), fall risk positive 14.8% (95% CI 13.5%–16.1%), fall risk negative 52.3% (95% CI 49.9%–54.8%), nutritional deficiency positive 4.0% (CI 3.0%–4.9%), and nutritional deficiency negative 71.8% (CI 67.7%–75.9%) (see Supplementary Tables S6 and S7 for complete PPV and F1 score results for all models).
Figure 4.
Precision-recall curves for the positive (orange), negative (red), and neutral (blue) class of respiratory impairment (A), musculoskeletal problem (B), fall risk (C), and nutritional deficiency (D) for the best performing elastic net model with word2vec embeddings.
In total, 216 patients (66%) identified as white and 101 (31%) identified as nonwhite, of whom 88 (87%) identified as Black or African American. About 9 patients (3%) did not have race recorded. For the best elastic net regression model using word2vec embeddings, the point estimates for SBS were better for white patients compared to nonwhite patients for all predictions (Table 4). Differences in performance were small for respiratory impairment and musculoskeletal problems, the 2 most common labels, and larger for risk and nutritional deficiencies, the 2 least common labels. Across most threshold values, PPV was worse for nonwhite compared to white patients for fall risk and nutritional deficiencies, and PPV was similar for respiratory impairment and musculoskeletal problems (Supplementary Figure S8). Model performance was similar for male and female patients. PPV for positive class predictions at most classification thresholds were better for male patients in respiratory impairment and nutritional deficiencies and better for female patients in fall risk and musculoskeletal problems (Supplementary Figure S9).
Table 4.
Performance of the best model selected by cross-validation, an elastic net regression model using word2vec embeddings, stratified by race and sex
| Frailty aspect | Group | Multiclass SBS (95% CI) | Negative class SBS (95% CI) | Neutral class SBS (95% CI) | Positive class SBS (95% CI) |
|---|---|---|---|---|---|
| Respiratory impairment | White | 0.57 (0.56–0.59) | 0.60 (0.59–0.62) | 0.54 (0.52–0.56) | 0.56 (0.53–0.59) |
| Nonwhite | 0.54 (0.52–0.57) | 0.57 (0.55–0.59) | 0.51 (0.48–0.53) | 0.54 (0.5–0.59) | |
| Male | 0.57 (0.55–0.59) | 0.60 (0.57–0.62) | 0.55 (0.52–0.57) | 0.54 (0.5–0.58) | |
| Female | 0.56 (0.54–0.58) | 0.59 (0.57–0.61) | 0.51 (0.48–0.54) | 0.56 (0.53–0.59) | |
| Musculo-skeletal | White | 0.44 (0.41–0.47) | 0.46 (0.43–0.5) | 0.40 (0.36–0.45) | 0.43 (0.39–0.47) |
| Problem | Nonwhite | 0.41 (0.38–0.44) | 0.44 (0.41–0.47) | 0.39 (0.35–0.43) | 0.37 (0.32–0.43) |
| Male | 0.38 (0.35–0.42) | 0.41 (0.38–0.45) | 0.33 (0.28–0.38) | 0.41 (0.35–0.46) | |
| Female | 0.45 (0.42–0.48) | 0.47 (0.45–0.5) | 0.43 (0.39–0.46) | 0.41 (0.37–0.45) | |
| Fall risk | White | 0.53 (0.5–0.55) | 0.55 (0.52–0.58) | 0.32 (0.28–0.36) | 0.61 (0.58–0.65) |
| Nonwhite | 0.43 (0.39–0.47) | 0.46 (0.42–0.5) | 0.19 (0.12–0.27) | 0.52 (0.48–0.57) | |
| Male | 0.50 (0.47–0.54) | 0.53 (0.5–0.57) | 0.26 (0.19–0.33) | 0.60 (0.56–0.64) | |
| Female | 0.49 (0.46–0.51) | 0.51 (0.48–0.53) | 0.29 (0.24–0.33) | 0.57 (0.54–0.6) | |
| Nutritional deficiency | White | 0.64 (0.6–0.69) | 0.67 (0.63–0.72) | 0.14 (0.01–0.3) | 0.7 (0.66–0.75) |
| Nonwhite | 0.49 (0.42–0.57) | 0.50 (0.43–0.59) | 0.10 (−0.05 to 0.3) | 0.59 (0.51–0.68) | |
| Male | 0.66 (0.6–0.72) | 0.70 (0.65–0.75) | 0.37 (0.25–0.5) | 0.68 (0.61–0.75) | |
| Female | 0.55 (0.5–0.6) | 0.57 (0.52–0.62) | −0.03 (−0.17 to 0.14) | 0.66 (0.61–0.72) |
Note: Each scaled Brier score is the macro average across all 4 frailty aspects. The multiclass scaled Brier score is our primary performance metric. We also report the scaled Brier score for each class (positive, negative, or neutral) separately and evaluated as a binary outcome against the other 2 classes.
