Abstract
Currently, monoclonal antibodies (mAbs) are the most used biopharmaceuticals for human therapy. One of the key aspects in their development is the control of effector functions mediated by the interaction between fragment crystallizable (Fc) and Fcγ receptors, which is a secondary mechanism of the action of biotherapeutics. N-glycosylation at the Fc portion can regulate these mechanisms, and much experimental evidence suggests that modifications of glycosidic chains can affect antibody binding to FcγRIIIa, consequently impacting the immune response. In this work, we try to elucidate via in silico procedures the structural role exhibited by glycans, particularly fucose, in mAb conformational freedom that can potentially affect the receptor recognition. By using adalimumab, a marketed IgG1, as a general template, after rebuilding its three-dimensional (3D) structure through homology modeling approaches, we carried out molecular dynamics simulations of three differently glycosylated species: aglycosylated, afucosylated, and fucosylated antibody. Trajectory analysis showed different dynamical behaviors and pointed out that sugars can influence the overall 3D structure of the antibody. As a result, we propose a putative structural mechanism by which the presence of fucose introduces conformational constraints in the whole antibody and not only in the Fc domain, preventing a conformation suitable for the interaction with the receptor. As secondary evidence, we observed a high flexibility of the antibodies that is translated into an asymmetric behavior of Fab portions shown by all the simulated biopolymers, making the dynamical asymmetry a new, to our knowledge, molecular aspect that may be further investigated. In conclusion, these findings can help understand the contribution of sugars on the structural architecture of mAbs, paving the way to novel strategies of pharmaceutical development.
Significance
Monoclonal antibodies are biotechnological drugs used in the treatment of several diseases, and their function is in part regulated by N-glycosylation. In particular, fucosylated antibodies weakly bind the receptor involved in the immune response activation, inducing a downregulation of the immune system. Based on published experimental data, in our study we try to describe via computational methods the conformational behavior of a marketed antibody, chosen as a case study, to better understand why fucosylation provokes a decrease in binding affinity of antibodies for their receptor. Our results lead us to hypothesize that the fucose is responsible for a conformational change of the whole antibody structure that reduces the affinity to the receptor.
Introduction
In the last decades, monoclonal antibodies (mAbs) have been recognized as the most widely used therapeutic biomolecules. According to the Antibody Society, 118 therapeutic mAbs are on the market or in review in EU and US, and the annual “Antibodies to watch in 2021” reports over 800 molecules in clinical phases (1). Their ability to specifically recognize unique targets, restraining the therapeutic action to specific tissues and reducing the risk of side effects, makes mAbs very interesting as biotherapeutics.
Among immunoglobulin G (IgG) subclasses, IgG1s is the most used in therapeutic treatments (2). From a structural point of view, IgG1 is composed of four polypeptidic chains, two light chains (LCs; λ or κ) with a molecular mass of ∼25 kDa and two heavy chains (HCs) of ∼50 kDa, that are connected by a disulfide bond between the conserved Cys216 (LC) and Cys220 (HC), according to the standard human γG1 immunoglobulin (Eu) numbering (3). The two HC are paired by other two disulfide bonds, located in a very flexible portion named the “hinge,” leading to a tetrameric Y-shaped molecule.
mAbs are organized in three functional domains: two fragment antigen binding (Fab) domains and one fragment crystallizable (Fc) that are linked by the hinge (Fig. 1 A) (2).
Figure 1.
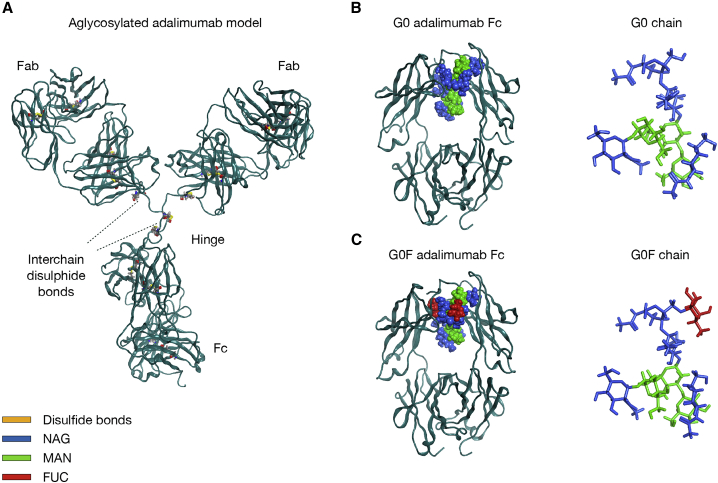
The general IgG1 architecture and the comparison between human and CHO cell glycosylation patterns. (A) IgG1 structural organization: in blue, HCs, divided into variable (VH) and constant (CH1, CH2, CH3) domains; in gray, LCs, organized into one variable (VL) and one constant (CL) domain; interchain disulphide bonds are represented as yellow sticks. (B) Glycosylation patterns expressed in human versus those expressed in CHO cells, including G0 and G0F glycans used in this study. In both panels, sugars are represented according to the Symbol Nomenclature for Glycans (4). To see this figure in color, go online.
Fab domains are responsible for epitopes recognition (5, 6, 7), whereas the Fc triggers the antibody effector functions and antibody recycling by interacting with specific receptors (8, 9, 10). According to published literature, Fc effector functions are regulated and influenced by N-glycosylation that is added as a post-translational modification to the conserved Asn297 (CH2 domain, standard EU numbering (3)), which is located in the N-X-S/T motif (where X is not Pro) (11,12). N-glycosylation patterns mainly depend upon the host cell type used to produce the molecule. Among mammalian cell lines, Chinese hamster ovary (CHO) cells are the most used expression system (60% of marketed products) because they can produce glycosylation patterns compatible with those expressed by humans (13). CHO and human cell lines share a common sugar backbone that is composed of a complex-type biantennary structure, consisting of a mannose (MAN) core and N-acetyl-glucosamine (NAG) units, named G0 (4,14,15). The backbone can be further modified by the addition of fucose, galactose, and sialic acid residues to form more complex species (13). In Fig. 1 B, a comparison between N-glycans highly expressed in humans and in CHO cells is reported, together with the composition of G0 and G0F glycans used in this study.
It is well known that an altered antibody functionality is associated with glycan modifications, whereas a full ablation of the Fc glycosylation affects binding between Fc and its receptors or complement proteins (16,17). However, up to now, the published literature about the impact of glycosylation on the antibody structure and function is still controversial, and a clear explanation of the molecular mechanisms that influence the conformational behavior of IgG1 has not been produced yet (18).
Among the most common modifications, several studies demonstrated that the presence of a core fucose, consisting of a fucose unit attached to the first NAG residue, can dramatically decrease the antibody-dependent cell-mediated cytotoxicity (ADCC) response by affecting the interaction between the antibody and Fcγ receptor IIIa (FcγRIIIa or CD16a) and being responsible for a decrease in ADCC activity of at least two orders in humans, suggesting the crucial role of this sugar in the immune response of therapeutic IgG1 (19, 20, 21, 22, 23, 24, 25, 26).
Starting from crystallographic data of fucosylated and afucosylated Fc in complex with FcγRIIIa, Ferrara and colleagues (27) showed that the fucose makes a steric hindrance effect on the FcγRIIIa glycans involved in the interaction with Fc sugars, proposing that the decrease of binding affinity to the FcγRIIIa, and consequently of ADCC activity, is mainly due to the masking by the fucose of a specific carbohydrate-carbohydrate interaction between Fc and the receptor glycans. However, because of the lack of a whole atomistic structure of an IgG1 in complex with FcγRIIIa, the role of the hinge region and the Fab portions in these recognition mechanisms still remains unknown, and the effect of sugars on the entire antibody structure cannot be elucidated. As a very flexible region, in fact, the hinge drives the conformational exploration of Fab arms, making the antibody able to adopt several conformational states (28, 29, 30). So, an impact of glycosylation on the dynamics of full-length antibodies cannot be neglected and should be adequately investigated.
In addition, to the best of our knowledge, only a few computational studies focused on mAbs conformational behavior via extensive molecular dynamics (MD) simulations of full-length antibodies have been published. In detail, two studies carried out on trastuzumab, a marketed humanized mAb, have been reported, both suggesting a very high flexibility of IgG1 mainly due to different types of contacts and interactions between domains (31,32). More studies are related to specific portions of the antibody (i.e., Fab-antigen complexes, Fc-receptor complexes, etc.) or to isolated glycan chains (33), which, despite their significant scientific contribution, cannot give a complete picture of the molecular mechanisms of IgG. So, the lack of both full x-ray structures and computational studies focused on elucidating the contribution of glycans to the whole conformational behavior of the antibody makes it very difficult to clarify the mechanisms that drive the biological activity of this class of biotherapeutics.
On this basis, the scope of this study is to try to fill the gap between structural information and functional aspects of IgG1 by merging different in silico structural approaches and paving the way to explain new molecular mechanisms of IgG1 functionality. Because of the key role of ADCC activity in immune system activation, both for the endogenous antibodies and for the recombinant mAbs, as a secondary mechanism of action of biotherapeutics, we try to address with structural evidence why a fucosylated antibody exhibits less propensity to interact with FcγRIIIa.
Adalimumab, a well-known therapeutic human IgG1 produced in CHO cells, was used as a model antibody for this study. We are confident that this molecule can be a representative case study because it is well described and deeply characterized in published experimental works and because its amino acid composition is highly conserved with respect to classical IgG1. Three different models of adalimumab have been built by homology: aglycosylated, glycosylated afucosylated (G0), and glycosylated fucosylated (G0F) (Fig. 1 B). Then, MD simulations were carried out per each antibody to evaluate the effects of glycosylation on the antibody tertiary and quaternary structures.
Materials and methods
Homology modeling
The three-dimensional (3D) structure of adalimumab was built by a chimeric homology modeling approach through the “Homology model” tool of the “Protein” module included in the Molecular Operating Environment 2019.01 (MOE, Chemical Computing Group, Montreal, QC, Canada) (34). Fab domains were modeled according to the x-ray structure of adalimumab Fab in complex with TNF-α (PDB: 3WD5) (35), and the hinge and Fc portions were built on the anti-HIV1 antibody (PDB: 1HZH) (36), selected from the MOE antibody templates library. Both templates were optimized and refined with the MOE “Structure preparation” tool (34) to correct any crystallographic issues and add missing hydrogens. Residues’ protonation states were adjusted by the “Protonate 3D” tool at pH 7. The last three C-terminal residues in LC (Gly212, Glu213, and Cys214) of 3WD5.pdb were modeled on 1HZH.pdb to preserve the orientation of both the C-terminal Cys and of the interchain (LC-HC) disulfide bond. Sequence alignment and structural superposition of 3WD5.pdb and 1HZH.pdb were performed to correctly orient the Fab domains with respect to the hinge and Fc and to build each half of the antibody independently, adopting the “Override template” function of the MOE “Homology model” tool. The interchain disulfide bonds (HC-HC) were modeled on 1HZH.pdb.
10 intermediate models were obtained by the homology modeling procedure, and the final structure was selected basing on the best-scoring intermediate one. The score was computed according to the generalized Born-volume integral methodology that calculates the free energy of hydration as the sum of the electrostatic energy term and a cavitation energy based on a volume integral London dispersion energy and not on a surface area, as in the classical generalized Born-surface area function (37). An energy minimization step was carried out until the root mean-square (RMS) gradient reached a value of 0.5 kcal/mol/Å2 by choosing the “medium” option to operate only a moderate relaxation and relieve steric strain. For each heterodimer, free cysteines were manually bonded through the “Builder” tool to assemble the whole molecule. A further energy minimization (RMS gradient 0.01 kcal/mol/Å2) was carried out to refine the final structure. Starting from the optimized aglycosylated antibody, two glycosylated structures of adalimumab were modeled. G0F and G0 glycan chains were added to obtain fucosylated and afucosylated species, respectively. Sugars were manually attached unit by unit to Asn301 (Asn297 according to the IgG1 standard numbering) on both HCs by the MOE “Carbohydrate builder.” A final minimization step was carried out on entire glycosylated models until the RMS gradient reached 0.01 kcal/mol/Å2. All the calculations in the modeling procedure were performed with the AMBER10:EHT force field (38), and the reaction field was applied to treat electrostatics contribution (39,40).
MD simulation of glycosylated hCG
Two MD simulations of glycosylated human chorionic gonadotropin (hCG) were carried out in parallel by NAMD 2.13 package (41) handled by the MOE graphical user interface (GUI) (34) and GROMACS (Groningen Machine for Chemical Simulations) 2020.1 (42). The structure of hCG was downloaded from the Protein Data Bank (PDB; PDB: 1HD4) and optimized by the “Protein preparation” tool included in MOE 2019.01 (34). MD simulations were carried out in transferable intermolecular potential with 3 points (TIP3P) explicit water model and NaCl (0.1 M) for 200 ns in an isothermal-isobaric (NPT) ensemble (p = 101.3 kPa; T = 300 K). In the NAMD simulation, the MOE “Solvate” tool was used to add water and ions to the system. hCG was centered and oriented in a rectangular water box with XYZ sides of 79.5 × 70.9 × 60.7 Å, and periodic boundary conditions were enabled. The solvated system was then prepared for the simulation by an energy minimization run for 5000 steps. In this simulation, the AMBER10:EHT force field (38) was used to parameterize the system. NPT conditions have been maintained by using the Langevin piston Nosé-Hoover method (43,44) for constant pressure and the Langevin thermostat for constant temperature.
The CHARMM-GUI toolbox (45, 46, 47) was instead used to generate GROMACS input files to run a simulation with the CHARMM36 force field (48). The protein was centered in a cubic periodic water box of XYZ side dimensions of 90 Å, and the system was minimized for 5000 steps. An equilibration step 125 ps long was run in a canonical (NVT) ensemble using the Nosé-Hoover thermostat to set constant temperature. The production phase was performed in an NPT ensemble using the Parrinello-Rahman barostat and the Nosé-Hoover thermostat for pressure and temperature control, respectively.
In both simulations, coordinates and velocities were saved every 10 ps, and the integration time step was set to 2 fs. The analysis of glycosidic bond distances over trajectory was performed by the MDTraj tool (49).
MD simulation of antibodies
A 1 μs MD simulation was carried out for each adalimumab model (aglycosylated, G0, and G0F) with the AMBER10:EHT force field (38) in TIP3P explicit water and NPT ensemble (p = 101.3 kPa; T = 300 K). All the three simulations were performed by NAMD 2.13 package (41), and systems were configured by the MOE GUI (34). Each antibody was capped at the N- and C-terminus of each chain with acetyl and N-methyl amide groups, respectively, and centered and oriented in a rectangular periodic box with XYZ side dimensions of 186.801 × 155.314 × 85.0184 Å (aglycosylated), 186.627 × 160.304 × 90.2284 Å (G0), and 185.041 × 161.404 × 90.1505 Å (G0F). Solvation and counterion addition (NaCl 0.1 M) to neutralize the system were performed by the MOE “Solvate” tool. Systems were minimized to an RMS gradient of 0.001 kcal/mol/Å2. The Langevin piston Nosé-Hoover method (43,44) was used to set constant pressure, the Langevin thermostat was applied for temperature control, and the sample time was set to 10 ps and the integration time step to 2 fs.
Analysis of trajectories
Trajectory analyses were performed by MDTraj (49), MOE 2019.01 (34) and in-house Python scripts. To visualize the movement of Fab domains, a reference frame, jointed to Fc and centered in the hinge “H” (as defined by Cys230, Pro231, Pro232, and Cys233), was defined as follows. Asp316 residues in mid-Fc (CH2 regions) were selected as R1 and R2, and the x axis of the reference frame was oriented parallel to the direction defined by the line R1-R2. The y axis was then oriented orthogonal to the plane R1R2H. Finally, the z axis was oriented to obtain a right-handed reference frame. Thus, the computed angles θ and φ are the spherical polar representation of vectors going from H to Met34 on Fab1 (F1) and Met34 on Fab2 (F2), representing the Fab movement with respect to Fc and the Fab rotation around the y axis, respectively. According to this, 0° < θ < 180° and 0° < φ < 360°, meaning that for θ > 90°, the Fab is collapsed onto the Fc. Cluster analysis was performed with an in-house Python script according to the Gromos algorithm (50), and a RMS deviation (RMSD)-based threshold of 7 Å was chosen to discriminate the groups. Hydrogen bond analysis was carried out by MDTraj according to the Baker-Hubbard criterion (51), with a cutoff distance of 0.35 nm, a cutoff angle of 120°, and a cutoff occupancy of 10%.
ED
For each trajectory (aglycosylated, G0, and G0F adalimumab), an essential dynamics (ED) was computed by using the covariance analysis tool included in GROMACS 2020.1 (42). First of all, the mass-weighted covariance matrix of Cα atoms was computed and diagonalized per each antibody with respect to an average structure by the “gmx covar” function. Then, an eigenvalue decomposition of each matrix was applied to generate a set of eigenvectors sorted in a descending eigenvalue index to determine the principal components (PCs) of each system by “gmx anaeig.” The trajectories were filtered along the three most prevalent eigenvectors associated with high eigenvalues using the “-filt” option. The two extreme projections of the components on the average structure were computed per each system to visualize the type of motion described and to deeply analyze the conformational behavior of the molecules.