SBS: scaled Brier Score.
In our error analysis of false positive and false negative cases (Supplementary Figure S10), manual review revealed that the model struggled to appropriately classify complex phrasing and negations. We also identified errors in annotation that were correctly classified by the model. Compute time, measured in wall clock time, was lower for elastic net regression and random forest models compared to neural networks (Supplementary Table S8).
DISCUSSION
We built several classification models that leveraged structured and unstructured EHR data to identify actionable aspects of frailty in a community-dwelling population with chronic lung disease. Our best performing model, an elastic net regression using pretrained, clinically relevant word2vec embeddings and structured data, was well calibrated across the positive, negative, and neutral classes for fall risk, respiratory impairment, and musculoskeletal problems. Discrimination was also adequate to recognize relevant sentences which would allow trained clinical personnel to rapidly identify patients for interventions to address frailty, consistent with the proposed use case.
Several of our findings have important implications for researchers building text-based classification models to support population health management programs. First, regression models consistently outperformed more complex neural network models at a substantially lower computational cost. Second, clinically trained language models consistently outperformed nonclinical language models. These findings are consistent with prior work demonstrating the advantages of these approaches in the clinical domain.56,62,75,76 Third, iterative development of an annotation guide with input from a multidisciplinary team led to imperfect but reliable labels that were adequate for training and testing. Finally, these findings confirm and extend previous work to detect frailty using EHR data, and text data in particular, by identifying aspects of frailty for which there are evidenced-based interventions, making such a model clinically actionable.33–38,77
We observed small and inconsistent differences in performance by patient sex and larger and consistent differences by patient race, with the models generally performing better for white compared to Black patients. Thus, further work is needed to ensure algorithmic equity in order to avoid reinforcing known disparities in the care of patients living with chronic lung disease.78 These performance differences have several potential explanations including differences in sample size, clinician documentation patterns that may vary by patient subgroup, and factors that affect sharing or disclosure of relevant medical and social history to clinicians. Future approaches should consider recalibration or refitting methods and decomposition of error into bias and variance components to inform further data gathering strategies, including alternative data sources, prior to final deployment of any model.79
This article has several strengths. First, we relied on a team of clinical experts to develop an annotation guide focused on labels that have clear implications for patient care. Second, we used active learning to efficiently acquire training labels until performance plateaued in the training set, and we used a power calculation for the test set in order to make inferences about the relative performance of several models. Third, we evaluated a wide array of model specifications and featurization approaches and used cross-validation to carefully select appropriate tuning parameters and model architectures.
This article should be interpreted in light of several limitations. First, calibration and discrimination were poor for the positive class of nutritional deficiencies, the least common label. For this frailty aspect, performance is not sufficient for deployment. Second, although we observed plateaus in the learning curves for neural network models between the fourth and fifth round of active learning, we cannot exclude the possibility that much larger training corpora may have led to improved performance. Third, models were trained and tested on data from a single health system. However, we have provided reproducible code and methods to allow other systems to fit models on local data. Fourth, we classified sentences within notes rather than making predictions for patients. Therefore, the model will require prospective patient-level validation before implementation. Finally, performance may not be equivalent in a pragmatic setting. Thus, in future work, these models will likely require iterative updating after deployment.
CONCLUSION
This study offers insights into the identification of actionable aspects of frailty using structured and unstructured data found in the EHR. The top-performing NLP-based classification models exhibited good calibration and discrimination, thereby offering a promising approach to support a population health management program in patients with chronic lung disease. This approach also provides a blueprint for similar programs targeted towards other aspects of frailty and for patients living with other chronic diseases. The tools presented here warrant future testing in CDS systems that might overcome logistical and clinical barriers to facilitate the use of interventions that are known to improve patient outcomes.