In silico solvent analysis
The solvent analysis was performed by the “Solvent analysis” tool included in MOE 2019.01 to estimate the solvent contribution to the glycans-protein and glycans-glycans interaction. The program computes the distribution functions of the solvent moieties and the involved free energy. The calculation was carried out on medoid structures with the AMBER10:EHT force field and using the 3D reference interaction site model (3D-RISM) (52, 53, 54). The “solute” mode was used, adding a concentration of NaCl 100 mM. For the representation, a cutoff distance of 5 Å with respect to the selected atoms (CH2 domains and glycans) was chosen, and only water densities with negative hydration free energy (dG) values were displayed.
Cutoff values were set as fourfold denser than bulk, and a multiplicator factor of 1 was applied to water density grid isolevels to improve the graphical representation.
Results
Force field parameter validation for description of glycans
The main challenge in studying glycosylated proteins through MD simulations is the treatment of glycans, whose experimental parameters are fully characterized only in a few force fields. Nowadays, GLYCAM (55) is the only force field exclusively dedicated to the computational analysis of sugars, but it is not suitable to parameterize glycoproteins if not in combination with other force fields. For this reason, to carry out our simulations, we decided to use the AMBER10:EHT (38,56) generalist force field, which is suitable for both proteins and small molecules. AMBER10:EHT was validated for glycans parametrization by comparing in silico MD results with experimental data. Specifically, two MD simulations of a di-galactosylated small protein, hCG, for which the NMR structure was solved (PDB: 1HD4) (57), were carried out in explicit water for 200 ns (Fig. S1 A).
First, a simulation of N-glycosylated hCG was run with NAMD 2.13 software (41) and the AMBER10:EHT force field (38), and the glycosidic bond distances computed in silico, including the one between the ND2 atom of glycosylated Asn78 and the C1 atom of the first NAG unit, were compared to experimental nuclear Overhauser effect distances reported in the structure. This analysis showed that distance values are strongly comparable and suggests that the AMBER10:EHT (38) force field can be considered good for parameterization of N-glycosylated systems investigated in this study. However, to doubly validate our results, another simulation was carried out with the GROMACS 2020.1 package using the CHARMM36 force field (48) that includes specific parameters for carbohydrates.
Data analysis showed that bond distances are conserved over time in both simulations, with values spanning the same range of experimental nuclear Overhauser effect distances (Fig. S1, B and C; Table S1). This confirms a suitable parameterization of the glycan chain and of N-glycosylated proteins and validates the use of the AMBER10:EHT force field for the glycans studied in our research.
Homology modeling and MD simulations of aglycosylated, G0, and G0F adalimumab
The 3D model of aglycosylated adalimumab was obtained by a chimeric homology modeling approach, as described in previous sections (see Homology modeling) and presents all the features described in the the schematic representation in Fig. 1. The structured domains share a conserved secondary structure with respect to crystallographic templates, and as expected, all domains are characterized by β-sheets connected by loops, whereas the hinge region is completely not structured (Fig. 2 A). The Ramachandran plot of the final model showed 15 outliers, all located in loop regions (data not shown). The model was then modified by the addition of glycan chains leading to three different species overall: aglycosylated, G0, and G0F adalimumab (Fig. 2, B and C).
Figure 2.
Homology model of aglycosylated adalimumab and glycosylated Fc portions. (A) The chimeric homology model of adalimumab rendered as ribbons and the structural representation of afucosylated (B) and fucosylated (C) Fc portion of adalimumab, together with G0 and G0F glycan chains represented as sticks, colored according to the Symbol Nomenclature for Glycans system (4). To see this figure in color, go online.
Three independent MD simulations, 1 μs long, were performed in parallel for the three mAbs to estimate the effects of different glycosylation patterns and the impact of glycosylation abrogation on the 3D structure of the protein.
To estimate the convergence of simulations, both the autocorrelation of potential energy and RMSD analysis were computed. More specifically, the autocorrelation plots (Fig. S2) show that in all systems, the potential energy reaches a convergence within the first 5000 timeframes (corresponding to 50 ns of MD), suggesting that the systems equilibrize.
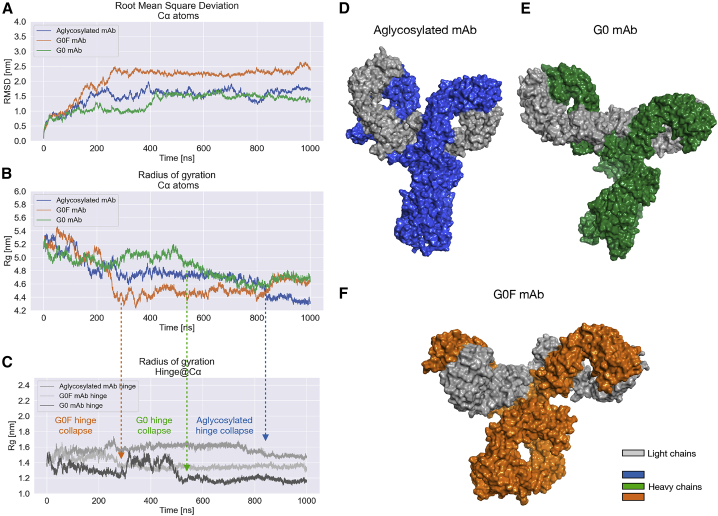
On the other hand, the RMSD of atomic positions was computed for Cα atoms with respect to the homology model, showing that all systems get out of the equilibration phase at different points. Together with this, three different RMSD profiles were recognized, indicating a significant difference among the three mAbs in the explored conformational space within the simulated time window (Fig. 3 A). In detail, the G0 mAb reaches a first RMSD equilibrium in ∼100 ns with a maximal RMSD value of 1.25 nm, and then a transition is observed after 400 ns, leading to another equilibrium state around 1.5 nm; the G0F antibody shows an RMSD plateau only after 250 ns, reaching a maximal deviation of 2.5 nm; finally, the RMSD of the aglycosylated adalimumab, as for the G0F antibody, oscillates in the first part of simulation, with intermediate RMSD values with respect to the other systems (between 1 and 2 nm), but reaches an equilibrium around 1.5 nm after 400 ns. To further confirm system equilibration and convergence, the RMSD calculation was performed for single antibody domains, demonstrating that they are all intrinsically stable and preserve their secondary structure, as suggested also by the structural analysis (see Fig. S3 for the analysis of two Fab regions, Fig. 4 for the Fc one). However, the RMSD profile of Fab1 in the G0F antibody suggests that this domain can explore different orientations, as reported by the structural superposition in Fig. S3 A, and that this behavior is probably responsible for the initial instability observed for the G0F antibody. The convergence of simulations was estimated also computing all the structural and dynamical observables by the block analysis technique, as reported in the Appendix in the Supporting material.
Figure 3.
RMSD profiles, radius of gyration, and centroid structures isolated by cluster analysis. (A) RMSD of Cα positions computed over trajectory per all three adalimumab forms: aglycosylated (blue) shows an intermediate profile with respect to other antibodies, with some RMSD oscillation during the simulation; G0 (green) reaches a plateau state in 100 ns with a shift around 200 ns; and G0F (orange) reaches an equilibrium only after 250 ns. (B) The radius of gyration analysis showed three different profiles describing three different conformations. The lowest Rg value (∼4.4 nm) has been detected for the G0F mAb (in orange), suggesting a compact conformation, whereas both the aglycosylated (in blue) and the G0 (in green) show higher values, likely corresponding to extended conformations. (C) Rg of hinge shows that in all the proteins, there is a collapse in this region. (D–F) Medoid structures identified by cluster analysis and rendered as gray (LC) and blue, green, and orange (HC) molecular surface. Glycans are not displayed in this picture. To see this figure in color, go online.
Figure 4.
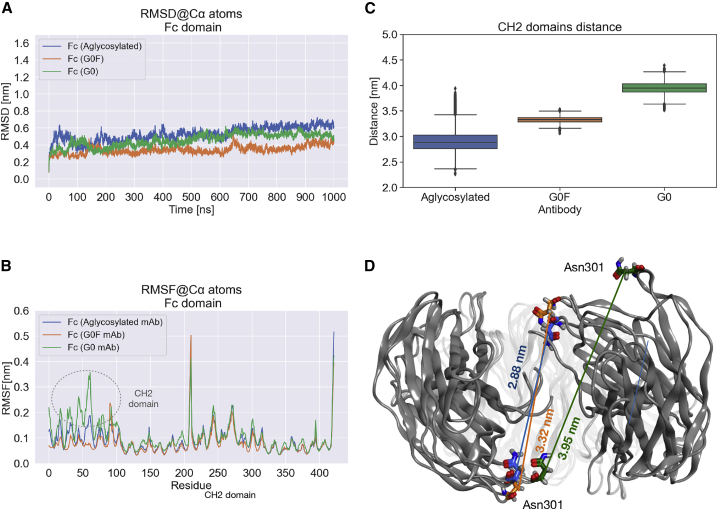
RMSD and RMSF of Fc and distances distribution between CH2 domains. (A) The RMSD computed for Cα atoms of Fc portions and (B) the RMSF analysis show quite overlapping trends among the three Fc, with a higher fluctuation in the first CH2 region of G0 mAb. (C) Box plots of Asn301-Asn301 Cα distances computed in the last 6000 timeframes, showing the higher opening of G0 Fc. On the y axis, distances are reported in nanometers, and on the x axis, the antibody species is given. Outliers are shown as diamonds. (D) Structural superposition of medoid structures with median values of each distribution. To see this figure in color, go online.