FUNDING
This work was supported by NIH K23HL141639, NIH R38HL143613-01, and an Accelerator Grant from the University of Pennsylvania Center for Precision Medicine.
AUTHOR CONTRIBUTIONS
GEW and JAM conceived of the study and wrote the annotation guide. ACD, GEW, and JAM wrote code and performed statistical analyses. FCL, JCP, TEK, and JAS performed annotation and consulted on the annotation guide. LHU, BEH, RAH, GEW, ACD, and JAM provided statistical expertise. BPK, JMD, VA, MWS, SDH, GEW, and JAM provided clinical expertise. All authors contributed to refinement of the study protocol and approved the final manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article cannot be shared publicly for the privacy of individuals that participated in the study. The data will be shared on reasonable request to the corresponding author.
Supplementary Material
REFERENCES
- 1. Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001; 56: M146–56. [DOI] [PubMed] [Google Scholar]
- 2. Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging. Sci World J 2001; 1: 323–36. doi:10.1100/tsw.2001.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bone AE, Hepgul N, Kon S, et al. Sarcopenia and frailty in chronic respiratory disease. Chron Respir Dis 2017; 14 (1): 85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaz Fragoso CA, Enright PL, McAvay G, et al. Frailty and respiratory impairment in older persons. Am J Med 2012; 125: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mittal N, Raj R, Islam EA, et al. The frequency of frailty in ambulatory patients with chronic lung diseases. J Prim Care Commun Health 2016; 7: 10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kennedy CC, Novotny PJ, LeBrasseur NK, et al. Frailty and clinical outcomes in chronic obstructive pulmonary disease. Ann Am Thorac Soc 2018; 16: 217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muscedere J, Waters B, Varambally A, et al. The impact of frailty on intensive care unit outcomes: a systematic review and meta-analysis. Intensive Care Med 2017; 43: 1105–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marengoni A, Vetrano DL, Manes-Gravina E, et al. The relationship between COPD and frailty: a systematic review and meta-analysis of observational studies. Chest 2018; 154: 21–40. [DOI] [PubMed] [Google Scholar]
- 9. Lahousse L, Ziere G, Verlinden VJA, et al. Risk of frailty in elderly with COPD: a population-based study. J Gerontol A Biol Sci Med Sci 2016; 71: 689–95. [DOI] [PubMed] [Google Scholar]
- 10. McCarthy B, Casey D, Devane D, et al. Pulmonary rehabilitation for chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2015; (2): CD003793. doi:10.1002/14651858.CD003793.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puhan MA, Gimeno-Santos E, Scharplatz M, et al. Pulmonary rehabilitation following exacerbations of chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2016; 12 (12): CD005305. doi:10.1002/14651858.CD005305.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferreira IM, Brooks D, White J, et al. Nutritional supplementation for stable chronic obstructive pulmonary disease. Cochrane Database Syst Rev 2012; 12: CD000998. [DOI] [PubMed] [Google Scholar]
- 13. Maddocks M, Kon SSC, Canavan JL, et al. Physical frailty and pulmonary rehabilitation in COPD: a prospective cohort study. Thorax 2016; 71: 988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Torres-Sánchez I, Valenza MC, Cabrera-Martos I, et al. Effects of an exercise intervention in frail older patients with chronic obstructive pulmonary disease hospitalized due to an exacerbation: a randomized controlled trial. COPD 2017; 14: 37–42. [DOI] [PubMed] [Google Scholar]
- 15. Franssen FME, Broekhuizen R, Janssen PP, et al. Effects of whole-body exercise training on body composition and functional capacity in normal-weight patients with COPD. Chest 2004; 125: 2021–8. [DOI] [PubMed] [Google Scholar]
- 16. Abizanda P, López MD, García VP, et al. Effects of an oral nutritional supplementation plus physical exercise intervention on the physical function, nutritional status, and quality of life in frail institutionalized older adults: the ACTIVNES Study. J Am Med Dir Assoc 2015; 16: 439.e9–439.e16. [DOI] [PubMed] [Google Scholar]
- 17. Sherrington C, Fairhall NJ, Wallbank GK, et al. Exercise for preventing falls in older people living in the community. Cochrane Database Syst Rev 2019; 1: CD012424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson MC, Campbell AJ, Gardner MM, et al. Preventing injuries in older people by preventing falls: a meta-analysis of individual-level data. J Am Geriatr Soc 2002; 50: 905–11. [DOI] [PubMed] [Google Scholar]
- 19. Liu-Ambrose T, Davis JC, Best JR, et al. Effect of a home-based exercise program on subsequent falls among community-dwelling high-risk older adults after a fall: a randomized clinical trial. JAMA 2019; 321: 2092–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Latham NK, Harris BA, Bean JF, et al. Effect of a home-based exercise program on functional recovery following rehabilitation after hip fracture: a randomized clinical trial. JAMA 2014; 311: 700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harrison SL, Robertson N, Graham CD, et al. Can we identify patients with different illness schema following an acute exacerbation of COPD: a cluster analysis. Respir Med 2014; 108: 319–28. [DOI] [PubMed] [Google Scholar]
- 22. Jones SE, Green SA, Clark AL, et al. Pulmonary rehabilitation following hospitalisation for acute exacerbation of COPD: referrals, uptake and adherence. Thorax 2014; 69: 181–2. [DOI] [PubMed] [Google Scholar]
- 23. Jones AW, Taylor A, Gowler H, et al. Systematic review of interventions to improve patient uptake and completion of pulmonary rehabilitation in COPD. ERJ Open Res 2017; 3 (1): 00089–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones SE, Barker RE, Nolan CM, et al. Pulmonary rehabilitation in patients with an acute exacerbation of chronic obstructive pulmonary disease. J Thorac Dis 2018; 10: S1390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jordan RE, Adab P, Enocson A, et al. Interventions to promote referral, uptake and adherence to pulmonary rehabilitation for people with chronic obstructive pulmonary disease (COPD). Cochrane Database Syst Rev 2017; 2017 (10): CD012813. [Google Scholar]
- 26. Burns ER, Haddad YK, Parker EM. Primary care providers’ discussion of fall prevention approaches with their older adult patients—DocStyles, 2014. Prev Med Rep 2018; 9: 149–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crowe B, Eckstrom E, Lessing JN. Missed opportunity for fall prevention: a teachable moment. JAMA Intern Med 2021; 181 (5): 689–90. [DOI] [PubMed] [Google Scholar]
- 28. Kim DH, Schneeweiss S, Glynn RJ, et al. Measuring frailty in medicare data: development and validation of a claims-based Frailty Index. J Gerontol A Biol Sci Med Sci 2018; 73 (7): 980–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kinosian B, Wieland D, Gu X, et al. Validation of the JEN frailty index in the National Long-Term Care Survey community population: identifying functionally impaired older adults from claims data. BMC Health Serv Res 2018; 18 (1): 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gilbert T, Neuburger J, Kraindler J, et al. Development and validation of a Hospital Frailty Risk Score focusing on older people in acute care settings using electronic hospital records: an observational study. Lancet Lond Engl 2018; 391: 1775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howlett SE, Rockwood MR, Mitnitski A, et al. Standard laboratory tests to identify older adults at increased risk of death. BMC Med 2014; 12: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ellis HL, Wan B, Yeung M, et al. Complementing chronic frailty assessment at hospital admission with an electronic frailty index (FI-Laboratory) comprising routine blood test results. CMAJ Can 2020; 192 (1): E3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koleck TA, Dreisbach C, Bourne PE, et al. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc 2019; 26 (4): 364–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gensheimer MF, Aggarwal S, Benson KRK, et al. Automated model versus treating physician for predicting survival time of patients with metastatic cancer. J Am Med Inform Assoc 2021; 28 (6): 1108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kharrazi H, Anzaldi LJ, Hernandez L, et al. The value of unstructured electronic health record data in geriatric syndrome case identification. J Am Geriatr Soc 2018; 66 (8): 1499–507. [DOI] [PubMed] [Google Scholar]