As a further validation of the data reported above and of those reported below, two replicas of each system were carried out as described in Supporting materials and methods. According to the results of the other two simulations, the convergence of the systems and the different dynamical behavior were further confirmed. Specifically, RMSD profiles computed for replicas (Fig. S4) confirmed the presence of three different trends among antibody species, and especially the difference between the two glycosylated ones, thus suggesting that glycosylation can potentially influence the conformational exploration of these biomolecules. Considering the RMSDs of the aglycosylated antibody, three different trends are observed among the replicas, suggesting a higher propensity of this species to explore multiple conformational states. The RMS fluctuation (RMSF) of Cα atoms was computed excluding the first 200 ns of trajectory to assess domain flexibility, showing comparable trends across the replicas and between the species and suggesting in another way the system convergence observed by RMSD (Fig. S5).
Radius of gyration analysis
The radius of gyration (Rg) of the three molecules was calculated over trajectories to estimate the preferred conformation for the antibodies (Fig. 3 B). In fact, Rg is defined as the mass-weighted RMS distance of a set of atoms from their common center of mass (58). This analysis can provide an insight of the overall dimensions of the protein and an idea of how much the protein conformation is globular rather than extended. According to this parameter, the aglycosylated and G0 adalimumab show a more extended and relaxed conformation than the G0F antibody, which, during the dynamics, seems to collapse in a more compact assembly. This is inferred from the higher values of Rg observed for aglycosylated (∼4.7 nm) and G0 (∼5 nm) and the lower value showed by the G0F antibody (∼4.4 nm). However, G0 mAb also shows a decrease of its Rg value after 500 ns of simulation, leading to a trend overlapping that of aglycosylated species. This change in Rg profile is mainly due to a collapse of the hinge region, as demonstrated by the Rg calculated for this portion (Fig. 3 C). With Rg potentially being affected by both the hinge collapse, that occurs in all antibodies, and the rotation of domains, a cluster analysis (Fig. S6) was performed to identify a reference structure for each antibody to use for visually comparing the conformations and for illustrating analysis results. To this purpose, the medoid structure of the most populated cluster was selected per each mAb, showing three completely different conformations, which are reported in Fig. 3, D–F. As expected from Rg analysis, aglycosylated and G0 antibodies show a comparable conformation, in which Fab portions are positioned far away from Fc and the hinge presents an extended orientation, with a limited effect of hinge behavior on the overall conformation of the G0 antibody (please see Structural analysis for further details).
Looking at the G0F form, a more compact assembly can be recognized, in which Fab domains, especially one of the two, are collapsed on the Fc and the hinge region is highly compacted.
Rg analysis was also performed for replicas, and the trends suggest that there is a significant conformational change in G0F antibody not occurring in the G0 one, which maintains a Y-shaped architecture, as demonstrated by an Rg plateau around 5 nm. These data further confirm that the Rg oscillation observed for G0 mAb is a sporadic event, restricted only to the first simulation. Considering overall the behavior of the aglycosylated antibody, in some cases its results are comparable to G0 and in others to G0F. These data, together with RMSD profiles, suggest that the absence of glycans can contribute to a higher flexibility of the molecule that results in it being freer to explore different states. In Fig. S7, the Rg profiles computed for each species in each replica are reported together with their medoid structures computed by cluster analysis. The identified structures are different among the species but conserved across the replicas except for the aglycosylated mAb, which, as mentioned before, assumes different orientations. Specifically, as expected from the Rg analysis, aglycosylated and G0 antibodies show a comparable Y-shaped conformation only in the first replica. In the other two simulations, whereas the G0 antibody confirmed the Y-shaped form, the aglycosylated one reached a T-shaped conformation. Looking at the G0F mAb, the more T-shaped compact assembly was observed in all the replicas, further confirming preliminary observations.
Structural analysis
A structural analysis was computed specifically for the Fc portion to highlight local differences among the three antibody forms. The RMSD contribution of this domain to the whole adalimumab dynamics was isolated, showing very similar trends among the three species (Fig. 4 A). RMSF calculation showed overlapped fluctuation trends and confirmed that, despite the different glycosylation pattern, the dynamical behavior of the three Fc domains is almost conserved (Fig. 4 B). However, a higher fluctuation was observed in the first CH2 domain of G0 antibody than in the others, suggesting a structural rearrangement of this portion. Of note, CH2 domains contain the glycosylated Asn301 (Asn297 according to the standard EU numbering (3)), and in principle, their dynamics can exert a key role in modulating the behavior of antibodies. To further assess this change, the distribution of the distances between Cα atoms of the two Asn301 residues was computed for the last 6000 timeframes (Fig. 4 C). As a result, the G0 Fc presents the highest distance value corresponding to an “open” conformation, as shown also by the representative structure reported in Fig. 4 D. Considering the aglycosylated and G0F adalimumab, they present more comparable distance distributions, with median values lower than those of G0. These data, together with the structural representation, suggest a “closed” Fc conformation in these two species. The presence of a closed structure in aglycosylated Fc has been already observed in many published works (59), pointing out that an open conformation of CH2 domains, mediated by the presence of sugars, is needed to allow FcγR recognition. Because a closed conformation has also been observed for the fucosylated antibody, although with slightly higher distance, a similar behavior of this mAb to the aglycosylated one in terms of receptor recognition can be hypothesized. Furthermore, the same analysis was performed for replicas, confirming the higher opening of CH2 domains in the G0 mAb with respect to G0F (Fig. S8) and showing a wider distance distribution for the aglycosylated antibody, suggesting in another way the major conformational variability of this species.
We also decided to investigate the behavior of Fab domains by computing the angles that they describe with respect to the hinge and the Fc. In particular, two angles were considered: the first one was called “θ” and explains how much the Fab comes closer to the Fc; the second one was called “φ” and defines the rotation of Fab around a central axis that passes across the hinge (please see Materials and methods for further details and Fig. 5 A for a schematic representation). The two angles can be considered as coordinates that define the position of Fab domains in space. So, as a result, we identified the most preferred positions of the two Fabs for each antibody. To simplify data discussion, we named the two domains Fab1 and Fab2, according to the numbering used in the input files generated for the MD.
Figure 5.
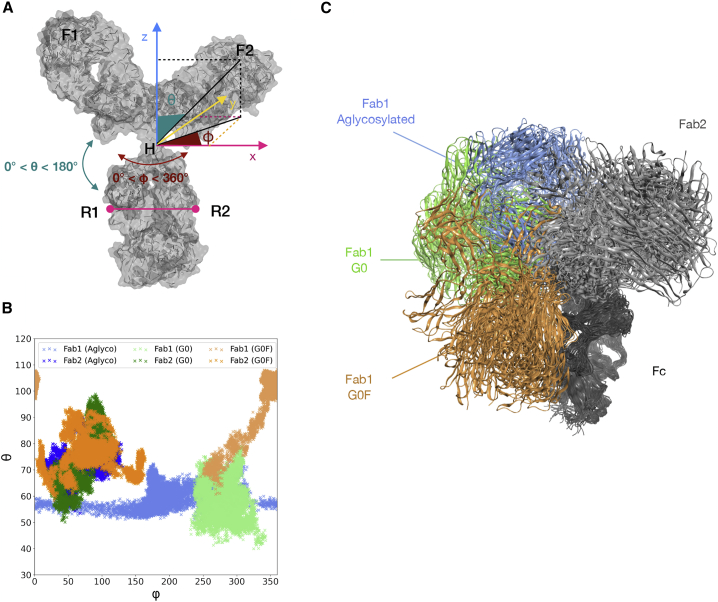
Schematic representation and scatter plots of φ and θ angles. (A) A schematic representation of the angles considered for calculation. H, the hinge region, is the center of a system defined by x (parallel to R1R2), y (perpendicular to x and defined by the plane R1R2H), and z. φ and θ are the spherical polar representation of vectors going from H to Met34 on Fab1(F1) and Met34 on Fab2 (F2). (B) Scatter plots of the φ and θ angles computed for aglycosylated, G0, and G0F antibodies, pointing out the very similar behavior of Fab2 domains and the different exploration performed by Fab1. (C) Structural superposition of 10 conformations per mAb sampled during the MD, showing comparable orientations of Fab2 domains and different orientations of Fab1. In the plots, φ- and θ-values of Fab1 are colored in light blue (aglycosylated), light green (G0), and light orange (G0F); values of Fab2 are colored in dark blue (aglycosylated), dark green (G0), and dark orange (G0F). Structures are rendered as ribbons colored with the same color code. To see this figure in color, go online.