- 36. Doing-Harris K, Bray BE, Thackeray A, et al. Development of a cardiac-centered frailty ontology. J Biomed Semantics 2019; 10 (1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen T, Dredze M, Weiner JP, et al. Identifying vulnerable older adult populations by contextualizing geriatric syndrome information in clinical notes of electronic health records. J Am Med Inform Assoc 2019; 26 (8-9): 787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anzaldi LJ, Davison A, Boyd CM, et al. Comparing clinician descriptions of frailty and geriatric syndromes using electronic health records: a retrospective cohort study. BMC Geriatr 2017; 17: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gill TM, Baker DI, Gottschalk M, et al. A program to prevent functional decline in physically frail, elderly persons who live at home. N Engl J Med 2002; 347: 1068–74. [DOI] [PubMed] [Google Scholar]
- 40. Speechley M, Tinetti M. Falls and injuries in frail and vigorous community elderly persons. J Am Geriatr Soc 1991; 39: 46–52. [DOI] [PubMed] [Google Scholar]
- 41. Rockwood K, Song X, MacKnight C, et al. A global clinical measure of fitness and frailty in elderly people. CMAJ Can 2005; 173: 489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rockwood K, Stadnyk K, MacKnight C, et al. A brief clinical instrument to classify frailty in elderly people. Lancet Lond Engl 1999; 353: 205–6. [DOI] [PubMed] [Google Scholar]
- 43. Saliba D, Elliott M, Rubenstein LZ, et al. The Vulnerable Elders Survey: a tool for identifying vulnerable older people in the community. J Am Geriatr Soc 2001; 49: 1691–9. [DOI] [PubMed] [Google Scholar]
- 44. Abellan van Kan G, Rolland Y, Bergman H, et al. The I.A.N.A Task Force on frailty assessment of older people in clinical practice. J Nutr Health Aging 2008; 12: 29–37. [DOI] [PubMed] [Google Scholar]
- 45. Winograd CH, Gerety MB, Chung M, et al. Screening for frailty: criteria and predictors of outcomes. J Am Geriatr Soc 1991; 39: 778–84. [DOI] [PubMed] [Google Scholar]
- 46. Buta BJ, Walston JD, Godino JG, et al. Frailty assessment instruments: systematic characterization of the uses and contexts of highly-cited instruments. Ageing Res Rev 2016; 26: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ter Beek L, van der Vaart H, Wempe JB, et al. Coexistence of malnutrition, frailty, physical frailty and disability in patients with COPD starting a pulmonary rehabilitation program. Clin Nutr Edinb Scotl 2020; 39: 2557–63. [DOI] [PubMed] [Google Scholar]
- 48. Ensrud KE, Ewing SK, Taylor BC, et al. Frailty and risk of falls, fracture, and mortality in older women: the study of osteoporotic fractures. J Gerontol A Biol Sci Med Sci 2007; 62: 744–51. [DOI] [PubMed] [Google Scholar]
- 49. Ensrud KE, Ewing SK, Cawthon PM, et al. A comparison of frailty indexes for the prediction of falls, disability, fractures and mortality in older men. J Am Geriatr Soc 2009; 57: 492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nowak A, Hubbard RE. Falls and frailty: lessons from complex systems. J R Soc Med 2009; 102: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spruit MA, Singh SJ, Garvey C, et al. An official American Thoracic Society/European Respiratory Society statement: key concepts and advances in pulmonary rehabilitation. Am J Respir Crit Care Med 2013; 188: e13–64. [DOI] [PubMed] [Google Scholar]
- 52. Goldstein RS, Gort EH, Guyatt GH, et al. Economic analysis of respiratory rehabilitation. Chest 1997; 112: 370–9. [DOI] [PubMed] [Google Scholar]
- 53. Griffiths TL, Phillips CJ, Davies S, et al. Cost effectiveness of an outpatient multidisciplinary pulmonary rehabilitation programme. Thorax 2001; 56: 779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoogendoorn M, Wetering C V, Schols AM, et al. Is INTERdisciplinary COMmunity-based COPD management (INTERCOM) cost-effective? Eur Respir J 2010; 35: 79–87. [DOI] [PubMed] [Google Scholar]
- 55. Neumann M, King D, Beltagy I, et al. ScispaCy: fast and robust models for biomedical natural language processing. In: Proceedings of the 18th BioNLP Workshop and Shared Task; 2019; 319–27. [Google Scholar]
- 56. Flamholz ZN, Crane-Droesch A, Ungar LH, Weissman GE. Word embeddings trained on published case reports are lightweight, effective for clinical tasks, and free of protected health information. J Biomed Inform 2021. doi: 10.1101/19013268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eckart de Castilho R, Mújdricza-Maydt É, Yimam SM, et al. A web-based tool for the integrated annotation of semantic and syntactic structures. In: Proceedings of the Workshop on Language Technology Resources and Tools for Digital Humanities (LT4DH); Osaka, Japan: The COLING 2016 Organizing Committee; 2016; 76–84. https://www.aclweb.org/anthology/W16-4011 Accessed April 21, 2021.