In all the antibodies, both θ and φ explored for Fab2 a more restricted range of values than for Fab1, suggesting a limited conformational space sampling for this domain (Fig. 5 B). Looking at Fab1 instead, especially in aglycosylated and G0F antibodies, there is a strong variability of the φ angle, suggesting a certain rotation of this domain. Then, an exploration of θ was mainly observed for G0F mAb, in which this angle assumes values ≥90°. This suggests a conformational transition, and more specifically the collapse of this domain onto the Fc. Considering the G0 antibody, very stable values of both θ and φ were observed, suggesting the higher conformational stability of this species and a lower rotation propensity of its Fabs, as already shown by previous analyses. Moreover, the structural superposition of 10 conformations isolated for each antibody with respect to Fc better showed that, whereas the position of Fab2 is almost conserved, Fab1 explores different states according to the glycosylation pattern of the antibody (Fig. 5 C). This analysis was also computed for replicas as reported in Fig. S9, confirming the trends already observed and suggesting that most of the conformational variability is related to the Fab1 domain. Because a certain variability can be observed among replicas, it is necessary to clarify that this calculation has been performed assuming the Fc as a rigid body, without considering its rotation and vibration modes that, of course, may influence the results. Moreover, a cumulative statistical distribution was computed for these angles considering the last 6000 timeframes of the three simulations. According to these data (Fig. S10), the propensity of the G0F mAb to assume a compact T-shaped conformation and at the same time the propensity of G0 to maintain an extended Y-shaped form are confirmed. Surely, the aglycosylated antibody instead is more prone to explore different states around both the θ and φ angles than the others, suggesting that its behavior could be due to the absence of the modulatory effect mediated by glycosylation.
Overall, this analysis suggests that a single antibody molecule can show a huge number of degrees of freedom that allow the protein to explore different conformations. This mechanism is essentially due to the high flexibility of hinge portion that, because it is a nonstructured region, can assume many orientations driving the exploration of Fab domains. However, according to our data, the orientation assumed by the hinge and consequently by Fab arms can be influenced by the presence of glycans and in particular by the fucose. These results are aligned with the Rg analysis described above that shows 1) a significant conformational change in G0F mAb that reaches a compact conformation during the dynamics, 2) an intermediate dynamical behavior of the aglycosylated antibody, and 3) a stable conformational trend for the G0 one, confirming the different conformational freedom of the molecules. In conclusion, the presence of fucose seems to be responsible for the loss of the typical antibody structural architecture, and we hypothesize that this is the cause of the lower affinity to FcγRIIIa.
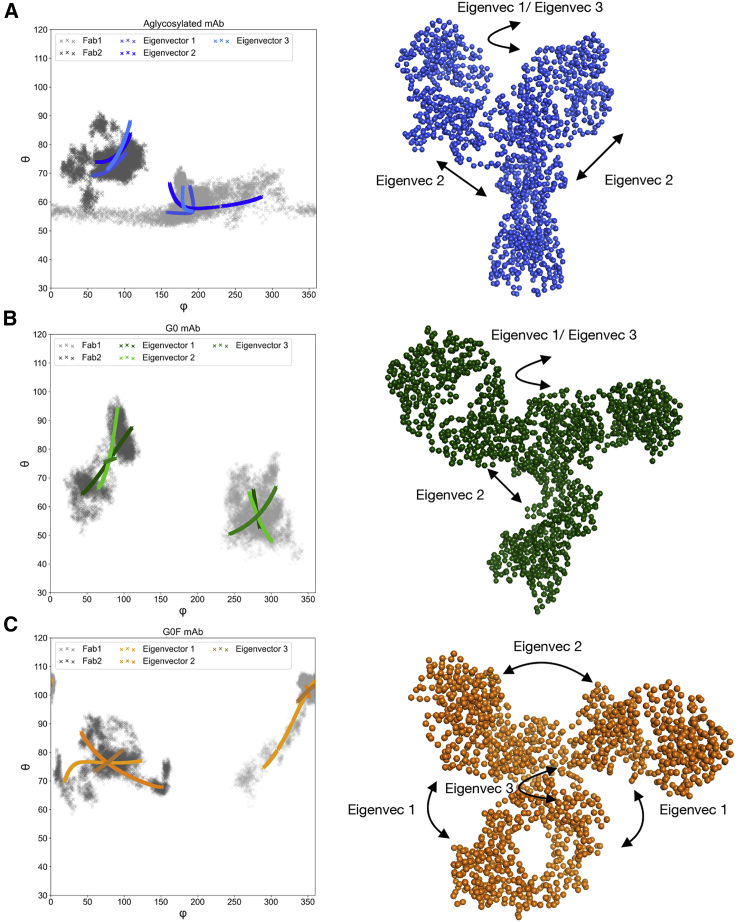
PC analysis
ED was carried out by GROMACS 2020.1 (42) to identify principal coordinated motions of the protein in the three systems and to better explain the three observed conformations. The first three PCs were considered for this analysis. The statistical significance of observing the first three PCs is confirmed by the normalized PC plot (Fig. S11), in which the first 15 eigenvectors of each replica are plotted against their percent contribution to the overall motion of the molecules. According to these data, the sum of the first three components represents more than 85% of the motion for all adalimumab species, thus suggesting that considering these eigenvectors is sufficient to define the most of protein dynamics in all the simulations. As a representative case, in this section we discuss only the results related to the first replica. Trajectories were filtered along the first three PCs, and the calculation of the Fab angles previously discussed was repeated on these trajectories. This was useful to understand whether there is a correlation between the observed motions and the Fab exploration. According to the results, the first three modes can describe the motion of Fab domains. In fact, looking at the three scatter plots reported in Fig. 6, the exploration of both θ and φ angles is described by the three isolated PCs in all the systems. In particular, in the aglycosylated mAb, eigenvector 1 and eigenvector 2 describe the variation of the φ angle on Fab1 and the θ angle on Fab2, and eigenvector 3 represents a variation of the θ angle on both Fab domains (Fig. 6 A). In the G0 antibody, all three eigenvectors essentially describe the θ variation on both Fabs, suggesting that in this case, their rotation is not a principal mode and confirming what was suggested by the Rg analysis (Fig. 6 B). Finally, in G0F adalimumab, eigenvector 1 is responsible for the θ variation on Fab1 and φ variation on Fab2, whereas eigenvector 2 and eigenvector 3 consist mainly of the θ variation on Fab2 (Fig. 6 C). As a conclusion of this analysis, all the species are more prone to explore around the θ angle, suggesting that their dynamics is characterized by breathing motions in which the Fab domains can come closer to or farther away from Fc. However, a rotational component (φ) is still present, meaning that because of the hinge flexibility, domains can explore a wide 3D space along several directions. Furthermore, this analysis confirmed the wider exploration of the θ angle on both Fabs of G0F antibody than in the others, better showing the motions that induce a compact T-shaped conformation. Moreover, extreme eigenvector positions were projected on the trajectory to further describe the type of motion. For aglycosylated and G0 mAbs, the isolated motions are very similar, essentially consisting of Fab rotations (eigenvectors 1 and 3) and hinge stretching (eigenvector 2). In the case of G0F antibody instead, eigenvector 1 is a contraction of the entire structure that induces Fab domains to collapse on the Fc, as described by the variation of the θ angle; eigenvector 2 includes the in-and-out movement of Fab domains with each other; eigenvector 3 is a rotation of all domains around a hinge, as described by the variation of the φ angle. Overall, these findings further confirm that the aglycosylated and G0 mAbs can show comparable dynamics, whereas the G0F one is characterized by a completely different conformational variability, suggesting the key role of fucose in modulating the structural behavior of IgG1 and indicating that the three different dynamical behaviors are likely responsible for different biological functions.
Figure 6.
Principal component analysis. Bidimensional projection of first three PCs identified by ED on the same subspace of φ and θ angles and schematic representation of the isolated motions of aglycosylated (A), G0 (B), and G0F (C) mAbs. Scatter plots show how the first three eigenvectors describe the exploration of φ and θ dimensions in all the systems, with a particular effect on θ in G0F antibody, which can explain the T-shaped conformation. In all mAbs, a rotational component is present, as shown by the exploration on φ; this exploration is less evident in the G0 antibody. The schematic representation of the modes shows that the aglycosylated and the G0 mAbs are characterized by a comparable behavior, with a rotational mode (eigenvectors 1 and 3) and hinge stretching (eigenvector 2), whereas G0F shows a different dynamics, including breathing motions that allow the approaching of Fabs to Fc (eigenvectors 1 and 2) and a rotational component (eigenvector 3) that affects all domains. To see this figure in color, go online.