- 58. Schein AI, Ungar LH. Active learning for logistic regression: an evaluation. Mach Learn 2007; 68: 235–65. [Google Scholar]
- 59. Figueroa RL, Zeng-Treitler Q, Kandula S, et al. Predicting sample size required for classification performance. BMC Med Inform Decis Mak 2012; 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lybarger K, Ostendorf M, Yetisgen M. Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. J Biomed Inform 2021; 113: 103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In: NAACL-HLT, vol. 1; January 1, 2019: 4171–86; Stroudsburg, PA.
- 62. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020; 36 (4): 1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop; June 2019: 72–8. [Google Scholar]
- 64. Liu Y, Ott M, Goyal N, et al. RoBERTa: A Robustly Optimized BERT Pretraining Approach. ArXiv190711692 Cs Published Online First: 26 July 2019.http://arxiv.org/abs/1907.11692 Accessed May 12, 2021.
- 65. Ramos J. Using tf-idf to determine word relevance in document queries. In: Proceedings of the First Instructional Conference on Machine Learning. Citeseer 2003; 29–48.
- 66. Steyerberg EW. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. Cham: Springer International Publishing; 2019. [Google Scholar]
- 67. Gneiting T, Raftery AE. Strictly proper scoring rules, prediction, and estimation. J Am Stat Assoc 2007; 102: 359–78. [Google Scholar]
- 68. Johansson U, König R, Niklasson L. Genetic rule extraction optimizing brier score. In: Proceedings of the 12th Annual Conference on Genetic and Evolutionary Computation. New York, NY: Association for Computing Machinery 2010; 1007–14. doi: 10.1145/1830483.1830668.
- 69. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiol Camb Mass 2010; 21 (1): 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brier G. Verification of forecasts expressed in terms of probability. Mon Weather Rev 1950; 78: 1–3. [Google Scholar]
- 71. Canty A, Ripley BD. boot: Bootstrap R (S-Plus) Functions. 2020. https://CRAN.R-project.org/package=boot [Google Scholar]
- 72. Obermeyer Z, Powers B, Vogeli C, et al. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019; 366: 447–53. [DOI] [PubMed] [Google Scholar]
- 73. Corbett-Davies S, Goel S. The measure and mismeasure of fairness: a critical review of fair machine learning. ArXiv180800023 Cs Published Online First: 14 August 2018.http://arxiv.org/abs/1808.00023 Accessed May 21, 2021.
- 74. Weissman GE, Teeple S, Eneanya ND, et al. Effects of neighborhood-level data on performance and algorithmic equity of a model that predicts 30-day heart failure readmissions at an urban academic medical center. J Card Fail 2021; 27 (9): 965–73. doi:10.1016/j.cardfail.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. Npj Digit Med 2018; 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Christopoulou F, Tran TT, Sahu SK, et al. Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. J Am Med Inform Assoc 2020; 27 (1): 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tarekegn A, Ricceri F, Costa G, et al. Predictive modeling for frailty conditions in elderly people: machine learning approaches. JMIR Med Inform 2020; 8 (6): e16678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mamary AJ, Stewart JI, Kinney GL, et al. Race and gender disparities are evident in COPD underdiagnoses across all severities of measured airflow obstruction. Chronic Obstr Pulm Dis J COPD Found 5: 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen IYJohansson FD, Sontag D. Why is my classifier discriminatory? In: Proceedings of the 32nd International Conference on Neural Information Processing Systems; December 3, 2018: 3543–54.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly for the privacy of individuals that participated in the study. The data will be shared on reasonable request to the corresponding author.