Sugars analysis
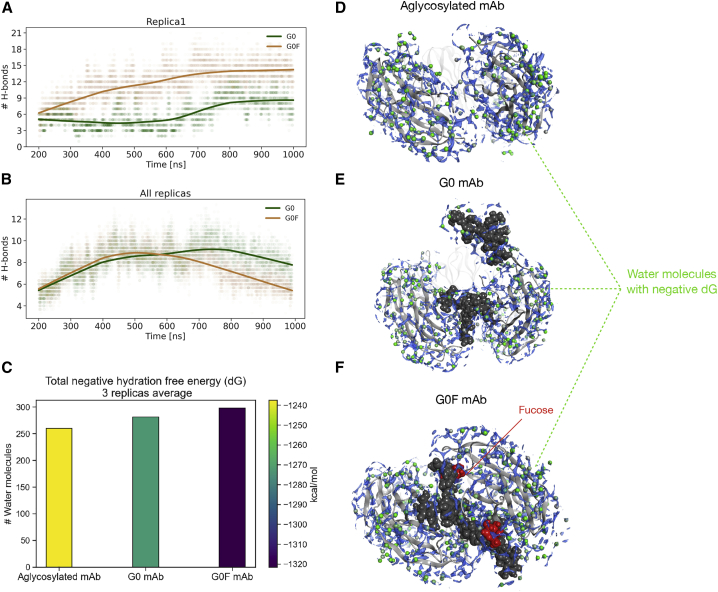
An analysis of dihedral angles that define the glycosidic bonds among glycan residues was performed to estimate the conformational space explored by glycans and to assess their contribution to the variability of the dynamic behavior in the two glycosylated molecules (Fig. S12). To facilitate data discussion, sugars were numbered progressively, and the two chains attached to each half of Fc were named chain A and chain B. This analysis was performed for all replicas, excluding from calculation the first 200 ns, which were considered as an equilibration step. As a result, a prevalently unimodal distribution of angles was observed in both G0 and G0F chains with some exceptions represented by terminal residues, i.e., MAN5-NAG7 in G0-chain B and G0F-chain A, thus suggesting that sugars have also reached conformational stability. An analysis of the hydrogen bond (H-bond) network between sugars and antibodies was performed to further assess the role of carbohydrates in stabilizing or destabilizing the antibody 3D structure. The total number of H-bonds computed over time per each glycan chain (intended as the sum of chain A and chain B) is reported in Fig. 7 A. The lowess nonparametric interpolation shows the propensity of G0F glycans in replica 1 to form more bonds with the protein than G0. However, considering the mean number of bonds computed for the three simulations (Fig. 7 B), the trends of the two chains are comparable, suggesting that this analysis can provide only a partial description of what happens in the Fc region. Although many crystallographic structures available report carbohydrate-carbohydrate interactions within the Fc core (60), H-bonds analysis did not identify interactions between the two glycan chains. Thus, we supposed that this could be due to the presence of water bridges in the pocket that coordinate the interactions between glycans. According to the hydrophilic nature of sugars, in fact, water molecules may play a key role in stabilizing the interaction network, and to further assess this feature, a solvent analysis was performed with the 3D-RISM. As a result, the time-averaged distribution of water H and O densities, along with free-energy maps for analyzing solvent stability and solvation contributions to binding free energy, is computed. The calculation was performed on medoid structures of all the replicas (Fig. S13), and Fig. 7 C contains a bar chart that quantitatively reports the mean number of water molecules predicted per each mAb in CH2 domains and associated with a negative hydration free energy (dG) together with the average dG of the three replicas. In Fig. 7, D–F, water densities with negative dG values located in the Fc cavity are displayed, considering medoids of replica 1. These results show that the solvent contribution in the Fc can change according to glycosylation. Specifically, in G0 and aglycosylated Fc, the presence of water seems to be disfavored. In the first case, this is mainly due to the position of G0 sugar chains that are placed far from each other, recruiting water molecules from outside the cavity. In the case of the aglycosylated mAb, the low water density is due to the absence of sugars and to the hydrophobicity of the Fc. Finally, in the case of G0F chains, the water position within the Fc cavity seems to be energetically favored, as the water molecules are placed between the sugars and the protein and between the two sugar chains, coordinating the interactions and stabilizing the closed conformation of Fc. The calculation was performed also on medoid structures of other replicas, confirming the observed results (Fig. S13). Overall, this analysis suggests that water can play a key role in stabilizing the antibody conformation and its interaction with glycans, especially in the presence of fucosylated chains. According to the cluster analysis, in fact, G0F glycans tend to stay inside the Fc cavity in all the replicas, recruiting water molecules that may coordinate the interactions with the protein, promoting and stabilizing the T-shaped conformation.
Figure 7.
H-bond number and solvent analysis. (A) The H-bond number between G0 or G0F chains (A and B) and the antibody versus simulation time. The first 200 ns were excluded from calculation. (B) The mean H-bond number between G0 or G0F chains (A and B) and the antibody computed for replicas. For both the scatter plots, a lowess nonparametric interpolation was used. (C–F) Solvent analysis performed by 3D-RISM on aglycosylated, G0, and G0F mAbs. In (C), a bar chart showing the mean number of predicted water molecules associated with a negative dG and the average negative dG of three replicas is given. Bars are colored according to the energy colormap. In (D)–(F), the top view of CH2 domains is shown as gray ribbons and glycans as dark gray spheres, with fucose in red. The dG isolevel density is shown in blue, and water molecules with a negative dG value are shown as green spheres. To see this figure in color, go online.
Discussion
N-glycosylation of mAbs exerts an important role in regulating their structure and function. In particular, it has been experimentally demonstrated that the presence of a core fucose can downregulate the ADCC response that is a secondary mechanism of action of these biotherapeutics (61). In fact, fucosylated IgG1 binds the FcγRIIIa with at least 50-fold lower binding affinity than afucosylated antibodies (20). Despite the multitude of published studies (21, 22, 23, 24, 25, 26, 27), a clear explanation of why the presence of fucose is detrimental for mAb binding to FcγRIIIa has not been given yet. On this basis, the scope of this work was to elucidate via in silico approaches the structural role of fucose in modulating IgG1 ADCC activity. The study was focused on the 3D structure prediction of the whole adalimumab molecule, chosen as a typical therapeutic IgG1 case study for which the crystal structure of Fab portion is published, and on MD simulations of the aglycosylated and two differently glycosylated adalimumab forms, afucosylated (G0) and fucosylated (G0F).
According to reported data, a wide range of conformations are explored by antibodies in all the simulated systems, both in the aglycosylated form, in which the impact of glycans is not considered, and in the two glycosylated ones. However, three different dynamics were observed for the three species, suggesting that antibodies are very flexible molecules able to adopt a huge number of reasonable conformations and that Fc N-glycosylation, particularly the fucosylation, extensively affects their whole conformational behavior and not only the Fc structure.
This was confirmed by the geometric parameters such as RMSD, RMSF, and Rg and by the PC analysis. In fact, according to our data and looking at the RMSD profile of the whole molecules, three different conformational behaviors were detected, suggesting that antibodies do not explore the same conformational space. Despite the strong discrepancy, the RMSD of the single antibody regions showed that their structure is highly conserved, suggesting that the difference is mainly related to the relative rearrangement of domains in the space. In particular, a clear difference was observed between the glycosylated species, indicating the G0 mAb as the one exploring a relative limited conformational space and the G0F antibody as the one most subject to conformational changes. Concerning the aglycosylated form, it shows an intermediate behavior in all the computed parameters, hence suggesting that the absence of glycans makes the molecule able to explore many states, some similar to the G0 antibody and others more comparable to the G0F one. This observation was inferred by the Rg and cluster analysis, which suggested a compact T-shaped conformation for G0F mAb, an extended Y-shaped orientation for G0, and both conformation types for the aglycosylated antibody.
Focusing on the dynamics of the Fc fragment, which according to literature should be the domain mostly influenced by glycosylation (59), comparable geometric parameters among the three investigated mAbs were observed, suggesting that globally, this portion does not undergo significant conformational changes. However, the distribution of the distances between the corresponding glycosylated Asn301 residues showed that in the G0 antibody, the Fc domain is in an “open” conformation, whereas in the other forms, it presents a “closed” one, with only slight differences between aglycosylated and G0F species. These data suggest that Fc can adopt different orientations depending on glycosylation, and this is perfectly aligned to what is reported in published studies that demonstrated a role of glycans in modulating the Fc orientation, particularly the glycosylated loops and CH2 domains (17,62, 63, 64). Published studies also suggested that the presence of an open Fc structure, as observed for G0 and G0F mAbs, is a necessary, but not sufficient, condition to allow receptor binding. Actually, aglycosylated IgG1, which presents a closed Fc conformation, does not bind FcγRIIIa (10), whereas both G0 and G0F species are able to form complexes, albeit with different binding affinities. Also in this case, a certain variability was observed for the aglycosylated form that, according to our simulations, can adopt both closed and open conformations, suggesting that in any case, an exploration of all possible orientations is performed by the molecule.
Despite the importance of Fc domain in binding the FcγRIIIa, the main finding of this work is that glycans can influence the orientation of Fab domains, as inferred by the analysis of angles that they describe. This analysis shows how much the position of each Fab with respect to the other and the Fc can be different among the species, suggesting not only the modulatory role of glycans in this mechanism but also the ability of these molecules to explore a very large 3D space.
The typical flexibility of antibodies was already experimentally demonstrated by Zhang and colleagues (28), who, even not considering the glycosylation contribution, identified ∼120 different 3D structures from each IgG1 antibody particle that they analyzed via individual-particle electron tomography, and by Zhao et al., who analyzed the conformational sampling of Fab portions in “apo” and “holo” mAbs (29). According to these studies and to our data, the intrinsic flexibility of antibodies allows a huge conformational exploration of Fab arms, leading us to hypothesize that there can be an undefined number of different conformations. This flexibility is mainly due to the hinge region, which is completely unstructured and can adopt a great number of orientations, influencing the antibody tertiary structure and the system global convergence. Although we demonstrated that a local convergence is reached by structured domains, considering the simulated time and the system dimensions, we cannot state that the desired global convergence is reached in our simulations, and we hypothesize that this effect is mainly due to the high flexibility of these biomolecules. However, within each structured domain, we observed more than enough convergence, and so we propose that the structural equilibrium of whole mAbs as highly flexible molecules resides in the continuous oscillation of the hinge and Fab regions.
In addition, our results showed that the type of Fab orientation can be strongly influenced by the presence of glycans, particularly the fucose, which determines the conformational freedom of the molecule and its ability to explore the conformational space. In fact, considering the aglycosylated mAb, it can assume a wide variety of conformations, ranging from Y to T shaped. If instead we consider the glycosylated antibodies, in the case of G0, the preferred conformation is always the Y-shaped one, whereas for G0F, a clear preference for the T-shaped orientation was observed. These data were confirmed by all the performed replicas, further suggesting that glycans can modulate the conformational freedom of IgG1. Moreover, considering both the work by Zhao et al. (29) and other published full-length IgG structures, antibodies can adopt both of these two orientations (Y-shaped like: i.e., PDB: 1IGY, 5DK3; T-shaped like: i.e., PDB: 1HZH, 1IGT), and a structure-dependent discrimination mechanism by antibody partners can be hypothesized. Zhao and colleagues also found out that the T-shaped form represents the hFcγRI-bound structure, but they hypothesized that other receptors could prefer the Y-shaped one, and this is in line with our findings.
In addition, we observed that in all simulations and in all species, most of the conformational exploration is restricted to one of the two Fab domains. Even though this effect could be due to a simulation or starting model bias, it suggests that the structural symmetry of the molecule might not necessarily correspond to symmetric dynamics, and this aspect needs to be further investigated in the near future.
By PC analysis, it was demonstrated that each antibody is characterized by different principal motions, significantly influenced by its glycosylation pattern, and transposed in different antibody conformations, which are likely responsible for different biological functions. Moreover, a direct correlation between the computed Fab angles and the principal motion projections was established, showing that the dynamics of G0F antibody is characterized by different modes with respect to others that drive the different exploration of Fab domains.
The modulatory role of fucose was further confirmed by the solvent analysis performed on the three molecules. According to this prediction, a critical role of solvent was identified especially in the G0F antibody, in which sugars are positioned inside the Fc cavity and the water seems to stabilize the interactions with the protein and between glycans. In other species, the solvent cannot enter the hydrophobic pocket because of the absence of sugars or their orientation outside the Fc, thus suggesting that the conformation assumed by G0F glycans is responsible for the different dynamics of this antibody.
So, according to the data shown above, we can assume that sugars act as structural and dynamical modulators of IgG1 driving their conformational sampling, whereas in the absence of carbohydrates, the antibody is able to explore several states in a nonregulated manner. Specifically, the presence of fucose has an impact on the overall antibody conformation because of some structural constraints that can change both the sugar conformational freedom and the protein dynamics.
Furthermore, the flexibility that we observed for all the simulated proteins confirmed that MD studies can be a very useful tool to elucidate the dynamics of antibodies and to sample the huge variety of possible conformations explored by the molecules. Because of this feature, in fact, strong efforts are required to experimentally investigate the conformational behavior of mAbs. Specifically, to determine the whole atomistic structure of these molecules via crystallography or other experimental techniques is a big challenge; thus, up to now, only one structure of a whole human IgG1 is available in the PDB (36). Furthermore, investigating the entire conformational space explored by proteins, especially antibodies, is very hard when using experimental methods (i.e., X-ray) because by crystallography, only a snapshot of the molecule in specific experimental conditions can be observed.
Taken together, all these data can pose the structural basis of why fucosylated antibodies are less prone to interact with FcγRIIIa, as reported by published experimental data (21, 22, 23, 24, 25, 26, 27). Hence, we propose that the G0F antibody is blocked in a conformation that could be less compatible with accommodating the receptor and that the fucose, modulating both glycan and antibody behavior, is the main factor responsible for this. On the other hand, in the G0 mAb, the lack of fucose confers more conformational freedom to the molecule, allowing it to adopt an arrangement suitable for binding FcγRIIIa. Thus, the changes observed in the Fc conformation, which are mainly related to CH2 domains, are probably a necessary, but not sufficient, condition to stabilize the receptor binding, and there is a mechanism of interaction regulated by the dynamics of the whole antibody, likely influenced by glycans.
The conformational change as a mechanism of interaction control between the mAb and the receptor has been already proposed by Kiyoshi and colleagues (62), who, starting from an x-ray structure of the Fc-FcγRI complex, suggested that Fab portions may adopt a specific conformation to allow the antibody-receptor interaction, highly supporting our findings. In fact, even if the study by Kiyoshi et al. were performed for the high-affinity receptor, a mechanism based on conformational changes could be also more critical for the recognition of low-affinity receptors such as FcγRIIIa. For this reason, the impact of fucose on the whole antibody conformation may be more relevant for the binding to this receptor.
Conclusions
In this work, starting from published experimental data, we tried to elucidate by in silico approaches the role of mAb N-glycosylation, especially of fucose, in modulating the antibody structure and, as a result, the Fc effector functions. Globally, our data suggest that N-glycans act as structural modulators of IgG1 and play a crucial role in their conformational behavior. The presence or the absence of glycans can lead to different antibody conformations that may influence its functions. Based on our results, a structural explanation of the attitude of fucosylated antibodies to be less prone to interact with FcγRIIIa is given. In particular, we propose that the presence of fucose can introduce structural constraints that force the antibody into a state not suitable for receptor recognition, in which Fab portions are collapsed onto the Fc in a T-shaped conformation. On the other hand, the lack of fucose allows the three antibody domains to assume a Y-shaped form that, in principle, can better fit to the receptor. As secondary evidence for this study, an asymmetric behavior of Fab portions was observed in all the simulated species, showing that the type of conformational space explored by these domains strongly depends on both the intrinsic flexibility of the molecule and glycosylation and suggesting that the orientation of Fab portions can influence the receptor recognition.
Considering the role of carbohydrates in the modulation of the Fc effector functions and consequently of the mechanisms of action of biotherapeutics, this study can help understand the contribution of N-glycosylation on the structural organization of mAbs, paving the way for novel strategies in the development of new biotechnological products.
Author contributions
S.S. performed the research, analyzed data, and wrote the manuscript. C. Parravicini and U.G. helped and supported data analysis and interpretation. C. Pergola and M.R. discussed the data and revised the manuscript. I.E. and F.C. designed the study, checked the quality of the simulations and data, discussed the results, and revised the manuscript.
Acknowledgments
The authors acknowledge Dr. Enrico Guarnera, Dr. Luca Palazzolo, and Dr. Tommaso Laurenzi for technical support and fruitful scientific discussion.
This study was supported by a Department of Excellence grant from the Italian Ministry of University and Research 2018–2022. I.E. gratefully acknowledges the departmental grant “Linea 2 – Azione A 2019” from the University of Milan.
Editor: Yuji Sugita.
Footnotes
Fabio Centola and Ivano Eberini contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.10.026.
Supporting material
▪▪▪
References
- 1.Kaplon H., Reichert J.M. Antibodies to watch in 2021. MAbs. 2021;13:1860476. doi: 10.1080/19420862.2020.1860476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edelman G.M., Cunningham B.A., et al. Waxdal M.J. The covalent structure of an entire gammaG immunoglobulin molecule. Proc. Natl. Acad. Sci. USA. 1969;63:78–85. doi: 10.1073/pnas.63.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A., Cummings R.D., et al. Kornfeld S. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potter M. Structural correlates of immunoglobulin diversity. Surv. Immunol. Res. 1983;2:27–42. doi: 10.1007/BF02918394. [DOI] [PubMed] [Google Scholar]
- 6.Kabat E.A., Wu T.T., Bilofsky H. Variable region genes for the immunoglobulin framework are assembled from small segments of DNA--a hypothesis. Proc. Natl. Acad. Sci. USA. 1978;75:2429–2433. doi: 10.1073/pnas.75.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu T.T., Johnson G., Kabat E.A. Length distribution of CDRH3 in antibodies. Proteins. 1993;16:1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- 8.Nimmerjahn F., Ravetch J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 9.Meyer S., Leusen J.H.W., Boross P. Regulation of complement and modulation of its activity in monoclonal antibody therapy of cancer. MAbs. 2014;6:1133–1144. doi: 10.4161/mabs.29670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Mathieu M., Brezski R.J. IgG Fc engineering to modulate antibody effector functions. Protein Cell. 2018;9:63–73. doi: 10.1007/s13238-017-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart G.W. Glycosylation. Curr. Opin. Cell Biol. 1992;4:1017–1023. doi: 10.1016/0955-0674(92)90134-x. [DOI] [PubMed] [Google Scholar]
- 12.Wada R., Matsui M., Kawasaki N. Influence of N-glycosylation on effector functions and thermal stability of glycoengineered IgG1 monoclonal antibody with homogeneous glycoforms. MAbs. 2019;11:350–372. doi: 10.1080/19420862.2018.1551044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh J.B., Ng S.K. Impact of host cell line choice on glycan profile. Crit. Rev. Biotechnol. 2018;38:851–867. doi: 10.1080/07388551.2017.1416577. [DOI] [PubMed] [Google Scholar]
- 14.Harvey D.J., Merry A.H., et al. Rudd P.M. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- 15.Varki A., Cummings R.D., et al. Etzler M.E. Symbol nomenclature for glycan representation. Proteomics. 2009;9:5398–5399. doi: 10.1002/pmic.200900708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnold J.N., Wormald M.R., et al. Dwek R.A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 17.Krapp S., Mimura Y., et al. Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi Y., Barb A.W. A synopsis of recent developments defining how N-glycosylation impacts immunoglobulin G structure and function. Glycobiology. 2020;30:214–225. doi: 10.1093/glycob/cwz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothman R.J., Perussia B., et al. Warren L. Antibody-dependent cytotoxicity mediated by natural killer cells is enhanced by castanospermine-induced alterations of IgG glycosylation. Mol. Immunol. 1989;26:1113–1123. doi: 10.1016/0161-5890(89)90055-2. [DOI] [PubMed] [Google Scholar]
- 20.Shields R.L., Lai J., et al. Presta L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 21.Shinkawa T., Nakamura K., et al. Shitara K. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003;278:3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- 22.Li H., Sethuraman N., et al. Gerngross T.U. Optimization of humanized IgGs in glycoengineered Pichia pastoris. Nat. Biotechnol. 2006;24:210–215. doi: 10.1038/nbt1178. [DOI] [PubMed] [Google Scholar]
- 23.Iida S., Misaka H., et al. Satoh M. Nonfucosylated therapeutic IgG1 antibody can evade the inhibitory effect of serum immunoglobulin G on antibody-dependent cellular cytotoxicity through its high binding to FcgammaRIIIa. Clin. Cancer Res. 2006;12:2879–2887. doi: 10.1158/1078-0432.CCR-05-2619. [DOI] [PubMed] [Google Scholar]
- 24.Masuda K., Kubota T., et al. Nakamura K. Enhanced binding affinity for FcgammaRIIIa of fucose-negative antibody is sufficient to induce maximal antibody-dependent cellular cytotoxicity. Mol. Immunol. 2007;44:3122–3131. doi: 10.1016/j.molimm.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Liu L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J. Pharm. Sci. 2015;104:1866–1884. doi: 10.1002/jps.24444. [DOI] [PubMed] [Google Scholar]
- 26.Pereira N.A., Chan K.F., et al. Song Z. The “less-is-more” in therapeutic antibodies: afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs. 2018;10:693–711. doi: 10.1080/19420862.2018.1466767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrara C., Grau S., et al. Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. USA. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Zhang L., et al. Ren G. 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci. Rep. 2015;5:9803. doi: 10.1038/srep09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J., Nussinov R., Ma B. Antigen binding allosterically promotes Fc receptor recognition. MAbs. 2019;11:58–74. doi: 10.1080/19420862.2018.1522178. Published online October 5, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright D.W., Elliston E.L.K., et al. Perkins S.J. Atomistic modeling of scattering curves for human IgG1/4 reveals new structure-function insights. Biophys. J. 2019;117:2101–2119. doi: 10.1016/j.bpj.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandt J.P., Patapoff T.W., Aragon S.R. Construction, MD simulation, and hydrodynamic validation of an all-atom model of a monoclonal IgG antibody. Biophys. J. 2010;99:905–913. doi: 10.1016/j.bpj.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kortkhonjia E., Brandman R., et al. Swartz T.E. Probing antibody internal dynamics with fluorescence anisotropy and molecular dynamics simulations. MAbs. 2013;5:306–322. doi: 10.4161/mabs.23651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbison A.M., Brosnan L.P., et al. Fadda E. Sequence-to-structure dependence of isolated IgG Fc complex biantennary N-glycans: a molecular dynamics study. Glycobiology. 2019;29:94–103. doi: 10.1093/glycob/cwy097. [DOI] [PubMed] [Google Scholar]
- 34.MOE . 2019. Molecular Operating Environment (MOE), 2019.01; Chemical Computing Group ULC, Montreal, Canada. [Google Scholar]
- 35.Hu S., Liang S., et al. Lou Z. Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor α-associated diseases from a molecular view. J. Biol. Chem. 2013;288:27059–27067. doi: 10.1074/jbc.M113.491530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saphire E.O., Parren P.W., et al. Wilson I.A. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 37.Labute P. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 2008;29:1693–1698. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 38.Case D.A., Darden T.A., et al. Merz K.M. San Francisco: University of California; 2008. AMBER 10. [Google Scholar]
- 39.Barker J.A., Watts R.O. Monte Carlo studies of the dielectric properties of water-like models. Mol. Phys. 1973;26:789–792. [Google Scholar]
- 40.Watts R.O. Monte Carlo studies of liquid water. Mol. Phys. 1974;28:1069–1083. [Google Scholar]
- 41.Phillips J.C., Braun R., et al. Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Der Spoel D., Lindahl E., et al. Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 43.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 44.Feller S.E., Zhang Y., et al. Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 45.Lee J., Cheng X., et al. Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks B.R., Brooks C.L., III, et al. Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo S., Kim T., et al. Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 48.Best R.B., Zhu X., et al. Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGibbon R.T., Beauchamp K.A., et al. Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daura X., Gademann K., et al. Fischer J. Peptide folding: when simulation meets experiment. Angew. Chem. Int.Engl. 1998;31:1387–1404. [Google Scholar]
- 51.Baker E.N., Hubbard R.E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 52.Hansen J.-P., McDonald I.R. Elsevier, Amsterdam, the Netherlands; 2006. Theory of Simple Liquids: with Applications to Soft Matter. [Google Scholar]
- 53.Beglov D., Roux B. An integral equation to describe the solvation of polar molecules in liquid water. J. Phys. Chem. B. 1997;101:7821–7826. [Google Scholar]
- 54.Kovalenko A., Hirata F. Self-consistent description of a metal-water interface by the Kohn-Sham density functional theory and the three-dimensional reference interaction site model. J. Chem. Phys. 1999;110:10095–10112. [Google Scholar]
- 55.Kirschner K.N., Yongye A.B., et al. Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerber P.R., Müller K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided Mol. Des. 1995;9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- 57.Erbel P.J.A., Haseley S.R., et al. Vliegenthart J.F.G. Studies on the relevance of the glycan at Asn-52 of the α-subunit of human chorionic gonadotropin in the alphabeta dimer. Biochem. J. 2002;364:485–495. doi: 10.1042/BJ20011482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar C.V., Swetha R.G., et al. Ramaiah S. Computational analysis reveals the association of threonine 118 methionine mutation in PMP22 resulting in CMT-1A. Adv. Bioinformatics. 2014;2014:502618. doi: 10.1155/2014/502618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pincetic A., Bournazos S., et al. Ravetch J.V. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 2014;15:707–716. doi: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagae M., Yamaguchi Y. Function and 3D structure of the N-glycans on glycoproteins. Int. J. Mol. Sci. 2012;13:8398–8429. doi: 10.3390/ijms13078398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiner L.M., Surana R., Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 2010;10:317–327. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiyoshi M., Caaveiro J.M.M., et al. Tsumoto K. Structural basis for binding of human IgG1 to its high-affinity human receptor FcγRI. Nat. Commun. 2015;6:6866. doi: 10.1038/ncomms7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsumiya S., Yamaguchi Y., et al. Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J. Mol. Biol. 2007;368:767–779. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 64.Oganesyan V., Damschroder M.M., et al. Dall’Acqua W.F. Structural insights into neonatal Fc receptor-based recycling mechanisms. J. Biol. Chem. 2014;289:7812–7824. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
▪▪▪